Abstract
We report the prevalence of the O139 serogroup in Calcutta, India, after its reemergence in August 1996 and the spread of the reemerged clone to other parts of the country by using previously established molecular markers. Phenotypically, the reemerged Vibrio cholerae O139 displayed a difference compared to those that appeared in late 1992 and 1993 in that the current O139 strains are sensitive to co-trimoxazole. Ribotyping with the enzyme BglI produced two rRNA restriction patterns in the O139 strains isolated after August 1996, and these patterns were identical to those exhibited by strains of O139 isolated in 1992. Three clones of V. cholerae O139 are currently prevailing in the country, with strains exhibiting three bands after HindIII digestion and hybridization with a ctxA probe being dominant. The reemergence of V. cholerae O139 in Calcutta after a 32-month quiescent period reestablishes the O139 serogroup as an entity which is likely to play a crucial role in the temporal antigenic variations among the serogroups of V. cholerae causing cholera.
Vibrio cholerae is classified into more than 155 serogroups based on the heat-stable somatic O antigen (21, 22). The disease cholera is, however, caused by only two serogroups, namely, O1 and O139. The O139 serogroup is a recent addition which appeared abruptly in September 1992 in southern India (17) and rapidly spread to virtually all areas where cholera is endemic in India (14) and in neighboring countries (15). The extent and rapidity of the spread of the O139 serogroup led us to conclude that this event was the beginning of the eighth pandemic of cholera (14). In February 1994, the O1 serogroup, which had reappeared sporadically in July 1993, again replaced the O139 serogroup and became the dominant serogroup causing cholera in Calcutta, India (11). Subsequent studies showed that the O1 serogroup which replaced the O139 serogroup was a new clone of O1 ElTor biotype (7, 19, 27).
A quiescent period followed in the brief history of V. cholerae O139, and it was thought that the appearance of O139 was a one-time event. However, an upsurge of V. cholerae O139 was observed in Calcutta in August 1996 (10), and the O139 serogroup again became the dominant serogroup causing cholera by September 1996. Molecular studies showed that the O139 strains which reemerged in August 1996 were indistinguishable from the O139 strains isolated in 1992 and 1993 by ribotyping but showed a unique change in the structure and organization of the CTX genetic element (20). In this study, the prevalence of the O139 serogroup in Calcutta after its reemergence and the spread of the reemerged clone to other parts of the country were investigated by using previously established molecular markers.
The present study is a part of the continuing surveillance program of the National Institute of Cholera and Enteric Diseases, Calcutta, on cholera. Stool specimens were obtained from patients admitted to the Infectious Diseases Hospital, Calcutta, the only hospital which admits cholera patients from the metropolitan city and suburban areas. Methods adopted for collection, transport, bacteriological examination of stool samples, identification, and serotyping of V. cholerae have been described in detail previously (11). The National Institute of Cholera and Enteric Diseases, India’s national reference laboratory for cholera, and a World Health Organization Reference Centre for Training and Research on Diarrhoeal Diseases, confirms and characterizes V. cholerae strains submitted from all over the country and from neighbouring countries. Strains received are characterized by using a panoply of tests as previously described (14). A total of 17 strains of V. cholerae O139 isolated between August 1996 and July 1997 from Calcutta and other parts of the country (Table 1) were included in this study for extensive molecular characterization. Another two strains of V. cholerae O139, one isolated in 1992 in Calcutta (SG24) and the other isolated in the same year in Madras (MO45 [ATCC 51394]), were included in this study for comparison.
TABLE 1.
