Abstract
The purpose of the present study was to determine if the quantity of herpes simplex virus (HSV) DNA in the cerebrospinal fluid (CSF) of patients with herpes encephalitis would be useful in establishing the prognosis of the disease and to determine the effect of antiviral therapy on the clearance of viral DNA from the CSF. Quantitation of HSV DNA was done by constructing an internal standard (IS) from the glycoprotein B amplicon which had a 25-bp deletion between primer annealing sites. Each CSF specimen was coamplified with the IS and the ratio of the amount of HSV/amount of IS was compared to the ratios on a standard curve constructed with the same IS plus known amounts of HSV DNA. CSF specimens were available from 16 patients who were treated with intravenous acyclovir, and the amount of HSV DNA ranged from <25 to 18,000 copies per μl in CSF obtained before or within 4 days of the initiation of acyclovir therapy. Patients with >100 copies of HSV DNA per μl were older, were found by computed tomography to have lesions, and had poorer outcomes than patients with <100 copies. Follow-up CSF specimens were available from seven patients. In six of these seven patients, the HSV DNA levels decreased during therapy. One patient had a twofold increase in HSV DNA levels after 1 week of therapy and died on day 8. The application of this assay may be helpful in establishing the prognosis and in the monitoring of patients with herpes simplex encephalitis.
Herpes simplex encephalitis (HSE) is associated with high rates of mortality and morbidity (35). Antiviral therapy is highly effective in reducing the rate of mortality from HSE (31); however, fewer than one-half of the patients return to normal following brain biopsy and treatment (34). Several clinical features, including age over 30 years, late introduction of antiviral treatment, low Glasgow score, and detection by computed tomography (CT) of a focal lesion at the onset of therapy have been associated with a poor prognosis of the disease (31, 23). A previous study which titrated the amount of infectious virus in brain biopsy tissue suggested that larger amounts of virus were predictive of more severe disease (24). Herpes simplex virus (HSV) DNA detection by PCR of cerebrospinal fluid (CSF) samples is very efficient in establishing the diagnosis of HSE (1, 3, 6, 18, 20, 22, 29). The use of a semiquantitative PCR suggested that the severity of disease and the age could influence the amount of HSV DNA in CSF (27). A quantitative PCR assay that assesses the effect of acyclovir therapy on the number of HSV type 1 (HSV-1) DNA copies in CSF has been described previously (2). The limitation of such assays is that they were unable to control the differences in amplification efficiency caused by sample interference, including the presence of Taq polymerase inhibitors. Therefore, the development of a quantitative assay which could estimate the extent of viral replication in the central nervous systems (CNS) of HSE patients would be helpful as a prognostic marker and in monitoring HSE treatment. In this report, a quantitative PCR (QC-PCR) which uses an internal standard (IS) that has the same primer binding sites as HSV-1 DNA is described. The amount of HSV-1 DNA in CSF samples from 16 HSE patients was measured. The ages, genders, severities of disease, durations of disease, durations of acyclovir treatment, and outcomes for patients with high and low levels of HSV-1 DNA were compared. Also, the effect of acyclovir therapy on the levels of HSV DNA is shown for six patients. Patients with larger amounts of HSV-1 DNA tended to have a more severe disease and a poor outcome. Moreover, this method showed potential utility for monitoring the treatment of HSE with antiviral agents.
MATERIALS AND METHODS
Patients and clinical specimens.
All 16 patients were prospectively admitted to seven different medical institutions in São Paulo and Campinas, Brazil. All of them were diagnosed as having HSE and were treated intravenously with acyclovir (30 mg/kg of body weight/day) for 10 to 21 days. Age, gender, historical findings, neurological examination, and neurodiagnostic data were collected on case record forms. Glasgow Coma Scale scores from the following three areas were totaled: verbal output, eye opening, and best motor response. Normal responses score 4, 6, and 5 points, respectively; no response gives a score of 1. Clinical outcome was assessed for 3 months after the completion of the treatment. Morbidity was defined as reported previously (32): normal; mild impairment, for patients with minor neuropsychological deficits; moderate impairment, for patients possessing limitations due to motor, speech, memory, or seizure disorder; severe impairment, for patients requiring supportive care; and death. CSF specimens were stored at −20°C. All PCR and QC-PCR assays were performed with neat CSF, which was boiled for 10 min and spun at 4°C for 5 min. The diagnosis of HSE was established by a PCR with two different sets of primers, one that amplifies a 179-bp region of the DNA polymerase gene and another that amplifies a 148-bp region of the glycoprotein B gene. The HSV typing was carried out by restriction enzyme digestion of DNA polymerase PCR products with the enzyme HhaI.
Viral DNA quantitation.
The QC-PCRs were performed in a solution containing buffer II (10 mM Tris [pH 8.3], 50 mM KCl), 2.5 mM MgCl2, each primer at a concentration of 1 μM, dATP, dCTP, dTTP, and dGTP each at a concentration of 80 μM and 160 μM dUTP, Taq polymerase at 2.5 U/reaction, uracil N-glycosylase at 0.04 U/reaction, 10 μl of the target (HSV-1 DNA or CSF sample), and 5 μl of IS. The cycles used were 1 cycle of 50°C for 2 min and 1 cycle of 95°C for 5 min, followed by 40 cycles at 95, 62, and 72°C for 45 s, with a 3-s extension added to each 72°C cycle.
