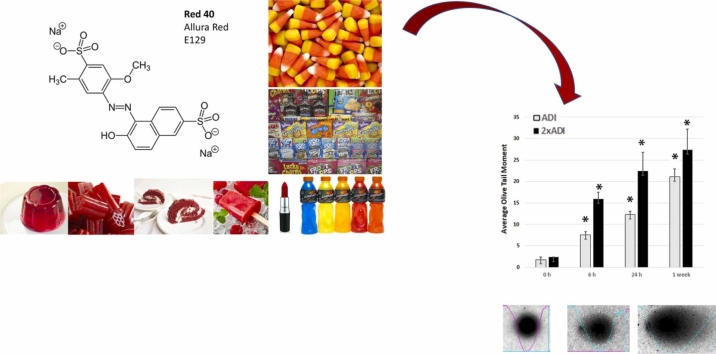
Abstract
The incidence of colorectal cancer (CRC) among young people has been on the rise for the past four decades and its underlying causes are only just starting to be uncovered. Recent studies suggest that consuming ultra-processed foods and pro-inflammatory diets may be contributing factors. The increase in the use of synthetic food colors in such foods over the past 40 years, including the common synthetic food dye Allura Red AC (Red 40), coincides with the rise of early-onset colorectal cancer (EOCRC). As these ultra-processed foods are particularly appealing to children, there is a growing concern about the impact of synthetic food dyes on the development of CRC. Our study aimed to investigate the effects of Red 40 on DNA damage, the microbiome, and colonic inflammation. Despite a lack of prior research, high levels of human exposure to pro-inflammatory foods containing Red 40 highlight the urgency of exploring this issue. Our results show that Red 40 damages DNA both in vitro and in vivo and that consumption of Red 40 in the presence of a high-fat diet for 10 months leads to dysbiosis and low-grade colonic inflammation in mice. This evidence supports the hypothesis that Red 40 is a dangerous compound that dysregulates key players involved in the development of EOCRC.
Keywords: Red 40, DNA damage, Colorectal cancer, Microbiome, P53
Graphical Abstract
Highlights
-
•
Red-40 damages DNA both in vitro and in vivo.
-
•
Red-40 consumption in the presence of a high-fat diet leads to dysbiosis in mice.
-
•
Red-40 causes inflammation in the distal colon and rectum of the A/J mouse model.
-
•
Red-40 dysregulates key players involved in the development of EOCRC.
1. Introduction
Colorectal cancer (CRC) in the young (<50 years old) - termed Early Onset Colorectal Cancer (EOCRC) - has been on the rise over the past 40 years [1], [2], [3], [4], [5], [6]. Although standard risk factors such as obesity, alcohol, and smoking are linked to EOCRC in epidemiological studies [1], [2], [3], [5], [7], [8], [9]; there remains a gap in our understanding as to why we are seeing a rise in CRC in otherwise healthy young people in their 20’s and 30’s.
Diet plays a pivotal role in influencing the risk of colorectal cancer, a significant public health concern. Research suggests that a diet high in processed meats, red meats, and saturated fats may elevate the risk of developing colorectal cancer [10]. Conversely, diets rich in fiber, whole grains, fruits, and vegetables offer protective benefits by promoting regular bowel movements, maintaining gut health, and reducing inflammation. Furthermore, certain nutrients like calcium, vitamin D, and antioxidants found in various foods have been associated with a decreased risk of colorectal cancer. These findings underscore the crucial link between dietary choices and colorectal cancer development, emphasizing the potential for preventive strategies and dietary modifications to contribute significantly to public health efforts aimed at reducing the burden of this disease.
The prevalence of ultra-processed, “westernized” diets that are typically high in fat and simple carbohydrates has increased significantly in the past 40 years [11], [12], [13], concomitant with the rise in EOCRC. This diet consists of foods that are heavily processed, include synthetic chemicals; are low in fiber, vitamins, minerals, and phytochemicals; and are often high in added sugar [14], [15]. Indeed, rigorous science has consistently shown that a highly processed, high-fat, westernized diet can drive inflammation and gut dysbiosis [16], [17], [18], and may increase risk of EOCRC [9], [19], [20], [21], [22], [23], [24]. There remains a gap, however, to the specific ingredients and their mechanisms toward EOCRC. Because of the prevalence of Red 40 in highly processed, westernized diets; it is reasonable to investigate the impact of Red 40 - that often comes with the consumption of a high-fat, westernized diet - on colon health in relation to carcinogenesis.
Synthetic dyes are prevalent in the global food supply chain. Three dyes [Allura Red (a.k.a. “Red 40”), Tartrazine (“Yellow-5”), and Sunset Yellow (“Yellow-6”)] account for 90 % of all dyes used in food in the USA; and Red 40 is by far the most common [25]. Alarmingly, 94 % of people over 2 years old in the USA consume Red 40 [26]; and over 40 % of foods marketed toward children in the USA contain such dyes [27]. It is used extensively in processed foods as a coloring for beverages, frozen treats, powder mixes, gelatin products, candies, icings, jellies, spices, dressings, sauces, and baked goods. It is critical we better understand the interaction of Red 40 with players involved in carcinogenesis.
The lack of rigorous prior research combined with high levels of human exposure to westernized diets containing Red 40, underscores the importance of researching this subject with current technologies, platforms, and appropriate animal models. We posit that the synthetic dye Red 40, acting as a foreign substance, induces a subtle and low-grade inflammatory response specific to the colon and rectum. This chronic inflammation may contribute to the development of CRC, particularly in the distal colon and rectum. Supporting the premise of this study, there evidence shows that Red 40 interacts with inflammatory components, impacts the microbiome, and can be perceived as foreign by the body [28], [29], [30], [31], [32], [33]. Indeed, it has recently been shown that Red 40 drives colitis under experimental conditions by several groups [34], [35], [36], and synthetic food dyes elevate inflammatory cytokines and modulate protein and gene expression related to inflammation [28], [37], [38]. Here, we present data consistent with the hypothesis that Red 40 damages DNA in vitro and in vivo; and that a westernized diet combined with Red 40 causes dysbiosis, functional mutations, and low-grade inflammation in the distal colon and rectum.
2. Materials and methods
2.1. Cell line and reagents
Human colorectal carcinoma cell line HCT 116 were maintained in media recommended by ATCC supplemented with 10 % New Born Calf serum (NBCS) (Biofluids, Rockville, MD), penicillin (10 U/ml, Biofluids) and streptomycin (10 μg/ml, Biofluids) at 37 °C in a humidified chamber with 5 % CO2 atmosphere. Experiments with Red 40 were carried out by treating the cells with indicated concentrations of Red 40 dissolved in media. Red 40 (Allura Red AC) was purchased from Sigma-Aldrich.
2.2. Mouse experiments
We chose the A/J mice mouse model because these mice tend to acquire more distal colon and rectal cancers with a carcinogen [39], [40] (as is the case for EOCRC) and are sensitive to chemical carcinogenesis [39]. No carcinogen was used in our experiments because we wanted to examine the impact of a HFD and/or Red 40 on the normal colon. Three-week old, female mice of the A/J strain (Stock No: 000646, Jackson Lab) were purchased from Jackson Laboratory (Bar Harbor, ME). Animals were cared for in accordance with protocols approved by the Institutional Animal Care and Use Committees of the University of South Carolina. Care and use of animals were overseen by the Department of Laboratory Animal Resources (DLAR) of the University of South Carolina under the direction of a veterinarian. The DLAR is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International, is registered with the U.S. Department of Agriculture (56-R-003) and has an active letter of Assurance of Compliance on file at the NIH. Mice were maintained at 22 ° C, with 12 h dark, 12 h light cycle. Female mice were maintained on rodent chow for one week and the experiment was started when the mice were four weeks old. Mice were randomly assigned to experimental groups. For the 10-month experiment, Red 40 was delivered in the drinking water ad libitum; and volume consumed was measured every other day. The mouse-equivalent of the human Acceptable Daily Intake (ADI, 7 mg/kg daily) is 86 mg/kg daily. With the calculation that mice consume 2 ml daily, mice were treated with the ADI and 2x ADI Red 40 and/or a LFD or HFD for 10 months. For the in vivo experiment on DNA damages, Red 40 was administered orally (e.g., oral gavage) as one time dose (either ADI or 2xADI).
2.3. Diets
We designed a diet that resembles a western-style diet with a high fat content (‘high-fat diet, or HFD’). The HFD (Bio Serv® #F3282) contained 20.5 % protein, 36.0 % fat, 35.8 % carbohydrate, 141 g/kg total saturated fatty acids, 202.2 g/kg total unsaturated fatty acids, and 5.49 kcal/g total calories) (Supplemental Table 1). The corresponding low-fat diet (LFD) was the control diet; has the same protein content; and is matched in micronutrients (Bio Serv® #F4031, protein 20.5 %, fat 7.1 %, carbohydrate 61.5 %, total saturated fatty acids 27 g/kg, total unsaturated fatty acids 40 g/kg, total caloric 3.39 kcal/g) (Supplemental Table 1). Animal diets were kept at 4 ° C. Food was re-filled every other day for all the animals, which keep the food fresh and available for the animals’ appetite. The drinking bottles were changed bi-weekly. Animal diets and water were checked daily; and mice weight, diarrhea, and hemoccult (fecal blood) were monitored twice a week. Food and drink consumption were monitored twice a week; fecal samples were collected once a month. Animal weight loss, diarrhea, hemoccult indicate the disease condition in the intestinal tract. For example, hemoccult and anemia suggesting pathological problems in the mucosa, which may reflect early age of onset, with carcinogenesis potentially starting in the mucosa.
