Summary
During eukaryotic DNA replication, Pol α-primase generates primers at replication origins to start leading-strand synthesis and every few hundred nucleotides during discontinuous lagging-strand replication. How Pol α-primase is targeted to replication forks to prime DNA synthesis is not fully understood. Here, by determining cryoelectron microscopy (cryo-EM) structures of budding yeast and human replisomes containing Pol α-primase, we reveal a conserved mechanism for the coordination of priming by the replisome. Pol α-primase binds directly to the leading edge of the CMG (CDC45-MCM-GINS) replicative helicase via a complex interaction network. The non-catalytic PRIM2/Pri2 subunit forms two interfaces with CMG that are critical for in vitro DNA replication and yeast cell growth. These interactions position the primase catalytic subunit PRIM1/Pri1 directly above the exit channel for lagging-strand template single-stranded DNA (ssDNA), revealing why priming occurs efficiently only on the lagging-strand template and elucidating a mechanism for Pol α-primase to overcome competition from RPA to initiate primer synthesis.
Keywords: DNA replication, replisome, priming, Pol α-primase, genome stability, helicase, DNA polymerase, CMG, cryo-EM
Graphical abstract
Highlights
-
•
Structures of DNA engaged yeast and human replisomes containing Pol α-primase
-
•
A conserved mechanism targets Pol α-primase to the replisome for priming
-
•
A multisite interaction network targets primase to the lagging-strand template
-
•
Direct interactions between CMG and Pol α-primase are critical for priming
By determining cryo-EM structures of budding yeast and human replisomes containing the Pol α-primase complex, Jones et al. reveal a conserved mechanism for the coordination of nascent-strand priming in the eukaryotic replisome. The mechanism explains why priming by Pol α-primase is highly efficient on the lagging-strand but not leading-strand template.
Introduction
Following replisome assembly and template unwinding at bi-directional origins of DNA replication, Pol α-primase is recruited to the two advancing replisomes where it primes the lagging-strand template.1 To start coupled leading-strand replication, these primers are extended across the origin by the main lagging-strand polymerase, Pol δ,2 before a polymerase switch transfers the nascent strand to the principal leading-strand polymerase, Pol ε.1,3,4,5,6 As replication forks progress primers are synthesized every few hundred nucleotides to support discontinuous lagging-strand replication. If leading-strand synthesis is interrupted due to DNA damage, biochemical reconstitution experiments have demonstrated that S. cerevisiae (budding yeast) replisomes continue lagging-strand replication but do not frequently reinitiate leading-strand replication due to a failure to support efficient primer synthesis on this strand.7,8,9,10 Collectively, these observations indicate that the replisome efficiently targets Pol α-primase to the lagging-strand template but not the leading-strand template, which likely explains why some eukaryotes, including humans, encode a second primase-polymerase, PRIMPOL, to restart leading-strand replication.11,12,13,14 Currently the mechanistic basis underlying the preference of Pol α-primase for lagging- rather than leading-strand priming is unknown.
Pol α-primase is a constitutive heterotetramer composed of a dimeric primase (PRIM1 and PRIM2 in H. sapiens [human], Pri1 and Pri2 in budding yeast) and a dimeric DNA polymerase (POLA1 and POLA2 in human, Pol1 and Pol12 in budding yeast) (Figure 1A). Primase synthesizes 8–10 nucleotides (nt) of RNA that are transferred to the Pol α DNA polymerase for limited extension to a total primer length about 20–35 nt.15,16,17,18 In vitro, the ability of budding yeast7 and human19,20,21 Pol α-primase to initiate primer synthesis on single-stranded DNA (ssDNA) templates is blocked when the template is saturated with RPA, indicating that a mechanism exists to target primase to ssDNA at the eukaryotic replication fork. Consistent with this idea, Pol α-primase interacts with several core components of the replisome including AND-1 (Ctf4 in budding yeast), MCM10, GINS, and CMG.22,23,24,25,26,27
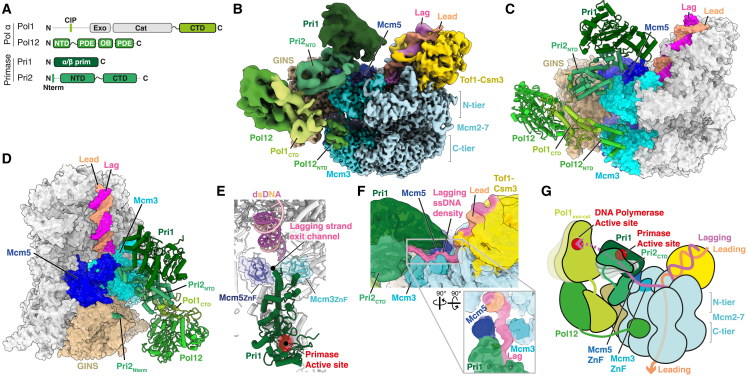
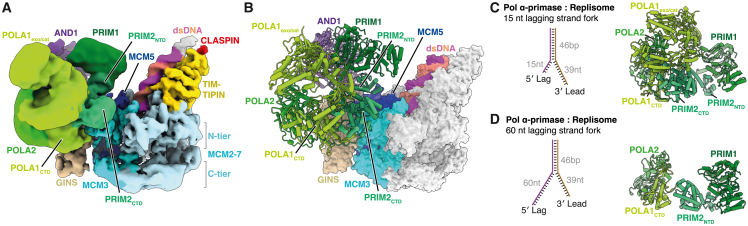
Figure 1.
Structure of Pol α-primase in the budding yeast replisome
(A) Domain architecture of yeast Pol α-primase. exo, exonuclease domain; cat, catalytic domain; CIP, Ctf4-interacting peptide; NTD, N-terminal domain; CTD, C-terminal domain; PDE, phosphodiesterase domain; OB, oligonucleotide/oligosaccharide-binding domain.
(B) Composite cryo-EM map of the budding yeast Pol α-primase associated replisome bound to replication fork DNA containing a 60 nucleotide 5′ flap. Density for Ctf4 is not observed in this map. The map was derived from combining individual focused refinements and is colored according to chain occupancy.
(C and D) Atomic model of the budding yeast Pol α-primase associated replisome lacking Ctf4 derived from cryo-EM data displayed in (B). Regions of CMG that physically interact with Pol α-primase are colored.
(E) Focused view of the Pri1 catalytic subunit of primase, showing how it is positioned above the exit channel for lagging-strand template ssDNA.
(F) Cryo-EM reconstruction displaying continuous density for lagging-strand template ssDNA extending from the point of dsDNA strand separation toward the active site region of Pri1. Map colored by chain occupancy with the density assigned to the lagging-strand template post-strand separation colored manually.
(G) Schematic illustrating the organization of Pol α-primase in the budding yeast replisome. The path of lagging-strand template ssDNA visualized in the structure immediately following strand separation is illustrated (solid pink line). The putative path of the lagging-strand template between the Pri1 and Pol1 active sites is also illustrated (dashed pink line).
In budding yeast, Pol α-primase is tethered to replisome progression complexes (RPCs) via interaction between its Pol1 subunit and Ctf4, a trimeric scaffold protein that binds directly to CMG.28,29,30,31 Similarly, human Pol α-primase associates with AND-1 but does so primarily via the N-terminal domain (NTD) of POLA2, which binds a C-terminal HMG-box in AND-1.32,33 However, considerable evidence indicates that AND-1/Ctf4 does not provide a pivotal link between Pol α-primase and the replisome to support priming: Ctf4 is a non-essential protein in budding yeast; disruption of the Pol1:Ctf4 interaction does not result in obvious DNA replication defects in vivo34; Pol α-primase localizes to replisomes in yeast cells lacking Ctf435; depletion of AND-1 in DT40 cells does not prevent the completion of bulk replication36; AND-1 and Ctf4 are dispensable for lagging-strand synthesis in DNA replication reactions reconstituted with purified proteins.27,37,38,39 Similarly, reconstituted budding yeast and human replisomes perform lagging-strand replication in the absence of MCM10.27,39,40
Accumulating evidence suggests that Pol α-primase is recruited to the replisome for priming via direct interactions with CMG. Yeast Pol α-primase can execute lagging-strand replication when functioning only with CMG and RPA.39 We recently found that minimal human replisomes consisting of CMG, Pol α-primase, Pol ε, CTF18-RFC, PCNA, and RPA support lagging-strand replication and that human Pol α-primase comigrates with CMG in glycerol gradient sedimentation experiments.27 However, because there are no structures of Pol α-primase in the eukaryotic replisome, we do not know how Pol α-primase binds to CMG, how these putative interactions contribute to lagging-strand DNA replication, how Pol α-primase might overcome competition with RPA for exposed ssDNA, where in the replisome Pol α-primase is positioned, and why Pol α-primase efficiently primes the lagging-strand but not leading-strand template. To address these questions, we have determined cryoelectron microscopy (cryo-EM) structures of budding yeast and human replisomes bound to Pol α-primase and replication fork DNA.
Results
Yeast Pol α-primase replisome structure
To assemble budding yeast replisomes associated with Pol α-primase for cryo-EM analysis, CMG was bound to a model replication fork containing a 39 nt 3′ flap, onto which CMG loads, and a 60 nt 5′ ssDNA flap to mimic the unwound lagging-strand template (Figure S1A). Fork-bound CMG was incubated with Pol α-primase and the replisome accessory factors Mrc1 and Tof1-Csm3. Tof1-Csm3 binds to the leading edge of CMG where it engages and stabilizes the parental DNA duplex and fork junction,30 which we reasoned might be important to aid the visualization of template DNA in cryo-EM reconstructions. After glycerol gradient sedimentation, complexes were isolated containing all replisome and Pol α-primase subunits and used to prepare grids for cryo-EM data collection and analysis (Figures S1 and S2; Table 1).
Table 1.
Cryo-EM statistics
| Budding yeast | Budding yeast | Budding yeast | Human | Human | |
|---|---|---|---|---|---|
| replisome:Pol α-primase | replisome:Pol α-primase | replisome:Pol α-primase | replisome:Pol α-primase | replisome:Pol α-primase | |
| 60 nt 5′ flap (−) Ctf4 | 60 nt 5′ flap (+) Ctf4. CIP #1 | 60 nt 5′ flap (+) Ctf4. CIP #2 | 60 nt 5′ flap | 15 nt 5′ flap | |
| (EMDB-16322) | (EMDB-15902) | (EMDB-15902) | (EMDB-15341) | (EMD-15922) | |
| (PDB: 8B9C) | (PDB: 8B9A) | (PDB: 8B9B) | (PDB: 8B9D) | – | |
| Data collection and processing | |||||
| Magnification | 81,000 × | 81,000× | 81,000× | 81,000× | 81,000× |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 40.184 | 40.184 | 40.184 | 37.8 | 88.5 |
| Defocus range (μm) | 1.5–3 | 1.5–3 | 1.5–3 | 1.5–3 | 3.5–0.9 |
| Pixel size (Å) (super resoution) | 0.86 | 0.86 | 0.86 | 1.23 | 1.07 |
| Symetry imposed | none | none | none | none | none |
| Movies collected | 12,819 | 12,819 | 12,819 | 7,355 | 6,718 |
| Initial particle images (no.) | 2,003,322 | 2,003,322 | 2,003,322 | 1,535,548 | 724,557 |
| Final particle images (no.) | 100,179 | 54,970 | 44,970 | 174,696 | 258,339 |
| Map resolution (Å) (0.143 FSC threshold) | 3.34 | 3.5 | 3.5 | 3.4 | 3.3 |
| Map resolution range (Å) | 2.8–12 | 2.9–12 | 2.9–12 | 2.8–12 | 3.0–12 |
| Refinement | |||||
| Initial model used (PDB code) | PDB: 6SKL | PDB: 6SKL | PDB: 6SKL | PDB: 7PFO | – |
| Model resolution (Å) (0.5 FSC threshold) | 4.1 | 4.2 | 4.2 | 4.1 | – |
| Map sharpening B factor (Å2) | −20 to −50 | −20 to −50 | −20 to −50 | −20 to −50 | – |
| Model composition | |||||
| Non-hydrogen atoms | 59,119 | 69,886 | 69,866 | 66,207 | – |
| Protein residues | 7,193 | 8,528 | 8,528 | 8,108 | – |
| Ligands | 4 AMP-PNP | 4 AMP-PNP | 4 AMP-PNP | 3 AMP-PNP | – |
| 4 Mg2+, 4 Zn2+ | 4 Mg2+, 4 Zn2+ | 4 Mg2+, 4 Zn2+ | 3 Mg2+, 4 Zn2+ | – | |
| RMSDs | |||||
| Bond lengths (Å) | 0.023 | 0.011 | 0.01 | 0.027 | – |
| Bond angles (°) | 1.913 | 1.396 | 1.101 | 2.66 | – |
| Validation | |||||
| MolProbity score | 0.73 | 0.75 | 0.77 | 0.78 | – |
| Clashscore | 0.32 | 0.62 | 0.69 | 0.32 | – |
| Poor rotamers (%) | 0.29 | 0.5 | 0.53 | 0.66 | – |
| Ramachandran plot | |||||
| Favored (%) | 97.42 | 97.81 | 97.81 | 97.12 | – |
| Allowed (%) | 2.58 | 2.19 | 2.19 | 2.88 | – |
| Disallowed (%) | 0 | 0 | 0 | 0 | – |
Three-dimensional (3D) reconstructions revealed well resolved cryo-EM density for CMG and Tof1-Csm3. Multiple additional densities were also apparent extending from the N-tier face of CMG atop Mcm3 and Mcm5 toward the MCM C-tier beside Mcm3 and GINS (Figure 1B). The resolution of these densities was typically lower than for CMG and displayed considerable variability (Figure S1E), indicating large conformational flexibility. Nonetheless, following extensive focused classification and refinement (see Figure S2), these densities could be unambiguously attributed to Pol α-primase (Figures 1B, S1E–S1O, and S3B–S3D), enabling us to construct a model of a DNA engaged yeast replisome encompassing CMG, Tof1-Csm3, several small sections of Mrc1 (Figure S3A) and regions of all four Pol α-primase subunits (Figures 1C and 1D; Video S1). The Pol α-primase model comprises the primase catalytic subunit Pri1 aside from its flexible N and C termini, the N terminus (residues 1–5) and NTD (residues 44–177 and 181–299) of the primase accessory subunit Pri2, the C-terminal domain (CTD) of the Pol α catalytic subunit Pol1 (Pol1CTD) (residues 1,271–1,468) and the majority of the Pol α accessory subunit Pol12 (residues 1–79, 203–582, and 604–705) (Figures 1C, 1D, and S3B–S3D).
First, individual replisome components are sequentially colored and labeled to aid in their visualisation and the interpretation of the overall structure. The video then highlights each pol α-primase interface with the replisome (sites a-e) using different views. Regions of the model forming an interaction interface are displayed using transparent surface rendering.
Pri1 is positioned close to the incoming parental double-stranded DNA (dsDNA) above a channel between the Mcm3 and Mcm5 zinc-finger (ZnF) domains, through which lagging-strand template ssDNA is extruded after strand separation (Figure 1E).41,42,43,44 It is localized to the replisome through its interaction with the Pri2 NTD (Pri2NTD), which sits on the periphery of the MCM N-tier straddling Mcm3 and Mcm5 (Figures 1C and 1D). Pol α (Pol1CTD and Pol12) is situated between the CMG N- and C-tiers, close to Mcm3 and GINS (Figures 1C and 1D) and is coupled to primase via an interaction between Pol1CTD and Pri2NTD.45 Pol12 interacts extensively with Pol1CTD and is anchored to the MCM C-tier through an interface involving its flexibly tethered NTD and Mcm3 (Figure 1C). Although Ctf4 was not included in replisome reconstitutions, we identified a 3D class containing both Pol α-primase and Ctf4 (Figure S3E). The presence of Ctf4 likely resulted from endogenous Ctf4 co-purifying with CMG due to the extensive interface between the two complexes.30,31 Comparison of the structures with and without Ctf4 revealed no substantial changes in the conformation of Pol α-primase (Figures S3E and S3F).
Clear densities for the Pol1 exonuclease-catalytic (exo-cat) domain (Pol1exo-cat) and Pri2 CTD (Pri2CTD) were not observed in our consensus refinement (Figures 1B and S1E), indicating that neither domain adopts a single stable conformation when Pol α-primase is bound to the replisome. This behavior contrasts with the crystal structure of human apo Pol α-primase, where both domains were well ordered.46 However, we recovered several rare 3D classes with low-resolution densities of the appropriate shape and volume to accommodate Pol1exo-cat and Pri2CTD, although the precise orientation of each domain could not be assigned (Figures S3G and S3H). In these reconstructions, Pri2CTD is adjacent to Pri1 close to the primase active site, while Pol1exo-cat sits above Pri2NTD on the periphery of the replisome adjacent to the Pri2CTD. This configuration more closely resembles the architecture of human Pol α-primase bound to CST (CTC1-STN1-TEN1) and telomeric ssDNA47 (Figure S3I), and a very recent structure of a human Pol α-primase elongation complex,48 than the human Pol α-primase apo structure.46 This led us to consider that Pol α-primase conformation in the yeast replisome might be modulated by protein-protein interactions and/or DNA engagement. Because DNA binding was heterogeneous across the dataset, we obtained a 3D replisome reconstruction lacking DNA (Figure S3J). Here, the positioning of Pol1exo-cat and Pri2CTD resembled the human apo crystal structure46 (Figure S3K), indicating that Pol α-primase undergoes DNA-dependent conformational changes when associated with CMG in the budding yeast replisome.
