Abstract
Within a 1-year period, six surgical-site infections (SSI) caused by Staphylococcus schleiferi were observed in the department of cardiac surgery of Ignatius Hospital, Breda, The Netherlands. Since outbreaks caused by this species of coagulase-negative staphylococci have not been described before, an extensive environmental survey and a case control study were performed in combination with molecular typing of the causative microorganism in order to identify potential sources of infection. Variability, as detected by four different genotyping methods (random amplification of polymorphic DNA [RAPD], conventional and PCR-mediated ribotyping, and pulsed-field gel electrophoresis [PFGE] of DNA macro restriction fragments), appeared to be limited both among the clinical isolates and among several control strains obtained from various unrelated sources. Among unrelated strains, RAPD and PCR-mediated ribotyping identified two types only, whereas seven different types were identified in a relatively concordant manner by conventional ribotyping and PFGE. The latter two procedures proved to be the most useful tools for tracking the epidemiology of S. schleiferi. Four of the outbreak-related strains were identical by both methods, and two isolates showed limited differences. In the search for a potential source of S. schleiferi infection, two slightly different PFGE types were encountered on several occasions in the nose of a single surgeon. These strains were, however, clearly different from the outbreak type. In contrast, S. schleiferi cultures remained negative for two persons identified on the basis of case control analysis. It was demonstrated that SSI caused by S. schleiferi had a clinical impact for patients comparable to that of a wound infection caused by Staphylococcus aureus. This report describes the first well-documented outbreak of S. schleiferi infection. A source of the outbreak was not detected.
Staphylococcus schleiferi was recognized in the late 1980s as a new species of coagulase-negative staphylococci (CoNS) (5). Since then, this pathogen has been recovered from several kinds of infections in humans, e.g., brain empyema, surgical-site infections (SSI), intravascular device-related bacteremia, infections of implanted prosthetic material (including pacemakers [3]), and endocarditis (4, 13). Its involvement in urinary tract infections was considered to be proven in 0.7% of 404 infections caused by CoNS (18). The pathogenicity of S. schleiferi was confirmed in a model study of abscess formation in mice (14). S. schleiferi was shown to be more virulent than, for instance, Staphylococcus warneri or Staphylococcus hominis. Moreover, all S. schleiferi strains produce beta-hemolysin, lipase, and esterase as putative virulence factors.
Little is known of the epidemiology of S. schleiferi; for this reason, S. schleiferi strains from diverse sources have been studied by various genotyping methods in order to define the genetic diversity within the species. Plasmid typing appeared to be unsuccessful because extrachromosomal elements were present in only a small fraction of strains (8). DNA restriction analysis with five different restriction enzymes showed no divergence in a diverse group of 31 strains. Ribotyping appeared to be more adequate in detecting genetic polymorphisms among these isolates (8). In a preliminary pulsed-field gel electrophoresis (PFGE) trial, a single strain of S. schleiferi was included (20). The PFGE fingerprint obtained for this strain clearly separated it from isolates of other species of CoNS. A subsequent study, including five S. schleiferi strains, once again revealed the genetic homogeneity of the species: only minor variation was observed upon SmaI digestion of genomic DNA and separation of the macro restriction fragments, either by PFGE or by field inversion gel electrophoresis (15, 21, 30). However, despite the availability of technically adequate typing technology, the precise clinical epidemiology of S. schleiferi remained unknown, since major outbreaks of infection due to this species had not been described.
In our department of cardiac surgery (Ignatius Hospital, Breda, The Netherlands), six patients nursed within the department developed SSI with S. schleiferi in 1 year. All infections involved the sternotomy site. Since outbreaks of S. schleiferi infection have not been reported before, an investigation into the source of these infections was performed. This investigation involved environmental sampling, a case control study, and molecular typing of the outbreak-related and environmental strains.
MATERIALS AND METHODS
Setting.
In the cardiac surgery department of Ignatius Hospital in Breda, approximately 1,500 cardiac surgical procedures are performed each year. The department consists of an operating theater, a postoperative intensive care unit, and a general postoperative ward. There is an active infection control policy which includes continuous surveillance of postoperative sternal wound infections. Overall, the deep SSI rate was approximately 1% during the years 1991 to 1996, and approximately half of these infections were caused by Staphylococcus aureus.
Bacteriology.
