Abstract
STUDY QUESTION
How does an altered maternal hormonal environment, such as that seen during superovulation with gonadotropins in ART, impact human uterine immune cell distribution and function during the window of implantation?
SUMMARY ANSWER
Hormonal stimulation with gonadotropins alters abundance of maternal immune cells including uterine natural killer (uNK) cells and reduces uNK cell ability to promote extravillous trophoblast (EVT) invasion.
WHAT IS KNOWN ALREADY
An altered maternal hormonal environment, seen following ART, can lead to increased risk for adverse perinatal outcomes associated with disordered placentation. Maternal immune cells play an essential role in invasion of EVTs, a process required for proper establishment of the placenta, and adverse perinatal outcomes have been associated with altered immune cell populations. How ART impacts maternal immune cells and whether this can in turn affect implantation and placentation in humans remain unknown.
STUDY DESIGN, SIZE, DURATION
A prospective cohort study was carried out between 2018 and 2021 on 51 subjects: 20 from natural cycles 8 days after LH surge; and 31 from stimulated IVF cycles 7 days after egg retrieval.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Endometrial biopsies and peripheral blood samples were collected during the window of implantation in subjects with regular menstrual cycles or undergoing superovulation. Serum estradiol and progesterone levels were measured by chemiluminescent competitive immunoassay. Immune cell populations in blood and endometrium were analyzed using flow cytometry. uNK cells were purified using fluorescence-activated cell sorting and were subjected to RNA sequencing (RNA-seq). Functional changes in uNK cells due to hormonal stimulation were evaluated using the implantation-on-a-chip (IOC) device, a novel bioengineered platform using human primary cells that mimics early processes that occur during pregnancy in a physiologically relevant manner. Unpaired t-tests, one-way ANOVA, and pairwise multiple comparison tests were used to statistically evaluate differences.
MAIN RESULTS AND THE ROLE OF CHANCE
Baseline characteristics were comparable for both groups. As expected, serum estradiol levels on the day of biopsy were significantly higher in stimulated (superovulated) patients (P = 0.0005). In the setting of superovulation, we found an endometrium-specific reduction in the density of bulk CD56+ uNK cells (P < 0.05), as well as in the uNK3 subpopulation (P = 0.025) specifically (CD103+ NK cells). In stimulated samples, we also found that the proportion of endometrial B cells was increased (P < 0.0001). Our findings were specific to the endometrium and not seen in peripheral blood. On the IOC device, uNK cells from naturally cycling secretory endometrium promote EVT invasion (P = 0.03). However, uNK cells from hormonally stimulated endometrium were unable to significantly promote EVT invasion, as measured by area of invasion, depth of invasion, and number of invaded EVTs by area. Bulk RNA-seq of sorted uNK cells from stimulated and unstimulated endometrium revealed changes in signaling pathways associated with immune cell trafficking/movement and inflammation.
LIMITATIONS, REASONS FOR CAUTION
Patient numbers utilized for the study were low but were enough to identify significant overall population differences in select immune cell types. With additional power and deeper immune phenotyping, we may detect additional differences in immune cell composition of blood and endometrium in the setting of hormonal stimulation. Flow cytometry was performed on targeted immune cell populations that have shown involvement in early pregnancy. A more unbiased approach might identify changes in novel maternal immune cells not investigated in this study. We performed RNA-seq only on uNK cells, which demonstrated differences in gene expression. Ovarian stimulation may also impact gene expression and function of other subsets of immune cells, as well as other cell types within the endometrium. Finally, the IOC device, while a major improvement over existing in vitro methods to study early pregnancy, does not include all possible maternal cells present during early pregnancy, which could impact functional effects seen. Immune cells other than uNK cells may impact invasion of EVTs in vitro and in vivo, though these remain to be tested.
WIDER IMPLICATIONS OF THE FINDINGS
These findings demonstrate that hormonal stimulation affects the distribution of uNK cells during the implantation window and reduces the proinvasive effects of uNK cells during early pregnancy. Our results provide a potential mechanism by which fresh IVF cycles may increase risk of disorders of placentation, previously linked to adverse perinatal outcomes.
STUDY FUNDING/COMPETING INTEREST(S)
Research reported in this publication was supported by the University of Pennsylvania University Research Funding (to M.M.), the Eunice Kennedy Shriver National Institute of Child Health and Human Development (P50HD068157 to M.M., S.S., and S.M.), National Center for Advancing Translational Sciences of the National Institutes of Health (TL1TR001880 to J.K.), the Institute for Translational Medicine and Therapeutics of the Perelman School of Medicine at the University of Pennsylvania, the Children’s Hospital of Philadelphia Research Institute (to S.M.G.), and the National Institute of Allergy and Infectious Diseases (K08AI151265 to S.M.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. All authors declare no conflict of interest.
TRIAL REGISTRATION NUMBER
N/A.
Keywords: IVF, ART, uterine immune cells, extravillous trophoblast, placenta
Introduction
Perinatal complications result in over 300000 maternal deaths and 15 million premature deliveries worldwide every year (Maternal Mortality, 2015). Multiple adverse pregnancy outcomes including pre-eclampsia, intrauterine fetal growth restriction, preterm birth, and placenta accreta, have been associated with disordered placental development (Goldman-Wohl and Yagel, 2002; Kaufmann et al., 2003; Kim et al., 2003; Tantbirojn et al., 2008). After the fertilized embryo implants into a receptive endometrium, successful establishment of pregnancy requires invasion of specialized fetal cells known as extravillous trophoblasts (EVTs) into the uterus, and remodeling of uterine spiral arteries into low-resistance vessels. Endometrial cells including vascular endothelial cells (ECs), stromal cells, and immune cells regulate processes essential to early placentation, including trophoblast invasion and arterial remodeling (Pollheimer et al., 2018; Park et al., 2022).
Work from our lab and others has demonstrated links between changes to the maternal environment and placental abnormalities. Specifically, we showed in both human and animal models that the supraphysiologic hormonal environment created during IVF cycles, with significant changes in estradiol levels, progesterone levels, and other factors secreted by the corpus luteum, can lead to changes in placentation (de Waal et al., 2012; Song et al., 2015; Weinerman et al., 2017; Senapati et al., 2018; Stuart et al., 2018; Sullivan-Pyke et al., 2020; Vrooman et al., 2020). When compared to unassisted conception or frozen embryo transfer, in which the embryo is transferred to the uterus under more physiologic hormonal conditions, fresh IVF cycles result in increased risk of fetal growth restriction and preterm birth (Kalra et al., 2011; Pandey et al., 2012). Our findings in a mouse model support these observations in humans, namely that an altered maternal hormonal environment during early placentation can lead to disordered placentation and fetal growth (Weinerman et al., 2017; Sullivan-Pyke et al., 2020). The mechanisms responsible for these changes in placentation remain incompletely understood.
Uterine natural killer (uNK) cells are the most abundant immune cell type in the preimplantation endometrium and in the decidua during early pregnancy (Lee et al., 2011). Kinetics of uNK cell abundance corresponds to cyclic hormonal changes seen during the normal menstrual cycles, and these cells reach a peak during the secretory phase, when early embryo implantation occurs (Flynn et al., 2000). They remain abundant through the first trimester of pregnancy but diminish in number once EVT invasion slows (King et al., 1989; Bulmer et al., 1991; Ho et al., 1996; Lee et al., 2011). Further, immunohistochemical studies show that uNK cells cluster near spiral arteries, and in vitro experiments indicate that uNK cells secrete cytokines, chemokines, and other factors that regulate EVT invasion and facilitate maternal artery remodeling (King et al., 1989; Smith et al., 2009; Lash et al., 2010; Robson et al., 2012; Tilburgs et al., 2015; Ma et al., 2017). Taken together, these data support essential roles for uNK cells in the establishment of pregnancy and normal placentation (Kanter et al., 2021).
By examining changes in gene expression using tissue-level microarray in endometrial biopsies obtained during the window of implantation, we found that hormonal stimulation disrupted the expression of several genes associated with natural killer (NK) cell signaling (Senapati et al., 2018). However, examining the function of human uNK cells during early pregnancy is challenging. Common animal models do not fully recapitulate the depth of invasion and cellular complexity seen in human placentation (Carter, 2007; Carter and Mess, 2014; Swanson and David, 2015; Grigsby, 2016; Silva and Serakides, 2016). Human and animal uNK cells exhibit important differences in development, phenotype, and function (Gaynor and Colucci, 2017). Studies of NK cells from human term placentae or from hysterectomized uteri, which often have underlying pathology, only partially model biology unique to the peri-implantation period or early in pregnancy (James et al., 2016; Silva and Serakides, 2016). In vitro studies of uNK cells are commonly performed in isolation or with one other cell type, and incompletely mimic cellular interactions at the maternal–fetal interface (James et al., 2016).
