Abstract
M. tuberculosis, an etiological agent of tuberculosis, requires a long treatment regimen due to its ability to respond to stress and persist inside the host. The second messenger (p)ppGpp-mediated stress response plays a critical role in such long-term survival, persistence, and antibiotic tolerance which may also lead to the emergence of multiple drug resistance. In mycobacteria, (pp)pGpp molecules are synthesized predominantly by two bifunctional enzymes-long RSH-Rel and short SAS-RelZ. The long RSH-Rel is a major (p)ppGpp synthetase and hydrolase. How it switches its activity from synthesis to hydrolysis remains unclear. RelMtb mutant has been reported to be defective in biofilm formation, cell wall function, and persister cell formation. The survival of such mutants has also been observed to be compromised in infection models. In M. smegmatis, short SAS-RelZ has RNase HII activity in addition to (pp)Gpp synthesis activity. The RNase HII function of RelZ has been implicated in resolving replication–transcription conflicts by degrading R-loops. However, the mechanism and regulatory aspects of such a regulation remain elusive. In this article, we have discussed (p)ppGpp metabolism and its role in managing the stress response network of mycobacteria, which is responsible for long-term survival inside the host, making it an important therapeutic target.
Introduction
Microorganisms live in constantly changing hostile environments that threaten their survival and existence. The stringent response (SR) is an evolutionarily conserved mechanism that allows bacteria to thrive and persist in adverse environments. Most bacteria under stressful conditions such as nutritional limitation produce guanosine 5′-diphosphate 3′-diphosphate (ppGpp) and guanosine 5′-triphosphate 3′-diphosphate (pppGpp)—collectively known as alarmone molecules or (p)ppGpp.1 These “alarmones” are the master regulators of the stringent response, a universal stress response, classically shown to be induced by amino acid deprivation. In 1969, Cashel and Gallant reported the appearance of a magic spot-(p)ppGpp over the thin layer chromatography sheet in the cell extract derived from starved bacterial cells (of Escherichia coli) while analyzing the nucleotide content.2 Notably, (p)ppGpp and a corresponding stringent response have emerged as a crucial master regulator of not only the bacterial response to stress but also several aspects of bacterial physiology, including growth rate, phase transition, sporulation, motility, competence, biofilm formation, toxin production, and a wide range of other virulence associations.3−5
Additionally, mounting evidence links (p)ppGpp, mediated stringent response to antibiotic tolerance, and the emergence of multidrug resistance.6 It is plausible that targeting these pathways related to (p)ppGpp could represent an alternative to conventional therapies.7,8 The lack of amino acids has been shown to trigger the activation of the stringent response in E. coli upon accumulation of uncharged tRNAs at the A site of ribosomes. RelA/Rel senses stalled ribosomes and responds by synthesizing (p)ppGpp from GTP/GDP and ATP. SpoT, a protein that functions as an accompanying hydrolase in E. coli, degrades the (p)ppGpp once the amino acid deprivation has been resolved. The overall effect of (p)ppGpp during the stringent response is that it reduces the transcription of most metabolic genes involved in the exponential growth phase while increasing the transcription of genes involved in amino acid biosynthesis and stress responses in E. coli.1 The Cashel group showed that ppGpp and pppGpp differentially regulate transcription in E. coli, but the mode of action was not clear. Syal and Chatterji showed that ppGpp and pppGpp exhibit differential binding to RNA polymerase, which could explain their different modes of transcriptional regulation in E. coli.9 Interestingly, (p)ppGpp does not bind to RNA polymerase in mycobacteria, and it mediates stress response through various mechanisms, as elaborated in other sections. Evidently, the basal levels of (p)ppGpp have been shown to be essential for viability in different classes of bacteria.10
The bacterial classes differ in (i) the mechanisms by which (p)ppGpp affects transcription and translation11 and (ii) the configuration and number of (p)ppGpp-producing enzymes they possess.12 As a highly charged species, (p)ppGpp has structural similarities to its precursor (GTP), which allows it to bind to diverse binding partners. The identification and characterization of the biologically relevant interactions remains a challenge.13−15 Since humans do not produce second messengers (p)ppGpp, targeting their synthesis or associated pathways in bacteria will not be harmful. The recent discoveries further demonstrate that it plays an essential role in virulence and antibiotic tolerance, making it an excellent therapeutic target.16,17
In this paper, we have focused on stringent response in mycobacteria. Two decades back, Ojha and Chatterji discovered (p)ppGpp in Mycobacterium smegmatis,18 and later in the same year, their findings were confirmed in Mycobacterium tuberculosis (Mtb).19 The Rel-bifunctional (p)ppGpp synthetase and hydrolase were characterized as the principal mediator of stringent response in all mycobacterium species.20M. tuberculosis experiences various stresses inside the host, including oxidative, nitrosative, and nutrient deprivation stress. However, it successfully overcomes these potentially lethal stresses and establishes chronic infection.20,21 Adapting to such stress conditions requires large-scale reprogramming of signaling cascades, allowing M. tuberculosis to infect macrophages and survive in granulomas for years.20,21M. tuberculosis has a bifunctional Rel enzyme that produces most (p)ppGpp in response to nutrient deprivation. Additionally, Rel can hydrolyze (p)ppGpp as well; however, triggering factors for hydrolysis are not well understood. (p)ppGpp expression indirectly tunes the expression of nearly 159 genes in M. tuberculosis, many of which encode important antigens, proteins, and virulence factors involved in persistence.22
This review elaborates the function of two bifunctional (p)ppGpp synthetases and their role in mycobacteria. Notably, the novel protein RelZ has RNAase H function (in M. smegmatis) in addition to (pp)pGpp synthetase activity; however, its physiological significance remains unclear. We further discussed the role of (p)ppGpp synthetase in biofilm formation, long-term survival, GTP homeostasis, and antibiotic tolerance in mycobacteria.
Family of (p)ppGpp Synthetase
Genes encoding enzymes for (p)ppGpp metabolism have been found in all sequenced bacterial genomes—except Planctomycetes, Chlamydia, Verrucomomicrobia, and some obligate intracellular bacterial species—which makes stringent response a nearly universal phenomenon in bacteria.12 The RelA-SpoT homologue (RSH) protein family members regulate the cellular pool of (p)ppGpp. The RSH family includes small alarmone synthetases (SASs) containing a synthetic domain, small alarmone hydrolases (SAHs) with a hydrolytic domain, and multidomain proteins containing both a synthetase and hydrolase domain. The standard nomenclature for multidomain RSH proteins is long RSHs.12 Several bacteria, including mycobacteria, also encode homologues of RSH proteins that are shorter in length. These are typically single-domain proteins with either synthetase or hydrolase activity. Hence, they are called small alarmone synthetases (SASs) or small alarmone hydrolases (SAH).20,23 Jimmy et al. gave the most recent classification of SAS and identified 30 subfamilies.24 Different SAS have distinct roles, and various signals trigger their activity.24
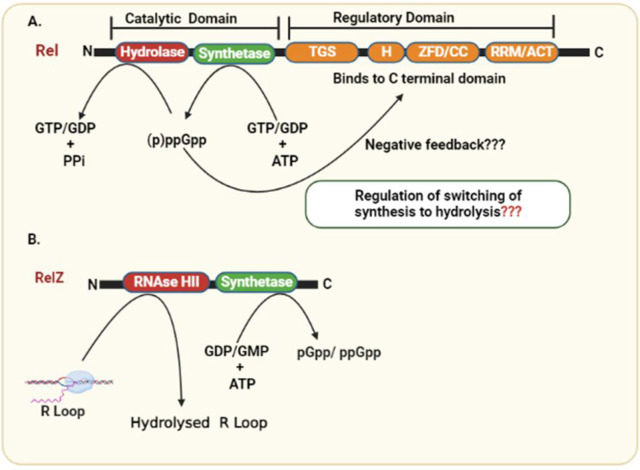
The stringent response in Gram-negative bacteria is governed by two enzymes: RelA and SpoT. The relA gene encodes the RelA protein, a monofunctional synthetase responsible for synthesizing (p)ppGpp. The bifunctional SpoT, encoded by the spoT gene, primarily functions as a hydrolase for degradation of (p)ppGpp. In response to specific stress signals, such as fatty acid starvation, SpoT can also synthesize (p)ppGpp. RelA and SpoT descend from the same ancestral Rel protein, and the hydrolase domain in RelA has been rendered inactive. Thus, RelA-SpoT in E. coli, and Rel proteins in mycobacteria have similar domain architecture and have been classified as members of the RelA-SpoT Homology (RSH) superfamily of proteins.12 Ojha et al. reported that M. smegmatis accumulates the stringent factor ppGpp under nutrient starvation, indicating a link between persistors and the stringent response.25 In mycobacteria alarmones are synthesized and degraded by Rel, a bifunctional enzyme.26 The gene rv2583c in M. tuberculosis encodes bifunctional Rel, a 738 amino acid long multidomain protein with a catalytic N-terminal domain [1–394 amino acids (aa)] and a regulatory C-terminal domain (395–738 aa). The transfer of the 5′-β,γ-pyrophosphate group from ATP to the 3′ OH group of GDP or GTP is catalyzed by the synthetase domain, which results in the formation of ppGpp or pppGpp, respectively. The hydrolysis domain catalyzes the reverse reaction, namely, the hydrolysis of the pyrophosphate from (p)ppGpp, which results in the formation of GDP or GTP.27 Mn2+ or Mg2+ cations are required as cofactors for both enzymatic activities.28 Half of the C-terminal protein harbors two regulatory domains: TGS (ThrRS, GTPase, and SpoT) and ACT (aspartate kinase, chorismate mutase, and TyrA). The TGS domain has a ligand binding function, while the ACT domain is found in proteins regulated by amino acid concentration.14 The TGS and ACT domains are linked by an intrinsically disordered intermediate region of roughly 200 amino acids, resulting in increased flexibility in the region, thereby facilitating domain–domain interaction.29 This intermediate region (INT) between TGS and ACT domains is conserved in several other distantly related Rel protein29 (Figure 1). Experimentally, it has been demonstrated that the deletion of regulatory region from C-terminal domain increases Rel synthetase activity and renders it independent of the accessory components needed for activation.14,25
Figure 1.
