Summary
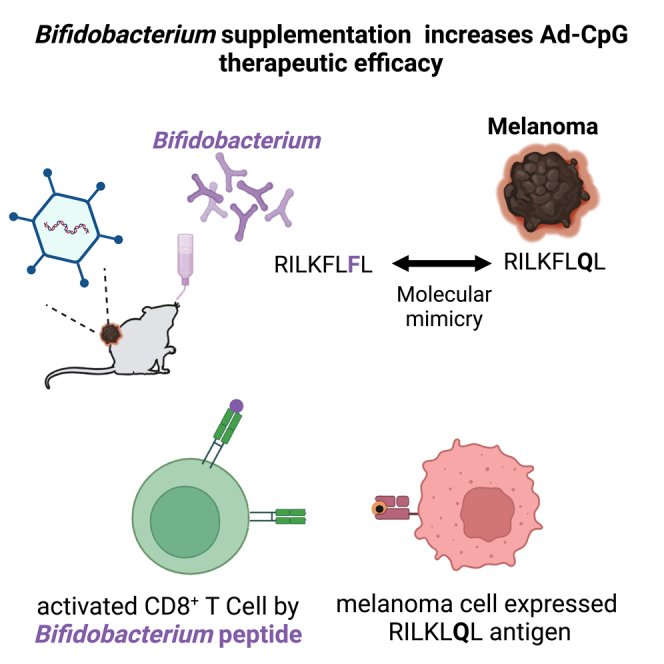
Gut microbiota plays a key role in modulating responses to cancer immunotherapy in melanoma patients. Oncolytic viruses (OVs) represent emerging tools in cancer therapy, inducing a potent immunogenic cancer cell death (ICD) and recruiting immune cells in tumors, poorly infiltrated by T cells. We investigated whether the antitumoral activity of oncolytic adenovirus Ad5D24-CpG (Ad-CpG) was gut microbiota-mediated in a syngeneic mouse model of melanoma and observed that ICD was weakened by vancomycin-mediated perturbation of gut microbiota. Ad-CpG efficacy was increased by oral supplementation with Bifidobacterium, reducing melanoma progression and tumor-infiltrating regulatory T cells. Fecal microbiota was enriched in bacterial species belonging to the Firmicutes phylum in mice treated with both Bifidobacterium and Ad-CpG; furthermore, our data suggest that molecular mimicry between melanoma and Bifidobacterium-derived epitopes may favor activation of cross-reactive T cells and constitutes one of the mechanisms by which gut microbiota modulates OVs response.
Subject areas: Virology, Microbiome, Cancer
Graphical abstract
Highlights
-
•
Therapeutic antitumor efficacy of Ad-CpG is gut microbiota mediated
-
•
Bifidobacterium supplementation increases the antitumoral efficacy of Ad-CpG s in vivo
-
•
Enrichment of species belonging to Firmicutes in the microbiota of Ad-CpG-treated mice
-
•
Immunogenicity of a Bifidobacterium peptide, highly similar to a melanoma peptide
Virology; Microbiome; Cancer
Introduction
In the era of incredible successes achieved by immunotherapy, many efforts are focusing on the identification of the sources of the variability of therapies, observed in clinical outcomes.1,2 The basis for variable patients’ responses to antibodies targeting programmed cell death protein 1 (PD-1) and cytotoxic T lymphocyte antigen-4 (CTLA-4/B7.1/B7.2) involves many biological factors, including genetic variability, differences in tumor mutational loads, and microbiota composition. The role of gut microbiota in the modulation of response to antitumoral therapies is becoming evident even considering new studies on its impact on the response to both chemotherapy3 and immunotherapy.4,5 In addition, tumor growth is modulated by intestinal microbiota: in fact, bacterial depletion by aggressive antibiotic treatment induces faster tumor progression and reduces the efficacy of many treatments, as shown in murine model of breast and colorectal cancer.6,7 Multiple studies show that microbiota plays a pivotal role in modulating response to immune-checkpoint inhibitors (ICIs) in mice models and patients, promoting its ability to affect treatment outcomes.8,9 Strong evidence in support of the immunomodulatory activity exerted by the gut microbiota was confirmed by the loss of therapeutic activity of recombinant monoclonal antibody anti-CTLA-4 (9D9) against sarcoma in mice housed under specific pathogen-free (SPF) conditions and then treated with multiple broad-spectrum antibiotics to sterilize the gut.10 Sivan et al. demonstrated that members of the Bifidobacteriales were abundant in mice that exhibited reduced growth of melanomas and improved cytotoxic T lymphocytes (CTLs)-mediated immunosurveillance. Supplementation with Bifidobacterium breve or Bifidobacterium longum into Bifidobacteriales-free mice was sufficient to reduce melanoma growth and restore anti-melanoma CTL response.11 Several studies indicate that gut microbiota affects the efficacy of cancer immunotherapies by engaging innate and/or adaptive immune systems9,12,13,14; for instance, commensal bacterium Bifidobacterium breve stimulates antitumoral immune responses by amplifying specific T cells; the latter cross-react with model neoantigen expressed in melanoma tumor.15,16,17 Indeed, specific antigens, derived from peptides of intracellular bacteria peptides, can be presented within human leukocyte antigen (HLA)-I and HLA-II molecules expressed on the surface of melanoma cells and elicit a tumor-infiltrating T cell immune response. In particular, bacterial peptides that invade cancer cells can be presented on the tumor cell surface and recognized by the immune system.18 The intratumoral bacteria repertoire identified in melanoma patients can affect T cell immune reactivity in the tumor microenvironment (TME).19 All these discoveries consistently support the idea that the gut microbiota plays a pivotal role in shaping antitumoral immune responses elicited by several immunotherapies, even if, different scenarios have been identified according to the type of tumor and therapy.20
Oncolytic virotherapy is a promising approach in cancer immunotherapy: working as an active immunizing agent, oncolytic viruses (OVs) convert immunologically “cold” tumors into “hot” ones. OVs are very attractive for potential synergistic combination with ICIs; indeed, their ability to recruit CTLs can be combined with the ICIs' skill to avoid tumor immune evasion and therefore induce a sustained response.21,22,23 The immune-mediated eradication of tumors through oncolytic adenoviruses makes them a promising strategy for combination therapy21,24,25,26 to address the limitations of ICIs. Actually, multiple ongoing clinical trials are aiming to investigate the combined therapeutic efficacy of ICIs and oncolytic adenoviruses.27 Despite the fact that the role of the gut microbiota in modulating immunotherapy efficacy has been widely considered in many types of cancers, its role in the oncolytic virotherapy of solid tumors has never been investigated to date. Given that gut microbiota influences ICIs efficacy, it is possible to hypothesize that it could also affect the antitumoral activity of oncolytic adenoviruses. In addition, the eligibility of OVs as a valuable candidate for combination therapy with ICIs, whose activity is gut microbiome mediated, is relevant to elucidate the possible role of microbiota in the modulation of response to OV treatment. Here, we decided to investigate whether the efficacy of oncolytic adenovirus Ad5D24-CpG (Ad-CpG) treatment24 could be affected by the gut microbiota, in a syngeneic mouse model of melanoma. Ad-CpG is a modified version of the oncolytic adenovirus Ad5D24 virus that includes the insertion of 18 immuno-stimulatory CpG islands.24 We observed that gut microbiota depletion by treatment with antibiotics10 impaired the response of subcutaneous tumors to Ad-CpG therapy, reducing tumor-infiltrating interferon (IFN)-γ CD8 (cluster of differentiation 8) T cells, confirming the role of intestinal bacteria in the antitumoral responses elicited by Ad-CpG. Considering the ability of Bifidobacterium to improve the effector function of tumor-specific CD8+ T cells,11 we evaluated tumor progression in the same model of melanoma after treatment with a combination of Ad-CpG and Bifidobacterium spp. cocktail (Bifidus), used as single agents or in combination, and we observed a reduced tumor progression in mice treated with the combination therapy. To better understand the mechanism responsible for this synergistic effect,28 we identified all Bifidobacterium-derived peptides highly similar to murine melanoma epitopes and evaluated their possible contribution to the antitumoral effect.
Results
Gut microbiota affects the response to Ad-CpG in a syngeneic mouse model of melanoma
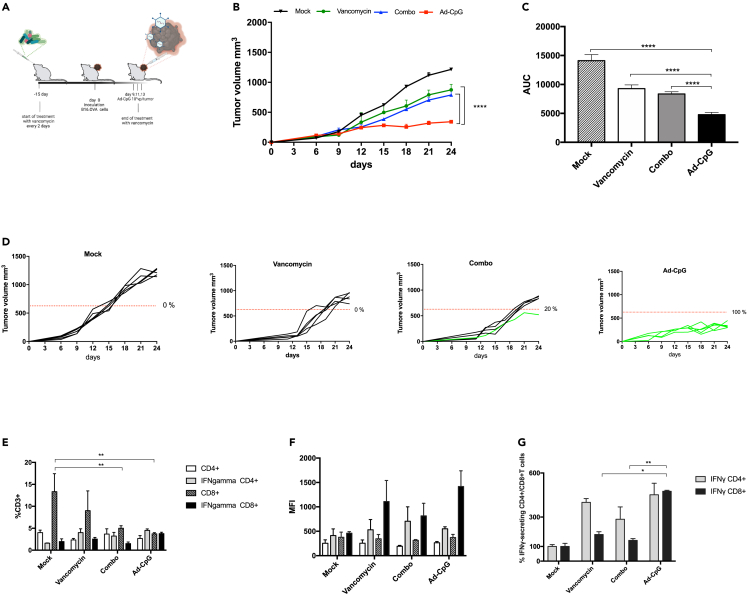
The observation that cancer immunotherapy with CTLA-4 and PD-L1 blockade is affected by gut microbiota composition10,11 prompted us to investigate whether oncolytic virotherapy efficacy could also be influenced by intestinal bacteria. At first, we planned to evaluate whether gut microbiota depletion and unbalance after long-term antibiotic use could affect the response to Ad-CpG therapy. To this end, we treated a group of mice with vancomycin starting two weeks before B16.OVA melanoma cells inoculation until the first intratumoral administration of Ad-CpG, as indicated in the experimental design (Figure 1, panel A). The group of mice treated with vancomycin and Ad-CpG had significantly faster tumor growth compared to the group treated with Ad-CpG alone (Figure 1, panel B); interestingly, tumor progression in the group treated with a combined regimen (Ad-CpG + vancomycin) was comparable to what was observed in mice treated with vancomycin alone. Therefore, vancomycin treatment seems to abrogate the effect of virotherapy. The Ad-CpG-treated tumors showed significantly slower growth kinetics as represented by the area under the curve (AUC) analysis (Figure 1, panel C). Furthermore, the reduced response observed in mice treated with Ad-CpG combined with vancomycin can be better appreciated considering the single tumor growth curves for each mouse treated (20% responders, Figure 1, panel D). Vancomycin treatment however reduced tumor growth compared to mock-treated mice: this observation is consistent with the current literature.29 To assess how vancomycin pre-treatment influences the immune response induced by Ad-CpG, we analyzed the phenotype of tumor-infiltrating lymphocytes (TILs) in treated mice and controls. Both CD4+ and CD8+ T cells levels were unaffected by vancomycin pre-treatment (Figure 1, panels E and F); on the other hand, the percentage of IFN-γ-producing CD8+ T cells was significantly lower in mice that received the combined regimen compared to mice treated only with Ad-CpG (Figure 1, panel G). The quadrant gating strategy that has been used for all samples is reported in supplemental data (Figure S1). These data suggest that antibiotic pre-treatment causes a reduction of the IFN-γ-secreting CD8+ T cells infiltrating the tumor, negatively affecting the anticancer Ad-CpG efficacy.
Figure 1.
Perturbation of gut microbiome reduces the efficacy of Ad-CpG treatment in a syngeneic mouse model of melanoma
(A) Experimental design: vancomycin was administered by oral gavage every 2 days, starting 15 days before tumor implantation until the first Ad-CpG injection. At day 0, 3.5 × 105 B16.OVA cells were injected in the right flank of female C57BL/6J mice (n = 5 per group). Ad-CpG was administered intratumorally on days 9, 11, and 13.
