Abstract
Deep convolutional neural networks (DCNNs) have shown promise in brain tumor segmentation from multi-modal MRI sequences, accommodating heterogeneity in tumor shape and appearance. The fusion of multiple MRI sequences allows networks to explore complementary tumor information for segmentation. However, developing a network that maintains clinical relevance in situations where certain MRI sequence(s) might be unavailable or unusual poses a significant challenge. While one solution is to train multiple models with different MRI sequence combinations, it is impractical to train every model from all possible sequence combinations. In this paper, we propose a DCNN-based brain tumor segmentation framework incorporating a novel sequence dropout technique in which networks are trained to be robust to missing MRI sequences while employing all other available sequences. Experiments were performed on the RSNA-ASNR-MICCAI BraTS 2021 Challenge dataset. When all MRI sequences were available, there were no significant differences in performance of the model with and without dropout for enhanced tumor (ET), tumor (TC), and whole tumor (WT) (p-values 1.000, 1.000, 0.799, respectively), demonstrating that the addition of dropout improves robustness without hindering overall performance. When key sequences were unavailable, the network with sequence dropout performed significantly better. For example, when tested on only T1, T2, and FLAIR sequences together, DSC for ET, TC, and WT increased from 0.143 to 0.486, 0.431 to 0.680, and 0.854 to 0.901, respectively. Sequence dropout represents a relatively simple yet effective approach for brain tumor segmentation with missing MRI sequences.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-023-00860-7.
Keywords: Brain tumor segmentation, 3D U-Net, Sequence dropout, Multi-contrast MRI, Deep learning
Introduction
Brain tumors are extremely heterogeneous, exhibiting different degrees of aggressiveness and variable prognosis that can influence treatment approaches. They contain heterogeneous histological subregions including necrotic core, peritumoral edema, and enhancing and non-enhancing tumor core. This heterogeneity can be evidenced by varying radiographic phenotypes. Due to heterogeneity in shape and appearance, it is very challenging to automatically segment these subregions accurately. T1 without contrast (T1), T1 with gadolinium contrast (T1Gd), T2, fluid-attenuated inversion recovery (FLAIR), and diffusion-weighted imaging are standard MRI sequences when evaluating such tumors.
Deep learning methods have shown success in various lesion segmentation tasks, including brain tumor segmentation. Different MRI sequences/contrasts are sensitive to different brain tumor subregions. They can thus be combined to improve network performance through either early fusion, in which different sequences are concatenated as different input channels, or late fusion, in which sequences are concatenated in a late stage of the network. Such fusion is a classic example of multiple knowledge representation prevalent in artificial intelligence where separate components of the model focus on different aspects of the image to produce a final segmentation [1]. For example, fusion of sequences allows for the model to excel in both its contrast-enhancing region segmentation by utilizing the specific features encoded from the T1Gd input and peritumor edema segmentation by utilizing the features from the FLAIR input. While combining all available sequences ensures that networks exploit all information provided by training data, it presents potential drawbacks when sequences are missing. With imaging acquisition protocols varying across clinical sites, the same orders may include different MRI sequences. Even if all required sequences are acquired, it is possible that one or more are unusable due to severe motion artifacts [2]. Furthermore, significant heterogeneity in acquisition parameters may exist, such as the degree of T2 weighting. Statistically, the likelihood of “at least one unusable” sequence is greatly increased when multiple sequences are needed for brain tumor automatic segmentation, even if the failure rate of an individual sequence is low. Unlike human readers who can take full advantage of and synthesize all available information when a case has missing/unusable sequence(s), a network trained conventionally with a fixed number of sequences as input may not be able to extract all information or may fail completely if it is trained to be reliant on the coexistence of different input sequences. A straightforward solution to this issue may be to train a network for each combination of MRI sequences. However, this is impractical due to the large number of possible combinations and the high cost of training DCNNs.
Several approaches have been investigated to compensate for missing imaging modalities [3]. Original methods include Hetero-Modal Image Segmentation (HeMIS) and Hetero-Modal Variational Encoder-Decoder (U-HVED), which translate the available modalities to a common latent space and calculate the mean and variance of available features to address missing modalities [4, 5]. These two methods unfortunately fail when key sequences are unavailable as they treat each modality equally during tumor segmentation. Alternatively, Wang et al. use Adversarial Co-Training Networks (ACN), in which independent, related models are trained for situations with all modalities and missing modalities to supplement each other and recover missing information [6]. ACNs perform considerably better than HeMIS and U-HVED, especially when segmenting enhanced tumor (ET), which includes the enhancing tumor core, when T1Gd was missing, and when segmenting whole tumor (WT), which includes necrotic core, peritumoral edema, invaded tissue, and enhancing and non-enhancing tumor core, when FLAIR was missing. However, the authors mention significant training costs with their design having a multimodal and unimodal path with three separate adversarial learning modules to connect the two paths. Some use knowledge distillation networks, in which a larger, more complicated teacher model is used to transfer knowledge to a smaller student network [7]. Others determine similarity metrics and optimize the common information across modalities [8, 9]. Chen et al. use RobustSeg where they use feature disentanglement to train a model to isolate independent features from each modality so that it is not dependent on weights from a single modality [10]. Generative adversarial networks (GAN) have also been used to generate the missing modalities [11]. Azad et al. utilize a style matching U-Net (SMU-Net). Style transfer networks became popular with their application of being able to alter a photo to match the style of another (e.g., a landscape photo is transformed to resemble the style of Van Gough’s Starry Night) [12]. SMU-Net has two learning pathways, one where all modalities are available and another with missing modalities, and uses style transfer to cull informative features from the full learning pathway to the missing modality one [13]. They show that robustness is superior to U-HeMIS and HVED (e.g., U-HeMIS Dice similarity coefficient [DSC] 0.249, HVED 0.248, SMU 0.461 on ET when only FLAIR is available; 0.680, 0.703, 0.773, respectively, on ET when only FLAIR and T1GD are available). Similarly to the above works though, SMU-Net relies on having multiple pathways during training that can burden computing resources and make generalization to other segmentation models challenging.