Domain architecture of Rel and RelZ proteins involved in the synthesis and hydrolysis of alarmones (pp)pGpp. Rel is composed of a catalytic and regulatory domain with opposite (hydrolase and synthetase) enzymatic activities. (p)ppGpp binds to the regulatory domain at CTD and may show negative feedback loop. RelZ has N-terminal RNaseHII domain and the (pp)pGpp synthetase domain. Synthetase domain of RelZ are responsible for the synthesis of (pp)pGpp and hydrolyze RNA: DNA hybrid in addition to R-loops.
Rel—Bifunctonal (p)ppGpp Synthetase/Hydrolase
As discussed, M. tuberculosis has a bifunctional (p)ppGpp synthetase-RelMtb conserved in most species of mycobacteria.28 As part of the relA-spoT family of genes, RelMtb mediates a global stringent response in mycobacteria.12 Interestingly, Rel from M. tuberculosis is a single-gene-encoding for a bifunctional enzyme capable of catalyzing both synthesis and hydrolysis of (p)ppGpp (Figure 2). The N-terminal region of the Rel enzyme has dedicated synthesis and hydrolysis domains with mutually exclusive activities. The regulation and switching of such opposite activities are not well understood. The (p)ppGpp hydrolysis domain stretches from 1 to 181 amino acids (aa) residues, whereas the (p)ppGpp synthetase domain is formed by 87–394 aa residues. It also includes an overlapping three-helix bundle (87–181) between the two activities that is conserved across RSH proteins.27,30 RelMtb also harbors a regulatory C-terminal domain (CTD, 395–738 aa), which include the TGS-domain as present in ThrRS (threonyl tRNA synthetase), GTPase (Obg family GTPases), and SpoT, as well as the ACT-domain (as seen in aspartate kinase, chorismate mutase, and TyrA). Jain et al. showed that the TGS subdomains (residues 400 to 459) and the ACT subdomains (residues 657 to 723) have regulatory functions in NTDs.14,30 The N-terminal’s hydrolase domain (1–181) consists of helices 1–11 and forms a catalytic domain structurally conserved among HD proteins. The four of 11 helices that are α1 to α4 form a helix bundle that facilitates the hydrolytic activity of enzymes.30 The binding pocket of the substrate (p)ppGpp is present in regions 41–53 that connect α2 to α3.30 The HD motif, which consists of conserved histidine (H80) and aspartate (D81) residues, is present in the β-turn that connects α4 and α5. The α8, α9/α10, and α11 (three-helice bundle) are common among hydrolase and the synthetase domain (136–197). Singal et al. concluded that the catalytic tetrad (H-Xn-HD-Xn-D) is bound to the divalent ions among the hydrolase domains and is conserved in RelMtb NTD. The divalent cations are predicted to be coordinated by the histidine and aspartate residues, as most of the conserved residues in this domain are either aspartates or histidine. The coordination of divalent metal ions has been reported to be essential for the domain’s hydrolase activity.31 Avarbock et al. observed that alanine substitution of H80 or D81 in RelMtb abolishes hydrolase activity, while leaving pppGpp synthesis activity unaffected in an in vitro study. The (p)ppGpp synthetase domain consists of five β-sheets surrounded by five α-helices.30 The (p)ppGpp synthetase domain was found to be structurally homologous to DNA polymerase β (pol β), and the D190 and D256 residues in pol β that are crucial for coordinating the Mg2+ cofactor correspond to D265 and E325 in RelMtb. Both of these residues of RelMtb are necessary for (p)ppGpp synthesis.26,32 The positively charged residues R242, K244, and K252 are most likely involved in the RelMtb ATP binding pocket. These residues coordinate the ATP’s phosphates at a location close to the GDP binding pocket and the Mg2+ binding site.26,30 RelMtb has strong cation cofactor requirements, including Mg2+ or Mn2+ for (p)ppGpp synthesis and Mn2+ for (p)ppGpp hydrolysis.27 The (p)ppGpp synthetase domain transfers the 5-β,γ-pyrophosphate from ATP to the 3′-OH of GDP or GTP to synthesize ppGpp and pppGpp, respectively.28 The transferase activity of both RelA and RelMtb depends on Mg2+. RXKD and EXDD are conserved motifs in the Rel protein. A charge reversal in a conserved motif within the synthesis subdomain has been shown to inhibit bifunctional RelMtb (p)ppGpp synthesis.33,34 For an optimal synthesis of (p)ppGpp, Mg2+ and Mn2+ concentrations must equal the combined concentrations of substrates ATP and GTP.27 Due to a highly conserved RXKD motif in the synthetase domain of bifunctional Rel enzymes, the activity of RelMtb synthetase is inhibited by amounts of Mg2+ (or Mn2+) that are greater than those of the GTP and ATP substrates concentration.34 Sajish et al. reported that monofunctional RelA in E. coli uses GDP as the primary pyrophosphate acceptor, whereas bifunctional RelMtb utilizes GTP. Here, the EXDD and RXKD motifs determine this specificity. They also discovered that an RXKD motif promoted cooperative nucleotide binding, whereas EXDD did not. Surprisingly, substituting RXKD for EXDD (in RelMtb) significantly diminished (p)ppGpp synthesis in a bifunctional protein. A similar reversal in a monofunctional protein, on the other hand, resulted in increased synthesis in E. coli. Importantly, RXKD to EXDD substitution in the bifunctional RelMtb resulted in synthesizing a novel molecule identified as pGpp.33,34 Sajish et al. also concluded that the C-terminal region negatively regulates (p)ppGpp synthesis by interaction mediated by these motifs in the N-terminal domain. Synthesis is tuned by the interactions between the C-terminal region and the EXDD and RXKD motifs.33,34
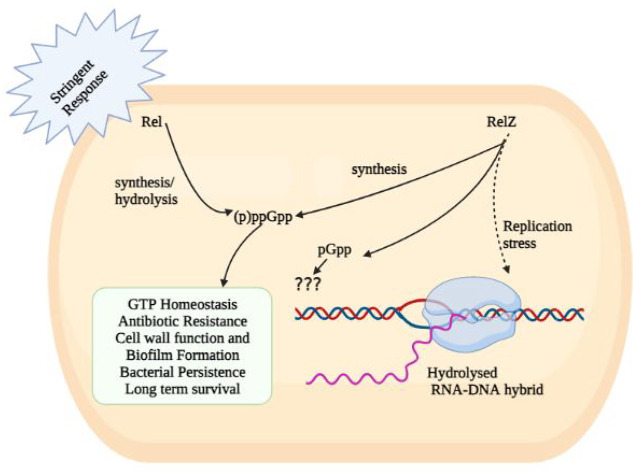
Figure 2.
Rel, RelZ, and their pleiotropic role in the management of bacterial stress response and housekeeping functions.
The relMtb gene is constitutively expressed at basal levels, possibly through a (−10) promoter element upstream of the gene recognized by the housekeeping factor σA (σ-factor).35 The CTD represses RelMtb synthetase activity in the absence of stress or stimuli, resulting in a low level of (p)ppGpp production.25,36 Even though the level of (p)ppGpp is low in normal conditions, it is still vital for growth.36 It is best understood that (p)ppGpp synthesis is induced during amino acid starvation by a Rel enzyme associated with ribosomes upon entry of uncharged tRNAs into the A site of the ribosome.37 Ribosomes, uncharged tRNAs, and cognate mRNA (RAC) tune the (p)ppGpp synthesis upon binding to the RelMtb enzyme. Together RAC enables RelMtb to alter its synthesis and hydrolysis rates. In the abundance of nutrients, the uncharged tRNAs are not present, thereby keeping both synthesis and hydrolysis at basal levels. Here, uncharged tRNA mimics starvation conditions, which further increases (p)ppGpp synthesis rate as well as synthetase affinity for its GTP/GDP/ATP substrates.27
The basal level of the (p)ppGpp synthesis is not significantly affected by the deletion of the C-terminal end (residues 1–394 or 87–394) of full-length RelMtb. A further investigation of the CTD domain in M. smegmatis, a nonpathogenic species often used as a model organism of M. tuberculosis, revealed its direct role in regulating (p)ppGpp synthesis. The CTD domain has been implicated in sensing uncharged tRNA.25 Out of six cysteine residues, four cysteine residues are conserved across RSH, and mutation of cysteine at 692 positions to even a very closely related amino acid-like serine makes it unresponsive to uncharged tRNA. Jain et al. through FRET and anisotropy measurement showed that cysteine at position 692 moves away from the NTD to form a more compact CTD when uncharged tRNA binds to RelMsm, thus allowing more space for substrates to enter the catalytic site.38 The presence of a flexible conserved linker region between the TGS and ACT supports this conclusion.29 In addition to the uncharged tRNA-induced conformational change within the CTD, alternate ligand binding to this portion of RelMsm can unfold the protein and repress its synthetic functions.14 Syal et al. reported that (p)ppGpp binds to the CTD region between the TGS and ACT domains of the RelMsm protein, resulting in a negative feedback loop.14 It has been shown that binding of pppGpp to RelMsm CTD represses (p)ppGpp synthesis and increases (p)ppGpp hydrolysis. Consequently, RelMsm-mediated pppGpp synthesis is reduced at saturating concentrations of pppGpp.14,38 In 2013, Weiss and Stallings concluded that (p)ppGpp production by RelMtb is necessary for efficient growth and biofilm formation in culture and for maintaining titers in a mouse model of infection.36
Mutant relMtb (ΔrelMtb) is unable to survive long-term starvation in the culture.19 The survival of relMtb mutant has also been reported to be compromised in mice models.19 RelMtb mediated (p)ppGpp synthesis regulates more than 80 genes and is critical for establishing a persistent Mtb infection in mice.19,22 (p)ppGpp synthesis by RelMtb is essential for chronic Mtb infection in mice and guinea pigs, especially when the immune system is impeding the bacteria’s growth. The RelMtb mutant could not grow in THP-1 macrophages in cell culture, suggesting survival during chronic in vivo infection depends on the stringent response.19,22 The differential expression of several genes was observed through microarray analysis of H37Rv and H37RvΔrelMtb mutant strains upon starvation for 6 h.22 The downregulation of 54 genes that encode ribosomal proteins was reported in the parental H37Rv strain, in comparison to the H37RvΔrelMtb mutant. Late-log phase cultures of H37RvΔrelMtb exhibited a minimum of 5-fold increase in ribosomes per unit protein in comparison to the H37Rv wild-type strain. Certain aspects of the stringent response are unique to mycobacteria including regulation through CarD and inorganic polyphosphate (polyP).20 Here, CarD is an essential protein in mycobacteria responsible for controlling rRNA transcription, and its depletion has been shown to impair the stringent response.57
The H37RvΔrelMtb mutant failed to survive the oxygen limitation and increased temperature of 42 °C, and lost viability sooner than the parental M. tuberculosis H37Rv strain.27 A subsequent study revealed that H37RvΔrelMtb exhibited significantly reduced levels of heat-shock protein HspX, which helps in adapting to heat shock. The observed low expression of HspX in H37RvΔrelMtb explains its inability to adapt to heat shock.10 Likewise, it has been observed that in M. smegmatis, the absence of the rel gene results in reduced viability during nutrient deprivation and sluggish growth under cold shock.39,40 Together, the presence of Rel offers a survival advantage for M. tuberculosis under stress conditions. Multiple genes linked to mycobacterial pathogenicity and antigens were also differently expressed in mutant as determined by transcriptomic analysis.22 The expression of groEL2, groES, LpqH (lipoprotein), and the PE_PGRS3 was also affected in mutant. Here, groEL2 is a heat shock protein, whereas groES is a chaperone protein. A recent demonstration has revealed that PE_PGRS3, situated on the surface of mycobacterial cells, is expressed under phosphate limitation.41 Secreted antigens like esat6 (early secretory antigen target), the antigen 85 complex, mpt83, and cfp7 (essential for the pathogenesis of Mtb) were dysregulated in the H37RvrelMtb mutant. Lipoprotein LpqH expression is also downregulated in the H37RvrelMtb mutant.42 PE_PGRS3, which is dependent on (p)ppGpp, is required for M. tuberculosis and host cell contact and infection.43 This suggests the involvement of stringent response in both M. tuberculosis and M. smegmatis.
