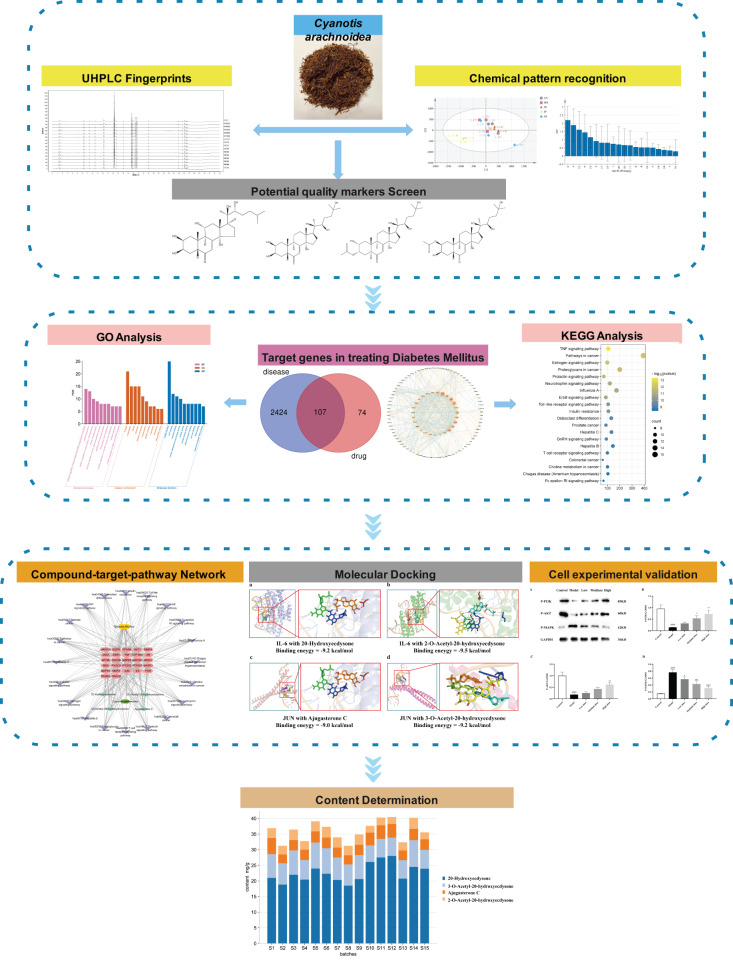
Abstract
Cyanotis arachnoidea C. B. Clarke is a traditional Chinese medicinal herb that has a limited clinical use in the treatment of diabetes mellitus (DM) in minority areas of Guizhou in China. However, few prior reports are available on the quality control of Cyanotis arachnoidea, and its quality markers and hypoglycemic mechanism are still unclear. The purpose of this study is to explore the quality markers (Q-markers) of Cyanotis arachnoidea and predict its hypoglycemic mechanism. In this study, ultra-high-performance liquid chromatography (UHPLC) fingerprint combined with chemical pattern recognition were performed, and four differential components were screened out as quality markers, including 20-Hydroxyecdysone, 3-O-acetyl-20-hydroxyecdysone, Ajugasterone C, and 2-O-acetyl-20-hydroxyecdysone. Network pharmacology analysis revealed 107 therapeutic target genes of Cyanotis arachnoidea in DM treatment, and the key targets were Akt1, TNF, IL-6, MAPK3, and JUN. The hypoglycemic mode of action of Cyanotis arachnoidea may be mediated by tumor necrosis factor (TNF) signaling, cancer, insulin resistance, and JAK-STAT pathways. Molecular docking analysis disclosed that the foregoing quality markers effectively bound their key target genes. An in vitro experiment conducted on pancreatic islet β-cells indicated that the forenamed active components of Cyanotis arachnoidea had hypoglycemic efficacy by promoting PI3K/Akt and inhibiting MAPK signaling. UHPLC also accurately quantified the quality markers. The identification and analysis of quality markers for Cyanotis arachnoidea is expected to provide references for the establishment of a quality control evaluation system and clarify the material basis and hypoglycemic mechanisms of this traditional Chinese medicine (TCM).
Keywords: Cyanotis arachnoidea, Quality markers, UHPLC, Chemical pattern recognition, Network pharmacology, Hypoglycemic, Ultra-high-performance liquid chromatography
Introduction
Diabetes mellitus (DM) is a complex and prevalent chronic metabolic condition. Its global incidence is increasing annually. DM is described as a disorder of glucose metabolism resulting from absolute or relative insulin insufficiency (Mustapha et al., 2021). According to the International Diabetes Federation, DM affects up to 463 million people in the age range of 20–79 years globally. China has approximately 114 million DM patients, and this number is expected to rise to 120 million by 2045 (Hu et al., 2019). DM is a disease associated with the most complications. The long-term increases in blood glucose associated with DM can cause damage to macro- and micro-vessels and adversely affect heart, brain, kidney, peripheral nerves, eyes, and feet (Xiaopeng et al., 2016). Consequently, extremely high disability and mortality rates are associated with DM, and it seriously threatens physical health and imposes a substantial socioeconomic burden on the patients afflicted with it.
Chinese herbal medicine has been administered to prevent and treat DM for thousands of years and has attracted research attention as it has multi-target and multi-pathway characteristics. The glycosides (Aswal et al., 2020), polyphenols (Riyaphan et al., 2021), phytosterones (Gu et al., 2021), flavones, and isoflavones (Xu et al., 2020) in Chinese herbal medicines provide efficacious glycemic control and ameliorate insulin resistance through various mechanisms. Cyanotis arachnoidea is a traditional Chinese herbal and ethnic medicine that is widely distributed in India, Sri Lanka, and the Yunnan Province of China. Ancient Chinese medicine texts reported that it has been used to treat nephritic edema, diabetes, rheumatoid arthritis, and other diseases (Lan & Su, 2017; Wang, 1975; Guizhou Institute of TCM, 1970; Bu, Zhang & Guo, 2013; Tan et al., 2002). Modern research has demonstrated that Cyanotis arachnoidea contains several phytosterones and is particularly rich in 20-Hydroxyecdysone (Nie & Qiu, 1987). This compound promotes de novo nucleic acid and protein synthesis (Omanakuttan et al., 2016), has pharmacological effects on glucose and lipid metabolism (Marschall et al., 2021), and has anti-inflammatory (Tang et al., 2020), antioxidant (Baev et al., 2022), neuroprotectant (Lim et al., 2020), and antitumor (Romaniuk-Drapała et al., 2021) activity. Previous studies (Chen, Xia & Qiu, 2006; Chen et al., 2017) showed that 20-Hydroxyecdysone ameliorated insulin resistance in HepG2 cells by activating the IRS-1/Akt pathway and upregulating GLUT4 and GLUT2 proteins. Hence, it has potential hypoglycemic efficacy. A clinical trial (Yi et al., 2018) demonstrated that a combination of Cyanotis arachnoidea and metformin significantly reduced the glycosylated hemoglobin and serum C-peptide levels in patients with type 2 diabetes mellitus (T2DM), thereby improving their typical symptoms. Nevertheless, active ingredients in Cyanotis arachnoidea and their hypoglycemic mechanisms remain unclear, and the adoption of this herb in clinical practice is severely limited. Furthermore, differences in the sources, harvest seasons, and processing methods of Cyanotis arachnoidea result in variability of its components, and effective quality control methods for Cyanotis arachnoidea are lacking.
Traditional Chinese medicine (TCM) herbs have complex compositions and multiple components, targets, and pathways. Thus, it is difficult to elucidate their modes of action. In 2016, quality markers (Q-markers) were proposed to evaluate and control TCM quality (Zhao et al., 2019; Bai et al., 2018). A Q-marker is a unique chemical component that is either inherent in TCM or forms when TCM herbs, decoctions, extracts, and patent medicines are prepared. It is closely related to the functional attributes of TCM, should be qualitatively and quantitatively determined, and could serve as a marker of TCM safety and efficacy (Liu, Guo & Liu, 2018). To search for Q-markers and hypoglycemic constituents in Cyanotis arachnoidea, and examine their hypoglycemic molecular mechanisms of action through the analysis of Q-markers and biomarkers, chemical pattern recognition and network pharmacology were performed to screen differential components, identify active ingredients and predict their potential targets. The TCM fingerprint is a comprehensive and quantifiable identification method that has been widely used for TCM quality control in combination with chemical pattern recognition techniques, such as hierarchical cluster analysis (HCA) and principal component analysis (PCA) (Jiang et al., 2022; Zhang et al., 2021). Network pharmacology is systematic and holistic and can be widely used to predict and research the multi-target mechanisms of TCM in the treatment of various diseases (Pan et al., 2022; Li et al., 2021; Wang, Cui & Li, 2019).
Molecular docking technology is a type of computer-aided drug design (CADD). This new drug research and development technology has rapidly developed to explore rational drug design. It uses information technology and exploits existing knowledge of related drugs and their targets to guide research in drug discovery and drug action mechanisms via calculations, simulations, and predictions (Baig et al., 2016). Digital CADD technology significantly shortens the time and lowers the cost of research. It decreases blindness, increases accuracy, and facilitates the discovery of high-activity, high-affinity drug molecules while targeting multiple sites. For these reasons, the advent of digital CADD technology is an important breakthrough in TCM development. Nevertheless, the models have low prediction accuracy, omit biologically active drug candidates, and do not consider component/drug toxicity (Kaur & Khatik, 2021). Molecular docking technology is important for further identifying the key targets of the active ingredients in TCMs by docking the ligands to their receptors and evaluating the stability of the docking system (Zhu et al., 2022).
In the present study, we used ultra-high-performance liquid chromatography (UHPLC) to establish a fingerprint reflecting the integrity of the chemical composition of Cyanotis arachnoidea. We screened potential quality markers and identified them by chemical pattern recognition technology. We used network pharmacology to predicte the hypoglycemic mechanisms of the components based on their quality markers and verified the latter by molecular docking and in vitro experiments. Finally, we quantitatively analyzed the quality markers by UHPLC. In conclusion, the present work devised a quality control evaluation method for Cyanotis arachnoidea and provided a reference for clarifying the pharmacodynamic mechanisms of the active ingredients of this TCM in DM. The experimental design is shown in Fig. 1.
