Abstract
Epidermal growth factor receptor (EGFR) is one of the most well-studied oncogenes with roles in proliferation, growth, metastasis, and therapeutic resistance. This intense study has led to the development of a range of targeted therapeutics including small-molecule tyrosine kinase inhibitors (TKIs), monoclonal antibodies, and nanobodies. These drugs are excellent at blocking the activation and kinase function of wild-type EGFR (wtEGFR) and several common EGFR mutants. These drugs have significantly improved outcomes for patients with cancers including head and neck, glioblastoma, colorectal, and non-small cell lung cancer (NSCLC). However, therapeutic resistance is often seen, resulting from acquired mutations or activation of compensatory signaling pathways. Additionally, these therapies are ineffective in tumors where EGFR is found predominantly in the nucleus, as can be found in triple negative breast cancer (TNBC). In TNBC, EGFR is subjected to alternative trafficking which drives the nuclear localization of the receptor. In the nucleus, EGFR interacts with several proteins to activate transcription, DNA repair, migration, and chemoresistance. Nuclear EGFR (nEGFR) correlates with metastatic disease and worse patient prognosis yet targeting its nuclear localization has proved difficult. This review provides an overview of current EGFR-targeted therapies and novel peptide-based therapies that block nEGFR, as well as their clinical applications and potential for use in oncology.
Keywords: Triple negative breast cancer, epidermal growth factor, mucin-1, retrotranslocation
Introduction
Discovered in 1965 by Cohen [1], the epidermal growth factor receptor (EGFR) is a member of the transmembrane tyrosine kinase human EGFR (HER)/ErbB family. The HER family of receptors are potent drivers of growth, proliferation, and migration [2] and consist of EGFR, ErbB2, ErbB3, and ErbB4. While essential for the development and maintenance of normal epithelial tissue, changes to EGFR biology including overexpression/amplification [3, 4], mutation [5, 6], and altered trafficking [7, 8], allow aberrant signaling and functions leading to cancer and other diseases. As a transmembrane tyrosine kinase, therapeutic interventions for EGFR-driven cancers have largely focused on targeting via monoclonal antibodies and tyrosine kinase inhibitors (TKIs). These therapies have shown some success, as in EGFR overexpressing head and neck [9], and lung adenocarcinoma [10]. Although these treatments are initially effective, the majority of tumors eventually acquire resistance via several mechanisms [11].
Monoclonal antibodies
Cetuximab was the first EGFR targeted therapeutic antibody to gain FDA approval in 2004 and acts by competitively inhibiting the ligand binding pocket to prevent activation (Figure 1) [12]. In phase 3 studies, when given in combination with chemotherapy or radiation, the inclusion of cetuximab has increased median overall survival by ~3 months for colorectal cancer [13], ~5 months in head and neck [14], and ~8 months for non-small cell lung cancer (NSCLC) [15]. Since then, several other antibodies have been developed including panitumumab, nimotuzumab, necitumumab, and zalutumumab [16, 17]. These antibodies have shown efficacy in clinical trials for colorectal, head and neck, non-small cell lung, and biliary cancers. Most of these antibodies act by blocking the EGFR ligand binding pocket. However, nimotuzumab induces natural killer (NK) cell activation and T-cell expansion in addition to blocking ligand binding. This immune activation allows nimotuzumab to be more effective than cetuximab despite having a lower binding efficiency to EGFR [18]. Despite promising efficacy in multiple tumors, these antibodies have had little effect in triple negative breast cancer (TNBC) [19], even though these cancers express high levels of EGFR.
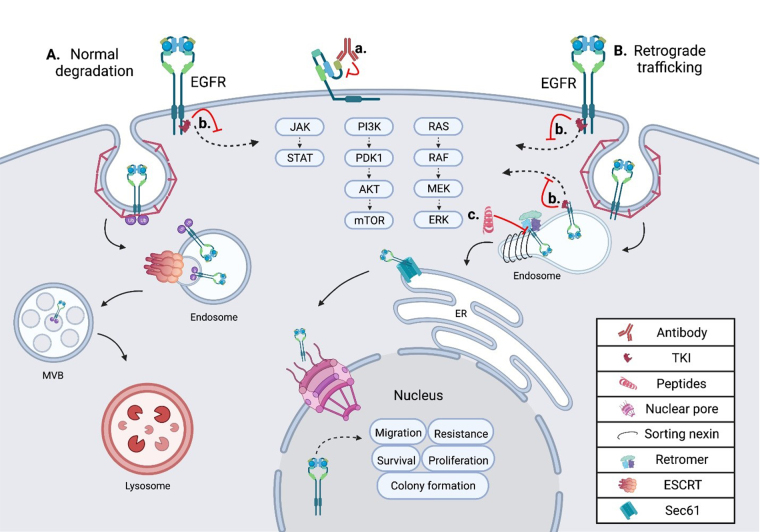
Figure 1.