To quantitate the viral DNA, a standard curve was obtained for each experiment by coamplification of known amounts of HSV-1 DNA (Sigma Chemical Co., St. Louis, Mo.) with 250 copies of the IS. Four consecutive dilutions of HSV-1 containing 100 (102.0), 200 (102.3), 500 (102.7), and 1,000 (103) copies/reaction yielded double bands when HSV-1 was coamplified with the IS (see Fig. 2). The CSF samples were coamplified with the same amount of IS. In cases in which the amount of HSV-1 DNA per reaction was greater than 2,000 copies per reaction (200 copies/μl), additional amplifications were performed with serial twofold dilutions of the sample in order to plot data for at least two different dilutions of the sample onto the standard curve. Twenty microliters of the products was applied to a 3% agarose gel stained with ethidium bromide (0.8 mg/ml). After electrophoresis, the intensities of the bands were determined by densitometric analysis of the gels with Collage software (FOTODYNE Incorporated, New Berlin, Wis.) for Apple Macintosh. To construct the standard curve the ratios of the amount of HSV/amount of IS were plotted on a logarithmic scale against the number of HSV copies. The log amounts of HSV DNA in the samples were obtained by plotting the ratio of the amount of HSV-1 DNA in the sample/amount of IS (sample/IS ratio) onto the standard curve.
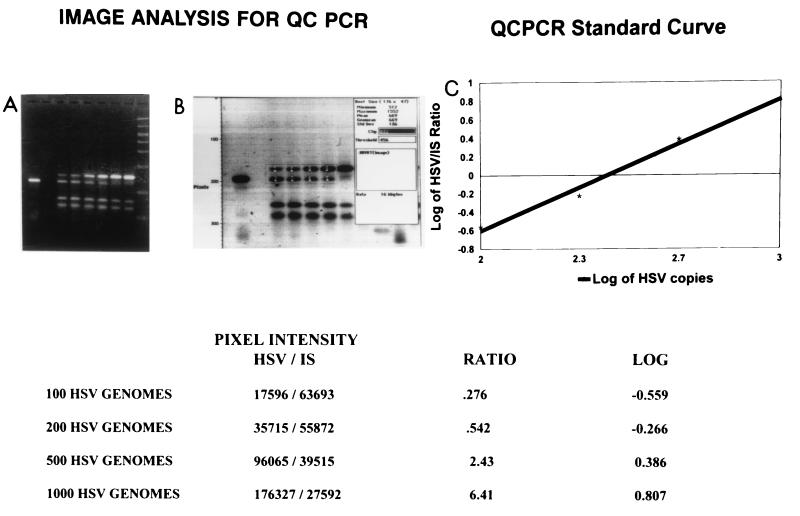
FIG. 2.
Construction of a standard curve for quantitation of HSV-1. (A) Agarose-ethidium bromide gel; (B) image analysis of the gel in panel A; (C) standard curve plotted from the data in the table at the bottom. In the gels, the first band is 250 copies of the 123-bp IS. The next six lanes contain 100, 200, 500, 1,000, 2,000, and 5,000 copies of HSV DNA, respectively, plus 250 copies of IS. The last lane in the agarose gel contains a molecular size marker (1,000, 700, 500, 400, 300, 200, 100, and 50 bp). The table shows the pixel intensities of the amount of HSV/amount of IS. The double bands are the 148-bp product of HSV DNA and the 123-bp product of the IS and are seen in the lanes containing 100, 200, 500, and 1,000 copies of HSV DNA.
Statistical analysis.
The patients were divided into two groups according to whether their CSF samples had amounts of HSV DNA that were smaller or larger than 102 copies/μl. The ages, genders, Glasgow scores, the presence of lesions detected by CT, durations of disease, durations of acyclovir therapy, and outcomes were compared between these two groups. All the statistical comparisons were carried out by the Wilcoxon rank sum test or the Fisher’s exact two-tailed test.
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of HSV-1 glycoprotein B is K01760.
RESULTS
Patients.
The mean age of the patients was 36.6 ± 21 years, with females accounting for 56% of the study population. All patients were in good health prior to the onset of HSE symptoms. HSV DNA was detected in the initial CSF samples from all 16 patients with both sets of primers complementary to the DNA polymerase gene and the glycoprotein B gene. Both diagnostic PCR protocols were able to detect 10 copies of the HSV-1 DNA-positive control per μl. The restriction endonuclease analysis of the DNA polymerase products for all 16 patients revealed fragments of ∼135 and ∼44 bp, indicative of HSV-1 infection.
QC-PCR primers and IS DNA.