Figure 1. Detailed diagram of the overall study design.
*: Compared with the model group, P < 0.05; **: Compared with the model group, P < 0.01; ***: Compared with the model group, P < 0.001; ****: Compared with the model group, P < 0.0001; ####: Compared with the blank control group, P < 0.0001.
Materials and Methods
Materials and reagents
Chinese medicinal materials
Fifteen batches of Cyanotis arachnoidea were purchased from China Shineway Pharmaceutical Group Ltd., Shijiazhuang, Hebei, China and authenticated by Professor Huixiang Yu of the Traditional Chinese Medicine Hospital of Chuxiong Yi Autonomous Prefecture, Yunnan, China. The dried roots were used in the experimental research, and the samples were deposited at the Hebei Traditional Chinese Medicine Formula Granule Engineering & Technology Innovate Center of the Hebei University of Chinese Medicine, Shijiazhuang, China.
Reference standards
20-Hydroxyecdysone (PU0362-0025MG), Ajugasterone C (PS0716-0010MG), 3-O-acetyl-20-hydroxyecdysone (PS210615-78-0005MG), and 2-O-acetyl-20-hydroxyecdysone (JOT-12130-0020MG) were obtained from Chengdu Push Biotechnology Co., Ltd. and Chengdu Pufei De Biotech Co., Ltd. (Sichuan, China). The purity of all the chemicals were greater than 98%. HPLC-grade methanol was obtained from Thermo Fisher Scientific (Waltham, MA, USA). Ultrapure water was generated by a Millipore Milli-Q Reagent Water System (EMD Millipore, Billerica, MA, USA). Chromatography-grade acetonitrile and formic acid were acquired from Sigma-Aldrich Corp. (St. Louis, MO, USA).
Cell lines
MIN6 islet β-cells (Lot No. CL-0674) were purchased from Procell Life Science & Technology Co., Ltd., (Wuhan, Hubei, China).
Reagents
The Roswell Park Memorial Institute (RPMI 1640) culture medium, the glucose assay kit and the radioimmunoprecipitation assay (RIPA) lysis solution were bought from Beijing Solarbio Science & Technology Co., Ltd., (Beijing, China). Protein bands were visualized with an enhanced chemiluminescence (ECL) detection kit (Thermo Fisher Scientific, Waltham, MA, USA). Anti-P-PI3K, anti-P-Akt, anti-P-MAPK and anti-GAPDH primary antibodies were acquired from bioWorld Technology Inc., (Dublin, OH, USA). Secondary antibodies were purchased from Zhongshanjinqiao Biotech (Beijing, China). Polyvinylidene fluoride (PVDF) transfer membranes were obtained from EMD Millipore.
Preparation of the herbal extract and the reference substances
The herbs were pulverized and sifted, and the 15 Cyanotis arachnoidea sample batches are listed in Table S1. Herb powder (0.2 g) was dissolved in 25 mL of 70% (v/v) methanol in an Erlenmeyer flask and sonicated at constant ultrasound frequency (40 kHz; 250 W) for 45 min. Then 70% (v/v) methanol was added after the extraction to compensate for weight loss. The supernatant was removed, passed through a 0.22-μm filter membrane, and used in the subsequent analyses.
The reference substances were accurately weighed and dissolved in 10 mL chromatographic methanol to prepare stock solutions, and then were mixed at concentrations of 0.1723, 0.0251, 0.0539, and 0.0256 mg/mL. All solutions were stored at 4 °C until further use.
UHPLC fingerprint analysis
UHPLC chromatography conditions
Chromatographic separation was performed in a Phenomenex Kinetex 2.6-μm C18 chromatographic column (100 mm × 3 mm; 2.6 μm) (Phenomenex, Torrance, CA, USA). The mobile phases were pure comprised pure acetonitrile (A) and 0.1% (v/v) formic acid in water (B), and the flow rate was 0.3 mL/min. The gradient elution program was as follows: 0–3 min, 5–13% A; 3–8 min, 13–15% A; 8–20 min, 15–35% A; 20–25 min, 35–95% A; 25–27 min, 95% A; 27–30 min, 95–5% B, and 30–33 min, 5% B. The column temperature was kept at 35 °C; the injection volume was 2.0 μL, and the detection wavelength was 248 nm.
Method validation
The same sample solution (S1) was measured sixfold in parallel to determine instrument precision. The relative standard deviation (RSD) values of the relative retention times and the peak areas under the common peaks were calculated using the peak of 20-Hydroxyecdysone as the reference. Repeatability was evaluated by analyzing nine independently prepared triplicate sample solutions at low (0.1 g), medium (0.2 g), and high (0.3 g) concentrations, each in triplicates. The stability of the method of analyzing the sample solutions was assessed at 0, 2, 4, 8, 12, and 24 h on the same day. Each sample solution was simultaneously assessed twice.
Fingerprint establishment and similarity analysis
Test solutions were prepared from powdered materials derived from the 15 sample batches and subjected to UHPLC analysis. The chromatograms generated were exported as CDF (computable document format) files and imported to the Similarity Evaluation System for Chromatographic Fingerprint of TCM (Version 2012) (Gu, Hall & Miles, 2016). The chromatogram for Batch 1 (S1) served as a reference and its common characteristic peaks were marked by multi-point correlation and automated Mark peak matching. A reference chromatogram (R) was generated by the median method. The degrees of similarity among the chromatograms for the 15 batches and that of R were calculated. Common peaks were identified by alignment with the reference compounds.
Chemical pattern recognition analysis
HCA analysis
HCA was performed using the Ward method in SPSS 23.0 (IBM Corp., Armonk, NY, USA). The areas of the common peaks of the 15 batches were the variables and the Euclidean distance (Qin et al., 2022) was the metric.
PCA and PLS-DA analysis
PCA was performed in SIMCA v. 14.1 (https://umetrics-simca.software.informer.com/14.0/) to distinguish the quality of the materials from different regions and was based on the peak areas of the common peaks of the fifteen batches. Sample quality and discrepancy were judged according to the distances between sample points (Li et al., 2021). PLS-DA was conducted to screen out the differential components based on their variable importance in projection (VIP) values.
Network pharmacology analysis
Prediction of target genes for the differential components and disease
Components contributing to the regional difference in material quality were identified by PLS-DA and used as candidate quality markers. The “SMILES” and “InChI” representations of the components were retrieved from the PubChem Compound database (https://www.ncbi.nlm.nih.gov/pubmed/). Standard 3D molecular structures were exported in structure-data file (SDF) format. Component targets were predicted using the BATMAN-TCM database (http://bionet.ncpsb.org.cn/batman-tcm/) (Liu et al., 2016) and based on their “InChI” representations (score > 20; P < 0.05). BATMAN-TCM is a bioinformatics analysis tool that elucidates the molecular mechanism of TCMs. It simultaneously analyzes multiple medicines and components and clarifies their mechanism at the molecular and systemic levels. Cytoscape v. 3.7.2 (https://cytoscape.org/download.html) was used to construct a component-target network.
Diabetes mellitus (DM)-associated target genes were collected from the GeneCards (Barshir et al., 2021) (https://www.genecards.org/), DrugBank (Wishart et al., 2018) (https://go.drugbank.com/), DisGeNET (Piñero et al., 2020) (https://www.disgenet.org/), and OMIM (Amberger et al., 2019) (https://omim.org/) databases by using the keyword/search term “diabetes mellitus”. The target genes were standardized by the UniProt database (UniProt Consortium, 2021) (https://www.uniprot.org/). The intersection target genes of component and DM were obtained on the Draw Venn Diagram platform (http://bioinformatics.psb.ugent.be/webtools/Venn/).
PPI network analysis and core target identification
A PPI network analysis of the intersecting target genes was constructed on the STRING database platform (Szklarczyk et al., 2021) (https://cn.string-db.org/). The confidence level for the interaction analysis in the network was >0.9. A topological analysis of the PPI network was performed using the “Analyze Network” function in Cytoscape v. 3.7.2, and the core targets were identified based on the degree, betweenness centrality (BC) and closeness centrality (CC) of each node.
GO and KEGG pathway enrichment analysis
GO annotation and KEGG pathway enrichment analyses were performed by importing the core targets to the DAVID v. 6.8 database (Jiao et al., 2021) (https://david.ncifcrf.gov). The biological processes and pathways potentially involved in the treatment of DM with Cyanotis arachnoidea were obtained at P ≤ 0.01 and Benjamini-Hochberg correction ≤0.01. Data for the active compounds and their core targets were imported into Cytoscape v. 3.7.2 to build a “drugs-active compounds-target genes-pathways” network.
Molecular docking analysis
Molecular docking was performed on the four potential quality markers and the top five core targets according to their degree values. The crystal structures of the targets proteins were downloaded from the PDB database (Burley et al., 2022) (https://www.rcsb.org/) and saved in protein databank (PDB) file format. The PDB IDs of the top five target proteins (Akt1, TNF, IL-6, MAPK3, and JUN) were 4EKL, 2AZ5, 4NI9, 2ZOQ, and 5T01. Solvents and water molecules were removed with PyMOL software (https://www.pymol.org/). AutoDockTools v. 1.5.6 (https://autodocksuite.scripps.edu/adt/) was used to add hydrogens, calculate the atomic charges for the target proteins, and export the results in a pdbqt file format. Molecular docking was performed in AutoDock Vina (http://vina.scripps.edu/) after adjustment for the binding pocket radius size and the corresponding coordinate site to obtain reliable docking models for the core targets. Forty was the grid size of the docking box in blind docking, and the center X, Y, and Z axes of the top five key targets were (21.003, 2.523, 16.167), (−13.69, 71.605, 27.0), (−11.213, 33.474, 11.162), (15.007, −7.839, 24.613), and (−23.326, 17.31, 20.956), respectively.
