Summary
Type 2 immune responses are critical in tissue homeostasis, anti-helminth immunity, and allergy. T helper 2 (Th2) cells produce interleukin-4 (IL-4), IL-5, and IL-13 from the type 2 gene cluster under regulation by transcription factors (TFs) including GATA3. To better understand transcriptional regulation of Th2 cell differentiation, we performed CRISPR-Cas9 screens targeting 1,131 TFs. We discovered that activity-dependent neuroprotector homeobox protein (ADNP) was indispensable for immune reactions to allergen. Mechanistically, ADNP performed a previously unappreciated role in gene activation, forming a critical bridge in the transition from pioneer TFs to chromatin remodeling by recruiting the helicase CHD4 and ATPase BRG1. Although GATA3 and AP-1 bound the type 2 cytokine locus in the absence of ADNP, they were unable to initiate histone acetylation or DNA accessibility, resulting in highly impaired type 2 cytokine expression. Our results demonstrate an important role for ADNP in promoting immune cell specialization.
Keywords: immunity, ADNP, T helper 2 cells, Th2, type 2 cytokines, IL-13, asthma, histone remodeling, AP-1, GATA3
Graphical abstract
Highlights
-
•
CRISPR screen identifies ADNP-mediated regulation of type 2 cytokine expression
-
•
ADNP is required to promote type 2 cytokine production
-
•
ADNP recruits the CHD4-BRG1 complex to remodel chromatin at the Th2 cytokine locus
-
•
ADNP-deficient mice display impaired antigen-specific Th2 cell responses
Th2 cells orchestrate type 2 immune responses by producing specific cytokines, but the mechanism underlying their regulation is not fully understood. Ferreira et al. discover that the neuroprotective protein ADNP is required for effective IL-13 production by Th2 cells and is indispensable for immune reactions to allergens.
Introduction
To perform their roles, T lymphocytes and innate lymphoid cells (ILCs) must be able to differentiate in response to external stimuli to initiate specialized protective immune responses. For example, T cells and ILCs express interferon-γ promote so-called type 1 defenses against bacteria, viruses, and cancer, whereas fungi induce type 3 immunity.1 Type 2 immune responses are critical for tissue homeostasis, tissue repair, and protection of the host from infections such as parasitic helminths.2,3,4 However, inappropriate type 2 immunity can lead to asthma and allergy.5,6 The key cellular orchestrators of these responses are ILC2s and adaptive T helper 2 (Th2) cells, which produce the secreted type 2 effector cytokines interleukin-4 (IL-4), IL-5, and IL-13.2,3,4,5 These cytokines act as messengers to induce the widespread activation of immune effector cells such as eosinophils, mucus-producing goblet cells, B cells, T cells, and macrophages.7 Consequently, the precise regulation of type 2 lymphocytes and ILs is critical in health and disease.6
The type 2 cytokine gene locus comprises a region of ∼150 kb on mouse chromosome 11 (and human chromosome 5), which harbors the Il4, Il5, and Il13 gene cluster, encoding IL-4, IL-5, and IL-13. During Th2 cell differentiation, this locus undergoes extensive chromatin remodeling and architectural reorganization.8,9,10 The pioneer transcription factor (TF) GATA3 is the “master regulator” of Th2 cell differentiation and type 2 IL expression11 and acts synergistically with other pioneer TFs such as the activator protein 1 (AP-1) factors JUNB and BATF,12,13 and TFs from the NF-κB and NFAT families.14 By binding gene regulatory regions, these factors promote the recruitment of histone-modifying enzymes (e.g., histone acetyl transferases [HATs]) and chromatin remodeling complexes, which reshape the local chromatin landscape.15,16,17 These processes increase chromatin accessibility for the recruitment of additional factors and promote transcription. Indeed, components of the ATPase-dependent BAF (BRG/BRM associated factor, mSWI/SNF) chromatin remodeling complex, for example, Brahma-related gene-1 (BRG1, encoded by Smarca4), play key roles in Th2 cell differentiation and type 2 cytokine production.18 Contributing to this activated Th2 cell phenotype, regulators of genome architecture and long-range chromosome interactions also play roles in Th2 cell differentiation.8,19 The DNA-binding protein CCCTC-binding factor (CTCF) which insulates topologically associated domains (TADs), and cooperates with the cohesin complex to form chromatin loops to bring together enhancers and/or silencers to regulate gene expression, is known to be required for type 2 cytokine expression.20 Further, the TF Ying Yang (YY1) was found to bind multiple regions in the type 2 cytokine locus before the recruitment of GATA3, to which it associates physically and induce DNA loop formation.21
Despite our knowledge that GATA3 is necessary for type 2 cytokine expression,2,22 in combination with TFs such as NF-κB, AP-1 and NFAT, and chromatin modifiers, we lack insight into how these transitions are coordinated and maintained in a Th2-cell-specific manner. To address this question, we performed a CRISPR-Cas9 screen in differentiating Th2 cells to identify factors that selectively regulated type 2 cytokine gene expression. This approach identified a mechanism by which activity-dependent neuroprotector homeobox protein (ADNP) has a critical role in promoting the Th2 cell phenotype.
Results
Identification of ADNP as a regulator of IL-13 expression by Th2 cells
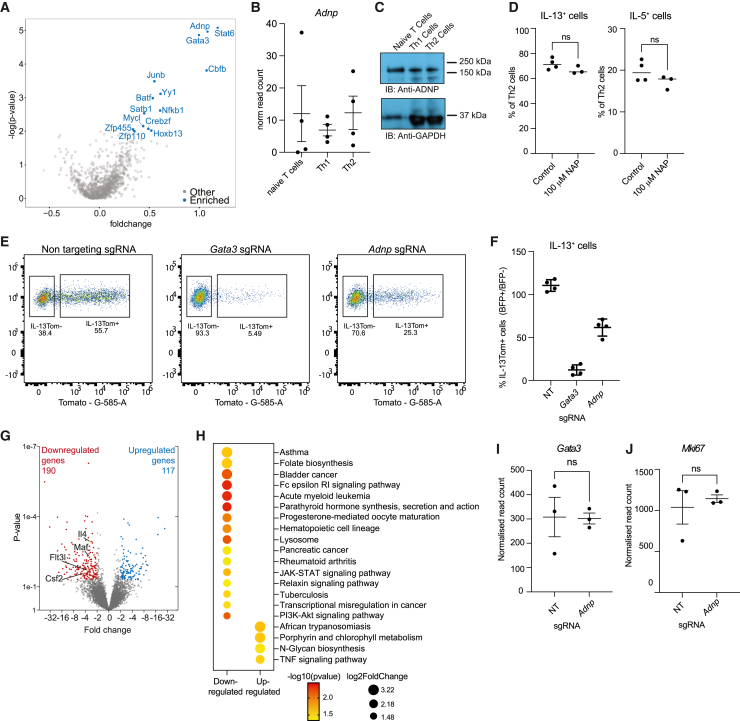
To identify additional regulators of Il13 gene expression in mouse Th2 cells from IL-13-reporter mice (Rosa26Cas9EGFPIl13tdTomato), we undertook an unbiased retrovirus-mediated CRISPR-Cas9 screening approach using a sgRNA library targeting 1,131 TFs. Naive primary splenic CD4+ T cells were retrovirally transduced and differentiated into Th2 cells by initiating T cell receptor and IL-4 signaling pathways.23 After 3 days in the CRISPR culture conditions, transduced cells were sorted by IL-13Tom expression (BFP+IL-13Tom+ and BFP+IL-13Tom−) and purified for next-generation sequencing (NGS) of integrated sgRNA inserts. Consistent with the known roles of GATA3 and STAT6 in Th2 cell differentiation, sgRNAs targeting Gata3 and Stat6 were highly enriched in IL-13Tom− populations (Figure 1A; Table S1) as were Yy1, Nfkb1, Junb, and Batf. Notably, Adnp was identified as a previously unappreciated candidate for the regulation of IL-13 expression (Figure 1A).
Figure 1.
Identification of ADNP as a regulator of IL-13 expression by Th2 cells
(A) Volcano plot showing positive regulators of Th2 cell differentiation (blue). Data are pooled from 2 independent screens.
(B) Adnp gene expression (from RNA-seq analysis) in naive T cells, Th1, and Th2 cells. Mean ± SD.
(C) Detection of ADNP (150 kDa) and GAPDH (36 kDa) proteins in T cell lysates.
(D) Flow cytometric analysis of cytokine expression by Th2 cells cultured in the presence of vehicle (PBS) or NAP. Data are representative of 2 independent experiments; unpaired two-sided t test; not significant (ns).
(E) Representative flow cytometry gating strategy for the analysis of IL-13Tom expression by Th2 cells transduced with sgRNAs.
(F) Flow cytometric analysis of IL-13Tom expression by Th2 cells transduced with sgRNAs. NT, non-targeting. Mean ± SD.
(G) Volcano plot showing RNA sequencing analysis of Adnp sgRNA-targeted versus non-targeted Th2 cells.
(H) KEGG pathway analysis of genes in Adnp sgRNA-targeted versus non-targeted Th2 cells. All shown pathways were enriched (p < 0.05).
(I and J) (I) Gata3 and (J) Mik67 gene expression (from RNA-seq analysis) in Adnp sgRNA-targeted and non-targeted cells. Mean ± SD; unpaired two-sided t test; not significant (ns).
There is little-to-no characterization of ADNP in the context of immune cells. Deletion of the Adnp in mice leads to compromised brain formation and embryonic lethality,24 and mutations in ADNP underlie the complex neurological and developmental disorder ADNP syndrome (Helsmoortel-Van der Aa autism syndrome, ADNP-related disorder).25,26 ADNP is reported to be expressed ubiquitously and have pleiotropic roles including repressive functions during embryonic cell (EC) development.27,28,29 We found that Adnp gene and protein expression were equivalent in naive T, Th1, and Th2 cells (Figures 1B and 1C). ADNP contains 9 zinc-finger domains and a C-terminal homeobox domain mediating DNA binding, and an 8 amino acid neuroprotective peptide called NAP (NAPVSIPQ) which mediates neuroprotection when secreted.30 However, treatment with NAP had no effect on Th2 cell differentiation (Figure 1D).
To confirm ADNP as a regulator of IL-13 expression in Th2 cells we used sgRNAs to individually genetically ablate Adnp in vitro, compared with non-targeting and Gata3 sgRNAs. Adnp disruption reduced the number of IL-13+ cells by approximately 50% (Figures 1E and 1F). We next performed bulk RNA-seq to compare gene expression in cultured Th2 cells following Adnp deletion as compared with non-deleted cells. Targeting Adnp resulted in the downregulation of 190 genes (including Maf, Il4, and Flt3l) and upregulation of 117 genes (Figure 1G; Table S2). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that the differentially expressed genes (DEGs) downregulated in the absence of ADNP were associated with “Asthma” pathways, which correlates with the roles of IL-13 and Th2 cells in allergic asthma (Figure 1H). Importantly, the effect of targeting Adnp was independent of GATA3 expression, which was essential for IL-13 production (Figure 1I). Additionally, the expression of Mki67 indicated that the proliferation of ADNP-deficient Th2 cells was not perturbed (Figure 1J). Thus, ADNP represents a previously unappreciated regulator of immune gene activation.
ADNP-deficient mice have reduced antigen-specific Th2 cell responses
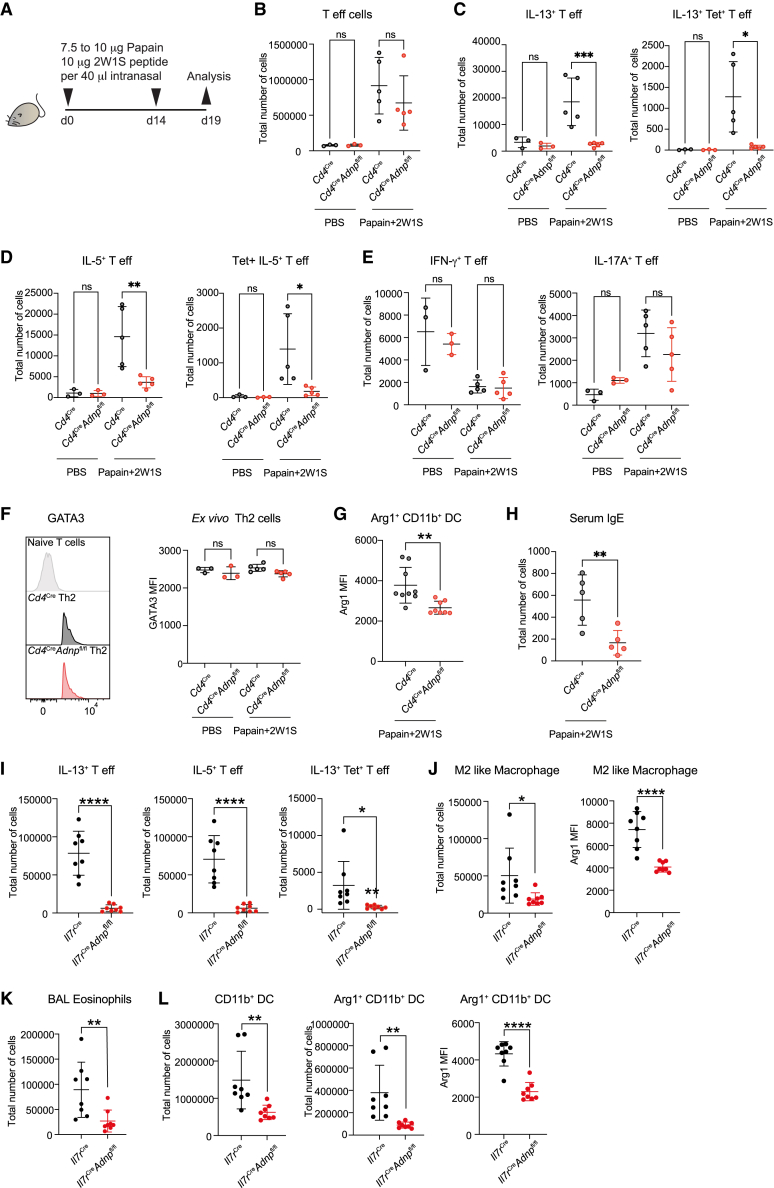
To investigate the potential impact of ADNP in primary cells and Th2 cell responses in vivo, we established Adnpfl/fl mice intercrossed with Cd4Cre mice in which ADNP was deleted preferentially in T cells (Cd4CreAdnpfl/fl mice, Figures S1A and S1B). To induce robust type 2-cell-mediated allergic immune responses in the lungs of these mice, we primed and re-challenged animals intranasally with the protease-allergen papain.31,32 The 2W1S peptide immunogen was co-administered with papain to track antigen-specific CD4+ T cells using the 2W1S:I-Ab MHCII tetramer33 (Figures 2A, S1C, and S1D). We found that although the total numbers of Th effector cells (viable CD45+CD3+CD4+CD44+ cells) were unchanged (Figure 2B), there was a reduction in IL-13 and IL-5 expression by total T effector cells and 2W1S peptide-specific T cells in the Cd4CreAdnpfl/fl mice as compared with controls in the lung (Figures 2C and 2D). In contrast, we observed no changes in IFN-γ+ or IL-17+ cells (Figure 2E). Numbers of IL-4+ Th effector cells in the lung and in the mediastinal lymph nodes (medLNs) were not impacted by ADNP deletion, though they were relatively rare (Figure S2A). The compromised IL-13 and IL-5 production was not associated with changes in GATA3 expression (Figure 2F). Total numbers of NK cells, B cells, natural killer T (NKT) cells, regulatory T (T reg) cells, CD4+ naive T cells and CD4+ Th cells were not altered at homeostasis or after antigen challenge in the lungs from Cd4CreAdnpfl/fl mice (Figures S2B and S2C). However, we observed a reduction in lung CD8+ T cells (Figure S2D). We analyzed the thymus of Cd4CreAdnpfl/fl mice to determine if there was a defect in the genesis of CD8+ T cells. We found comparable numbers of CD4 single-positive (SP) and CD8 SP cells (Figure S2E), but an increased number of CD4/CD8 double-positive (DP) cells and a decrease in NKT cells (Figures S2E and S2F). These results suggested that ADNP may play unappreciated roles at specific stages of lymphocyte development from DP to NKT cells, but not from DP to SP CD4 and CD8 cells. By performing combined adoptive transfer of equal numbers of Cd4CreAdnpfl/fl (CD45.2) and control bone marrow cells (CD45.1) into lethally irradiated recipient mice (expressing both CD45.1and CD45.2), we assessed the T cell-intrinsic versus extrinsic effects of ADNP deficiency (Figure S2G). Reconstitution was equivalent (Figures S2H and S2I). Co-reconstitution with wild-type T cells was sufficient to reverse the CD8+ T cell deficit observed above, indicating that it was not intrinsic to Cd4CreAdnpfl/fl T cells (Figure S2J). By contrast, the defects in IL-13, IL-5, and IL-4-producing Th effector cells were intrinsic to the Cd4CreAdnpfl/fl cells as they were not reversed by the presence of wild-type T cells (Figure S2K).