To address some of these significant limitations, we recently developed a microengineered ‘implantation-on-a-chip’ (IOC) device to model directional invasion of EVTs toward maternal vessels in vitro (Park et al., 2022). In this system, primary human EVTs and endometrial ECs are seeded into parallel microchannels separated by a mock extracellular matrix (ECM) composed of a Matrigel-Collagen mixture within which other maternal cells can be seeded. The ECs form a vessel-like structure in their microchannel and induce directional migration of EVTs. After migrating through the ECM, EVTs interact with ECs to recapitulate such aspects of uterine spiral artery remodeling as disruption of EC junctions and induction of EC apoptosis. The IOC device offers several advantages, including individually addressable microchannels with distinct media feeds to allow for control and manipulation of microenvironmental cues, imaging and analysis capabilities, and a compartmentalized design populated by primary human cells that closely mimics the 3D microarchitecture of the maternal–fetal interface. Using the IOC device, we recently showed that when uNK cells isolated from endometrium during the window of implantation were seeded into the ECM, EVT invasion in the presence of ECs was significantly enhanced. These data support that uNK cells from secretory phase endometrium promote the invasion of EVTs (Park et al., 2022).
In this study, we examine the effects of the maternal hormonal environment on the immune cell landscape of the receptive endometrium. We then use the novel IOC device to study potential impacts of hormonal stimulation on uNK cell function. This study represents the first described practical application of a newly developed organ-on-a-chip device that is able to model the maternal–fetal interface during early placentation. We demonstrate that superovulation with gonadotropins in preparation for ART results in decreased density of uNK cells. Further, we show that supraphysiologic levels of maternal hormones impair the ability of uNK cells to promote invasion of EVTs on the IOC device. These data demonstrate essential roles for the maternal hormonal environment and uNK cells in early human placentation and suggest potential modifiable factors that may be able to reduce the risks of adverse pregnancy outcomes.
Materials and methods
Sample collection
Endometrial biopsies to obtain endometrial immune cells, as well as blood samples, were obtained from patients and volunteers at Penn Fertility Care. The University of Pennsylvania Institutional Review Board (IRB) approved the collection of endometrial tissue (IRB#828613). Written informed consent was obtained from all subjects. Two separate groups of women were recruited: patients receiving exogenous gonadotropins for oocyte or embryo cryopreservation (stimulated group) and naturally cycling volunteers (unstimulated control group). All women for both groups were of reproductive age, between the ages of 18 and 43, with no significant medical history and regular menstrual cycles (between 25 and 35 days over the previous 3 months). Stimulated patients (n = 31) underwent gonadotropin stimulation per standard clinic practice using either luteal phase leuprolide or an antagonist protocol with administration of hCG or hCG and leuprolide co-trigger to induce follicle maturation per clinical protocol 36 h prior to oocyte retrieval. In these women, endometrial biopsy was performed 7 days following oocyte retrieval. Endometrial biopsies were obtained from nonstimulated controls (n = 20) 8 days after an LH surge as detected in the urine by the BFP® Ovulation Test (Fairhaven Health, USA). Biopsies were obtained using a Pipelle® Endometrial Suction Curette (CooperSurgical, USA). At this study visit, venous blood draw was also performed for subsequent testing of serum hormone levels and flow cytometry.
Serum hormone analysis
Serum estradiol (E2) and progesterone (P4) levels were measured by chemiluminescent competitive immunoassay using the Roche E170 immunoassay analyzer (Roche Diagnostic).
Endometrial tissue digestion
Single-cell suspensions of endometrial tissue for flow cytometry and cell sorting were prepared as previously described (Gordon et al., 2020). Briefly, endometrial tissue was washed in phosphate-buffered saline (PBS) and finely minced before being digested in a shaking 37°C water bath for 15 min in digestion medium (sterile serum-free Roswell Park Memorial Institute (RPMI) medium, 0.28 WU/ml Liberase TM (Roche 05401127001, Switzerland), 30 µg/ml DNase (Roche 10104159001, Switzerland)). A large-bore transfer pipet was used to pipet tissue up and down during digestion to breakdown tissue. Digested tissue was filtered through a 70-micron filter and pelleted at 300×g for 5 min. To remove red blood cell contamination, the cell pellet was resuspended in 5 ml Ammonium-Chloride-Potassium (ACK) lysis buffer and incubated on ice for 5 min. After lysis, ice cold PBS was added, and cells spun down at 300×g for 5 min. Cells were resuspended and prepared for flow cytometry staining.
Flow cytometry of immune cells
Flow cytometry was performed on either a MacsQuant Analyzer 10 (Miltenyi Biotech, Germany) or a CytoFLEX LX (Beckman Coulter, USA). Raw data were analyzed using FlowJo 10. Data were aggregated, and statistical analysis with two-way ANOVA or unpaired t-tests was performed with Graphpad Prism. All antibody staining was performed at 4°C for 30 min in the dark and all antibodies (Supplementary Table SI) were used at a dilution of 1:100. LIVE/DEAD blue or aqua (Thermo Fisher, USA) and 1:100 human Fc block (BD, USA) were used prior to staining with fluorescently-labeled antibodies.
Fluoresence-activated cell sorting of uNK cells
Cell sorting was performed on a FACSAria Fusion (BD, USA). Gating of live, singlet, CD45+ lymphocytes was performed first. Bulk uNK cells were further defined as CD3−CD19−CD14−CD56 bright cells. uNK cells were sorted into pure fetal bovine serum (FBS), pelleted, and frozen at −80°C for RNA isolation or sorted into RPMI with 10% FBS for use on the IOC device.
Implantation-on-chip device
Each experimental run consisted of three cellular environments, and each chip was run in triplicate: (i) co-culture of ECs and EVTs, (ii) triculture of ECs, EVTs, and addition of hormonally stimulated uNKs to the ECM, and (iii) triculture of ECs, EVTs, and addition of uNKs from naturally cycling subjects to the ECM. In addition, three biological replicates were performed each with different human subjects.
Extravillous trophoblast isolation and culture
EVTs were collected from placental villi obtained from first-trimester pregnancy terminations performed at the Penn Family and Pregnancy Loss Center (IRB#827072). Written informed consent was obtained from all subjects. Patients with pre-existing medical conditions or any pregnancy complications were excluded from the study. Collected tissue was kept on ice and cell isolation was carried out within 1 h of obtaining tissue. Primary EVTs were isolated from these samples based on an EVT-outgrowth-based protocol established by Graham et al. (1992) and well-documented by other investigators (Graham et al., 1992; Getsios et al., 1998; Anton et al., 2012; 2019). Briefly, villous tissue was finely minced and cultured at 37°C in RPMI 1640 medium containing 20% charcoal-stripped FBS. After villous fragment attachment, EVT outgrowth occurred and cells were separated from tissue during washing and passaging of the cells. Isolated EVTs were maintained in RPMI 1640 medium containing 20% FBS and 1% penicillin (100 U/ml)/streptomycin (100 U/ml) solution. EVT identity was confirmed by immunostaining for cytokeratin-7 and HLA-G. Cells were used within the first three passages.
Human endothelial cells and culture
Human endometrial microvascular cells (ScienCell, Catalog #7010) were cultured in Endothelial Cell Growth Medium MV2 (EGM-MV2, Promocell, Catalog # C-22022). They were used between passages three and five.
Implantation-on-chip model preparation and cell seeding
IOC devices were fabricated using standard soft lithography protocols as reported (Park et al., 2022). After device production, mock ECM was first injected into the middle lane. To make the ECM hydrogel precursor solution, rat tail collagen Type-1 (8 mg/ml, Corning) solution was prepared by mixing 10× PBS and 1 N NaOH to achieve physiological pH, and then Matrigel (10 mg/ml, Corning, USA) was mixed with Col-1 solution at a ratio of 1:1 (v/v). ECM precursor solution was injected and the device was incubated 37°C for 20 min to form an ECM hydrogel. After gelation, fibronectin solution (0.1 mg/ml in PBS) was introduced into the vascular chamber and treated for 2 h at 37°C to facilitate EC attachment. Maternal ECs (10 million/ml) were injected into the vascular chamber and incubated for 1 h at 37°C. EVTs (8 million cells/ml) were then seeded, and the device was tilted so that EVTs could adhere to the exposed surface of ECM hydrogel. EVTs and uNK cells used in this study were fluorescently labeled by preincubating them with 5 μg/ml of CellTracker Green CMFDA (Thermo Fisher Scientific, USA) and 1 μg/ml of CellTracker Deep Red (Thermo Fisher Scientific, USA) in DMEM supplemented 2% (v/v) FBS for 15 min at 37°C, respectively. To produce uNK cell-containing devices, sorted uNK cells were suspended in an ECM precursor solution at a density of 2 million cells/ml and injected into the middle lane of the device. Following incubation for 20 min at 37°C, a polydimethylsiloxane slab containing media reservoirs was conformally bonded to the top surface of the device and filled with 2% FBS supplemented RPMI and EGM2 medium. Four hundred microliters of culture medium were added to the medium reservoirs connected to each chamber.