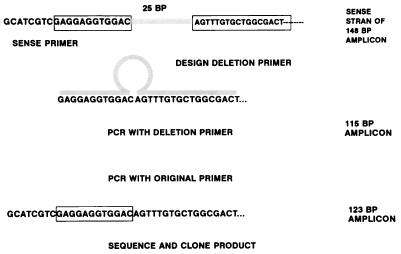
The sequences of the primers used in the QC-PCR assay were 5′-GCATCGTCGAGGAGGTGGAC-3′ (sense) and 5′-TTGAAGCGGTCGGCGGCGTA-3′ (antisense). These oligonucleotides amplify a 148-bp region of the glycoprotein B gene and are complementary to bases 1359 to 1378 and bases 1487 to 1506 of this gene, respectively (22). The procedure used to construct the IS is outlined in Fig. 1. The IS was obtained by amplification of HSV-1 DNA in a PCR in which the normal sense primer was replaced by a primer composed of the last 12 bases plus 17 bases located 25 bases downstream. This 29-base primer is complementary to bases 1367 to 1378 and bases 1404 to 1420 of the glycoprotein B gene and loops out a 25-bp region from bases 1379 to 1403 of this gene. This PCR was performed with the usual reagent concentrations and cycles. The 115-bp product of this reaction was reamplified with sense and antisense primers in order to reincorporate the region of the glycoprotein B binding site from bases 1359 to 1366. The last PCR resulted in a 123-bp amplicon containing sites complementary to the sense and antisense primers. These products were cloned into pCRII (TA Cloning Kit; Invitrogen, San Diego, Calif.) and were assessed with an automated sequencer (Applied Biosystems, Foster City, Calif.). The only differences from the original glycoprotein B gene sequence were the deletions from bases 1379 to 1403.
FIG. 1.
Procedure used to construct IS. The top line depicts the locations of the sequences used to construct the IS. The normal glycoprotein B primer is shown at the left; the IS primer was composed of the last 12 bases of this primer plus 17 bases located downstream, as shown in the box. The resulting primer shown in the second line loops out 25 bases resulting in a 115-bp product. This product was reamplified to reconstruct the original primer site and produce a 123-bp amplicon. BP, base pairs.
Quantitation of viral DNA.
The construction of a standard curve for the quantitation of HSV-1 is shown in Fig. 2. Double bands, the 148-bp product of HSV DNA and the 123-bp product of the IS, are seen in the lanes containing 100, 200, 500, and 1,000 copies of HSV DNA. The linear correlation shown in Fig. 2 indicates that this assay was efficient in measuring the amount of target HSV-1 DNA. Quantitation of the HSV-1 DNA in the clinical samples was highly reproducible when considering the log10 copy number. For patients for whom the amount of DNA was greater than 2 × 102 copies/μl, data for two consecutive dilutions were plotted onto the standard curve, and no differences in the final log10 copy numbers were found for different dilutions of the same sample. For patients with <2.5 copies/μl, it was not possible to determine the log10 number of copies because no band of HSV DNA was detected in the presence of competing IS. As shown in Table 1, the number of copies ranged from less than 2.5 × 101 to 1.8 × 104 copies/μl.
TABLE 1.
QC-PCR and clinical dataa
| Patient no. | Sex | Age (yr) | No. of HSV-1 DNA copies/μl | Time of disease (days) | Time of acyclovir (days) | Glasgow score | Detection of lesion by CT | Impairment after 3 mo |
|---|---|---|---|---|---|---|---|---|
| 1 | F | 48 | 1.8 × 104 | 7 | 2 | 14 | Yes | Moderate |
| 2 | M | 61 | 1.2 × 104 | 6 | 2 | 13 | Yes | Mild |
| 3 | F | 48 | 8.0 × 103 | 12 | 0 | 5 | Yes | Severe |
| 4 | M | 48 | 7.9 × 103 | 6 | 3 | 13 | Yes | Mild |
| 5 | M | 15 | 7.9 × 103 | 4 | 1 | 13 | Yes | Moderate |
| 6 | F | 59 | 3.1 × 103 | 11 | 0 | 13 | Yes | Moderate |
| 7 | M | 59 | 2.3 × 103 | 5 | 2 | 14 | Yes | Moderate |
| 8 | F | 59 | 1.9 × 103 | 5 | 1 | 6 | Yes | Death |
| 9 | M | 12 | 1.5 × 103 | 3 | 0 | 7 | Yes | Mild |
| 10 | F | 41 | 3.2 × 101 | 2 | 1 | 15 | No | Normal |
| 11 | F | 4 | 3.1 × 101 | 6 | 0 | 12 | No | Normal |
| 12 | F | 47 | <2.5 × 101 | 4 | 3 | 15 | Yes | Normal |
| 13 | M | 17 | <2.5 × 101 | 3 | 1 | 11 | No | Normal |
| 14 | F | 1.5 | <2.5 × 101 | 1 | 0 | 15 | No | Moderate |
| 15 | F | 44 | <2.5 × 101 | 10 | 4 | 15 | No | Normal |
| 16 | M | 22 | <2.5 × 101 | 1 | 0 | 14 | No | Normal |
Time of disease and time of acyclovir refer to the duration of disease and duration of therapy, respectively, at the time that the CSF was obtained. Glasgow scores refer to the level of consciousness at the time of enrollment. Patients were categorized as follows: 15, alert; 12 to 14, lethargic; 7 to 11, semicomatose; <7, comatose. F, female; M, male.