Schematic showing the intracellular trafficking and pharmacological inhibitors of EGFR. (A) Shows the internalization and degradation of EGFR, seen in normal polarized epithelium; EGFR is first internalized through clathrin-mediated endocytosis, then undergoes trafficking through the endosomal network before being degraded in the lysosome; (B) the retrograde trafficking of EGFR is depicted, beginning with clathrin-mediated endocytosis; EGFR is then bound by sorting nexins (SNXs) and the retromer within the endosome, inducing further retrograde trafficking to the nucleus; within the nucleus EGFR drives oncogenic pathways through several mechanisms; (a) an EGFR targeted monoclonal antibody is depicted binding EGFR at the cell surface, inhibiting ligand binding and subsequent activation; (b) an EGFR specific TKI is depicted inhibiting EGFR signal transduction from the plasma membrane and the endosome in both normal cells and tumor cells; (c) the EGFR retrograde trafficking inhibitor cSNX1.3 is shown blocking the interaction of EGFR and cSNX1.3 which blocks the nuclear accumulation of EGFR. AKT: protein kinase B; cSNX1.3: capped sorting nexin peptide 1.3; ER: endoplasmic reticulum; ERK: extracellular signal-regulated kinase; ESCRT: endosomal sorting complex required for transport; JAK: Janus kinase; MEK: mitogen-activated protein kinase kinase; mTOR: mechanistic target of rapamycin; MVB: multivesicular body; PDK1: 3-phosphoinositide-dependent kinase 1; PI3K: phosphatidylinositol 3-kinase; RAF: rapidly accelerated fibrosarcoma; RAS: rat sarcoma virus; Sec61: transmembrane translocon channel; STAT: signal transducer of activators of transcription
Conjugated antibodies
Antibody-drug conjugates (ADCs) targeting EGFR have also shown promise in treating certain cancers. One of the first ADCs was an EGFR targeted antibody (mAb108), which was conjugated to the DNA intercalating agent, doxorubicin, in 1989 [20]. This ADC, mAb108-dextran-doxirubicin, showed a significantly improved anti-tumor effect when compared to free doxorubicin in mouse xenografts. Since their introduction, several next generation ADCs have been developed which use more tumor specific antibodies and stronger payloads [21]. Depatuxizumab mafodotin (ABT-414) is an EGFR targeted ADC currently in phase 3 clinical trials for glioblastoma [22]. ABT-414 uses the EGFR targeted mAb806 which has a higher affinity for EGFRvIII and greater efficacy against glioblastoma than cetuximab [23]. This antibody is conjugated to the anti-microtubule agent monomethyl auristatin F (MMAF) which is highly toxic as a single agent yet becomes tolerable with the tumor specificity granted by the conjugated antibody. In the phase 2 clinical trial for ABT-414 in glioblastoma patients, a hazard ratio of 0.66 was found for ABT-414 plus temozolamide compared to the temozolamide alone control arm [24]. Another ADC, M1231, uses a bispecific antibody that binds EGFR and mucin-1 (MUC1), proteins that are isolated to separate membrane domains in healthy tissue and colocalize during tumorigenesis [25]. A recent phase 1/1b clinical trial of the anti-EGFR antibody-monomethyl auristatin E, MRG003, found manageable toxicity with some efficacy in head and neck and nasopharyngeal carcinoma [26].
Another group has generated an antibody conjugate for combination therapy by creating a multilayered sphere called, anti-EGFR-PTX-TCS-GNS. The layers from the innermost are a silica sphere, gold shell, thiol chitosan, paclitaxel, and EGFR directed antibodies [27]. This complex drug simultaneously optimizes several therapies to create a first-in-class combinatorial therapeutic. Tumor directed gold particles increase the efficacy of radiotherapy by absorbing the near-infrared (NIR) wavelengths within the tumor, increasing the necrotic effect [28]. Thiol chitosan is a biopolymer that is nontoxic and biodegradable, as it absorbs heat from the NIR waves it melts releasing the paclitaxel payload.
Nanobodies
Nanobodies are camel-derived antibodies that lack the antibody light chain. One of the first successful nanobodies targeted EGFR to block epidermal growth factor (EGF) binding and EGFR phosphorylation and showed promised in xenograft models, primarily the A431 xenograft model [29]. Since their initial development, nanobodies have become increasingly complicated and effective [30]. Shortly after the first EGFR targeted nanobody was created, the same group created several bivalent (2 conjugated nanobodies which bind the same epitope) and biparatopic (2 conjugated nanobodies which bind different epitopes of the same protein) nanobodies and tested their efficacy again in A431 xenografts [31]. They found that while bivalent nanobodies had a modest increase, biparatopic nanobodies showed a significant improvement in efficacy.
Subsequently, the creation of bispecific nanobodies (2 fused nanobodies which bind different proteins) with an αEGFR-αEGFR-αAlbumin nanobody, which binds albumin in the blood to reduce renal clearance and increase tumor delivery [32]. This nanobody showed greater tumor uptake and deeper tumor penetration than cetuximab. In 2021, a bispecific nanobody was created, using αEGFR nanobody (7D12) fused to αCD16 nanobody (C21) to bind NK cells [33]. The authors reported that in in vitro and ex vivo studies, this bispecific nanobody induced degranulation of NK cells.
TKIs
TKIs are widely used in cancers with EGFR tyrosine kinase activating mutations as drivers, most effectively in lung adenocarcinoma, specifically NSCLC [34]. In NSCLC, EGFR TKIs significantly improve progression-free survival, as recently reviewed in [35]. EGFR-directed TKIs are designated into 3 generations of approved therapies with fourth generations currently being developed (Figure 1). First generation EGFR TKIs, including gefitinib and erlotinib, reversibly bind the ATP binding pocket within the kinase domain [36]. Second generation TKIs covalently bind the ATP pocket and showed improvement in PFS in head to head trials [37, 38]. However, these second-generation inhibitors also had slightly higher toxicity and side effects.
While first and second generation TKIs have shown significant clinical impact, the majority of tumors tend to acquire resistance to these drugs within a year [39]. While some tumors develop resistance through upregulation of other receptor tyrosine kinases (RTKs) or activating mutation in signaling intermediates, the majority of resistance arises with the EGFR T790M mutation [40]. This is a mutation to the ATP binding pocket that increase EGFRs affinity for ATP which outcompetes first and second generation TKIs [41]. A third generation EGFR TKI which overcome this mutation are currently available in clinic [42]. Osimertinib, an oral third generation irreversible EGFR TKI was shown in a pivotal phase 3 clinical trial to show superior efficacy to first and second generation EGFR TKIs when given as first-line therapy [43]. Similar to antibodies, these TKIs while having shown significant clinical impact in many cancers including; head and neck, NSCLC, and colon are ineffective in TNBC [44].
Alternative trafficking as a mechanism of EGFR-driven oncogenesis
In TNBC, EGFR is frequently found amplified and overexpressed, but not mutated. In these tumors, it is observed in intracellular locations in addition to the plasma membrane [45]. Several protein interactions and regulatory events are required for altered intracellular localization of EGFR to occur, including the complexing of EGFR and MUC1 (model of trafficking; Figure 1). MUC1 is comprised of a C-terminal cytoplasmic tail, including a transmembrane and small extracellular domain, that is bound through cysteine bonds to the heavily O-glycosylated N-terminal extracellular domain [46]. In healthy epithelium, MUC1 is localized to the apical domain and acts as a protective barrier against infection and inflammation [47]. However, during cancer progression MUC1 regulation is altered, resulting in its overexpression and gain of several oncogenic functions [48]. Concurrent with MUC1 overexpression during tumorigenesis, a loss of cellular polarity across the epithelium occurs. Loss of polarity allows traditionally apical and basolateral localized proteins to interact, as is the case with EGFR and MUC1. The interaction of MUC1 and EGFR at the plasma membrane inhibits the ubiquitylation of EGFR [49]. This interaction increases the rate of receptor internalization into the early endosome (EE) and is paired with a loss in degradation. Under these conditions, EGFR is found to interact with the SNX, specifically SNX1 which directly binds EGFR and regulates its intracellular trafficking [50] (Figure 1).
