Summary
With COVID-19 becoming endemic, there is a continuing need to find biomarkers characterizing the disease and aiding in patient stratification. We studied the relation between COVID-19 and cholesterol biosynthesis by comparing 10 intermediates of cholesterol biosynthesis during the hospitalization of 164 patients (admission, disease deterioration, discharge) admitted to the University Medical Center of Ljubljana. The concentrations of zymosterol, 24-dehydrolathosterol, desmosterol, and zymostenol were significantly altered in COVID-19 patients. We further developed a predictive model for disease severity based on clinical parameters alone and their combination with a subset of sterols. Our machine learning models applying 8 clinical parameters predicted disease severity with excellent accuracy (AUC = 0.96), showing substantial improvement over current clinical risk scores. After including sterols, model performance remained better than COVID-GRAM. This is the first study to examine cholesterol biosynthesis during COVID-19 and shows that a subset of cholesterol-related sterols is associated with the severity of COVID-19.
Subject areas: Assessment in health technology, Lipid, Bioinformatics
Graphical abstract

Highlights
-
•
Intracellular synthesis of cholesterol is significantly modified during COVID-19
-
•
A handful of clinical variables outperform current patient stratification tools
-
•
A subset of cholesterol-related sterols was associated with COVID-19 severity
Assessment in health technology; Lipid; Bioinformatics
Introduction
COVID-19 is still very much present and, according to epidemiologists, its causative agent SARS-CoV-2 is likely to become endemic with new variants recurring seasonally.1 Therefore, searching for new biomarkers for disease course and outcome prediction remains of high importance.
Viral infections can trigger changes in the host organism’s lipid profile, which could serve as a biomarker. In fact, dyslipidemia seems to be a hallmark of viral infections, as previously shown in hepatitis C virus (HCV), human immunodeficiency virus (HIV), and dengue virus infection.2 In 2020, Hu et al.3 were among the first to report an altered lipid profile in COVID-19 patients. Serum total cholesterol as well as HDL- and LDL-cholesterol levels were significantly lowered in patients suffering from COVID-19. Reports about dyslipidemia linked to SARS-CoV-2 infection from other groups soon followed,4,5,6,7,8,9,10,11,12,13,14,15,16,17 but findings were not always concordant. Furthermore, increased serum levels of the liver enzymes alkaline phosphatase (ALP), alanine aminotransferase (ALT), and aspartate aminotransferase (AST) seen in COVID-19 patients were suggested to be explained by liver damage as a result of the infection.2,5 Chen et al.11 showed that liver-specific proteins that regulate sterol and cholesterol transport were downregulated in COVID-19 patients. Although altered blood cholesterol levels have been reported in patients suffering from COVID-19, the effect of COVID-19 on intracellular biosynthesis of cholesterol remains to be determined.
Besides being a fundamental lipid component of vertebrate cell membranes, primarily as a lipid building block of ordered membrane micro-domains – lipid rafts2,18,19 – cholesterol also modulates their permeability, signaling, and transport. Furthermore, it can be integrated into lipoproteins, and stored in lipid droplets and cholesteryl esters.2,20 Importantly, its metabolic pathways branch out in several ways, resulting in physiologically important, active compounds (e.g., bile acids, oxysterols, steroid and glucocorticoid hormones, vitamin D, coenzyme Q).2,20,21,22 As cholesterol is an important molecule with versatile functions in numerous physiological processes and an excess of non-esterified cholesterol may potentially be toxic, maintaining its homeostasis is pivotal.2,23 Cholesterol biosynthesis is a tightly regulated housekeeping pathway and takes place mainly in the liver. The pre-squalene part of de novo cholesterol biosynthesis starts with acetyl-CoA and terminates with the enzymatic conversion of farnesyl-PP to squalene (Figure 1). A more detailed description of the pre-squalene pathway is given elsewhere.24 Conversion of squalene to lanosterol is the link between the pre- and post-squalene parts of the biosynthesis. Lanosterol is converted through a series of enzymatic reactions to cholesterol via the Bloch and/or Kandutsch-Russell (K-R) pathway.22,24,25 Both pathways are enzymatically identical, except for the first and the last steps (Figure 1). In the Bloch pathway, CYP51A1 catalyzes the conversion of lanosterol to FF-MAS, while in the K-R pathway, sterol-Δ24-reductase (DHCR24) catalyzes the same sterol intermediate to 24,25-dihydrolanosterol. In the last enzymatic step of the Bloch pathway desmosterol is converted to cholesterol by DHCR24, while the final reaction of the K-R branch is the conversion of 7-dehydrocholesterol to cholesterol by DHCR7. Since all sterol intermediates (referred to as sterol intermediates or sterols throughout the manuscript) from lano-to desmosterol in the Bloch branch contain Δ24 double bonds, DHCR24 can in principle metabolize any of them, and both branches, therefore, intertwine. Nonetheless, an in vitro study showed that 24-dehydrolathosterol is the most plausible switching point between both branches, indicating that cholesterol biosynthesis preferentially starts via Bloch and later shifts to the K-R pathway.26 Depending on tissue type, one or an interplay of both pathways dominates de novo cholesterol biosynthesis.
Figure 1.
The cholesterol biosynthesis pathway
The cholesterol biosynthesis pathway is divided into pre- and post-squalene parts. In the pre-squalene part, acetyl-CoA is converted to squalene through a series of enzymatic reactions. The conversion of squalene to lanosterol represents a linking point between the two parts of the biosynthesis. After lanosterol, the biosynthesis branches into the Bloch and Kandutsch-Russell (K-R) pathways. In the Bloch pathway, lanosterol is converted to FF-MAS by CYP51A1, while in the K-R pathway DHCR24 converts it to 24,25-dihydrolanosterol. The final reaction in the Bloch pathway is a conversion of desmosterol to cholesterol by DHCR24, whilst in the K-R pathway DHCR7 catalyzes the conversion of 7-dehydrocholesterol to cholesterol. Both pathways can intertwine but data show that 24-dehydrolathosterol is the most plausible substrate where switching occurs, indicating that cholesterol biosynthesis preferentially starts via the Bloch and later shifts to the K-R pathway. Enzymes catalyzing each reaction are shown in gray. Sterol chemical names are listed in Table S1.
The aim of this study was to evaluate the effects of COVID-19 on intracellular cholesterol biosynthesis, to develop a predictive model for the severity of COVID-19 course based on simple clinical parameters obtained at hospital admission using machine learning models, and to investigate the potential benefits of measuring metabolic pathways for disease monitoring. Including metabolic biomarkers into clinical diagnostics is attracting growing interest, emphasizing the importance of investigating their potential use in this context. Previous research has focused on altered blood cholesterol levels in COVID-19 patients, but detailed knowledge of endogenous cholesterol biosynthesis in patients with SARS-CoV-2 infection is scarce. In addition, existing prognostic models have limitations and lack reproducibility across different populations. Thus, we sought to identify readily available clinical variables and reliable methods to predict disease severity beyond the established COVID-GRAM risk score, as well to investigate biomarker potential of sterols. We performed targeted lipidomics to monitor changes in blood sterol intermediates – indicators of de novo intracellular cholesterol biosynthesis – in hospitalized COVID-19 patients. In addition, machine learning techniques were used to retrospectively estimate disease severity based on clinical parameters alone and also to investigate the biomarker potential of cholesterol-related sterols measured at hospital admission.
Results
Cohort description
164 adult patients admitted to the Department of Infectious Diseases of the University Medical Center Ljubljana (Slovenia) from July 2020 to July 2021 suffering from a severe course of COVID-19 were enrolled in this study. Their basic clinical characteristics are shown in Tables 1 and S2.
Table 1.
Basic demographic, clinical, and biochemical parameters of hospitalized COVID-19 patients
| Number of patients | 164 |
|---|---|
| Age | 61.0 ± 14.0 |
| BMI | 31.9 ± 6.7 |
| Disease category | |
| Mild | 7 (4.27%) |
| Moderate | 23 (14.02%) |
| Severe | 100 (60.98%) |
| Critical | 34 (20.73%) |
| Comorbidities | |
| Yes | 125 |
| No | 39 |
| Other parameters | |
| Reason for admissiona | need for oxygen: 119 subjective feeling of difficulty breathing: 17 worsening of the underlying diseases: 6 other: 23 |
| Oxygen saturation measured with or without oxygen supplementationa | oxygen: 95 air: 69 |
| Oxygen saturation [%]a | 94.0 ± 3.0 |
| CRP [mg/L]a | 104.4 ± 75.1 |
| Ferritin [μg/L]a | 1008.7 ± 839.4 |
| LDH [μkat/L]a | 5.72 ± 1.99 |
| X-ray – lung abnormalitiesa | yes: 140 |
| no: 22 | |
| X-ray – lung opacities locationsa | unilateral: 16 |
| bilateral: 121 | |
| In-hospital outcome | |
| Survival | 159 |
| Deaths | 5 |
The comorbidities section contains the presence of diabetes, hyperlipidemia, thyroid disease, arterial hypertension, heart failure, chronic disease of lung, liver, and kidney, rheumatic disease, active malignant disease, and/or transplantation. See also Table S2. Disease category data are number (%); other values are given as mean ± SD. BMI, body mass index; CRP, C-reactive protein; LDH, lactate dehydrogenase; SD, standard deviation.
At admission to hospital.
