Abstract
Cryptococcus neoformans serotypes A and D are responsible for the overwhelming majority of infections in patients with AIDS. The genetic relationship between the serotypes is poorly understood, but there are significant differences in the epidemiology and clinical presentation of serotype A and D infections. We evaluated the genetic relationship between reference C. neoformans strains belonging to serotypes A and D by analyzing their URA5 sequences and restriction fragment length polymorphisms (RFLPs) with the C. neoformans repetitive element 1 (CNRE-1) probe. The results were compared to those previously obtained for isolates from Brazil and New York City by the same typing methods, and dendrograms were generated. Serotype A and D strains produced distinct RFLP patterns consistent with their separation into two major clusters in the dendrogram generated on the basis of RFLP data. Similarly, serotype A and D strains clustered independently on the basis of the nucleotide sequences of their URA5 genes. Pairwise comparisons revealed average numbers of nucleotide differences within serotypes A and D of 3.0 ± 1.7 and 7.2 ± 3.4, respectively (P < 0.0001), and between serotypes A and D of 41.9 ± 2.7. In summary, our results indicate phylogenetic differences between the two serotypes of C. neoformans var. neoformans and suggest that these serotypes could probably be considered different varieties of C. neoformans.
Cryptococcus neoformans is an encapsulated yeast that can cause severe meningoencephalitis in immunocompromised patients, especially those with AIDS. On the basis of the antigenic composition of the capsular polysaccharide, C. neoformans has been divided into four serotypes: serotypes A, B, C, and D. The four serotypes have been further divided into two varieties, C. neoformans var. neoformans (serotypes A and D) and C. neoformans var. gattii (serotypes B and C), on the basis of biochemical, morphological and genetic characteristics (22, 24). C. neoformans var. neoformans is primarily the etiologic agent of cryptococcosis in patients with AIDS, and serotype A comprises the overwhelming majority of clinical isolates. Infections due to strains belonging to serotype D are more prevalent in certain geographic areas, including France, Italy, and Denmark (2, 12, 24). In France, serotype D causes 21% of cases of cryptococcosis (11, 13). Serotype D infections are more likely than serotype A infections to occur in older patients, to result in skin involvement, and to be associated with corticosteroid therapy (11, 13).
The genetic relationship between isolates classified as serotypes A and D is uncertain. Guého et al. (17) found a relatively large phylogenetic distance between serotypes A and D by analysis of partial 26S rRNA sequences. Meyer et al. (23) used PCR fingerprinting analysis to demonstrate that strains of serotypes A and D could be distinguished from each other. Similarly, Varma and Kwon-Chung (31) reported the isolation of a DNA probe (UT-4p) that was able to discriminate between these serotypes. Brandt et al. (4) demonstrated that serotypes A and D could be distinguished by their multilocus enzyme electrophoresis profile. Serotypes A and D also show consistent differences in their electrophoretic karyotypes (26, 27). Hence, there is evidence that serotypes A and D belong to genetically distinct groups, but they remain within a single varietal classification because occasional strains of A and D isolates have been successfully mated (20). The uncertainty regarding the genetic relationship between serotype A and D strains is compounded by extensive genetic heterogeneity for strains grouped within a serotype.
The field of cryptococcal research is relatively small, and independent groups tend to work with different strains. This raises the concern that findings with a particular strain may not be generalizable. In this study we used two molecular typing techniques, DNA fingerprinting with the C. neoformans repetitive element 1 (CNRE-1) probe and analysis of the nucleotide sequence of the URA5 gene, to investigate the genetic relationship between reference C. neoformans strains belonging to serotypes A and D. For the purpose of this study we defined a reference strain as one that has been used in more than one study and/or more than one laboratory. Our results indicate phylogenetic differences between the two serotypes of C. neoformans var. neoformans and the genetic relationship between commonly used laboratory strains.
MATERIALS AND METHODS
C. neoformans strains.
A detailed list of the reference strains used in this study is presented in Table 1. Clinical and environmental isolates from Brazil (designated C and E followed by a number, respectively) were described previously (16). Clinical strains from New York (J isolates) were also reported earlier (6). All strains were kept in 50% glycerol in a freezer at −80°C and were grown overnight on Sabouraud’s broth at 30°C for DNA isolation.
TABLE 1.