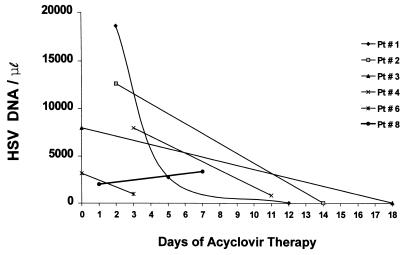
Consecutive CSF samples for QC-PCR was obtained from six patients. In order to avoid the effect of small variations in this analysis, all the samples obtained from the same patient were repeated in the same experiment. As shown in Fig. 3, five of six patients had a decline in HSV-1 DNA levels with treatment with acyclovir. The most rapid decline was observed in patient 1, from whose CSF 6.8 times fewer HSV-1 DNA copies were collected after 4 days of acyclovir therapy. Despite acyclovir therapy, the amount of HSV-1 DNA in the second CSF sample from patient 8 was higher than that in the first CSF sample. This patient died 8 days after antiviral treatment was introduced.
FIG. 3.
Quantity of HSV-1 DNA per microliter of CSF from patients (Pt) 1, 2, 3, 4, 6, and 8 according to duration of acyclovir therapy.
Correlation with severity of disease and outcome.
Table 2 correlates the clinical data and HSV-1 DNA levels in the first CSF sample. There was no significant difference in the gender distribution between patients with HSV-1 DNA amounts less than or greater than 102 copies/μl (P = 0.358; Fisher’s exact test). Patients with high levels of HSV-1 DNA received acyclovir for 1.22 ± 1.09 days, while the time of treatment among patients with low levels of viral DNA was 1.28 ± 1.6 days. The time of acyclovir therapy was not significantly different between these two groups (P = 0.869; Wilcoxon rank sum test). The mean duration of symptoms was 6.55 ± 3.04 days for patients with more than 102 HSV-1 DNA copies/μl and 3.85 ± 3.23 days for patients with fewer viral DNA copies (P = 0.0624; Wilcoxon rank sum test). The mean age was 45.44 ± 18.9 years among patients with high levels of HSV-1 DNA and 25.21 ± 19 years among patients with low levels of HSV-1 DNA (P = 0.0221; Wilcoxon rank sum test). A reduced level of consciousness was found for all patients with large amounts of HSV-1 DNA but was found for only three (42.86%) of the patients with low levels of viral DNA (P = 0.019; Fisher’s exact test). Lesions were found by CT in all patients with more than 102 HSV-1 DNA copies/μl but in only one (14.29%) patient in the group with smaller amounts of viral DNA (P = 0.000874; Fisher’s exact test). The outcome was also significantly different between these groups. No patient with more than 102 viral DNA copies/μl regained normal function, seven (77.78%) patients had mild or moderate levels of impairment, and two (22.22%) patients died or had severe levels of impairment. In contrast, six (85.71%) of the patients with smaller amounts of HSV-1 DNA returned to normal activities, and only one (14.29%) patient had moderate levels of impairment (P = 0.0021; Fisher’s exact test).
TABLE 2.
Correlation between HSV-1 DNA level, severity of disease, and outcomea
| DNA level | Age (yr) | Duration of disease (days) | Duration of acyclovir treatment (days) | No. (%) of patients with lesion de- tected by CT | No. (%) of patients with reduced level of consciousness | No. (%) of patients with the following outcomeb
|
||
|---|---|---|---|---|---|---|---|---|
| Healthy | Mild or moderate level of im- pairment | Severe level of impairment or death | ||||||
| >100 copies/μl | 45.4 ± 18.9 | 6.5 ± 3 | 1.2 ± 1 | 9 (100) | 9 (100) | 0 | 7 (77.8) | 2 (22.2) |
| <100 copies/μl | 25.2 ± 19 | 3.8 ± 3.2 | 1.3 ± 1.6 | 1 (14.86) | 3 (42.9) | 6 (85.7) | 1 (14.3) | 0 |
| P value | 0.0221c | 0.0624c | 0.869c | 0.000874d | 0.019d | |||
Duration of disease is the number of days that the patients were ill prior to obtaining the CSF specimen. Time of acyclovir treatment is the number of days that the patient had been treated prior to obtaining the CSF specimen. Reduced level of consciousness indicates the number of patients with a Glasgow Coma Scale score of <15 (see footnote a of Table 1).
The P value for outcome was 0.0021 (Fisher’s exact test).
Based on Wilcoxon rank sum test.
Based on Fisher’s exact test.
DISCUSSION
Quantitative analyses of viral load in vivo have been shown to be useful in the evaluation of several human viral infections (4, 5, 14, 16, 21, 28, 36). QC-PCR has been tested as a means of evaluating antiviral therapy and assessing disease progression, and for some infections, the quantitation of viral DNA can be necessary to distinguish between dormant and active infection (4, 5, 14, 16, 21, 30). This is not the case for HSV infection of the central nervous system (CNS), in which the qualitative detection of the HSV genome in CSF samples has a high correlation with active disease and is presently the best noninvasive method for the diagnosis of HSE. Nevertheless, few studies have addressed the potential utility of quantitation of HSV DNA in CSF samples from HSE patients as a prognostic marker and for assessing the response to antiviral therapy.