Surgical sites are routinely monitored for signs of infection, and wound sampling with sterile cotton swabs is performed whenever an infection is suspected. Prior to sampling, the surface of the wound is cleaned with a disinfecting agent. Swabs are transported to the microbiology laboratory; for all different morphotypes of staphylococci growing in the resulting cultures, a slide agglutination test (Staphaurex Plus; Murex Diagnostics, Breukelen, The Netherlands) and a test for the presence of heat-stable thermonuclease are routinely performed. If these two tests are both positive the isolate is considered to be S. aureus, and if the tests are both negative the isolates are considered to be CoNS. If the tests are discordant, a tube coagulase test and a biochemical identification test with Api ID32 Staph (bioMerieux, Lyon, France) are performed. After the outbreak of S. schleiferi was recognized, this procedure was modified by adding the tube coagulase test to the routinely performed tests. Based on these procedures, S. schleiferi was identified and isolated from clinical samples of six patients from September 1995 to September 1996.
Environmental sampling.
From all surgeons, anesthetists, nurses of the operating theater and the postoperative wards, and technicians for extracorporeal circulation, nasal swabs were obtained for culture. From the surgeons, hands were sampled as well. After actively moving one hand in a sterile surgical glove containing sterile broth for one minute, the broth was used for culture. Both the swabs and the broths were inoculated on blood agar plates and incubated at 37°C for 48 h. The outbreak-related isolates of S. schleiferi showed characteristic beta-hemolysis after this period (10). Furthermore, during 10 surgical sessions, environmental samples were collected while a surgical procedure was being performed. Sampling methods included air settling plates and active air sampling.
Case control study.
Prospective surveillance for SSI has routinely been performed in the department of cardiothoracic surgery since 1991. Criteria for the presence of SSI are those of the Centers for Disease Control and Prevention (11). The six patients with SSI caused by S. schleiferi served as experimental subjects. Two control groups were selected. First, 24 patients were randomly selected from among patients who had had operations during the time period in which the six S. schleiferi SSI cases were identified. Second, all patients who had developed SSI with S. aureus in 1995 and 1996 were selected as a control group. From S. schleiferi SSI patients and controls, the following variables were recorded: patient identification, age, sex, height, weight, underlying diseases, immunosuppressive drugs, diabetes mellitus, smoking habits, New York Heart Association (NYHA) score (a score for the severity of cardiovascular disease, with a range in increasing severity of 1 to 4), date of admission, date of surgery, date of discharge, surgeons (first and assistant), anesthetist, operating room nurses (first assistant and second assistant), technician for extracorporeal circulation, perioperative antibiotic prophylaxis, duration of surgery, duration of extracorporeal circulation, operating room, volume of perioperative loss of blood, size of blood transfusion, emergency procedure, rethoracotomy, postoperative infections at other sites, and outcome (survival). The results were analyzed with Statistical Package for the Social Sciences software. S. schleiferi SSI patients were compared with controls, and crude odds ratios were determined. Statistical significance was determined with Fisher’s exact test for categorical variables or the t test for continuous variables. To find out if risk factors were independent, multiple logistic regression was performed. Statistical significance was accepted at P of <0.05.
Molecular typing.
The six outbreak-related isolates and a collection of S. schleiferi isolates from the environmental samples were included in the analysis (see Table 3 for a description of the strains). For comparison and validation, a reference collection consisting of 10 epidemiologically unrelated strains of diverse geographical origin was included in all studies. All typing procedures were performed and analyzed without knowledge of the origin of the isolates (blind).
TABLE 3.
Survey of origins and genotypes of S. schleiferi strains
| Strain | Origina | Isolation dateb | PCR ribotype | Compound RAPD codec | Conventional ribotype | PFGE type |
|---|---|---|---|---|---|---|
| Epidemiologically unrelated international isolates | ||||||
| 1 | U.K. | 1993 | A | A/A | A | A |
| 2 | U.K. | 1993 | A | A/A | B | B |
| 3 | U.S. | 1988 | A | A/A | C | C |
| 4 | France | 1989 | A | A/A | D | D |
| 5 | France | 1989 | A | A/A | E | E |
| 6 | New Zealand | 1989 | A | A/A | C | C |
| 7 | France | 1989 | A | A/A | F | F |
| 8 | France | 1988 | A | A/A | B | B |
| 9 | France | 1986 | A | A/A | A | A |
| 10 | France | 1984 | A | A/A | E | B |
| Strains from patients involved in outbreak | ||||||
| 12 | Pat. 5 | 8-96 | A | A/A | E | B |
| 13 | Pat. 3 | 11-95 | A | A/A | G | B |
| 14 | Pat. 2 | 10-95 | A | A/A | E | B |
| 15 | Pat. 1 | 10-95 | A | A/A | C | C |
| 16 | Pat. 4 | 8-96 | A | A/A | E | B |
| 17 | Pat. 6 | 9-96 | A | A/A | E | B |
| Strains from surgeon A | ||||||
| 11 | Nose | 10-96 | A | A/A | C | G |
| 19 | Shedding | 10-96 | A | A/A | C | G |
| 20 | Nose | 10-96 | A | A/A | C | H |
| 21 | Shedding | 10-96 | A | A/A | C | H |
| 22 | Shedding | 10-96 | A | A/A | C | G |
| 23 | Shedding | 10-96 | A | A/A | C | H |
| 24 | Nose | 10-96 | A | A/A | C | H |
| 25 | Shedding | 10-96 | A | A/A | C | H |
| Strains not involved in outbreak | ||||||
| 18 | 95-1-2cd | 1995 | A | A/A | A | C |
| 26 | Pat. nose | 1997 | B | B/B | E | I |
U.K., United Kingdom; U.S., United States; Pat., patient.
