Summary
Nasopharyngeal carcinoma (NPC) is known for high curability during early stage of the disease, and early diagnosis relies on nasopharyngeal endoscopy and subsequent pathological biopsy. To enhance the early diagnosis rate by aiding physicians in the real-time identification of NPC and directing biopsy site selection during endoscopy, we assembled a dataset comprising 2,429 nasopharyngeal endoscopy video frames from 690 patients across three medical centers. With these data, we developed a deep learning-based NPC detection model using the you only look once (YOLO) network. Our model demonstrated high performance, with precision, recall, mean average precision, and F1-score values of 0.977, 0.943, 0.977, and 0.960, respectively, for internal test set and 0.825, 0.743, 0.814, and 0.780 for external test set at 0.5 intersection over union. Remarkably, our model demonstrated a high inference speed (52.9 FPS), surpassing the average frame rate (25.0 FPS) of endoscopy videos, thus making real-time detection in endoscopy feasible.
Subject areas: Cancer systems biology, Machine learning
Graphical abstract

Highlights
-
•
We employed the YOLO network to develop an NPC diagnostic model
-
•
Datasets from three clinical centers were used to develop and validate the model
-
•
The proposed model demonstrated outstanding performance and robustness
-
•
Real-time detection for NPC during nasopharyngeal endoscopic videos can be achieved
Cancer systems biology; Machine learning
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor originating from the mucosal epithelium of the nasopharynx. In 2020, there were 96,371 new cases and 58,094 deaths worldwide, with over 70% of new cases occurring in East and Southeast Asia, revealing a highly uneven global distribution.1,2
Patients with early stage NPC exhibit a high overall survival rate after treatment.3 However, due to the atypical symptoms often associated with early stage NPC and the possibility of asymptomatic cases, the majority (>70%) of patients are diagnosed at an advanced stage of NPC. For the early screening of NPC, endoscopy is considered to be indispensable. The ultimate gold standard for diagnosing NPC is nasopharyngeal endoscopy-guided biopsy of abnormal nasopharyngeal lesions.4 Hence, it is particularly important for endoscopists to observe the morphological characteristics of masses through endoscopy and make preliminary judgments. However, distinguishing nasopharyngeal inflammation, lymphoid hyperplasia, adenoid hypertrophy, and residual adenoid tissue from early NPC under endoscopy can be challenging, resulting in false-negative outcomes.5,6,7 Atypical and small lesions may require multisite and repeated biopsies to improve detection rates. Repeated biopsies increase patient trauma and may delay treatment. Therefore, accurately identifying lesions and localizing biopsy sites are critical for early tumor diagnosis. However, not all endoscopists possess the necessary training, experience, or equipment to adequately identify and localize nasopharyngeal lesions, particularly early stage, insidious lesions. In addition, repeatedly reviewing endoscopic images of NPC can be time-consuming and mentally exhausting for endoscopists, as the human eye and brain are less sensitive to identifying lesions. Consequently, developing automatic computer-aided detection (CADe) and diagnosis (CADx) systems to support physicians in diagnosing NPC is crucial.
Computer-aided systems employing machine learning (ML) and deep learning (DL) techniques, such as convolutional neural networks (CNNs), can enhance disease detection and diagnostic accuracy and efficiency. By learning feature information from input images, CNN models can recognize specific patterns and correlate them with predefined results or parameters (output detection or diagnosis) to train network parameters. In recent years, CNNs have emerged as a promising method for image recognition or classification, serving as the foundation for automated image perception, processing, and decision-making. CNNs have proven highly beneficial in endoscopy and have been applied in various medical endoscopy imaging areas. For example, a large-dataset DL model was developed for the detection of upper gastrointestinal tumors in digestive endoscopy, an ear endoscopic image classification model based on DL was developed and validated, a DL model was applied to laryngoscopy for real-time laryngeal cancer detection, and a real-time system using DL was applied to detect and track ureteral orifices during urinary endoscopy.8,9,10,11,12,13,14 In the field of endoscopic NPC recognition, several studies have developed artificial intelligence models based on static endoscopic images, demonstrating their feasibility and recognition performance. However, these models often require a balance of high accuracy and fast inference speed, impeding real-time dynamic detection in video nasopharyngeal endoscopy.15,16,17,18,19 Furthermore, existing studies have mainly focused on the recognition and classification of endoscopic images of NPC, which makes it difficult to accurately locate lesions in images. This is obviously unfavorable for guiding inexperienced doctors to perform biopsy site selection. In addition, as these studies rely on single-center datasets, the actual performance, generalization, and robustness of the models still need to be investigated.
The First Affiliated Hospital of Sun Yat-sen University, Macau Kiang Wu Hospital, and Guangzhou First People’s Hospital are located in southeastern China, an area with a high global prevalence of NPC. By leveraging artificial intelligence technology and our extensive nasopharyngeal endoscopy data, we utilized the you only look once (YOLO) network to develop a fast and accurate real-time object detection model for NPC. In this study, we assessed the model’s performance, determined the optimal configuration, and validated the feasibility and effectiveness of the model for real-time automated NPC detection in nasopharyngeal endoscopy using both internal and external datasets. The proposed model represents a novel approach to assist physicians with NPC identification and guide biopsy site selection during nasopharyngeal endoscopy. In addition, the model’s predictive capabilities can validate a physician’s clinical judgment. Our contributions are as follows:
-
1.
We have harnessed the power of the YOLO network to create a real-time NPC diagnostic model designed specifically for video nasopharyngeal endoscopy. This model excels in terms of diagnostic accuracy and inference speed, thereby enabling rapid and precise localization of NPC.
-
2.
We use a dataset from three different clinical centers to develop and validate our model. This unique approach facilitates a more realistic assessment of our model’s performance and generalizability in real-world clinical environments.
-
3.
We have developed a robust model that maintains consistent performance across a wide array of scenarios, including variations in video brightness, hue, contrast, video quality, and lens stability. This level of robustness in varied conditions is an advancement in the field.
Results
Detection results
The purpose of this experiment was to evaluate the detection performance of various algorithms for NPC lesions of varying size, shape, and appearance in different datasets. The experimental results are displayed in Table 1. The results demonstrated that the object detection performance of the YOLOv8l model was superior to that of YOLOv6m, YOLOv7, Faster-RCNN, Cascade-RCNN, and SSD (single shot multibox detector) for the internal test set, with precision, recall, F1-score, and mAP (mean average precision) values of 0.977, 0.943, 0.960, and 0.977, respectively. In the comparison test for the external test set, YOLOv7 exhibited higher precision than the remaining five models, with a value of 0.862. Furthermore, the recall of YOLOv6m was the highest among all models, with a value of 0.750. However, the F1-score can more comprehensively reflect the model’s performance. The F1-score of YOLOv8l for the external test set was 0.780, higher than that of YOLOv7, YOLOv6m, and the remaining three models. In terms of inference speed, YOLOv7 demonstrated the fastest speed and the least delay in the comparative experiments among the six models. Except for Faster-RCNN, Cascade-RCNN, and SSD, the inference speed of YOLOv6m, YOLOv7, and YOLOv8l exceeded the average frame rate (25 FPS) of the nasopharyngeal endoscopy videos, enabling real-time detection in endoscopy. Based on the performance comparison of the six models, we selected the top three performing models for further comparative analysis: YOLOv6m, YOLOv7, and YOLOv8l. The detailed results of this comparative analysis can be found in Figure 1. In summary, these comparative experiments provided compelling evidence that the YOLOv8l model exhibited exceptional stability and accuracy across both internal and external datasets. Notably, YOLOv8l demonstrated remarkable accuracy and real-time lesion detection capabilities.