Experimental validation
Recovered MiN6 islet β-cells were inoculated into RPMI 1640 medium and incubated under a 5% CO2 atmosphere and at 37 °C. Cells in insulin solution with the concentration of 1 × 10−7 mol/L comprised the model group. Cells in buffer solution (no insulin) comprised the control group. The cells in both groups were subjected to 0.1 mL of 10 mM glucose solution. After continued culture for 24 h, the glucose concentration in the supernatant of the model group was significantly higher than in that of the control group. Hence, the cellular insulin resistance model was successfully established. The cells were then collected and their density was adjusted to 1 × 106/L. Then 0.1 mL cell suspension was added to a 96-well culture plate. Chinese herbal extract (64 mg) was weighed out and dissolve in 10 mL of serum-free medium to obtain a 6,400 mg/L storage solution. The latter was diluted to 100, 200, and 400 mg/L for administration to the low, medium and high dose groups, respectively. Each group had triplicate wells. 0.1 mL of 10 mmol/L glucose was simultaneously added to all cells in each group. The cells were then incubated for 24 h, and the glucose concentrations in the cell culture media were measured by the glucose oxidase method (Xuguang et al., 2019).
The expression levels of the phosphorylated PI3K (p-PI3K), Akt (p-Akt), and MAPK (p-MAPK) proteins were detected by western blot. After 24 h cell culture, the supernatants were removed, 200 μL cell lysate was added, and the cells were incubated at 4 °C overnight and centrifuged at 4 °C and 12,000 rpm for 10 min. The UV method and 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were used to determine the protein content in the supernatants. to determine the protein content by UV method, and carried out 10% SDS-PAGE electrophoresis. The polyvinylidene fluoride (PVDF) membrane was then subject to a constant 250 mA for 60 min, and blocked with skim milk at 37 °C for 2 h. The membrane was then washed with phosphate-buffered saline (PBS), and a mixture of PBS-diluted primary antibodies (anti-p-PI3K, 1:3,000; anti-p-MAPK, 1:1,500; anti-p-Akt, 1:1,200; and anti-GADPH, 1:2,000) was added. The membrane was incubated overnight at 4 °C, washed again, and subjected to horseradish peroxidase (HRP)-labeled secondary antibody. Then the enhanced chemiluminescence (ECL) method was used to expose and develop the membrane with a gel imager. Finally, GAPDH was used as the internal reference and the protein bands were scanned and analyzed in Tanon Gis software to determine their grayscale values. The relative protein expression level equals the grayscale values of the target protein bands divided by those of the GAPDH protein bands.
Content determination of the potential quality markers
Method validation
The mixed reference solutions were serially diluted to obtain gradient samples at six different concentrations (n = 6). The mixed reference solutions (0.2, 0.5, 1, 2, 3, and 5 μL) were placed in 10-mL volumetric flasks, diluted to the mark with 70% (v/v) methanol, and shaken. In this manner, a concentration series of mixed reference solutions for the four components were obtained. Two microliters of the reserve solution were subjected to UHPLC analysis as previously described. The reference substance concentrations were plotted on the abscissa (x-axis; X) while the peak areas were plotted on the ordinate (y-axis; Y). Standard curves were then drawn and the regression equations and linear ranges were calculated for each component.
An appropriate amount of the mixed reference solution was diluted with an equal volume of 70% (v/v) methanol. Then 2 μL diluted mixed reference solution was accurately pipetted and injected into the UHPLC. The chromatographic analysis was performed in duplicate. The precision, repeatability and stability of the determination method in methodological validation was refered to the UHPLC fingerprint. The peak area of each compound was recorded and the results of the RSD were calculated to determine sample precision.
Quantitative analysis
The 15 sample batches were accurately weighed and prepared as test solutions. UHPLC was then performed to establish the peak areas. The contents of the four potential quality markers were calculated and determined by measuring the peak areas.
Statistical analysis
All data were expressed as ( ± s) and the statistical analyses were performed in GraphPad Prism v. 9.4.0 (GraphPad Software, La Jolla, CA, USA). One-way ANOVA was used to compare data among different groups. Multiple comparisons were made using Tukey’s test. P < 0.05 was considered statistically significant.
Results and discussion
Establishment of UHPLC fingerprints
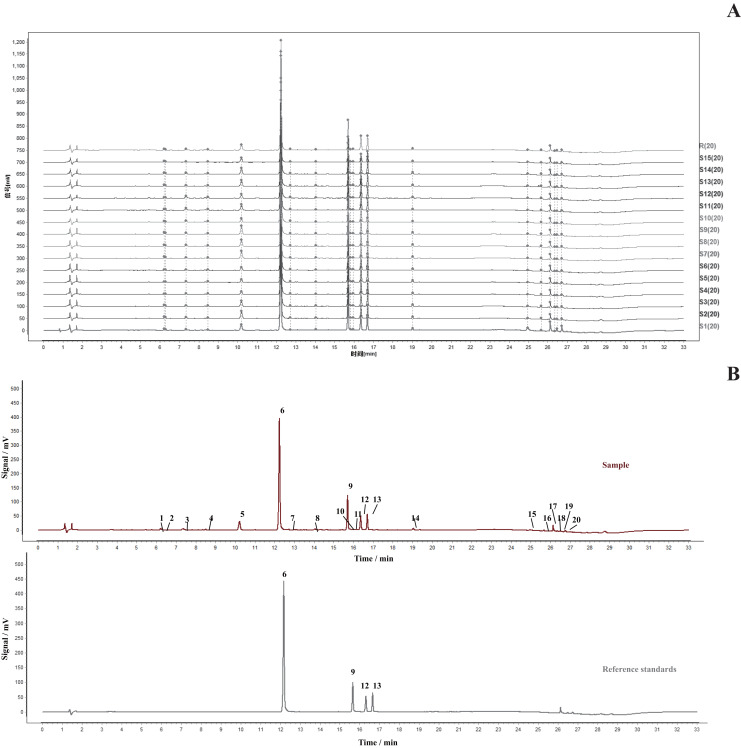
The fifteen Cyanotis arachnoidea samples batches were collected in five different regions of Yunnan Province including Anning (S1–S3), Menghai (S4–S6), Jinning (S7–S9), Jinping (S10–S12), and Jinghong (S13–S15), and their fingerprints were established by UHPLC. Twenty common characteristic peaks were automatically selected as the fingerprints. Peak-6 served as the reference for a standard R chromatogram that was determined by the median method and superimposed on the fingerprints (Fig. 2A). Comparison of the UHPLC chromatograms and the peak retention times of the samples and the mixed control solution, four components were identified, including 20-Hydroxyecdysone (peak-6), 3-O-Acetyl-20-hydroxyecdysone (peak-9), Ajugasterone C (peak-12), and 2-O-Acetyl-20-hydroxyecdysone (peak-13) (Fig. 2B).
Figure 2. Identification and fingerprints of 15 sample batches of Cyanotis arachnoidea.
UHPLC fingerprints of 15 Cyanotis arachnoidea samples batches (S1–S15) and reference fingerprint (S1). Twenty common characteristic peaks were selected (A). Comparison of the UHPLC chromatograms for the samples and reference standards identified four components. Peaks 6, 9, 12, and 13 correspond to 20-Hydroxyecdysone, 3-O-Acetyl-20-hydroxyecdysone, Ajugasterone C, and 2-O-Acetyl-20-hydroxyecdysone, respectively (B).
According to the Similarity Evaluation System for Chromatographic Fingerprint of TCMs (Version 2012), the similarity values of the S1–S15 samples were in the range of 0.996–1.000 and generally remained at high levels (Table S1). Hence, the overall quality of 15 batches of Cyanotis arachnoidea was relatively stable, but the content of each component might be different. For this reason medicinal materials from different sources might vary in terms of their quality stability and homogeneity. Systematic studies of the chemical components of TCM indicate that their fingerprints could be used in their analysis and the determination of their overall quality control.
The precision, repeatability, and stability of the foregoing method were validated, and the RSDs of the average retention time and the peak area were used to evaluate it. According to the precision, repeatability, and stability tests, the RSD values for the average retention times and peak areas were all <0.2% and <2.89%, <0.14% and <2.81%, and <0.28% and <2.96%, respectively (Table S2). Validation of the method demonstrated good instrument precision, excellent repeatability, and sample solutions that were stable for 24 h. Therefore, the method could be used in the qualitative identification of Cyanotis arachnoidea.
Chemical pattern recognition analysis
Hierarchical cluster analysis
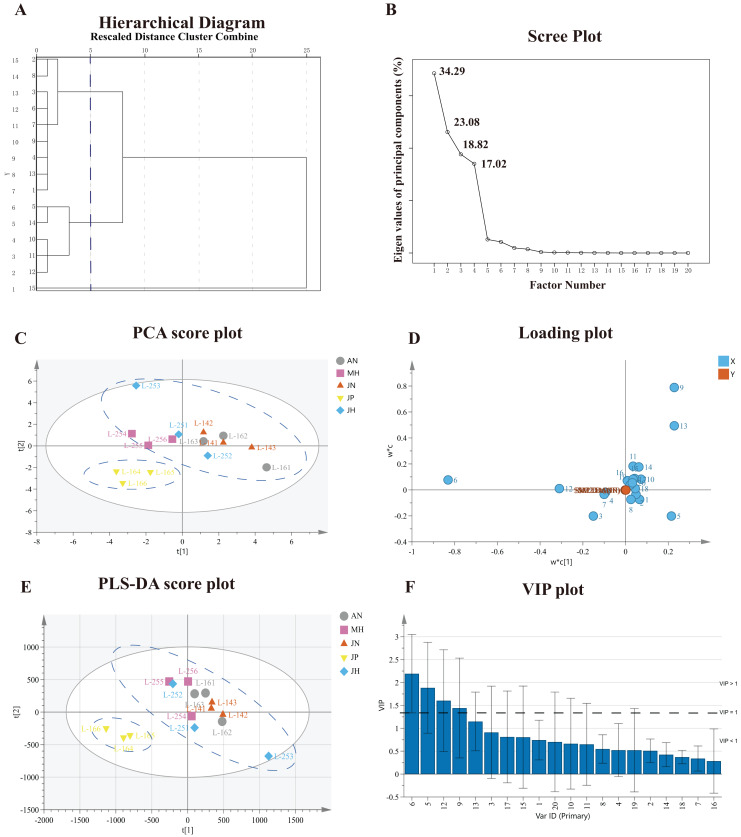
The twenty common characteristic peaks were subjected to HCA in SPSS v. 23.0 to validate the results of the similarity assessment. The average distance between groups served as the measurement standard. At a distance of five, 15 sample batches were divided into two main clusters (Fig. 3A). S1–S4, S6–S9, and S13 were assigned to Cluster І while all others were assigned to Cluster ІІ. After standardized processing and operation of the original data matrix, the four main components with eigenvalues greater than 1 were extracted, and their cumulative contribution rate has reached to be 93.21%. The components and their corresponding eigenvalues were shown in a scree plot (Fig. 3B). Curve steepness increases with the components contribution rate. In accordance with similarity analysis, these results indicated that the chemical constituents in Cyanotis arachnoidea samples were different although there were high similarities among the samples collected in various areas.