Ding et al. capitalize on the different information provided by each modality with RFNet. For example, enhancing regions would be better visualized on T1Gd and edema on FLAIR. They create a separate encoder for each modality to segment each modality individually. A vowel-based probability map of each tumor region is generated and used to strategically weight the contributions from each encoder during fusion based on which sequence is ideal for the likely tumor region [14]. They further augment the handling of missing sequences by incorporating a segmentation-based regularizer so that the model is forced to identify discriminating features from each modality rather than becoming reliant on a subset of modalities. Their implementation consistently outperforms HeMIS, U-HVED, and RobustSeg with any permutation of missing sequences, achieving a DSC of 0.759 on ET when FLAIR and T1 are missing and 0.780 when FLAIR and T2 are missing.
Such prior works are cleverly designed and robust to missing modalities but share a common limitation of difficult generalizability onto existing and future segmentation models. They require altering the network architecture, complicated mathematical principles, and heavy computing resources that may prevent missing modality robustness from becoming the general expectation of all segmentation models. The objective of this study is to develop a computationally resourceful deep learning-based method robust to missing imaging modalities for automatic segmentation of brain tumor subregions that can be easily generalized to any existing segmentation model. We propose a relatively simple sequence dropout framework that only changes the training implementation by randomly “dropping” imaging modalities or replacing them with all-zero arrays throughout training while preserving the model network.
Methods
Patient Cohort
We utilized the publicly available RSNA-ASNR-MICCAI Brain Tumor Segmentation (BraTS) Challenge 2021, which comprises multi-institutional pre-operative multi-parametric brain MRI scans of GBM/high-grade glioma (HGG) and low-grade glioma (LGG) [15]. Average age was 61.2 (STD 11.9) with mean survival days 445.8 (STD 355.9). Sequences include T1, T1Gd, T2, and FLAIR volumes. A total of 1251 cases from different patients with annotated gadolinium-enhancing tumor, peritumoral edema, and necrotic tumor core were provided, where all sequences were employed for delineation of each tumor volume.
In our study, brain tumors were classified into three types: (1) ET; (2) tumor core (TC), including necrotic core, enhancing and non-enhancing tumor core; and (3) WT. The dataset was randomly split into training and testing cohorts in a 2:1 ratio. Due to the computational limitations of having to test on 15 different combinations from 4 sequences, we then randomly sampled 50 cases from the testing cohort to use for performance evaluation. The images within the testing set that were not sampled were not used anywhere else in the study.
Furthermore, we used an external dataset for additional validation/testing, where we randomly sampled 30 cases from a public dataset from The Cancer Genome Atlas Program (TCGA). Similar to the BraTS Challenge 2021 dataset, the TCGA dataset comprises multi-sequence MRI brain scans of glioblastoma (GBM) and low-grade glioma (LGG). The MR sequences include T1, T1Gd, T2, and FLAIR, with some potential missing sequences. The manual annotations (ground truth) include delineated ET volume on T1Gd, TC volume on T2, and WT volume on FLAIR. If T1Gd sequence is missing, ET volume is delineated on T1 sequence.
Image Pre-Processing
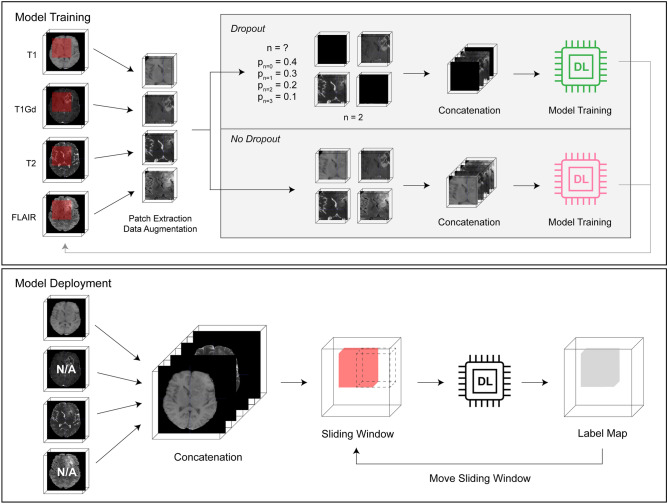
The sequence dropout framework consisted of (1) image pre-processing, (2) training the 3D nnU-Net model, and (3) deployment and validation (Fig. 1).
Fig. 1.
Framework for sequence dropout model training and deployment
The dataset already included standard pre-processing, such as co-registration to the same anatomical template, resampling to isotropic 1 mm3 resolution, and skull-stripping [15]. Each 3D image was then normalized to 0-mean, unit variance by subtracting the mean value and dividing by the standard deviation for all voxels of the 3D image. The normalized images of all sequences for each subject were then concatenated, resulting in a whole input image size of MxNxPx4, where M,N,P correlate to the whole image dimensions, in the following order: T1, T1Gd, T2, FLAIR. Corresponding multi-class ground truth label maps were generated, with background labeled “0,” necrosis and non-enhancing tumor core “1,” enhancing tumor core “2,” and peritumoral edema/infiltrative tumor “3.”
Model Architecture and Training
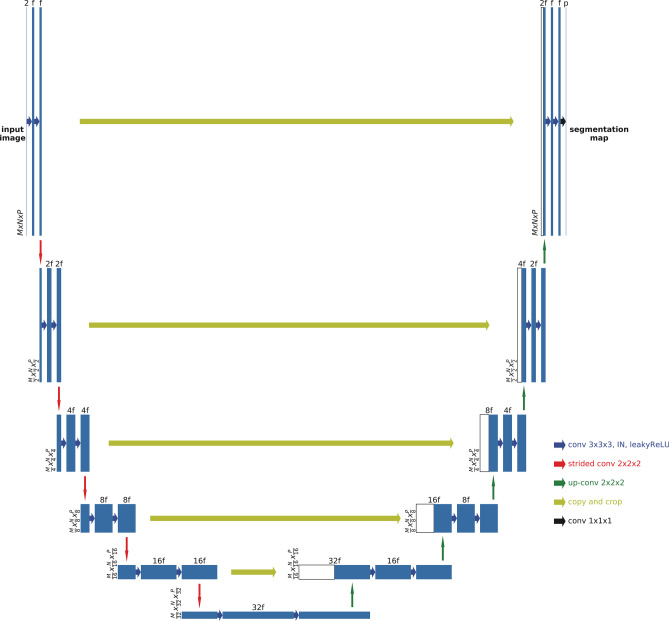
The model used an nnU-Net architecture, which uses the traditional 3D U-Net in an adaptive way, with 6 levels and Tensorflow backend (Fig. 2) [16, 17]. Each encoding block consists of two consecutive 3D convolutional layers, followed by instance normalization and leaky rectified linear activation layers. For the decoding blocks, symmetric blocks were used with skip connections from corresponding encoding blocks, with 3D convolutional layers replaced by 3D transposed convolutional layers. Features were concatenated to the de-convolution outputs, and the segmentation map of the input patch was expanded to the multi-class (3 foreground classes and background) ground truth labels.