Dates for strains isolated from patients involved in the outbreak and from surgeon A are given as month-year.
The first letter in the compound RAPD code identifies the ERIC1-ERIC2 result; the second letter identifies the RAPD1-RAPD7 score.
Strain from a quality control study conducted by the Dutch Foundation for Quality Assessment of Medical Microbiological Laboratories (SKMM).
RAPD.
Random amplification of polymorphic DNA (RAPD) was performed essentially as described previously (26–28). Bacteria were treated with lysostaphin (35 mg/ml; Sigma, Zwijndrecht, The Netherlands) and subsequently lysed by the addition of guanidinium containing lysis buffer. DNA was purified by affinity chromatography onto Celite (0.2 g/ml; Jansen Pharmaceuticals, Beerse, Belgium) according to established protocols (1). The DNA concentration in the resulting eluates was determined by agarose gel electrophoresis (Hispanagar; Sphaero Q, Leiden, The Netherlands). Samples of the DNA preparations were coelectrophoresed with known amounts of lambda DNA. After staining, the amounts were estimated by comparison of fluorescence intensities upon UV transillumination of the gel. For RAPD, SuperTaq DNA polymerase (Sphaero Q) was used, and cycling was performed in BioMed (Theres, Germany) PCR machines (model 60). Primers used were ERIC1 and ERIC2 (32) or RAPD1 and RAPD7 (25) in single amplification reactions. Amplicons were analyzed on agarose gels, and the resulting fingerprints were photographed with a charge-coupled device coupled to a thermoprinter (Progress Control; Mitsubishi, Waalwijk, The Netherlands).
PCR ribotyping.
PCR ribotyping was performed according to an optimized protocol (16) based on prior publications (2, 9). The 16S-to-23S intergenic region was amplified with primers sp1 and sp2, and the amplicons were separated on agarose gels. The ribopatterns were visualized by UV transillumination after ethidium bromide staining.
Conventional ribotyping.
Ribotyping was performed as described previously (8). In short, DNA was isolated from staphylococcal cultures by standard procedures (19), and HindIII digests were prepared according to recommendations of the manufacturer of the restriction enzyme (Boehringer, Mannheim, Germany). DNA fragments were size separated by electrophoresis and blotted onto nylon membranes. Plasmid pKK3535, containing the rrnB ribosomal operon of Escherichia coli, was used as a probe (8). The probe was labeled with digoxigenin-11-dUTP by random priming, and chemiluminescence was generated with Lumigen PPD as the substrate (Boehringer). Ribotypes were finally visualized by exposure of X-ray films.
PFGE.
PFGE was carried out based on protocols previously described for DNA from S. aureus or other species of CoNS (17, 24, 29). A suspension of bacteria was mixed in a 1:1 ratio with 1% InCert agarose (FMC Bioproducts, Rockland, Maine). Agarose plugs were prepared with Bio-Rad (Veenendaal, The Netherlands) casting forms and incubated with lysostaphin (Sigma). Spheroplasts were lysed by incubating the plugs in buffer containing 1% sodium dodecyl sulfate and 1 mg of proteinase K (Boehringer) per ml. Plugs were washed six times for 30 min each in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA and stored at 4°C. DNA within half a plug was digested by SmaI (Boehringer), and PFGE was carried out in 1% SeaKem GTG agarose gels (FMC Bioproducts). The buffer consisted of 0.5× Tris-borate-EDTA. Electrophoresis was performed in a Bio-Rad CHEF Mapper. Running time was 22 h with linear ramping from 2.16 to 44.69 s at an angle of 120° (60°/−60°). The voltage was 6 V/cm, gel dimensions were 120 by 140 by 5 mm, and the temperature was set at 14°C. Gels were stained with ethidium bromide and photographed with instant Polaroid equipment. Differences in banding patterns were documented by at least two independent observers. Strains belonging to a single PFGE type should display electropherograms that differ in a maximum of three bands. If more differences in DNA restriction fragment migration are observed, this is considered to be a new type. This interpretation is according to general guidelines issued by an American working party (22, 23).
RESULTS
Patients and case control studies.