Interestingly, the group of Ojha from Wadsworth Centre-New York has reported the induction of ribosome hibernation in M. smegmatis by a zinc-limiting growth condition involving stringent response. They reported a novel role of this intracellular RelA|SpoT homologue (Rsh) in constitutive scanning of translating ribosomes, and detection of the deacylated A-site tRNA in the first cycle of elongation and in consequent triggering of the stringent response.44,45 The ribosome hibernation results in the depletion of this intracellular RelA/SpoT homologue via a Clp protease-dependent mechanism. Ojha et al. through cryo-EM structure showed that the ACT domain of Rsh engages in a constitutive interaction with translating ribosomes during the intricate process of initiation complex formation and its subsequent transition to the pre-elongation stage. Together, they proposed a surveillance role for RelA/SpoT homologue.44
Evidently, Rel mutant of M. smegmatis still showed detectable (p)ppGpp which lead to the discovery of another (p)ppGpp synthetase23 as described in the next section.
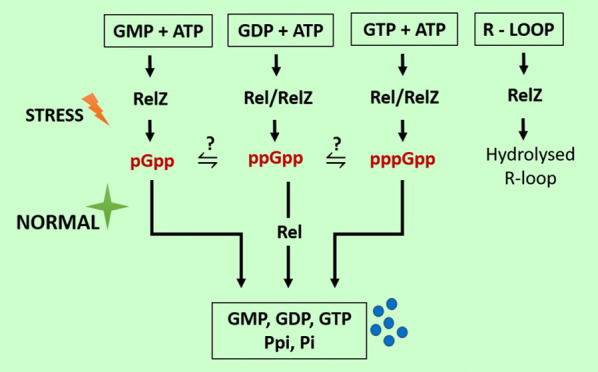
RelZ: (pp)pGpp synthetase with RNase H Domain
Interestingly, Murdeshwar and Chatterji reported MS_RHII-RSD, also known as RelZ, that has both RNase HII and (p)ppGpp synthesis activity in a single polypeptide chain in M. smegmatis.23 Here, MSMEG 5849 gene in M. smegmatis encodes for RelZ, which is similar to other SAS in terms of its C-terminal RSD domain, but distinct from them due to the presence of an N-terminal RNase HII domain in the same polypeptide chain.23 A homologue of bifunctional Rel (RelMtb) and SAS that is Rv1366 has been reported in the pathogen M. tuberculosis.28 Here, Rv1366 lacks the RHII domain and cannot synthesize (p)ppGpp invitro.46 It has been demonstrated that RelZ from M. smegmatis exhibits a preference for GDP substrate, Mg2+ ion-independent (p)ppGpp synthesis, and lack of (p)ppGpp hydrolysis activity.23 Along with these functions, RelZ also has the ability to synthesize pGpp. GMP is a preferred substrate for RelZ. The role of pGpp remains elusive. Given that the levels of pGpp and pppGpp synthesis were highest in the cases of RelZ and RelMsm, respectively, it was clear that the two enzymes Rel and RelZ have different patterns of substrate consumption. GMP > GDP > GTP is the substrate preference hierarchy for RelZ, while GTP > GDP is the preference hierarchy for RelMsm.47 Notedly, RelMsm does not make pGpp.47 According to Petchiappan et al., RNA and (p)ppGpp subtly alter the RelZ-mediated synthesis of pGpp. RelZ hydrolyzes RNA/DNA hybrids and R-loops; therefore, alarmone synthesis would not be necessary once the RHII domain had degraded them. It seems conceivable that the hydrolyzed RNA would prevent RelZ from producing pGpp.47 They also hypothesized that the inhibition of RelZ by (p)ppGpp may regulate the cell’s total alarmone levels. It may be advantageous when the cells no longer need to synthesize alarmone.47 RelZ hydrolyzes the RNA moiety of RNA:DNA heteroduplexes in the presence of Mn2+, and its amino-terminal region resembles the structural properties of bacterial RNase HII proteins.23 The function of RelZ’s RNase H domain led to the discovery of its role in the R-loop-induced stress response. R-loops play a significant role in the replication-transcription conflicts responsible for stalled RNA polymerase arrays and promoting replication stress.23,48 On one hand, RNase HII removes R-loops,49 and on the other hand, (p)ppGpp synthesis destabilizes stalled RNA polymerase.1,50 RelZ possesses these important activities (RNase HII and (pp)pGpp synthetase) in a single polypeptide.48 Krishnan et al. reported the upregulation of relZ expression and consequent removal of R-loops induced by UV stress.48 RelZ active site mutational studies have shown that the inactivation of one domain of RelZ did not alter the activity of the other domain, but the purified subdomains have been observed to be inactive. This domain interdependence suggests that full-length RelZ is essential for its function. Krishnan et al. also reported the altered cell surface properties of ΔrelZ strain suggesting that RelZ plays a important role in cell wall metabolism.47,48 The soil bacteria M. smegmatis is exposed to highly variable hostile conditions, and additional (ppp)Gpp synthetase RelZ enables better tuning of (p)ppGpp mediated stringent response in the cell. Also, formation of R-loops due to the UV stress and other hostile conditions may be more prevalent in M. smegmatis, which may explain the potential role of additional RNase HII domain of RelZ which is not the case with M. tuberculosis.
Biofilm
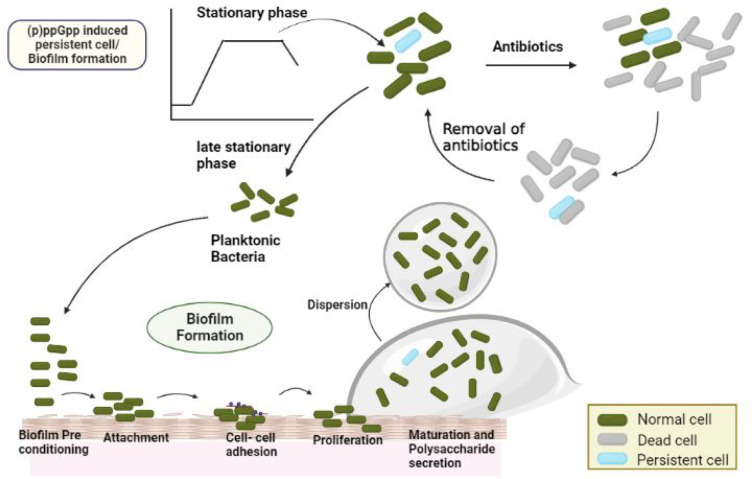
Bacteria use various adaptive strategies, including (p)ppGpp-mediated biofilm formation, to survive stressful external conditions. The biofilm is an aggregation of microbial cells encased in an extracellular polymeric matrix with the surface attachment57,58 (Figure 3). Microorganisms in the form of colonies survive better than those in the planktonic form. The formation of biofilms protects microorganisms from hostile environmental conditions such as heat shock, nutrient deprivation, antibiotics, and other environmental stresses.57,59 The alarmone (p)ppGpp regulates biofilm formation in both Gram-negative bacteria, like Vibrio cholerae and E. coli, and Gram-positive bacteria, such as Streptococcus mutans and Enterococcus faecalis. The ability of bacteria to develop three-dimensionally stable, multicellular communities, known as biofilms, has been strongly linked with their survival in the human host.60,61
Figure 3.
(p)ppGpp enables biofilm formation and mediates bacterial persistence. Persistent cell survives lethal antibiotic treatment and replenish the population upon return of favorable conditions.
Gupta et al. and Syal et al. investigated defects in biofilm formation in rel mutants. They compared M. smegmatis mc2155 (the wild type/WT) and its isogenic variants relMsm (rel gene removed) and the rel complemented strain (rel comp).14 They found that the knockout strains where the rel gene was removed were defective in biofilm formation and exhibited altered surface properties. These phenotypes are directly correlated with various glycopeptidolipids present in the cell wall of M. smegmatis.14 It was found that the rel comp strain where rel gene was added in the knock out strain formed more biofilms than the WT strain; however, its primary adherence values were less than those of the WT.