Figure 2.
ADNP-deficient mice have reduced antigen-specific Th2 cell responses
(A) Schematic of the experimental induction of type 2 inflammation in the mouse lung with papain and 2W1S peptide.
(B–H) Flow cytometric analysis of lung (B) total Th effector cells, (C) total and 2W1S-tetramer-specific IL-13-producing Th effector cells, (D) total and 2W1S-tetramer-specific IL-5-producing Th effector cells, (E) total IFN-γ and IL-17A-producing Th effector cells, (F) GATA3 expression in Th2 cells, (G) Arg1 expression in Arg1+CD11b+ dendritic cells (DCs), and (H) serum IgE quantified by ELISA. Data are representative of 2 independent experiments; mean ± SD; one-way ANOVA with Tukey’s post-hoc test. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, not significant (ns).
(I–L) Flow cytometric analysis of lung (I) total and 2W1S-tetramer-specific IL-13-producing Th effector cells and total IL-5-producing Th effector cells, (J) total M2 macrophage and Arg1 expression in M2 macrophage, (K) total bronchoalveolar eosinophils, (L) total CD11b+ and Arg1+CD11b+ DCs and Arg1 expression in Arg1+CD11b+ DCs. Data are representative of 2 independent experiments; mean ± SD; one-way ANOVA with Tukey’s post-hoc test. ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p < 0.05.
See also Figures S1 and S2.
In the lungs of Cd4CreAdnpfl/fl mice, we did not detect major reductions in M2 macrophages or eosinophils that are dependent on IL-13 and IL-5, respectively, but did observe reduced arginase 1 (Arg1) expression by CD11b+ dendritic cells (DCs) (Figure 2G) and a reduction in serum IgE, which are dependent on IL-13 and IL-4 (Figure 2H). We speculated whether the residual type 2 immunity observed could result from the partial functional redundancy between Th2 cells and ILC2s, with ILC2 acting as additional ADNP-dependent sources of type 2 cytokine production in Cd4CreAdnpfl/fl mice (Figure S2L). Therefore, we intercrossed Adnpfl/fl mice with Il7raCre mice (Il7raCreAdnpfl/fl mice) to delete ADNP in all lymphocytes. These mice were challenged as above with papain and 2W1S peptide. In Il7raCreAdnpfl/fl mice very few effector Th and 2W1S peptide-specific T cells or ILC2 expressed IL-13 or IL-5 (Figures 2I and S2M). Deletion of Adnp in all lymphocytes resulted in a notable reduction in B cells and NK cells (Figure S2N), and a modest decrease in CD4+ and CD8+ T cells (Figure S2O), further suggesting that ADNP may have additional roles in lymphocyte development or proliferation. Allied to the impairment in lymphocyte-derived type 2 cytokine production in Il7raCreAdnpfl/fl mice, we observed reduced M2-like macrophage polarization (Figure 2J) and eosinophilia (Figure 2K). In addition, ADNP-deficient mice presented notable reductions in CD11b+ and Arg1+CD11b+ DC recruitment and lower expression of Arg1 (Figure 2L), indicative of inefficient type 2 cytokine stimulation. Together, these results confirm a critical role for ADNP in Th2 cell polarization and ILC2 type 2 cytokine expression and type 2 immunity. Indeed, deletion of Adnp in all lymphocytes almost totally abrogated the type 2 immune response to lung allergen.
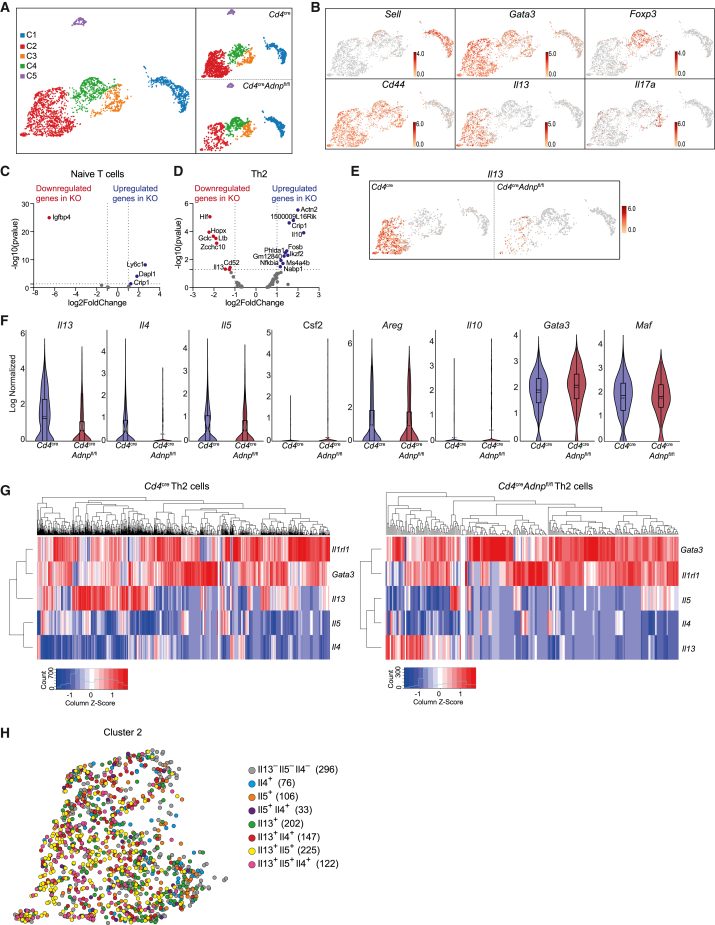
To better understand how ADNP modulates Th2 cell responses to allergen we performed single-cell RNA sequencing (scRNA-seq) analysis of naive CD4+CD44−CD62L+CD25− T cells and CD4+CD44+CD62L−ST2+ effector T cells purified from the lungs of papain-challenged Cd4Cre and Cd4CreAdnpfl/fl mice (Figure 2A). The cells formed 5 distinct clusters in both control and ADNP-deficient mice (Figure 3A). Cells in cluster 1 expressed genes characteristic of naive T cells, e.g., CD62L (encoded by Sell) while lacking expression of Cd44 (Figures 3B and S3A). Cluster 2 was characterized by the expression of Th2-cell-associated genes, including Il13, Gata3, and Il1rl1 (encoding ST2) (Figures 3B and S3A). Cluster 3 represented a minor population expressing Il17a and Il18r1, typical Th17 cell-associated genes (and were probably purified due to background anti-ST2 antibody staining) (Figures 3B and S3A). Cells in cluster 4 expressed Foxp3, characteristic of Treg cells (Figures 3B and S3A). Cluster 5 was composed of mitotic cells characterized by expression of genes associated with cell proliferation and cell cycle regulation (Mki67, Birc5, and Ccna2) (Figure S3B) and was removed from further analysis for simplification.
Figure 3.
Single-cell gene expression analysis confirms the importance of ADNP for IL-13 expression
(A) UMAP plot of single-cell gene expression analysis of naive CD4+CD44−CD62L+CD25− T cells and CD4+CD44+CD62L−ST2+ effector T cells purified from the lungs of papain-challenged Cd4Cre and Cd4CreAdnpfl/fl mice.
(B) UMAP plot with expression (log2 expression) of indicated genes per individual cell.
(C and D) Volcano plot comparing genes from naive T cells (cluster 1) (C) and Th2 cells (cluster 2) (D) from Cd4Cre and Cd4CreAdnpfl/fl (KO) mice.
(E) UMAP plot with expression (log2 expression) of Il13 per individual cell.
(F) Violin plots with expression (log2 expression) of indicated genes. Rectangle, solid line, and dashed line represent the interquartile range, the median and the mean, respectively.
(G) Heatmap showing the expression of indicated genes in cells from cluster 2 from Cd4Cre (left) and Cd4CreAdnpfl/fl (right) mice. Columns represent cluster 2 individual cells and rows represent the different genes.
(H) UMAP plot showing cluster 2. Cells are colored by their cytokine expression pattern as shown, with number of cells indicated in parentheses.
See also Figure S3.
Comparison of control naive T cells with ADNP-deficient naive T cells indicated only four DEGs (Figure 3C). The only downregulated gene was Igfbp4, which is associated with inhibition of cell proliferation and the stimulation of apoptosis but does not alter lymphocyte development.34 The absence of ADNP resulted in very few DEG in Treg or Th17 cells (Figure S3C). However, Hopx and Hlf gene expression was notable in all the Th cell populations analyzed, but not in naive T cells (Figures 3C, 3D, and S3C). Hopx is expressed in T cells where it can regulate IL-2 expression,35,36 which can promote T cell proliferation. Hlf has not been associated with T cell function. Comparing the DEG from control and ADNP-deficient Th2 cells demonstrated that Il13 expression was reduced in the absence of ADNP (Figures 3D–3G) indicating that ADNP is required for the expression of Il13 following the differentiation of naive T cells. Il4 and Il5 expression was not dysregulated, though cell numbers were small (Figures 3D, 3F, and 3G). Furthermore, the scRNA-seq data also confirmed previous reports37,38,39 that the expression of the Il13, Il4, and Il5 are not always synchronized, with individual cells expressing one, two, or all these cytokines (Figures 3G and 3H). Additionally, Il13 expression was more associated with Gata3 and Il1rl1 expression (Figure 3G), and this association was reduced in the absence of ADNP (Figure 3G). We also confirmed that Gata3 expression was not affected by ADNP deletion (Figures 3F and 3G). Other type 2 cytokines such as GM-CSF (encoded by Csf2) and Areg, were not impacted by ADNP deficiency or were not sufficiently expressed to allow detection by scRNA-seq analysis (Figure 3F). Il10 was one of the upregulated genes in Cd4CreAdnpfl/fl Th2 cells but was only detected in a few cells (Figure 3F). In contrast to what we observed in vitro, Maf expression was not reduced in ADNP-deficient Th2 cells (Figure 3F).
These data indicate that ADNP is required for Il13 expression following the differentiation of naive T cells, even in the context of normal GATA3 expression.
