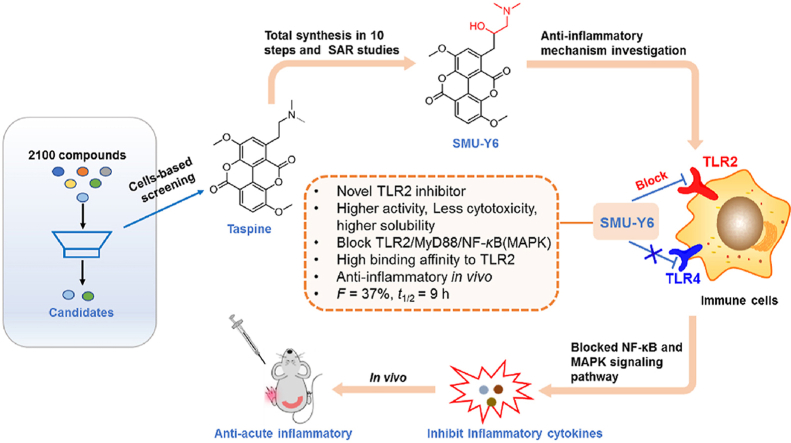
Abstract
Toll-like receptor 2 (TLR2) mediated macrophages regulate the protective immune response to infectious microorganisms, but the aberrant activation of macrophages often leads to pathological inflammation, including tissue damage. In this study, we identified antagonists of TLR2 by screening 2100 natural products and subsequently identified Taspine, an aporphine alkaloid, as an excellent candidate. Furthermore, analysis of the 10 steps chemical synthesis route and structural optimization yielded the Taspine derivative SMU-Y6, which has higher activity, better solubility, and improved drug-feasible property. Mechanistic studies and seq-RNA analysis revealed that SMU-Y6 inhibited TLR2 over other TLRs, hindered the formation of TLR2/MyD88 complex, and blocked the downstream NF-κB and MAPK signaling pathway, thus suppressing the release of inflammatory cytokines. SMU-Y6 could stabilize TLR2 and bind to TLR2 protein with a Kd of 0.18 μmol/L. Additionally, SMU-Y6 could efficiently reverse the M1 phenotype macrophage polarization, reduce the production of cytokines as well as infiltration of neutrophiles and alleviate the local inflammation in mice with acute paw edema and colitis. Collectively, we reported the first aporphine alkaloid derivative that selectively inhibits TLR2 with high binding affinity and superior drug-feasible property, thus providing an urgently-needed molecular probe and potential drug candidate for inflammatory and autoimmune disease therapy.
Key words: Taspine derivative; TLR2 inhibitor; MyD88, NF-κB signaling pathway; Macrophage polarization; Anti-acute inflammatory
Graphical abstract
A novel TLR2 inhibitor, SMU-Y6, inhibits the TLR2/MyD88/NF-κB and MAPK signaling pathways and reduces the inflammatory cytokines release, macrophage polarization, as well as neutrophil infiltration, exhibiting excellent anti-inflammatory effects in vivo.
1. Introduction
The human immune system is made up of innate and adaptive immune. Compared with innate immune, the adaptive immune produces specific antibodies or effector T lymphocytes to respond to foreign antigens as well as immune memory cells that prepare for any future infection1. Before the production of the former, most B or T lymphocytes need to receive co-stimulated signaling from antigen-presenting cells (APCs), even though some antigens are recognizable on their own2,3. In addition, these APCs are able to recognize and eliminate “perceived threats” by either phagocytosis or via secreting immune factors; thus, playing a key role in the innate immune response. Theoretically, APCs contribute to both innate and adaptive immunity through pattern recognition receptors (PRRs)2,4. PRRs are able to recognize a series of targets, which share similar characteristics, which can mainly be classified as pathogen-associated molecular patterns (PAMPs) or endogenous danger-associated molecular patterns (DAMPs)5,6. Thus, given their importance to the immune response, PPRs have been increasingly popular as a subject of investigation, with one of the most well-studied subgroups of this receptor family being the TLRs.
TLRs were first discovered in Drosophila melanogaster and were the first mammalian PRR to be cloned7. To date, 13 subtypes have been identified. Of these, TLR4 was the first to be identified as a receptor that recognized lipopolysaccharide and bacterial wall-related components from gram-negative bacteria, whereas TLR2 was shown to recognize lipopeptides or bacterial wall-related components from gram-positive bacteria8. Both TLR2 and TLR4 are extracellular and work together to form a line of defense against invading pathogens. Once TLR2 binds to its ligand, it recruits myeloid differentiation factor 88 (MyD88) to activate specific protein kinases (such as mitogen-activated protein kinase (MAPK) and inhibitory subunit of NF-κB (IκB) kinase) and activates certain transcription factors, such as NF-κB, which all lead to the production of inflammatory cytokines9. TLR2 agonist became a potential target for vaccine-adjuvant and tumor immunity because it binds with TLR2 and activates APCs, which could enhance the ability of up-taking and processing antigens of APCs and promote the subsequent activation of cytotoxic T lymphocytes and B cells against “threats” and tumor cells10,11. Our group has previously reported several novel TLR1/2 specific agonists, capable of activating cytotoxic T lymphocytes that target tumor cells, which are considered to have potential applications in cancer immunotherapy9,11,12 and TLR3 specific antagonists that reduce the uptake of oxidized low-density lipoproteins (oxLDLs) by macrophages, decreasing the formation of foam cells13. Unfortunately, the over-activation of TLR2 is shown to be associated with the development of inflammation. This conclusion has been supported by several studies that have revealed that the over-activation of TLR2 was found in inflammatory diseases and that the blockade of TLR2 slows down the progression of diseases, such as arthritis14, Alzheimer's disease15, and hepatitis16. Clanchy et al.17 found that TLR2 was over-expressed in human RA synovium isolated from RA patients. Moreover, the RA patients who had no effect in anti-TNF treatment were detected to perform more TLR2 expression than that anti-TNF treatment worked17. Some studies revealed that the expression of IL-10 was significantly reduced after TLR2 activation in systemic lupus erythematosus (SLE) patients18. IL-10 is a common immunonegative regulatory cytokine, which antagonizes the effect of pro-inflammatory cytokines. Rangasam et al.15 found that a high level of TLR2 and MyD88 was found in AD patients’ prefrontal cortex, compared to no cognitive impairment. Those suggest TLR2 inhibitors are likely to be potent anti-inflammatory agents. To date, some progress has been made in the development of small-molecule TLR2 inhibitors; however, none of these studies have progressed to an approved drug. These identified TLR2 inhibitors tend to have a moderate activity or unstable structures (based on Schiff-base and polyphenol configurations―see Supporting Information Fig. S1)19, 20, 21, 22, 23, 24, 25, 26. Given that these characteristics make existing TLR2 inhibitors unsuitable for clinical application, we have devoted our research to discovering novel small molecule inhibitors of TLR2 that will lend themselves to being used in a therapeutic capacity.
Natural products are an important source of structurally novel and pharmacologically active compounds; libraries of which can be screened during the early stages of the drug discovery process. The success rate from using such libraries is relatively high, with 8/40 and 5/36 of chemical drugs approved by US Food and Drug Administration in 2020 and 2021, respectively, being derived from natural products27,28. Given the significance of natural products as a reservoir of drug-feasible bioactive compounds, we commenced this study by screening a library of 2100 natural compounds. From these, we identified an aporphine alkaloid, Taspine, that successfully inhibited TLR2 activation in HEK-Blue hTLR2 cells. Previous studies have already reported that Taspine and its derivatives have: anti-tumor activity29,30, anti-neuropathic pain properties31, and can act as a fluorescent probe32. However, the potential anti-inflammation targets for Taspine and its derivatives are still poorly understood. Moreover, those reported derivatives were limited, mainly focusing on either removing the side-chain amino group29,32 and/or opening the lactone ring30,32 to simplify synthesis. However, we found that the lactone ring was the key to Taspine's anti-TLR2 activity. Nevertheless, retaining the lactone ring, whilst modifying its side-chain amino group proved challenging, as the synthesis of any such compound was complex. Thus, in order to improve the anti-TLR2 activity and drug feasibility, but reduce the toxicity of Taspine, we modified its chemical structure. In this study, we kept the lactone ring and mainly focused on the modification of its side-chain amino group, including (I) replacing dimethylamine with different cyclic aliphatic amines, (II) introducing hydrophilic hydroxyl group and lengthening the carbon chain (III), replacing the amine group with various amides (Supporting Information Fig. S1). The preliminary structure–activity relationship (SAR) studies highlighted SMU-Y6 as being the most promising target candidate. Following, we identified SMU-Y6 targeted TLR2/MyD88/NF-κB and MAPK signaling pathway to regulate inflammation. Protein and molecular binding experiments revealed that SMU-Y6 selectively targeted TLR2 but not TLR4. Finally, SMU-Y6 exhibited superior anti-acute inflammation properties, high bioavailability and long-term anti-inflammation protection in vivo. All these suggest that TLR2 inhibitor is a promising anti-inflammation agent in clinical investigation and we hope to provide a feasible method in inflammation therapy.
2. Results and discussion
2.1. Screening of potential anti-inflammatory agent and chemical synthesis
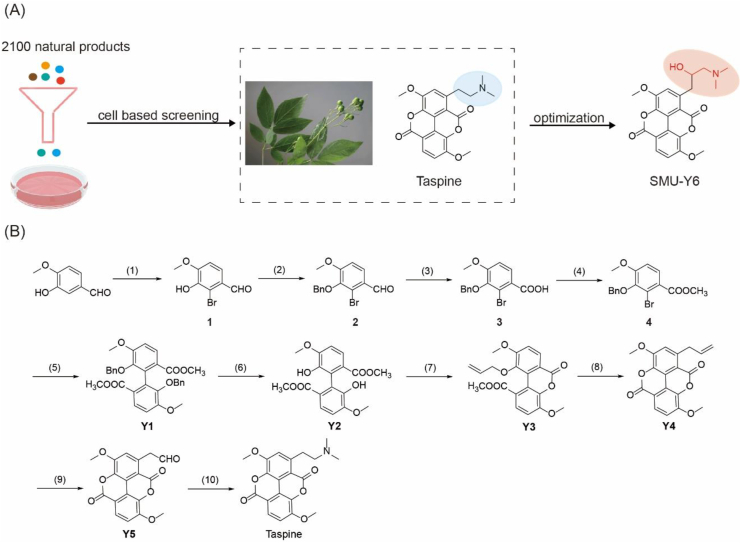
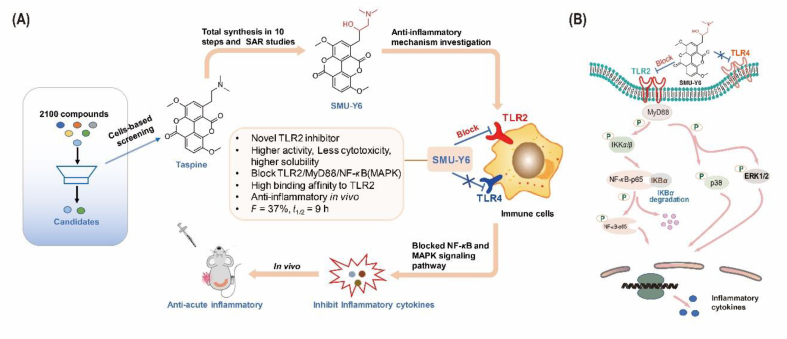
By screening 2100 nature compounds (Target Mol and NCI Nature product library), we determined that the aporphine alkaloid, Taspine (NSC76022), showed the best TLR2 inhibitory activity in HEK-Blue hTLR2 cells (Fig. 1A, Supporting Information Table S1). Taspine is a natural product found in several plants including Caulophyllum, which belongs to the Berberidaceae family. Caulophyllum is used in traditional Chinese medicine to treat injury following a fall and rheumatism, suggesting that Taspine may have anti-inflammatory properties. Although Taspine shows significant anti-TLR2 activity, it also exhibits a certain level of toxicity and poor water solubility, which limits its use as a therapeutic drug. Therefore, we made a series of structurally modified derivatives with the aim of identifying a superior candidate drug. Despite being a natural product, it is important to be able to synthesize this compound for the sake of subsequent structure modification. Therefore, we looked to optimize the synthesis methodologies that had already been reported31 and were able to completely synthesize Taspine in 10 steps, including oxidation, reduction and coupling reaction (Fig. 1B).
Figure 1.
Screening and synthesis of potential TLR2 inhibitors. (A) Cell-based screening of 2100 natural products and structure optimization of Taspine. (B) Total synthesis route of Taspine and derivatives. (1) Br2, Fe, CH3COONa, CH3COOH, rt, 3 h, 70%; (2) BnBr, K2CO3, CH3CH2OH, rt, 4 h, 95%; (3) NaClO2, H2O2, NaH2PO4, THF, H2O, rt, 4 h, 90%; (4) CH3COCl, CH3OH, rf, 4 h, 80%; (5) Cu, anhyd DMF, 155 °C, 4 h, 70%; (6) Pd/C, H2, THF, rt, 95%; (7) allyl bromide, K2CO3, DMF, 60 °C, 4 h, 43%; (8) 215 °C, 1 h, 70%; (9) OsO4, NaIO4, NMO, CH2Cl2, t-BuOH, H2O, rt, 20 h, 40%; (10) dimethylamine, NaBH3CN, CH2Cl2, 4 h, rt, 45%.
2.2. Structure–activity relationship (SAR) studies
To effectively design the Taspine derivatives, it was necessary to identify the key structural element(s) responsible for anti-TLR2 activity. Thus, we tested the anti-TLR2 activity of each intermediate involved in the synthesis of Taspine. The results indicated that the lactone ring and amino side chain were essential to Taspines’ activity (Supporting Information Table S2). When creating the various derivatives, we kept the basic skeleton of Taspine, including the lactone ring, but looked to modify the amino side chain. Firstly, we experimented with replacing the dimethylamine with different aliphatic amines to investigate whether steric hindrance plays a role in anti-TLR2 activity (Supporting Information Scheme S1). Limited by commercial availability, we obtained some derivative compounds Y6–Y10 (Supporting Information Table S2). We concluded that cyclic aliphatic amines maintain the anti-TLR2 activity, but those with six-membered rings had the best activity of this group. These results suggest that the relevant protein binding site of the side chain amino group had good spatial tolerance.