Table 1.
Quantification of organoids derived in different culture environments. Data are shown as mean ± SD.
| Culture Medium | Treatment | Percentage (%) |
|---|---|---|
| WENRAS (control media) |
LFD | 100 |
| WENRAS + Nutlin (only p53 mutant survive) |
LFD LFD + Red 40 |
20 ± 2.3 52 ± 5.1* |
| ENRAS (only APC mutant survive) |
LFD LFD + Red 40 |
1 ± 0.4 1 ± 0.5 |
| WENRAS (control media) |
HFD | 100 |
| WENRAS + Nutlin (only p53 mutant survive) |
HFD HFD + Red 40 |
23 ± 2.5 46 ± 4.8** |
| ENRAS (only APC mutant survive) |
HFD HFD + Red 40 |
16 ± 1.8 20 ± 2.2 |
* indicates a significant difference from LFD/WENRAS+Nutlin
** indicates a significant difference from HFD/WENRAS+Nutlin
2.4. Macroscopic foci counting
Colon macroscopic foci were counted under a dissecting microscope. The number and location of foci in the GI tract were recorded and imaged. After all the necessary observations were made, the colon was swiss-rolled and fixed with 10 % neutral buffered formalin for 20 h and embedded in paraffin. Tissue sections were prepared and stained with hematoxylin and eosin (H&E) for histological examination.
2.5. Histology and quantifying inflammation
H&E staining was performed on 5-µm frozen sections, followed by bright field microscopy image analysis and scored for inflammation by our pathologist in a blinded fashion. Scores were assigned to each slide that reflects three histological features by the percent area of involvement. Inflammation severity was scored as following: 0 for none, 1 for minimal, 2 for moderate, and 3 for severe; inflammation extent as following: 0 for none, 1 for mucosa, 2 for mucosa and submucosa, and 3 for transmural; crypt damage as following: 0 for none, 1 for one-third of crypt damaged, 2 for two-thirds of crypt damaged, 3 for crypt loss and surface epithelium intact, 4 for crypt loss and surface epithelium loss. Percent area involvement was scored as following: 0 for 0 %, 1 for 1–25 %, 2 for 26–50 %, 3 for 51–75 %, and 4 for 76–100 %. Therefore, the minimal score is 0, and the maximal score is 40. We have used this method of method extensively in previous publications [41], [42], [43]. For immunohistochemical staining, formalin-fixed, paraffin-embedded serial sections of mouse colon tissues were incubated overnight with antibodies against iNOS (rabbit polyclonal, cat# 160862, diluted 1:3500; Cayman Chemical, Ann Arbor, MI) by slow rocking using the Antibody Amplifier (ProHisto, Columbia, SC) to ensure even staining and reproducible results. After incubation with primary antibodies, sections were processed using EnVision+ System-HRP kits (DakoCytomation, Carpinteria, CA) according to kit protocols. The chromogen was diaminobenzidine, and sections were counterstained with 1 % methyl green. Intensity and degree of staining were evaluated independently by three blinded investigators (QZ, AC and IC). For each tissue section, the percentage of positive cells was scored on a scale of 0–5 for the percentage of tissue stained: 0 (0 % positive cells), 1 (<10 %), 2 (11–25 %), 3 (26–50 %), 4 (51–80 %), or 5 (>80 %). Staining intensity was scored on a scale of 0–3: 0 (negative staining), 1 (weak staining), 2 (moderate staining), or 3 (strong staining). The two scores were multiplied, resulting in an immunoreactivity score (IRS) value ranging from 0 to 15 [43], [44], [45], [46].
2.6. Comet assay (DNA damage) analysis
The single-cell gel electrophoresis Comet assay was performed on HCT 116 cells, treated with 0–1500 μM, as well as on colon epithelial cells of mice treated with human equivalent of the acceptable daily intake (ADI) of Red 40 or double acceptable daily intake (2xADI) for 6 h, 24 h, or 1 week. The Comet Assay has been described by our group previously [47]. Briefly, following treatment of mice with Red 40, we dissected out the colon, flushed it out with 1 × PBS, opened it longitudinally and cut the colon into two small pieces, which were incubated in 10 % fetal bovine serum/5 mM ethylenediaminetetraacetic acid in 1 × Ca2+/Mg2+-free PBS for 15 min at room temperature. Colon tissues were then shaken to dislodge the epithelial layer into single-cells suspensions. Cell viability was checked by trypan blue exclusion and > 95 % cells were viable. The single-cell suspension was centrifuged (200 g.m. for 5 min), and the pellet was brought up in freezing media and frozen at –80 °C until Comet analysis. An alkali Comet assay was performed according to instructions provided by the kit manufacturer (CometAssay™, Trevigen, Gaithersburg, MD). Cells treated with hydrogen peroxide (200 μM, 20 min) were used as positive controls. A minimum of 75 Comets per treatment were quantified after capturing with and quantified by the Automated Comet Assay Analysis System CometAssay™, Trevigen, Gaithersburg, MD). Olive tail moment was used to evaluate DNA damage. The tail moment, expressed in arbitrary units, is calculated by multiplying the percent of DNA (fluorescence) in the tail by the length of the tail in μm. The tail length is measured between the edge of Comet head and the end of the Comet tail. An advantage of using the tail moment as an index of DNA damage is that both the amount of DNA damage and the distance of migration of the genetic material in the tail are represented by a single number. Three mice per treatment condition were used and the average of three Comet assays was plotted. For in vitro assay the average of the tree independent experiments were plotted.
2.7. Colon organoids
Colonic tissue organoids developed from experimental mice crypts were studied. Our understanding of the molecular and cellular mechanisms that mediate the stem cell microenvironment have been leveraged to assess mutations in colon cancer driver genes: APC and p53. To quantify effects, the mouse colon was opened lengthwise and cut into 2–4 cm pieces, placed in a 50 ml conical tube filled with ice-cold 1X RPMI 1640 (Gibco) buffer. The tissue was transferred into 20 ml ice-cold DPBS (Gibco) for cleaning. Tubes were inverted gently four times, then the tissue was transferred to sterile 50 ml tubes containing 30 mM EDTA, 1.5 mM DTT, diluted into 1X DPBS and incubated on ice for 20 min. After dissociation, tissues were placed into a pre-warmed (37 °C) 30 mM EDTA, diluted into 1X DPBS and incubated at 37 °C for 10 min; then vigorously shook for 30 s to detach the epithelium from the basement membrane. Suspended cells were transferred into a sterile 15 ml conical tubes and pelleted by centrifugation at 500 g for 10 min at 4 °C. Supernatant was decanted, and washed 2x with the same buffer, then centrifuged at 500 g for 5 mins to pellet. Advanced DMEM/F12 was supplemented with penicillin/streptomycin, 10 mM HEPES, 2 mM GlutaMAX, 1 × B27 (Life Technologies), 10 nM gastrin I (Sigma), and 1 mM N-acetylcysteine (Wako). The following niche factors were used: 50 ng/ml mouse recombinant EGF (Life Technologies), 100 ng/ml mouse recombinant Noggin (Peprotech), 10 % R-spondin-1 conditioned medium, 50 % Wnt-3A conditioned medium, 500 nM A83–01 (Tocris), and 10 μM SB202190 (Sigma). After matrigel polymerization, the cells were overlaid with culture media representing different niche factor conditions. Crypts were observed under the microscope and the final pellet was plated into matrigel (Corning Life Sciences, Durham, NC) which supports 3D structural growth. The minimum amount of matrigel used as 50/50 ratio of the pellet size, with 75 µL/well in a 12-well plate. 75 µL of organoids/matrigel were placed in the center of each well forming a dome. We maintained the plate in the incubator until the matrigel solidified. Then, we added 1 ml of WENRAS media with ROCK and GSK inhibitors in each well. After several days, organoids were placed in standard media, or WENRAS+nutlin (surviving clones are p53 mutant), or ENRAS (media minus Wnt-3a; surviving clones are APC-mutant). Media was changed every other day; and cultured for 3 weeks. Organoids were counted under a 40x microscope at the end of the experiment.