Fig. 2.
3D U-Net structure with 5 encoding and 5 decoding blocks
To accommodate GPU memory limitations and focus training onto target lesions, patches of 96×96x96×4 with 50% bias to foreground voxels were randomly extracted from each image at each epoch during training. Data augmentation was then performed onto the patches by randomly incorporating left–right flip, rotation, scaling, gaussian noise, contrast, brightness, and scaling transformations before being input into model training.
Deep supervision was applied to the network by computing loss at each decoding block (except the bottleneck layer and first decoding block). This approach allows for gradients to be injected deeper into the network and facilitates training at each layer [18]. The final loss was calculated as the sum of the weighted loss computed at each decoding block. Stochastic gradient descent with polynomial learning rate decay and momentum of 0.99 were used, and models optimized the sum of Dice loss and cross-entropy loss with a batch size of 1. Model training and testing were performed with a NVIDIA Titan Xp 12 GB GPU.
Stabilizing Model for Missing Imaging Sequences
In order to train a model capable of segmenting the ground truth with missing sequences, we implemented a novel technique called “sequence dropout,” inspired by the dropout method widely used in training neural networks [19]. While the dropout method randomly drops hidden units in certain layers to reduce overfitting, our sequence dropout method randomly drops MRI sequences when forming training inputs to prevent complex co-adaptation between sequences in training.
Before sequence concatenation, we randomly drop n sequences by replacing them with an all-zero voxel array. By replacing the dropped-out sequence with an image of zeros, we essentially created a pure background, preventing any unwanted interference with the network. The relative probabilities of dropping n random sequences were pn = 0 = 0.4, pn = 1 = 0.3, pn = 2 = 0.2, and pn = 3 = 0.1, respectively. After determining n, the specific sequences that were dropped were chosen randomly with uniform distribution.
To assess how sequence dropout affects the ability of a 4-sequence-input model to accurately segment regions of interest (ROI) despite missing sequences, we trained one model without and another with sequence dropout. The non-dropout model was trained for 4000 epochs, while we increased the dropout model training to 6000 epochs given the large number of input variability introduced by sequence dropout.
Given that most state-of-the-art segmentation models currently do not incorporate sequence dropout and must have all sequences the model was trained with, we also compared our dropout model capable of handling one to four sequences with models specifically trained on a subset of sequences. For instance, we trained a model that only accepts T1Gd and T2 and compared its performance to that of our dropout model if it were only given T1Gd and T2.
Model Performance Evaluation
Due to similar GPU memory limitations during model training, a sliding window approach was used to generate ROI segmentations. The deployment window size was chosen to be the same as the training patch size, and the stride was designated as 1/2 of the window size. For each window, the original image and left–right flipped image were both predicted. After flipping back the output of the flipped input, the average probability was used as the output. Therefore, each voxel, except for those on the edges, was predicted 16 times when sliding in all directions. Although smaller stride sizes could be used to further improve accuracy, the deployment time would increase by 8 times for every 1/2 reduction of the window size and quickly become unmanageable.
To evaluate the performance of our segmentation models, DSC was computed using ground truth and our model’s segmentation. Along with DSC, absolute volume error was also calculated between ground truth and predicted segmentations. We then compared the differences in performance between the dropout and non-dropout models when a certain permutation of imaging sequences was removed using paired t-tests (α = 0.05), applying the Holm-Bonferroni method to address the multiple comparisons problem.
External Validation
For evaluation with an external dataset, we used the TCGA dataset as described in the “Patient Cohort” section. After further investigation of the training dataset (BraTS Challenge 2021), we found out that 166 cases of the BraTS dataset were from the TCGA dataset. To solve the issue, we retrained a separate model on the training dataset without samples from TCGA to perform external validation calculations.
Results
Overall Comparison of Sequence Non-Dropout and Dropout Model
The dropout model performed significantly better than the non-dropout model for most MRI sequence combinations across ET, TC, and WT. With only T1Gd input, the DSC on ET, TC, and WT was significantly larger with the dropout model (0.748 vs 0.241, p ≤ 0.001; 0.771 vs 0.247, p< 0.001; 0.762 vs 0.093, p< 0.001, respectively). With only T1Gd and T1, the dropout model again achieved higher dice for segmentations of all tumor types (0.761 vs 0.401, p< 0.001; 0.717 vs 0.407, p< 0.001; 0.744 vs 0.191, p< 0.001, respectively). The dropout model was most robust to missing sequences when T1Gd was included. For instance, the DSC for ET with just a single T1Gd input was 0.748 ± 0.222, whereas the DSC with three sequences of T1, T2, and FLAIR was 0.486 ± 0.199 (p< 0.001). For all segmentations, it took about 1 min to generate a multi-class label map per subject.
Table 1 shows the DSC of the two models deployed on different combinations of the four MRI sequences to segment different tumor types (ET, TC, and WT). Similarly, Supplementary Table 1 shows the absolute volume error when the two models are deployed on different combinations of the four MRI sequences to segment ET, TC, and WT. Average tumor sizes were 18,366 mm [3]; 31,011 mm [3]; and 99,119 mm [3] for ET, TC, and WT, respectively.
Table 1.
