Abstract
Nested PCR assays were used for the direct identification of Coxiella burnetii plasmids in human sera. A total of 81 serum samples from 81 patients with Q fever were tested by nested PCR with four sets of primers. The first set of primers was used to detect the genomic sequences. The second set of primers was used to detect the conserved sequences of the plasmids. Another two sets of primers were used to identify the QpH1 and QpRS plasmids. QpH1 and QpRS plasmid-specific sequences were identified in 40 (49.4%) and 24 (29.6%) of the serum samples, respectively. Both of the QpH1 and QpRS plasmid-specific sequences were detected in 5 (8.6%) of the serum samples but were not found in 12 (20.7%) of the serum samples. Furthermore, all of the 23 acute-phase serum samples were positive for the QpH1 plasmid and negative for the QpRS plasmid. Nested PCR with plasmid-specific primers appears to be a useful method for the direct typing of C. burnetii plasmids in human sera.
Coxiella burnetii is an obligate intracellular bacterium that causes acute Q fever and chronic endocarditis in humans (1). Acute Q fever is a flu-like illness which is self-limiting or easily treated with antibiotics, when an appropriate diagnosis is made. Chronic Q fever is a severe disease that requires prolonged antibiotic therapy, because the infection can result in endocarditis (8, 15) or granulomatous hepatitis (21). Rapid differentiation of C. burnetii in clinical specimens is very important for the control of Q fever, because prompt antibiotic therapy may lead to a better prognosis for individuals.
Routine diagnosis of Q fever is usually based on serological tests (2, 13, 14), since isolation of C. burnetii from patients is time-consuming, difficult, and hazardous. However, serological tests offer only a retrospective diagnosis and are useless for the treatment of the afflicted patients, because antibodies cannot be detected during the early stage of the infection and it is difficult to discriminate between current and past infection by a test with a single serum sample. Also, serological tests cannot provide the ability to predict whether the patient has acute or chronic disease because they do not detect differences in C. burnetii isolates. Although the C. burnetii isolates derived from patients with acute and chronic cases of Q fever could be differentiated by their lipopolysaccharide (3), restriction fragment length polymorphism (4, 5), and plasmid (17, 20) types, these methods are difficult to perform with clinical samples, and therefore, they are useless for the early differential diagnosis of acute and chronic Q fever and for epidemiological investigations. Thus, there have been no valid methods that could be used for the differentiation of C. burnetii in clinical samples.
Five plasmid types (QpH1, QpRS, QpDG, QpDV, and plasmidless) have been found in C. burnetii. The QpH1 plasmid was first obtained from a tick isolate (16) and was also detected in most isolates originating from ticks, domestic animals (cows, goats, and sheep), and acute Q fever patients (17). The QpRS plasmid was first detected in an isolate from an aborted goat and was then found in most isolates from patients with chronic Q fever (17). The QpDG plasmid was found in only a few isolates from wild rodents (17, 22). In several isolates from humans with endocarditis, separate plasmid DNA was not isolated, but the plasmid sequences were integrated into the chromosomes of these isolates (17). A new plasmid, QpDV, was discovered in an isolate from cow’s milk and an isolate from a human with pneumonia and was also found in three isolates from human patients with acute Q fever, an aortic aneurysm, and chronic endocarditis (20). Thus, identification of C. burnetii plasmids may provide important information for the differential diagnosis of Q fever and for epidemiological investigations.
Because of the extensive serological and bacteriological evidence of Coxiella infection in animals and humans in Japan (6, 7, 11), C. burnetii was assumed to be widespread. We have cultured many isolates of C. burnetii from samples from cattle, ticks, and humans with acute Q fever (6, 7, 11). Our recent serological investigation also showed a high prevalence of antibodies to C. burnetii in humans (12). However, the plasmid type of C. burnetii has not yet been examined in Japan.
The highly sensitive PCR has become a useful tool for the detection of C. burnetii (7, 18, 23). This report describes the results of the nested PCR assays for the direct identification of C. burnetii plasmids in human sera.
MATERIALS AND METHODS
Microorganisms.