2.8. Stool and microbiome analysis
Blood in stool was detected using Hemoccult (Beckman Coulter) fecal immunochemical test. Immediately before sacrifice, stool consistency (0-fully formed stool; 2-loose stool; 3-diarrhea) and blood in the stool (0-no blood; 2-detected using Hemoccult; 3-rectal bleeding) were scored, and these measurements were used along with the weight difference in mice from the beginning to the end of the experiment (0 =no weight loss; 1 = 0–5 % weight loss; 2 = 6–10 % weight loss; 3 =11–15 % weight loss; 4 =16–20 % weight loss), to calculate a cumulative disease index (CDI). Fecal samples were freshly collected from 1 month old, 7 months old, and 11 months old. During fecal sample collection, mice were put in a large empty tip box, which have been punched with multiple holes to maintain airflow. After approximately 20 min, mice produced enough fecal drops in the box, then collected fecal samples in a clean 1.5 ml microcentrifuge tube. Fecal samples were freshly frozen in – 80 ° C for microbial analysis. DNA extraction was performed, and the eluted DNA was divided into 2 plates; one of which was used to prepare 16 S libraries. After extraction, samples were quantified; then the V4 region of the 16 s rRNA gene was amplified from each sample using the Dual indexing sequencing strategy. Sequencing was done on the Illumina MiSeq platform. PCR was performed and products were visualized. Libraries were normalized using SequalPrep Normalization Plate Kit (Life technologies) following the manufacturer’s protocol for sequential elution. The concentration of the pooled samples was determined and the sizes of the amplicons in the library was determined. The final library consisted of equal molar amounts from each of the plates, normalized to the pooled plate at the lowest concentration. Library Preparation for Sequencing and Sequencing Libraries were prepared according to Illumina’s protocol for Preparing Libraries for Sequencing on the MiSeq. PhiX and genomes were added in 16 s amplicon sequencing to create diversity. A mock community was used for error analysis.
2.9. Statistical analysis
The bioinformatics pipeline includes comparisons of each group's phylogenetic alpha diversity in Shannon vector and evenness vector, which indicates diversity of microbial communities. To evaluate phylogenetic and possible metabolomic alterations within samples, QIIME reads processing with chimera removal, open reference, and SILVA rRNA database (Silva_99) options were used. For PiCRUSt application, a closed reference against the Greengenes database selection was used. Operational Taxonomic Unit (OTU) tables were generated from Nephele and were subjected to Linear Discrimination Analysis of effect size (LEfSe). Data for each group presented as mean with standard deviation. Multiple comparisons between the groups were conducted by one-way analyses of variance (ANOVAs), followed by Newman-Keuls' post hoc tests. The p-value chosen for significance was 0.05. All statistical analyses were performed in GraphPad PRISM 8.0 (GraphPad Software, San Diego, CA).
3. Results
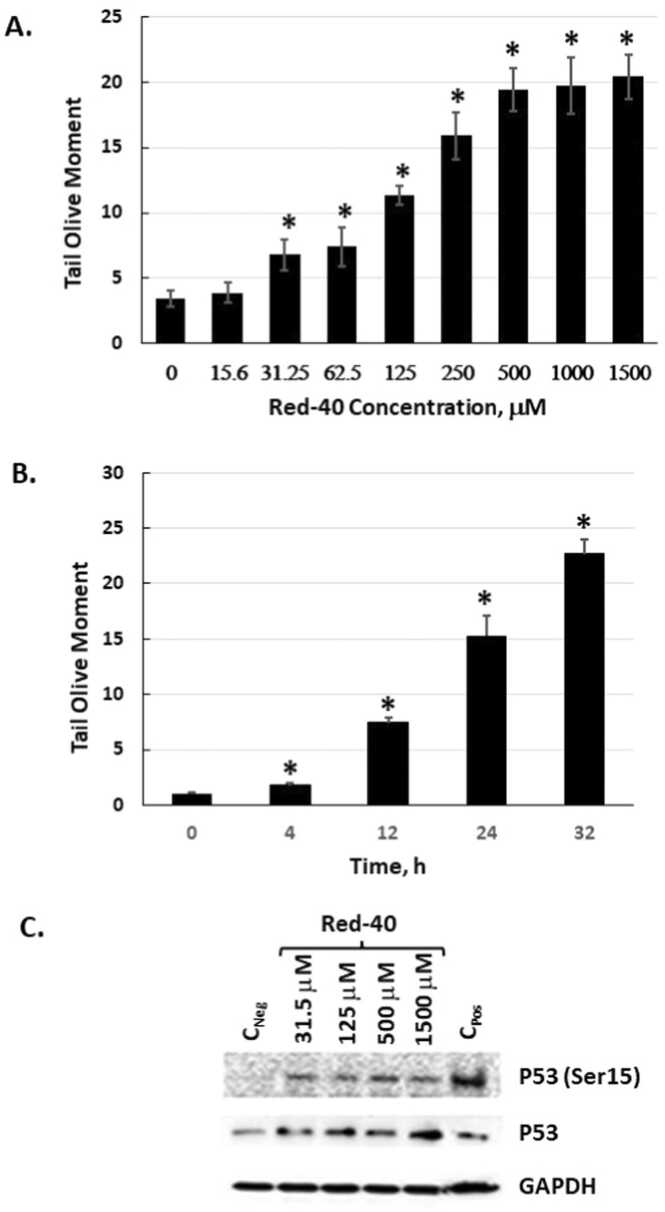
3.1. Red 40 causes DNA damage in vitro
In vitro experiments were conducted to examine the impact of Red 40 on human HCT 116 colon cancer cells. To this, we carried out dose- and time-course experiments with physiologically relevant doses of Red 40. Fig. 1 shows that Red 40 causes DNA damage (determined by the comet assay) in a dose (Fig. 1A) and time (Fig. 1B) dependent manner. Consistent with these findings, Red 40 also causes a dose-dependent increase in the DNA damage markers, p53 and phosphorylated p53 (on serine 15) (Fig. 1C). It is important to mention that Red 40 does not have significant cytostatic or cytotoxic effect under these experimental conditions as the cell grows rate /count and viability (94–96 %, n = 3) for Red 40 treated cells have not changed in comparison to non-treated cells (viability of 96 %, n = 3).
Fig. 1.
Red 40 induces DNA damage in vitro. A. HCT116 cells were exposed to Red 40 as indicated by dose then harvested after 24 h exposure. B. Time course of exposure to 1 mM Red 40. C. Red 40 causes phosphorylation of P53 at serine 15 and stabilizes P53. For the comet assay (A, B), an alkaline Comet assay was carried out, which detects both double and single stranded DNA breaks. Tail moment is the product of the tail length and the fraction of total DNA in the tail; and a longer tail moment indicates higher DNA damage. Error bars represent the standard error (n = 3).
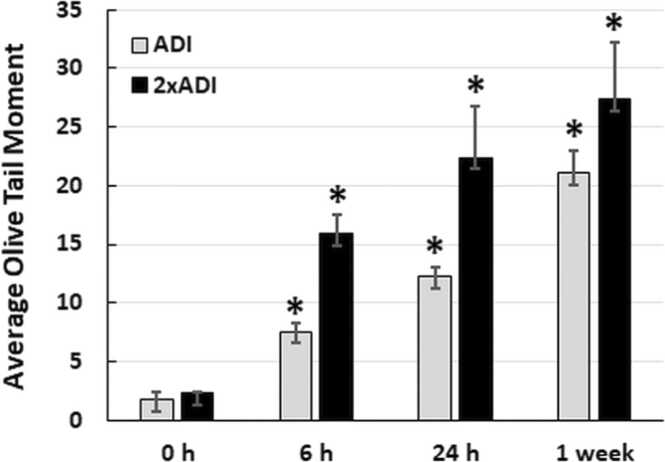
3.2. Red 40 causes DNA damage in vivo
Although previous studies from the early 2000’s have shown that Red 40 causes DNA damage to the colon) [48], [49], [50]; others have shown a lack of DNA damage [51]. Because this is an unresolved issue, and it makes sense to confirm our in vitro results; here we evaluated the impact of Red 40 on DNA damage in vivo. Fig. 2 shows that Red 40 – at doses equivalent to the Accepted Daily Allowance (ADI) or twice the ADI, causes DNA damage in the colon. Mice were given a bolus of Red 40 at indicated doses, then colon cells collected at indicated times (0. 6, 24 hrs; and 1 week) by scraping and examined by the Comet Assay.
Fig. 2.
Red 40 induces DNA damage in vivo. A. Red 40 given at a human equivalent dose of 7 mg/kg (ADI) and 14 mg/kg (2x ADI) for 6 h, 24 h or 1 week causes DNA damage in cells collected from the colon after indicated time points. Female A/J mice were given Red 40 orally (e.g., oral gavage) at indicated doses (ADI or 2xADI), then colon cells collected at indicated times (0. 6, or 24 h; and 1 week) by scraping and examined by the Comet Assay. Error bars represent the standard deviation (n = 3).