Details for some C. neoformans var. neoformans strains used in this studya
| Strain | Source | Serotype | Original source or reference | GenBank accession no. (URA5 gene) |
|---|---|---|---|---|
| 3501 | ATCC (ATCC 34873) | D | K. J. Kwon-Chung, NIH | M34606 |
| 3502 | ATCC (ATCC 34874) | D | K. J. Kwon-Chung, NIH | AF032430 |
| 24067 | ATCC | D | Clinical isolate (32) | AF032431 |
| 24064 | ATCC | A | Clinical isolate (32) | AF032432 |
| 145 | S. Levitz (BU) | A | Clinical isolate | AF032433 |
| 184 | J. Murphy (UOklahoma) | A | Clinical isolate (25) | AF032434 |
| H99 | J. Perfect, DUMC | A | Clinical isolate from a lymphoma patient | AF032436 |
| SB4 | E. D. Spitzer, SUNY | A | Clinical isolate from an AIDS patient (30) | AF032435 |
| J9 | A. Casadevall (AECOM) | D | Clinical isolate from an AIDS patient (30) | —b |
| J11 | A. Casadevall (AECOM) | A | Clinical isolate from an AIDS patient (30) | —c |
| J21 | A. Casadevall (AECOM) | D | Clinical isolate from an AIDS patient (10) | L38585 |
| J22 | A. Casadevall (AECOM) | D | Clinical isolate from an AIDS patient (10) | —d |
Abbreviations: ATCC, American Type Culture Collection (Rockville, Md.); NIH, National Institutes of Health (Bethesda, Md.); BU, Boston University (Boston, Mass.); UOklahoma, University of Oklahoma (Oklahoma City); DUMC, Duke University Medical Center (Durham, N.C.); SUNY, State University of New York (Stony Brook); AECOM, Albert Einstein College of Medicine (Bronx, N.Y.).
—, the URA5 sequence is the same as that of strain 24067.
—, the URA5 sequence is the same as that of strain H99.
—, the URA5 sequence same as that of strain J21.
Serotyping.
The serotype classifications for the reference strains are listed in Table 1. Recent isolates from Brazil (isolates C5, C7, C24, C25, C31, C33, RJ1, RJ2, E3, E4, E5, E6, E9, and E12) and New York (isolates J15, J17, J19, J24, J25, and J26) were serotyped by the slide agglutination test with sera containing cryptococcal antigen factors 1, 5, 6, 7, and 8 (Iatron Laboratories, Inc., Tokyo, Japan). On the basis of the patterns of agglutination, the results were interpreted as follows: serotypes A, B, C, D, and AD reacted with antigen factors 1, 7; 1, 5; 1, 6; 1, 8; and 1, 7, 8, respectively.
The reactivity of monoclonal antibody (MAb) 13F1 with the recent isolates from Brazil and New York was also examined by indirect immunofluorescence (IF). Previously, we reported that IF with MAb 13F1 discriminates between most serotype A and D strains by producing annular and punctate binding patterns, respectively (9). IF studies were performed as described previously (9). Briefly, stationary-phase cells were washed and incubated with MAb 13F1 at 10 μg/ml for 2 h at room temperature. MAb binding was detected with fluorescein isothiocyanate-labeled goat anti-mouse immunoglobulin M (Southern Biotechnology, Birmingham, Ala.). Samples were viewed with a Zeiss (Thornwood, N.Y.) Axiophot microscope equipped with a fluorescein isothiocyanate filter. All the reference strains listed in Table 1 were previously examined by IF (9).
CNRE-1 RFLP analysis.
All strains were typed by Southern blot analysis with CNRE-1 as described previously (16). Briefly, genomic DNA was extracted from protoplasts (16) and was digested with SacI (Boehringer Mannheim, Indianapolis, Ind.), and the resulting fragments were probed with CNRE-1 labeled with [α-32P]dCTP. The bands were visualized by autoradiography.