To further explore the putative DNA engagement state of Pol α-primase, we performed additional classification focusing on regions of primase close to the replication fork junction (Figure S2). Strikingly, this strategy revealed a 3D class with continuous density extending from the parental DNA duplex at the point of strand separation, through the channel between the Mcm3 and Mcm5 ZnF domains and alongside Pri1 in the direction of the primase active site (Figure 1F). The density between the Mcm3 and Mcm5 ZnF domains is in an equivalent position to the previously identified path of the lagging-strand template following strand separation in the human replisome,41,42 strongly suggesting that it corresponds to lagging-strand template ssDNA. Moreover, the close proximity of the density to Pri1 and its continuation beyond the Mcm3-Mcm5 ZnF channel—which has not been observed in prior human and yeast replisome structures lacking Pol α-primase—indicate that Pri1 engages lagging-strand template ssDNA in the yeast replisome structure. We hypothesize that this configuration functions to ensure a minimal length of ssDNA is required for the lagging-strand template to reach the primase active site, thereby enabling primase to outcompete RPA for access to the template to initiate primer synthesis. Moreover, the positioning of Pri1 and Pol1exo-cat arranges the primase and DNA polymerase catalytic centers in synthesis order along the template (Figures 1G and S3L), suggesting a possible mechanism for transfer of the RNA primer to the Pol α DNA polymerase as the replisome advances, similar to the mechanism proposed for human Pol α-primase during telomere C strand fill-in.47
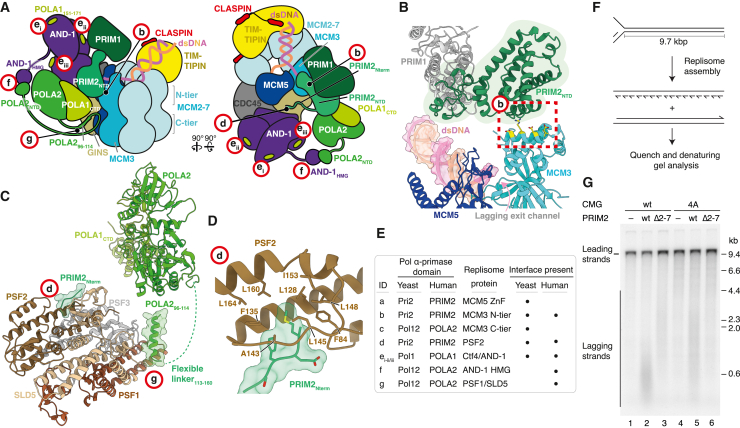
Pol α-primase replisome interactions
Four small interaction sites, labeled sites a–d in Figure 2A, tether Pol α-primase directly to CMG and position primase to engage lagging-strand template ssDNA (Video S1). Pri2NTD forms electrostatic interfaces with both the Mcm5 ZnF domain (site a) and the Mcm3 N-terminal helical domain (site b) (Figure 2B). The interface with the Mcm5 ZnF is mediated by a small insertion in Pri2NTD that appears confined to a subset of fungal species (Figure S4A), while the interface with Mcm3 involves three flexible loops within the Pri2NTD (between helices α3-4, α4-5, an α6-7) that are positioned to interact with conserved surface-exposed charged residues on the first alpha helix (α1) of Mcm3 (Figures 2B, 2C, and S4B). 3D variability analysis49 shows that Pri2NTD adopts a continuum of rotational states with respect to Mcm3 while remaining engaged, likely due to the electrostatic nature of the interface (Figure S4C).
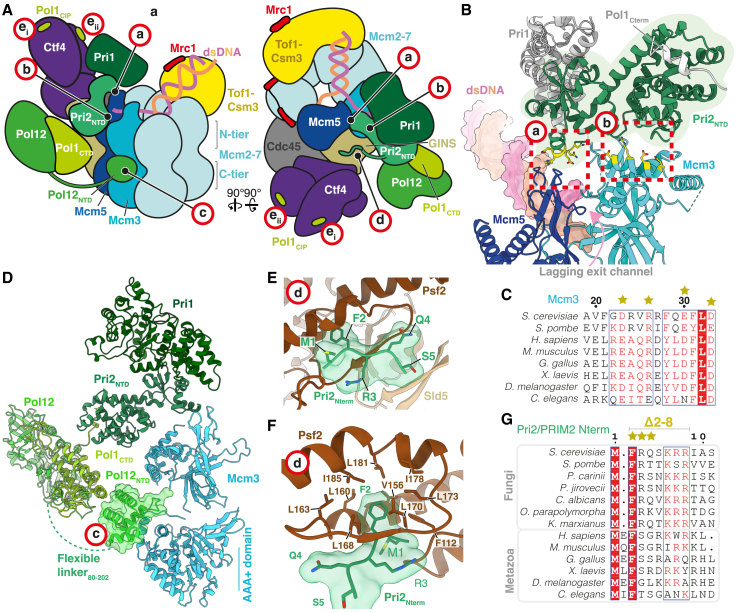
Figure 2.
The structural basis for Pol α-primase recruitment to the budding yeast replisome
(A) Schematic of the budding yeast replisome highlighting Pol α-primase-binding sites (red circles labeled a–e).
(B) Atomic model highlighting the interfaces between Pri2NTD (green) and the Mcm5 (blue) zinc finger (site a) and Mcm3 (cyan) N-terminal helical domain (site b). Residues colored yellow with side chains displayed represent those targeted for mutational analysis.
(C) Multiple sequence alignment indicating the conservation of Mcm3 residues contacting Pri2NTD (site b), colored according to conservation. Stars correspond to the Mcm3 residues colored yellow in (B) that were mutated.
(D) Atomic model highlighting the interface between the Pol12NTD (green) and the Mcm3 (cyan) AAA+ domain in the MCM C-tier (site c).
(E) Atomic model highlighting the interface between the Pri2Nterm (green) and the Psf2 subunit of GINS (brown) (site d).
(F) Atomic model showing how Pri2-F2 projects into a hydrophobic pocket on Psf2, colored as in (E).
(G) Multiple sequence alignment of Pri2Nterm residues contacting Psf2. The alignment is grouped into fungal and metazoan sequences and colored according to conservation. Stars indicate residues mutated to alanine in the Pri2-AAA mutant.
The remaining two interfaces between Pol α-primase and CMG (sites c and d) involve regions of the Pri2 and Pol12 subunits situated at the ends of regions of polypeptide predicted to be unstructured,50 indicating that they form flexible tethering points (Figures 1A, 1D, 2A, and 2D–2F). The 79 amino acid (aa) Pol12 NTD (Pol12NTD) adopts a compact helical fold connected to the phosphodiesterase (PDE) domain via a 130 aa unstructured linker region. Pol12NTD binds the Mcm3 AAA+ domain in the MCM C-tier (site c), where helices α8 and α18 of Mcm3 form an electrostatic cradle into which α4 of the Pol12NTD docks (Figures 2D and S4D). During ATP-dependent DNA translocation the MCM C-tier adopts multiple conformational states dependent on nucleotide occupancy and DNA engagement.30,44 While we observe only one C-tier conformation when Pol12NTD is bound to Mcm3, the Pol12NTD can be docked without clashes onto Mcm3 via the same interface through a range of C-tier configurations, indicating it might remain associated with Mcm3 throughout active replication (Figure S4E). Site d involves the N-terminal 5 amino acids of Pri2 (Pri2Nterm), where Pri2-F2—invariant in fungal species—docks into a surface-exposed hydrophobic pocket on the GINS subunit Psf2 (Figures 2E–2G). Pri2Nterm is connected to Pri2NTD via a 40 aa linker and, although this linker is predicted to be unstructured, at low map thresholds, continuous density is visible between the last modeled residue of Pri2Nterm (S5) and the first modeled residue of the Pri2NTD (S44), indicating that a section of the linker might adopt a structured conformation (Figure S4F).
In reconstructions containing Ctf4, local refinement revealed the presence of Pol α-primase-dependent density on the surface of the C-terminal α-helical domains of Ctf4 at the previously identified Pol1 binding site,29 into which the Pol1 Ctf4-interacting peptide (CIP box) (Pol1CIP) can be docked (PDB: 4C93) (Figures 2A, S4G, and S4H). Although we see no evidence for the presence of multiple copies of Pol α-primase in our dataset, density for Pol1CIP was observed on two Ctf4 monomers (the third monomer is bound to the Sld5 CIP box30,31), suggesting mixed occupancy within our reconstructions. Thus, Pol1CIP can bind to either Ctf4 monomer while Pol α-primase is associated with CMG (labeled sites ei and eii in Figure 2A), presumably because the CIP box is linked to Pol1exo via ∼200 aa of largely unstructured polypeptide.
Although we obtained 3D reconstructions where Pol α-primase was bound to CMG at all 4 sites, a substantial fraction of the dataset lacking Ctf4 displayed binding at just the Mcm5 ZnF and GINS interfaces (sites a and d), demonstrating that only a subset of binding sites are necessary to anchor Pol α-primase to CMG (Figures S4I–S4K). Inspection of the cryo-EM density in reconstructions where only sites a and d were engaged reveals that Pri2NTD and Pol1CTD-Pol12 are less well resolved, indicating that these regions are stabilized by the binding of Pri2NTD and Pol12NTD to Mcm3 (sites b and c, respectively). These data indicate that Pol α-primase can utilize only a subset of interaction sites for replisome association, which might be important to permit conformational changes during the priming cycle.18,46
Pol α-primase interaction mutants
To examine the contributions of the Pol α-primase:CMG interfaces during DNA replication, we purified Pol α-primase mutants and truncations designed to disrupt the Pri2:Mcm5 (Pri2-5A), Pol12:Mcm3 (Pol12-ΔN) and Pri2:GINS (Pri2-Δ2-8) interfaces (sites a, c, and d, respectively) and a Cdt1-Mcm2-7 charge reversal (CR) mutant (Mcm3-CR) designed to disrupt the Pri2NTD:Mcm3 binding site (site b) (Figures 3A and S5A). We also purified a Pol α-primase complex in which the Pol1 CIP box was mutated to abrogate its interaction with Ctf429 (Pol1-4A) (targeting sites ei and eii) (Figures 3A and S5A). Figure S5B shows that all Pol α-primase mutants displayed similar priming and DNA synthesis activities to the wild-type protein on ssDNA templates. Origin-dependent DNA replication reactions that generate leading- and lagging-strand products were reconstituted with purified budding yeast proteins on a 10.1 kbp linear DNA template with the origin positioned roughly at its center (Figure 3B).1,37,38 In reconstituted replication reactions in which the lagging-strand maturation machinery is omitted, the length distribution of lagging-strand products is dependent on Pol α-primase concentration, with less frequent priming resulting in the synthesis of longer lagging strands.37,39,51
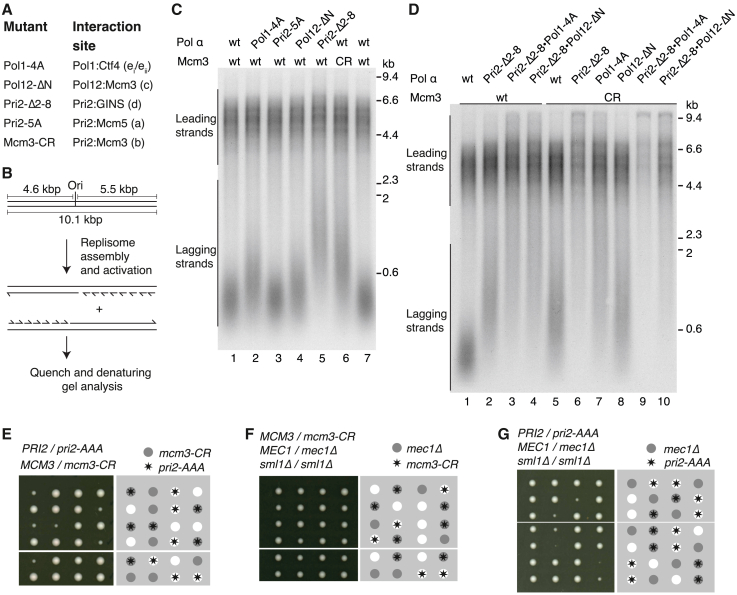
Figure 3.
Pol α-primase CMG binding sites are critical for DNA replication
(A) Summary of Pol α-primase and Cdt1-Mcm2-7 mutants and the interaction sites that are targeted. CR, charge reversal.
(B) Schematic of the DNA template and anticipated products for origin-dependent budding yeast in vitro DNA replication reactions.
(C and D) Denaturing agarose gel analysis of origin-dependent DNA replication reactions performed as illustrated in (B) for 20 min.
(E–G) Diploid budding yeast cells of the indicated genotype were sporulated and the resulting tetrads were dissected and grown on YPD medium for 3 days at 25°C. Dissections that displayed abnormal segregation patterns were cropped from plate images.
Replication with wild-type proteins produced a population of ∼4.5–6.5 kb leading strands and lagging strands of less than 0.6 kb (Figure 3C, lanes 1 and 7). Figure 3C shows that all mutant proteins with a single binding site targeted were competent for leading and lagging-strand DNA replication, demonstrating that no single interface is essential for Pol α-primase function. However, the distribution of lagging-strand products varied considerably among mutants, indicating that each interface does not contribute equally to productive primer synthesis. Surprisingly, mutations designed to target the Pri2:Mcm5 interface (Pri2-5A, site a) did not affect the length of lagging-strand products (Figure 3C, lane 3). Disruption of the Pol12:Mcm3 (Pol12-ΔN, site c) and Pol1:Ctf4 (Pol1-4A, sites ei/eii) binding sites resulted in slightly longer lagging-strand products than reactions containing wild-type Pol α-primase under these conditions (Figures 3C, lanes 1, 2, and 4 and S5C), indicating that these interfaces make relatively minor contributions to primer synthesis in the replisome. Strikingly, there was a marked lengthening of lagging-strand products when the Pri2:Mcm3 (Mcm3-CR, site b) and Pri2:GINS (Pri2-Δ2-8, site d) interfaces were perturbed (Figure 3C, lanes 5 and 6). Loss of the Pri2:GINS interaction had the most pronounced effect, with lagging-strand products displaying a broad length distribution of between 0.6 and 2 kb (Figure 3C, lane 5). These data indicate that the Pri2:GINS (site d) and Pri2:Mcm3 (site b) interfaces are the most important interaction sites for priming in the budding yeast replisome.
To gain further insight into the hierarchy of Pol α-primase:replisome interactions during DNA replication, we purified additional complexes harboring combinations of mutations/truncations (Figures S5A and S5B). Figures 3D, S5C, and S5D show that, in almost all cases, disrupting multiple Pol α-primase binding sites resulted in further increases in the length of lagging-strand products. Notably, lagging-strand synthesis was all but abolished in a reaction where both the Pri2:Mcm3 and Pri2:GINS interfaces (sites b and d) were disrupted, and there was a reduction in intensity and subtle lengthening of leading-strand products (Figure 3D, lane 6), which is indicative of delayed synthesis of the primers used to start leading-strand replication.1 Leading-strand replication was further compromised in reactions where the Pri2:Mcm3, Pri2:GINS, and Pol1:Ctf4 interfaces (sites b, d, and ei/eii) were targeted simultaneously (Figures 3D, lane 9 and S5D, lane 7). Similar defects were observed when the Pri2:GINS interface was disrupted together with both Pol12:Mcm3 and Pol1:Ctf4 (Figure S5D, lane 5) and when the four sites that individually contribute to DNA replication were simultaneously disrupted (Figure S5D, lane 10). In contrast, we observed robust leading-strand replication and some long lagging-strand products when the Pri2:Mcm3, Pol12:Mcm3, and Pol1:Ctf4 interfaces were simultaneously targeted (Figures S5C, lane 8 and S5D, lane 9), indicating that the interaction between Pri2Nterm and GINS is sufficient to support the necessary priming to start leading-strand replication. These data demonstrate that four distinct interfaces between Pol α-primase and the replisome contribute to nascent-strand priming and that collectively they are essential for efficient in vitro DNA replication. Importantly, the data also indicate that the contribution of each interface is not equal: disruption of the interface between Pri2Nterm and GINS (site d) is most deleterious for lagging-strand replication followed by the interface between Pri2NTD and Mcm3 (site b), whereas the interactions between Pol1 and Ctf4 (sites ei/eii) and Pol12NTD and Mcm3 (site c) make more minor contributions.
Pol α-primase mutants in vivo
Priming at replication forks is an essential function of Pol α-primase and therefore the key interactions we have identified should be critical for cell growth. To test this, we generated budding yeast strains with mutations targeting the Pri2:GINS (Pri2-AAA) and Pri2:Mcm3 (Mcm3-CR) interfaces (Figure 2A, sites b and d). In the Pri2-AAA allele, amino acids F2, R3, and Q4 are substituted to alanine. Figure S5E shows that Pol α-primase complexes containing Pri2-AAA and Pri2-Δ2-8 displayed almost indistinguishable behavior in in vitro replication assays. Colony growth of both pri2-AAA and mcm3-CR cells was comparable to control cells, indicating that priming was occurring at sufficient levels to permit relatively normal DNA replication (Figure S5F). We therefore combined the pri2-AAA and mcm-CR mutations. This resulted in a profound reduction in colony size relative to control cells, consistent with these cells having DNA replication defects (Figures 3E and S5G), which is concordant with the near absence of lagging-strand products in in vitro reactions when these interfaces are disrupted (Figures 3D, lane 6 and S5E, lanes 5 and 6).
Although the lack of obvious growth defects for pri2-AAA and mcm3-CR cells was somewhat surprising, previous work has shown that budding yeast are reasonably tolerant of reduced Pol α-primase levels.52 Moreover, colony growth of pol1-F1463A cells in which the interaction between primase and the Pol1 C terminus is disrupted, was comparable to control cells.45 However, pol1-F1463A is synthetic lethal with deletion of the gene encoding the apical checkpoint kinase Mec1, the ortholog of ATR, indicating that these cells do in fact have DNA replication defects.45 We therefore wondered if pri2-AAA and mcm3-CR might have subtle DNA replication defects that render cells dependent on checkpoint activation. Figure 3F shows that deletion of MEC1 in combination with mcm3-CR had minimal effect on colony growth. In contrast there was a notable reduction in colony size when mec1Δ was combined with pri2-AAA (Figure 3G), revealing that tethering of Pol α-primase to CMG via the Pri2:GINS interface is essential for unperturbed DNA replication in budding yeast.