Quantification of EVT invasion
Fluorescence images of EVTs were obtained from the ECM matrix region between the maternal vascular and fetal compartments of the IOC device. High-magnification images were collected from three separate devices per experimental group. EVT invasion was quantified by (i) the number of invading EVTs, (ii) the depth of EVT invasion, and (iii) the area of EVT invasion. To evaluate the cell number, EVTs in the ECM compartment were manually counted, and the average of total cell counts was plotted. The depth of invasion was determined by averaging the vertical distance that EVTs had traveled in the ECM scaffold. Analysis of invasion area was achieved by using the Analyze pixels function of ImageJ (NIH) with appropriate thresholding to measure the area of ECM hydrogel covered by invading EVTs.
RNA-sequencing of bulk uterine NK cells
A total of 10 uNK samples, 5 from naturally cycling secretory endometrium and 5 following gonadotropin stimulation, were selected for RNA sequencing (RNA-seq). Only 4 naturally cycling samples met quality control metrics, thus one was excluded from analysis. RNA library preparations, sequencing reactions, and initial bioinformatics analysis were conducted at GENEWIZ, LLC. (South Plainfield, NJ, USA). Total RNA was extracted following the Trizol Reagent User Guide (Thermo Fisher Scientific, USA). RNA was quantified using Qubit Fluorometer (Life Technologies, USA), and RNA integrity was checked with TapeStation (Agilent Technologies, USA). SMART-Seq v4 Ultra Low Input Kit for Sequencing was used for full-length complementary DNA (cDNA) synthesis and amplification (Clontech, USA), and Illumina Nextera XT library was used for sequencing library preparation. Briefly, cDNA was fragmented, and adaptor was added using Transposase, followed by limited-cycle PCR to enrich and add index to the cDNA fragments. The final library was assessed with Agilent TapeStation.
The sequencing libraries were multiplexed and clustered on one lane of a flowcell. After clustering, the flowcell was loaded on the Illumina HiSeq instrument according to manufacturer’s instructions. The samples were sequenced using a 2 × 150 Paired End (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq were converted into fastq files and demultiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification. After investigating the quality of the raw data, sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the Homo sapiens reference genome available on ENSEMBL using the STAR aligner v.2.5.2b. The STAR aligner is a splice aligner that detects splice junctions and incorporates them to help align the entire read sequences. BAM files were generated as a result of this step. Unique gene hit counts were calculated by using feature Counts from the Subread package v.1.5.2. Only unique reads that fall within exon regions were counted. After extraction of gene hit counts, the gene hit counts table was used for downstream differential expression analysis. Using DESeq2, a comparison of gene expression between the groups of samples was performed. The Wald test was used to generate P-values and log2 fold changes. Genes with P-values <0.01 were called as differentially expressed genes for each comparison.
To place proteins secreted by uNKs in a biological context, differentially regulated genes between uNKs from hormonally stimulated versus naturally cycling subjects were analyzed using a bioinformatics pathway analysis tool, Ingenuity Pathway Analysis (IPA), to predict related signals and their biofunctions (www.qiagen.com/ingenuity). Filter terms were set to molecules/relationships specific to Humans, and experimentally observed direct/indirect relationships. Top canonical pathways, upstream regulators, disease, and physiological functions were examined for biological significance.
Statistical analysis
Unpaired t-tests were performed using Prism to examine differences in baseline characteristics and to evaluate differences in immune cell distribution. RNA-seq data were analyzed using DESeq2 and Wald test was used to generate P-values and log2 fold changes. For IOC models, one-way ANOVA followed by multiple comparison testing was performed using Prism, with a minimum of three independent devices for each experimental group (n = 3).
Study approval for collection of human tissue
Human tissue samples were obtained following approval of the University of Pennsylvania IRB (828613 and 827072). All patients provided written informed consent.
Results
Hormonal stimulation changes the abundance of uNK cells and B cells during the window of implantation
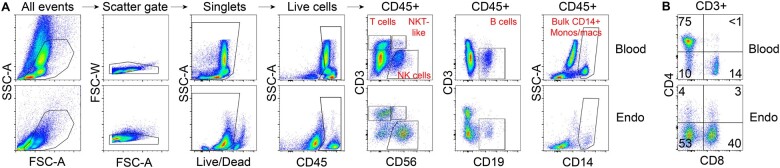
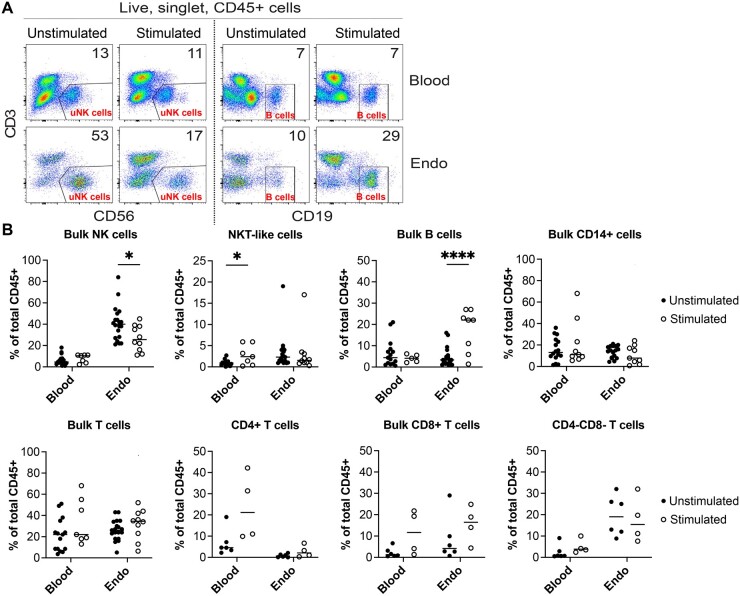
We first investigated how gonadotropin stimulation impacts the abundance of maternal immune cells in secretory phase endometrium during the window of implantation. We performed flow cytometry on endometrial biopsies from naturally cycling women (n = 20) and from women following hormonal stimulation for either oocyte or embryo cryopreservation (n = 31). Baseline demographic characteristics were similar for both groups except for serum estradiol levels on the day of biopsy, which were significantly higher in gonadotropin-stimulated patients, as expected (P = 0.0005) (Table I). Immune cells profiled were chosen based on prior evidence of involvement in pregnancy, and included uNK cells (CD56+CD3−); T cells (CD3+CD56−), including: CD4+ T cells, CD8+ T cells, CD4−CD8− T cells; B cells (CD19+CD3−); NKT-like cells (CD3+CD56+); and monocytes and macrophages (CD14+). (Van Nieuwenhoven, 2003; Liu et al., 2017) Figure 1 shows our flow cytometry gating strategy using representative panels from one naturally cycling patient. We found significant differences specifically in the endometrium of two immune cell populations profiled, uNK cells and B cells. uNK cells of hormonally stimulated subjects were significantly reduced in the endometrium, but not in blood (natural n = 20, stimulated n = 10, P < 0.05) (Fig. 2A and B). We also found that the percentage of B cells was significantly increased in the endometrium (natural n = 19, stimulated n = 8, P < 0.0001) but not in blood of subjects undergoing superovulation (natural n = 15, stimulated n = 8) (Fig. 2A and B). We did not see any statistically significant changes in bulk CD14+ cells (endometrium natural n = 18, stimulated n = 9; blood natural n = 16, stimulated n = 7), encompassing monocytes, dendritic cells, and uterine macrophages (Thiruchelvam et al., 2013), bulk T cells (endometrium natural n = 19, stimulated n = 10; blood natural n = 15, stimulated n = 7), CD4+ (natural n = 6, stimulated n = 4), CD8+ (natural n = 6, stimulated n = 4), or CD4−CD8− T cells (natural n = 6, stimulated n = 4) in either endometrium or blood samples (Fig. 2B). NKT-like cells showed a modest but statistically significant increase in blood samples (natural n = 15, stimulated n = 7, P < 0.05), but not endometrium (natural n = 19, stimulated n = 10), but these cells represented a very low overall proportion (<5%) of the immune cell population, so the clinical significance of this change is unclear (Fig. 2B).