The CNRE-1 restriction fragment length polymorphism (RFLP) patterns obtained for each strain were compared by using the Molecular Analyst/PC Fingerprinting software (Bio-Rad, Hercules, Calif.). Band positions were normalized by equating the HindIII-digested bacteriophage λ DNA molecular weight marker (Boehringer Mannheim), and Dice coefficients of similarity (number of shared bands × 2/total number of fragments in the two strains) were calculated for each pair of strains compared, generating a matrix of similarity coefficients. Dendrograms based on these matrices were then generated by the unweighted pair-group method of average linkage (28). When two patterns were compared, a match was recorded if the normalized molecular size of the fragment in the first pattern was within a window of ±1.5% of the molecular size of a fragment in the second pattern. The CNRE-1 RFLP patterns of Brazilian and New York City isolates reported earlier (10, 16) were also scanned and compared to those of the reference strains.
URA5 nucleotide sequencing.
URA5 DNA was amplified from genomic DNA by PCR as described previously (16) and was cloned into the pCR 2.1 vector of the TA cloning system (Invitrogen, San Diego, Calif.). Escherichia coli transformants were selected on plates containing 50 μg of kanamycin per ml, and plasmid DNA was purified with Midi-Prep columns (Qiagen, Chatsworth, Calif.). The insert was sequenced in the DNA Sequencing Facility of the Albert Einstein College of Medicine with automated sequencing instrument models ABI373A and ABI377 (Perkin-Elmer, Foster City, Calif.). Samples were analyzed by fluorescent cycle sequencing with dye-labeled primers.
The URA5 sequences obtained from the reference strains were compared with sequences previously obtained for seven New York City clinical isolates (GenBank accession no. L38582 to L38858, respectively), 10 Brazilian clinical and environmental isolates (GenBank accession no. U67723 to U67732, respectively), isolate B-3501 (GenBank accession no. M34606), and C. neoformans var. gattii (GenBank accession no. M93026). All sequences were first aligned by using Clustal V (18), and phylogenies were then estimated by use of the DNAPENNY (branch and bound parsimony), CONSENSE, and SEQBOOT programs of the PHYLIP package, version 3.5c (15).
Nucleotide sequence accession numbers.
The URA5 DNA sequences of the reference strains have been deposited in GenBank, and the accession numbers are listed in Table 1.
RESULTS
Serotyping.
Of the 20 recent isolates from Brazil and New York, all but isolates J25 and J26 presented agglutination patterns consistent with their assignment to serotype A. Isolates J25 and J26 showed positive reactions with antigenic factors 1, 7, and 8 characteristic of serotype AD. IF with MAb 13F1 produced an annular pattern of binding typical of serotype A strains for all recent isolates, thus confirming the results obtained with the sera containing cryptococcal antigen factors.
CNRE-1 RFLP analysis.
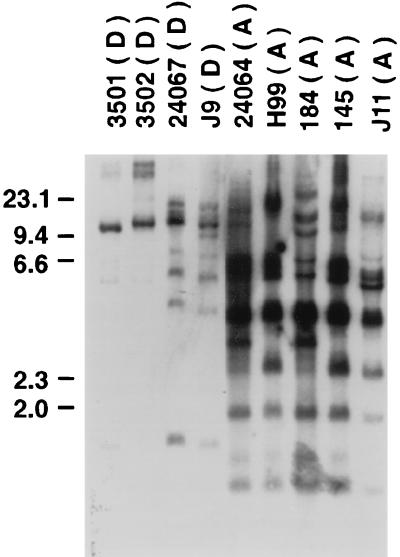
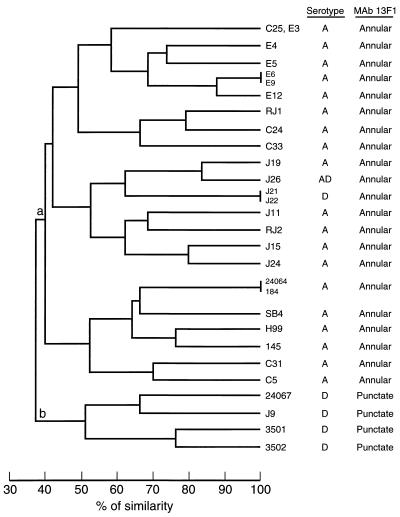
To determine whether the DNA fingerprints of the reference strains correlated with their serotypic status, Southern hybridization with the CNRE-1 probe was performed with serotype A and D isolates of C. neoformans var. neoformans. As seen in Fig. 1, hybridization of CNRE-1 to SacI-digested genomic DNA from serotype A isolates generated complex patterns of bands ranging from 11 to 16 restriction fragments with various intensities. A strongly hybridizing band at approximately 3.5 kb was present in all serotype A isolates. No two serotype A isolates had identical CNRE-1 RFLPs, although the dendrogram generated on the basis of the RFLP data grouped isolates 24064 and 184 together because it considered a band position tolerance of 1.5% (Fig. 2). This dendrogram also showed that isolates H99 and 145 were highly related, with 76% similarity. CNRE-1 hybridized to fewer bands and with a lower intensity for serotype D isolates (Fig. 1). The number of restriction fragments produced ranged from 5 to 11. Isolates 3501 and 3502 were very similar, clustering together with 76% similarity. Overall, serotype A and D isolates produced distinct RFLP patterns consistent with their separation into two major clusters (clusters a and b), as demonstrated in the dendrogram presented in Fig. 2. All recent isolates from Brazil and New York were placed among the reference serotype A strains.