ADNP is associated with genes involved in lymphocyte differentiation
We next sought to identify the mechanism by which ADNP regulates IL-13 production and Th2 cell polarization. Studying ES cell differentiation, Buhler and colleagues reported that the CHD4, ADNP, HP1γ complex (called ChAHP) impairs chromatin accessibility around its DNA-binding site preventing access to transcriptional activators,28 in part by competing for CTCF sites and thus modifying TADs.27,28,29,40 Further, an ADNP complex with the chromatin remodeling regulators CHD4 and BRG1 is known to repress gene expression in mouse ES cells.27,41
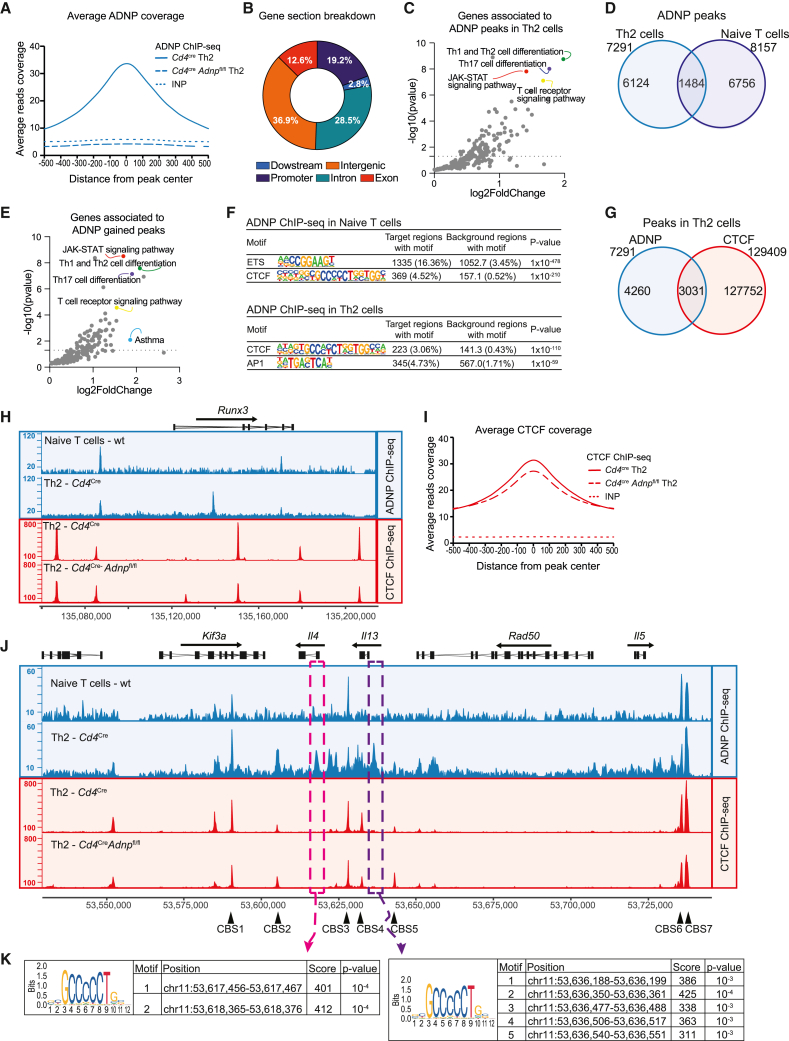
We therefore sought to define the molecular interactions of ADNP in naive T cells and in-vitro-differentiated Th2 cells using chromatin immunoprecipitation followed by sequencing (ChIP-seq). The in-vitro-differentiated Cd4CreAdnpfl/fl Th2 cells showed reduced IL-13 expression (Figures S4A and S4B), similar to in vivo Th2 cells (Figure 2C). However, no change was observed in IL-5 expression in in-vitro-differentiated cells (in contrast to in vivo) (Figures S4A and S4B). IFN-γ expression by Th1 cells was also unaffected, and we observed no deficit in T cell proliferation as assessed by expression of Ki67 or the activation marker CD25 (Figure S4B). To validate the assay, we performed ChIP-seq using ADNP-deficient Cd4CreAdnpfl/fl Th2 cells to establish assay background (Figure 4A). As in ES cells,28 around 60% of ADNP-binding sites in Th2 cells were associated with protein-coding genes and almost 20% were located at promoters (Figure 4B). Analysis of enriched KEGG pathways revealed that ADNP target genes included those associated with Th2 cell differentiation, JAK-STAT, and T cell receptor signaling, and asthma-related pathways (Figure 4C; Tables S3 and S4). Only 20% of ADNP-binding sites in Th2 cells were already occupied by ADNP in naive T cells, indicating the dynamic regulation of ADNP binding during Th2 cell differentiation (Figure 4D). By comparing ADNP binding in naive T cells and Th2 cells, we observed that those sites newly occupied by ADNP in differentiated T cells were enriched for “Th1 and Th2 cell differentiation” pathways including the genes encoding IL-4, IL-13, and cMaf (Figure 4E), whereas those originally present in the naive cells were not (Figure S4C).
Figure 4.
ADNP is associated with genes involved in lymphocyte differentiation
(A) Average ChIP-seq signal over all ADNP peaks in Th2 cells. Data are representative of 3 biological replicates.
(B) Pie chart displaying distribution of 7,291 ADNP peaks across genomic features. Promoter includes peaks between 1-kb upstream and 1-kb downstream of the transcription start site (TSS). Downstream includes peaks up to 1-kb downstream transcription termination site (TTS).
(C) KEGG pathway analysis of the genes associated to ADNP peaks. All pathways shown were enriched (p < 0.05).
(D) Venn diagram showing the overlap between ADNP ChIP-seq peaks in Th2 and naive T cells. Peak list was generated using two biological replicates.
(E) KEGG pathway analysis of the genes associated with ADNP gained peaks (peaks present in Th2 cells but not in naive T cells). All pathways shown were enriched (p < 0.05).
(F) Top two enriched motifs of ADNP peaks in naive T cells and Th2 cells. Enrichment was assessed using a one-sided cumulative binomial distribution in HOMER.
(G) Venn diagram showing the overlap between ADNP and CTCF ChIP-seq peaks in Th2 cells. Peak list was generated using two biological replicates.
(H) Representative binding profiles of ADNP in wild-type naive T cells and Cd4Cre Th2 cells and CTCF in Cd4Cre or Cd4CreAdnpfl/fl Th2 cells at the Runx3 locus. Data are representative of 3 biological replicates.
(I) Average ChIP-seq signal over all CTCF peaks in Cd4Cre or Cd4CreAdnpfl/fl Th2 cells. Data are representative of 2 biological replicates.
(J) Representative binding profiles of ADNP in wild-type naive T cells and Cd4Cre Th2 cells; and CTCF in Cd4Cre or Cd4CreAdnpfl/fl Th2 cells at the type 2 cytokine locus. Black arrows indicate CTCF-binding sites (CBSs). Data are representative of 3 biological replicates.
(K) Predicted CTCF motifs within the ADNP ChIP-seq peaks at the Il4 and Il13 promoter regions (magenta and purple rectangles in J). Prediction was performed using Jaspar transcription factor database.
Motif enrichment analysis of ADNP-associated peaks from naive T cells identified the CTCF and ETS motifs, whereas ADNP was associated with CTCF and AP-1 sites in Th2 cells (Figure 4F). This raised the possibility that in some locations ADNP might bind directly to sites in the vicinity of AP-1 sites, or directly to the AP-1 motif, or that it may associate with other proteins, such as AP-1 factors to interact with chromatin indirectly or in combination. Notably, we had identified the Junb and Batf genes, which encode the AP-1 factors JUNB and BATF, in our CRISPR-Cas9 screen (Figure 1A). The association of ADNP with CTCF motifs was confirmed using anti-CTCF ChIP-seq, with around 40% of ADNP-binding sites overlapping with CTCF binding (Figure 4G), similar to previously published ES cell data.29 However, in Th2 cells in contrast to ES cells, ADNP and CTCF did not appear to compete for the CTCF-binding sites, since CTCF binding was not affected by the absence of ADNP (Figures 4H and 4I). Moreover, the ADNP peaks identified in the naive T cells showed a greater association with CTCF than the ADNP peaks identified in the Th2 cells (Figure S4D). This suggested that an alternative mechanism may underlie ADNP-mediated gene regulation in Th2 cells.
At the Th2 cell cytokine locus, confirming previous reports, we found that CTCF bound predominantly to CTCF-binding sites (CBSs) 1, CBS3, CBS6, and CBS720 (Figure 4J). Super-imposition with ADNP binding showed that ADNP colocalized with CTCF at CBS1, CBS6, and CBS7 (Figure 4J), and that ADNP was already present at these locations in naive T cells. However, we did not observe that the deletion of ADNP influenced CTCF binding at these loci either (Figure 4J). Unexpectedly, we observed that ADNP bound to the Il13 (purple rectangle) and Il4 (magenta rectangle) promoter regions. ADNP was recruited to these locations during Th2 cell differentiation and was not present in naive T cells (Figure 4J). These regions contain predicted CTCF-binding motifs, which were not bound by CTCF in T cells, providing a platform for ADNP interaction (Figure 4K). The Il5 promoter region was not bound by ADNP (Figure 4J).
Together, these results demonstrate that under Th2 cell differentiation conditions ADNP binds to the type 2 cytokine locus in a region known to regulate Il13 gene expression, which includes CTCF and AP-1 motifs.
ADNP associates with chromatin remodeling factors and AP-1 factors in Th2 cells
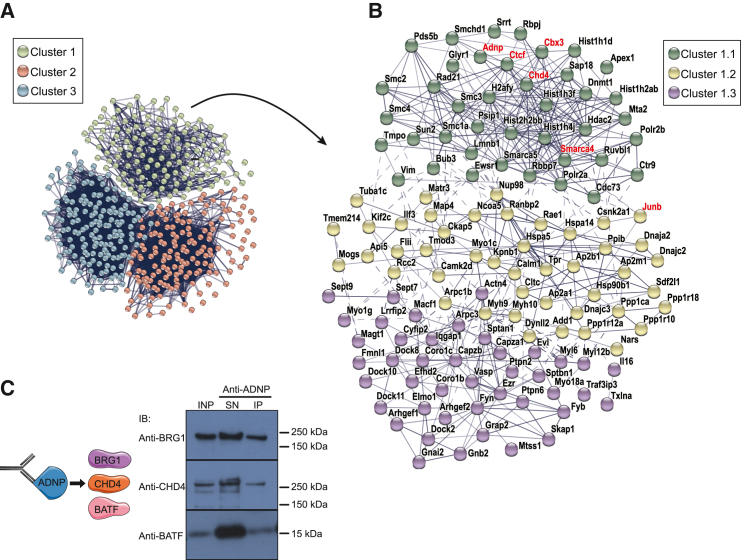
Next, to investigate possible ADNP-binding partners in Th2 cells, we performed anti-ADNP immunoprecipitation followed by mass spectrometry (MS) analysis. We identified 457 proteins as compared with isotype control (Table S5). STRING protein interaction analysis (https://string-db.org/) showed the complexity of ADNP interactions in Th2 cells with 3 major clusters: cluster 1 chromatin-binding (GO:0003682) and TF-binding (GO:0008134) factors; cluster 2, RNA binding (GO:0003723), and cluster 3, mRNA binding molecular functions (GO:0003729) (Figure 5A). We focused on cluster 1 which contains ADNP and can be subdivided into 3 subclusters (Figure 5B). Cluster 1.1 includes ADNP, histones, and components of the chromatin remodeling complexes (NuRD, SWI/SNF, and ChAHP complexes), for example, CHD4, HP1γ (CBX3), and BRG1 (SMARCA4), which associate with ADNP in ESC.28,29 CTCF and cohesins were also present in this cluster. Gene ontology analysis indicated enrichment of chromosome organization (GO:0051276), chromatin organization (GO:0006325), regulation of gene expression (GO:0010468), and histone modification (GO:0016570) biological processes. Of note, cluster 1.2 contained the AP-1 TF JUNB suggesting an association with ADNP (Figure 5B).
Figure 5.
ADNP associates with chromatin remodeling factors and AP-1 factors in Th2 cells
(A) STRING analysis of the 457 proteins immunoprecipitated with anti-ADNP from Th2 cells and identified by mass spectrometry. Connecting lines between proteins denote the confidence of the interactions (line thickness indicates the strength of data support). K means clustering was performed to identify three clusters represented by individual colors. Co-expression and co-occurrence were removed from active interaction sources.
(B) STRING analysis of cluster 1 in (A).
(C) Protein associations confirmed by co-immunoprecipitation experiments. Detection of BRG1 (181 kDa), CHD4 (226 kDa), and BATF (14 kDa) proteins in immunocomplexes generated with Th2 cell nuclear lysate co-immunoprecipitated with anti-ADNP antibody.
INP, input nuclear extract; SN, supernatant; IP, immunoprecipitation elution. Data are representative of 2 independent experiments. IB refers to immunoblotting antibody.
We performed co-immunoprecipitation (coIP) experiments to verify ADNP interaction partners. We confirmed that endogenous ADNP co-immunoprecipitated with CHD4 and BRG1 (Figure 5C). Anti-BRG1 immunoprecipitation also pulled down ADNP and CHD4 (Figure S5A). Although ADNP did not directly co-precipitate JUNB (Figure S5B), it did co-precipitate BATF (Figure 5C), which binds to JUNB to form a dimeric AP-1 TF. Further, endogenous BATF pulled down ADNP, BRG1, and JUNB (Figure S5C). These data demonstrate that ADNP can interact with CHD4 and BRG1 as well as the pioneer TF AP-1, composed of JUNB and BATF. Both BATF and JUNB are known to be important for Th2 cell differentiation and regulation of the Th2 cytokine locus.12,42
ADNP is required for efficient recruitment of CHD4 and BRG1 to the Th2 cytokine locus
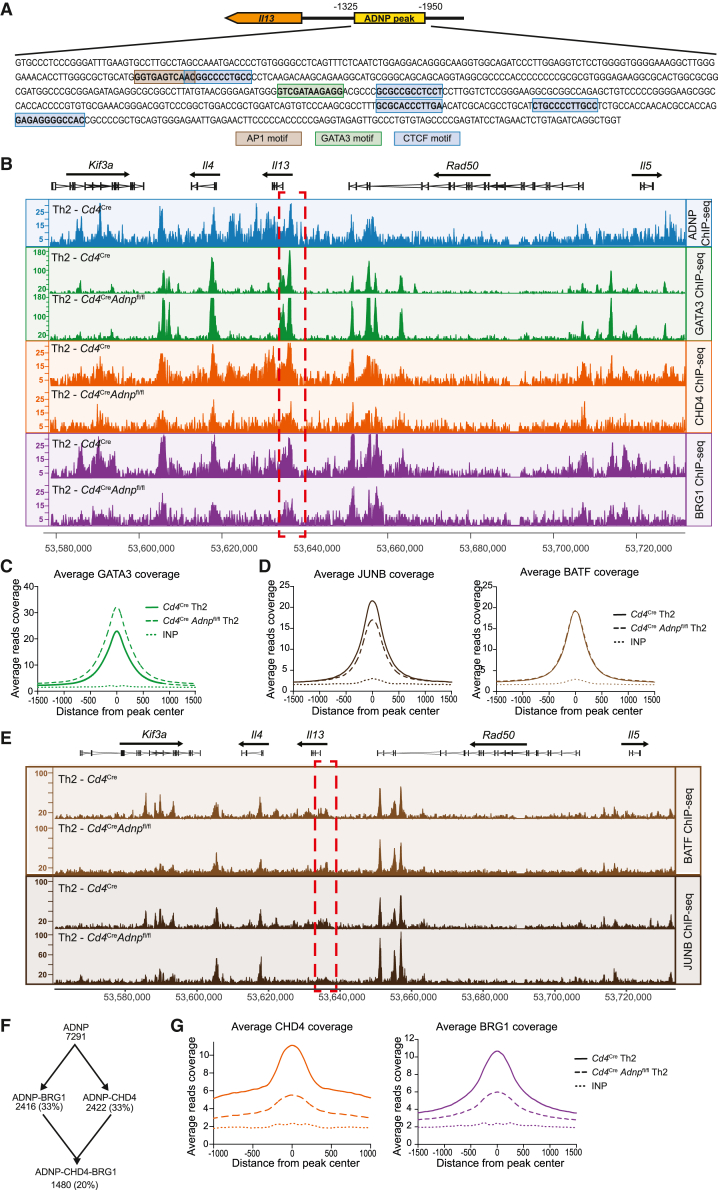
Given the interaction of ADNP with CHD4, BRG1, and BATF, we performed ChIP-seq to investigate their localization within the Th2 locus. As indicated by anti-ADNP ChIP-seq analysis, ADNP binds upstream of the Il13 promoter in a region harboring one AP-1 motif followed by a GATA3 motif12,43 and five predicted CTCF motifs (Figure 6A). Using anti-GATA3 ChIP-seq, we confirmed that this region aligns with the conserved GATA3 response element (CGRE) (Figure 6B), which has been postulated to regulate Th2 cell cytokine production via GATA3 binding.43 We determined that GATA3 binding at the CGRE preceded ADNP binding, since GATA3 was present at this location in naive T cells, before ADNP recruitment (Figures 4J and S6A). We observed a modest increase of GATA3 binding in the CGRE region and across the genome of ADNP-deficient Th2 cells (Figures 6B and 6C), which may arise from the ADNP-deficient Th effector cells becoming arrested and unable to progress beyond the GATA3-binding stage. Anti-JUNB and anti-BATF ChIP-seq indicated that JUNB and BATF binding also occurred independently of ADNP (Figures 6D and 6E), suggesting that they, similar to GATA3, can access and bind DNA before or without prior binding of ADNP. However, although ADNP co-precipitated with BATF (and BATF co-precipitated JUNB), it is currently unclear whether this AP-1 complex binds to the AP-1 site present in the CGRE where AP-1 enrichment was only modest (Figure 6E).12
Figure 6.