Table I.
Patient demographics.
| Unstimulated (n = 20) | Stimulated (n = 31) | P-value | |
|---|---|---|---|
| Age | 31.7 ± 7.0 | 33.5 ± 4.9 | 0.21 |
| BMI (kg/m2) | 23.8 ± 3.8 | 26.0 ± 5.8 | 0.15 |
| No. of prior pregnancies | 1.0 ± 1.5 | 0.8 ± 1.3 | 0.57 |
| Cycle day when OPK positive | 14.7 ± 2.5 | N/A | N/A |
| Biopsy day estradiol (pg/ml) | 144.7 ± 69.9 | 595.8 ± 468.1 | 0.0005 |
| Biopsy day progesterone (ng/ml) | 14.0 ± 6.3 | 23.5 ± 23.5 | 0.09 |
| Stimulation length (days) | N/A | 11.1 ± 1.6 | N/A |
| Trigger day estradiol (pg/ml) | N/A | 2316.2 ± 949.8 | N/A |
| Trigger day LH (IU/l) | N/A | 4.1 ± 8.2 | N/A |
| Trigger day progesterone (ng/ml) | N/A | 1.37 ± 0.7 | N/A |
| Trigger days EMS (mm) | N/A | 10.3 ± 3.3 | N/A |
| Post-trigger estradiol (pg/ml) | N/A | 2351.9 ± 1024.3 | N/A |
| Post-trigger LH (IU/l) | N/A | 26.9 ± 37.4 | N/A |
| Post-trigger progesterone (ng/ml) | N/A | 7.6 ± 4.4 | N/A |
Baseline demographic characteristics of subjects cycling naturally (‘unstimulated’) or following hormonal stimulation (‘stimulated’). P-values shown were calculated with unpaired t-test. OPK, ovulation prediction kit; EMS, endometrial stripe measurement; N/A: not applicable. Bold typeface indicates statistically significant values.
Figure 1.
Raw flow cytometry gating strategy used to identify basic immune populations. Blood and endometrial (Endo) profiles reveal tissue-specific effects on immune composition. Data shown are from matched blood and endometrial biopsy from the same patient cycling naturally. Samples were processed and stained with the identical antibody cocktails at the same time. (A) Gating scheme for live, singlet, and CD45+ cells. Among CD45+ cells, we show T cells (CD3+CD56−), natural killer T-like (NKT-like) cells (CD3+CD56+), NK cells (CD56+CD3-), B cells (CD19+CD3−), bulk CD14+ monocytes (blood), and bulk CD14+ monocytes/macrophages (endometrium). (B) Subsetting of CD45+CD3+ T cells by expression of CD4 and CD8. CD4+ T cells are rare in the endometrium, while CD4/CD8 double-negative T cells emerge in the endometrium. Numbers on plot denote percent of cells within indicated gates.
Figure 2.
Tissue-specific changes in density of innate and adaptive lymphocytes in response to hormonal stimulation. (A) Reduced density of bulk natural killer (NK) cells and increased density of B cells in endometrium (Endo), but not blood, of hormonally stimulated subjects. Flow cytometry plots gated on live, singlet, and CD45+ cells from matched blood and endometrium from two subjects whose samples were collected and run simultaneously are shown. One subject was cycling naturally, and one was hormonally stimulated. Numbers on plot denote percent of cells within indicated gates. (B) Quantification of indicated immune cells as a percentage of total live, singlet, and CD45+ cells. Maximum n per tissue per group was 20, minimum n per tissue per group was 4. Each point represents a single tissue sample and bar represents the mean of each set of samples. Multiple unpaired t-tests were performed in order to determine effects of hormonal treatment on indicated immune cells within blood or within endometrium. *P < 0.05; ****P < 0.0001. NKT-like, natural killer T-like.
Since hormonal stimulation leads to supraphysiologic levels of estradiol, we next examined the relationship between serum estradiol levels and the abundance of uNK cells and B cells. Using linear regression analysis, we did not find any correlation between uNK cell abundance and serum estradiol level on day of trigger (stimulated n = 10) and day of sample collection (natural n = 14, stimulated n = 9) (Supplementary Fig. S1A and B). Similar to uNK cells, B-cell abundance was not correlated with serum estradiol levels (natural n = 18, stimulated n = 7) (Supplementary Fig. S1C).
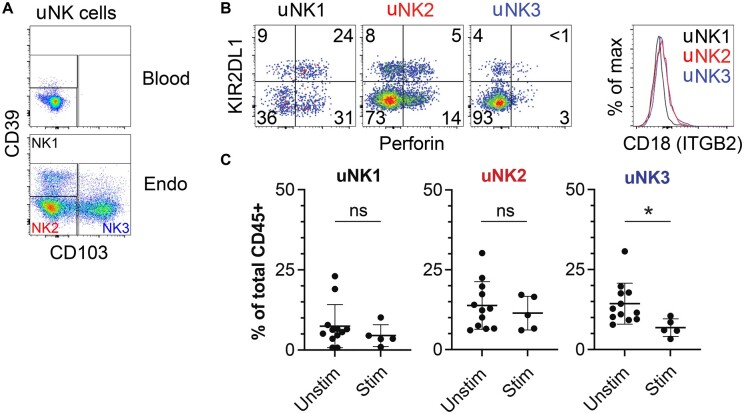
As percentages of bulk uNK cells were reduced after hormonal stimulation, we next sought to investigate whether all known subtypes of uNK cells were affected or whether one subset of uNK cells was disproportionately affected. Recent work profiling the maternal–fetal interface using single-cell RNA-seq in first-trimester tissue found three subsets of decidual NK (dNK) cells, denoted dNK1, dNK2, and dNK3 (Vento-Tormo et al., 2018). These transcriptionally distinct clusters of NK cells can be distinguished by flow cytometry with the surface proteins CD39 and CD103: dNK1 as CD103−CD39+ NK cells, dNK2 as CD103−CD39− NK cells, and dNK3 as CD103+ cells. Further, dNK1 cells are enriched for Perforin and killer cell immunoglobulin-like receptor 2DL1 (KIR2DL1), while dNK2 and dNK3 cells express CD18 (integrin subunit beta 2 (ITGB2)). We identified similar subsets in pre-pregnancy endometrial tissue, hereafter called uNK1, uNK2, and uNK3 cells (Fig. 3A and B). We analyzed the abundance of these subpopulations of NK cells in naturally cycling endometrium (n = 12) and hormonally stimulated endometrium (n = 5). We found that hormonal stimulation led to a significant decrease in the abundance of uNK3 cells as a proportion of all leukocytes (P = 0.025), while the abundance of uNK1 and uNK2 cells as a proportion of all leukocytes were not significantly changed (Fig. 3C). In contrast to uNK cells subsets in the endometrium, NK cells in the blood were unaffected by hormonal stimulation. Of note, blood NK cells exhibited minimal surface levels of CD39 and CD103 in our study (Fig. 3A).
Figure 3.
Specific reduction in endometrial uterine natural killer 3 (uNK3) cells after hormonal stimulation. (A) Raw flow cytometry gating strategy to identify uterine natural killer 1 (uNK1) (CD39+CD103−), uNK2 (CD39−CD103−), and uNK3 (CD39−CD103+) cells in the endometrium (‘endo’). Events shown are live, singlet, and CD45+CD3−CD56+ NK cells. (B) uNK1, uNK2, and uNK3 cells found in endometrium phenocopy previously described decidual NK1, NK2, and NK3 cells based on expression of KIR2DL1, Perforin, and CD18 (integrin beta 2 (ITGB2)). (C) Reduction in density of endometrial NK3 cells after hormonal stimulation. Each point represents a single tissue sample and bar represents the mean of each set of samples. Naturally cycling subjects (n = 11), hormonally stimulated subjects (n = 5). *P = 0.025 by unpaired t-test.
These data demonstrate that the maternal hormonal environment uniquely affects the composition of bulk uNK cells driven by changes in the uNK3 subset, and B cells in the maternal endometrium during the window of implantation. Due to our prior work demonstrating changes in the expression of genes associated with NK cell signaling due to hormonal stimulation in whole endometrial samples (Senapati et al., 2018), and our findings of a significant role for uNK cells in directing EVT invasion (Park et al., 2022), we focused our investigation on effects of maternal hormonal stimulation on uNK cell function during early placentation.
