Abstract
Catalytic asymmetric α-alkylation of carbonyl compounds represents a long-standing challenge in synthetic organic chemistry. Herein, we advance a dual biocatalytic platform for the efficient asymmetric alkylation of α-keto acids. First, guided by our recently obtained crystal structures, we develop SgvMVAV as a general biocatalyst for the enantioselective methylation, ethylation, allylation and propargylation of a range of α-keto acids with total turnover numbers (TTNs) up to 4,600. Second, we mine a family of bacterial HMTs from Pseudomonas species sharing less than 50% sequence identities with known HMTs and evaluated their activities in SAM regeneration. Our best performing HMT from P. aeruginosa, PaHMT, displays the highest SAM regeneration efficiencies (TTN up to 7,700) among HMTs characterized to date. Together, the synergistic use of SgvMVAV and PaHMT affords a fully biocatalytic protocol for asymmetric methylation featuring a record turnover efficiency, providing a solution to the notorious problem of asymmetric alkylation.
Subject terms: Biocatalysis, Synthetic chemistry methodology, Biocatalysis
Catalytic asymmetric a-alkylation of carbonyl compounds represents a longstanding challenge in synthetic organic chemistry but general methods for the catalytic symmetric alkylation of carbonyl compounds remain elusive. Here, the authors mine and engineer methyltransferases as synthetically useful biocatalysts for the catalytic asymmetric alkylation reactions of carbonyl compounds.
Introduction
As catalysts capable of exerting exquisite control over the stereochemical outcome of chemical reactions under mild and benign conditions, enzymes hold the potential to revolutionize the practice of asymmetric synthesis. Over the past three decades, the advent of powerful protein engineering technologies1–3 has enabled the rapid development of customized enzymes with excellent activity and stereoselectivity toward non-native substrates, furnishing exciting biocatalytic solutions to synthetic problems4. However, compared to the powerful toolbox of abiotic chemistry discovered and optimized by synthetic chemists, the reaction type and substrate scope of biocatalysts still remain limited5. In particular, in contrast to the widespread application of enzymatic functional group manipulation, synthetically useful biocatalytic methods to form C–C bonds6,7—an essential process to forge the backbone of organic compounds—remains largely underdeveloped.
Among the various C–C bond forming reactions, the α-alkylation of carbonyl compounds is a fundamentally important process taught in introductory organic chemistry courses8,9. These reactions lead to the formation of a stereogenic center adjacent to the synthetically versatile carbonyl group, and are widely utilized in the construction of complex molecular architectures, including bioactive natural products and clinically important therapeutics10. Despite the apparent simplicity and fundamental importance, general methods for the catalytic asymmetric alkylation of carbonyl compounds have eluded synthetic chemists for decades9. Previously developed and widely implemented approaches for the asymmetric alkylation of carboxylic acid derivatives typically hinge on the use of a stoichiometric amount of chiral auxiliaries, such as the Evans8,11 and the Myers auxiliaries12–15, or Koga16–19 and Zakarian’s20–22 chiral polydentate amines to achieve excellent stereocontrol (Fig. 1a–c). Furthermore, the enantioselective incorporation of the smallest alkyl group, namely the methyl group, into the α-position of carbonyls represents a disproportionately large hurdle facing synthetic chemists23. As the introduction of a methyl substituent into medicinal agents can dramatically enhance their bioactivity due to the “magic methyl effect”24,25, methods allowing for selective methylation are particularly valuable to medicinal chemists.
Fig. 1. Asymmetric α-methylation and α-alkylation of carboxylic acid derivatives: state-of-the-art methods based on stoichiometric chiral auxiliaries and chiral controllers, and our dual biocatalytic platform for asymmetric α-alkylation.
a Use of chiral auxiliaries: Evans’s oxazolidinone. b Use of chiral auxiliaries: Myers’s pseudoephedrine. c Use of chiral controllers: Koga/Zakarian’s chiral polydentate amine-supported organolithium reagents. d Development of a dual biocatalytic platform consisting of optimized CMT and HMT for asymmetric α-alkylation using alkyl halides as the alkyl donor. CMT carbon methyltransferase, HMT halogen methyltransferase.
To address these challenges, we initiated a research program to mine and engineer methyltransferases as synthetically useful biocatalysts for the catalytic asymmetric α-methylation and other alkylation reactions of carbonyl compounds (Fig. 1d). Nature employs methylation as a central mechanism to regulate biology26,27. Thus, a large variety of methyl transfer enzymes has been evolved in the biological world. Our inspiration came from a largely underutilized enzyme family using S-adenosylmethionine (SAM), an organic cofactor bearing a highly electrophilic and stereogenic sulfonium ion, to catalyze selective methyl transfer28–31. We envisioned a fully biocatalytic platform consisting of an engineered SAM-dependent carbon methyltransferase (CMT)28 and a highly efficient halogen methyltransferase (HMT)32–34 to facilitate the asymmetric α-alkylation of carbonyl compounds using readily available alkyl electrophiles as the terminal alkyl donor. In this biocatalytic asymmetric alkylation, the CMT enzyme catalyzes the stereocontrolled delivery of the alkyl group using a sulfonium cofactor shuttle. The HMT enzyme then regenerates the sulfonium cofactor from S-adenosylhomocysteine (SAH) using alkyl (pseudo)halides, as recently elegantly demonstrated by Seebeck, Bornscheuer and Hammer35–40, and completes the catalytic cycle. Herein, we describe the implementation of this proposal through the mining, structural analysis, and protein engineering of carbon methyltransferases and halogen methyltransferases. First, through structure-guided enzyme engineering, we developed a general, selective, and highly efficient biocatalyst SgvMVAV for asymmetric alkylation. Furthermore, through the mining of a family of bacterial HMTs from Pseudomonas species, we discovered one of the best SAM regeneration enzymes, PaHMT, allowing record turnover efficiencies to be accomplished.