Given that Taspine only has minor hydrophilic groups, we hoped to improve the solubility of the derivatives by extending the carbon chain (which connects the main, planar structure with the R group) and introducing hydrophilic hydroxyl groups to these additional carbons. Our rationale was the hope that the introduction of extra hydrophilic groups would improve the lipid-water partition coefficient, thus improving drug feasibility. Consequently, we epoxidized the olefin of compound Y4 and opened it under alkaline conditions, resulting in the compounds Y11–Y15 (Scheme S1, Table S2). Our results showed that lengthening the carbon chain and introducing hydroxyl groups did not affect anti-TLR2 activity, giving us more latitude to make modifications to improve the lipid‒water partition coefficient through the addition of a hydrophilic hydroxyl group.
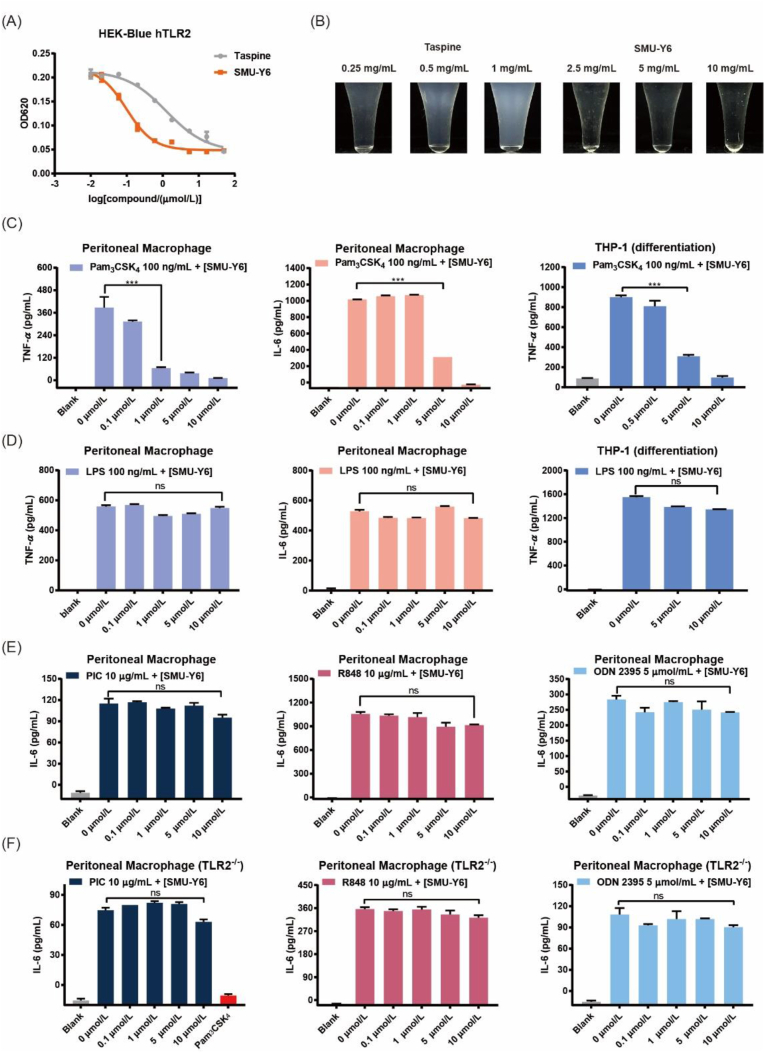
Because hydroxyl and carbonyl groups are isosteres, we designed and synthesized amide derivatives Y16–Y22 (Scheme S1). As shown in Table S2, amide compounds exhibited little anti-TLR2 activity, so no further synthesis of this type of derivative was carried out and the SAR was discontinued. We concluded that the side chain required a certain level of alkalinity for the derivative to have anti-TLR2 activity. Based on SAR studies, we found that out of a panel of tested compounds, SMU-Y6 exhibited the best inhibition of TLR2 (IC50 = 0.11 ± 0.04 μmol/L), which represented a 10-fold improvement compared with Taspine. This optimized derivative also exhibited reduced toxicity and improved water solubility that was 20 times greater than Taspine (Fig. 2A and B, Supporting Information Fig. S2). In short, we discovered that the lactone ring and the side chain of amino group were essential to Taspine's anti-TLR2 activity and that extending the carbon side chain and introducing hydrophilic groups yielded an optimized compound (SMU-Y6) that had enhanced anti-TLR2 activity, improved water solubility and reduced toxicity.
Figure 2.
Comparison between Taspine and SMU-Y6. (A) SEAP signaling of Taspine and SMU-Y6 in HEK-Blue hTLR2 cells. HEK-Blue hTLR2 cells were treated with Pam3CSK4 100 ng/mL and Taspine or SMU-Y6 for 24 h. The supernatant was collected for SEAP signaling. (B) Comparison of water solubility between Taspine and SMU-Y6. (C) TNF-α or IL-6 in supernatants of primary murine peritoneal macrophage cells after treatment with indicated Pam3CSK4 (100 ng/mL) and differing concentrations of SMU-Y6 for 24 h. TNF-α in supernatants of THP-1 cells after treatment with Pam3CSK4 (100 ng/mL) and differing concentrations of SMU-Y6 for 24 h. (D) TNF-α or IL-6 in supernatants of primary murine peritoneal macrophage cells after treatment with indicated LPS (100 ng/mL) and differing concentrations of SMU-Y6 for 24 h. TNF-α in supernatants of THP-1 cells after treatment with indicated LPS (100 ng/mL) and differing concentrations of SMU-Y6 for 24 h. (E) IL-6 in supernatants of primary murine peritoneal macrophage cells after treatment with indicated Poly I:C (10 μg/mL), R848 (10 μg/mL), ODN 2395 (5 μmol/L) and differing concentrations of SMU-Y6 for 24 h. (F) IL-6 in supernatants of primary murine peritoneal macrophage cells (TLR2−/−) after treatment with indicated Poly I:C (10 μg/mL), R848 (10 μg/mL), ODN 2395 (5 μmol/mL) and differing concentrations of SMU-Y6 for 24 h. Data presented is mean ± SD and the figures shown are representative of three independent experiments.
2.3. SMU-Y6 inhibits TLR2 preferentially to other TLRs in vitro
TLR2 can form a dimer with either TLR1 or TLR6, so we conjectured whether SMU-Y6 exhibited selectivity in binding with TLR1/2 and TLR2/6. The results of our experimentation showed that SMU-Y6 suppressed the SEAP signaling induced by Pam2CSK4 (TLR2/6 ligand) or Pam3CSK4 (TLR1/2 ligand) in HEK-Blue hTLR2 cells, which indicated that SMU-Y6 inhibits both TLR1/2 and TLR2/6 (Supporting Information Fig. S3A). In addition, TLR4 recognizes lipopolysaccharide or bacterial wall-related components of Gram-negative bacteria, whose ligand and location are both similar to TLR2, so it is promising for it to exhibit selectivity between TLR2 and TLR4. Interestingly, as shown in Fig. 2C and D, SMU-Y6 reduces the production of the TLR2-induced inflammatory cytokines, TNF-α and IL-6, in human and murine cell lines; whereas it exhibits little or even no inhibitory activity with regards to TLR4, which suggested that SMU-Y6 tend to block TLR2 rather than TLR4. In addition, TLR3, TLR7 and TLR9 are all intracellular and mainly recognize DNA or RNA from bacteria and viruses. Thus, we measured the release of IL-6 in peritoneal macrophage which was activated by TLR3, TLR7 or TLR9 agonist. The results indicated that SMU-Y6 has a negative effect on TLR3, TLR7 and TLR9 (Fig. 2E). In summary, compound SMU-Y6 not only showed selectivity between TLR2 and TLR4, but also no significant inhibition to other TLRs (TLR3, TLR7 and TLR9). Furthermore, we employed the peritoneal macrophage from TLR2−/− knockout mice and measured the effect of SMU-Y6 on IL-6 after being stimulated by TLR3, TLR7 or TLR9 agonist. The results indicated that SMU-Y6 exhibited almost no inhibition on TLR3, TLR7 and TLR9, which showed strong specific to TLR2 (Fig. 2F). Thus, we determined that SMU-Y6 was a TLR2 inhibitor that shows excellent selectivity between TLR2 and other TLRs in vitro. The further selectivity, anti-inflammatory mechanism, as well as anti-inflammatory activity studies of SMU-Y6 are discussed as follows.
2.4. SMU-Y6 inhibited the release of inflammatory cytokines in human and murine primary cell lines
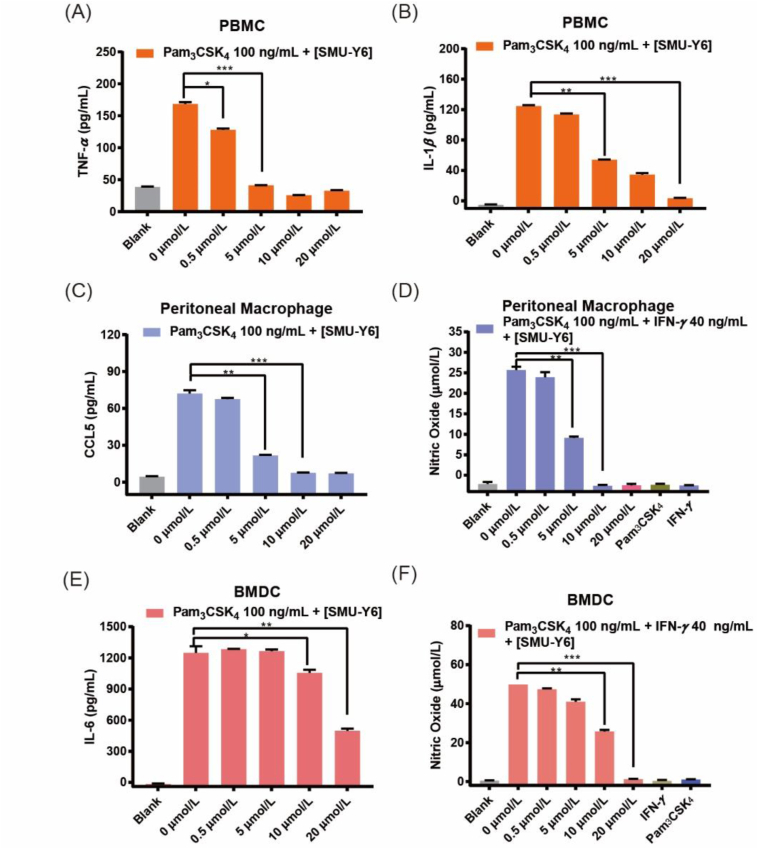
Inflammatory cytokines play an important role in inflammation development and are commonly used as indicators when evaluating the progression of inflammation, such as TNF, interleukin, chemokines, NO and so on. Blocking the production of inflammatory cytokines is beneficial to alleviating inflammation. We have revealed that SMU-Y6 inhibited the release of Pam3CSK4-induced TNF-α and IL-6 in peritoneal macrophages, as well as TNF-α in THP-1 cells (Fig. 2C). Meanwhile, we were more interested in the effect of SMU-Y6 on healthy human immune cells for it is much more closely mimic the real immune condition in vivo. In healthy human peripheral blood mononuclear cells (PBMCs), 5 μmol/L SMU-Y6 completely inhibited the release of TNF-α and IL-1β, but in a dose-dependent manner, after the cells were treated by Pam3CSK4 (Fig. 3A and B). The chemokine, regulated upon activation of normal T cell expressed and secreted (RANTES), also called CCL5, acts as an agonist of CCR1 and CCR5 (which are widely distributed in T cell, DC cell, NK cell and basophilic granulocytes), activating these cells and consequently the immune response33. As shown in Fig. 3C, levels of CCL5, induced by Pam3CSK4 in primary murine peritoneal macrophages, decreased significantly after treatment with SMU-Y6. In addition, the production of pro-inflammatory nitric oxide (NO) dilates the blood vessels local to the inflamed tissue and facilitates the recruitment and infiltration of immune cells34,35. In other words, inhibiting the production of pro-inflammatory NO could reduce the infiltration of immune cells at the inflammation site. In primary murine peritoneal macrophages, 10 μmol/L SMU-Y6 significantly reduced the production of NO induced by Pam3CSK4 (Fig. 3D). Dendritic cells also play an important role as APCs, affecting some autoimmune diseases due to their strong antigen uptake and presentation abilities. Similarly, they also produce inflammatory cytokines during activation. As shown in Fig. 3E, SMU-Y6 inhibited the release of IL-6 in bone marrow-derived dendritic cells (BMDCs). Similarly, NO induced by Pam3CSK4 decreased significantly after SMU-Y6 intervention (Fig. 3F). Inflammatory cytokines, induced by Pam2CSK4 were also blocked by SMU-Y6 (Supporting Information Fig. S3B and C) and these results were consistent with previous observations. Thus, we can summarize that SMU-Y6 reduced the release of TLR2-induced inflammatory cytokines in both human and murine primary cell lines to carry out its anti-inflammation effect. However, the underlying anti-inflammatory mechanisms need further investigation.
Figure 3.
Inhibitory effect of SMU-Y6 on inflammatory cytokines in human and murine cell lines. (A) TNF-α and (B) IL-1β in supernatants of primary PBMCs after treatment with Pam3CSK4 (100 ng/mL) and different concentrations of SMU-Y6 for 24 h. (C) CCL5 in the supernatants of murine peritoneal macrophage after treatment with indicated Pam3CSK4 (100 ng/mL) and differing concentrations of SMU-Y6 for 24 h. (D) Pro-inflammatory NO in murine peritoneal macrophage after treatment with Pam3CSK4 (100 ng/mL), IFN-γ (40 ng/mL) and differing concentrations of SMU-Y6 for 24 h. (E) IL-6 in supernatants of BMDC after treatment with indicated Pam3CSK4 (100 ng/mL) and differing concentrations of SMU-Y6 for 24 h. (F) Pro-inflammatory NO in murine BMDCs after treatment with indicated Pam3CSK4 (100 ng/mL), IFN-γ (40 ng/mL) and different concentrations of SMU-Y6 for 24 h. Data presented is mean ± SD and the figures shown are representative of three independent experiments.