(B) tumor-bearing mice (n = 5 per group) were treated with saline solution (Mock), 100 μL of vancomycin (10 mg/mL) by oral gavage, 1 × 10E9 vp/tumor of Ad-CpG and with a combination of Ad-CpG + vancomycin. Tumor size was determined for each mouse with a digital caliper; results are graphed as mean for each treatment groups +/− standard single error of the mean (SEM); statistical difference has been determined with two-way ANOVA (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; p < 0.0001).
(C) Average area under the curves relative to tumor growth in the different experimental groups indicated as mean ± SEM. The statistical significance was evaluated by unpaired t test with Welch’s correction and the asterisks indicate statistical significance (∗∗∗∗p < 0.0001).
(D) Growth curves for each tumor are reported as a single graph for each group (n = 5 animals per group). The percentage of responders (mice with an absolute tumor volume lower than 627.25 mm3) for each group is indicated. Volumes of tumors in responders are represented in green, whereas tumors volumes in non-responder are represented by black curves.
(E) Percentages of total CD3+ CD4+ T lymphocytes, IFN-γ CD4+ CD3+, CD3+ CD8+ T lymphocytes and IFN-γ CD8+ CD3+ T cells present in the tumors. The statistical significance was evaluated by two-way ANOVA test using Tukey’s multiple comparisons test and the asterisks indicate statistical significance (∗∗p < 0.01) compared to CD8+CD3+ of tumors in mock-treated mice.
(F) Mean Fluorescent Intensity (MFI) of T cells represented in plot E as average +/− SEM.
(G) Percentages of IFN-γ+ CD4+ CD3+ and IFN-γ+ CD8+ CD3+ T cells calculated as ratio of IFNγ+ CD4+ CD3+/CD4+ CD3+ and ratio IFNγ+ CD8+ CD3+/CD8+ CD3+ and normalized on ratio value of mock group. The statistical significance was evaluated by paired student’s t test and the asterisks indicate statistical significance (∗p < 0.05; ∗∗p < 0.01) compared to the percentage of IFNγ+ CD8+ CD3+ of Ad-CpG-treated tumor. The data are represented as mean ± SEM.
Restoring gut microbiota balance reverses the effect of vancomycin treatment on the efficacy of Ad-CpG treatment
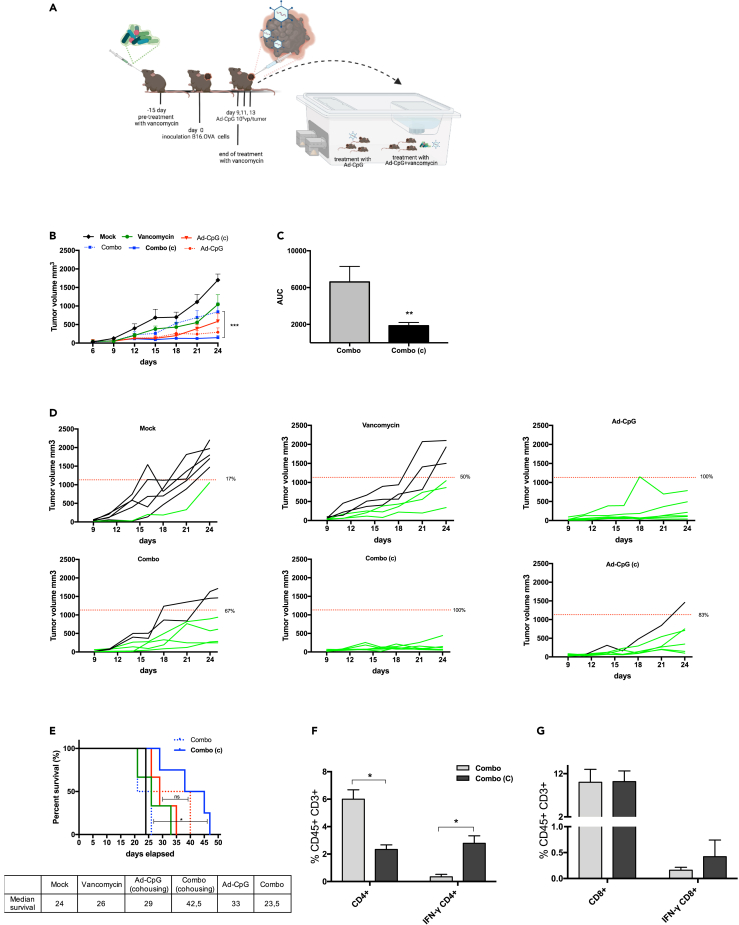
In order to confirm that the reduced efficacy of Ad-CpG was due to gut microbiota alterations, we cohoused a group of mice treated with a combined regimen (Ad-CpG + vancomycin) with mice treated with Ad-CpG, as indicated in the experimental design (Figure 2, panel A). We compared melanoma growth in the cohoused group to mice treated with Ad-CpG alone and to an additional group, treated with the same combined regimen but kept isolated. We observed that cohousing significantly reduces differences in tumor growth between mice treated with a combined regimen and those treated with Ad-CpG alone, indicating that a positive perturbation and likely a restoration of gut microbiota occurred (Figure 2, panel B). In addition, tumors of mice treated with the combined regimen and cohoused showed significantly slower growth kinetics compared to the isolated mice as represented by the AUC analysis (Figure 2, panel C). In fact, 100% of responders to Ad-CpG therapy was observed in the cohoused mice treated with the combined regimen while 67% of responders was observed in control group kept isolated (Figure 2, panel D). Overall survival of mice pre-treated with vancomycin was significantly improved by cohousing (median survival 42.5 days) compared to isolated mice (median survival 23.5 days, p < 0.05, Figure 2, panel E). We then investigated the levels of IFN-γ-producing CD4+ and CD8+ T cells in tumors: interestingly, IFN-γ CD4+/CD4+ T cells ratio in tumors in mice treated with the combined regimen and cohoused was 2.78 compared to a 0.34 ratio observed in tumors from mice treated with the combined regimen and kept isolated (Figure 2, panel F). Similarly, IFN-γ CD8+/CD8+ T cells ratio in tumors from mice treated with a combined regimen and cohoused was significantly higher compared to what was observed in mice treated with a combined regimen and kept isolated (Figure 2, panel G). These data suggest that the perturbation of gut microbiota caused by vancomycin affects levels of IFN-γ-producing CD8+ and CD4+ T cells and that subsequent cohousing with mice that did not receive antibiotic treatment influences microbiota composition, restoring oncolytic virotherapy efficacy.
Figure 2.
Cohousing with antibiotic-untreated mice can restore Ad-CpG efficacy in vancomycin-treated mice
(A) Experimental design of the cohousing experiment: vancomycin was administered by oral gavage every two days, 15 days before tumor implantation. At day 0, 3.5 × 105 B16.OVA cells were injected in the flank of female C57BL/6J mice (n = 6 per group). Ad-CpG was administered intratumorally on days 9, 11, and 13. After the third injection of the virus, we cohoused the group treated with the combined regimen with mice treated with only Ad-CpG. The groups involved in the cohousing were indicated as (Ad-CpG (c) and (Combo (c). Tumor-bearing mice (n = 6 per group) were treated with saline solution (Mock), 100 μL of 10 mg/mL vancomycin (Vancomycin), 1 × 10E9 vp/tumor of adenovirus (Ad-CpG) and with a combination of Ad-CpG and vancomycin (Combo).
(B) Tumor size was measured with a digital caliper and results are graphed as mean ± SEM; statistical difference has been determined with two-way ANOVA (∗∗∗p < 0.001).
(C) Average area under the curves relative to tumor growth in the different experimental groups is indicated as mean ± SEM; the statistical significance was evaluated by unpaired t test with Welch’s correction and the asterisks indicate statistical significance (∗∗p < 0.01).
(D) Growth curves for each tumor are reported as a single graph for each group (n = 6 animals per group). Percentage of responders (mice with an absolute tumor volume lower than 1132.22 mm3) for each group is indicated. The threshold level to evaluate tumor-bearing mice responder to Ad-CpG therapy was defined by the median value of volumes of the saline solution-treated tumors on day 21. Volumes of tumors in responders are represented in green, whereas tumors volumes in non-responders are represented by black curves.
(E) Survival curves relative to the experimental groups represented in panel B. The median survival of each group is reported in the table below the graph. The log rank Mantel-Cox analysis was used to calculate the p value (∗p < 0.05) of the survival curves.
(F) Percentage of IFN-γ CD4+/CD4+ T cells ratio. The statistical significance was examined by paired student’s t test and the asterisks indicate statistical significance (∗p < 0.05) compared to control group, treated with combined regimen and kept isolated (Combo).
(G) Percentage of IFN-γ CD8+/CD8+ T cells ratio. Results are graphed as mean for each treatment group +/− SEM.
Bifidobacterium spp. improves Ad-CpG efficacy by reducing tumor-infiltrating T-regs lymphocytes
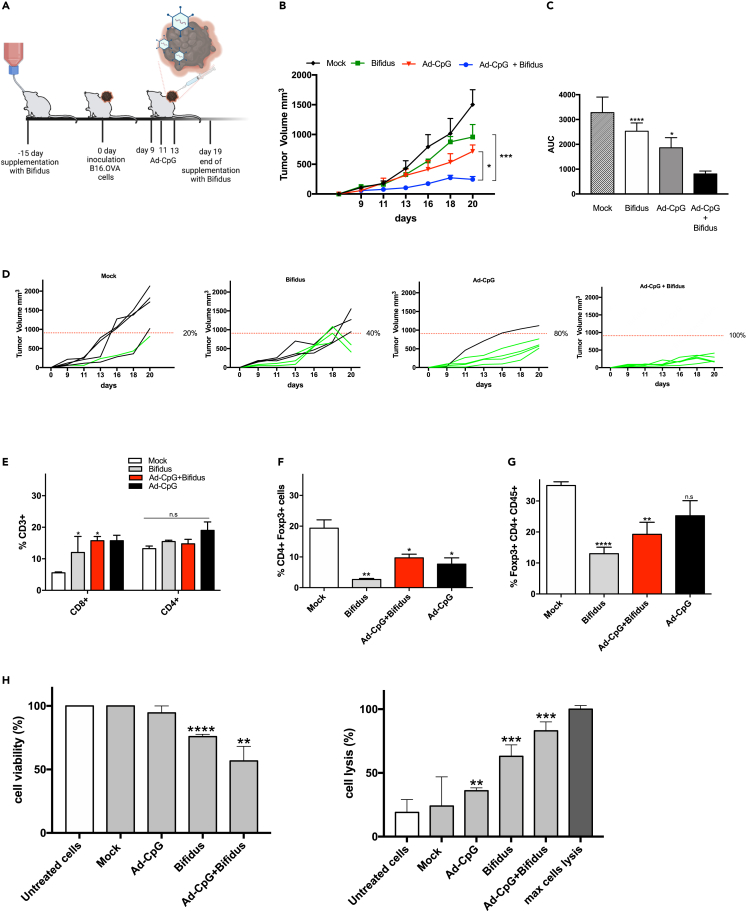
Recently, investigators have linked abundance of specific bacteria to immunotherapy success.8,9,10,11 In addition, commensal bacteria, such as Bifidobacterium, have been shown to modulate dendritic cells (DCs) function resulting in enhanced antitumor T cell activity.11 We then decided to inoculate syngeneic B16-OVA melanoma cells in female C57BL/6J and treat them with Ad-CpG alone or in combination with a Bifidobacterium cocktail (Bifidus) (Figure 3, panel A). We observed an increased response to Ad-CpG therapy in mice that received also Bifidus treatment (Figure 3, panel B) compared to groups treated with Ad-CpG alone and PBS. We observed significantly slower growth kinetics, as represented by the AUC analysis, in mice treated with the combination therapy (Ad-CpG + Bifidus) compared to those treated with Ad-CpG alone (Figure 3, panel C). Interestingly, oral administration of Bifidus by itself resulted in a slightly slower tumor progression compared to mock-treated mice; a different growing trend can be appreciated considering the tumor growth curves for single mouse treated (40% responders, Figure 3, panel D). These surprising results confirmed that Bifidus affects tumor progression and shows a synergistic action with Ad-CpG. Considering the potent immunogenicity of Ad-CpG and the limitation of human serotype 5-based-Ads that do not induce significant oncolysis in murine cells due to species-specific incompatibility, the observed effect most probably relies on the stimulation of the immune system. We analyzed lymphocytes in spleens and tumors collected from all experimental groups. We observed a consistent increase in CD8+ T cells in spleens from mice treated with Bifidus, Ad-CpG, and the combination therapy compared to controls (Figure 3, panel E). No significant differences were appreciated for CD4+ T cells between different treatments; however, a reduced percentage of T-regs lymphocytes was observed in the spleen (CD4+ Foxp3+) and in tumor samples (CD45+CD4+Foxp3+) from mice treated with the combination therapy (Figure 3, panels F–G, respectively) compared to mock group. This reduction is even more significant in mice that received only Bifidus supplementation, suggesting that Bifidus shapes the composition of the gut microbiota and is able to alter the T-regs lymphocytes subset intratumoral infiltration. The quadrant gating strategy that has been used for all samples is reported in supplementary materials (Figures S2–S5). In order to evaluate the presence of immunological memory against the tumor, we collected spleen samples from each group and performed a co-culture assay with B16-OVA cells and preimmunized murine lymphocytes to assess their ability to induce tumor cell death, as previously reported for co-culture-based assays set up to test immunomodulatory agents.30,31 This experiment is a preliminary assessment of antitumor effect of lymphocytes from mice treated with Ad-CpG, Bifidus, and their combination on cancer cells. The viability of B16-OVA treated with splenocytes derived from mice treated with the combination therapy was significantly lower (56.92% of untreated cells) compared to cells co-cultured with splenocytes collected from mice that received the other treatments (Figure 3, panel H, left plot). This reduced cell survival can be appreciated by images of B16-OVA acquired after splenocytes removal (Figure S6). Cytotoxic effect revealed an increased cell death in cells co-cultured with splenocytes derived from combination therapy-treated mice (Figure 3, panel H, right plot). Despite these preliminary results, we cannot exclude the possibility that the activation of splenocytes collected from Ad-CpG and Bifidobacterium-treated mice could be due to a non-specific killing mechanism by T cells activated by the combined regimen.