COVID-19 impact on de novo intracellular cholesterol biosynthesis
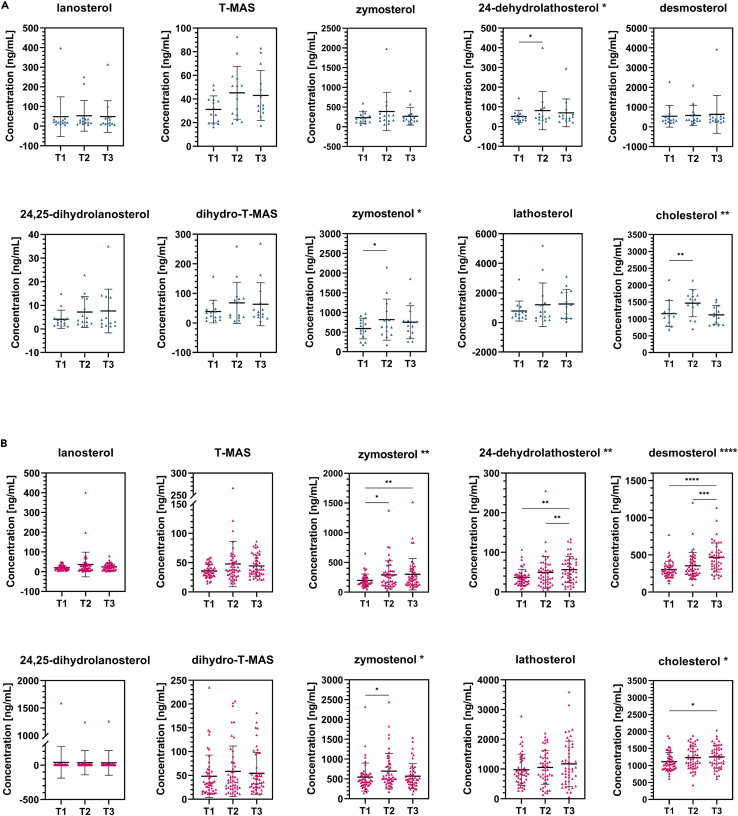
As the concentration of blood cholesterol depends on different factors, e.g., de novo biosynthesis and dietary cholesterol, the rate of de novo intracellular cholesterol biosynthesis can only be estimated according to the presence of sterol intermediates. We used liquid chromatography with tandem mass spectrometry (LC-MS/MS)-based targeted lipidomics to provide insight into cholesterol intermediates during COVID-19. Ten sterols from the post-squalene part of cholesterol biosynthesis were measured in serum samples at three different time points during hospitalization of 62 COVID-19 patients, i.e., lanosterol, 24,25-dihydrolanosterol, T-MAS, dihydro-T-MAS, zymosterol, zymostenol, 24-dehydrolathosterol, lathosterol, desmosterol, and cholesterol. Samples were collected upon admission to the hospital care (T1), in case of severe deterioration or in the middle of treatment (T2), and upon discharge (T3). An additional 102 COVID-19 patients serum samples were collected only at T1 and sterol intermediates were measured. Concentrations of sterol intermediates at all time points are shown in Table 2. Statistical significance was tested using the nonparametric Friedman test (Figures 2A and 2B) comparing three time points of sterol concentrations. For multiple comparisons, the adjusted p-values were determined using Dunn’s test. Results show more significant alterations in cholesterol biosynthesis in patients with severe (Figure 2B), compared to those with mild disease course (Figure 2A). In the latter, statistically significant changes during the course of the disease were found in the concentrations of 24-dehydrolathosterol (T1 vs. T2), zymostenol (T1 vs. T2), and cholesterol (T1 vs. T2), whereas in patients with severe course of COVID-19 statistically significant changes were observed in zymosterol (T1 vs. T2, T1 vs. T3), 24-dehydrolathosterol (T1 vs. T3, T2 vs. T3), desmosterol (T1 vs. T3, T2 vs. T3), zymostenol (T1 vs. T2) and finally cholesterol (T1 vs. T3), most of them being representatives of the Bloch pathway. Findings are shown in Figures 2A and 2B, Tables 2 and S3.
Table 2.
Concentration of sterol intermediates in patients with mild (N = 14) and severe (N = 48) COVID-19 measured at hospital admission
| Sterol name | Concentration [ng/mL] |
|||||
|---|---|---|---|---|---|---|
| T1 |
T2 |
T3 |
||||
| Mild | Severe | Mild | Severe | Mild | Severe | |
| lanosterol | 48.34 ± 100.98 | 19.95 ± 11.77 | 52.65 ± 77.73 | 36.21 ± 62.40 | 48.76 ± 80.54 | 24.42 ± 16.60 |
| 24,25-dihydrolanosterol | 4.15 ± 3.83 | 40.46 ± 228.38 | 7.18 ± 6.54 | 36.09 ± 178.55 | 7.63 ± 9.27 | 33.92 ± 180.78 |
| T-MAS | 31.18 ± 11.46 | 36.13 ± 11.16 | 45.17 ± 22.25 | 47.65 ± 38.81 | 43.02 ± 21.14 | 44.32 ± 18.36 |
| dihydro-T-MAS | 38.83 ± 38.33 | 48.17 ± 44.41 | 68.10 ± 69.70 | 58.52 ± 53.27 | 63.56 ± 72.67 | 54.53 ± 43.83 |
| zymosterol | 234.29 ± 146.37 | 196.70 ± 107.56 | 387.69 ± 487.68 | 291.07 ± 234.56 | 266.49 ± 224.34 | 300.75 ± 262.96 |
| zymostenol | 593.27 ± 263.01 | 546.95 ± 352.53 | 819.32 ± 521.05 | 696.03 ± 446.04 | 752.53 ± 419.62 | 569.46 ± 314.30 |
| 24-dehydrolathosterol | 51.84 ± 30.73 | 36.94 ± 20.31 | 81.06 ± 96.69 | 49.50 ± 40.53 | 69.65 ± 70.39 | 56.50 ± 32.96 |
| lathosterol | 781.71 ± 677.21 | 974.93 ± 524.65 | 1200.01 ± 1475.55 | 1050.77 ± 565.37 | 1258.76 ± 979.63 | 1171.96 ± 764.83 |
| desmosterol | 538.41 ± 548.88 | 302.72 ± 112.58 | 577.32 ± 516.68 | 352.64 ± 180.33 | 630.24 ± 957.82 | 465.34 ± 195.47 |
| cholesterol | 1156.96 ± 385.43 | 1112.98 ± 271.54 | 1442.01 ± 398.03 | 1233.75 ± 335.29 | 1107.95 ± 267.97 | 1253.15 ± 336.29 |
Data are represented as mean ± SD. See also Table S3. SD, standard deviation.
Figure 2.
Changes in serum lipid profile during COVID-19
(A and B) Changes in serum lipid profile during COVID-19 in patients with (A) mild and (B) severe course of the disease tested with the nonparametric Friedman’s test followed by Dunn’s post hoc test for multiple comparisons testing. Statistical significance of Friedman’s and Dunn’s tests is indicated beside the sterol name and inside the graph, respectively. Data are represented as mean ± SD. See also Table S3. SD, standard deviation. ∗ ≤0.05, ∗∗ ≤0.01, ∗∗∗ ≤0.001, ∗∗∗∗ ≤0.0001.
Developing machine learning models for COVID-19 course prediction
The specific aims of the study were (1) to predict disease severity based on clinical parameters using machine learning models better than the currently established COVID-19 clinical risk score (i.e., COVID-GRAM27), and (2) to evaluate whether the inclusion of sterol intermediates could strengthen the prediction performance.
The cohort utilized in this study recorded more than 70 clinical variables upon admission and measured concentrations of 10 sterols at T1. Using all clinical variables to estimate disease severity would lead to overfitting of machine learning models, resulting in poor generalizability. Therefore, to identify a smaller subset of meaningful variables, we conducted an unsupervised variable selection to identify important predictors for disease severity among clinical, as well as sterol intermediates measurements. This yielded a total of 8 clinical and 4 sterol variables (Table S4), which included the reason for hospital admission, information on how oxygen saturation was measured and its level, conclusions drawn from X-ray analyses, and concentrations of ferritin, LDH, CRP, 24,25-dihydrolanosterol, zymostenol, 24-dehydrolathosterol, and desmosterol. An overview of these variables can be found in Table 3, with a more detailed description in Table S5.
Table 3.
Summary overview on variables, measured at admission to hospital, contained in each variable set (clinical, T1 sterols, clinical + T1 sterols)
| Variable | Scenario | |
|---|---|---|
| Reason for admission | clinical | clinical + T1 sterols |
| Oxygen saturation measured with or without oxygen supplementation | ||
| Oxygen saturation [%] | ||
| CRP | ||
| Ferritin | ||
| LDH | ||
| X-ray – lung abnormalities | ||
| X-ray – lung opacities locations | ||
| 24,25-dihydrolanosterol | T1 sterols | |
| zymostenol | ||
| 24-dehydrolathosterol | ||
| desmosterol | ||
To evaluate the predictive power of the selected clinical variables, T1 sterols, and their combination, we first trained eight different machine learning models on each scenario using leave-one-out-cross-validation. Their performances were compared with several metrics and the best performing model selected based on highest scores across all metrics, model simplicity, and interpretability (Figure S1; Table S6). The best performing models of each scenario were subsequently compared to each other (Figure 3A; Table 4). A model trained on clinical variables alone showed excellent predictive power (“clinical”, AUC = 0.96). Based on T1 sterol measurements alone, disease severity can be estimated with moderate confidence (“T1 sterols”, AUC = 0.66). The performance of the model remained excellent when both groups of variables were combined (“clinical + T1 sterols”, AUC = 0.95).