Human Pol α-primase replisome structure
Because priming is fundamental for genome duplication, we considered it likely that key features of the mechanism targeting Pol α-primase to prime DNA synthesis were conserved. To examine this directly we determined the cryo-EM structure of a human replisome containing CMG, TIMELESS-TIPIN, AND-1, CLASPIN, Pol α-primase and a DNA replication fork with a 60 nt 5′ ssDNA flap (Figures S6A–S6D; Table 1). Similar to the yeast replisome, in addition to well resolved cryo-EM density for CMG, TIMELESS-TIPIN, and AND-1, poorly resolved density extended from the N-tier face of CMG atop MCM3 (Figures S6E–S6J). Following focused classification and refinement (Figure S6C) this density could be unambiguously assigned to Pol α-primase enabling assignment of PRIM1 (residues 9–349 and 386–408), the N terminus (residues 1–5) and NTD (residues 17–252) of PRIM2, the CTD of POLA1 (residues 1,279–1,445 and 1,448–1,458) and the majority POLA2 (residues 96–114 and 170–598) (Figures 4A, 4B, and S7A–S7C; Video S2).
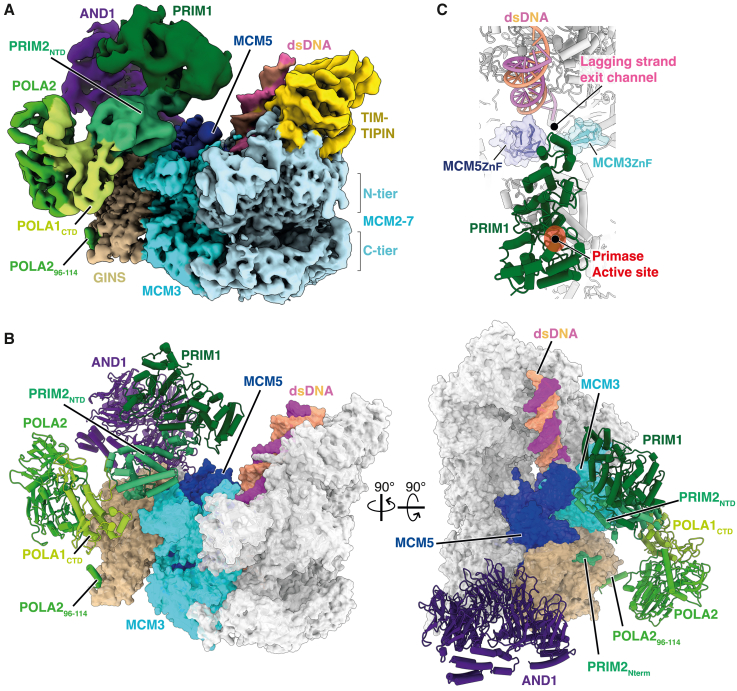
Figure 4.
Structure of Pol α-primase in a human replisome assembled on fork DNA with a 60-nt 5′ flap
(A) Composite cryo-EM map of the human replisome containing Pol α-primase, assembled on forked DNA containing a 60 nucleotide 5′ flap (Figure S1A). The map was derived from combining individual focused refinements and is colored according to chain occupancy.
(B) Atomic model for the human Pol α-primase associated replisome, derived from cryo-EM data displayed in (A). Regions of CMG that interact directly with Pol α-primase are colored.
(C) Focused view of PRIM1 showing its position at the mouth of the exit channel for lagging-strand ssDNA.
First, individual replisome components are sequentially colored and labeled to aid in their visualisation and the interpretation of the overall structure. The video then highlights each pol α-primase interface with the replisome (sites b, d, g, and e) using different views. Regions of the model forming an interaction interface are displayed using transparent surface rendering.
The conformation of human Pol α-primase and its positioning in the replisome are remarkably similar to yeast (Figures S7D and S7E), with the catalytic PRIM1 subunit again positioned above the mouth of the lagging-strand template exit channel (Figures 4A–4C). Similar to yeast, POLA1exo-cat is invisible when CMG is bound to replication fork DNA but is visualized stably engaging Pol α-primase in reconstructions lacking DNA, where it adopts the conformation observed for apo human Pol α-primase46 (Figures S7F and S7G). This suggested that the conformation of Pol α-primase in the human replisome might represent a DNA engaged state. To investigate this further, we repeated our human replisome sample preparation and cryo-EM analysis as before, but with a replication fork containing a 15 nt 5′ ssDNA flap that we reasoned would be too short to fully engage Pol α-primase (Figures S8A–S8H; Table 1). In the resulting 3D reconstructions, clear density is observed for the POLA1exo-cat domain, both when CMG is bound to the replication fork and when CMG is not engaging DNA (Figures 5, S8I, and S8J). In both situations, Pol α-primase adopts the same conformation as observed for the human apo structure,46 strongly suggesting that Pol α-primase is bound to lagging-strand template ssDNA when the human replisome is assembled on a replication fork with a 60 nt 5′ flap.
Figure 5.
Structure of Pol α-primase in a human replisome assembled on fork DNA with a 15-nt 5′ flap
(A) Cryo-EM reconstruction of the Pol α-primase associated human replisome engaged on a DNA fork containing a 15 nucleotide 5′ flap. Map colored according to subunit occupancy.
(B) Atomic model for the human Pol α-primase associated replisome, derived from cryo-EM data displayed in (A). Regions of CMG that interact directly with Pol α-primase are colored.
(C and D) Comparison of Pol α-primase from human replisomes bound to forked DNA with a 15 nt 5′ flap (C) and 60 nt 5′ flap (D) as illustrated.
Conservation of Pol α-primase tethering
Human Pol α-primase is tethered directly to CMG via three small interfaces that are all occupied independently of DNA engagement state (Figures 6A–6D and S9A–S9C; Video S2). PRIM2 binds to MCM3 (site b) and GINS (site d) in an analogous manner to Pri2 in the budding yeast replisome (Figures 6B–6D, S9D, and S9E), consistent with these binding sites being the most important for priming in yeast (Figures 3C and 3D). The PRIM2:MCM3 interface involves charged residues on α4 and the α3-4 linker of the PRIM2NTD that form electrostatic contacts with four conserved residues on α1 of MCM3 (Figures 2C, 6B, and S9D). Binding of PRIM2 to GINS is mediated by the N terminus of PRIM2, with amino acids M1 and F3—invariant in Metazoa—projecting into a surface-exposed hydrophobic pocket on PSF2 in a comparable manner to Pri2-F2 in yeast (Figure 6D). Continuous cryo-EM density links the last modeled residue of PRIM2Nterm (G5) and the first modeled residue of PRIM2NTD (Q17) indicating this interface spatially constrains the position of the PRIM2NTD and PRIM1 (Figure S9E). In addition to sites b and d, a flexibly tethered helix in POLA2 (residues 96–114) interacts with PSF1 and SLD5 (site g, Figures 6A, 6C, and S9F). Although this helix is predicted to be absent from Pol12, we note the presence of low-resolution Pol α-primase-dependent density on the surface of Psf1 in our budding yeast replisome maps, suggesting a similar binding site could be present in the yeast replisome (Figure S9G). We also note that binding of the POLA2 helix to PSF1 and SLD5 will localize the POLA2 NTD—that binds to the C-terminal AND-1 HMG-box32 (labeled site f in Figure 6A)—to this region of the replisome because the POLA2 helix and NTD are separated by a short (12 aa) linker. In contrast to the budding yeast replisome, we find no evidence of PRIM2NTD binding to the MCM5 ZnF, indicating it is not a conserved mode of interaction and perhaps explaining why the yeast Pri2-5A mutant did not display a lagging-strand replication defect (Figure 3C). Finally, consistent with reports that AND-1 binds the unstructured POLA1 N terminus (residues 151–171),32,33 we observe a small region of Pol α-primase dependent density on the C-terminal α-helical domain of each AND-1 monomer (labeled sites ei–iii, Figures S9H and S9I), indicating that Pol1 can access all available binding sites on the AND-1 trimer.
Figure 6.
Structural basis for Pol α-primase recruitment to the human replisome for priming
(A) Schematic of the human replisome engaged by Pol α-primase. Red circled labels indicate protein-protein interaction sites between Pol α-primase and the replisome.
(B) Atomic model highlighting the interface between PRIM2NTD (green) and the MCM3 (cyan) N-terminal helical domain (site b). Residues colored yellow with side chains displayed are those targeted for mutational analysis.
(C) Atomic model highlighting the interfaces between PRIM2Nterm and the PSF2 subunit of GINS (site d) and the POLA2 N-terminal helix (residues 96–114) and both PSF1 and SLD5 (site g).
(D) Zoomed in view of the PRIM2Nterm:PSF2 interface (site d).
(E) Table summarizing the protein-protein interfaces between Pol α-primase and the replisome in both budding yeast and human. Each discrete site is assigned a letter identifier corresponding to the labeling in (A) and Figure 2A.
(F) Schematic of the forked DNA template and anticipated products of in vitro DNA replication with purified human proteins.27
(G) Denaturing agarose gel analysis of an in vitro DNA replication reaction performed as in (A) with the indicated proteins for 20 min.
The conservation of the PRIM2:GINS and PRIM2:MCM3 interfaces (Figure 6E) suggested they would be important for priming in the human replisome. To test this directly we purified a Pol α-primase complex lacking the PRIM2 N terminus (PRIM2-Δ2-7) and a CMG complex where four conserved residues on helix α1 of MCM3 were mutated to alanine (MCM3-4A) (Figures S9J–S9L) and analyzed them in an in vitro human DNA replication system that we recently developed (Figure 6F).27 Here, replisomes assembled around purified CMG at model replication forks perform leading and lagging-strand DNA replication at rates comparable to those measured in cultured human cells. Figures 6G and S9M show that lagging-strand products distributed around ∼0.6 kb in length were synthesized with wild-type proteins. These products were substantially longer when the PRIM2:GINS interface (PRIM2-Δ2-7) was disrupted (Figure 6G, lanes 2 and 6). While disruption of the PRIM2:MCM3 (MCM3-4A) interface was less severe, there was still a notable increase in the length of lagging-strand products, which were longer compared with when the AND-1HMG:POLA2 interface was abolished (AND-1-ΔHMG) (Figures 6G and S9M). Disruption of multiple interfaces in the same reaction further compromised lagging-strand replication compared with single site mutants (Figure S9M). Collectively, these data indicate that key anchor points for attaching Pol α-primase to CMG to facilitate primer synthesis at replication forks are structurally and functionally conserved between yeast and human.
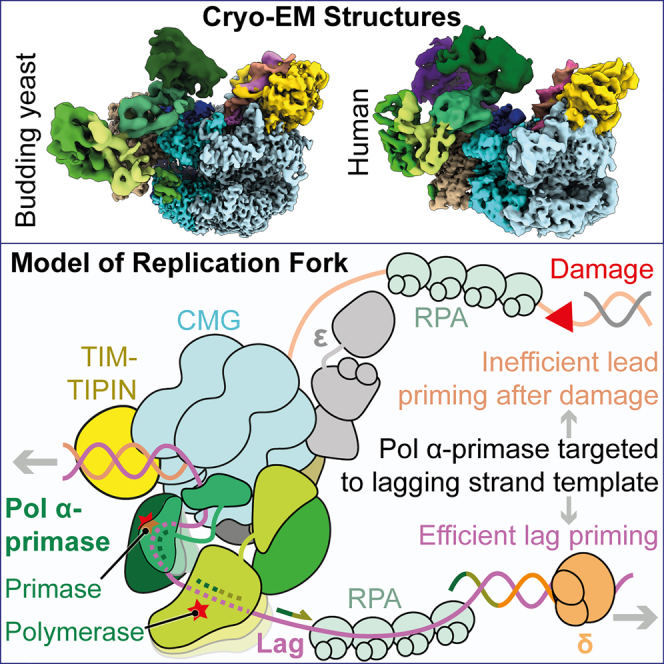
Discussion
By determining cryo-EM structures of budding yeast and human replisomes that are poised to initiate primer synthesis, we have elucidated a conserved mechanism for targeting Pol α-primase to replication forks for priming. The positioning of the catalytic Pri1/PRIM1 subunit at the mouth of the exit channel for lagging-strand template ssDNA explains how Pol α-primase functions so efficiently on this template strand and reveals a mechanism for primase to overcome competition with RPA for access to the DNA template. By contrast, the unwound leading-strand template exits CMG ∼150 Å away from Pri1/PRIM1 on the opposite side of the replisome, which is presumably not conducive for leading-strand priming, thus explaining why the core yeast replisome cannot efficiently restart leading-strand replication by repriming downstream of DNA damage7,8 or secondary structures10 and why lagging-strand primers are used to start leading-strand replication.1,3,4
Pol α-primase is targeted to the lagging-strand template for priming via a complex multisite interaction network involving several direct interactions with CMG. These interactions explain why Pol α-primase tethering by Ctf4/AND-1 is dispensable for DNA replication.27,34,36,37 The primary function of Ctf4/AND-1-dependent tethering of Pol α-primase is to facilitate the transfer of parental histones to the lagging strand.34,53 It will be interesting to discover why Ctf4/AND-1-dependent tethering is required for this activity and why the interactions between Pol α-primase and CMG that we have identified cannot fulfill this role. In both the yeast and human replisomes, the majority of Pol α-primase docking sites—including the crucial interaction between Pri2/PRIM2 and GINS—are mediated by regions of Pol α-primase situated at the end of, or within, unstructured linker regions, thereby providing flexible tethering points. Of the interactions that contribute to priming, only binding of the Pri2/PRIM2 NTD to MCM3 involves the association of two large rigid bodies. However, this interface is frequently disengaged and, due to its electrostatic nature, permits considerable motion between the two domains. This suggests that, although the positioning of Pol α-primase at the mouth of the lagging-strand template exit channel is crucial for priming, it is also important that primase is not rigidly fixed in this location. We hypothesize that flexible tethering of Pol α-primase in the replisome is required to allow other proteins access to key binding sites on CMG. For example, the E3 ubiquitin ligase that regulates replisome disassembly (Cul2LRR1 in human and SCFDia2 in budding yeast) binds across the lagging-strand template exit channel42 and this binding site is inaccessible when Pri2/PRIM2 is bound to MCM3. Flexible tethering may also function to enable Pol α-primase to remain associated with the replisome while it undergoes conformational changes during the primer synthesis reaction.
Our structures indicate that incorporation of Pol α-primase into the replisome does not induce conformational changes in CMG that are likely to modulate helicase activity. Consequently, concomitant primer synthesis and template unwinding will result in increasing lengths of ssDNA being formed between the primase/DNA polymerase active sites and the point of template unwinding, thereby generating what has been termed a “priming loop.”54,55 Currently, we do not know whether Pol α-primase remains fully engaged with CMG throughout the priming reaction, or whether the multiple docking sites are utilized dynamically. We consider it likely that Pol α-primase remains associated with CMG via at least one docking site for the entirety of the priming cycle given the prolonged replisome association kinetics that have been observed in single molecule experiments.35,56 The conformational dynamics of Pol α-primase and its utilization of docking sites during primer synthesis are interesting subjects for future investigation that we anticipate will also influence the disposition of priming loops in the replisome.
Considerable recent progress has been made in delineating the mechanisms of primer synthesis including the molecular basis for DNA primer initiation,57 how Pol α-primase activity is coordinated by the CST complex during telomeric C strand fill-in.47,58,59 Our work represents another important advance by revealing a conserved mechanism for targeting Pol α-primase to replisomes to prime eukaryotic DNA replication and also provides a platform to visualize additional key intermediates during this fundamental process.