Figure 3.
Co-administration of Ad-CpG and Bifidobacterium spp. reduces melanoma growth in a syngeneic mouse model by reducing the activity of CD4+ regulatory T cells
(A and B) Experimental design: Bifidobacterium spp. cocktail (Bifidus) was added to the drinking water, every 2 days, starting 15 days before the cancer cell inoculation and until day 19. At day 0, 3.5 × 105 B16.OVA cells were injected in the right flank of female C57BL/6J mice (n = 5 per group). Ad-CpG was administered intratumorally on days 9, 11 and 13; B) tumor-bearing mice (n = 5 per group) were treated with saline solution (Mock), 10E9 CFU/mL of Bifidus (Bifidus), 1 × 10E9 vp/tumor of adenoviral therapy (Ad-CpG) and with a combination of the two monotherapies (Ad-CpG + Bifidus). Tumor size was determined for each mouse; results are graphed as mean for each treatment group +/− SEM; the statistical difference has been determined with two-way ANOVA (∗p < 0.05; ∗∗∗p < 0.001).
(C) Average area under the curves relative to the tumor growth in the different experimental groups is indicated as mean ± SEM. The statistical significance was evaluated by unpaired t test with Welch’s correction and the asterisks indicate statistical significance (∗p < 0.05; ∗∗∗p < 0.001).
(D) Growth curves for each tumor reported as a single graph for each group (n = 5 animals per group). The percentage of responders (mice with an absolute tumor volume lower than 907.91 mm3) for each group is indicated. Volumes of tumors in responders are represented in green, whereas tumors volume in non-responders are represented by black curves.
(E) Percentage of total CD3+ CD4+ T-lymphocytes present in the spleen of treated mice.
(F) Percentage of total CD3+ CD4+ Foxp3+ T-lymphocytes present in the spleens.
(G) Percentage of total CD45+ CD4+ Foxp3+ T lymphocytes present in the tumors. The statistical significance was evaluated by Student’s t test and the asterisks indicate statistical significance (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001) compared to the relative lymphocytes’ population of spleens or tumors in mock-treated mice.
(H) B16-OVA cells were incubated with or without murine splenocytes isolated from spleens of mice, previously treated with PBS (mock), Ad-CpG, Bifidus or a combination of virus and Bifidobacterium for 48 h. Left plot: cell viability is reported as a percentage of viable cells compared to untreated B16-OVA cancer cells (white bar). Right plot: tumor cell lysis was determined by measurement of the LDH levels released in the culture medium and expressed as a percentage with respect to max lysis control represented by cells treated with 10% Triton X-100. The experiments were performed in triplicate and statistical significance (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001) was assessed by the Student’s t test by comparing the cells, treated as indicated, to untreated cells. The data are represented as mean +/− standard deviation (SD).
Treatments with Ad-CpG alone and in combination with Bifidus cause perturbations in mice’s fecal microbiome
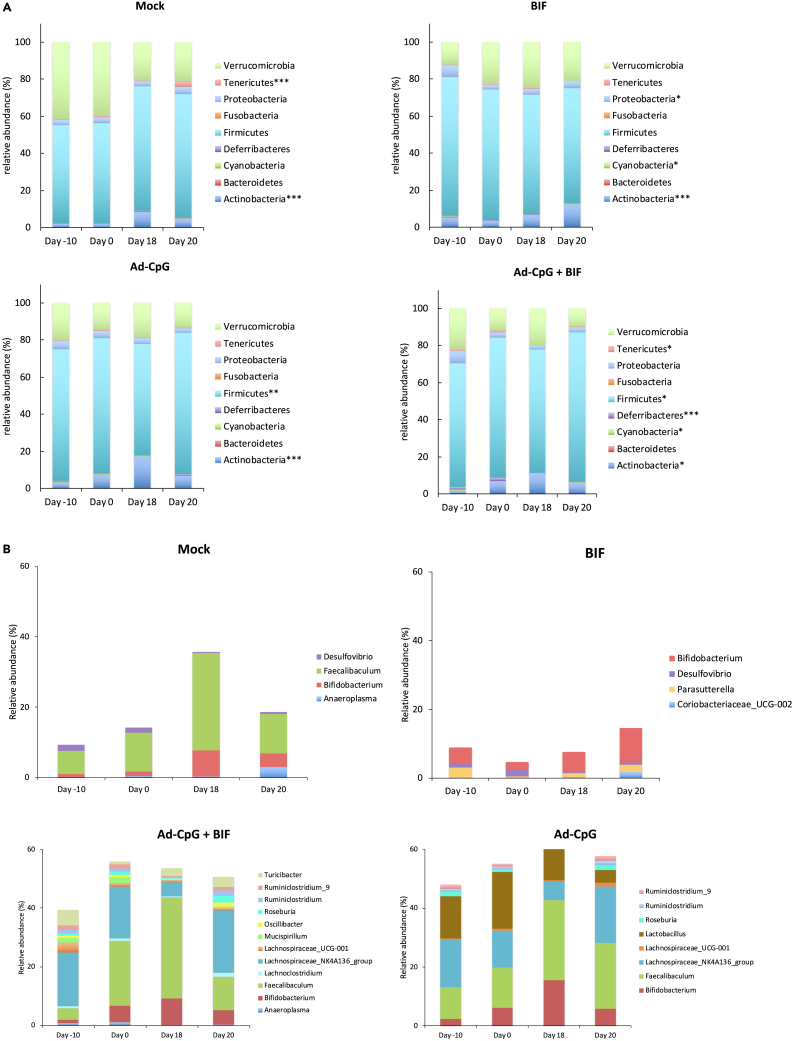
In order to better characterize the effect of Ad-CpG alone and in combination with Bifidus, we decided to investigate the fecal microbiome profiles of treated mice. We monitored fecal bacterial content over time in treated mice with 16S ribosomal RNAs (rRNAs). Ninety-three fecal samples were collected and processed for microbiome evaluation in a single next-generation sequencing (NGS) run. In order to investigate a possible link between gut microbiome composition and the different therapeutic regimens, we sought to investigate differences between the microbial profiles of groups. Alpha diversity plots showed that the microbial richness, measured by observed operational taxonomic unit (OTU), Chao1, and Shannon indices, does not differ between treatments or time points (Figure S7). Beta diversity analysis performed to test the inter-group variability showed that the combination of time and treatment explained a significant proportion of the variation in the microbial community. The statistical difference was evaluated by PERMANOVA (Adonis) statistical test using both weighted (p = 0.001, R2 = 0.449) and unweighted (p = 0.001, R2 = 0.31) UniFrac distance. (Figure S8, panels A–B). According to the Kruskal-Wallis test among the four times points for each treatment, distinct phyla were identified such as Verrucomicrobia, Tenericutes, Proteobacteria, Fusobacteria, Firmicutes, Deferribacteres, Cyanobacteria, Bacteroidetes, and Actinobacteria (Figure 4, panel A). In the untreated group (Mock), Tenericutes (p < 0.001) and Actinobacteria (p < 0.001) were significantly different between the four time points whereas, in the Bifidus-treated group, significant differences were observed in Proteobacteria, Cyanobacteria, and Actinobacteria. On the other hand, in the group treated with Ad-CpG, Firmicutes and Actinobacteria were significantly different between the four time points; in the group treated with Ad-CpG and Bifidus, additional phyla Tenericutes, Deferribacteres, and Cyanobacteria were identified (Figure 4, panel A). Interestingly, the fecal microbiota of both groups (Ad-CpG and Ad-CpG and Bifidus) treated with Ad-CpG was characterized by a significant abundance of phyla Firmicutes and Actinobacteria; specifically considering the time point Day 18, the relative abundance of Firmicutes phylum was about 63% in both groups treated with adenoviral therapy (Figure 4, panel A).
Figure 4.
Treatment of tumor with Ad-CpG increases Firmicutes in murine fecal microbiome
(A) The bar plots show Kruskal-Wallis test results on amplicon sequence variants (ASVs) grouped in phyla for each study group over the time points (Day-10, Day 0, Day 18, and Day 20). The treatments were indicated as Mock: untreated tumor-bearing-mice; BIF: Bifidobacterium oral supplementation-receiving mice; Ad-CpG: Ad5-CpG-treated mice; Ad-CpG + BIF: combined regimen-treated mice. Each column in the plot represents a time point and each color in the column represents the percentage of relative abundance. The phyla significantly (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001) different between the four time points and for each treatment, were shown in each panel alongside each bar plot graph.
(B) The bar plots show the relative abundances of differentially abundant genera (p < 0.05) according to the Kruskal-Wallis test among the four time points for each treatment. The color legends show the identified genera.
