Abstract
The formation of learning and memory is regulated by synaptic plasticity in hippocampal neurons. Here we explored how gestational exposure to dexamethasone, a synthetic glucocorticoid commonly used in clinical practice, has lasting effects on offspring's learning and memory. Adult offspring rats of prenatal dexamethasone exposure (PDE) displayed significant impairments in novelty recognition and spatial learning memory, with some phenotypes maintained transgenerationally. PDE impaired synaptic transmission of hippocampal excitatory neurons in offspring of F1 to F3 generations, and abnormalities of neurotransmitters and receptors would impair synaptic plasticity and lead to impaired learning and memory, but these changes failed to carry over to offspring of F5 and F7 generations. Mechanistically, altered hippocampal miR-133a-3p-SIRT1-CDK5-NR2B signaling axis in PDE multigeneration caused inhibition of excitatory synaptic transmission, which might be related to oocyte-specific high expression and transmission of miR-133a-3p. Together, PDE affects hippocampal excitatory synaptic transmission, with lasting consequences across generations, and CDK5 in offspring's peripheral blood might be used as an early-warning marker for fetal-originated learning and memory impairment.
Key words: Dexamethasone, Early-warning marker, Hippocampus, Histon acetylation, Learning and memory impairment, MicroRNA, Synaptic transmission, Transgenerational inheritance
Graphical abstract
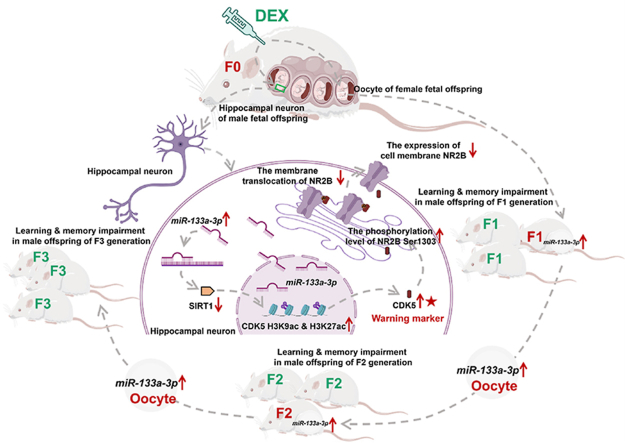
Hippocampal miR-133a-3p–SIRT1–CDK5–NR2B signaling axis mediates transgenerational effects on learning and memory in PDE offspring. CDK5 in offspring's blood may be an early-warning marker for fetal-originated learning and memory impairment.
1. Introduction
Dexamethasone (DEX) is a synthetic glucocorticoid commonly used clinically for the treatment of conditions associated with preterm birth1. Studies have shown that prenatal application of DEX has a “double-edged sword” effect, which can not only effectively prevent and treat preterm birth-related diseases, but also cause a series of developmental toxicities such as fetal or intrauterine growth restriction (FGR/IUGR) and impaired neurogenesis in offspring2, 3, 4. Clinical studies have shown that IUGR fetuses display intrauterine neurodevelopmental delay, low postnatal intelligence index, and altered executive functions related to long-term and short-term memory5, 6, 7. Children treated with dexamethasone before birth also have shown some cognitive deficits8,9. Animal studies in our and other laboratories have also demonstrated that prenatal dexamethasone exposure (PDE) can induce neurobehavioral changes in adult offspring10,11. However, the mechanistic underpinnings linking early-life DEX exposure and later behavioral outcomes in life has lagged these findings.
The hippocampus is a highly sensitive brain region that is primarily responsible for the production and transformation of learning and memory. Numerous animal models of brain injury or related disorders have shown that sustained learning and memory impairment is associated with damaged excitatory synaptic transmission in the hippocampal region12,13. NR2B, an important subtype of N-methyl-d-aspartate receptor (NMDAR), is highly expressed in hippocampal pyramidal and granule cells and plays an important role in hippocampal excitatory synaptic transmission; inhibition of its activity can reduce excitatory postsynaptic currents and inhibit excitatory synaptic function14. Cyclin-dependent kinase 5 (CDK5) is a proline-guided serine/threonine kinase that plays a variety of biological roles by phosphorylating different substrates, including participating in synaptogenesis and transmission, neuronal migration, and so on15. Abnormal activation of CDK5 in a variety of different conditions has been shown to be involved in hippocampal synaptic injury and learning and memory impairment16,17. CDK5-dependent phosphorylation of NR2B has been reported to reduce the expression of cell membrane NR2B, thereby attenuating synaptic transmission in stressed rats18.
Given that PDE may have lasting effects on learning and memory in offspring, we investigated whether fetal exposure to DEX affected synaptic transmission in hippocampal neurons. We determined that hippocampal neurons were particularly sensitive to PDE and showed that impaired synaptic transmission in hippocampal neurons had lasting effects on learning and memory that persisted across multi-generation. As an important epigenetic regulatory mechanism, microRNA (miRNA) plays a vital role in mediating multigenerational inheritance19. We further revealed the molecular mechanism of hippocampal CDK5/NR2B signaling changes induced by miR-133a-3p overexpression in mediating the multi-generation of learning and memory impairment in PDE offspring, which might be associated with oocyte-specific miR-133a-3p overexpression and transmission. At the same time, it is proposed that the detection of CDK5 level in the peripheral blood of offspring may be used as an early warning marker for PDE-related fetal-originated learning and memory impairment.
2. Materials and methods
2.1. Experimental design
This study was designed to assess the effects of gestational exposure to clinically relevant doses of DEX delivered via maternal rat injection to mimic human exposure levels, on the developing hippocampus of offspring. All animal and experimental protocols were approved by the Animal Experiment Ethics Committee of Wuhan University School of Medicine (Permit: 2017-0018) and followed the Guidelines for the Care and Use of Laboratory Animals of the Chinese Animal Welfare Committee. The number of samples was determined based on the experimental methods, availability, and feasibility required to obtain results and was described in the detailed methods or results below. Male animals were used for all behavioral experiments. Where feasible, all data points were shown in the graphs. To reduce bias in animal experiments, rats were housed by one technician, while different co-authors were in charge of rats’ treatment, sample collection, detection, and data analysis respectively.
2.2. Rat husbandry and breeding offspring
Rats were housed and maintained at the Center for Animal Experiments of Wuhan University (Wuhan, China). Specific pathogen-free Wistar rats from Hubei Provincial Academy of Preventive Medicine (Wuhan, Hubei, China) were paired. The breeding success of females was monitored by vaginal smear every morning, and the day when sperm was observed was the gestational Day (GD) 0. From GD9 to GD20, pregnant rats in the experimental group were subcutaneously injected with 0.2 mg/kg·day of DEX (Shuanghe Pharmaceutical Company, Wuhan, China), while the control group was given an equal volume of normal saline. On GD20, part of the pregnant rats was euthanized under 2% isoflurane anesthesia (Baxter Healthcare Co., Deerfield, IL, USA). Pregnant rats with litter sizes of 8–14 were qualified. Fetal blood and hippocampus were collected, and serum was separated. The hippocampus and blood of all male fetuses in each litter were combined into one independent sample respectively, and reserved in −80 °C for follow-up experiment.
The remaining pregnant rats were raised to normal delivery and produced the F1 generation of offspring. The number of pregnant rats in each group was assigned to 12 (the litter size was comprised from 8 to 14 at birth and the male/female ratio was approximately 1:1). After weaning, randomly 1 male and 2 female offspring of F1 generation from each litter were left to feed ad libitum. Part of the offspring were sacrificed at postnatal Week (PW) 28 after behavioral tests, and the samples including the hippocampus, serum, and peripheral blood mononuclear cells (PBMC) were collected.
The female offspring of F1 generation were outcrossed to wild-type males at PW12 to generate F2 generation for analysis of behavior or treated with pregnant mare serum gonadotropin (PMSG, 40 IU, Head Biotechnology Co., Ltd., Beijing, China) and human chorionic gonadotropin (hCG, 40 IU, ProSpec-Tany TechnoGene Ltd., Ness-Ziona, Israel) to obtain oocytes. The female offspring of F2 generation were given the same treatment to obtain the offspring of F3 generation and the oocytes of F2 generation. Similarly, F4–F7 generations were obtained in sequence according to the above method. The mating method was shown briefly as in Fig. 1A1.
Figure 1.
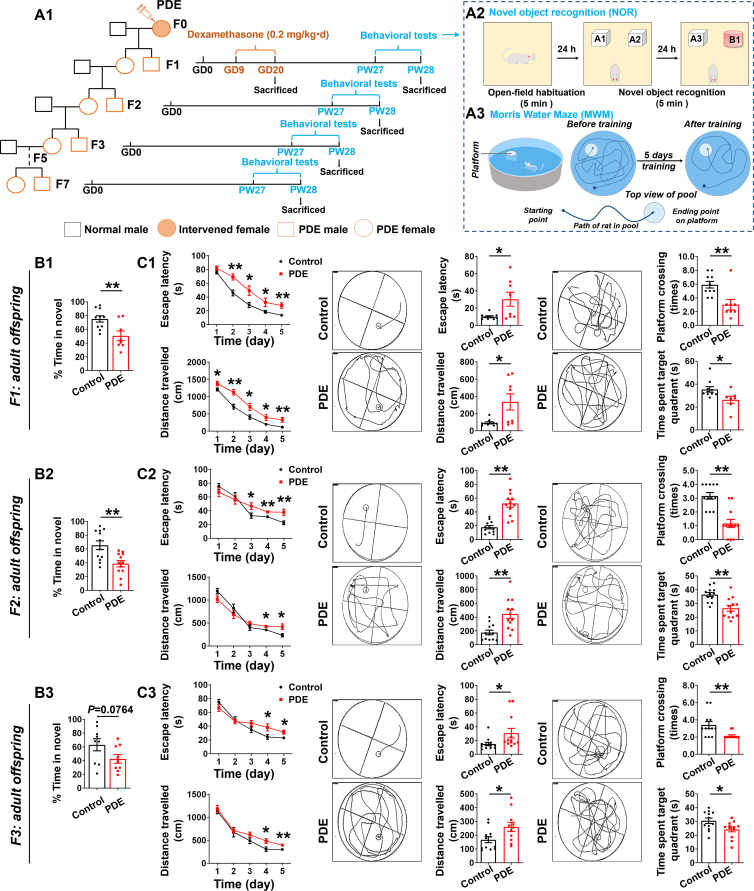
Prenatal dexamethasone exposure (PDE) caused learning and memory impairment in male adult offspring rats with transgenerational inheritance effects. (A1) PDE treatment model and multigenerational transmission via maternal-line. GD, gestational day; PW, postnatal week. (A2, A3) Novel object recognition (NOR) and Morris water maze (MWM) pattern diagram. (B1–B3) The discrimination ratio in NOR test (n = 9–12). (C1–C3) (left) The escape latency and movement distance to find the hidden platform during the five consecutive days of the training phase. (middle) Representative swim patterns of rats for spatial orientation during the hidden platform trials. The escape latency and movement distance to find the hidden platform during the hidden platform trials. (right) Representative swim patterns of rats for spatial orientation during the probe trials. The number of swim crossings over the previous platform location and time spent in the previous platform location during the probe trials (n = 11–12). Mean ± SEM; ∗P < 0.05, ∗∗P < 0.01 vs. respective controls. Using unpaired t test.
2.3. CDK5-RNAi by AAV vector injection
The AAV vector used was derived from adenovirus serotype 9. Vectors containing sequences encoding rat CDK5-RNAi (AAV9-CDK5-RNAi: 4.08E12v.g/mL) and relevant control vectors (AAV-EMPTY: 4.08E12v.g/mL) were generated, produced, and purified by Genechem Technology (Shanghai, China). Rats were anesthetized with a mixture of isoflurane and air (5% isoflurane induction, 2.5% isoflurane maintenance), placed in a stereotaxic frame on a heated plate at 37 °C, and the scalp was cut after local disinfection. According to the rat brain in stereotaxic coordinates20, the anterior fontanel (anterior and posterior = 0 mm, beside the sagittal line = 0 mm, under the dura mater = 0 mm) was used as the origin point. 2 μL viral carrier suspension and 1 μL mannitol mixture were injected into the bilateral hippocampus (anterior-posterior, −3.3 mm; Medial-lateral, 1.8 mm; Dorsal-Ventral, −2.6 mm). The pipette was left in place for 5 min after injection to prevent the return of virus particles through the injection tract, and then was slowly exited for the next 5 min. The rats were further subjected to topical penicillin disinfection and skin suture, and then returned to the feeding room. For the ventricular localization injection experiment, two offspring of F1 generation from the same litter were randomly divided into the control AAV group and the CDK5-RNAi group, and a total of four groups of offspring were obtained, with 8–10 animals in each group. Behavioral tests were performed 3–4 weeks after the viral injection.