Limitations of the study
Our structures of budding yeast and human Pol α-primase bound to the replisome likely only represent a small subset of conformations that Pol α-primase adopts during the priming cycle. Moreover, while our data strongly support the conclusion that Pol α-primase is engaging ssDNA in both the yeast and human replisomes, it is not possible to determine precisely which step of the priming cycle the structures represent. Although the structures provide important insights into how Pol α-primase is targeted to the replisome for priming, including identifying key protein:protein interaction sites, additional proteins that were not included in our replisome preparations might also modulate Pol α-primase activity at replication forks. For example, subunits of Pol α-primase have been reported to interact directly with Mcm10, RPA, and Pol δ. Therefore, an important future goal will be to determine structures of more complete replisomes performing lagging-strand replication to visualize intermediates along the primer synthesis pathway, the handoff of primers from Pol α-primase to Pol δ, and gain insights into how proteins such as RPA modulate the activity of Pol α-primase in the context of the replisome.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli 5-alpha Competent (High Efficiency) | New England Biolabs | Cat# C2987H |
| E. coli: Rosetta™ 2(DE3) strain: F-ompT hsdSB(rB- mB-) gal dcm (DE3) pRARE2 (CamR) | Novagen / Merck Millipore | Cat# 71400 |
| E. coli DH10 EMBacY | Geneva Biotech | https://geneva-biotech.com/product_category/insect-cell-expression/multibac/ |
| Chemicals, peptides, and recombinant proteins | ||
| 3X FLAG peptide | Sigma | Cat# F4799 |
| Adenosine 5’-(β,γ-imido)triphosphate lithium salt hydrate (AMP-PNP) | Sigma | Cat# A2647 |
| dNTP set | Invitrogen | Cat# 10297018 |
| NTP set | Invitrogen | Cat# R0481 |
| [alpha-P32]dCTP | Hatmann analytic | Cat# SRP-205 |
| Glutaraldehyde | Sigma | Cat# G5882 |
| Nonidet P-40 substitute (NP-40-S) | Roche | Cat# 11754599001 |
| Glutathione Sepharose 4B | GE Healthcare | Cat# 17-0756-01 |
| Suberic acid bis(3-sulfo-N-hydroxysuccinimide ester) sodium salt (BS3) | Sigma | Cat# S5799 |
| TWEEN® 20 | Sigma | Cat# P8341 |
| Biotin | Sigma | Cat# B4501 |
| cOmplete™, Mini, EDTA-free protease inhibitor cocktail | Sigma | Cat# 11873580001 |
| FuGENE® HD | Promega | Cat# E2311 |
| Insect-XPRESS protein-free insect cell media with L-glutamine | Lonza | Cat# BELN12-730Q |
| Sf-900™ II serum-free media | GIBCO | Cat# 10902-088 |
| SilverQuest™ staining kit | Invitrogen | Cat# LC6070 |
| Bovine Serum Albumin | Invitrogen | Cat# AM2616 |
| Phusion® High-Fidelity DNA Polymerase | New England Biolabs | Cat# E0553 |
| SphI | New England Biolabs | Cat# R0182 |
| SapI | New England Biolabs | Cat# R0569 |
| FspI | New England Biolabs | Cat# R0135 |
| BsaHI | New England Biolabs | Cat# R0556 |
| β-Glucuronidase | Sigma | Cat# G7017 |
| TEV protease | Nagai laboratory | N/A |
| Proteinase K | New England Biolabs | P8107 |
| Recombinant proteins | ||
| (see also Table S2) | ||
| Budding yeast (S. cerevisiae) | ||
| Cdt1-Mcm2-7 | Coster et al.60 | N/A |
| ORC | Frigola et al.61 | N/A |
| Cdc6 | Frigola et al.61 | N/A |
| DDK | On et al.62 | N/A |
| Sld3/7 | Yeeles et al.38 | N/A |
| Cdc45 | Yeeles et al.38 | N/A |
| Dpb11 | Yeeles et al.38 | N/A |
| Sld2 | Yeeles et al.38 | N/A |
| GINS | Yeeles et al.38 | N/A |
| Pol ε | Yeeles et al.38 | N/A |
| S-CDK | Yeeles et al.38 | N/A |
| Mcm10 | Yeeles et al.38 | N/A |
| Pol α | Yeeles et al.38 | N/A |
| Ctf4 | Yeeles et al.38 | N/A |
| RPA | Baretić et al.30 | N/A |
| Mrc1 | Baretić et al.30 | N/A |
| Tof1-Csm3 | Baretić et al.30 | N/A |
| RFC | Yeeles et al.37 | N/A |
| PCNA | Yeeles et al.37 | N/A |
| Pol δ | Yeeles et al.37 | N/A |
| CMG | Baretić et al.30 | N/A |
| Cdt1-Mcm2-7: Mcm3-CR | This study | N/A |
| Pol α-primase: Pol1-4A | This study | N/A |
| Pol α-primase: Pol12ΔN | This study | N/A |
| Pol α-primase: Pol1-4A⋅Pol12ΔN | This study | N/A |
| Pol α-primase: Pri2-Δ2-8 | This study | N/A |
| Pol α-primase: Pri2-5A | This study | N/A |
| Pol α-primase: Pri2-AAA | This study | N/A |
| Pol α-primase: Pol1-4A⋅Pri2-Δ2-8 | This study | N/A |
| Pol α-primase: Pol1-4A⋅Pol12-ΔN⋅Pri2-Δ2-8 | This study | N/A |
| Pol α-primase: Pol12-ΔN⋅Pri2-Δ2-8 | This study | N/A |
| Human (H. sapiens) | ||
| CMG | Jones et al.41 | N/A |
| CLASPIN | Jones et al.41 | N/A |
| TIMELESS-TIPIN | Jones et al.41 | N/A |
| AND-1 | Jones et al.41 | N/A |
| Pol ε | Jones et al.41 | N/A |
| Pol α-primase | Baris et al.27 | N/A |
| PCNA | Jones et al.41 | N/A |
| RPA | Jones et al.41 | N/A |
| Ctf18-RFC | Baris et al.27 | N/A |
| Pol δ | Baris et al.27 | N/A |
| Pol α-primase: PRIM2-Δ2-7 | This study | N/A |
| CMG: MCM3-4A | This study | N/A |
| AND1: ΔHMG (Δ1017) | Baris et al.27 | N/A |
| Deposited data | ||
| Budding yeast (S. cerevisiae) | ||
| Co-ordinate file for the Pol α-primase associated replisome in the absence of Ctf4 | This study | 8B9C |
| Co-ordinate file for the Pol α-primase associated replisome in the presence of Ctf4, CIP box site #1 | This study | 8B9A |
| Co-ordinate file for the Pol α-primase associated replisome in the presence of Ctf4, CIP box site #2 | This study | 8B9B |
| Pol α-primase associated replisome consensus refinement in the absence of Ctf4 (binned) | This study | EMD-16320 |
| Pol α-primase associated replisome consensus refinement in the absence of Ctf4 (un-binned) | This study | EMD-16322 |
| Tof1-Csm3 local refinement | This study | EMD-15304 |
| Mcm2-7 C-tier local refinement | This study | EMD-15305 |
| Pol12, Pol1CTD, Pri2NTD local refinement | This study | EMD-15306 |
| Pol12, Pol1CTD local refinement | This study | EMD-16885 |
| Pol α-primase associated replisome consensus refinement in the presence of Ctf4 (binned) | This study | EMD-15309 |
| Pol α-primase associated replisome consensus refinement in the presence of Ctf4 (un-binned) | This study | EMD-15902 |
| Ctf4 local refinement | This study | EMD-15310 |
| Pri1, Pri2CTD local refinement | This study | EMD-16247 |
| Pol α-primase associated replisome consensus refinement containing density for the Pol1exo/cat and Pri2CTD domains | This study | EMD-16248 |
| Pol α-primase associated replisome consensus refinement containing density for the lagging strand DNA template and the Pri1CTD (binned) | This study | EMD-15924 |
| Pol α-primase associated replisome consensus refinement containing density for the lagging strand DNA template and the Pri1CTD (un-binned) | This study | EMD-15303 |
| Pol α-primase associated replisome consensus refinement where only the Pri2:Mcm5ZnF interface is engaged | This study | EMD-16323 |
| Human (H. sapiens) | ||
| Co-ordinate file for the Pol α-primase associated replisome | This study | 8B9D |
| Pol α-primase associated replisome consensus refinement (un-binned) | This study | EMD-15341 |
| MCM2-7 C-tier local refinement | This study | EMD-15340 |
| AND-1 local refinement | This study | EMD-15342 |
| Pol α-primase associated replisome consensus refinement containing strong PRIM1 density (binned) | This study | EMD-15349 |
| PRIM1, POLA2, PolA1CTD, Pri2NTD local refinement | This study | EMD-15351 |
| TIMELESS-TIPIN local refinement | This study | EMD-15356 |
| Composite map assembled from EMD-15342:15341:15340:15349:15351:15356 | This study | EMD-15904 |
| Pol α-primase associated replisome consensus refinement, not engaged on DNA derived from dataset including a 15 nucleotide 5ʹ-flap DNA fork | This study | EMD-15918 |
| Pol α-primase associated replisome consensus refinement, not engaged on DNA derived from dataset including a 60 nucleotide 5ʹ-flap DNA fork | This study | EMD-15923 |
| Pol α-primase associated replisome consensus refinement, engaged on a 15 nucleotide 5ʹ-flap DNA fork | This study | EMD-15922 |
| Experimental Models: Cell Lines | ||
| Hi5 | Thermo Fisher | B85502 |
| Experimental models: Organisms/strains | ||
| S. cerevisiae strains (See also Table S3 for additional details of strains constructed as part of this study) | ||
| yAM33 (Cdt1-Mcm2-7 purification) | Coster et al.60 | N/A |
| ySDORC (ORC purification) | Frigola et al.61 | N/A |
| ySDK8 (DDK purification) | On et al.62 | N/A |
| yTD6 (Sld3/7 purification) | Yeeles et al.38 | N/A |
| yTD8 (Sld2 purification) | Yeeles et al.38 | N/A |
| yJY13 (Cdc45 purification) | Yeeles et al.38 | N/A |
| yJY26 (Dpb11 purification) | Yeeles et al.38 | N/A |
| yAJ2 (Pol epsilon purification) | Yeeles et al.38 | N/A |
| yAE88 (S-CDK purification) | Jake et al.63 | N/A |
| yAE95 (Pol alpha purification) | Jake et al.63 | N/A |
| yAE40 (Ctf4 purification) | Yeeles et al.38 | N/A |
| yJY106 (RPA purification) | Baretić et al.30 | N/A |
| yJY32 (Mrc1 purification) | Yeeles et al.37 | N/A |
| yAE48 (Tof1-Csm3 purification) | Yeeles et al.37 | N/A |
| yAE41 (RFC purification) | Yeeles et al.37 | N/A |
| yAE34 (Pol delta purification) | Yeeles et al.37 | N/A |
| yJY197 (CMG purification) | Jenkyn-Bedford et al.42 | N/A |
| yVA87 (Cdt1-Mcm2-7: Mcm3-CR) | This study | N/A |
| yVA96 (Pol α-primase: Pol1-4A) | This study | N/A |
| yJY239 (Pol α-primase: Pol12-ΔN) | This study | N/A |
| yJY232 (Pol α-primase: Pri2-5A) | This study | N/A |
| yJY241 (Pol α-primase: Pri2-Δ2-8) | This study | N/A |
| yJY242 (Pol α-primase: Pri2-AAA) | This study | N/A |
| yJY381 (Pol α-primase: Pol1-4A⋅Pol12ΔN) | This study | N/A |
| yMJ12 (Pol α-primase: Pol1-4A⋅Pri2-Δ2-8) | This study | N/A |
| yMJ13 (Pol α-primase: Pol1-4A⋅Pol12-ΔN⋅Pri2-Δ2-8) | This study | N/A |
| yMJ18 (Pol α-primase: Pol12-ΔN⋅Pri2-Δ2-8) | This study | N/A |
| yJY244 | This study | N/A |
| yJY297 | This study | N/A |
| yJY321 | This study | N/A |
| yJY300 | This study | N/A |
| yJY301 | This study | N/A |
| yJY365 | This study | N/A |
| yJY367 | This study | N/A |
| yJY345 | This study | N/A |
| yJY350 | This study | N/A |
| yJY351 | This study | N/A |
| yJY356 | This study | N/A |
| yJY357 | This study | N/A |
| yJY255 | This study | N/A |
| yJY302 | This study | N/A |
| yJY326 | This study | N/A |
| yJY328 | This study | N/A |
| yJY313 | This study | N/A |
| yJY315 | This study | N/A |
| yJY317 | This study | N/A |
| yJY352 | This study | N/A |
| yJY354 | This study | N/A |
| Oligonucleotides | ||
| Leading strand: 5ʹ-(Cy3)-TAGAGTAGGAAG TGAGGTAAGTGATTAGAGAATTGGAGAGT GTG(T)34T∗T∗T∗T∗T∗T – 3ʹ (∗ - phosphorothioate) |
Integrated DNA Technologies (IDT) | N/A |
| 15 Nucleotide 5ʹ-flap lagging strand: 5ʹ-GGCAGG CAGGCAGGCACACACTCTCCAATTCTCTAATCA CTTACCACACTTCCTACTCTA – 3ʹ |
Integrated DNA Technologies (IDT) | N/A |
| 60 nucleotide 5ʹ-flap lagging strand: (T)60ACACAC TCTCCAATTCTCTAATCACTTACCATCACTTCCT ACTCTA – 3ʹ |
Integrated DNA Technologies (IDT) | N/A |
| MT096: 5′phos/GCTATGTGGTAGGA AGTGAGAATTGGAGAGTGTGTTTTT TTTTTTTTTTTTTTTTTTTTTTTTTTTT TTTTTTTGAGGAAAGAATGTTGGTG AGGGTTGGGAAGTGGAAGGATGG GCTCGAGAGGTTTTTTTTTTTTTTTT TTTTTTTTTTTTTTTTTT |
Integrated DNA Technologies (IDT) | N/A |
| JY197: 5′- TTTTTTTTTTTTTTTTTTTTCACA CTCTCCAATTCTCACTTCCTACCACAT |
Integrated DNA Technologies (IDT) | N/A |
| JY195: 5′ - CCTCTCGAGCCCATC CTTCCACTTCCCAACCCTCACC |
Integrated DNA Technologies (IDT) | N/A |
| JY104: 5′ - GAATTGCGCTCTATGAAGTTGAC | Merck | N/A |
| JY105: 5′ - GAACTGCGGCTTGATAATGG | Merck | N/A |
| JY370: 5′ - GGACTAGGATGAGTAGCAGC | Merck | N/A |
| JY491: 5′ - GAGTCAGACAACCAGCAAGC | Merck | N/A |
| JY604: 5′ - GGTTGAAGAGCAGGCCAAGG | Merck | N/A |
| JY609: 5′ - TCAGGCCAAAGGTGATACGAC | Merck | N/A |
| VA212: 5′ - GACCTGTCGAATTCTCTCAA | Merck | N/A |
| Recombinant DNA | ||
| vVA20 | Aria and Yeeles1 | N/A |
| M13mp18 ssDNA | New England Biolabs | Cat# N4040S |
| ZN3 | Taylor and Yeeles7 | N/A |
| pJFDJ5 (yeast GINS purification) | Yeeles et al.38 | N/A |
| vJY19 (yeast PCNA purification) | Yeeles et al.37 | N/A |
| pAM3 (yeast Cdc6 purification) | Coster et al.60 | N/A |
| pET28a-Mcm10 (yeast Mcm10 purification) | Yeeles et al.38 | N/A |
| YB_X1 (human RPA purification) | This study | N/A |
| MT_EB1 (human PCNA purification) | Jones et al.41 | N/A |
| YB_2 (human CMG purification) | Jones et al.41 | N/A |
| YB_1 (human CMG purification) | Jones et al.41 | N/A |
| MT_01 (human CMG purification) | Jones et al.41 | N/A |
| MT_BF1 (AND-1 purification) | Jones et al.41 | N/A |
| MT_DB1 (CLASPIN purification) | Jones et al.41 | N/A |
| MT_DF1 (TIMELESS-TIPIN purification) | Jones et al.41 | N/A |
| MT_BD1 (TIMELESS-TIPIN purification) | Jones et al.41 | N/A |
| MT_BH1 (human RFC purification) | Jones et al.41 | N/A |
| MT_BJ1 (human RFC purification) | Jones et al.41 | N/A |
| MT_BK1 (human RFC purification) | Jones et al.41 | N/A |
| MT_BL1 (human RFC purification) | Jones et al.41 | N/A |
| MT_BI1 (human RFC purification) | Jones et al.41 | N/A |
| YB_7 (CTF18-RFC purification) | Baris et al.27 | N/A |
| YB_5 (CTF18-RFC purification) | Baris et al.27 | N/A |
| YB_6 (CTF18-RFC purification) | Baris et al.27 | N/A |
| YB_4 (CTF18-RFC purification) | Baris et al.27 | N/A |
| MT_CF1 (human Pol delta purification) | Baris et al.27 | N/A |
| MT_CH1 (human Pol delta purification) | Baris et al.27 | N/A |
| YB_3 (human Pol delta purification) | Baris et al.27 | N/A |
| MT_FC1 (human Pol delta purification) | Baris et al.27 | N/A |
| MT_BC3 (human Pol alpha-primase purification) | Baris et al.27 | N/A |
| MT_AE1 (human Pol alpha-primase purification) | Baris et al.27 | N/A |
| MT_AF1 (human Pol alpha-primase purification) | Baris et al.27 | N/A |
| MT_AG1 (human Pol alpha-primase purification) | Baris et al.27 | N/A |
| MT_U2 (human Pol epsilon purification) | Baris et al.27 | N/A |
| MT_L1 (human Pol epsilon purification) | Baris et al.27 | N/A |
| MT_M1 (human Pol epsilon purification) | Baris et al.27 | N/A |
| MT_N1 (human Pol epsilon purification) | Baris et al.27 | N/A |
| YB_8 (AND-1-ΔHMG purification) | Baris et al.27 | N/A |
| YB_X2 (MCM3-4A construction) | This study | N/A |
| YB_X3 (CMG: MCM3-4A purification) | This study | N/A |
| vVA62 (CMG: MCM3-4A purification) | This study | N/A |
| vMJ9 (human Pol alpha-primase: PRIM2-Δ2-7) | This study | N/A |
| vVA52 (Cdt1-Mcm2-7: Mcm3-CR strain construction) | This study | N/A |
| vVA58 (yeast Pol alpha-primase: Pol1-4A strain construction) | This study | N/A |
| vJY186 (yeast Pol alpha-primase: Pol12-ΔN) strain construction) | This study | N/A |
| vJY187 (yeast Pol alpha-primase: Pol1-4A⋅Pol12-ΔN) strain construction) | This study | N/A |
| vJY196 (yeast Pol alpha-primase: Pri2-Δ2-8) strain construction) | This study | N/A |
| vJY183 (yeast Pol alpha-primase: Pri2-5A) strain construction) | This study | N/A |
| vJY199 (yeast Pol alpha-primase: Pri2-AAA strain construction) | This study | N/A |
| vJY177 (construction of Mcm3-CR (Ura3) strain) | This study | N/A |
| vJY206 (construction of Pri2-AAA (Ura3) strain) | This study | N/A |
| Software and algorithms | ||
| Chimera (v1.13) | UCSF Resource for Biocomputing, Visualization, and Informatics | https://www.cgl.ucsf.edu/chimera/ |
| ChimeraX (v1.52) | UCSF Resource for Biocomputing, Visualization, and Informatics | https://www.cgl.ucsf.edu/chimerax/ |
| Coot (v1.0) | Paul Emsley (Medical Research Council Laboratory of Molecular Biology) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| EPU (v2.0) | ThermoFisher Scientific (FEI) | https://www.fei.com/software/epu-automated-single-particles-software-for-life-sciences |
| ESPript (v3.0.7) | Patrice Gouet (Lyon University); Xavier Robert (Centre national de la recherche scientifique) | http://espript.ibcp.fr/ESPript/ESPript/ |
| FIJI (v1.0) | National Institute of Health | https://imagej.net/Fiji/Downloads |
| Gautomatch (v0.53) | Kai Zhang (Medical Research Council Laboratory of Molecular Biology) | https://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/ |
| ImageJ (v1.50i) | National Institute of Health | https://imagej.nih.gov/ij/ |
| ISOLDE (v1.4) | Tristan Croll (Cambridge Institute for Medical Research) | https://isolde.cimr.cam.ac.uk/ |
| Phenix (v1.20-4459) | Cambridge University; Duke University; Lawrence Berkeley National Laboratory; Los Alamos National Laboratory | https://www.phenix-online.org/ |
| Photoshop 2020 | Adobe | https://www.adobe.com/uk/products/photoshop.html |
| Prism (v9.0.0) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| RELION (v2.1 & v3.1) | Sjors Scheres (Medical Research Council Laboratory of Molecular Biology) | https://www3.mrc-lmb.cam.ac.uk/relion/ |
| MUSCLE | European Molecular Biology Laboratory -European Bioinformatics Institute (EMBL-EBI) | https://www.ebi.ac.uk/Tools/msa/muscle/ |
| cryoSPARC (v3.2, v4.0 & v4.1) | Structura Biotechnology | https://cryosparc.com/updates |
| CTFFIND-4.1 | The Grigorieff Lab | https://grigoriefflab.umassmed.edu/ctffind4 |
| AlphaFold (v2.0) | DeepMind | https://www.deepmind.com/open-source/alphafold |
| AlphaFold-multimer (v2.0) | DeepMind | https://github.com/deepmind/alphafold |
| ColabFold (v1.5.2) | Ovchinnikov & Steinegger Labs | https://github.com/sokrypton/ColabFold |
| Epson Scan 3.9.3.0EN | Seiko Epson Corporation | https://www.epson.co.uk |
| Amersham Typhoon (1.1.0.7) | Cytiva | |
| Other | ||
| Amicon Ultra Centrifugal Filter Units | Millipore | Cat# UFC901096 |
| QUANTIFOIL Copper 400 mesh R2/2 holey carbon TEM grids | Electron Microscopy Sciences | Cat# Q450CR2 |
| HiTrap Blue HP | GE Healthcare | Cat# 17-0412-01 |
| HiTrap DEAE Fast Flow | GE Healthcare | Cat# 17-5055-01 |
| HiTrap Heparin HP | GE Healthcare | Cat# 17-0406-01 |
| HiTrap SP HP | GE Healthcare | Cat# 29-0513-24 |
| HiTrap SP FF | GE Healthcare | Cat# 29-0513-24 |
| IgG Sepharose Fast Flow | GE Healthcare | Cat# 17-0969-01 |
| StrepTactin Superflow high-capacity resin | IBA life sciences | Cat# 2-1208-002 |
| MonoQ PC 1.6/5 | GE Healthcare | Cat# 17-0671-01 |
| MonoS 5/50 GL | GE Healthcare | Cat# 17-5168-01 |
| Ni-NTA Agarose | QIAGEN | Cat# 30210 |
| Superdex 200 Increase 10/300 GL | GE Healthcare | Cat# 28-9909-44 |
| Superose™ 6 Increase 10/300 GL | GE Healthcare | Cat# 29-0915-96 |
| Sepharose 4B | Sigma | Cat# 4B200 |
| Microspin G-50 columns | GE Healthcare | Cat# GE27-5330-02 |
| Anti-FLAG M2 affinity gel | Sigma | Cat# A2220 |
| Bio-Gel HT (Hydrated) Hydroxyapatite | Bio-Rad | Cat# 130-0150 |
| Calmodulin-Sepharose 4B | GE Healthcare | Cat# 17-0529-01 |
| Criterion XT 4-12% Bis-Tris precast gels | BioRad | Cat# 3450124 |
| NuPAGE™ 4-12% Bis-Tris precast gels | Thermo Fisher | Cat# NPO323box |
| Whatman 3 MM paper | Cytivia | Cat# 11895375 |
| BAS-IP MS phosphor screen | Cytivia | Cat# 28956474 |
| Amersham Hyperfilm MP | Cytivia | Cat# 28906842 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Joseph Yeeles (jyeeles@mrc-lmb.cam.ac.uk).