Antibiogram and RFLP analyses of the CTX genetic element of the 17 clinical V. cholerae O139 strains isolated from different parts of India after September 1996
| Strain no. | Place of isolation | Region | Antibiogram | Restriction patterna of CTX genetic element | Ribotypeb |
|---|---|---|---|---|---|
| AS258 | Calcutta | Eastern | AP-FZ-NM-SM | HindIII: 14.0, 9.0, 7.2; BglII: 23.0 | 3a |
| MD1 | Midnapur | Eastern | AP-FZ-NM-SM | HindIII: 14.0, 9.0, 7.2; BglII: 23.0 | 5a |
| MD3 | Midnapur | Eastern | AP-FZ-NM-SM | HindIII: 14.0, 9.0, 7.2; BglII: 23.0 | 3a |
| NPO549 | Nagpur | Deccan | AP-FZ-NM-SM | HindIII: 14.0, 9.0, 7.2; BglII: 23.0 | 3a |
| YO29 | Yavatmal | Deccan | AP-FZ-SM | HindIII: 14.0, 9.0, 7.2; BglII: 23.0 | 3a |
| MO585 | Madras | Southern | AP-FZ-NM-SM | HindIII: 14.0, 9.0, 7.2 | 3a |
| NPO554 | Nagpur | Deccan | AP-FZ-SM | HindIII: 14.0, 9.0, 7.2 | 3a |
| NPO564 | Nagpur | Deccan | AP-FZ-NM-SM | HindIII: 14.0, 9.0, 7.2 | 3a |
| YO30 | Yavatmal | Deccan | AP-FZ-SM | HindIII: 14.0, 9.0, 7.2 | 3a |
| SO58 | Sewagram | Deccan | AP-FZ-SM | HindIII: 14.0, 9.0, 7.2 | 3a |
| APO16 | Amravati | Deccan | AP-FZ-NM-SM | HindIII: 14.0, 9.0, 7.2 | 3a |
| VO82 | Vellore | Southern | AP-FZ-NA-SM | HindIII: 20.0; BglII: 8.2, 7.2 | 3a |
| VO83 | Vellore | Southern | AP-FZ-NA-SM | HindIII: 20.0; BglII: 8.2, 7.2 | 3a |
| PO42 | Pune | Western | AP-FZ-NA-SM | HindIII: 20.0; BglII: 8.2, 7.2 | 5a |
| WSO10 | Solapur | Deccan | AP-FZ-NA-SM | HindIII: 20.0; BglII: 8.2, 7.2 | 5a |
| BOM6 | Bombay | Western | AP-FZ-NM-NA-SM | HindIII: 15; BglII: 7.2 | 5a |
| MO579 | Madras | Southern | AP-FZ-SM | HindIII: 15 | 3a |
Restriction enzyme and sizes (in kilobases) of the fragments of the CTX genetic element of V. cholerae O139 which hybridized with the ctxA gene probe.
Ribotype designations are the same as those of Popovic et al. (16).
Representative strains of V. cholerae O139 isolated from hospitalized patients in Calcutta in 1992 and 1993 (101 strains) and 1996 and 1997 (89 strains) and all 17 representative strains of V. cholerae O139 included in the detailed molecular characterization study were examined for resistance to ampicillin (AP) (10 μg), chloramphenicol (30 μg), co-trimoxazole (25 μg), ciprofloxacin (5 μg), furazolidone (FZ) (100 μg), gentamicin (10 μg), neomycin (NM) (30 μg), nalidixic acid (NA) (30 μg), norfloxacin (10 μg), streptomycin (SM) (10 μg), and tetracycline (30 μg) by using commercial discs (Hi Media, Bombay, India) as described previously (11). Characterization of strains as susceptible or resistant was based on size of the inhibition zone around each disc according to the manufacturer’s instructions, which matched the interpretive criteria recommended by the World Health Organization (25). Strains showing an intermediate zone of inhibition were interpreted as resistant to that drug on the basis of previous MIC studies conducted with V. cholerae (26).
Genomic DNA extraction and Southern hybridization were performed as described earlier (19). Genomic DNA was digested with BglI for ribotyping and with HindIII and BglII for CTX restriction fragment length polymorphism (RFLP) following the manufacturer’s instructions (Boehringer Mannheim GmbH, Mannheim, Germany). Digested fragments were separated by agarose gel electrophoresis (0.8% gel) and Southern hybridized on nylon membrane (Hybond-N+, Amersham Life Science, Little Chalfont, Buckinghamshire, England). The rRNA gene probe was a 7.5-kb BamHI fragment of pKK3535, which is a pBR322-derived plasmid containing an Escherichia coli rRNA operon consisting of one copy each of the genes coding for 5S rRNA, 16S rRNA, 23S rRNA, and tRNAGlu (5). The gene probe for cholera toxin was a 0.5-kb EcoRI fragment of pCVD27, which is a pBR325-derived plasmid containing an XbaI-ClaI fragment representing 94% of the gene encoding the A subunit of cholera toxin (ctxA) cloned with EcoRI linkers (9). Labelling of the probes, hybridization, and detection of the bands were performed with the ECL detection system (Amersham Life Science) and Kodak Biomax Film (Eastman Kodak Co., Rochester, N.Y.).