Between September 1995 and September 1996, six patients developed SSI with S. schleiferi (Table 1). This accounted for 40% of the number of SSI observed in this particular period, which strongly underscores the potential outbreak relatedness. The SSI rate in the year of the outbreak was 1.0%. This was due to 15 SSI, 12 of which were deep SSI. The deep SSI rate in this year was 0.8%. Eight SSI were caused by S. aureus, one by Staphylococcus epidermidis. All patients with an S. schleiferi SSI were males; their average age was 63 years. S. schleiferi SSI patients were comparable with both control groups with regard to all preoperative variables included in the case control study (Table 2). For the perioperative variables, a significant association was observed with the presence during surgery of an anesthetist (A) and an operating room nurse (A). Both anesthetist A and nurse A were repeatedly sampled for S. schleiferi carriage, but this species was never cultured. From one of the surgeons (A), S. schleiferi was isolated repeatedly. In Table 2 it is shown that, contrary to expectations, he was not significantly associated with the S. schleiferi SSI patients. To control for possible confounding, logistic regression analysis was performed. The presence of anesthetist A, operating room nurse A, or surgeon A and age, gender, NYHA score, and smoking habits were entered into the model. Anesthetist A (P = 0.041) and operating room nurse A (P = 0.018) were both identified as independent risk factors for development of SSI with S. schleiferi when noninfected patients were the control group. When patients with an S. aureus SSI were considered controls, only anesthetist A (P = 0.002) was identified as an independent risk factor. The pathogenicity of S. schleiferi can be deduced from Table 2. The median postoperative length of stay was significantly longer for patients with S. schleiferi (27 days) than for patients without SSI (11 days) and was comparable to that of patients suffering from S. aureus infection (26 days). The mean highest value of C-reactive protein was higher in the group of S. schleiferi-infected individuals than in the noninfected group; this difference approached statistical significance.
TABLE 1.
S. schleiferi SSI patient data included in the case control studya
| Patient no. | Date of surgery (mo/day/yr) | Age (yr) | Length of stay (days) | S | A | N | OR | Kind of SSI | Days before onset of SSI | PFGE type | Ribotype |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 9/27/95 | 57 | 29 | Yes | Yes | No | 4 | Deep | 11 | C | C |
| 2 | 10/03/95 | 62 | 22 | Yes | No | Yes | 3 | Deep | 45 | B | E |
| 3 | 10/27/95 | 64 | 22 | No | No | No | 2 | Deep | 11 | B | G |
| 4 | 7/15/96 | 55 | 29 | Yes | Yes | Yes | 1 | Superficial | 10 | B | E |
| 5 | 7/31/96 | 73 | 28 | No | Yes | Yes | 2 | Deep | 12 | B | E |
| 6 | 8/29/96 | 69 | 25 | No | Yes | No | 3 | Superficial | 8 | B | E |
All patients were male. Abbreviations: S, surgeon A; A, anesthetist A; N, nurse A; OR, operating room.
TABLE 2.
Case control dataa
| Variable | S. schleiferi-infected patients | Uninfected patients | P | S. aureus-infected patients | P |
|---|---|---|---|---|---|
| No. of patients | 6 | 24 | 15 | ||
| Preoperative variables | |||||
| No. (%) of males | 6 (100.0) | 20 (83.3) | 0.56 | 12 (80.0) | 0.53 |
| Mean age (SD) | 63.3 (6.9) | 60.5 (11.4) | 0.45 | 63.6 (12.3) | 0.96 |
| Mean NYHA score (SD) | 3.0 (1.1) | 2.8 (1.3) | 0.70 | 3.1 (0.9) | 0.89 |
| Mean body mass index (SD) | 24.8 (2.3) | 26.6 (3.1) | 0.15 | 26.5 (3.4) | 0.20 |
| No. of diabetes mellitus patients | 0 | 4 | 0.88 | 2 | 1.00 |
| No. of patients taking immunosuppressive drugs | 0 | 1 | 1.00 | 1 | 1.00 |
| Perioperative variables | |||||
| Mean duration of surgery (min) (SD) | 217.2 (52.5) | 219.9 (56.9) | 0.91 | 243 (98.9) | 0.44 |
| Mean duration of extracorporeal circulation (min) (SD) | 112.0 (28.2) | 113.1 (47.1) | 0.94 | 135.7 (68.1) | 0.29 |
| Mean amount of blood transfused (ml) (SD) | 66.7 (163.3) | 268.2 (464.4) | 0.10 | 270.1 (139.2) | 0.20 |
| Mean blood loss (ml) (SD) | 550.0 (63.2) | 569.1 (84.6) | 0.56 | 743.5 (513.0) | 0.17 |
| Mean no. of bypasses (SD) | 3.7 (1.5) | 2.7 (1.8) | 0.21 | 3.7 (1.6) | 1.00 |
| No. (%) of procedures with surgeon A | 3 (50.0) | 4 (16.6) | 0.12 | 2 (13.3) | 0.115 |
| No. (%) of procedures with anesthetist A | 4 (66.6) | 4 (16.6) | 0.029 | 1 (6.6) | 0.011 |
| No. (%) of procedures with OR nurse Ab | 3 (50.0) | 1 (4.2) | 0.018 | 1 (6.6) | 0.053 |
| Postoperative variables | |||||
| Mean highest value of C-reactive protein (SD) | 125.8 (61.2) | 68.1 (54.0) | 0.07 | 182.2 (122.8) | 0.18 |
| Median length of stay (days) | 27 | 11 | 0.003 | 26 | 0.98 |
P values represent comparisons between S. schleiferi-infected patients and uninfected patients and between S. schleiferi-infected patients and S. aureus-infected patients.