Figure 3.
Computational models for estimating COVID-19 severity
(A) Performance evaluation of machine learning classification models for predicting disease severity in a cohort of COVID-19 patients (N = 164). This utilized three different sets of variables, namely clinical parameters (F = 8), sterol intermediates measured at T1 (F = 4), and their combination (F = 12). Only the best performing models for each variable set are displayed. F, number of features; N, number of patients.
(B) Variable importance in different scenarios. Feature importance for all three scenarios is shown: upper left - clinical parameters; upper right - sterol intermediates in T1; lower: combination of both clinical and T1 sterols. ∗Oxygen saturation measured with or without oxygen supplementation upon admission.
(C) Receiver operating characteristic (ROC) curve of all three scenarios, COVID-GRAM, and clinical baseline model. The area under the curve (AUC) for each set is displayed in the legend. The dashed diagonal line reflects the performance of a diagnostic test that is no better than chance level. See also Tables S5 and S6.
Table 4.
Evaluation metrics for binary classification for best performing classifiers in each scenario
| Dataset | Classifier | Performance metric |
||||
|---|---|---|---|---|---|---|
| precision | recall | f1 | accuracy | AUC | ||
| clinical | GNB | 0.976 | 0.910 | 0.942 | 0.909 | 0.955 |
| T1 sterols | RFC | 0.849 | 0.963 | 0.902 | 0.829 | 0.664 |
| clinical + T1 sterols | RFC | 0.926 | 0.940 | 0.933 | 0.890 | 0.950 |
| MCL | 0.817 | 1.000 | 0.899 | 0.817 | 0.500 | |
See also Table S6. AUC, area under the ROC curve; f1, F1-score; GNB, Gaussian Naive Bayes; MCL, Majority Classifier; RFC, Random Forest.
To gain more insight into the decision making of each model, feature importance for each scenario was evaluated using a permutation-based feature importance method (see STAR methods, classification models), revealing sets of features with the highest importance in the prediction of COVID-19 severity (Figure 3B). We observe that although variables were selected in a data-driven manner, not all of them display equal importance during the predictive task. While the primary reason for hospital admission (“reason for admission”), CRP, and the measurement method of oxygen saturation (“Oxygen saturation measured with or without oxygen supplementation”) remain highly important across scenarios, we see a shift in the importance of, e.g., X-ray measurements or 24,25-dihydrolanosterol upon combining clinical and T1 sterols information. This may indicate that the selected sterols correlate with the respective clinical measurements, resulting in an overlap of information (Figure S2).
In order to put our model performances in the context of established clinical practices, we compared them to the COVID-GRAM risk score, as well as a “clinical baseline” model we developed using three variables commonly used to stratify patients into disease severity groups, namely concentrations of LDH, ferritin, and CRP upon admission (Figure 3C).
It is evident that the “clinical” as well as combined “clinical + T1 sterols” models greatly outperform the COVID-GRAM and clinical baseline estimates. We can also observe that while the clinical baseline model can predict disease severity fairly accurately, the inclusion of only a handful of additional variables would greatly benefit model accuracy. Sterol measurements alone show moderate performance similar to COVID-GRAM risk estimates. In general, the validity of COVID-GRAM for this cohort is to be questioned, as although it uses a similar set of variables to the “clinical” and “clinical baseline” model it seems to be unable to delineate patient trajectories.
Discussion
Viral infections lead to changes in the lipid metabolism of the host
It is well known that host undergoes a number of physiological changes during viral infection. Studies have previously demonstrated significant alterations in lipid metabolism driven by bacterial and viral infection,2,3,28,29,30,31,32,33,34 including SARS-CoV-2.3,5,35,36,37,38,39,40,41,42,43,44,45,46,47,48 The degree of hypolipidemia was shown to inversely correlate with disease severity and fatality rate in COVID-19 patients.8,36,44,49,50 Additionally, a few studies reported progressive changes in levels of different phospholipids43 and sphingolipids,47 suggesting lipidome signature as a putative biomarker of disease severity and outcome.
A handful of papers have reported viral infection affecting levels of sterol intermediates. Mercorelli et al.51 have recently shown that human cytomegalovirus (HCMV) increased CYP51 gene expression in the U-373 MG cell line, whose protein product is in charge of the enzymatic conversion of lanosterol to FF-MAS. Furthermore, CYP51 expression is also induced by HIV-1 Nef protein in order to upregulate cholesterol biosynthesis and its transport to lipid rafts, resulting in increased virion infectivity.52 In contrast, significantly decreased levels of serum lathosterol were seen in HCV genotype 3 patients53 while metabolite profiling of JFH1 cell culture infected with HCV demonstrated an approximately 10-fold accumulation of desmosterol.54,55 However, to our knowledge there is no report that provides a detailed characterization of endogenous cholesterol biosynthesis during viral infection. The results of our study show that in patients suffering from severe COVID-19, the Bloch pathway (three of five sterol intermediates – zymosterol, 24-dehydrolathosterol, desmosterol) and, to a lesser extent, the K-R pathway (one of four sterol intermediates – zymostenol) of endogenous cholesterol biosynthesis are significantly impaired (Figures 1 and 2B). In patients with mild/moderate disease course only 24-dehydrolathosterol from the Bloch and zymostenol from the K-R pathway were significantly altered (Figures 1 and 2A). However, serum concentrations of cholesterol were significantly elevated in both patient groups (Figures 2A and 2B). At hospital discharge, an increase in the concentrations of T-MAS, zymosterol, zymostenol, 24-dehydrolathosterol, lathosterol, and desmosterol was noted, indicating that their levels started to recover. This is consistent with previous reports on other biochemical parameters.5,37,56,57 However, we cannot say with certainty that they reached their basal level. Accordingly, some studies report that alterations of some metabolic parameters persist in recovered patients for a longer period of time, suggesting a long-term systemic effect on the hosts’ metabolism following SARS-CoV-2 infection.48,58,59,60 Different durations and stages of illness at admission to hospital, and the fact that T2 samples were collected either in the case of severe deterioration or in the middle of hospitalization might explain higher standard deviations seen in several sterol intermediates (i.e., 24,25-dihydrolanosterol, T-MAS, zymostenol, and lathosterol; Figures 2A and 2B). However, the broad distribution of sterol concentrations in T2 could also indicate a different course of the disease between individual patients.
Plasma levels of sterol intermediates reflect liver function. Liver injury seen in COVID-19 patients, especially in those with severe or critical course of the disease, may be a result of a variety of mechanisms including direct viral damage, systemic inflammation, immune injury, hypoxia and ischemia, drug-induced liver injury as well as worsening of underlying liver diseases.61,62,63
Data-driven variable selection identifies a set of clinical variables highly predictive of disease severity
Patients suffering from severe COVID-19 may experience rapid deterioration and admission to the intensive care unit, which represents, on the one hand, a threat to the patient’s life and, on the other, a considerable burden on the medical system, as experienced during the first epidemic waves.64 Timely recognition and correct prognosis of the disease are essential for the optimal use of available resources. Therefore, we investigated whether we could better predict the development of severe COVID-19 in our patient cohort using machine learning models based on clinical parameters measured at hospital admission than using the currently available clinical risk score COVID-GRAM. In the second part, we aimed to delineate a possible biomarker potential of sterols in predicting disease severity.
Our analyses revealed that a set of 8 clinical measurements is sufficient to predict disease severity with high accuracy (Tables 3 and S5). Among the most important clinical variables were those found to be the reason for admission to hospital, such as the need for oxygen treatment or difficulties breathing, and “oxygen saturation measured with or without oxygen supplementation”, which detailed whether patients at the time of saturation measurement evidently need supplemental oxygen (and were therefore receiving oxygen therapy) or not (they were breathing normal air). These findings are not surprising, as dyspnea has been reported as an important factor determining COVID-19 course.65 Hentsch et al.66 showed that perceived breathlessness usually occurs in an advanced stage of the disease, which suggests that the reason for admission might be an indicator on how far the disease has progressed prior to hospital admission. Also, several biochemical parameters, namely concentrations of LDH, ferritin, and CRP, were deemed important for severity predictions. Increased concentration of CRP, a type I acute phase response protein synthetized in the liver and regulated by the pro-inflammatory cytokines IL-6, IL-1, and TNF-α, correlates with disease severity and predicts a need for ICU treatment, as well as mechanical ventilation. Patients with a critical course of COVID-19 also show elevated levels of ferritin, D-dimer, and lactate dehydrogenase (LDH), which are associated with poor prognosis and outcome.65,67,68 Our analyses further found chest X-ray information to be relevant, which is supported by recent publications showing the usefulness of X-rays in risk stratification for clinical worsening and prediction of fatality rate in COVID-19 patients.69,70
In addition, we found that a subset of four sterols was associated with disease severity, namely 24,25-dihydrolanosterol, zymostenol, 24-dehydrolathosterol, and desmosterol (Table 3). The latter three were also significantly altered during the course of the disease (Figures 2A and 2B). We believe that this is not a coincidence, since oxygen is needed for several enzymatic reactions in cholesterol biosynthesis and COVID-19 patients suffer from hypoxemia.26 Three molecules of oxygen are needed for conversion of lanosterol to FF-MAS (or 24,25-dihydrolanosterol to dihydro-FF-MAS) by CYP51A1 and for conversion of T-MAS to zymosterol (or dihydro-T-MAS to zymostenol) by SC4MOL, while one molecule of oxygen is required for transformation of 24-dehydrolathosterol to 7-dehydrodesmosterol (not measured within this study) and further to desmosterol by SC5DL and DHCR7, respectively (Figure 1). The most significant change during COVID-19 was seen in desmosterol concentrations in severe COVID-19 patients (Figure 2B) which is not surprising as it is the last precursor before cholesterol and the effect of oxygen deprivation accumulates through the biosynthesis chain. However, the significant changes in cholesterol biosynthesis could also be a result of promoting viral infectivity. Namely, host-derived lipids are required for the viral life cycle (i.e., entry, replication, and assembly) and could therefore also be a potential target for antiviral drug development.69,71 Higher level of plasma membrane cholesterol increases viral infection rate by promoting membrane fusion,71,72 while its depletion disrupts virion membrane composition.73 Additionally, Costello et al.55 showed the importance of desmosterol for HCV replication by increasing membrane fluidity, while the inhibition of its biosynthesis resulted in an antiviral effect.