A semiquantitative assay that measured the intensity of a radioactively labeled probe found a correlation between the signal intensity and age as well as the clinical severity of disease (27). Ando et al. (2) have described a quantitative PCR assay for HSV-1 DNA which used known amounts of HSV DNA as an external control. However, such methods provide only a relative measurement and are susceptible to error due to sample variability, which affects amplification efficiency (26). The coamplification of an unrelated DNA sequence with the target DNA has been studied in the quantitation of other viruses (12); however, this method can also be inaccurate due to differences in the melting temperature and amplification efficiency between the target DNA and heterologous target sequences. The coamplification of an IS containing the same primer binding sites as the target DNA can overcome these problems because any interference within the reaction equally affects the amplification of the target DNA and the IS and does not affect the sample/IS ratio. Several studies have used an internal standard to quantitate HSV, cytomegalovirus, and human herpesvirus 6 (7, 13, 17). Those studies were able to quantitate viral DNA over a range of 3 to 6 logs by using radioactive labels or postamplification hybridization. The method used in the study described in this report was able to quantitate viral DNA only over a range of 10- to 20-fold and required dilution of the sample to determine the number of genomes present in the original CSF specimen. We assume that the lack of a detectable IS band in reactions with greater than a 20-fold excess of HSV DNA is due to the relatively high amount of DNA required for visualization in ethidium bromide gels. In this study, the logarithms of the ratio of sample/IS were plotted against the logarithms of known amounts of HSV-1 DNA. The results obtained with two controls suggest that this method is accurate for the quantitation of HSV DNA. First, the ratios on all standard curves were zero at approximately 2.4 logs, or 250 copies, of HSV DNA. A ratio of zero indicates equivalency between the amounts of IS and HSV DNA, and the amount of IS was known to be 250 copies. Second, when CSF was diluted two- or fourfold, the results obtained from the extrapolation of the standard curve indicated two- or fourfold less HSV DNA.
The data presented in Tables 1 and 2 support and extend previous observations regarding the diagnosis of HSE and the prognosis of patients with HSE. Among the 16 patients in this study, only 10 (63%) had evidence of a brain lesion by CT. Morawetz et al. (23) found that 8 of 10 patients with biopsy-proven HSE had some abnormality in preoperative CT scans. Thus, while CT scans fail to detect all patients with HSE, there is a striking correlation between patient outcome and evidence of lesions by CT. Among the 18 patients with abnormal CT scans, 9 had mild to moderate impairment, and 7 either died or had severe neurological sequelae; only 2 of 18 patients in whom lesions were detected by CT returned to normal. The quantitative results suggest that lesions result from higher levels of viral replication. Magnetic resonance imaging has also been shown to be useful in the evaluation of presumed HSE (9).
In this study gender, age, duration of disease, severity of disease, and outcome were compared between patients with amounts of HSV-1 DNA smaller or larger than 102 copies/μl. Although the duration of acyclovir treatment was previously shown to affect the intensity of product bands (3), in this study the duration of antiviral therapy was not significantly different between these two groups. Different doses of acyclovir could be another bias; however, all patients received the same therapeutic regimen. A more severe disease was found in the group of patients with greater than 102 HSV-1 copies/μl. The numbers of patients with reduced levels of consciousness and in whom focal lesions were detected by CT were significantly higher in this group (P = 0.019 and 0.000874, respectively; Fisher’s exact test). Also, this group of patients was significantly older. These findings are in agreement with those of a previous study in which the signal intensity obtained after hybridization was correlated with age and severity of disease (27). Patients with higher HSV-1 DNA levels also had a poor outcome compared with patients with low viral DNA levels (P = 0.0021; Fisher’s exact test). Although the difference was not statistically significant (P = 0.0624; Fisher’s exact test), the duration of disease was almost two times longer in the group of patients with greater than 102 HSV-1 DNA copies/μl. These data confirm the need for the early introduction of antiviral treatment in order to reduce the amount of virus within the CNS and improve the clinical outcome (33). It is apparent for this group of patients that viral loads above 102 copies of HSV-1 DNA/μl in the first CSF specimen correlates with a poor prognosis.
While the method described here is reasonably rapid, is not labor intensive, and does not require radioactivity or postamplification enhancement techniques, it does require expensive image analysis software. Until rapid, quantitative assays for HSV become available, the current methods are primarily restricted to research laboratories and retrospective studies. The data presented here suggest that a simple method might be useful in establishing the prognosis of HSE. All patients with >100 copies/μl were positive when the initial CSF was diluted 1/100 and had a lesion: that was detected by CT; therefore, the combination of these two observations would strongly suggest a poor outcome. We performed the assay with CSF obtained prior to antiviral therapy from five biopsy-positive patients enrolled in the Collaborative Antiviral Study Group study (22). Of the five patients, three were positive when the diluted CSF was tested and all had lesions on CT. Two patients were negative by PCR and did not have focal findings on CT. Follow-up for the first three patients indicated that two patients died within 2 months and that the third patient was classified as moderately impaired. Of the two patients with negative PCRs, one had a mild level of impairment and the other had a moderate level of impairment. Studies have shown that temporal abnormalities detected by CT scans increase the likelihood of a positive PCR; however, patients with such findings can be negative by PCR and some PCR-positive patients with less severe forms of HSE have normal CT scans (8).