Table 1.
Performance Evaluation of various models
| Model | Parameters of the Model (Millions) | Internal Test Set |
External Test Set |
Frame Rate (FPS) | Delay (ms) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P@.5iou | R@.5iou | F1@.5iou | mAP@.5 | P@.5iou | R@.5iou | F1@.5iou | mAP@.5 | ||||
| YOLOv8l | 43.7 | 0.977 | 0.943 | 0.960 | 0.977 | 0.825 | 0.743 | 0.780 | 0.814 | 52.9 | 18.9 |
| YOLOv7 | 36.9 | 0.944 | 0.924 | 0.930 | 0.944 | 0.862 | 0.621 | 0.730 | 0.634 | 57.1 | 17.5 |
| YOLOv6m | 35.9 | 0.946 | 0.937 | 0.941 | 0.946 | 0.746 | 0.750 | 0.758 | 0.705 | 43.0 | 23.3 |
| Faster-RCNN | 41.3 | 0.563 | 0.391 | 0.406 | 0.563 | 0.235 | 0.377 | 0.290 | 0.235 | 8.0 | 125.0 |
| Cascade-RCNN | 69.2 | 0.930 | 0.621 | 0.742 | 0.930 | 0.676 | 0.415 | 0.544 | 0.676 | 6.3 | 158.7 |
| SSD | 24.4 | 0.858 | 0.683 | 0.759 | 0.858 | 0.779 | 0.454 | 0.573 | 0.779 | 10.4 | 96.2 |
RCNN = Region Convolutional neural network; SSD = Single Shot MultiBox Detector; P@.5iou = Precision with an Intersection over Union threshold of 0.5; R@.5iou = Recall with an Intersection over Union threshold of 0.5; F1@.5iou = F1 Score with an Intersection over Union threshold of 0.5; mAP@.5 = Mean Average Precision with an Intersection over Union threshold of 0.5; FPS = Frames Per Second; ms = Millisecond.
Figure 1.
YOLO models performance metrics on the internal test set and the external test set for NPC detection
(A and E) Precision curves.
(B and F) Recall curves.
(C and G) F1-score curves.
(D and H) Precision-Recall curves.
Visualization of DL model prediction
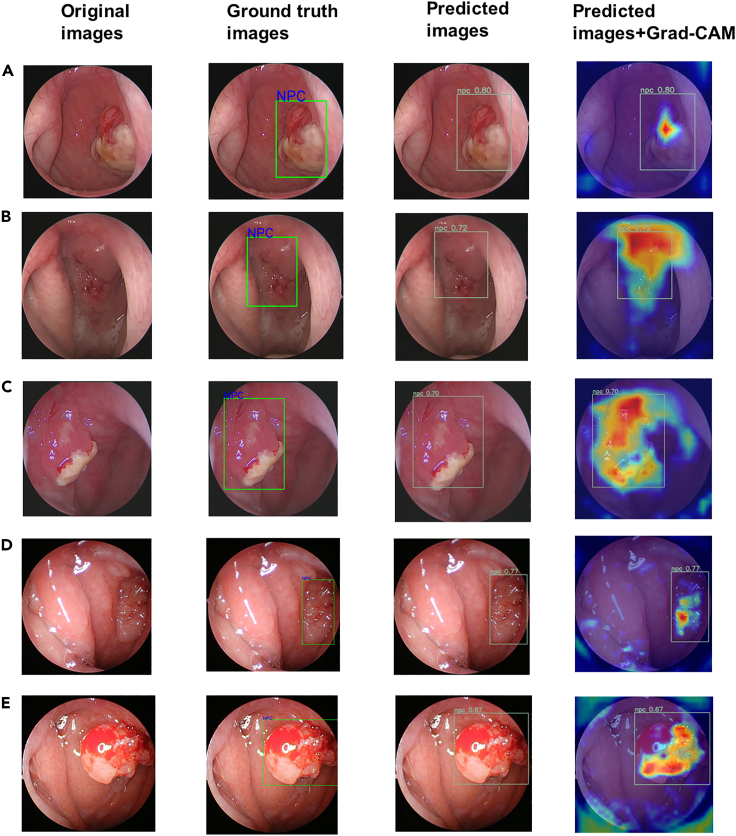
In the field of medical image processing, ensuring the interpretability of a model is of utmost importance. Providing doctors with an understanding of the reasoning behind the model’s predictions allows for enhanced trust and utilization in clinical practice. To address this, we incorporated gradient-weighted class activation mapping (Grad-CAM) into our methodology.20 Grad-CAM aids to generate activation maps specific to the predicted class by producing a weighted linear sum of visual patterns across different spatial locations. By employing Grad-CAM, we are able to determine which regions of an image the model relies on for its predictions. Our analysis of the Grad-CAM results revealed that the model consistently directs its attention toward lesion areas, with a particular focus on regions exhibiting distinct elevation and more intricate blood vessels, as visually depicted in Figure 2. This observation suggests that these areas hold crucial information for identifying and diagnosing potential cases of NPC. The focus on these regions indicates their significance in contributing to the model’s accurate predictions. As a result, the Grad-CAM technique shows promise as a reference tool for guiding precise lesion biopsies, potentially improving the diagnostic accuracy and treatment planning in clinical settings.
Figure 2.
Examples of automatic NPC prediction provided by the model (YOLOv8l)
The first column on the left contains the original images. The second column contains images with ground truth bounding boxes. The third column contains images with YOLO-predicted bounding boxes. The fourth column contains images with predicted bounding boxes and heat maps. Cases A–E are nasopharyngeal endoscopic images of different NPC patients. Grad-cam: Gradient-weighted Class Activation Mapping.
Verification of real-time detection
To evaluate the model’s suitability for real-time detection in video streams, the focus of this study was primarily on the running time. The YOLOv8l model was chosen for its superior performance, as evidenced by the aforementioned results. As shown in Table 2, the inference speed of a model that processed six videos was greater than the videos' frame rate. To illustrate the detection effect, we selected some original video frames and their corresponding frames processed by the model, which are displayed in Figure 3. Furthermore, to provide a more comprehensive demonstration, three sample videos processed by the model, referred to as Videos S1, S2, and S3 in supplemental information, were made available. The aforementioned experimental results validated the efficacy and feasibility of utilizing YOLOv8l for the real-time NPC detection of nasopharyngeal endoscopy videos. The division of the dataset, the research process, and the definition of IoU are shown in Figures 4, 5 and 6.
Table 2.
Characteristics and Computation Times of the Testing Videos After Applying the Model (YOLOv8l) for NPC Detection
| Video ID | Size (Mb) | Video Format | Video Resolution | Video Frame Rate (FPS) | Total Frame Count | NPC | Average Computation Time Per Frame (s) | Model Frame Rate (FPS) |
|---|---|---|---|---|---|---|---|---|
| 1 | 18.3/89.6 | MP4/MKV | 1920x1080 | 25.00 | 306 | Y | 0.0183 | 54.64 |
| 2 | 18.9/81.2 | MP4/MKV | 1920x1080 | 25.00 | 272 | Y | 0.0176 | 56.82 |
| 3 | 9.92/42.7 | MP4/MKV | 1920x1080 | 25.00 | 148 | Y | 0.0176 | 56.82 |
| 4 | 20.2/82.3 | MP4/MKV | 1920x1080 | 25.00 | 286 | Y | 0.0175 | 57.14 |
| 5 | 26.6/113 | MP4/MKV | 1920x1080 | 25.00 | 390 | Y | 0.0174 | 57.47 |
| 6 | 19.2/89.4 | MP4/MKV | 1920x1080 | 25.00 | 311 | N | 0.0170 | 58.82 |
NPC = nasopharyngeal carcinoma; Mb = megabytes; FPS = frame per second.