OR, operating room.
Molecular typing.
Molecular typing data are categorized into two groups, based on the observed and overlapping resolution of RAPD and PCR ribotyping versus Southern hybridization-based ribotyping and PFGE.
PCR ribotyping and RAPD.
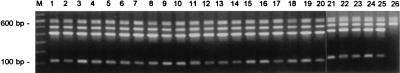
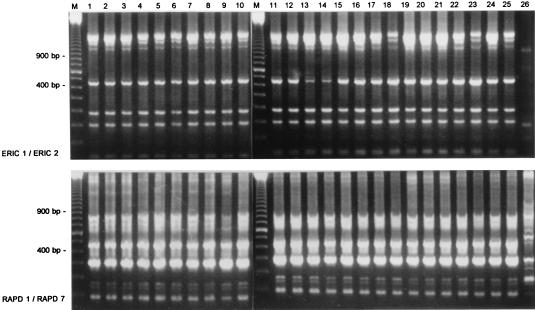
PCR ribotyping and RAPD experimental data are depicted in Fig. 1 and 2. PCR ribotyping reveals homogeneity among the strains. Only strain 26, encountered as a noninvasive colonizer in the nose of a surgical patient, differed from the other strains, in the sense that the two smaller amplicons were lacking. RAPD analysis employing two combinations of primers (ERIC1-ERIC2 and RAPD1-RAPD7) in two separate assays was performed. The strain grouping deduced from the PCR ribotyping data was confirmed by the RAPD tests. The only deviating fingerprint, again, was acquired for strain 26. Furthermore, all PCR fingerprints are identical, except for some minor differences in band staining intensity.
FIG. 1.
PCR-mediated ribotyping of S. schleiferi strains collected during the present study. Strain numbers are indicated at the top of the lanes and correspond with those given in Table 3. Note that only for strain 26 is an aberrant DNA banding pattern observed. On the left (lane M), a 100-bp length marker is displayed; fragments with sizes of 600 and 100 bp are highlighted.
FIG. 2.
RAPD analysis of S. schleiferi strains collected during the present study. The top panel shows results obtained by the combined application of primers ERIC1 and ERIC2; the fingerprints in the bottom panel were generated with the RAPD1-RAPD7 combination. On the left and between lanes 10 and 11 (lanes M), a 100-bp length marker is displayed; fragments with sizes of 900 and 400 bp are highlighted.
PFGE and ribotyping.
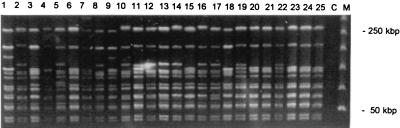
The experimental results of PFGE analysis are depicted in Fig. 3. The numbers of DNA macro restriction fragments vary from 13 to 15; of these, 12 are universally present. If these results were interpreted on the basis of guidelines issued previously (22, 23), all isolates would have been considered clonally related. However, if single band differences are taken into consideration as well, different types can be distinguished. Table 3 surveys all of the typing data, including those based on single band differences in the PFGE electropherograms. It is comforting to note that this classification is largely corroborated by the conventional ribotyping data (Table 3). A schematic representation of the seven different ribotyping patterns is shown in Fig. 4.
FIG. 3.
PFGE data obtained for S. schleiferi strains collected during the present study. On the right an array of bacteriophage lambda DNA concatemers is shown; the sizes of two of the fragments are indicated; fragments differ in length by units of 50 kbp. Note that the result obtained for strain 26 is not depicted; this strain was analyzed on a separate gel, but the fingerprint strongly resembled the basic patterns shown here.
FIG. 4.