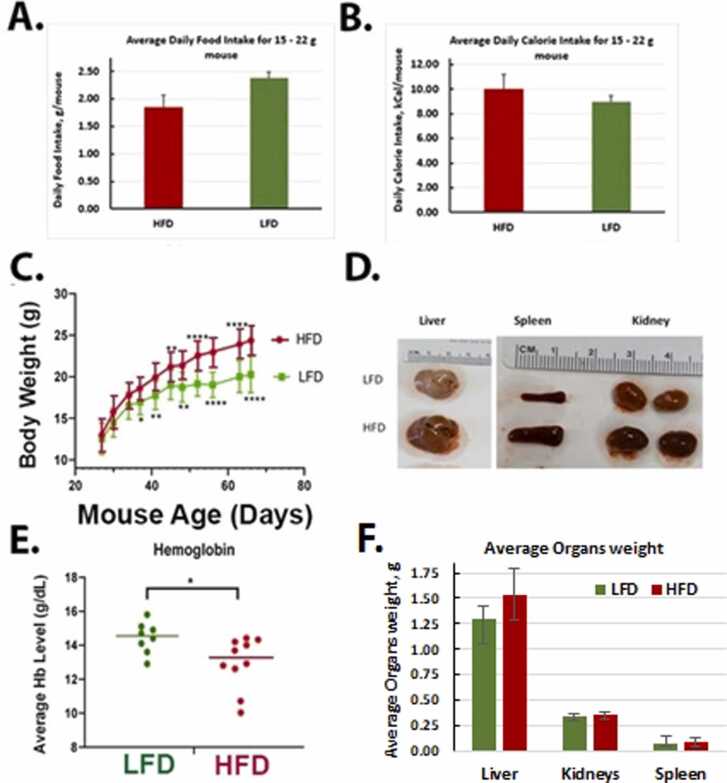
3.3. Impact of a HFD and Red 40 on health and the colon
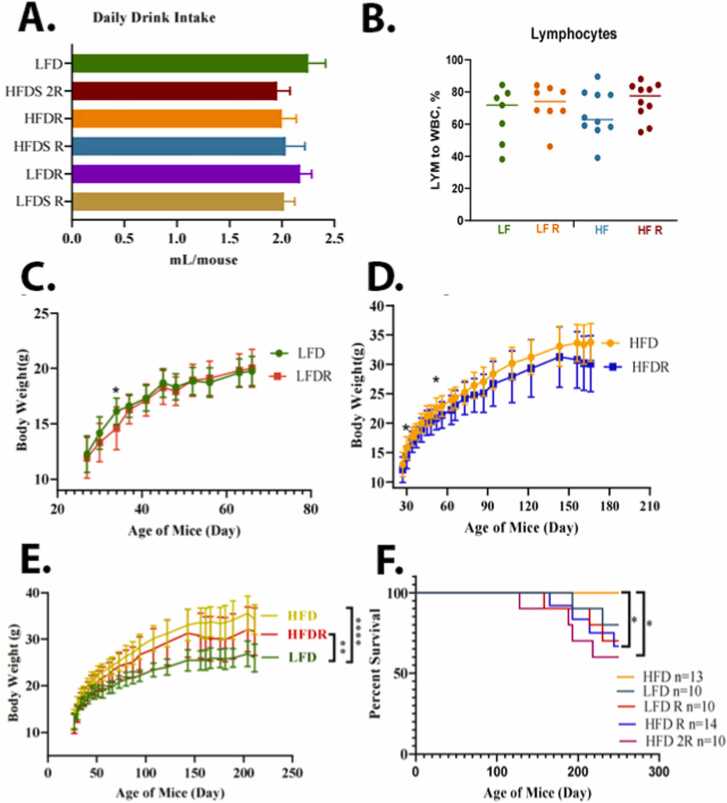
We compared the food intake, body weight changes, and other endpoints of health in the different treatment groups. Although the average amount of food intake was higher in the LFD mice (Fig. 3A), the weight gain was higher in mice consuming the HFD (Fig. 3C). The HFD animals’ overall caloric intake was 10.29 Kcal/mouse/day; while the LFD caloric intake was 8.88 Kcal/mouse/day (Fig. 3B). After consumption of the HFD for 10 months, there was an increase in the size of the spleen, liver, and kidney (Fig. 3D); and a decrease in hemoglobin levels (Fig. 3E). For LFD group, liver, kidney, and spleen weights were 1.3 ± 0.12 g, 0.34 ± 0.03 g, and 0.08 ± 0.06 g, respectively (Fig. 3F). A HFD group, liver, kidney, and spleen weights were 1.25 ± 0.35 g, 0.04 ± 0.09 g, and 0.09 ± 0.04 g, respectively (Fig. 3F). After an addition of the Red 40 to diets, a LFD group liver, kidney, and spleen weights were 1.46 ± 0.13 g, 0.34 ± 0.04 g, 0.09 ± 0.02 g, respectively, and HFD group liver, kidney, and spleen weights were 1.27 ± 0.24 g, 0.34 ± 0.06 g, and 0.08 ± 0.03 g, respectively. Colon length decreased (although non-significant) also with consumption of the HFD (9.26 ± 0.6 cm for LFD and 8.7 ± 0.82 cm for HFD), and even further with an addition of Red 40–8.75 ± 1.38 cm for LFD and 7.79 ± 1.27 cm. Daily liquid and food intake were monitored for all the groups; with no significant difference in the amount consumed (Fig. 4A). In comparing the impact of Red 40 on body weight, we saw that within the first several weeks after Red 40 consumption, the LFD with Red 40 group had significant lower body weight than control group, (p < 0.05) (Fig. 4C, D, E), which indicate toxicity of Red 40 in mice fed a HFD. Indeed, survival was lower in mice consuming Red 40 (Fig. 4F). Another observation was that the HFD overlaid with Red 40, increased lymphocytes percentage in peripheral blood (Fig. 4B).
Fig. 3.
Impact of consuming a low-fat diet (LFD) vs. high-fat diet (HFD) on food intake (A), calorie intake (B), body weight changes (C), organ impact (D), systemic hemoglobin (E) and organ weight (F). Female A/J mice were fed LFD or HFD starting at weaning. The average amount of food intake was higher in the LFD mice (Fig. 3A); although the average daily calorie intake was lower in this group (Fig. 3B). Weight gain was higher in mice consuming the HFD (Fig. 3C). There was an increase in the size of the spleen, liver, and kidney in the HFD group (Fig. 3D, F); and a decrease in hemoglobin levels in the HFD group (Fig. 3E).
Fig. 4.
Impact of consuming a LFD vs. HFD on liquid intake (A), systemic lymphocyte count (B), body weight changes (C, D, E), and percent survival (F). Female A/J mice were exposed to Red 40 in the drinking water (mouse equivalent of 7 mg/kg/d (HFDR) or 14 mg/kg/d (HFD2R)) beginning at 3 months of age and continuing until 11 months of age.
*, indicates significant difference (p < 0.05).
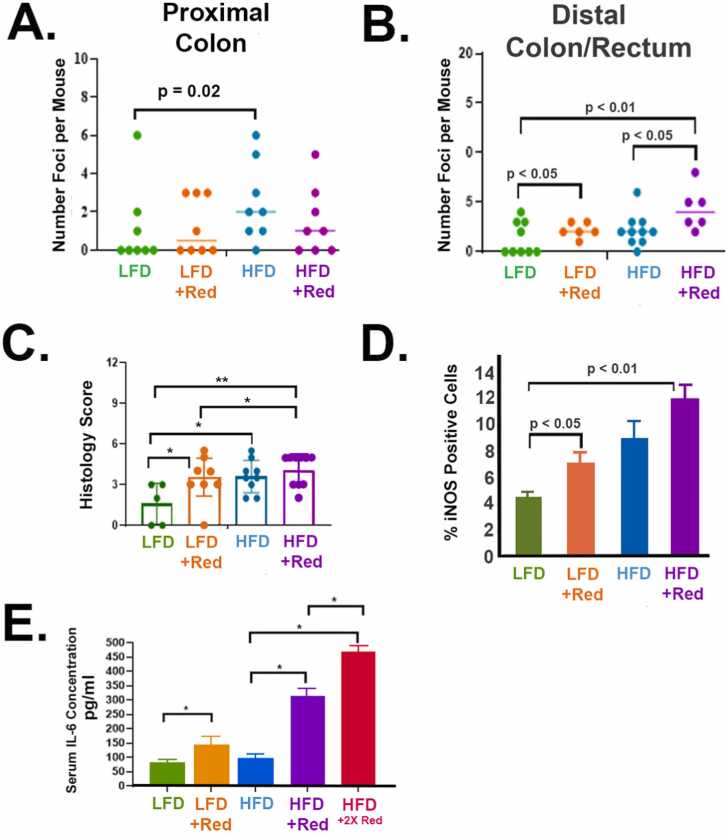
Next, colon macroscopic foci were counted. All groups had foci by 11 months old, which indicates that aging is also a parameter for developing foci anomalies. Of note, mice consuming Red 40 had their intestine content and tissue dyed red.
Consumption of a HFD with Red 40 increases foci number and size in the colon compared to other groups. Flat structures, resembling aberrant crypt foci, were counted under a dissecting microscope in the proximal colon (Fig. 5A) and distal (Fig. 5B) colon/rectum. Red 40 increases the number of foci in the distal colon in the presence or absence of the HFD. A HFD appears to increase the number of foci in the proximal colon; and Red 40 does not impact this endpoint in this part of the colon. We then scored the inflammatory “histological” changes in the distal colon and rectum (Fig. 5C); which shows the degree of pathological inflammation increasing with the HFD ± Red 40; and Red 40 alone (+LFD) also increases distal colon inflammation. We also stained for inducible nitric oxide synthase (iNOS) in the distal colon/rectum as a marker of inflammation (Fig. 5D). iNOS – a marker of inflammation – is also induced by Red 40 ± HFD. Finally, to see if Red 40 affects systemic inflammatory load, we measured serum IL-6; and saw that Red 40 indeed elevates the levels of IL-6 in the presence of a HFD (Fig. 5E).