FIG. 1.
CNRE-1 RFLP patterns obtained from reference strains of C. neoformans of serotypes A and D. The serotype of each strain is indicated in parentheses. The numbers on the left are in kilobases.
FIG. 2.
Dendrogram generated from the Dice coefficients computed from the CNRE-1 patterns for reference strains of C. neoformans and recent isolates from Brazil (C and E isolates) and New York City (J isolates). The serotypes and binding patterns obtained by IF with MAb 13F1 are presented on the right.
URA5 sequence analysis.
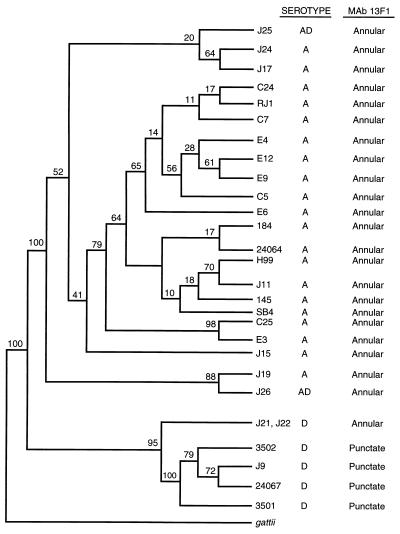
Partial sequences of the URA5 gene for serotype A and D strains are presented in Fig. 3 along with the partial sequence of strain 3501 (14). Isolates 24067 and J9 had identical sequences, as did isolates J11 and H99. Among the isolates belonging to serotype D, the number of nucleotide substitutions averaged 7.2 ± 3.4 (n = 15 pairwise comparisons). The URA5 genes from strains J21 and J22 showed the greatest number of base differences (between 9 and 10 substitutions) compared to the numbers for the other serotype D isolates. The nucleotide sequences of the serotype A isolates differed from each other by an average of 3.0 ± 1.7 base differences (n = 15 pairwise comparisons). The average number of URA5 base differences obtained by pairwise comparisons between serotype A and D isolates was 41.9 ± 2.7 (n = 30). Parsimony analysis (with bootstrapping) of these sequences identified a consensus tree that confirmed the dendrogram obtained from the CNRE-1 RFLP data, indicating that isolates of serotypes A and D clustered independently (Fig. 4). Remarkably, the separate clusters obtained for these serotypes occurred in 100% of the bootstrap replicates. Again, recently isolated strains from Brazil and New York were placed in the cluster that contained reference serotype A isolates.
FIG. 3.
Partial nucleotide sequence of the URA5 genes from reference strains of C. neoformans serotypes A and D. Serotypes are indicated in parentheses. A hyphen indicates that the base is identical to that in strain 3501 (14). A space implies that the base was not present in the allele. Lowercase letters indicate synonymous substitutions. The complete sequences are available in GenBank under the accession numbers listed in Table 1.
FIG. 4.
Relationship of reference strains of C. neoformans serotypes A and D and recent isolates from Brazil and New York City obtained from phylogenetic analysis of URA5 sequence data. The tree was obtained by use of DNAPENNY and CONSENSE of the PHYLIP program. The numbers at each branch point were generated by use of SEQBOOT and indicate the percentage of bootstrap replications. C. neoformans var. gattii was designated the outgroup. The serotypes and binding patterns obtained by IF with MAb 13F1 are presented on the right. The sequences of isolates C31, E5, and RJ2 were identical to the sequence of isolate C7 (16). Isolate C33 had the same sequence as isolate C5 (16).