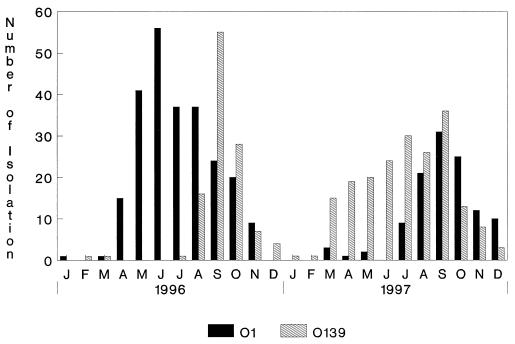
The monthly incidence of V. cholerae O1 and O139 in patients admitted to the Infectious Diseases Hospital, Calcutta, from January 1996 to December 1997 is shown in Fig. 1. The resurgence of V. cholerae O139 in Calcutta began in August 1996, and by September 1996, the O139 serogroup dominated over the O1 serogroup after a lapse of 32 months. After September 1996, O139 strains were also received from different parts of the country. The O139 serogroup dominated until September 1997. From October to December 1997, the O1 serogroup again became the dominant serogroup associated with cholera in Calcutta.
FIG. 1.
Monthly isolation of various serogroups of V. cholerae from patients hospitalized because of acute secretory diarrhea at the Infectious Diseases Hospital, Calcutta, from January 1996 to December 1997 (J to D, respectively).
Comparison of the drug resistance patterns between O139 strains isolated in 1992 and 1993 and those isolated in 1996 and 1997 showed that the strains from 1996 and 1997 were susceptible to co-trimoxazole, unlike the O139 strains from 1992 and 1993, which were resistant to this drug, corroborating our previous observation (10). Interestingly, the O139 strains isolated in 1996 and 1997 are becoming increasingly resistant to ampicillin and neomycin but increasingly susceptible to chloramphenicol and, to a certain extent, streptomycin. The drug resistance pattern of the O139 strains isolated from different parts of the country was similar to the drug resistance pattern of the newly emerged O139 strains in Calcutta with the exception of five strains which were additionally resistant to nalidixic acid (Table 1).
Ribotyping with the enzyme BglI produced two distinct rRNA restriction patterns in the O139 strains isolated after August 1996 which were similar to the ribotype patterns of the O139 strains isolated in 1992 and 1993 and were designated as 3a and 5a by Popovic et al. (16). The ribotypes 3a and 5a differed from each other by a single 6-kb band. Southern blot analysis of HindIII-digested genomic DNA of 11 of the 17 O139 strains showed that three bands of 14, 9, and 7.2 kb hybridized with the ctxA probe, as reported previously (20). The strains demonstrating this pattern were widely distributed in the country and dominated in the eastern and Deccan regions in India. Two other HindIII patterns, one in which a 15-kb fragment (shown by two strains) and one in which a 20-kb fragment (shown by four strains) hybridized with the ctxA probe, were found sporadically among strains from Southern and Western regions and from a single place in the Deccan region (Table 1). The HindIII digest showing the 15-kb fragment resembled the pattern shown by SG24 and MO45, the O139 strains which appeared in 1992 (data not shown).
We further analyzed representative strains showing the three different HindIII patterns with BglII to understand the structure of the CTX genetic element. Strains which showed three bands after HindIII digestion showed a single band of about 23 kb when digested with BglII and probed with ctxA, indicating that in these strains the BglII site resides outside the CTX genetic element, which corroborates our earlier findings (20). Southern blot hybridization of BglII-digested genomic DNA by using a ctxA probe of a representative strain which gave the 15-kb band upon HindIII digestion showed a single fragment of 7.2 kb. This indicates the presence of a single copy of the CTX genetic element. Southern blot hybridization of BglII-digested genomic DNA of representative strains which showed a 20-kb HindIII fragment with a ctxA probe demonstrated two fragments of 7.2 and 8.2 kb. This indicates that in these strains there is a tandem duplication of the CTX genetic element.