2.5. SMU-Y6 blocked the activation of NF-κB and MAPK signaling pathway
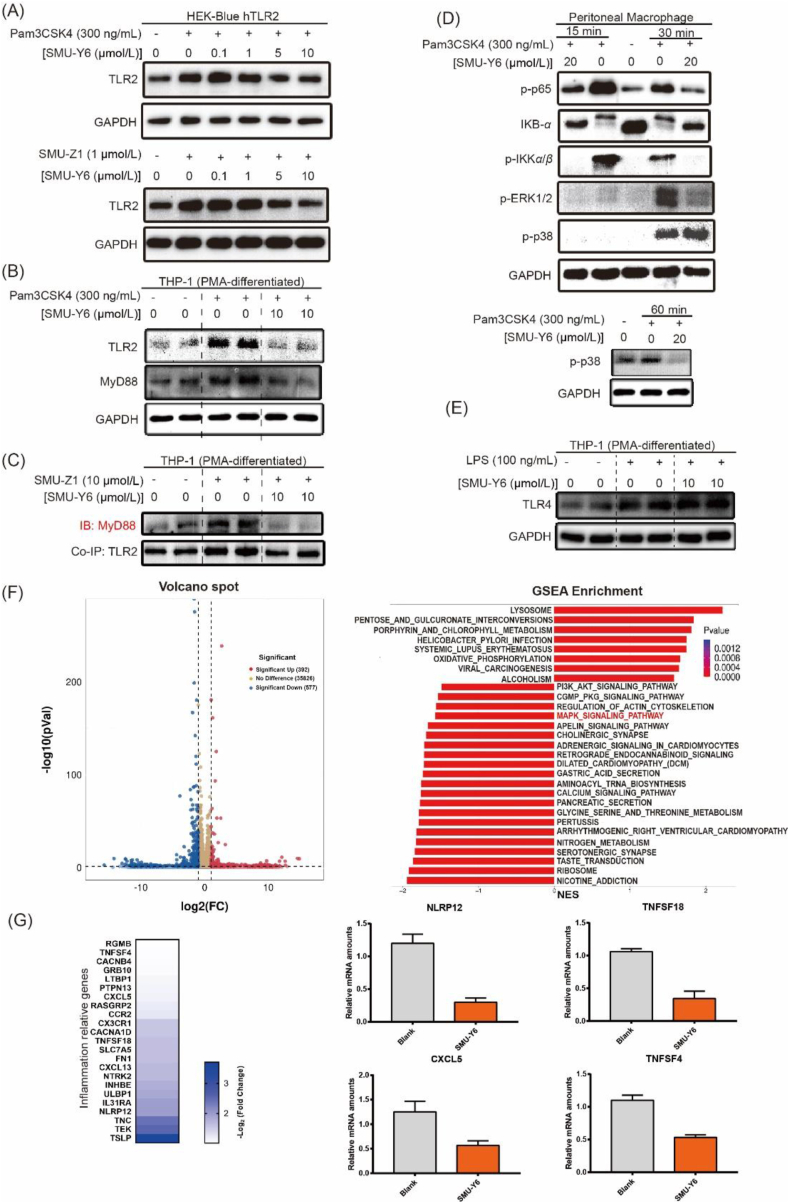
We have revealed that SMU-Y6 inhibited the release of TLR2-induced inflammatory cytokines. However, the underlying mechanism of action and signaling pathway remains to be elucidated. Therefore, the next step was to confirm at a protein level whether SMU-Y6 regulates inflammation through TLR2 and its downstream adaptor protein. As illustrated in Fig. 4A, the up-regulation of TLR2 protein by Pam3CSK4 in HEK-Blue-hTLR2 cells was reversed by SMU-Y6 in a dose-dependent manner. Similar results were obtained when we used the specific TLR1/2 agonist SMU-Z19, as an alternative positive control. Considering that TLR2 is highly expressed in HEK-Blue-hTLR2 cells, we also used THP-1 cells, which have more normal levels of TLR2 expression. Once again, SMU-Y6 reduced the up-regulation of TLR2 and MyD88 in THP-1 cells (Fig. 4B). It has already been documented that TLR2 recruits MyD88 to form a complex that activates its downstream signaling pathway19. We observed that the formation of TLR2/MyD88 complex increased after treatment with SMU-Z1 in THP-1 cells, whereas this formation decreased after the intervention of SMU-Y6 (Fig. 4C, Supporting Information Fig. S4). TLR2/MyD88 complex performs its pro-inflammatory function through activating downstream NF-κB and MAPK signaling pathway19. Our results show that SMU-Y6 reduces the recruitment of MyD88 to TLR2. Thus, our next focus was whether the activation of NF-κB and MAPK signaling pathways were affected. As shown in Fig. 4D, as a result of stimulation by Pam3CSK4, IKKα/β is phosphorylated, which promotes the degradation of IKB-α and increases the phosphorylation of p65. This cascade accounts for the activation of the NF-κB signaling pathway. Significantly, SMU-Y6 reverses the activation of the NF-κB signaling pathway by reducing the phosphorylation of IKKα/β, preventing IKB-α from being degraded, thus reducing the phosphorylation of p65 (Fig. 4D). With regards to the MAPK signaling pathway, phosphorylation of ERK1/2 and p38 are shown to be significantly increased at 30 min after treatment with Pam3CSK4. We determined that 30 min post treatment, SMU-Y6 had suppressed the activation of the ERK-MAPK signaling pathway. SMU-Y6 had little effect on the p38-MAPK signaling pathway during this time, however, we detected a decrease in phosphorylation of p38 at 60 min post SMU-Y6 treatment (Fig. 4D). Therefore, SMU-Y6 can inhibit the MAPK signaling pathway by blocking the phosphorylation of ERK and p38. In contrast, SMU-Y6 did not reverse the LPS-induced up-regulation of TLR4 (Fig. 4E), which may account for why SMU-Y6 failed to inhibit the TLR4-regulated release of inflammatory cytokines, providing further evidence to illustrate its selectivity between TLR2 and TLR4. We also carried out RNA-seq bioinformatics analysis to investigate the potential anti-inflammatory mechanism of SMU-Y6. After the treatment of SMU-Y6, there are 392 genes that significantly up-regulated (fold change >2 and P value < 0.05), while 577 genes are down-regulated (fold change >2 and P value < 0.05) (Fig. 4F). We analyzed the signaling pathway in which these genes were located through GSEA enrichment and found that MAPK signaling pathway was involved (Fig. 4F). These results are consistent with our previous observations that SMU-Y6 blocked MAPK signaling pathway, via interaction with p38 and ERK. Among those signaling pathway, some genes with significant differences in expression and associated with inflammation were list out, such as NLRP12, TNFSF18, CXCL5 and TNFSF4 (Fig. 4G). Overall, we determined that SMU-Y6 reduced the recruitment of TLR2 to MyD88 and blocked the activation of the NF-κB and MAPK signaling pathways through reducing the phosphorylation of IKKα/β, p65, p38 and ERK1/2, as well as the degradation of IKB-α.
Figure 4.
SMU-Y6 blocked the activation of TLR2/MyD88/NF-κB and MAPK signaling pathway and differential gene expression of seq-RNA analysis. (A) SMU-Y6 inhibited the upregulation of TLR2 induced by Pam3CSK4 (300 ng/mL) and SMU-Z1 (1 μmol/L) in HEK-blue hTLR2 cells. (B) SMU-Y6 inhibited the up-regulation of TLR2 and MyD88 induced by Pam3CSK4 (300 ng/mL) in THP-1 cells. (C) Co-immunoprecipitation showed that SMU-Y6 inhibited the formation of TLR2 and MyD88 complex induced by TLR2 agonist. (D) Murine peritoneal macrophages were pre-treated for 1 h with medium (or SMU-Y6) and treated with Pam3CSK4 (300 ng/mL) for 15, 30, and 60 min in the presence of medium or SMU-Y6. IB was performed using whole-cell lysates. (E) SMU-Y6 did not inhibit the up-regulation of TLR4 induced by LPS (100 ng/mL) in THP-1 cells. (F) Volcano plot exhibited genes differentially expressed (determined by RNA-seq analysis) in untreated THP-1 cells compared to SMU-Y6 treated cells and GSEA enrichment analysis shows variation in signaling pathway after SMU-Y6 treatment, noting that MAPK signaling pathway was involved. (G) Heatmap showed the expression of inflammation-associated genes on the treatment of SMU-Y6 among GSEA enrichment signaling pathway. The gene expression data were assessed by RNA-seq array.
2.6. SMU-Y6 targeted and competitive combined with TLR2
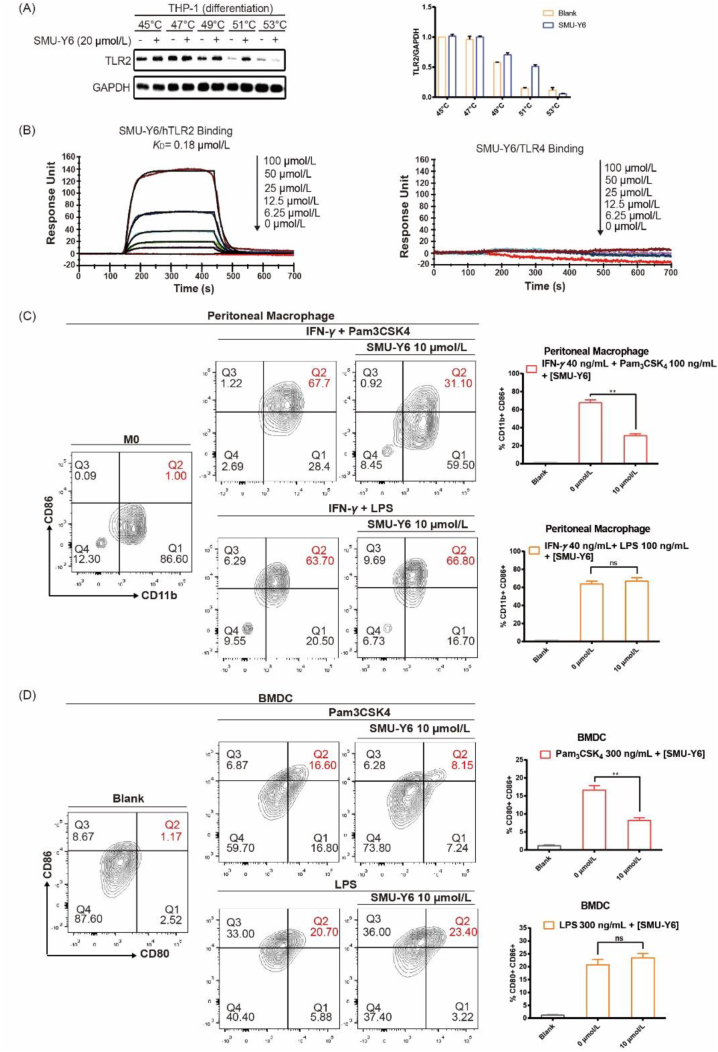
The interaction between the drug candidate and the target protein is the basis of its pharmacological activity. Therefore, evaluation of this ligand‒protein binding represents an important task in the drug development process. Cellular thermal shift assay (CETSA), which is based on the ligand-induced thermal stabilization of target proteins after combination, was used to evaluate drug binding to the target protein in cells and tissue samples36,37. We used CETSA to investigate whether SMU-Y6 increased TLR2 protein thermal stabilization and confirm whether SMU-Y6 interacted directly with TLR2 protein. As shown in Fig. 5A, the TLR2 protein was gradually degraded as the temperature increased from 45 to 53 °C. However, the TLR2 protein exhibited higher thermal stabilization after the addition of SMU-Y6, especially at 51 °C, which indicated that SMU-Y6 interacted with and stabilized the TLR2 protein. On the contrary, SMU-Y6 failed to induce the thermal stabilization of TLR4 (Supporting Information Fig. S5A). In addition, we carried out an experiment, using surface plasmon resonance (SPR), to investigate the binding affinity between SMU-Y6 and hTLR2 protein. As shown in Fig. 5B, SMU-Y6 bound with recombinant hTLR2 protein with a KD value of 0.18 μmol/L. We also demonstrated that the interaction between SMU-Y6 and TLR2 protein sets up a state of dynamic equilibrium with the molecules quickly binding and dissociating. SMU-Y6 completely dissociates from TLR2 protein after 500 s, indicating that SMU-Y6 does not covalently bind with TLR2 protein. However, we did not detect a binding signal between SMU-Y6 and recombinant hTLR4 protein (Fig. 5B); whereas the TLR4-specific inhibitor TAK-242, exhibited the ability to bind with recombinant hTLR4 protein (Fig. S5B). Moreover, we further investigated the potential binding sites of SMU-Y6 to TLR2. Pam3CSK4 is the natural ligand of TLR2 and its binding mode to TLR2 has been reported38. The competitive binding experiment between SMU-Y6 and Rhodamine-Pam3CSK4 demonstrated that SMU-Y6 reduced the fluorescence intensity of Rhodamine-Pam3CSK4 at 1 μmol/L (Supporting Information Fig. S6). Thus, we hypothesized that the binding site of SMU-Y6 to TLR2 is similar to Pam3CSK4. When SMU-Y6 binds with TLR2, Pam3CSK4 fails to combine with TLR2 in the way that it is supposed to be. In summary, we determined that SMU-Y6 competitively binds with the binding sites where Pam3CSK4 binds with TLR2 and had no influence on TLR4.
Figure 5.