The efficacy of acyclovir treatment is well established by large clinical trials (9, 31). Nonetheless, the rates of mortality and morbidity among HSE patients treated with acyclovir remain high. Both age and duration of disease when antiviral therapy is introduced are known to influence mortality and morbidity; however, other factors may also have a role in the clinical response to antiviral therapy. The primary virological factor is the resistance of HSV strains to acyclovir. The majority of cases of disease attributed to resistant HSV strains have occurred in immunocompromised patients, primarily AIDS patients, and is normally restricted to ulcerative mucocutaneous lesions (10, 11). However, there has been one report of the isolation of an acyclovir-resistant HSV strain from the CSF of a patient with a fatal CNS disease (15). The establishment of a method to estimate prospectively the levels of viral DNA in patients with HSE would be desirable since no isolate is available for sensitivity testing. In this study, late CSF specimens from six patients were available for assessment of the response to antiviral therapy by QC-PCR. A decline in the number of HSV-1 copies was observed in five patients, and all of them clinically responded to acyclovir therapy. One patient had almost twice as many HSV-1 DNA copies in the second CSF and died 8 days after the onset of acyclovir therapy. Although the number of patients prospectively evaluated by QC-PCR was small, these data suggest that this assay can be helpful in monitoring the response to antiviral therapy. One of the unknowns in the diagnosis of HSE by PCR is the effect of acyclovir on a positive signal. It is not uncommon to initiate therapy prior to obtaining the CSF specimen for PCR or to assay a CSF specimen obtained after the completion of therapy. Our previous experience suggested that a duration of antiviral therapy of less than 1 week does not negate a positive PCR result; however, the patients enrolled in this study received different antiviral agents at different doses. While it is impossible to draw valid conclusions from the results for the limited number of patients in this study, two points are warranted. First, all six patients received the currently recommend dosage of acyclovir for 14 days. Positive PCR results were obtained after 2, 3, and 8 days of antiviral therapy. Importantly, all six of the patients had high levels of HSV in their initial CSF and none of these patients returned to normal. Viral drug resistance should be suspected and studied for patients with no decline in viral DNA levels with treatment. Indeed, this method can be useful in the evaluation of new antiviral drugs for the treatment of HSE.
In conclusion, the competitive PCR described here is a reliable method for the quantitation of HSV-1 DNA in CSF samples from patients with HSE. This assay can be used as prognostic marker since patients with higher viral loads in their CSF are more likely to develop more severe neurological impairment. Also, this method was shown to be of potential utility for monitoring antiviral therapy in patients with HSE. Further studies should address the use of QC-PCR in anticipating the clinical relapse of encephalitis permitting the prompt reintroduction of antiviral therapy (19, 25).
ACKNOWLEDGMENTS
This work was supported by the following institutions and grants: contract 201330/95-4 from CNPq, São Paulo, Brazil; grant 94/1776-0 from FAPESP, São Paulo, Brazil; contracts N01-AI-15113, N01-AI-62554, and N01-AI-12667 from the Antiviral Research Branch of the National Institute of Allergy and Infectious Diseases; a grant from the Division of Research Resources (RR-032) from the National Institutes of Health; and a grant from the state of Alabama.
We gratefully acknowledge collaboration with the following physicians and institutions: Paulo E. Marchiori, Getúlio Rabello, and Milberto Scaff Department of Neurology, University of São Paulo, and Vicente Amato Neto.
REFERENCES
- 1.Anderson N E, Powell K F, Croxson M C. A polymerase chain reaction assay of cerebrospinal fluid in patients with suspected herpes simplex encephalitis. J Neurol Neurosurg Psychiatry. 1993;56:520–525. doi: 10.1136/jnnp.56.5.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando Y, Kimura H, Miwata H, Kuto T, Shibata M, Morishima T. Quantitative analysis of herpes simplex virus DNA in cerebrospinal fluid of children with herpes simplex encephalitis. J Med Virol. 1993;41:170–173. doi: 10.1002/jmv.1890410214. [DOI] [PubMed] [Google Scholar]