2.4. Behavioral tests
The offspring of F1, F2, F3, F5, and F7 generations were raised to PW28 and then underwent behavioral testing for 7–10 days. The rats were moved into the behavioral room a week early for adaptive feeding. All behavioral tests were conducted between 8:00 a.m. and 6:00 p.m. The novel object recognition (NOR) experiment was conducted on an open field (80 cm × 80 cm × 40 cm) to evaluate the cognitive ability of rats. The resolution in new object recognition = TB1/(TA3+TB1) (TA3 = the time spent in exploring the object A3; TB1 = the time spent in exploring object B1). A 180 cm diameter and 60 cm height circular black pool were used in the Morris water maze (MWM) test, which was divided into four hypothetical, equal quadrants. A platform (diameter: 10 cm) was placed 1 cm below the water surface in the middle of the target quadrant. The water temperature was controlled at 22 ± 1 °C. As previously described10, for the training test, each rat was placed in a slot from one of four virtual quadrants to find the platform, which was hidden 1 cm below the surface of the water. The rats need to reach the platform within 60 s and stay on the platform for 15 s. The escape incubation period and escape distance were recorded using a video tracking program for analysis. If the rats failed to find the platform within the specified time, they were guided to the target and kept for 15 s, and the escape incubation period was recorded as 60 s. The training tests were carried out twice a day for five consecutive days. For the exploration test, on the 6th day, the platform was removed, and each rat was allowed 60 s to search the pool for the platform. The incubation period of escape and the time spent in the target quadrant of the platform were recorded by the Smart V3.0 intelligent video tracking system (Panlab, Spain) and analyzed. For all behavioral data analyses, there were 8–12 male rats in each group, all from different litters.
2.5. Cell culture and treatment
The H19-7 fetal rat hippocampal neuronal cell line (No. CRL-2526™) was obtained from American Type Culture Collection (ATCC). The details of the H19-7 cell line culture have been described in our previous study21. The cells were treated with 0, 20, 100, and 500 nmol/L DEX for 3 days and then collected for further analysis. To confirm the role of the glucocorticoid receptor (GR), miR-133a-3p and CDK5, the cells were cultured for 3 days with 500 nmol/L DEX and (or) 2.5 μmol/L RU486 (HY-13683, MedchemExpress Biotechnology Inc., USA, a GR antagonist), 500 nmol/L DEX and (or) 10 μmol/L miR-133a-3p inhibitor (RiboBio Technology, Guangzhou, China), or 500 nmol/L DEX and (or) 25 μmol/L Roscovitine (HY-B0231, Medchem Express Biotechnology Inc., a CDK5 inhibitor), respectively, and harvested for subsequent analysis. The miR-133a-3p oligonucleotides were as follows, miR-133a-3p inhibitor: 5′-CAGCUGGUUGAAGGGGACCAAA-3′; inhibitor NC: 5′-CAGUACUUUUGUGUAGUACAAA-3′.
2.6. Zebrafish rearing, microinjection, and detection
Sexually mature zebrafish were reared in a constant temperature environment at 28 °C. One male and two female zebrafish were placed in separate tanks one day in advance. After 12 h, the dividers were removed to fertilize the males and females. The fertilized eggs were collected after 30 min. Morpholino oligonucleotides (MOs) were purchased from Gene Tools (Philomath, OR, USA). The MOs sequences used were as follows, miR-133a MO: 5′-TGATTTGGTTCCATTTTACCAGCTT-3′, control MO: 5′-CCTCTTACCTCAGTTACAATTTATA-3′. MOs were stored in RNase-free water diluted to a concentration of 1 mmol/L and diluted to the appropriate concentration before use22. MOs were injected into the yolk of zebrafish at the single-cell stage using a glass microinjection needle (0.07 mm inner diameter at the tip). The injection volume was 1.5–15 nL. The spontaneous tail curl rate of zebrafish embryos was measured at 24 h post-injection, and embryo death and hatching were observed and recorded every 24 h. At 120 hpf, zebrafish brains were collected. 10–15 zebrafish brains were combined to form an independent sample.
2.7. Human subjects, blood collection, and PBMC isolation
Human blood samples were obtained from Renmin Hospital of Wuhan University, China (Ethics Permit No. Whu2021-jc015). According to medication use during pregnancy, 39 infants were recruited: control male infants who did not receive dexamethasone (n = 18) and male infants whose mothers received a single course of dexamethasone at 24–37 weeks of gestation (n = 21). We compared maternal age, corrected gestational age and birth weight of the infants between the two groups. Patients with any of the following conditions were considered ineligible for the study: (i) Receiving other synthetic glucocorticoids during pregnancy, such as betamethasone and prednisone; (ii) Hereditary disease; (iii) Any other clinical conditions that, in the opinion of the investigator, would render the subject unsuitable for the study. Umbilical cord blood or peripheral blood samples (2–3 mL) were obtained from infants aged 0–1 years with ethylene diamine tetraacetic acid vacuum catheters, and PBMC were immediately separated by peripheral blood lymphocyte isolation solution (Catalog # 17-1440-02, GE) or serum was collected after centrifugation. All the above analyses were performed in a single sample.
2.8. Real-time quantitative PCR (RT-qPCR) assays
Total RNA was extracted from rat hippocampus, H19-7 cells, zebrafish brain and PBMC, using TRIzol reagent (Invitrogen, USA). cDNA was prepared from total RNA using the Reverse Transcription Kit (Takara Biotechnology, Japan). For miRNA, Qiagen miRNA Reverse Transcription Kit (miScript II RT Kit, Qiagen Bio, Germany) was used. All oligonucleotide primers were synthesized by Tianyihuiyuan Biotechnology Co. Primers for miR-133a-3p were designed and synthesized by RiboBio Technology Co. RNU6A (u6) primers were provided by QIAGEN Bio Co. (miScript Primer Assays). The results were calculated using the 2−ΔΔCt method by normalizing with glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and beta-actin (β-actin), or u6 gene. The corresponding primers are shown in Supporting Information Table S1.
2.9. Western blotting assays
Rat hippocampus, H19-7 cells, and zebrafish brain were lysed in RIPA buffer (Beyotime Biotechnology, China) containing phenylmethanesulfonyl fluoride (PMSF, Beyotime Biotechnology, China) and phosphatase inhibitors. Lysates were centrifuged and protein concentrations were determined using the bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology, China). The boiled samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad Lab., Hercules, CA, USA). After soaking in the blocking solution at room temperature for 2 h, the membranes were incubated overnight at 4 °C with the following primary antibodies, including rabbit anti-SIRT1 (dilution at 1:1000, ab189494, Abcam, USA), rabbit anti-CDK5 (dilution at 1:1000, ab40773, Abcam, USA), rabbit anti-NR2B (dilution at 1:1000, ab65783, Abcam, USA), rabbit anti-NR2B Ser1303 (dilution at 1:1000, ab81271, Abcam, USA), mouse anti-GR antibody (dilution at 1:50, sc-12763, Santa Cruz, USA) and rabbit anti-β-actin (dilution at 1:50,000, AC026, ABclonal, China), respectively. Then incubation with appropriate secondary antibodies was performed for 2 h at room temperature and the signal was detected using an enhanced chemiluminescence (ECL) kit (Bridgen, China) and visualized by an imaging system (Bio-ID VL, Conn France). The grayscale of the target protein bands was analyzed with Image-Pro Plus 6.0 (Media Cybernetics, Silver Spring, MD, USA).
2.10. Immunofluorescence analysis
Cells were fixed with 4% paraformaldehyde at 4 °C for 15 min and then were blocked at room temperature for 2 h by adding 10% goat serum-PBS blocking solution. Cells were then incubated with rabbit anti-NR2B antibody (dilution at 1:100, ab65783; Abcam, USA) at 4 °C in a wet box overnight. After washing 5 times with deionized water, a cy3-labeled goat anti-rabbit antibody (B100802, Baiqiandu Inc., China) was added and the dark box was incubated at room temperature for 40–60 min. 4′,6-diamidino-2-phenylindole (DAPI, B0011, Baiqiandu Inc., China) was added and incubated at room temperature in the dark for 20 min. Photographs were taken after washing with deionized water for 15 min.
Brain tissues were fixed in 4% paraformaldehyde for 24 h, paraffin embedded and cut into 2 μm thick sections. Sections were dewaxed and washed in PBS. After antigen repair, the sections were blocked with blocking solution for 2 h. The sections were subsequently incubated with rabbit anti-CDK5 antibody (dilution at 1:100, ab40773, Abcam, USA) or mouse anti-GR antibody (dilution at 1:50, sc-12763, Santa Cruz, USA) at 4 °C overnight, washed and incubated with cy3-labeled goat anti-rabbit antibody (B100802, Baiqiandu Inc., China) or FITC-labeled goat anti-mouse antibody (AS001, Abclonal, China). DAPI (B0011, Baiqiandu Inc., China) was added and incubated at room temperature in the dark for 20 min. The sections were then washed with deionized water for 15 min before filming. Images were collected using inverted fluorescence microscopy for both cells and tissues, and fluorescence intensity was determined by measuring 3 random average optical densities for each sample and quantified using Image-Pro Plus 6.0 (Media Cybernetics, Silver Spring, MD, USA).
2.11. Luciferase reporter assays
H19-7 cells were co-transfected with luciferase reporter plasmids containing wild-type (WT) or mutant (MUT) Sirt1 3′UTR binding sites and miR-133a-3p mimic or negative control of mimic NC. The luciferase reporter plasmids were constructed by GenePharma Technology (Shanghai, China). After transfection, the Firefly/Renilla Luciferase Reporter Assay Kit (MA0518-1; Dalian Meilun Co., Ltd., China) and the Glomax 20/20Luminometer (Promega, USA) were used to measure the Firefly/Renilla Luciferase activity ratio. miR-133a-3p mimic and mimic NC were provided by RiboBio Technology (Guangzhou, China), and the oligonucleotides were as follows, miR-133a-3p mimic: 5′-UUUGGUCCCCUUCAACCAGCUG-3′, 3′-AAACCAGGGGAAGUUGGUCGAC-5′; mimic NC: 5′-UUUGUACUACACAAAAGUACUG-3′, 3′-AAACAUGAUGUGUUUUCAUGAC-5′.
2.12. Chromatin immunoprecipitation (ChIP) assay
Homogenates of hippocampal tissue or H19-7 cells were fixed with 1% formaldehyde for 15 min to cross-link DNA and its associated proteins. Glycine (final concentration of 0.125 mol/L) was added, and the reaction was terminated at 4 °C. Then after shearing the DNA in the lysate to a size of 200–800 bp using ultrasound, the samples were collected by centrifugation and dilution buffer was added. After mixing, 10 μL of supernatant was aspirated as Input and used for Input chromatin normalization. The remaining solution was transferred to a new Eppendorf tube containing Protein G agar beads at 200 μL of solution per tube. The samples were incubated with the following antibodies: 4 μg of acetyl-histone H3 at lys9 (H3K9ac, A7255, ABclonal, China), or 4 μg of H3K27ac (A7253, ABclonal, China), or 4 μg of normal rabbit IgG (AC005, ABclonal, China), respectively, spun at 4 °C overnight, and then eluted in stages. Samples were incubated with 200 μg/mL proteinase K at 65 °C overnight. DNA was subsequently purified and collected using the DNA Purification Kit (639549, Tiangen Biotech Co., China), and the purified DNA was detected by q-PCR. The IgG negative control values were used as the background and the Input values were quantified. The primer sequences were as follows: Forward primers: 5′-TAGGGTGAGGTAAAGGCTGC-3′, Reverse primer: 5′-GGCAGGTCAGTTACTTCCCT-3′.
2.13. miRNA sequencing
The hippocampal tissues of male fetal rats from three different litters in each group and provided them to Genergy (Shanghai, China). The subsequent miRNA sequencing and results analysis were completed by Genergy.
2.14. CDK5 ELISA assays
Serum samples were collected from male fetal rats at GD20, male offspring rats at PW28, and male infants aged 0–1 years old, and CDK5 levels in serum were measured according to the kit instructions (HY31782, HY13509, HYCEZMBIO, China). A four-parameter logistic curve fitting (4-pl) was used to create a standard curve equation, and the concentration values of the samples were calculated from the absorbance (450 nm) of the samples. All the above analyses were performed in a single sample.
2.15. Golgi staining
The Golgi staining kit (FD RapidGolgiStain Kit, PK401) was purchased from FD NeuroTechnologies, lnc. (USA). The rat brains were washed with clean PBS and placed in a mixture of solution A and solution B prepared 24 h in advance. After soaking for 6 h, replaced with new dipping solution. After 2 weeks at room temperature and away from light, the tissues were transferred to solution C. After soaking for 24 h, replaced with new liquid C, and stored at room temperature away from light for 7 days. The frozen microtome prepared brain tissue into slices of 100–200 μm and transferred the slices to gelatin coated slides containing solution C. After drying at room temperature for 3 days, the slides were rinsed with distilled water twice for 4 min each time. The mixture of solution D, solution E and distilled water (1:1:2) was proportioned and the slides were placed in the mixture for 10 min. The slides were then rinsed with distilled water twice for 4 min each time. The sections were dehydrated in 50%, 75%, and 95% ethanol for 4 min per concentration gradient. Then the slices were dehydrated in anhydrous ethanol 4 times for 4 min each time. The sections were transparent in xylene 3 times for 4 min each time. After the film was sealed with tablet sealer, the image of neurons in the hippocampus was collected by orthographic microscopy, and the Image-Pro Plus 6.0 (Media Cybernetics, Silver Spring, MD, USA) was used for quantitative analysis.