The ΔrelMsm strain has reduced glycopeptidolipid (GPLs) levels in its cell wall compared to the parental mc2155 strain. The stringent response may regulate biofilm formation and colony morphology in M. smegmatis by regulating GPL synthesis.20,62−65 In addition, both relMtb and relMsm strains exhibit differential expression of several genes involved in the synthesis of cell envelopes.22,40,66 The typical thick and wrinkled surface appearance was missing in the ΔrelZ mutant in comparison to the wild-type strain. The double knockout (rel-relZ) shows the most robust inhibition of biofilm formation in M. smegmatis.47
As discussed, Weiss et al. described the role of RelMtb in both growth in culture and pathogenesis in mice. They studied a point mutation that specifically abolished (p)ppGpp synthesis by RelMtb and compromised biofilm formation in M. tuberculosis.36 ΔrelMsm strain has reduced sliding motility and possesses rough colony morphology. Together, the ΔrelMsm strain has been shown to be defective in biofilm formation, exhibit compromised sliding motility, and possess rough colony morphology.67
Persistence/Long-Term Survival
The persistence is a pervasive phenomenon adopted by most bacteria involving formation of a dormant or a slow-growing state that transiently leads to the multidrug tolerant phenotype.5 Soon after the discovery of penicillin, Bigger reported the existence of a small persistent surviving fraction of Staphylococcus aureus that survived the treatment with penicillin.68 The persistence can be vividly observed in M. tuberculosis, as well. The routine treatment for these stubborn bacteria is a combination of drugs such as rifampicin, isoniazid, pyrazinamide, and ethambutol for a minimum period of six months. It is crucial to study persistence, and its underlying mechanisms, as treating M. tuberculosis has become a difficult task due to a commonly observed relapse of infection.69 The persistence has been implicated in recurrent and chronic infections and is a bet-hedging strategy that ensures survival under fluctuating hostile environmental conditions.68 The antibiotics affect bacteria in two phases: a rapid killing phase in which most bacteria are killed and another stagnant phase in which few bacteria persist. Without antibiotics, these bacterial persisters again began multiplying in the host, leading to delayed clearance and recurrent bacterial infections.69 In the case of M. tuberculosis, to survive in hostile conditions, they rapidly downregulate ribosome biogenesis to match the declining translational need. This response requires coordinated transcriptional regulation of all ribosome components and entry into a dormancy state. It is a global regulatory mechanism in which transcription of stable RNAs is inhibited, in part by the production of the hyperphosphorylated guanine nucleotides (p)ppGpp in mycobacteria.70 The details of the mechanism are still under investigation. (p)ppGpp aids in elevating the stationary phase sigma factor σS, which increases persister formation due to its role in stress-related pathways. Their absence often leads to impaired ability to survive antibiotic insult suggesting a crucial role of (p)ppGpp in antibiotic tolerance/resistance.22
Further, a few environmental cues (like stress in phagocytic vacuoles) can potentially trigger persister formation via (p)ppGpp.22,69 Evidently, persistence of M. tuberculosis within host granulomas is directly linked with the RelMtb gene function.71 In addition, (p)ppGpp promotes polyP accumulation, which results in an overhaul of M. tuberculosis metabolism that arrests growth and facilitates its persistence. In turn, polyP promotes RelMtb expression and the production of (p)ppGpp through a signaling cascade involving the two-component system MprAB and the alternative σ-factor E (σE).72 Together, PolyP accumulation and stringent response are linked to M. tuberculosis persistence. The potential of M. tuberculosis to persist in the host is attributed to the formation of persister cells that exhibit decreased replication, altered metabolism, increased antibiotic tolerance, and increased stress resistance.26,19,10,39,22
The Δrel strain of M. tuberculosis was incompetent to persist in mice22 and unable to form tubercle lesions in guinea pigs,73 demonstrating the importance of (p)ppGpp in virulence and the long-term survival of mycobacteria. Syal et al. also observed significant inhibition of long-term survival in the presence of the (p)ppGpp inhibitors (AC and AB-ppGpp analogs) in comparison to the wild-type untreated controls in M. smegmatis (Table 1). Both AC and AB compounds showed considerable inhibition.74 Δrel mutant showed no further inhibition of long-term survival in the presence of these compounds, indicating that Rel was the target. Their compounds targeted Rel and inhibited (p)ppGpp synthesis, thereby affecting the long-term survival in M. smegmatis. Vitamin C’s ability to prevent the production of (p)ppGpp raises another possibility of discovering a viable treatment. This argument is further supported by the correlation between the inhibition of (p)ppGpp synthesis and the impairment in long-term survival. As Vitamin C affects numerous pathways,75,76 other pathways should also be studied for abnormalities in long-term survival.74,77 A cellular (p)ppGpp concentration can be fine-tuned through Rel in response to nutrients or other stresses, ensuring survival in growth-limited conditions for M. tuberculosis.22 Dahl et al. also reported that the role of relMsm would contribute to M. smegmatis survival under prolonged nutrient or oxygen starvation conditions.10
Table 1. Compounds That Inhibit a Bacterial Stringent Response in Mycobacteria.
| compound name | species | target | mode of action | author and year |
|---|---|---|---|---|
| DMNP | M. smegmatis | RelMsm and RelZ | Binds to adjacent of GTP/GDP active sites (H177) in the catalytic domain of proteins, and inhibits GTP/GDP binding. | Tkachenko (2021)51 |
| X9 | M. tuberculosis | RelMtb | Binds to active site of protein but it is unknown whether the compound binds to amino acid D265 and/or E325, which are essential for ppGpp synthesis. | Dutta (2019)52 |
| Pyrazinoic acid | M. tuberculosis | RelMtb | Results in the conformational changes of protein by binding to Asp67 of Rv2783 and inhibit its catalytic activities. | Njire (2017)53 |
| Acetylated and acetylated (AC) benzoylated Relacin (AB) compound | M. smegmatis, M. tuberculosis | RelMsm | It may inhibit by binding to active site or CTD of Rel protein as suggested by enzyme kinetics resulting in impaired biofilm formation and the emergence of elongated cells. | Syal (2017)54 |
| Vitamin C | M. smegmatis | RelMsm | GTP analogue; suggested to bind to the active site of Rel enzyme and inhibit its catalytic activities. | Syal (2017)55 |
| NSC9037 and NSC35676 | M. tuberculosis | PPK2 | Mechanism is unclear. | Singh (2016)56 |
Role of Rel and RelZ in Antibiotic Tolerance
Antibiotic tolerance is the ability of bacteria to withstand the presence of antibiotics (up to a specific concentration), thus contributing toward antibiotic treatment failure. Also, it acts as a precursor for antibiotic resistance. It describes the ability of bacteria to survive by slowing down metabolic cascades, and it is often referred to as phenotypic resistance.78,79 Antibiotic tolerance and persistence may lead to the emergence of antibiotic resistance as persistence provides a viable group of cells with time in which the resistant mutants can emerge by de novo chromosomal mutations or horizontal gene transfer.80,81 A recent de novo study demonstrated that M. tuberculosis cells exposed to lethal concentrations of antibiotics would generate antibiotic tolerance, and such cells could become resistant to the same antibiotics.82 Most antibiotics target active metabolic processes including replication and translation, and decreased growth rate may leads to multidrug tolerance.83 The stringent response has also been implicated in the downregulation of the genes required for growth in bacteria, including ribosome and cell wall synthesis.84,85 The inhibition of growth by stringent response indirectly protects the cells from stress and antibiotics that usually target pathways involved in growth and metabolism. In most species, stringent response upregulates genes like stress-specific transcription factors, and heat shock proteins which help cells to survive during stress conditions.86,87M. tuberculosis lacking Rel has been shown to lose its ability to become quiescent. Targeting Rel may enhance the capacity of isoniazid drug to target M. tuberculosis by limiting formation of persister cells in infected mice and starvation conditions.52 Our previous work has showed that targeting stringent response is a promising approach to overcome persistence, and it may potentially shorten the tuberculosis treatment.54 In M. smegmatis, the high throughput microarray technique has been used to study the relationship between a stringent response and antibiotic tolerance. It has been shown that the ΔrelMSm strain outgrew the mc2155 wild-type strain in the presence of multiple classes of antibiotics which is surprising and may be due to the inability to detect hostile conditions and respond to stress.88 The antibiotic sensitivity for a knockout strain of relZ was performed by Murdeshwar et al. in M. smegmatis.89 The ΔrelZ strain has shown sensitivity to most antibiotics, including rifampin, ofloxacin, and bleomycin, which target RNA polymerase and DNA gyrase compared to the ΔrelMsm strain. Similarly, a double knockout strain ΔrelΔrelZMSm has been shown to be even more sensitive to antibiotics than the ΔrelZ strain.90 Deletion of RelZ induces slow growth, which confers protection against antibiotics. It has been hypothesized that knockout strains are sensitive to antibiotics because of the alteration in the influx/efflux of antibiotics due to defective (p)ppGpp homeostasis. Due to the presence of dual bifunctional (p)ppGpp synthetase, the relationship between antibiotic tolerance and (p)ppGpp levels appears more complex in mycobacteria in comparison to the other bacteria.91
Toxin–Antitoxin Systems in Mycobacteria—Are They Linked to the Stringent Response?