Figure 3. Screening of potential quality markers via chemical pattern recognition techniques.
(A) Hierarchical clustering analysis (HCA) of fifteen Cyanotis arachnoidea sample batches. (B) Scree plot of Cyanotis arachnoidea samples. Curve steepness increases with the component contribution rate. (C) PCA score plot of Cyanotis arachnoidea samples. (D) Loading plot of Cyanotis arachnoidea samples. Variable influence increases with the distance from the origin. (E) PLS-DA score plot of Cyanotis arachnoidea samples and cluster separation. (F) Importance variable in projection (VIP) scores of the peak area variables of the samples in the PLS-DA model.
Principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA)
Principal component analyses (PCA) and partial least squares discriminant analyses (PLS-DA) were conducted in Simca-P v. 14.1 to screen components as potential quality markers and identify the differences in the chemical constituents and quality of the Cyanotis arachnoidea samples. The areas of the twenty common characteristic peaks generated by the fifteen sample batches were used as the original variables.
The clusters containing similar samples and the separation of the sample classes can be displayed in score plost (Yu et al., 2022). The PCA score plot (Fig. 3C) showed that the 15 batches of samples were divided into two clusters. The S10–S13 samples from Jinping were in Cluster І, all others were in Cluster ІІ, and the results were roughly consistent with those of the HCA. The influences of the variables on the principal component weight increased with the distance from the origin. The eigenvalues of the four principal components extracted were 8.82, 3.17, 1.49, and 1.34, respectively. The cumulative variance contribution rate for the first four principal components have reached was 98.9%, indicating that the four principal components can reflect the basic characteristics and most of the information for Cyanotis arachnoidea. Loading plots showed that the similarities between the variables and their effects on the scores. Loading and influence on the scores increase with the distance of the variables from the origin (Pan et al., 2022). Thus, the chromatographic peaks 3, 5, 6, 9, 12 and 13 contributioned the most to the principal components (Fig. 3D). These six components might mainly account for the observed differences among samples of different origins.
PLS-DA was also performed to demonstrate the differences in Cyanotis arachnoidea of different origins. The PLS-DA also divided the 15 sample batches into two clusters (Fig. 3E). Cross-validation disclosed that the model interpretation rate parameter R2X was 0.932, and the prediction ability parameters Q2 was 0.574, indicated that the model was both stable, reliable, and with high predictive abilities. Then the variable importance in projection (VIP) values of the 20 sample peak area variables in the PLS-DA model were extracted to screen the components explaining the difference among the fifteen sample batches. The components 20-Hydroxyecdysone, 3-O-acetyl-20-hydroxyecdysone, Ajugasterone C and 2-O-acetyl-20-hydroxyecdysone all had VIP scores > 1 (Jiang et al., 2022). For this reason, they were deemed potential quality markers distinguishing Cyanotis arachnoidea samples from different origins (Fig. 3F).
Network pharmacology analysis
Target gene screening and interaction network construction
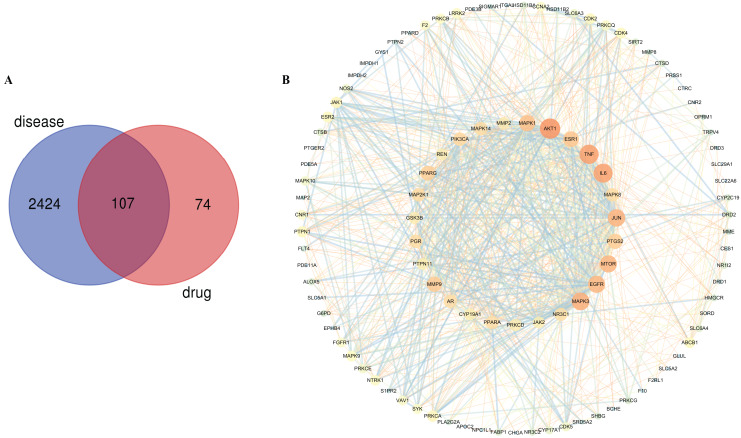
A network pharmacological method was performed to predict the pharmacodynamic activity of the quality markers, explore the underlying hypoglycemic mechanisms of Cyanotis arachnoidea, and identify the potential targets and corresponding pathways of Cyanotis arachnoidea in the treatment of DM. Chemical information for four potential quality markers was obtained from the PubChem database and 181 predictive targets were obtained from BATMAN-TCM. DM-related targets were identified by searching the DisGeNET, GeneCards, DrugBank, and OMIM databases using the keyword “diabetes mellitus”. We obtained 2,571 genes related to DM deleting duplicated targets. The Cyanotis arachnoidea and DM targets were uploaded to the online tool Venny v. 2.8 (http://bioinfogp.cnb.csic.es/tools/venny/) for mapping and intersection. A total of 107 intersection target genes were obtained and they were considered to be the potential targets of Cyanotis arachnoidea in DM treatment (Fig. 4A).
Figure 4. Target genes predcition of Cyanotis arachnoidea in DM treatment.
After intersection of 181 compounds target genes and 2,571 disease target genes, 107 target genes associated with DM treatment by Cyanotis arachnoidea were obtained (A). The PPI network of the intersecting target genes was established and 27 core target genes were selected according to their degree value (B). The nodes in the inner circle represented the core target genes and the node size and color represented the degree. The node from tiny to large is represented by the degree from low to high, and the color is orange to cyan. Edge betweenness was expressed by edge size and color. The border between low and high symbolized the transition from thin to thick and the change in hue from orange to cyan. The darker the color, the more important of the targets in DM treatment.
The intersecting target genes were imported into the STRING database for the construction of a protein-protein interaction (PPI) network. There were 107 nodes (target proteins) and 878 edges (interactions) in the network (Fig. 4B). Cytoscape v. 3.7.2 software was used to visualize the protein interactions and the topological analysis was performed by the NetworkAnalyzer plugin. Based on the degree value, the betweenness centrality (BC) and the closeness centrality (CC) of each node in the network, take more than double median as the standard, 27 target genes were selected with degree ≥ 24, BC ≥ 0.0056 and CC ≥ 0.9722. These were the core targets implicated in DM treatment by Cyanotis arachnoidea (Table 1).
Table 1. Detailed diagram of the overall study design.
| No. | Name | Betweenness centrality | Closeness centrality | Degree |
|---|---|---|---|---|
| 1 | AKT1 | 0.1443 | 0.7047 | 61 |
| 2 | TNF | 0.0696 | 0.6731 | 56 |
| 3 | IL6 | 0.0712 | 0.6731 | 55 |
| 4 | MAPK3 | 0.0390 | 0.6442 | 50 |
| 5 | JUN | 0.0296 | 0.6364 | 48 |
| 6 | EGFR | 0.0574 | 0.6325 | 47 |
| 7 | MTOR | 0.0597 | 0.6325 | 47 |
| 8 | MAPK1 | 0.0183 | 0.6105 | 43 |
| 9 | ESR1 | 0.0288 | 0.6034 | 41 |
| 10 | MMP9 | 0.0379 | 0.5966 | 39 |
| 11 | PTGS2 | 0.0308 | 0.5966 | 38 |
| 12 | PPARG | 0.0343 | 0.5866 | 37 |
| 13 | PIK3CA | 0.0091 | 0.5526 | 32 |
| 14 | MAPK8 | 0.0076 | 0.5738 | 31 |
| 15 | MAPK14 | 0.0153 | 0.5645 | 31 |
| 16 | MAP2K1 | 0.0058 | 0.5615 | 31 |
| 17 | PGR | 0.0211 | 0.5556 | 31 |
| 18 | AR | 0.0233 | 0.5615 | 31 |
| 19 | PPARA | 0.0348 | 0.5707 | 31 |
| 20 | NR3C1 | 0.0202 | 0.5585 | 30 |
| 21 | MMP2 | 0.0069 | 0.5469 | 26 |
| 22 | PRKCD | 0.0176 | 0.5224 | 26 |
| 23 | REN | 0.0319 | 0.5497 | 25 |
| 24 | GSK3B | 0.0093 | 0.5440 | 25 |
| 25 | PTPN11 | 0.0032 | 0.5198 | 24 |
| 26 | CYP19A1 | 0.0129 | 0.5385 | 24 |
| 27 | JAK2 | 0.0085 | 0.5303 | 24 |
Pathway analysis and construction of the compound-target-pathway network
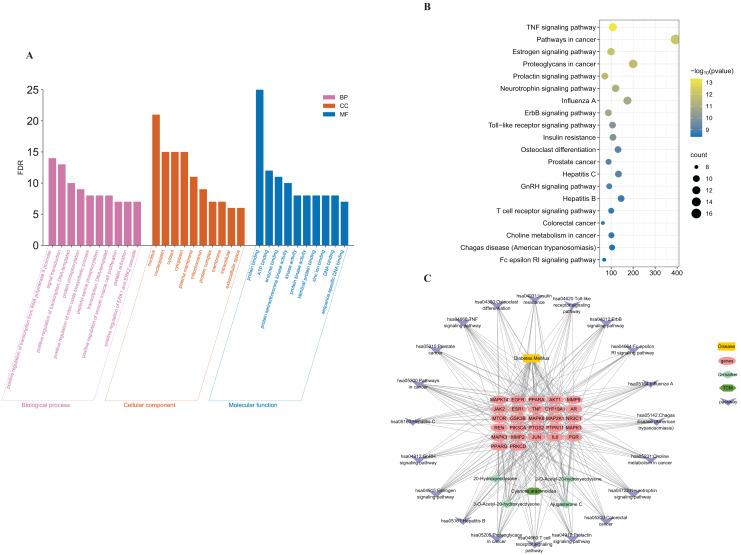
In order to predict the key pathways of Cyanotis arachnoidea for DM treatment, GO analysis and KEGG pathway enrichment analyses of 27 core targets were conducted using the DAVID database. For the GO analysis, a total of 295 GO entries with P < 0.05 were obtained, including 230 biological process (BP), 42 cell component (CC), and 23 molecular function (MF). The top 10 BP, CC, and MF entries were selected for visualization (Fig. 5A). The GO analysis of the core target genes showed that the “biological” process term included mainly RNA polymerase II promoter transcription, protein phosphorylation, protein kinase activity, protein serine/threonine kinase activity, biological membrane protein interaction, and sequence-specific DNA binding.
