Summary
The cochlea consists of diverse cellular populations working in harmony to convert mechanical stimuli into electrical signals for the perception of sound. Otic mesenchyme cells (OMCs), often considered a homogeneous cell type, are essential for normal cochlear development and hearing. Despite being the most numerous cell type in the developing cochlea, OMCs are poorly understood. OMCs are known to differentiate into spatially and functionally distinct cell types, including fibrocytes of the lateral wall and spiral limbus, modiolar osteoblasts, and specialized tympanic border cells of the basilar membrane. Here, we show that OMCs are transcriptionally and functionally heterogeneous and can be divided into four distinct populations that spatially correspond to OMC-derived cochlear structures. We also show that this heterogeneity and complexity of OMCs commences during early phases of cochlear development. Finally, we describe the cell-cell communication network of the developing cochlea, inferring a major role for OMC in outgoing signaling.
Subject areas: Cell biology, Developmental biology, Transcriptomics
Graphical abstract

Highlights
-
•
OMCs are the most numerous cell type in the cochlea during development
-
•
OMCs are heterogeneous, with spatially and functionally distinct subpopulations
-
•
OMC heterogeneity is present early and continues to refine during development
-
•
OMCs express outgoing signals predicated to be necessary for cochlear development
Cell biology; Developmental biology; Transcriptomics
Introduction
Currently, over 5% of the worldwide population suffers from hearing loss with this number expected to increase to 10% by 2050 (World Health Organization, 2021). Unfortunately, to date, no therapeutics are available to treat the underlying molecular mechanisms of hearing loss, leaving amplification (e.g., hearing aids) and cochlear implantation as the only forms of treatment. The development of novel therapeutics that address the underlying causes of sensorineural hearing loss (SNHL) requires a detailed understanding of the diverse cochlear cell types and their role in the development and maintenance of auditory function. The transcriptional heterogeneity of the developing cochlear sensory epithelium, auditory neurons, stria vascularis, and developing hair cells has now been described.1,2,3,4,5,6,7,8,9,10 However, the developmental profiles of otic mesenchyme cells (OMCs), the most abundant cell type in the developing inner ear, remain only partially resolved.
OMCs are a specialized type of neural crest and cranial paraxial mesoderm.11 OMCs express the transcription factors Tbx1, Tbx18, and Pou3f4, all of which are also known deafness genes, highlighting their importance in auditory function.12,13,14 OMCs have been shown to be necessary for the development of essential cochlear processes such as spiral ganglion neuron (SGN) axonal guidance and survival, endocochlear potential generation, modiolar ossification, extracellular matrix organization, and overall cochlear structure.14,15,16,17,18,19 In addition, OMCs terminally differentiate into a variety of crucial cochlear cell types, including specialized tympanic border cells, fibrocytes of the lateral wall and spiral limbus, basal cells of the stria vascularis, and modiolar osteoblasts. Nonetheless, although OMCs have diverse roles in cochlear development well before terminal differentiation, their molecular distinctness and functionality remain largely unresolved.
In this study, we utilize single-cell RNA-sequencing (scRNA-seq) at embryonic day (E) 15 and postnatal day (P) 7, followed by in situ marker gene validation to reveal four subpopulations of OMCs that are transcriptionally, functionally, and spatially distinct with divergent gene regulatory networks. While the four OMC subpopulations are spatially distinct as early as E15, their spatial distribution is further refined by P7. Ultimately, each OMC subpopulation corresponds to one of the four major cochlear structures that arise from OMCs: the basilar membrane, spiral limbus, modiolus, and lateral wall. Finally, as OMCs are known to influence surrounding cell types, we constructed the cell-cell communication network at both E15 and P7 to elucidate possible signaling pathways impacting developmental processes within the cochlea. In conclusion, these data suggest that OMCs diversification occurs early during development, resulting in heterogeneous subpopulations that are predicted to be responsible for the majority of outgoing cochlear signaling.