SMU-Y6 targeted TLR2 and flow cytometry analysis of peritoneal macrophage and BMDC. (A) CETSA experiment indicates SMU-Y6 (20 μmol/L) reduces the degradation of TLR2 when heated to different temperatures. THP-1 cells were stimulated with PMA for 24 h, then cultured in a fresh medium for another 24 h. Cells were collected, suspended in PBS, and incubated with 20 μmol/L SMU-Y6 or medium for 0.5 h. Cells were heated at indicated temperature for 3 min. The proteins were extracted for Western blot. (B) Surface plasmon resonance experiment of SMU-Y6 binding to recombinant hTLR2 and hTLR4 protein. (C) Flow cytometry analysis of CD86+ and CD11b+ in mouse primary peritoneal macrophage. Mouse primary Peritoneal macrophage was treated with medium, IFN-γ (40 ng/mL) plus Pam3CSK4 (100 ng/mL), IFN-γ (40 ng/mL) plus Pam3CSK4 (100 ng/mL) and SMU-Y6, IFN-γ (40 ng/mL) plus LPS (100 ng/mL), IFN-γ (40 ng/mL) plus LPS (100 ng/mL) and SMU-Y6 for 24 h. (D) Flow cytometry analysis of CD86 and CD80 in mouse primary BMDCs. BMDCs were treated with medium, Pam3CSK4 (300 ng/mL), Pam3CSK4 (300 ng/mL) plus SMU-Y6, LPS (300 ng/mL) and LPS (300 ng/mL) plus SMU-Y6 for 24 h. Figures presented are mean ± SD, derived from three independent experiments.
2.7. SMU-Y6 reverses the polarization of M1 phenotype in macrophage and activation in dendritic cells
TLR2 was wildly distributed in APCs, given that APCs played an important role in innate and adaptive immunity, however, the over-activation of APCs could cause some inflammatory diseases39. Research suggests that resting macrophages (M0s) can polarize into a pro-inflammatory phenotype (M1) following TLR2 activation, which resulted in autoimmune rheumatic diseases40. We had revealed that SMU-Y6 blocked TLR2/MyD88/NF-κB and MAPK signaling pathway to reduce the production of cytokines in macrophages. Hence, we hypothesize that reducing the M1 polarization of macrophage induced by TLR2 may impair inflammation. Studies indicate that CD86, a surface marker of macrophages, is up-regulated when macrophages are polarized to their M1 phenotype40. As shown in Fig. 5C, after treatment of Pam3CSK4 and IFN-γ, the expression of CD86 is up-regulated significantly compared to the control. The percentage of CD11b+ CD86+ macrophage cells decreased from 51% to 20.7% after treatment with 10 μmol/L SMU-Y6, while the treatment had no effect on the polarization induced by LPS and IFN-γ (Fig. 5C). We were also interested in another important APCs, dendritic cells. Similarly, when BMDCs triggered by Pam3CSK4 are treated with 10 μmol/L SMU-Y6, the percentage of CD80+ CD86+ cells more than halved from 16.6% to 8.1% (Fig. 5D). SMU-Y6 failed to reverse the LPS-induced activation of BMDCs (Fig. 5D). In summary, these results indicated that SMU-Y6 reverses the polarization of M1 phenotype macrophage and the activation of dendritic cell induced by TLR2, but not TLR4, thus having potential application in selective inflammation suppression.
2.8. Evaluation of the in vivo anti-inflammatory activity of SMU-Y6
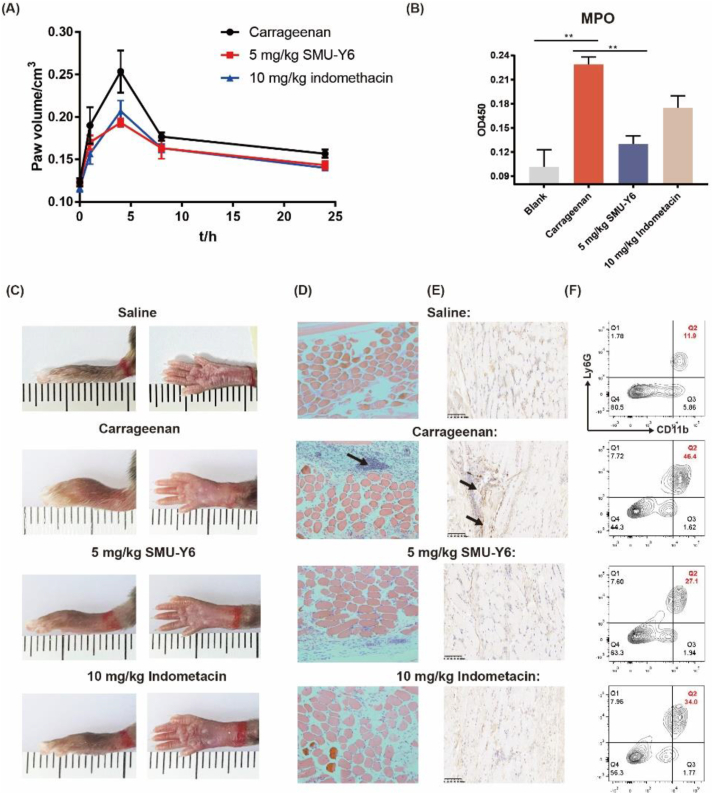
Given that our in vitro experiments demonstrated that SMU-Y6 exhibits excellent anti-inflammatory potential, through binding with TLR2, blocking its downstream signaling pathway, thus reducing the production of inflammatory cytokines, we looked to investigate if the same were true in vivo. The anti-inflammatory effect of SMU-Y6 in acute inflammation was investigated with the carrageenan-induced paw edema model41. As shown in Fig. 6A, after the mice received a subcutaneous injection in their paws of 1% carrageenan, the resulting edema lasted for 24 h, peaking at 4 h. When the mice were intraperitoneally injected with SMU-Y6 or indomethacin (used as an anti-inflammatory agent in the clinic) prior to the injection with carrageenan, the paw swelling (measured by volume) was significantly reduced at 4 h (Fig. 6A). In addition, SMU-Y6 not only impaired the severity of paw edema at 4 h, but the mice receiving this treatment also experienced a better overall prognosis (Fig. 6A). We measured the activity of myeloperoxidase in paw tissue to indicate the development of inflammation42. As shown in Fig. 6B, the activity of local myeloperoxidase (MPO) was significantly up-regulated after carrageenan treatment, compared with the control. However, this inflammation could be reversed with SMU-Y6 or indomethacin treatment. Histopathologic sections indicated that the mouse group injected with carrageenan presented higher aggregation of immune cells (black arrow), compared to the group injected with saline; however, treatment with SMU-Y6 or indomethacin could effectively reduce this aggregation (Fig. 6D). In addition, we detected M1 phenotype macrophage through immunohistochemical staining to iNOs. As shown in Fig. 6E, we concluded that iNOs was identified at local tissue significantly when treated with carrageenan, suggesting that those M0 macrophage was recruited and polarized into M1 phenotype. On the contrary, iNOs was not detected significantly in local tissue after SMU-Y6 intervention. The rapid infiltration of neutrophiles and secretion of MPO contributed to the formation of inflammation. We have identified SMU-Y6 reduced the production of MPO, so we wondered whether SMU-Y6 reduced the infiltration of neutrophile in mice paw to impair inflammation. We identified neutrophiles through anti-CD11b/Ly6G antibodies as reported43. As shown in Fig. 6F, we found that the infiltration of neutrophile was signficantly increased in local tissue after carrageenan treatment, while SMU-Y6 could reverse it. We also detected the expression of TLR2 protein in mouse paw, which was significantly up-regulated in the carrageenan group, compared with the control group. Once again, the presence of SMU-Y6 effectively reversed this over-expression of TLR2 observed in the paw tissue (Supporting Information Fig. S7A and S7B).
Figure 6.
The anti-inflammatory effect of SMU-Y6 in mice. (A) Mouse paw edema volume at different time points. (B) Myeloperoxidase activity in the inflamed foot pads at 4 h. Before mice paws received a subcutaneous injection of 1% carrageenan, mice were treated with saline, 5 mg/kg carrageenan, 10 mg/kg indomethacin. (C) Photograph of mouse paw edema at 4 h taken after treatment with either saline, carrageenan plus saline, carrageenan plus SMU-Y6 (5 mg/kg), carrageenan plus indomethacin (10 mg/kg). (D) HE staining and (E) immunohistochemical analysis of iNOs of mouse paws treated with either saline, carrageenan plus saline, carrageenan plus SMU-Y6 (5 mg/kg), carrageenan plus indomethacin (10 mg/kg) at 4 h injection. (F) Flow cytometry analysis of neutrophiles with Ly6G+ and CD11b+ in local mice paws.
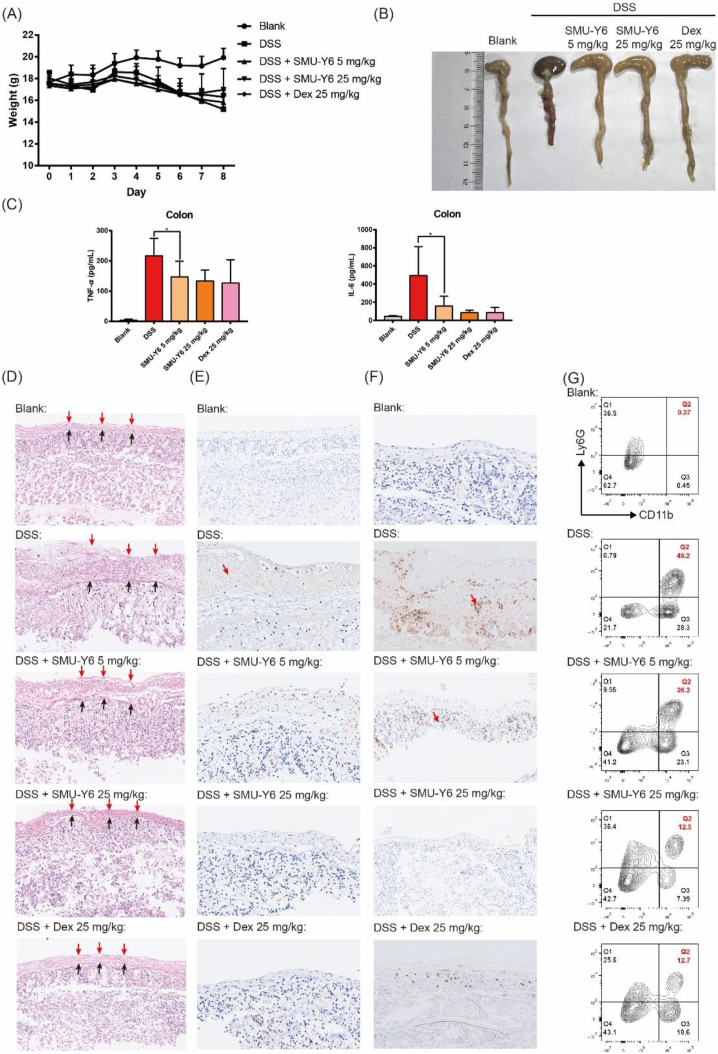
There are a large number of Gram-positive bacteria and their secretion of lipid polypeptide in the gut. Moreover, the incidence of enteritis gradually increases with the acceleration of the pace of life and irregular diet. It had been reported that TLR signaling pathway regulated inflammation in colitis44,45, so we further investigated the anti-inflammation effect of SMU-Y6 in dextran sulfate sodium-induced colitis. As shown in Fig. 7A, the weight of mice declined gradually since day 5 with oral administration of 3.5% dextran sulfate sodium (DSS), which indicated that the mice colitis model had been established. Oral administration of SMU-Y6 or dexamethasone reduced weight loss on Day 8, suggesting that SMU-Y6 may perform anti-inflammatory effects in colitis. On Day 8, the mice were sacrificed and the colon were collected. We noticed that the colon from the DSS model displayed shorted length and dark red than the control (Fig. 7B). Contrastly, the intervention of SMU-Y6 or dexamethasone could reverse it. Inflammatory cytokines were secreted in local colonic tissue to promote inflammation during colitis, so we measured the representative inflammatory factors TNF-α and IL-6 in local colonic tissue to evaluate the progress of colitis. Both TNF-α and IL-6 were increased in DSS model group, while SMU-Y6 reduced the production of TNF-α and IL-6 (Fig. 7C). As shown in Fig. 7D, histopathological analysis indicated that the mice with DSS treatment exhibited incompleted supermucous (red arrow) and submucous (black arrow), lysis of muscular cells (between supermucous and submucous) and irregular colonic recesses. On the contrary, SMU-Y6 reduced the injury of the mucous layer, lysis of muscular cells and maintained the function of colonic recesses in a dose-dependent way. We were also interested in whether M1 phenotype macrophage played a role in the formation of colitis, so we detected them through immunohistochemical analysis of iNOs. We could conclude that when treated with DSS, most iNOs (red arrow) were expressed in the mucosa layer (Fig. 7E), but the intervention of SMU-Y6 could reverse it in a dose-dependent manner. The over-production of NO increases the permeability of local tissue, which may lead to loss of interstitial fluid and give rise to severe diarrhea. In addition, the increasing permeability of local tissue was beneficial to the infiltration of immune cells, which produced TNF-α and IL-6 to arise inflammation and damage muscular cells. Meanwhile, we detected the expression of MPO in local tissue. Results revealed that the expression of MPO was significantly up-regulated in DSS treated group, which suggested the infiltration of neutrophiles in the inflammatory tissue (Fig. 7F). Notably, SMU-Y6 could reduce the production of MPO in a dose-dependent way. In addition, we analyzed the neutrophils in colon tissue by flow cytometry. Results showed that SMU-Y6 significantly reduced neutrophil infiltration (Fig. 7G), which is consistent with the situation observed in MPO. Thus, it could be seen that our TLR2 inhibitor has potential anti-inflammatory effects both in acute paw edema and colitis model. TLR2 was initially designed as a membrane receptor to recognize Gram-positive bacteria to avoid infection, so we were also interested in whether SMU-Y6 performed protection effect in infectious disease like sepsis. We constructed the sepsis model through cecum ligation and puncture. Surprisingly, SMU-Y6 seemed to perform protection effect on sepsis, as we found that SMU-Y6 significantly reduced the production of TNF-α and IL-6 (Supporting Information Fig. S8). In short, our results showed that SMU-Y6 could be a potential anti-inflammatory agent in acute inflammation treatment through reducing the infiltration of neutrophils, production of MPO, polarization of macrophage and secretion of inflammatory cytokines.