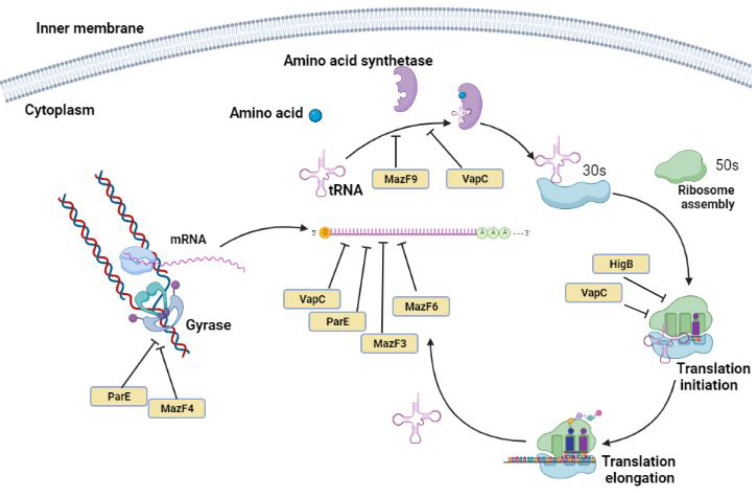
Toxin–antitoxin (TA) systems are small genetic modules initially discovered on bacterial plasmids. It is well-known that TA systems are widely distributed in prokaryotic genomes and have been proposed to play a crucial role in several cellular functions, including persistence. TA systems consist of two gene operons encoding toxins and antitoxins. Toxin (protein or RNA) targets various cellular functions and inhibits growth, whereas antitoxin (protein or RNA) counters and neutralizes toxin’s effects.92 The M. tuberculosis genome wide scan suggest the existence of nearly 88 putative TA modules, whereas nontubercular mycobacteria such as M. ulcerans, M. smegmatis, or M. marinum possess less than five TA module.93,94 A high number of TA systems present in M. tuberculosis in comparison to their nonpathogenic counterparts suggests that TA systems may play a crucial role in its survival and pathogenesis. TA modules may also detect and integrate environmental stimuli during bacterial infection, enabling mycobacteria to develop or maintain the dormant state of latent tuberculosis.95,96 The TA systems are involved in different processes such as bacterial persistence, biofilm formation, control of stress response, and defense against phage infection.97 Bioinformatics and phylogenomic studies have revealed that the genome of M. tuberculosis predominantly encodes a type II TA system, which includes 51 VapBC, 10 MazEF, 3 HigBA, 2 RelBE, 1 YefM/YoeB, and 2 ParDE family members. Here, the VapBC (virulence associated protein) TA modules are the most abundant loci encoded by the genome of M. tuberculosis.98In vitro experiments suggest that most of the VapC (VapC1, VapC2, VapC5, VapC11, VapC20, and VapC29) from M. tuberculosis exhibit ribonuclease activity.99 VapBC TA loci mediate functions in different stages of infection and persistence. M. tuberculosis genome encodes for 10 MazEF TA loci, and overexpression of MazF3, MazF6, and MazF9 has been shown to arrest growth.100M. tuberculosis also encodes for two ParDE, three RelBE, and three HigBA systems.94 ParDE/RelBE TA superfamily inhibits translation by cleaving RNA. Here, ParE1 has been shown to be necessary for the survival of mycobacteria in activated macrophages. ParE homologues interact with DNA gyrase and block replication in cells. HigB toxins bind to 50S subunits of 70S ribosomes and target translation by cleaving AAA sequences of mRNAs101 (Figure 4). HigA1 and HigA2 antitoxin neutralizes the activity of HigB toxin.102 Together, TA systems augment survival by regulating growth rate, metabolism, and cell division and inducing slow growth phenotype. It is plausible that the TA system works in synergy with a stringent response to induce persister cell formation in mycobacteria. Both ppGpp and the TA systems are vital for persistence. However, the direct evidence that may associate TA systems with ppGpp is yet to be reported in mycobacteria.
Figure 4.
Schematic representation of the regulation of the TA system influencing vital processes in mycobacteria. The pathways associated with replication (left) and translation (right) are targeted predominantly by free active toxins in stress conditions in mycobacteria.
GTP Homeostasis
The purine nucleotides are key molecules involved in DNA replication, energy processes, and different metabolic cascades. GTP levels across species are critical for fitness, and any dysregulation may lead to genomic instability.103 The GTP homeostasis has not been studied in detail in mycobacteria but is explored well in different Gram-positive and Gram-negative bacteria. GTP is a major contributor to metabolism and is essential for multiple cellular processes.104 In some cases, the levels of GTP lie within a limited, narrow range, like in some Gram-positive bacteria (Bacillus subtilis), and excess GTP is also severely detrimental to cell growth and survival.103 In contrast, reduced GTP levels lower transcription of rRNA and trigger sporulation in Gram-positive bacteria like B. subtilis resulting in slow growth.104 Interestingly, cell growth is inhibited at high GTP levels in E. coli as well.103 (p)ppGpp levels in a cell are associated with moderation of GTP levels. In B. subtilis, under limiting nutrition, cells produce (p)ppGpp and inhibit GTP production by regulating several enzymes involved in its synthesis pathway such as Gmk.105B. subtilis (p)ppGpp-null strains, when exposed to an amino acid-depleted medium for 10 min, resulted in cell death, and it has been partly attributed to the increased GTP concentrations. Interestingly, inhibiting (p)ppGpp hydrolysis has an effect not only on (p)ppGpp levels but also on ATP and GTP levels within bacteria.104 GTP homeostasis has been shown to be disrupted in (p)ppGpp0 cells with GTP levels uncontrollably rising to 10 mM or higher, resulting in toxicity and cell death. This dysregulation shows that (p)ppGpp is a master regulator of GTP homeostasis and not only a contributor.104 GTP levels reduce upon induction of (p)ppGpp synthesis, which could be due to the utilization of GTP as a substrate for pppGpp synthesis. In addition, the GTP biosynthesis enzymes IMP dehydrogenase-GuaB and guanylate kinase-Gmk are both inhibited by (p)ppGpp.104,106 Although B. subtilis RNAP lacks ppGpp binding motifs, it mounts a stringent response via an indirect mechanism that alters GTP homeostasis. In B. subtilis, GTP is one of the initiating nucleotides, and increasing (p)ppGpp synthesis decreases the GTP pool, modulating rRNA promoter activity.107 Gross GTP dysregulation occurs without (p)ppGpp, suggesting a crucial housekeeping role for (p)ppGpp. During amino acid deprivation, (p)ppGpp is made from GTP/GDP and ATP, and its synthesis coincides with a decrease in cellular GTP levels. Kriel et al. demonstrated that (p)ppGpp lowers GTP levels during starvation by directly repressing the activity of two enzymes, Gmk and HprT. This (p)ppGpp-mediated control also stops GTP from rising to toxic high levels even in the absence of starvation.104 Evidently, different modes regulate GTP homeostasis. First, the most understood mechanisms involve regulation of the de novo pathway, where (p)ppGpp prevents de novo and salvages GTP biosynthesis. The second mode is direct transcriptional feedback loop of the control genes responsible for GTP production.108,109 The third mode includes the maintenance of GTP homeostasis; (p)ppGpp is an off-pathway product made from GTP, and it shows a negative feedback loop. (p)ppGpp enables GTP homeostasis by (1) buffering GTP against fluctuations at lower (p)ppGpp levels and (2) modulating high GTP levels to stabilize metabolism in response to external stress signal.104 It is unclear how excessive GTP levels cause cell death; however, (p)ppGpp’s mediated regulation of GTP is essential for survival of B. subtilis.103 Together, (p)ppGpp regulates GTP homeostasis in response to extrinsic stress and intrinsic cell status, thereby preventing death-by-GTP and preserving metabolic stability in B. subtilis. It also suggests an important and pleiotropic role for (p)ppGpp as a global player in the metabolome. Here, interconversion of GTP and (p)ppGpp may fine-tune GTP homeostasis and it may be a common strategy employed by many bacteria including mycobacteria.
Discussion
Two decades back, the (p)ppGpp-mediated stringent response was discovered in mycobacteria. The (p)ppGpp family has now emerged as the master regulator of stress response that helps mycobacteria survive hostile conditions such as the presence of antibiotics. Earlier, Rel, a bifunctional enzyme capable of synthesis and hydrolysis of (p)ppGpp, was primarily held responsible for maintaining levels of (p)ppGpp. With an aim to understand the function of Rel, it was knocked out in M. smegmatis. Surprisingly, Rel mutant still had detectable (p)ppGpp, which led to the discovery of RelZ. Like Rel, RelZ also possesses dual activities including (pp)pGpp synthetase and RNase HII both in a single polypeptide chain. Here the function of the RNase HII domain coupled with (pp)pGpp synthesis is still not completely understood and is under investigation. Both Rel and RelZ have been implicated in biofilm formation, persistence, antibiotic tolerance, GTP homeostasis, and virulence. Together, these key enzymes control the cell’s defense network, allowing it to survive hostile conditions. RelZ also the ability to synthesize pGpp.23 The function of pGpp remains unclear.47,48 Interestingly, double knockout of Rel and RelZ in M. smegmatis still showed detectable (p)ppGpp (unpublished data). The family of alarmone molecules has now been broadened to include a variety of molecules such as pppGpp, ppGpp, pGpp, and (pp)pApp as distinct members. These molecules may aid in fine-tuning stress responses under hostile conditions. How are these molecules interconverted? Do these molecules work synergistically or compete with each other? How they perform different functions or augment each other’s function is unclear. Ahmad et al. discovered the Tas1 enzyme in Pseudomonas aeruginosa, that produces (p)ppApp and not (p)ppGpp.110 Like (p)ppGpp, excessive amounts of (p)ppApp is also toxic and can be reversed by the corresponding hydrolase;111 however, function remains elusive. (pp)pGpp family mediated biofilms help bacteria persist against antibiotic treatment and confer protection from the host immune system and other environmental disturbances.57,112 Current antibiotics are becoming increasingly ineffective against the slow-growing persister state that may result in revival of the infection. In this regard, (pp)pGpp synthetase inhibitors constitute a new line of antimicrobial agents that can inhibit the persister cell formation and block phenotypes such as biofilm formation and long-term survival. The possibility of combining (pp)pGpp synthetase inhibitors with antibiotics and establishing potential synergies with an aim to reduce the duration of the antitubercular antibiotic regimen should be further investigated. Such strategies will place us one step closer to treating people more effectively while concomitantly preventing the emergence of multidrug resistance in M. tuberculosis.
Acknowledgments
KS acknowledges DBT, Government of India for RLS fellowship and Research funding. Authors acknowledge BITS-Pilani-Hyderabad for additional CRG and RIG grants, and for fellowship to NRS. YD acknowledges CSIR-UGC, Delhi for Junior Research Fellowship. Authors acknowledge Biorender and Canva online web tools which were used to prepare schematics. The authors of this manuscript apologize to the colleagues whose research papers are not cited because of space limitations.
The authors declare no competing financial interest.
References
- Cashel M., Gentry D. R., Hernandez V. J., Vinella D.. “The stringent response”, in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, eds Neidhardt F. C.; Curtiss III R.; Ingraham J. L.; Lin E. E. C.; Low K. B.; Magasanik B., et al. ASM Press: Washington, DC, 1996, 1458–1496.