Figure 3.
Panel of testing videoframes extracted from six nasopharyngeal endoscopic videos
Each row represents a different video: the first two pictures of every row are extracted from the original videos, the second two and the last two images are the same frames extracted after the prediction of the model (YOLOv8l).
Figure 4.
The flowchart of dataset creation
Figure 5.
The flowchart of research & YOLOv8l architecture
Figure 6.
Evaluation of whether model was correctly diagnosed. The red rectangle is marked by the model as a predicted NPC
The green rectangle is the location of NPC, which is manually marked by the expert physicians. Intersection over the union (IoU) is the area of overlap divided by area of union.
Validation of robustness
The robustness of a model may have significant implications for its practical application in the real world, manifested by its ability to exhibit outstanding detection performance in diverse environments. A series of rigorous tests were conducted to validate the robustness of the proposed model. We applied image corruption methods to the test sets to simulate various image characteristics in different scenes, including noise, blur, fog, and changes in brightness.21 The experimental results are displayed in Table 3. The experiments demonstrated that the model exhibited reliable detection performance when dealing with different video qualities, blurry visuals caused by lens shake, foggy scenes resulting from patient respiration, and changes in device brightness for both the internal and external datasets. It is worth mentioning that, in terms of the F1-score, in addition to the zoom blur and fog methods, which significantly decreased the model performance, the average impact on the performance metrics of the other methods for the internal and external test sets was −0.017 and −0.018, respectively. In summary, the proposed model demonstrated exceptional performance and reasonable generalizability for a diverse range of clinical scenarios.
Table 3.
Robustness validation of the model
| Model and Methods of Image corruption | Internal Test Set |
External Test Set |
||||||
|---|---|---|---|---|---|---|---|---|
| P@.5iou | R@.5iou | F1@.5iou | mAP@.5 | P@.5iou | R@.5iou | F1@.5iou | mAP@.5 | |
| YOLOv8l | 0.977 | 0.943 | 0.960 | 0.977 | 0.825 | 0.743 | 0.780 | 0.814 |
| Gaussian noise | 0.956 | 0.911 | 0.933 | 0.959 | 0.805 | 0.711 | 0.755 | 0.795 |
| Shot noise | 0.948 | 0.911 | 0.929 | 0.960 | 0.792 | 0.710 | 0.749 | 0.797 |
| Impulse noise | 0.954 | 0.889 | 0.920 | 0.949 | 0.805 | 0.684 | 0.739 | 0.787 |
| Defocus blur | 0.965 | 0.932 | 0.948 | 0.971 | 0.812 | 0.735 | 0.771 | 0.807 |
| Zoom blur | 0.810 | 0.758 | 0.783 | 0.837 | 0.680 | 0.587 | 0.630 | 0.705 |
| Motion blur | 0.966 | 0.966 | 0.966 | 0.969 | 0.818 | 0.732 | 0.772 | 0.803 |
| Fog | 0.945 | 0.661 | 0.778 | 0.830 | 0.796 | 0.493 | 0.610 | 0.689 |
| Brightness + | 0.977 | 0.943 | 0.960 | 0.977 | 0.824 | 0.743 | 0.780 | 0.815 |
| Brightness - | 0.956 | 0.932 | 0.944 | 0.971 | 0.804 | 0.731 | 0.766 | 0.803 |
P@.5iou = Precision with an Intersection over Union threshold of 0.5; R@.5iou = Recall with an Intersection over Union threshold of 0.5; F1@.5iou = F1 Score with an Intersection over Union threshold of 0.5; mAP@.5 = Mean Average Precision with an Intersection over Union threshold of 0.5.
Discussion
In this study, we successfully employed a YOLO network to develop an NPC diagnostic model for video nasopharyngeal endoscopy. The model demonstrated outstanding accuracy and inference speed with both internal and external datasets. By providing real-time and precise lesion localization during the early screening of NPC, our model has the potential to significantly assist clinicians in their decision-making processes. Furthermore, the model shows promising prospects in guiding biopsy procedures for NPC, ultimately contributing to more accurate diagnoses and improving patient outcomes.
In areas where NPC is endemic, early screening for high-risk individuals currently includes serum Epstein-BarrVirus (EBV) DNA testing combined with nasopharyngeal endoscopy and magnetic resonance imaging (MRI). NPC exhibits characteristics that are distinct from those of most other tumors, and nasopharyngeal endoscopy plays a vital role in the early screening and auxiliary diagnosis of NPC that MRI cannot replace.22 Presently, there are two main types of nasopharyngeal endoscopy: white light imaging (WLI) and narrow-band imaging (NBI). The former mainly identifies the overall characteristics of a lesion, and the latter mainly identifies the microvascular morphology of a lesion.23 While NBI is more beneficial in identifying occult NPC, its clinical applicability is hampered by the intensive training and expertise required for optical image interpretation. Furthermore, NBI endoscopic equipment is more expensive than WLI endoscopic equipment. Given that many areas with high NPC prevalence in China and Southeast Asia are situated in rural or remote locations, WLI endoscopes are the most prevalent endoscopic equipment in local hospitals. Thus, the prevalence of NBI endoscopy is limited.24 After considering these factors, we primarily focused on detecting and diagnosing NPC in the WLI mode of nasopharyngeal endoscopy.