Computational models for estimating COVID-19 severity
Our machine learning models using a unique set consisting of only clinical information measured at hospital admission to predict COVID-19 disease course showed excellent performances (AUC: 0.96; Table 4; Figure 3A) comparable to other machine learning models published (Figure 3C).74,75 Other biostatistical tools attempting to predict COVID-19 course have found sets of relevant variables and models comparable to our study. The neural network developed by Statsenko et al.76 has demonstrated the ability to estimate the risk of patients being administered to the ICU, with ferritin, LDH, and CRP being powerful predictors. In an impressive and early work, Yan et al.77 showed the stratification of patients at high and low risk of mortality by a tree-based model utilizing only CRP, LDH, and lymphocyte measurements. Xiong et al.74 successfully trained several machine learning models to predict COVID-19 severity identifying a set of laboratory and imaging features as relevant. To put our model performances directly in context with established clinical practice, we compared them to the COVID-GRAM risk score,27 as well as a clinical baseline model we developed using three variables commonly used to stratify patients into disease severity groups (i.e., CRP, LDH, ferritin) (Figure 3C). It is evident that all of our models using clinical information significantly outperformed current clinical patient stratification strategies (AUC: 0.96 vs. AUCbaseline: 0.74 and AUCCOVID-GRAM: 0.68). Although the baseline model performed well despite only using three measurements, it is clear that by adding a few variables that are easy to assess and routinely measured, for example the reason for hospital admission or oxygen saturation levels, performances could be greatly improved. Unfortunately, the usefulness of COVID-GRAM as a risk score predicting critical illness among patients hospitalized with COVID-19 could not be confirmed in our cohort. This finding supports the concerns initially raised by Moreno-Pérez et al.78 who found similar limitations in their Spanish cohort (AUC: 0.72). The inability to validate COVID-GRAM in European cohorts reflects the limitations encountered when applying risk prediction tools in new populations. In contrast to this, Sebastian et al.79 have reported good correlation of COVID-GRAM derived risk scores with mortalities in a Polish cohort. This may indicate the usefulness of COVID-GRAM tool assessing the risk of fatality rate rather than disease severity.
A subset of 4 sterol measurements alone shows similar performance to COVID-GRAM risk estimates (AUCT1sterols: 0.66; AUCCOVID-GRAM: 0.68; Table 4; Figures 3A and 3C). However, when clinical variables were combined with sterol measurements, the performance of the model remained excellent without improving (AUCclinical: 0.96; AUCclinical+T1sterols: 0.95; Table 4; Figures 3A and 3C). We do not find it surprising since the clinical model already shows exceptional accuracies, reflecting a lack of performance improvement upon combining sterol measurements and clinical information. Although we do not find sterols to be indicative of disease severity, they should not be discarded as potentially useful biomarkers, as there are a number of other clinical outcomes highly relevant for optimal COVID-19 patient care. One of them gaining relevance with the rising number of long-COVID cases are the long-term adversarial effects of a SARS-CoV-2 infection, including on the liver. Independent of pre-existing chronic liver diseases, abnormalities in liver enzymes are common in COVID-19 patients, reflecting a dysregulation of hepatic function. In most patients without previous liver conditions hepatic injury is mild and transient,80 while a profound deterioration of hepatic injury was reported in patients with severe courses of COVID-19.81 Interestingly, studies also show elevation of the liver fibrosis index FIB-4 and serum hyaluronic acid in acute COVID-19 patients upon admission to hospital care, suggesting liver fibrogenesis.82 Moreover, patients with chronic liver diseases and COVID-19 present with higher risk of long-term morbidity and fatality rate, where approximately 30% of those patients present with symptoms consistent with long COVID-19.83 Also, hospitalized patients had significantly lower HDL-cholesterol values at a follow-up.84 A recent computational study shows that acute liver injury is a common complication in COVID-19 (∼39.9%) with patients unable to fully recover until hospital discharge. The average time to recover may take up to two months, but can be reliably estimated using statistical models and measurements taken upon patients leaving the hospital.85 All of this evidence suggests that the liver function, especially of metabolically compromised patients and those most susceptible for complications, should be monitored even after being discharged from hospital care.
Conclusion
In conclusion, infection-associated dyslipidemia in COVID-19 patients has been widely reported. However, most of the clinical studies concentrate only on lipoproteins6,7,56 while little is known about the underlying mechanism of cholesterol metabolism. Herein we focused in depth on de novo intracellular biosynthesis of cholesterol in hospitalized COVID-19 patients and showed statistically significant differences in sterol concentrations over the course of COVID-19. A greater number of sterols were significantly altered in patients with a severe disease course (i.e., zymosterol, zymostenol 24-dehydrolathosterol, desmosterol, cholesterol), than in patients with a mild disease course (i.e., zymostenol, 24-dehydrolathosterol, cholesterol).
Furthermore, SARS-CoV-2 is known to be rapidly evolving, with symptoms and disease courses changing according to the most prevalent variant. The inability to validate risk scores in populations outside their original development, and the fact that much is still unknown about the long-term adverse effects on the health of recovered COVID-19 patients, warrants a continuing search for novel biomarkers characterizing this disease. Our machine learning models provided a unique set of 8 clinical variables sufficient to predict disease severity with excellent accuracy (AUC = 0.96). This proved to be a substantial improvement over currently used clinical risk scores. Although the concentrations of sterol intermediates have not improved our already exceptional clinical model, we have shown that their concentrations change during disease course and that the changes differ between mild and severe COVID-19 cases. Accordingly, we strongly believe that their biomarker potential should be further explored, as they may have prognostic value for clinical outcomes other than disease severity prediction, such as the adverse effects of a SARS-CoV-2 infection (including on the liver).
To our knowledge, this is the first study relating to COVID-19 to examine the blood sterol intermediates that arise from the endogenous biosynthesis of cholesterol in detail, which contributes to our understanding of the SARS-CoV-2 pathogenesis and disease course. The second contribution of our study is a unique set of readily available clinical variables capable of predicting COVID-19 course with excellent accuracy. Finally, we believe that our study will also serve as inspiration for future studies aimed at investigating potential biomarkers outside the routine clinical setting.
Limitations of the study
Because our study was based on hospitalized patients, the majority of the patients had severe disease. Thus, the distribution of patients in the present study reflected the actual situation in hospitalized patients but not in outpatients in whom mild(er) illness prevailed. In addition, since patients were admitted to hospital with different pre-hospital durations of illness and at different disease stages, baseline as well as consecutive blood samples were obtained over a considerable time span. Furthermore, the T2 samples were collected either at the occurrence of severe deterioration or in the middle of the hospitalization. Another limitation of our study is that some clinical variables could not be included because of the high proportion of missing data in patients. Furthermore, COVID-19 deterioration may not only be a result, of course, of SARS-CoV-2 infection, but is usually a consequence of a rather complex condition including underlying disease worsening, secondary infections, or noninfectious complications, that were not considered. Finally, it is essential to test our models in an external cohort and, although we found evidence of the potential value of sterols as biomarkers for COVID-19 and discussed their potential importance in the manuscript, we were not able to test these hypotheses in the context of this study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human serum from patients with COVID-19 | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| lanosterol | Avanti Polar Lipids | Cat#700063 |
| 24,25-dihydrolanosterol | Avanti Polar Lipids | Cat#700067 |
| T-MAS | Avanti Polar Lipids | Cat#700073 |
| dihydro-T-MAS | Avanti Polar Lipids | Cat#700173 |
| zymosterol | Avanti Polar Lipids | Cat#700068 |
| zymostenol | Avanti Polar Lipids | Cat#700118 |
| 24-dehydrolathosterol | Avanti Polar Lipids | Cat#700114 |
| lathosterol | Avanti Polar Lipids | Cat#700069 |
| lathosterol-D7 | Avanti Polar Lipids | Cat#700056 |
| desmosterol | Avanti Polar Lipids | Cat#700060 |
| cholesterol | Merck | Cat# C3045 |
| Software and algorithms | ||
| scikit-learn | https://scikit-learn.org/stable/ | |
| Code for model development and validation; available upon request | This paper | https://github.com/sonjakatz/covid_sterols_ML |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Damjana Rozman (damjana.rozman@mf.uni-lj.si).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Human participants
165 adult patients (53 females and 112 males, aged 23 to 93 years) admitted to the Department of Infectious Diseases of the University Medical Center Ljubljana (Slovenia) from July 2020 to July 2021 with COVID-19, were enrolled in the prospective study. In all patients, infection with SARS-CoV-2 was demonstrated by the presence of the virus in nasopharyngeal swabs using real-time PCR.