Figure 7.
The anti-inflammatory effect of SMU-Y6 in mice colitis induced by DSS. (A) Body weight of mice of each group. (B) Photograph of mice colon on Day 8. (C) TNF-α and IL-6 were measured in serum and local colonic tissue after mice treated with vehicle, 5 mg/kg SMU-Y6, 25 mg/kg SMU-Y6 and 25 mg/kg dexamethasone. (D) HE staining results and immunohistochemical analysis of iNOs (E) and MPO (F) in colon tissue. Results of mice were treated with vehicle, 5 mg/kg SMU-Y6, 25 mg/kg SMU-Y6 and 25 mg/kg dexamethasome. (G) Flow cytometry analysis of neutrophile in local colon tissue.
2.9. Pharmacokinetics studies and safety evaluation in vivo
To further validate the potential of SMU-Y6 as a therapeutic agent, we also investigated its metabolism in vivo. Pharmacokinetics studies indicated that SMU-Y6 persisted in rat plasma, with t1/2 of almost 9 h. This may account for the sustained anti-inflammatory effect of SMU-Y6 in mouse paw edema model. The drug concentration in the plasma peaked at 3 h post-oral administration, but was still detectable at 24 h, indicating it was unnecessary to maintain blood concentration by frequent dosing. The plasma concentration–time curve with different administration procedures is shown in Supporting Information Fig. S9. In addition, SMU-Y6 exhibits a high bioavailability (F∼37%), which mitigated having to administer high doses of SMU-Y6 to achieve the required blood concentration (Table 1). In order to evaluate the safety of SMU-Y6 in vivo, KM mice were given 10 times the therapeutic dose of SMU-Y6 (250 mg/kg) in oral administration every other day. The body weight of mice were recorded every day. Compared with the control group, frequent oral administration of SMU-Y6 with large doses did not seem to exhibit immediate toxicity for no mouse sacrificed on Day 7 (Supporting Information Fig. S10A). We performed a pathological analysis of the local tissue of mice. Results showed that there was no obvious organic damage to the local tissue in mice which was treated with SMU-Y6, including heart, liver, spleen, lung and kidney (Fig. S10B). In summary, SMU-Y6 has a long half-life, high bioavailability and hypotoxicity in vivo, making it an excellent long-term anti-inflammatory drug candidate.
Table 1.
Pharmacokinetic parameters of SMU-Y6 in rat plasma following intraperitoneal injection (n = 5) and oral administration (n = 5).
| Parameters | Units | Intraperitoneal injection | Oral administration |
|---|---|---|---|
| Tmax | h | 0.25 ± 0.01 | 3.00 ± 1.00 |
| Cmax | μg/L | 208.00 ± 31.11 | 52.33 ± 5.83 |
| t1/2 | h | 9.16 ± 1.11 | 11.54 ± 2.70 |
| AUC0‒t | μg/L·h | 1787.22 ± 409.66 | 619.21 ± 84.17 |
| AUC0‒∞ | μg/L·h | 2159.02 ± 600 | 844.08 ± 184.14 |
| F | % | – | 37 ± 10 |
3. Conclusions
TLR2 is an important immunoregulatory protein in blocking infections, removing cancerous cells and maintaining the homeostasis of the internal environment. However, the over-activation of TLR2 is closely related to the progression of inflammation-related diseases, including arthritis, Alzheimer's, and sepsis14, 15, 16. Hence, the discovery of new TLR2 inhibitor is particularly interesting, as currently, there is no marketed drug that acts as an TLR2 inhibitor, and existing TLR2 inhibitors have shortcomings, such as easy oxidation and hydrolysis, which limit their feasibility as drugs. Natural products as a reservoir of feasible drugs have been demonstrated by numerous marketed drugs. In our works, after screening of 2100 natural compounds, one compound in particular, Taspine, stood out as an excellent drug candidate, as it inhibited TLR2 in HEK-Blue hTLR2 cells. However, it too had limitations, including poor solubility and unacceptable levels of toxicity. Thus, we looked at ways to overcome these problems and enhance Taspine's anti-inflammatory properties. We looked to optimize the 10 steps of chemical synthesis of Taspine, then carried out structure–activity relationship studies. The results of these analyses yielded a novel compound (SMU-Y6), which has an IC50 value of 0.11 ± 0.04 μmol/L; exhibited selective inhibition TLR2 as opposed to other TLRs, including TLR3, TLR4, TLR7 and TLR9; improved anti-inflammatory activity by 10-fold, and water solubility by a factor of 20, as well as exhibiting reduced toxicity. SAR studies revealed that it is Taspine's lactone ring and aliphatic side chain (terminating with a basic amino group) that are essential for its activity. As for the anti-inflammation mechanism, we identified a potential mechanism in vitro as follow. On the one hand, SMU-Y6 reduced the recruitment of MyD88 to TLR2, and impaired the phosphorylation of IKKα/β, p65, p38, ERK1/2 as well as the degradation of IKBα (Fig. 8A). As a result, TLR2 and its downstream NF-κB/MAPK signaling pathway were blocked and the release of inflammatory cytokines (such as TNF-α, IL-1β, IL-6) were reduced in both human and murine cell lines. On the other hand, SMU-Y6 reduces the polarization of macrophage into M1 phenotype and activation of dendritic cells, suggesting that SMU-Y6 inhibits the progress of inflammation, induced by APCs, in the innate and adaptive immune system. Potentially, SMU-Y6 might have advantages in the treatment of TLR2-related autoimmune diseases. In addition, CETSA and SPR assays suggested that SMU-Y6 targets TLR2 rather than TLR4 with a Kd value of 0.18 μmol/L. Competitive experiment showed that SMU-Y6 competitively binds with the binding sites where Pam3CSK4 binds with TLR2. In mice paw edema model, we found that SMU-Y6 reduced the local infiltration of neutrophiles, production of MPO and inhibited the polarization of M1 macrophages. In vivo colitis model, SMU-Y6 reduced the release of local cytokines, reversed the polarization of the M1 phenotype of macrophages and reduced the infiltration of neutrophiles, thereby alleviating colon damage. In summary, we first highlight the aporphine alkaloid TLR2 inhibitor (SMU-Y6) that was optimized from Taspine, by retaining the latter's lactone ring, but modifying its side chain amine, with better anti-TLR2 activity, superior water solubility and less toxicity. We reveal SMU-Y6's anti-inflammatory effect through blockage of TLR2/MyD88/NF-κB and MAPK signaling pathway to reduce the release of cytokines, the infiltration of neutrophiles and the polarization of macrophages in vitro and in vivo (Fig. 8B). As a result, we have identified a much-needed potential drug candidate for inflammatory and autoimmune disease therapy as well as lay a foundation for the anti-inflammatory research of TLR2 inhibitors.
Figure 8.
Cell-based screening and structural modification of Taspine yields SMU-Y6. (A) SMU-Y6 exhibited less cytotoxicity, higher solubility and better activity in blocking TLR2 in preference to TLR4. It exerts its anti-inflammatory effect through the TLR2/MyD88/NF-κB and MAPK signaling pathway. (B) SMU-Y6 blocked TLR2/MyD88/NF-κB and MAPK signaling pathways to reduce the production of inflammatory cytokines and play an anti-inflammatory role.
4. Experimental
4.1. Biology
4.1.1. QUANTI-blue SEAP assay
HEK-blue hTLR2 cells were cultured in 96-well plates (4 × 104 cells per well) (Thermo Scientific) at 37 °C with the condition of 5% CO2 for 24 h in 200 mL DMEM, supplemented with 10% FBS and 1% penicillin/streptomycin. After 24 h, no adherent cells and medium were removed and replaced with supplemented fresh DMEM medium. The cells were treated with different concentrations of compounds and agonist (100 ng/mL Pam3CSK4, InvivoGen) in 200 mL DMEM total. Cells were cultured for another 24 h. The supernatant (50 μL) was collected and transferred to another 96-well plate. QUANTI-Blue (50 μL, InvivoGen) buffer was added to each well and the 96-well plate was shaken in the dark. The measure was conducted by a plate reader at an absorbance of 620 nm (OD620) at 15–30 min.
4.1.2. Cell toxicity assays
HEK-hTLR2 cells (3 × 104 cells per well) were seeded in a 96-well plate with 100 μL DMEM medium (with 10% FBS and 1% pen/strep) and incubated at 37 °C overnight. After that, different concentrations of SMU-Y6 were added to 200 μL total and incubated at 37 °C for another 24 h. Cell counting kit-8 (CCK-8) (Beyotime, C0038) was added into each well of above 96-well plate for 20 μL and incubated at 37 °C for 1–4 h until it turned into orange. The absorbance was measured at an absorbance of 450 nm through a plate reader.
4.1.3. Cytokines ELISA assays
Murine peritoneal macrophage, BMDCs or human PBMC cells were seeded in 12-well plates at a density of 0.5 × 106 cells per well with 1 mL of RMPI 1640 medium supplemented with 10% FBS, and 1% penicillin/streptomycin. The cells were treated with different concentrations of SMU-Y6 and Pam3CSK4 (100 ng/mL, Invivogen) or LPS (100 ng/mL, Invivogen) as positive control and incubated for 24 h at 37 °C in a 5% CO2 humidified incubator. The cell culture supernatants were collected and frozen at −80 °C until measurement. THP-1 cells at a density of 2.5 × 105 cells/mL were differentiated by treatment with 100 nmol/L PMA (Sigma) in RPMI 1640 cell culture medium, containing 10% FBS, 1% penicillin and streptomycin for 24 h at 37 °C in a 5% CO2 humidified incubator. After that, cells were washed with PBS and cultured in fresh RPMI 1640 cell culture medium (containing 10% FBS, 1% pen/strep) for another 24 h. The cell culture supernatant was replaced with new fresh culture medium and treated with 100 ng/mL Pam3CSK4 or 100 ng/mL LPS and different concentrations of compound for 24 h. The cell culture supernatants were collected and frozen at −80 °C until measurement. The level of cytokines was determined using recombinant human-cytokine standards, cytokine-specific capture antibodies and detection antibodies according to the commercially available ELISA kit (BD Biosciences) with each sample for triplicate.
4.1.4. Immunoblot analysis
HEK-Blue hTLR2 cells were seeded in 6-well plate (Thermo Scientific, 145380) at density of 1.5 × 106 per well in 3 mL DMEM (supplemented with 10% FBS, 10 × penicillin/streptomycin, and 10 × glutamine) and incubated for 24 h. The supernatant was removed and replaced with fresh DMEM medium to 3 mL totally and treated with different concentrations of Pam3CSK4 (InvivoGen, 112208-00-1), SMU-Z1 and SMU-Y6 for 24 h. The cells were harvested and lysed with 150 μL cell lysis buffers (RIPA mixed with protease inhibitor and protein phosphatase inhibitor, Booster). Cell lysates of equal amounts were denatured, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membrane (Millipore). Then use skim milk (BD, 6342932) to block the membrane for 2 h, and then incubated with the TLR2 (cell signaling technology, 12276s), GAPDH (Boster, BM1623) antibody overnight at 4 °C, followed with incubating by a horseradish peroxidase conjugated secondary antibody. The immunoblots were visualized by enhanced exposure machine. Peritoneal macrophages were seeded in 6-well plate (Thermo Scientific, 145380) at density of 1.5 × 106 per well in 3 mL RPMI 1640 (supplemented with 10% FBS, 10 × penicillin/streptomycin, and 10 × glutamine) and incubated for 24 h. The supernatant was removed and replaced with fresh RPMI 1640 medium to 3 mL totally and treated with different concentrations of Pam3CSK4 (InvivoGen, 112208-00-1), SMU-Y6 for 15 min, 30 min. The cells were harvested and lysed with 100 μL cell lysis buffers (RIPA mixed with protease inhibitor and protein phosphatase inhibitor, Booster). Cell lysates of equal amount were denatured, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membrane (Millipore). Then use skim milk (BD, 6342932) to block the membrane for 2 h, and then incubated with the p-p65 (Cell Signaling Technology, 3033s), p-p38 (Cell Signaling Technology, 4511s), IKBα (Cell Signaling Technology, 44D4), p-IKKα/β (Cell Signaling Technology, 16A6), p-ERK (Cell Signaling Technology, 4370s), GAPDH (Boster, BM1623) antibody overnight at 4 °C, followed with incubating by a horseradish peroxidase conjugated secondary antibody. The immunoblots were visualized by an enhanced exposure machine.
4.1.5. Co-immunoprecipitation assay
THP-1 cells were seeded in 6-well plate (Thermo Scientific) at a density of 3 × 106 per well in 3 mL RPMI 1640 (supplemented with 10% FBS, 10 × penicillin/streptomycin) and treated with 100 nmol/L PMA (Sigma) for 24 h. The supernatant was removed and cells were washed with PBS twice, then fresh 3 mL RPMI 1640 (supplemented with 10% FBS, 1% penicillin/streptomycin) was added in. The cells were incubated for another 24 h. The supernatant was removed and cells were pre-treated with indicated SMU-Y6 for 0.5 h. Then SMU-Z1, as a TLR2 agent, was added in. An hour later, the cells were harvested and lysed with 100 μL cell lysis buffers (RIPA mixed with protease inhibitor and protein phosphatase inhibitor, Booster). The cell lysates were centrifuged at 13,000×g for 10 min, and the supernatant was collected. 1 μL TLR2 anti-body (cell signaling technology, 12276s) was added in 80 μL cell lysates and shaken overnight. After that, 30 μL protein A + G beads (EarthOx, PA001-01) were added in and shaken for 6 h. Later, the beads were collected through centrifuging at 300×g for 5 min and washed twice with cell lysis. Loading Buffer (Solarbio, PC0020) was added in and the samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membrane. Than 5% skim milk (BD, 6342932) was used to block the membrane for 1 h at room temperature. Then incubated with the primary antibody of TLR2 (Cell Signaling Technology, 12276s) and MyD88 (Proteintech, 23230-1-AP) overnight at 4 °C, followed with incubating by a horseradish peroxidase conjugated secondary antibody [diluted by 5% skim milk solution, goat anti-rabbit (Booster, BA1056). The immunoblots were visualized by an enhanced exposure machine.