Results and discussion
We focused our initial efforts on a family of SAM-dependent CMTs, whose native function is to enantioselectively introduce a methyl unit onto the α-position of keto acids (Figs. 1d and 2a). In particular, pioneering research from Müller and coworkers demonstrated the utility of wild-type SgvM41 in effecting asymmetric α-methylation and ethylation42. From a synthetic perspective, the α-keto acid moiety provides an excellent functional group handle, allowing convenient access to various valuable products bearing an α- and/or β-stereogenic center. Additionally, the α-keto acid substrates are easily accessible from the corresponding carboxylic acids using Bode’s methods43–45. Despite the synthetic potential of SAM-dependent CMTs from this class, previously characterized wild-type (wt) enzymes only accept native substrates or a narrow range of α-keto acids which are sterically and electronically similar to the native substrate41,42,46–51. Thus, we set out to develop general and synthetically useful biocatalysts for asymmetric alkylation through the mining and engineering of CMTs.
Fig. 2. Protein structure and engineering of SgvM toward a general enantioselective methylation biocatalyst.
a Phylogenetic analysis and evaluation of wt CMTs. Original organism: MarI: Streptomyces sp. B9173, MppJ: S. hygroscopicus, SgvM: S. griseoviridis, CdpJ: Chondromyces crocatus. b Overview of the structure of wt SgvM. c SgvM in complex with α-ketoleucine and methylthioadenosine (MTA). A simulated annealing omit map with ligand coordinates omitted is superimposed at 3σ above background in blue wire-mesh. d SgvM in complex with α-ketoleucine and S-adenosylhomocysteine (SAH). A simulated annealing omit map with ligand coordinates omitted is superimposed at 3σ above background in blue wire-mesh. e Superposition of the SAH- and MTA-bound structures of SgvM. f Structure-guided engineering of SgvM: development of the triple mutant SgvM F329V T331A M144V (SgvMVAV). Reactions were performed in triplicates and data are shown as mean ± SD (n = 3).
To commence our investigation, we performed phylogenetic analysis and assembled a library of CMTs, including MarI46, MppJ47,48, SgvM41,42, CdpJ49, MrsA50, and GlmT51 with distinct native ketoacid substrates (see Supplementary Fig. 2). While all these CMTs could be successfully expressed using E. coli as the microbial host, MppJ and SgvM were produced at higher expression levels ( > 100 mg/L TB culture). To develop generally applicable alkylation biocatalysts, we assayed the activity of these CMTs using 2-oxo-4-phenylbutanoic acid (1a), a non-native α-keto acid bearing a large pedant phenyl substituent, as the model substrate (Fig. 2a). In our initial studies, we used methionine adenosyltransferase from E. coli (EcMAT) or Bacillus subtilis (BsMAT) to generate SAM from ATP and 5′-methylthioadenosine/S-adenosylhomocysteine (MTA/SAH) nucleosidase from E. coli (EcMTAN) to degrade SAH. Among all the wild-type CMTs we examined (Supplementary Table 1), MarI (Fig. 2a, entry 1) and SgvM (entry 3) furnished measurable activities, with SgvM providing the highest yield of 2a (5%). SgvM is found in the biosynthetic gene cluster of the antibiotic viridogrisein and catalyzes methyl transfer to the C3 (i.e., β) position of α-ketoleucine (4-methyl-2-oxovalerate)41.
In light of its highest initial activity and substantially better expression level (ca. 150 mg/L TB culture) than MarI, we focused our efforts on the engineering of SgvM to further enhance its efficiency to catalyze a wider range of enantioselective biocatalytic methylation processes. To facilitate this protein engineering, we first determined the structure of wild-type SgvM by heavy atom phasing and solved several co-crystal structures with bound cofactors and substrates (Fig. 2b–e). The structure of SgvM captured in a non-productive complex with 5’-methylthioadenosine (MTA), likely derived from SAM hydrolysis, and α-ketoleucine has been reported in a published thesis but the atomic coordinates have not been deposited in any data bank52. Hence, we first determined the 2.13 Å resolution structure of wt SgvM in complex with MTA and α-ketoleucine. The overall structure consists of a two-domain fold that assembles into homodimer in both the solution and the crystal (Fig. 2b). The N-terminal domain (I6 through H147) consists of helices that mediate dimerization, while the C-terminal domain (F149 through H338) consists of a Rossman fold.
A single metal ion is located at the base of the C-terminal domain and is ligated by H244 and H296 (Fig. 2c). Anomalous difference Fourier maps calculated from data collected at the zinc absorption edge, and the tetrahedral coordination are consistent with a bound zinc ion. The zinc is likely integral to the enzyme as no exogenous metals were added during purification or crystallization. When modeled as zinc ion at full occupancy, the thermal (B) factors of the metal are consistent with those of the rest of the polypeptide. Presumably, the metal ion acts as a Lewis acid to trigger the soft enolization of the ketoacid substrate and facilitates electrophilic methylation. Density corresponding to the α-ketoleucine substrate can be observed directly bound to the metal. However, SAM could not be observed in the structure but rather density for MTA, likely derived from SAM hydrolysis, is seen in the near vicinity of the metal. The distance from the MTA sulfur atom to C3 position of α-ketoleucine is ca. 7 Å, suggesting that this represents a nonproductive complex. This non-productive complex resembles that observed in the previously reported structure of SgvM.
We next determined the structure of wt SgvM in complex with SAH and α-ketoleucine (2.2 Å resolution, Fig. 2d). A superposition with the MTA bound structure shows that the N-terminal domain has undergone a marked shift resulting in an overall closed conformation (Fig. 2e). The cofactor now resides in proximity to the substrate (3.7 Å distance between the sulfur of SAH and C3 of α-ketoleucine). The (Re)-face of the zinc enolate is shielded by the protein scaffold, while the (Si)-face is exposed to the methylating cofactor, thereby allowing methyl transfer to occur with excellent π-facial selectivity (Fig. 2d and Fig. 3c).