Hormonal stimulation impairs the function of uNK cells isolated from endometrial tissue
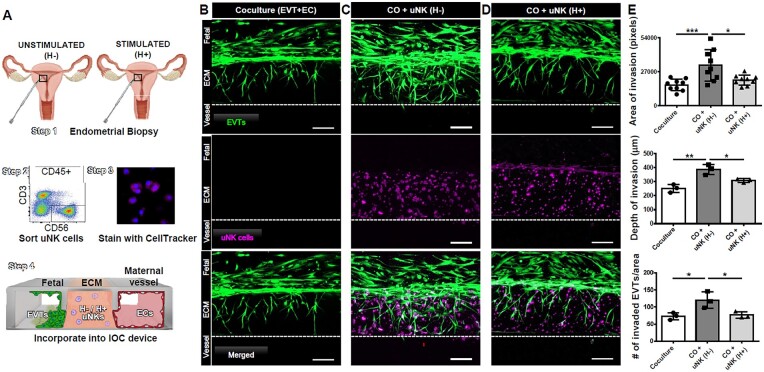
We next sought to investigate if hormonal stimulation can impact the function of uNK cells present in implantation window endometrium prior to pregnancy. In vitro experiments indicate that uNK cells can regulate invasion of trophoblasts (Kanter et al., 2021). We recently used the novel IOC device to study invasion of EVTs through the ECM toward maternal endometrial ECs and found that uNK cells had a proinvasive effect on EVTs (Park et al., 2022). Following a similar strategy, we obtained endometrial biopsies from hormonally unstimulated (n = 3) or stimulated (n = 3) endometrium during the mid-secretory phase and isolated bulk CD3−CD56+ uNK cells using fluorescence-activated cell sorting. We then incorporated the sort-purified uNK cells into the mock ECM of devices already seeded with EVTs in the fetal channel and maternal endometrial ECs seeded into the vascular channel (Fig. 4A). After 6 days of invasion, when EVTs breach the EC compartment, invasion was compared between the following devices: (i) coculture, containing EVTs and ECs; (ii) coculture devices with uNK cells from naturally cycling endometrium (H-); and (iii) coculture devices with uNK cells from hormonally stimulated endometrium (H+) (Fig. 4B–D). In the presence of maternal ECs only, EVTs migrated through the ECM and breached the vascular compartment in 6 days (Fig. 4B). Adding uNK cells isolated from naturally cycling women markedly increased EVT invasion (P = 0.01 by area of invasion in pixels, P = 0.03 by depth of invasion, P = 0.03 by number of invaded EVTs), confirming our previous data (Fig. 4C). In contrast, uNK cells isolated from women undergoing hormonal stimulation for IVF led to EVT invasion similar to that seen in coculture conditions, negating the proinvasive effect of uNK cells (Fig. 4D). Quantification of area invaded by EVTs, depth of invasion, and number of invaded EVTs by area in replicates using uNK cells isolated from different subjects confirmed these findings, showing a significant decrease in EVT invasion in devices with hormonally stimulated uNK cells compared to unstimulated uNK cells (P = 0.001 by area of invasion in pixels, P = 0.003 by depth of invasion, P = 0.02 by number of invaded EVTs) (Fig. 4E). These data demonstrate that superovulation impacts both abundance and function of uNK cells during the window of implantation.
Figure 4.
Hormonal stimulation alters the impact of uterine natural killer (uNK) cells on extravillous trophoblast (EVT) invasion. (A) Endometrial biopsies were obtained from unstimulated or stimulated endometrial biopsies (Step 1); uNK cells were identified as live, singlet, and CD45+ cells that express CD56 and lack CD3 (identifying T-lineage cells) by flow cytometry and sorted (Step 2); cells were stained with Cell Tracker (Step 3) and incorporated into gelated ECM within implantation-on-a-chip (IOC) devices containing EVTs and ECs (Step 4). (B) EVTs (green) reached the vascular compartment in 6 days in the presence of ECs (not shown). (C) Addition of unstimulated uNK cells (purple) increased EVT invasion compared to coculture devices. (D) uNK cells from hormonally stimulated endometrium reduced the proinvasive effect of uNK cells. Scale bars = 200 µm. (E) Quantification of area of EVT invasion by pixels (P = 0.001; P = 0.01; n = 9), depth of invasion (P = 0.003; P = 0.03; n = 3), and # of invaded EVTs by area (P = 0.02; P = 0.03; n = 3). *P < 0.05; **P < 0.01;***P < 0.001. Each point represents a single experimental run. Bars represent the standard deviation of the data (P < 0.05). EVTs, extravillous trophoblasts; ECs, endothelial cells; uNK, uterine natural killer; ECM, extracellular matrix; H−, unstimulated; H+, stimulated; CO, coculture.
Genes and pathways associated with cell migration and immune response are perturbed by hormonal stimulation
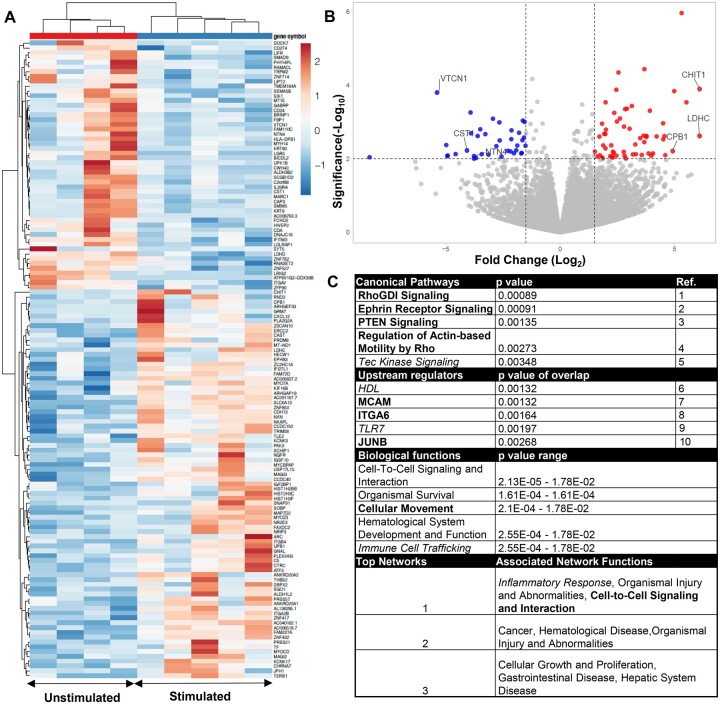
To identify potential candidate genes and pathways that are involved in uNK cell regulation of EVT invasion, we next examined how hormonal stimulation affects the gene expression profile of sorted bulk uNK cells. Samples were obtained from five hormonally stimulated and five naturally cycling subjects and were examined by RNA-seq following fluorescence-activated cell sorting (FACS). Principal component analysis demonstrated that one of the naturally cycling subjects was a remote outlier, data for which was excluded from subsequent analyses. Removal of the outlier resulted in subsequent clustering of stimulated and unstimulated subjects via hierarchical clustering and similarities in gene expression profiles were seen within each group (Fig. 5A). We found 196 genes with significant changes in expression (P-value < 0.01), of which 133 genes were protein-coding (Supplementary Table SII). Of these genes, 80 were upregulated (60%), while 53 were downregulated (40%) in stimulated uNK cells, and upregulated samples showed a greater magnitude of expression change as seen when represented by a volcano plot (Fig. 5B).
Figure 5.
Transcriptomic changes in bulk uterine natural killer (uNK) cells following hormonal stimulation. (A) Heat map showing differentially expressed genes based on bulk RNAseq of sort-purified uNK cells. Upregulated genes expressed in red with downregulation in blue. (B) Volcano plot of protein-coding genes examined via RNAseq. (C) Ingenuity pathway analysis of protein-coding genes showing top canonical pathways, upstream regulators, and biological function, and networks. In bold are pathways/genes/functions associated with immune cell trafficking and movement, and inflammation associated pathways, genes, and functions are italicized. 1: Tkachenko et al., 2011; 2: Sharfe et al., 2002; 3: Leong et al., 2015; 4: Wang et al., 2016; 5: Horwood et al., 2012; 6: Barter et al, 2004; 7: Guezguez et al., 2007; 8: Shannon and Mace, 2021; 9: Chatterjee et al., 2012; 10: Fontana et al., 2015. VTCN1, V-set domain containing T-cell activation inhibitor 1; CST1, cystatin SN; NTN4, netrin 4; CHIT1, chitinase 1; LDHC, lactate dehydrogenase C; CPB1, carboxypeptidase B1.
To place changes in gene expression seen in biological context, we next performed ingenuity pathway analysis (IPA) on differentially regulated protein-coding genes. Examining canonical pathways, upstream regulators, biological functions, and associated functions of potential networks, we found several pathways and genes with reported function in immune cell trafficking and migration (Fig. 5C, bold). Additionally, we found pathways involved in the regulation of inflammation (Fig. 5C, italicized).