The C. burnetii isolates used in this study comprised five QpH1 plasmid-containing strains (Nine Mile, Bangui, California 76, Ohio 314, and Henzerling) from the American Type Culture Collection, one QpRS plasmid-containing strain (Priscilla), and three plasmidless strains (GQ212, SQ217, and KoQ229) kindly provided by L. P. Mallavia of Washington State University, Pullman. All strains of C. burnetii were propagated in buffalo green monkey (BGM) cell cultures as described elsewhere (6).
Sera.
The 81 serum samples used in this study were obtained from 81 patients diagnosed with Q fever by an immunofluorescence (IF) test and PCR in our previous studies (7, 12, 24). Twenty-three acute-phase serum samples were taken between 1982 and 1983 from 23 children with acute atypical pneumonia in Gifu Prefecture. These children were aged 2 to 10 years and were diagnosed with Q fever pneumonia by an IF test, nested PCR, and isolation in our laboratory (7). The other 58 serum samples were collected routinely between September and December 1995 from 58 patients at Gifu University Medical Faculty Hospital and were demonstrated to be positive for C. burnetii by an IF test and PCR (12, 24). In addition, 50 serum samples from patients with pneumonia of viral or bacterial origin (influenza virus, parainfluenza virus, respiratory syncytial virus, Chlamydia psittaci, Legionella pneumophila, or Mycoplasma pneumoniae) served as negative serum controls for PCR.
Nested PCR. (i) Primers used.
The sequences of the primers used in the study and the PCR conditions are presented in Table 1. The primers OMP1-OMP2 and OMP3-OMP4 were designed from the nucleotide sequence of the com-1 gene, encoding a 27-kDa outer membrane protein, and were used to detect C. burnetii genomic sequences (24). The primers HFrag1-HFrag2 and HF1-HF2 were designed to amplify a fragment of a conserved region of the plasmid sequences. This region is present in all types of plasmids examined and is used to detect C. burnetii plasmid sequences (22). Two sets of the modified primers were used to detect C. burnetii plasmid-specific sequences. The first set of primers, CB5-CB6 and CB3-CB4, were designed from a specific gene of the QpH1 plasmid, cbhE′ (10, 19). The second set of primers, QpRS1-QpRS2 and QpRS3-QpRS4, were designed from a unique gene of the QpRS plasmid, cbbE′ (9, 22). The nested PCR was performed with serially diluted total DNA from various strains of C. burnetii to determine the minimum level of DNA detectable by the assay.
TABLE 1.
Sequences of primers and PCR conditions
| Target | Primer | Sequence | Gene detected | Amplimer length (bp) | PCR conditions (°C/no. of s)
|
No. of PCR cycles | ||
|---|---|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | ||||||
| Genome | OMP1 | 5′-AGTAGAAGCATCCCAAGCATTG-3′ | 501 | 94/60 | 54/60 | 72/60 | 36 | |
| OMP2 | 5′-TGCCTGCTAGCTGTAACGATTG-3′ | com-1 | ||||||
| OMP3 | 5′-GAAGCGCAACAAGAAGAACAC-3′ | 438 | 94/30 | 54/20 | 72/60 | 36 | ||
| OMP4 | 5′-TTGGAAGTTATCACGCAGTTG-3′ | |||||||
| Conserved region of plasmidsa | HFrag1 | 5′-ATTGCTATCACTGAGGGTGACG-3′ | 508 | 94/120 | 55/1 | 77/75 | 3 | |
| HFrag2 | 5′-CTGACGAAGAAGCAGCATTAGC-3′ | Conserved region of plasmids | 93/4 | 55/1 | 77/75 | 32 | ||
| HF1 | 5′-TCCTAAACAAGTGATGGTCTCC-3′ | 183 | 94/30 | 55/20 | 75/75 | 35 | ||
| HF2 | 5′-TTCGCAGAAAGTCAGCTATCG-3′ | |||||||
| QpH1 plasmid | CB5 | 5′-ATAATGAGATTAGAACAACCAAGA-3′ | 977 | 94/120 | 53/60 | 72/120 | 35 | |
| CB6 | 5′-TCTTTCTTGTTCATTTTCTGAGTC-3′ | cbhE′ | ||||||
| CB3 | 5′-TAATAGAACGTGTTAATCG-3′ | 266 | 94/60 | 50/60 | 72/60 | 35 | ||
| CB4 | 5′-GCTGGCAATCTGCTCGGC-3′ | |||||||
| QpRS plasmid | QpRS1 | 5′-CTCGTACCCAAAGACTATGAATATATCC-3′ | 693 | 94/60 | 54/60 | 72/120 | 36 | |
| QpRS2 | 5′-AACACCGATCAATGCGACTAGCCC-3′ | cbbE′ | ||||||
| QpRS3 | 5′-ACTTTACGTCGTTTAATTCGC-3′ | 309 | 94/30 | 51/20 | 72/90 | 36 | ||
| QpRS4 | 5′-CACATTGGGTATCGTACTGTCCCT-3′ | |||||||
As described previously (22).