Fig. 3. Engineered triple mutant alkyl transfer enzyme SgvMVAV: substrate scope and structural insights.
a SgvMVAV-catalyzed enantioselective methylation and various other alkylations of α−keto acids. b Crystal structure of triple mutant SgvMVAV in complex with model substrate 1a. A simulated annealing omit map with ligand coordinates omitted is superimposed at 3σ above background in blue wire-mesh. c Enantioinduction mechanism with SgvMVAV.
With these enzyme structures, we embarked on structure-guided protein engineering of SgvM (Fig. 2f). In wt SgvM-SAH-α-ketoleucine complex, the isopropyl chain of the substrate is in close contact with two residues, including S141 and F329. While S141 is located in an α-helix, the bulky aromatic amino acid residue F329 resides in a flexible loop. Upon further examining the crystal structure of SgvM, we found that the F329-containing flexible loop encapsulates the substrate binding pocket. We thus posited that flexible loop engineering53,54 can dramatically broaden the substrate scope of SgvM and furnish a general biocatalyst for enantioselective methylation. Additionally, multiple sequence alignment showed that this aromatic residue of SgvM (F329) is not conserved in the flexible loop of CMTs of this family (Supplementary Fig. 3), further supporting our envisioned flexible loop engineering approach.
Indeed, introducing a single F329A mutation into SgvM led to a 57-fold increase in total turnover number (TTN) for the biocatalytic transformation of 1a, confirming the critical role of residue 329 in modulating protein-substrate interaction (Fig. 2f, wt SgvM: TTN = 6, SgvM F329A: TTN = 340). We subsequently replaced F329 with residues possessing varying side chain sterics (Fig. 2f, Supplementary Fig. 6), including glycine (G), leucine (L), isoleucine (I) and valine (V), leading to the identification of SgvM F329V with the highest activity (TTN = 780). Encouraged by these results, we next evaluated the role of T331 in the same flexible loop (Supplementary Fig. 5). It was found that T331A and T331V had similar activating effects on the conversion of 1a, giving rise to TTNs of 32 and 70, respectively. Although the recombination of F329V and T331V led to inferior results, double mutant SgvM F329V T331A displayed a higher catalytic activity relative to the corresponding single mutants (TTN = 810). Further active site engineering showed that a single M144V mutation significantly improved enzyme activity (TTN = 1080). M144 resides in an α-helix proximal to the phenyl group of 1a (Supplementary Fig. 5). Ultimately, triple mutant SgvM F329V T331A M144V (SgvMVAV) exhibited the highest activity, furnishing excellent yield and enantiomeric ratio (e.r.) of 1a (TTN = 1,410, 93% yield and 99:1 e.r.). Compared to wt SgvM, SgvMVAV provided a 235-fold improvement in TTN, demonstrating its substantially enhanced catalytic efficiency.
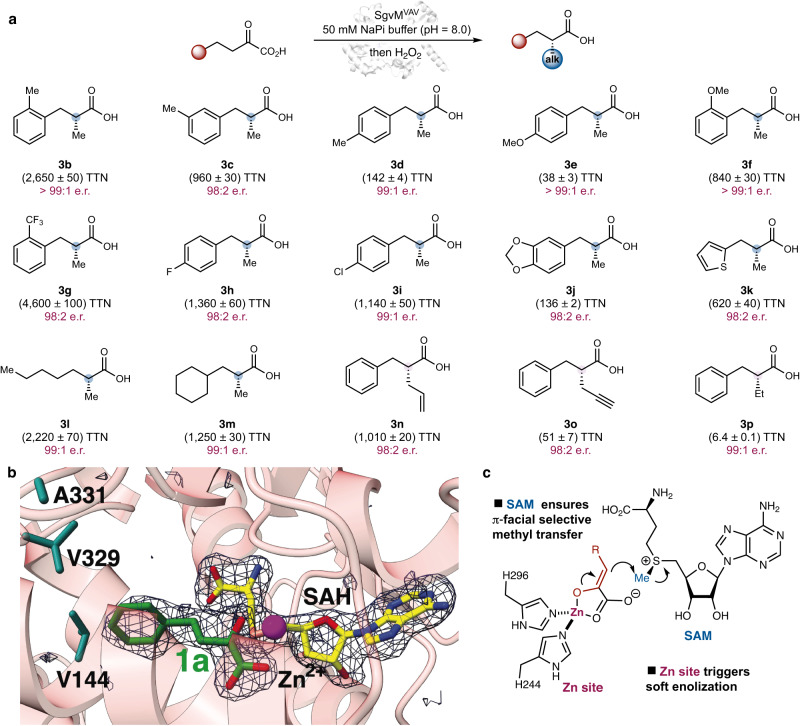
With this engineered SgvMVAV variant, we examined the substrate scope of this enantioselective α-methylation process (Fig. 3a). In this study, alkylated α-ketoacids (2a−2p) were converted into the corresponding carboxylic acids (3a−3p) (vide infra). It was found that keto acids bearing an ortho- and a meta-substituent (1b and 1c) were excellent substrates, affording methylated products 3b and 3c with good to excellent TTN and e.r. For para-substituted substrates 1d and 1e, SgvMVAV displayed reduced activities but high levels of enantioselectivity (3d and 3e). Electron-donating methoxy (3f) and electron-withdrawing trifluoromethyl (3g) groups of the aromatic ring were found to be compatible. Other functional groups, including a fluorine (3h) and a chlorine (3i), were also tolerated. Additionally, heterocycles such as a benzodioxole (3j) and a thiophene (3k) were successfully converted with excellent enantiocontrol. Finally, other aliphatic substrates underwent smooth enzymatic transformation to furnish methylated products (3l and 3m) in excellent enantioselectivity. To our knowledge, the total turnover numbers of our engineered SgvM triple mutant described herein (up to 4,600 TTN, 3g) represented a record high for SAM-dependent carbon methyltransferases.