For physicians, identifying NPC by nasopharyngeal endoscopy is a significant challenge. Li et al. reported that the accuracy, sensitivity, specificity, and positive prediction value (PPV) of experts with five years of experience in identifying nasopharyngeal malignant and benign lesions in WLI images were 80.5%, 89.5%, 70.8%, and 76.6%, respectively, and these metrics were significantly lower for less experienced physicians.15 As many areas with high NPC prevalence in China and Southeast Asia are located in rural or remote locations, local doctors might lack sufficient experience and advanced endoscopic equipment for NPC detection. Consequently, we believe that primary care hospitals need a CADe model based on DL to assist physicians in the early diagnosis of NPC more than national hospitals, and this model can help bridge the cancer diagnosis gap between them. Li et al.'s DL model demonstrated higher recognition accuracy and faster recognition speed than those of professional clinicians, proving that CADe technology can serve as a powerful assistant for clinicians.15
Over the past five years, the advent of the big data era has driven rapid development in the DL network represented by CNNs. In the processes of data acquisition, preprocessing, feature extraction, and data classification, CNNs have been widely used in tumor classification, detection, and segmentation due to their outstanding spatial feature extraction function and classification accuracy.11,13,25,26 Currently, numerous studies have developed object detection models based on YOLO, particularly in the field of video endoscopy, which requires real-time lesion recognition. With the exceptional accuracy and speed of the YOLO network, dynamic, real-time, and precise lesion recognition can be achieved in video endoscopy.11,27,28,29,30,31
To the best of our knowledge, only a few studies have employed artificial intelligence networks to construct NPC endoscopic diagnosis models. Li et al. retrospectively used 28,966 white light images of nasopharyngeal endoscopy to train and develop a CNN-based diagnostic model to identify endoscopic nasopharyngeal malignant tumors and guide biopsies.15 The accuracy, sensitivity, specificity, and PPV values of the model for the retrospective test set were 88.7%, 91.3%, 83.1%, and 92.2%, respectively. Simultaneously, for the prospective test set, these values were 88.0%, 90.2%, 85.5%, and 86.9%, respectively. It took approximately 40 s to process 1,430 images. The model exhibited excellent segmentation performance and could accurately outline tumor boundaries. However, since the model can only make diagnoses based on preacquired endoscopic images rather than real-time video, it was challenging to achieve real-time detection in video endoscopy. Mohammed et al. used 381 endoscopic images of NPC to construct an ML model that can handle classification and segmentation tasks. By employing a genetic algorithm for feature selection and an artificial neural network (ANN) for image classification, the model achieved precision, sensitivity, and specificity values of 95.15%, 94.80%, and 95.20%, respectively. In addition, the segmentation accuracy of the model was 92.65%.18 Moreover, Mohammed’s research team used the same NPC dataset to develop a diagnostic model using support vector machine (SVM)-based decision-level fusion of three image texture (local binary patterns, first-order statistics histogram properties, and grayscale histograms) schemes. The classifier approaches achieved an accuracy of 94.07%, a sensitivity of 92.05%, and a specificity of 93.07%.19 For the detection of endoscopic images of NPC, Mohammed’s team then developed an NPC detection model using a genetic algorithm and ANN based on Haar feature fear. The proposed model achieved accuracy, sensitivity, and specificity values of 96.22%, 95.35%, and 94.55%, respectively.17 Xu et al. developed a CNN-based NPC diagnostic model using 4,783 nasopharyngeal endoscopic images by combining the optical characteristics of NPC with WL and NB images and used cross-validation to expand the test set sample size. They reported an accuracy of 94.9%, a sensitivity of 94.8%, a specificity of 95.0%, a PPV of 95.2%, and an AUC of 0.986. Additionally, the processing time for 2,000 images was 39.04 s, enabling real-time diagnosis during nasopharyngeal endoscopy.16 However, their models primarily perform image classification tasks and cannot accurately obtain the location, size, and other important information of an NPC lesion. While some of the aforementioned models exhibit fast inference speeds, there has been no verification of their performance in video-based applications. Therefore, further investigation is necessary to evaluate and optimize the real-time performance of these models. Moreover, since these studies use single-center datasets, the generalizability and applicability of the proposed models in the real world remain to be further discussed. The comparison of our proposed model with previous studies is summarized in Table 4.
Table 4.
A review of AI diagnosis of NPC based on endoscopic images
| Authors, Year and Country | Site, No. of Cases (Data Type) | AI Subfield (Application) | AI Methods and its Application | Performance Metric (s) |
|---|---|---|---|---|
| Li et al.15 (2018) (China) |
NPC 28966 (Endoscopic images, white light imaging) |
Deep learning (Auto-contouring/Diagnosis) | 1. Detection: Fully CNN |
1. Detection performance - AUC: 0.930 - Sensitivity: 0.902 [CI:0.878–0.922] - Specificity: 0.855 [CI: 0.827–0.880] - Accuracy: 0.880 [CI: 0.861–0.896] - PPV: 0.869 [CI: 0.843–0.892] - NPV: 0.892 [CI: 0.865–0.914] - Time taken: 0.67 min (1430 images) 2. Segmentation performance - DSC: 0.75 ± 0.26 |
| Mohammed et al.18 (2018) (Malaysia, Iraq and India) |
NPC 381 (Endoscopic images, white light imaging) |
Machine learning (Auto-contouring/Diagnosis) | 1. Feature selection: Genetic algorithm 2. Classification: ANN & SVM |
1. Segmentation performance - Accuracy: 0.9265 2. Classification performance - Sensitivity: 0.9480 - Specificity: 0.9520 - Precision: 0.9515 |
| Abd Ghani MK et al.19 (2018) (Malaysia, Iraq and India) | NPC 381 (Endoscopic images, white light imaging) |
Machine learning (Diagnosis) | 1. Classification: SVM, ANN, KNN |
1. Classification performance - Sensitivity: 0.9205 - Specificity: 0.9307 - Accuracy: 0.9407 |
| Mohammed et al.17 (2018) (Malaysia, Iraq and India) |
NPC 381 (Endoscopic images, white light imaging) |
Machine learning (Diagnosis) | 1. Feature selection: Genetic algorithm 2. Classification: ANN |
1. Classification performance - Sensitivity: 0.9535 - Specificity: 0.9455 - Accuracy: 0.9622 |
| Xu et al.16 (2021) (China) |
NPC 4783 (Endoscopic images, white light imaging & narrow-band imaging) |
Deep learning (Diagnosis) | 1. Feature extraction: Xception 2. Classification: Deep CNN |
1. Classification performance - AUC: 0.986 [CI:0.982–0.992] - Sensitivity: 0.948 [CI:0.930–0.966] - Specificity: 0.950 [CI: 0.937–0.964] - Accuracy: 0.949 [CI: 0.933–0.965] - PPV: 0.952 [CI: 0.936–0.968] - NPV: 0.946 [CI: 0.933–0.960] 2. Inference Time: 39.4 s (1000 pairs of images) |
| Our proposed model | NPC 2429 (Endoscopic images, white light imaging) |
Deep learning (Object detection/Diagnosis) | 1. Object detection: YOLOv6, YOLOv7, YOLOv8, Faster-RCNN, Cascade-RCNN, SSD |
1. Object detection performance Internal dataset -Precision: 0.977 -Recall: 0.943 -F1-score: 0.960 -mAP: 0.977 External dataset -Precision: 0.825 -Recall: 0.743 -F1-score: 0.780 -mAP: 0.814 2. Inference speed: 52.9 FPS |
NPC = Nasopharyngeal carcinoma; AI = Artificial intelligence; CNN = Convolutional neural network; AUC = Area under curve; PPV = Positive prediction value; NPV = Negative prediction value; DSC = Dice similarity coefficient; ANN = Artificial neural network; SVM = Support vector machines; KNN = k-nearest neighbors’ algorithm; RCNN = Region Convolutional neural network; SSD = Single Shot MultiBox Detector; mAP = Mean average precision; FPS = Frames per second.