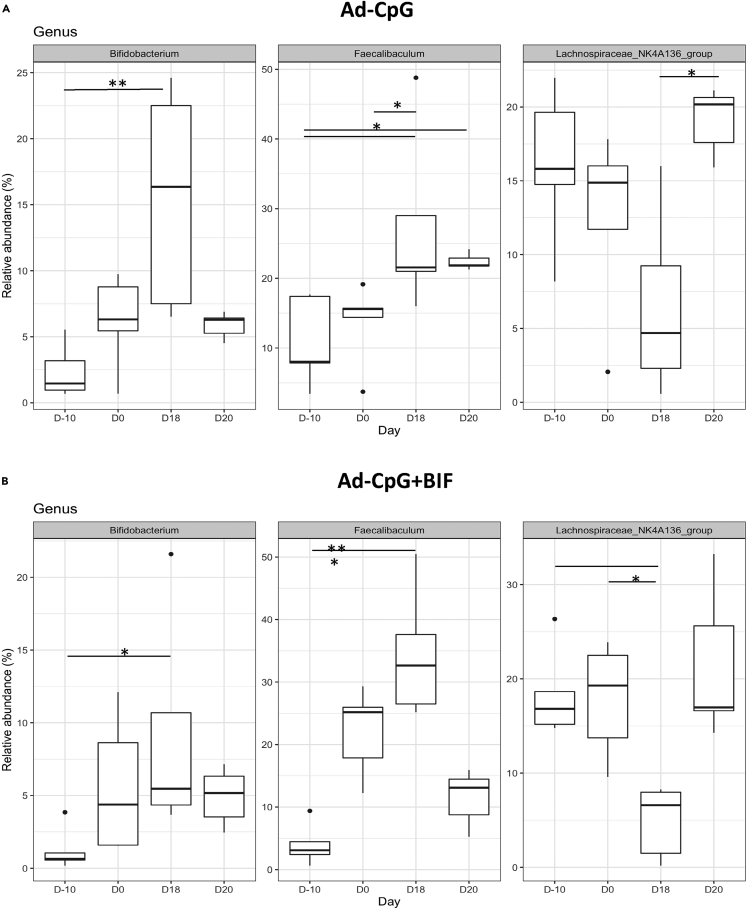
By using Dunn’s test, a post hoc non-parametric test applied to the Kruskal-Wallis significant amplicon sequence variants (ASVs), we performed multiple pairwise tests in order to identify between which time pairs the phyla differentially abundant were significant. As reported in Table S1, Tenericutes and Actinobacteria phyla abundances resulted significantly different between the earlier (Day −10) and later (Day 18 and Day 20) time points suggesting that their relative abundance was independent of tumor growth both in mock and Bifidus-treated groups. In both groups treated with Ad-CpG, the abundance of Actinobacteria was significantly different between Day −10 to Day 18, indicating that the pre-treatment with Bifidus did not affect its richness. In the Ad-CpG-treated group, Firmicutes phylum was differentially abundant between Day 0 and Day 18, when differences between tumor growth curves were well appreciated, as shown in Figure 3, panel B. A similar result was confirmed in the group treated with Ad-CpG and Bifidus, in which the relative abundance of the Firmicutes was different also between Day −10 and the endpoint (Day 20) (Table S1). These data confirmed that melanoma-bearing mice treated with Ad-CpG were characterized by an enrichment of Firmicutes in their fecal microbiota. The relative abundance of differentially abundant genera (p < 0.05) for each study group, according to the Kruskal-Wallis test among the four time points, was shown in Figure 4, panel B.
In fecal bacterial content of mice treated with Ad-CpG, the genera identified were Ruminiclostridium_9, Ruminiclostridium, Roseburia, Lachnospiraceae_UCG-001, Lachnospiraceae_NK4A136, and Faecalibaculum. All these bacterial genera belong to the Firmicutes phylum. In addition to these genera, Turicibacter, Oscillibacter, and Lachnoclostridium were identified in the group treated with Ad-CpG plus Bifidus. Bifidobacterium, belonging to the Actinobacteria phylum, was identified in all groups, but with a relative abundance of about 10% in the group treated with Bifidus (Day 20), compared to a relative abundance of 4.9% observed in the group treated with Ad-CpG and Bifidus (Day 20) (Figure 4, panel B).
At genus level, we have identified a specific fecal microbiota profile in both groups treated with Ad-CpG and with Ad-CpG and Bifidus (Figure 5, panels A and B). Specifically, Bifidobacterium, Faecalibaculum, and Lachnospiraceae_NK4A136 genera showed a relative abundance greater than 2% in fecal bacterial composition in groups treated with Ad-CpG. These data support the hypothesis that groups treated with Ad-CpG alone and in combination with Bifidus, for which we obtained higher antitumoral efficacy, were characterized by a specific fecal microbiota composition.
Figure 5.
Treatment of tumor with Ad-CpG increases the abundance of three specific genera in murine fecal microbiome
(A and B) Over the time points (Day-10, Day 0, Day 18, and Day 20), the three genera with a relative abundance >2% shown in the boxplots were in common between Ad-CpG and Ad-CpG + BIF group and belong to phyla Actinobacteria (Bifidobacterium) and Firmicutes (Faecalibaculum and Lacknospiraceae NK4A136). The treatments were indicated as Ad-CpG: Ad5-CpG-treated mice; Ad-CpG + BIF: combined regimen-treated mice. Middle line in boxes represents the median; lower box bounds the first quartile; upper box bounds the 3rd quartile. Whiskers represent the 95% confidence interval of the mean. The significance of distribution differences has been calculated by applying the Kruskal-Wallis test, Dunn’s post-hoc test and Benjamini-Hochberg p value adjustment (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.005).
Bifidobacterium-derived peptides stimulate antitumor responses via T cell cross-reactivity with melanoma-derived antigens
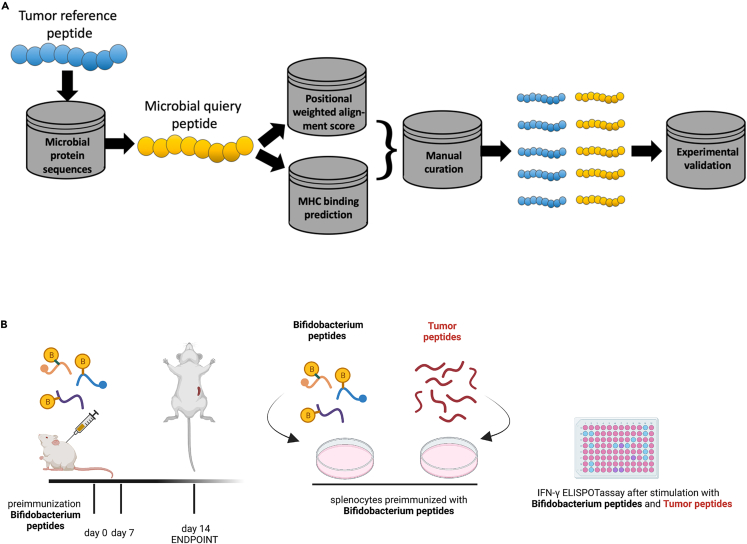
Considering that the supplementation with Bifidus in melanoma-bearing mice resulted in a slightly slower tumor progression compared to the control group, and in an increased response to Ad-CpG treatment, we decided to investigate a possible underlying mechanism that could explain the immunomodulatory activity of Bifidus linked to tumor regression. We hypothesized that several Bifidobacterium-derived peptides highly similar to melanoma epitopes could trigger a more effective antitumoral CTL response.
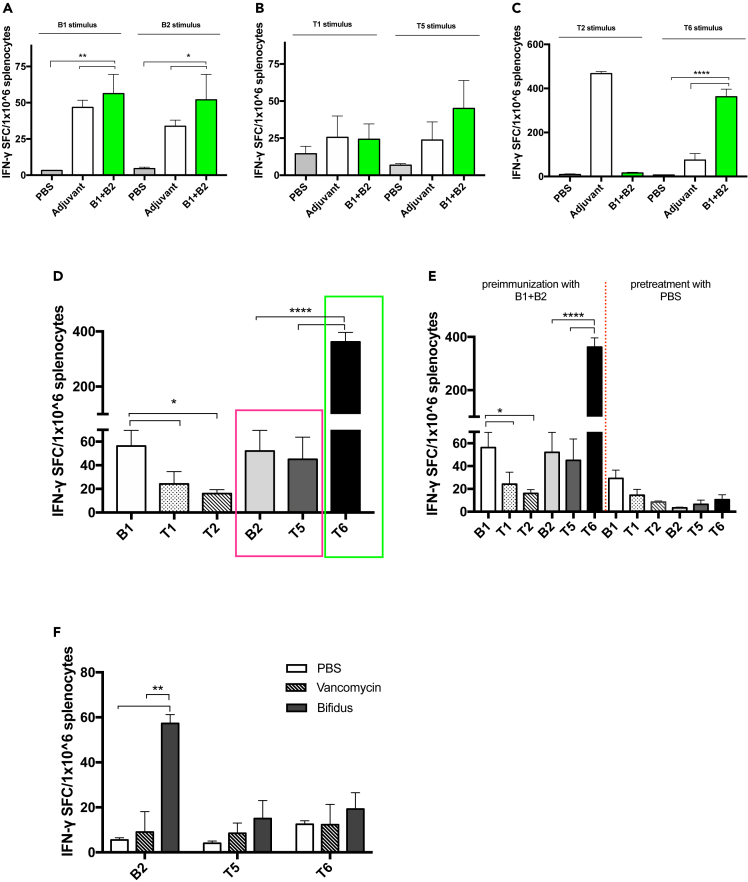
To this end, we compared the peptidome of the B16.F10 melanoma cell line with that of all Bifidobacterium species, including those present in the commercial cocktail used for in vivo supplementation, using Hamming similarity measure and Homology Evaluation Xenopeptides (HEX) platform, as represented by Figure 6, panel A. We selected Bifidobacterium peptides differing by up to three amino acid residues from the melanoma peptides, and after refinement, using the alignment and affinity binding scores by HEX, we obtained a list of ten Bifidobacterium peptides and fourteen melanoma peptides (Table 1). First, all Bifidobacterium peptides were tested in vivo, and in particular, each group of mice received a primary and a boosting dose of two Bifidobacterium peptides (B) complexed with the poly (I:C) adjuvant. Secondly, the corresponding highly similar melanoma peptides (T) to peptides B were tested in vitro by the IFN-γ ELISpot assay, as shown in the schematic representation of the immunization regimen (Figure 6, panel B). Splenocytes collected from mice preimmunized with peptides RQYGFIVL (B1) and RILKFLFL (B2) (indicated as B1+B2 group in the plot) showed a significant increase of IFN-γ CD8 T cells when re-stimulated singularly with peptide B1 (p < 0.01) and B2 (p < 0.05), compared to control groups treated with PBS or poly (I:C) adjuvant (Figure 7, panel A), confirming their stimulatory activity (legend of peptides indicated in Table 1).To assess the levels of immunogenicity of tumor peptides, we pulsed the preimmunized splenocytes of the B1+B2 group, with corresponding similar tumor peptides: T1, T5, T2, and T6. The first induced an IFN-γ production similar to that observed in the poly (I:C) control group; instead, stimulation with tumor peptides T5 and T6 increased the IFN-γ production more than that in control groups and for T2 no response was detected (Figure 7, panels B–C).Then, splenocytes harvested from preimmunized mice pulsed with B2 and T5 (bars in the magenta box) (Figure 7, panel D) exerted similar immunological activity in vitro and in vivo, confirming that the molecular mimicry between the bacterial peptides and tumor antigens can increase antitumor immunity. Additionally, a significantly increased immune activation was observed in pulsing splenocytes with tumor peptide T6 (p < 0.0001) (bar in the green box) than peptides investigated and control group preimmunized with PBS (Figure 7, panels D–E). To confirm the level immunogenicity of described peptide B2 and evaluate whether tumor peptides T5 and T6 were indeed immunogenic as B2, we harvested splenocytes from groups treated with vancomycin and with oral supplementation of Bifidobacterium spp. (Bifidus) and pulsed with peptides B2, T5, and T6. An additional group treated with PBS was used as a control group. Vancomycin-mediated microbiota perturbation significantly affected the response to peptides B2 compared to the Bifidus-treated group (p < 0.01), as shown by reduced immune activation (Figure 7, panel F). Therefore, we confirmed that gut microbiota perturbation negatively affects response to Ad-CpG therapy, as described in Figure 3, but also to B2 peptide. Splenocytes from mice pre-treated with Bifidus and stimulated with tumor peptides T5 and T6 showed a slightly increased trend of activation, although not achieving full statistical significance, compared to the group treated with vancomycin (Figure 7, panel F). Finally, we preimmunized mice only with peptide B1 and pulsed the splenocytes in vitro with the last two peptides, SQYRFIVF (T3) and RGYEFIVR (T4). Peptide T3 was able to elicit a significantly major immune response than other peptides (Figure S9, panels A and B).