4.1.6. Cellular thermal shift assay
THP-1 cells at a density of 1.5 × 106 cells per well were differentiated by treatment with 100 nmol/L PMA (Sigma) in RPMI 1640 cell culture medium, containing 10% FBS, 1% penicillin and streptomycin for 24 h at 37 °C in 6 well-plate. Cells were collected, washed with PBS in three times and suspended in PBS with protease inhibitor. The cells were divided into two aliquots with one aliquot being treated with indicated SMU-Y6 and the other aliquot with equivalent solvent. After 30 min incubation at room temperature, the respective cell suspensions were divided into smaller (40 μL) aliquots and heated individually at different temperatures for 3 min followed by cooling for 3 min at room temperature. Then, half volume of cell lysis buffer (20 μL) was added in. The whole cell suspensions were vortexed, frozen and thawed at −80 °C for three times. The suspensions were centrifuged at 16,000×g for 30 min at 4 °C. The supernatants were transferred to new microtubes and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis.
4.1.7. Surface plasmon resonance assay
(1) h-TLR2 and h-TLR4 protein coating: Human TLR2 protein was expressed in insect H5 cells as described earlier11. The TLR2 protein was diluted with PBS to a concentration of 1 mg/mL and the TLR4 protein (10146-H08B, SinoBiological) was diluted with PBS to a concentration of 0.1 mg/mL. The 3D dextran chip was washed in PBS three times and dried with N2. The solution of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (Macklin, N915145, 0.765 g dissolved in H2O for 10 mL) and N-hydroxy succinimide (Macklin, H6231, 0.115 g, dissolved in H2O to 10 mL) were mixed and the 3D dextran chip was immersed in the mixture for 15–20 min and then washed with PBS for three times before drying with N2. The TLR2 protein solution (1 μL) was dropped to the surface of the 3D dextran chip with a pipette, more gently. Then the chip was incubated at 4 °C overnight with more than 60% of humidity. On Day 2, the chip was washed three times with PBS and blocked by 1 mol/L ethanolamine aqueous solution (pH = 8.4). The chip was stored at 4 °C until used. (2) Preparation of loading buffer: 8.0 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4 and 0.24 g KH2PO4 were dissolved into 800 mL distilled water, and con. hydrochloric acid was used for pH adjustment (pH = 7.4). Then, the solution was constant to 1 L. (3) Preparation of recycling buffer: Glycine (Macklin, G800883, 1.965 g) was dissolved in 480 mL of deionized water in a volumetric flask. Con. hydrochloric acid was used to adjust the pH to 2.0. Finally, the volume of solution was made up to 500 mL. (4) Sample testing: SMU-Y6 was dissolved in the loading buffer at a final concentration of 100, 50, 25, 12.5, 6.26 and 0 μmol/L. All the samples were tested by PlexArray HT A100 (plexera) and data were analyzed using BIA evaluation Software.
4.1.8. Nitric oxide (NO) assay
Nitrite, the end product of NO metabolism, was measured using the Griess reagent. Briefly, murine primary peritoneal macrophages cells were seeded in a 12 well plastic plate at a density of 1 × 106 per well, pretreated with IFN-γ (40 ng/mL, peprotech) and Pam3CSK4 (100 ng/mL) and then stimulated with for 48 h. To measure the nitrite concentration, the culture supernatant (50 μL) was mixed with an equal volume of Griess reagent for 10 min. The nitrite concentration in each culture supernatant was measured at 570 nm with reference to the standard curve using sodium nitrite.
4.1.9. Preparation of peritoneal macrophage
Peritoneal macrophages were obtained by peritoneal lavage from 6 to 8 weeks C57BL/6 mice (purchased from the Laboratory Animal Center of Southern Medical University) 4 day after ip injection of 3 mL 4% sterile thioglycolate. Cells were washed with PBS three times and cultured in RPMI 1640 [supplemented with 10% FBS, penicillin (100 U/mL), and streptomycin (100 mg/mL)]. The cells were used in subsequent experiments.
4.1.10. Preparation of bone marrow-derived dendritic cells (BMDC)
Total cells were harvested by flushing cells from femurs and tibiae of C57BL/6 mice and cultured with RPMI 1640 [supplemented with 10% FBS, penicillin (100 U/mL), and streptomycin (100 mg/mL)] for 2 h in petri dish. The supernatant was transferred to new petri dish to remove adherent cells and murine IL-4 (10 ng/mL, peprotech), murine GM-CSF (40 ng/mL, peprotech) were added in. On day three, the cells were dissociated slightly to discard the strongly adherent cells. The suspended cells were transferred to a new petri dish, then murine IL-4 (10 ng/mL, peprotech), and murine GM-CSF (40 ng/mL, peprotech) were added in. On day seven, BMDC were collected for subsequent experiments.
4.1.11. Peritoneal macrophage phenotype assay
Murine Peritoneal Macrophage cells were seeded in 12-well plate (Thermo Scientific) at density of 1 × 106 per well in 1 mL RPMI 1640 [supplemented with 10% FBS, penicillin (100 U/mL), and streptomycin (100 mg/mL)] and incubated overnight. Peritoneal macrophage was treated with medium, Pam3CSK4 (InvivoGen, 100 ng/mL) plus murine IFN-γ (40 ng/mL, Peprotech), LPS (InvivoGen, 100 ng/mL) plus murine IFN-γ (40 ng/mL, Peprotech) and indicated SMU-Y6 for 24 h. Cells were collected into centrifuge tube after trypsinization and carefully aspirated the supernatant after centrifugation at 300×g for 5 min. The cells were washed twice with PBS and stained with a mixture of antibodies for 30 min, including anti-mouse CD86-PE-Cy7 (BD Biosciences), anti-mouse CD11b-FITC (BD Biosciences). After incubation, the cells were centrifuged at 300×g for 5 min and washed twice with PBS. Stained cells were analyzed by flow cytometer (FACScanto II, BD) and the flow cytometry data were analyzed using FlowJo software.
4.1.12. BMDCs activation assay
BMDCs were seeded in 12-well plate (Thermo Scientific) at density of 5 × 105 per well in 1 mL RPMI 1640 [supplemented with 10% FBS, penicillin (100 U/mL), and streptomycin (100 mg/mL)] and incubated overnight. BMDCs were treated with Pam3CSK4 (InviviGen, 300 ng/mL), LPS (InviviGen, 100 ng/mL) and indicated SMU-Y6 for 24 h. Cells were collected into centrifuge tube after trypsinization and carefully aspirated the supernatant after centrifugation at 300×g for 5 min. The cells were washed twice with PBS and stained with a mixture of antibodies for 30 min, including anti-mouse CD86-PE-Cy7 (BD Biosciences), anti-mouse CD80-PerCP-Cy5.5 (BD Biosciences) and CD11c-APC-Cy7 (BD Biosciences). After incubation, the cells were centrifuged at 300×g for 5 min and washed twice with PBS. Stained cells were analyzed by flow cytometer (FACScanto II, BD) and the flow cytometry data were analyzed using FlowJo software.
4.1.13. Paw edema experiment
All animal studies were conducted according to institutional animal care and use regulations approved by the Animal Research Center of Southern Medical University (Permit number SCKK 2016-0041). The anti-inflammatory activity was investigated using the carrageenan-induced paw edema model. The male C57Bl/6 mice were purchased from the Laboratory Animal Center of Southern Medical University at 6–8 weeks old and were randomly divided into four groups of six animals each. Prior to the injection of carrageenan, the mice's pedal volume up to the ankle joint was recorded using plethysmometer (Taimeng PV-200 7500, Chengdu, China). Before subcutaneous injection of carrageenan, mice were treated with saline, indomethacin (10 mg/kg, ip) or SMU-Y6 (5 mg/kg, ip) for 1 h. Later, 50 μL of 1% carrageenan suspended in saline or saline was injected into the plantar side of right hind paw. The paw volume was measured at 1, 4, 8 and 24 h after injection using a plethysmometer. The right hind paws of mice were surgically dissected at 4 h post-injection for the histological analysis and myeloperoxidase activity assay.
4.1.14. Histological analysis
Tissue slices were fixed in 10% formalin for 3 days, decalcified overnight and embedded in paraffin and sectioned into 4 μm tissue sections. Tissue sections were stained with hematoxylin and eosin before being examined under a BX60 microscope (Olympus, Melville, NY, USA) for pathological changes.
4.1.15. Tissue immunoblot analysis
The male C57Bl/6 mice were purchased from the Laboratory Animal Center of Southern Medical University at 6–8 weeks old and were randomly divided into three groups. Before subcutaneous injection of carrageenan, mice were treated with saline or SMU-Y6 (5 mg/kg, ip) for 1 h. Later, 50 μL of 1% carrageenan suspended in saline or saline was injected into the plantar side of right hind paw. Four hours later, the right hind paws of mice were surgically dissected. The skin surface of each paw was removed with surgical scissors and local tissue was isolated. Then the tissue was lysed with RIPA lysis buffers (containing protease inhibitor) under ultrasonic treatment for 0.5 h. The samples were centrifuged at 12,000×g for 10 min at 4 °C. The supernatants were collected. The subsequent treatment was followed by immunoblot analysis.
4.1.16. Myeloperoxidase activity assay
Tissue myeloperoxidase activity assay was performed as an index of granulocyte recruitment, as previously described by Kotsyfakis. Briefly, the injected paws obtained from the above experiments were weighed and homogenized in 2 mL of 0.5% hexadecyl-trimethylammonium bromide phosphate buffered solution (pH 6.0). The homogenate was then centrifuged at 12,000 rpm for 5 min. Three aliquots of each supernatant were then transferred into 96-well plates before the addition of a solution containing 3,3′-dimethoxybenzidine and 1% hydrogen peroxide. In parallel, dilutions of pure myeloperoxidase (Sigma–Aldrich, St. Louis, MO, USA) were used for the construction of a standard curve (OD as a function of units of enzyme activity). OD readings at 450 nm were taken at 1 min (time point corresponding to the linear portion of the enzymatic reaction) using a microplate reader (TECAN Company, Männedorf).
4.1.17. RNA-seq analysis
THP-1 cells at a density of 1.5 × 106 cells per well were differentiated by treatment with 100 nmol/L PMA (Sigma) in RPMI 1640 cell culture medium, containing 10% FBS, 1% penicillin and streptomycin for 24 h at 37 °C in 6 well-plate. The cells were washed with PBS three times and were cultured in new fresh RPMI 1640 cell culture medium for 24 h. Total RNA were collected through Trizol method (LS1040, Gibco), then analyzed RNA integrity assessment by Bioanalyzer2100 (Agilent) and samples with RIN value ≥ 7 were used. Poly (A) RNA is purified by using Dynabeads Oligo (dT) (2561005, ThermoFisher) and broken into pieces by Magnesium RNA Fragmentation Module (e6150, NEB) under 94 °C for 5–7 min. Then cDNA was obtained through the reverse transcription of cleaved RNA by SuperScript™II Reverse Transcriptase (1896649, Invitrogen) and indicated to synthesize U-labeled second-stranded DNAs by E. coli DNA polymerase I (m0209, NEB), RNaseH (m0297, NEB) and dUTP Solution (R0133, ThermoFisher). A-base is then ligated to the blunt ends of each strand for index adapters ligation containing a T-base overhang, and AMPureXP beads performed size selection. Under the treatment of heat-labile UDG enzyme (NEB, cat. m0280, USA), the ligated samples are amplified through PCR by the following steps: the denaturation of samples at 95 °C for 3 min; denaturation at 98 °C for 15 s for 8 cycles, annealing at 60 °C for 15 s, and extension at 72 °C for 30 s; and the last extension at 72 °C for 5 min. For the cDNA library in RNA-Seq, the average insert size was 300 ± 50 bp. We finally displayed the paired-end sequencing (PE150) on an Illumina Novaseq™ 6000 (LC-BioTechnology Co., Ltd., Hangzhou, China) following the recommended protocol. FPKM were calculated by StringTie and ballgown, and were applied to evaluate the expression of all mRNA transcripts. The significant expression of mRNAs was identified with fold change>2 or fold change<0.5 and P value < 0.05 by R package edgeR or DESeq2, and were used for KEGG enrichment analysis.
4.1.18. Establishment of mice colontis
The male C57Bl/6 mice were purchased from the Laboratory Animal Center of Southern Medical University at 6–8 weeks old and were randomly divided into five groups (control group; DSS induced-colitis group treated with vehicle; DSS induced-colitis group treated with 5 mg/kg SMU-Y6; DSS induced-colitis group treated with 25 mg/kg SMU-Y6; DSS induced-colitis group treated with 25 mg/kg dexamethasome). All mice were given 3.5% DSS dissolved in daily drinking water for 5 days except for the control group. After the fifth day, 3.5% DSS solution was removed and all mice were given normal water. From Day 2, 5 mg/kg SMU-Y6, 25 mg/kg SMU-Y6, and 25 mg/kg dexamethasome were given through oral administration. The body weight of each mice were collected for each day. On Day 8, all mice were sacrificed and blood samples as well as colonic tissue were collected.
4.1.19. Seurm elisa assay
All blood samples from mice were placed in room temperature for 1 h. The samples were centrifuged for 15 min at 12,000×g. After that, the serum was collected and IL-6 was tested by using recombinant mouse-cytokine standards, cytokine-specific capture antibodies and detection antibodies according to the commercially available ELISA kit (BD Biosciences).