Fig. 5.
Red 40 causes inflammation in the distal colon and rectum of the A/J mouse model. A/J mice were exposed to Red 40 in the drinking water (mouse equivalent of 7 mg/kg/d or 14 mg/kg/d) beginning at 3 months of age and continuing until 11 months of age. Because of the use of Red 40 in a high-fat, westernized diet; we also included groups consuming both a low-fat diet (LFD) and a high-fat diet (HFD). Flat structures, resembling aberrant crypt foci, were counted under a dissecting microscope in the proximal colon (A) and distal (B) colon/rectum. We then scored the inflammatory “histological” changes in the distal colon and rectum as we have done many times previously (C). Foci count in the distal colon and rectum were also quantified (D). We also stained for inducible nitric oxide synthase (iNOS) in the distal colon/rectum as a marker of inflammation (E). Finally, to see if Red 40 affects systemic inflammatory load, we measured serum IL-6; and saw that Red 40 indeed elevates the levels of IL-6 in the presence of a HFD (F). HFDR = High-Fat Diet + 1x Acceptable Daily Intake (ADI) Red 40; HFD2R = HFD + 2x ADI Red 40.
*, indicates significant difference (p < 0.05).
3.4. HFD and Red 40 impacts the microbiome in vivo
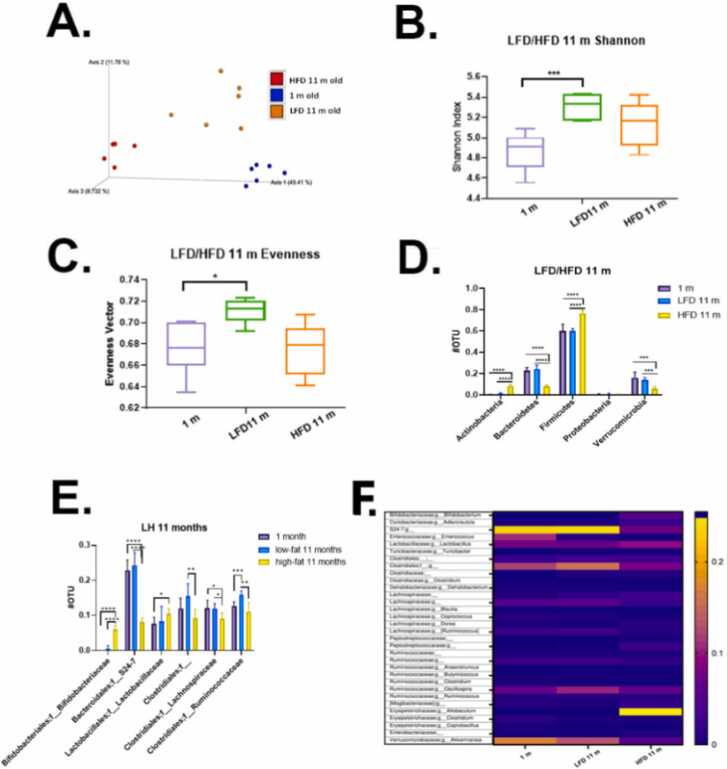
Because the microbiome is a key regulator of colonic health [52], we wanted to examine how a Red 40 impacts the microbiome with and without HFD. While alpha-diversity represents the diversity within each sample, beta-diversity represents the difference between samples. In other words, it describes how similar or different the two ecosystems are. First, we asked how the fecal microbiome changes after 10 months of consumption of the HFD vs. LFD. Fig. 6A shows a Bray-Curtis dissimilarity between samples; and that diet significantly changes the beta-diversity of the samples. Also, after 10 months of consuming a HFD, there is a decreased phylogenetic alpha diversity in Shannon vector (Fig. 6B) and evenness vector (Fig. 6C), which indicate lower diversity of microbial communities, and less healthy microbial environment. As expected, 1-month old mice microbiota – still influenced by mother - had less diverse and richness (Fig. 6B, C). Quantification of bacteria based on Phyla (Fig. 6D) shows that mice fed a HFD over 10 months had significantly increased Actinobacteria and Firmicutes; and significantly lower quantities of protective Bacteroidetes and Verrucomicrobia. Operational taxonomic units at the family level were also compared (Fig. 6E). The 10-month consumption of the HFD results in fecal samples having significantly increased levels of Bifidobacteriaceae and Lactobacillaceae; and decreased levels of S24–7, Clostridiales, Lachnospiraceae, Ruminococcaceae and one undefined family. Finally, we present microbial communities through heatmap plots (Fig. 6F). Mice consuming a HFD had elevated levels of Bifidobacteriaceae;g__Bifidobacterium, Lactobacillaceae;g__Lactobacillus; and especially Erysipelotrichaceae; g_Allobaculum. There also was a decreased S24–7, Verrucomicrobiaceae Akkermansia, and Enterococcus.
Fig. 6.
Microbial communities’ analysis: age and LFD vs HFD. A. Phylogenetic based beta diversity-principal component analysis (PCA) plot of fecal microbiota was examined. Plots based on unweighted UniFrac distance matrices of microbial communities in fecal samples, three separated clusters were displayed. PC1 (x‐axis) explained 49.41 %, PC2 (y‐axis) explained 11.78 % of variability, PC3 (z‐axis) explained 8.73 % of the variability. B. Phylogenetic based alpha diversity-the Shannon index. Lower Shannon vectors indicate the lower diversity of the microbial communities. LFD mice had the highest diversity than the other two groups. 1-month old mice majority microbial communities came from the mother and less diverse and richness influenced by diet. Microbiota in young mice tend to be less diverse than that of older mice. C. Alpha diversity-the evenness was compared in three groups. The LFD group had the highest richness and evenness than the other two groups, indicating LFD fed mice microbiome community has a small disparity between the number of individuals within each species. D. Quantitative phyla levels of operational taxonomic unit (OTU) was compared. E. Family level of operational taxonomic unit (OTU) was compared. The HFD mice samples increased in Bifidobacteriaceae, Lactobacillaceae; decreased in S24–7, Clostridiales, Lachnospiraceae, Ruminococcaceae and one undefined family. Significance, *p < 0.05, * *p < 0.01, * **p < 0.001, * ** * p < 0.0001. F. Microbial communities’ analysis. Heatmap plots of 11 months mice fecal samples showed the gut microbiota composition between samples at the genus level. Relative abundances of individual taxa (rows) in each sample (columns) are indicated in the associated color scale.
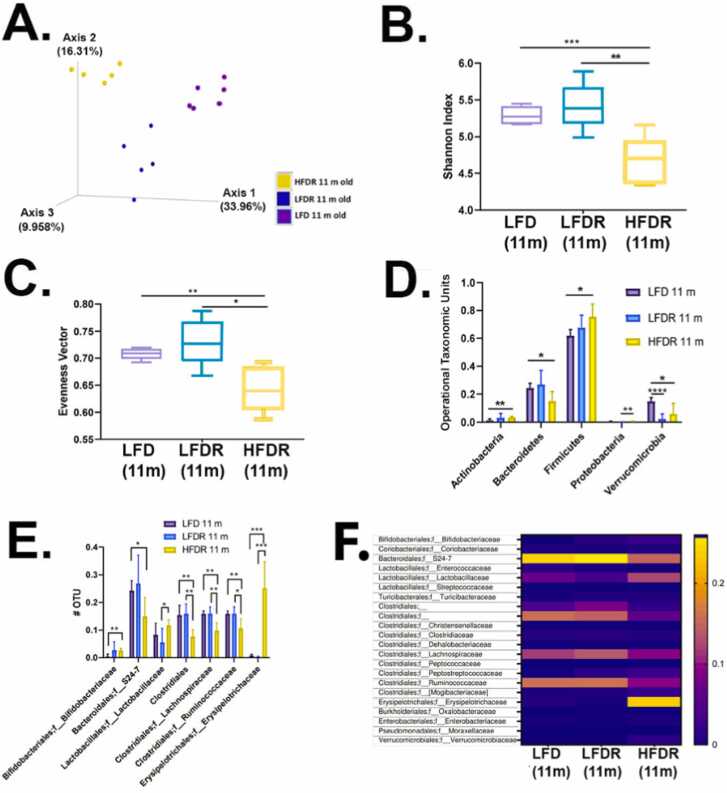
We next asked how Red 40 affects the microbiome. Bray-Curtis dissimilarity between samples shows that the beta-diversity is different amongst the three groups; including the control LFD group with and without Red 40 (Fig. 7A). However, Red 40 - alone - does not appear to change the alpha diversity (Fig. 7B, C); a result consistent with another recent study [35]. The introduction of the HFD with Red 40 however significantly decreases beneficial microbial communities and increases harmful microbial communities in A/J mice (Figs. 7D, 7E). Operational taxonomic units at the phyla level were quantified and showed that the consumption of a HFD+Red 40 causes a significant increase in Actinobacteria, Firmicutes; and Proteobacteria; and a decrease in Bacteroidetes (Fig. 7D). The beneficial species, Verrucomicrobia, was significantly reduced by Red 40 alone (Fig. 7D). Finally, heatmap plots show the gut microbiota composition between samples at the family level. Consumption of Red 40 in the backdrop of a LFD causes an increase in S24–7 and clostridiales; and a decrease in Lactobacillaceae (Fig. 7F). Also, our HFD + Red 40 causes an increase in Erysipelotrichaceae compared with consumption of the LFD (Fig. 7F).