All participants provided written informed consent, and the study was approved by the Medical Ethics Committee of the Republic of Slovenia (No. 0120–211/2020/7 and No. 0120-33/2022/3).
Method details
Materials
All sterol standards (See Table S1 and key resources table) were bought from Avanti Polar Lipids (Alabaster, AL, USA), except cholesterol, which was purchased from Merck (Darmstadt, Germany). LC-MS grade cyclohexane, methanol, and 1-propanol were purchased from Honeywell (Charlotte, NC, USA). Formic acid was from Fluka (Honeywell) and sodium hydroxide from Merck (Darmstadt, Germany).
Methods
Sample collection
Blood samples were collected at 3 different time points during the hospitalization of 62 COVID-19 patients: upon admission to hospital care due to a severe course of COVID-19, in case of severe deterioration or in the middle of treatment, and upon discharge from the hospital. In 103 COVID-19 patients, blood samples were only collected upon hospitalization. Samples were collected in vacutainer tubes and centrifuged at 1811 × g (Eppendorf 5810R) at room temperature for 10 min to obtain serum samples which were stored at −80°C until further use.
Assessment of clinical parameters
Information was obtained using a questionnaire. More than 200 clinical parameters were recorded, including patient demographics, comorbidities and associated diseases, regular therapies, COVID-19 vaccination status, symptoms and signs of the disease and their intensity, laboratory and X-ray findings, information regarding specific treatments, as well as a patient condition upon discharge. According to disease severity, patients were classified into 4 groups – mild, moderate, severe, and critical. Grouping was done according to NIH recommendations (Table S7).86 Demographic data and selected clinical and biochemical parameters are listed in Tables 1 and S2.
Sterol isolation
200 ng of lathosterol-D7 (Avanti Polar Lipids, Cat. No. 700056P) and 1 mL of hydrolysis solution (4g NaOH dissolved in 10 mL Milli-Q water and 90 mL 99.5% ethanol) were added to 250 μL of serum sample and mixed well. After 1 h of incubation in a water bath with shaking at 65°C, 500 μL of Milli-Q water and 3 mL of cyclohexane were added to the solution, vortexed, and centrifuged at 1301 × g (5810 R centrifuge, Eppendorf, Germany) for 10 min at room temperature. The upper organic phase was transferred to a new 15 mL glass vial and the extraction step with cyclohexane was repeated. Extracts were then combined and the organic solvent was evaporated at 45°C using Eppendorf concentrator 5301. Lipid films were dissolved in 100 μL of LC-MS grade methanol, transferred to HPLC vials, purged with N2, and stored at −20°C until LC-MS/MS analysis.
LC-MS/MS analysis
The analysis was carried out according to a modified method from our previous study.87 Briefly, chromatographic separation using two pentafluorophenyl columns Phenomenex Luna 3 μm (Phenomenex, USA) was performed on a Shimadzu Nexera XR HPLC (Shimadzu, Japan), with an oven temperature of 40°C, mobile phase composition of methanol/1-propanol/formic acid/water (v/v/v/v, 80:10:0.05:9.95), and isocratic flow 200 μL/min, except for cholesterol 300 μL/min. The injected volume of standard or sample was 5 μL, except for cholesterol the injection volume was 1 μL.
Detection was performed on an SCIEX Triple Quad 3500 mass spectrometer (AB Sciex LLC, USA) with APCI ionization in a positive mode. Detailed information about sterols and mass spectrometry detection conditions are listed in Table S1. A standard solution consisting of the same concentration of each sterol was used for their quantification in serum samples. Analyst software 1.6.3 (AB Sciex LLC, USA) was used for data evaluation.
Data preprocessing
The selected target variable for predictions was the degree of disease severity, which was categorized into 4 classes, according to increasing severity and in consensus with NIH guidelines (Table S7).86 Class 1 represented the mildest and class 4 the most severe course of the disease. Due to the small number of members in classes 1, 2, and 4, patients were combined into two groups, namely those with mild illness (classes 1 and 2) and those with severe illness (classes 3 and 4) cases. Patients with missing disease severity annotations were discarded (N = 1), resulting in a sample of 30 and 134 patients for the mild and severe groups, respectively.
159 variables (also referred to as features throughout the manuscript) with the potential to be used for prediction were included in the information available upon hospital admission, namely patient demographics, vaccination status, comorbidities and associated diseases, regular therapies upon admission, symptoms and/or signs of the disease and their intensity, and other clinical and laboratory findings. Irrelevant or potentially biasing variables (e.g., dates, patient IDs, some clinical endpoints) were omitted during preprocessing. 77 variables were used as an input for variable selection.
Imputation was performed for variables if their degree of missingness did not exceed 15%, as imputation accuracy cannot be guaranteed in the case of higher missingness. Variables exceeding this threshold were removed from the analysis. During imputation, we accounted for the mixed data types present in the collected data by differentiating between continuous, binary, and categorical variables. Continuous features were treated using the IterativeImputer from scikit-learn88 and scaled through min-max normalization. Binary information was completed using the K-Nearest Neighbors method. Missing categorical features were imputed by the most frequently occurring value and encoded to numerical representation by an ordinal encoder.
Variable selection
To combat overfitting of the machine learning models, variables relevant for disease severity prediction were selected prior to model training using a variant of the unsupervised feature selection method proposed by Kursa et al.,89 called Boruta. Our variant of Boruta, referred as iterative Boruta, included the recording of relevant variables over 100 iterations of the feature selection process. Only variables occurring in at least 50% of iterations were deemed relevant and kept.
This feature selection process was conducted separately for the clinical ("clinical") and sterol ("T1 sterols") datasets. Selected variables from both datasets were subsequently merged into a combined set ("clinical + T1 sterols").
Classification models
Eight predictive models were trained, namely Random Forest,90 Gaussian Processes,91 AdaBoost,92 Logistic Regression,93 K-Nearest Neighbors,94 Multilayer Perceptron,95 Gaussian Naive Bayes,96 and Quadratic Discriminant Analysis.97 To ensure optimal performance, model hyperparameters were optimized during training through an exhaustive search, with balanced accuracy as a scoring metric.
The predictive power of each classification model was assessed by leave-one-out cross-validation (Figure S3) and evaluated using several metrics including balanced accuracy, precision, recall, F1-score, and ROC-AUC score. Their definition is based on a confusion matrix consisting of four elements (TP, true positive; TN, true negative; FP, false positive; FN, false negative). For additional information about each metric please refer to Note S1. Within each cross-validation split, the importance of individual variables for prediction was measured by applying an iterative permutation-based feature importance assessment90 with 100 iterations. For analyses, the feature importance of all 100 iterations was averaged.
Comparison to COVID-GRAM and clinical baseline
COVID-GRAM, developed by Liang et al.,27 is a risk score for COVID-19 patients used to predict patients' risk of developing a critical illness. It utilizes 10 clinical variables available at the time of admission including age, the presence of hemoptysis, dyspnea, or abnormalities in the X-ray, whether patients arrive unconscious, the number of comorbidities, records of cancer history, the neutrophil to lymphocyte ratio, and the concentrations of lactate dehydrogenase (LDH) and bilirubin. As the COVID-GRAM score was not assessed during patient recruitment, we calculated it post-hoc for each patient. Individual risk scores were subsequently translated into probabilities and used for further analyses. For respective formulas, please refer to the Note S2.
To make a comparison to current clinical practices, we selected three variables commonly used to stratify patients into disease severity groups, namely concentrations of LDH, ferritin, and C-reactive protein (CRP) upon admission. Classification models were subsequently trained using only these three variables (as outlined in section classification models) - we refer to those as “clinical baseline” models.
Quantification and statistical analysis
Statistical significance was tested comparing three time points of serum sterol intermediate concentrations with a nonparametric Friedman’s test since the collected data did not follow a normal distribution. For multiple comparisons, the adjusted p-values were determined using Dunn’s test. The adjusted p-value ≤ 0.05 was considered statistically significant. Statistical analysis was carried out using GraphPad Prism 9 software (Dotmatics, California, USA).
Acknowledgments
We would like to thank Tanja Blagus and prof. dr. Vita Dolžan (Pharmacogenetics Laboratory, Institute of Biochemistry and Molecular Genetics, Faculty of Medicine, University of Ljubljana), Petra Nassib, Jaka Brzin, and Jadranka Stojnić for the help with sample collection. Finally, we thank Benjamin Bajželj for critical input and carefully reading the manuscript, and John Hancock for appraisal of the manuscript.
Funding: This work was funded by the Slovenian Research and Innovation Agency (ARIS) program grants P1-0390, P2-0359, P3-0296, and the Ph.D. grant for young researchers to E.K.; S.K. was supported by the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 860895 TranSYS. We would also like to acknowledge the support by the Network of Research and Infrastructure Centres of the University of Ljubljana (MRIC-UL-CFGBC, IP-0022), financed by the ARIS, and infrastructure program ELIXIR-SI RI-SE-2 financed by the European Regional Development Fund and by the Ministry of Education, Science and Sport of Republic of Slovenia. The funding sources had no role in the design of the study and collection, analysis, and interpretation of data nor in writing the manuscript.
Author contributions
Conceptualization: D.R., F.S., E.K., and C.S. Methodology: E.K., S.K., Ž.P., and M.Mo. Software: S.K., Ž.P., and M.Mo. Formal Analysis: E.K., S.K., Ž.P., and M.Mo. Resources: E.K., S.K., Ž.P., M.Mo., P.B., G.T., and F.S. Data curation: S.K., E.K., G.T., P.B., and F.S. Writing – Original Draft: E.K. and S.K. Writing – Review and Editing: D.R., M.Mo., C.S., T.R., F.S., V.M.d.S. Visualization: E.K. and S.K. Supervision: D.R., M.Mo., F.S., M.Mr., and V.M.d.S. Project Administration: D.R., M.Mo., E.K., and S.K. Funding Acquisition: D.R., M.Mr., F.S., and V.M.d.S.