ADNP is required for efficient recruitment of CHD4 and BRG1 to the Th2 cytokine locus
(A) Predicted AP-1, GATA3, and CTCF motifs within ADNP ChIP-seq peak upstream of Il13 promoter. Prediction was performed using Jaspar transcription factor database.
(B) Representative binding profiles of ADNP, GATA3, CHD4, and BRG1 in Th2 cells from Cd4Cre or Cd4CreAdnpfl/fl mice at the type 2 cytokine locus. Data are representative of 3 biological replicates.
(C) Average ChIP-seq signal over all GATA3 peaks in Cd4Cre or Cd4CreAdnpfl/fl Th2 cells. Data are representative of 2 biological replicates.
(D) Average ChIP-seq signal over all JUNB and BATF peaks in Cd4Cre or Cd4CreAdnpfl/fl Th2 cells. Data are representative of 3 biological replicates.
(E) Representative binding profiles of BATF and JUNB in Th2 cells from Cd4Cre or Cd4CreAdnpfl/fl mice at the type 2 cytokine locus. Data are representative of 2 biological replicates.
(F) Number and percentages of all ADNP, ADNP-BRG1, ADNP-CHD4, and ADNP-CHD4-BRG1 overlapping ChIP-seq peaks in Th2 cells. Peak list was generated using two biological replicates.
(G) Average ChIP-seq signal over all CHD4 and BRG1 peaks in Cd4Cre or Cd4CreAdnpfl/fl Th2 cells. Data are representative of 3 biological replicates.
See also Figure S6.
Anti-BRG1 and anti-CHD4 ChIP-seq demonstrated that these chromatin remodeling proteins also colocalized with ADNP binding at the CGRE (Figure 6B), with ∼20% of ADNP-binding sites in Th2 cells coincident with BRG1 and CHD4 binding (Figure 6F), but not HP1γ binding (described as a repressor in the ChAHP complex in ES cells28) (Figure S6B). In the absence of ADNP, we observed a strong reduction in the binding of CHD4 and BRG1 to the CGRE, and at other ADNP-binding regions across the genome (Figure 6G), indicating a critical role for ADNP in recruiting these chromatin modifiers. ADNP also colocalizes with GATA3, BRG1, and CHD4 at the Il4 promoter region where there was a reduction in BRG1 and CHD4 binding in ADNP-deficient cells (Figure 6B).
These data demonstrate that ADNP is not required for the binding of the pioneer factors GATA3 and AP-1 to the Th2 locus but is necessary for recruitment of BRG1 and CHD4 to chromatin.
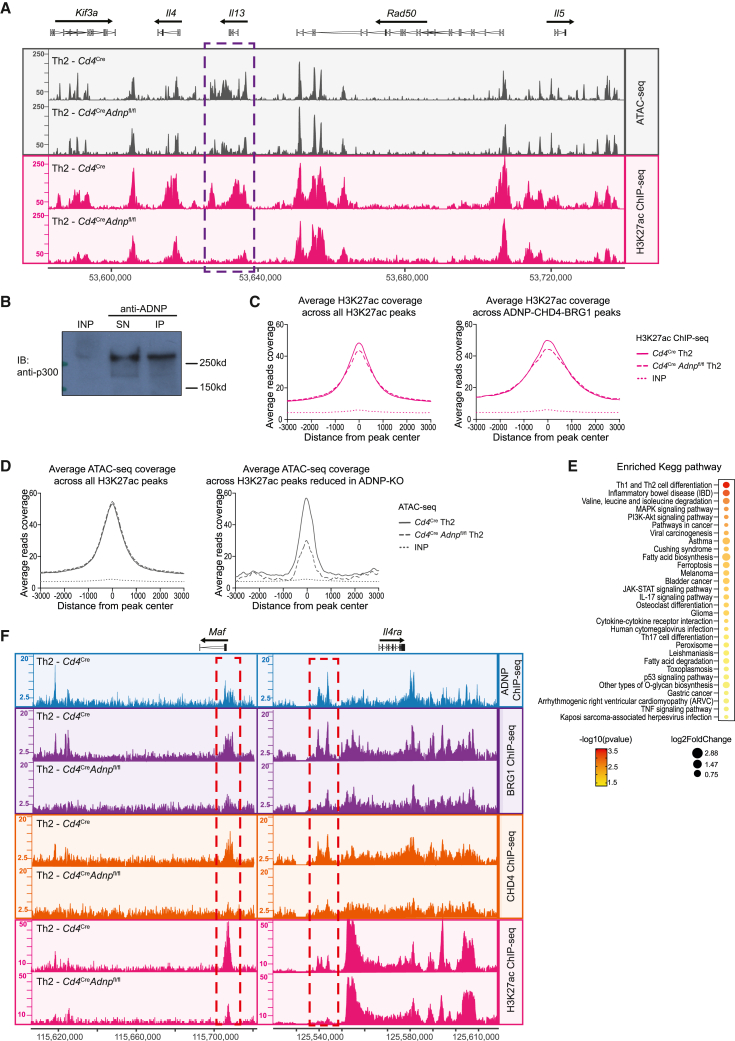
The absence of ADNP at the Il13 promoter reduces local H3K27 acetylation and DNA accessibility
Next, we investigated how ADNP, with CHD4 and BRG1, altered gene expression. We found using assay for transposase-accessible chromatin sequencing (ATAC-seq) assays that without ADNP full accessibility to the Il13 locus was not initiated (Figure 7A). To investigate this defect in accessibility, we assessed the acetylation status of the Il13 locus in the presence and absence of ADNP. Histone H3 lysine 27 acetylation (H3K27ac) is known to shape active promoters and enhancers by opening chromatin, thereby allowing the transcriptional machinery to assemble.44,45 Indeed, the Il13 locus in Th2 cells was marked by H3K27ac (Figure 7A, purple rectangle). We found that the absence of ADNP led to a profound localized deficit in H3K27 acetylation at the Il13 locus (Figure 7A, purple rectangle). Since CHD4 is important for the recruitment of the HAT P300 to the Th2 cytokine locus, including to the CGRE,46 we assessed the relationship of P300 with ADNP and found that P300 also associated with ADNP in Th2 cells (Figure 7B). Although we identified co-localization of ADNP, GATA3, BRG1, and CHD4 at the Il4 promoter, similar to the Il13 promoter, we did not observe a pronounced or reproducible change in gene accessibility or H3K27 acetylation at this locus in the absence of ADNP (Figure 7B). As the proportion of Th2 cells that express IL-4 is relatively small when compared with IL-13-expressing cells (Figures 3F–3H and S2A), it is likely that these IL-4-positive cells become diluted within whole-population analysis such as ATAC-seq and ChIP-seq, making it difficult to determine the modulation of this locus.
Figure 7.
Absence of ADNP at the Il13 promoter reduces local H3K27 acetylation and DNA accessibility
(A) Representative ATAC-seq and H3K27ac ChIP-seq tracks from Th2 cells from Cd4Cre or Cd4creAdnpfl/fl mice in the type 2 cytokine locus. Data are representative of 2 biological replicates.
(B) Detection of p300 protein (218 kDa) in immunocomplex generated with Th2 cell nuclear lysate co-immunoprecipitated with anti-ADNP antibody. INP, input nuclear extract; SN, supernatant; IP, immunoprecipitation elution. Data are representative of 2 independent experiments. IB refers to immunoblotting antibody.
(C) Average ChIP-seq signal over all H3K27ac peaks or over ADNP-CHD4-BRG1 peaks in Cd4Cre or Cd4CreAdnpfl/fl Th2 cells. Data are representative of 3 biological replicates.
(D) Average ATAC-seq signal over all H3K27ac peaks or over H3K27ac peaks that were reduced in Cd4CreAdnpfl/fl Th2 cells. Data are representative of 2 biological replicates.
(E) KEGG pathway analysis of genes associated to the genomic regions where H23K27ac was reduced in Cd4CreAdnpfl/fl Th2 cells. All shown pathways were enriched (p < 0.05).
(F) Representative binding profiles of ADNP, CHD4, BRG1, and H3K27ac in Th2 cells of Cd4Cre or Cd4CreAdnpfl/fl mice at the Maf and Il4ra loci. Data are representative of 2 biological replicates.
The co-localization of ADNP-CHD4-BRG1 was highly associated with H3K27ac regions (Figure 7C). As expected, regions where H3K27 acetylation was reduced in ADNP-deficient Th2 cells displayed a reduction in DNA accessibility (Figure 7D). Pathway enrichment analyses of the genes associated with those regions revealed Th cell differentiation, JAK-STAT, and asthma, among the enriched pathways (Figure 7E). Comparing those genes with the genes associated to ADNP-CHD4-BRG1 peaks, we found 228 genes that were present in both lists (Table S6), and 8 of those were related to the Th1 and Th2 cell differentiation pathway. In addition to the Il13 locus, the Il4ra and Maf loci also feature convergent binding of the chromatin modifiers and a coincident reduction in H3K27 acetylation in ADNP-deficient Th2 cells (Figure 7F). Notably, the Maf and Il4ra genes play key roles in Th2 differentiation.47,48,49
These results support a key role for ADNP in focusing Th2 gene expression patterns to potentiate Th2 cell differentiation and activate type 2 cytokine production, acting as a critical component in the recruitment of chromatin remodeling proteins CHD4, BRG1, and P300.
Discussion
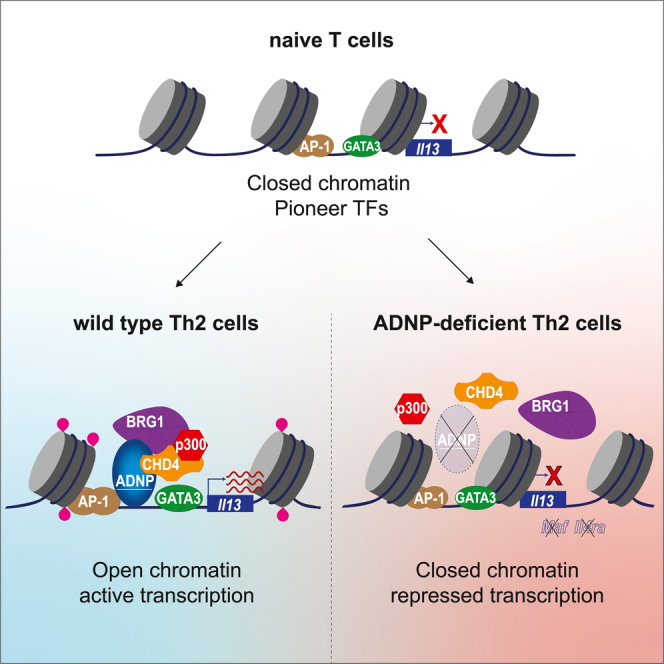
Our CRISPR-Cas9 screen identified ADNP as a factor, which has not been characterized previously in immune cells, as well as several TFs already known to play roles in Th2 cell differentiation and type 2 cytokine production. Conditional deletion of ADNP from T cells, or all lymphocytes, revealed that ADNP was required for efficient type 2 immune responses to allergic challenge and also in lymphocyte development. Mechanistically, ADNP bound to unoccupied CTCF motifs within the CGRE of the type 2 cytokine locus upstream of the Il13 coding region and was critical for localized histone acetylation and gene accessibility, leading to IL-13 expression. ADNP performs this function by recruiting a complex of chromatin remodeling factors including BRG1, CHD4, and P300 to gene regulatory regions.