Fig. 7.
Microbial diversity of after 10 months exposure to Red 40. A. Phylogenetic based beta diversity, a qualitative principal component analysis (PCA) plot of fecal microbiota. Compare LFD 11-month old mice fecal samples, LFD/HFD with Red 40 11 months old mice fecal sample. Unweighted UniFrac distance matrices of microbial communities in fecal samples, PC1 (x‐axis) explained 33.96 %, PC2 (y‐axis) explained 16.31 % of variability, PC3 (z‐axis) explained 9.96 % of the variability. B. Phylogenetic based alpha diversity, the Shannon index. Lower Shannon vectors indicate the lower diversity of the microbial communities. Comparing HFD with Red 40 11 months old mice to LFD with or without Allura Red AC 11 months old mice Shannon vector significantly decreased. *p < 0.05, * *p < 0.01, * **p < 0.001. C. Comparing the alpha diversity evenness. Comparing the HFD with Red 40 11 months old mice to LFD with or without Red 40 11 months old mice samples, diversity of evenness significantly decreased. Significant, D. Operational taxonomic unit (OTU) of the Phyla level were compared: LFD and HFD with and without Red 40 11 months mice microbiota. The significantly increased in Actinobacteria, Firmicutes; and Proteobacteria. Decreased in Bacteroidetes only in HFD with Red 40 samples, and decreased Verrucomicrobia in both LFD and HFD with Red 40 samples. E. Comparing the OTU of the family level: Increased in Bifidobacteriaceae and Erysipelotrichaceae, especially in overlaid with HFD and Red 40, decreased in Bacteroidales family level in S24–7, especially in overlaid with HFD and Red 40; Clostridiales family level in Ruminococcaceae and Lachnospiraceae, especially in overlaid with HFD and Red 40. F. Heatmap of family level. Heatmap plots show the gut microbiota composition between samples at the family level. Relative abundances of individual taxa (rows) in each sample (columns) are indicated in the associated color scale. *p < 0.05, * *p < 0.01, * **p < 0.001, * ** * p < 0.0001.
3.5. Impact of a HFD and Red 40 on functional p53 and APC mutations
Organoids were cultured according to well established mouse organoid protocols [53]. Indeed, we have extensive experience establishing, culturing, and testing colon organoids in our college core [54], [55]. Organoids analyzed ex-vivo were functionally tested for the acquisition of mutations to the gene by plating in the absence of ligands, as described previously [56]. Briefly, organoids are selected for TP53 and APC mutants by adding Nutlin-3a (TP53) or removing Wnt-3A (APC) to the media. Adding Nutlin-3a or Wnt-3A to the growth media will kill cells with wildtype TP53 or APC, respectively, thus enriching for organoids containing mutations. We observed that organoids derived from HFD colonic culture are 2-fold larger than LFD colonic organoids. We also observed that p53 is functionally mutated by Red 40 in the backdrop of consuming either a LFD and a HFD. Although APC was functionally mutated by consuming either the LFD or the HFD; Red 40 did not induce further functional mutations (Table 1).
4. Discussion
Diseases such as Inflammatory Bowel Disease (IBD) are characterized by persistent, cyclic, and heightened inflammation, often associated with an elevated cancer risk within the affected tissue, such as the increased risk of CRC due to IBD. While this demonstrates a robust connection between intrinsic, recurrent, high-grade inflammation and cancer, emerging evidence highlights that even subtle, chronic inflammation can contribute to colorectal cancer development. In this study, we postulated that Red 40, a synthetic food dye, might incite a concealed, low-grade, tissue-specific inflammation in the colon and rectum, thereby laying the groundwork for colorectal carcinogenesis. With ubiquitous exposure to synthetic chemicals through daily diets, particularly in highly processed, Western-style high-fat diets (HFDs), which are often rich in synthetic food dyes, such as Red 40, the potential impact is concerning. Disturbingly, the escalating consumption of Red 40 parallels the rising incidence of early-onset colorectal cancer (EOCRC).
As our study investigates the effects of Red 40 on CRC-related endpoints—DNA damage, low-grade inflammation, and the gut microbiome—we present foundational evidence substantiating its deleterious influence. Notably, our findings align with recent research indicating that Red 40 triggers colitis in experimental settings [34], [35]. He and Chen et al. [35], [36] showed Red 40 (7 mg/kg/d) triggers experimental colitis in mice expressing IL-23; and that this was dependent on CD4 + T-cell and IFN-γ signaling [54]. Kwon et al. [34] showed that chronic (100 ppm in diet ad libitum for 12 weeks) exposure to Red 40 prior to experimental induction of colitis exacerbates this condition [34]; while intermittent Red 40 exposure (1 day per week for 12 week) does not. Additionally, early life exposure to Red 40 primed mice to a heightens susceptibility to DSS-induced colitis [34]. So, multiple studies – including this one - demonstrate that chronic exposure to Red 40 provokes mild colitis, akin to our results following a ten-month chronic exposure (Fig. 5). Importantly, Red 40 induces DNA damage and activates the p53 pathway, a significant revelation considering p53's pivotal role in colorectal carcinogenesis (Fig. 1, Fig. 2). Our finding that p53 is functionally mutated by Red 40 in the backdrop of consuming either a LFD and a HFD (Table 1) is novel and of significance given the key role that p53 plays in colorectal carcinogenesis [57]. Interestingly, Red 40 does not – in itself – functionally mutate APC. This impact of Red 40 on p53 but not APC parallels the findings that human EOCRC has a relatively high p53 mutation load; but low APC mutation load [7], [58], [59]. Because Red 40 is metabolized by the gut microbiome to cresidine-4-sulfonic acid (CSA-Na) and 1-amino-2-naphthol-6-sulfonic acid (ANSA-Na), future studies will test whether these metabolites also damage DNA. Indeed, previous studies have shown that ANSA-Na can trigger colitis [35]; and that Red 40 and its’ metabolites can impact the DNA and have pro-inflammatory properties [28], [29], [60], [61].
In studying the impact of Red 40 on the microbiome, our data show that Red 40 does not appear to change the general composition of the bacterial community - indicated by the lack of change in alpha diversity. This result is consistent with other recent studies [35], [36]. However, in the presence of a HFD, the consumption of Red 40 contributes to a change in the alpha and beta diversities of the fecal microbiome (Fig. 6, Fig. 7). Although we have evidence of specific species of bacteria impacted by Red 40, further studies are needed to delineate which species mechanistically contribute to CRC because of Red 40 exposure. Only two studies have previously explored which bacteria species metabolize Red 40 and contribute to colon anomalies [35], [62]. He et al. showed that Red 40 is metabolized by B. ovatus and E. faecalis; but not E. coli [35]. Chung et al. [62] showed high metabolism by Anaerobes, Fusobacterium; modest metabolism by Bacteroides, Bifidobacterium, and Citrobacter; and that Red 40 is not metabolized by Acidamiococcus, Peptostreptococcus [62]. Given the significance of the gut microbiome to health, and that Red 40 is consumed by a diverse and large number of people, a further understanding of the interactions between Red 40 and the gut microbiome will be of high importance to public health.
Because of the tight control of dietary exposures and careful measurement of CRC-related outcomes, our study has added to the foundation of growing scientific evidence that Red 40 and its metabolites target endpoints that control the genesis of CRC: DNA damage, functional mutation to p53, low-grade inflammation in the distal colon, and the microbiome. The use of in vitro and in vitro models has allowed us to dive into mechanisms relevant to CRC. Despite its strengths, this study has some weaknesses, including a lack of mechanistic insight into these observational changes. Further studies will be needed to build on the foundation that Red 40 causes low-grade inflammation; and ask how it does this, and which microbial species are most relevant to this process. Until then, limitations remain to extrapolating results from cell culture and animal studies to humans. Studies in humans will be needed to deepen understanding of the role of Red 40 in the natural history EOCRC. These studies will require careful planning and execution to address practical and ethical concerns. Interventions will need to be designed with temporal limitation in mind – e.g., using the same kinds of assays employed in this study and taking advantage of methodologic innovations that we have discussed earlier [63]. As with most epidemiologic advancements, this also will require careful design, planning, execution, and analyses of observational studies that do not share the constraints of trials that are constrained to studying small study groups over relatively short periods of time (i.e., the weeks or months needed to see changes in intermediate biomarkers as opposed to the years or decades needed to see CRC [64]. Future laboratory studies, conducted in tandem with human studies, should elaborate relevant mechanisms of action.