4.1.20. Colonic tissue elisa assay
All colonic tissue samples from mice were collected and extracted with 200 μL cell lysis buffers (RIPA mixed with protease inhibitor and protein phosphatase inhibitor, Booster) under an ultrasonic cell disruptor at 4 °C. The samples were centrifuged for 30 min at 12,000×g. The supernatant was collected and the protein quantification using BCA method. The lysis with the same concentration of protein were tested IL-6 as well as TNF-α by using recombinant mouse-cytokine standards, cytokine-specific capture antibodies and detection antibodies according to the commercially available ELISA kit (BD Biosciences).
4.1.21. Toxicity studies in vivo assay
The male and female KM mice were purchased from the Laboratory Animal Center of Southern Medical University at 6–8 weeks old and were randomly divided into two groups (the control group and SMU-Y6 group). Each group contained 10 KM mice (half male). The mice in SMU-Y6 group were given 250 mg/kg SMU-Y6 through oral administration, while those in the control group were given the same volume of solvent on Day 1, 3 and 5. The body weight of each mouse was recorded every day and mice were given ample food and water. On Day 7, all mice were sacrificed and local tissue were collected for HE pathological analysis.
4.1.22. Competitive combination assay
HEK-hTLR2 cells (3 × 104 cells per well) were seeded in 96-well plate (Cellvis, P96-1-N) with 100 μL DMEM medium (with 10% FBS and 1% pen/strep) and incubated at 37 °C overnight. The supernatant was removed and 50 μL fresh DMEM medium which contained 800 ng/mL Rhodamine-Pam3CSK4 (InvivoGen, tlr-rpms) and 1 μmol/L SMU-Y6 was added in for 4 h incubation. The supernatant was removed and the cells were washed with PBS for three times. DAPI (Beyotime, C1005) was added in and incubated for 5 min. Later, The DAPI was removed and the cells were washed with PBS for three times. The cells were observed under confocal microscopy (Carl Zeiss, LSM 880 with Airyscan).
4.1.23. Flow cytometry analysis of neutrophile
The epidermis of mice paw was removed and tissue was collected through the scalpel. Mice colon was collected and the contents were removed. Part of colon was separated and washed with PBS three times. The mice paw tissue or colon tissue were placed in a centrifuge tube, RPMI 1640 medium (containing collagenase V, 2 mg/mL) was added in. The tissue was cut into pieces through surgical scissors and was digested at 37 °C for 2 h. The cell suspension was filtered through a 40 μm filter membrane. The cells were collected after centrifugation at 300×g for 5 min. The cells were washed twice with PBS and stained with a mixture of antibodies for 30 min, including anti-mouse Ly6G-APC (Elabscience, E-AB-F1108E), anti-mouse CD11b-FITC (BD Biosciences). After incubation, the cells were centrifuged at 300×g for 5 min and washed twice with PBS. Stained cells were analyzed by flow cytometer (FACScanto II, BD) and the flow cytometry data were analyzed using FlowJo software.
4.1.24. Cecum ligation and puncture model
The male C57Bl/6 mice were purchased from the Laboratory Animal Center of Southern Medical University at 6–8 weeks old. The polymicrobial sepsis was carried out by CLP. After the mice were anesthetized, the cecum was exposed. The cecum was ligated with a surgical suture and a through-and-through puncture was made into the cecum by an 18-gauge needle. A portion of the cecal content was extruded and the cecum was reset and sutured. Sham-operated mice underwent laparotomy without CLP. All mice were sacrificed 8 h after surgery and seurm and lung tissue were collected for Elisa assay. 5 and 25 mg/kg dose of SMU-Y6 was oral administrated to mice 1 h before CLP. The sham-operated mice were received the same volume of saline.
4.1.25. Statistical analysis
Data are displayed as mean ± standard deviation (SD). Statistical analysis was carried out using an unpaired two-tailed Student's t-test. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 and ns denotes not significant. P value < 0.05 was considered as significant.
4.2. Chemistry: General information and synthetic procedures
2-Bromo-3-hydroxy-4-methoxybenzaldehyde (compound1). To a mixture of 3-hydroxy-4-methoxybenzaldehyde (25.0 g, 0.164 mol), NaOAc (27.0 g, 0.325 mol), and Fe powder (0.755 g, 0.015 mol) were added in acetic acid (100 mL). The mixture was stirred at room temperature for 15 min Br2 (9.9 mL, 0.18 mol) was dissolved in acetic acid (50 mL) and dropped into the above mixture. The mixture was stirred at room temperature for 4 h. Ice water (150 mL) was added to the mixture and stirred for another 1 h. The solid was obtained by filtration and washed with EtOH (40 mL) to get compound 1 (26.5 g, 70%) as a gray solid. 1H NMR (400 MHz, DMSO-d6): δ = 10.11 (s, 1H), 9.90 (s, 1H), 7.42 (d, J = 7.4 Hz, 1H), 7.39 (d, J = 7.4 Hz, 1H), 3.92 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 190.8, 153.3, 144.0, 126.6, 122.0, 113.3, 110.4, 56.4. ESI-HRMS m/z: calculated C8H8BrO3[M+H]+ 230.9651, found 230.9647.
3-Benzyloxy-2-bromo-4-methoxybenzaldehyde (compound2). Compound 1 (26.5 g, 0.114 mol) was dissolved in dehydrated alcohol (200 mL). Anhydrous K2CO3 (47.7 g, 0.346 mol) and benzyl bromide (19.8 mL, 0.172 mol) was added in above mixture. The mixture was refluxed for 4 h. The alcohol was removed in a vacuum after filtration. The residue was dissolved in EtOAc (100 mL). The organic layers were washed with H2O (30 mL), 1 mol/L NaOH (30 mL), 2 mol/L HCl (30 mL), and brine (20 mL). Then, the organic layer was concentrated and purified by column chromatography to give compound 2 (38.6 g, 95%) as a white solid. 1H NMR (400 MHz, CDCl3): δ = 10.27 (s, 1H), 7.76 (d, J = 8.4 Hz, 1H), 7.55 (d, 2H), 7.40–7.35 (d, J = 9.2 Hz, 3H), 6.97 (d, J = 8.4, 1H), 5.05 (s, 2H), 3.96 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 190.6, 156.9, 145.4, 136.4, 128.6, 128.5, 125.4, 122.5, 121.0, 117.2, 74.9, 56.6.
3-Benzyloxy-2-bromo-4-methoxybenzoic acid (compound3). To a solution of compound 2 (38.6 g, 0.120 mol) in THF (200 mL) was added distilled H2O (80 mL) and NaH2PO4 (8.65 g, 0.072 mol). The mixture was stirred for 5 min at room temperature. NaClO2 (36.08 g, 0.4 mol) and 30% H2O2 (27.52 mL, 0.265 mol) in distilled H2O (80 mL) were added into the above mixture at ice-water bath. The mixture was stirred for 3 h. THF was evaporated under vacuum and the residue was extracted with EtOAc (200 mL). The organic layers were washed with H2O (50 mL) and the product was extracted with 2 mol/L NaOH (3 × 50 mL). The aqueous phase was collected and acidified with con HCl. The white solid was obtained by filtration to give compound 3 (36.52 g, 90%). 1H NMR (400 MHz, CDCl3): δ = 7.88 (d, J = 8.8 Hz, 1H),7.57 (d, J = 6.9, 2H), 7.43–7.32 (m, 3H), 6.93 (d, J = 8.8 Hz, 1H), 5.03 (s, 2H), 3.94 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 170.6, 157.5, 146.3, 136.9, 129.4, 128.6, 128.5, 128.3, 123.0, 120.1, 110.4, 74.7, 56.3. ESI-HRMS m/z: calculated C15H12BrO4[M+H]+ 338.0070, found 337.0068.
Methyl 3-benzyloxy-2-bromo-4-methoxybenzoate (compound4). Acetyl chloride (15.2 mL, 0.213 mol) was added in MeOH (150 mL) at 0 °C and stirred at room temperature for 15 min. Compound 3 (36 g, 0.107 mol) was added to the above mixture and refluxed for 4 h. MeOH was removed under vacuum and the residue was extracted with EtOAc (150 mL). The organic extracts were washed with H2O (50 mL), saturated NaHCO3 solution (2 × 50 mL), brine (50 mL), and concentrated. The oily liquid was purified by chromatographed on silica gel (PE:EtOAc = 5:1) to give compound 4 (29.9 g, 80%) as an oil. 1H NMR (400 MHz, CDCl3): δ = 7.64 (d, J = 8.7 Hz, H), 7.56 (d, J = 6.9, 1H)7.42–7.31 (m, 3H), 6.89 (d, J = 8.7 Hz, 1H), 5.01 (s, 2H), 3.91 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 166.3, 156.5, 146.0, 136.9, 128.5, 128.4, 128.2, 127.8, 124.8, 119.0, 110.4, 74.6, 56.2, 52.3. ESI-HRMS m/z: calculated C16H14BrO4[M+H]+ 351.0226, found 351.0019.
Dimethyl 6,6-dibenzyloxy-5,5-dimethoxybiphenyl-2,2-dicar-boxylate (Y1). Compound 4 (29 g, 0.083 mol) was dissolved in drying DMF (50 mL), added activated Cu (53 g, 0.83 mol) under N2 protection, and the solution was heated at 156 °C for 4 h. The mixture was filtered and the residue was washed with CHCl3 (3 × 100 mL). The organic layers were washed with aq 2 mol/L HCl (3 × 50 mL) and brine (2 × 50 mL), and concentrated. The residue was purified by chromatographed on silica gel (PE: EtOAc = 5:1) to give Y1 (15.6 g, 70%) as a white solid; 1H NMR (400 MHz, CDCl3): δ = 7.88 (d, J = 8.6 Hz, 2H),7.16–6.93 (m, 12H), 4.85 (d, J = 11.1 Hz, 2H), 4.71 (d, J = 11.1 Hz, 2H), 3.92 (s, 6H), 3.58 (s, 6H). 13C NMR (101 MHz, CDCl3) δ 166.5, 156.3, 145.3, 138.1, 135.0, 127.9, 127.5, 127.3, 122.6, 110.4, 74.1, 55.9, 51.6. ESI-HRMS m/z: calculated C32H31O8[M+H]+ 543.2013, found 543.2000.
Dimethyl 6,6-dihydroxy-5,5-dimethoxybiphenyl-2,2-dicar-boxylate (Y2). Compound Y1 (15 g) was dissolved in THF (100 mL), added 10% Pd/C (4.5 g) under and stirred under H2 atmosphere at room temperature for 6 h. The precipitate was filtered and washed with CH2Cl2 until there was no distinct gray solids and only Pd/C left. The combined organic layers were evaporated under vacuum to give Y2 (9.6 g, 95%) as a gray solid. 1H NMR (400 MHz, DMSO-d6): δ = 7.43 (d, J = 8.5 Hz, 2H),6.96 (d, J = 8.6 Hz, 2H), 3.87 (s, 6H), 3.46 (s, 6H). 13C NMR (101 MHz, DMSO-d6) δ = 166.1, 150.2, 143.2, 126.6, 122.4, 121.3, 108.9, 55.6, 51.0. ESI-HRMS m/z: calculated C18H19O8[M+H]+ 363.1074, found 363.1071.
Methyl 10-(allyloxy)-4,9-dimethoxy-6-oxo-6H-benzo[c]chromene-1-carboxylate (Y3). Compound Y2 (9.6 g, 0.026 mol) was dissolved in DMF (40 mL), added K2CO3 (5.56 g, 0.04 mol), ally bromide (2.78 mL, 0.04 mol) and stirred at 60 °C for 12 h under N2 protection. The mixture was filtered and the residue was washed with CH2Cl2 (120 mL). The organic layers were extracted were washed with H2O (2 × 50 mL), 2 mol/L HCl (2 × 30 mL), brine (2 × 30 mL) and concentrated. The residue was purified by chromatographed on silica gel (PE:EtOAc = 5:1) to give Y3 (4.2 g, 43%) as a white solid. 1H NMR (400 MHz, CDCl3): δ = 8.19 (d, J = 8.7 Hz, 1H),7.63 (d, J = 8.5 Hz, 1H), 7.20 (d, J = 8.7, 1H), 7.02 (d, J = 8.5Hz, 1H), 5.80 (dd, J = 17, 10.4 Hz, 1H), 5.07–4.98 (m, 2H), 4.23 (d, J = 6.5 Hz, 2H), 4.01 (s, 3H),3.99 (s, 3H), 3.75 (s, 3H). 13C NMR (101 MHz, CDCl3): δ = 168.6, 160.2, 157.9, 149.8, 144.1, 141.3, 133.1, 129.0, 127.4, 126.1, 124.6, 119.1, 116.0, 115.7, 113.6, 110.6, 75.1, 56.4, 52.1. ESI-HRMS m/z: calculated C20H19O7[M+H]+ 371.1125, found 371.1118.
3,8-Dimethoxy-1-(prop-2-enyl)[1]benzopyrano[5,4,3-cde][1]benzopyran-5,10-dione (Y4). Compound Y3 (4.2 g) was stirred at 210–220 °C for 50 min under N2 protection. The residue was purified by column dichloromethane (CH2Cl2) to give Y4 (2.68 g, 70%) as a white solid. 1H NMR (400 MHz, CDCl3): δ = 8.21 (d, J = 8.7 Hz, 1H),7.31 (d, J = 8.7 Hz, 1H), 7.14 (s, 1H), 6.12–6.06 (m, 1H),5.17–5.10 (m, 2H), 4.13–4.12 (m, 2H), 4.11 (s, 3H), 4.10 (s, 3H). 13C NMR (101 MHz, CDCl3): δ = 151.4, 144.0, 136.1, 127.1, 117.0, 115.8, 113.8, 111.8, 56.7, 56.6, 38.9. ESI-HRMS m/z: calculated C19H15O6[M+H]+ 339.0863, found 339.0855.