Figure 6.
Experimental characterization of peptides
(A) Flowchart of the bioinformatics workflow for peptides discovery.
(B) Schematic representation of the schedule followed during the preimmunization procedure. The mice have been subcutaneously injected with two Bifidobacterium peptides (such as B1+B2) at days 0 and 7. PBS or poly (I:C) were used as controls as well. The spleens were harvested at day 14 and IFN-γ ELISpot was performed on harvested splenocytes and individual response to Bifidobacterium peptides and the corresponding similar tumor peptides were evaluated for each mouse.
Table 1.
Bifidobacterium and melanoma peptides
| B16.F10 peptides (Reference) | Microbial peptides (Query) | Bifidobacterium species | Hamming distance | weighted alignment score | H-2Kb binding affinity (nM) |
|---|---|---|---|---|---|
| AVLKYYKV | ASLKYYKV | bifidum | 1 | 269 | 40.4 |
| TAYEFAKL | TMYEFAKL | longum | 1 | 259 | 2.5 |
| SGYIPARL | SKYIPARL | breve, longum, bifidum | 1 | 256 | 181.9 |
| SNYERLESL | SNYERLEFL | breve, bifidum | 1 | 254 | 54.4 |
| ISLEFRNL | IALEFRNL | longum | 1 | 246 | 3 |
| KILTFDQL | KGLTFDQL | longum | 1 | 244 | 136.7 |
| SGYKFGVL | SGYKFDVL | lactis | 1 | 225 | 12.4 |
| VSTKFEHL | VSTKFSHL | longum | 1 | 220 | 4.8 |
| RQYIFSKL T1 |
RQYGFIVL B1 |
breve | 3 | 86 | 156.65 |
| RQYMFSSL T2 |
RQYGFIVL B1 |
breve | 3 | 99 | 156.65 |
| SQYRFIVF T3 |
RQYGFIVL B1 |
breve | 3 | 168 | 156.65 |
| RGYEFIVR T4 |
RQYGFIVL B1 |
breve | 3 | 158 | 156.65 |
| RILKFLKL T5 |
RILKFLFL B2 |
breve, longum | 1 | 226 | 67.2 |
| RILKFLQL T6 |
RILKFLFL B2 |
breve, longum | 1 | 226 | 67.2 |
Tables 1.List of ten Microbial peptides (query) differing by up to three amino acid residues from the fourteen B16.F10 peptides (reference). The different residue between reference sequence and query sequence is indicated in bold font. The legend of peptides is indicated below the reference and query sequences. The query peptide B1 is similar to four reference peptides: T1, T2, T3, and T4 (Hamming distance = 3). The query peptide B2 is similar to two reference peptides: T5 and T6 (Hamming distance = 1). The microbial peptides belong to different Bifidobacterium species, indicated in the third column. The distance of amino acid residues between query and reference are indicated by the hamming distance value; the weighted alignment score and the H-2Kb affinity binding scores were refined by software HEX.
Figure 7.
Bifidobacterium-derived peptides can resemble murine melanoma epitopes and mediate strong CTL response
(A–E) IFN-γ ELISpot was performed on harvested splenocytes from mice preimmunized with PBS, adjuvant poly (I:C) or with two Bifidobacterium peptides (B1+B2) and individual response to B1 stimulus and B2 (B) to T1 and T5 stimulus (C) to T2 and T6 stimulus (D) to stimulus B1, T1, T2, B2, T5, and T6 for each group of mice (n = 3) is reported as the value of IFN-γ spot-forming cells (SFC) on 1 × 106 splenocytes. Comparison of number of spots IFN-γ detected between B2 and T5, in magenta box, and comparison between B2 and T6 in the green box (E) IFN-γ ELISpot was performed on harvested splenocytes from mice preimmunized with two Bifidobacterium peptides (B1+B2) and with PBS and stimulated with peptides B1, T1, T2, B2, T5, and T6.
(F) IFN-γ ELISpot was performed on harvested splenocytes from mice treated with PBS, with vancomycin and from mice received oral supplementation with Bifidobacterium spp. (Bifidus) and stimulated with peptides B2, T5, and T6. For each group of mice (n = 3) number of spots (IFN-γ) is reported. All data are depicted as bar plots and represented as mean ± SEM. The statistical analysis was performed with ordinary one-way ANOVA test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
All these results strongly confirmed our initial hypothesis, according to which a T cell cross-reactivity occurs between highly immunogenic Bifidobacterium-derived epitope and melanoma antigens restricted to major histocompatibility complex (MHC)-I. These shared peptides could be deeply exploited to design powerful and cancer-specific OVs-based therapeutic vaccines, increasing their antitumoral effect.
Discussion
Despite the clinical impact of ICIs in cancer therapy, accumulating evidence suggests that these therapeutic agents show some limitations: only a fraction of patients benefit from ICIs treatment that is ineffective against immunologically “cold” tumors characterized by low TILs count.21 To this end, OVs represent attractive therapeutic agents that can be combined with ICIs, increasing their response blockade in poorly immunogenic tumors.21,23,25 Variability in the observed response rate is also due to environmental and genetic factors: intestinal microbiota has been indicated as a cause of variability in response to ICIs because of its general immunomodulatory action. Considering OVs therapy as a possible complement to increase the success of ICIs immunotherapy, we decided to investigate whether intestinal microbiota could affect OVs therapy efficacy.
We used an oncolytic adenovirus enriched with CpG island as OV for this study.24 Ad-CpG has been successfully evaluated in the past using a syngeneic melanoma model based on the inoculation of B16-OVA cells in C57BL/6J mice; this melanoma model responds to ICIs therapy and is sensitive to microbiota perturbation.32,33 In order to evaluate whether gut microbiota was necessary to the antitumoral activity of the Ad-CpG, we reduced intestinal bacterial diversity, pre-treating mice with the antibiotic vancomycin, which has been previously used for the same purpose by Vetizou et al.10,34
We found that pre-treatment with vancomycin reduced the efficacy of Ad-CpG therapy: mice had a faster tumor progression compared to mice treated only with Ad-CpG (Figure 1, panel B). We can therefore conclude that bacterial diversity in general is necessary for the efficacy of Ad-CpG therapy. In fact, vancomycin-based bacterial depletion induced an alteration of antitumoral immune response leading to a reduced count of IFN-γ+CD8+ TILs. In order to further confirm that bacterial depletion was the cause of the poor efficacy of Ad-CpG in vancomycin-treated mice, we cohoused a group of mice pre-treated with vancomycin and then treated with Ad-CpG (combined regimen) with a group of mice that did not receive any antibiotics. Cohousing is supposed to reduce or abrogate microbiota differences in the two groups because of mice coprophagy and grooming behavior35; transmission of the gut microbiota, by cohousing, significantly reduced differences efficacy of Ad-CpG therapy in mice pre-treated with vancomycin and mice that did not receive any antibiotics (Figure 2, panels B–C). These results confirmed that microbial diversity is relevant for the efficacy of Ad-CpG treatment in this melanoma model; in fact, mice pre-treated with vancomycin and kept isolated showed reduced survival (Figure 2). Cohousing-mediated improvement of Ad-CpG efficacy was also accompanied by an increase of tumor-infiltrating IFN-γ CD4+ and IFN-γ CD8+ T cells, underlining the effect of intestinal microbiota on the immune response against melanoma in this model.
Mounting evidence has demonstrated the effects of the gut microbiota on the efficacy of ICIs in humans and mice;4,8,36 specifically favorable gut bacteria increase the population of CD8+ T cells and decrease T-regs within the tumor bed.36 As reported, beneficial commensal Bifidobacterium boosts the efficacy of the PD-1/PD-L1 blockade by activating DCs and tumor-specific CD8+ T cells.11 Therefore, we evaluated a possible immunomodulatory activity of Bifidus supplementation, combined with Ad-CpG therapy, in our melanoma mouse model. Our data strongly indicate that Bifidus supplementation increased the response to Ad-CpG compared to mice that did not receive this probiotic. We observed fewer tumor-infiltrating T-regs in mice that received Bifidus supplementation; these lymphocytes are well known to exert an immunosuppressive action. Thus, Bifidus supplementation increases the efficacy of Ad-CpG treatment by modulating antitumoral immune response and, more specifically, by reducing the presence of immune suppressive lymphocytes and increasing intratumoral CTLs compared to the control group treated with virus alone. Therefore, Bifidus supplementation was able to boost the immunovirotherapy efficacy (Figure 3). As proof of this, splenocytes harvested from vaccinated mice with Ad-CpG and Bifidus cause extensive T cell-mediated killing of melanoma cells. We believe that this preliminary assessment of T cell-mediated tumor killing and then of immunological memory could not be directly connected to the presence of viral therapy combined with Bifidus supplementation.
Based on these observations, our data indicate that the immune-mediated eradication of melanoma, exerted by Ad-CpG, was affected by intestinal microbiota composition. As previously shown, antibiotic-mediated gut microbiota alteration reduces some beneficial bacteria taxa altering intestinal anti-inflammatory and immunomodulatory properties.10 These bacterial communities contribute to a healthy host-microorganism balance useful to optimally perform metabolic and immune functions and reduce disease development. The phylogenetic composition of the bacterial communities in both humans and mice seems to be similar at the phylum level, where the two main bacterial phyla of the murine intestinal tract are the Bacteroidetes and the Firmicutes,37,38 the most predominant phyla in a human healthy gut.39 For this reason, we compared fecal microbiome of mice treated with a combined regimen (Ad-CpG + Bifidus) to that of mice that received a single treatment and a control group, by DNA sequencing of the 16S rRNA gene in bacteria. Ad-CpG and combined regimen-recipient mice showed a predominance of the Firmicutes and Actinobacteria phyla in their fecal bacterial content showing, respectively, seven and nine genera belonging to the Firmicutes phylum (Figure 4). Specifically, groups treated with Ad-CpG showed, in their fecal bacterial content, three genera Lachnospiraceae_NK4A136 (14,5%), Faecalibaculum (18%), and Bifidobacterium (13%) with a relative abundance >2% belonging to phyla Actinobacteria and Firmicutes, respectively (Figure 5).
Therefore, we had evidence that Ad-CpG treatment, as a stand-alone and in combination with Bifidus, affects fecal microbiota composition. The enrichment of the phylum Firmicutes in the gut microbiota of Ad-CpG-treated groups is likely to create a favorable and immunologically permissive gut microbiota that contributes to the antitumoral immune responses.
In order to understand how gut microbiota exerts antitumor effects, we hypothesized that the molecular mimicry between antigens expressed by melanoma cells and antigens derived from peptides of Bifidobacterium could be responsible for the synergistic effect observed in groups treated with a combined regimen. To this end, we compared the peptidome of melanoma cell line B16.F10 with that of all Bifidobacterium species, as described in the schematic representation of the immunization regimen (Figure 6). Fourteen MHC I-restricted melanoma murine peptides extremely similar to peptides expressed in proteins from Bifidobacterium species were identified and then tested in vivo and in vitro. As reported by IFN-γ ELISpot assay, re-stimulation of Bifidobacterium (B1+B2) preimmunized splenocytes with peptides B1 and B2 induced an increased IFN-γ CD8 T cells, confirming the immunomodulatory potential of both peptides (Figure 7, panel A). Despite the high similarity between Bifidobacterium B1 and melanoma peptides T1 and T2, no significant differences were appreciated in the number of spots IFN-γ. Surprisingly, stimulation of B1+B2 preimmunized splenocytes, with specific melanoma epitopes T5 and T6, induced an increased IFN-γ CD8 T cells, indicating a mechanism of T cells cross-reactivity, based on possible molecular mimicry mechanism (Figure 7, panels D magenta and green boxes). One of the most interesting findings is that the specific epitope B2 (RILKFLFL) derived from peptides of B.breve and B.longum is efficiently presented on cell surface and triggers specific T cell responses; in addition, melanoma epitope T6 (RILKFLQL) is highly immunogenic and induces in vitro a greater amount of effector T cells that are cross-reactive with the B2 antigen. In the present study, for the first time to our knowledge, a possible association between the gut microbiota and the OVs therapy has been explored. The identification and use of specific antigens of Bifidobacterium, similar to melanoma antigens, for immunovirotherapy, has to be carefully considered. Antigens selection could be useful to optimize the manipulation of OVs, expanding OVs potency and reducing the variability of clinical outcomes. The effect of these modifications could be further enhanced by supplementation with Bifidobacterium species related to antigens selected, favoring a targeted bacterial colonization that positively affects the response to cancer immunotherapy. Then, results obtained could open up new perspective to design powerful anticancer OVs-based vaccines obtaining even more durable clinical benefits.