Until September 1997, the O139 serogroup dominated as the causal serogroup of cholera among hospitalized patients admitted to the Infectious Diseases Hospital, Calcutta. As shown previously (20), the recent O139 strains show substantial reorganization in the structure of the CTX genetic element compared to the 1992 and 1993 O139 strains. In this study, we exploited the unusual HindIII RFLP of the CTX genetic element of strains of the reemergent O139 serogroup to monitor the spread of this clone to other parts of India. From the results, it is clear that strains of O139 exhibiting three bands after HindIII digestion of the genomic DNA and subsequently hybridized with ctxA probe constitute the dominant O139 clone currently prevailing in the country. This new O139 clone appears to have originated in Calcutta because the representative strain (MO579) of O139 isolated after September 1996 in Madras showed a single 15-kb band after HindIII digestion, while the representative 1997 Madras strain (MO585) showed three bands like those shown by the Calcutta O139 strains after HindIII digestion. Therefore, it appears that the new O139 clone may have spread from Calcutta to Madras, which is in contrast to the 1992 situation, when O139 spread from Madras to Calcutta (14, 17).
This study also shows that O139 strains similar to a 1992 clone of O139, which showed a single band of around 15 kb after HindIII digestion, are prevalent in certain areas, like Bombay. Further restriction analysis with BglII revealed that this strain had only a single copy of the CTX genetic element. In the 1992 epidemic strains of O139, most strains had two tandemly duplicated copies of the CTX genetic element, while in some a single copy of the element was detected (3, 24). The strains BOM6 and MO579, therefore, appear to be remnants of the O139 clone from 1992 and 1993, which continue to prevail in or represent areas where the new O139 clone has not yet spread. A third HindIII restriction pattern among O139 strains from Vellore, Pune, and Solapur, India, was observed. Although in these strains there was a duplication of the CTX genetic element, as evident from BglII digestion, the HindIII 20-kb band does not resemble the 1992 O139 pattern. Therefore, it is evident that three clones of V. cholerae O139 are currently prevailing in the country, with strains exhibiting three bands after HindIII digestion being dominant.
Phenotypically, the reemerged V. cholerae O139 displayed a difference compared to those that appeared in late 1992 and 1993 in that the current O139 strains are sensitive to co-trimoxazole. There was an apparent correlation between antibiogram and HindIII restriction patterns of the CTX genetic element in the O139 strains examined in this study. The dominant antibiogram of O139 strains displaying three bands after HindIII digestion and hybridization with a ctxA probe was AP-FZ-NM-SM or AP-FZ-SM, while the prominent antibiogram of O139 strains displaying a single 20- or 15-kb HindIII band was AP-FZ-NA-SM (Table 1). Careful scrutiny of data on multidrug resistance of V. cholerae over the past few years indicates that the recent rapid changes being witnessed may be a consequence of the rapid changes in drug resistance of V. cholerae (12). In fact, at the time of the genesis of the O139 serogroup, two principal features which distinguished O139 from O1 ElTor strains were the novel O139 serogroup antigen and the distinct pattern of antibiotic resistance (1, 17). While examining the spread of the novel clone of O1 ElTor, we recently observed that the emergence of a new clone of V. cholerae O1 was preceded by a change in the antibiogram (2).
Although there were differences in the antibiogram and CTX RFLP between the O139 strains isolated in 1992 and 1993 and in 1996 and 1997, the ribotypes displayed by these temporally spaced-out O139 strains were similar to the ribotypes of the strains isolated in 1992 and 1993. No new ribotype was detected in this study. In contrast, in Bangladesh, differences in ribotypes of O139 strains isolated between 1993 and 1996 have been reported (8, 16). Despite the geographical proximity of and the movement of population between Calcutta and Bangladesh, the epidemiology of cholera can be quite different in the two areas and may be dependent on, among other things, ecological conditions. An outstanding example was the reappearance of the classical biotype of V. cholerae in 1983 in Bangladesh (18). The classical biotype of V. cholerae O1, however, did not spread to Calcutta or to any other areas where cholera is endemic in India. Within Bangladesh itself, there appears to be a selective distribution of biotypes, with the classical biotype clustering in the southern region while the ElTor biotype prevails in the other regions (23).