Figure 5. Enrichment of GO and KEGG pathway analysis and compound-target-pathway network establishment.
GO enrichment analysis including the top 10 BP, MF, and CC entries. Histogram height is proportional to the number of biological processes expressed by the enrichment score (A). In the KEGG pathway analysis (top 20), the node size represents the number of enriched target genes, and the node color progression from green to red indicates a decrease in P value (B). The compound-target-pathway network was constructed by linking the components, and their potential targets, and biological pathways. The yellow, red, dark green, light green, and purple nodes represent the disease, core target genes, herb, four quality markers, and pathways, respectively (C).
A total of 101 KEGG pathways were obtained from the DAVID database and the top 20 were selected based on their-log P values (Fig. 5B, Table 2). The KEGG pathway enrichment analysis showed that the 27 core target genes were related mainly to the tumor necrosis factor (TNF), estrogen signaling, insulin resistance, JAK-STAT, and other pathways. Hence, Cyanotis arachnoidea may treat DM by acting on multiple signaling pathways. Moreover, there may be complex interactions among these pathways.
Table 2. The KEGG enrichment analysis information.
| ID | Term | Count | P value | Genes |
|---|---|---|---|---|
| hsa04668 | TNF signaling pathway | 12 | 4.87E−14 | IL6/MAP2K1/JUN/MAPK8/PIK3CA/MAPK1/AKT1/MAPK14/PTGS2/TNF/MMP9/MAPK3 |
| hsa05200 | Pathways in cancer | 16 | 7.53E−13 | GSK3B/MAP2K1/JUN/MMP2/PTGS2/MMP9/EGFR/MTOR/AR/IL6/MAPK8/PIK3CA/AKT1/MAPK1/PPARG/MAPK3 |
| hsa04915 | Estrogen signaling pathway | 11 | 1.05E−12 | MAP2K1/JUN/PIK3CA/MMP2/PRKCD/MAPK1/AKT1/ESR1/MMP9/EGFR/MAPK3 |
| hsa05205 | Proteoglycans in cancer | 13 | 1.78E−12 | MAP2K1/MMP2/PTPN11/MAPK14/ESR1/TNF/MMP9/EGFR/MTOR/PIK3CA/AKT1/MAPK1/MAPK3 |
| hsa04917 | Prolactin signaling pathway | 10 | 2.14E−12 | GSK3B/MAP2K1/MAPK8/PIK3CA/MAPK1/AKT1/JAK2/MAPK14/ESR1/MAPK3 |
| hsa04722 | Neurotrophin signaling pathway | 11 | 7.51E−12 | GSK3B/MAP2K1/JUN/MAPK8/PIK3CA/PRKCD/MAPK1/AKT1/PTPN11/MAPK14/MAPK3 |
| hsa05164 | Influenza A | 12 | 1.10E−11 | GSK3B/IL6/MAP2K1/JUN/MAPK8/PIK3CA/MAPK1/AKT1/JAK2/MAPK14/TNF/MAPK3 |
| hsa04012 | ErbB signaling pathway | 10 | 1.42E−11 | GSK3B/MAP2K1/JUN/MAPK8/PIK3CA/MAPK1/AKT1/EGFR/MTOR/MAPK3 |
| hsa04620 | Toll-like receptor signaling pathway | 10 | 8.73E−11 | IL6/MAP2K1/JUN/MAPK8/PIK3CA/MAPK1/AKT1/MAPK14/TNF/MAPK3 |
| hsa04931 | Insulin resistance | 10 | 1.04E−10 | GSK3B/IL6/MAPK8/PIK3CA/PRKCD/AKT1/PTPN11/PPARA/TNF/MTOR |
| hsa04380 | Osteoclast differentiation | 10 | 5.94E−10 | MAP2K1/JUN/MAPK8/PIK3CA/MAPK1/AKT1/PPARG/MAPK14/TNF/MAPK3 |
| hsa05215 | Prostate cancer | 9 | 6.72E−10 | GSK3B/AR/MAP2K1/PIK3CA/MAPK1/AKT1/EGFR/MTOR/MAPK3 |
| hsa05160 | Hepatitis C | 10 | 6.81E−10 | GSK3B/MAPK8/PIK3CA/MAPK1/AKT1/MAPK14/PPARA/TNF/EGFR/MAPK3 |
| hsa04912 | GnRH signaling pathway | 9 | 8.83E−10 | MAP2K1/JUN/MAPK8/MMP2/PRKCD/MAPK1/MAPK14/EGFR/MAPK3 |
| hsa05161 | Hepatitis B | 10 | 1.48E−09 | IL6/MAP2K1/JUN/MAPK8/PIK3CA/MAPK1/AKT1/TNF/MMP9/MAPK3 |
| hsa04660 | T cell receptor signaling pathway | 9 | 1.89E−09 | GSK3B/MAP2K1/JUN/PIK3CA/MAPK1/AKT1/MAPK14/TNF/MAPK3 |
| hsa05210 | Colorectal cancer | 8 | 1.96E−09 | GSK3B/MAP2K1/JUN/MAPK8/PIK3CA/MAPK1/AKT1/MAPK3 |
| hsa05231 | Choline metabolism in cancer | 9 | 2.05E−09 | MAP2K1/JUN/MAPK8/PIK3CA/MAPK1/AKT1/EGFR/MTOR/MAPK3 |
| hsa05142 | Chagas disease (American trypanosomiasis) | 9 | 2.59E−09 | IL6/JUN/MAPK8/PIK3CA/MAPK1/AKT1/MAPK14/TNF/MAPK3 |
| hsa04664 | Fc epsilon RI signaling pathway | 8 | 3.81E−09 | MAP2K1/MAPK8/PIK3CA/MAPK1/AKT1/MAPK14/TNF/MAPK3 |
Compound-target-pathway network construction
The compound-target-pathway network contained four potential quality markers, 27 core targets, and 20 pathways, and was constructed with Cytoscape v. 3.7.2 (Fig. 5C). The network contained 282 edges and 53 nodes representing 27 core target genes, four active components, one disease, one compound prescription, and the top 20 KEGG pathways. The results indicated that the potential quality markers in Cyanotis arachnoidea might play roles in the treatment of DM by regulating multiple target proteins and acting on multiple pathways.
The compound-target-pathway network revealed that all four quality markers were phytosterones. According to modern pharmacology, 20-Hydroxyecdysone has antioxidant, anti-microbial, anti-inflammatory, and glucose-regulating activity. DM is closely associated with oxidative stress (Lima, Moreira & Sakamoto-Hojo, 2021), inflammation (Takahashi et al., 2022) and cancer (Carstensen et al., 2016). The disorders of glucose and lipid metabolism associated with DM activate the immune system, and macrophages synthesize and secrete various proinflammatory factors such as TNF-α, IL-1β, and IL-6 (Oliver et al., 2010). These substances affect the blood and the paracrine, the sensitivity of target tissues to insulin, and the function of islet β-cells (Kitade et al., 2012). Analyses of the KEGG enrichment and the core target genes revealed that the hypoglycemic efficacy of Cyanotis arachnoidea may be attributed to the amelioration of insulin resistance, antioxidant and anti-inflammatory activity, and impact on the TNF-α signaling pathway.
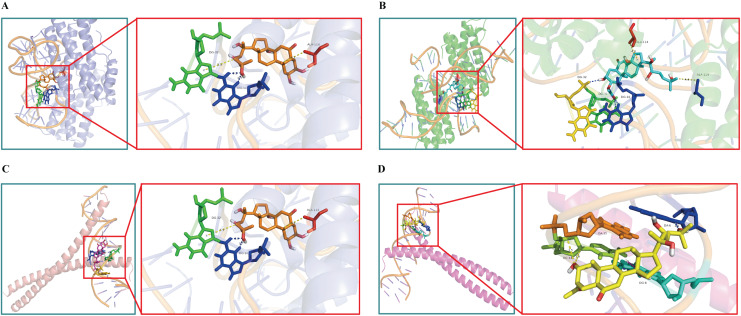
Molecular docking verification
Molecular docking strategy was used to further verify the activity of the quality markers and disclose the binding pattern between the active components and the core genes. AutoDock Vina was used to dock the four potential quality markers with the top five target proteins. The latter were selected according to their degree values and included Akt1 (PDB ID: 4EKL), TNF (PDB ID: 2AZ5), IL-6 (PDB ID: 4NI9), MAPK3 (PDB ID: 2ZOQ), and JUN (PDB ID: 5T01). The binding energy docking results were listed in Table 3. Docking groups with the strongest target binding energies were selected for each component and visualized in PyMOL (Fig. 6). The results show that all the potential quality markers had molecular binding energies less than −6.7 kcal/mol with their corresponding core targets, indicating that each compound had a good affinity for its target proteins. The four quality markers could be the major active ingredients of Cyanotis arachnoidea for DM treatment. Moreover, their antihyperglycemic effect may be mediated by regulating the activity of Akt1, TNF, IL-6, MAPK3, and JUN activity.