Limitations of the study
The present study is limited to an oncolytic adenovirus in a specific syngeneic mouse model of melanoma; therefore, further studies will be necessary to evaluate the role of intestinal microbiota and its possible perturbations in other tumor models and with additional OVs and, finally, in clinical settings. In addition, role of metabolites and other bacterial components has to be taken into account and will be part of a further investigation. Therefore, exploitation of the potential of the OV immunotherapy-gut microbiome axis has to be further examined to further improve the efficiency of this therapeutic strategy.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD3 PerCP-Cy5.5 | eBioscience, Invitrogen | Cat# 45-0031-82; RRID: AB_1107000 |
| anti-mouse CD8 FITC | eBioscience, Invitrogen | Cat# 11-0083-82; RRID: AB_657764 |
| anti-mouse CD4 PeCy7 | eBioscience, Invitrogen | Cat# 25-0041-82; RRID: AB_469576 |
| anti-mouse CD45R/B220 APC | Biolegend, San Diego, California | Cat# 103211; RRID: AB_312996 |
| anti-mouse CD45 APC-Cy7 | Sony Biotechnology, San Jose, California | Cat# 1153580; RRID: N/A |
| anti-mouse IFN-γ PE | Biolegend, San Diego, California | Cat# 505807; RRID: AB_315401 |
| anti-mouse Foxp3 PE | eBioscience, Invitrogen | Cat# 12-5773-82; RRID: AB_465936 |
| FC-Block | Biolegend, San Diego, California | Cat# 101301; RRID: AB_312800 |
| Bacterial and virus strains | ||
| Ad5D24-CpG | Prof.Vincenzo Cerullo | Cerullo et al.24 |
| Chemicals, peptides, and recombinant proteins | ||
| Cell stimulation cocktail | eBioscience, Invitrogen | Cat# 00-4970-03 |
| Brefeldin A | eBioscience, Invitrogen | Cat# 00-4506-51 |
| Foxp3/Transcription Factor Staining Buffer Set | eBioscience, Invitrogen | Cat# 00-5523-00 |
| Critical commercial assays | ||
| CyQUANTTMLDH Cytotoxicity Assay | Invitrogen | Cat# C20300 |
| Mouse IFN-γ 96-well White Precoated | Immunospot, CTL Europe GmbH | Cat# SKU: hIFNgp-1M |
| QIAamp DNA mini kit | Qiagen, Venlo, The Netherlands | Cat# 56304 |
| Agencourt AMPure XT beads | Beckman Coulter, Brea, CA, USA | Cat# A63881 |
| Deposited data | ||
| Sample metadata and raw data Release data: 2023-10-31 |
NCBI Sequence Read Archive (SRA) https://www.ncbi.nlm.nih.gov/sra/PRJNA999658 | BioProject accession number PRJNA999658 |
| Experimental models: Cell lines | ||
| B16.OVA | Prof. Vincenzo Cerullo | Sigma-Aldrich Cat# SCC420 |
| Experimental models: Organisms/strains | ||
| Female C57BL/6J 6–8 weeks old | Charles Rivers Italy RMS | Cat# 632C57BL/6J |
| Authorization number by the Ministry of Health | Provincial Government of Italy | n°331/2019-PR-protocol D5A89.35 |
| Software and algorithms | ||
| DADA2 v. 1.15.0 | Callahan et al.40 | (https://github.com/benjjneb/dada2 |
| Phyloseq 1.28.0 R packages | McMurdie et al.41 | R version 3.6.1 |
| SILVA reference database v.128 | https://zenodo.org/record/824551#.XmIcO5NKhuU | N/A |
| DECIPHER 2.12.0 R package | Wright et al.42 | N/A |
| phangorn 2.5.5 R package | Schliep et al.43 | N/A |
| FSA 0.8.27 package | Dunn et al.44 | N/A |
| 909 H-2Kb-specific peptides from B16-F10 murine melanoma cell line | Caron et al.45 | ProteomeXchange (Accession no: PXD001872) |
| Microbial peptides database | NCBI protein databases | https://www.ncbi.nlm.nih.gov/protein/ |
| in silico tool Homology Evaluation Xenopeptides (HEX) | Chiaro et al.28 | https://whalleyt.shinyapps.io/homology-evalutation-xenopeptides/. |
| NetMHC | ||
| GraphPad Prism 7 software | GraphPad Software, Inc., La Jolla, CA, USA | D4106509D18 |
| Other | ||
| Standard diet mice | Diete standard | Cat# 4RF25 |
| Vancomycin | Mylan | Cat# MINSAN 041220070 |
| ProBiotaTM Bifido Seeking health 10 Billion Bifidobacterium | Sivan et al.11 | Probiotic Blend (10 billion CFU) in a 280 mg∗ base of inulin (derived from chicory root) |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lucio Pastore (lucio.pastore@unina.it)
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The datasets used and/or analyzed during the current study is publicly accessible. Information specific to these datasets are listed in the key resources table.
-
•
Sample metadata and raw data obtained from 16S bacterial rRNA sequencing of fecal samples from mice enrolled in the experiment in vivo are reported in NCBI Sequence Read Archive (SRA), the BioProject accession number is PRJNA999658.
-
•
This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study partecipant details
Animal experiments
All animal experiments were reviewed and approved by the body responsible for animal welfare (OPBA) and by experimental animal committee of the CEINGE Biotecnologie Avanzate Franco Salvatore and the Provincial Government of Italy (n°331/2019-PR-protocol D5A89.35) national regulations guidelines in accordance with EU Directive 2010/63/EU. We used the ARRIVE 1 reporting guidelines.46 Female C57BL/6J 6–8 weeks old were obtained from Charles Rivers and used as syngeneic mouse melanoma model. Animals were kept in individually ventilated cages under standard conditions (12 h light:dark, temperature-controlled and humidity-controlled conditions) and received ad libitum access to water and food. Animals were fed with diet standard (Mucedola) and housed in Animal Biosafety Level 2 (ABSL-2) in an animal facility at CEINGE Biotecnologie Avanzate Franco Salvatore. Drinking water was sterilized in an autoclave and changed every two days. Animals were monitored daily for symptoms related to distress and pain including hunched posture, overall activity/ability to move and roughness of the hair coat. 3.5 × 105 B16.OVA cells were engrafted subcutaneously in right flank of all animals. After 7–9 days from inoculation, we ensured that all the tumors were implanted and have reached a diameter of 5 mm and then were randomized into treated and control groups. Blinding from the people running the study and analysing the data was adopted during some stages of animal experiments: during animal’s allocation to a group, the conduct of the experiment and during the outcome assessment. Some animals have been dropped from the study and euthanized before the endpoint, when the size of the largest diameter of the subcutaneous tumor exceeded 18 mm. To guarantee a sample-size for each experimental group equal to 5 and 6 animals useful for statistical analyses, as described in figure legends, experimental units in each group at the start of the study were respectively equal to 7 and 8 animals. Animal experiments were not replicated. In the present study, we collected spleen and tumor samples for flow cytometry analyses and feces samples from all enrolled animals in experimental research of treatment with Bifidus and Ad-CpG. Spleen and tumor samples were triturated into single-cell suspension in a sterile freezing medium with 90% FBS and 10% DMSO, then filtered by a cell strainer 70 μm and stored at −80°C. Viable cells were counted using an automated cell counter. Feces samples were immediately cooled in dry ice and stored at −80°C until DNA extraction for microbiome analysis. For the preimmunization experiment, 5-6 weeks-old female C57BL/6J (n = 3 per group) were allocated in different groups according to the treatment and each mouse was subcutaneously injected with 25 μg of Bifidobacterium peptide complexed with 25 μg of poly (I:C) adjuvant. The prime and boosting were done respectively on days 0 and 7 and mice were sacrificed on day 14.
Tumor implantation and tumor growth measurement
At day 0, 3.5 × 105 B16.OVA cells were diluted in 200 μL of PBS and injected subcutaneously into the right flank of each mouse. 9 days after cell inoculation, tumors were measured with an electronic digital caliper (largest tumor diameter and perpendicular tumor diameter) every two days, starting on the day cancer cell inoculation. Tumors volume was calculated as length × width × height (mm3). Mice were sacrificed when the tumor volume was greater than 1.800 mm3 or when they were in poor condition and expected to die shortly. Tumors and spleens were collected and used for further experiments. In the In vivo experiment related to restoring Ad-CpG efficacy in vancomycin-treated mice (Figure 2, panel E), the survival of treated mice has been followed until they were sacrificed or naturally died. Additional specific details of each animal experiment are well described in figure legends and by experimental design representations.
Virus administration
Mice were anesthetized in an isoflurane chamber and then 1 × 10E9 vp of Ad-CpG was injected intratumorally at days 9, 11 and 13 after cell inoculation. The protocol schedule of tumor, Ad-CpG injection is shown in Figure 1, panel A.
Antibiotic and bifidobacterium treatment
Mice were treated with vancomycin starting two weeks before tumor cells inoculation until the first injection of Ad-CpG. Vancomycin (0.25 mg/mL Mylan) was administered by oral gavage every two days. A cocktail of lyophilized Bifidobacterium spp. (B. bifidum, B. longum, B. lactis and B. breve- Seeking Health) was resuspended in PBS at a 5 × 10E9 CFU/mL concentration. Each mouse was treated with 200 μL of Bifidobacterium spp. (1 × 10E9 CFU/mouse) added to the drinking water starting 15 days before tumor inoculation and until the end of the experiment.
Method details
Cell lines and reagents
B16.OVA, a mouse melanoma cell line expressing chicken ovalbumin (OVA) derived from C57BL/6J, was kindly provided by Prof. Vincenzo Cerullo. Cells were cultured according to ATCC recommendations in RPMI 1640 medium. Medium was supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco), penicillin (50 U/mL), streptomycin (500 μg/mL), and glutamine (4 mmol/L) in a humidified atmosphere with 5% CO2 at 37°C. 1% of Geneticine (GIBCO) was added to the cells. At the endpoint of in vivo experiments, peripheral blood mononuclear cells (PBMCs) were isolated from the spleens of treated mice, to perform the IFN-γ ELISpot.
Virus production
Oncolytic adenovirus Ad5D24-CpG was amplified and characterized using standard protocols, as previously described.24 For a first step of amplification, A549 cells infected with virus Ad5D24-CpG at MOI (multiplicity of infection) equal to 10 pfu/cell. After 72h, the cell lysate was collected and stored at −80°C. Subsequently, to induce cell lysis, the sample was subjected to three freeze-thaw cycles and then centrifuged at 4,000 rpm for 25 min at 25°C. The supernatant containing the virus was collected and purified from the lysate by a first step in an ultracentrifuge at 27,000 rpm for 1-2h at 4°C on a cesium chloride (CsCl) gradient. The band containing the oncolytic adenovirus was subjected to a further ultracentrifugation step under the same conditions described above for a time exceeding 18h. Successively, the sample was dialyzed by placing it in TM solution (10 mM Tris-HCl pH 8.0, 2 mM MgCl2) for 2h under stirring at 4°C. After 2h, the dialysis cassette was transferred to the Freezing solution (10 mM Tris-HCl pH 8.0, 2 mM MgCl2, 4% sucrose) overnight at 4°C under stirring. The virus was collected, aliquoted and stored at −80°C. Ad5D24-CpG was titrated by spectrophotometric reading, obtaining a titer of 4.4 × 10E11 vp/mL.