Materials availability
Budding yeast strains and protein expression plasmids will be made available on request.
Experimental model and study participant details
S. cerevisiae strains constructed for genetic experiments were based on the W303 genetic background. Comprehensive information regarding the genotypes of these S. cerevisiae strains can be found in Table S4.
Method details
Protein expression and purification
Details of protein expression plasmids and strains made during this study can be found in Tables S1 and S2. An overview of the purification strategy for each protein is provided in Table S3. All wild type budding yeast proteins were expressed and purified as described previously.30,37,38,42 Cdt1-Mcm2-7 and Pol α-primase mutants / truncations were purified using the same procedure as for the wild type proteins. Human proteins were expressed and purified as described previously.27,41,42 Human Pol α-primase (Pri2-Δ2-7) was expressed and purified as described for the wild type protein.27 Human CMG (MCM3-4A) was expressed by coinfecting Hi5 cells at a density of 1 x 106 cells/ml with four viruses (generated as previously described42) expressing: MCM2, MCM5, MCM3-4A (vVA62); MCM7, MCM4, MCM6 (YB_X3); Cdc45 (MT_O1)41; PSF1, PSF2, PSF3, SLD5 (YB_1).41 Cells were grown for 72 hours before harvest by centrifugation. Protein purification was performed as described previously for the wild type protein.41
TIMELESS-TIPIN purification
Cells from a 1L culture were resuspended in lysis buffer (25 mM HEPES-KOH pH 7.2, 150 mM KCl, 5% glycerol, 0.5 mM TCEP, 0.01% NP-40-S) + protease inhibitors (cOmplete, EDTA-free, one tablet per 50 ml buffer) and lysed by dounce homogenization. Insoluble material was cleared by centrifugation (235,000 g, 4°C, 45 min) and 0.5 ml Strep-Tactin XT superflow high capacity resin was added to the lysate. Following a 30 min incubation at 4°C the resin was collected in a 20-ml column and was washed with 50 ml lysis buffer. The resin was resuspended in ∼ 2 ml lysis buffer and TEV protease was added to 100 ug/ml. The sample was incubated at 4°C overnight with gentle rotation. The sample was collected and applied to a 1 ml HiTrap Q HP column (GE Healthcare) equilibrated in 25 mM HEPES-KOH pH 7.2, 150 mM KCl, 5% glycerol, 0.5 mM TCEP, 0.01% NP-40-S. Proteins were eluted with a 20 column volume gradient from 150 to 1,000 mM KCl and peak fractions containing TIMELESS-TIPIN were pooled, concentrated to ∼ 500 μl in an Amicon Ultra-15 30 kDa MWCO concentrator and applied to a Superdex 200 Increase 10/300 gel filtration column (GE Healthcare) equilibrated in 25 mM Tris–HCl pH 7.2, 5% glycerol, 0.01% NP-40-S, 1 mM DTT, 150 mM NaCl. Peak fractions were pooled, frozen in liquid nitrogen and stored at −80°C.
Preparation of fork DNA for cryo-EM
To prepare forked DNA for cryo-EM sample preparation, leading and lagging strand oligonucleotides (Integrated DNA Technologies) were mixed at equimolar ratios in annealing buffer (25 mM HEPES-NaOH, pH 7.5, 150 mM NaOAc, 0.5 mM TCEP, 2 mM Mg(OAc)2) and gradually cooled from 80°C to room temperature. Leading strand oligo: 5′-(Cy3)-TAGAGTAGGAAGTGAGGTAAGTGATT
AGAGAATTGGAGAGTGTG(T)34 T∗T∗T∗T∗T∗T – 3′
∗ Denotes a phosphorothioate backbone linkage. 15 Nucleotide 5′-flap lagging strand oligo:
5′-GGCAGGCAGGCAGGCACACACTCTCCAATTCTCTAATCACTTACCACACTTCCTACT
CTA – 3′. 60 nucleotide 5′-flap lagging strand sequence:
5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAC
ACACTCTCCAATTCTCTAATCACTTACCATCACTTCCTACTCTA – 3′
Replisome assembly for cryo-EM
Reconstitution reactions were set up to yield a final volume of 300 μl, containing 150 nM CMG with a 1.5-fold molar excess of replisome proteins and fork DNA in reconstitution buffer (25 mM HEPES-NaOH pH 7.6, 150 mM NaOAc, 0.5 mM TCEP, 500 μM AMP-PNP, 10 mM Mg(OAc)2). Firstly, CMG was incubated with fork DNA for 10 min on ice in an 80 μl reaction. Next, the additional proteins were added in the following order: Ctf4/AND-1, Tof1-Csm3/TIMELESS-TIPIN, Mrc1/CLASPIN and Pol α-primase. The reaction volume was then adjusted to 300 μl using reconstitution buffer before being incubated for 20 min on ice. Following incubation, 132 μl of the reconstitution reaction was loaded separately onto two 10-30% glycerol gradients, each containing crosslinker. The remaining 36 μl of the reconstitution reaction was diluted in reconstitution buffer to 132 μl and this sample loaded onto a glycerol gradient prepared in the absence of crosslinker. Glycerol gradients were prepared as previously described30: Buffer A (40 mM HEPES-NaOH, pH 7.5, 150 mM NaOAc, 0.5 mM TCEP, 10% v/v glycerol, 0.5 mM AMP-PNP and 3 mM Mg(OAc)2) was layered on top an equal volume of Buffer B (Buffer A, except 30% v/v glycerol, 0.16% glutaraldehyde [Sigma] and 2mM bis(sulfosuccinimidyl)suberate (BS3, ThermoFisher Scientific)) in a 2.2 mL TLS-55 tube (Beranek Laborgerate) and gradients made using a gradient-making station (Biocomp Instruments, Ltd.) before cooling on ice. The sample was separated by centrifugation (200,000g, 4°C, 2 h) prior to manual fractionation. SDS-PAGE gel analysis was used to identify two peak fractions from each gradient containing crosslinker (total volume 368 μl) as previously described.41 These fractions were then buffer exchanged and concentrated prior to being immediately used for cryo-EM grid preparation as previously described.41
Cryo-EM data collection
Budding yeast replisome + Pol α-primase + 60 nucleotide 5′-flap DNA fork
A total of 12,819 raw movies were acquired using a 300 keV Titan Krios microscope (FEI) equipped with a K3 direct electron detector (Gatan) operated in electron counting mode using the EPU automated acquisition software (ThermoFisher) with “Faster Acquisition” mode (AFIS) enabled. A slit width of 20 eV was used for the BioQuantum energy filter. Data were collected in super-resolution mode bin 2 at an effective pixel size of 0.86 Å/pixel over a defocus range of -1.8 to -3.5 μm. Movies were dose-fractionated into 39 fractions over a 4 s exposure, resulting in a total dose of 39.2 e-/Å2.
Human replisome + Pol α-primase + 60 nucleotide 5′-flap DNA fork
A total of 7,355 raw movies were acquired using a 300 keV Titan Krios microscope (FEI) equipped with a K3 direct electron detector (Gatan) operated in electron counting mode using the EPU automated acquisition software (ThermoFisher) with “Faster Acquisition” mode (AFIS) enabled. A slit width of 20 eV was used for the BioQuantum energy filter. Data were collected at a pixel size of 0.86 Å/pixel using a defocus range of -2.1 to -3.5 μm. Movies were dose-fractionated into 39 fractions over a 4 s exposure resulting in a total dose of 47.4 e-/Å2.
Human replisome + Pol α-primase + 15 nucleotide 5′-flap DNA fork
A total of 6,718 raw movies were acquired using a 300 keV Titan Krios microscope (FEI) equipped with a Falcon III direct electron detector (Thermo) operated in linear mode using the EPU automated acquisition software (ThermoFisher). Data were collected at a pixel size of 1.07 Å/pixel using a defocus range of -0.9 to -3.5 μm. Movies were dose-fractionated into 39 fractions over a 1 s exposure resulting in a total dose of 88.5 e-/Å2.
Cryo-EM data processing
Budding yeast replisome + Pol α-primase + 60 nucleotide 5′-flap DNA fork
The data processing pipeline outlined here is schematised in Figure S2. Data were processed using either RELION-364,65,66,67 (henceforth referred to as RELION) or cryoSPARC-368 (henceforth referred to as cryoSPARC) unless otherwise stated. 12,819 39-fraction movies were aligned and dose-weighted (1.00513 e-/Å2/fraction, 5 x 5 patches, 150 Å2 B-factor) using RELION’s implementation of a MotionCor2-like program.69 CTF parameters were estimated using CTFFIND-4.170 and 112 poor-quality micrographs excluded from future processing. Particles were picked using RELION’s Laplacian-of-Gaussian (LoG) function providing a minimum diameter of 200 Å and maximum of 350 Å. 2,003,322 picked particles were extracted using a box size of 430 Å. During extraction the data were down-sampled to a pixel size of 3.44 Å/pixel. One round of RELION 2D classification was carried out and 1,623,209 particles were selected for further classification. Four successive rounds of RELION 3D classification (regularisation parameter, T = 4), each generating 6 classes, were carried out using a previously obtained map of the budding yeast replisome as a reference (EMD-10227).30 Class selection was based upon the presence of secondary structure features within CMG. The first two round of 3D classification were performed with a 250 Å diameter circular mask to focus classification on the CMG, with the subsequent two rounds using a more dilated mask of 380 Å to incorporate signal from Pol α-primase.
202,655 particles containing poor density for dsDNA were selected for an additional two rounds of 3D classification in RELION. This resulted in the selection of 44,871 particles in classes displaying density for both Pol α-primase and CMG in the absence of dsDNA. These particles were refined using 3D auto-refinement in RELION generating a reconstruction, after post-processing, at 7.4 Å resolution. In order to enrich for replisomes stably bound by Pol α-primase in the absence of dsDNA, signal subtraction was carried out in RELION focussing on the interface between Pol α-primase and Mcm3. The refined reconstruction was low-pass filtered to 10 Å in UCSF Chimera71 and a soft mask was generated covering density for both Pol α-primase and the Mcm3 N-terminal helical domain. Subtracted particles were re-centred on the mask and sub-classified using 3D classification without alignment in RELION. 18,412 particles in classes containing strong density for both Pol α-primase and the Mcm3 N-terminal helical domain were selected and reverted to the original (non-subtracted) particles prior to refinement using 3D auto-refine in RELION. This generated a reconstruction, following postprocessing, in RELION at 6.8 Å resolution (Figure S3J).
Returning to the results of the fourth overall round of 3D classification, 884,301 particles were selected for further processing to enrich for replisome complexes bound by Pol α-primase engaged on DNA. These particles were re-extracted using an un-binned pixel size of 0.86 Å in a 450 Å box and submitted for refinement using RELION 3D auto-refine, yielding a reconstruction at 3.5 Å following postprocessing. These data were submitted for two rounds of iterative per-particle motion correction using dataset-trained particle polishing in RELION72 and RELION CTF-refinement64 (beamtilt and trefoil correction, anisotropic magnification correction, and per-particle defocus and astigmatism CTF correction). These data were refined to an improved resolution of 3.0 Å following postprocessing in RELION. At this stage of processing the density for CMG, Ctf4, Tof1-Csm3 and DNA was of high quality yet the Pol α-primase density was disordered and fragmented with only the Mcm5ZnF contact preserved at appropriate map thresholds.
In order to identify classes in which Pol α-primase was stably engaged with the replisome, the strategy previously described to enrich for Pol α-primase stably engaged on replisomes in the absence of DNA was employed. Signal subtraction was carried out using a mask encompassing both Pol α-primase and the Mcm3 N-terminal helical domain. These subtracted particles were than sub-classified using 3D classification without alignment, resulting in 614,228 particles in classes with improved Pol α-primase density. These data were reverted to the original non-subtracted particles and refined using RELION 3D auto-refine. The subtraction and subclassification process was then iterated to select for 3D classes representing 588,597 particles with improved Pol α-primase density. These particles were imported into cryoSPARC and subsequently down sampled to a pixel size of 1.72 Å/pixel to boost the signal-to-noise for spatial frequencies describing secondary structure elements. These data were then classified in 3D via heterogeneous refinement using five different replisome reference maps (composition indicated in Figure S2). Classes in which Pol α-primase was only engaged at both the Mcm5ZnF and Psf2 sites were selected representing 434,311 particles. These particles were refined using non-uniform refinement,73 with a pixel size of 0.86 Å, to 3.0 Å resolution (Figure S1N). To aid interpretation, all non-uniform and local refinements completed in cryoSPARC were subsequently locally filtered using their respective local resolution maps.
Returning to the previous heterogeneous refinement, classes with improved Pol α-primase density representing 140,426 particles were selected and further classified via heterogeneous refinement with six identical 3D references, the results of which are used as the input for processing strategies 1-4:
Strategy 1: Classes representing 84,628 particles were selected based on the presence of both strong Ctf4 and Pol α-primase density. A soft mask was generated in UCSF Chimera71 covering Pol α-primase and masked 3D classification without alignment carried out using five identical 3D references in cryoSPARC (target resolution 8 Å, initialisation mode PCA). Masks generated for use in cryoSPARC were binarised using the vop_threshold command in UCSF Chimera71 and a soft padding width applied using the Volume Tools in cryoSPARC (mask softness=5∗resolution(Å)/pixel-size(Å)). Classes representing 54,970 particles were selected based upon Pol α-primase Pri2NTD engagement with the Mcm3 N-terminal helical domain. A consensus refinement for these unmasked particles was carried out using non-uniform refinement in cryoSPARC, with an binned pixel size, resulting in a reconstruction at 4.6 Å resolution in which the MCM C-tier adopts conformation II. This process was iterated to generate a reconstruction, with an un-binned pixel size of 0.86, at 3.5 Å resolution (Figure S1M). In this reconstruction Pol α-primase engages the replisome via contacts with Mcm5ZnF, Psf2, Mcm3 N-terminal helical domain, Mcm3 AAA+ domain and Ctf4. Reconstructions in which these sites are all engaged (+/- Ctf4) we define as the “fully engaged” complex. Soft masks were generated covering both the visible regions of Ctf4/Cdc45/GINS in the resulting map, and the remainder of the density. These maps were used to subtract the non Ctf4/Cdc45/GINS density from the particle images and subsequently carry out masked local refinement of the Ctf4/Cdc45/GINS region in cryoSPARC. Local refinement was carried out using the default parameters and a fulcrum point defined by the mask centre, resulting in a reconstruction at 3.3 Å resolution for Ctf4/Cdc45/GINS (Figure S1H).