Schematic representation of the different ribotyping patterns (ribo) obtained from strains included in the present study. The designations listed on the left correspond to the ribotype patterns given in Tables 1 and 3. Patterns H through L are patterns which have been observed in other S. schleiferi strains. At the top, a 100-bp length marker is indicated.
DISCUSSION
This report describes the first documented outbreak of S. schleiferi infection. In the laboratory, S. schleiferi is difficult to differentiate from S. aureus: the species are morphologically similar, and S. schleiferi subsp. schleiferi produces both clumping factor and heat-stable thermonuclease. For these and maybe other reasons, this pathogen may be underreported. If only slide coagulase testing and assessment of thermostable nuclease were performed diagnostically, the present outbreak would have been considered to be due to S. aureus. However, a negative result in the tube coagulase test performed by one of the technicians revealed that the isolate was S. schleiferi, which was confirmed by biochemical analysis with API ID32 Staph. Subsequent reexamination of all of the clinical S. aureus strains from the preceding year (1995) revealed two more patients infected with S. schleiferi (patients 1 and 2). This illustrates how a potentially misidentified bacterial species can turn into an “emerging” pathogen. According to standard microbiology laboratory procedures, performance of a slide agglutination test for clumping factor combined with a test for thermonuclease is sufficient to discriminate S. aureus from CoNS. However, S. schleiferi subsp. schleiferi would also be considered S. aureus by this approach. Therefore, the magnitude of S. schleiferi infection in clinical practice may be underestimated. This observation underscores the usefulness of including a tube coagulase test in routine procedures to differentiate S. aureus from other staphylococcal species.
A variety of accurate procedures for correct identification of clinical strains of CoNS have been described lately, although potential experimental pitfalls have been demonstrated for “difficult” isolates (31). Gas-liquid chromatography of fatty acids, though not yet within reach of the routine diagnostic laboratory, appears to be reliable and robust (21). Automated Microscan identification (Pos ID and Rapid Pos ID panels) of species was shown to be less reliable for the infrequently occurring species of CoNS (7). The procedure we used during the present study for definitive identification of S. schleiferi (API ID32 Staph) was demonstrated to be an accurate means of identification. In a multicenter study, it led to 100% efficient characterization of this species (12). This reliability was underscored in the present study: all S. schleiferi strains included shared several genotypic characteristics as well, most clearly demonstrated by the homogeneity of the RAPD and PCR ribotyping data. This implies that the current collection of strains (n = 26) could serve the purpose of validating novel species-specific identification assays, such as heat shock protein-based species identification tests (6).
It was shown previously that PFGE performed on DNA isolated from strains of S. schleiferi detected limited genetic heterogeneity among the strains (30). In the present study, however, PFGE together with conventional ribotyping turned out to be the typing combination of choice for analysis of S. schleiferi strains. PCR ribotyping was inadequate, as was RAPD employing the current combinations of random or ERIC primers. The latter two procedures have been applied successfully for other species, which indicates that S. schleiferi may be genetically a rather homogeneous species. This putative clonality is further corroborated by strikingly similar PFGE fingerprints (Fig. 3). Other studies have tried to optimize typing methods (among them PFGE) for S. schleiferi without satisfactory results (8, 15, 20, 30). The conclusion is that the results to date all point toward S. schleiferi being a rather homogeneous species genetically. Further studies should reveal whether alternative approaches yield better results. The question of whether minor differences are relevant can be answered only after more profound investigation into this relatively rare species. In view of the higher heterogeneity of unrelated strains by PFGE (six different patterns among 10 strains) than of outbreak-related strains (two patterns among 6 strains), we consider the outbreak-related strains with identical types to be clonally related.
Four of the six S. schleiferi isolates encountered in the cardiac surgery patients were identical with respect to the parameters examined. Five of the six isolates had the same PFGE pattern. This provided proof of an ongoing outbreak, and, theoretically, a source should have been present during the outbreak period. However, neither microbiological screening nor detailed case control studies revealed a potential reservoir for the outbreak-related genotype. In conclusion, it can be stated that S. schleiferi can cause significant infections in a clinically persistent fashion: in our case a single type was encountered regularly within a period of nearly a year. Unfortunately, the source of infection remained enigmatic. An apparent source, one of the surgeons who persistently carried S. schleiferi in the nose, was ruled out by typing of the strain and by the outcome of the case control study: the nasal inhabitant differed from the outbreak strain. On the other hand, the individuals implicated as possible sources of infection by the case control analysis (anesthetist A and operating room nurse A) did not carry S. schleiferi despite repeated culture assays. Consequently, causal involvement could not be proven. This may have been a consequence of the insensitivity of bacteriological detection of S. schleiferi, in which experience is limited. Experience is limited not only with bacteriological techniques but also with the epidemiology of S. schleiferi. The ecological niches of this microorganism are at present unknown. Also, it is not known if carriage is persistent or transient. The observation of this outbreak strongly suggests that there has been a persistent source in the department, either human or in the inanimate environment. Unfortunately, the source of the outbreak was not identified.