Tumor cell lysis assays
Tumor cell lysis was determined through the quantitative analysis of Lactate dehydrogenase (LDH) by using an LDH Detection Kit (Cyquant, Invitrogen), according to the manufacturer’s instructions. The B16-OVA were plated and incubated for 16 h at 37°C, and successively pre-immunized murine splenocytes were added at ratio of 5:1. After 48 h of incubation at 37°C, images of each experimental point, before and after the removal of the murine splenocytes were captured by a phase-contrast microscopy. The dosage of LDH activity was performed on the supernatants collected 48-h of co-culture assay, by spectrophotometric analysis at 490 nm.
Isolation of immune cells from mice spleen and tumor
After the collection, spleen and tumor samples were kept in sterile RPMI 1640 medium, supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco), and transferred into a 70 μm cell strainer fitted on a 50 mL tube. Then spleen and tumor samples were gently cut into small pieces using a 5 mL syringe plunger and pressed through a cell strainer. Once the single-cell suspensions were collected, up to 25–30 mL of media was added and cells were centrifuged at 2,000 rpm, 10 min, at +4°C. Splenocytes and tumor cells were centrifuged at 2,000 rpm, 10 min, at +4°C and filtered through a new 70 μm cell strainer, to remove dead cell clumps and other debris. Some cells were used directly for cell assays and IFN-γ ELISpot while the remaining cells were frozen in a medium supplemented with 90% of FBS and 10% of DMSO.
Flow cytometry analysis
Surface and intracellular staining were performed using the following anti-mouse antibodies: CD3 PerCP-Cy5.5 (eBioscience, Invitrogen); CD8 FITC (eBioscience, Invitrogen); CD4 PeCy7 (eBioscience, Invitrogen); CD45R/B220 APC (Biolegend, San Diego, California); CD45 APC-Cy7 (Sony Biotechnology, San Jose, California); IFN-γ PE (Sony Biotechnology); Foxp3 PE (eBioscience, Invitrogen). Cell stimulation cocktail (eBioscience, Invitrogen); Brefeldin A (eBioscience, Invitrogen). Cells were initially stained with surface markers (CD3, CD8, CD4, CD45R/B220 and CD45) and then stained for Foxp3 (Foxp3/Transcription Factor Staining Buffer Set, eBioscience, Invitrogen) using a protocol for nuclear detection, according to manufacturers' instructions. T-cells were stimulated to produce IFN-γ in vitro using a cell stimulation cocktail (eBioscience, Invitrogen) and its secretion was blocked using protein transport inhibitors (Brefeldin A Solution) (eBioscience, Invitrogen). Subsequently, cells were stained with surface markers (CD3, CD8, CD4, CD45R/B220 and CD45) and then stained for IFN-γ using a protocol for cytoplasmatic detection, according to manufacturers' instructions. All stained cells were acquired by FACS Canto II cytometer (BD Biosciences Franklin Lakes, NJ) and analyzed with FACS Diva software (BD Biosciences).
IFN-γ ELISpot
IFN-γ ELISpot assays were performed using a commercially available mouse ELISpot reagent set (Mouse IFN-γ 96-well White Precoated, ImmunoSpot, Bonn Germany) and 0.02 μg/μL of each peptide was tested in vitro stimulations of 3 × 105 splenocytes for each well at 37°C for 72h. The experiments were performed in duplicate in each 96-well. Spots were counted using an ELISpot reader system (ImmunoSpot, Bonn Germany).
Graphics
The graphical representations, schematics, and timelines used in the figures were created using BioRender.com.
Quantification and statistical analysis
16S rRNA microbiome sequencing
Five fecal samples were collected from every group (n = 5) at different time points: before B16.OVA cells inoculation (day-10), when tumor has reached a diameter of 5 mm and before treatment (day 0), the day before the last feeding integration with the Bifidobacterium spp. mix (day 18), and at the end of the experiment (day 20) as described in the related paragraph. DNA was extracted from fecal samples using a QIAamp DNA mini kit (Qiagen, Venlo, The Netherlands). All extractions were performed in a pre-PCR designated room. To deeply investigate microbiome composition, we used a multiplexed 16S rDNA amplicon-based approach coupled with the NGS system MiSeq (Illumina, San Diego, CA, USA).47,48 In particular, 500 bp amplicons, spanning the V4-V6 hyper-variable regions of the 16S rRNA gene, were obtained. Each sample was individually amplified and purified (Agencourt AMPure XT beads, Beckman Coulter, Brea, CA, USA). Primers used in the first round of PCR contained the overhang sequences with Illumina adapters:
forward primer, 5′- TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCAGCAGCCGCGGTAA -3’; reverse primer, 5′- GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGGTTGCGCTCGTTGC - 3’. PCR conditions were 95°C for 10 min; 30 cycles of 95°C for 30 s, 59°C for 30 s, 72°C for 1 min; 72°C for 7 min and 4°C to the end. A second round of PCR was used to add the Illumina index to the amplicons for the library preparation according to the Nextera XT protocol (Illumina). PCR conditions were 72°C for 3 min; 95°C for 30 s; 12 cycles of 95°C for 10 s, 55°C for 30 s, and 72°C for 30 s; 72°C for 5 min and 10°C to the end. After appropriate quality assessment (TapeStation, Agilent Technologies, Santa Clara, CA, USA), the amplification products from different DNA samples were pooled in equimolar ratios. The obtained multiple amplicon libraries were quality assessed (TapeStation, Agilent Technologies) and quantified (Qubit dsDNA BR assay, Thermo Fisher, Waltham, MA, USA), according to the manufacturer’s instructions, in order to obtain a pool of equimolar libraries, so ensuring a normalization across the different samples sequenced in the same run. All libraries were sequenced with the Illumina MiSeq System, PE 300 × 2 protocol, according to the specifications of the manufacturer.
Microbiome data processing
Quality of microbial sequences was assessed using MultiQC v1.5 [1]. Reads adapters and low-quality reads/ends were removed using Trimmomatic v0.38. To analyze the taxonomic composition of samples, DADA2 v. 1.15.040 and Phyloseq 1.28.041 R packages were used (R version 3.6.1). As a scarce overlapping between paired-end reads was observed, we chose to analyze only the forward reads to have a more reliable alignment. Before aligning reads, the forward primer was trimmed out from reads and reads were filtered according to the following parameters: maxEE = 2; minLen = 50; maxN = 0; truncQ = 2. After chimeric sequences removal, taxonomy was assigned to ASVs (Amplicon Sequence Variants) by using the SILVA reference database v.128, formatted for being used by DADA2 software and available at the link https://zenodo.org/record/824551#.XmIcO5NKhuU. The phylogenetic tree was constructed by performing a multiple-alignment using the DECIPHER 2.12.0 R package.42 The phangorn 2.5.5 R package43 was then used to first construct a neighbor-joining tree, and then fit a GTR+G+I (Generalized time-reversible with Gamma rate variation) maximum likelihood tree using the neighbor-joining tree as a starting point. Statistical analyses of the dataset were carried out through combining all the data (cleaned ASVs, taxa assignment, phylogenetic tree, and metadata) into a phyloseq object. The α- and β-diversity were computed on the counts normalized with the respect to the total and the ANOSIM tests were performed. Wilcoxon rank-sum test (Mann-Whitney) was applied to test significance in terms of richness measured by the several α-diversity distances. The significance of differential abundance among groups at different taxonomic levels was assessed by the Kruskal-Wallis Rank-Sum Test in R environment.49 The pairwise comparison was then performed by Dunn’s test44 (FSA 0.8.27 package) on the significant Kruskal-Wallis tests and the p value corrected for multiple comparisons by the Benjamini-Hochberg adjustment.
Discovery and characterization of microbial peptides
We collected 909 H-2Kb-specific peptides from B16-F10 murine melanoma cell line obtained from Caron et al.45 These 909 murine peptides were used to search microbial peptides from Bifidobacterium bifidum, Bifidobacterium longum, Bifidobacterium breve and Bifidobacterium lactis NCBI protein databases (https://www.ncbi.nlm.nih.gov/protein/) based on Hamming similarity measure. We identified 62 unique pairs of melanoma-microbial peptides (58 pairs with Hamming distance 1 and 4 pairs with Hamming distance 3). These were analyzed using the in silico tool Homology Evaluation Xenopeptides (HEX), published in28,50 and available at https://whalleyt.shinyapps.io/homology-evalutation-xenopeptides/. HEX compares the similarity between melanoma peptides (reference peptides) and microbial peptides (query peptides) by calculating the weighted alignment score, using the BLOSUM substitution matrix, and predicting the H-2Kb binding affinity, using NetMHC.51
Fourteen unique murine-microbial peptide pairs (comprising fourteen B16.F10 peptides and ten microbial peptides) were selected based on their alignment and binding affinity scores (Table 1) for in vivo and in vitro experiments. The schematic of the bioinformatics workflow is shown in Figure 6, panel A. All peptides identified and tested in vitro and in vivo were purchased from Twin Helix. 14mg of each peptide was ordered with a guaranteed purity of >90%. The immunogenic peptides tested are reported in Table 1.
Statistical analysis
Statistical analyses were performed using the GraphPad Prism 7 software (GraphPad Software, Inc., La Jolla, CA, USA). Data were represented as mean +/− standard error of the mean (SEM) or +/−standard deviation (SD) as well defined in the figure legends. Significant differences of tumor growth between different groups was analyzed by two-way analysis of variance (ANOVA), and a p value <0.05 was considered statistically significant for all test used (∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). For average area under the curves (AUC) relative to tumor growth in the different experimental groups indicated as mean ± SEM, the statistical significance was evaluated by Unpaired T-test with Welch’s correction. The log rank Mantel-Cox analysis was used to calculate the p value (p < 0.05) of the survival curves. The statistical significance of percentages of T lymphocytes between experimental groups was evaluated by two-way ANOVA test using Tukey’s multiple comparison test. The statistical significance of percentages of IFN-γ+ T-cells between groups was evaluated by paired student’s T-test. The relative abundances of differentially abundant genera (p < 0.05) among the four-time points for each treatment was evaluated according to the Kruskal-Wallis test. Regarding the significance of distribution differences, between the three genera with a relative abundance >2%, has been calculated by applying the Kruskal-Wallis test, Dunn’s post-hoc test and Benjamini-Hochberg p value adjustment. Comparison between levels of immunogenicity between Bifidobacterium-derived peptides, the statistical analysis was performed by ordinary one-way ANOVA test. Further details about the statistical tests for each experiment can be found in the corresponding figure legends.
Acknowledgments
The authors thank Dr. Silvia Esposito, veterinary doctor responsible for the welfare of animals of animal facility of CEINGE Biotecnologie Avanzate Franco Salvatore, and Dr. Flavio Starnone for their valuable help and for sharing their extensive professional expertise. Professor Lucio Pastore is supported by funding of Italian Natural Recovery and Resilience Plan (PNRR) and POR Campania FESR 2014/2020: Sviluppo di Approcci Terapeutici INnovativi per patologie neoplastiche resistenti ai trattamenti (SATIN). Professor Vincenzo Cerullo is supported by funding of Italian Natural Recovery and Resilience Plan (PNRR), the European Research Council under the European Union's Horizon 2020 Framework program (H2020)/ERC-CoG-2015 Grant Agreement No. 681219, the Helsinki Institute of Life Science (HiLIFE), the Jane and Aatos Erkko Foundation (decision 19072019), the Cancer Society of Finland (Syöpäjärjestöt).
Authors’ contributions
LT, SF, MP, CC, LC, and MV performed in vitro and in vivo experiments; GS and LG contributed to flow cytometry analyses; IG, MRG, and VDA contributed to 16S rRNA microbiome sequencing and data processing and interpreted the results; CDL, GC, and VDA contributed to experiment design; TW and BS contributed to discovery peptide analyses; LT, SF, VC, and LP designed the study, interpreted the results, and contributed to manuscript writing. All authors read, corrected, and approved the final manuscript.