Dice similarity coefficient (DSC) for different combinations of MRI sequences
| MRI sequences | Enhancing tumor | Tumor core | Whole tumor | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Flair | T1 | T1Gd | T2 | No dropout | Dropout | p | No dropout | Dropout | p | No dropout | Dropout | p |
| ✔ | 0.085 | 0.363 | < 0.001 | 0.140 | 0.596 | < 0.001 | 0.315 | 0.812 | < 0.001 | |||
| ✔ | 0.241 | 0.748 | < 0.001 | 0.247 | 0.771 | < 0.001 | 0.093 | 0.762 | < 0.001 | |||
| ✔ | 0.005 | 0.411 | < 0.001 | 0.002 | 0.592 | < 0.001 | 0.036 | 0.682 | < 0.001 | |||
| ✔ | 0.309 | 0.349 | 0.664 | 0.454 | 0.583 | < 0.001 | 0.805 | 0.864 | < 0.001 | |||
| ✔ | ✔ | 0.731 | 0.803 | 0.002 | 0.740 | 0.844 | < 0.001 | 0.813 | 0.879 | < 0.001 | ||
| ✔ | ✔ | 0.401 | 0.761 | < 0.001 | 0.407 | 0.717 | < 0.001 | 0.191 | 0.744 | < 0.001 | ||
| ✔ | ✔ | 0.057 | 0.489 | < 0.001 | 0.391 | 0.685 | < 0.001 | 0.774 | 0.866 | < 0.001 | ||
| ✔ | ✔ | 0.003 | 0.464 | < 0.001 | 0.216 | 0.656 | < 0.001 | 0.651 | 0.868 | < 0.001 | ||
| ✔ | ✔ | 0.262 | 0.394 | < 0.001 | 0.441 | 0.626 | < 0.001 | 0.833 | 0.898 | < 0.001 | ||
| ✔ | ✔ | 0.793 | 0.807 | 0.313 | 0.832 | 0.848 | 0.535 | 0.860 | 0.882 | 0.012 | ||
| ✔ | ✔ | ✔ | 0.800 | 0.807 | 1.000 | 0.861 | 0.861 | 1.000 | 0.846 | 0.882 | < 0.001 | |
| ✔ | ✔ | ✔ | 0.143 | 0.486 | < 0.001 | 0.431 | 0.680 | < 0.001 | 0.854 | 0.901 | 0.007 | |
| ✔ | ✔ | ✔ | 0.803 | 0.808 | 1.000 | 0.846 | 0.857 | 0.664 | 0.902 | 0.907 | 0.686 | |
| ✔ | ✔ | ✔ | 0.798 | 0.810 | 0.664 | 0.802 | 0.859 | 0.018 | 0.782 | 0.886 | < 0.001 | |
| ✔ | ✔ | ✔ | ✔ | 0.813 | 0.815 | 1.000 | 0.866 | 0.871 | 1.000 | 0.904 | 0.907 | 0.799 |
Mean DSC with bolded text indicate statistically significant superior performance
P-values are from paired t-tests comparing the dropout and no dropout models and are adjusted for multiple comparisons with the Holm-Bonferroni method
When tested on individual MRI sequences only, the dropout model outperformed the non-dropout model in a statistically significant way for all sequences when segmenting ET, TC, and WT, except with FLAIR for ET where there was no significant difference between models (p = 0.664). This improvement was least notable when deployed on FLAIR alone, with DSC increasing from 0.454 to 0.583 (p< 0.001) and 0.805 to 0.864 (p< 0.001) for TC and WT, respectively. Interestingly, the non-dropout model achieved the highest DSC for segmentations for all tumor types when deployed on FLAIR among all the individual sequences.
When all four MRI sequences were available, there was no significant difference between the dropout and non-dropout models across segmentations of all tumor types (p = 1.000, 1.000, 0.799 for ET, TC, and WT, respectively). Average performance between the two models for ET, TC, and WT was 0.814, 0.868, and 0.906.
Dropout Model Performance with Missing Sequences
When one sequence was missing—out of the four different combinations of remaining available sequences—the DSC of the dropout model was statistically different to its DSC with no missing sequences for only one combination for ET (FLAIR, T1, T2), one combination for TC (FLAIR, T1, T2), and two combinations for WT (FLAIR, T1, T1Gd and T1, T1Gd, T2).
When two sequences were missing—out of the six different combinations of remaining available sequences—the DSC of the dropout model was statistically different to its DSC with no missing sequences for four, four, and five combinations for ET, TC, and WT, respectively.
When only one sequence was available, the dropout model performed significantly worse than its performance with no missing sequences. Specific p-values of comparisons between the performance of the dropout model with missing sequences versus with no missing sequences are shown in Supplementary Table 2. These p-values were determined using paired t-tests (α = 0.05) and applying the Holm-Bonferroni method for multiple comparisons.
Evaluation of T1Gd for Segmentation of ET and TC
When T1Gd was missing, the dropout model performed significantly better. For example, when tested on only T1, T2, and FLAIR sequences together, DSC for ET, TC, and WT increased from 0.143 to 0.486 (p< 0.001), 0.431 to 0.680 (p< 0.001), and 0.854 to 0.901 (p = 0.007), respectively.
For ET, the absence of T1Gd sequence in deployment was significantly detrimental to segmentation performance, with DSC decreasing from 0.798 (T1, T1Gd, T2) to 0.143 (T1, T2, FLAIR) for the non-dropout model (p< 0.001). With dropout, DSC still decreased from 0.810 (T1, T1Gd, and T2) to 0.486 (T1, T2, FLAIR) (p< 0.001). The combination of T1Gd and either T2 or FLAIR with dropout achieved DSC of 0.803 and 0.807, respectively, which was not significantly different to performance with all sequences (DSC = 0.815, p = 0.294 and 0.899).
T1Gd was also the most important sequence in segmenting TC, with DSC decreasing from 0.802 (T1, T1Gd, T2) to 0.431 (T1, T2, FLAIR) (p< 0.001) with the non-dropout model when T1Gd was missing. For the dropout model, DSC decreased from 0.859 (T1, T1Gd, T2) to 0.680 (T1, T2, FLAIR) (p< 0.001).
When deployed solely on T1Gd, the model with sequence dropout achieved relatively high performance for segmentations of all tumor types, achieving DSC of 0.748, 0.771, and 0.762 for ET, TC, and WT, respectively.
Evaluation of T2 and FLAIR for WT Segmentation
For WT segmentation, the dropout model achieved the highest DSC of 0.812 when deployed on T2 alone and 0.864 on FLAIR alone when comparing single input sequence performance. Without dropout, the DSC for T2 and FLAIR were 0.315 and 0.805, respectively, compared to its performance of only 0.191 when deployed on both T1 and T1Gd. The highest DSC for a two-sequence combination was 0.898, deployed on T2 and FLAIR with dropout.
Comparing DSC and Absolute Volume Error Results
Absolute volume error results are generally in agreement with DSC where the sequence dropout method improved segmentation performance. Among instances where absolute volume error for ET was greater in the dropout model (T1Gd, FLAIR; T1, T1Gd, FLAIR; T1Gd, T2, FLAIR; T1, T1Gd, T2, FLAIR), there was no significant difference in DSC between both models.