The situation at the Il4 and Il5 loci appears to be more complex. At the Il4 locus ADNP colocalizes with GATA3, BRG1, and CHD4 indicating that an ADNP-dependent mechanism may be directly involved in Il4 regulation and in vitro Th2 cell differentiation assays indicated a reliance upon ADNP for Il4 expression, which was shown to be T cell intrinsic in the context of in vivo bone marrow transfer and antigen challenge. However, we observed inconsistency in ADNP-dependent IL-4 expression in some in vivo experiments, potentially due to the relatively small numbers of IL-4-positive cells. By contrast, we could not identify direct ADNP binding at the Il5 locus, despite the expression of IL-5 being impaired in a Th2 cell-intrinsic ADNP-dependent manner following allergen challenge. This suggests that Il5 gene regulation may not be due to direct ADNP binding to the Il5 gene, but that ADNP influences Il5 transcription by an alternative mechanism, perhaps by contributing to dysregulated 3D chromatin architecture.37,50 The prominent role for ADNP in regulating IL-13 expression was further illustrated by single-cell gene expression analysis of Th2 cells following allergen challenge which showed that Il13 expression was predominant within a mix of Il4, Il5, and Il13 expressing cells (representing single, double, and triple-positive Th2 cells), and that these were almost undetectable in the absence of ADNP. These data support previous observations indicating that the cytokine genes in this cluster can be independently regulated by discrete control mechanisms and argue for a role for ADNP in this process.38,39
Mutations in the gene encoding ADNP underlie ADNP syndrome, which is characterized by neurological and developmental abnormalities.25,51 More recent studies have focused on the ability of ADNP to act genome-wide to locally restrict gene expression in ES cells to prevent spontaneous cell differentiation.27,28,29,40 This effect results from ADNP associating with CHD4 and the transcriptional repressor HP1γ to form ChAHP complexes, which locally restrict chromatin accessibility by competing for CBSs and thereby modifying TADs.27,28,29,40 A similar role for ADNP in gene repression in ES cells was reported by Sun and colleagues, who found ADNP associated with CHD4 and BRG1.27,28,29,40,41 In contrast to these repressive roles for ADNP in ES cells, our data from primary Th2 cells demonstrated that ADNP can also play a fundamental and previously unappreciated role in directly promoting gene expression. This suggested a mechanism distinct from the repression model previously proposed in ES cells.27,28,29,40 Indeed, when we analyzed the DNA binding of CTCF and ADNP in primary wild-type or ADNP-deficient Th2 cells, we found no evidence that ADNP competed for CBSs within the type 2 cytokine locus, or across the genome. Instead, we determined that unoccupied CTCF motifs in the Il13 promoter were bound by ADNP during the differentiation of naive T cells to cytokine-producing Th2 cells.
The deletion of ADNP in T cells resulted in a pronounced defect in the recruitment of the histone-modifying factors CHD4 and BRG1 to the CGRE upstream of the Il13 promoter, and coIP verified that in Th2 cells ADNP associated with CDH4, BRG1, and P300. Although CHD4, which induces ATP-dependent distortion of nucleosomal DNA during chromatin remodeling52 is commonly viewed as a component of the repressive NuRD complex,53 it can also act as a transcriptional activator. Indeed, CHD4 is required for Cd4 gene transcription in T cells through its association with the HAT P300 at the Cd4 enhancer.54 A similar association was reported in Th2 cells where CHD4 was found to bind to GATA3, and was proposed to recruit P300 to the CGRE.46 The amino-terminal region of CHD4 has also been demonstrated to associate with BRG1 to activate transcription,55 which in turn has also been reported to bind to the Il13 promoter in Th2 cells.56 Thus, although an ADNP-CHD4-BRG1 complex can be repressive in ES cells,27 our results are congruent with BRG1 hydrolyzing ATP to drive chromatin accessibility and transcription at the type 2 gene cluster in T cells.57
In ES cells ADNP maintains multipotency with an overarching requirement to suppress cell differentiation programs. In this situation, the competition between ADNP and CTCF helps control TAD formation and repression predominates.29 In Th2 cells, ADNP is predominantly associated with activation during the polarization of cytokine production in response to cytokine and T cell receptor signaling. Although the mechanism underlying these differences is not clear, it is possible that the interactions that we observed between ADNP and the pioneer factors AP-1 and GATA3 may help focus ADNP to active loci in T cells in response to IL-4 signaling. GATA3 is essential for Th2 development2,58,59 and AP-1 factors (BATF and JUNB) also play key roles in this cellular program and cytokine expression.12,13,42,60,61 Our results indicate that GATA3 and AP-1 bind to the CGRE independently of ADNP but that ADNP appears to form a complex that includes BATF as well as BRG1, CHD4, and P300. In addition, BATF binds JUNB, and CHD4 and JUNB have been reported to bind GATA3.46 This suggests that a larger complex can form at genomic locations where there is a conjunction of GATA3, AP-1, and ADNP binding to their juxtaposed DNA motifs to facilitate the ADNP-dependent recruitment of BRG1, CHD4, and P300 and provide specific locus activation. Indeed, when we looked beyond the type 2 gene cluster for loci with GATA3, AP-1, ADNP, BRG1, and CHD4 protein co-localization, which also showed a deficit in acetylation and histone accessibility in ADNP-deficient in-vitro-cultured Th2 cells, we found that these loci encoded MAF and the IL-4 receptor alpha (IL-4Rα). The MAF proto-oncogene was one of the first TFs reported to activate the Il4 promoter, leading to IL-4 expression and Th2 cell differentiation.47 Furthermore, IL-4Rα is the primary binding chain for IL-4, which stimulates Th2 cell differentiation through the activation of STAT6 and the induction of GATA3 expression.2,49 These results suggest that ADNP (with associated factors) may be capable of promoting a Th2 cell feedback loop that reinforces the Th2 cell phenotype. However, we did not observe a reduction in MAF expression in vivo, suggesting that there are alternative ADNP-independent pathways that can regulate MAF expression in vivo. By contrast, our data indicate no such ADNP-independent regulation of the Il13 locus.
The critical biological role of this ADNP bridge is vividly revealed by the incapacity of mice with ADNP-deficient T cells or ADNP-deficient lymphocytes to respond efficiently to allergen challenge. These mice displayed impaired production of type 2 cytokines by Th2 and ILC2 and failed to mount a robust type 2 immune response. Although ADNP has not been identified as a common factor in asthma susceptibility, unlike IL-4, IL-5, and IL-13, it was reported among a subset of genes that were differentially expressed by CD4+ lymphocytes and that predicted more atopic from less atopic children.62 However, there are currently no reports of children with ADNP syndrome presenting specific immunological disorders, though recurring infections (∼50% of cases) are reported.26
Together, our results are consistent with ADNP functioning as an adapter or bridge to specifically localize CHD4 and BRG1 to ADNP-bound CTCF motifs forming a complex that can include the acetylase P300 and drive histone acetylation and genome accessibility. This ADNP-dependent mechanism is essential to promote Th2 cell differentiation and type 2 cytokine production in response to IL-4 signaling. Our results also raise the possibility that ADNP may play additional key roles in hematopoiesis and immunity.
Limitations of the study
For the future, manipulation of the CTCF and/or ADNP motifs within the whole cytokine locus and in the vicinity of individual cytokine genes will help to define ADNP-binding specificity. This could be attempted in primary cells or gene-modified mice to allow in vitro and in vivo readouts of ADNP function. Furthermore, the application of single-cell ChIP-seq and ATAC-seq could also provide improved resolution of the chromatin changes which are dependent on ADNP. Moreover, our study raised the involvement of ADNP in pan-lymphocyte development within the bone marrow, and of NKT cell differentiation in the thymus, and additional studies will be required to identify the mechanisms of action of ADNP in these processes. In addition, it will be important to assess the relevance of ADNP on human Th2 cell function, as our study focused on the mouse model.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD3e (145-2C11) BV510 | BioLegend | Cat#100353; RRID: AB_2565879 |
| Anti-mouse CD4 (GK1.5) PerCP/Cy5.5 | BioLegend | Cat#100434; RRID: AB_893324 |
| Anti-mouse CD8a (53-6.7) Alexa Fluor700 | BioLegend | Cat#100730; RRID: AB_493703 |
| Anti-mouse/human CD11b (M1/70) PE-Cy7 | BioLegend | Cat#101216; RRID: AB_312799 |
| Anti-mouse CD19 (6D5) Alexa Fluor700 | BioLegend | Cat#115528; RRID: AB_493735 |
| Anti-mouse CD25 (PC61) PE | BioLegend | Cat#102008; RRID: AB_312857 |
| Anti-mouse CD25 (PC61) BV510 | BioLegend | Cat#102042; RRID: AB_2562270 |
| Anti-mouse/human CD44 (IM7) PerCP/Cy5.5 | BioLegend | Cat#103032; RRID: AB_2076204 |
| Anti-mouse/human CD44 (IM7) BV605 | BioLegend | Cat#103047; RRID: AB_2562451 |
| Anti-mouse CD62L (MEL-14) BV421 | BioLegend | Cat#104436; RRID: AB_2562560 |
| Anti-mouse CD127 (SB/199) biotin | BioLegend | Cat#121104; RRID: AB_493502 |
| Anti-mouse F4/80 (BM8) BV785 | BioLegend | Cat#123141; RRID: AB_2563667 |
| Anti-mouse FceR1 (MAR-1) Alexa Fluor700 | BioLegend | Cat#134324; RRID: AB_2566734 |
| Anti-mouse/human IL-5 (TRFK5) APC | BioLegend | Cat#504306; RRID: AB_315330 |
| Anti-mouse IFN-γ (XMG1.2) BV785 | BioLegend | Cat#505838; RRID: AB_2629667 |
| Anti-mouse Arginase 1 (A1exF5) PE | eBioscience | Cat#12-3697-82; RRID: AB_2734839 |
| Anti-mouse CD3 (17A2) Alexa Fluor700 | eBioscience | Cat#56-0032-82; RRID: AB_529507 |
| Anti-mouse CD4 (GK1.5) ef450 | eBioscience | Cat#48-0041-82; RRID: AB_10718983 |
| Anti-mouse CD4 (GK1.5) Alexa Fluor700 | eBioscience | Cat#56-0041-82; RRID: AB_493999 |
| Anti-mouse CD4 (GK1.5) FITC | eBioscience | Cat#11-0041-82; RRID: AB_464892 |
| Anti-mouse CD8a (53-6.7) PE-Cy5 | eBioscience | Cat#15-0081-82; RRID: AB_468706 |
| Anti-mouse CD11b (M1/70) Alexa Fluor700 | eBioscience | Cat#56-0112-82; RRID: AB_657585 |
| Anti-mouse CD11c (N418) Alexa Fluor700 | eBioscience | Cat#56-0114-82; RRID: AB_493992 |
| Anti-mouse/human CD44 (IM7) APC | eBioscience | Cat#17-0441-82; RRID: AB_469390 |
| Anti-mouse CD45.1 (A20) PE-Cy5 | eBioscience | Cat#15-0453-82; RRID: AB_468759 |
| Anti-mouse CD45.2 (104) Alexa Fluor700 | eBioscience | Cat#56-0454-82; RRID: AB_657752 |
| Anti-mouse/human Gata-3 (TWAJ) eFluor660 | eBioscience | Cat#50-9966-42; RRID: AB_10596663 |
| Anti-mouse GR-1/Ly-6G/C (RB6-8C5) Alexa Fluor700 | eBioscience | Cat#56-5931-82; RRID: AB_494007 |
| Anti-mouse IL-13 (eBio13A) PE | eBioscience | Cat#12-7133-82; RRID: AB_763559 |
| Anti-mouse IL-13 (eBio13A) PE-Cy7 | eBioscience | Cat#25-7133-82; RRID: AB_2573530 |
| Anti-mouse Ki67 (SolA15) PE-Cy7 | eBioscience | Cat#25-5698-82; RRID: AB_11220070 |
| Anti-mouse KLRG1 (2F1) PerCP-eFluor710 | eBioscience | Cat#46-5893-82; RRID: AB_10670282 |
| Anti-mouse Ly-6G (1A8-Ly6G) PerCP-eFluor710 | eBioscience | Cat#46-9668-82; RRID: AB_2573893 |
| Anti-mouse MHCII (M5/114.15.2) eFluor450 | eBioscience | Cat#48-5321-82; RRID: AB_1272204 |
| Anti-mouse NK1.1 (PK136) Alexa Fluor700 | eBioscience | Cat#56-5941-82; RRID: AB_2574505 |
| Anti-mouse TCR beta (H57-597) eFluor450 | eBioscience | Cat#48-5961-82; RRID: AB_11039532 |
| Anti-mouse TER-119 (TER-119) Alexa Fluor700 | eBioscience | Cat#56-5921-82; RRID: AB_2815252 |
| Fixable Viability Dye eFluor 780 | eBioscience | Cat#65-0865-18 |
| Anti-mouse CD45 (30-F11) BUV395 | BD Biosciences | Cat#564279; RRID: AB_2651134 |
| Anti-mouse NK1.1 (PK136) BUV395 | BD Biosciences | Cat#564144; RRID: AB_2738618 |
| Anti-mouse SiglecF (E50-2440) Alexa Fluor647 | BD Biosciences | Cat#562680; RRID: AB_2687570 |
| Streptavidin BUV737 | BD Biosciences | Cat#612775; RRID: AB_2870104 |
| Anti-mouse ST2 (DJ8) FITC | MD bioproducts | Cat#101001F; RRID: AB_947549 |
| 2W1S-tetramer PE | NIH Tetramer Facility | N/A |
| anti-ADNP | Novus Biologicals | Cat#NBP1-89236; RRID: AB_11008573 |
| anti-CTCF | Cell Signaling | Cat#2899; RRID: AB_2086794 |
| anti-GATA3 | Cell Signaling | Cat#5852; RRID: AB_10835690 |
| anti-CHD4 | Cell Signaling | Cat#12011; RRID: AB_2734702 |