Declaration of interests
Authors declare no competing interests.
Published: August 31, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107799.
Contributor Information
Miha Moškon, Email: miha.moskon@fri.uni-lj.si.
Damjana Rozman, Email: damjana.rozman@mf.uni-lj.si.
Supplemental information
Data and code availability
-
•
Data used for analysis in this study cannot be deposited in a public repository because of patient privacy concerns. If you would like to request access for academic purposes, contact the lead contact for more information.
-
•
For all preprocessing steps and classification algorithms the implementations available in the scikit-learn Python library (version 1.1.3)88 were used. Source code and fitted models are available on a private GitHub repository and will be released upon request by the lead contact.
-
•
Any additional information required to re-analyze the data reported in this work is available upon request from the lead contact.
References
- 1.Antia R., Halloran M.E. Transition to endemicity: Understanding COVID-19. Immunity. 2021;54:2172–2176. doi: 10.1016/j.immuni.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kočar E., Režen T., Rozman D. Cholesterol, lipoproteins, and COVID-19: Basic concepts and clinical applications. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2021;1866 doi: 10.1016/j.bbalip.2020.158849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu X., Chen D., Wu L., He G., Ye W. Low Serum Cholesterol Level Among Patients with COVID-19 Infection in Wenzhou, China. Lancet. 2020 doi: 10.2139/ssrn.3544826. [DOI] [Google Scholar]
- 4.Wei C., Wan L., Zhang Y., Fan C., Yan Q., Yang X., Gong J., Yang H., Li H., Zhang J., et al. Cholesterol Metabolism--Impact for SARS-CoV-2 Infection Prognosis, Entry, and Antiviral Therapies. medRxiv. 2020 doi: 10.1101/2020.04.16.20068528. Preprint at. [DOI] [Google Scholar]
- 5.Wei X., Zeng W., Su J., Wan H., Yu X., Cao X., Tan W., Wang H. Hypolipidemia is associated with the severity of COVID-19. J. Clin. Lipidol. 2020;14:297–304. doi: 10.1016/j.jacl.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Zhang Y., Lu R., Dai M., Shen M., Zhang J., Cui Y., Liu B., Lin F., Chen L., et al. Lipid metabolism changes in patients with severe COVID-19. Clin. Chim. Acta. 2021;517:66–73. doi: 10.1016/j.cca.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi D., Yan R., Lv L., Jiang H., Lu Y., Sheng J., Xie J., Wu W., Xia J., Xu K., et al. The serum metabolome of COVID-19 patients is distinctive and predictive. Metabolism. 2021;118:154739. doi: 10.1016/j.metabol.2021.154739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yue J., Xu H., Zhou Y., Liu W., Han X., Mao Q., Li S., Tam L.S., Ma J., Liu W. Dyslipidemia Is Related to Mortality in Critical Patients With Coronavirus Disease 2019: A Retrospective Study. Front. Endocrinol. 2021;12:611526. doi: 10.3389/fendo.2021.611526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabre B., Fernandez Machulsky N., Olano C., Jacobsen D., Gómez M.E., Perazzi B., Zago V., Zopatti D., Ferrero A., Schreier L., Berg G. Remnant cholesterol levels are associated with severity and death in COVID-19 patients. Sci. Rep. 2022;12:17584–17586. doi: 10.1038/s41598-022-21177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aladağ N., Şipal A., Atabey R.D., Akbulut T., Asoğlu R., Özdemir M. Containment measures established during the COVID-19 outbreak and its impact on lipid profile and neutrophil to lymphocyte ratio. Eur. Rev. Med. Pharmacol. Sci. 2020;24:12510–12515. doi: 10.26355/eurrev_202012_24047. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y.M., Zheng Y., Yu Y., Wang Y., Huang Q., Qian F., Sun L., Song Z.G., Chen Z., Feng J., et al. Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage. EMBO J. 2020;39:e105923. doi: 10.15252/embj.2020105896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu X., Chen D., Wu L., He G., Ye W. Declined Serum High Density Lipoprotein Cholesterol Is Associated with the Severity of COVID-19 Infection. Clin. Chim. Acta. 2020 doi: 10.1016/j.cca.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka S., De Tymowski C., Assadi M., Zappella N., Jean-Baptiste S., Robert T., Peoch K., Lortat-Jacob B., Fontaine L., Bouzid D., et al. Lipoprotein concentrations over time in the intensive care unit COVID-19 patients: Results from the ApoCOVID study. PLoS One. 2020;15:e239615. doi: 10.1371/journal.pone.0239573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D., Li R., Wang J., Jiang Q., Gao C., Yang J., Ge L., Hu Q. Correlation analysis between disease severity and clinical and biochemical characteristics of 143 cases of COVID-19 in Wuhan, China: A descriptive study. BMC Infect. Dis. 2020;20:519. doi: 10.1186/s12879-020-05242-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G., Zhang Q., Zhao X., Dong H., Wu C., Wu F., Yu B., Lv J., Zhang S., Wu G., et al. Low high-density lipoprotein level is correlated with the severity of COVID-19 patients: An observational study. Lipids Health Dis. 2020;19:204–207. doi: 10.1186/s12944-020-01382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alcántara-Alonso E., Molinar-Ramos F., González-López J.A., Alcántara-Alonso V., Muñoz-Pérez M.A., Lozano-Nuevo J.J., Benítez-Maldonado D.R., Mendoza-Portillo E. High triglyceride to HDL-cholesterol ratio as a biochemical marker of severe outcomes in COVID-19 patients. Clin. Nutr. ESPEN. 2021;44:437–444. doi: 10.1016/j.clnesp.2021.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aparisi Á., Iglesias-Echeverría C., Ybarra-Falcón C., Cusácovich I., Uribarri A., García-Gómez M., Ladrón R., Fuertes R., Candela J., Tobar J., et al. Low-density lipoprotein cholesterol levels are associated with poor clinical outcomes in COVID-19. Nutr. Metabol. Cardiovasc. Dis. 2021;31:2619–2627. doi: 10.1016/j.numecd.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 19.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 20.Rezen T., Rozman D., Pascussi J.M., Monostory K. Interplay between cholesterol and drug metabolism. Biochim. Biophys. Acta. 2011;1814:146–160. doi: 10.1016/j.bbapap.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Kovač U., Skubic C., Bohinc L., Rozman D., Režen T. Oxysterols and gastrointestinal cancers around the clock. Front. Endocrinol. 2019;10:1–19. doi: 10.3389/fendo.2019.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skubic C., Rozman D. Sterols from the Post-Lanosterol Part of Cholesterol Synthesis: Novel Signaling Players. Mamm. Sterols. 2020;1–22:1–22. doi: 10.1007/978-3-030-39684-8_1. [DOI] [Google Scholar]
- 23.Rodríguez-Acebes S., de la Cueva P., Fernández-Hernando C., Ferruelo A.J., Lasunción M.A., Rawson R.B., Martínez-Botas J., Gómez-Coronado D. Desmosterol can replace cholesterol in sustaining cell proliferation and regulating te SREBP pathway in a sterol-Δ24-reductase deficient cell line. Biochem. J. 2009;420:305–315. doi: 10.1042/BJ20081909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown A.J., Sharpe L.J. In: Lipoproteins and Membranes. Sixth Edition. Ridgway N.D., McLeod R., editors. Elsevier; 2016. Chapter 11 - Cholesterol Synthesis; pp. 327–358. [DOI] [Google Scholar]
- 25.Kandutsch A.A., Russell A.E. Preputial Gland Tumor Sterols. J. Biol. Chem. 1960;235:2256–2261. doi: 10.1016/s0021-9258(18)64608-3. [DOI] [PubMed] [Google Scholar]
- 26.Belič A., Pompon D., Monostory K., Kelly D., Kelly S., Rozman D. An algorithm for rapid computational construction of metabolic networks: A cholesterol biosynthesis example. Comput. Biol. Med. 2013;43:471–480. doi: 10.1016/j.compbiomed.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 27.Liang W., Liang H., Ou L., Chen B., Chen A., Li C., Li Y., Guan W., Sang L., Lu J., et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern. Med. 2020;180:1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka S., Couret D., Tran-Dinh A., Duranteau J., Montravers P., Schwendeman A., Meilhac O. High-density lipoproteins during sepsis: From bench to bedside. Crit. Care. 2020;24:134. doi: 10.1186/s13054-020-02860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cirstea M., Walley K.R., Russell J.A., Brunham L.R., Genga K.R., Boyd J.H. Decreased high-density lipoprotein cholesterol level is an early prognostic marker for organ dysfunction and death in patients with suspected sepsis. J. Crit. Care. 2017;38:289–294. doi: 10.1016/j.jcrc.2016.11.041. [DOI] [PubMed] [Google Scholar]
- 30.Chien J.-Y., Jerng J.-S., Yu C.-J., Yang P.-C. Low serum level of high-density lipoprotein cholesterol is a poor prognostic factor for severe sepsis. Crit. Care Med. 2005;33:1688–1693. doi: 10.1097/01.ccm.0000171183.79525.6b. [DOI] [PubMed] [Google Scholar]
- 31.Barlage S., Gnewuch C., Liebisch G., Wolf Z., Audebert F.X., Glück T., Fröhlich D., Krämer B.K., Rothe G., Schmitz G. Changes in HDL-associated apolipoproteins relate to mortality in human sepsis and correlate to monocyte and platelet activation. Intensive Care Med. 2009;35:1877–1885. doi: 10.1007/s00134-009-1609-y. [DOI] [PubMed] [Google Scholar]