External Validation Showing Some Generalizability
Model performance (trained without TCGA cases) on the external dataset was 0.525 for ET, 0.636 for TC, and 0.797 for WT. The model was deployed on all available MR sequences, where there were some missing sequences in some cases. As mentioned in the “Patient Cohort” section, there were some differences in annotation protocol between the training dataset (BraTS) and external validation dataset (TCGA) leading to inter-observer variability. In the BraTS dataset, the annotation was produced by employing multiple MR sequences (T1, T1Gd, T2, and FLAIR) for each tumor volume. With the TCGA dataset, on the other hand, annotation was produced for each tumor volume using specific MR sequence only (T1Gd for ET, T2 for TC, and FLAIR for WT). The differences in annotation protocol account for the significant decrease in performance on external validation dataset. While the model performance seems lower compared to the internal split validation dataset, there is some generalizability where the tumor volumes for WT can be segmented reliably, and the volumes for TC can be segmented somewhat reliably.
Qualitative Comparison of Model Outputs
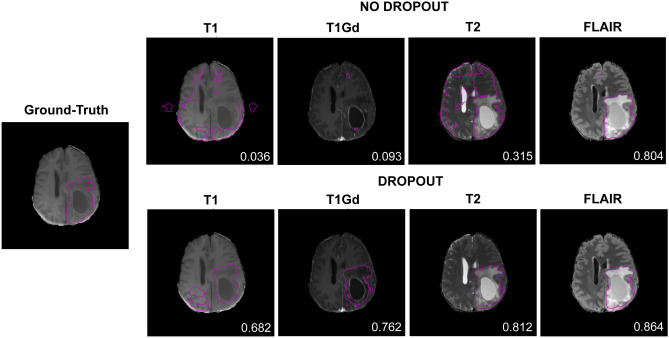
Figure 3 shows the results of segmentation when the models (no dropout and dropout) were deployed on one MRI sequence from the BraTS dataset. The results from the model with sequence dropout were visibly improved over the model without dropout, except when deployed on FLAIR where segmentations appear similar.
Fig. 3.
Segmentation results of models without and with sequence dropout on individual MRI sequences. Contoured regions represent whole tumor (WT). Dice similarity coefficient (DSC) is indicated in each image
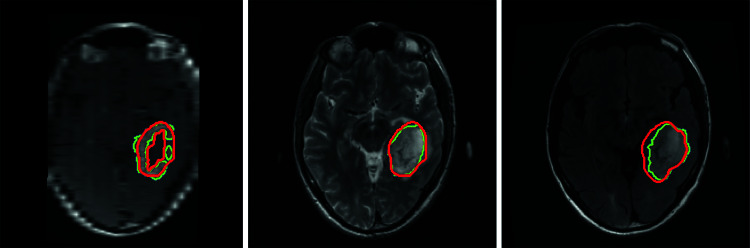
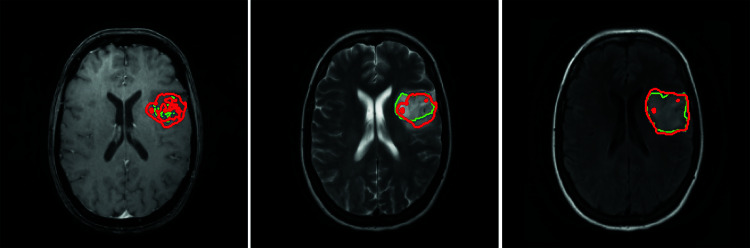
Figure 4 shows the results of segmentation when the final model was deployed on a case from an external (TCGA) dataset. For visualization, ET, TC, and WT are shown on T1Gd, T2, and FLAIR, respectively.
Fig. 4.
Segmentation results of the final model on a case from external dataset. Contoured regions represent enhanced tumor (ET) on T1Gd (left), tumor core (TC) on T2 (middle), and whole tumor (WT) on FLAIR (right). Green contour represents ground truth, and red contour represents model output. DSC 0.833 (left), 0.884 (middle), and 0.806 (right)
Figure 5 shows the results of segmentation when the final model was deployed on a case with missing T1Gd sequence from the external (TCGA) validation dataset. In this scenario, manual annotation was performed on T1 sequence. With T1Gd missing, the segmentation model suffers and performs poorly for ET. However, the performance on TC is fairly well despite missing an important sequence (T1Gd) for TC. Furthermore, the manual annotation for ET on T1 sequence we believe might also not be fully reliable since the T1 does not provide the contrast enhancement necessary to identify the enhanced region of the tumor.
Fig. 5.
Segmentation results of final model on a case from external dataset with T1Gd sequence missing. Contoured regions represent enhanced tumor (ET) on T1 (left), tumor core (TC) on T2 (middle), and whole tumor (WT) on FLAIR (right). Green contour represents ground truth, and red contour represents model output. DSC 0.168 (left), 0.719 (middle), and 0.878 (right)
Comparison of Dropout Model with Missing Sequences to Model Trained Specifically with Just the Available Sequences
Table 2 compares the DSC results for the dropout model versus a fixed, limited input model trained only on T1Gd as well as the dropout model versus a fixed, limited input model trained only on T1Gd and T2 across all tumor subtypes. The p-values for these comparisons were determined using paired t-tests (α = 0.05). The differences in performance are statistically significant for all of these combinations, except for TC when trained on T1Gd and T2 (p = 0.405). However, these differences have minimal clinical significance, as the differences in DSC across all tumor subtypes when trained on T1Gd only are around 0.07–0.08 and the differences across all tumor subtypes when trained only on T1Gd and T2 are around 0.02–0.03.
Table 2.
DSC between models trained on fixed, limited subset of inputs versus dropout model
| MRI sequences | Enhancing tumor (ET) | Tumor core (TC) | Whole tumor (WT) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Flair | T1 | T1Gd | T2 | Fixed input | Dropout | Fixed input | Dropout | Fixed input | Dropout |
| ✔ | 0.822 ± 0.198 | 0.748 ± 0.222 | 0.853 ± 0.203 | 0.771 ± 0.227 | 0.839 ± 0.150 | 0.762 ± 0.157 | |||
| ✔ | ✔ | 0.829 ± 0.169 | 0.803 ± 0.186 | 0.861 ± 0.190 | 0.844 ± 0.177 | 0.896 ± 0.097 | 0.879 ± 0.106 | ||
T1Gd p-values comparing ET, TC, and WT, respectively:< 0.001, 0.002,< 0.001. T1Gd, T2 p-values: 0.017, 0.405, 0.017
Discussion
Missing sequences are common in clinical practice, whether that be due to a certain sequence not being ordered, significant motion artifact, or data loss. Given that many of the current state-of-the-art models expect a certain series of sequences, they can be unusable or suffer in performance in these scenarios. Our study addresses this by randomly dropping MRI sequences when forming training inputs and thus preventing complex co-adaptation between sequences in training. This ensures that a network can learn intrinsic information from each sequence and any subsequent combination. This technique has the potential to improve the robustness of a single model trained on multi-modal MRI sequences in deployment on either single-modality images or multi-modality MR images as needed.