2.16. Statistical analysis
SPSS22 (SPSS Science Inc., Chicago, IL, USA) and Prism 8.0 (GraphPad Software, La Jolla, CA, USA) were used to analyze experimental data. All data are expressed as mean ± standard error of mean (SEM). Independent Student's t-test compared means between the control and treated group. Multiple groups comparison analyzed by one-way ANOVA test, and then use Dunnett-t-test to identify significant differences between the two groups. The Pearson correlation analysis was used to analyze the correlation between two indicators. A value of P < 0.05 was considered statistically significant.
3. Results
We established a rat model of gestational DEX exposure based on clinical medication practice, and the F1–F7 multigenerational offspring were obtained through maternal transmission (Fig. 1A1). Due to the difficulty of early diagnosis of preterm birth, pregnant women who do not respond to a single course of DEX treatment are switched from preventive medication to continuous medication, resulting in multiple courses of treatment, sometimes even more than 11 courses23. Combined with the present situation of the treatment, and to avoid the occurrence of early embryo abortion and stillbirth, we chose to use 0.2 mg/kg·day DEX to treat pregnant mice at GD9–GD20. According to the dose conversion relationship between humans and rats (1:6.17), 0.2 mg/kg·day DEX is equivalent to a crowd dose of 0.03 mg/kg·day24, and does not exceed the clinical DEX dosing criteria of 0.05–0.2 mg/kg·day25. The fetal blood DEX concentration of 267 nmol/L was measured in a 0.2 mg/kg·day PDE rat model in our previous study26, so 20, 100, and 500 nmol/L DEX were used to treat H19-7 fetal hippocampal neuron cell line in vitro. Therefore, the timing, dose, or concentration of DEX used in this study in both in vivo and in vitro experiments were reasonable and had clinical practical significance. At the same time, considering that female individuals are more prone to emotional problems under the influence of hormones and sexual cycles, we chose male offspring as the object of this study to clarify the mechanism of learning and memory impairment.
3.1. PDE caused learning and memory impairment in offspring rats and had transgenerational effects
To test whether PDE offspring exhibited hippocampal-dependent changes in learning and memory, we examined the behavior of offspring rats in NOR experiments and MWM tests (Fig. 1A2–A3). In the F1 generation, PDE male offspring at PW28 showed a decline in cognitive ability for novel objects, as assessed by a significant reduction in the time spent in the novelty (red cylinder) in NOR experiments (Fig. 1B1). Further MWM tests showed significantly longer escape latencies and movement distances for the PDE offspring both over 5 consecutive days of training and in place navigation test, indicating their reduced spatial learning and memory ability (Fig. 1C1). In spatial probe test, the reduced number of platform crossings and time spent in the target quadrant of the PDE offspring further confirmed their reduced spatial learning and memory abilities (Fig. 1C1). We went on to examine the behavior of F2 generation at PW28. Compared with the control group, the PDE male offspring of F2 generation spent less time on novel objects in NOR experiments (Fig. 1B2).
In MWM tests for 5 consecutive days of training and place navigation test, the PDE offspring of F2 generation showed significantly prolonged escape latencies and movement distances, as well as reduced number of platform crossings and time spent in the target quadrant in spatial probe test (Fig. 1C2). These suggested that, like the F1 generation, the PDE offspring of F2 generation showed reduced cognitive and memory ability for novelty and spatial learning.
Under PDE, the mother, fetus, and its primordial germ cells were all directly exposed to DEX, so the persistent changes in the F1 and F2 generations caused by PDE are only intergenerational changes. There was no direct DEX exposure in the F3 generation, so the effect of PDE on the F3 generation and subsequent generations was a transgenerational inheritance effect. Therefore, we continued to examine the behavioral changes of F3, F5, and F7 generations at PW28. Compared with the control group, the PDE offspring of F3 generation spent less time staying on novel objects in NOR experiments (Fig. 1B3) and showed prolonged escape latencies and movement distances in training test and place navigation test, as well as reduced number of platform crossings and time spent in the target quadrant of spatial probe test in MWM experiments (Fig. 1C3). However, in the F5 and F7 generations, we did not observe changes for the PDE group in various behavioral tests (Supporting Information Fig. S1A1–S1D1 and S1A2–S1D2). It was suggested that the learning and memory impairment of male adult offspring caused by PDE could persist to the F2 generation and could be transmitted across generations to the F3 generation, but not to the F5 and F7 generations.
3.2. PDE induced changes in hippocampal CDK5/NR2B signaling and synaptic damages in multigenerational adult offspring rats
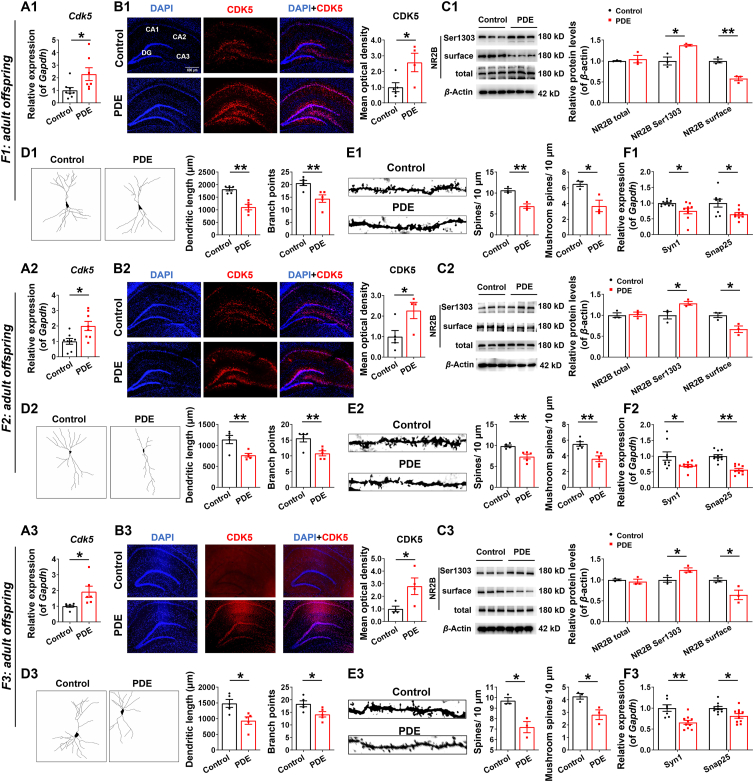
In the offspring of F1 generation at PW28, the mRNA expression of Cdk5 and P35 in hippocampus of PDE group was significantly increased compared with that of control group (Fig. 2A1 and Supporting Information Fig. S2A). Immunofluorescence detection and semi-quantitative analysis showed that CDK5 (red) was mainly expressed in neurons in different regions of the hippocampus, and the expression of CDK5 in hippocampal neurons in the PDE group was higher than that in the control group (Fig. 2B1), suggesting that CDK5 was overactivated in hippocampal neurons of PDE offspring. Protein detection revealed that compared with the control group, the total protein of NR2B in the hippocampus of the PDE group had no change, but the phosphorylation level of NR2B Ser1303 was increased and the expression of the cell membrane NR2B was decreased (Fig. 2C1). The NetPhos-3.1 website was used to predict and found that kinases such as CDK5, protein kinase A (PKA), ribosomal protein S6 kinase (RSK), calcium/calmodulin dependent protein kinase II alpha (CaMK2a), protein kinase C (PKC), and death associated protein kinase 1 (DAPK1), could act at the Ser1303 site of NR2B27,28. It was further found that the mRNA expression of the above kinases (Pka, Rsk, Camk2a, Pkc, and Dapk1) did not change in the hippocampus of PDE offspring (Fig. S2B), except for the increased expression of CDK5, suggesting that CDK5 overactivation might mediate the change of the phosphorylation level of NR2B Ser1303. Subsequent Golgi staining showed that, compared with the control group, the dendritic complexity and the dendritic length of hippocampal neurons were reduced, the number of branching points of neurons was decreased, the density of total dendritic spines and the number of mushroom-type dendritic spines (mature dendritic spines) were also decreased in PDE offspring (Fig. 2D1 and E1). It was also accompanied by a significant inhibition of mRNA expression of hippocampal synaptic proteins-synapsin I (Syn1) and synaptosome-associated protein 25 (Snap25) in PDE offspring (Fig. 2F1).
Figure 2.
Prenatal dexamethasone exposure (PDE) induced changes in hippocampal CDK5/NR2B signaling and synaptic damages in multigenerational adult offspring rats. (A1–A3, F1–F3) mRNA expression of hippocampal cyclin-dependent kinase 5 (Cdk5), synapsin I (Syn1) and synaptosome-associated protein 25 (Snap25) (n = 8–12). (B1–B3) Immunofluorescence co-labeling and quantification for hippocampal CDK5 (red) and 4′,6-diamidino-2-phenylindole (DAPI) (blue) (n = 4–5). Scale bar, 600 μm. (C1–C3) Protein levels of hippocampal N-methyl-d-aspartate receptor (NR2B) (n = 3). (D1–D3, E1–E3) Golgi staining of hippocampal neurons, the neuronal dendrite length, the number of branch points, the density of total dendritic spines and the number of mushroom spines in adult offspring (n = 3–5). Mean ± SEM; ∗P < 0.05, ∗∗P < 0.01 vs. respective controls. Using unpaired t test.
Like the F1 generation, the mRNA expression and immunofluorescence semi-quantification of CDK5, as well as the mRNA expression of P35 in the hippocampus of PDE adult offspring of F2 and F3 generations were significantly increased (Fig. 2A2, B2, A3 and B3 and Fig. S2A). Protein detection also showed no changes in the total protein of hippocampal NR2B, but the phosphorylation level of NR2B Ser1303 was increased and the expression of the cell membrane NR2B was decreased (Fig. 2C2 and C3). Moreover, the dendritic complexity and the dendritic length of hippocampal neurons were reduced, the number of branching points of neurons was decreased, while the density of total dendritic spines and the number of mushroom-type dendritic spines (mature dendritic spines) were also decreased (Fig. 2D2, E2, D3 and E3) in the hippocampus of PDE adult offspring of F2 and F3 generations, and accompanied by a significant decrease in hippocampal Syn1 and Snap25 mRNA expression (Fig. 2F2 and F3). We did not observe changes in the expression of hippocampal Syn1 and Snap25 in the offspring of F5 and F7 generations (Supporting Information Fig. S1E1 and S1E2). These results suggest that PDE could induce hippocampal CDK5/NR2B signaling changes and synaptic damages in male adult offspring and persisted to F2 and F3 generations, but not to F5 and F7 generations.
3.3. Aberrant epigenetic modification of CDK5 was involved in intrauterine origin and multigenerational programming effects of altered hippocampal CDK5/NR2B signaling in PDE offspring rats
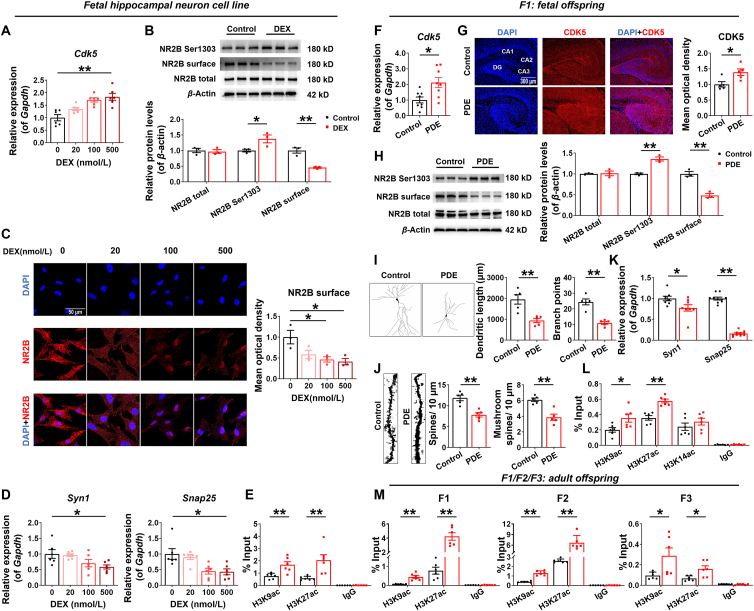
To confirm the direct regulation of DEX on CDK5/NR2B signaling in the fetal hippocampus, we first treated the H19-7 rat fetal hippocampal neuron cell line with different concentrations of DEX (0, 20, 100, 500 nmol/L) for 3 days. Compared with the control group, the mRNA expression of Cdk5 in fetal hippocampal neurons in the DEX-treated group was increased in a concentration-dependent manner (Fig. 3A). The total protein of NR2B was not altered in the 500 nmol/L DEX-treated group, but the phosphorylation level of NR2B Ser1303 was increased and the expression of the cell membrane NR2B was decreased (Fig. 3B). Immunofluorescence assay further showed a DEX concentration-dependent decrease in NR2B membrane expression in fetal hippocampal neurons (Fig. 3C). The mRNA expression of Syn1 and Snap25 also decreased in a DEX concentration-dependent manner (Fig. 3D).