(ii) Preparation of samples for PCR.
DNA of C. burnetii was extracted from the isolates as described previously (5). Briefly, the purified organisms from BGM cell cultures were suspended in TNE buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA) and were digested with proteinase K in the presence of 0.1% sodium dodecyl sulfate at 55°C for 60 min. DNA was extracted with phenol, phenol-chloroform, and chloroform, precipitated with ethanol, dried under vacuum, and resuspended in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The DNA concentration and purity were determined by measuring the optical density at both 260 and 280 nm with a DNA calculator (GeneQuant II; Pharmacia Biotech). The DNA solution was kept at −20°C.
Samples for PCR from sera were prepared as described previously (7). Ten microliters of each serum sample was mixed with 40 μl of sample buffer (1% Nonidet P-40, 1% Tween 20, 10 mM Tris-HCl [pH 8.0]), the mixture was boiled for 10 min and then centrifuged at 12,000 × g for 5 min, and the supernatant was directly used for the PCR analysis.
(iii) DNA amplification.
The first amplification for the nested PCR was performed in a total volume of 50 μl containing 5 μl of DNA sample, 2 U of Taq DNA polymerase (Takara Shuzo, Co., Ltd., Shiga, Japan), and final concentrations of 50 mM KCl; 10 mM Tris-HCl (pH 8.3); 1.5 mM MgCl2; dATP, dCTP, dGTP, and dTTP at a concentration of 200 μM each; and the primers at a concentration of 0.5 μM each. Five microliters of the first amplification product was then subjected to the second amplification with the nested primers. A positive control with 5 pg of C. burnetii DNA as the template and a negative control without DNA template were included in each PCR run. The mixtures were overlaid with 2 drops of mineral oil and were amplified in a DNA thermal cycler (Perkin-Elmer GeneAmp PCR system 9600; Takara Biomedicals, Kyoto, Japan).
Restriction endonuclease digestion.
The specificity of the nested PCR with primers QpRS1-QpRS2 and QpRS3-QpRS4 was confirmed by digesting the amplification products of the reference strain with the restriction enzyme MspI. One MspI site is present in the amplified region of the cbbE′ gene sequence.
Detection of PCR products.
The amplified products of PCR and the restricted products were examined by electrophoresis in a 1.5% agarose gel, stained with ethidium bromide (0.5 μg/ml), visualized under UV illumination (TM-20; UVP, Inc.) at 320 nm, and photographed.
RESULTS
Specificity and sensitivity of the nested PCR assays.