Results
OMCs are transcriptionally and spatially heterogeneous at P7
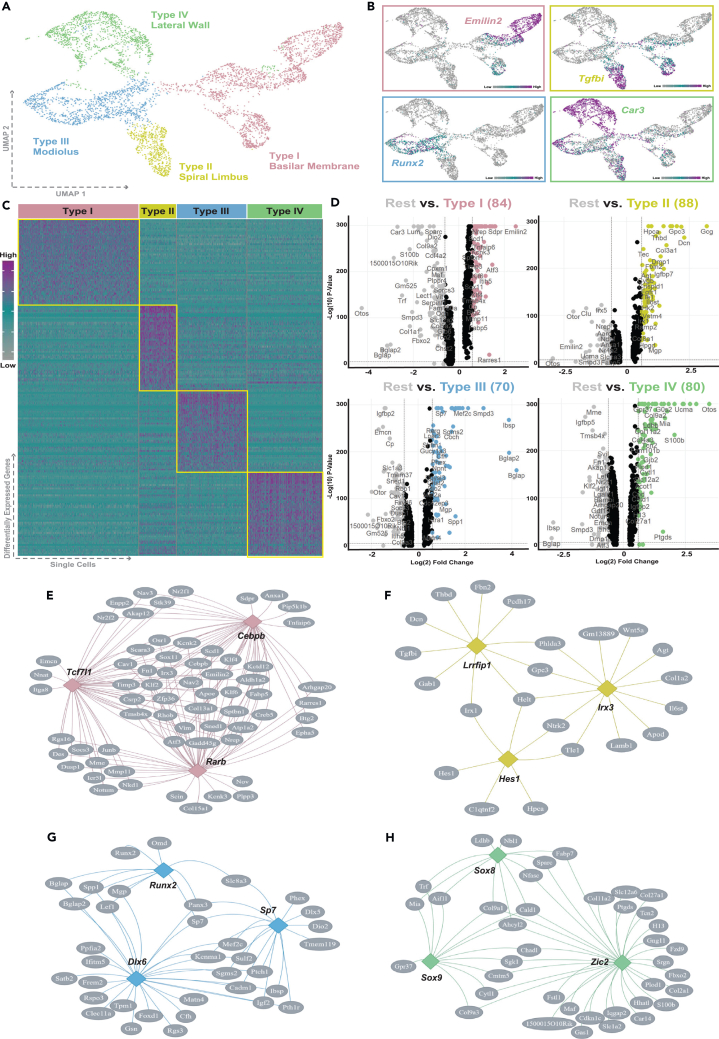
OMC-derived structures include the basilar membrane, found between the organ of Corti and tympanic border cells, the spiral limbus anchoring Reisner’s membrane and the tectorial membrane, the modiolar bone that surrounds the SGNs, and the lateral wall adjacent to the stria vascularis (Figures 1A–1D). To fully determine the transcriptional heterogeneity of OMCs in early postnatal cochlear development, we obtained cochlear ducts, including all OMC-derived structures, from P7 male mice and measured gene expression using scRNA-seq. After filtering out low complexity droplets, the dataset contained 7,703 cochlear cells (Figure S1A). Graph-based unbiased clustering using Seurat identified 15 groups of cells with unique patterns of expression (Figures 1E and 1F). Cellular identities were established by examination of the top 25 genes defining each cluster as well as established marker genes from the literature (Figures 1F and S1B; Table S1).
Figure 1.
OMCs are transcriptionally and spatially heterogeneous at P7
(A–D) Immunohistochemistry of P0 mouse cochleae cross sections stained with TUJ1 (neurons and their projections), CD326 (epithelium), MYO6 (hair cells), or KCNQ1 (marginal cells of SV) in yellow, OMC marker POU3F4 in magenta and DAPI in cyan. Scale bar: 50μm.
(E) Identification of P7 cochlear cell types, including 4 OMC subpopulations labeled type I–IV, visualized in UMAP space. Neurons are contaminated with glial cells and marked with (C). One unknown cluster (colored by auburn) not labeled.
(F) Heatmap of the top 25 genes of each cell cluster labeled by respective color. Cell types expressing universal OM markers labeled with cell identification above heatmap. Gene expression represented by gray-cyan-magenta color scale. Full differential expression shown in Table S1.
(G–J) Table of the top 9 differentially expressed genes for each OMC subpopulation, including the log2 fold change between OMC subpopulation and rest of cell types, percent expression in the OMC subpopulation and all other cell populations, and adjusted p value. Marker gene chosen for spatial localization highlighted in OMC subpopulation color and visualized on UMAP. Arrows indicate OMC subpopulation corresponding to selected marker gene.