Figure 3.
Aberrant epigenetic modification of CDK5 was involved in intrauterine origin and multigenerational programming effects of altered hippocampal CDK5/NR2B signaling and synaptic damages in male offspring rats of prenatal dexamethasone exposure (PDE). (A, D, F, K) mRNA expression of hippocampal cyclin-dependent kinase 5 (Cdk5), synapsin I (Syn1), synaptosome-associated protein 25 (Snap25) in H19-7 cell line (n = 6) or in fetal rats (n = 8–12). (B, H) Protein levels of hippocampal N-methyl-d-aspartate receptor (NR2B) in H19-7 cell line or fetal rats (n = 3). (C) Immunofluorescence co-labeling and quantification for NR2B (red) and 4′,6-diamidino-2-phenylindole (DAPI) (blue) in H19-7 cell line (n = 3). Scale bar, 50 μm. (E, L, M) Levels of acetyl-histone H3 at lys9 (H3K9ac), H3K14ac, and H3K27ac in the promoter of hippocampal Cdk5 in fetal and adult offspring rats, as well as in H19-7 cell line (n = 6–8). (G) Immunofluorescence co-labeling and quantification for hippocampal CDK5 (red) and DAPI (blue) in fetal rats (n = 5). Scale bar, 300 μm. (I, J) Golgi staining of hippocampal neurons, the neuronal dendrite length, the number of branch points, the density of total dendritic spines and the number of mushroom spines in fetal rats (n = 3–5). Mean ± SEM; ∗P < 0.05, ∗∗P < 0.01 vs. respective controls. Using unpaired t-test compared means between the control and treated groups. The comparisons among multiple groups were analyzed by one-way ANOVA test, followed by Dunnett-t test.
We further examined the changes of CDK5/NR2B signaling in fetal rat hippocampus of F1 generation at GD20. Compared with the control group, the mRNA expression of Cdk5 and P35 in the hippocampus of PDE male fetal rats was significantly increased (Fig. 3F and Fig. S2A), which suggested that CDK5 was also overactivated in hippocampal neurons of PDE fetal rats. Immunofluorescence results showed that the level of CDK5 was higher in the PDE group than in the control group (Fig. 3G). Protein detection also revealed the total protein of NR2B in the fetal hippocampus of the PDE group had no change, but the phosphorylation level of NR2B Ser1303 was increased and the expression of the cell membrane NR2B was decreased (Fig. 3H). Moreover, the dendritic complexity and the dendritic length of fetal hippocampal neurons were reduced, the number of branching points of neurons, the density of total dendritic spines and the number of mushroom-type dendritic spines (mature dendritic spines) were also decreased in PDE group (Fig. 3I and J), accompanied by the inhibited mRNA expression of series hippocampal synaptic proteins, including Syn1 and Snap25 (Fig. 3K). These changes are consistent with multi-generation of adult offspring.
Critical periods of intrauterine development are most susceptible to epigenetic regulation29, and epigenetic mechanisms may well explain the programming and multigenerational transmission of phenotypes associated with intrauterine growth restriction30. Studies have suggested that histone acetylation-mediated regulation of CDK5 expression played a key role in determining neurite length in embryonic neurons31. We screened for altered acetylation modifications at different histone sites in the hippocampal Cdk5 promoter region of fetal rats at GD20 and found increased levels of H3K9ac and H3K27ac in the Cdk5 promoter region of the PDE group, while H3K14ac did not change (Fig. 3L). Simultaneously the increased levels of H3K9ac and H3K27ac persisted until after birth and continued into the F2 and F3 generations (Fig. 3M). In vitro experiments on H19-7 cell line also confirmed a marked increase of H3K9ac and H3K27ac levels in the Cdk5 promoter region in the 500 nmol/L DEX-treated group (Fig. 3E). These effects were consistent with high expression of CDK5, altered CDK5/NR2B signaling and synaptic damages in the in vivo and in vitro experiments. It was suggested that persistent alterations of H3K9ac and H3K27ac in the Cdk5 promoter region might be essential in mediating the intrauterine origin and multigenerational programming effects of altered hippocampal CDK5/NR2B signaling and synaptic damages in PDE offspring.
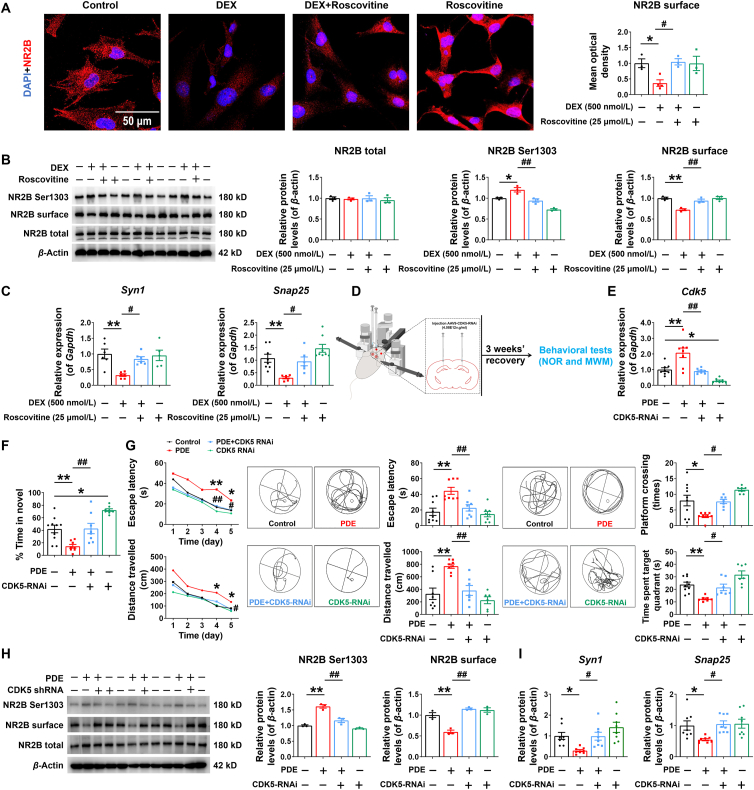
3.4. Intervention of CDK5 could reverse the changes of hippocampal NR2B phosphorylation and learning and memory impairment in PDE offspring rats
To confirm the key regulatory role of CDK5, we first observed the effect of roscovitine, a CDK5 inhibitor, on DEX-induced synaptic damages in H19-7 cell line in vitro. Immunofluorescence assay displayed that the expression of the cell membrane NR2B was lower in the DEX group than of the control group, but it was significantly higher in the DEX + roscovitine group than of the DEX group after 3 days of roscovitine treatment (Fig. 4A). The Western blotting assay showed that the total protein of NR2B was not changed in all groups, but roscovitine reversed the increase of the phosphorylation level of NR2B Ser1303 and the decrease of the expression of cell membrane NR2B induced by DEX (Fig. 4B). Also, roscovitine reversed the DEX-induced reduction in the mRNA expression of Syn1 and Snap25 in fetal hippocampal neurons (Fig. 4C). It was suggested that in vitro intervention of CDK5 reversed the altered NR2B phosphorylation and synaptic damages induced by DEX in fetal hippocampal neurons.
Figure 4.
Intervention of CDK5 could reverse the changes of hippocampal NR2B phosphorylation and learning and memory impairment in male offspring rats of prenatal dexamethasone exposure (PDE). (A–C) The H19-7 cell line was treated with 500 nmol/L DEX and (or) 25 μmol/L roscovitine. (A) Immunofluorescence co-labeling and quantification for N-methyl-d-aspartate receptor (NR2B) (red) and 4′,6-diamidino-2-phenylindole (DAPI) (blue) (n = 3–4). Scale bar, 50 μm. (B) Protein levels of NR2B (n = 3). (C) mRNA expression of Synapsin I (Syn1) and Synaptosome-associated protein 25 (Snap25) (n = 6). (D) Diagram summarizing the experimental procedure. 3 weeks after the appropriate adeno-associated virus (AAV) injection, rats were undergone novel object recognition (NOR) experiments and Morris water maze (MWM) tests. (E) mRNA expression of hippocampal cyclin-dependent kinase 5 (Cdk5) 3 weeks after AAV injection (n = 8). (F) The discrimination ratio in NOR experiment (n = 8–10). (G) (left) The escape latencies and movement distances to find the hidden platform during the five consecutive days of the training phase. (middle) Representative swim patterns of rats for spatial orientation during the hidden platform trials. The escape latencies and movement distances to find the hidden platform during the hidden platform trials. (right) Representative swim patterns of rats for spatial orientation during the probe trials. The number of swim crossings over the previous platform location and time spent in the previous platform location during the probe trials (n = 8–9). (H) Protein levels of hippocampal NR2B (n = 3). (I) mRNA expression of hippocampal Syn1 and Snap25 (n = 8). Mean ± SEM; ∗P < 0.05, ∗∗P < 0.01 vs. Control; #P < 0.05, ##P < 0.01 vs. DEX or PDE. Using one-way ANOVA.
We continued to intervene CDK5 expression by ventricular localization injection of AAV9-CDK5-RNAi in PDE male offspring rats at PW24 and detected behavioral and hippocampus-related indexes three weeks later (Fig. 4D). Compared with the control group, the mRNA expression of hippocampal Cdk5 in the CDK5-RNAi group was decreased (Fig. 4E), confirming the effectiveness of ventricular localization injection of AAV9-CDK5-RNAi. In NOR experiments, the time spent in the novelty of the PDE offspring decreased, while the PDE + CDK5-RNAi group spent considerably longer time in the novelty compared to the PDE group, and the CDK5-RNAi group also showed a significant increase in the dwell time (Fig. 4F). In the training test, the place navigation test and the spatial probe test of MWM tests, PDE offspring showed longer escape latencies and movement distances, as well as reduced number of platform crossings and time spent in the target quadrant, while CDK5-RNAi reversed these changes caused by PDE (Fig. 4G). It was suggested that CDK5-RNAi treatment notably ameliorated the reduced novelty cognition ability and spatial learning and memory impairment in PDE offspring. Subsequently, the Western blotting assay revealed that compared with the PDE group, the total protein of NR2B in the hippocampus of the PDE + CDK5 RNAi group had no change, but the phosphorylation level of NR2B Ser1303 was decreased and the expression of the cell membrane NR2B was increased (Fig. 4H), accompanied by the upregulation of the mRNA expression of hippocampal Syn1 and Snap25 (Fig. 4I).
Combined with the results of CDK5 intervention both in vivo and in vitro, it was suggested that CDK5 could mediate the change of the phosphorylation level of NR2B Ser1303, and silencing of hippocampal CDK5 could significantly reverse the changes of NR2B phosphorylation and learning and memory impairment in offspring induced by PDE.
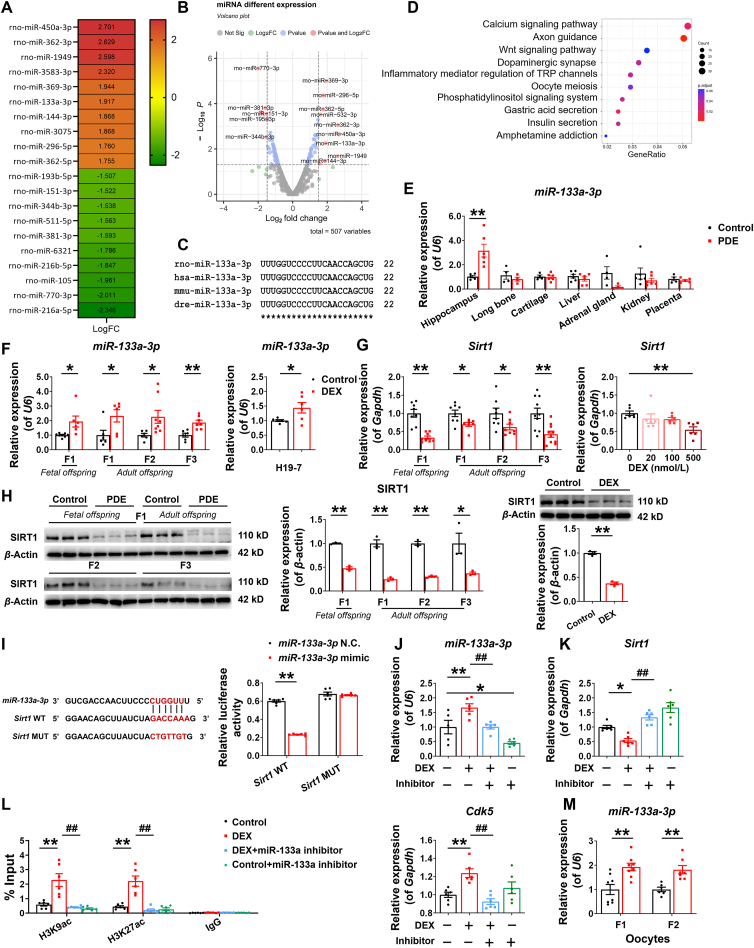
3.5. miR-133a-3p mediated the altered epigenetic modifications of hippocampal CDK5 in multi-generation of PDE offspring rats, which might be related to the high expression and transmission of miR-133a-3p in oocytes
miRNAs are abundantly expressed, target a wide variety of genes, and are highly genetically conserved. To determine whether miRNAs are involved in the regulation of epigenetic modifications of hippocampal CDK5 in the multi-generation of PDE offspring, we sequenced miRNAs in the hippocampus of fetal rats of F1 generation at GD20. We found that 259 miRNAs were up-regulated, and 248 miRNAs were down-regulated in the PDE group, and 97 significantly differentially expressed miRNAs were obtained by calculation. Conservativeness analysis of the top ten differentially paired up-regulated miRNAs by using the miRBase database32 and the Clustal Omega website33 revealed that miR-450a-3p, miR-362-3p, miR-369-3p, miR-133a-3p, miR-144-3p, miR-296-5p, miR-362-5p are well conserved with high sequence homology and are expressed in human and rat, as well as in mouse, where miR-133a-3p, miR-144-3p are also highly conserved in zebrafish (Fig. 5A–C and Fig. S2C).