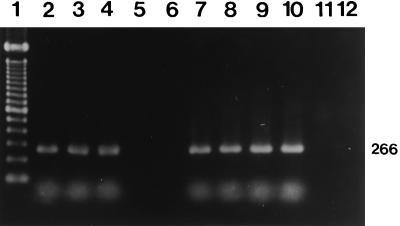
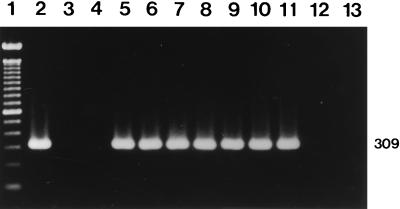
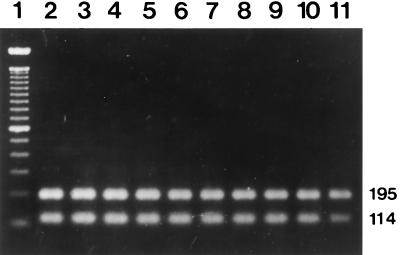
Primers OMP1-OMP2 and OMP3-OMP4 and primers HFrag1-HFrag2 and HF1-HF2 amplified the predicted products from all of the reference strains. The primers CB5-CB6 and CB3-CB4 amplified expected products of 977 and 266 bp, respectively, in the first and the second PCRs from the Nine Mile, Bangui, California 76, Ohio 314, and Henzerling strains containing the QpH1 plasmid (Fig. 1). The primers QpRS1-QpRS2 and QpRS3-QpRS4 also yielded predicted products of 693 and 309 bp in the first and the second PCRs from the Priscilla strain containing the QpRS plasmid (Fig. 2). The GQ212, SQ217, and KoQ229 strains with plasmid sequences integrated into the genome did not react with either set of nested primers (primers CB5-CB6 and CB3-CB4 or primers QpRS1-QpRS2 and QpRS3-QpRS4). Also, the four sets of primers did not amplify any products from negative serum controls or the reagent control. The specificity of the nested PCR with primers QpRS1-QpRS2 and QpRS3-QpRS4 was also confirmed by digesting the amplified products with MspI, which yielded 195- and 114-bp fragments (Fig. 3).
FIG. 1.
Identification of the QpH1 plasmid by nested PCR with primers CB5-CB6 and CB3-CB4. An agarose gel electrophoretogram of the 266-bp amplification products after the nested PCR and ethidium bromide staining is shown. Lane 1, molecular size markers (100-bp DNA ladder); lanes 2 to 4, three reference strains (Nine Mile, Henzerling, and Ohio, respectively); lane 5, reference strain Priscilla; lane 6, reference strain GQ212; lanes 7 to 10, human serum samples; lane 11, negative serum control; lane 12, reagent-negative control. The number on the right is in base pairs.
FIG. 2.
Identification of the QpRS plasmid by nested PCR with primers QpRS1-QpRS2 and QpRS3-QpRS4. An agarose gel electrophoretogram of the 309-bp amplification products after the nested PCR and ethidium bromide staining is shown. Lane 1, molecular size markers (100-bp DNA ladder); lane 2, reference strain Priscilla; lane 3, reference strain Nine Mile; lane 4, reference strain GQ212; lanes 5 to 11, human serum samples; lane 12, negative serum control; lane 13, reagent-negative control. The number on the right is in base pairs.
FIG. 3.
Specificity of the nested PCR with primers QpRS1-QpRS2 and QpRS3-QpRS4 demonstrated by digesting the amplified products with MspI. The 309-bp amplification products were digested with MspI, electrophoresed on agarose gels, and stained with ethidium bromide. Lane 1, molecular size markers (100-bp DNA ladder); lane 2, reference strain Priscilla; lanes 3 to 11, human serum samples. The numbers on the right are in base pairs.
Identification of C. burnetii plasmids in human sera.
The usefulness of the nested PCR was first evaluated for the direct identification of C. burnetii plasmids in human sera. Initially, all of the samples were PCR positive when primers OMP1-OMP2 and OMP3-OMP4 and primers HFrag1-HFrag2 and HF1-HF2 were used. The genomic and plasmid sequences were detected in all of the samples. In addition, the plasmid types of C. burnetii were directly identified in the sera by the nested PCR with primers targeted to the QpH1 and QpRS plasmids. Among the 81 serum samples tested, 40 (49.4%) were positive for the QpH1 plasmid, 24 (29.6%) were positive for the QpRS plasmid, and 5 (6.2%) and 12 (14.8%) were positive and negative for both of the QpH1 and QpRS plasmids, respectively (Table 2). Furthermore, all of the 23 acute-phase serum samples were positive for the QpH1 plasmid and were negative for the QpRS plasmid.
TABLE 2.