| anti-JUNB | Cell Signaling | Cat#3753; RRID: AB_2130002 |
| anti-H3K27ac | Cell Signaling | Cat#8173; RRID: AB_10949503 |
| anti-BRG1 | abcam | Cat#ab110641; RRID: AB_10861578 |
| anti-BATF | abcam | Cat#ab236876; |
| anti-HP1g | abcam | Cat#ab217999; RRID: AB_217999 |
| anti-p300 | abcam | Cat#ab275378 |
| anti-CD3 | 2B Scientific | Cat#Ab00105-1.1 |
| anti-CD28 | 2B Scientific | Cat#AGEL0759 |
| anti-IFNg | 2B Scientific | Cat#AGEL2200 |
| anti-IL-4 | BioLegend | Cat#504101 |
| Chemicals, peptides, and recombinant proteins | ||
| 2W1S peptide | Designer Bioscience | N/A |
| papain | Sigma-Aldrich | Cat#76216 |
| rmIL-2, carrier-free | Biolegend | Cat#575406 |
| rmIL-4, carrier-free | Biolegend | Cat#574306 |
| rmIL-12, carrier-free | Biolegend | Cat#577006 |
| PMSF | Sigma Aldrich | Cat#P7626 |
| collagenase I | Gibco | Cat#17100017 |
| DNase I, from bovine pancreas | Sigma-Aldrich | Cat#D5025/DN25 |
| Percoll | GE Healthcare | Cat#17-0891-01 |
| cOmplete protease inhibitor | Roche | Cat#4693116001 |
| Pierce 660nm protein assay reagent | ThermoFisher, | Cat#22660 |
| A/G dynabeads) | Thermo Scientific | Cat#88802 |
| 1X NuPage LDS sample buffer | Invitrogen | Cat#NP0008 |
| ECL western blotting detection reagent | GE Healthcare | Cat#RPN2106 |
| protein A Dynabeads | ThermoFisher | Cat#10002D |
| NAP | Biotechne | Cat#6779 |
| Critical commercial assays | ||
| Mouse IgE Uncoated ELISA kit | Invitrogen | Cat#88-504460-88 |
| Ovation RNA-seq System V2 | Tecan | Cat#7102-32 |
| Ovation Ultralow Library Systems | Tecan | Cat#0344NB-32 |
| Fixable Dye eFluor 780 | Invitrogen | Cat#65-0865-14 |
| BD Cytofix/Cytoperm Plus reagents | BD Biosciences | Cat#555028 |
| Protein Transport Inhibitor Cocktail | eBioscience | Cat#00-4980-9 |
| Gibson assembly | New England BioLabs | Cat#E5510S |
| NEB T4 DNA ligase | New England BioLabs | Cat#M0202S |
| RPMI 1640 + GlutaMAX | GIBCO | Cat#61870-010 |
| Fetal Calf Serum | GIBCO | Cat#10270-106 |
| 2-mercaptoethanol | Sigma-Aldrich | Cat#M6250 |
| DMEM | GIBCO | Cat#10564011 |
| Fugene HD Transfection Reagent | Promega | Cat#E2311 |
| OPTI-MEM | GIBCO | Cat#31985062 |
| DNeasy Blood & Tissue Kits | QIAGEN | Cat#69504 |
| Herculase II Fusion DNA polymerase | Agilent | Cat#600675 |
| KAPA library quantification kit | Roche | Cat#KK4824 |
| RNeasy Plus Micro kit | Qiagen | Cat#74034 |
| truChIP Chromatin Shearing kit | Covaris | Cat#520154 |
| Foxp3 Staining kit reagents | eBioscience | Cat#00-5523-00 |
| Qiagen MinElute kit | Qiagen | Cat#28004 |
| Kappa HiFi HotStart Ready mix | Roche | Cat#KK2601 |
| Illumina Tagment DNA Enzyme and Buffer | Illumina | Cat#20034197 |
| SPRI Ampure XP beads | Beckman Coulter | Cat#A63881 |
| Deposited data | ||
| scRNAseq | This paper | GEO: GSE218017 |
| ATACseq | This paper | GEO: GSE218017 |
| ChIPseq | This paper | GEO: GSE218017 |
| Experimental models: Cell lines | ||
| Platinum-E retroviral packaging cells | Cell biolabs | Ca#RV-101 |
| Experimental models: Organisms/strains | ||
| Mouse: Rosa26Cas9EGFP | The Jackson Laboratory | JAX 026179 |
| Mouse: Il13tdTom | Barlow et al.38 | N/A |
| Mouse: Il7rCre | Schlenner et al.63 | MGI:4441349 |
| Mouse: CD4Cre | Taconic | Ca#4196 |
| Mouse: Adnptm1a(KOMP)Wtsi | KOMP repository | RRID:MMRRC_051854-UCD |
| Mouse: C57BL/6JOla | Jackson Labs (Bred in LMB) | Cat#000664; RRID: IMSR_JAX:000664 |
| Oligonucleotides | ||
| Primer for sgRNA-insert amplification (Forward) AATGGACTATCATATG CTTACCGTAACTTGAAAGTATTTCG |
Sigma-Aldrich | N/A |
| Primer for sgRNA-insert amplification (Reverse) CTTTAGTTTGTATGTCTGTT GCTATTATGTCTACTATTCTTTCC |
Sigma-Aldrich | N/A |
| Software and algorithms | ||
| FlowJo | FlowJo, LLC | RRID: SCR_008520 |
| Prism 9 | GraphPad Prism | RRID: SCR_002798 |
| Cutadapt | N/A | https://journal.embnet.org/index.php/embnetjournal/article/view/200 |
| Trim Galore | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| DESeq2 | Love et al.64 | https://bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html |
| 10x Cell Ranger | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| 10x Genomics Loupe Browser | 10x Genomics | https://www.10xgenomics.com/products/loupe-browser |
| Scaffold programme (Proteome Software Inc., USA)65 | Keller et al.65 | Proteome Software Inc., USA |
| Bowtie2 (version 2.3.5.1) | Langmead et al.66 | https://bowtie-bio.sourceforge.net/bowtie2/manual.shtml |
| HOMER | Heinz et al.67 | http://homer.ucsd.edu/homer/index.html |
| SeqMonk software (v1.48.0) | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/ |
| FACSDiva software | BD Biosciences | https://www.bdbiosciences.com/en-gb/products/software/instrument-software/bd-facsdiva-software |
| Macs2 (v2.1.2) | Zhang et al.68 | https://github.com/macs3-project/MACS/wiki |
| STAR (version 2.6.0a) | N/A | https://github.com/alexdobin/STAR |
| Other | ||
| Mouse Brie CRISPR knockout pooled library | Addgene | Ca#73633 |
| MSCV-pU6-(BbsI)-CcdB-(BbsI)-Pgk-Puro-T2A-BFP | Addgene | Ca#86457 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andrew McKenzie (anm@mrc-lmb.cam.ac.uk).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mice
Rosa26Cas9EGFP (JAX 026179),69 Il13tdTom,38 Il7rCre 63 and CD4Cre (Taconic, model #4196) mice were on the C57BL/6 background. C57BL/6 controls were bred in-house. Adnpflox targeted ES cells (to delete exon 5) were purchased from KOMP repository (Adnptm1a(KOMP)Wtsi) and Adnpfl mouse line was generated by methods previously described.70 All mice were maintained in the Medical Research Council ARES animal facility under specific pathogen-free conditions, at 19-23oC with a 12-h light-dark cycle. In individual experiments, mice were matched for age, sex and background strain and all experiments undertaken in this study were done so with the approval of the LMB Animal Welfare and Ethical Review Body (AWERB) and of the UK Home Office.
Method details
Antibodies
Antibodies against the following proteins were used in immunoprecipitation and ChIP-seq experiments: ADNP (Novus Biologicals NBP1-89236), CTCF (Cell Signaling, #2899), GATA3 (Cell Signaling, #5852), CHD4 (Cell Signaling, #12011), BRG1 (abcam, ab110641), JUNB (Cell Signaling, #3753), BATF (abcam, ab236876), H3K27ac (Cell Signaling, #8173), HP1g (abcam, ab217999). In flow cytometry experiments we used antibodies from BioLegend (CD3e (BV510, 145-2C11, 1:300 dilution), CD4 (ef450, GK1.5, 1:500 dilution) or (BV785, RM-4-5, 1:500 dilution), CD8a (Alexa Fluor700, BV421 or BV785, 53-6.7, 1:500 dilution), CD11b (PE-Cy7, M1/70, 1:1000 dilution), CD19 (Alexa Fluor700 or BV605, 6D5, 1:500 dilution), CD25 (PE or BV510, PC61, 1:300 dilution), CD44 (PerCP/Cy5.5 or BV605, IM7, 1:500 dilution), CD45 (BV510, 30-F11, 1:500 dilution), CD62L (BV421, MEL-14, 1:500 dilution), CD127 (biotin, SB/199, 1:500 dilution), F4/80 (BV785, BM8, 1:500 dilution), FceR1 (Alexa Fluor700, MAR-1, 1:500 dilution), IL-5 (APC, TRFK5, 1:300 dilution), IFN-g (BV785, XMG1.2, 1:300 dilution), eBioscience (Arginase 1 (PE, A1exF5, 1:300 dilution), CD3e (Alexa Fluor 700, 17A2, 1:300 dilution), CD4 (Alexa Fluor700 or FITC, GK1.5, 1:500 dilution), CD8a (FITC or PE-Cy7, 53-6.7, 1:500 dilution), CD11b (Alexa Fluor700, M1/70, 1:500 dilution), CD11c (Alexa Fluor 700 or PE-Cy7, N418, 1:500 dilution), CD19 (PerCP-Cy5.5 or PE-Cy7, eBio1D3, 1:500 dilution), CD44 (FITC or APC, IM7, 1:500 dilution), CD45 (FITC, 30-F11, 1:500 dilution), FceR1 (PE-Cy7, MAR-1, 1:500 dilution), Gata-3 (eFluor 660, TWAJ, 1:300 dilution), GR-1/Ly-6G/C (Alexa Fluor700 or PE-Cy7, RB6-8C5, 1:500 dilution), IL-13 (PE or PE-Cy7, eBio13A, 1:300 dilution), KLRG1 (PerCP-eFluor710, 2F1, 1:500 dilution), Ly-6G (PerCP-eFluor710, 1A8-Ly6G, 1:500 dilution), MHCII (eFluor450, M5/114.15.2, 1:1000 dilution), NK1.1 (Alexa Fluor700 or PE-Cy7, PK136, 1:500 dilution), TER-119 (Alexa Fluor700 or PE-Cy7, TER-119, 1:500 dilution)), BD Biosciences (CD45 (BUV395, 30-F11, 1:500 dilution), NK1.1 (BUV395, PK136, 1:300 dilution), SiglecF (Alexa Fluor 647, E50-2440, 1:500 dilution), Streptavidin (BUV737, 1:500 dilution)), MD bioproducts (ST2 (FITC, DJ8, 1:500 dilution)) and the NIH Tetramer Facility (2W1S-tetramer, PE, 1:500 dilution). ‘Lineage’ staining included antibodies specific for CD3, CD4, CD8, CD11b, CD11c, CD19, FceRI, GR-1, NK1.1 and TER-119. All samples were co-stained with a cell viability dye (Fixable Dye eFluor 780, Invitrogen).
Adoptive transfers
Bone marrow cells were purified by flow cytometry from wildtype control mice (CD45.1) and from Cd4CreAdnpfl/fl (CD45.2). Cells from both sources were mixed at a ratio of 1:1 and implanted via tail vein injection into lethally irradiated (600 rad) CD45.1/CD45.2 recipients. After 6 weeks of cell transfer, mice were challenged with papain or PBS.
In vivo stimulation
Mice were anesthetized by isoflurane inhalation followed by the intranasal injection of papain (7.5 mg for males, 5 mg for females, Sigma-Aldrich #76216) with or without 2W1S peptide (50 mg, Designer Bioscience) in 40 μl PBS on days 0 and 14. Mice were sacrificed for analysis on day 19.
Tissue preparation
Cell suspensions from spleen, lymph nodes, and thymus tissue were obtained by passing the tissues through a 70-mm strainer. Lung tissue was predigested with 750 U ml−1 collagenase I (Gibco) and 0.3 mg ml−1 DNaseI (Sigma-Aldrich) and cell suspensions were obtained by passing the tissues through a 70-mm strainer. Red blood cells were removed by incubating with RBC lysis solution (140 mM NH4Cl, 17 mM Tris, pH 7.2). Lung lymphocytes were further enriched by centrifugation in 30% Percoll at 800g (GE Healthcare). Serum samples were obtaied following blood coagulation and centrifugation of coagulated cells at 3,300 x g. Serum IgE was quantified using the Invitrogen Mouse IgE Uncoated ELISA kit (88-504460-88) following the manufacturer’s instructions.
Flow cytometry
Single-cell suspensions were incubated with fluorochrome- or biotin-conjugated antibodies in the presence of anti-CD16/CD32 (Fc block, clone 2.4G2) as indicated. All samples were co-stained with a cell viability dye (Fixable dye eFluor780, Invitrogen). For cell sorting an iCyt Synergy (70-μm nozzle, Sony Biotechnology) was used. Intracellular cytokine staining was performed using BD Cytofix/Cytoperm Plus reagents (BD Biosciences) following pre-culture with RPMI, supplemented with 50 ng ml-1 phorbol 12-myristate 13-acetate (PMA), 500 ng ml-1 ionomycin and Protein Transport Inhibitor Cocktail (eBioscience), for 4 h at 37°C. Intracellular TF staining was performed using Foxp3 Staining kit reagents (eBioscience). Analysis was performed on an LSRFortessa system (BD Biosciences) with FACSDiva software (version 6.2, BD Biosciences). For cell sorting, an iCyt Synergy system (70-μm nozzle, Sony Biotechnology) was used. Data were analyzed with FlowJo software (version 10).
sgRNA cloning into retroviral expression vector
MSCV-pU6-(BbsI)-CcdB-(BbsI)-Pgk-Puro-T2A-BFP was a gift from Ralf Kuehn (Addgene plasmid # 86457; http://n2t.net/addgene:86457; RRID:Addgene_86457).71 Mouse Brie CRISPR pooled library was a gift from David Root and John Doench (Addgene #73633).72 Custom sgRNA libraries were synthesised by Twist Bioscience. sgRNA libraries were cloned into the retroviral vector by Gibson assembly. sgRNA library representation was verified by next generation sequencing to contain > 90% perfectly matching sgRNAs, < 0.5% undetected sgRNAs and a skew ratio of less than 10. sgRNA oligo pairs were purchased from Sigma-Aldrich. Individual CRISPR sequences were inserted into the retroviral vector by ligation (NEB T4 DNA ligase). Sequences of individual sgRNA-expressing constructs were confirmed by Sanger sequencing.
Th2 cell culture for CRISPR screening
Splenic naïve CD4+ T cells were sorted as Live CD4+CD44loCD62LhiCD25– cells. Cells were maintained in RPMI1640, 10% FCS with penicillin-streptomycin and 2-mercaptoethanol. Naïve CD4+ T cells were isolated from Rosa26Cas9EGFP x Il13tdTom mice and cultured on anti-CD3 coated plates (2B Scientific, 145-2C11, 5 mg ml-1, 37oC, 1 h), supplemented with anti-CD28 (2B Scientific, 37.51, 2 mg ml-1) and IL-2 (10 ng ml-1) for 24 hr. Cells were collected and mixed with retroviruses and spinoculated on retronectin-coated plates (Takara, 4 mg cm-2, non-TC-treated plate) at 37oC for 1 h. Cells were incubated further for 3 h at 37oC before transfer to fresh TC-treated plates until day 6. Fresh media containing 10 ng ml-1 IL-2 was supplemented at day 3. On day 6, cells were transferred to anti-CD3 coated plates and cultured in the presence of anti-CD28 (2 mg ml-1), IL-2 (10 ng ml-1), IL-4 (10 ng ml-1) and anti-IFNγ neutralising antibody (2B Scientific, 1 mg ml-1). After 3 days of differentiation, GFP+ BFP+ cells were sorted into IL13Tom+ and IL13Tom- populations.