(K–N) Immunohistochemistry of P7 mouse cochlea cross sections stained with identified OMC-specific markers EMILIN2 (type I), TGFBI (type II), RUNX2 (type III), and CAR3 (type IV). Arrows indicate areas of expression corresponding to OMC subtype-specific expression. Dotted arrow represents overlap of expression in basilar membrane. Scale bar: 50μm.
To identify OMCs, we used known markers including: Pou3f4, Tbx18, Tbx1, and Pou3f3 (Figure S1C).12,13,14,20 A total of 4,742 OMCs were identified and divided into four clusters, each with a unique transcriptional profile. To further identify OMC subpopulations, we applied a differential gene expression analysis using a non-parametric Wilcoxon rank-sum test. OMC clusters were numbered I–IV arbitrarily where type I OMCs were the most distinct in terms of cell cluster separation. Marker genes for each cluster were selected based on multiple criteria including: (1) log fold change between the OMC subpopulation and the rest of cochlear cell types, (2) adjusted p value, and (3) specificity of RNA expression. From the top 9 marker genes, Emilin2, Tgfbi, Runx2, and Car3 were selected based on known biological function and antibody availability to determine the spatial distribution of each OMC subpopulation via immunohistochemistry (Figures 1G–1J and S2C). EMILIN2 (elastin microfiber interfacer 2) is an extracellular matrix protein found in the basilar membrane and shown to be expressed by OMC-derived tympanic border cells.21,22 Indeed, expression of EMILIN2 marked type I OMCs, which localize to the basilar membrane (Figure 1K). TGFBI (transforming growth factor β induced) is a secreted protein known to play a role in cell migration and adhesion.23 TGFBI marked type II OMCs, which localize to the spiral limbus, with some expression in the basilar membrane (Figure 1L). Type III OMCs were marked by RUNX2 (runt-related transcription factor 2), an essential regulator of osteoblast differentiation and a known deafness gene.24,25,26 RUNX2 expression was confined to the modiolar bone OMCs which include OMCs that surround Rosenthal’s canal (location of SGN cell bodies, Figure 1M). Finally, CAR3 (carbonic anhydrase 3), a zinc-containing metalloenzyme important for pH and CO2 homeostasis, especially in vasculature, localized to the lateral wall/spiral ligament and marked type IV OMCs (Figure 1N).27 Overall, our data show that OMCs consist of four transcriptionally distinct groups of cells at P7, which each mark spatially distinct OMC-derived cochlear structures.
Analysis of embryonic OMCs demonstrates early diversification of OMC subpopulations
The developmental time course of OMC differentiation has not been well defined. To describe the temporal specification of OMC subpopulation throughout development we used the P7 OMC marker genes identified and extended our analysis to E15 until the onset of hearing (P14; Figures 2A–2L and S2). At E15 RUNX2 is not expressed, consistent with the non-osseous state of the modiolus at this stage in development (Figure 2C). EMILIN2, TGFBI, and CAR3 are expressed at E15 in different locations surrounding the developing cochlear epithelium, including below the organ of Corti (EMILIN2, type I OMCs, Figure 2A), medial to Kölliker’s organ or the greater epithelial ridge (TGFBI, type II OMCs, Figure 2B), and directly above the developing stria vascularis (CAR3, type IV OMCs; Figure 2D). OMC marker genes at P2 show similar expression patterns as P7, although TGFBI expression extends to the modiolar bone OMCs and RUNX2 expression does not extend to the OMCs that surround Rosenthal’s canal (Figures 2E–2H). After the onset of hearing (P14), both EMILIN2 and CAR3 continue to be expressed by type I and type IV OMCs, respectively (Figures 2I and 2L). In contrast, RUNX2 can no longer be detected in the modiolar bone, possibly correlating with the conclusion of ossification, and TGFBI is no longer detected in the spiral limbus but becomes sequestered to Rosenthal’s canal (Figures 2J and 2K). To determine if the division of OMC subpopulations is specific to the C57BL/6J mouse strain, we repeated our immunohistochemical analysis using the outbred mouse strain CD-1/ICR (Figure S3). This analysis showed similar results, although the loss of expression of TGFBI from the spiral limbus was noted as early as P7, consistent with a possible earlier maturation of CD-1/ICR mice in comparison to C57BL/6J (Figure S3F).28
Figure 2.