In summary, we found several protein-coding genes perturbed in bulk uNK cells isolated from endometrial tissue during the implantation window, and preliminary pathway analysis suggests potential roles for hormonal stimulation used during IVF in the regulation of uNK cell migration and in the mediation of inflammation.
Discussion
In this study, we survey immune cell populations in secretory phase endometrium during the window of implantation in natural cycles and following gonadotropin stimulation. We find significant changes in abundance, function, and gene expression of uNK cells following gonadotropin stimulation. These changes may be a result of supraphysiologic estrogen levels but may also be related to direct or indirect effects of progesterone or other factors secreted by the corpus luteum. uNK cells are a unique population of NK cells present in the endometrium prior to and during pregnancy. They are phenotypically and functionally distinct from their peripheral blood counterparts. uNK cells drastically increase in abundance during the implantation window, becoming the most prominent lymphocyte population during early pregnancy. An emerging body of work using in vitro cell culture and explant models points to an active role for uNK cells in the regulation of EVT invasion and maternal artery remodeling through the secretion of soluble factors (Kanter et al., 2021). Mouse models deficient in uNK cells additionally demonstrate contributions to spiral artery remodeling and placentation, which are rescued when uNK cells are restored (Croy et al., 2010). However, their role in adverse pregnancy outcomes and disorders of human placentation remains incompletely defined. Epidemiologic studies demonstrate that pregnancies achieved following IVF with fresh embryo transfer, when the embryo is transferred immediately following ovarian stimulation with gonadotropins, are at increased risk for disordered placentation compared to unassisted pregnancies. Our findings thereby offer a potential etiology which may contribute to these observed adverse outcomes through alteration in the abundance, function, and gene expression of uNK cells. Our study examines immune cells during the window of implantation, when populations such as the uNK cells first undergo dramatic increases in abundance. Additionally, using our novel physiologically relevant IOC model, we are able to examine the functional repercussions of these changes on processes essential to early pregnancy. While we survey a large number of immune cell types, the study is biased toward those that have shown some involvement in pregnancy. Additionally, the implantation-on-chip device, while improving significantly on existing in vitro models, does not contain all maternal cell types of interest, and there is scope for increasing the complexity of the device. Finally, sourcing of cells from the endometrium of subjects preparing for both natural and stimulated frozen embryo transfer cycles is needed to understand if these approaches mitigate the changes in uNK cells.
Several reports attempting to correlate uNK cell abundance to adverse pregnancy outcomes have emerged with conflicting conclusions, nd how uNK cells function during normal and abnormal pregnancy is an active area of investigation (Faas and de Vos, 2017; Kanter et al., 2021). In this study, we show that hormonal stimulation, a ubiquitous intervention utilized as a part of IVF, results in significant changes in bulk uNK cell abundance. We find that these changes are driven by changes in the uNK3 subset first described by Vento-Tormo et al. (2018). This uNK cell subtype is believed to be a powerful producer of cytokines and chemokines that may support the invasive functions of EVTs (Vento-Tormo et al., 2018; Huhn et al., 2020). Studies examining uNK cells in the context of pregnancy complications frequently utilize uNK cells isolated from first-trimester maternal tissue, and assume homogeneity as a population. As immune cells responsible for promoting EVT invasion are likely those that accumulate prior to implantation of the embryo into the uterus, a particular strength of this study is use of uNK cells from endometrial biopsies rather than from first-trimester decidua. In line with this, evidence exists that shows phenotypic and gene expression differences between bulk endometrial NK cells and bulk first-trimester decidual NK cells (Manaster et al., 2008). Analysis of first-trimester tissue by Vento-Tormo et al. (2018) revealed multiple uNK cell subtypes, and subsequent studies have described other subsets distinct from those described by this group (Fu et al., 2017; Gamliel et al., 2018; Suryawanshi et al., 2018; Huhn et al., 2020) highlighting the complexity of uNK cells present during pregnancy. Examination of prepregnancy endometrial tissue in this study and identification of NK1, 2, and 3 subsets suggests that uNK cell subsets exist prior to arrival of the embryo. These findings highlight the need to consider and examine uNK cells as phenotypically and functionally heterogeneous. Future work is ongoing and will examine functional differences within these populations, which might then allow us to alter the course of pregnancies threatened by functionally impaired uNK cells.
Fetal–maternal crosstalk is essential for appropriate regulation of the directionality and extent of EVT invasion. Current studies investigating the contribution of individual maternal cell types to early pregnancy rely on animal models and in vitro culture systems that do not adequately capture the complex cellular interactions involved in embryo implantation and early placentation. Additionally, functional readouts relevant to nonuterine NK cells, such as traditional cytotoxicity assays, do not translate appropriately to uNK cells (Kopcow et al., 2005). With our IOC device, we have demonstrated that uNK cells play a critical role in regulating EVT invasion (Park et al., 2022). In this study, we found that, unlike uNK cells from naturally cycling subjects, uNK cells exposed to supraphysiologic levels of hormones did not significantly enhance EVT invasion. This change in the ability of uNK cells obtained from hormonally stimulated women to promote EVT invasion could lead to abnormalities in placentation and, in turn, to adverse fetal and maternal outcomes. Indeed, women undergoing IVF suffer from pre-eclampsia and intrauterine growth restriction much more commonly than women with naturally conceived pregnancies (Shevell et al., 2005; Zhu et al., 2016). It has been suggested that such outcomes are characterized by insufficient invasion of EVTs (Kaufmann et al., 2003). uNK cells are just one of many cell types regulating EVT invasion, however. Prior work from our lab demonstrates influence of other cell types, including decreased EVT invasion in the presence of decidualized stromal cells, however, proinvasive effects of uNK cells are persistent in the presences of decidualized stromal cells (Park et al., 2022). Future work examining interactions among numerous immune cell types including macrophages and dendritic cells, as well as other maternal cells including vascular smooth muscle cells and decidualized stromal cells, as well as the influence of hormonal stimulation on these cell types, are important avenues for future investigation on the IOC device.
In order to identify potential factors that may be regulating EVT invasion, we identified significant differences in gene expression via RNA-seq of sorted prepregnancy uNK cells from stimulated and unstimulated subjects. IPA analysis indicated that genes and pathways involved in immune cell migration and inflammation are affected by hormonal stimulation.
uNK cell populations are dynamic and go through significant enrichment during the window of implantation and when EVT invasion occurs. How this enrichment occurs is an ongoing area of investigation, but it is thought to involve proliferation of existing uNK cells, as well as recruitment and migration of peripheral NK cells (Kanter et al., 2021). Our data showing a lower abundance of uNK cells and defective uNK cell function suggests that hormonal stimulation could lead to abnormalities in uNK cell migration to their site of action, though this remains to be tested. Another emerging theme from IPA on our dataset was the mediation of inflammation. Pregnancy is a unique inflammatory environment, and studies have found that proinflammatory signals are necessary for normal pregnancy, especially during the first trimester, and during parturition (Mor et al., 2017). In line with this, conditions associated with systemic inflammation, such as lupus, result in several pregnancy complications (Peart and Clowse, 2014). However, how inflammation contributes to pregnancy is commonly studied in the context of maternal tolerance toward the fetus, while understanding how inflammation could participate in normal processes involved in pregnancy establishment remains unknown. Further work investigating the role of particular genes involved in these pathways in the context of uNK cells and pregnancy, and examination of differentially expressed genes within uNK cells subsets is required. Furthermore, in order to garner a better understanding of effects on other cell types present within the maternal endometrium, we plan to pursue a comprehensive single-cell RNAseq in order to assess these changes.
The use of IVF is continuing to rise and new interventions and treatment options continue to emerge. However, we still do not understand the mechanisms by which these interventions may increase the risk of adverse outcomes. Mitigation of the increased risks for adverse outcomes associated with IVF, seen even in singleton pregnancies, depends upon a more comprehensive understanding of mechanisms underlying adverse clinical outcomes. Findings from this study provide a possible contributing factor in changes in uNK cell number and function seen early in gestation and influence on EVT invasion, thought to be a factor in complications that are not clinically apparent until later in gestation. Here we demonstrate for the first time, the applicability of a novel research platform, the IOC device, to dissecting processes critical to the understanding of normal and abnormal pregnancy in humans. Future work utilizing this novel device will be critical to developing diagnostic and therapeutic options for the treatment of the multiple adverse pregnancy outcomes associated with abnormal placentation.