In conclusion, our study, distinguished by meticulous dietary control and selected CRC-related outcome measurements, underscores Red 40's adverse effects. The combination of in vitro and in vivo models facilitates nuanced exploration of CRC-relevant mechanisms. Nonetheless, our study does possess limitations, including the need for more detailed mechanistic insights into observed changes. As we continue to examine how Red 40 triggers low-grade inflammation, with emphasis on relevant microbial species, we acknowledge the challenges in extrapolating results from animal models to human contexts. To this, the current study advances our understanding of Red 40's detrimental effects on health, highlighting the potential of chronic exposure to elevate CRC risk. We demonstrate that Red 40 inflicts DNA damage, particularly in the presence of a HFD, which leads to altered gut microbiota and subsequent inflammation in the distal colon. These findings contribute to the growing body of evidence illustrating Red 40's adverse impact on colorectal carcinogenesis. Future endeavors should incorporate human studies to deepen insights into Red 40's role in EOCRC's natural history, alongside further laboratory investigations to elucidate underlying mechanisms.
Author contributions
QZ, AAC, and LJ conceived and designed the experiments; QZ, N, AAC, NS, JH, MK, and QH performed the experiments; IC examined pathology; PJB and CEB oversaw organoid experiments. All contributed to writing the manuscript.
Funding
This work was supported by the National Institutes of Health, grants 1R01CA246809 and 1U01CA272977 issued to Lorne J. Hofseth.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Lorne Hofseth reports financial support was provided by National Institutes of Health.
Acknowledgements
We thank Dr. Vitali Sikirzhytski, a Director of the Microscopy Core of the University of South Carolina Center for Targeted Therapeutics for his assistance with Comet assay imaging, and image analysis.
Handling Editor: Dr. L.H. Lash
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.toxrep.2023.08.006.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
No data was used for the research described in the article.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Colorectal cancer mortality rates in adults aged 20 to 54 years in the United States, 1970-2014. JAMA. 2017;318(6):572–574. doi: 10.1001/jama.2017.7630. Aug 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fedewa S.A., et al. Colorectal cancer statistics, 2017. CA: Cancer J. Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. May 6. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Fedewa S.A., Anderson W.F., et al. Colorectal cancer incidence patterns in the United States, 1974-2013. J. Natl. Cancer Inst. 2017;109(8) doi: 10.1093/jnci/djw322. Aug 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA: Cancer J. Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. (Jan) [DOI] [PubMed] [Google Scholar]
- 5.Siegel R.L., Jemal A. Percentage of colorectal cancer diagnosed in adults aged younger than 50 years. Cancer. 2016;122(9):1462–1463. doi: 10.1002/cncr.29980. May 1. [DOI] [PubMed] [Google Scholar]
- 6.Venugopal A., Carethers J.M. Epidemiology and biology of early onset colorectal cancer. Excli J. 2022;21:162–182. doi: 10.17179/excli2021-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofseth L.J., Hebert J.R., Chanda A., et al. Early-onset colorectal cancer: initial clues and current views. Nat. Rev. Gastroenterol. Hepatol. 2020 doi: 10.1038/s41575-019-0253-4. Feb 21. Feb 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen L.H., Liu P.H., Zheng X., et al. Sedentary behaviors, TV viewing time, and risk of young-onset colorectal cancer. JNCI Cancer Spectr. 2018;2(4):pky073. doi: 10.1093/jncics/pky073. (Nov) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng X., Hur J., Nguyen L.H., et al. Comprehensive assessment of diet quality and risk of precursors of early-onset colorectal cancer. J. Natl. Cancer Inst. 2020 doi: 10.1093/jnci/djaa164. Nov 2. Nov 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hebert J.R., Hofseth L.J. Elsevier Science,; 2022. Diet, Inflammation, and Health. [Google Scholar]
- 11.Srour B., Kordahi M.C., Bonazzi E., Deschasaux-Tanguy M., Touvier M., Chassaing B. Ultra-processed foods and human health: from epidemiological evidence to mechanistic insights. Lancet Gastroenterol. Hepatol. 2022;7(12):1128–1140. doi: 10.1016/s2468-1253(22)00169-8. (Dec) [DOI] [PubMed] [Google Scholar]
- 12.Kliemann N., Al Nahas A., Vamos E.P., et al. Ultra-processed foods and cancer risk: from global food systems to individual exposures and mechanisms. Br. J. Cancer. 2022;127(1):14–20. doi: 10.1038/s41416-022-01749-y. (Jul) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baygi F., Mohammadi-Nasrabadi F., Zyriax B.C., et al. Global overview of dietary outcomes and dietary intake assessment methods in maritime settings: a systematic review. BMC Public Health. 2021;21(1):1579. doi: 10.1186/s12889-021-11593-z. Aug 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popkin B.M., Adair L.S., Ng S.W. Global nutrition transition and the pandemic of obesity in developing countries. Nutr. Rev. 2012;70(1):3–21. doi: 10.1111/j.1753-4887.2011.00456.x. (Jan) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Giovannucci E.L. Ultra-processed foods and health: a comprehensive review. Crit. Rev. Food Sci. Nutr. 2022:1–13. doi: 10.1080/10408398.2022.2084359. Jun 6. [DOI] [PubMed] [Google Scholar]
- 16.Statovci D., Aguilera M., MacSharry J., Melgar S. The impact of western diet and nutrients on the microbiota and immune response at mucosal interfaces. Front. Immunol. 2017;8:838. doi: 10.3389/fimmu.2017.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Keefe S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016;13(12):691–706. doi: 10.1038/nrgastro.2016.165. (Dec) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benninghoff A.D., Hintze K.J., Monsanto S.P., et al. Consumption of the total western diet promotes colitis and inflammation-associated colorectal cancer in mice. Nutrients. 2020;12(2) doi: 10.3390/nu12020544. Feb 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen L.H., Cao Y., Hur J., et al. The sulfur microbial diet is associated with increased risk of early-onset colorectal cancer precursors. Gastroenterology. 2021 doi: 10.1053/j.gastro.2021.07.008. Jul 14. Jul 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ugai T., Liu L., Tabung F.K., et al. Prognostic role of inflammatory diets in colorectal cancer overall and in strata of tumor-infiltrating lymphocyte levels. Clin. Transl. Med. 2022;12(11) doi: 10.1002/ctm2.1114. (Nov) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hang D., Wang L., Fang Z., et al. Ultra-processed food consumption and risk of colorectal cancer precursors: results from three prospective cohorts. J. Natl. Cancer Inst. 2022 doi: 10.1093/jnci/djac221. Dec 7. Dec 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joh H.K., Lee D.H., Hur J., et al. Simple sugar and sugar-sweetened beverage intake during adolescence and risk of colorectal cancer precursors: Adolescent sugar intake and colorectal polyp. Gastroenterology. 2021 doi: 10.1053/j.gastro.2021.03.028. Mar 19. Mar 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hur J., Otegbeye E., Joh H.K., et al. Sugar-sweetened beverage intake in adulthood and adolescence and risk of early-onset colorectal cancer among women. Gut. 2021 doi: 10.1136/gutjnl-2020-323450. May 6. May 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue Y., Hur J., Cao Y., et al. Prospective evaluation of dietary and lifestyle pattern indices with risk of colorectal cancer in a cohort of younger women. Ann. Oncol.: Off. J. Eur. Soc. Med. Oncol. 2021 doi: 10.1016/j.annonc.2021.03.200. Mar 31. Mar 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potera C. The artificial food dye blues. Environ. Health Perspect. 2010;118(10):A428. doi: 10.1289/ehp.118-a428. (Oct) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doell D.L., Folmer D.E., Lee H.S., Butts K.M., Carberry S.E. Exposure estimate for FD&C colour additives for the US population. Food Addit. Contam. Part A, Chem., Anal., Control, Expo. risk Assess. 2016;33(5):782–797. doi: 10.1080/19440049.2016.1179536. (May) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batada A., Jacobson M.F. Prevalence of artificial food colors in grocery store products marketed to children. Clin. Pediatr. 2016;55(12):1113–1119. doi: 10.1177/0009922816651621. (Oct) [DOI] [PubMed] [Google Scholar]
- 28.Leo L., Loong C., Ho X.L., Raman M.F.B., Suan M.Y.T., Loke W.M. Occurrence of azo food dyes and their effects on cellular inflammatory responses. Nutr. (Burbank, Los Angel Cty., Calif. ) 2018;46:36–40. doi: 10.1016/j.nut.2017.08.010. (Feb) [DOI] [PubMed] [Google Scholar]