The dropout model performed significantly better than the non-dropout model for most MRI sequence combinations across ET, TC, and WT especially when the number of missing sequences increased.
For ET and TC, T1Gd was an essential sequence for accurate segmentation. This inherently makes sense as the contrasting enhancing portion comprises much of the lesion perimeter for both ET and TC and would only be visible on T1Gd. Despite this, even when T1Gd was missing, the dropout model was able to utilize some information from other sequences to identify the ROI as we can see almost a four-fold improvement in DSC by the dropout model for segmenting the ET when only given T1, T2, and FLAIR. The robustness of the dropout model despite missing sequences and especially with missing T1Gd in segmenting ET is further relevant with the current gold standard for response assessment by the Response Assessment in Neuro-Oncology relying on contrast-enhancing lesions for tumor size calculation [20].
For WT, FLAIR and T2 were the most important sequences. Even when the dropout model was missing 75% of the sequences, it was able to achieve DSCs of 0.812 with T2 alone and 0.864 on FLAIR alone. The importance of FLAIR is further seen as it was able to achieve a DSC of 0.805 with the non-dropout model, indicating predictive models naturally learn to zero out the weights from the input channels of the other sequences. This also makes sense given that peritumoral edema and infiltrative neoplasm would comprise much of the WT perimeter. With FLAIR and T2 making fluid hyperintense and FLAIR attenuating CSF such that only fluid abnormalities remain bright, the WT border would be most visible on these sequences.
While most of this study focuses on the degree to which the sequence dropout method stabilizes performance despite missing sequences, we were also interested in the performance of the dropout model with a limited subset of input sequences versus a model specially trained on this limited subset. Given that the current gold standard for segmentation models expect the full subset of sequences it was trained on and are vulnerable to missing sequences, such comparison would allow us to determine if a dropout model capable of accepting a variable number of input sequences can be the new gold standard without performance compromise. While there are statistically significant differences between the limited, fixed input model and the dropout model for T1Gd only and T1Gd and T2 inputs across ET, TC, and WT, these differences had minimal clinical significance since the differences in DSC were relatively low. This minimal clinical difference supports the application of dropout models in the context of missing sequences.
Notably, the above findings of important sequences do not necessarily indicate that a single T1Gd sequence input model for TC and a single FLAIR sequence input model for WT can replace a four-sequence input model robust to missing sequences. Given that glioblastomas and gliomas are one of the most common central nervous tumors, other brain tumor datasets are likely to identify similar key sequences but may differ if the tumor subtype distribution is different [15]. HGGs, for example, tend to disrupt the blood–brain barrier with subsequent contrast leakage more so than LGG. Furthermore, peritumoral edema is also more characteristic of HGG compared to LGG due to greater disruption and infiltration [15]. Accordingly, certain MRI sequences may purposefully not be ordered due to lower utility, further accentuating the importance of having a model capable of both single and multiple sequence inputs.
The dropout model demonstrates similar performance to previously published works that also explore brain tumor segmentation with missing MRI sequences. Table 3 shows both our results and the results of ACN, SMU-Net, and RFNet. These prior studies are recent works that have demonstrated superior performance to previous state-of-the-art methods, like U-HeMIS and HVED, on BraTS datasets [6, 13, 14]. The prior study results are from their respective publications and do not use the same BraTS 2021 dataset version as our study does. For gross standardization of results, Table 3 also shows the performance of the model given a certain permutation of missing modalities as a percentage to the DSC when all modalities are available. Because the studies use different datasets, no claims of superiority among models can be made. However, given the similarity of datasets, Table 3 suggests that our methods achieve similar performance despite its marked simplicity. Interestingly, the performance dependence of a particular tumor region segmentation mentioned above may suggest that dropout may inherently take advantage of certain modalities being more sensitive to a particular tumor region similarly to the design of RFNet. As mentioned previously, RFNet creates a voxel-based probability map for each tumor region. It then uses separate encoders for each modality and assigns higher weights during fusion to the features returned from the encoder of the modality that is most sensitive to the highest probably tumor region. For example, RFNet achieves DSC of 0.749 for ET and 0.873 for WT on the BraTS 2020 dataset when only T1Gd and FLAIR are available, respectively. Likewise, dropout achieves a DSC of 0.748 for ET and 0.864 for WT, respectively, on the BraTS 2021 dataset.
Table 3.