The sorting nexin family is a diverse group of peripheral membrane proteins that are classified based on the SNX-phox homology (PX) domain [51]. The PX domain of the SNX family is a phosphatidylinositol 3-phosphate (PI3P) binding motif, with minimal affinity for PI2P, that allows recruitment to membranes throughout the endosomal network and plasma membrane [52]. Initial studies suggested that SNX1 enhanced the degradation of EGFR when expressed in cells that did not normally express SNX1 [50]. This is likely due to the ability of SNX to elongate tubule vesicles out of domains such as the EE [53]. SNX, including SNX1, can associate their cargo with the retromer complex, a heterotrimer of proteins required for sorting (Vps26, Vps29, and Vps35). This complex is involved in cargo recognition at the endosomal network and sorting for retrograde trafficking. Unlike the SNX which directly binds the membrane via its PX domains, the retromer has no membrane binding domains. Instead of directly binding the membrane, the retromer complex binds sorting proteins like Rab7 or the SNX, as they are recruited to late endosomes [54] (Figure 1). Together, SNX proteins recognize and bind cargo proteins such as EGFR, induce the formation of vesicles, associate with the retromer complex, and route cargo for retrograde trafficking.
EGFR has several roles in the nucleus that can be either kinase dependent or more often kinase independent (Figure 2). While EGFR has no DNA binding domain, it can act as a co-transcriptional regulator for a growing list of oncogenes through associations with several nuclear proteins [55] (Figure 2). Nuclear EGFR (nEGFR) has been shown to interact with the transcription factors E2F1, STAT3, and STAT5 as well as RNA helicase A [56–59]. nEGFR can induce the expression of cyclinD1 [60], nitric oxide synthase (iNOS) [57], B-Myb [56], Aurora kinase A (Aurora-A) [58], cyclooxygenase-2 (COX-2) [61], c-Myc [62], and the breast cancer resistance protein, breast cancer resistance protein (BCRP) [63].
Figure 2.
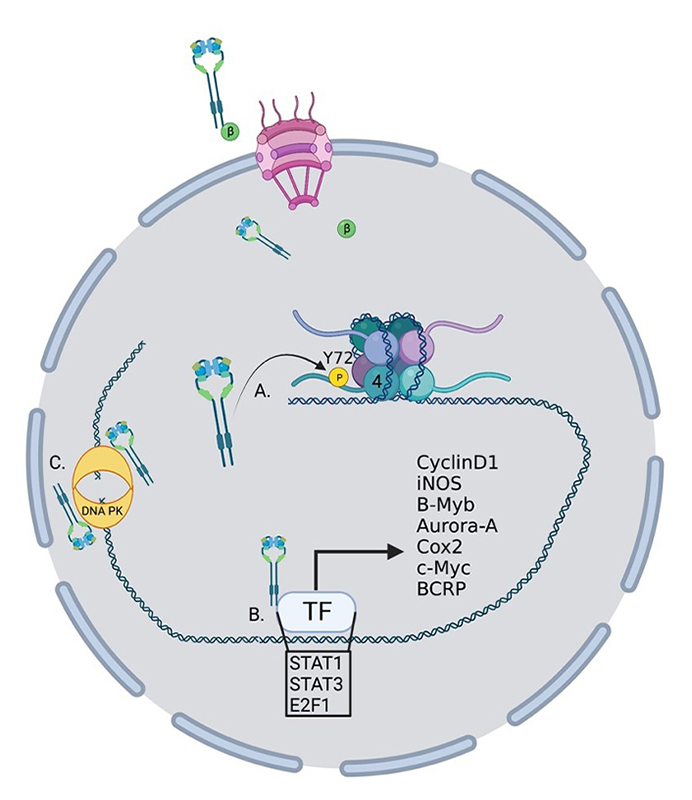
Oncogenic roles for nucelar EGFR. EGFR performs several functions within the nucleus to drive the transcription of oncogenes. (A) EGFR phosphorylates histone 4 Y72 which induces the recruitment of histone methyl transferaces and the methylation of histone 4 K20, enhancing DNA replication and repair; (B) EGFR interacts with several transcription factors to induce the transcription of a growing list of oncogenes; (C) EGFR interacts with DNA-dependent protein kinase (DNA-PK) to induce its translocation to the nucleus and activation of DNA repair. TF: transcription factor; P: phosphorylation; β: importin β
nEGFR has also been linked to resistance to radiotherapy through the activation of DNA repair pathways [64]. Infrared (IR) radiation can induce nuclear localization and interaction with DNA-PK and induction of DNA repair [65] and DNA repair can be eliminated by blocking retrograde trafficking and the nuclear accumulation of EGFR [66] (Figure 2). In another study, EGFR was shown to be recruited to the nucleus and aid in DNA repair in response to cisplatin treatment [67]. nEGFR has also been shown to modulate the epigenetic landscape by phosphorylating histone 4Y72 [68] (Figure 2). This phosphorylation increases the recruitment of the histone methyltransferases SET8 and SUV 4-20H leading to an increase in DNA transcription and repair.
The non-canonical trafficking and activities of EGFR may drive resistance to conventional therapies in several ways. The presence of EGFR within intracellular endosomes or the nucleus would prevent cell impermeable antibodies from reaching the receptor. Further, antibodies like cetuximab which act by competitively inhibiting the ligand binding pocket would not be effective as the ligand is already bound, and receptor activated and internalized [12]. Antibodies, like nimotuzumab, that act by recruiting immune cells would also be ineffective as they need to be on the cell surface to recruit immune cells [18]. TKIs on the other hand are freely cell permeable and inhibit EGFR activation through competitive inhibition of the ligand binding pocket. However, the noncanonical roles of nEGFR also evade TKI therapies as many of these roles may not involve EGFR’s kinase function. Instead, nEGFR can directly bind transcription factors within the nucleus and alter their DNA binding profiles to induce a novel suite of oncogenic protein expression. The shortcomings of conventional therapies to target intracellular and nEGFR highlight the need for new and different therapies that can inhibit the nuclear roles of EGFR in TNBC.