3,8-Dimethoxy-1-(2-oxoethyl)[1]benzopyrano[5,4,3-cde][1]ben-zopyran-5,10-dione (Y5). To a suspension of compound Y4 (240 mg, 0.7 mmol) in a mixture of CH2Cl2, H2O, and t-BuOH (50 mL, 3:1:1) was added OsO4 (20 μL, 1 g/mL), 4-methylmorpholine N-oxide (100 μL, 50% w/w). The mixture was stirred at room temperature for 2 h in dark. NaIO4 (600 mg, 2.8 mmol) was added to the above mixture and stirred at room temperature for another 20 h in dark. The mixture was evaporated under vacuum to remove the organic solvent, and then filtration. The residue was purified by column chromatography (CH2Cl2: MeOH) to Y5 (96 mg, 40%) as a white solid. 1H NMR (400 MHz, CF3COOD): δ = 11.43 (s, H), 9.90 (d, J = 8.7 Hz, 1H), 9.04 (d, J = 8.7 Hz, 2H), 6.09 (s, 2H), 5.66 (s, 6H). 13C NMR (101 MHz, CF3COOD): δ = 206.6, 154.2, 153.9, 138.4, 138.3, 130.2, 121.4, 119.6, 117.7, 116.7, 116.0, 114.9, 113.9, 111.5, 111.1, 110.2, 57.5, 57.4. ESI-HRMS m/z: calculated C18H13O7[M+H]+ 341.0655, found 341.0651.
General procedure for the synthesis of compounds Y6–Y22, 1H NMR, 13C NMR spectrum and HRMS (ESI) of all the compounds please find in Supporting Information Note S1.
Acknowledgments
This study was supported by Foundation National Natural Science Foundation of China (Nos. 82073689, 82273762), National Key Research and Development Program of China (No. 2022YFC2304203), Key Program of National Natural Science Foundation of China (No. 82130101), National Natural Science Foundation of Guangdong Province (No. 2018B030312010, China) and Science and Technology Program of Guangzhou (No. 201904010380, China).
Author contributions
Conceptualization: Junjie Yang, Yue Pan, Shuwen Liu, Zhipeng Chen and Kui Cheng; Designed and performed research: Junjie Yang, Yue Pan, Shuwen Liu, Zhipeng Chen and Kui Cheng; Data analysis: Junjie Yang, Yue Pan and Kui Cheng; Manuscript writing, review and editing: Junjie Yang, Yue Pan, Shuwen Liu, Xiaoshan Zeng, Zhipeng Chen and Kui Cheng.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.05.034.
Contributor Information
Shuwen Liu, Email: Liusw@smu.edu.cn.
Zhipeng Chen, Email: czpwyq@smu.edu.cn.
Kui Cheng, Email: Chengk@smu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Netea M.G., Joosten L.A., Latz E., Mills K.H., Natoli G., Stunnenberg H.G., et al. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khader S.A., Divangahi M., Hanekom W., Hill P.C., Maeurer M., Makar K.W., et al. Targeting innate immunity for tuberculosis vaccination. J Clin Invest. 2019;129:3482–3491. doi: 10.1172/JCI128877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messina N.L., Zimmermann P., Curtis N. The impact of vaccines on heterologous adaptive immunity. Clin Microbiol Infect. 2019;25:1484–1493. doi: 10.1016/j.cmi.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Netea M.G., van Crevel R. BCG-induced protection: effects on innate immune memory. Semin Immunol. 2014;26:512–517. doi: 10.1016/j.smim.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Zindel J., Kubes P. DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu Rev Pathol. 2020;15:493–518. doi: 10.1146/annurev-pathmechdis-012419-032847. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patra M.C., Choi S. Recent progress in the development of toll-like receptor (TLR) antagonists. Expert Opin Ther Pat. 2016;26:719–730. doi: 10.1080/13543776.2016.1185415. [DOI] [PubMed] [Google Scholar]
- 8.Kawai T., Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z., Zhang L., Yang J., Zheng L., Hu F., Duan S., et al. Design, synthesis, and structure‒activity relationship of N-aryl-N′-(thiophen-2-yl)thiourea derivatives as novel and specific human TLR1/2 agonists for potential cancer immunotherapy. J Med Chem. 2021;64:7371–7389. doi: 10.1021/acs.jmedchem.0c02266. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Su L., Morin M.D., Jones B.T., Mifune Y., Shi H., et al. Adjuvant effect of the novel TLR1/TLR2 agonist Diprovocim synergizes with anti-PD-L1 to eliminate melanoma in mice. Proc Natl Acad Sci U S A. 2018;115:E8698–E8706. doi: 10.1073/pnas.1809232115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cen X., Zhu G., Yang J., Yang J., Guo J., Jin J., et al. TLR1/2 specific small-molecule agonist suppresses leukemia cancer cell growth by stimulating cytotoxic T lymphocytes. Adv Sci. 2019;6 doi: 10.1002/advs.201802042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng K., Gao M., Godfroy J.I., Brown P.N., Kastelowitz N., Yin H. Specific activation of the TLR1‒TLR2 heterodimer by small-molecule agonists. Sci Adv. 2015;1 doi: 10.1126/sciadv.1400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cen X., Wang B., Liang Y., Chen Y., Xiao Y., Du S., et al. Small molecule SMU-CX24 targeting toll-like receptor 3 counteracts inflammation: a novel approach to atherosclerosis therapy. Acta Pharm Sin B. 2022;12:3667–3681. doi: 10.1016/j.apsb.2022.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arjumand S., Shahzad M., Shabbir A., Yousaf M.Z. Thymoquinone attenuates rheumatoid arthritis by downregulating TLR2, TLR4, TNF-α, IL-1, and NF-κB expression levels. Biomed Pharmacother. 2019;111:958–963. doi: 10.1016/j.biopha.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Rangasamy S.B., Jana M., Roy A., Corbett G.T., Kundu M., Chandra S., et al. Selective disruption of TLR2–MyD88 interaction inhibits inflammation and attenuates Alzheimer's pathology. J Clin Invest. 2018;128:4297–4312. doi: 10.1172/JCI96209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z., Trippler M., Real C.I., Werner M., Luo X., Schefczyk S., et al. Hepatitis B virus particles activate toll-like receptor 2 signaling initially upon infection of primary human hepatocytes. Hepatology. 2020;72:829–844. doi: 10.1002/hep.31112. [DOI] [PubMed] [Google Scholar]
- 17.Clanchy F.I.L., Borghese F., Bystrom J., Balog A., Penn H., Hull D.N., et al. TLR expression profiles are a function of disease status in rheumatoid arthritis and experimental arthritis. J Autoimmun. 2021;118 doi: 10.1016/j.jaut.2021.102597. [DOI] [PubMed] [Google Scholar]
- 18.Durcan L., O'Dwyer T., Petri M. Management strategies and future directions for systemic lupus erythematosus in adults. Lancet. 2019;393:2332–2343. doi: 10.1016/S0140-6736(19)30237-5. [DOI] [PubMed] [Google Scholar]
- 19.Cheng K., Wang X., Zhang S., Yin H. Discovery of small molecule inhibitors of the TLR1–TLR2 complex. Angew Chem Int Ed. 2012;51:12246–12249. doi: 10.1002/anie.201204910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mistry P., Laird M.H., Schwarz R.S., Greene S., Dyson T., Snyder G.A., et al. Inhibition of TLR2 signaling by small molecule inhibitors targeting a pocket within the TLR2 TIR domain. Proc Natl Acad Sci U S A. 2015;112:5455–5460. doi: 10.1073/pnas.1422576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bi J., Wang W., Du J., Chen K., Cheng K. Structure‒activity relationship study and biological evaluation of SAC-Garlic acid conjugates as novel anti-inflammatory agents. Eur J Med Chem. 2019;179:233–245. doi: 10.1016/j.ejmech.2019.06.059. [DOI] [PubMed] [Google Scholar]
- 22.Grabowski M., Murgueitio M.S., Bermudez M., Rademann J., Wolber G., Weindl G. Identification of a pyrogallol derivative as a potent and selective human TLR2 antagonist by structure-based virtual screening. Biochem Pharmacol. 2018;154:148–160. doi: 10.1016/j.bcp.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 23.Wietzorrek G., Drexel M., Trieb M., Santos-Sierra S. Anti-inflammatory activity of small-molecule antagonists of Toll-like receptor 2 (TLR2) in mice. Immunobiology. 2019;224:1–9. doi: 10.1016/j.imbio.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Zhong Z., Liu L.J., Dong Z.Q., Lu L., Wang M., Leung C.H., et al. Structure-based discovery of an immunomodulatory inhibitor of TLR1-TLR2 heterodimerization from a natural product-like database. Chem Commun. 2015;51:11178–11181. doi: 10.1039/c5cc02728d. [DOI] [PubMed] [Google Scholar]
- 25.Liang Q., Wu Q., Jiang J., Duan J., Wang C., Smith M.D., et al. Characterization of sparstolonin B, a Chinese herb-derived compound, as a selective Toll-like receptor antagonist with potent anti-inflammatory properties. J Biol Chem. 2011;286:26470–26479. doi: 10.1074/jbc.M111.227934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J., Hu F., Guo C., Liang Y., Song H., Cheng K. Discovery of isoliquiritigenin analogues that reverse acute hepatitis by inhibiting macrophage polarization. Bioorg Chem. 2021;114 doi: 10.1016/j.bioorg.2021.105043. [DOI] [PubMed] [Google Scholar]
- 27.Torre B.G., Albericio F. The pharmaceutical industry in 2020. An analysis of FDA drug approvals from the perspective of molecules. Molecules. 2021;26:627. doi: 10.3390/molecules26030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torre B.G., Albericio F. The pharmaceutical industry in 2021. An analysis of FDA drug approvals from the perspective of molecules. Molecules. 2022;27:1075. doi: 10.3390/molecules27031075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Zhang Y., Pan X., Wang S., He L. Synthesis and cytotoxic evaluation of novel symmetrical taspine derivatives as anticancer agents. Med Chem. 2011;7:286–294. doi: 10.2174/157340611796150914. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J., Zhang Y., Zhang S., Wang S., He L. Discovery of novel taspine derivatives as antiangiogenic agents. Bioorg Med Chem Lett. 2010;20:1797. doi: 10.1016/j.bmcl.2009.11.073. [DOI] [PubMed] [Google Scholar]
- 31.Nadzirin I.B., Fortuny-Gomez A., Ngum N., Richards D., Ali S., Searcey M., et al. Taspine is a natural product that suppresses P2X4 receptor activity via phosphoinositide 3-kinase inhibition. Br J Pharmacol. 2021;178:4859–4872. doi: 10.1111/bph.15663. [DOI] [PubMed] [Google Scholar]
- 32.He H., Zhan Y., Zhang Y., Zhang J., He L. Synthesis of novel taspine diphenyl derivatives as fluorescence probes and inhibitors of breast cancer cell proliferation. Luminescence. 2012;27:310–314. doi: 10.1002/bio.1331. [DOI] [PubMed] [Google Scholar]
- 33.Li M., Sun X., Zhao J., Xia L., Li J., Xu M., et al. CCL5 deficiency promotes liver repair by improving inflammation resolution and liver regeneration through M2 macrophage polarization. Cell Mol Immunol. 2020;17:753–764. doi: 10.1038/s41423-019-0279-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsolmongyn B., Koide N., Jambalganiin U., Odkhuu E., Naiki Y., Komatsu T., et al. A Toll-like receptor 2 ligand, Pam3CSK4, augments interferon-γ-induced nitric oxide production via a physical association between MyD88 and interferon-γ receptor in vascular endothelial cells. Immunology. 2013;140:352–361. doi: 10.1111/imm.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller E., Christopoulos P.F., Halder S., Lunde A., Beraki K., Speth M., et al. Toll-like receptor ligands and interferon-γ synergize for induction of antitumor M1 macrophages. Front Immunol. 2017;8:1383. doi: 10.3389/fimmu.2017.01383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 37.Hart E.M., Mitchell A.M., Konovalova A., Grabowicz M., Sheng J., Han X., et al. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc Natl Acad Sci U S A. 2019;116:21748–21757. doi: 10.1073/pnas.1912345116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin M.S., Kim S.E., Heo J.Y., Lee M.E., Kim H.M., Paik S.G., et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Arebro J., Tengroth L., Razavi R., Kumlien Georén S., Winqvist O., Cardell L.O. Antigen-presenting epithelial cells can play a pivotal role in airway allergy. J Allergy Clin Immunol. 2016;137:957–960. doi: 10.1016/j.jaci.2015.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cutolo M., Campitiello R., Gotelli E., Soldano S. The role of M1/M2 macrophage polarization in rheumatoid arthritis synovitis. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.867260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng B., Chai J., Deng Z., Ye T., Chen W., Li D., Chen X., et al. Functional characterization of a novel lipopolysaccharide-binding antimicrobial and anti-inflammatory peptide in vitro and in vivo. J Med Chem. 2018;61:10709–10713. doi: 10.1021/acs.jmedchem.8b01358. [DOI] [PubMed] [Google Scholar]
- 42.Kolaczkowska E., Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen C.T., Furuya H., Das D., Marusina A.I., Merleev A.A., Ravindran R., et al. Peripheral γδ T cells regulate neutrophil expansion and recruitment in experimental psoriatic arthritis. Arthritis Rheumatol. 2022;74:1524–1534. doi: 10.1002/art.42124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.lantis K., Polykratis A., Welz P.S., van Loo G., Pasparakis M., Wullaert A. TLR-independent anti-inflammatory function of intestinal epithelial TRAF6 signalling prevents DSS-induced colitis in mice. Gut. 2016;65:935–943. doi: 10.1136/gutjnl-2014-308323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irizarry-Caro R.A., McDaniel M.M., Overcast G.R., Jain V.G., Troutman T.D., Pasare C. TLR signaling adapter BCAP regulates inflammatory to reparatory macrophage transition by promoting histone lactylation. Proc Natl Acad Sci U S A. 2020;117:30628–30638. doi: 10.1073/pnas.2009778117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.