Strategy 2: Classes representing 73,884 particles were selected following heterogeneous refinement displaying strong Pol α-primase density in the absence of Ctf4 with the Mcm2-7 C-tier in conformation II. Masked 3D classification without alignment using ten 3D references was carried out in cryoSPARC, as described in strategy 1, to enrich for classes where the Pol α-primase Pri2NTD is engaged with the Mcm3 N-terminal helical domain. Classes representing 53,964 particles were selected for two non-uniform refinement jobs in cryoSPARC using either a pixel size of 0.86 Å or 2.27 Å, resulting reconstructions at both 3.5 Å and 4.6 Å resolution respectively for the fully engaged complex in the absence of Ctf4 (Figure S1E). Signal subtraction and masked local refinement was carried out in cryoSPARC using a pixel size of 0.86 Å, as described in Strategy 1, focussing on both the Pol12/Pol1CTD (Figure S1I) and Pol12/Pol1CTD/Pri2NTD (Figure S1J) regions of the complex, yielding reconstructions at 4.8 Å and 5.0 Å resolution respectively. In order to improve the noisy density adjacent to Pri1, a soft mask covering this region and Pri1 was generated in UCSF Chimera71 based upon a low-pass filtered map derived from the consensus non-uniform refinement at 3.5 Å resolution. Masked 3D classification of this region (10 classes, target resolution 12 Å) resulted in the selection of 39,555 particles that were unmasked and refined using non-uniform refinement, resulting in a reconstruction at 4.6 Å resolution at a pixel size of 2.27 Å (Figure S1K). The improved resolution in this region resulted in its assignment as the Pri2CTD.
Strategy 3: Classes representing 100,178 particles with strong density for both the Mcm2-7 C-tier in conformation II and Pol α-primase regardless of Ctf4 occupancy. These data were refined using non-uniform refinement in cryoSPARC, using a pixel size of 0.86 Å, to a resolution of 3.5 Å. Signal subtraction and masked local refinement was carried out in cryoSPARC using a pixel size of 0.86 Å, as described in Strategy 1, focussing on the Mcm2-7 C-tier yielding a reconstruction at 3.2 Å resolution (Figure S1F).
Strategy 4: Classes representing 84,350 particles with strong density for Tof1-Csm3/dsDNA and Pol α-primase regardless of Ctf4 occupancy. These data were refined using non-uniform refinement in cryoSPARC, using a pixel size of 0.86 Å, to a resolution of 3.4 Å. Signal subtraction and masked local refinement was carried out in cryoSPARC using a pixel size of 0.86 Å, as described in Strategy 1, focussing on Tof1-Csm3/DNA yielding a reconstruction at 3.9 Å resolution (Figure S1G).
In order to enrich for particles in which the Pri2CTD was well resolved, the 588,597 particle subset initially imported into cryoSPARC following processing in RELION was re-processed using a 3D reference obtained from refinement of the 39,555 particle subset containing density for the Pri2CTD. Four rounds of iterative heterogeneous refinement were carried out using four 3D references containing Pri2CTD domain density resulting in an 87,540 particle subset. The non-Pol α-primase density was subtracted from these particles and 3D classification without alignment was carried out within a mask encompassing all of the visible regions of Pol α-primase. Classes were selected based on clear Pri2CTD domain density resulting in a 45,111 particle subset. The corresponding un-subtracted particles were then refined using non-uniform refinement to a resolution of 4.6 Å prior to being further classified using 3D variability analysis49 using 3 modes and a filter resolution of 9 Å. The results were displayed using clustering analysis using 6 clusters and a filter resolution of 9 Å. Inspection of the resulting reconstructions revealed the presence of additional density corresponding to the Pol1exo/cat domain. An additional round of 3D variability analysis was carried out using the same parameters with a new mask additionally encompassing the Pol1exo/cat domain density. This procedure identified a subset of 9,633 particles which were then refined to a resolution of 4.6 Å that displayed density for both the Pri2CTD and the Pol1exo/cat domains (Figures S3G and S3H).
Human replisome + Pol α-primase + 60 nucleotide 5′-flap DNA fork
The data processing pipeline is illustrated by the schematic in Figure S6C. 7,355 39-fraction movies were aligned and dose-weighted (1.22 e-/Å2/fraction, 5 x 5 patches, 150 Å2 B-factor) using RELION’s implementation of a MotionCor2-like program.69 CTF parameters were estimated using CTFFIND-4.170 and 44 poor-quality micrographs excluded from future processing. Particles were picked using RELION’s Laplacian-of-Gaussian (LoG) function providing a minimum diameter of 180 Å and maximum of 330 Å. 1,535,548 particles were extracted using a box size of 380 Å. During extraction the data were down-sampled to a pixel size of 3.78 Å/pixel. Two successive rounds of RELION 3D classification (regularisation parameter, T = 4), using 6 classes, were carried out using a previously obtained map of the human core replisome as a 3D reference (EMD-13375).41 Class selection was based upon the presence of secondary structure features within CMG. 3D classification was performed with a 250 Å diameter circular mask to focus classification on CMG. A resulting 550,340 particles were subsequently refined using 3D auto-refinement in RELION generating a reconstruction at 7.6 Å resolution following postprocessing. These data were submitted for per-particle motion correction using dataset-trained particle polishing in RELION72 using a pixel size 0.86 Å in a 450 Å box. RELION CTF-refinement64 (beamtilt and trefoil correction, anisotropic magnification correction, and per-particle defocus and astigmatism CTF correction) was then carried out and the data refined to an improved resolution of 3.6 Å following postprocessing in RELION. A further round of 3D classification was carried out with a dilated circular mask of 380 Å and classes with significant Pol α-primase density representing 359,677 particles selected for subclassification. Signal subtraction and masked 3D classification without alignment was carried out in RELION as described in the budding yeast data processing methods to enrich for replisomes stably associated with Pol α-primase using a mask covering both Pol α-primase and the MCM3 N-terminal helical domain.
3D classes in which Pol α-primase adopted the previously reported autoinhibited primosome conformation (PDB:5EXR)46 were selected, representing 74,940 particles. Following reversion to the original non-subtracted particles.star file these data were imported into cryoSPARC and refined using non-uniform refinement73 to 3.6 Å resolution (Figure S7G). Density for DNA was not observed within this reconstruction.
Returning to the previous masked 3D classification without alignment in RELION, classes comprising 174,696 particles were selected in which Pol α-primase adopted a conformation distinct from that of the primosome. Following reversion to the original non-subtracted particles.star file these data were refined via 3D auto-refinement in RELION and postprocessed to a resolution of 4.1 Å. These data were then submitted to an additional round of particle polishing and CTF-refinement in RELION using the same parameters as the previous round. Particles were imported into cryoSPARC and a consensus refinement carried out using non-uniform refinement generating a reconstruction at a resolution of 3.4 Å (Figure S6E). Local refinements were carried out in cryoSPARC, as described in the budding yeast data processing methods, for regions encompassing both TIMELESS-TIPIN/DNA (Figure S6F) and the MCM2-7 C-tier (Figure S6H) resulting in reconstructions at 4.1 Å and 3.7 Å respectively.
In order to improve the quality of the AND-1 density, particle subtraction followed by masked 3D classification without alignment was carried out as described in the budding yeast data processing methods. 3D classification was carried out on the total dataset imported into cryoSPARC, in a conformation distinct from the primosome, focussing on the AND-1/CDC45/GINS region of the map. 3D classes were selected, consisting of 63,393 particles, based on the presence of continuous strong AND-1 density. These data were then locally refined to generate a reconstruction at 3.3 Å resolution (Figure S6G).
In order to improve the quality of the Pol α-primase density, particle subtraction followed by masked 3D classification without alignment was carried out using a mask covering the POLA1CTD/POLA2/PRIM1/PRIM2NTD region of the map. 3D classes were selected, consisting of 148,103 particles which were locally refined to generate a reconstruction at 4.4 Å resolution (Figure S6I). A single 3D class was selected from this procedure, containing 23,758 particles, with particularly strong PRIM1 density. The non-subtracted particles comprising this class were subjected to consensus non-uniform refinement generating a reconstruction at 4.3 Å resolution.
Human replisome + Pol α-primase + 15 nucleotide 5′-flap DNA fork
The data processing pipeline is illustrated by the schematic in Figure S8D. 6,718 39-fraction movies were aligned and dose-weighted (2.27 e-/Å2/fraction, 5 x 5 patches, 150 Å2 B-factor) using RELION’s implementation of a MotionCor2-like program.69 CTF parameters were estimated using CTFFIND-4.170 and 109 poor-quality micrographs excluded from future processing. Particles were picked using Gautomatch v0.56 (https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/#gauto) leading to extraction of 724,557 particles using a box size of 380 Å and pixel size of 4.28 Å/pixel (raw pixel size 1.07 Å/pixel). Two successive rounds of RELION 3D classification (regularisation parameter, T = 4), using 6 classes, were carried out using a previously obtained map of the human core replisome as a 3D reference (EMD-13375).41 Class selection was based upon the presence of secondary structure features within CMG. 3D classification was performed with a 250 Å diameter circular mask to focus classification on CMG. A resulting 584,362 particles were subsequently refined using 3D auto-refinement in RELION generating a reconstruction at 8.8 Å resolution. These data were submitted for per-particle motion correction using dataset-trained particle polishing in RELION72 using a pixel size 1.02 Å in a 450 Å box. RELION CTF-refinement64 (beamtilt and trefoil correction, anisotropic magnification correction, and per-particle defocus and astigmatism CTF correction) was then carried out and the data refined to an improved resolution of 3.4 Å following postprocessing in RELION. A further round of 3D classification was carried out with a dilated circular mask of 380 Å. 28,202 particles in 3D classes lacking DNA were imported into cryoSPARC and refined using non-uniform refinement to 3.6 Å resolution. 492,011 particles in 3D classes with significant Pol α-primase and DNA density were selected for further subclassification. Signal subtraction and masked 3D classification without alignment was carried out in RELION as described in the budding yeast data processing methods to enrich for replisomes stably associated with Pol α-primase using a mask covering both Pol α-primase and the MCM3 N-terminal helical domain. The signal subtraction followed by 3D classification without alignment procedure was iterated resulting in the selection of 258,339 particles in 3D classes with strong Pol α-primase density. These data were imported into cryoSPARC and refined using non-uniform refinement to 3.3 Å resolution.
Cryo-EM model building
Budding yeast replisome + Pol α-primase + 60 nucleotide 5′-flap DNA fork
To begin model building, structures of the budding yeast core replisome (PDB:6SKL)30 with the MCM C-tier removed and the MCM C-tier in conformation II (PDB:6SKO),30 were rigid body docked into the cryo-EM map of the budding yeast replisome fully engaged by Pol α-primase in the absence of Ctf4 at 4.6 Å resolution (binned pixel size of 2.27 Å) using ChimeraX.74 The atomic model for Ctf4 contained within 6SKO was manually removed at this stage. The structure of the human primosome (PDB:5EXR)46 was then rigid body docked into the region of the cryo-EM map that remained unassigned, guided by the presence of secondary structure features within density for the Pri2NTD. Inspection of the fit-to-density following 5EXR docking revealed a lack of strong cryo-EM density corresponding to both the POLA1 exonuclease and catalytic domains and the PRIM2CTD, therefore these were subsequently removed from the model. The quality of the fit-to-density for the remaining human Pol α-primase model was then improved via manual manipulation followed by automated docking for both the PRIM1 and PRIM2NTD domains and the POLA1CTD/POLA2 module respectively. The positions of these human Pol α-primase subunits provided a reference to which the AlphaFold50 models for budding yeast Pri2NTD (residues S44-T299), PRIM1 (residues S12-D402) and the crystal structure the Pol1CTD/Pol12 dimer (PDB:3FLO) were aligned prior to being rigid body fit into the density. An AlphaFold multimer75 model for the Pol12 (residues 203-705) / Pol1CTD (residues 1260-1468) complex was aligned to the Pol12 subunit of 3FLO, rigid-body fit into the cryo-EM density and the most N-terminal residue of Pol1 trimmed to I1271. The Pol1 C-terminus was remodelled based on an AlphaFold-Multimer75 result indicating complex formation between the Pol1 C-terminus and Pri2NTD, in an analogous fashion to the POLA1C-term in the primosome structure 5EXR.
The quality of the fit for the model into the fully engaged cryo-EM map lacking Ctf4 at 4.6 Å resolution was optimised via an all-atom simulation in ISOLDE,27 using adaptive distance restraints for the dsDNA model (kappa=100). The resulting model was further refined using Phenix76 real-space-refine, utilising the input model as a reference to generate restraints with sigma=0.1 and global minimisation with nonbonded_weight=2000 and weight=0.5. Regions of the model that fit poorly to the density were the manually refined in Coot77 using the local refinement and regularisation tools incorporating stereochemical restraints. The model for the Mcm2-ZnF (residues 338–378) was truncated due to the absence of well resolved density in this region of the map.
At this stage in the modelling process, regions of cryo-EM density that remained unmodelled were identified for further analysis to determine their identity. Density for a small four-helical bundle bound to the Mcm3 AAA+ domain was assigned as the flexibly tethered Pol12NTD (residues M1-I79). An AlphaFold-Multimer prediction for the Pol12NTD interacting with the AAA+ domain of Mcm3 was used to dock the Pol12NTD into the cryo-EM density.
Two regions of disconnected helical density were identified in analogous positions to CLASPIN in the core human replisome (PDB:7PFO).41 This led us to speculate that these represented regions of the budding yeast homologue of CLASPIN, Mrc1. Furthermore, AlphaFold-Multimer modelling predicted multiple high-confidence interactions between Mrc1 and the replisome. Predictions were validated by the presence of corresponding side-chain density for Mrc1, present in the un-binned cryo-EM reconstruction of the fully engaged Pol α-primase complex at 3.5 Å resolution. Mrc1 residues S339-K323 interact with the α-solenoid of Tof1, whilst D468-Q483 contacts Mcm2 in the N-tier and the Tof1 N-terminus. An additional region of disconnected density, on the same side of the replisome to the two modelled regions of Mrc1, was also assigned to Mrc1 based on AlphaFold-Multimer prediction and the presence of clear side-chain density. Alphafold-Multimer predicts an interaction between Mrc1 residues L815-E858 spanning both Cdc45 and the Mcm2 N-tier, for which there is cryo-EM density present in the fully engaged reconstruction at 4.6 Å resolution. However, there is only cryo-EM density of sufficient resolution to enable unambiguous assignment, in the 3.5 Å resolution fully engaged map, for Mrc1 residues N842-E858, therefore these are the only residues deposited in the final model for this particular Mrc1 interface.
A region of unmodelled density bound to the GINS subunit Psf2 was ascribed to Pri2Nterm (residues M1-S5). Assignment was based upon the close proximity of the otherwise most N-terminal modelled residue of Pri2 (S44) and the presence of continuous density between S5-S44 present at low map thresholds. Clear side chain density was present for residues M1-Q4. As the second residue in Pri2 is a large phenylalanine residue the first methionine will not be removed78 and is likely to be acetylated.79
At this stage the model was inspected residue-by-residue in Coot,77 docked into the highest resolution cryo-EM map (consensus and focused refinements) for the corresponding region of the model. Focussed refinements were rigid-body docked into the consensus and resampled onto the same origin. The model was manually refined against the map in Coot77 and both backbone Ramachandran and rotamer outliers corrected. A final global run in ISOLDE80 was carried out to minimise the clash-score using distance and torsion restraints prior to Phenix76 real-space-refinement using the same restraints as described above. Model validation was carried out using the Molprobity server,81 Phenix76 validation and the wwPDB OneDep validation server.
In order to model the budding yeast replisome fully engaged by Pol α-primase in the presence of Ctf4, the structure of trimeric Ctf4 (PDB:6SKL)30 was rigid body docked into the corresponding density. The fit-to-density was subsequently refined via local ISOLDE80 simulation followed by Phenix76 real-space-refinement and manual adjustment in Coot.77 Inspection of the locally refined Ctf4/GINS/Cdc45 cryo-EM map revealed the presence of unmodelled density bound to the helical bundles of two Ctf4 monomers in the correct position to accommodate the Pol1 CIP-box (a.a. F140-S149).29 The Pol1 CIP box was subsequently modelled into each discrete density based on the crystal structure of the Ctf4CTD-Pol1 CIP-box (PDB: 4C93).29 It was not possible to sub-classify the Ctf4 density to generate reconstructions with only one-site occupied at any time, therefore two models were deposited to the PDB with the Pol1 CIP-box interacting with a different monomer of Ctf4 in each.
Human replisome + Pol α-primase + 60 nucleotide 5′-flap DNA fork
A previously determined structure of the core human replisome (PDB: 7PFO)41 was rigid body docked into the locally filtered cryo-EM map of the human replisome fully engaged by Pol α-primase on a DNA fork containing a 60 nucleotide 5ʹ-flap at 3.4 Å resolution using ChimeraX.74 The atomic model for Pol ε contained within 7PFO41 was manually removed at this stage, as were MCM2 residues 324-368 which comprise its zinc-finger motif due to the lack of corresponding density. Using the same strategy employed for the modelling of the budding yeast Pol α-primase-replisome structure, a previously determined model for the human primosome (PDB:5EXR) was rigid body docked into the remaining unassigned density. The 5EXR model was then manually edited to remove both the POLA1 exonuclease and catalytic domains and the PRIM2CTD due to a lack of corresponding cryo-EM density. The fit-to-density for the Pol α-primase subunits was improved by docking each module: PRIM1, PRIM2 and the POLA1CTD/POLA2 dimer, individually as a rigid body. AlphaFold multimer75 structure predications for the PRIM1- PRIM2NTD complex and the POLA2-POLA1CTD complex were aligned to the corresponding subunits derived from 5EXR, replacing the crystal structure subunits. Using this strategy atomic models were generated for the following regions of sequence: PRIM2 residues Q17-H252, PRIM1 residues M9-T349 and T386-G408, POLA1 residues Q1279-G1445 and E1448-C1458 and POLA2 residues I96-T114 and V170-I598.