DSC results in comparison to recent works that use BraTS data for brain tumor segmentation [6, 13, 14]. Prior model results are taken directly from respective publication and displayed here for ease of comparison. ACN and SMU use the BraTS 2018 dataset, and RFNet uses the 2020 dataset. Our study dropout (bolded) uses the 2021 dataset. The value in parentheses represents the percent of the DSC result with respect to that using all MRI sequences. With the displayed works using different datasets, direct comparisons for superiority should not be made, but given the similarity of datasets, a baseline performance regarding studies that address missing modalities may be appreciated
| MRI sequences | Enhancing tumor (ET) | Tumor core (TC) | Whole tumor (WT) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Flair | T1 | T1Gd | T2 | ACN | SMU | RFNet | Dropout | ACN | SMU | RFNet | Dropout | ACN | SMU | RFNet | Dropout |
| ○ | ○ | ○ | ● | 0.430 (55.8%) | 0.431 (54.4%) | 0.463 (59.4%) | 0.363 (44.5%) | 0.679 (79.8%) | 0.672 (77.0%) | 0.710 (83.3%) | 0.596 (68.4%) | 0.856 (95.9%) | 0.857 (96.4%) | 0.861 (94.4%) | 0.812 (89.5%) |
| ○ | ○ | ● | ○ | 0.781 (101.3%) | 0.783 (98.7%) | 0.749 (96.0%) | 0.748 (91.8%) | 0.842 (98.8%) | 0.841 (96.3%) | 0.815 (95.7%) | 0.771 (88.5%) | 0.805 (90.2%) | 0.803 (90.3%) | 0.768 (84.3%) | 0.762 (84.0%) |
| ○ | ● | ○ | ○ | 0.415 (53.9%) | 0.428 (54.0%) | 0.373 (47.8%) | 0.411 (50.4%) | 0.712 (83.6%) | 0.695 (79.6%) | 0.660 (77.5%) | 0.592 (68.0%) | 0.793 (88.9%) | 0.786 (88.4%) | 0.772 (84.7%) | 0.682 (75.2%) |
| ● | ○ | ○ | ○ | 0.428 (55.5%) | 0.461 (58.1%) | 0.382 (49.0%) | 0.349 (42.8%) | 0.677 (79.5%) | 0.718 (82.2%) | 0.692 (81.2%) | 0.583 (66.9%) | 0.873 (97.8%) | 0.875 (98.4%) | 0.873 (95.8%) | 0.864 (95.3%) |
| ○ | ○ | ● | ● | 0.757 (98.2%) | 0.757 (95.5%) | 0.759 97.3%) | 0.803 (98.5%) | 0.844 (99.1%) | 0.850 (97.4%) | 0.835 (97.9%) | 0.844 (96.9%) | 0.864 (96.9%) | 0.861 (96.9%) | 0.877 (96.3%) | 0.879 (96.9%) |
| ○ | ● | ● | ○ | 0.752 (97.6%) | 0.751 (94.7%) | 0.780 (100.0%) | 0.761 (93.4%) | 0.846 (99.3%) | 0.844 (96.7%) | 0.834 (97.9%) | 0.717 (82.3%) | 0.801 (89.7%) | 0.803 (90.3%) | 0.811 (89.0%) | 0.744 (82.0%) |
| ● | ● | ○ | ○ | 0.437 (56.7%) | 0.440 (55.5%) | 0.410 (52.6%) | 0.489 (60.0%) | 0.713 (83.7%) | 0.712 (81.6%) | 0.731 (85.8%) | 0.685 (78.6%) | 0.875 (98.1%) | 0.873 (98.2%) | 0.897 (98.5%) | 0.866 (95.5%) |
| ○ | ● | ○ | ● | 0.474 (61.5%) | 0.477 (56.4%) | 0.457 (58.6%) | 0.464 (56.9%) | 0.733 (86.0%) | 0.735 (84.2%) | 0.731 (85.8%) | 0.656 (75.3%) | 0.855 (95.8%) | 0.856 (96.3%) | 0.877 (96.3%) | 0.868 (95.7%) |
| ● | ○ | ○ | ● | 0.460 (59.6%) | 0.460 (58.0%) | 0.493 63.2%) | 0.394 (48.3%) | 0.716 (84.1%) | 0.712 (81.6%) | 0.741 (87.0%) | 0.626 (71.9%) | 0.878 (98.4%) | 0.879 (98.9%) | 0.899 (98.6%) | 0.898 (99.0%) |
| ● | ○ | ● | ○ | 0.775 (100.5%) | 0.773 (97.5%) | 0.767 (98.3%) | 0.807 (99.0%) | 0.834 (97.9%) | 0.841 (96.3%) | 0.847 (99.3%) | 0.848 (97.4%) | 0.883 (98.9%) | 0.884 (99.4%) | 0.899 (98.7%) | 0.882 (97.2%) |
| ● | ● | ● | ○ | 0.762 (98.8%) | 0.762 (96.1%) | 0.768 (98.5%) | 0.807 (99.0%) | 0.843 (98.9%) | 0.842 (96.4%) | 0.851 (99.8%) | 0.861 (98.9%) | 0.890 (99.7%) | 0.882 (99.2%) | 0.907 (99.5%) | 0.882 (97.2%) |
| ● | ● | ○ | ● | 0.421 (54.6%) | 0.431 (54.4%) | 0.499 (64.0%) | 0.486 (59.6%) | 0.679 (79.7%) | 0.679 (77.8%) | 0.752 (88.2%) | 0.680 (78.1%) | 0.884 (99.0%) | 0.883 (99.3%) | 0.906 (99.4%) | 0.901 (99.3%) |
| ● | ○ | ● | ● | 0.760 (98.6%) | 0.754 (95.1%) | 0.771 (98.8%) | 0.808 (99.1%) | 0.829 (97.3%) | 0.825 (94.5%) | 0.850 (99.7%) | 0.857 (98.4%) | 0.883 (99.0%) | 0.882 (99.2%) | 0.907 (99.5%) | 0.907 (100.0%) |
| ○ | ● | ● | ● | 0.761 (98.8%) | 0.762 (96.1%) | 0.770 (98.7%) | 0.810 (99.4%) | 0.847 (99.4%) | 0.844 (96.7%) | 0.835 (98.0%) | 0.859 (98.6%) | 0.869 (97.4%) | 0.865 (97.3%) | 0.883 (96.9%) | 0.886 (97.7%) |
| ● | ● | ● | ● | 0.771 (-) | 0.793 (-) | 0.780 (-) | 0.815 (-) | 0.852 (-) | 0.873 (-) | 0.852 (-) | 0.871 (-) | 0.892 (-) | 0.889 (-) | 0.911 (-) | 0.907 (-) |
The major strength of this study is the simplicity of the sequence dropout implementation. By solely replacing an input sequence with an all-zero array and not altering the out-of-the-box neural network architecture, we avoid complex strategies, such as artificially generating the missing sequence, and allow generalizability to future studies. Despite its straightforward design, our comparisons with prior studies suggest a similar degree of robustness to current state-of-the-art methods that utilize a more complicated approach.
By using an external validation dataset separate from the training dataset, some generalizability among different datasets has been established. The model performance on the external dataset seems to be lower compared to the internally split validation dataset. However, accounting for the significant difference in annotation protocol among the datasets and the fact that TC and WT volumes could be segmented reliably, our model shows great potential in generalizability.
One limitation of this study is that it does not investigate the model performance in a real-world setting. Given that every sequence was available for all patients, we artificially assign different permutations of sequences as missing to generate performance metrics. In the clinical setting, certain sequences may be more readily available than others. Future studies may investigate which sequences provide the most clinical utility to determine which portion of our results would be most applicable in the real-world setting. Another limitation is that in clinical practice, the concept of “missing” is less straightforward unless a sequence was never captured initially. For example, the degree of motion artifact or differences in optimal image acquisition time for contrast studies that would deem a sequence unusable or missing is unclear. With our results demonstrating that certain sequences are more informative, the model may benefit in including a suboptimal image or be robust enough to drop it despite minimal artifact based on the sequence. Further studies may incorporate an automatic classifier in the usability of available sequences to increase the clinical utility of sequence dropout models.