Targeting EGFR retrograde trafficking
The nuclear accumulation of EGFR represents an ideal target for therapeutic intervention as it appears to rarely occur outside the context of cancer, yet drives tumor progression, therapeutic resistance, and correlates with worse patient outcomes [69–71]. nEGFR has been shown to drive a growing list of oncogenes when associated with transcription factors in the nucleus, as well as modulate the epigenetic landscape to favor tumor progression [56–58, 60–62, 65, 66, 68]. The nuclear localization of EGFR has been shown to correlate with worse prognosis in an ever growing list of cancer including, breast [71], colorectal [70], ovarian [72], lung [73], oropharyngeal [74] and laryngeal [75]. Importantly, these studies demonstrate that nEGFR levels are a better predictor of outcomes than surface or whole cell EGFR levels. This presents an opportunity to develop therapeutics to block the nuclear functions of EGFR, to make clinical impact in TNBC and other tumors where EGFR undergoes retrograde trafficking.
Several therapeutic agents have been developed to target the retrograde trafficking of EGFR, primarily by blocking one of several protein interactions which drive this trafficking. At the plasma membrane in unpolarized epithelia during tumor progression, EGFR forms a complex with MUC1. This complex initiates a series of events that drive EGFR retrograde trafficking and has been targeted with several peptide-based inhibitors. These therapeutic peptides typically mimic some portion of the binding interface to act as a competitive inhibitor once introduced to the cell. However, therapeutic peptides are typically too large and charged or polar to be readily cell permeable. To allow cell penetration, a protein transduction domain is added to these peptides, allowing them to freely enter the cell [76].
The Schroeder lab and others have previously produced therapeutic cell penetrating peptides that target the interaction of EGFR and MUC1. The first peptide generated, PTD4-MUC1-inhibitory-peptide (PMIP), based on the cytoplasmic domain of MUC1 which interacts with EGFR, initiating its retrograde trafficking [77] (Table 1). PMIP was shown to reduce EGFR levels in cell culture and in the mouse mammary tumor virus-driven polyoma middle T-antigen (MMTV-PyV MT) mouse model of breast cancer. Along with the reduction of EGFR levels, a reduction in tumor growth and an induction of caspase3 cleavage was observed, suggesting these tumors were undergoing apoptotic cell death. Clinical translation of this peptide was not possible, as it proved to be unstable in high concentrations (data not shown). Another series of peptides were generated to target the cytoplasmic domain of MUC1 called GO-201 [78], GO-202 [78], GO-203 [79] (Table 1). These peptides mimicked regions of the intracellular domain and induced tumor regression in several xenograph models of breast, prostate cancer [80], and NSCLC [81]. The GO-203 peptide was then reformulated into a nanoparticle containing several 203 peptides called GO-203-NP which increases efficacy of GO-203 by increasing the cellular uptake of GO-203 [82]. Phase 1/2 clinical trial results demonstrate efficacy for GO-203-2C, a peptide that blocks MUC1 dimerization, in combination therapy with decitabine in acute myeloid leukemia (AML) patients [83].
Table 1.
Theraputic peptides targeting protiens required for EGFR retrograde trafficking
| Name | Peptide mimetic | Mechanism of action | Reference |
|---|---|---|---|
| PMIP | MUC1 c-tail | EGFR-Muc1 binding | Bitler et al. [77] |
| GO-201 | MUC1 c-tail | Binds Muc1 c-tail | Raina et al. [78] |
| GO-202 | MUC1 c-tail | Binds Muc1 c-tail | Raina et al. [78] |
| GO-203 | MUC1 c-tail | EGFR-Muc1 binding | Kharbanda et al. [79] |
| GO-203-NP | MUC1 c-tail nano particle | Increased cellular uptake of GO-203 peptide | Hasegawa et al. [82] |
| EJ-1 | EGFR JXM domain | EGFR dimerization, nuclear localization, calmodulin binding, basolateral targeting, mitochondrial localization | Hart et al. [84] |
| SAH5 | Hydrocarbon stapled EGFR JXM domain | EGFR dimerization, nuclear localization, calmodulin binding, basolateral targeting, mitochondrial localization | Maisel et al. [85] |
| cSNX1.3 | SNX1 Bar domain | EGFR-SNX1 binding | Atwell et al. [86] |
Peptides derived from MUC1 cytoplasmic tail, EGFR juxtamembrane (JXM) domain, and SNX1 have demonstrated anti-tumor effects in models of breast, prostate, glioblastoma, and lung cancer. PMIP, EGFR JXM-1 (EJ-1), stapled aromatic hydrocarbon EJ1-5 (SAH5), and cSNX1.3 have all been shown to directly inhibit the retrograde trafficking of EGFR. The GO peptides have been shown to inhibit the nuclear accumulation of MUC1 and inhibit the retrograde trafficking of EGFR
Alternatively, an EJ-1 was generated to interfere directly with the nuclear localization sequence of EGFR [84] (Table 1). The JXM, region of EGFR, is a significant regulator of EGFR biology as it contains a nuclear localization sequence [87], calmodulin-binding domain [88], dimerization domain [89], and basolateral targeting domain [90]. Not only is this region critical for EGFR regulation, but it is also structured as an alpha-helix, and the resulting EJ-1 peptide was far more stable than PMIP. To increase the stability further, this peptide was stapled by substituting 2 residues with non-natural amino acids that were covalently linked through a hydrocarbon linker arm [85]. The resulting stapled peptide, SAH5, had greater stability and was more potent than EJ-1 (Table 1). SAH5 significantly reduced the growth of SUM149, an inflammatory TNBC cell line, xenographs in severe combined immunodeficiency (SCID) mice. However, this peptide was shown to have toxic side effects that included injection site irritation, constriction of veins upon injection, and the induction of seizures in animals shortly after injection (data not shown). Cell culture analysis demonstrated that this peptide induced the opening of calcium channels and altered subsequent calcium signaling (data not shown). These off-target effects increased the toxicity to a level that prevented clinical translation of this peptide.