Table 3. Binding energies of the quality markers with top five core targets.
| Number | Compound | Binding energies (kcal/mol) | ||||
|---|---|---|---|---|---|---|
| AKT1 | TNF | IL-6 | MAPK3 | JUN | ||
| 1 | 20-Hydroxyecdysone | −8.2 | −8.1 | −9.2 | −8.6 | −9.1 |
| 2 | 3-O-Acetyl-20-hydroxyecdysone | −7.7 | −7.2 | −9.1 | −8.3 | −9.2 |
| 3 | Ajugasterone C | −7.7 | −8.5 | −8.6 | −8.3 | −9.0 |
| 4 | 2-O-Acetyl-20-hydroxyecdysone | −8.2 | −7.5 | −9.5 | −8.7 | −9.5 |
Figure 6. Molecular docking diagrams.
(A) 20-Hydroxyecdysone with IL-6 (Binding eneygy = −9.2 kcal/mol); (B) 2-O-Acetyl-20-hydroxyecdysone with IL-6 (Binding eneygy = −9.5 kcal/mol); (C) Ajugasterone C with JUN (Binding eneygy = −9.0 kcal/mol); (D) 3-O-Acetyl-20-hydroxyecdysone with JUN (Binding eneygy = −9.2 kcal/mol). Proteins shown in cartoon mode are represented by different colors. Hydrogen bonds are depicted as yellow dotted lines, and the ligands are represented by sticks.
In vitro validation
It can be seen from Table S3 that the cell supernatant glucose concentration was significantly higher for insulin resistance than the control group (P < 0.05). It shows that the Cyanotis arachnoidea extract increased the glucose uptake and utilization by the islet β-cells, effectively lowered glucose concentration, alleviated insulin resistance, and exhibited hypoglycemic activity.
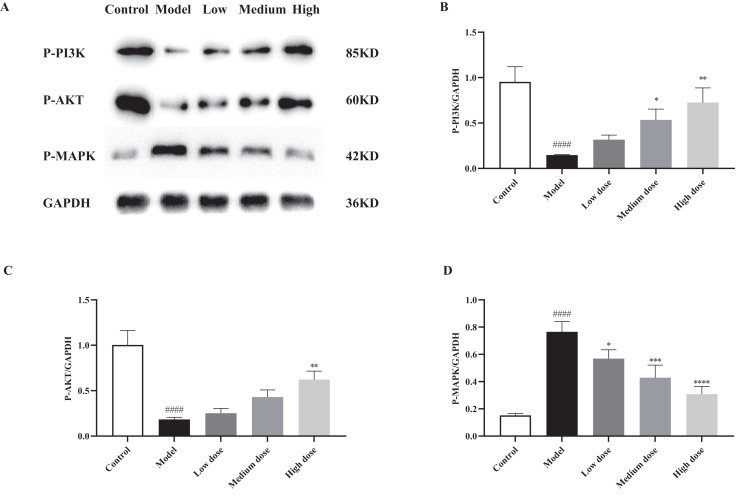
p-PI3K/GAPDH and p-Akt/GAPDH were 0.15 ± 0.01 and 0.18 ± 0.02 in insulin resistance group, and were significantly lower than those in the blank control group (P < 0.05). p-MAPK/GAPDH was 0.77 ± 0.06 which was significantly higher than that of the blank control group (P < 0.05). Compared with the insulin resistance group, the p-PI3K and p-Akt protein expression levels significantly increased while that of p-MAPK significantly decreased with increasing Cyanotis arachnoidea extract dose (P < 0.05; Figs. 7A–7B and Table S4). The PI3K/Akt signaling pathway plays an important role in blood glucose regulation. Insulin resistance is related to the dysregulated signal transduction of the PI3K/Akt pathway and the overexpression of MAPK. The results of this study were consistent with those previously reported (Zheng et al., 2022). The expression levels of the phosphorylated PI3K and Akt proteins were higher in the 200 and 400 mg/L Cyanotis arachnoidea extract groups than in the insulin resistance group (P < 0.05). The glucose concentration was significantly lower in the cell culture medium than in the insulin resistance group. Thus, Cyanotis arachnoidea upregulates PI3K and Akt proteins, strengthens the signal transduction barrier of the PI3K/Akt signal pathway, inhibits the MAPK signal pathway, enhances glucose intake, improves insulin resistance, and play a hypoglycemic role in insulin-resistant pancreatic islet cells.
Figure 7. Protein expression results (±s, n = 3).
(A) Electrophoregram of the p-PI3K, p-Akt and p-MAPK proteins in the control, insulin resistance model, and various Cyanotis arachnoidea extract intervention groups. (B) Relative phosphorylated PI3K protein expression. (C) Relative phosphorylated Akt protein expression. (D) Relative phosphorylated MAPK protein expression. *: Compared with the model group, P < 0.05; **: Compared with the model group, P < 0.01; ***: Compared with the model group, P < 0.001; ****: Compared with the model group, P < 0.0001; ####: Compared with the blank control group, P < 0.0001.
Quantitative analysis
To ensure the efficacy and consistency, the content of quality markers should be monitored. UHPLC was uesd for the simultaneous quantification of the four quality marker compounds. The UHPLC analysis was quantitatively validated in terms of linearity, precision, repeatability, stability, and recovery.
According to method validation results of linearity, all calibration curves showed good linear ranges (r2 > 0.999) and simultaneously quantified the four quality markers in the samples (Table 4). Standard curves for the four components are shown in Fig. S1.
Table 4. Calibration curves for four potential active ingredient markers (n = 6).
| Analyte | Calibration curve | r 2 | Linear range (µg/mL) |
|---|---|---|---|
| 20-Hydroxyecdysone | Y = 5.49 × 106 X − 2.33 × 104 | 0.9997 | 0.0344–0.8602 |
| 3-O-Acetyl-20-hydroxyecdysone | Y = 4.02 × 106 X − 6.01 × 103 | 1.0000 | 0.0092–0.2300 |
| Ajugasterone C | Y = 4.38 × 106 X − 3.91 × 103 | 0.9996 | 0.0050–0.1256 |
| 2-O-Acetyl-20-hydroxyecdysone | Y = 4.61 × 106 X − 4.35 × 103 | 0.9998 | 0.0051–0.1281 |
The relative standard deviations (RSDs) for the precision, repeatability, and stability analyses of the four potential quality markers were all <3%. The result indicated that the instrument precision, method repeatability, and sample stability were good. Recovery of all four components was in the range of 98.72–101.44% which proved that the method was accurate (Table S5).
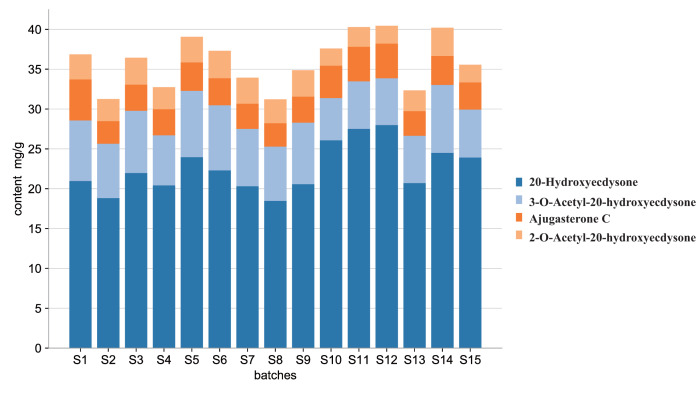
The contents of the four quality markers in fifteen batches were quantified, and the results were shown in Fig. 8. The most abundant component in Cyanotis arachnoidea was 20-Hydroxyecdysone (18.47–27.99 mg/g) followed by 3-O-Acetyl-20-hydroxyecdysone (5.30–8.54 mg/g), Ajugasterone C (2.84–5.16 mg/g) and 2-O-Acetyl-20-hydroxyecdysone (2.16–3.54 mg/g) (Table S6). The relative differences in the amounts of the components among the various batches indicate that the Cyanotis arachnoidea collected from different producing areas vary in composition and quality. The established method provided a reference for the control and evaluation of the quality of Cyanotis arachnoidea. The four components including 20-Hydroxyecdysone, 3-O-Acetyl-20-hydroxyecdysone, Ajugasterone C and 2-O-Acetyl-20-hydroxyecdysone could be considered as the quality markers of Cyanotis arachnoidea.
Figure 8. Histogram of quality markers content.
Content of the four quality markers in 15 sample batches of Cyanotis arachnoidea collected from different origins. Different colors represent different components and 20-Hydroxyecdysone had the highest content of all four quality markers.
Conclusion
In this study, we used chemical pattern recognition and network pharmacology to design and test a method of evaluating the overall quality of the TCM Cyanotis arachnoidea. We explored its quality markers and predicted the molecular mechanism in treating diabetes mellitus. UHPLC was used to characterize the chemical constituents of Cyanotis arachnoidea and analyze 15 sample batches collected from five different origins in Yunnan Province, China. The fingerprints of medicinal materials were established. The chromatographic peaks, HCA, PCA, and PLS-DA identified and screened 20-Hydroxyecdysone, 3-O-Acetyl-20-hydroxyecdysone, Ajugasterone C and 2-O-Acetyl-20-hydroxyecdysone as potential quality markers of Cyanotis arachnoidea. We predicted their biological activity via network pharmacology and molecular docking. We determined that Cyanotis arachnoidea may mediate its hypoglycemic efficacy by regulating its target genes encoding Akt1, TNF, IL-6, MAPK3, and JUN. The hypoglycemic property might also be explained by its impact on the PI3K/Akt, MAPK, TNF, TLR, and estrogen signaling, cancer, insulin resistance, and JAK-STAT pathways. In vitro experiments on pancreatic islet β-cells confirmed that Cyanotis arachnoidea enhances glucose uptake, improves insulin resistance and play a hypoglycemic role by promoting the PI3K/Akt and inhibiting the MAPK signal pathways. The foregoing results suggest that the quality markers in Cyanotis arachnoidea may act on the entire biological network rather than a single target gene. The present work demonstrated the integrity and complexity of Cyanotis arachnoidea function and also explained the mechanism of Cyanotis arachnoidea in treating diabetes mellitus with multiple components, gene targets, and signal pathways.