DISCUSSION
To better understand the genetic relationship between serotypes A and D of C. neoformans var. neoformans, we have determined the CNRE-1 RFLP profiles and the nucleotide sequences of the URA5 genes from a set of reference strains. The CNRE-1 RFLP profiles of serotype A strains were more complex than those of serotype D strains, with average band numbers of 14 ± 2.1 and 8.2 ± 2.4 bands, respectively. Spitzer and Spitzer (29) also observed that CNRE-1 hybridized less intensely and to fewer bands for a serotype D isolate. In our study the CNRE-1 RFLP patterns for serotype D strains were also less intense than those for serotype A strains. This difference in intensity most likely represents a quantitative difference in CNRE-1 copy number in the genome and suggests that serotype D isolates contain fewer repetitive elements within one restriction fragment.
Genetic differences between serotypes A and D were also demonstrated by the nucleotide sequence of the URA5 gene. Within a serotype, the base replacements occurred mostly at the same nucleotide position. The only exceptions were isolates J21 and J22, which showed a few substitutions (5 of 22) that were more typical of serotype A isolates than D isolates (Fig. 3). The RFLP patterns of strains J21 and J22 also contained more bands than those of the other serotype D isolates tested (data not shown). Interestingly, these strains are classified as serotype D, but it was later shown for isolate J22 that it has a novel GXM triad structure not found in serotype A or D isolates (8). Studies with a MAb that discriminates between serotype A and D isolates on the basis of the fluorescence pattern revealed that strain J22 is like serotype A isolates (9). Hence, strain J22 appears to be an unusual strain with certain unique characteristics.
Overall, our results indicate that serotypes A and D segregated into two groups which appear to be phylogenetically distant from each other, as demonstrated by the independent clusters observed in the trees generated from RFLP and sequencing data. Our findings are in agreement with those of Guého et al. (17), who also found a relative phylogenetic difference between serotypes A and D on the basis of the sequence of the 26S large subunit of rRNA. We were also able to detect phylogenetic distance between serotypes A and D by investigating a gene that has been shown to be highly polymorphic among cryptococcal isolates (5, 6). In addition, a comparison between the reference isolates and those from Brazil and New York suggested that the observed phylogenetic distance of serotype A and serotype D isolates is independent of the geographic background of the isolate.
Analysis of several commonly used C. neoformans laboratory strains revealed significant phylogenetic differences. Among the reference serotype A strains, strains H99 and 145 and strains 24064 and 184 were most closely related by both URA5 sequence and CNRE-1 RFLPs. Nevertheless, each of the isolates was distinguishable by DNA typing, suggesting the possibility of different biological traits. Considering the existence of significant genetic differences among isolates classified within a serotype and among reference strains, one should be cautious in generalizing conclusions on the basis of data obtained for one strain. Our results suggest that it may be prudent to include both serotype A and D strains when conducting biological studies with C. neoformans.
The taxonomy of C. neoformans and its teleomorph, Filobasidiella neoformans, has been the subject of several studies. Initially, two different species were recognized: C. neoformans (serotypes A and D) and its sexual state, F. neoformans (20), and C. bacillispora (serotypes B and C) and its teleomorph, F. bacillispora (21). Further taxonomic studies with Cryptococcus species and their teleomorphs reclassified these two species into C. neoformans var. neoformans and C. neoformans var. gattii on the basis of successful interspecific crosses and DNA relatedness (1, 22). Although the current classification of C. neoformans remains at the varietal status, more recent studies have brought into question this proposed scheme. Boekhout et al. (3) demonstrated that C. neoformans var. neoformans and C. neoformans var. gattii differ in their genetic makeups and may represent separate species. Genetic differences between serotypes A and D, as described in this report and by others (see the introduction), provide additional evidence that the taxonomy of C. neoformans may need to be reinvestigated. Consistent with this, phenotypic differences between serotypes A and D have also been reported. These differences include the chemical structure of the capsular polysaccharide (7, 19) and qualitative and quantitative antigenic dissimilarities among serotype A and D strains (9). In one study, it was suggested that serotypes D and A may not have the same virulence, tropism, and/or ecological niche (13).