Recently, the Schroeder lab targeted the endosomal trafficking protein SNX1 to generate a peptide that mimics the interaction domain between EGFR and SNX1, called cSNX1.3 (Figure 1, Table 1) [86]. This end-capped peptide directly binds EGFR with a similar binding efficiency as SNX1 and results in a significant reduction of nEGFR in TNBC cells with amplified EGFR. cSNX1.3 treatment induces an EGFR-dependent impact on oncogenic phenotypes including proliferation, survival, migration, and mammosphere formation while having no impact on immortalized breast epithelial cells. In the transgenic mouse model whey acidic protein-transforming growth factor α (WAP-TGFα), which develops EGFR-dependent mammary carcinomas, 1/3 of animals showed partial regression, 1/3 no progression and 1/3 demonstrated a complete regression with no observable tumor upon necropsy. Further analysis demonstrated no toxicity and circulation of cSNX1.3 was found up to 24 h after injection, indicating this may be a viable therapeutic (data not shown).
Of note, in cell culture experiments, cSNX1.3 had no effect on EGFR signal transduction, internalization, or degradation. This indicates that results observed were not due to an effect on canonical activities of EGFR. Rather, cSNX1.3 seemed to only effect the retrograde trafficking of EGFR, further highlighting the importance of this pathway for the understanding and treatment of cancer. These and other peptide-based therapeutics that block protein-protein interactions hold promise for future clinical therapeutics.
Conclusions
EGFR is a complex protein that plays roles in many aspects of tumor growth and metastasis. Many drugs have been designed to target well characterized aspects of EGFR biology like ligand binding and kinase signaling. TKIs and antibody-based therapies have significantly improved many patient outcomes over several decades. However, tumors often develop resistance to these drugs either through activation of other signal transduction pathways or through additional mutations. In cases where there is an abundance of nEGFR these drugs don’t often work. nEGFR is a major driver of tumor progression, metastasis, and resistance to anti-EGFR therapies and has been suggested to be a better biomarker than total cell EGFR levels.
The tumor specificity of nEGFR makes it a potentially valuable therapeutic target as nEGFR is rarely seen outside the context of cancer. This tumor specificity is in stark contrast to the roles of EGFR’s kinase domain which are critical in a majority of human tissues at nearly all developmental stages. The reliance on EGFR to develop and maintain human tissues, including the skin and gastrointestinal (GI) tract, explains many side effects seen with anti-EGFR therapies including rash, nausea, and diarrhea.
As has been demonstrated with several peptide inhibitors of EGFR retrograde trafficking, nEGFR is a druggable target. The trafficking of EGFR to the nucleus requires several protein interactions that alter the posttranslational modification seen on EGFR, leading to a switch from lysosomal degradation to nuclear trafficking. The interactions of EGFR with MUC1 and SNX1 have both been targeted and shown efficacy in animal mouse tumor models. The interaction of EGFR and MUC1 is an upstream step in the nuclear trafficking of EGFR and might induce the degradation of EGFR in addition to the loss off nuclear localization making it an ideal anti-nEGFR therapy. In contrast targeting a nEGFR trafficking protein, like SNX1, is further downstream and is unlikely to induce EGFR degradation. However, targeting SNX1 might have additional benefits that have not been fully characterized. SNX1 binds EGFR on the kinase domain and induces the retrograde trafficking of EGFR. SNX1 also binds several other RTKs that have also been shown to drive oncogenesis with their nuclear activities. The peptide cSNX1.3 which was designed to block the interaction of EGFR and SNX1 inhibited 2 dimensional (2D) cell migration driven by EGFR. cSNX1.3 also inhibited migration driven by HER3 and c-Met, which are both RTKs shown to interact with SNX1. They undergo nuclear translocation and drive oncogenesis from within the nucleus suggesting the nuclear localization of RTKs more broadly could be a target for therapeutic intervention and targeting sorting proteins could be an avenue to a broad acting anti-RTK retrograde trafficking therapy.
Abbreviations
- ADCs
antibody-drug conjugates
- cSNX1.3
capped sorting nexin peptide 1.3
- EGFR
epidermal growth factor receptor
- EJ-1
epidermal growth factor receptor juxtamembrane-1
- HER
human epidermal growth factor receptor
- MUC1
mucin-1
- JXM
juxtamembrane
- nEGFR
nuclear epidermal growth factor receptor
- NK
natural killer
- NSCLC
non-small cell lung cancer
- PMIP
PTD4-mucin-1-inhibitory-peptide
- PX
phox homology
- RTKs
receptor tyrosine kinases
- SAH5
stapled aromatic hydrocarbon epidermal growth factor receptor juxtamembrane-1
- SNXs
sorting nexins
- STAT
signal transducer of activators of transcription
- TKIs
tyrosine kinase inhibitors
- TNBC
triple negative breast cancer
Declarations
Author contributions
BA and JS: conceptualization, Writing—original draft, Writing—review & editing. PC: Writing—review & editing.
Conflicts of interest
The authors declare that they has no conflicts of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
Not applicable.
Copyright
© The Author(s) 2023.