This study preliminarily identified the quality markers and potential pharmacological components of Cyanotis arachnoidea. Nevertheless, the hypoglycemic efficacy and modes of action of each active ingredient in this TCM must be explored and elucidated. This information will facilitate the utilization and optimization of Cyanotis arachnoidea for the clinical treatment of diabetes mellitus.
Supplemental Information
Funding Statement
This work was supported by the Hebei Provincial Key Research and Development Projects (No. 23372502D); and the Natural Science Foundation of Hebei Province (Key Program) (Grant/Award Number: H2022423335). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Liying Niu, Email: niuliyingyy@163.com.
Xinguo Wang, Email: wangxinguozy@163.com.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Jingnan Hu performed the experiments, authored or reviewed drafts of the article, and approved the final draft.
Yu Feng performed the experiments, authored or reviewed drafts of the article, and approved the final draft.
Baolin Li analyzed the data, prepared figures and/or tables, and approved the final draft.
Fengxia Wang analyzed the data, prepared figures and/or tables, and approved the final draft.
Qi Qian performed the experiments, authored or reviewed drafts of the article, and approved the final draft.
Wei Tian performed the experiments, authored or reviewed drafts of the article, and approved the final draft.
Liying Niu conceived and designed the experiments, prepared figures and/or tables, and approved the final draft.
Xinguo Wang conceived and designed the experiments, prepared figures and/or tables, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental File.
References
- Amberger et al. (2019).Amberger JS, Bocchini CA, Scott AF, Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Research. 2019;47(D1):D1038–D1043. doi: 10.1093/nar/gky1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswal et al. (2020).Aswal S, Kumar A, Chauhan A, Semwal RB, Kumar A, Semwal DK. A molecular approach on the protective effects of mangiferin against diabetes and diabetes-related complications. Current Diabetes Reviews. 2020;16(7):690–698. doi: 10.2174/1573399815666191004112023. [DOI] [PubMed] [Google Scholar]
- Baev et al. (2022).Baev AY, Charishnikova OS, Khasanov FA, Nebesnaya KS, Makhmudov AR, Rakhmedova MT, Khushbaktova ZA, Syrov VN, Levitskaya YV. Ecdysterone prevents negative effect of acute immobilization stress on energy metabolism of rat liver mitochondria. The Journal of Steroid Biochemistry and Molecular Biology. 2022;219(2011):106066. doi: 10.1016/j.jsbmb.2022.106066. [DOI] [PubMed] [Google Scholar]
- Bai et al. (2018).Bai G, Zhang T, Hou Y, Ding G, Jiang M, Luo G. From quality markers to data mining and intelligence assessment: a smart qualityevaluation strategy for traditional Chinese medicine based on quality markers. Phytomedicine. 2018;44:109–116. doi: 10.1016/j.phymed.2018.01.017. [DOI] [PubMed] [Google Scholar]
- Baig et al. (2016).Baig MH, Ahmad K, Roy S, Ashraf JM, Adil M, Siddiqui MH, Khan S, Kamal MA, Provazník I, Choi I. Computer aided drug design: success and limitations. Current Pharmaceutical Design. 2016;22(5):572–581. doi: 10.2174/1381612822666151125000550. [DOI] [PubMed] [Google Scholar]
- Barshir et al. (2021).Barshir R, Fishilevich S, Iny-Stein T, Zelig O, Mazor Y, Guan-Golan Y, Safran M, Lancet D. GeneCaRNA: a comprehensive gene-centric database of human non-coding RNAs in the GeneCards suite. Journal of Molecular Biology. 2021;433(11):166913. doi: 10.1016/j.jmb.2021.166913. [DOI] [PubMed] [Google Scholar]
- Bu, Zhang & Guo (2013).Bu YH, Zhang XD, Guo WS. Determination of ecdysterone in Cyanotis arachnoidea capsules by HPLC. Chinese Traditional Patent Medicine. 2013;35(10):2301–2302. doi: 10.3969/j.issn.1001-1528.2013.10.058. [DOI] [Google Scholar]
- Burley et al. (2022).Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV, Duarte JM, Dutta S, Fayazi M, Feng Z, Flatt JW, Ganesan SJ, Goodsell DS, Ghosh S, Kramer Green R, Guranovic V, Henry J, Hudson BP, Lawson CL, Liang Y, Lowe R, Peisach E, Persikova I, Piehl DW, Rose Y, Sali A, Segura J, Sekharan M, Shao C, Vallat B, Voigt M, Westbrook JD, Whetstone S, Young JY, Zardecki C. RCSB protein data bank: celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D. Protein Science. 2022;31(1):187–208. doi: 10.1002/pro.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstensen et al. (2016).Carstensen B, Read SH, Friis S, Sund R, Keskimäki I, Svensson AM, Ljung R, Wild SH, Kerssens JJ, Harding JL, Magliano DJ, Gudbjörnsdottir S, Diabetes and Cancer Research Consortium Cancer incidence in persons with type 1 diabetes: a five-country study of 9,000 cancers in type 1 diabetic individuals. Diabetologia. 2016;59(5):980–988. doi: 10.1007/s00125-016-3884-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Xia & Qiu (2006).Chen Q, Xia YP, Qiu ZY. Effect of ecdysterone on insulin sensitivity and glucose metabolism in insulin-resistant cell model. Chinese Pharmacological Bulletin. 2006;22(4):460–464. doi: 10.3321/j.issn:1001-1978.2006.04.018. [DOI] [Google Scholar]
- Chen et al. (2017).Chen L, Zheng SJ, Huang M, Ma XH, Yang J, Deng SH, Huang Y, Wen YZ, Yang XZ. β-ecdysterone from Cyanotis arachnoidea exerts hypoglycemic effects through activating IRS-1/Akt/GLUT4 and IRS-1/Akt/GLUT2 signal pathways in KK-Ay mice. Journal of Functional Foods. 2017;39(6821):123–132. doi: 10.1016/j.jff.2017.09.061. [DOI] [Google Scholar]
- Gu, Hall & Miles (2016).Gu F, Hall P, Miles NJ. Performance evaluation for composites based on recycled polypropylene using principal component analysis and cluster analysis. Journal of Cleaner Production. 2016;115(Mar.1):343–353. doi: 10.1016/j.jclepro.2015.12.062. [DOI] [Google Scholar]
- Gu et al. (2021).Gu Y, Yang X, Shang C, Thao TTP, Koyama T. Inhibition and interactions of alpha-amylase by daucosterol from the peel of Chinese water chestnut (Eleocharis dulcis) Food & Function. 2021;12(18):8411–8424. doi: 10.1039/d1fo00887k. [DOI] [PubMed] [Google Scholar]
- Guizhou Institute of TCM (1970).Guizhou Institute of TCM . Guizhou herbal medicine (episode 2) Guiyang: Guizhou People’s Publishing House; 1970. [Google Scholar]
- Hu et al. (2019).Hu H, Peng L, Jiang H, Shen H, Zhou P, Gao Y. Silenced CHOP protects pancreatic B-cell function by targeting peroxisome proliferator-activated receptor-γ coactivator-1α through nuclear factor-κB signaling pathway in diabetes mellitus. Journal of Cellular Biochemistry. 2019;120(8):12595–12603. doi: 10.1002/jcb.28526. [DOI] [PubMed] [Google Scholar]
- Jiang et al. (2022).Jiang M, Cao J, Zhang C, Su B, Wang S, Ning N, Lei T, Li P. A comprehensive strategy for quality evaluation of Wushe Zhiyang pills by integrating UPLC-DAD fingerprint and multi-ingredients rapid quantitation with UPLC-MS/MS technology. Journal of Pharmaceutical and Biomedical Analysis. 2022;210:114556. doi: 10.1016/j.jpba.2021.114556. [DOI] [PubMed] [Google Scholar]
- Jiang et al. (2022).Jiang JL, Kang A, Li Q, Song MM, Han HF, Cai YQ, Xu ZC, Cheng HB, Sun DD. Multi-index components of Scutellariae Barbatae Herba according to UHPLC-QTRAP-MS coupled with chemometrics. China Journal of Chinese Materia Medica. 2022;47(2):437–443. doi: 10.19540/j.cnki.cjcmm.20210901.201. [DOI] [PubMed] [Google Scholar]
- Jiao et al. (2021).Jiao X, Sherman BT, da Huang W, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2021;28(13):1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur & Khatik (2021).Kaur P, Khatik G. An overview of computer-aided drug design tools and recent applications in designing of anti-diabetic agents. Current Drug Targets. 2021;22(10):1158–1182. doi: 10.2174/1389450121666201119141525. [DOI] [PubMed] [Google Scholar]
- Kitade et al. (2012).Kitade H, Sawamoto K, Nagashimada M, Inoue H, Yamamoto Y, Sai Y, Takamura T, Yamamoto H, Miyamoto K, Ginsberg HN, Mukaida N, Kaneko S, Ota T. CCR5 plays a critical role in obesity-induced adipose tissue inflammation and insulin resistance by regulating both macrophage recruitment and M1/M2 status. Diabetes. 2012;61(7):1680–1690. doi: 10.2337/db11-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan & Su (2017).Lan M, Su GY. Diannan materia medica. Kunming: Yunnan People’s Publishing House; 2017. [Google Scholar]
- Li et al. (2021).Li T, Yang WZ, Song TX, Liu CJ, Jiang MM. Integrating chemical profiling and network pharmacology analysis based on anti-inflammatory effects for quality control of Scutellaria barbata. Phytochemical Analysis. 2021;32(6):1141–1151. doi: 10.1002/pca.3055. [DOI] [PubMed] [Google Scholar]
- Li et al. (2021).Li ZT, Zhang FX, Fan CL, Ye MN, Chen WW, Yao ZH, Yao XS, Dai Y. Discovery of potential Q-marker of traditional Chinese medicine based on plant metabolomics and network pharmacology: periplocae cortex as an example. Phytomedicine. 2021;85:153535. doi: 10.1016/j.phymed.2021.153535. [DOI] [PubMed] [Google Scholar]
- Lim et al. (2020).Lim HS, Moon BC, Lee J, Choi G, Park G. The insect molting hormone 20-hydroxyecdysone protects dopaminergic neurons against MPTP-induced neurotoxicity in a mouse model of Parkinson’s disease. Free Radical Biology & Medicine. 2020;159(1):23–36. doi: 10.1016/j.freeradbiomed.2020.07.010. [DOI] [PubMed] [Google Scholar]