Moreover, we found that without additional protein engineering, SgvMVAV readily allowed asymmetric α-allylation to occur with excellent activity and enantiocontrol using the S-allyl analog of SAM (3n)28. In light of the synthetic versatility of the allyl moiety and the central role of enantioselective allylation in asymmetric synthesis55,56, this result represented a synthetically useful advance. Furthermore, enantioselective α-propargylation and ethylation were accomplished with excellent enantioselectivity, although with lower activity. Together, these results demonstrated the excellent potential of SgvMVAV variant for diverse asymmetric α-alkylative functionalization processes. Further engineering of alkyl transfer biocatalysts derived from SgvMVAV with enhanced catalytic activities is currently underway in our laboratory.
In order to provide further insights into the broader substrate scope and enhanced total turnover number of SgvMVAV, we determined its crystal structure in complex with SAH and 1a (2.04 Å resolution, Fig. 3b). Protein crystallography showed that although the three amino acid substitutions led to limited alterations in the overall structure, they resulted in a substantially widening of the binding pocket to accommodate larger substrates (Supplementary Fig. 7). Meanwhile, interactions between the protein, the metal ion and the bound SAH cofactor are sufficient to accommodate these dramatic changes in the active site contours with minimal perturbations to the overall scaffold. Taken together, the expanded substrate binding pocket and the retained overall enzyme scaffold allowed for a broad range of substrates to be effectively transformed, leading to a generally useful enantioselective alkylation biocatalyst.
Next, we sought to couple this engineered CMT with a halogen methyl transferase (HMT)-based cofactor regeneration system to develop a fully biocatalytic asymmetric methylation system using methyl iodide as the methyl donor (Fig. 4). Based on sequence similarity network (SSN) analysis, we focused on the mining of previously unexplored putative halogen methyl transferases from Pseudomonas species (Fig. 4a). Annotated as thiopurine S-methyltransferase (TPMT), these SAM-dependent methyltransferases display less than 50% sequence identities with other previously mined and biochemically characterized HMTs and are thus distinct SAM-dependent enzymes (Fig. 4a, see Supplementary Fig. 4 for a phylogenetic tree). We cloned five putative HMTs from this subfamily, and expressed them in our engineered E. coli BL21(DE3)Δmtn strain (Supplementary Fig. 1). Compared to previously investigated bacterial, fungal and plant HMTs, these Pseudomonas HMTs showed higher production levels when heterologously expressed in E. coli, thus indicating their potential as practical biocatalysts in biotechnology.
Fig. 4. Development of a fully biocatalytic platform for asymmetric α-methylation with a record turnover efficiency.
a Mining of Pseudomonas HMTs: sequence similarity network (SSN) of selected putative HMTs. b Dual biocatalytic enantioselective methylation using SgvMVAV and HMTs mined from Pseudomonas species. PaHMT: P. aeruginosa HMT, PeHMT: P. entomophila HMT, PfHMT: P. fluorescens HMT, PpHMT: P. putida HMT, PsHMT: P. syringae HMT. a0.01 mol% PaHMT was used. c SAM regeneration efficiency with HMTs from Pseudomonas sp.
Furthermore, it was found that our Pseudomonas HMTs exhibited promising activities toward SAM regeneration from SAH and methyl iodide (Fig. 4b, c). In particular, HMT from P. aeruginosa (Fig. 4b, entry 1) showed the highest SAM regeneration efficiency in the dual enzyme system among mined HMTs of this family (entries 1−5) as well as previously studied bacterial, fungal and plant HMTs (entries 6−9)35,37,39,40. Notably, in the presence of 0.1 mol% SgvMVAV, 0.02 mol% PaHMT, 0.1 mol% SAH and 2 equiv of methyl iodide, enantioenriched 2a formed in 95% yield and 99:1 e.r. (entry 1). This corresponds to a TTN of 4,750 for PaHMT and a TTN of 950 for SgvMVAV. By further lowering the PaHMT loading to 0.01 mol%, its TTN could be further increased to 7,700 (entry 1). To our knowledge, these TTNs represented the highest turnover efficiencies for dual biocatalytic methylation processes of this type. Additionally, with 0.1 mol% engineered SgvMVAV and 0.02 mol% PaHMT, the total turnover number of the SAH cofactor approached 1,000, which is of the same order of magnitude for practical NAD(P)H-regeneration systems57. Furthermore, PaHMT also showed the highest relative activity in SAM regeneration from SAH and methyl iodide among HMTs studied herein (Fig. 4c). Taken together, the excellent catalytic efficiency and enantioselectivity of our optimized dual biocatalytic system consisting of SgvMVAV and PaHMT showcased the synthetic potential of SAM-dependent methyl transfer enzymes for asymmetric C−C bond formation.