Recently, we have proposed that the population structure of C. neoformans is clonal on the basis of analysis of URA5 sequences, electrophoretic karyotypes, and CNRE-1 RFLPs (16). The results of this study are consistent with that proposal and extend it by suggesting the divergence of serotype A and D strains along different lineages. C. neoformans mating types a and α have been described for serotype D strains, but no serotype A a strain has ever been recovered from clinical or environmental sources. C. neoformans strains which are efficient mating pairs have all been serotype D. The divergence of serotype A and D lineages combined with a high mating efficiency restricted to serotype D suggests that future population structure studies should consider strains of serotype A and D separately.
In summary, our findings revealed genetic unrelatedness between serotypes A and D of C. neoformans and indicate that these serotypes could probably be considered different varieties of C. neoformans. Our results suggest that C. neoformans var. gattii and C. neoformans var. neoformans could be regarded as separate species, as originally proposed, and serotypes A and D and serotypes B and C would then represent different varieties of each species. Additional genetic studies will be necessary to further define the exact genetic relationship between the serotypes of C. neoformans, including those belonging to serotypes B and C.
ACKNOWLEDGMENTS
A.C. is supported by a Burroughs Wellcome Fund Development Therapeutics Award and National Institutes of Health grants AI-22774 and AI-13342.
We thank J. W. Taylor for careful review of and helpful comments on the manuscript.
REFERENCES
- 1.Aulakh H S, Straus S E, Kwon-Chung K J. Genetic relatedness of Filobasidiella neoformans (Cryptococcus neoformans) and Filobasidiella bacillispora (Cryptococcus bacillispora) as determined by DNA base composition and sequence homology studies. Int J Syst Bacteriol. 1981;31:97–103. [Google Scholar]
- 2.Bennett J E, Kwon-Chung K J, Howard D. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1984;105:582–586. doi: 10.1093/oxfordjournals.aje.a112423. [DOI] [PubMed] [Google Scholar]
- 3.Boekhout T, van Belkum A, Leenders A C, Verbrugh H A, Mukamurangwa P, Swinne D, Scheffers W A. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int J Syst Bacteriol. 1997;47:432–442. doi: 10.1099/00207713-47-2-432. [DOI] [PubMed] [Google Scholar]
- 4.Brandt M E, Bragg S L, Pinner R W. Multilocus enzyme typing of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2819–2823. doi: 10.1128/jcm.31.10.2819-2823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall A, Freundlich L F, Marsh L, Scharff M. Extensive allelic variation in Cryptococcus neoformans. J Clin Microbiol. 1992;30:1080–1084. doi: 10.1128/jcm.30.5.1080-1084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen F, Currie B P, Chen L C, Spitzer S G, Spitzer E D, Casadevall A. Genetic relatedness of Cryptococcus neoformans clinical isolates with the repetitive DNA probe CNRE-1. J Clin Microbiol. 1995;33:2818–2822. doi: 10.1128/jcm.33.11.2818-2822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherniak R, Sundstrom J B. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect Immun. 1994;62:1507–1512. doi: 10.1128/iai.62.5.1507-1512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherniak R, Morris L C, Belay T, Spitzer E D, Casadevall A. Variation in the structure of glucuronoxylomannan in isolates from patients with recurrent cryptococcal meningitis. Infect Immun. 1995;63:1899–1905. doi: 10.1128/iai.63.5.1899-1905.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleare W, Casadevall A. The different binding patterns of two immunoglobulin M monoclonal antibodies to Cryptococcus neoformans serotype A and D strains correlate with serotype classification and differences in functional assays. Clin Diagn Lab Immunol. 1998;5:125–129. doi: 10.1128/cdli.5.2.125-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Currie B P, Freundlich L F, Casadevall A. Restriction fragment length polymorphism analysis of Cryptococcus neoformans isolates from environmental (pigeon excreta) and clinical sources in New York City. J Clin Microbiol. 1994;32:1188–1192. doi: 10.1128/jcm.32.5.1188-1192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dromer F, Varma A, Ronin A, Mathoulin S, Dupont B. Molecular typing of Cryptococcus neoformans serotype D clinical isolates. J Clin Microbiol. 1994;32:2364–2371. doi: 10.1128/jcm.32.10.2364-2371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dromer F, Mathoulin S, Dupont B, Laporte A the French Cryptococcosis Study Group. Epidemiology of cryptococcosis in France: a 9-year survey (1985–1993) Clin Infect Dis. 1996;23:82–90. doi: 10.1093/clinids/23.1.82. [DOI] [PubMed] [Google Scholar]