To the best of our knowledge, we are the first to develop a DL model for real-time NPC object detection in video endoscopy. We used YOLOv6m, YOLOv7, YOLOv8l, Faster-RCNN, Cascade-RCNN, and SSD to construct object detection models and compared their results, ultimately finding that YOLOv8l provided the best accuracy and speed for tumor detection. The model demonstrated a good balance between precision and recall, with F1-scores of 0.960 and 0.780 for the internal and external test sets, respectively, which maximized the detection rate and reduced the misdiagnosis rate. The CADe’s ability to detect small lesions has been shown to be comparable or even superior to that of professional doctors, which can help less experienced doctors accurately detect lesions during endoscopy.28 The YOLOv8l model required only 17 ms to analyze a video frame. Additionally, the model’s average frame rate was 57.6 FPS, while the average frame rate of nasopharyngeal endoscopy video was 25–30 FPS, indicating that the model was fully capable of real-time lesion detection in nasopharyngeal endoscopy. In addition, our endoscopic video verification experiment also confirmed the real-time performance of the model. Most importantly, the proposed model demonstrated outstanding stability during robustness testing. We believe that the model can accurately detect NPC across various settings of nasopharyngoscopy examinations, differences in hospital equipment, and instances where the examination field may be blurred due to inexperienced doctor’s operation. Similar to those used by Xu et al., we used interpretive tools to visually explain which regions of images the model focuses on to make predictions, which had important implications.16 On the one hand, when the model is significantly weaker than endoscopists in terms of NPC detection, the goal of explanations is to identify the failure modes, thereby helping researchers focus their efforts on the most fruitful research directions. On the other hand, when the model is significantly stronger than endoscopists in terms of NPC detection, the goal of explanations is in machine teaching, i.e., a machine teaching an endoscopist about how to make better decisions in detecting NPC during nasopharyngeal endoscopy. Unlike CAM, Grad-CAM can extract the heatmap of any layer of the feature map without modifying the network structure of the model. It can be applied to the network structure of nonglobal average pooling connections to provide more accurate visualization results.20 Furthermore, due to the lightweight structure of the YOLO network, this model can be widely used and promoted in grassroots or community hospitals.
Since DL models typically perform well for internal datasets and poorly for extrapolation, we incorporated data from other medical centers as external test sets to validate the models' generalizability across various patient populations and healthcare systems. The dataset from multiple clinical medical centers included characteristics of different populations and different endoscopic systems, which could make our sample population more consistent with the actual population and more accurately reflect the model’s performance in practical applications. Often, the metric values of a model for an external test set may be less than or equal to those for an internal test set because the external test set contains more unknown data, which may have different distributions from the data in the internal test set. This could be due to a variety of factors, including differences in the equipment used at different centers, variations in the skills of the technical staff, and differences in the types and stages of diseases among patient populations. In the process of model training, the model will attempt to adapt to the data distribution of the training set and the validation set but may overfit these data distributions, resulting in degraded performance with the external test set. Additionally, there may be some selection bias in the internal test set data, resulting in slightly higher model performance for the internal test set than in reality. However, despite these differences, our model demonstrated good generalizability, maintaining reasonable performance across different centers and patient populations. This outcome gives us confidence in the application of our model in the real world and provides directions for improvements to our model. Consequently, future research should focus on further validating and optimizing the model’s practicality and generalizability in real-world scenarios. Increasing the diversity and quantity of data used for training or employing cross-validation methodologies in limited data should be considered. Furthermore, it is crucial to test the model’s performance across different sex, age, region, and disease stage subgroups and assess its robustness when dealing with various video qualities, lighting conditions, and lens angles. However, it should be noted that achieving perfect generalization across all datasets is often unattainable, so it may be necessary to strike a balance between increased performance with additional data and the possibility of overfitting the model.
Limitations of the study
There are limitations in this study. First, the model only recognizes NPC and non-NPC. Non-NPC includes benign and malignant lesions, such as hypertrophic adenoids, tuberculosis, lymph node hyperplasia, cysts, lymphoma, olfactory neuroblastoma, malignant melanoma, and adenoid cystic carcinoma. Among them, the number of video frames of lymphoma is small, and its endoscopic morphology is similar to that of NPC, so the model is prone to mistakenly consider lymphoma as NPC. Although NPC is the most common malignant tumor in the nasopharynx, further research is necessary. The next stage of this study will focus on expanding the number of other pathological types to enrich the dataset and build a reliable algorithm. Second, the number of video frames of NPC growing under the mucosa in the dataset, that is, atypical lesions and small lesions, is small, which leads to insufficient training of the model for this type of NPC. Therefore, the model is prone to missed diagnosis. In nasopharyngeal endoscopy, the local resolution can be improved by making the lens close to the lesion, thereby improving the detection rate. Third, the dataset consists of manually selected video frames rather than continuous video frames. Continuous video frames provide a more comprehensive representation of the dynamic changes and progression of lesions over time. This may cause the model to fail to learn effectively and adapt to a wider range of scenarios. Fourth, the whole dataset was collected retrospectively, which might have led to a certain level of selection bias. Finally, the accuracy of our model’s detection needs to be compared with different ranks of doctors, which in turn validates the model’s suitability for true clinical practice.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Nasopharyngeal Endoscopic Image Datasets | This paper | N/A |
| Software and algorithms | ||
| YOLOv6 | Li et al.32 | https://github.com/meituan/YOLOv6 |
| YOLOv7 | Wang et al.33 | https://github.com/WongKinYiu/yolov7 |
| YOLOv8 | Ultralytics company | https://github.com/ultralytics/ultralytics |
| Single Shot MultiBox Detector (SSD) | Liu et al.34 | https://github.com/weiliu89/caffe |
| Faster-RCNN | Girshick et al.35 | https://github.com/rbgirshick/py-faster-rcnn |
| Cascade-RCNN | Cai et al.36 | https://github.com/zhaoweicai/cascade-rcnn |
| Image Corruptions | Hendrycks et al.21 | https://github.com/bethgelab/imagecorruptions |
| Gradient-weighted Class Activation Mapping (Grad-CAM) | Selvaraju et al.20 | https://github.com/jacobgil/pytorch-grad-cam |
| PyTorch | Version 1.11.0 | https://pytorch.org/docs/1.11/ |
| Matplotlib | Version 3.7.1 | https://pypi.org/project/matplotlib/ |
| Python | Version 3.8.13 | https://www.python.org/downloads/release/python-3813/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Jian Li (lijianent@hotmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
This study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Ethics Committees of the First Affiliated Hospital of Sun Yat-sen University, the Kiang Wu Hospital and the Guangzhou First People's Hospital. Due to the retrospective nature of the study and the negligible risk to subjects, informed consent was waived. All patients underwent examination in the endoscopy room using a high-definition video nasopharyngeal endoscope (KARL STORZ-endoskope, Tuttlingen, Germany) in white light mode after local anesthesia with bupivacaine and mucosal shrinkage with epinephrine.
All images were video frames captured from nasopharyngeal endoscopy videos. The inclusion criteria for images were: (1) a minimum resolution of 400x400 pixels; (2) a minimum size of 60 kb; (3) acquired during the initial diagnosis; (4) nasopharyngeal images without nasal structure; (5) clearly visible nasopharyngeal mucosa without overlying material; (6) clear focus; (7) standard white light used during inspection and image capture, with white balance correction performed before inspection; (8) definitive pathological diagnosis. The exclusion criteria for images included: (1) missing pathological information; (2) missing endoscopic images; (3) images that were out of focus, too low in brightness, or had motion artifacts.
We retrospectively collected 2,429 nasopharyngeal endoscopic video frame images, clinicopathological data, imaging reports, and medical records from 690 patients at three medical centers from January 1, 2020, to December 1, 2021. This included 2,000 images from 519 patients at the First Affiliated Hospital of Sun Yat-sen University and 429 images from 171 patients at the Kiang Wu Hospital in Macau and the First People's Hospital in Guangzhou. All patients were Chinese. In the internal dataset, we recorded a total of 369 male patients and 150 female patients. Similarly, in the external dataset, our findings showed 112 male patients and 59 female patients. The average age of the entire dataset was 41.05 years old. All images were anonymized and reconstructed in random order. Images pathologically confirmed as other than NPC, according to the World Health Organization histopathological classification, were considered to be in the non-NPC category, which included nasopharyngeal cysts, lymphomas, tuberculosis, fibrovascular tumors, malignant melanoma, etc. The ratio of NPC to non-NPC was approximately 1:1.