- 3.Aurelius E, Johansson B, Sköldenberg B, Staland A, Forsgren M. Rapid diagnosis of herpes simplex encephalitis by nested polymerase chain reaction assay of cerebrospinal fluid. Lancet. 1991;337:189–192. doi: 10.1016/0140-6736(91)92155-u. [DOI] [PubMed] [Google Scholar]
- 4.Boivin G, Chou S, Quirk M R, Erice A, Jordan M C. Detection of ganciclovir resistance mutations and quantitation of cytomegalovirus (CMV) DNA in leukocytes of patients with fatal disseminated CMV disease. J Infect Dis. 1996;173:523–528. doi: 10.1093/infdis/173.3.523. [DOI] [PubMed] [Google Scholar]
- 5.Boivin G, Olson C A, Quirk M R, Kringstad B, Hertz M I, Jordan M C. Quantitation of cytomegalovirus DNA and characterization of viral gene expression in bronchoalveolar cells of infected patients with and without pneumonitis. J Infect Dis. 1996;173:1304–1312. doi: 10.1093/infdis/173.6.1304. [DOI] [PubMed] [Google Scholar]
- 6.Caparros-Lefebvre D, Dewilde A, Guffond T, Berier A, Vallee L, Wattre P, Petit H. Valeur de l’amplification génetique du virus herpétique dans le diagnostic et le traitment des encéphalites aiguës virales. Rev Neurol. 1995;151:124–128. [PubMed] [Google Scholar]
- 7.Clark D A, Ait-Khaled M, Wheeler A C, Kidd I M, McLaughlin J E, Johnson M A, Griffiths P D, Emery V C. Quantification of human herpesvirus 6 immunocompetent persons and post-mortem tissues from AIDS patients by PCR. J Gen Virol. 1996;77:2271–2275. doi: 10.1099/0022-1317-77-9-2271. [DOI] [PubMed] [Google Scholar]
- 8.Domingues R B, Tsanaclis A M C, Pannuti C S, Mayo M S, Lakeman F D. Evaluation of the range of clinical presentations of herpes simplex encephalitis by using polymerase chain reaction assay of cerebrospinal fluid samples. Clin Infect Dis. 1997;25:86–91. doi: 10.1086/514494. [DOI] [PubMed] [Google Scholar]
- 9.Domingues, R. B., M. C. D. Fink, A. M. C. Tsanaclis, C. C. de Castro, G. G. Cerri, M. S. Mayo, and F. D. Lakeman. Diagnosis of herpes simplex encephalitis by magnetic resonance imaging and polymerase chain reaction assay of cerebrospinal fluid. J. Neurol. Sci., in press. [DOI] [PubMed]
- 10.Englund J A, Zimmerman M E, Swierkosz E M, Goodman J L, Scholl D R, Balfour H H. Herpes simplex virus resistant to acyclovir. Ann Intern Med. 1990;112:416–422. doi: 10.7326/0003-4819-76-3-112-6-416. [DOI] [PubMed] [Google Scholar]
- 11.Erlich K S, Mills J, Chatis P, Mertz G J, Busch D F, Follansbe S E, Grant R M, Crumpacker C S. Acyclovir-resistant herpes simplex virus infections in patients with the acquired immunodeficiency syndrome. N Engl J Med. 1989;320:293–296. doi: 10.1056/NEJM198902023200506. [DOI] [PubMed] [Google Scholar]
- 12.Ferre, F., A. L. Marchese, S. L. Griffin, A. E. Daigle, S. P. Richieri, F. C. Jensen, and D. J. Carlo. 1993. Development and validation of a polymerase chain reaction method for the precise quantitation of HIV-1 DNA in blood cells from subjects undergoing a 1-year immunotherapeutic treatment. AIDS 7(Suppl. 2):S21–S27. [DOI] [PubMed]
- 13.Fox J C, Griffiths P D, Emery V C. Quantification of human cytomegalovirus DNA using the polymerase chain reaction. J Gen Virol. 1992;73:2405–2408. doi: 10.1099/0022-1317-73-9-2405. [DOI] [PubMed] [Google Scholar]
- 14.Fox J C, Kidd I M, Griffiths P D, Sweny P, Emory V C. Longitudinal analysis of cytomegalovirus load in renal transplant recipients using a quantitative polymerase chain reaction: correlation with disease. J Gen Virol. 1995;76:309–319. doi: 10.1099/0022-1317-76-2-309. [DOI] [PubMed] [Google Scholar]
- 15.Gateley A, Gander R M, Johnson P C, Kit S, Otsuka H, Kohl S. Herpes simplex type 2 meningoencephalitis resistant to acyclovir in a patient with AIDS. J Infect Dis. 1990;161:711–715. doi: 10.1093/infdis/161.4.711. [DOI] [PubMed] [Google Scholar]
- 16.Gerna G, Baldanti F, Sarasini A, Furione M, Percivalle E, Revello M G, Zipeto D, Zella D. Effect of foscarnet treatment on quantitation of human cytomegalovirus (HCMV) DNA in peripheral blood polymorphonuclear leukocytes and aqueous humor of AIDS patients with HCMV retinitis. Antimicrob Agents Chemother. 1994;38:38–44. doi: 10.1128/aac.38.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobson A, Wald A, Wright N, Corey L. Evaluation of a quantitative competitive PCR assay for measuring herpes simplex virus DNA content in genital tract secretions. J Clin Microbiol. 1997;35:548–552. doi: 10.1128/jcm.35.3.548-552.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kessler H H, Pierer K, Weber B, Sakrauski B, Santner B, Stuenzner D, Gergely E, Marth E. Detection of herpes simplex virus DNA from cerebrospinal fluid by PCR and a rapid, nonradioactive hybridization technique. J Clin Microbiol. 1994;32:1881–1886. doi: 10.1128/jcm.32.8.1881-1886.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura H, Aso K, Kuzushima K, Hanada N, Shibata M, Morishima T. Relapse of herpes simplex encephalitis in children. Pediatrics. 1992;89:891–894. [PubMed] [Google Scholar]