The reemergence of V. cholerae O139 in Calcutta after a 32-month quiescent period reestablishes the O139 serogroup as an entity which is likely to play a crucial role in the temporal antigenic variations among the cholera-causing serogroups of V. cholerae. The periodic shift between the O1 and O139 serogroups is reminiscent of the shifts from the Ogawa to the Inaba serotypes periodically witnessed among O1 V. cholerae in earlier years, possibly mediated by the immune pressure in the population. For some inexplicable reason, the Inaba serotype of the O1 serogroup has disappeared, with the last Inaba dominance being recorded in 1989 in Calcutta (13). The complete disappearance of the Inaba serotype from Calcutta strangely coincides with the genesis of the O139 serogroup. Molecular studies have now shown that the O139 serogroup originated from an O1 biotype ElTor strain by acquisition of novel DNA which replaced the rfb genes encoding the O1 antigen (4, 6). Epidemiologically, it appears that the O139 serogroup has appeared as an alternate to the Inaba serotype of the O1 serogroup to aid the persistence and perpetuate the spread of cholera.
REFERENCES
- 1.Albert M J, Siddique A K, Islam M S, Faruque A S G, Ansaruzzaman M, Faruque S M, Sack R B. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. doi: 10.1016/0140-6736(93)90481-u. [DOI] [PubMed] [Google Scholar]
- 2.Bag, P. K., S. Maiti, C. Sharma, A. Ghosh, A. Basu, R. Mitra, S. K. Bhattacharya, S. Nakamura, S. Yamasaki, Y. Takeda, and G. B. Nair. Rapid spread of the new clone of Vibrio cholerae O1 biotype ElTor in cholera endemic areas in India. Epidemiol. Infect., in press. [DOI] [PMC free article] [PubMed]
- 3.Bhadra R K, Roychoudhury S, Banerjee R K, Kar S, Majumdar R, Sengupta S, Chatterjee S, Khetawat G, Das J. Cholera toxin (CTX) genetic element in Vibrio cholerae O139. Microbiology. 1995;141:1977–1983. doi: 10.1099/13500872-141-8-1977. [DOI] [PubMed] [Google Scholar]
- 4.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of Escherichia coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 6.Comstock L E, Johnson J A, Michalski J M, Morris J G, Kaper J B. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol Microbiol. 1996;19:815–828. doi: 10.1046/j.1365-2958.1996.407928.x. [DOI] [PubMed] [Google Scholar]
- 7.Faruque S M, Ahmed K M, Alim A R M A, Qadri F, Siddique A K, Albert M J. Emergence of a new clone of toxigenic Vibrio cholerae O1 biotype El Tor displacing V. cholerae O139 Bengal in Bangladesh. J Clin Microbiol. 1997;35:624–630. doi: 10.1128/jcm.35.3.624-630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque S M, Ahmed K M, Siddique A K, Zaman K, Alim A R M A, Albert M J. Molecular analysis of toxigenic Vibrio cholerae O139 Bengal strains isolated in Bangladesh between 1993 and 1996: evidence for emergence of a new clone of the Bengal vibrios. J Clin Microbiol. 1997;35:2299–2306. doi: 10.1128/jcm.35.9.2299-2306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaper J B, Jr, Morris J G, Nishibuchi M. DNA probes for pathogenic Vibrio species. In: Tenover F C, editor. DNA probes for infectious disease. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 65–77. [Google Scholar]
- 10.Mitra R, Basu A, Dutta D, Nair G B, Takeda Y. Resurgence of Vibrio cholerae O139 Bengal with altered antibiogram in Calcutta, India. Lancet. 1996;348:1181. doi: 10.1016/s0140-6736(05)65326-3. [DOI] [PubMed] [Google Scholar]