This study shows the outbreak potential of S. schleiferi. Environmental sources may be of crucial importance in the acquisition of S. schleiferi infections; future studies should reveal whether this is an exception or the rule.
REFERENCES
- 1.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cartwright C P, Stock F, Beekmann S E, Williams E C, Gill V J. PCR amplification of rRNA intergenic spacer regions as a method for epidemiologic typing of Clostridium difficile. J Clin Microbiol. 1995;33:184–187. doi: 10.1128/jcm.33.1.184-187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celard M, Vandenesch F, Darbas H, Grando J, Jean-Pierre J, Kirkorian G, Etienne J. Pacemaker infection caused by Staphylococcus schleiferi, a member of the human preaxillary flora: four case reports. Clin Infect Dis. 1997;24:1014–1015. doi: 10.1093/clinids/24.5.1014. [DOI] [PubMed] [Google Scholar]
- 4.Fleurette J, Bes M, Brun Y, Freney J, Forey F, Coulet M, Reverdy M E, Etienne J. Clinical isolates of Staphylococcus lugdunensis and Staphylococcus schleiferi: bacteriological characteristics and susceptibility to antimicrobial agents. Res Microbiol. 1989;140:107–118. doi: 10.1016/0923-2508(89)90044-2. [DOI] [PubMed] [Google Scholar]
- 5.Freney J, Brun Y, Bes M, Meugnier H, Grimont F, Grimont P A D, Nervi C, Fleurette J. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int J Syst Bacteriol. 1988;38:168–172. [Google Scholar]
- 6.Goh S H, Potter S, Wood J O, Hemmingsen S M, Reynolds R P, Chow A W. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J Clin Microbiol. 1996;34:818–823. doi: 10.1128/jcm.34.4.818-823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant C E, Sewell D L, Pfaller M A, Bumgardner R V, Williams J A. Evaluation of two commercial systems for identification of coagulase negative staphylococci to species level. Diagn Microbiol Infect Dis. 1994;18:1–5. doi: 10.1016/0732-8893(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 8.Grattard F, Etienne J, Pozzetto B, Tardy F, Gaudin O G, Fleurette J. Characterization of unrelated strains of Staphylococcus schleiferi by using ribosomal DNA fingerprinting, DNA restriction patterns, and plasmid profiles. J Clin Microbiol. 1993;31:812–818. doi: 10.1128/jcm.31.4.812-818.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurtler V. Typing of Clostridium difficile strains by PCR amplification of variable length 16S-23S rDNA spacer regions. J Gen Microbiol. 1993;139:3089–3097. doi: 10.1099/00221287-139-12-3089. [DOI] [PubMed] [Google Scholar]
- 10.Hebert G A. Hemolysins and other characteristics that help differentiate and biotype Staphylococcus lugdunensis and Staphylococcus schleiferi. J Clin Microbiol. 1990;28:2425–2431. doi: 10.1128/jcm.28.11.2425-2431.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horan T C, Gaynes R P, Martone W J, Jarvis W R, Emori T G. CDC definitions of nosocomial surgical site infections, 1992: a modification of CDC definitions of surgical wound infections. Am J Infect Control. 1992;20:271–274. doi: 10.1016/s0196-6553(05)80201-9. [DOI] [PubMed] [Google Scholar]
- 12.Ieven M, Verhoeven J, Pattyn S R, Goossens H. Rapid and economical method for species identification of clinically significant coagulase-negative staphylococci. J Clin Microbiol. 1995;33:1060–1063. doi: 10.1128/jcm.33.5.1060-1063.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jean-Pierre H, Darbas H, Jean-Roussenq A, Boyer G. Pathogenicity in two cases of Staphylococcus schleiferi, a recently described species. J Clin Microbiol. 1989;27:2110–2111. doi: 10.1128/jcm.27.9.2110-2111.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambe D W, Ferguson K P, Keplinger J L, Gemmell C G, Kalbfleisch J H. Pathogenicity of Staphylococcus lugdunensis, S. schleiferi and three other coagulase negative staphylococci in a mouse model and possible virulence factors. Can J Microbiol. 1990;36:455–463. doi: 10.1139/m90-080. [DOI] [PubMed] [Google Scholar]
- 15.Lina B, Vandenesch F, Etienne J, Kreiswirth B, Fleurette J. Comparison of coagulase negative staphylococci by pulsed field gel electrophoresis. FEMS Microbiol Lett. 1992;71:133–138. doi: 10.1016/0378-1097(92)90501-e. [DOI] [PubMed] [Google Scholar]