- 29.Khayyat L.I., Essawy A.E., Sorour J.M., Soffar A. Sunset yellow and allura red modulate Bcl2 and COX2 expression levels and confer oxidative stress-mediated renal and hepatic toxicity in male rats. PeerJ. 2018;6 doi: 10.7717/peerj.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(JECFA) JFWECoFA . World Health Organization,; 2017. Safety evaluation of certain food additives. [Google Scholar]
- 31.Raposa B., Ponusz R., Gerencser G., et al. Food additives: sodium benzoate, potassium sorbate, azorubine, and tartrazine modify the expression of NFkappaB, GADD45alpha, and MAPK8 genes. Physiol. Int. 2016;103(3):334–343. doi: 10.1556/2060.103.2016.3.6. (Sep) [DOI] [PubMed] [Google Scholar]
- 32.Meyer S.K., Probert P.M.E., Lakey A.F., et al. Hepatic effects of tartrazine (E 102) after systemic exposure are independent of oestrogen receptor interactions in the mouse. Toxicol. Lett. 2017;273:55–68. doi: 10.1016/j.toxlet.2017.03.024. May 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown J.P. Reduction of polymeric azo and nitro dyes by intestinal bacteria. Appl. Environ. Microbiol. 1981;41(5):1283–1286. doi: 10.1128/aem.41.5.1283-1286.1981. (May) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon Y.H., Banskota S., Wang H., et al. Chronic exposure to synthetic food colorant Allura Red AC promotes susceptibility to experimental colitis via intestinal serotonin in mice. Nat. Commun. 2022;13(1):7617. doi: 10.1038/s41467-022-35309-y. Dec 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Z., Chen L., Catalan-Dibene J., et al. Food colorants metabolized by commensal bacteria promote colitis in mice with dysregulated expression of interleukin-23. Cell Metab. 2021 doi: 10.1016/j.cmet.2021.04.015. May 11. May 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L., He Z., Reis B.S., et al. IFN-γ(+) cytotoxic CD4(+) T lymphocytes are involved in the pathogenesis of colitis induced by IL-23 and the food colorant Red 40. Cell Mol. Immunol. 2022;19(7):777–790. doi: 10.1038/s41423-022-00864-3. (Jul) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan I.S., Ali S., Dar K.B., et al. Toxicological analysis of synthetic dye orange red on expression of NFκB-mediated inflammatory markers in Wistar rats. Drug Chem. Toxicol. 2021:1–11. doi: 10.1080/01480545.2021.1979579. Sep 24. [DOI] [PubMed] [Google Scholar]
- 38.Vojdani A., Vojdani C. Immune reactivity to food coloring. Alter. Ther. Health Med. 2015;21(Suppl 1):52–62. [PubMed] [Google Scholar]
- 39.Guda K., Marino J.N., Jung Y., Crary K., Dong M., Rosenberg D.W. Strain-specific homeostatic responses during early stages of azoxymethane-induced colon tumorigenesis in mice. Int. J. Oncol. 2007;31(4):837–842. (Oct) [PubMed] [Google Scholar]
- 40.Rosenberg D.W., Giardina C., Tanaka T. Mouse models for the study of colon carcinogenesis. Carcinogenesis. 2009;30(2):183–196. doi: 10.1093/carcin/bgn267. (Feb) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaparala A., Tashkandi H., Chumanevich A.A., et al. Molecules from American ginseng suppress colitis through nuclear factor erythroid-2-related factor 2. Nutrients. 2020;12(6) doi: 10.3390/nu12061850. Jun 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaparala A., Poudyal D., Tashkandi H., et al. Panaxynol, a bioactive component of American ginseng, targets macrophages and suppresses colitis in mice. Oncotarget. 2020;11(22):2026–2036. doi: 10.18632/oncotarget.27592. Jun 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chumanevich A.A., Chaparala A., Witalison E.E., et al. Looking for the best anti-colitis medicine: a comparative analysis of current and prospective compounds. Oncotarget. 2017;8(1):228–237. doi: 10.18632/oncotarget.13894. Jan 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chumanevich A.A., Causey C.P., Knuckley B.A., et al. Suppression of colitis in mice by Cl-amidine: a novel peptidylarginine deiminase inhibitor. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;300(6):G929–G938. doi: 10.1152/ajpgi.00435.2010. (Jun) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chumanevich A.A., Witalison E.E., Chaparala A., et al. Repurposing the anti-malarial drug, quinacrine: new anti-colitis properties. Oncotarget. 2016;7(33):52928–52939. doi: 10.18632/oncotarget.10608. Aug 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaked H., Hofseth L.J., Chumanevich A., et al. Chronic epithelial NF-κB activation accelerates APC loss and intestinal tumor initiation through iNOS up-regulation. Proc. Natl. Acad. Sci. USA. 2012;109(35):14007–14012. doi: 10.1073/pnas.1211509109. Aug 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin Y., Kotakadi V.S., Ying L., et al. American ginseng suppresses inflammation and DNA damage associated with mouse colitis. Carcinogenesis 2008. 2008;29(12):2351–2359. doi: 10.1093/carcin/bgn211. (Epub) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimada C., Kano K., Sasaki Y.F., Sato I., Tsudua S. Differential colon DNA damage induced by azo food additives between rats and mice. J. Toxicol. Sci. 2010;35(4):547–554. doi: 10.2131/jts.35.547. (Aug) [DOI] [PubMed] [Google Scholar]
- 49.Sasaki Y.F., Kawaguchi S., Kamaya A., et al. The comet assay with 8 mouse organs: results with 39 currently used food additives. Mutat. Res. 2002;519(1–2):103–119. doi: 10.1016/s1383-5718(02)00128-6. Aug 26. [DOI] [PubMed] [Google Scholar]
- 50.Tsuda S., Murakami M., Matsusaka N., Kano K., Taniguchi K., Sasaki Y.F. DNA damage induced by red food dyes orally administered to pregnant and male mice. Toxicol. Sci.: Off. J. Soc. Toxicol. 2001;61(1):92–99. doi: 10.1093/toxsci/61.1.92. (May) [DOI] [PubMed] [Google Scholar]
- 51.Bastaki M., Farrell T., Bhusari S., Pant K., Kulkarni R. Lack of genotoxicity in vivo for food color additive Allura Red AC. Food Chem. Toxicol.: Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2017;105:308–314. doi: 10.1016/j.fct.2017.04.037. (Jul) [DOI] [PubMed] [Google Scholar]
- 52.Yu I., Wu R., Tokumaru Y., Terracina K.P., Takabe K. The role of the microbiome on the pathogenesis and treatment of colorectal cancer. Cancers. 2022;14(22) doi: 10.3390/cancers14225685. Nov 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato T., Clevers H. Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 2013;945:319–328. doi: 10.1007/978-1-62703-125-7_19. [DOI] [PubMed] [Google Scholar]
- 54.Liu C., Banister C.E., Weige C.C., et al. PRDM1 silences stem cell-related genes and inhibits proliferation of human colon tumor organoids. Proc. Natl. Acad. Sci. USA. 2018;115(22):E5066–e5075. doi: 10.1073/pnas.1802902115. May 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saatci O., Kaymak A., Raza U., et al. Targeting lysyl oxidase (LOX) overcomes chemotherapy resistance in triple negative breast cancer. Nat. Commun. 2020;11(1):2416. doi: 10.1038/s41467-020-16199-4. May 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Canli O., Nicolas A.M., Gupta J., et al. Myeloid cell-derived reactive oxygen species induce epithelial mutagenesis. Cancer Cell. 2017;32(6):869–883 e5. doi: 10.1016/j.ccell.2017.11.004. Dec 11. [DOI] [PubMed] [Google Scholar]
- 57.Liebl M.C., Hofmann T.G. The role of p53 signaling in colorectal cancer. Cancers. 2021;13(9) doi: 10.3390/cancers13092125. Apr 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xicola R.M., Manojlovic Z., Augustus G.J., et al. Lack of APC somatic mutation is associated with early-onset colorectal cancer in African Americans. Carcinogenesis. 2018;39(11):1331–1341. doi: 10.1093/carcin/bgy122. Dec 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Figer A., Irmin L., Geva R., Flex D., Sulkes A., Friedman E. Genetic analysis of the APC gene regions involved in attenuated APC phenotype in Israeli patients with early onset and familial colorectal cancer. Br. J. Cancer. 2001;85(4):523–526. doi: 10.1054/bjoc.2001.1959. Aug 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kobylewski S., Jacobson M.F. Toxicology of food dyes. Int. J. Occup. Environ. Health. 2012;18(3):220–246. doi: 10.1179/1077352512z.00000000034. (Jul-Sep) [DOI] [PubMed] [Google Scholar]
- 61.Jabeen H.S., ur Rahman S., Mahmood S., Anwer S. Genotoxicity assessment of amaranth and allura red using Saccharomyces cerevisiae. Bull. Environ. Contam. Toxicol. 2013;90(1):22–26. doi: 10.1007/s00128-012-0870-x. (Jan) [DOI] [PubMed] [Google Scholar]
- 62.Chung K.T., Fulk G.E., Egan M. Reduction of azo dyes by intestinal anaerobes. Appl. Environ. Microbiol. 1978;35(3):558–562. doi: 10.1128/aem.35.3.558-562.1978. (Mar) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hebert J.R., Frongillo E.A., Adams S.A., et al. Perspective: randomized controlled trials are not a panacea for diet-related research. Adv. Nutr. (Bethesda, Md) 2016;7(3):423–432. doi: 10.3945/an.115.011023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hebert J.R., Miller D.R. Methodologic considerations for investigating the diet-cancer link. Am. J. Clin. Nutr. 1988;47:1068–1077. doi: 10.1093/ajcn/47.6.1068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
No data was used for the research described in the article.