Supplementary Material
Contributor Information
J Kanter, Division of Reproductive Endocrinology and Infertility, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
S M Gordon, Division of Neonatology, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Pediatrics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
S Mani, Division of Reproductive Endocrinology and Infertility, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
A Sokalska, Division of Reproductive Endocrinology and Infertility, Stanford University, Stanford, CA, USA.
J Y Park, Department of Bioengineering, University of Pennsylvania, Philadelphia, PA, USA.
S Senapati, Division of Reproductive Endocrinology and Infertility, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
D D Huh, Department of Bioengineering, University of Pennsylvania, Philadelphia, PA, USA; NSF Science and Technology Center for Engineering Mechanobiology, University of Pennsylvania, Philadelphia, PA, USA.
M Mainigi, Division of Reproductive Endocrinology and Infertility, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA.
Data availability
The data underlying this article are available in the Gene Expression Omnibus (GEO) database at https://www.ncbi.nlm.nih.gov/geo/ under accession number GSE222892.
Authors’ roles
S.M., J.K., S.M.G., and M.M. conceived and planned the data analyses and experiments. S.M., J.K., J.Y.P., and S.M.G. carried out the experiments and performed the data analysis. All authors contributed to interpretation of the results. S.M., J.K., S.M.G., and M.M. wrote the article, with all authors critically editing the article.
Funding
Research reported in this publication was supported by the University of Pennsylvania University Research Funding (to M.M.), the Eunice Kennedy Shriver National Institute of Child Health and Human Development (P50HD068157 to M.M., S.S., and S.M.), National Center for Advancing Translational Sciences of the National Institutes of Health (TL1TR001880 to J.K.), the Institute for Translational Medicine and Therapeutics of the Perelman School of Medicine at the University of Pennsylvania, the Children’s Hospital of Philadelphia Research Institute (to S.M.G.), and the National Institute of Allergy and Infectious Diseases (K08AI151265 to S.M.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest
All authors declare no conflict of interest.
References
- Anton L, Brown AG, Parry S, Elovitz MA. Lipopolysaccharide induces cytokine production and decreases extravillous trophoblast invasion through a mitogen-activated protein kinase-mediated pathway: possible mechanisms of first trimester placental dysfunction. Hum Reprod 2012;27:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton L, DeVine A, Polyak E, Olarerin-George A, Brown AG, Falk MJ, Elovitz MA. HIF-1α stabilization increases miR-210 eliciting first trimester extravillous trophoblast mitochondrial dysfunction. Front Physiol 2019;10:699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter PJ, Nicholls S, Rye KA, Anantharamaiah GM, Navab M, Fogelman AM. Antiinflammatory properties of HDL. Circ Res 2004;95:764–772. [DOI] [PubMed] [Google Scholar]
- Bulmer JN, Morrison L, Longfellow M, Ritson A, Pace D. Granulated lymphocytes in human endometrium: histochemical and immunohistochemical studies. Hum Reprod 1991;6:791–798. [DOI] [PubMed] [Google Scholar]
- Carter AM. Animal models of human placentation—a review. Placenta 2007;28(Suppl A):S41–S47. [DOI] [PubMed] [Google Scholar]
- Carter AM, Mess AM. Mammalian Placentation: Implications for Animal Models. In McManus LM, Mitchell RN (eds). Pathobiology of Human Disease: A Dynamic Encyclopedia of Disease Mechanisms. Waltham, MA, USA: Elsevier, 2014, 2423–2442. [Google Scholar]
- Chatterjee P, , WeaverLE, , DoerschKM, , KoprivaSE, , ChiassonVL, , AllenSJ, , NarayananAM, , YoungKJ, , JonesKA, , Kuehl TJ et al. Placental Toll-like receptor 3 and Toll-like receptor 7/8 activation contributes to preeclampsia in humans and mice. PLoS One 2012;7:e41884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croy BA, Zhang J, Tayade C, Colucci F, Yadi H, Yamada AT. Analysis of uterine natural killer cells in mice. Methods Mol Biol 2010;612:465–503. [DOI] [PubMed] [Google Scholar]
- de Waal E, Yamazaki Y, Ingale P, Bartolomei MS, Yanagimachi R, McCarrey JR. Gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice. Hum Mol Genet 2012;21:4460–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faas MM, de Vos P. Uterine NK cells and macrophages in pregnancy. Placenta 2017;56:44–52. [DOI] [PubMed] [Google Scholar]
- Flynn L, Byrne B, Carton J, Kelehan P, O’Herlihy C, O’Farrelly C. Menstrual cycle dependent fluctuations in NK and T-lymphocyte subsets from non-pregnant human endometrium. Am J Reprod Immunol 2000;43:209–217. [DOI] [PubMed] [Google Scholar]
- Fontana MF, , BaccarellaA, , PancholiN, , PufallMA, , HerbertDR, , Kim CC. JUNB is a key transcriptional modulator of macrophage activation. J Immunol 2015;194:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B, Zhou Y, Ni X, Tong X, Xu X, Dong Z, Sun R, Tian Z, Wei H. Natural killer cells promote fetal development through the secretion of growth-promoting factors. Immunity 2017;47:1100–1113.e6. [DOI] [PubMed] [Google Scholar]
- Gamliel M, Goldman-Wohl D, Isaacson B, Gur C, Stein N, Yamin R, Berger M, Grunewald M, Keshet E, Rais Y et al. Trained memory of human uterine NK cells enhances their function in subsequent pregnancies. Immunity 2018;48:951–962.e5. [DOI] [PubMed] [Google Scholar]
- Gaynor LM, Colucci F. Uterine natural killer cells: functional distinctions and influence on pregnancy in humans and mice. Front Immunol 2017;8:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getsios S, Chen GT, Huang DT, MacCalman CD. Regulated expression of cadherin-11 in human extravillous cytotrophoblasts undergoing aggregation and fusion in response to transforming growth factor beta 1. J Reprod Fertil 1998;114:357–363. [DOI] [PubMed] [Google Scholar]
- Goldman-Wohl D, Yagel S. Regulation of trophoblast invasion: from normal implantation to pre-eclampsia. Mol Cell Endocrinol 2002;187:233–238. [DOI] [PubMed] [Google Scholar]
- Gordon SM, Nishiguchi MA, Chase JM, Mani S, Mainigi MA, Behrens EM. IFNs drive development of novel IL-15-responsive macrophages. J Immunol 2020;205:1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CH, Lysiak JJ, McCrae KR, Lala PK. Localization of transforming growth factor-beta at the human fetal-maternal interface: role in trophoblast growth and differentiation. Biol Reprod 1992;46:561–572. [DOI] [PubMed] [Google Scholar]
- Grigsby PL. Animal models to study placental development and function throughout normal and dysfunctional human pregnancy. Semin Reprod Med 2016;34:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guezguez B, Vigneron P, Lamerant N, Kieda C, Jaffredo T, Dunon D. Dual role of melanoma cell adhesion molecule (MCAM)/CD146 in lymphocyte endothelium interaction: MCAM/CD146 promotes rolling via microvilli induction in lymphocyte and is an endothelial adhesion receptor. J Immunol 2007;179:6673–6685. [DOI] [PubMed] [Google Scholar]
- Ho HN, Chao KH, Chen CK, Yang YS, Huang SC. Activation status of T and NK cells in the endometrium throughout menstrual cycle and normal and abnormal early pregnancy. Hum Immunol 1996;49:130–136. [DOI] [PubMed] [Google Scholar]
- Horwood NJ, Urbaniak AM, Danks L. Tec family kinases in inflammation and disease. Int Rev Immunol 2012;31:87–103. [DOI] [PubMed] [Google Scholar]
- Huhn O, Ivarsson MA, Gardner L, Hollinshead M, Stinchcombe JC, Chen P, Shreeve N, Chazara O, Farrell LE, Theorell J et al. Distinctive phenotypes and functions of innate lymphoid cells in human decidua during early pregnancy. Nat Commun 2020;11:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James J, Tun W, Clark A. Quantifying trophoblast migration: in vitro approaches to address in vivo situations. Cell Adh Migr 2016;10:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra SK, Ratcliffe SJ, Coutifaris C, Molinaro T, Barnhart KT. Ovarian stimulation and low birth weight in newborns conceived through in vitro fertilization. Obstet Gynecol 2011;118:863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanter JR, Mani S, Gordon SM, Mainigi M. Uterine natural killer cell biology and role in early pregnancy establishment and outcomes. F S Rev 2021;2:265–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann P, Black S, Huppertz B. Endovascular trophoblast invasion: implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol Reprod 2003;69:1–7. [DOI] [PubMed] [Google Scholar]