- 20.Klapper P E, Cleator G M, Dennet C, Lewis A G. Diagnosis of herpes encephalitis via Southern-blotting of cerebrospinal fluid DNA amplified by polymerase chain reaction. J Med Virol. 1990;32:261–264. doi: 10.1002/jmv.1890320413. [DOI] [PubMed] [Google Scholar]
- 21.Kojima, E., T. Shirasaka, B. Anderson, S. Chokekijchai, S. Sei, R. Yarchoan, and H. Mitsuya. 1993. Monitoring the activity of antiviral therapy for HIV infection using a polymerase chain reaction method coupled with reverse transcription. AIDS 7(Suppl. 2):S101–S105. [DOI] [PubMed]
- 22.Lakeman F D, Whitley R J. Diagnosis of herpes simplex encephalitis: application of polymerase chain reaction to cerebrospinal fluid from brain-biopsed patients and correlation with disease. J Infect Dis. 1995;171:857–863. doi: 10.1093/infdis/171.4.857. [DOI] [PubMed] [Google Scholar]
- 23.Morawetz R B, Whitley R J, Murphy D M. Experience with brain biopsy for suspected herpes encephalitis: a review of forty consecutive cases. Neurosurgery. 1983;12:654–657. doi: 10.1227/00006123-198306000-00011. [DOI] [PubMed] [Google Scholar]
- 24.Nahmias A J, Whitley R J, Visintine A N, Takei Y, Alford C A. Herpes simplex virus encephalitis laboratory evaluations and their diagnostic significance. J Infect Dis. 1982;145:829–836. doi: 10.1093/infdis/145.6.829. [DOI] [PubMed] [Google Scholar]
- 25.Nicolaidou P, Iacovidou N, Youroukos S, Liancopopou-Tsitsipi T, Kattanis C. Relapse of herpes simplex encephalitis after acyclovir therapy. Eur J Pediatr. 1993;152:737–738. doi: 10.1007/BF01953988. [DOI] [PubMed] [Google Scholar]
- 26.Piatak M, Jr, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 27.Puchhammer-Stöckl E, Heinz F X, Kund I M, Popow-Kraupp T, Grimm G, Millner M M, Kunz C. Evaluation of polymerase chain reaction for diagnosis of herpes simplex virus encephalitis. J Clin Microbiol. 1993;31:146–148. doi: 10.1128/jcm.31.1.146-148.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen L, Sipeto D, Wolitz R A, Dowling A, Dfron B, Merigan T C. Risk for retinitis in patients with AIDS can be assessed by quantitation of threshold levels of cytomegalovirus DNA burden in blood. J Infect Dis. 1997;176:1146–1155. doi: 10.1086/514106. [DOI] [PubMed] [Google Scholar]
- 29.Rowley A H, Whitley R J, Lakeman F D, Wolinsky S M. Rapid detection of herpes simplex virus DNA in cerebrospinal fluid of patients with herpes simplex encephalitis. Lancet. 1990;35:440–441. doi: 10.1016/0140-6736(90)90667-t. [DOI] [PubMed] [Google Scholar]
- 30.Secchiero P, Zella D, Crowley R W, Gallo R C, Russo P. Quantitative PCR for human herpesviruses 6 and 7. J Clin Microbiol. 1995;33:2124–2130. doi: 10.1128/jcm.33.8.2124-2130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitley R J, Alford C A, Hirsh M S, Schooley R T, Luby J P, Aoki F Y, Hanley D, Nahmias A J, Soong S-J NIAID Collaborative Antiviral Study Group. Vidarabine versus acyclovir therapy in herpes simplex encephalitis. N Engl J Med. 1986;314:144–149. doi: 10.1056/NEJM198601163140303. [DOI] [PubMed] [Google Scholar]
- 32.Whitley R J, Cobbs C G, Alford C A, Soong S-J, Morawetz R, Benton J W, Hirsch M S, Reichman R C, Aoki F Y, Connor J, Oxman M, Corey L, Hanley D F, Wright P F, Levin M, Nahmias A, Powell D A NIAID Collaborative Antiviral Study Group. Diseases that mimic herpes simplex encephalitis-diagnosis, presentation and outcome. JAMA. 1989;262:234–239. [PubMed] [Google Scholar]
- 33.Whitley R J, Gnann J W. Acyclovir: a decade later. N Engl J Med. 1992;327:782–789. doi: 10.1056/NEJM199209103271108. [DOI] [PubMed] [Google Scholar]
- 34.Whitley R J, Lakeman F D. Herpes simplex virus infections of the central nervous system therapeutic and diagnostic considerations. Clin Infect Dis. 1995;20:414–420. doi: 10.1093/clinids/20.2.414. [DOI] [PubMed] [Google Scholar]
- 35.Whitley R J, Soong S-J, Linneman C, Liu C, Pazin G, Alford C A. Herpes simplex encephalitis—clinical assessment. JAMA. 1982;247:317–320. [PubMed] [Google Scholar]
- 36.Zaia J A, Gallez-Hawkins G M, Tegtmeier B R, ter Vee R A, Li X, Niland J C, Forman S J. Late cytomegalovirus disease in marrow transplantation is predicted by virus load in plasma. J Infect Dis. 1997;176:782–785. doi: 10.1086/517301. [DOI] [PubMed] [Google Scholar]