To further demonstrate the synthetic utility of this fully biocatalytic asymmetric alkylation system, we carried out preparative scale biocatalytic synthesis and cascades (Fig. 5). Enantioenriched keto acid 2a was prepared on a 100 mg scale with 91% yield and 99:1 e.r., showcasing the synthetic utility of this enzymatic process. On the basis of this result, a range of derivatization reactions was successfully carried out. First, oxidative decarboxylation of α-keto acid 2a using hydrogen peroxide furnished carboxylic acid 3a in quantitative yield and 99:1 e.r.. Moreover, taking advantage of the synthetically versatile ketoacid moiety, biocatalytic cascades were successfully carried out to afford stereochemical dyads with excellent diastereo- and enantioselectivities (4a and 4b). By coupling the SgvMVAV-PaHMT dual enzyme system with a ketoacid reductase58, α-hydroxy-β-methyl acid 4a with adjacent stereogenic centers formed in 76% yield, 19:1 d.r. and >99:1 e.r.. Additionally, cascade enantioselective methylation/diastereoselective transamination59 furnished α-amino-β-methyl acid 4b with vicinal stereocenters in 68% yield, >20:1 d.r. and >99:1 e.r. We note that using chemical methods, low levels of diastereoselectivity (d.r. = ca. 1:1) were observed for hydroxyl acid and amino acid synthesis, thus highlighting the power of biocatalytic cascades to access well-defined stereochemical dyads. Together, these chemical and biocatalytic transformations demonstrated the value of the α-keto acid functional group handle to access a wide range of synthetically and medicinally valuable scaffolds.
Fig. 5. Preparative-scale biotransformations and biocatalytic cascade reactions of α-keto acids.
Conditions: a SgvMVAV, PaHMT, MeI. b H2O2, then catalase. c KmAT, L-glutamate, NaPi buffer (pH = 8.0), 30 °C, 24 h. d YiaE, NADP+, NaPi buffer (pH = 7.5), Codex cofactor recycling mix, 30 °C, 24 h.
In conclusion, we developed a dual biocatalytic platform composed of our engineered SAM-dependent CMT and mined HMT for the efficient catalytic asymmetric alkylation of α-keto acids. First, SgvMVAV was engineered as a general biocatalyst for the asymmetric α-alkylation of α-keto acids, displaying record total turnover numbers (TTN up to 4,600) and excellent enantioselectivities across a diverse range of substrates. In this effort, the crystal structures of both the wild-type and engineered biocatalysts were solved, revealing the presence of two essential catalytic components, including a Lewis acidic Zn site to trigger substrate enolization and an adjacent SAM to ensure stereoselective methyl transfer. Second, we mined, cloned, and assessed a family of HMTs from Pseudomonas species that share less than 50% sequence identity to previously evaluated HMTs. In particular, HMT from P. aeruginosa exhibited the highest catalytic efficiency in the dual biocatalytic asymmetric methylation, allowing a record turnover efficiency (TTN of HMT up to 7,700, TTN of SAH up to 1,000) to be accomplished, thereby demonstrating the synthetic potential of this fully biocatalytic platform. Furthermore, based on this dual enzyme platform, a range of valuable enantioenriched scaffolds, including α-alkyl carboxylic acids, β-alkyl,α-hydroxy acids and β-alkyl,α-amino acids, could be conveniently prepared through biocatalytic cascades. Collectively, these enzyme mining and engineering efforts set the stage for advancing a general biocatalytic platform for catalytic asymmetric α-alkylation using easily available alkyl (pseudo)halides, providing a solution to this long-standing challenging facing synthetic organic chemists.
Methods
Analytical scale biocatalytic asymmetric methylation of α-ketoacids using SgvMVAV
To a 2 mL vial were added NaPi buffer (50 mM, pH 8.0), SAM (80 μL of 100 mM stock solution in 50 mM NaPi buffer, pH 8.0), SgvMVAV, EcMTAN, and substrate 1 (250 μL of 10 mM stock solution in 50 mM NaPi buffer, pH 8.0) in succession. Final reaction volume was 1 mL; final concentrations were 2.5 mM 1, 8.0 mM SAM, 0.4 – 1.6 μM SgvMVAV, and 2.5 μM EcMTAN. The vials were sealed and shaken at 30 °C and 250 rpm for 10 – 14 h and analyzed by HPLC as detailed in the Supplementary Information.
Analytical scale biocatalytic asymmetric methylation with methyl iodide as the terminal methyl donor using SgvMVAV and HMT
To a 2 mL vial were added NaPi buffer (50 mM, pH 8.0), SAH (2.5 μL of 1 mM stock solution in DMSO), methyl iodide (25 μL of 200 mM stock solution in DMSO), SgvMVAV, HMT, and substrate 1a (250 μL of 10 mM stock solution in 50 mM NaPi buffer, pH 8.0) in succession. Final reaction volume was 1 mL; final concentrations were 2.5 mM 1a, 2.5 μM SAH, 5.0 mM methyl iodide, 0.25 μM SgvMVAV, 0.25–2.5 μM HMT. The vials were sealed and shaken at 30 °C and 250 rpm for 6–24 h. The reactions were then analyzed by HPLC as detailed in the Supplementary Information.
Preparative scale biocatalytic asymmetric methylation with methyl iodide as the terminal methyl donor using SgvMVAV and PaHMT
To a 100 mL flask were added NaPi buffer (50 mM, pH 8.0), SAH (60 μL of 10 mM stock solution in DMSO), methyl iodide (3 mL of 400 mM stock solution in DMSO), SgvMVAV, PaHMT, and substrate 1a (15 mL of 40 mM stock solution in NaPi buffer) in succession. Final reaction volume was 30 mL; final concentrations were 20 mM 1a, 20 μM SAH, 40 mM methyl iodide, 20 μM SgvMVAV, 20 μM PaHMT. The reaction flask was sealed and shaken at 30 °C and 250 rpm for 14 h.
Upon the completion of the reaction, the reaction mixture was then treated with H2O2 (30% w/w) and allowed to stir at room temperature for 2 h. Excess H2O2 was decomposed with catalase. The resulting mixture was concentrated in vacuo, and the pH of the aqueous layer was adjusted to 3–4 using 1.0 M HCl (aq.). This mixture was then shaken vigorously and centrifuged (12,000 × g, 20 min).