Retroviral production
Platinum-E retroviral packaging cells (Cell biolabs, #RV-101) were maintained in DMEM, 10% FCS with penicillin-streptomycin, supplemented with puromycin (1 mg ml-1) and blasticidin (10 mg ml-1). On the day before transfection, 3 million cells were seeded in a 100 mm culture dish in 10 ml of media without antibiotics. Cells were transfected at 70% confluency using Fugene HD Transfection Reagent (Promega). For each 100 mm culture dish, 950 ml OPTI-MEM (GIBCO) was mixed with 11 mg pCl-Eco, 22 mg library plasmid and 99 ml Fugene HD. The transfection mixture was incubated for 10 min at room temperature prior to addition. At 18 h post-transfection, the media was replaced with 10 ml fresh media, and viral supernatant was harvested at 48 and 72 h post-transfection. Cells were removed by filtering through a 0.45 mm syringe filter.
Genomic extraction and sequencing library preparation
Genomic DNA from sorted cells were extracted using the QIAGEN DNeasy Blood & Tissue Kits following the manufacturer’s protocol, with the exception of DNA elution in water instead of buffer AE. sgRNA-insert was first PCR-amplified using Herculase II Fusion DNA polymerase (Agilent) with primers (Forward) AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG and (Reverse) CTTTAGTTTGTATGTCTGTTGCTATTATGTCTACTATTCTTTCC, using up to 2 mg genomic DNA per 50 μl reaction. Equal volumes from each reaction were pooled and used for a further PCR amplification step to attach Illumina sequencing adaptors and Illumina P7 barcodes, using Herculase II Fusion DNA polymerase. The 330 bp library was gel purified and quantified using KAPA library quantification kit (Roche). Libraries were pooled and sequenced with a HiSeq 4000 at the CRUK Cambridge NGS facility.
Analysis of CRISPR screen results
20 nt sgRNA sequences were trimmed from backbone sequences using Cutadapt (version 1.4.1) (5’ GACGAAACACCG, 3’ GTTTTAGAGCTA). sgRNA sequences were aligned to reference sgRNA libraries using Bowtie2 (version 1.2.3). sgRNAs with counts less than 20 (genome-wide screens) or 50 (all other screens) in either of the populations were excluded from the analysis. The stat.wilcox function from the caRpools package (version 0.83) was applied to each screen separately. The function was modified to return the non-adjusted p-values. The stat.wilcox function collapses the sgRNAs to genes returning an enrichment score and a p-value for each gene. NT sgRNAs were used as a reference population. To combine data from screen replicates, the mean of enrichment score for each gene was calculated, and Fisher’s method was used to combine the p-values.
In vitro mouse Th cell culture
Splenic naïve T cells were sorted (CD4+CD44-CD25-CD62L+) and were cultured (250,000 cells /well) on anti-CD3 coated plates (5 mg ml-1), supplemented with anti-CD28 (2 mg ml-1) and IL-2 (10 ng ml-1). The following cytokines and neutralising antibodies were additionally supplemented in different Th conditions. Th1: IL-12 (10 ng ml-1) and anti-IL-4 neutralising antibody (BioLegend, 11B11, 1 mg ml-1). Th2: IL-4 (10 ng ml-1) and anti-IFNγ neutralising antibody (1 mg ml-1). Cells were passaged on day 2 or day 3, then analysed by flow cytometry on day 5. Where appropriate, 100 mM of the neuroprotective peptide called NAP (NAPVSIPQ) was supplemented during the in vitro differentiation.
RNA-sequencing
Cells were sorted by flow cytometry into PBS, 50% FCS, and RNA was extracted using the RNeasy Plus Micro kit (Qiagen). After assessment using a Bioanalyzer (Agilent), RNA was processed for RNA-seq using an Ovation RNA-seq System V2 (Nugen), fragmented using the Covaris M220 ultrasonicator and bar-coded using Ovation Ultralow Library Systems (Nugen). Samples were sequenced using an Illumina HiSeq 4000, by running a single-read 50-bp protocol (Cancer Research UK Cambridge Institute). Sequence data were trimmed to remove adaptors and sequences with a quality score below 30 using Trim Galore (version 0.50, Babraham Bioinformatics) and then aligned to the mouse genome (GRCm38) using STAR (version 2.6.0a), and differential expression was calculated using DESeq2 (version 1.18.1).64
Single-cell RNA sequencing
10x single-cell library preparation was performed using the 10x Genomics technology platform. The 10x Genomics Chromium Single Cell 3′ v3 protocol was followed to obtain 3′ libraries for subsequent sequencing. The reads were aligned to the mouse transcriptome (GRCm38), and expression was calculated using the 10x Cell Ranger (version 3.0.2) wrapper for the STAR aligner (version 2.60a). Separate libraries were generated using cells purified from the lungs of papain-challenged Cd4Cre and Cd4CreAdnpfl/fl mice (7,500 CD4+CD44+CD62L–ST2+ effector T cells and 500 naïve CD4+CD44–CD62L+CD25– T cells) and then combined using Cell Ranger. Analysis and statistical calculations were performed using the 10x Genomics Loupe Browser (https://support.10xgenomics.com/single-cell-gene-expression/software/visualization/latest/what-is-loupe-cell-browser).
Immunoprecipitation
Th2 cells were lysed in lysis buffer (50 mM Tris pH 8.0, 0.1% NP40, 10% glycerol and 2 mM EDTA), supplemented with 1x cOmplete protease inhibitor (Roche) and PMSF (Sigma Aldrich). After 10 min incubation on ice with intermittent mixing the lysates were centrifuged at 1,700 g at 4oC for 5 min and the supernatant was collected. The pelleted nuclei were resuspended in nuclear extraction buffer (50 mM Tris pH 8.0, 1 mM EDTA, 150 mM NaCl, 1% NP40 and 5% glycerol) supplemented with protease inhibitor cocktail and PMSF, and incubated on ice for 1 hour. Nuclear extract was collected by centrifugation at 13,000 x g at 4oC for 10 min. Protein concentration was quantified using the Pierce 660nm protein assay reagent (ThermoFisher, #22660). Lysates were incubated with antibodies (2 mg antibody per 100 mg protein) overnight at 4oC on a rotator. Immunocomplexes were precipitated with protein A/G dynabeads (Thermo Scientific #88802), washed three times with lysis buffer and once with TE buffer (10 mM Tris and 0.1 mM EDTA, pH 8). For western blot analysis, cell lysates or immunocomplexes were denatured by boiling at 95oC for 5 min in 1X NuPage LDS sample buffer (#NP0008) supplemented with 1% 2-mercaptoethanol. Proteins were resolved with Novex Tris-Glycine gels and transferred to PVDF membranes. Membranes were sequentially blocked with 5% BSA in PBST, incubated with primary and HRP-conjugated secondary antibodies and ECL western blotting detection reagent (GE Healthcare #RPN2106). For mass spectrometry analysis, the immunocomplexes were resuspended in 50mM NH4HCO3 followed by reduction with 10 mM DTT and alkylation with 55mM iodoacetamide. Then, proteins were digested (50 mM (NH₄)HCO₃ pH 8.0, 1μg trypsin, overnight, 37°C). Digestion was terminated by the addition of formic acid to a final concentration of 2% v/v. After separation (C18 Acclaim PepMap100 3 μm, 75 μm x 150 mm nanoViper, ThermoScientific Dionex, San Jose, USA), peptides were eluted with a gradient of acetonitrile. The analytical column outlet was directly interfaced via a modified nano-flow electrospray ionisation source, with a hybrid dual pressure linear ion trap mass spectrometer (Orbitrap Velos, ThermoScientific, San Jose, USA). Data dependent analysis was carried out, using a resolution of 30,000 for the full MS spectrum, followed by ten MS/MS spectra in the linear ion trap. MS spectra were collected over a m/z range of 300–2000. MS/MS scans were collected using a threshold energy of 35 for collision induced dissociation. LC-MS/MS data were then searched against a protein database (UniProt KB) using the Mascot search engine programme (Matrix Science, UK).73 Database search parameters were set with a precursor tolerance of 5 ppm and a fragment ion mass tolerance of 0.8 Da. Two missed enzyme cleavages were allowed and variable modifications for oxidized methionine, carbamidomethyl cysteine, pyroglutamic acid, phosphorylated serine, threonine and tyrosine were included. MS/MS data were validated using the Scaffold programme (Proteome Software Inc., USA).65 All data were additionally interrogated manually.
ChIP-seq using ChIPmentation
Chromatin extracts from in vitro cultured Th2 cells (1.0 × 107 cells) were prepared using the truChIP Chromatin Shearing kit (Covaris), with 5 min of crosslinking and optimized shearing conditions (peak power, 75; duty factor, 10.0; cycles per burst, 200; duration, 300 s), to obtain fragments of ∼500 bp. Extracts were exposed to 1% SDS and diluted 10x with dilution buffer (5.5 mM EDTA, 55 mM Tris-HCl, pH 8, 200 mM NaCl, 0.5% NP-40). Chromatin extracts were incubated overnight at 4 °C with 2 μg of antibody. In addition, 25 μl protein A Dynabeads (Thermo Fisher Scientific) per immunoprecipitation were blocked in PBS containing 0.1% BSA (Sigma) by incubating overnight at 4 °C. The next day, beads were added to the chromatin extracts, followed by incubating for 1 h at 4 °C. Beads were collected and washed twice with low-salt buffer (0.1% SDS, 1% Triton X-100, 1 mM EDTA, 10 mM Tris-HCl, pH 8, 140 mM NaCl, 0.1% sodium deoxycholate), twice with high-salt buffer (0.1% SDS, 1% Triton X-100, 1 mM EDTA, 10 mM Tris-HCl, pH 8, 500 mM NaCl, 0.1% sodium deoxycholate), twice with LiCl buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate) and once with 10 mM Tris-HCl, pH 8. Chromatin–antibody–bead complexes were then subjected to tagmentation, followed by the elution of DNA, and libraries were amplified and purified as described previously.74 Pooled libraries were sequenced using an Illumina Novaseq 6000, running a pair-read 150-bp protocol (Cancer Research UK Cambridge Institute). Sequenced reads were aligned to the mouse genome (GRCm38) using Bowtie2 (version 2.3.5.1) with default parameters, and reads that could not be uniquely mapped were removed from further analyses. Aligned reads were visualised using the SeqMonk software (v1.48.0). HOMER66 (v4.10.4) software was used for motif find analysis. Peak calling analysis was performed using Macs2 (v2.1.2) and the target genes were defined by the closest gene from each peak (bedtools closest). Only target genes identified in two independent experiments were used in further analysis.
ATAC-seq
ATAC-seq was performed as previously described.67 20,000 to 50,000 FACS purified cells were lysed using cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.1% NP-40) to obtain nuclei extract. Nuclei were immediately used in the transposase reaction (25 μl 2× TD buffer, 2.5 μl transposase (Illumina) and 22.5 μl nuclease- free water) for 30 min at 37 °C, followed by sample purification (Qiagen MinElute kit). Then, we amplified library fragments using Kappa HiFi HotStart Ready mix and 1.25 M of custom Nextera PCR primers as previously described.68 Libraries were purified using dual (0.5x-0.7x) SPRI Ampure XP beads (Beckman Coulter), pooled and were subjected to high-throughput sequencing. ATAC-seq data was aligned to the genome using the same pipeline as the ChIP-seq data.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism version 9 software. Statistical significance was calculated by unpaired Student’s t-test (two-tailed), one-way or two-way ANOVA. ∗∗∗∗: P<0.0001, ∗∗∗: P<0.001, ∗∗: P<0.01, ∗: P<0.05, ns: not significant. No samples were excluded from the analysis.
Acknowledgments
We are grateful to the Ares, genotyping, mass spectroscopy, and flow cytometry core facilities for their technical assistance. We acknowledge Hans-Reimer Rodewald for the Il7ra-Cre mice and Balaji Santhanam for assistance with collation of the TF library. This work was supported by the Medical Research Council as part of United Kingdom Research and Innovation (MRC grant U105178805). For the purpose of open access, the MRC Laboratory of Molecular Biology has applied a CC BY public copyright license to any Author Accepted Manuscript version arising. This study was also supported by the Wellcome Trust (100963/Z/13/Z and 220223/Z/20/Z), and A.C.H.S. was supported by a Croucher Cambridge International Scholarship.
Author contributions
A.C.F.F. and A.C.H.S. designed and performed experiments and wrote the paper. P.A.C., A.C., H.E.J., and P.K. performed experiments, provided advice on experimental design and interpretation, and commented on the manuscript. A.N.J.M. supervised the project, designed the experiments, and wrote the paper.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: June 6, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2023.05.010.
Contributor Information
Ana C.F. Ferreira, Email: ferreira@mrc-lmb.cam.ac.uk.
Andrew N.J. McKenzie, Email: anm@mrc-lmb.cam.ac.uk.