The quality of the fit-to-density was optimised via an all-atom simulation in ISOLDE,80 using adaptive distance restraints for the dsDNA model (kappa=100). The resulting model was further refined using Phenix76 real-space-refine, utilising the input model as a reference to generate restraints with sigma=0.1 and global minimisation with nonbonded_weight=2000 and weight=0.5. Regions of the model that fit poorly to the density were then manually refined in Coot77 using the local refinement and regularisation tools incorporating stereochemical restraints.
Inspection of the cryo-EM density following the modelling procedure outlined revealed a short region of unmodelled helical density adjacent to the PSF1 subunit. AlphaFold-Multimer analysis predicted a helix within the flexible N-terminus of POLA2, residues I96-T114, to bind at the location of the unmodelled density. Furthermore, clear side chain density for POLA2 Y113 and L109 corroborated the prediction in addition to stereo-chemically favourable contacts formed. This region of POLA2 was subsequently incorporated into the final model.
A region of unmodelled density bound to the GINS subunit PSF2 was assigned to the PRIM2Nterm, residues M1-G5, and incorporated into the final model. Assignment was based upon the close proximity of the otherwise most N-terminal modelled residue of Pri2, Q17, and the presence of continuous density between G5-Q17 present at low map thresholds. Clear side chain density was present for residues M1-S4. Furthermore, the PRIM2Nterm – PSF2 contact was predicted by AlphaFold-Multimer at high confidence.
At this stage the model was inspected residue-by-residue in Coot,77 docked into the highest resolution cryo-EM map (consensus and focused refinements) for the corresponding region of the model. The model was manually refined against the map in Coot77 and both backbone Ramachandran and rotamer outliers corrected, followed by real-space refinement in both ISOLDE80 and Phenix.76 Each focussed refinement was rigid-body docked into the consensus refinement (EMD-15341) and resampled onto the same map origin. Only density at the AND-1 – CDC45/GINS interface was used to align the CDC45/GINS/AND-1 local refinement (EMD-15342) to the consensus (EMD-15341). However, due to subtle differences between the density at this interface between the local and consensus refinements it was not possible to build a model that perfectly satisfied both maps. A final global run in ISOLDE80 including distance and torsion restraints was carried out to minimise the clash-score prior to Phenix76 real-space-refinement using the same restraints as described above into the consensus refinement with additional reference model restraints. Model validation was carried out using the Molprobity server,81 Phenix76 validation and the wwPDB OneDep validation server. Following this procedure, the model for CLASPIN residues E299-E310 was removed due to the lack of corresponding cryo-EM density.
Multiple sequence alignments
Amino acid sequences were retrieved from UniProt and protein sequence alignments carried out using Clustal Omega.82 Alignments were rendered using ESPript3.0 (http://espript.ibcp.fr).83
AlphaFold and AlphaFold-Multimer
AlphaFold models were obtained from the AlphaFold Protein Structure Database50 (https://alphafold.ebi.ac.uk). AlphaFold-Multimer structure predictions were determined using the Colabfold84 implementation (version 1.2.0) using the following parameters: num_models=5, num_recycles=3, use_amber=false, use_templates=false. Colabfold sequence alignments were performed using Mmseqs2. Below is a description of the AlphaFold-Multimer input sequences, including protein names and the residue ranges included as input: Pri2NTD (44-299) / Pol1C-term (1140-1468); Pol12N-term (1-74) / Mcm3 AAA+ domain (337-750); Pol12 (203-705) / Pol1CTD (1260-1468); Mrc1site #1 (300-350) / Tof1 (1-779); Mrc1site #2 (420-550) / Tof1 (1-300) / Mcm2 (150-458); Mrc1site #3 (600-1000) / Mcm2 (150-458) / Cdc45 (1-468); Pri2Nterm (1-30) / Psf2 (1-213); PRIM2Nterm (1-14) / PSF2 (1-185); POLA2 (1-200) / PSF1 (1-196) / SLD5 (1-223); POLA2 (100-598) / POLA1CTD (1260-1458); PRIM1 (1-420) / Pri2 (1-509).
Budding yeast primase-polymerase assay on M13mp18 ssDNA
Reactions (10 μl) were performed at 30oC in a buffer containing: 25 mM HEPES-KOH (pH 7.6); 100 mM potassium glutamate; 10 mM Mg(OAc)2; 1 mM DTT; 0.01 % NP-40 substitute (NP-40-S) (Roche #11754599001), 0.1 mg/ml bovine serum albumin (BSA); 0.5 nM M13mp18 ssDNA (NEB #N4040S); 30 μM dC, dG, dT, dATP; 200 μM G, C, UTP; 33 nM α-[32P]-dCTP (Hartmann Analytic #SCP-205); 3 mM ATP; 40 nM RPA; 20 nM Pol α-primase. RPA was prebound to the ssDNA template for 10 min and then reactions were started by addition of Pol α-primase. Reactions were quenched with 10 μl 100 mM EDTA and unincorporated radiolabelled nucleotide was removed using a Microspin G-50 column (Cytiva). 1/10 volume loading buffer (10% w/v sucrose; 500 mM NaOH; ∼0.25% w/v xylene cyanol) was added to the sample before analysis on 0.7% alkaline agarose gels run in 30 mM NaOH, 2 mM EDTA for 16 hours at 25 volts. After electrophoresis gels were incubated at 4oC in 5% trichloroacetic acid for 30 min with one buffer change after 15 min. The gel was then incubated in 500 mM Tris-Cl (pH 8) for 15 mins before being dried at 75oC under vacuum onto Whatman 3 MM paper (Cytiva). The dried gel was imaged using a BAS-IP MS phosphor screen (Cytiva) and an Amersham Typhoon phosphorimager and on Amersham Hyperfilm MP (Cytiva).
Origin-dependent budding yeast DNA replication assay
Origin-dependent budding yeast DNA replication reactions were based on a previously established protocol.1,7,37,38 First, Mcm2-7 double hexamers were loaded onto the DNA template in an MCM loading reaction (10-40 μl) containing: 25 mM HEPES-KOH (pH 7.6); 100 mM potassium glutamate; 10 mM Mg(OAc)2; 1 mM DTT; 0.01 % NP-40-S, 0.1 mg/ml bovine serum albumin (BSA); 40 mM KCl, 3 mM ATP, 3 nM SphI-linearised vVA201; 75 nM Cdt1-Mcm2-7, 40 nM Cdc6; 20 nM ORC; 25 nM DDK. The reaction was incubated at 24oC for 10 mins, at which point S-CDK was added to a final concentration of 80 nM and incubation continued at 24oC for a further 5 min. The MCM loading reaction was diluted 4-fold into a replication buffer to give a final replication reaction buffer consisting of: 25 mM HEPES-KOH (pH 7.6); 250 mM potassium glutamate; 1 mM DTT; 0.01 % NP-40-S, 0.1 mg/ml BSA; 10 mM KCl, 3 mM ATP, 30 μM dC, dG, dT, dATP; 200 μM G, C, UTP, 33 nM α-[32P]-dCTP (Hartmann Analytic #SCP-205); 0.75 nM SphI-linearised vVA20; 18.75 nM Cdt1-Mcm2-7, 10 nM Cdc6; 5 nM ORC; 6.25 nM DDK; 20 nM S-CDK. Pol α-primase was added to a final concentration of 10 nM and reactions (10 μl) were equilibrated at 30oC. DNA replication was initiated by addition of replication proteins to final concentrations of: 30 nM Dpb11; 100 nM GINS; 30 nM Cdc45; 10 nM Mcm10; 20 nM Pol ε; 20 nM Ctf4; 100 nM RPA; 20 nM RFC; 20 nM Tof1-Csm3; 20 nM PCNA; 10 nM Pol δ; 12.5 nM Sld3/7; 20 nM Sld2; 10 nM Mrc1. Reactions were incubated at 30oC for 20 mins before quenching, processing and analysis as described for budding yeast primase-polymerase assays on M13mp18 ssDNA.
Budding yeast tetrad dissection
Details of yeast strains can be found in Table S4. Diploid yeast cells were patched onto sporulation plates (0.25% yeast extract; 0.1% glucose; 1.5% potassium acetate; 2% agar; 5 μg/ml Arginine, 10 μg/ml Adenine, 10 μg/ml Uracil; 5 μg/ml Histidine; 5 μg/ml Leucine; 5 μg/ml Lysine; 5 μg/ml Tryptophan, 2 μg/ml Tyrosine; 25 μg/ml Phenylalanine; 5 μg/ml Methionine; 1 μg/ml Proline) and incubated for 3-5 days at 30oC. Sporulated cells were picked using sterile toothpicks and resuspended in 75 μl sterile Milli-Q water and 5 μl β-Glucuronidase (Sigma #G7017). Following incubation at room temperature for 15-20 min, 150 μl sterile Milli-Q water was added and 10 μl of this mix was streaked onto YPD plates (1.1% yeast extract; 2.2% peptone; 2% glucose; 55 μg/ml Adenine; 2.5% agar). Tetrads were dissected using a micromanipulator (Singer Instruments) and the resulting cells were grown for 3 days at 25oC. Plates were imaged on an Epson Perfection V850 Pro scanner.
Cells were genotyped by analysing growth on appropriate selective plates or, when two alleles were associated with the same auxotrophic marker, by PCR from purified genomic DNA. For Pri2, primers JY105 and JY609 were used to generate a 608 bp PCR product. Products were digested with BsaHI, which digests the Pri2-AAA allele but not the wild type. For Mcm3, primers JY604 and VA212 were used to generate a 728 bp PCR product. Products were digested with FspI, which digests the wild type allele but not Mcm3-CR. Alternatively, primers JY370 / JY491 were used to amplify a region of the Mcm3 allele linked to the Ura3 marker. Primers JY370 / JY104 were used to amplify a region of the Pri2 allele linked to the Ura3 marker.
Human primase-polymerase assay on M13mp18 ssDNA
Reactions were performed at 37°C in a buffer containing 25 mM HEPES-KOH (pH 7.6), 100 mM potassium glutamate, 0.01% NP-40-S, 1 mM DTT, 10 mM Mg(OAc)2, 0.1 mg/ml BSA, 5 mM ATP, 200 μM CTP, GTP, UTP, 30 μM dATP, dCTP, dGTP, dTTP, and 33 nM α-[32P]-dCTP. 0.5 nM M13mp18 single-stranded DNA (New England Biolabs) was pre-incubated with 40 nM RPA for 10 min. Reactions were initiated by the addition of 20 nM Pol α-primase. After 20 min, reactions were quenched by addition of EDTA to 50 mM. Post reaction processing and analysis were performed as described for budding yeast primase-polymerase assays on M13mp18 ssDNA.
Human DNA replication assays
Replication reactions were performed on a 9.7 kbp forked linear DNA template (made from SapI-linearized ZN3 plasmid) as previously described27 with minor modifications. Reactions were conducted at 37°C in a replication buffer consisting of 25 mM HEPES-KOH (pH 7.6), 0.01% NP-40-S, 100 mM potassium glutamate, 1 mM DTT, 10 mM Mg(OAc)2 and 0.1 mg/ml BSA. The final concentration of potassium glutamate in reactions was increased to 230 mM when replication was initiated. Protein and nucleotide concentrations in the final reactions were: 25 nM CMG, 15 nM Pol ε, 20 nM RFC, 7 nM Pol δ, 20 nM PCNA, 20 nM AND-1, 10 nM Pol α-primase, 100 nM RPA, 20 nM CLASPIN, 20 nM TIMELESS–TIPIN, 20 nM CTF18–RFC, 4 mM ATP, 30 μM dC/dT/dG/dATP, 200 μM C/G/UTP and 33 nM α-[32P]-dCTP. Reactions were set up as follows: 2 nM linear forked DNA template was pre-incubated with 50 nM CMG for 10 min in replication buffer. The reaction was diluted twofold by the addition of replication buffer containing 60 μM dA/dCTP, PCNA, Pol ε, Pol α-primase, Pol δ, CLASPIN, TIMELESS–TIPIN, AND-1 and CTF18–RFC. Replication was initiated by addition of a 10X solution containing ATP, RFC, dTTP, dGTP, GTP, CTP, UTP, α-[32P]-dCTP and RPA. Reactions were stopped by addition of 50 mM EDTA and unincorporated α-[32P]-dCTP was removed with illustra MicroSpin G-50 columns (GE Healthcare). Reactions were run on 0.7% alkaline agarose gels in 2 mM EDTA and 30 mM NaOH and for 16 h at 26 V. Post reaction processing and analysis were performed as described for budding yeast primase-polymerase assays on M13mp18 ssDNA.
Quantification and statistical analysis
No quantification or statistical analysis were performed in this manuscript.
Acknowledgments
We thank K. Labib for sharing budding yeast strains. We thank J. Shi for operation of the LMB baculovirus facility; LMB media preparation for budding yeast plates; S. Chen, G. Sharov, G. Cannone, A. Yeates, and B. Ahsan for smooth running of the MRC LMB EM facility; and J. Grimmett, T. Darling, and I. Clayson for maintenance of scientific computing facilities. We are grateful to F. Abid-Ali and members of the Yeeles lab for comments on the manuscript. This work was supported by the MRC as part of UK Research and Innovation (MRC grant MC_UP_1201/12 to J.T.P.Y.).
Author contributions
Investigation—performed all cryo-EM experiments, data analysis, and model building, methodology—purified proteins, writing – original draft, review & editing, M.L.J.; investigation—performed preliminary DNA replication assays and Pol α-primase:CMG interaction studies, methodology—generated expression vectors and yeast strains and purified proteins, V.A.; investigation—performed human DNA replication assays, methodology—purified proteins, Y.B.; conceptualization, supervision, funding acquisition, investigation—performed biochemistry and genetic experiments, methodology—DNA template preparation, protein purification, yeast strains, writing – original draft, review & editing, J.T.P.Y.
Declaration of interests
The authors declare no competing interests.
Published: July 27, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2023.06.035.
Supplemental information
Data and code availability
-
•Cryo-EM density maps used in model building have been deposited in the Electron Microscopy Data Bank (EMDB) https://www.ebi.ac.uk/pdbe/emdb, under the following accession numbers:
- Budding yeast. EMD-16320 (binned), EMD-16322 (un-binned), Pol α-primase associated replisome consensus refinement in the absence of Ctf4, 60 nucleotide 5ʹ-flap DNA fork. EMD-15304, Tof1-Csm3 local refinement. EMD-15305, Mcm2-7 C-tier local refinement, conformation II. EMD-15306, Pol12, Pol1CTD, Pri2NTD local refinement. EMD-16885, Pol12, Pol1CTD local refinement. EMD-15309 (binned), EMD-15902 (un-binned), Pol α-primase associated replisome consensus refinement in the presence of Ctf4, 60 nucleotide 5ʹ-flap DNA fork. EMD-15310, Ctf4 local refinement. EMD-16247, Pri1, Pri2CTD local refinement. EMD-16248, Pol α-primase associated replisome consensus refinement including density for the Pol1exo/cat and Pri2CTD domains. EMD-15924 (binned), EMD-15303 (un-binned), Pol α-primase associated replisome consensus refinement with continuous density for the lagging strand DNA template extending towards the Pri1 active site and density for the Pri1CTD, 60 nucleotide 5ʹ-flap DNA fork. EMD-16323, consensus refinement of Pol α-primase associated with the replisome only via the Pri2:Mcm5ZnF and Pri2Nterm:Psf2 interfaces, 60 nucleotide 5ʹ-flap DNA fork.
- Human. EMD-15341, Pol α-primase associated replisome un-binned consensus refinement, 60 nucleotide 5ʹ-flap DNA fork. EMD-15342, AND-1 local refinement. EMD-15340, MCM2-7 C-tier local refinement. EMD-15351, PRIM1, POLA2, PolA1CTD, Pri2NTD local refinement. EMD-15356, TIMELESS-TIPIN local refinement. EMD-15904. EMD-15349, Pol α-primase associated replisome binned consensus refinement, strong PRIM1 density. composite map assembled from EMD-15342: 15341:15340:15349:15351:15356. EMD-15918, Pol α-primase associated replisome consensus refinement, not engaged on DNA derived from dataset including a 15 nucleotide 5ʹ-flap DNA fork. EMD-15923, Pol α-primase associated replisome consensus refinement, not engaged on DNA derived from dataset including a 60 nucleotide 5ʹ-flap DNA fork. EMD-15922, Pol α-primase associated replisome consensus refinement, engaged on a 15 nucleotide 5ʹ-flap DNA fork.
- Atomic coordinates have been deposited in the Protein Data Bank (PDB), http://www.pdb.org, with the following accession numbers:
- Budding yeast. 8B9C - Pol α-primase associated replisome in the absence of Ctf4. 8B9A - Pol α-primase associated replisome in the presence of Ctf4, CIP box site #1. 8B9B - Pol α-primase associated replisome in the presence of Ctf4, CIP box site #2.
- Human. 8B9D – Pol α-primase associated replisome.
- Unprocessed images of the data featured in this manuscript have been deposited at Mendeley Data and are publicly available as of the date of publication (https://doi.org/10.17632/n2wm36mrmw.1).