Figure 5.
The high expression and transmission of oocyte miR-133a-3p might mediate the epigenetic regulation of hippocampal Cdk5 in multi-generation of male offspring rats of prenatal dexamethasone exposure (PDE). (A, B) Heat map and volcano map showing the top 20 differentially expressed miRNAs in the hippocampus of PDE male fetal rats on gestational day 20 (GD20). (C) Sequence conservation of miR-133a-3p in rat, human, mouse, and zebrafish. (D) Signaling enrichment map of miR-133a-3p targeted genes. (E) Relative expression of miR-133a-3p in the hippocampus, long bones, cartilage, liver, adrenal gland, kidney, and placenta in male fetal rats (n = 4–6). (F, G) Relative expression of miR-133a-3p and mRNA expression of Sirt1 in the hippocampus of male fetal and adult offspring rats of F1, F2, and F3 generations, as well as in H19-7 cell line (n = 6–8). (H) Protein levels of SIRT1 in the hippocampus of male fetal and adult offspring rats of F1, F2, and F3 generations, as well as in H19-7 cell line (n = 3). (I) (left) The complementary sequences of miR-133a-3p were discovered in 3′UTR of Sirt1 mRNA using Target Scan; (right) Relative luciferase activity was analyzed after wild-type or mutant 3′UTR reporter plasmids were co-transfected with different plasmids in H19-7 cell line (n = 6). (J–L) Relative expression of miR-133a-3p, mRNA expression of Sirt1 and Cdk5, and the levels of H3K9ac and H3K27ac in the promoter of Cdk5 in the H19-7 cell line treated with 500 nmol/L DEX and (or) 10 μmol/L miR-133a-3p inhibitor (n = 6). (M) Expression of miR-133a-3p in oocytes in adult offspring rats of F1 and F2 generations (n = 6–8). Mean ± SEM; ∗P < 0.05, ∗∗P < 0.01 vs. Control; #P < 0.05, ##P < 0.01 vs. DEX. Using unpaired t test compared means between the control and treated groups. Multiple groups comparison was analyzed by one-way ANOVA test, followed by Dunnett-t-test.
Axon guidance is the process by which neurons send out axons and form synapses at the correct location. We found that only miR-133a-3p was highly associated with neuronal axon guidance among the dozens of significantly variable and highly conserved miRNAs by GO and KEGG pathway enrichment analysis (Fig. 5D and Fig. S2D)34,35. We detected the expression of miR-133a-3p in multiple tissues of fetal rats of F1 generation at GD20 and found that the basal expression of miR-133a-3p was high in hippocampus, long bones, cartilage, and adrenal gland, and low in liver, kidney, and placenta (Fig. S2E). At the same time, the expression of miR-133a-3p in fetal hippocampus was specifically increased under the PDE model (Fig. 5E), but there was no change in other tissues (Fig. 5E and Fig. S2E). The expression of miR-133a-3p in the hippocampus of fetal rats of F1 generation and adult offspring of F1, F2, and F3 generations, were further detected, which were significantly higher than that of the respective control groups (Fig. 5F). In vitro experiments also confirmed that 500 nmol/L DEX could significantly up-regulate the expression of miR-133a-3p in H19-7 neurons (Fig. 5F).
However, is the highly expressed miR-133a-3p involved in the regulation of histone acetylation in the Cdk5 promoter region? Sirtuin (Sirt), a nicotinamide adenosine dinucleotide-dependent deacetylase, is an important enzyme family mediating deacetylation and is involved in the regulation of cell cycle and development, cellular senescence and neuroprotection36,37. We found that the mRNA expression and protein levels of SIRT1 in the hippocampus of PDE fetal rats were decreased (Fig. 5G and H), but the mRNA expression of Sirt2, Sirt3, Sirt4, Sirt5, Sirt6, Sirt7 were not changed (Fig. S2F). At the same time, the expression of hippocampal SIRT1 of adult offspring of F1, F2, and F3 generations also showed consistent inhibitory changes with that of fetal rats of F1 generation (Fig. 5G and H). The expression of SIRT1 in H19-7 cell line also decreased after DEX treatment (Fig. 5G and H). The Target Scan website38 predicted that miR-133a-3p has a binding site for Sirt1 3′UTR (Fig. 5I). Therefore, a dual luciferase reporter gene was used to detect the interaction between Sirt1 3′UTR and miR-133a-3p in H19-7 cell line. It was found that the luciferase activity was decreased in fetal hippocampal neurons transfected with the wild-type Sirt1 construct, compared with the negative control group (Fig. 5I), whereas the luciferase activity remained unchanged in fetal hippocampal neurons transfected with the mut-type Sirt1 construct. These results indicated that miR-133a-3p and Sirt1 had a direct negative regulatory effect. It was further found that intervention of miR-133a-3p (Fig. 5J) in fetal hippocampal neurons could reverse the DEX-induced low expression of Sirt1, the increase of H3K9ac and H3K27ac levels in the Cdk5 promoter region, and the high expression of Cdk5 (Fig. 5K and L). It was preliminary suggested that high expression of miR-133a-3p could mediate the regulation of epigenetic modification and expression of hippocampal Cdk5 in multi-generation of PDE offspring by targeted inhibition of Sirt1.
Substantial evidence suggests that epigenetically induced phenotypic changes can persist for generations39, which means that the memory of these epigenetic changes must be retained during germ cell development and passed on to the next generation. Studies have shown that parental stress-induced changes in the miR-200 family in offspring's uterus extended to the F2 generation, and these changes appeared in both the uterus and the cerebral cortex of the F2 generation40. Our previous studies found that miR-320a-3p and miR-17-5p were inhibited in the oocytes of the F1 and F2 generations directly and indirectly exposed to DEX, which caused abnormal development of ovarian follicles, inhibition of estrogen synthesis function and decreased ovarian reserve, and could persist until F3 generation19,41. To determine whether the persistently high expression of miR-133a-3p in the hippocampus of F2 and F3 generations was associated with oocyte germ cells, we detected the expression of miR-133a-3p in the oocytes of F1 and F2 generations, and found that their expressions were dramatically increased compared to the respective control groups (Fig. 5M), and were consistent with the results in hippocampus. These results suggest that the high expression of miR-133a-3p in offspring induced by PDE is stably inherited in maternal germ cells and may be transmitted to the hippocampus, thereby causing the sustained high expression of miR-133a-3p in the hippocampus of multi-generation, and then participating in the regulation of CDK5 epigenetic modification.
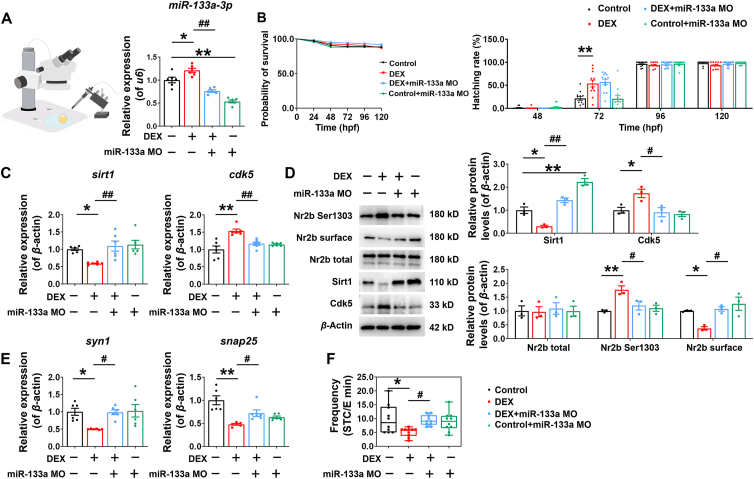
3.6. miR-133a-3p was confirmed to mediate dexamethasone-induced changes in brain SIRT1/CDK5/NR2B signaling axis by using zebrafish embryos
Zebrafish is a powerful model organism for studying vertebrate embryonic development, but the developmental process is accelerated in zebrafish. Zebrafish embryos begin to develop ganglia at 10 h post fertilization (hpf), hatch at 48–72 hpf, and open for foraging at 120 hpf42. Spontaneous tail contraction (STC) in zebrafish is a good parameter reflecting neurodevelopmental and neurobehavioral changes43. We treated zebrafish embryos with 0, 25, 50, and 100 μmol/L DEX at 10–72 hpf and found no changes in the survival of zebrafish embryos in the 25–100 μmol/L DEX treatment group, compared with the 0 μmol/L DEX treatment group. The hatchability increased in a DEX concentration-dependent manner at 48 hpf. The hatchability of the DEX-treated group was higher than that of the control group at 72 hpf, and there was no difference between the groups at 96 hpf (Supporting Information Fig. S3A). At 120 hpf, the expression of miR-133a-3p in the brain of DEX-treated zebrafish was increased, while the expression of sirt1 and cdk5 were inhibited or up-regulated respectively, with the most significant in the 50 μmol/L group (Fig. S3B–S3E). At this time, the phosphorylation level of Nr2b Ser1303 was increased and the expression of the cell membrane Nr2b was decreased (Fig. S3E). Meanwhile, the mRNA expression of brain-derived neurotrophic factor (bdnf), syn1, and snap25 were inhibited (Fig. S3F), and the STC of zebrafish embryos at 24 hpf treated with 50 μmol/L DEX was decreased (Fig. S3G). It was suggested that although DEX could promote the hatching of zebrafish embryos to some extent, the neurotoxicity of 50 μmol/L DEX on zebrafish embryos was like that of PDE rats.
Like humans, early developmental events in fish are regulated by maternally supplied gene products44. The developmental process of zebrafish embryos is like that of humans and rodents. To confirm the transgenerational effect mediated by miR-133a-3p alterations in germ cell, we inhibited miR-133a-3p expression in fertilized eggs by microinjection of miR-133a MO into zebrafish embryos at the zygote stage. Compared with the control group, the expression of miR-133a-3p in zebrafish brain was decreased at 120 hpf (Fig. 6A), confirming that the microinjection of miR-133a MO was effective. At the same time, after injection of miR-133a MO, there was no difference in embryo survival rate among all treatment groups (Fig. 6B). Subsequently, it was found that compared with the DEX group, the expression of sirt1 and cdk5 were inhibited or up-regulated respectively (Fig. 6C and D), the total protein of Nr2b was not changed, the phosphorylation level of Nr2b Ser1303 was decreased while the expression of the cell membrane Nr2b was increased in the DEX + miR-133a MO group (Fig. 6D). In CON + miR-133a MO group, the protein expression of Sirt1 was increased, while the protein expression of Cdk5 and Nr2b did not change (Fig. 6D). In addition, miR-133a MO could reverse DEX-induced down-regulation of syn1 and snap25 mRNA expression (Fig. 6E), as well as the inhibition of STC at 24 hpf (Fig. 6F), while there was no change in CON + miR-133a MO group (Fig. 6F). These results suggest that the intervention of miR-133a-3p in zebrafish zygote could reverse the DEX-induced CDK5/NR2B signaling changes and synaptic function by regulating Sirt1.
Figure 6.
miR-133a-3p was confirmed to mediate dexamethasone-induced changes in brain SIRT1/CDK5/NR2B signaling axis by using zebrafish embryos. (A) (left) Schematic diagram of the microinjection of miR-133a Morpholinos (miR-133a MO) into zebrafish embryos at zygote stage. After microinjection, the zebrafish embryos were treated with 50 μmol/L dexamethasone (DEX) or not; (right) Relative expression of miR-133a-3p in zebrafish embryos at 120-h post fertilization (hpf) (n = 6). (B) Probability of survival and hatching rate of zebrafish embryos at 24, 48, 72, 96, and 120 hpf (n = 12). (C, E) mRNA expression of silent information regulator 1 (sirt1), cyclin-dependent kinase 5 (cdk5), synapsin I (syn1) and synaptosome-associated protein 25 (snap25) in zebrafish at 120 hpf (n = 6). (D) Protein levels of Sirt1, Cdk5 and N-methyl-d-aspartate receptor (Nr2b) in zebrafish at 120 hpf (n = 3). (F) The spontaneous tail contractions (STC) of zebrafish embryos at 24 hpf (n = 10). Mean ± SEM; ∗P < 0.05, ∗∗P < 0.01 vs. Control; #P < 0.05, ##P < 0.01 vs. DEX. Using one-way ANOVA.