- Cashel M.; Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 1969, 221, 838–841. 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- Dalebroux Z.D.; Svensson S.L.; Gaynor E.C.; Swanson M.S. ppGpp conjures bacterial virulence. Microbiology Molecular Biology Reviews 2010, 74 (2), 171–199. 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte C. C.; Crosson S. Bacterial lifestyle shapes stringent response activation. Trends in microbiology 2013, 21 (4), 174–180. 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca A. O.; Colomer-Winter C.; Lemos J. A. Many means to a common end: the intricacies of (p) ppGpp metabolism and its control of bacterial homeostasis. Journal of bacteriology 2015, 197 (7), 1146–1156. 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs J. K.; Boraston A. B. (p) ppGpp and the stringent response: an emerging threat to antibiotic therapy. ACS infectious diseases 2019, 5 (9), 1505–1517. 10.1021/acsinfecdis.9b00204. [DOI] [PubMed] [Google Scholar]
- Syal K.; RS N.; Reddy M V N J. The extended (p) ppGpp family: New dimensions in Stress response. Current Research in Microbial Sciences 2021, 2, 100052. 10.1016/j.crmicr.2021.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Salazar C.; Calero P.; Espinosa-Portero R.; Jiménez-Fernández A.; Wirebrand L.; Velasco-Domínguez M. G.; López-Sánchez A.; Shingler V.; Govantes F. The stringent response promotes biofilm dispersal in Pseudomonas putida. Sci. Rep. 2017, 7 (1), 1–13. 10.1038/s41598-017-18518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K.; Chatterji D. Differential binding of ppGpp and pppGpp to E. coli RNA polymerase: photo-labeling and mass spectral studies. Genes to Cells 2015, 20 (12), 1006–1016. 10.1111/gtc.12304. [DOI] [PubMed] [Google Scholar]
- Dahl J. L.; Arora K.; Boshoff H. I.; Whiteford D. C.; Pacheco S. A.; Walsh O. J.; Lau-Bonilla D.; Davis W. B.; Garza A. G. The relA homolog of Mycobacterium smegmatis affects cell appearance, viability, and gene expression. Journal of bacteriology 2005, 187 (7), 2439–2447. 10.1128/JB.187.7.2439-2447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V.; Atkinson G. C.; Murakami K. S.; Tenson T.; Gerdes K. Recent functional insights into the role of (p) ppGpp in bacterial physiology. Nature Reviews Microbiology 2015, 13 (5), 298–309. 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson G. C.; Tenson T.; Hauryliuk V. The RelA/SpoT Homolog (RSH) Superfamily: Distribution and Functional Evolution of ppGpp Synthetases and Hydrolases across the Tree of Life. PLoS One 2011, 6 (8), e23479 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L.; Keck J. L. Magic spots cast a spell on DNA primase. Cell 2007, 128 (5), 823–824. 10.1016/j.cell.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Syal K.; Joshi H.; Chatterji D.; Jain V. Novel pppGpp binding site at the C-terminal region of the Rel enzyme from Mycobacterium smegmatis. FEBS journal 2015, 282 (19), 3773–3785. 10.1111/febs.13373. [DOI] [PubMed] [Google Scholar]
- Wexselblatt E.; Oppenheimer-Shaanan Y.; Kaspy I.; London N.; Schueler-Furman O.; Yavin E.; Glaser G.; Katzhendler J.; Ben-Yehuda S. Relacin, a novel antibacterial agent targeting the stringent response. PloS Pathogens 2012, 8 (9), e1002925 10.1371/journal.ppat.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatnaparat T.; Li Z.; Korban S. S.; Zhao Y. The bacterial alarmone (p) ppGpp is required for virulence and controls cell size and survival of P seudomonas syringae on plants. Environmental microbiology 2015, 17 (11), 4253–4270. 10.1111/1462-2920.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley C. L.; Zhang X.; Fortney K. R.; Ellinger S.; Johnson P.; Baker B.; Liu Y.; Janowicz D. M.; Katz B. P.; Munson R. S. Jr DksA and (p) ppGpp have unique and overlapping contributions to Haemophilus ducreyi pathogenesis in humans. Infection immunity 2015, 83 (8), 3281–3292. 10.1128/IAI.00692-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha A. K.; Mukherjee T. K.; Chatterji D. High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infection immunity 2000, 68 (7), 4084–4091. 10.1128/IAI.68.7.4084-4091.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primm T. P.; Andersen S. J.; Mizrahi V.; Avarbock D.; Rubin H.; Barry C. E. III The stringent response of Mycobacterium tuberculosis is required for long-term survival. Journal of bacteriology 2000, 182 (17), 4889–4898. 10.1128/JB.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K. R.; Arora G.; Mattoo A.; Sajid A. Stringent Response in Mycobacteria: From Biology to Therapeutic Potential. Pathogens (Basel, Switzerland) 2021, 10 (11), 1417. 10.3390/pathogens10111417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai M.; Behr M.A.; Dowdy D.; Dheda K.; Divangahi M.; Boehme C.C.; Ginsberg A.; Swaminathan S.; Spigelman M.; Getahun H. Tuberculosis (Primer). Nature Reviews: Disease Primers 2016, 2 (1), 1. 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- Dahl J. L.; Kraus C. N.; Boshoff H. I.; Doan B.; Foley K.; Avarbock D.; Kaplan G.; Mizrahi V.; Rubin H.; Barry C. E. III The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (17), 10026–10031. 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdeshwar M. S.; Chatterji D. MS_RHII-RSD, a dual-function RNase HII-(p) ppGpp synthetase from Mycobacterium smegmatis. Journal of bacteriology 2012, 194 (15), 4003–4014. 10.1128/JB.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimmy S.; Saha C. K.; Kurata T.; Stavropoulos C.; Oliveira S. R. A.; Koh A.; Cepauskas A.; Takada H.; Rejman D.; Tenson T. A widespread toxin– antitoxin system exploiting growth control via alarmone signaling. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (19), 10500–10510. 10.1073/pnas.1916617117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V.; Saleem-Batcha R.; China A.; Chatterji D. Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 2006, 15 (6), 1449–1464. 10.1110/ps.062117006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusa J.; Zhu D. X.; Stallings C. L. J. The stringent response and Mycobacterium tuberculosis pathogenesis. Pathogens disease 2018, 76 (5), fty054. 10.1093/femspd/fty054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarbock A.; Avarbock D.; Teh J.-S.; Buckstein M.; Wang Z.-m.; Rubin H. Functional regulation of the opposing (p) ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 2005, 44 (29), 9913–9923. 10.1021/bi0505316. [DOI] [PubMed] [Google Scholar]
- Avarbock D.; Salem J.; Li L.-s.; Wang Z.-m.; Rubin H. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 1999, 233 (1–2), 261–269. 10.1016/S0378-1119(99)00114-6. [DOI] [PubMed] [Google Scholar]
- Ekal L.; Ganesh B.; Joshi H.; Lama D.; Jain V. Evidence of a conserved intrinsically disordered region in the C-terminus of the stringent response protein Rel from mycobacteria. FEBS letters 2014, 588 (9), 1839–1849. 10.1016/j.febslet.2014.03.048. [DOI] [PubMed] [Google Scholar]
- Singal B.; Balakrishna A. M.; Nartey W.; Manimekalai M. S. S.; Jeyakanthan J.; Grüber G. Crystallographic and solution structure of the N-terminal domain of the Rel protein from Mycobacterium tuberculosis. FEBS letters 2017, 591 (15), 2323–2337. 10.1002/1873-3468.12739. [DOI] [PubMed] [Google Scholar]
- Aravind L.; Koonin E. V. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends in biochemical sciences 1998, 23 (12), 469–472. 10.1016/S0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- Hogg T.; Mechold U.; Malke H.; Cashel M.; Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p) ppGpp metabolism during the stringent response. Cell 2004, 117 (1), 57–68. 10.1016/S0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- Sajish M.; Kalayil S.; Verma S. K.; Nandicoori V. K.; Prakash B. The significance of EXDD and RXKD motif conservation in Rel proteins. J. Biol. Chem. 2009, 284 (14), 9115–9123. 10.1074/jbc.M807187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajish M.; Tiwari D.; Rananaware D.; Nandicoori V. K.; Prakash B. A charge reversal differentiates (p) ppGpp synthesis by monofunctional and bifunctional Rel proteins. J. Biol. Chem. 2007, 282 (48), 34977–34983. 10.1074/jbc.M704828200. [DOI] [PubMed] [Google Scholar]
- Jain V.; Sujatha S.; Ojha A. K.; Chatterji D. Identification and characterization of rel promoter element of Mycobacterium tuberculosis. Gene 2005, 351, 149–157. 10.1016/j.gene.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Weiss L. A.; Stallings C. L. Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p) ppGpp. Journal of bacteriology 2013, 195 (24), 5629–5638. 10.1128/JB.00759-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarbock D.; Avarbock A.; Rubin H. Differential Regulation of Opposing RelMtb Activities by the Aminoacylation State of a tRNA⊙ Ribosome⊙ mRNA⊙ RelMtb Complex. Biochemistry 2000, 39 (38), 11640–11648. 10.1021/bi001256k. [DOI] [PubMed] [Google Scholar]
- Jain V.; Saleem-Batcha R.; Chatterji D. Synthesis and hydrolysis of pppGpp in mycobacteria: a ligand mediated conformational switch in Rel. Biophys. Chem. 2007, 127 (1–2), 41–50. 10.1016/j.bpc.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Mathew R.; Ojha A. K.; Karande A. A.; Chatterji D. Deletion of the rel gene in Mycobacterium smegmatis reduces its stationary phase survival without altering the cell-surface associated properties. Curr. Sci. 2004, 149–153. [Google Scholar]