Identification of C. burnetii plasmids in sera of 81 patients with Q fever by nested PCR
| Plasmid | No. (%) of positive sera
|
||
|---|---|---|---|
| Acute | Nonacute | Total | |
| QpH1a | 23 (100) | 17 (29.3) | 40 (49.4) |
| QpRSb | 0 | 24 (41.4) | 24 (29.6) |
| QpH1 and QpRS | 0 | 5 (8.6) | 5 (6.2) |
| Undefined | 0 | 12 (20.7) | 12 (14.8) |
| Total | 23 (100) | 58 (100) | 81 (100) |
Identified by nested PCR with primers CB5-CB6 and CB3-CB4.
Identified by nested PCR with primers QpRS1-QpRS2 and QpRS3-QpRS4.
DISCUSSION
Although the PCR technique has previously been used for the differentiation of C. burnetii plasmid types (19, 22), our investigation is the first to evaluate the nested PCR assays for use in the direct identification of C. burnetii plasmids in human sera. False-positive PCR results as a result of contamination did not appear to be a problem during the present study, as evidenced by repeatedly negative results for the negative controls. The nested PCR assays were demonstrated to be highly specific for the QpH1 and QpRS plasmids and useful for the direct identification of these plasmids in human sera.
Several serological and bacteriological studies have suggested that Q fever is distributed widely in Japan (6, 7, 11, 12). C. burnetii has been isolated from arthropods, animals, and humans (6, 7, 11). However, it is still unclear whether the C. burnetii organism that causes acute Q fever is different from the C. burnetii organism that causes chronic Q fever. In the present study, the QpH1- and QpRS-specific sequences were detected in 40 and 24 patients with Q fever, respectively. This result indicates that different strains of C. burnetii have spread in humans in Japan. We also demonstrated that 59 isolates originating from ticks, cattle, and humans possessed the QpH1 plasmid (unpublished data). These data suggest that C. burnetii strains possessing the QpH1 or QpRS plasmid are the most prevalent strains in Japan. Samuel et al. (17) demonstrated that the isolates originating from patients with acute Q fever contained the QpH1 plasmid, while the isolates originating from patients with chronic Q fever possessed the QpRS plasmid or plasmid sequences integrated into the chromosome. In the present study, we found that all of the 23 acute-phase serum samples possessed the QpH1 plasmid-specific gene. These sera were taken from 23 children with acute atypical pneumonia who were diagnosed with Q fever pneumonia by an IF test, nested PCR, and isolation of C. burnetii (7). This result agreed with the results of Samuel et al. (17) and suggests that the QpH1 plasmid is associated with acute Q fever. Also, the QpRS plasmid-specific gene was identified in 24 patients with Q fever. This is the first report of a QpRS plasmid-containing C. burnetii isolate occurring in humans in Japan. However, because Q fever is still not diagnosed routinely in Japan, we have been unable to obtain the detailed clinical data for these patients. We know only that these patients were diagnosed with hepatitis, cerebellitis, lymphangitis, arthritis, or cancer. The presence of C. burnetii antibodies and specific genes in these patients suggests that C. burnetii is a causative agent of hepatitis, cerebellitis, lymphangitis, arthritis, or cancer. To our knowledge, endocarditis is the most common manifestation of chronic Q fever, but vascular infection, bone infection, chronic hepatitis, and osteomyelitis are other manifestations of chronic Q fever. At present, the occurrence of QpRS plasmid-containing C. burnetii in patients with hepatitis, cerebellitis, lymphangitis, arthritis, or cancer may suggest that these diseases are also manifestations of chronic Q fever in Japan. Recently, chronic endocarditis associated with Q fever was also shown in a retrospective study by Yuasa et al. (23) in which C. burnetii DNA was detected by nested PCR in paraffin-embedded endocardial tissues from 4 of 56 patients with chronic endocarditis. These studies suggest that chronic Q fever is not uncommon in Japan and that there is a diversity of clinical forms of chronic Q fever.
It is noteworthy that both of the QpH1- and QpRS-specific sequences were found in five patients. This observation may be explained by the following possibilities: (i) these patients were complicatedly infected with the QpH1 or QpRS plasmid-containing C. burnetii, or (ii) these patients were infected with a single strain of C. burnetii that possessed both QpH1 and QpRS plasmids. The finding of both plasmids in single serum samples may be confirmed by extracting the plasmids after the bacteria are isolated from the samples.