Subsequently, three expert physicians manually labeled the images using LabelImg software. In an image with histopathological evidence of NPC, a bounding box, defined as Ground Truth (GT), was outlined along the largest boundary of the tumor that surrounded the entire tumor area and was labeled as "NPC" according to the label. The accuracy of the GT bounding boxes profile was cross-checked by the three expert physicians. We classified the NPC images into three categories based on relative bounding-box size of lesion in proportion to the image. The classification criteria were as follows: small: GT bounding box of lesion occupying equal or less than 10% of the image; medium: GT bounding box of lesion occupying more than 10% but equal or less than 30% of the image; large: GT bounding box of lesion occupying more than 30% of the image. Among them, small, medium and large bounding boxes accounted for 3.6 %, 36.2 % and 60.2 % respectively. The size distribution of ground-truth bounding boxes for different datasets is shown in the Table S1 and Figure S1 in supplemental information.
We randomly divided the dataset of the main center into a training set, a validation set, and an internal test set in an 8:1:1 ratio. The dataset from the remaining two centers was used as an external test set to evaluate the model's generalization ability. Lastly, six unedited videos of nasopharyngeal endoscopy were selected to validate the real-time NPC detection performance of the model. The dataset division is shown in Figure 4.
Method details
Data augmentation
Data augmentation is a technique to improve model generalization and reduce model overfitting, aiming to increase the number and diversity of data in the training set by transforming and expanding the original data to enhance the model's generalization and robustness. The data augmentation techniques we employed included adjusting image brightness, contrast, saturation, noise, random scaling of images, cropping, flipping, rotating, copy-paste, mixup, and mosaic data augmentation.33,37,38
DL model training and testing
We utilized the YOLO network of open-source CNNs as the object detection model. YOLO is a single-stage DL object detector capable of identifying objects by framing them in a bounding box while simultaneously classifying the object based on probability. At the time of our analysis, the latest version of YOLO was YOLOv8, which demonstrated excellent accuracy and inference speed. In the backbone network of YOLOv8, additional branches have been introduced during feature extraction. These additional branches help enhance the model's training accuracy by capturing and leveraging more diverse and informative features from the input images. Importantly, during the inference process, these additional branches are not involved in the computations. This optimization ensures that the inference speed is not compromised, allowing for efficient real-time or near-real-time object detection. The head section adopts the popular decoupled head structure, separating the classification and regression heads. Moreover, it transitions from an anchor-based approach to an anchor-free approach. In the classification head, YOLOv8 utilizes the Binary Cross Entropy (BCE) Loss for efficient and accurate object classification. The regression head, on the other hand, incorporates the concepts from the Distribution Focal Loss (DFL) and Complete Intersection over Union (CIoU) Loss. DFL is a loss function proposed to address the class imbalance issue in object detection tasks while CIoU Loss is a localization-based loss function that measures the geometric similarity between predicted and GT bounding boxes.
YOLOv8 consists of five different models that vary in terms of the number of parameters, trainable weight sizes, and computation time. Models range from small to extra-large versions (YOLOv8n, YOLOv8s, YOLOv8m, YOLOv8l, and YOLOv8x). In this study, we chose YOLOv8l as the target algorithm model. The flowchart of the research and YOLOv8l architecture are depicted in Figure 5. Additionally, to verify the NPC detection performance of the YOLOv8l model, we selected the other five algorithms, YOLOv7, YOLOv6m, Faster-RCNN, Cascade-RCNN and SSD for comparison tests.32,33,34,35,36
To further assess the model's robustness in various environmental conditions, including different video brightness, hue, contrast, video quality, and lens stability, we conducted a series of rigorous robustness tests. We employed the image corruption methods in test sets to simulate various image characteristics under different scenes, such as noise, blur, fog and brightness changes.
Gaussian noise was added to the images, simulating random variations with a Gaussian distribution. This noise introduces randomness and decreases in intensity as the distance from the center increases. It can be represented mathematically as follows:
For each pixel (i, j) in the image:
where I'(i, j) represents the corrupted pixel, I(i, j) is the original pixel, and N(0, σ) represents Gaussian noise with zero mean and standard deviation σ.
Shot noise was applied to simulate the random fluctuations in pixel intensities caused by variations in light. This type of noise is typically associated with uncertainties in light intensity and results in visible artifacts at areas of brightness changes. Shot noise can be modeled using the Poisson distribution:
For each pixel (i, j) in the image:
where I'(i, j) represents the corrupted pixel, I(i, j) is the original pixel, and λ controls the intensity of the noise.
Impulse noise, also known as salt-and-pepper noise, introduces sudden, isolated changes in brightness or color values. This type of noise is commonly observed due to sensor malfunctions or transmission errors, resulting in the appearance of bright and dark pixels. The corruption process involves randomly replacing a certain percentage of pixels with either the maximum intensity or minimum intensity.
For each pixel (i, j) in the image:
with a probability of p,
, if a random number less than p/2;
, if a random number exceeds p/2 and is less than p.where I'(i, j) represents the corrupted pixel, I(i, j) is the original pixel, and p represents the corruption ratio.
Defocus blur simulates the blurring effect caused by inaccurate focus settings in the endoscopic lens. It results in the loss of sharpness in image details, blurring of edges, or overall image blurriness. This effect is achieved by convolving the image with a given blur kernel, which represents the defocused point spread function.
For each pixel (i, j) in the image:
where I'(i, j) represents the blurred pixel, I(i, j) is the original pixel, and ⊗ denotes convolution with the blur kernel K.
Zoom blur mimics the blur effect caused by endoscopic lens or lesions movement during exposure. This blur results in the diffusion and blurring of details in the image. The process involves cropping and scaling the image, which enlarge or reduces the edges of the original image, leading to blurred edge information.
For each pixel (i, j) in the image:
where I'(i, j) represents the zoom-blurred pixel, I_zoomed(k)(i', j') represents the pixel value from the k-th zoomed-in image at coordinates (i', j'), N represents the total number of zoomed-in images.
Motion blur replicates the blurring effect caused by the movement of the endoscopic lens or the lesions. It manifests as a blurred trajectory of moving objects or overall image blurring. The blur effect is achieved by shifting and weighting the input image based on given parameters and a randomly chosen angle.
For each pixel (i, j) in the image:
where I'(i, j) represents the blurred pixel, I(i - dx, j - dy) are the shifted pixels, W(dx, dy) represents the motion blur kernel weights, and the sum is taken over the motion blur kernel.
Fog simulation introduces a decrease in lens clarity caused by the patient 's breathing fog. This degradation method results in a blurry appearance, faded colors, and loss of fine details. The fog function adds a generated plane fractal image to the input image, creating the desired foggy effect. The plane fractal image is a complex structure with self-similarity generated through a mathematical algorithm or process.
For each pixel (i, j) in the image:
where I'(i, j) represents the corrupted pixel, I(i, j) is the original pixel, and F(i, j) is the plane fractal image.
The brightness+ operation increases the overall brightness level of the image, resulting in a brighter appearance. This enhancement is achieved by adjusting the pixel values in the V (Value) channel of the HSV (Hue, Saturation, Value) color space.
Convert the image to HSV color space:
where V'(i, j) represents the modified pixel value in the V channel, V(i, j) is the original pixel value, and c controls the brightness increment.
The brightness- operation decreases the overall brightness level of the image, resulting in a darker appearance. Similar to brightness+, it adjusts the pixel values in the V channel of the HSV color space.