- Kim YM, Bujold E, Chaiworapongsa T, Gomez R, Yoon BH, Thaler HT, Rotmensch S, Romero R. Failure of physiologic transformation of the spiral arteries in patients with preterm labor and intact membranes. Am J Obstet Gynecol 2003;189:1063–1069. [DOI] [PubMed] [Google Scholar]
- King A, Wellings V, Gardner L, Loke YW. Immunocytochemical characterization of the unusual large granular lymphocytes in human endometrium throughout the menstrual cycle. Hum Immunol 1989;24:195–205. [DOI] [PubMed] [Google Scholar]
- Kopcow HD, Allan DS, Chen X, Rybalov B, Andzelm MM, Ge B, Strominger JL. Human decidual NK cells form immature activating synapses and are not cytotoxic. Proc Natl Acad Sci USA 2005;102:15563–15568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash GE, Otun HA, Innes BA, Percival K, Searle RF, Robson SC, Bulmer JN. Regulation of extravillous trophoblast invasion by uterine natural killer cells is dependent on gestational age. Hum Reprod 2010;25:1137–1145. [DOI] [PubMed] [Google Scholar]
- Lee JY, Lee M, Lee SK. Role of endometrial immune cells in implantation. Clin Exp Reprod Med 2011;38:119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong JW, Schneider SE, Sullivan RP, Parikh BA, Anthony BA, Singh A, Jewell BA, Schappe T, Wagner JA, Link DC et al. PTEN regulates natural killer cell trafficking in vivo. Proc Natl Acad Sci USA 2015;112:E700–E709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Diao L, Huang C, Li Y, Zeng Y, Kwak-Kim JYH. The role of decidual immune cells on human pregnancy. J Reprod Immunol 2017;124:44–53. [DOI] [PubMed] [Google Scholar]
- Ma L, Li G, Cao G, Zhu Y, Du MR, Zhao Y, Wang H, Liu Y, Yang Y, Li YX et al. dNK cells facilitate the interaction between trophoblastic and endothelial cells via VEGF-C and HGF. Immunol Cell Biol 2017;95:695–704. [DOI] [PubMed] [Google Scholar]
- Manaster I, Mizrahi S, Goldman-Wohl D, Sela HY, Stern-Ginossar N, Lankry D, Gruda R, Hurwitz A, Bdolah Y, Haimov-Kochman R et al Endometrial NK cells are special immature cells that await pregnancy. J Immunol 2008;181:1869–1876. [DOI] [PubMed] [Google Scholar]
- Maternal Mortality. Gl obal Health Observatory Data. Switzerland: World Health Organization, 2015. [Google Scholar]
- Mor G, Aldo P, Alvero AB. The unique immunological and microbial aspects of pregnancy. Nat Rev Immunol 2017;17:469–482. [DOI] [PubMed] [Google Scholar]
- Pandey S, Shetty A, Hamilton M, Bhattacharya S, Maheshwari A. Obstetric and perinatal outcomes in singleton pregnancies resulting from IVF/ICSI: a systematic review and meta-analysis. Hum Reprod Update 2012;18:485–503. [DOI] [PubMed] [Google Scholar]
- Park JY, Mani S, Clair G, Olson HM, Paurus VL, Ansong CK, Blundell C, Young R, Kanter J, Gordon S et al. A microphysiological model of human trophoblast invasion during implantation. Nat Commun 2022;13:1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart E, Clowse ME. Systemic lupus erythematosus and pregnancy outcomes: an update and review of the literature. Curr Opin Rheumatol 2014;26:118–123. [DOI] [PubMed] [Google Scholar]
- Pollheimer J, Vondra S, Baltayeva J, Beristain AG, Knofler M. Regulation of placental extravillous trophoblasts by the maternal uterine environment. Front Immunol 2018;9:2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson A, Harris LK, Innes BA, Lash GE, Aljunaidy MM, Aplin JD, Baker PN, Robson SC, Bulmer JN. Uterine natural killer cells initiate spiral artery remodeling in human pregnancy. FASEB J 2012;26:4876–4885. [DOI] [PubMed] [Google Scholar]
- Senapati S, Wang F, Ord T, Coutifaris C, Feng R, Mainigi M. Superovulation alters the expression of endometrial genes critical to tissue remodeling and placentation. J Assist Reprod Genet 2018;35:1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon MJ, , Mace EM. Natural killer cell integrins and their functions in tiss residency. Front Immunol 2021;12:647358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharfe N, Freywald A, Toro A, Dadi H, Roifman C. Ephrin stimulation modulates T cell chemotaxis. Eur J Immunol 2002;32:3745–3755. [DOI] [PubMed] [Google Scholar]
- Shevell T, Malone FD, Vidaver J, Porter TF, Luthy DA, Comstock CH, Hankins GD, Eddleman K, Dolan S, Dugoff L et al. Assisted reproductive technology and pregnancy outcome. Obstet Gynecol 2005;106:1039–1045. [DOI] [PubMed] [Google Scholar]
- Silva JF, Serakides R. Intrauterine trophoblast migration: a comparative view of humans and rodents. Cell Adh Migr 2016;10:88–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Dunk CE, Aplin JD, Harris LK, Jones RL. Evidence for immune cell involvement in decidual spiral arteriole remodeling in early human pregnancy. Am J Pathol 2009;174:1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Ghosh J, Mainigi M, Turan N, Weinerman R, Truongcao M, Coutifaris C, Sapienza C. DNA methylation differences between in vitro- and in vivo-conceived children are associated with ART procedures rather than infertility. Clin Epigenetics 2015;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart TJ, O’Neill K, Condon D, Sasson I, Sen P, Xia Y, Simmons RA. Diet-induced obesity alters the maternal metabolome and early placenta transcriptome and decreases placenta vascularity in the mouse. Biol Reprod 2018;98:795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan-Pyke C, Mani S, Rhon-Calderon EA, Ord T, Coutifaris C, Bartolomei MS, Mainigi M. Timing of exposure to gonadotropins has differential effects on the conceptus: evidence from a mouse model. Biol Reprod 2020;103:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max KEA, Garzia A, Kustagi M, Tuschl T, Williams Z. A single-cell survey of the human first-trimester placenta and decidua. Sci Adv 2018;4:eaau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson AM, David AL. Animal models of fetal growth restriction: considerations for translational medicine. Placenta 2015;36:623–630. [DOI] [PubMed] [Google Scholar]
- Tantbirojn P, Crum CP, Parast MM. Pathophysiology of placenta creta: the role of decidua and extravillous trophoblast. Placenta 2008;29:639–645. [DOI] [PubMed] [Google Scholar]
- Thiruchelvam U, Dransfield I, Saunders PT, Critchley HO. The importance of the macrophage within the human endometrium. J Leukoc Biol 2013;93:217–225. [DOI] [PubMed] [Google Scholar]
- Tilburgs T, Evans JH, Crespo AC, Strominger JL. The HLA-G cycle provides for both NK tolerance and immunity at the maternal-fetal interface. Proc Natl Acad Sci USA 2015;112:13312–13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachenko E, Sabouri-Ghomi M, Pertz O, Kim C, Gutierrez E, Machacek M, Groisman A, Danuser G, Ginsberg MH. Protein kinase A governs a RhoA-RhoGDI protrusion-retraction pacemaker in migrating cells. Nat Cell Biol 2011;13:660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nieuwenhoven ALV, Heineman MJ, Faas MM. The immunology of successful pregnancy. Hum Reprod Update 2003;9:347–357. [DOI] [PubMed] [Google Scholar]
- Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polanski K, Goncalves A et al Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018;563:347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrooman LA, Rhon-Calderon EA, Chao OY, Nguyen DK, Narapareddy L, Dahiya AK, Putt ME, Schultz RM, Bartolomei MS. Assisted reproductive technologies induce temporally specific placental defects and the preeclampsia risk marker sFLT1 in mouse. Development 2020;147:dev186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun J, Ma C, Gao W, Song B, Xue H, Chen W, Chen X, Zhang Y, Shao Q et al. Reduced expression of galectin-9 contributes to a poor outcome in colon cancer by inhibiting NK cell chemotaxis partially through the Rho/ROCK1 signaling pathway. PLoS One 2016;11:e0152599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinerman R, Ord T, Bartolomei MS, Coutifaris C, Mainigi M. The superovulated environment, independent of embryo vitrification, results in low birthweight in a mouse model. Biol Reprod 2017;97:133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Liu Y, Zhang R, Wu Y, Huang Y, Liu F, Li M, Sun S, Xing L et al. Maternal and live-birth outcomes of pregnancies following assisted reproductive technology: a retrospective cohort study. Sci Rep 2016;6:35141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the Gene Expression Omnibus (GEO) database at https://www.ncbi.nlm.nih.gov/geo/ under accession number GSE222892.