- 32.van Leeuwen H.J., Heezius E.C.J.M., Dallinga G.M., van Strijp J.A.G., Verhoef J., van Kessel K.P.M. Lipoprotein metabolism in patients with severe sepsis. Crit. Care Med. 2003;31:1359–1366. doi: 10.1097/01.CCM.0000059724.08290.51. [DOI] [PubMed] [Google Scholar]
- 33.Drobnik W., Liebisch G., Audebert F.X., Fröhlich D., Glück T., Vogel P., Rothe G., Schmitz G. Plasma ceramide and lysophosphatidylcholine inversely correlate with mortality in sepsis patients. J. Lipid Res. 2003;44:754–761. doi: 10.1194/jlr.M200401-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Lima W.G., Souza N.A., Fernandes S.O.A., Cardoso V.N., Godói I.P. Serum lipid profile as a predictor of dengue severity: A systematic review and meta-analysis. Rev. Med. Virol. 2019;29:e2056. doi: 10.1002/rmv.2056. [DOI] [PubMed] [Google Scholar]
- 35.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masana L., Correig E., Ibarretxe D., Anoro E., Arroyo J.A., Jericó C., Guerrero C., Miret M.L., Näf S., Pardo A., et al. Low HDL and high triglycerides predict COVID-19 severity. Sci. Rep. 2021;11:7217–7219. doi: 10.1038/s41598-021-86747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barman H.A., Pala A.S., Dogan O., Atıcı A., Yumuk M.T., Alici G., Sit O., Gungor B., Dogan S.M. Prognostic significance of temporal changes of lipid profile in COVID-19 patients. Obes. Med. 2021;28 doi: 10.1016/j.obmed.2021.100373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caterino M., Gelzo M., Sol S., Fedele R., Annunziata A., Calabrese C., Fiorentino G., D’Abbraccio M., Dell’Isola C., Fusco F.M., et al. Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Sci. Rep. 2021;11:2941. doi: 10.1038/s41598-021-82426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dei Cas M., Ottolenghi S., Morano C., Rinaldo R., Roda G., Chiumello D., Centanni S., Samaja M., Paroni R. Link between serum lipid signature and prognostic factors in COVID-19 patients. Sci. Rep. 2021;11:21633. doi: 10.1038/s41598-021-00755-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J.T., Chen Z., Nie P., Ge H., Shen L., Yang F., Qu X.L., Ying X.Y., Zhou Y., Wang W., et al. Lipid Profile Features and Their Associations With Disease Severity and Mortality in Patients With COVID-19. Front. Cardiovasc. Med. 2020;7:584987. doi: 10.3389/fcvm.2020.584987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T., Cao Y., Zhang H., Wang Z., Man C.H., Yang Y., Chen L., Xu S., Yan X., Zheng Q., Wang Y.P. COVID-19 metabolism: Mechanisms and therapeutic targets. MedComm. 2022;3:e157. doi: 10.1002/mco2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masoodi M., Peschka M., Schmiedel S., Haddad M., Frye M., Maas C., Lohse A., Huber S., Kirchhof P., Nofer J.R., Renné T. Disturbed lipid and amino acid metabolisms in COVID-19 patients. J. Mol. Med. 2022;100:555–568. doi: 10.1007/s00109-022-02177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciccarelli M., Merciai F., Carrizzo A., Sommella E., Di Pietro P., Caponigro V., Salviati E., Musella S., Sarno V.d., Rusciano M., et al. Untargeted lipidomics reveals specific lipid profiles in COVID-19 patients with different severity from Campania region (Italy) J. Pharm. Biomed. Anal. 2022;217:114827. doi: 10.1016/j.jpba.2022.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aydın S.Ş., Aksakal E., Aydınyılmaz F., Gülcü O., Saraç İ., Kalkan K., Aydemir S., Doğan R., Aksu U., Tanboğa İ.H. Relationship Between Blood Lipid Levels and Mortality in Hospitalized COVID-19 Patients. Angiology. 2022;73:724–733. doi: 10.1177/00033197211072346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janneh A.H., Kassir M.F., Dwyer C.J., Chakraborty P., Pierce J.S., Flume P.A., Li H., Nadig S.N., Mehrotra S., Ogretmen B. Alterations of lipid metabolism provide serologic biomarkers for the detection of asymptomatic versus symptomatic COVID-19 patients. Sci. Rep. 2021;11:14232. doi: 10.1038/s41598-021-93857-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ballout R.A., Kong H., Sampson M., Otvos J.D., Cox A.L., Agbor-Enoh S., Remaley A.T. The NIH Lipo-COVID Study: A Pilot NMR Investigation of Lipoprotein Subfractions and Other Metabolites in Patients with Severe COVID-19. Biomedicines. 2021;9 doi: 10.3390/biomedicines9091090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torretta E., Garziano M., Poliseno M., Capitanio D., Biasin M., Santantonio T.A., Clerici M., Lo Caputo S., Trabattoni D., Gelfi C. Severity of covid-19 patients predicted by serum sphingolipids signature. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai Y., Huang W., Li Y., Lai C., Huang S., Wang G., He Y., Hu L., Chen C. Lipidomic alteration of plasma in cured COVID-19 patients using ultra high-performance liquid chromatography with high-resolution mass spectrometry. Biosci. Rep. 2021;41 doi: 10.1042/BSR20204305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., Pan Y., Yin Y., Chen W., Li X. Association of dyslipidemia with the severity and mortality of coronavirus disease 2019 (COVID-19): a meta-analysis. Virol. J. 2021;18:157. doi: 10.1186/s12985-021-01604-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y., Zhang J., Li H., Kong W., Zheng J., Li Y., Wei Q., Li Q., Yang L., Xu Y., et al. Prognostic Value of Leucocyte to High-Density Lipoprotein-Cholesterol Ratios in COVID-19 Patients and the Diabetes Subgroup. Front. Endocrinol. 2021;12:727419. doi: 10.3389/fendo.2021.727419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mercorelli B., Luganini A., Celegato M., Palù G., Gribaudo G., Lepesheva G.I., Loregian A. The Clinically Approved Antifungal Drug Posaconazole Inhibits Human Cytomegalovirus Replication. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00056-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Y.H., Plemenitas A., Fielding C.J., Peterlin B.M. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proc. Natl. Acad. Sci. USA. 2003;100:8460–8465. doi: 10.1073/pnas.1437453100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheridan D.A., Shawa I.T., Thomas E.L., Felmlee D.J., Bridge S.H., Neely D., Cobbold J.F., Holmes E., Bassendine M.F., Taylor-Robinson S.D. Infection with the hepatitis C virus causes viral genotype-specific differences in cholesterol metabolism and hepatic steatosis. Sci. Rep. 2022;12:5562. doi: 10.1038/s41598-022-09588-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodgers M.A., Villareal V.A., Schaefer E.A., Peng L.F., Kathleen E., Chung R.T., Yang P.L. Lipid Metabolite Profiling Identifies Desmosterol Metabolism as a New Antiviral Target for Hepatitis C Virus. J. Am. Chem. Soc. 2013;134:6896–6899. doi: 10.1021/ja207391q.Lipid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costello D.A., Villareal V.A., Yang P.L. Desmosterol increases lipid bilayer fluidity during hepatitis C virus infection. ACS Infect. Dis. 2016;2:852–862. doi: 10.1021/acsinfecdis.6b00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang S., Zhou C., Yuan Z., Xiao H., Wu X. The clinical value of high-density lipoprotein in the evaluation of new coronavirus pneumonia. Adv. Clin. Exp. Med. 2021;30:153–156. doi: 10.17219/ACEM/130606. [DOI] [PubMed] [Google Scholar]
- 57.Li G., Du L., Cao X., Wei X., Jiang Y., Lin Y., Nguyen V., Tan W., Wang H. Follow-up study on serum cholesterol profiles and potential sequelae in recovered COVID-19 patients. BMC Infect. Dis. 2021;21:299. doi: 10.1186/s12879-021-05984-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He X., Liu C., Peng J., Li Z., Li F., Wang J., Hu A., Peng M., Huang K., Fan D., et al. COVID-19 induces new-onset insulin resistance and lipid metabolic dysregulation via regulation of secreted metabolic factors. Signal Transduct. Target. Ther. 2021;6:427. doi: 10.1038/s41392-021-00822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bizkarguenaga M., Bruzzone C., Gil-Redondo R., SanJuan I., Martin-Ruiz I., Barriales D., Palacios A., Pasco S.T., González-Valle B., Laín A., et al. Uneven metabolic and lipidomic profiles in recovered COVID-19 patients as investigated by plasma NMR metabolomics. NMR Biomed. 2022;35:e4637. doi: 10.1002/nbm.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Q., Zhou L., Sun X., Yan Z., Hu C., Wu J., Xu L., Li X., Liu H., Yin P., et al. Altered Lipid Metabolism in Recovered SARS Patients Twelve Years after Infection. Sci. Rep. 2017;7:9110–9112. doi: 10.1038/s41598-017-09536-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian D., Ye Q. Hepatic complications of COVID-19 and its treatment. J. Med. Virol. 2020;92:1818–1824. doi: 10.1002/jmv.26036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhong P., Xu J., Yang D., Shen Y., Wang L., Feng Y., Du C., Song Y., Wu C., Hu X., Sun Y. COVID-19-associated gastrointestinal and liver injury: clinical features and potential mechanisms. Signal Transduct. Target. Ther. 2020;5:256. doi: 10.1038/s41392-020-00373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saviano A., Wrensch F., Ghany M.G., Baumert T.F. Liver Disease and Coronavirus Disease 2019: From Pathogenesis to Clinical Care. Hepatology. 