- Gupta K. R.; Baloni P.; Indi S. S.; Chatterji D. Regulation of growth, cell shape, cell division, and gene expression by second messengers (p) ppGpp and cyclic Di-GMP in Mycobacterium smegmatis. Journal of bacteriology 2016, 198 (9), 1414–1422. 10.1128/JB.00126-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio F.; Battah B.; Palmieri V.; Petrone L.; Corrente F.; Salustri A.; Palucci I.; Bellesi S.; Papi M.; Rubino S. PE_PGRS3 of Mycobacterium tuberculosis is specifically expressed at low phosphate concentration, and its arginine-rich C-terminal domain mediates adhesion and persistence in host tissues when expressed in Mycobacterium smegmatis. Cellular Microbiology 2018, 20 (12), e12952 10.1111/cmi.12952. [DOI] [PubMed] [Google Scholar]
- Post F. A.; Manca C.; Neyrolles O.; Ryffel B.; Young D. B.; Kaplan G. Mycobacterium tuberculosis 19-kilodalton lipoprotein inhibits Mycobacterium smegmatis-induced cytokine production by human macrophages in vitro. Infection immunity 2001, 69 (3), 1433–1439. 10.1128/IAI.69.3.1433-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio F.; Salustri A.; Battah B.; Palucci I.; Marchionni F.; Bellesi S.; Palmieri V.; Papi M.; Kramarska E.; Sanguinetti M. PE_PGRS3 ensures provision of the vital phospholipids cardiolipin and phosphatidylinositols by promoting the interaction between M. tuberculosis and host cells. Virulence 2021, 12 (1), 868–884. 10.1080/21505594.2021.1897247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Majumdar S.; Treen R.; Sharma M. R.; Corro J.; Gamper H. B.; Manjari S. R.; Prusa J.; Banavali N. K.; Stallings C. L.; Hou Y. M.; Agrawal R. K.; Ojha A. K. Starvation sensing by mycobacterial RelA/SpoT homologue through constitutive surveillance of translation. Proc. Natl. Acad. Sci. U.S.A. 2023, 120 (22), e2302006120 10.1073/pnas.2302006120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Corro J. H.; Palmer C. D.; Ojha A. K. Progression from remodeling to hibernation of ribosomes in zinc-starved mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (32), 19528–19537. 10.1073/pnas.2013409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bag S.; Das B.; Dasgupta S.; Bhadra R. K. Mutational analysis of the (p) ppGpp synthetase activity of the Rel enzyme of Mycobacterium tuberculosis. Archives of microbiology 2014, 196, 575–588. 10.1007/s00203-014-0996-9. [DOI] [PubMed] [Google Scholar]
- Petchiappan A.; Naik S. Y.; Chatterji D. RelZ-mediated stress response in Mycobacterium smegmatis: pGpp synthesis and its regulation. Journal of bacteriology 2020, 202 (2), e00444–19. 10.1128/JB.00444-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S.; Petchiappan A.; Singh A.; Bhatt A.; Chatterji D. R-loop induced stress response by second (p) ppGpp synthetase in Mycobacterium smegmatis: functional and domain interdependence. Molecular microbiology 2016, 102 (1), 168–182. 10.1111/mmi.13453. [DOI] [PubMed] [Google Scholar]
- Aguilera A.; García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 2012, 46, 115–124. 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Ross W.; Vrentas C. E.; Sanchez-Vazquez P.; Gaal T.; Gourse R. L. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Molecular cell 2013, 50 (3), 420–429. 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko A. G.; Kashevarova N. M.; Sidorov R. Y.; Nesterova L. Y.; Akhova A. V.; Tsyganov I. V.; Vaganov V. Y.; Shipilovskikh S. A.; Rubtsov A. E.; Malkov A. V. A synthetic diterpene analogue inhibits mycobacterial persistence and biofilm formation by targeting (p)ppGpp synthetases. Cell chemical biology 2021, 28 (10), 1420–1432. 10.1016/j.chembiol.2021.01.018. [DOI] [PubMed] [Google Scholar]
- Dutta N. K.; Klinkenberg L. G.; Vazquez M.-J.; Segura-Carro D.; Colmenarejo G.; Ramon F.; Rodriguez-Miquel B.; Mata-Cantero L.; Porras-De Francisco E.; Chuang Y.-M.; Rubin H.; Lee J. J.; Eoh H.; Bader J. S.; Perez-Herran E.; Mendoza-Losana A.; Karakousis P. C. Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Science Advances 2019, 5 (3), eaav2104. 10.1126/sciadv.aav2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njire M.; Wang N.; Wang B.; Tan Y.; Cai X.; Liu Y.; Mugweru J.; Guo J.; Hameed H. M. A.; Tan S.; Liu J.; Yew W. W.; Nuermberger E.; Lamichhane G.; Liu J.; Zhang T. Pyrazinoic Acid Inhibits a Bifunctional Enzyme in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2017, 61 (7), e00070–17. 10.1128/AAC.00070-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K.; Flentie K.; Bhardwaj N.; Maiti K.; Jayaraman N.; Stallings C. L.; Chatterji D. Synthetic (p)ppGpp Analogue Is an Inhibitor of Stringent Response in Mycobacteria. Antimicrob. Agents Chemother. 2017, 61 (6), e00443–17. 10.1128/AAC.00443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K.; Bhardwaj N.; Chatterji D. Vitamin C targets (p)ppGpp synthesis leading to stalling of long-term survival and biofilm formation in Mycobacterium smegmatis. FEMS Microbiol Lett. 2017, 364 (1), fnw282. 10.1093/femsle/fnw282. [DOI] [PubMed] [Google Scholar]
- Singh M.; Tiwari P.; Arora G.; Agarwal S.; Kidwai S.; Singh R. Establishing Virulence Associated Polyphosphate Kinase 2 as a drug target for Mycobacterium tuberculosis. Sci. Rep. 2016, 6, 26900. 10.1038/srep26900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddam A. D.; Zaslow S. J.; Wang Y.; Phillips K. S.; Silverman M. D.; Regan P. M.; Amarasinghe J. J. Characterization of Biofilm Formation by Mycobacterium chimaera on Medical Device Materials. Front Microbiol 2021, 11, 586657. 10.3389/fmicb.2020.586657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarakonda L.; RS N.; Bhalerao M; Syal K.. (p)ppGpp Mediated Biofilm Formation and Estimation on Chip. In Miniaturized Electrochemical Devices; CRC Press, 2023; p 20. [Google Scholar]
- Potrykus K.; Cashel M. (p) ppGpp: still magical?. Annu. Rev. Microbiol. 2008, 62, 35–51. 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Richards J. P.; Cai W.; Zill N. A.; Zhang W.; Ojha A. K. Adaptation of Mycobacterium tuberculosis to biofilm growth is genetically linked to drug tolerance. Antimicrobial agents chemotherapy 2019, 63 (11), e01213–19. 10.1128/AAC.01213-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K. Novel Method for Quantitative Estimation of Biofilms. Curr. Microbiol. 2017, 74 (10), 1194–1199. 10.1007/s00284-017-1304-0. [DOI] [PubMed] [Google Scholar]
- Maiti K.; Syal K.; Chatterji D.; Jayaraman N. Synthetic Arabinomannan Heptasaccharide Glycolipids Inhibit Biofilm Growth and Augment Isoniazid Effects in Mycobacterium smegmatis. Chembiochem 2017, 18 (19), 1959–1970. 10.1002/cbic.201700247. [DOI] [PubMed] [Google Scholar]
- Syal K.; Maiti K.; Naresh K.; Avaji P. G.; Chatterji D.; Jayaraman N. Synthetic arabinomannan glycolipids impede mycobacterial growth, sliding motility and biofilm structure. Glycoconj J. 2016, 33 (5), 763–77. 10.1007/s10719-016-9670-6. [DOI] [PubMed] [Google Scholar]
- Syal K.; Maiti K.; Naresh K.; Chatterji D.; Jayaraman N. Synthetic glycolipids and (p)ppGpp analogs: development of inhibitors for mycobacterial growth, biofilm and stringent response. Adv. Exp. Med. Biol. 2015, 842, 309–27. 10.1007/978-3-319-11280-0_20. [DOI] [PubMed] [Google Scholar]
- Naresh K.; Avaji P. G.; Maiti K.; Bharati B. K.; Syal K.; Chatterji D.; Jayaraman N. Synthesis of beta-arabinofuranoside glycolipids, studies of their binding to surfactant protein-A and effect on sliding motilities of M. smegmatis. Glycoconj J. 2012, 29 (2–3), 107–18. 10.1007/s10719-012-9369-2. [DOI] [PubMed] [Google Scholar]
- Arora K.; Whiteford D. C.; Lau-Bonilla D.; Davitt C. M.; Dahl J. L. Inactivation of lsr2 results in a hypermotile phenotype in Mycobacterium smegmatis. Journal of bacteriology 2008, 190 (12), 4291–4300. 10.1128/JB.00023-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K. R.; Kasetty S.; Chatterji D. Novel functions of (p) ppGpp and cyclic di-GMP in mycobacterial physiology revealed by phenotype microarray analysis of wild-type and isogenic strains of Mycobacterium smegmatis. Appl. Environ. Microbiol. 2015, 81 (7), 2571–2578. 10.1128/AEM.03999-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar A.; De Piano C.; Gelman E.; McKinney J. D.; Dhar N. Elucidating the role of (p) ppGpp in mycobacterial persistence against antibiotics. IUBMB life 2018, 70 (9), 836–844. 10.1002/iub.1888. [DOI] [PubMed] [Google Scholar]
- Petchiappan A.; Chatterji D. Antibiotic resistance: current perspectives. ACS omega 2017, 2 (10), 7400–7409. 10.1021/acsomega.7b01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallings C. L.; Stephanou N. C.; Chu L.; Hochschild A.; Nickels B. E.; Glickman M. S. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 2009, 138 (1), 146–159. 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakousis P. C.; Yoshimatsu T.; Lamichhane G.; Woolwine S. C.; Nuermberger E. L.; Grosset J.; Bishai W. R. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. Journal of experimental medicine 2004, 200 (5), 647–657. 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureka K.; Dey S.; Datta P.; Singh A. K.; Dasgupta A.; Rodrigue S.; Basu J.; Kundu M. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Molecular microbiology 2007, 65 (2), 261–276. 10.1111/j.1365-2958.2007.05814.x. [DOI] [PubMed] [Google Scholar]
- Klinkenberg L. G.; Lee J.-H.; Bishai W. R.; Karakousis P. C. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. Journal of infectious diseases 2010, 202 (9), 1397–1404. 10.1086/656524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K.; Flentie K.; Bhardwaj N.; Maiti K.; Jayaraman N.; Stallings C.L.; Chatterji D. Synthetic (p)ppGpp Analogue Is an Inhibitor of Stringent Response in Mycobacteria. Antimicrob. Agents Chemother. 2017, 61 (6), 1. 10.1128/AAC.00443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S.; Syal K.; Bhattacharyya R.; Banerjee D. Vitamin deficiency and tuberculosis: need for urgent clinical trial for management of tuberculosis. J. Nutrition Health Food Sci. 2014, 2 (2), 1–6. 10.15226/jnhfs.2014.00118. [DOI] [Google Scholar]