- Lima, Moreira & Sakamoto-Hojo (2021).Lima JEBF, Moreira NCS, Sakamoto-Hojo ET. Mechanisms underlying the pathophysiology of type 2 diabetes: from risk factors to oxidative stress, metabolic dysfunction, and hyperglycemia. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 2021;874–875(45):503437. doi: 10.1016/j.mrgentox.2021.503437. [DOI] [PubMed] [Google Scholar]
- Liu, Guo & Liu (2018).Liu C, Guo DA, Liu L. Quality transitivity and traceability system of herbal medicine products based on quality markers. Phytomedicine. 2018;44(1–2):247–257. doi: 10.1016/j.phymed.2018.03.006. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2016).Liu Z, Guo F, Wang Y, Li C, Zhang X, Li H, Diao L, Gu J, Wang W, Li D, He F. BATMAN-TCM: a bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Scientific Reports. 2016;6(1):21146. doi: 10.1038/srep21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall et al. (2021).Marschall MJM, Ringseis R, Gessner DK, Grundmann SM, Most E, Wen G, Maheshwari G, Zorn H, Eder K. Effect of ecdysterone on the hepatic transcriptome and lipid metabolism in lean and obese Zucker rats. International Journal of Molecular Sciences. 2021;22(10):5241. doi: 10.3390/ijms22105241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustapha et al. (2021).Mustapha S, Mohammed M, Azemi AK, Jatau AI, Shehu A, Mustapha L, Aliyu IM, Danraka RN, Amin A, Bala AA, Ahmad WANW, Rasool AHG, Mustafa MR, Mokhtar SS. Current status of endoplasmic reticulum stress in type II diabetes. Molecules. 2021;26(14):4362. doi: 10.3390/molecules26144362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie & Qiu (1987).Nie RL, Qiu MH. The ecdysterone in the ecdysone waste residue produced by Cyanotis arachnoidea industrial. Acta Botanica Yunnanica. 1987;9:253–256. [Google Scholar]
- Oliver et al. (2010).Oliver E, McGillicuddy F, Phillips C, Toomey S, Roche HM. The role of inflammation and macrophage accumulation in the development of obesity-induced type 2 diabetes mellitus and the possible therapeutic effects of long-chain n-3 PUFA. The Proceedings of the Nutrition Society. 2010;69(2):232–243. doi: 10.1017/S0029665110000042. [DOI] [PubMed] [Google Scholar]
- Omanakuttan et al. (2016).Omanakuttan A, Bose C, Pandurangan N, Kumar GB, Banerji A, Nair BG. Nitric Oxide and ERK mediates regulation of cellular processes by Ecdysterone. Experimental Cell Research. 2016;346(2):167–175. doi: 10.1016/j.yexcr.2016.07.019. [DOI] [PubMed] [Google Scholar]
- Pan et al. (2022).Pan MH, Su YF, Liu XJ, Xiang SQ, Ding YX, Li Q. Identification of Forsythia suspensa (Thunb.) Vahl in different harvest periods using intelligent sensory technologies, HPLC characteristic fingerprint coupled with chemometrics. Phytochemical Analysis. 2022;33(3):490–501. doi: 10.1002/pca.3104. [DOI] [PubMed] [Google Scholar]
- Pan et al. (2022).Pan HT, Xi ZQ, Wei XQ, Wang K. A network pharmacology approach to predict potential targets and mechanisms of “Ramulus Cinnamomi (cassiae)—Paeonia lactiflora” herb pair in the treatment of chronic pain with comorbid anxiety and depression. Annals of Medicine. 2022;54(1):413–425. doi: 10.1080/07853890.2022.2031268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero et al. (2020).Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Research. 2020;48(D1):D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin et al. (2022).Qin LL, Yu M, Zhang HX, Jia HM, Ye XC, Zou ZM. Quality markers of Baizhu dispensing granules based on multi-component qualitative and quantitative analysis combined with network pharmacology and chemometric analysis. Journal of Ethnopharmacology. 2022;288:114968. doi: 10.1016/j.jep.2022.114968. [DOI] [PubMed] [Google Scholar]
- Riyaphan et al. (2021).Riyaphan J, Pham DC, Leong MK, Weng CF. In silico approaches to identify polyphenol compounds as α-Glucosidase and α-Amylase inhibitors against type-II diabetes. Biomolecules. 2021;11(12):1877. doi: 10.3390/biom11121877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk-Drapała et al. (2021).Romaniuk-Drapała A, Lisiak N, Totoń E, Matysiak A, Nawrot J, Nowak G, Kaczmarek M, Rybczyńska M, Rubiś B. Proapoptotic and proautophagic activity of 20-hydroxyecdysone in breast cancer cells in vitro. Chemico-Biological Interactions. 2021;342(2):109479. doi: 10.1016/j.cbi.2021.109479. [DOI] [PubMed] [Google Scholar]
- Szklarczyk et al. (2021).Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Research. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi et al. (2022).Takahashi P, Xavier DJ, Lima JEBF, Evangelista AF, Collares CVA, Foss-Freitas MC, Rassi DM, Donadi EA, Passos GA, Sakamoto-Hojo ET. Transcript expression profiles and MicroRNA regulation indicate an upregulation of processes linked to oxidative stress, DNA repair, cell death, and inflammation in type 1 diabetes mellitus patients. Journal of Diabetes Research. 2022;2022:3511329. doi: 10.1155/2022/3511329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan et al. (2002).Tan CY, Wang JH, Zhang H, Li X. A new phytosterone from Cyanotis arachnoidea. Journal of Asian Natural Products Research. 2002;4(1):7–10. doi: 10.1080/10286020290019631. [DOI] [PubMed] [Google Scholar]
- Tang et al. (2020).Tang Y, Mo Y, Xin D, Zeng L, Yue Z, Xu C. β-ecdysterone alleviates osteoarthritis by activating autophagy in chondrocytes through regulating PI3K/AKT/mTOR signal pathway. American Journal of Translational Research. 2020;12(11):7174–7186. [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2021).UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Research. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang (1975).Wang G. Compilation of China’s medicinal herb. Beijing: People’s Medical Publishing House; 1975. [Google Scholar]
- Wang, Cui & Li (2019).Wang YL, Cui T, Li YZ. Prediction of quality markers of traditional Chinese medicines based on network pharmacology. Chinese Herbal Medicines. 2019;11(4):349–356. doi: 10.1016/j.chmed.2019.08.003. [DOI] [Google Scholar]
- Wishart et al. (2018).Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Research. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaopeng et al. (2016).Xiaopeng L, Yong S, Shuyi L, Bijun X, Zhida S. A-type procyanidins from litchi pericarp ameliorate hyperglycaemia by regulating hepatic and muscle glucose metabolism in streptozotocin (STZ)-induced diabetic mice fed with high fat diet. Journal of Functional Foods. 2016;27(9):711–722. doi: 10.1016/j.jff.2016.08.010. [DOI] [Google Scholar]
- Xu et al. (2020).Xu X, Niu L, Liu Y, Pang M, Lu W, Xia C, Zhu Y, Yang B, Wang Q. Study on the mechanism of Gegen Qinlian Decoction for treating type II diabetes mellitus by integrating network pharmacology and pharmacological evaluation. Journal of Ethnopharmacology. 2020;262(18):113129. doi: 10.1016/j.jep.2020.113129. [DOI] [PubMed] [Google Scholar]
- Xuguang et al. (2019).Xuguang H, Aofei T, Tao L, Longyan Z, Weijian B, Jiao G. Hesperidin ameliorates insulin resistance by regulating the IRS1-GLUT2 pathway via TLR4 in HepG2 cells. Phytotherapy Research. 2019;33(6):1697–1705. doi: 10.1002/ptr.6358. [DOI] [PubMed] [Google Scholar]
- Yi et al. (2018).Yi Q, Wang H, Shen A, Ma J. Clinical analysis of Cyanotis arachnoidea combined with metformin in the treatment of type 2 diabetes with blood glucose and C-peptide changes. Medicine and Pharmacy of Yunnan. 2018;39:463–465. [Google Scholar]
- Yu et al. (2022).Yu Y, Zhu Z, Xie M, Deng L, Xie X, Zhang M. Investigation on the Q-markers of Bushen Huoxue prescriptions for DR treatment based on chemometric methods and spectrum-effect relationship. Journal of Ethnopharmacology. 2022;285(4):114800. doi: 10.1016/j.jep.2021.114800. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2021).Zhang SJ, Zhang YG, Li DH, Wu HW, Niu JT, Si XL, Li YF. Prediction of Q-markers of Astragali Radix based on network pharmacology and fingerprint. Zhongguo Zhong Yao Za Zhi=China Journal of Chinese Materia Medica. 2021;46(11):2691–2698. doi: 10.19540/j.cnki.cjcmm.20200925.201. [DOI] [PubMed] [Google Scholar]
- Zhao et al. (2019).Zhao C, Liu H, Miao P, Wang H, Yu H, Wang C, Li Z. A strategy for selecting “Q-Markers” of Chinese medical preparation via components transfer process analysis with application to the quality control of Shengmai injection. Molecules. 2019;24(9):1811. doi: 10.3390/molecules24091811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng et al. (2022).Zheng S, Chen N, Kang X, Hu Y, Shi S. Irisin alleviates FFA induced β-cell insulin resistance and inflammatory response through activating PI3K/AKT/FOXO1 signaling pathway. Endocrine. 2022;75(3):740–751. doi: 10.1007/s12020-021-02875-y. [DOI] [PubMed] [Google Scholar]
- Zhu et al. (2022).Zhu W, Li Y, Zhao J, Wang Y, Li Y, Wang Y. The mechanism of triptolide in the treatment of connective tissue disease-related interstitial lung disease based on network pharmacology and molecular docking. Annals of Medicine. 2022;54(1):541–552. doi: 10.1080/07853890.2022.2034931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental File.