2021;74:1088–1100. doi: 10.1002/hep.31684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Popescu M., Ştefan O.M., Ştefan M., Văleanu L., Tomescu D. ICU-Associated Costs during the Fourth Wave of the COVID-19 Pandemic in a Tertiary Hospital in a Low-Vaccinated Eastern European Country. Int. J. Environ. Res. Publ. Health. 2022;19 doi: 10.3390/ijerph19031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skevaki C., Fragkou P.C., Cheng C., Xie M., Renz H. Laboratory characteristics of patients infected with the novel SARS-CoV-2 virus. J. Infect. 2020;81:205–212. doi: 10.1016/j.jinf.2020.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hentsch L., Cocetta S., Allali G., Santana I., Eason R., Adam E., Janssens J.P. Breathlessness and COVID-19: A call for research. Respiration. 2021;100:1016–1026. doi: 10.1159/000517400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q., Akdis C.A., Gao Y.D. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy Eur. J. Allergy Clin. Immunol. 2020;75:1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 69.Borghesi A., Zigliani A., Golemi S., Carapella N., Maculotti P., Farina D., Maroldi R. Chest X-ray severity index as a predictor of in-hospital mortality in coronavirus disease 2019: A study of 302 patients from Italy. Int. J. Infect. Dis. 2020;96:291–293. doi: 10.1016/j.ijid.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rubin G.D., Ryerson C.J., Haramati L.B., Sverzellati N., Kanne J.P., Raoof S., Schluger N.W., Volpi A., Yim J.-J., Martin I.B.K., et al. The Role of Chest Imaging in Patient Management During the COVID-19 Pandemic. Chest. 2020;158:106–116. doi: 10.1016/j.chest.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meher G., Bhattacharjya S., Chakraborty H. Membrane Cholesterol Modulates Oligomeric Status and Peptide-Membrane Interaction of Severe Acute Respiratory Syndrome Coronavirus Fusion Peptide. J. Phys. Chem. B. 2019;123:10654–10662. doi: 10.1021/acs.jpcb.9b08455. [DOI] [PubMed] [Google Scholar]
- 72.Abu-Farha M., Thanaraj T.A., Qaddoumi M.G., Hashem A., Abubaker J., Al-Mulla F. The Role of Lipid Metabolism in COVID-19 Virus Infection and as a Drug Target. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21103544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salimi H., Johnson J., Flores M.G., Zhang M.S., O’Malley Y., Houtman J.C., Schlievert P.M., Haim H. The lipid membrane of HIV-1 stabilizes the viral envelope glycoproteins and modulates their sensitivity to antibody neutralization. J. Biol. Chem. 2020;295:348–362. doi: 10.1074/jbc.RA119.009481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiong Y., Ma Y., Ruan L., Li D., Lu C., Huang L., National Traditional Chinese Medicine Medical Team Comparing different machine learning techniques for predicting COVID-19 severity. Infect. Dis. Poverty. 2022;11:19. doi: 10.1186/s40249-022-00946-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou K., Sun Y., Li L., Zang Z., Wang J., Li J., Liang J., Zhang F., Zhang Q., Ge W., et al. Eleven routine clinical features predict COVID-19 severity uncovered by machine learning of longitudinal measurements. Comput. Struct. Biotechnol. J. 2021;19:3640–3649. doi: 10.1016/j.csbj.2021.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Statsenko Y., Al Zahmi F., Habuza T., Gorkom K.N.V., Zaki N. Prediction of COVID-19 severity using laboratory findings on admission: Informative values, thresholds, ML model performance. BMJ Open. 2021;11:e044500. doi: 10.1136/bmjopen-2020-044500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan L., Zhang H.T., Goncalves J., Xiao Y., Wang M., Guo Y., Sun C., Tang X., Jing L., Zhang M., et al. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020;2:283–288. doi: 10.1038/s42256-020-0180-7. [DOI] [Google Scholar]
- 78.Moreno-Pérez Ó., Andrés M., León-Ramirez J.M., Sánchez-Payá J., Boix V., Gil J., Merino E. The COVID-GRAM Tool for Patients Hospitalized With COVID-19 in Europe. JAMA Intern. Med. 2021;181:1000–1001. doi: 10.1001/jamainternmed.2021.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sebastian A., Madziarski M., Madej M., Proc K., Szymala-Pędzik M., Żórawska J., Gronek M., Morgiel E., Kujawa K., Skarupski M., et al. The Usefulness of the COVID-GRAM Score in Predicting the Outcomes of Study Population with COVID-19. Int. J. Environ. Res. Publ. Health. 2022;19 doi: 10.3390/ijerph191912537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nardo A.D., Schneeweiss-Gleixner M., Bakail M., Dixon E.D., Lax S.F., Trauner M. Pathophysiological mechanisms of liver injury in COVID-19. Liver Int. 2021;41:20–32. doi: 10.1111/liv.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patel K.P., Patel P.A., Vunnam R.R., Hewlett A.T., Jain R., Jing R., Vunnam S.R. Gastrointestinal, hepatobiliary, and pancreatic manifestations of COVID-19. J. Clin. Virol. 2020;128:104386. doi: 10.1016/j.jcv.2020.104386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kolesova O., Vanaga I., Laivacuma S., Derovs A., Kolesovs A., Radzina M., Platkajis A., Eglite J., Hagina E., Arutjunana S., et al. Intriguing findings of liver fibrosis following COVID-19. BMC Gastroenterol. 2021;21:370. doi: 10.1186/s12876-021-01939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aby E.S., Moafa G., Latt N., Sultan M.T., Cacioppo P.A., Kumar S., Chung R.T., Bloom P.P., Gustafson J., Daidone M., et al. Long-term clinical outcomes of patients with COVID-19 and chronic liver disease: US multicenter COLD study. Hepatol. Commun. 2023;7 doi: 10.1097/01.hc9.0000897224.68874.de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barbara J.M., Gatt J., Xuereb R.A., Tabone Adami N., Darmanin J., Erasmi R., G Xuereb R., Barbara C., Stephen F., Jane Magri C. Clinical outcomes at medium-term follow-up of COVID-19. J. R. Coll. Physicians Edinb. 2022;52:220–227. doi: 10.1177/14782715221124617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu J.Y., Ho S.L., Buczek A., Fleysher R., Hou W., Chacko K., Duong T.Q. Clinical predictors of recovery of COVID-19 associated-abnormal liver function test 2 months after hospital discharge. Sci. Rep. 2022;12:17972. doi: 10.1038/s41598-022-22741-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.COVID-19 Treatment Guidelines Panel Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. National Institutes of Health. https://www.covid19treatmentguidelines.nih.gov/ [PubMed]
- 87.Skubic C., Vovk I., Rozman D., Križman M. Simplified LC-MS method for analysis of sterols in biological samples. Molecules. 2020;25 doi: 10.3390/molecules25184116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 89.Kursa M.B., Rudnicki W.R. Feature selection with the boruta package. J. Stat. Softw. 2010;36:1–13. doi: 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- 90.Breiman L. Random forest. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 91.Rasmussen C.E. Gaussian Processes in Machine Learning. Lect. Notes Comput. Sci. 2004;3176:63–71. doi: 10.1007/978-3-540-28650-9_4. [DOI] [Google Scholar]
- 92.Thongkam J., Xu G., Zhang Y. IEEE Int. Jt. Conf. Neural Networks (IEEE World Congr. Comput. Intell. 2008. AdaBoost algorithm with random forests for predicting breast cancer survivability; pp. 3062–3069. [DOI] [Google Scholar]
- 93.Dreiseitl S., Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J. Biomed. Inf. 2002;35:352–359. doi: 10.1016/S1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 94.Kramer O. 2013. K-Nearest Neighbors BT - Dimensionality Reduction with Unsupervised Nearest Neighbors; pp. 13–23. [DOI] [Google Scholar]
- 95.Gardner M.W., Dorling S.R. Artificial neural networks (the multilayer perceptron)—a review of applications in the atmospheric sciences. Atmos. Environ. X. 1998;32:2627–2636. doi: 10.1016/S1352-2310(97)00447-0. [DOI] [Google Scholar]
- 96.Rish I. IJCAI 2001 Work. Empir. methods Artif. Intell. 2001. An empirical study of the naive Bayes classifier; pp. 41–46. [Google Scholar]
- 97.Tharwat A. Linear vs. quadratic discriminant analysis classifier: a tutorial. Int. J. Appl. Pattern Recognit. 2016;3:145. doi: 10.1504/ijapr.2016.079050. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data used for analysis in this study cannot be deposited in a public repository because of patient privacy concerns. If you would like to request access for academic purposes, contact the lead contact for more information.
-
•
For all preprocessing steps and classification algorithms the implementations available in the scikit-learn Python library (version 1.1.3)88 were used. Source code and fitted models are available on a private GitHub repository and will be released upon request by the lead contact.
-
•
Any additional information required to re-analyze the data reported in this work is available upon request from the lead contact.