The supernatant containing the product was filtered and loaded onto a Biotage Sfär C18 cartridge (12 g) that had been equilibrated to 10% MeOH/H2O with 0.1% formic acid. The column was flushed with 2 column volumes (CV) of 10% MeOH/H2O (0.1% formic acid). Then, the product was eluted with a gradient from 10% to 100% methanol with 0.1% formic acid over 8 CV. Product-containing fractions were combined and concentrated in vacuo. The residual aqueous solution was lyophilized to afford the analytically pure product as a white powder in 81% yield (99:1 e.r.).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We are grateful to Profs. Armen Zakarian (UCSB), Tom Pettus (UCSB), Liming Zhang (UCSB) and Yiming Wang (University of Pittsburgh) for helpful discussions and critical reading of this manuscript. We acknowledge the NSF CAREER award (CHE-2145749 to Y.Y.) and the NIH (R01GM131347 to S.K.N.) for financial support. Transaminase study is partially supported by the ACS Herman Frasch Foundation (947-HF22 to Y.Y.). We thank the NSF BioPACIFIC MIP (DMR-1933487) and NSF MRSEC at UCSB (DMR-2308708) for access to instrumentation. We acknowledge Drs. Hongcheng Shen and Dian Li for assistance with substrate synthesis, Dr. Lei Cheng for assistance with Marfey’s analysis of 4b, and Spencer Anderson and the staff at LS-CAT (Sector 21, APS, Argonne IL) for facilitating data collection.
Author contributions
Y.Y. conceived and directed biocatalysis research. S.K.N. directed protein crystallography research. S.J. and J.W. designed and performed enzyme mining, cloning and engineering as well as all the biocatalytic reactions. S.J. and R.G. synthesized substrates and authentic racemic products. S.J. and B.K.-H. performed bioinformatic analysis of HMTs. K.P.K. carried out protein crystallography. Y.Y. and S.K.N. wrote the manuscript with the input of all other authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data are available in the main text and the Supplementary Information. Plasmids encoding carbon and halogen methyltransferases reported in this study are available for research purposes from Y.Y. under a material transfer agreement (MTA) with the University of California Santa Barbara.
Competing interests
Y.Y. and S.J. are inventors on a patent application submitted by the University of California Santa Barbara (UC Case No. 2023-879, US Prov. Appl. No. 63/482,527) that covers systems and constructs for biocatalytic alkyl transfer and cofactor regeneration. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shuyun Ju, Kaylee P. Kuzelka.
Contributor Information
Satish K. Nair, Email: snair@illinois.edu
Yang Yang, Email: yang@chem.ucsb.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-023-40980-w.
References
- 1.Bloom JD, Arnold FH. In the light of directed evolution: pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. USA. 2009;106:9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lutz S. Beyond directed evolution—semi-rational protein engineering and design. Curr. Opin. Biotechnol. 2010;21:734–743. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 4.Bornscheuer UT, et al. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 5.Renata H, Wang ZJ, Arnold FH. Expanding the enzyme universe: accessing non-natural reactions by mechanism-guided directed evolution. Angew. Chem. Int. Ed. 2015;54:3351–3367. doi: 10.1002/anie.201409470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zetzsche LE, Narayan AR. Broadening the scope of biocatalytic C–C bond formation. Nat. Rev. Chem. 2020;4:334–346. doi: 10.1038/s41570-020-0191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandoval BA, Hyster TK. Emerging strategies for expanding the toolbox of enzymes in biocatalysis. Curr. Opin. Chem. Biol. 2020;55:45–51. doi: 10.1016/j.cbpa.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoltz BM, et al. Alkylations of enols and enolates. Comprehens. Organic Synth. 2014;3:1–55. [Google Scholar]
- 9.Cano R, Zakarian A, McGlacken GP. Direct asymmetric alkylation of ketones: still unconquered. Angew. Chem. Int. Ed. 2017;56:9278–9290. doi: 10.1002/anie.201703079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farina V, Reeves JT, Senanayake CH, Song JJ. Asymmetric synthesis of active pharmaceutical ingredients. Chem. Rev. 2006;106:2734–2793. doi: 10.1021/cr040700c. [DOI] [PubMed] [Google Scholar]
- 11.Evans DA, Ennis MD, Mathre DJ. Asymmetric alkylation reactions of chiral imide enolates. A practical approach to the enantioselective synthesis of.alpha.-substituted carboxylic acid derivatives. J. Am. Chem. Soc. 1982;104:1737–1739. doi: 10.1021/ja00370a050. [DOI] [Google Scholar]
- 12.Myers AG, Yang BH, Chen H, Gleason JL. Use of pseudoephedrine as a practical chiral auxiliary for asymmetric synthesis. J. Am. Chem. Soc. 1994;116:9361–9362. doi: 10.1021/ja00099a076. [DOI] [Google Scholar]
- 13.Myers AG, et al. Pseudoephedrine as a practical chiral auxiliary for the synthesis of highly enantiomerically enriched carboxylic acids, alcohols, aldehydes, and ketones. J. Am. Chem. Soc. 1997;119:6496–6511. doi: 10.1021/ja970402f. [DOI] [Google Scholar]