Conclusion
In this study, we developed a self-adaptive network for brain tumor segmentation from multi-sequence MRI using the sequence dropout technique. This network can adapt to different combinations of input sequences and achieve maximal performance similar to training a new network using the corresponding input sequences.
T1Gd was critical to the segmentation of ET and TC, while FLAIR and T2 were important for WT. The conclusions drawn on the level of importance of different MRI sequences can translate to an efficient clinical workflow where informed decisions can be made on what MRI scans are essential for brain tumor segmentation.
Furthermore, based on our experiment with the external dataset, while the model performance seems lower compared to the internal split validation dataset, there is some generalizability where the tumor volumes can be segmented reliably, especially for TC and WT.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
All authors contributed to the study conception and design. Material preparation, data collection, and experimentation were done by XF, KG, QC, and SP. External dataset preparation and annotation were done by JP. Data analysis and the first draft of the manuscript were performed by XF, KG, DDK, RSC, and HZ. All authors commented on the previous version of the manuscript. All authors read and approved the final manuscript.
Funding
This project was funded by NCI R44CA254844 to XF, the Natural Science Foundation of China 81971696 to LY, and the Natural Science Foundation of Hunan 2022JJ30861 to LY.
Declarations
Ethics Approval
The study used a public dataset of MRI images, and informed consent was not applicable.
Consent to Participate
This study did not use human subjects. It used a public dataset of MRI images.
Consent for Publication
This study did not use individual person’s data.
Competing Interests
RSC reports personal fees from Roivant Sciences, personal fees from Sumitovant Biopharma, outside the submitted work. XF, KG, GH, and QC are employees of Carina Medical LLC. The rest of the authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Li Yang, Email: yangli762@csu.edu.cn.
Harrison Bai, Email: hbai7@jhu.edu.
References
- 1.Yang Y, Zhuang Y, Pan Y. Multiple knowledge representation for big data artificial intelligence: framework, applications, and case studies. Frontiers of Information Technology & Electronic Engineering. 2021/12/01 2021;22(12):1551–1558. 10.1631/FITEE.2100463
- 2.Havsteen I, Ohlhues A, Madsen KH, Nybing JD, Christensen H, Christensen A. Are Movement Artifacts in Magnetic Resonance Imaging a Real Problem?-A Narrative Review. Front Neurol. 2017;8:232. doi: 10.3389/fneur.2017.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad R, Khosravi N, Dehghanmanshadi M, Cohen-Adad J, Merhof D. Medical image segmentation on mri images with missing modalities: A review. arXiv preprint https://arxiv.org/abs/2203.06217. 2022
- 4.Havaei M, Guizard N, Chapados N, Bengio Y. Hemis: Hetero-modal image segmentation. Springer; 2016:469–477
- 5.Dorent R, Joutard S, Modat M, Ourselin S, Vercauteren T. Hetero-modal variational encoder-decoder for joint modality completion and segmentation. Springer; 2019:74–82
- 6.Wang Y, Zhang Y, Liu Y, et al. Acn: Adversarial co-training network for brain tumor segmentation with missing modalities. Springer; 2021:410–420
- 7.Wang Q, Zhan L, Thompson P, Zhou J. Multimodal Learning with Incomplete Modalities by Knowledge Distillation. Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. 2020:1828–1838. 10.1145/3394486.3403234
- 8.Zhou T, Canu S, Vera P, Ruan S. Conditional generator and multi-sourcecorrelation guided brain tumor segmentation with missing MR modalities. arXiv preprint https://arxiv.org/abs/2105.13013. 2021
- 9.Zhou T, Canu S, Vera P, Ruan S. Latent Correlation Representation Learning for Brain Tumor Segmentation With Missing MRI Modalities. IEEE Transactions on Image Processing. 2021;30:4263–4274. doi: 10.1109/TIP.2021.3070752. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Dou Q, Jin Y, Chen H, Qin J, Heng P-A. Robust Multimodal Brain Tumor Segmentation via Feature Disentanglement and Gated Fusion. Springer International Publishing; 2019:447–456
- 11.Sharma A, Hamarneh G. Missing MRI Pulse Sequence Synthesis Using Multi-Modal Generative Adversarial Network. IEEE Transactions on Medical Imaging. 2020;39(4):1170–1183. doi: 10.1109/TMI.2019.2945521. [DOI] [PubMed] [Google Scholar]
- 12.Gatys LA, Ecker AS, Bethge M. A neural algorithm of artistic style. arXiv preprint https://arxiv.org/abs/1508.06576. 2015
- 13.Azad R, Khosravi N, Merhof D. SMU-Net: Style matching U-Net for brain tumor segmentation with missing modalities. Medical Imaging with Deep Learning. 2022
- 14.Ding Y, Yu X, Yang Y. RFNet: Region-aware Fusion Network for Incomplete Multi-modal Brain Tumor Segmentation. IEEE/CVF International Conference on Computer Vision (ICCV). 2021;2021:3955–3964. doi: 10.1109/ICCV48922.2021.00394. [DOI] [Google Scholar]
- 15.Baid U, Ghodasara S, Mohan S, et al. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv preprint https://arxiv.org/abs/2107.02314. 2021
- 16.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. 2015: http://arxiv.org/abs/1505.04597. Accessed May 01, 2015. https://ui.adsabs.harvard.edu/abs/2015arXiv150504597R
- 17.Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods. 2021/02/01 2021;18(2):203–211. 10.1038/s41592-020-01008-z [DOI] [PubMed]
- 18.Bakas S, Akbari H, Sotiras A, et al. Data from: Segmentation Labels for the Pre-operative Scans of the TCGA-GBM collection. 2017. The Cancer Imaging Archive. 10.7937/K9/TCIA.2017.KLXWJJ1Q
- 19.Gal Y, Ghahramani Z. Dropout as a Bayesian Approximation: Representing Model Uncertainty in Deep Learning. 2015: http://arxiv.org/abs/1506.02142. Accessed June 01, 2015. https://ui.adsabs.harvard.edu/abs/2015arXiv150602142G
- 20.Lin NU, Lee EQ, Aoyama H, et al. Response assessment criteria for brain metastases: proposal from the RANO group. Lancet Oncol. 2015;16(6):e270–e278. doi: 10.1016/s1470-2045(15)70057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.