- 16.Martirosian G, Kuipers S, Verbrugh H, van Belkum A, Meisel-Mikolajczyk F. PCR ribotyping and arbitrarily primed PCR for typing strains of Clostridium difficile from a Polish maternity hospital. J Clin Microbiol. 1995;33:2016–2021. doi: 10.1128/jcm.33.8.2016-2021.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nur Y A, VandenBregh M F Q, Yusuf M A, van Belkum A, Verbrugh H A. Nasal carriage of multiresistant Staphylococcus aureus among health care workers and pediatric patients in two hospitals in Mogadishu, Somalia. Int J Infect Dis. 1997;1:186–191. [Google Scholar]
- 18.Ozturkeri H, Kocabeyoglu O, Yergok Y Z, Kosan E, Yenen O S, Keskin K. Distribution of coagulase negative staphylococci, including the newly described species Staphylococcus schleiferi, in nosocomial and community acquired urinary tract infections. Eur J Clin Microbiol Infect Dis. 1994;13:1076–1079. doi: 10.1007/BF02111833. [DOI] [PubMed] [Google Scholar]
- 19.Renaud F, Etienne J, Bertrand A, Brun Y, Greenland T B, Freney J, Fleurette J. Molecular epidemiology of Staphylococcus haemolyticus strains isolated in an Albanian hospital. J Clin Microbiol. 1991;29:1493–1497. doi: 10.1128/jcm.29.7.1493-1497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snopkova S, Gotz F, Doskar J, Rosypal S. Pulsed field gel electrophoresis of the genomic restriction fragments of coagulase negative staphylococci. FEMS Microbiol Lett. 1994;124:131–139. doi: 10.1016/0378-1097(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 21.Stoakes L, John M A, Lannigan R, Schieven B C, Ramos M, Harley D, Hussain Z. Gas-liquid chromatography of cellular fatty acids for identification of staphylococci. J Clin Microbiol. 1994;32:1908–1910. doi: 10.1128/jcm.32.8.1908-1910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover F C, Arbeit R D, Goering R V The Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect Control Hosp Epidemiol. 1997;18:426–439. doi: 10.1086/647644. [DOI] [PubMed] [Google Scholar]
- 23.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trzcinski K, van Leeuwen W, van Belkum A, Grzewiowski P, Kluijtmans J, Sijmons M, Verbrugh H, Witte W, Hryniewicz W. Two clones of methicillin resistant Staphylococcus aureus in Poland. Clin Microbiol Infect. 1997;3:198–207. doi: 10.1111/j.1469-0691.1997.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 25.van Belkum A, Kluytmans J, van Leeuwen W, Bax R, Quint W, Peters E, Fluit A, Vandenbroucke-Grauls C, van den Brule A, Koeleman H, Melchers W, Meis J, Elaichouni A, Vaneechoutte M, Moonens F, Maes N, Struelens M, Tenover F, Verbrugh H. Multicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J Clin Microbiol. 1995;33:1537–1547. doi: 10.1128/jcm.33.6.1537-1547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Belkum A, Bax R, Prevost G. Comparison of four genotyping assays for epidemiological studies of methicillin resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1994;13:420–424. doi: 10.1007/BF01972002. [DOI] [PubMed] [Google Scholar]
- 27.van Belkum A, Bax R, Peerbooms P, Goessens W, van Leeuwen N, Quint W G V. Comparison of phage typing and DNA fingerprinting for discrimination of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:798–803. doi: 10.1128/jcm.31.4.798-803.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Belkum, A., R. Bax, P. J. C. van der Straaten, W. G. V. Quint, and E. Veringa. PCR fingerprinting for epidemiological study of Staphylococcus aureus. J. Microbiol. Methods 20:235–247.
- 29.van Belkum A, van Leeuwen W, Verkooyen R, Can Sacilik S, Cokmus C, Verbrugh H. Dissemination of a single clone of methicillin-resistant Staphylococcus aureus among Turkish hospitals. J Clin Microbiol. 1997;35:978–981. doi: 10.1128/jcm.35.4.978-981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandenesch F, Lina B, Lebeau C, Greenland T B, Etienne J. Epidemiological markers of coagulase negative staphylococci. Intensive Care Med. 1993;19:311–315. doi: 10.1007/BF01694703. [DOI] [PubMed] [Google Scholar]
- 31.Vandenesch F, Lebeau C, Bes M, Lina G, Lina B, Greenland T, Benito Y, Brun Y, Fleurette J, Etienne J. Clotting activity in Staphylococcus schleiferi subspecies from human patients. J Clin Microbiol. 1994;32:388–392. doi: 10.1128/jcm.32.2.388-392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]