Convert the image to HSV color space:
where V'(i, j) represents the modified pixel value in the V channel, V(i, j) is the original pixel value, and c controls the brightness decrement. Figure S2 illustrates the schematic diagram of the image corruption methods.
The experimental platform for this study was based on Ubuntu, with an Intel(R) Xeon(R) CPU (2.40GHz) and Nvidia GeForce RTX3090 GPUs (24G). We implemented the YOLO model using the Pytorch deep learning framework, and the experimental environment was torch 1.11.0 + cu113 CUDA. The hyperparameters used to train the YOLO network were as follows: batch size 160; image size 640; number of epochs 100; optimizer Stochastic Gradient Descent (SGD); dropout rate 0.1; confidence threshold 0.25; intersection over union (IoU) threshold 0.5; initial learning rate (lr0) 0.01; learning rate decay factor (lrf) 0.1; momentum 0.937; weight decay 0.0005; warm-up epochs 3.0; warm-up momentum 0.8; warm-up bias learning rate 0.1; box loss gain 4.0; classification loss gain 4.5; DFL loss gain 1.5; focal loss gamma 0.0; label smoothing 0.0; nominal batch size (nbs) 64; HSV-Hue augmentation 0.015; HSV-Saturation augmentation 0.4; HSV-Value augmentation: 0.4.
Quantification and statistical analysis
We evaluated the model performance using precision, recall, mean average precision (mAP), F1-score, and frame rate. Intersection over Union (IoU) is the ratio of the intersection and the union of the predicted bounding box and the GT box, as shown in Figure 6. True positive (TP) was defined as IoU ≥ 0.5. A false positive (FP) was defined as an IoU < 0.5 or detection of a duplicate bounding box for the same GT box. False negative (FN) was defined as the model detecting the target object as a negative class in its presence. True negative (TN) indicates that the model detects the target object as a negative class in its absence. The object detection metric does not consider TN because it does not reflect the algorithm's performance in detecting the target object. All statistical analyses were conducted using Python (version 3.8.13 (Matplotlib library, version 3.7.1). Recall is the probability of correct identification in all positive samples and corresponds to the model’s sensitivity, expressed as:
Precision is the probability of correct detection among all detected targets and corresponds to the model’s positive predicted value (PPV), expressed as:
For the object detection model, mAP is the standard performance metric. mAP is the area under the precision-recall curve, defined by the following equation:
where Q is the number of queries in the set, and AveP(q) is the average precision for a given query, q. In our study, as we set the model threshold as 0.5 (at IoU = 0.5), mAP@.5 denotes that this value was achieved under the condition of IoU ≥ 0.5.
The F1-score takes into account both precision and recall of the object detection model and is the summed average of the two, which is expressed as:
To evaluate the inference speed of a model, frame rate is usually used, and its unit is FPS (frames per second), and the expression is:
where FrameNum is the number of video frames and elapsedTime is the time taken by the model to process the video frames. In addition, we recorded the time delay of the model in milliseconds (ms).
Acknowledgments
Study funding: This study was funded by the National Natural Science Foundation of China (81974141), the Provincial Natural Science Foundation of Guangdong Province (2022A1515010506).
Author contributions
Design or conceptualization of the study: Z.H., J.C., and J.L.; Acquisition of data: Y.W., Z.H., W.H., Q.M., and J.L.; Models training: K.Z. and N.Z.; Analysis or interpretation of the data: K.Z. and N.Z.; Drafting or revising the manuscript for intellectual content: Z.H., K.Z., N.Z., C.L., J.C., and J.L.; Grant proposal and funding acquisition: J.L. and J.C.; Supervision and mentoring: J.L. and J.C.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. We avoided “helicopter science” practices by including the participating local contributors from the region where we conducted the research as authors on the paper.
Published: July 24, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107463.
Contributor Information
Junzhou Chen, Email: chenjunzhou@mail.sysu.edu.cn.
Jian Li, Email: lijianent@hotmail.com.
Supplemental information
Data and code availability
-
•
Nasopharyngeal endoscopic image data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Chen Y.P., Chan A.T.C., Le Q.-T., Blanchard P., Sun Y., Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. doi: 10.1016/s0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Ca - Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Lee A.W.M., Ng W.T., Chan L.L.K., Hung W.M., Chan C.C.C., Sze H.C.K., Chan O.S.H., Chang A.T.Y., Yeung R.M.W. Evolution of treatment for nasopharyngeal cancer--success and setback in the intensity-modulated radiotherapy era. Radiother. Oncol. 2014;110:377–384. doi: 10.1016/j.radonc.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Bossi P., Chan A.T., Licitra L., Trama A., Orlandi E., Hui E.P., Halámková J., Mattheis S., Baujat B., Hardillo J., et al. Nasopharyngeal carcinoma: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up(dagger) Ann. Oncol. 2021;32:452–465. doi: 10.1016/j.annonc.2020.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Cengiz K., Kumral T.L., Yıldırım G. Diagnosis of pediatric nasopharynx carcinoma after recurrent adenoidectomy. Case Rep. Otolaryngol. 2013;2013:653963. doi: 10.1155/2013/653963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y.P., Cai P.Q., Tian L., Xu J.H., Mitteer R.A., Jr., Fan Y., Zhang Z. Hypertrophic adenoids in patients with nasopharyngeal carcinoma: appearance at magnetic resonance imaging before and after treatment. Chin. J. Cancer. 2015;34:130–136. doi: 10.1186/s40880-015-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D.H., Lee M.H., Lee S., Kim S.W., Hwang S.H. Comparison of Narrowband Imaging and White-Light Endoscopy for Diagnosis and Screening of Nasopharyngeal Cancer. Otolaryngol. Head Neck Surg. 2022;166:795–801. doi: 10.1177/01945998211029617. [DOI] [PubMed] [Google Scholar]
- 8.Luo H., Xu G., Li C., He L., Luo L., Wang Z., Jing B., Deng Y., Jin Y., Li Y., et al. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645–1654. doi: 10.1016/s1470-2045(19)30637-0. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z., Lin L., Wu C., Li C., Xu R., Sun Y. Artificial Intelligence for Assisting Cancer Diagnosis and Treatment in the Era of Precision Medicine. Cancer Commun. 2021;41:1100–1115. doi: 10.1002/cac2.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng X., Jiang Z., Luo W., Li H., Li H., Li G., Shi J., Wu K., Liu T., Lin X., et al. Efficient and accurate identification of ear diseases using an ensemble deep learning model. Sci. Rep. 2021;11:10839. doi: 10.1038/s41598-021-90345-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azam M.A., Sampieri C., Ioppi A., Africano S., Vallin A., Mocellin D., Fragale M., Guastini L., Moccia S., Piazza C., et al. Deep Learning Applied to White Light and Narrow Band Imaging Videolaryngoscopy: Toward Real-Time Laryngeal Cancer Detection. Laryngoscope. 2022;132:1798–1806. doi: 10.1002/lary.29960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu D., Peng X., Liu X., Li Y., Bao Y., Xu J., Bian X., Xue W., Qian D. A real-time system using deep learning to detect and track ureteral orifices during urinary endoscopy. Comput. Biol. Med. 2021;128:104104. doi: 10.1016/j.compbiomed.2020.104104. [DOI] [PubMed] [Google Scholar]