- 11.Mukhopadhyay A K, Garg S, Mitra R, Basu A, Rajendran K, Dutta D, Bhattacharya S K, Shimada T, Takeda T, Takeda Y, Nair G B. Temporal shifts in traits of Vibrio cholerae strains isolated from hospitalized patients in Calcutta: a 3-year (1993 to 1995) analysis. J Clin Microbiol. 1996;34:2537–2543. doi: 10.1128/jcm.34.10.2537-2543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukhopadhyay A K, Garg S, Nair G B, Kar S, Ghosh R K, Pajni S, Ghosh A, Shimada T, Takeda T, Takeda Y. Biotype traits and antibiotic susceptibility of Vibrio cholerae serogroup O1 before, during and after the emergence of the O139 serogroup. Epidemiol Infect. 1995;115:427–434. doi: 10.1017/s0950268800058581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair G B, Ramamurthy T, Garg S, Takeda T, Takeda Y. Characteristics of Vibrio cholerae isolated from hospitalized patients with acute diarrhoea in Calcutta, India: a four year analysis. LabMed Int. 1993;10:29–33. [Google Scholar]
- 14.Nair G B, Ramamurthy T, Bhattacharya S K, Mukhopadhyay A K, Garg S, Bhattacharya M K, Takeda T, Shimada T, Takeda Y, Deb B C. Spread of Vibrio cholerae O139 Bengal in India. J Infect Dis. 1994;169:1029–1034. doi: 10.1093/infdis/169.5.1029. [DOI] [PubMed] [Google Scholar]
- 15.Nair G B, Albert M J, Shimada T, Takeda Y. Vibrio cholerae O139 Bengal, the new serogroup causing cholera. Rev Med Microbiol. 1996;7:43–51. [Google Scholar]
- 16.Popovic T, Fields P I, Olsvik O, Wells J G, Evins G M, Cameron D N, Farmer III J J, Bopp C A, Wachsmuth K, Sack R B, Albert M J, Nair G B, Shimada T, Feeley J C. Molecular subtyping of toxigenic Vibrio cholerae O139 causing epidemic cholera in India and Bangladesh, 1992–93. J Infect Dis. 1995;171:122–127. doi: 10.1093/infdis/171.1.122. [DOI] [PubMed] [Google Scholar]
- 17.Ramamurthy T, Garg S, Sharma R, Bhattacharya S K, Nair G B, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. Emergence of a novel strain of V. cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 18.Samadi A R, Shahid N, Eusof A, Yunus M, Huq M I, Khan M U, Rahman A S M M, Faruque A S G. Classical Vibrio cholerae biotype displaces ElTor in Bangladesh. Lancet. 1983;i:805–807. doi: 10.1016/s0140-6736(83)91860-3. [DOI] [PubMed] [Google Scholar]
- 19.Sharma C, Nair G B, Mukhopadhyay A K, Bhattacharya S K, Ghosh R K, Ghosh A. Molecular characterization of V. cholerae O1 biotype El Tor strains isolated between 1992 and 1995 in Calcutta, India: evidence for the emergence of a new clone of the El Tor biotype. J Infect Dis. 1997;175:1134–1141. doi: 10.1086/516453. [DOI] [PubMed] [Google Scholar]
- 20.Sharma C, Maiti S, Mukhopadhyay A K, Basu A, Basu I, Nair G B, Mukhopadhyaya R, Das B, Kar S, Ghosh R K, Ghosh A. Unique organization of the CTX genetic element in Vibrio cholerae O139 strains which reemerged in Calcutta, India, in September 1996. J Clin Microbiol. 1997;35:3348–3350. doi: 10.1128/jcm.35.12.3348-3350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada, T. Unpublished data.
- 22.Shimada T, Arakawa E, Itoh K, Okitsu T, Matsushima A, Asai Y, Yamai S, Nakazato T, Nair G B, Albert M J, Takeda Y. Extended serotyping scheme for Vibrio cholerae. Curr Microbiol. 1994;28:175–178. [Google Scholar]
- 23.Siddique A K, Baqui A H, Eusof A, Haider K, Hossain M A, Bashier I, Zaman K. Survival of classic cholera in Bangladesh. Lancet. 1991;337:1125–1127. doi: 10.1016/0140-6736(91)92789-5. [DOI] [PubMed] [Google Scholar]
- 24.Waldor M K, Mekalanos J J. Emergence of a new cholera pandemic: molecular analysis of virulence determinants in Vibrio cholerae O139 and development of a live vaccine prototype. J Infect Dis. 1994;170:278–283. doi: 10.1093/infdis/170.2.278. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. Guidelines for cholera control. Geneva, Switzerland: World Health Organization; 1993. [Google Scholar]
- 26.Yamamoto T, Nair G B, Takeda Y. Emergence of tetracycline resistance due to multiple drug resistance plasmid in Vibrio cholerae O139. FEMS Immunol Med Microbiol. 1995;11:131–136. doi: 10.1111/j.1574-695X.1995.tb00099.x. [DOI] [PubMed] [Google Scholar]
- 27.Yamasaki S, Nair G B, Bhattacharya S K, Yamamoto S, Kurazono H, Takeda Y. Cryptic appearance of a new clone of Vibrio cholerae O1 biotype El Tor in Calcutta, India. Microbiol Immunol. 1997;41:1–6. doi: 10.1111/j.1348-0421.1997.tb01165.x. [DOI] [PubMed] [Google Scholar]