References
- 1.Cohen S. The stimulation of epidermal proliferation by a specific protein (EGF) Dev Biol. 1965;12:394–407. doi: 10.1016/0012-1606(65)90005-9. [DOI] [PubMed] [Google Scholar]
- 2.Roskoski R Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Sholl LM, Yeap BY, Iafrate AJ, Holmes-Tisch AJ, Chou YP, Wu MT, et al. Lung adenocarcinoma with EGFR amplification has distinct clinicopathologic and molecular features in never-smokers. Cancer Res. 2009;69:8341–8. doi: 10.1158/0008-5472.CAN-09-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Liang R, Song C, Xiang Y, Liu Y. Prognostic significance of epidermal growth factor receptor expression in glioma patients. Onco Targets Ther. 2018;11:731–42. doi: 10.2147/OTT.S155160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol Mech Dis. 2011;6:49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 6.Yoon HY, Ryu JS, Sim YS, Kim D, Lee SY, Choi J, et al. Clinical significance of EGFR mutation types in lung adenocarcinoma: a multi-centre Korean study. PLoS One. 2020;15:e0228925. doi: 10.1371/journal.pone.0228925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas R, Weihua Z. Rethink of EGFR in cancer with its kinase independent function on board. Front Oncol. 2019;9:800. doi: 10.3389/fonc.2019.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maisel SA, Schroeder J. Wrong place at the wrong time: how retrograde trafficking drives cancer metastasis through receptor mislocalization. J Cancer Metastasis Treat. 2019;5:7. doi: 10.20517/2394-4722.2018.82. [DOI] [Google Scholar]
- 9.Fasano M, Della Corte CM, Viscardi G, Di Liello R, Paragliola F, Sparano F, et al. Head and neck cancer: the role of anti-EGFR agents in the era of immunotherapy. Ther Adv Med Oncol. 2021;13:1758835920949418. doi: 10.1177/1758835920949418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah R, Lester JF. Tyrosine kinase inhibitors for the treatment of EGFR mutation-positive non-small-cell lung cancer: a clash of the generations. Clin Lung Cancer. 2020;21:e216–28. doi: 10.1016/j.cllc.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Hrustanovic G, Lee BJ, Bivona TG. Mechanisms of resistance to EGFR targeted therapies. Cancer Biol Ther. 2013;14:304–14. doi: 10.4161/cbt.23627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, Schmitz KR, Jeffrey PD, Wiltzius JJ, Kussie P, Ferguson KM. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–11. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Qin S, Li J, Wang L, Xu J, Cheng Y, Bai Y, et al. Efficacy and tolerability of first-line cetuximab plus leucovorin, fluorouracil, and oxaliplatin (FOLFOX-4) versus FOLFOX-4 in patients with RAS wild-type metastatic colorectal cancer: the open-label, randomized, phase III TAILOR trial. J Clin Oncol. 2018;36:3031–9. doi: 10.1200/JCO.2018.78.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch FR, Redman MW, Moon J, Agustoni F, Herbst RS, Semrad TJ, et al. EGFR high copy number together with high EGFR protein expression predicts improved outcome for cetuximab-based therapy in squamous cell lung cancer: analysis from SWOG S0819, a phase III trial of chemotherapy with or without cetuximab in advanced NSCLC. Clin Lung Cancer. 2022;23:60–71. doi: 10.1016/j.cllc.2021.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai WQ, Zeng LS, Wang LF, Wang YY, Cheng JT, Zhang Y, et al. The latest battles between EGFR monoclonal antibodies and resistant tumor cells. Front Oncol. 2020;10:1249. doi: 10.3389/fonc.2020.01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu MJ, Johnson DE, Grandis JR. EGFR-targeted therapies in the post-genomic era. Cancer Metastasis Rev. 2017;36:463–73. doi: 10.1007/s10555-017-9687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazorra Z, Lavastida A, Concha-Benavente F, Valdés A, Srivastava RM, García-Bates TM, et al. Nimotuzumab induces NK cell activation, cytotoxicity, dendritic cell maturation and expansion of EGFR-specific T cells in head and neck cancer patients. Front Pharmacol. 2017;8:382. doi: 10.3389/fphar.2017.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, et al. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol. 2012;30:2615–23. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aboud-Pirak E, Hurwitz E, Bellot F, Schlessinger J, Sela M. Inhibition of human tumor growth in nude mice by a conjugate of doxorubicin with monoclonal antibodies to epidermal growth factor receptor. Proc Natl Acad Sci USA. 1989;86:3778–81. doi: 10.1073/pnas.86.10.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Fang T, Yun C, Liu X, Cai X. Antibody-drug conjugates targeting the human epidermal growth factor receptor family in cancers. Front Mol Biosci. 2022;9:847835. doi: 10.3389/fmolb.2022.847835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Achenbach C, Silginer M, Blot V, Weiss WA, Weller M. Depatuxizumab mafodotin (ABT-414)-induced glioblastoma cell death requires EGFR overexpression, but not EGFRY1068 phosphorylation. Mol Cancer Ther. 2020;19:1328–39. doi: 10.1158/1535-7163.MCT-19-0609. [DOI] [PubMed] [Google Scholar]
- 23.Reilly EB, Phillips AC, Buchanan FG, Kingsbury G, Zhang Y, Meulbroek JA, et al. Characterization of ABT-806, a humanized tumor-specific anti-EGFR monoclonal antibody. Mol Cancer Ther. 2015;14:1141–51. doi: 10.1158/1535-7163.MCT-14-0820. [DOI] [PubMed] [Google Scholar]
- 24.Van Den Bent M, Eoli M, Sepulveda JM, Smits M, Walenkamp A, Frenel JS, et al. INTELLANCE 2/EORTC 1410 randomized phase II study of Depatux-M alone and with temozolomide vs temozolomide or lomustine in recurrent EGFR amplified glioblastoma. Neuro Oncol. 2020;22:684–93. doi: 10.1093/neuonc/noz222. Erratum in: Neuro Oncol. 2021;23:1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Codony-Servat J, Dotterweich J, Molina-Vila MA, Román R, Giménez-Capitán A, Aldeguer E, et al. Preclinical studies of the bispecific MUC1xEGFR antibody drug conjugate M1231 in EGFR mutant NSCLC. Eur J Cancer. 2022;174:S89. doi: 10.1016/S0959-8049(22)01035-8. [DOI] [Google Scholar]