- 14.Kummer DA, Chain WJ, Morales MR, Quiroga O, Myers AG. Stereocontrolled alkylative construction of quaternary carbon centers. J. Am. Chem. Soc. 2008;130:13231–13233. doi: 10.1021/ja806021y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morales MR, Mellem KT, Myers AG. Pseudoephenamine: a practical chiral auxiliary for asymmetric synthesis. Angew. Chem. Int. Ed. 2012;51:4568–4571. doi: 10.1002/anie.201200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirai R, Tanaka M, Koga K. Enantioselective deprotonation by chiral lithium amide bases: asymmetric synthesis of trimethylsilyl enol ethers from 4-alkylcyclohexanones. J. Am. Chem. Soc. 1986;108:543–545. doi: 10.1021/ja00263a051. [DOI] [PubMed] [Google Scholar]
- 17.Imai M, Hagihara A, Kawasaki H, Manabe K, Koga K. Catalytic asymmetric benzylation of achiral lithium enolates using a chiral ligand for lithium in the presence of an achiral ligand. J. Am. Chem. Soc. 1994;116:8829–8830. doi: 10.1021/ja00098a061. [DOI] [Google Scholar]
- 18.Matsuo JI, Koga K. Enantioselective α-alkylation of phenylacetic acid using a chiral bidentate lithium amide as a chiral auxiliary. Chem. Pharm. Bull. 1997;45:2122–2124. doi: 10.1248/cpb.45.2122. [DOI] [Google Scholar]
- 19.Yamashita Y, Odashima K, Koga K. Construction of chiral quaternary carbon centers by asymmetric alkylation of achiral lithium enolates mediated by chiral tetradentate ligands: stoichiometric and catalytic approaches. Tetrahedron Lett. 1999;40:2803–2806. doi: 10.1016/S0040-4039(99)00299-3. [DOI] [Google Scholar]
- 20.Stivala CE, Zakarian A. Highly enantioselective direct alkylation of arylacetic acids with chiral lithium amides as traceless auxiliaries. J. Am. Chem. Soc. 2011;133:11936–11939. doi: 10.1021/ja205107x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu K, et al. Lithium enolates in the enantioselective construction of tetrasubstituted carbon centers with chiral lithium amides as noncovalent stereodirecting auxiliaries. J. Am. Chem. Soc. 2017;139:527–533. doi: 10.1021/jacs.6b11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu K, Miao B, Wang W, Zakarian A. Direct enantioselective and regioselective alkylation of β,γ-unsaturated carboxylic acids with chiral lithium amides as traceless auxiliaries. Org. Lett. 2019;21:1930–1934. doi: 10.1021/acs.orglett.9b00587. [DOI] [PubMed] [Google Scholar]
- 23.Nagib DA. Catalytic desymmetrization by C−H functionalization as a solution to the chiral methyl problem. Angew. Chem. Int. Ed. 2017;56:7354–7356. doi: 10.1002/anie.201703311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barreiro EJ, Kümmerle AE, Fraga CA. The methylation effect in medicinal chemistry. Chem. Rev. 2011;111:5215–5246. doi: 10.1021/cr200060g. [DOI] [PubMed] [Google Scholar]
- 25.Schönherr H, Cernak T. Profound methyl effects in drug discovery and a call for new C-H methylation reactions. Angew. Chem. Int. Ed. 2013;52:12256–12267. doi: 10.1002/anie.201303207. [DOI] [PubMed] [Google Scholar]
- 26.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 27.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett MR, Shepherd SA, Cronin VA, Micklefield J. Recent advances in methyltransferase biocatalysis. Curr. Opin. Chem. Biol. 2017;37:97–106. doi: 10.1016/j.cbpa.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Abdelraheem E, et al. Methyltransferases: functions and applications. ChemBioChem. 2022;23:e202200212. doi: 10.1002/cbic.202200212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Q, Pavlidis IV, Badenhorst CP, Bornscheuer UT. From natural methylation to versatile alkylations using halide methyltransferases. ChemBioChem. 2021;22:2584–2590. doi: 10.1002/cbic.202100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKean IJ, Hoskisson PA, Burley GA. Biocatalytic alkylation cascades: recent advances and future opportunities for late-stage functionalization. ChemBioChem. 2020;21:2890–2897. doi: 10.1002/cbic.202000187. [DOI] [PubMed] [Google Scholar]
- 32.Wuosmaa AM, Hager LP. Methyl chloride transferase: a carbocation route for biosynthesis of halometabolites. Science. 1990;249:160–162. doi: 10.1126/science.2371563. [DOI] [PubMed] [Google Scholar]
- 33.Schmidberger JW, James AB, Edwards R, Naismith JH, O’Hagan D. Halomethane biosynthesis: structure of a SAM-dependent halide methyltransferase from Arabidopsis thaliana. Angew. Chem. Int. Ed. 2010;49:3646–3648. doi: 10.1002/anie.201000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayer TS, et al. Synthesis of methyl halides from biomass using engineered microbes. J. Am. Chem. Soc. 2009;131:6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 35.Liao C, Seebeck FP. S-adenosylhomocysteine as a methyl transfer catalyst in biocatalytic methylation reactions. Nat. Catal. 2019;2:696–701. doi: 10.1038/s41929-019-0300-0. [DOI] [Google Scholar]
- 36.Bengel LL, et al. Engineered enzymes enable selective N-alkylation of pyrazoles with simple haloalkanes. Angew. Chem. Int. Ed. 2021;60:5554–5560. doi: 10.1002/anie.202014239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Q, et al. Directed evolution of a halide methyltransferase enables biocatalytic synthesis of diverse SAM analogs. Angew. Chem. Int. Ed. 2021;60:1524–1527. doi: 10.1002/anie.202013871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng J, Liao C, Bauer C, Seebeck FP. Fluorinated S-adenosylmethionine as a reagent for enzyme-catalyzed fluoromethylation. Angew. Chem. Int. Ed. 2021;60:27178–27183. doi: 10.1002/anie.202108802. [DOI] [PubMed] [Google Scholar]