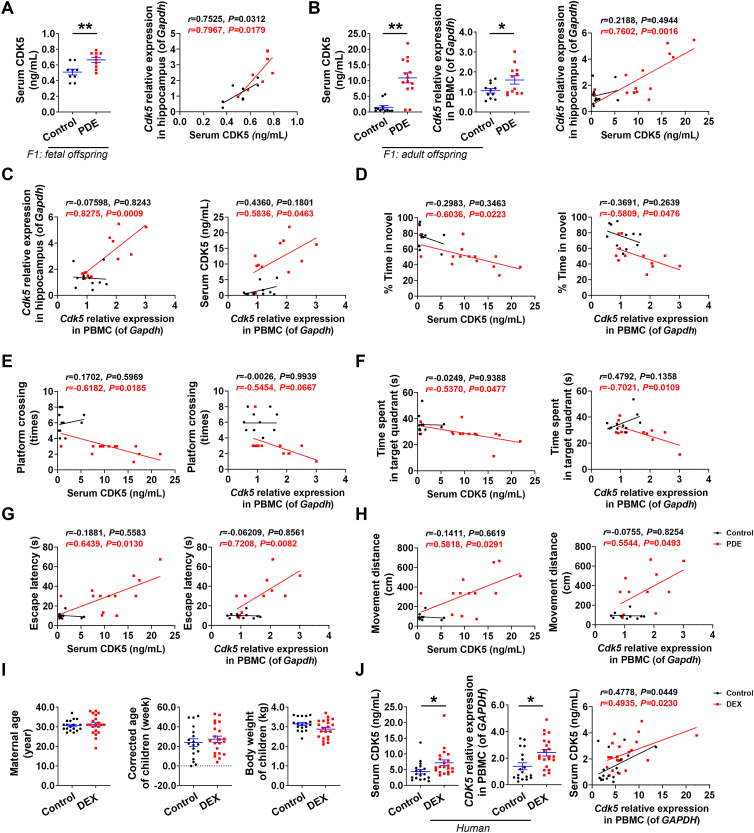
3.7. The level or expression of CDK5 in peripheral blood might serve as a potential early-warning marker for learning and memory impairment in PDE offspring
The exploration of biomarkers provides a direction for the prevention and treatment of complex neurological diseases. The discovery of highly specific peripheral blood biomarkers would greatly improve the early warning and diagnosis of neurological disorders. Based on the key regulatory role of CDK5 in learning and memory impairment in PDE offspring, we attempted to explore the peripheral blood biomarkers of fetal-originated learning and memory impairment. Compared with the control group, the level or expression of CDK5, in the serum of fetal rats at GD20 as well as in the serum and PBMC of offspring rats at PW28, were increased in the PDE group, and they were all positively correlated with the expression of Cdk5 in the hippocampus. The expression of CDK5 in the PBMC was also positively correlated with the level of CDK5 in the serum. The above correlation was more obvious in the PDE group (Fig. 7A–C). Further analysis revealed that the level or expression of CDK5 in the serum and PBMC of the PDE offspring at PW28 were negatively correlated with the time spent in novelty, the number of crossing platforms, and the time spent in the target quadrant, and positively correlated with the escape latencies and movement distances in the behavioral tests. However, there was no correlation in the control group (Fig. 7D–H). These results suggest that the level or expression of CDK5 in the serum and PBMC were well correlated with CDK5 expression in the hippocampus and could reflect the behavioral changes of the offspring with learning and memory impairment.
Figure 7.
The level or expression of CDK5 in peripheral blood might serve as a potential early-warning marker for learning and memory impairment in offspring of prenatal dexamethasone exposure (PDE). (A–C) Serum CDK5 levels and mRNA expression of CDK5 in peripheral blood mononuclear cell (PBMC) (n = 9–14), and the correlation analysis of CDK5 in serum or PBMC and in hippocampus of male offspring rats (n = 7–12). (D–H) The correlation analysis of CDK5 in serum or PBMC and novel object recognition (NOR) experiments or Morris water maze (MWM) tests of male adult offspring rats (n = 11–12). (I) Maternal age, corrected age, and birth weight of clinical infants (n = 18–21). (J) (left) Serum CDK5 levels and mRNA expression of CDK5 in PBMC of clinical male infants. (right) The correlation analysis of CDK5 in serum and in PBMC of clinical male infants (n = 18–21). Mean ± SEM; ∗P < 0.05, ∗∗P < 0.01 vs. Control. Using unpaired t test compared means between the control and treated groups. Pearson correlation analysis was used to analyze the correlation between two indicators.
To further explore the clinical translational potential of this peripheral blood biomarker, we collected plasma samples from 21 male infants under one year of age who received a single course of prenatal DEX and 18 age-matched control infants. Preterm infants perform worse on tests of cognitive and motor development than their full-term peers of similar chronological age45. To avoid underestimating their abilities in early assessments, age is usually adjusted using chronological age minus the number of weeks the child was born premature46. Our statistical analysis of relevant clinical information confirmed that there were no differences in maternal age (30.61 ± 2.73 in the control group, 31.00 ± 4.85 in the DEX group), corrected infant age (23.86 ± 16.23 in the control group, 27.20 ± 14.89 in the DEX group), or infant birth weight (3.14 ± 0.34 in the control group, 2.87 ± 0.47 in the DEX group) between the control and the DEX group, indicating that the basic information of the two groups was consistent (Fig. 7I and Supporting Information Table S2). Further detection of CDK5 level or expression in the serum and PBMC showed that they were higher in the DEX group than that of the control group, and the level or expression of CDK5 in serum and PBMC was positively correlated (Fig. 7J). These results indicate that the effect of PDE on CDK5 was consistent in rat models and humans, and the level or expression of CDK5 in peripheral blood might serve as an early warning marker for PDE-related fetal-originated learning and memory impairment.
4. Discussion
Here, we investigated the effects of clinically equivalent doses of DEX on hippocampal synaptic development and learning and memory function in offspring rats and reported the specific sensitivity of hippocampal excitatory synapses to this synthetic glucocorticoid. Specifically, we observed reduced cell membrane translocation of hippocampal excitatory glutamate receptor NR2B, damaged neuronal dendrites and dendritic spines, inhibited expression of synaptic proteins, and the onset of learning and memory impairments in offspring, using a rather clinically classical usage of prenatal DEX exposure. Despite the lack of sustained DEX exposure, hippocampal excitatory synaptic damage and learning and memory impairments persist over multiple generations, underscoring the lasting effects of DEX on hippocampal excitatory synapses in offspring. In addition, using rat, H19-7 fetal hippocampal neuron cell line, zebrafish model, and clinical peripheral blood samples, we showed that miR-133a-3p-mediated alterations in SIRT1/CDK5/NR2B signaling axis might be involved in hippocampal excitatory synaptic damage and learning and memory impairment in PDE offspring. The specific high expression of miR-133a-3p in oocytes could be an important reason for its transgenerational inheritance. Detection of CDK5 level in peripheral blood might forewarn the susceptibility of PDE offspring to learning and memory impairment. Taken together, these findings served as further evidence that PDE had transgenerational effects on brain development and revealed previously unknown cellular and molecular mechanisms underlying PDE-induced sustained hippocampal developmental damage, providing further evidence to re-examine the pros and cons of current DEX use during pregnancy.
With the deepening of research, the influence of adverse environment during pregnancy on offspring may not be limited to F1 generation, for example, mammalian embryos exposed to adverse environment can cause abnormal lipid metabolism in offspring until F3 generation47. Our previous studies also found that PDE could inhibit ovarian estrogen synthesis function and reduce ovarian reserve in offspring, which could extend to F3 generation19,41. PDE caused increased susceptibility to temporal lobe epilepsy in offspring rats, and lithium chloride-pilocarpine induced seizures accompanied by cognitive impairment could extend to the F2 generation48. These results suggest that PDE could cause persistent damage in offspring for multiple generations, thereby seriously impairing the quality of life of the offspring. We used NOR and MWM to assess the learning and memory abilities of rodents. NOR experiments showed that the PDE male offspring of the F1, F2 and F3 generations exhibited impairment in recognizing novel objects. MWM tests further show that the spatial learning and memory abilities of PDE offspring rats were weakened. In addition, the membrane translocation of the excitatory glutamate receptor NR2B in the hippocampus of the F1, F2 and F3 offspring was reduced, the neuronal dendrites and the dendritic spines were damaged, and the expression of the synaptic markers Syn1 and Snap25 was inhibited. This further refined our understanding of the hippocampus-dependent impairment of persistent learning and memory abilities in PDE offspring.
Various evidence highlighted the synaptic dysregulation in cognitive-related disorders49. Our study reveals a strong long-term effect of PDE on synaptic regulation of hippocampal excitatory neurons in offspring, which was directly related to CDK5. Notably, CDK5 can inhibit NR2B translocation to the cell membrane through direct or indirect phosphorylation, resulting in reduced synaptic plasticity18,50. Exposed of mice to propofol in the early postnatal period increased the phosphorylation of NR2B and decreased its membrane translocation in the hippocampus, accompanied by spatial memory deficits in pubertal period, whereas promoting the expression of the membrane NR2B in the hippocampal neurons improves their learning and memory functions18,51. Like previous results, our data indicate that NR2B phosphorylation and membrane translocation were abnormal in the hippocampus of PDE offspring. The downregulation of CDK5 by AAV not only reversed the expression changes of NR2B, but also alleviated the reduction of excitatory synaptic proteins and learning and memory impairment induced by PDE. Administration of CDK5 inhibitor roscovitine to H19-7 fetal hippocampal neurons similarly reversed the expression changes of NR2B and synaptic damage caused by DEX. These results suggest that CDK5 played an essential role in PDE-induced hippocampal excitatory synaptic inhibition and learning and memory impairment in offspring.
Previous studies have described that the expression of CDK5 in the rodent hippocampus is regulated by histone acetylation modifications in the promoter region52,53. Coincidentally, our data show that the expression of CDK5 and the levels of H3K9ac and H3K27ac in its promoter regions in the hippocampus of the PDE offspring increased continuously from in utero to adulthood in the F1 generation, and then to the F2 and F3 generations. SIRT1 is widely distributed in the rodent brain and is highly expressed in hippocampal neurons54. Sirt1 knockout mice develop hippocampus-dependent learning and memory impairment55. Interestingly, our results show a persistent decrease in hippocampal SIRT1 in PDE offspring from intrauterine to postnatal until F2 and F3 generations. H19-7 cells treated with DEX show similar results. This suggest that persistent changes of SIRT1 in the epigenetic profile may partially explain the persistent histone hyperacetylation status in the CDK5 promoter region and its high expression.
Biogenesis, activity, and degradation of specific miRNAs have been shown to be involved in the regulation of neuroplasticity responsible for learning and memory formation56. The hippocampus-specifically enriched miR-34c mediated learning impairment in Alzheimer's mice by suppressing Sirt1 expression57. Like miR-34c, we screened that the expression of SIRT1 was suppressed in the hippocampus of PDE offspring, which might be mediated by the specific miR-133a-3p transcriptional activation, and this effect could be extended to the F2 and F3 generations. The dual luciferase reporter gene further confirmed the direct negative regulatory effect of miR-133a-3p and Sirt1 in H19-7 cells, and miR-133a-3p inhibitor could reverse the expression changes of Sirt1, CDK5, NR2B and the inhibition of synaptic protein expression caused by DEX. As mentioned previously, spontaneous curly tail movement in zebrafish embryos is a commonly used parameter to assess the effects of exogenous agents on early development of the zebrafish nervous system58. Our data show that DEX induced a decrease in the rate of spontaneous curling in zebrafish embryos at 24 hpf. It was further found that miR-133a-3p is highly conserved in human, rat, mouse and zebrafish, and DEX exposure resulted in upregulation of miR-133a-3p expression and suppression of Sirt1 expression in zebrafish brain, along with aberrant activation of Cdk5, increased phosphorylation level of NR2B and decreased expression of the membrane NR2B, and these changes were consistent with the PDE rat model. These data suggest that altered miR-133a-3p/SIRT1/CDK5/NR2B signaling axis appeared to be an important mechanism for impaired learning and memory in PDE offspring.