Also, the QpH1- and QpRS-specific sequences were not detected in 12 patients. Because our nested PCR assays are not able to identify plasmids other than the QpH1 and QpRS plasmids, the failure to find either QpH1 or QpRS in some patients may be explained by the possibility that these patients were infected with C. burnetii strains which possessed other types of plasmids, such as QpDG or QpDV, which were of the plasmidless type, or which possessed a different plasmid. The C. burnetii plasmid types in these patients may be identified by PCR with primers specific for other plasmids when they are available.
The results of this study indicate that the nested PCR assays are useful for the direct typing of C. burnetii plasmids in human sera. Plasmid typing by PCR appears to be a more promising and useful method for the rapid differentiation of C. burnetii in clinical samples because of its sensitivity and specificity. Therefore, further studies are needed to validate the nested PCR assays for the early differentiation of acute Q fever from chronic Q fever.
ACKNOWLEDGMENT
This work was supported by a Grant-in-Aid for Developmental Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (grant 07306015).
REFERENCES
- 1.Baca O G, Paretsky D. Q fever and Coxiella burnetii: a model for host-parasite interactions. Microbiol Rev. 1983;47:127–149. doi: 10.1128/mr.47.2.127-149.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupuis G, Peter O, Peacock M, Burgdorfer W, Haller E. Immunoglobulin responses in acute Q fever. J Clin Microbiol. 1985;22:484–487. doi: 10.1128/jcm.22.4.484-487.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hackstadt T, Peacock M G, Hitchcock P J, Cole R L. Lipopolysaccharide variation in Coxiella burnetii: intrastrain heterogeneity in structure and antigenicity. Infect Immun. 1985;48:359–365. doi: 10.1128/iai.48.2.359-365.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinzen R, Stiegler G L, Whiting L L, Schmitt S A, Mallavia L P, Frazier M E. Use of pulsed field gel electrophoresis to differentiate Coxiella burnetii strains. Ann N Y Acad Sci. 1990;590:504–513. doi: 10.1111/j.1749-6632.1990.tb42260.x. [DOI] [PubMed] [Google Scholar]
- 5.Hendrix L R, Samuel J E, Mallavia L P. Differentiation of Coxiella burnetii isolates by analysis of restriction-endonuclease-digested DNA separated by SDS-PAGE. J Gen Microbiol. 1991;137:269–276. doi: 10.1099/00221287-137-2-269. [DOI] [PubMed] [Google Scholar]
- 6.Ho T, Htwe K K, Yamasaki N, Zhang G Q, Ogawa M, Yamaguchi T, Fukushi H, Hirai K. Isolation of Coxiella burnetii from dairy cattle and ticks, and some characteristics of the isolates in Japan. Microbiol Immunol. 1995;39:663–671. doi: 10.1111/j.1348-0421.1995.tb03254.x. [DOI] [PubMed] [Google Scholar]
- 7.Ho T, Kako N, Zhang G Q, Otsuka H, Ogawa M, Ochiai O, Nguyen S V, Yamaguchi T, Fukushi H, Nagaoka N, Akiyama M, Amano K, Hirai K. Q fever pneumonia in children in Japan. J Clin Microbiol. 1996;34:647–651. doi: 10.1128/jcm.34.3.647-651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimbrough R C D, Ormsbee R A, Peacock M, Rogers W R, Bennetts R W, Raaf J, Krause A, Gardner C. Q fever endocarditis in the United States. Ann Intern Med. 1979;91:400–402. doi: 10.7326/0003-4819-91-3-400. [DOI] [PubMed] [Google Scholar]
- 9.Minnick M F, Heinzen R A, Douthart R, Mallavia L P, Frazier M E. Analysis of QpRS-specific sequences from Coxiella burnetii. Ann N Y Acad Sci. 1990;590:514–522. doi: 10.1111/j.1749-6632.1990.tb42261.x. [DOI] [PubMed] [Google Scholar]