Supplemental information
Data and code availability
-
•
Single-cell RNAseq, ATACseq and ChIPseq data are deposited with the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under the accession number GSE218017.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Annunziato F., Romagnani C., Romagnani S. The 3 major types of innate and adaptive cell-mediated effector immunity. J. Allergy Clin. Immunol. 2015;135:626–635. doi: 10.1016/j.jaci.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Walker J.A., McKenzie A.N.J. TH2 cell development and function. Nat. Rev. Immunol. 2018;18:121–133. doi: 10.1038/nri.2017.118. [DOI] [PubMed] [Google Scholar]
- 3.Walker J.A., McKenzie A.N. Development and function of group 2 innate lymphoid cells. Curr. Opin. Immunol. 2013;25:148–155. doi: 10.1016/j.coi.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J., Paul W.E. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKenzie A.N. Type-2 innate lymphoid cells in asthma and allergy. Ann. Am. Thorac. Soc. 2014;11:S263–S270. doi: 10.1513/AnnalsATS.201403-097AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammad H., Lambrecht B.N. The basic immunology of asthma. Cell. 2021;184:1469–1485. doi: 10.1016/j.cell.2021.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Fallon P.G., Jolin H.E., Smith P., Emson C.L., Townsend M.J., Fallon R., Smith P., McKenzie A.N. IL-4 induces characteristic Th2 responses even in the combined absence of IL-5, IL-9, and IL-13. Immunity. 2002;17:7–17. doi: 10.1016/s1074-7613(02)00332-1. [DOI] [PubMed] [Google Scholar]
- 8.Spilianakis C.G., Flavell R.A. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 9.Fields P.E., Kim S.T., Flavell R.A. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J. Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 10.Fields P.E., Lee G.R., Kim S.T., Bartsevich V.V., Flavell R.A. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Kanhere A., Hertweck A., Bhatia U., Gökmen M.R., Perucha E., Jackson I., Lord G.M., Jenner R.G. T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat. Commun. 2012;3:1268. doi: 10.1038/ncomms2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao K., Carr T., Wu J., Barclay W., Jin J., Ciofani M., Reinhardt R.L. BATF modulates the Th2 locus control region and regulates CD4+ T cell fate during antihelminth immunity. J. Immunol. 2016;197:4371–4381. doi: 10.4049/jimmunol.1601371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schraml B.U., Hildner K., Ise W., Lee W.L., Smith W.A., Solomon B., Sahota G., Sim J., Mukasa R., Cemerski S., et al. The AP-1 transcription factor BATF controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwal S., Avni O., Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643–652. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- 15.Smale S.T., Fisher A.G. Chromatin structure and gene regulation in the immune system. Annu. Rev. Immunol. 2002;20:427–462. doi: 10.1146/annurev.immunol.20.100301.064739. [DOI] [PubMed] [Google Scholar]
- 16.Ansel K.M., Lee D.U., Rao A. An epigenetic view of helper T cell differentiation. Nat. Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 17.Michieletto M.F., Tello-Cajiao J.J., Mowel W.K., Chandra A., Yoon S., Joannas L., Clark M.L., Jimenez M.T., Wright J.M., Lundgren P., et al. Multiscale 3D genome organization underlies ILC2 ontogenesis and allergic airway inflammation. Nat. Immunol. 2023;24:42–54. doi: 10.1038/s41590-022-01295-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wurster A.L., Pazin M.J. BRG1-mediated chromatin remodeling regulates differentiation and gene expression of T helper cells. Mol. Cell. Biol. 2008;28:7274–7285. doi: 10.1128/MCB.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Schoonhoven A., Huylebroeck D., Hendriks R.W., Stadhouders R. 3D genome organization during lymphocyte development and activation. Brief. Funct. Genomics. 2020;19:71–82. doi: 10.1093/bfgp/elz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ribeiro de Almeida C., Heath H., Krpic S., Dingjan G.M., van Hamburg J.P., Bergen I., van de Nobelen S., Sleutels F., Grosveld F., Galjart N., Hendriks R.W. Critical role for the transcription regulator CCCTC-binding factor in the control of Th2 cytokine expression. J. Immunol. 2009;182:999–1010. doi: 10.4049/jimmunol.182.2.999. [DOI] [PubMed] [Google Scholar]
- 21.Hwang S.S., Kim Y.U., Lee S., Jang S.W., Kim M.K., Koh B.H., Lee W., Kim J., Souabni A., Busslinger M., Lee G.R. Transcription factor YY1 is essential for regulation of the Th2 cytokine locus and for Th2 cell differentiation. Proc. Natl. Acad. Sci. USA. 2013;110:276–281. doi: 10.1073/pnas.1214682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul W.E., Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat. Rev. Immunol. 2010;10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szeto A.C.H., Ferreira A.C.F., Mannion J., Clark P.A., Sivasubramaniam M., Heycock M.W.D., Crisp A., Jolin H.E., Kozik P., Knolle M.D., McKenzie A.N.J. An alphavbeta3 integrin checkpoint is critical for efficient T(H)2 cell cytokine polarization and potentiation of antigen-specific immunity. Nat. Immunol. 2023;24:123–135. doi: 10.1038/s41590-022-01378-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinhasov A., Mandel S., Torchinsky A., Giladi E., Pittel Z., Goldsweig A.M., Servoss S.J., Brenneman D.E., Gozes I. Activity-dependent neuroprotective protein: a novel gene essential for brain formation. Brain Res. Dev. Brain Res. 2003;144:83–90. doi: 10.1016/s0165-3806(03)00162-7. [DOI] [PubMed] [Google Scholar]
- 25.Helsmoortel C., Vulto-van Silfhout A.T., Coe B.P., Vandeweyer G., Rooms L., van den Ende J., Schuurs-Hoeijmakers J.H., Marcelis C.L., Willemsen M.H., Vissers L.E., et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat. Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Dijck A., Vulto-van Silfhout A.T., Cappuyns E., van der Werf I.M., Mancini G.M., Tzschach A., Bernier R., Gozes I., Eichler E.E., Romano C., et al. Clinical presentation of a complex neurodevelopmental disorder caused by mutations in ADNP. Biol. Psychiatry. 2019;85:287–297. doi: 10.1016/j.biopsych.2018.02.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun X., Yu W., Li L., Sun Y. ADNP controls gene expression through local chromatin architecture by association with BRG1 and CHD4. Front. Cell Dev. Biol. 2020;8:553. doi: 10.3389/fcell.2020.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostapcuk V., Mohn F., Carl S.H., Basters A., Hess D., Iesmantavicius V., Lampersberger L., Flemr M., Pandey A., Thomä N.H., et al. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature. 2018;557:739–743. doi: 10.1038/s41586-018-0153-8. [DOI] [PubMed] [Google Scholar]
- 29.Kaaij L.J.T., Mohn F., van der Weide R.H., de Wit E., Bühler M. The ChAHP complex counteracts chromatin looping at CTCF sites that emerged from SINE expansions in mouse. Cell. 2019;178:1437–1451.e14. doi: 10.1016/j.cell.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Gozes I., Morimoto B.H., Tiong J., Fox A., Sutherland K., Dangoor D., Holser-Cochav M., Vered K., Newton P., Aisen P.S., et al. NAP: research and development of a peptide derived from activity-dependent neuroprotective protein (ADNP) CNS Drug Rev. 2005;11:353–368. doi: 10.1111/j.1527-3458.2005.tb00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novey H.S., Marchioli L.E., Sokol W.N., Wells I.D. Papain-induced asthma--physiological and immunological features. J. Allergy Clin. Immunol. 1979;63:98–103. doi: 10.1016/0091-6749(79)90198-2. [DOI] [PubMed] [Google Scholar]
- 32.Halim T.Y., Steer C.A., Mathä L., Gold M.J., Martinez-Gonzalez I., McNagny K.M., McKenzie A.N., Takei F. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40:425–435. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon J.J., Chu H.H., Pepper M., McSorley S.J., Jameson S.C., Kedl R.M., Jenkins M.K. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou R., Flaswinkel H., Schneider M.R., Lahm H., Hoeflich A., Wanke R., Wolf E. Insulin-like growth factor-binding protein-4 inhibits growth of the thymus in transgenic mice. J. Mol. Endocrinol. 2004;32:349–364. doi: 10.1677/jme.0.0320349. [DOI] [PubMed] [Google Scholar]
- 35.Opejin A., Surnov A., Misulovin Z., Pherson M., Gross C., Iberg C.A., Fallahee I., Bourque J., Dorsett D., Hawiger D. A two-step process of effector programming governs CD4(+) T cell fate determination induced by antigenic activation in the steady state. Cell Rep. 2020;33:108424. doi: 10.1016/j.celrep.2020.108424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourque J., Kousnetsov R., Hawiger D. Roles of Hopx in the differentiation and functions of immune cells. Eur. J. Cell Biol. 2022;101:151242. doi: 10.1016/j.ejcb.2022.151242. [DOI] [PubMed] [Google Scholar]
- 37.Nagashima H., Petermann F., Pękowska A., Chaitankar V., Kanno Y., O’Shea J.J. 2022. Remodeling of Il4-Il13-Il5 locus underlies selective gene expression. [DOI] [Google Scholar]
- 38.Barlow J.L., Bellosi A., Hardman C.S., Drynan L.F., Wong S.H., Cruickshank J.P., McKenzie A.N. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J. Allergy Clin. Immunol. 2012;129:191–198.e1–e4. doi: 10.1016/j.jaci.2011.09.041. [DOI] [PubMed] [Google Scholar]
- 39.Liang H.E., Reinhardt R.L., Bando J.K., Sullivan B.M., Ho I.C., Locksley R.M. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol. 2011;13:58–66. doi: 10.1038/ni.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun X., Peng X., Cao Y., Zhou Y., Sun Y. ADNP promotes neural differentiation by modulating Wnt/beta-catenin signaling. Nat. Commun. 2020;11:2984. doi: 10.1038/s41467-020-16799-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandel S., Rechavi G., Gozes I. Activity-dependent neuroprotective protein (ADNP) differentially interacts with chromatin to regulate genes essential for embryogenesis. Dev. Biol. 2007;303:814–824. doi: 10.1016/j.ydbio.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 42.Li B., Tournier C., Davis R.J., Flavell R.A. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamashita M., Ukai-Tadenuma M., Kimura M., Omori M., Inami M., Taniguchi M., Nakayama T. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J. Biol. Chem. 2002;277:42399–42408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- 44.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang Y., Kim Y.W., Kang J., Kim A. Histone H3K4me1 and H3K27ac play roles in nucleosome eviction and eRNA transcription, respectively, at enhancers. FASEB J. 2021;35 doi: 10.1096/fj.202100488R. [DOI] [PubMed] [Google Scholar]
- 46.Hosokawa H., Tanaka T., Suzuki Y., Iwamura C., Ohkubo S., Endoh K., Kato M., Endo Y., Onodera A., Tumes D.J., et al. Functionally distinct Gata3/Chd4 complexes coordinately establish T helper 2 (Th2) cell identity. Proc. Natl. Acad. Sci. USA. 2013;110:4691–4696. doi: 10.1073/pnas.1220865110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho I.C., Lo D., Glimcher L.H. c-maf promotes T helper cell type 2 (Th2) and attenuates Th1 differentiation by both interleukin 4-dependent and -independent mechanisms. J. Exp. Med. 1998;188:1859–1866. doi: 10.1084/jem.188.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Junttila I.S. Tuning the cytokine responses: an update on interleukin (IL)-4 and IL-13 receptor complexes. Front. Immunol. 2018;9:888. doi: 10.3389/fimmu.2018.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul W.E. History of interleukin-4. Cytokine. 2015;75:3–7. doi: 10.1016/j.cyto.2015.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai S., Lee C.C., Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 51.Van Dijck A., Vandeweyer G., Kooy F. In: GeneReviews((R)) Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Mirzaa G.M., Amemiya A., editors. University of Washington; 1993. ADNP-related disorder. [Google Scholar]
- 52.Farnung L., Ochmann M., Cramer P. Nucleosome-CHD4 chromatin remodeler structure maps human disease mutations. eLife. 2020;9 doi: 10.7554/eLife.56178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagman J.R., Arends T., Laborda C., Knapp J.R., Harmacek L., O'Connor B.P. Chromodomain helicase DNA-binding 4 (CHD4) regulates early B cell identity and V(D)J recombination. Immunol. Rev. 2022;305:29–42. doi: 10.1111/imr.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams C.J., Naito T., Arco P.G., Seavitt J.R., Cashman S.M., De Souza B., Qi X., Keables P., Von Andrian U.H., Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Shimono Y., Murakami H., Kawai K., Wade P.A., Shimokata K., Takahashi M. Mi-2 beta associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J. Biol. Chem. 2003;278:51638–51645. doi: 10.1074/jbc.M309198200. [DOI] [PubMed] [Google Scholar]
- 56.De S., Wurster A.L., Precht P., Wood W.H., 3rd, Becker K.G., Pazin M.J. Dynamic BRG1 recruitment during T helper differentiation and activation reveals distal regulatory elements. Mol. Cell. Biol. 2011;31:1512–1527. doi: 10.1128/MCB.00920-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu N., Balliano A., Hayes J.J. Mechanism(s) of SWI/SNF-induced nucleosome mobilization. ChemBioChem. 2011;12:196–204. doi: 10.1002/cbic.201000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng W., Flavell R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 59.Zhu J., Yamane H., Cote-Sierra J., Guo L., Paul W.E. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 2006;16:3–10. doi: 10.1038/sj.cr.7310002. [DOI] [PubMed] [Google Scholar]
- 60.Sahoo A., Alekseev A., Tanaka K., Obertas L., Lerman B., Haymaker C., Clise-Dwyer K., McMurray J.S., Nurieva R. BATF is important for IL-4 expression in T follicular helper cells. Nat. Commun. 2015;6:7997. doi: 10.1038/ncomms8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller M.M., Patel P.S., Bao K., Danhorn T., O'Connor B.P., Reinhardt R.L. BATF acts as an essential regulator of IL-25-responsive migratory ILC2 cell fate and function. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.aay3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howrylak J.A., Moll M., Weiss S.T., Raby B.A., Wu W., Xing E.P. Gene expression profiling of asthma phenotypes demonstrates molecular signatures of atopy and asthma control. J. Allergy Clin. Immunol. 2016;137:1390–1397.e6. doi: 10.1016/j.jaci.2015.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlenner S.M., Madan V., Busch K., Tietz A., Läufle C., Costa C., Blum C., Fehling H.J., Rodewald H.R. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 64.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 66.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M., et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McKenzie G.J., Fallon P.G., Emson C.L., Grencis R.K., McKenzie A.N. Simultaneous disruption of interleukin (IL)-4 and IL-13 defines individual roles in T helper cell type 2-mediated responses. J. Exp. Med. 1999;189:1565–1572. doi: 10.1084/jem.189.10.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chu V.T., Graf R., Wirtz T., Weber T., Favret J., Li X., Petsch K., Tran N.T., Sieweke M.H., Berek C., et al. Efficient CRISPR-mediated mutagenesis in primary immune cells using CrispRGold and a C57BL/6 Cas9 transgenic mouse line. Proc. Natl. Acad. Sci. USA. 2016;113:12514–12519. doi: 10.1073/pnas.1613884113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 74.Schmidl C., Rendeiro A.F., Sheffield N.C., Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods. 2015;12:963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single-cell RNAseq, ATACseq and ChIPseq data are deposited with the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under the accession number GSE218017.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.