Declaration of interests
Vincenzo Cerullo is founder and shareholder at VALO therapeutics.
Published: August 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107668.
Contributor Information
Vincenzo Cerullo, Email: vincenzo.cerullo@helsinki.fi.
Lucio Pastore, Email: lucio.pastore@unina.it.
Supplemental information
References
- 1.Kim K., Kim H.S., Kim J.Y., Jung H., Sun J.-M., Ahn J.S., Ahn M.-J., Park K., Lee S.-H., Choi J.K. Predicting clinical benefit of immunotherapy by antigenic or functional mutations affecting tumour immunogenicity. Nat. Commun. 2020;11:951. doi: 10.1038/s41467-020-14562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sambi M., Bagheri L., Szewczuk M.R. Current Challenges in Cancer Immunotherapy: Multimodal Approaches to Improve Efficacy and Patient Response Rates. J. Oncol. 2019;2019 doi: 10.1155/2019/4508794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander J.L., Wilson I.D., Teare J., Marchesi J.R., Nicholson J.K., Kinross J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroentero. 2017;14:356–365. doi: 10.1038/nrgastro.2017.20. [DOI] [PubMed] [Google Scholar]
- 4.Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sepich-Poore G.D., Zitvogel L., Straussman R., Hasty J., Wargo J.A., Knight R. The microbiome and human cancer. Science. 2021;371 doi: 10.1126/science.abc4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKee A.M., Kirkup B.M., Madgwick M., Fowler W.J., Price C.A., Dreger S.A., Ansorge R., Makin K.A., Caim S., Le Gall G., et al. Antibiotic-induced disturbances of the gut microbiota result in accelerated breast tumor growth. iScience. 2021;24 doi: 10.1016/j.isci.2021.103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuczma M.P., Ding Z.-C., Li T., Habtetsion T., Chen T., Hao Z., Bryan L., Singh N., Kochenderfer J.N., Zhou G. The impact of antibiotic usage on the efficacy of chemoimmunotherapy is contingent on the source of tumor-reactive T cells. Oncotarget. 2017;8:111931–111942. doi: 10.18632/oncotarget.22953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 9.Iida N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A., Molina D.A., Salcedo R., Back T., Cramer S., et al. Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vétizou M., Pitt J.M., Daillère R., Lepage P., Waldschmitt N., Flament C., Rusakiewicz S., Routy B., Roberti M.P., Duong C.P.M., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.-L., et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanov I.I., Honda K. Intestinal Commensal Microbes as Immune Modulators. Cell Host Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper L.V., Littman D.R., Macpherson A.J. Interactions Between the Microbiota and the Immune System. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viaud S., Saccheri F., Mignot G., Yamazaki T., Daillère R., Hannani D., Enot D.P., Pfirschke C., Engblom C., Pittet M.J., et al. The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bessell C.A., Isser A., Havel J.J., Lee S., Bell D.R., Hickey J.W., Chaisawangwong W., Glick Bieler J., Srivastava R., Kuo F., et al. Commensal bacteria stimulate antitumor responses via T cell cross-reactivity. Jci Insight. 2020;5 doi: 10.1172/jci.insight.135597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tripodi L., Passariello M., D’Argenio V., Leggiero E., Vitale M., Colicchio R., Salvatore P., Cerullo V., Pastore C.D.L. Evaluation of the antiproliferative effect of Bifidobacterium longum BB-536 in solid tumor cell lines, co-cultured with murine splenocytes. Biochim. Clin. 2021;45:242–247. doi: 10.19186/bc_2021.021. [DOI] [Google Scholar]
- 17.Capasso C., Magarkar A., Cervera-Carrascon V., Fusciello M., Feola S., Muller M., Garofalo M., Kuryk L., Tähtinen S., Pastore L., et al. A novel in silico framework to improve MHC-I epitopes and break the tolerance to melanoma. OncoImmunology. 2017;6 doi: 10.1080/2162402x.2017.1319028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nejman D., Livyatan I., Fuks G., Gavert N., Zwang Y., Geller L.T., Rotter-Maskowitz A., Weiser R., Mallel G., Gigi E., et al. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalaora S., Nagler A., Nejman D., Alon M., Barbolin C., Barnea E., Ketelaars S.L.C., Cheng K., Vervier K., Shental N., et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature. 2021;592:138–143. doi: 10.1038/s41586-021-03368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zitvogel L., Daillère R., Roberti M.P., Routy B., Kroemer G. Anticancer effects of the microbiome and its products. Nat. Rev. Microbiol. 2017;15:465–478. doi: 10.1038/nrmicro.2017.44. [DOI] [PubMed] [Google Scholar]
- 21.Feola S., Capasso C., Fusciello M., Martins B., Tähtinen S., Medeot M., Carpi S., Frascaro F., Ylosmäki E., Peltonen K., et al. Oncolytic vaccines increase the response to PD-L1 blockade in immunogenic and poorly immunogenic tumors. OncoImmunology. 2018;7:e1457596. doi: 10.1080/2162402x.2018.1457596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vitale M., Scialò F., Patriciello T., Tripodi L., Coluccino L., Scognamiglio A., Pastore L. Oncolytic adenovirus treatment induces the reduction of tumor dimension in a model of mice lung adenocarcinoma multicellular spheroids. Biochim. Clin. 2023;47:251–258. doi: 10.19186/bc_2023.045. [DOI] [Google Scholar]
- 23.Vitale M., Scialò F., Passariello M., Leggiero E., D’Agostino A., Tripodi L., Gentile L., Bianco A., Castaldo G., Cerullo V., et al. Oncolytic Adenoviral Vector-Mediated Expression of an Anti-PD-L1-scFv Improves Anti-Tumoral Efficacy in a Melanoma Mouse Model. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.902190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerullo V., Diaconu I., Romano V., Hirvinen M., Ugolini M., Escutenaire S., Holm S.-L., Kipar A., Kanerva A., Hemminki A. An Oncolytic Adenovirus Enhanced for Toll-like Receptor 9 Stimulation Increases Antitumor Immune Responses and Tumor Clearance. Mol. Ther. 2012;20:2076–2086. doi: 10.1038/mt.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garofalo M., Bertinato L., Staniszewska M., Wieczorek M., Salmaso S., Schrom S., Rinner B., Pancer K.W., Kuryk L. Combination Therapy of Novel Oncolytic Adenovirus with Anti-PD1 Resulted in Enhanced Anti-Cancer Effect in Syngeneic Immunocompetent Melanoma Mouse Model. Pharm. Times. 2021;13:547. doi: 10.3390/pharmaceutics13040547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tripodi L., Vitale M., Cerullo V., Pastore L. Oncolytic Adenoviruses for Cancer Therapy. Int. J. Mol. Sci. 2021;22:2517. doi: 10.3390/ijms22052517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang J.K., Hong J., Yun C.-O. Oncolytic Viruses and Immune Checkpoint Inhibitors: Preclinical Developments to Clinical Trials. Int. J. Mol. Sci. 2020;21:8627. doi: 10.3390/ijms21228627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiaro J., Kasanen H.H.E., Whalley T., Capasso C., Grönholm M., Feola S., Peltonen K., Hamdan F., Hernberg M., Mäkelä S., et al. Viral Molecular Mimicry Influences the Antitumor Immune Response in Murine and Human Melanoma. Cancer Immunol. Res. 2021;9:981–993. doi: 10.1158/2326-6066.cir-20-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uribe-Herranz M., Rafail S., Beghi S., Gil-de-Gómez L., Verginadis I., Bittinger K., Pustylnikov S., Pierini S., Perales-Linares R., Blair I.A., et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J. Clin. Invest. 2020;130:466–479. doi: 10.1172/jci124332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasso E., D’Avino C., Passariello M., D’Alise A.M., Siciliano D., Esposito M.L., Froechlich G., Cortese R., Scarselli E., Zambrano N., et al. Massive parallel screening of phage libraries for the generation of repertoires of human immunomodulatory monoclonal antibodies. mAbs. 2018;10:1060–1072. doi: 10.1080/19420862.2018.1496772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Passariello M., Camorani S., Vetrei C., Cerchia L., De Lorenzo C. Novel Human Bispecific Aptamer–Antibody Conjugates for Efficient Cancer Cell Killing. Cancers. 2019;11:1268. doi: 10.3390/cancers11091268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rastrelli M., Tropea S., Rossi C.R., Alaibac M. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28:1005–1011. [PubMed] [Google Scholar]
- 33.Mellman I., Coukos G., Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strati F., Pujolassos M., Burrello C., Giuffrè M.R., Lattanzi G., Caprioli F., Troisi J., Facciotti F. Antibiotic-associated dysbiosis affects the ability of the gut microbiota to control intestinal inflammation upon fecal microbiota transplantation in experimental colitis models. Microbiome. 2021;9:39. doi: 10.1186/s40168-020-00991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ericsson A.C., Franklin C.L. Manipulating the Gut Microbiota: Methods and Challenges. ILAR J. 2015;56:205–217. doi: 10.1093/ilar/ilv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.-L., Luke J.J., Gajewski T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 38.Rawls J.F., Mahowald M.A., Ley R.E., Gordon J.I. Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-free Recipients Reveal Host Habitat Selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jandhyala S.M., Talukdar R., Subramanyam C., Vuyyuru H., Sasikala M., Nageshwar Reddy D. Role of the normal gut microbiota. World J. Gastroentero. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMurdie P.J., Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright E. Using DECIPHER v2.0 to Analyze Big Biological Sequence Data in R. R. J. 2016;8:352. doi: 10.32614/RJ-2016-025. [DOI] [Google Scholar]
- 43.Schliep K.P. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunn O.J. Multiple Comparisons Using Rank Sums. Technometrics. 1964;6:241–252. doi: 10.2307/1266041. [DOI] [Google Scholar]
- 45.Caron E., Espona L., Kowalewski D.J., Schuster H., Ternette N., Alpízar A., Schittenhelm R.B., Ramarathinam S.H., Lindestam Arlehamn C.S., Chiek Koh C., et al. An open-source computational and data resource to analyze digital maps of immunopeptidomes. Elife. 2015;4 doi: 10.7554/elife.07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kilkenny C., Browne W.J., Cuthill I.C., Emerson M., Altman D.G. Improving Bioscience Research Reporting: The ARRIVE Guidelines for Reporting Animal Research. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iaffaldano L., Granata I., Pagliuca C., Esposito M.V., Casaburi G., Salerno G., Colicchio R., Piccirillo M., Ciacci C., Del Vecchio Blanco G., et al. Oropharyngeal microbiome evaluation highlights Neisseria abundance in active celiac patients. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-29443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nardelli C., Granata I., D’Argenio V., Tramontano S., Compare D., Guarracino M.R., Nardone G., Pilone V., Sacchetti L. Characterization of the Duodenal Mucosal Microbiome in Obese Adult Subjects by 16S rRNA Sequencing. Microorg. 2020;8:485. doi: 10.3390/microorganisms8040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hollander M., Wolfe D.A., Chicken E. 2017. Nonparametric Statistical Methods: Hollander/Nonparametric Statistical Methods. [DOI] [Google Scholar]
- 50.Tripodi L., Sasso E., Feola S., Coluccino L., Vitale M., Leoni G., Szomolay B., Pastore L., Cerullo V. Systems Biology Approaches for the Improvement of Oncolytic Virus-Based Immunotherapies. Cancers. 2023;15:1297. doi: 10.3390/cancers15041297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreatta M., Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The datasets used and/or analyzed during the current study is publicly accessible. Information specific to these datasets are listed in the key resources table.
-
•
Sample metadata and raw data obtained from 16S bacterial rRNA sequencing of fecal samples from mice enrolled in the experiment in vivo are reported in NCBI Sequence Read Archive (SRA), the BioProject accession number is PRJNA999658.
-
•
This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.