- 26.Qiu MZ, Zhang Y, Guo Y, Guo W, Nian W, Liao W, et al. Evaluation of safety of treatment with anti-epidermal growth factor receptor antibody drug conjugate MRG003 in patients with advanced solid tumors: a phase 1 nonrandomized clinical Trial. JAMA Oncol. 2022;8:1042–6. doi: 10.1016/j.jconrel.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manivasagan P, Nguyen VT, Jun SW, Hoang G, Mondal S, Kim H, et al. Anti-EGFR antibody conjugated thiol chitosan-layered gold nanoshells for dual-modal imaging-guided cancer combination therapy. J Control Release. 2019;311–312:26–42. doi: 10.1016/j.jconrel.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Haume K, Rosa S, Grellet S, Śmiałek MA, Butterworth KT, Solov’yov AV, et al. Gold nanoparticles for cancer radiotherapy: a review. Cancer Nanotechnol. 2016;7:8. doi: 10.1186/s12645-016-0021-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roovers RC, Laeremans T, Huang L, De Taeye S, Verkleij AJ, Revets H, et al. Efficient inhibition of EGFR signaling and of tumour growth by antagonistic anti-EFGR nanobodies. Cancer Immunol Immunother. 2007;56:303–17. doi: 10.1007/s00262-006-0180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang EY, Shah K. Nanobodies: next generation of cancer diagnostics and therapeutics. Front Oncol. 2020;10:1182. doi: 10.3389/fonc.2020.01182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roovers RC, Vosjan MJ, Laeremans T, el Khoulati R, de Bruin RC, Ferguson KM, et al. A biparatopic anti-EGFR nanobody efficiently inhibits solid tumour growth. Int J Cancer. 2011;129:2013–24. doi: 10.1002/ijc.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tijink BM, Laeremans T, Budde M, Stigter-van Walsum M, Dreier T, de Haard HJ, et al. Improved tumor targeting of anti-epidermal growth factor receptor nanobodies through albumin binding: taking advantage of modular nanobody technology. Mol Cancer Ther. 2008;7:2288–97. doi: 10.1158/1535-7163.MCT-07-2384. [DOI] [PubMed] [Google Scholar]
- 33.Toffoli EC, Sheikhi A, Lameris R, King LA, van Vliet A, Walcheck B, et al. Enhancement of NK Cell antitumor effector functions using a bispecific single domain antibody targeting CD16 and the epidermal growth factor receptor. Cancers (Basel) 2021;13:5446. doi: 10.3390/cancers13215446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, Ke L, Zhang Z, Yu J, Meng X. Development of EGFR TKIs and options to manage resistance of third-generation EGFR TKI osimertinib: conventional ways and immune checkpoint inhibitors. Front Oncol. 2020;10:602762. doi: 10.3389/fonc.2020.602762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levantini E, Maroni G, Del Re M, Tenen DG. EGFR signaling pathway as therapeutic target in human cancers. Semin Cancer Biol. 2022;85:253–75. doi: 10.1016/j.semcancer.2022.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Karachaliou N, Fernandez-Bruno M, Bracht JWP, Rosell R. EGFR first- and second-generation TKIs—there is still place for them in EGFR-mutant NSCLC patients. Transl Cancer Res. 2019;8:S23–47. doi: 10.21037/tcr.2018.10.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu YL, Cheng Y, Zhou X, Lee KH, Nakagawa K, Niho S, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol. 2017;18:1454–66. doi: 10.1016/S1470-2045(17)30608-3. [DOI] [PubMed] [Google Scholar]
- 38.Paz-Ares L, Tan EH, O’Byrne K, Zhang L, Hirsh V, Boyer M, et al. Afatinib versus gefitinib in patients with EGFR mutation-positive advanced non-small-cell lung cancer: overall survival data from the phase IIb LUX-Lung 7 trial. Ann Oncol. 2017;28:270–7. doi: 10.1093/annonc/mdw611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–81. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 40.Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. 2018;17:38. doi: 10.1186/s12943-018-0777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaclova T, Grazini U, Ward L, O’Neill D, Markovets A, Huang X, et al. Clinical impact of subclonal EGFR T790M mutations in advanced-stage EGFR-mutant non-small-cell lung cancers. Nat Commun. 2021;12:1780. doi: 10.1038/s41467-021-22057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Cang S, Liu D. Third-generation inhibitors targeting EGFR T790M mutation in advanced non-small cell lung cancer. J Hematol Oncol. 2016;9:34. doi: 10.1186/s13045-016-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. FLAURA Investigators Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–25. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 44.Nakai K, Hung MC, Yamaguchi H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res. 2016;6:1609–23. [PMC free article] [PubMed] [Google Scholar]
- 45.Maisel S, Broka D, Schroeder J. Intravesicular epidermal growth factor receptor subject to retrograde trafficking drives epidermal growth factor-dependent migration. Oncotarget. 2017;9:6463–77. doi: 10.18632/oncotarget.23766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–57. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- 47.Dhar P, McAuley J. The role of the cell surface mucin MUC1 as a barrier to infection and regulator of inflammation. Front Cell Infect Microbiol. 2019;9:117. doi: 10.3389/fcimb.2019.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horm TM, Schroeder JA. MUC1 and metastatic cancer: expression, function and therapeutic targeting. Cell Adh Migr. 2013;7:187–98. doi: 10.4161/cam.23131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pochampalli MR, el Bejjani RM, Schroeder JA. MUC1 is a novel regulator of ErbB1 receptor trafficking. Oncogene. 2007;26:1693–701. doi: 10.1038/sj.onc.1209976. [DOI] [PubMed] [Google Scholar]
- 50.Kurten RC, Cadena DL, Gill GN. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science. 1996;272:1008–10. doi: 10.1126/science.272.5264.1008. [DOI] [PubMed] [Google Scholar]
- 51.Carlton JG, Cullen PJ. Sorting nexins. Curr Biol. 2005;15:R819–20. doi: 10.1016/j.cub.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 52.Cozier GE, Carlton J, McGregor AH, Gleeson PA, Teasdale RD, Mellor H, et al. The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J Biol Chem. 2002;277:48730–6. doi: 10.1074/jbc.M206986200. [DOI] [PubMed] [Google Scholar]
- 53.van Weering JR, Sessions RB, Traer CJ, Kloer DP, Bhatia VK, Stamou D, et al. Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 2012;31:4466–80. doi: 10.1038/emboj.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seaman MN. The retromer complex - endosomal protein recycling and beyond. J Cell Sci. 2012;125:4693–702. doi: 10.1242/jcs.103440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang YN, Hung MC. Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2012;2:13. doi: 10.1186/2045-3701-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45:10–7. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 57.Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–89. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Hung LY, Tseng JT, Lee YC, Xia W, Wang YN, Wu ML, et al. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008;36:4337–51. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huo L, Wang YN, Xia W, Hsu SC, Lai CC, Li LY, et al. RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc Natl Acad Sci USA. 2010;107:16125–30. doi: 10.1073/pnas.1000743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–8. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 61.Lo HW, Cao X, Zhu H, Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8:232–45. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gururaj AE, Gibson L, Panchabhai S, Bai M, Manyam G, Lu Y, et al. Access to the nucleus and functional association with c-Myc is required for the full oncogenic potential of ΔEGFR/EGFRvIII. J Biol Chem. 2013;288:3428–38. doi: 10.1074/jbc.M112.399352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, Tsai SL, et al. Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J Biol Chem. 2011;286:20558–68. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt-Ullrich RK, Mikkelsen RB, Dent P, Todd DG, Valerie K, Kavanagh BD, et al. Radiation-induced proliferation of the human A431 squamous carcinoma cells is dependent on EGFR tyrosine phosphorylation. Oncogene. 1997;15:1191–7. doi: 10.1038/sj.onc.1201275. [DOI] [PubMed] [Google Scholar]