- 10.Minnick M F, Small C L, Frazier M E, Mallavia L P. Analysis of the cbhE′ plasmid gene from acute disease-causing isolates of Coxiella burnetii. Gene. 1991;103:113–118. doi: 10.1016/0378-1119(91)90401-v. [DOI] [PubMed] [Google Scholar]
- 11.Nagaoka H, Akiyama M, Sugieda M, Nishio T, Akahane S, Hattori H, Ho T, Fukushi H, Hirai K. Isolation of Coxiella burnetii from children with influenza-like symptoms in Japan. Microbiol Immunol. 1996;40:147–151. doi: 10.1111/j.1348-0421.1996.tb03330.x. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen S V, Otsuka H, Zhang G Q, Ho T, Yamaguchi T, Fukushi H, Noma A, Hirai K. Rapid method for detection of Coxiella burnetii antibodies using high-density particle agglutination. J Clin Microbiol. 1996;34:2947–2951. doi: 10.1128/jcm.34.12.2947-2951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peter O, Dupuis G, Bee D, Luthy R, Nicolet J, Burgdorfer W. Enzyme-linked immunosorbent assay for diagnosis of chronic Q fever. J Clin Microbiol. 1988;26:1978–1982. doi: 10.1128/jcm.26.10.1978-1982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peter O, Dupuis G, Burgdorfer W, Peacock M. Evaluation of the complement fixation and indirect immunofluorescence tests in the early diagnosis of primary Q fever. Eur J Clin Microbiol. 1985;4:394–396. doi: 10.1007/BF02148690. [DOI] [PubMed] [Google Scholar]
- 15.Raoult D, Urvolgyi J, Etienne J, Roturier M, Puel J, Chaudet H. Diagnosis of endocarditis in acute Q-fever by immunofluorescence serology. Acta Virol. 1988;32:70–74. [PubMed] [Google Scholar]
- 16.Samuel J E, Frazier M E, Kahn M L, Thomashow L S, Mallavia L P. Isolation and characterization of a plasmid from phase I Coxiella burnetii. Infect Immun. 1983;41:488–493. doi: 10.1128/iai.41.2.488-493.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuel J E, Frazier M E, Mallavia L P. Correlation of plasmid type and disease caused by Coxiella burnetii. Infect Immun. 1985;49:775–779. doi: 10.1128/iai.49.3.775-779.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein A, Raoult D. Detection of Coxiella burnetii by DNA amplification using polymerase chain reaction. J Clin Microbiol. 1992;30:2462–2466. doi: 10.1128/jcm.30.9.2462-2466.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein A, Raoult D. Lack of pathotype specific gene in human Coxiella burnetii isolates. Microb Pathog. 1993;15:177–185. doi: 10.1006/mpat.1993.1068. [DOI] [PubMed] [Google Scholar]
- 20.Valková D, Kazár J. A new plasmid (QpDV) common to Coxiella burnetii isolates associated with acute and chronic Q fever. FEMS Microbiol Lett. 1995;125:275–280. doi: 10.1111/j.1574-6968.1995.tb07368.x. [DOI] [PubMed] [Google Scholar]
- 21.Weir W R C, Bannister B, Chambres S, Coke K D, Mistry H. Chronic Q fever associated with granulomatous hepatitis. J Infect. 1984;8:56–60. doi: 10.1016/s0163-4453(84)93354-1. [DOI] [PubMed] [Google Scholar]
- 22.Willems H, Thiele D, Krauss H. Plasmid based differentiation and detection of Coxiella burnetii in clinical samples. Eur J Epidemiol. 1993;9:411–418. doi: 10.1007/BF00157399. [DOI] [PubMed] [Google Scholar]
- 23.Yuasa Y, Yoshiie K, Takasaki T, Yoshida H, Oda H. Retrospective survey of chronic Q fever in Japan by using PCR to detect Coxiella burnetii DNA in paraffin-embedded clinical samples. J Clin Microbiol. 1996;34:824–827. doi: 10.1128/jcm.34.4.824-827.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G Q, Nguyen S V, Ho T, Ogawa M, Hotta A, Yamaguchi T, Fukushi H, Hirai K. Clinical evaluation of a new PCR assay for detection of Coxiella burnetii in human serum samples. J Clin Microbiol. 1998;36:77–80. doi: 10.1128/jcm.36.1.77-80.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]