Epigenetic modifications in germ cells are closely related to the transgenerational inheritance of altered phenotypic in offspring caused by adverse pregnancy environments59. Evidence suggested that epigenetic marks at specific genomic locations could evade reprogramming during germ cell maturation, thereby supporting transmission of epigenetic changes across generations60,61. Epigenetic information carried by non-coding RNAs, etc., could be tracked in germ cells and passed on to offspring62,63. For example, the offspring and even grandchildren of diabetic mothers exhibited a similar diabetic phenotype, which has been shown to be associated with altered epigenetic modifications and transmission in oocytes64. Mice injected with miR-1 in fertilized eggs developed cardiac hypertrophy in three generations of their offspring and all three generations of sperm showed elevated miR-165. In the maternal cross-generational inheritance of this study, the hippocampal-specific upregulation of miR-133a-3p in PDE male offspring continued from intrauterine to postnatal in F1 generation, until F2 and F3 generation. At the same time, the expression of miR-133a-3p in the oocytes of F1 and F2 generations of PDE offspring also showed a consistent upregulation. This suggested that the high expression of miR-133a-3p in zygotic oocytes of PDE offspring might escape reprogramming during oocyte formation and early embryonic development and was specifically transmitted to the hippocampus of multigeneration. Further, we microinjected miR-133a MO into zygotic zebrafish embryos to suppress miR-133a-3p expression and found that DEX-induced alterations in zebrafish brain miR-133a-3p/Sirt1/Cdk5/Nr2b signaling, synaptic protein expression and the suppression of spontaneous tail curl rate of zebrafish, were all reversed. It was suggested that intervention of miR-133a-3p expression in fertilized eggs could reverse DEX-induced abnormalities in the early neurodevelopment of the offspring. To sum up, PDE could increase the expression of miR-133a-3p in oocytes of offspring, and the epigenetic information carried by miR-133a-3p was transmitted to F2 and F3 generations via oocytes, which could mediate the change of SIRT1/CDK5/NR2B signaling axis by specifically upregulating miR-133a-3p in hippocampus. This might be the underlying molecular basis for the transgenerational genetic effect of learning and memory impairment in PDE offspring.
As a long-acting glucocorticoid, DEX exerts biological effects by activating GR. In this study, the expression of GR and its target genes in PDE fetal rats was significantly up-regulated, while H19-7 cells treated with DEX showed the same changes (Supporting Information Fig. S4A–S4D), suggesting the activation of GR in PDE fetal hippocampal neurons. Activated GR regulates transcription by acting on the glucocorticoid response element (GRE) of the target genes. Bioinformatics analysis revealed the presence of GRE sites in the promoter region of miR-133a-3p precursor transcripts (Fig. S4E). Meanwhile, GR antagonist RU486 could reverse the changes of miR-133a-3p/SIRT1/CDK5/NR2B signaling axis induced by DEX in fetal hippocampal neurons (Fig. S4F–S4J). These results suggest that PDE might upregulate the expression of miR-133a-3p in fetal hippocampus by activating GR, and then regulate the CDK5/NR2B pathway by targeting SIRT1.
Fetal-originated diseases originate from the in uterus, but most of them develop in adulthood, with complex mechanisms and unclear targets, making it difficult for early warning in clinical practice. Studies have demonstrated that altered expression of functionally regulated genes in early life can be used as biomarkers for long-term disease66,67. Unlike invasive methods such as brain biopsy or cerebrospinal fluid analysis, peripheral blood is a clinically accessible biological sample. Despite the influence of the blood–brain barrier, gene expression in peripheral blood can be used for the diagnosis of brain disorders including Alzheimer's disease, schizophrenia, and bipolar disorder68,69. CDK5 has highly specific kinase activity in the nervous system. Studies have confirmed the presence of CDK5 in human and rat monocytes70. Co-culture of HIV-infected monocyte-derived macrophages with primary neurons increases CDK5 activity and induces neuronal death71. This suggests that changes in CDK5 levels in PBMC may reflect neuronal states to some extent. In this study, we observed that the level or expression of CDK5 in peripheral blood serum and PBMC of PDE fetal rats and adult offspring rats were significantly increased, and it was significantly correlated with the expression of Cdk5 in the hippocampus and the behavioral changes related to learning and memory impairment in the offspring. Further, significant increases in CDK5 levels were also detected in serum and/or PBMC of male infants exposed to DEX in utero. It was suggested that the changes of CDK5 in peripheral blood were consistent in PDE offspring rats and infants, and the level or expression of CDK5 in peripheral blood might be an early warning marker for PDE-related fetal-originated learning and memory impairment.
Our study had certain limitations. In this study, it was not known how DEX led to the alteration of oocyte miR-133a-3p, and how miR-133a-3p in oocytes escaped the developing reprogramming and specifically transmitted to the hippocampus of offspring. Typically, differential methylation of imprinted genes can be erased in primordial germ cells and re-methylated de novo during gametogenesis. It has been shown that differential methylation changes in some imprinted genes such as H19 can be transmitted from oocytes to somatic cells72. Whether the transmission effect of oocyte miR-133a-3p escape reprogramming in this study is regulated by differential methylation of imprinted genes remains to be further explored. Genes have a natural self-repair function and can develop a corresponding repair remedy for possible genetic damage. In this study, the F5 and F7 generations of PDE offspring did not show obvious hippocampal synaptic damage and neurobehavioral changes, which might be caused by the repairment of the overexpression of miR-133a-3p, by gene recombination repair, excision repair, and other ways in the genetic process. In addition, although both epidemiological surveys and our studies had confirmed that PDE was an important cause of learning and memory impairment in offspring, and based on the PDE rat model and the peripheral blood samples of infants exposed to intrauterine DEX, we also proposed that the level of CDK5 in peripheral blood was expected to become an early warning marker for fetal-derived learning and memory impairment, but this still needs more clinical samples, including long-term neurobehavioral changes in subjects to be further validated.
5. Conclusions
We confirmed that PDE could cause learning and memory impairment in offspring and had transgenerational genetic effects, that is, PDE upregulated the expression of miR-133a-3p in oocytes and hippocampus in the offspring of F1 generation, while the epigenetic information carried by miR-133a-3p was transmitted to the offspring of F2 and F3 generations via oocytes. By specifically regulating the high expression of miR-133a-3p in the hippocampus, and further by targeting SIRT1 to mediate the increase of histone acetylation level in the CDK5 promoter region and the high expression of CDK5, the latter could increase the phosphorylation of NR2B Ser1303 and reduce the membrane translocation of NR2B, causing the inhibition of excitatory synaptic transmission in the hippocampus. Eventually, learning and memory impairment and transgenerational transmission occurred in the offspring. Detection of CDK5 level in peripheral blood of offspring might be an early warning of PDE-related fetal-originated learning and memory impairment.
Acknowledgments
This study was supported by grants from the National Key R&D Program of China No. 2020YFA0803900 (Hui Wang), and the National Natural Science Foundation of China No. 81973405 (Dan Xu), No. 82122071 (Dan Xu), and No. 82030111 (Hui Wang). We are grateful to the Demonstration Center for Experimental Basic Medicine Education of Wuhan University, for excellent help with the behavioral experiments. We are thankful to the Zebrafish Platform of Wuhan University Taikang Medical School (School of Basic Medical Sciences), for providing the zebrafish animals and microinjection technical support.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.05.013.
Author contributions
Mingcui Luo, Yiwen Yi, and Songqiang Huang conceived these experiments under the guidance of Hui Wang and Dan Xu. Mingcui Luo, Songqiang Huang, Yiwen Yi, and Shiyun Dai completed the behavioral test. Yiwen Yi, Tao Jiang, and Mingcui Luo completed the intraventricular localization injection and related analysis. Mingcui Luo, Yiwen Yi, Tao Jiang, and Lulu Xie completed the cell experiment. Lulu Xie and Baozhen Yao provided clinical samples. Mingcui Luo completed zebrafish-related experiments and clinical sample testing. Shuai Zhang and Mingcui Luo completed miRNA pathway enrichment. Mingcui Luo, Yiwen Yi, Shuai Zhang, Tingting Wang, and Kexin Liu analyzed the data. Mingcui Luo and Yiwen Yi wrote the manuscript with input from all authors. All authors discussed and commented on the results and the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Crowther C.A., McKinlay C.J., Middleton P., Harding J.E. Repeat doses of prenatal corticosteroids for women at risk of preterm birth for improving neonatal health outcomes. Cochrane Database Syst Rev. 2015;2015:Cd003935. doi: 10.1002/14651858.CD003935.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel J.P., Souza J.P., Gülmezoglu A.M., Mori R., Lumbiganon P., Qureshi Z., et al. Use of antenatal corticosteroids and tocolytic drugs in preterm births in 29 countries: an analysis of the WHO Multicountry Survey on Maternal and Newborn Health. Lancet. 2014;384:1869–1877. doi: 10.1016/S0140-6736(14)60580-8. [DOI] [PubMed] [Google Scholar]
- 3.Moisiadis V.G., Matthews S.G. Glucocorticoids and fetal programming part 1: outcomes. Nat Rev Endocrinol. 2014;10:391–402. doi: 10.1038/nrendo.2014.73. [DOI] [PubMed] [Google Scholar]
- 4.Lu Z., Guo Y., Xu D., Xiao H., Dai Y., Liu K., et al. Developmental toxicity and programming alterations of multiple organs in offspring induced by medication during pregnancy. Acta Pharm Sin B. 2023;13:460–477. doi: 10.1016/j.apsb.2022.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malhotra A., Allison B.J., Castillo-Melendez M., Jenkin G., Polglase G.R., Miller S.L. Neonatal morbidities of fetal growth restriction: pathophysiology and impact. Front Endocrinol. 2019;10:55. doi: 10.3389/fendo.2019.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacchi C., De Carli P., Mento G., Farroni T., Visentin S., Simonelli A. Socio-emotional and cognitive development in intrauterine growth restricted (IUGR) and typical development infants: early interactive patterns and underlying neural correlates. Rationale and methods of the study. Front Behav Neurosci. 2018;12:315. doi: 10.3389/fnbeh.2018.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhotra A., Ditchfield M., Fahey M.C., Castillo-Melendez M., Allison B.J., Polglase G.R., et al. Detection and assessment of brain injury in the growth-restricted fetus and neonate. Pediatr Res. 2017;82:184–193. doi: 10.1038/pr.2017.37. [DOI] [PubMed] [Google Scholar]
- 8.Maryniak A., Ginalska-Malinowska M., Bielawska A., Ondruch A. Cognitive and social function in girls with congenital adrenal hyperplasia—influence of prenatally administered dexamethasone. Child Neuropsychol. 2014;20:60–70. doi: 10.1080/09297049.2012.745495. [DOI] [PubMed] [Google Scholar]
- 9.Meyer-Bahlburg H.F., Dolezal C., Haggerty R., Silverman M., New M.I. Cognitive outcome of offspring from dexamethasone-treated pregnancies at risk for congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Eur J Endocrinol. 2012;167:103–110. doi: 10.1530/EJE-11-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S., Dong W., Jiao Z., Liu J., Li K., Wang H., et al. Prenatal dexamethasone exposure induced alterations in neurobehavior and hippocampal glutamatergic system balance in female rat offspring. Toxicol Sci. 2019;171:369–384. doi: 10.1093/toxsci/kfz163. [DOI] [PubMed] [Google Scholar]
- 11.Zeng Y., Brydges N.M., Wood E.R., Drake A.J., Hall J. Prenatal glucocorticoid exposure in rats: programming effects on stress reactivity and cognition in adult offspring. Stress. 2015;18:353–361. doi: 10.3109/10253890.2015.1055725. [DOI] [PubMed] [Google Scholar]
- 12.Wolf J.A., Johnson B.N., Johnson V.E., Putt M.E., Browne K.D., Mietus C.J., et al. Concussion induces hippocampal circuitry disruption in swine. J Neurotrauma. 2017;34:2303–2314. doi: 10.1089/neu.2016.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almeida-Suhett C.P., Prager E.M., Pidoplichko V., Figueiredo T.H., Marini A.M., Li Z., et al. GABAergic interneuronal loss and reduced inhibitory synaptic transmission in the hippocampal CA1 region after mild traumatic brain injury. Exp Neurol. 2015;273:11–23. doi: 10.1016/j.expneurol.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 14.Volianskis A., France G., Jensen M.S., Bortolotto Z.A., Jane D.E., Collingridge G.L. Long-term potentiation and the role of N-methyl-d-aspartate receptors. Brain Res. 2015;1621:5–16. doi: 10.1016/j.brainres.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi G.J., Chen Q., Chen L.J., Shu Y., Bu L.L., Shao X.Y., et al. Phosphorylation of connexin 43 by Cdk5 modulates neuronal migration during embryonic brain development. Mol Neurobiol. 2016;53:2969–2982. doi: 10.1007/s12035-015-9190-6. [DOI] [PubMed] [Google Scholar]