- 13.Dromer F, Mathoulin S, Dupont B, Letenneur L, Ronin O the French Cryptococcosis Study Group. Individual and environmental factors associated with infection due to Cryptococcus neoformans serotype D. Clin Infect Dis. 1996;23:91–96. doi: 10.1093/clinids/23.1.91. [DOI] [PubMed] [Google Scholar]
- 14.Edman J C, Kwon-Chung K J. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990;10:4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.5c. Distributed by the author. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 16.Franzot S P, Hamdan J S, Currie B P, Casadevall A. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J Clin Microbiol. 1997;35:2243–2251. doi: 10.1128/jcm.35.9.2243-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guého E, Improvisi L, Christen R, de Hoog G S. Phylogenetic relationships of Cryptococcus neoformans and some related basidiomycetous yeasts determined from partial large subunit rRNA sequences. Antonie Leeuwenhoek. 1993;63:175–189. doi: 10.1007/BF00872392. [DOI] [PubMed] [Google Scholar]
- 18.Higgins D G, Bleasby A J, Fuchs R. CLUSTALV: improved software for multiple sequence alignment. Comput Appl Biosci. 1997;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda R, Nishimura S, Nishikawa A, Shinoda T. Production of agglutinating monoclonal antibody against antigen 8 specific for Cryptococcus neoformans serotype D. Clin Diagn Lab Immunol. 1996;3:89–92. doi: 10.1128/cdli.3.1.89-92.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon-Chung K J. A new genus Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- 21.Kwon-Chung K J. A new species of Filobasidiella, the perfect state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:942–946. [Google Scholar]
- 22.Kwon-Chung K J, Bennett J E, Rhodes J C. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Leeuwenhoek. 1982;48:25–38. doi: 10.1007/BF00399484. [DOI] [PubMed] [Google Scholar]
- 23.Meyer W, Mitchell T G, Freedman E Z, Vilgalys R. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2274–2280. doi: 10.1128/jcm.31.9.2274-2280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy J W, Cozad G C. Immunological unresponsiveness induced by cryptococcal polysaccharide assayed by the hemolytic plate technique. Infect Immun. 1972;5:896–901. doi: 10.1128/iai.5.6.896-901.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perfect J R, Magee B B, Magee P T. Separation of chromosomes of Cryptococcus neoformans by pulsed-field gel electrophoresis. Infect Immun. 1989;57:2624–2627. doi: 10.1128/iai.57.9.2624-2627.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polacheck I, Lebens G A. Electrophoretic karyotype of the pathogenic yeast Cryptococcus neoformans. J Gen Microbiol. 1989;134:1037–1041. doi: 10.1099/00221287-135-1-65. [DOI] [PubMed] [Google Scholar]
- 28.Romesburg H C. Cluster analysis for researchers. Malabar, Fla: Robert E. Krieger Publishing Company; 1990. [Google Scholar]
- 29.Spitzer E D, Spitzer S G. Use of a dispersed repetitive DNA element to distinguish clinical isolates of Cryptococcus neoformans. J Clin Microbiol. 1992;30:1094–1097. doi: 10.1128/jcm.30.5.1094-1097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spitzer E D, Spitzer S G, Freundlich L F, Casadevall A. Persistence of initial infection in recurrent Cryptococcus neoformans meningitis. Lancet. 1993;341:595–596. doi: 10.1016/0140-6736(93)90354-j. [DOI] [PubMed] [Google Scholar]
- 31.Varma A, Kwon-Chung K J. DNA probe for strain typing of Cryptococcus neoformans. J Clin Microbiol. 1992;30:2960–2967. doi: 10.1128/jcm.30.11.2960-2967.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson D E, Bennett J E, Bailey J W. Serologic grouping of Cryptococcus neoformans. Proc Soc Exp Biol Med. 1968;127:820–823. doi: 10.3181/00379727-127-32812. [DOI] [PubMed] [Google Scholar]