-
•
This study does not report any original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Aria V., Yeeles J.T.P. Mechanism of bidirectional leading-strand synthesis establishment at eukaryotic DNA replication origins. Mol. Cell. 2019;73:199–211.e110. doi: 10.1016/j.molcel.2018.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nick McElhinny S.A., Gordenin D.A., Stith C.M., Burgers P.M., Kunkel T.A. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garbacz M.A., Lujan S.A., Burkholder A.B., Cox P.B., Wu Q., Zhou Z.X., Haber J.E., Kunkel T.A. Evidence that DNA polymerase delta contributes to initiating leading strand DNA replication in Saccharomyces cerevisiae. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-03270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Z.-X., Lujan S.A., Burkholder A.B., Garbacz M.A., Kunkel T.A. Roles for DNA polymerase δ in initiating and terminating leading strand DNA replication. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-11995-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daigaku Y., Keszthelyi A., Müller C.A., Miyabe I., Brooks T., Retkute R., Hubank M., Nieduszynski C.A., Carr A.M. A global profile of replicative polymerase usage. Nat. Struct. Mol. Biol. 2015;22:192–198. doi: 10.1038/nsmb.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pursell Z.F., Isoz I., Lundström E.B., Johansson E., Kunkel T.A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor M.R.G., Yeeles J.T.P. The initial response of a eukaryotic replisome to DNA damage. Mol. Cell. 2018;70:1067–1080.e12. doi: 10.1016/j.molcel.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor M.R.G., Yeeles J.T.P. Dynamics of replication fork progression following helicase–polymerase uncoupling in eukaryotes. J. Mol. Biol. 2019;431:2040–2049. doi: 10.1016/j.jmb.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guilliam T.A., Yeeles J.T.P. Reconstitution of translesion synthesis reveals a mechanism of eukaryotic DNA replication restart. Nat. Struct. Mol. Biol. 2020;27:450–460. doi: 10.1038/s41594-020-0418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casas-Delucchi C.S., Daza-Martin M., Williams S.L., Coster G. The mechanism of replication stalling and recovery within repetitive DNA. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-31657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guilliam T.A., Brissett N.C., Ehlinger A., Keen B.A., Kolesar P., Taylor E.M., Bailey L.J., Lindsay H.D., Chazin W.J., Doherty A.J. Molecular basis for PrimPol recruitment to replication forks by RPA. Nat. Commun. 2017;8 doi: 10.1038/ncomms15222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiavone D., Jozwiakowski S.K., Romanello M., Guilbaud G., Guilliam T.A., Bailey L.J., Sale J.E., Doherty A.J. PrimPol is required for replicative tolerance of G quadruplexes in vertebrate cells. Mol. Cell. 2016;61:161–169. doi: 10.1016/j.molcel.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Gómez S., Reyes A., Martínez-Jiménez M.I., Chocrón E.S., Mourón S., Terrados G., Powell C., Salido E., Méndez J., Holt I.J., Blanco L. PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell. 2013;52:541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan L., Lou J., Xia Y., Su B., Liu T., Cui J., Sun Y., Lou H., Huang J. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep. 2013;14:1104–1112. doi: 10.1038/embor.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellegrini L. The Pol alpha-primase complex. Subcell. Biochem. 2012;62:157–169. doi: 10.1007/978-94-007-4572-8_9. [DOI] [PubMed] [Google Scholar]
- 16.Perera R.L., Torella R., Klinge S., Kilkenny M.L., Maman J.D., Pellegrini L. Mechanism for priming DNA synthesis by yeast DNA polymerase alpha. eLife. 2013;2 doi: 10.7554/eLife.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg P., Burgers P.M.J. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 18.Baranovskiy A.G., Lisova A.E., Morstadt L.M., Babayeva N.D., Tahirov T.H. Insight into RNA–DNA primer length counting by human primosome. Nucleic Acids Res. 2022;50:6264–6270. doi: 10.1093/nar/gkac492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins K.L., Kelly T.J. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase alpha-primase. Mol. Cell. Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H., Weiner B.E., Zhang H., Fuller B.E., Gao Y., Wile B.M., Zhao K., Arnett D.R., Chazin W.J., Fanning E. Structure of a DNA polymerase alpha-primase domain that docks on the SV40 helicase and activates the viral primosome. J. Biol. Chem. 2010;285:17112–17122. doi: 10.1074/jbc.M110.116830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisshart K., Förster H., Kremmer E., Schlott B., Grosse F., Nasheuer H.P. Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J. Biol. Chem. 2000;275:17328–17337. doi: 10.1074/jbc.M000717200. [DOI] [PubMed] [Google Scholar]
- 22.Zhu W., Ukomadu C., Jha S., Senga T., Dhar S.K., Wohlschlegel J.A., Nutt L.K., Kornbluth S., Dutta A. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007;21:2288–2299. doi: 10.1101/gad.1585607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miles J., Formosa T. Protein affinity chromatography with purified yeast DNA polymerase alpha detects proteins that bind to DNA polymerase. Proc. Natl. Acad. Sci. USA. 1992;89:1276–1280. doi: 10.1073/pnas.89.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Falco M., Ferrari E., De Felice M., Rossi M., Hübscher U., Pisani F.M. The human GINS complex binds to and specifically stimulates human DNA polymerase α-primase. EMBO Rep. 2007;8:99–103. doi: 10.1038/sj.embor.7400870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricke R.M., Bielinsky A.-K. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol. Cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Chattopadhyay S., Bielinsky A.K. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol. Biol. Cell. 2007;18:4085–4095. doi: 10.1091/mbc.e06-12-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baris Y., Taylor M.R.G., Aria V., Yeeles J.T.P. Fast and efficient DNA replication with purified human proteins. Nature. 2022;606:204–210. doi: 10.1038/s41586-022-04759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R.C., Edmondson R.D., Calzada A., Labib K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon A.C., Zhou J.C., Perera R.L., van Deursen F., Evrin C., Ivanova M.E., Kilkenny M.L., Renault L., Kjaer S., Matak-Vinković D., et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature. 2014;510:293–297. doi: 10.1038/nature13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baretić D., Jenkyn-Bedford M., Aria V., Cannone G., Skehel M., Yeeles J.T.P. Cryo-EM structure of the fork protection complex bound to CMG at a replication fork. Mol. Cell. 2020;78:926–940.e13. doi: 10.1016/j.molcel.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan Z., Georgescu R., Santos R.L.A., Zhang D., Bai L., Yao N.Y., Zhao G., O'Donnell M.E., Li H. Ctf4 organizes sister replisomes and Pol alpha into a replication factory. eLife. 2019;8 doi: 10.7554/eLife.47405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilkenny M.L., Simon A.C., Mainwaring J., Wirthensohn D., Holzer S., Pellegrini L. The human CTF4-orthologue AND-1 interacts with DNA polymerase alpha/primase via its unique C-terminal HMG box. Open Biol. 2017;7 doi: 10.1098/rsob.170217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan C., Li J., Sun D., Liu Y., Liang H. The structure and polymerase-recognition mechanism of the crucial adaptor protein AND-1 in the human replisome. J. Biol. Chem. 2017;292:9627–9636. doi: 10.1074/jbc.M116.758524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evrin C., Maman J.D., Diamante A., Pellegrini L., Labib K. Histone H2A-H2B binding by Pol α in the eukaryotic replisome contributes to the maintenance of repressive chromatin. EMBO J. 2018;37 doi: 10.15252/embj.201899021. H2A–H2B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapadia N., El-Hajj Z.W., Zheng H., Beattie T.R., Yu A., Reyes-Lamothe R. Processive activity of replicative DNA polymerases in the replisome of live eukaryotic cells. Mol. Cell. 2020;80:114–126.e8. doi: 10.1016/j.molcel.2020.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Abe T., Kawasumi R., Giannattasio M., Dusi S., Yoshimoto Y., Miyata K., Umemura K., Hirota K., Branzei D. AND-1 fork protection function prevents fork resection and is essential for proliferation. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-05586-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeeles J.T.P., Janska A., Early A., Diffley J.F.X. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol. Cell. 2017;65:105–116. doi: 10.1016/j.molcel.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeeles J.T., Deegan T.D., Janska A., Early A., Diffley J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Georgescu R.E., Schauer G.D., Yao N.Y., Langston L.D., Yurieva O., Zhang D., Finkelstein J., O'Donnell M.E. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. eLife. 2015;4 doi: 10.7554/eLife.04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lõoke M., Maloney M.F., Bell S.P. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017;31:291–305. doi: 10.1101/gad.291336.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones M.L., Baris Y., Taylor M.R.G., Yeeles J.T.P. Structure of a human replisome shows the organisation and interactions of a DNA replication machine. EMBO J. 2021;40 doi: 10.15252/embj.2021108819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkyn-Bedford M., Jones M.L., Baris Y., Labib K.P.M., Cannone G., Yeeles J.T.P., Deegan T.D. A conserved mechanism for regulating replisome disassembly in eukaryotes. Nature. 2021;600:743–747. doi: 10.1038/s41586-021-04145-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan Z., Georgescu R., Bai L., Zhang D., Li H., O'Donnell M.E. DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-14577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eickhoff P., Kose H.B., Martino F., Petojevic T., Ali F.A., Locke J., Tamberg N., Nans A., Berger J.M., Botchan M.R., et al. Molecular basis for ATP-hydrolysis-driven DNA translocation by the CMG helicase of the eukaryotic replisome. Cell Rep. 2019;28:2673–2688.e8. doi: 10.1016/j.celrep.2019.07.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kilkenny M.L., De Piccoli G., Perera R.L., Labib K., Pellegrini L. A conserved motif in the C-terminal tail of DNA polymerase alpha tethers primase to the eukaryotic replisome. J. Biol. Chem. 2012;287:23740–23747. doi: 10.1074/jbc.M112.368951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baranovskiy A.G., Babayeva N.D., Zhang Y., Gu J., Suwa Y., Pavlov Y.I., Tahirov T.H. Mechanism of concerted RNA-DNA primer synthesis by the human primosome. J. Biol. Chem. 2016;291:10006–10020. doi: 10.1074/jbc.M116.717405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Q., Lin X., Chavez B.L., Agrawal S., Lusk B.L., Lim C.J. Structures of the human CST-Polα–primase complex bound to telomere templates. Nature. 2022;608:826–832. doi: 10.1038/s41586-022-05040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He Q., Baranovskiy A.G., Morstadt L.M., Lisova A.E., Babayeva N.D., Lusk B.L., Lim C.J., Tahirov T.H. Structures of human primosome elongation complexes. Nat. Struct. Mol. Biol. 2023;30:579–583. doi: 10.1038/s41594-023-00971-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Punjani A., Fleet D.J. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 2021;213 doi: 10.1016/j.jsb.2021.107702. [DOI] [PubMed] [Google Scholar]
- 50.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurat C.F., Yeeles J.T.P., Patel H., Early A., Diffley J.F.X. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell. 2017;65:117–130. doi: 10.1016/j.molcel.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porcella S.Y., Koussa N.C., Tang C.P., Kramer D.N., Srivastava P., Smith D.J. Separable, Ctf4-mediated recruitment of DNA polymerase α for initiation of DNA synthesis at replication origins and lagging-strand priming during replication elongation. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gan H., Serra-Cardona A., Hua X., Zhou H., Labib K., Yu C., Zhang Z. The Mcm2-Ctf4-Polalpha axis facilitates parental histone H3-H4 transfer to lagging strands. Mol. Cell. 2018;72:140–151.e3. doi: 10.1016/j.molcel.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson S.W., Kumar R., Benkovic S.J. RNA primer handoff in bacteriophage T4 DNA replication: the ROLE OF SINGLE-STRANDED DNA-BINDING PROTEIN AND polymerase ACCESSORY PROTEINS. J. Biol. Chem. 2008;283:22838–22846. doi: 10.1074/jbc.M802762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pandey M., Syed S., Donmez I., Patel G., Ha T., Patel S.S. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature. 2009;462:940–943. doi: 10.1038/nature08611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis J.S., Spenkelink L.M., Schauer G.D., Yurieva O., Mueller S.H., Natarajan V., Kaur G., Maher C., Kay C., O’Donnell M.E., van Oijen A.M. Tunability of DNA polymerase stability during eukaryotic DNA replication. Mol. Cell. 2020;77:17–25.e5. doi: 10.1016/j.molcel.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li A.W.H., Zabrady K., Bainbridge L.J., Zabrady M., Naseem-Khan S., Berger M.B., Kolesar P., Cisneros G.A., Doherty A.J. Molecular basis for the initiation of DNA primer synthesis. Nature. 2022;605:767–773. doi: 10.1038/s41586-022-04695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He Y., Song H., Chan H., Liu B., Wang Y., Sušac L., Zhou Z.H., Feigon J. Structure of Tetrahymena telomerase-bound CST with polymerase α-primase. Nature. 2022;608:813–818. doi: 10.1038/s41586-022-04931-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai S.W., Zinder J.C., Svetlov V., Bush M.W., Nudler E., Walz T., de Lange T. Cryo-EM structure of the human CST-Polα/primase complex in a recruitment state. Nat. Struct. Mol. Biol. 2022;29:813–819. doi: 10.1038/s41594-022-00766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coster G., Frigola J., Beuron F., Morris E.P., Diffley J.F. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol Cell. 2014;55:666–677. doi: 10.1016/j.molcel.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frigola J., Remus D., Mehanna A., Diffley J.F. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.On K.F., Beuron F., Frith D., Snijders A.P., Morris E.P., Diffley J.F. Prereplicative complexes assembled in vitro support origin-dependent and independent DNA replication. EMBO J. 2014;33:605–620. doi: 10.1002/embj.201387369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jake H., Patrik E., Lucy S.D., Alessandro C., John F.X.D. The eukaryotic replisome requires an additional helicase to disarm dormant replication origins. bioRxiv. 2020 doi: 10.1101/2020.09.17.301366. 2020.2009.2017.301366. [DOI] [Google Scholar]
- 64.Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scheres S.H. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 66.Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scheres S.H. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 69.Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rohou A., Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 72.Zivanov J., Nakane T., Scheres S.H.W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Punjani A., Zhang H., Fleet D.J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods. 2020;17:1214–1221. doi: 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 74.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Evans R., O’Neill M., Pritzel A., Antropova N., Senior A., Green T., Žídek A., Bates R., Blackwell S., Yim J., et al. 2022. Protein complex prediction with AlphaFold-Multimer. [DOI] [Google Scholar]
- 76.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wingfield P.T. N-terminal methionine processing. Curr. Protoc. Protein Sci. 2017;88:6.14.1–6.14.3. doi: 10.1002/cpps.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ree R., Varland S., Arnesen T. Spotlight on protein N-terminal acetylation. Exp. Mol. Med. 2018;50:1–13. doi: 10.1038/s12276-018-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Croll T.I. Isolde: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sievers F., Higgins D.G. In: Multiple Sequence Alignment Methods. Russell D.J., editor. Humana Press; 2014. Clustal Omega, accurate alignment of very large numbers of sequences; pp. 105–116. [DOI] [PubMed] [Google Scholar]
- 83.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat. Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
First, individual replisome components are sequentially colored and labeled to aid in their visualisation and the interpretation of the overall structure. The video then highlights each pol α-primase interface with the replisome (sites a-e) using different views. Regions of the model forming an interaction interface are displayed using transparent surface rendering.
First, individual replisome components are sequentially colored and labeled to aid in their visualisation and the interpretation of the overall structure. The video then highlights each pol α-primase interface with the replisome (sites b, d, g, and e) using different views. Regions of the model forming an interaction interface are displayed using transparent surface rendering.
Data Availability Statement
-
•Cryo-EM density maps used in model building have been deposited in the Electron Microscopy Data Bank (EMDB) https://www.ebi.ac.uk/pdbe/emdb, under the following accession numbers:
- Budding yeast. EMD-16320 (binned), EMD-16322 (un-binned), Pol α-primase associated replisome consensus refinement in the absence of Ctf4, 60 nucleotide 5ʹ-flap DNA fork. EMD-15304, Tof1-Csm3 local refinement. EMD-15305, Mcm2-7 C-tier local refinement, conformation II. EMD-15306, Pol12, Pol1CTD, Pri2NTD local refinement. EMD-16885, Pol12, Pol1CTD local refinement. EMD-15309 (binned), EMD-15902 (un-binned), Pol α-primase associated replisome consensus refinement in the presence of Ctf4, 60 nucleotide 5ʹ-flap DNA fork. EMD-15310, Ctf4 local refinement. EMD-16247, Pri1, Pri2CTD local refinement. EMD-16248, Pol α-primase associated replisome consensus refinement including density for the Pol1exo/cat and Pri2CTD domains. EMD-15924 (binned), EMD-15303 (un-binned), Pol α-primase associated replisome consensus refinement with continuous density for the lagging strand DNA template extending towards the Pri1 active site and density for the Pri1CTD, 60 nucleotide 5ʹ-flap DNA fork. EMD-16323, consensus refinement of Pol α-primase associated with the replisome only via the Pri2:Mcm5ZnF and Pri2Nterm:Psf2 interfaces, 60 nucleotide 5ʹ-flap DNA fork.
- Human. EMD-15341, Pol α-primase associated replisome un-binned consensus refinement, 60 nucleotide 5ʹ-flap DNA fork. EMD-15342, AND-1 local refinement. EMD-15340, MCM2-7 C-tier local refinement. EMD-15351, PRIM1, POLA2, PolA1CTD, Pri2NTD local refinement. EMD-15356, TIMELESS-TIPIN local refinement. EMD-15904. EMD-15349, Pol α-primase associated replisome binned consensus refinement, strong PRIM1 density. composite map assembled from EMD-15342: 15341:15340:15349:15351:15356. EMD-15918, Pol α-primase associated replisome consensus refinement, not engaged on DNA derived from dataset including a 15 nucleotide 5ʹ-flap DNA fork. EMD-15923, Pol α-primase associated replisome consensus refinement, not engaged on DNA derived from dataset including a 60 nucleotide 5ʹ-flap DNA fork. EMD-15922, Pol α-primase associated replisome consensus refinement, engaged on a 15 nucleotide 5ʹ-flap DNA fork.
- Atomic coordinates have been deposited in the Protein Data Bank (PDB), http://www.pdb.org, with the following accession numbers:
- Budding yeast. 8B9C - Pol α-primase associated replisome in the absence of Ctf4. 8B9A - Pol α-primase associated replisome in the presence of Ctf4, CIP box site #1. 8B9B - Pol α-primase associated replisome in the presence of Ctf4, CIP box site #2.
- Human. 8B9D – Pol α-primase associated replisome.
- Unprocessed images of the data featured in this manuscript have been deposited at Mendeley Data and are publicly available as of the date of publication (https://doi.org/10.17632/n2wm36mrmw.1).
-
•
This study does not report any original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.