- 16.Murphy J.A., Stein I.S., Lau C.G., Peixoto R.T., Aman T.K., Kaneko N., et al. Phosphorylation of Ser1166 on GluN2B by PKA is critical to synaptic NMDA receptor function and Ca2+ signaling in spines. J Neurosci. 2014;34:869–879. doi: 10.1523/JNEUROSCI.4538-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canet G., Zub E., Zussy C., Hernandez C., Blaquiere M., Garcia V., et al. Seizure activity triggers tau hyperphosphorylation and amyloidogenic pathways. Epilepsia. 2022;63:919–935. doi: 10.1111/epi.17186. [DOI] [PubMed] [Google Scholar]
- 18.Plattner F., Hernández A., Kistler T.M., Pozo K., Zhong P., Yuen E.Y., et al. Memory enhancement by targeting Cdk5 regulation of NR2B. Neuron. 2014;81:1070–1083. doi: 10.1016/j.neuron.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong X., Zhang J., Ge C., Yi Y., Dai S., Fan G., et al. miRNA320a-3p/RUNX2 signal programming mediates the transgenerational inheritance of inhibited ovarian estrogen synthesis in female offspring rats induced by prenatal dexamethasone exposure. Pharmacol Res. 2021;165 doi: 10.1016/j.phrs.2021.105435. [DOI] [PubMed] [Google Scholar]
- 20.Paxinos G., Watson C. 7th ed. Academic Press; 2013. The rat brain in stereotaxic coordinates. [Google Scholar]
- 21.He X., Lu J., Dong W., Jiao Z., Zhang C., Yu Y., et al. Prenatal nicotine exposure induces HPA axis-hypersensitivity in offspring rats via the intrauterine programming of up-regulation of hippocampal GAD67. Arch Toxicol. 2017;91:3927–3943. doi: 10.1007/s00204-017-1996-8. [DOI] [PubMed] [Google Scholar]
- 22.Gan S., Huang Z., Liu N., Su R., Xie G., Zhong B., et al. MicroRNA-140-5p impairs zebrafish embryonic bone development via targeting BMP-2. FEBS Lett. 2016;590:1438–1446. doi: 10.1002/1873-3468.12190. [DOI] [PubMed] [Google Scholar]
- 23.Elfayomy A.K., Almasry S.M. Effects of a single course versus repeated courses of antenatal corticosteroids on fetal growth, placental morphometry and the differential regulation of vascular endothelial growth factor. J Obstet Gynaecol Res. 2014;40:2135–2145. doi: 10.1111/jog.12466. [DOI] [PubMed] [Google Scholar]
- 24.Reagan-Shaw S., Nihal M., Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2008;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 25.Moisiadis V.G., Matthews S.G. Glucocorticoids and fetal programming part 1: outcomes. Nat Rev Endocrinol. 2014;10:391–402. doi: 10.1038/nrendo.2014.73. [DOI] [PubMed] [Google Scholar]
- 26.Lv F., Wan Y., Chen Y., Pei L., Luo D., Fan G., et al. Prenatal dexamethasone exposure induced ovarian developmental toxicity and transgenerational effect in rat offspring. Endocrinology. 2018;159:1401–1415. doi: 10.1210/en.2018-00044. [DOI] [PubMed] [Google Scholar]
- 27.Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 28.Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 29.Hussain N. Epigenetic influences that modulate infant growth, development, and disease. Antioxidants Redox Signal. 2012;17:224–236. doi: 10.1089/ars.2011.4484. [DOI] [PubMed] [Google Scholar]
- 30.Doan T.N.A., Briffa J.F., Phillips A.L., Leemaqz S.Y., Burton R.A., Romano T., et al. Epigenetic mechanisms involved in intrauterine growth restriction and aberrant kidney development and function. J Dev Orig Health Dis. 2021;12:952–962. doi: 10.1017/S2040174420001257. [DOI] [PubMed] [Google Scholar]
- 31.Lee J., Ko Y.U., Chung Y., Yun N., Kim M., Kim K., et al. The acetylation of cyclin-dependent kinase 5 at lysine 33 regulates kinase activity and neurite length in hippocampal neurons. Sci Rep. 2018;8 doi: 10.1038/s41598-018-31785-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manjula R., Anuja K., Alcain F.J. SIRT1 and SIRT2 activity control in neurodegenerative diseases. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.585821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng Y., Huang Y., Yu H., Wu C., Yan Q., Wang Y., et al. Nimbolide targeting SIRT1 mitigates intervertebral disc degeneration by reprogramming cholesterol metabolism and inhibiting inflammatory signaling. Acta Pharm Sin B. 2023;13:2269–2280. doi: 10.1016/j.apsb.2023.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGeary S.E., Lin K.S., Shi C.Y., Pham T.M., Bisaria N., Kelley G.M., et al. The biochemical basis of microRNA targeting efficacy. Science. 2019;366:eaav1741. doi: 10.1126/science.aav1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martos S.N., Tang W.Y., Wang Z. Elusive inheritance: transgenerational effects and epigenetic inheritance in human environmental disease. Prog Biophys Mol Biol. 2015;118:44–54. doi: 10.1016/j.pbiomolbio.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao Y., Robinson A.M., Zucchi F.C., Robbins J.C., Babenko O., Kovalchuk O., et al. Ancestral exposure to stress epigenetically programs preterm birth risk and adverse maternal and newborn outcomes. BMC Med. 2014;12:121. doi: 10.1186/s12916-014-0121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong X., Dai S., Wang T., Zhang J., Fan G., Luo M., et al. MiR-17-5p/FOXL2/CDKN1B signal programming in oocytes mediates transgenerational inheritance of diminished ovarian reserve in female offspring rats induced by prenatal dexamethasone exposure. Cell Biol Toxicol. 2023;39:867–883. doi: 10.1007/s10565-021-09645-6. [DOI] [PubMed] [Google Scholar]
- 42.Jarque S., Rubio-Brotons M., Ibarra J., Ordoñez V., Dyballa S., Miñana R., et al. Morphometric analysis of developing zebrafish embryos allows predicting teratogenicity modes of action in higher vertebrates. Reprod Toxicol. 2020;96:337–348. doi: 10.1016/j.reprotox.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Weichert F.G., Floeter C., Meza Artmann A.S., Kammann U. Assessing the ecotoxicity of potentially neurotoxic substances—evaluation of a behavioural parameter in the embryogenesis of Danio rerio. Chemosphere. 2017;186:43–50. doi: 10.1016/j.chemosphere.2017.07.136. [DOI] [PubMed] [Google Scholar]
- 44.Bertho S., Kaufman O., Lee K., Santos-Ledo A., Dellal D., Marlow F.L. A transgenic system for targeted ablation of reproductive and maternal-effect genes. Development. 2021;148 doi: 10.1242/dev.198010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gould J.F., Fuss B.G., Roberts R.M., Collins C.T., Makrides M. Consequences of using chronological age versus corrected age when testing cognitive and motor development in infancy and intelligence quotient at school age for children born preterm. PLoS One. 2021;16 doi: 10.1371/journal.pone.0256824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engle W.A. Age terminology during the perinatal period. Pediatrics. 2004;114:1362–1364. doi: 10.1542/peds.2004-1915. [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Dominguez X., Marco-Jiménez F., Peñaranda D.S., Diretto G., García-Carpintero V., Cañizares J., et al. Long-term and transgenerational phenotypic, transcriptional and metabolic effects in rabbit males born following vitrified embryo transfer. Sci Rep. 2020;10:11313. doi: 10.1038/s41598-020-68195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu S., Yi Y., Jiang T., Jiao Z., Dai S., Gong X., et al. Intrauterine RAS programming alteration-mediated susceptibility and heritability of temporal lobe epilepsy in male offspring rats induced by prenatal dexamethasone exposure. Arch Toxicol. 2020;94:3201–3215. doi: 10.1007/s00204-020-02796-1. [DOI] [PubMed] [Google Scholar]
- 49.Wang P., Wang F., Ni L., Wu P., Chen J. Targeting redox-altered plasticity to reactivate synaptic function: a novel therapeutic strategy for cognitive disorder. Acta Pharm Sin B. 2021;11:599–608. doi: 10.1016/j.apsb.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S., Edelmann L., Liu J., Crandall J.E., Morabito M.A. Cdk5 regulates the phosphorylation of tyrosine 1472 NR2B and the surface expression of NMDA receptors. J Neurosci. 2008;28:415–424. doi: 10.1523/JNEUROSCI.1900-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Han S., Han R., Su Y., Li J. Propofol-induced downregulation of NR2B membrane translocation in hippocampus and spatial memory deficits of neonatal mice. Brain Behav. 2017;7 doi: 10.1002/brb3.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guan J.S., Haggarty S.J., Giacometti E., Dannenberg J.H., Joseph N., Gao J., et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pascual M., Do Couto B.R., Alfonso-Loeches S., Aguilar M.A., Rodriguez-Arias M., Guerri C. Changes in histone acetylation in the prefrontal cortex of ethanol-exposed adolescent rats are associated with ethanol-induced place conditioning. Neuropharmacology. 2012;62:2309–2319. doi: 10.1016/j.neuropharm.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Zakhary S.M., Ayubcha D., Dileo J.N., Jose R., Leheste J.R., Horowitz J.M., et al. Distribution analysis of deacetylase SIRT1 in rodent and human nervous systems. Anat Rec. 2010;293:1024–1032. doi: 10.1002/ar.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herskovits A.Z., Guarente L. SIRT1 in neurodevelopment and brain senescence. Neuron. 2014;81:471–483. doi: 10.1016/j.neuron.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McNeill E., Van Vactor D. MicroRNAs shape the neuronal landscape. Neuron. 2012;75:363–379. doi: 10.1016/j.neuron.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zovoilis A., Agbemenyah H.Y., Agis-Balboa R.C., Stilling R.M., Edbauer D., Rao P., et al. microRNA-34c is a novel target to treat dementias. EMBO J. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He Y., Jia D., Du S., Zhu R., Zhou W., Pan S., et al. Toxicity of gabapentin-lactam on the early developmental stage of zebrafish (Danio rerio) Environ Pollut. 2021;287:117649. doi: 10.1016/j.envpol.2021.117649. [DOI] [PubMed] [Google Scholar]
- 59.Moisiadis V.G., Constantinof A., Kostaki A., Szyf M., Matthews S.G. Prenatal glucocorticoid exposure modifies endocrine function and behaviour for 3 generations following maternal and paternal transmission. Sci Rep. 2017;7 doi: 10.1038/s41598-017-11635-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perera F., Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reprod Toxicol. 2011;31:363–373. doi: 10.1016/j.reprotox.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei Y., Yang C.R., Wei Y.P., Zhao Z.A., Hou Y., Schatten H., et al. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A. 2014;111:1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bohacek J., Mansuy I.M. A guide to designing germline-dependent epigenetic inheritance experiments in mammals. Nat Methods. 2017;14:243–249. doi: 10.1038/nmeth.4181. [DOI] [PubMed] [Google Scholar]
- 63.Blake G.E., Watson E.D. Unravelling the complex mechanisms of transgenerational epigenetic inheritance. Curr Opin Chem Biol. 2016;33:101–107. doi: 10.1016/j.cbpa.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 64.Ge Z.J., Zhang C.L., Schatten H., Sun Q.Y. Maternal diabetes mellitus and the origin of non-communicable diseases in offspring: the role of epigenetics. Biol Reprod. 2014;90:139. doi: 10.1095/biolreprod.114.118141. [DOI] [PubMed] [Google Scholar]
- 65.Wagner K.D., Wagner N., Ghanbarian H., Grandjean V., Gounon P., Cuzin F., et al. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 66.Liu X., Zhang L., Cheng K., Wang X., Ren G., Xie P. Identification of suitable plasma-based reference genes for miRNAome analysis of major depressive disorder. J Affect Disord. 2014;163:133–139. doi: 10.1016/j.jad.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 67.Batistela M.S., Josviak N.D., Sulzbach C.D., de Souza R.L. An overview of circulating cell-free microRNAs as putative biomarkers in Alzheimer's and Parkinson's diseases. Int J Neurosci. 2017;127:547–558. doi: 10.1080/00207454.2016.1209754. [DOI] [PubMed] [Google Scholar]
- 68.Lewczuk P., Riederer P., O’Bryant S.E., Verbeek M.M., Dubois B., Visser P.J., et al. Cerebrospinal fluid and blood biomarkers for neurodegenerative dementias: an update of the consensus of the task force on biological markers in psychiatry of the world federation of societies of biological psychiatry. World J Biol Psychiatr. 2018;19:244–328. doi: 10.1080/15622975.2017.1375556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goldsmith D.R., Rapaport M.H., Miller B.J. A meta-analysis of blood cytokine network alterations in psychiatric patients: comparisons between schizophrenia, bipolar disorder and depression. Mol Psychiatr. 2016;21:1696–1709. doi: 10.1038/mp.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Studzinski G.P., Harrison J.S. The neuronal cyclin-dependent kinase 5 activator p35Nck5a and Cdk5 activity in monocytic cells. Leuk Lymphoma. 2003;44:235–240. doi: 10.1080/1042819021000030009. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y., White M.G., Akay C., Chodroff R.A., Robinson J., Lindl K.A., et al. Activation of cyclin-dependent kinase 5 by calpains contributes to human immunodeficiency virus-induced neurotoxicity. J Neurochem. 2007;103:439–455. doi: 10.1111/j.1471-4159.2007.04746.x. [DOI] [PubMed] [Google Scholar]
- 72.Poirier M., Smith O.E., Therrien J., Rigoglio N.N., Miglino M.A., Silva L.A., et al. Resiliency of equid H19 imprint to somatic cell reprogramming by oocyte nuclear transfer and genetically induced pluripotency. Biol Reprod. 2020;102:211–219. doi: 10.1093/biolre/ioz168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.