- 13.Min J.K., Kwak M.S., Cha J.M. Overview of Deep Learning in Gastrointestinal Endoscopy. Gut Liver. 2019;13:388–393. doi: 10.5009/gnl18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumiyama K., Futakuchi T., Kamba S., Matsui H., Tamai N. Artificial intelligence in endoscopy: Present and future perspectives. Dig. Endosc. 2021;33:218–230. doi: 10.1111/den.13837. [DOI] [PubMed] [Google Scholar]
- 15.Li C., Jing B., Ke L., Li B., Xia W., He C., Qian C., Zhao C., Mai H., Chen M., et al. Development and validation of an endoscopic images-based deep learning model for detection with nasopharyngeal malignancies. Cancer Commun. 2018;38:59. doi: 10.1186/s40880-018-0325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J., Wang J., Bian X., Zhu J.Q., Tie C.W., Liu X., Zhou Z., Ni X.G., Qian D. Deep Learning for nasopharyngeal Carcinoma Identification Using Both White Light and Narrow-Band Imaging Endoscopy. Laryngoscope. 2022;132:999–1007. doi: 10.1002/lary.29894. [DOI] [PubMed] [Google Scholar]
- 17.Mohammed M.A., Abd Ghani M.K., Arunkumar N., Hamed R.I., Abdullah M.K., Burhanuddin M.A. A real time computer aided object detection of nasopharyngeal carcinoma using genetic algorithm and artificial neural network based on Haar feature fear. Future Generat. Comput. Syst. 2018;89:539–547. doi: 10.1016/j.future.2018.07.022. [DOI] [Google Scholar]
- 18.Mohammed M.A., Abd Ghani M.K., Arunkumar N., Mostafa S.A., Abdullah M.K., Burhanuddin M.A. Trainable model for segmenting and identifying Nasopharyngeal carcinoma. Comput. Electr. Eng. 2018;71:372–387. doi: 10.1016/j.compeleceng.2018.07.044. [DOI] [Google Scholar]
- 19.Abd Ghani M.K., Mohammed M.A., Arunkumar N., Mostafa S.A., Ibrahim D.A., Abdullah M.K., Jaber M.M., Abdulhay E., Ramirez-Gonzalez G., Burhanuddin M.A. Decision-level fusion scheme for nasopharyngeal carcinoma identification using machine learning techniques. Neural Comput. Appl. 2018;32:625–638. doi: 10.1007/s00521-018-3882-6. [DOI] [Google Scholar]
- 20.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. 2017 IEEE International Conference on Computer Vision (ICCV) 2017. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. [Google Scholar]
- 21.Dan Hendrycks T.G.D. International Conference on Learning Representations. Vol. 2019. 2019. Benchmarking Neural Network Robustness to Common Corruptions and Surface Variations. [Google Scholar]
- 22.Li S., Deng Y.Q., Zhu Z.L., Hua H.L., Tao Z.Z. A Comprehensive Review on Radiomics and Deep Learning for Nasopharyngeal Carcinoma Imaging. Diagnostics. 2021;11:1523. doi: 10.3390/diagnostics11091523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen Y.H., Zhu X.L., Lei W.B., Zeng Y.H., Sun Y.Q., Wen W.P. Narrow-band imaging: a novel screening tool for early nasopharyngeal carcinoma. Arch. Otolaryngol. Neck Surg. 2012;138:183–188. doi: 10.1001/archoto.2011.1111. [DOI] [PubMed] [Google Scholar]
- 24.Ni X.G., Zhang Q.Q., Wang G.Q. Classification of nasopharyngeal microvessels detected by narrow band imaging endoscopy and its role in the diagnosis of nasopharyngeal carcinoma. Acta Otolaryngol. 2017;137:546–553. doi: 10.1080/00016489.2016.1253869. [DOI] [PubMed] [Google Scholar]
- 25.Pacal I., Karaboga D., Basturk A., Akay B., Nalbantoglu U. A comprehensive review of deep learning in colon cancer. Comput. Biol. Med. 2020;126:104003. doi: 10.1016/j.compbiomed.2020.104003. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki K. Overview of deep learning in medical imaging. Radiol. Phys. Technol. 2017;10:257–273. doi: 10.1007/s12194-017-0406-5. [DOI] [PubMed] [Google Scholar]
- 27.Lee J.Y., Jeong J., Song E.M., Ha C., Lee H.J., Koo J.E., Yang D.H., Kim N., Byeon J.S. Real-time detection of colon polyps during colonoscopy using deep learning: systematic validation with four independent datasets. Sci. Rep. 2020;10:8379. doi: 10.1038/s41598-020-65387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Z., Nemoto D., Zhu X., Li Q., Aizawa M., Utano K., Isohata N., Endo S., Kawarai Lefor A., Togashi K. Polyp detection algorithm can detect small polyps: Ex vivo reading test compared with endoscopists. Dig. Endosc. 2021;33:162–169. doi: 10.1111/den.13670. [DOI] [PubMed] [Google Scholar]
- 29.Pacal I., Karaboga D. A robust real-time deep learning based automatic polyp detection system. Comput. Biol. Med. 2021;134:104519. doi: 10.1016/j.compbiomed.2021.104519. [DOI] [PubMed] [Google Scholar]
- 30.Ku Y., Ding H., Wang G. Efficient Synchronous Real-Time CADe for Multicategory Lesions in Gastroscopy by Using Multiclass Detection Model. BioMed Res. Int. 2022;2022:8504149. doi: 10.1155/2022/8504149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pacal I., Karaman A., Karaboga D., Akay B., Basturk A., Nalbantoglu U., Coskun S. An efficient real-time colonic polyp detection with YOLO algorithms trained by using negative samples and large datasets. Comput. Biol. Med. 2022;141:105031. doi: 10.1016/j.compbiomed.2021.105031. [DOI] [PubMed] [Google Scholar]
- 32.Li C., Li L., Jiang H., Weng K., Geng Y., Liang L., Zaidan K., Li Q., Cheng M., Nie W., Li Y., et al. YOLOv6: A Single-Stage Object Detection Framework for Industrial Applications. arxiv. 2022 doi: 10.48550/arXiv.2209.02976. Preprint at. [DOI] [Google Scholar]
- 33.Wang C.Y., Bochkovskiy A., Liao Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. arxiv. 2022 doi: 10.48550/arXiv.2207.02696. Preprint at. [DOI] [Google Scholar]
- 34.Liu W., Anguelov D., Erhan D., Szegedy C., Reed S., Fu C.-Y., Berg A.C. 2016. SSD: Single Shot MultiBox Detector (Computer Vision – ECCV 2016) [DOI] [Google Scholar]
- 35.Ren S., He K., Girshick R., Sun J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39:1137–1149. doi: 10.1109/tpami.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 36.Zhaowei Cai N.V. Cascade R-CNN: Delving Into High Quality Object Detection. IEEE Conf. Comput. Vis. Pattern Recogn. 2018:6154–6162. [Google Scholar]
- 37.Ghiasi G., Cui Y., Srinivas A., Qian R., Lin T.Y., Cubuk E.D., Le Q.V., Zoph B. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2021. Simple Copy-Paste Is a Strong Data Augmentation Method for Instance Segmentation; pp. 2917–2927. [Google Scholar]
- 38.Zhang H., Cisse M., Dauphin Y.N., Lopez-Paz D. The International Conference on Learning Representations. Vol. 2018. 2018. mixup: Beyond empirical risk minimization. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Nasopharyngeal endoscopic image data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.