- Rs N.; Reddy M.; Batra S.; Srivastava S. K.; Syal K. Vitamin C and its therapeutic potential in the management of COVID19. Clin Nutr ESPEN 2022, 50, 8–14. 10.1016/j.clnesp.2022.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K.; Chatterji D. Vitamin C: A Natural Inhibitor of Cell Wall Functions and Stress Response in Mycobacteria. Adv. Exp. Med. Biol. 2018, 1112, 321–332. 10.1007/978-981-13-3065-0_22. [DOI] [PubMed] [Google Scholar]
- Corona F.; Martinez J. L. Phenotypic Resistance to Antibiotics. Antibiotics (Basel, Switzerland) 2013, 2 (2), 237–55. 10.3390/antibiotics2020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Reisman I.; Ronin I.; Gefen O.; Braniss I.; Shoresh N.; Balaban N. Q. Antibiotic tolerance facilitates the evolution of resistance. Science (New York, N.Y.) 2017, 355 (6327), 826–830. 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- Cohen N. R.; Lobritz M. A.; Collins J. J. Microbial persistence and the road to drug resistance. Cell host & microbe 2013, 13 (6), 632–42. 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels E. M.; Michiels J. E.; Fauvart M.; Wenseleers T.; Van den Bergh B.; Michiels J. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME Journal 2019, 13 (5), 1239–1251. 10.1038/s41396-019-0344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J.; Swaminath S.; Nair R.R.; Jakkala K.; Pradhan A.; Ajitkumar P. De Novo Emergence of Genetically Resistant Mutants of Mycobacterium tuberculosis from the Persistence Phase Cells Formed against Antituberculosis Drugs In Vitro. Antimicrob. Agents Chemother. 2017, 61 (2), 1. 10.1128/AAC.01343-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer A.; Wolz C. Role of (p)ppGpp in antibiotic resistance, tolerance, persistence and survival in Firmicutes. microLife 2023, 4, 1. 10.1093/femsml/uqad009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler M. F.; Summers S. M.; Nguyen H. T.; Zacharia V. M.; Hightower G. A.; Smith J. T.; Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Molecular microbiology 2008, 68 (5), 1128–48. 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield W. B.; Zimmermann-Kogadeeva M.; Zimmermann M.; Barry N. A.; Goodman A. L. The Stringent Response Determines the Ability of a Commensal Bacterium to Survive Starvation and to Persist in the Gut. Cell host & microbe 2018, 24 (1), 120–132. 10.1016/j.chom.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss L. A.; Stallings C. L. Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. Journal of bacteriology 2013, 195 (24), 5629–38. 10.1128/JB.00759-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer H.; Beckert B.; Frese C. K.; Steinchen W.; Nuss A. M.; Beckstette M.; Hantke I.; Driller K.; Sudzinová P.; Krásný L.; Kaever V.; Dersch P.; Bange G.; Wilson D. N.; Turgay K. The alarmones (p)ppGpp are part of the heat shock response of Bacillus subtilis. PLoS genetics 2020, 16 (3), e1008275 10.1371/journal.pgen.1008275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K. R.; Kasetty S.; Chatterji D. Novel Functions of (p)ppGpp and Cyclic di-GMP in Mycobacterial Physiology Revealed by Phenotype Microarray Analysis of Wild-Type and Isogenic Strains of Mycobacterium smegmatis. Appl. Environ. Microbiol. 2015, 81 (7), 2571–2578. 10.1128/AEM.03999-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdeshwar M. S.; Chatterji D. MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. Journal of bacteriology 2012, 194 (15), 4003–14. 10.1128/JB.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petchiappan A.; Naik S. Y.; Chatterji D. RelZ-Mediated Stress Response in Mycobacterium smegmatis: pGpp Synthesis and Its Regulation. Journal of bacteriology 2020, 202 (2), e00444–19. 10.1128/JB.00444-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K.R.; Arora G.; Mattoo A.; Sajid A. Stringent Response in Mycobacteria: From Biology to Therapeutic Potential. Pathogens (Basel, Switzerland) 2021, 10 (11), 1417. 10.3390/pathogens10111417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayden R.A.; Dawson C.C.; Cummings J.E. Toxin–antitoxin systems and regulatory mechanisms in Mycobacterium tuberculosis. Pathogens and Disease 2018, 76 (4), 1. 10.1093/femspd/fty039. [DOI] [PubMed] [Google Scholar]
- Boldrin F.; Provvedi R.; Cioetto Mazzabo L.; Segafreddo G.; Manganelli R. Tolerance and Persistence to Drugs: A Main Challenge in the Fight Against Mycobacterium tuberculosis. Frontiers in Microbiology 2020, 11, 1. 10.3389/fmicb.2020.01924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage H. R.; Connolly L. E.; Cox J. S. Comprehensive Functional Analysis of Mycobacterium tuberculosis Toxin-Antitoxin Systems: Implications for Pathogenesis, Stress Responses, and Evolution. PLoS genetics 2009, 5 (12), e1000767 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayden R.A.; Dawson C.C.; Cummings J.E. Toxin-antitoxin systems and regulatory mechanisms in Mycobacterium tuberculosis. Pathog Dis 2018, 76 (4), 1. 10.1093/femspd/fty039. [DOI] [PubMed] [Google Scholar]
- Georgiades K.; Raoult D. Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PLoS One 2011, 6 (3), e17962 10.1371/journal.pone.0017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K.; Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annual review of microbiology 2012, 66, 103–23. 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- Arcus V. L.; McKenzie J. L.; Robson J.; Cook G. M. The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng. Des Sel 2011, 24 (1–2), 33–40. 10.1093/protein/gzq081. [DOI] [PubMed] [Google Scholar]
- Sala A.; Bordes P.; Genevaux P. Multiple Toxin-Antitoxin Systems in Mycobacterium tuberculosis. Toxins (Basel) 2014, 6 (3), 1002–1020. 10.3390/toxins6031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M. V.; Dawson C. C.; Crew R.; England K.; Slayden R. A. MazF6 toxin of Mycobacterium tuberculosis demonstrates antitoxin specificity and is coupled to regulation of cell growth by a Soj-like protein. BMC microbiology 2013, 13, 240. 10.1186/1471-2180-13-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.; Nayyar N.; Chawla M.; Sitaraman R.; Bhatnagar R.; Banerjee N. The Chromosomal parDE2 Toxin-Antitoxin System of Mycobacterium tuberculosis H37Rv: Genetic and Functional Characterization. Front Microbiol 2016, 7, 886. 10.3389/fmicb.2016.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuessler D. L.; Cortes T.; Fivian-Hughes A. S.; Lougheed K. E.; Harvey E.; Buxton R. S.; Davis E. O.; Young D. B. Induced ectopic expression of HigB toxin in Mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mRNAs and cleavage of tmRNA. Molecular microbiology 2013, 90 (1), 195–207. 10.1111/mmi.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Justel D.; Marcos-Alcalde I.; Abascal F.; Vidana N.; Gomez-Puertas P.; Jimenez A.; Revuelta J. L.; Buey R. M. Diversity of mechanisms to control bacterial GTP homeostasis by the mutually exclusive binding of adenine and guanine nucleotides to IMP dehydrogenase. Protein Sci. 2022, 31 (5), e4314. 10.1002/pro.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriel A.; Bittner A. N.; Kim S. H.; Liu K.; Tehranchi A. K.; Zou W. Y.; Rendon S.; Chen R.; Tu B. P.; Wang J. D. Direct regulation of GTP homeostasis by (p) ppGpp: a critical component of viability and stress resistance. Molecular cell 2012, 48 (2), 231–241. 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrilero L.; Urwin L.; Ward E.; Choudhury N. R.; Monk I. R.; Turner C. E.; Stinear T. P.; Corrigan R. M. Stringent Response-Mediated Control of GTP Homeostasis Is Required for Long-Term Viability of Staphylococcus aureus. Microbiology Spectrum 2023, 11, e00447–23. 10.1128/spectrum.00447-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving S. E.; Choudhury N. R.; Corrigan R. M. The stringent response and physiological roles of (pp) pGpp in bacteria. Nature Reviews Microbiology 2021, 19 (4), 256–271. 10.1038/s41579-020-00470-y. [DOI] [PubMed] [Google Scholar]
- Kriel A.; Brinsmade S. R.; Tse J. L.; Tehranchi A. K.; Bittner A. N.; Sonenshein A. L.; Wang J. D. GTP dysregulation in Bacillus subtilis cells lacking (p) ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. Journal of bacteriology 2014, 196 (1), 189–201. 10.1128/JB.00918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky B. R.; Sonenshein A. L. CodY-mediated regulation of guanosine uptake in Bacillus subtilis. Journal of bacteriology 2011, 193 (22), 6276–6287. 10.1128/JB.05899-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole D. J.; Zalkin H. Bacillus subtilis pur operon expression and regulation. Journal of bacteriology 1989, 171 (4), 2136–2141. 10.1128/jb.171.4.2136-2141.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S.; Wang B.; Walker M. D.; Tran H.-K. R.; Stogios P. J.; Savchenko A.; Grant R. A.; McArthur A. G.; Laub M. T.; Whitney J. C. An interbacterial toxin inhibits target cell growth by synthesizing (p) ppApp. Nature 2019, 575 (7784), 674–678. 10.1038/s41586-019-1735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T.; Kästle B.; Gratani F. L.; Goerke C.; Wolz C. Two small (p) ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. Journal of bacteriology 2014, 196 (4), 894–902. 10.1128/JB.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maifreni M.; Frigo F.; Bartolomeoli I.; Buiatti S.; Picon S.; Marino M. Bacterial biofilm as a possible source of contamination in the microbrewery environment. Food Control 2015, 50, 809–814. 10.1016/j.foodcont.2014.10.032. [DOI] [Google Scholar]