- 65.Dittmann K, Mayer C, Kehlbach R, Rodemann HP. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol Cancer. 2008;7:69. doi: 10.1186/1476-4598-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dittmann K, Mayer C, Rodemann HP. Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother Oncol. 2005;76:157–61. doi: 10.1016/j.radonc.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 67.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011;71:1103–14. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chou RH, Wang YN, Hsieh YH, Li LY, Xia W, Chang WC, et al. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev Cell. 2014;30:224–37. doi: 10.1016/j.devcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brand TM, Iida M, Luthar N, Starr MM, Huppert EJ, Wheeler DL. Nuclear EGFR as a molecular target in cancer. Radiother Oncol. 2013;108:370–7. doi: 10.1016/j.radonc.2013.06.010. Erratum in: Radiother Oncol. 2019;130:195. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 70.Yang CC, Lin LC, Lin YW, Tian YF, Lin CY, Sheu MJ, et al. Higher nuclear EGFR expression is a better predictor of survival in rectal cancer patients following neoadjuvant chemoradiotherapy than cytoplasmic EGFR expression. Oncol Lett. 2019;17:1551–8. doi: 10.3892/ol.2018.9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–48. Erratum in: Cancer Res. 2005;65:2045. [PubMed] [Google Scholar]
- 72.Xia W, Wei Y, Du Y, Liu J, Chang B, Yu YL, et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2009;48:610–7. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Traynor AM, Weigel TL, Oettel KR, Yang DT, Zhang C, Kim K, et al. Nuclear EGFR protein expression predicts poor survival in early stage non-small cell lung cancer. Lung Cancer. 2013;81:138–41. doi: 10.1016/j.lungcan.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Psyrri A, Egleston B, Weinberger P, Yu Z, Kowalski D, Sasaki C, et al. Correlates and determinants of nuclear epidermal growth factor receptor content in an oropharyngeal cancer tissue microarray. Cancer Epidemiol Biomarkers Prev. 2008;17:1486–2. doi: 10.1158/1055-9965.EPI-07-2684. [DOI] [PubMed] [Google Scholar]
- 75.Marijić B, Braut T, Babarović E, Krstulja M, Maržić D, Avirović M, et al. Nuclear EGFR expression is associated with poor survival in laryngeal carcinoma. Appl Immunohistochem Mol Morphol. 2021;29:576–84. doi: 10.1097/PAI.0000000000000932. [DOI] [PubMed] [Google Scholar]
- 76.Bitler BG, Schroeder JA. Anti-cancer therapies that utilize cell penetrating peptides. Recent Pat Anticancer Drug Discov. 2010;5:99–108. doi: 10.2174/157489210790936252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bitler BG, Menzl I, Huerta CL, Sands B, Knowlton W, Chang A, et al. Intracellular MUC1 peptides inhibit cancer progression. Clin Cancer Res. 2009;15:100–9. doi: 10.1158/1078-0432.CCR-08-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raina D, Ahmad R, Joshi MD, Yin L, Wu Z, Kawano T, et al. Direct targeting of the mucin 1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res. 2009;69:5133–41. doi: 10.1158/0008-5472.CAN-09-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kharbanda A, Rajabi H, Jin C, Tchaicha J, Kikuchi E, Wong KK, et al. Targeting the oncogenic MUC1-C protein inhibits mutant EGFR-mediated signaling and survival in non-small cell lung cancer cells. Clin Cancer Res. 2014;20:5423–34. doi: 10.1158/1078-0432.CCR-13-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Joshi MD, Ahmad R, Yin L, Raina D, Rajabi H, Bubley G, et al. MUC1 oncoprotein is a druggable target in human prostate cancer cells. Mol Cancer Ther. 2009;8:3056–65. doi: 10.1158/1535-7163.MCT-09-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, et al. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Ther. 2011;10:806–16. doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hasegawa M, Sinha RK, Kumar M, Alam M, Yin L, Raina D, et al. Intracellular targeting of the oncogenic MUC1-C protein with a novel GO-203 nanoparticle formulation. Clin Cancer Res. 2015;21:2338–47. doi: 10.1158/1078-0432.CCR-14-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liegel J, Rosenblatt J, Stone RM, McMasters M, Levine JD, Myrna Nahas, et al. Phase I/Ib trial of the MUC1 inhibitor GO-203-2C alone and in combination with decitabine for acute myeloid leukemia. Blood. 2017;130:2659. doi: 10.1182/blood.V130.Suppl_1.2659.2659. [DOI] [Google Scholar]
- 84.Hart MR, Su HY, Broka D, Goverdhan A, Schroeder JA. Inactive ERBB receptors cooperate with reactive oxygen species to suppress cancer progression. Mol Ther. 2013;21:1996–2007. doi: 10.1038/mt.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maisel SA, Broka D, Atwell B, Bunch T, Kupp R, Singh SK, et al. Stapled EGFR peptide reduces inflammatory breast cancer and inhibits additional HER-driven models of cancer. J Transl Med. 2019;17:201. doi: 10.1186/s12967-019-1939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Atwell B, Chen CY, Christofferson M, Montfort WR, Schroeder J. Sorting nexin-dependent therapeutic targeting of oncogenic epidermal growth factor receptor. Cancer Gene Ther. 2023;30:267–76. doi: 10.1038/s41417-022-00541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–40. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- 88.Li H, Sánchez-Torres J, Del Carpio A, Salas V, Villalobo A. The ErbB2/Neu/HER2 receptor is a new calmodulin-binding protein. Biochem J. 2004;381:257–66. doi: 10.1042/BJ20040515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goldman R, Levy RB, Peles E, Yarden Y. Heterodimerization of the erbB-1 and erbB-2 receptors in human breast carcinoma cells: a mechanism for receptor transregulation. Biochemistry. 1990;29:11024–8. doi: 10.1021/bi00502a002. [DOI] [PubMed] [Google Scholar]
- 90.Hobert ME, Kil SJ, Medof ME, Carlin CR. The cytoplasmic juxtamembrane domain of the epidermal growth factor receptor contains a novel autonomous basolateral sorting determinant. J Biol Chem. 1997;272:32901–9. doi: 10.1074/jbc.272.52.32901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.