- 39.Schülke KH, Ospina F, Hörnschemeyer K, Gergel S, Hammer SC. Substrate profiling of anion methyltransferases for promiscuous synthesis of S-adenosylmethionine analogs from haloalkanes. ChemBioChem. 2022;23:e202100632. doi: 10.1002/cbic.202100632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen X, Leisinger F, Leopold V, Seebeck FP. Synthetic reagents for enzyme-catalyzed methylation. Angew. Chem. Int. Ed. 2022;61:e202208746. doi: 10.1002/anie.202208746. [DOI] [PubMed] [Google Scholar]
- 41.Xie Y, et al. Identification of the biosynthetic gene cluster and regulatory cascade for the synergistic antibacterial antibiotics griseoviridin and viridogrisein in Streptomyces griseoviridis. ChemBioChem. 2012;13:2745–2757. doi: 10.1002/cbic.201200584. [DOI] [PubMed] [Google Scholar]
- 42.Sommer-Kamann C, Fries A, Mordhorst S, Andexer JN, Müller M. Asymmetric C-alkylation by the S-adenosylmethionine-dependent methyltransferase SgvM. Angew. Chem. Int. Ed. 2017;129:4091–4094. doi: 10.1002/ange.201609375. [DOI] [PubMed] [Google Scholar]
- 43.Bode JW, Fox RM, Baucom KD. Chemoselective amide ligations by decarboxylative condensations of N-alkylhydroxylamines and α-ketoacids. Angew. Chem. Int. Ed. 2006;45:1248–1252. doi: 10.1002/anie.200503991. [DOI] [PubMed] [Google Scholar]
- 44.Ju L, Lippert AR, Bode JW. Stereoretentive synthesis and chemoselective amide-forming ligations of C-terminal peptide α-ketoacids. J. Am. Chem. Soc. 2008;130:4253–4255. doi: 10.1021/ja800053t. [DOI] [PubMed] [Google Scholar]
- 45.Harmand TJ, Murar CE, Bode JW. Protein chemical synthesis by α-ketoacid–hydroxylamine ligation. Nat. Protoc. 2016;11:1130–1147. doi: 10.1038/nprot.2016.052. [DOI] [PubMed] [Google Scholar]
- 46.Zou Y, et al. Stereospecific biosynthesis of β-methyltryptophan from L-tryptophan features a stereochemical switch. Angew. Chem. Int. Ed. 2013;52:12951–12955. doi: 10.1002/anie.201306255. [DOI] [PubMed] [Google Scholar]
- 47.Magarvey NA, Haltli B, He M, Greenstein M, Hucul JA. Biosynthetic pathway for mannopeptimycins, lipoglycopeptide antibiotics active against drug-resistant gram-positive pathogens. Antimicrob. Agents. Chemother. 2006;50:2167. doi: 10.1128/AAC.01545-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou XW, et al. Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase. Acta Crystallogr. D. 2014;70:1549–1560. doi: 10.1107/S1399004714005239. [DOI] [PubMed] [Google Scholar]
- 49.Surup F, et al. Crocadepsins-depsipeptides from the myxobacterium Chondromyces crocatus found by a genome mining approach. ACS Chem. Biol. 2018;13:267–272. doi: 10.1021/acschembio.7b00900. [DOI] [PubMed] [Google Scholar]
- 50.Braun SD, et al. Identification of the biosynthetic gene cluster for 3-methylarginine, a toxin produced by Pseudomonas syringae pv. syringae 22d/93. Appl. Environ. Microbiol. 2010;76:2500–2508. doi: 10.1128/AEM.00666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahlert C, Kopp F, Thirlway J, Micklefield J, Marahiel MA. Stereospecific enzymatic transformation of α-ketoglutarate to (2S,3R)-3-methyl glutamate during acidic lipopeptide biosynthesis. J. Am. Chem. Soc. 2007;129:12011–12018. doi: 10.1021/ja074427i. [DOI] [PubMed] [Google Scholar]
- 52.Kemper, F. Structural characterization of the methyltransferases SgvM and MrsA and the polyphosphate kinase MrPPK2-III. Doctoral dissertation, Dissertation, Albert-Ludwigs-Universität Freiburg (2016).
- 53.Nestl BM, Hauer B. Engineering of flexible loops in enzymes. ACS Catal. 2014;4:3201–3211. doi: 10.1021/cs500325p. [DOI] [Google Scholar]
- 54.Kreß N, Halder JM, Rapp LR, Hauer B. Unlocked potential of dynamic elements in protein structures: channels and loops. Curr. Opin. Chem. Biol. 2018;47:109–116. doi: 10.1016/j.cbpa.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 55.Trost BM, Crawley ML. Asymmetric transition-metal-catalyzed allylic alkylations: applications in total synthesis. Chem. Rev. 2003;103:2921–2944. doi: 10.1021/cr020027w. [DOI] [PubMed] [Google Scholar]
- 56.Cheng Q, et al. Iridium-catalyzed asymmetric allylic substitution reactions. Chem. Rev. 2019;119:1855–1969. doi: 10.1021/acs.chemrev.8b00506. [DOI] [PubMed] [Google Scholar]
- 57.Wang X, et al. Cofactor NAD(P)H regeneration inspired by heterogeneous pathways. Chem. 2017;2:621–654. doi: 10.1016/j.chempr.2017.04.009. [DOI] [Google Scholar]
- 58.Yun H, Choi H, Fadnavis NW, Kim BG. Stereospecific synthesis of (R)-2-hydroxy carboxylic acids using recombinant E. coli BL21 overexpressing YiaE from Escherichia coli K12 and glucose dehydrogenase from Bacillus subtilis. Biotechnol. Prog. 2005;21:366–371. doi: 10.1021/bp049694w. [DOI] [PubMed] [Google Scholar]
- 59.Zhou H, et al. Artificial biocatalytic cascade with three enzymes in one pot for asymmetric synthesis of chiral unnatural amino acids. Eur. J. Org. Chem. 2019;2019:6470–6477. doi: 10.1002/ejoc.201900828. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text and the Supplementary Information. Plasmids encoding carbon and halogen methyltransferases reported in this study are available for research purposes from Y.Y. under a material transfer agreement (MTA) with the University of California Santa Barbara.