Analysis of embryonic OMCs demonstrates early diversification of OMC subpopulations
(A–L) Immunohistochemistry of E15, P2, and P14 cochlea cross sections stained with OMC subtype-specific markers EMILIN2 (type I), TGFBI (type II), RUNX2 (type III), or CAR3 (type IV) in yellow, POU3F4 in magenta and DAPI in cyan. Arrows indicate areas of expression corresponding to OMC subtype-specific expression. Dotted arrows indicate overlap of expression. Scale bar: 50μm.
(M) Identification of E15 cochlear cell types, visualized in UMAP space. OMC subpopulations are not distinguishable at this resolution. Unknown clusters not labeled. Full differential expression shown in Table S2.
(N) Subcluster analysis of E15 OMCs, visualized in UMAP space, which includes the 4 OMC subpopulations found at P7 and two additional subpopulations: progenitors and type IE.
(O) Visualization of identified marker gene expression for the 4 main OMC subpopulations. OMC subpopulation cluster boundaries marked with colored dotted lines. Gene expression represented by gray-cyan-magenta color scale. Full differential expression shown in Table S3.
(P) E13 and E15 in situ hybridization images from the Allen Brain Atlas for the OM marker genes including Ccn3 (type I), Dcn (type II), Dkk2 (type III), and Car3 (type IV). Scale bar: 99μm.
(Q) Schematic of developmental time course of OM spatial localization between E15 and P7.
RM: Reissner’s Membrane, SV: Stria Vascularis, OS: Outer Sulcus, OC: Organ of Corti, PS: Prosensory Domain, IS: Inner Sulcus, KO: Kölliker’s organs, SGN: Spiral Ganglion Neurons, BM: Basilar Membrane, SL: Spiral Limbus, M: Modiolus, LW: Lateral Wall.
As OMC marker genes identified at P7 displayed an overlap in expression patterns at E15, we generated an E15 OMC enriched scRNA-seq dataset containing 4,521 high-quality OMCs to better characterize the OMC subpopulations in early cochlear development (Figures 2M and S4A; Table S2). Unbiased clustering of E15 OMCs revealed 6 subpopulations, each with unique transcriptional profiles, representing the 4 OMC subpopulations found at P7 and two additional subpopulations. One additional subpopulation at E15 are the progenitor OMCs, which express Top2a, a DNA topoisomerase essential transcription, and numerous histone genes, further suggesting proliferation as histone gene expression mark entry into the S-phase of the cell cycle.29 The other subpopulation, type IE OMCs, resembles a subpopulation of P7 type I OMCs (Figures 2N and S7A; Table S3). To further compare the OMCs at E15 and P7, the two datasets were integrated showing close alignment of clusters (Figure S4B). A reciprocal marker analysis was used to compare the datasets. P7 marker genes are expressed in their respective subpopulations at E15, with some overlap, except for TGFBI, which is broadly expressed in E15 OMCs (Figure S4C). Differential gene expression analysis identified marker genes for each OMC subpopulation at E15, including Ccn3 (also known as Nov) for type I, Dcn for type II, Dkk2 for type III, Car3 for type IV, Top2a for progenitors, and Kcnk2 for type IE (Figures 2O and S4C). Ccn3, Dcn, Car3, and Kcnk2 are expressed in the respective OMC subpopulations at P7, while Dkk2 and Top2a show little to no expression at P7 (Figure S4D). in situ hybridization analyses performed on E13 and E15 C57/B6J mice from the Allen Brain Atlas were used for spatial localization validation.30 Indeed, marker genes identified for E15 OMCs revealed spatially distinguishable OMC subpopulations with type I found below the developing organ of Corti (Ccn3 expression), type II medial to Kölliker’s organ (Dcn expression), type III oriented in the developing modiolus (Dkk2 expression), and type IV found above the developing stria vascularis (Car3 expression; Figure 2P). Furthermore, these OMC-subpopulation specific markers show some expression at E13, although with low expression (Figure 2P). Dcn and Dkk2 are also expressed in Kölliker’s organ and roof cells, respectively, confirmed by an E16 cochlear epithelium dataset as visualized by the gene expression analysis resource (UMgEAR.org; Figures 2P and S4E–S4F).1,31 Finally, to determine if OMCs are spatially distinct at E13, we analyzed a previously published dataset focused on the E13 otic/periotic region.32 Our analysis revealed a clear OMC progenitor population expressing Top2a, however, the other OMC subpopulations could not be deciphered using the previously identified E15 marker genes (Figures S5A and S5B; Ccn3 was not expressed in the E13 dataset, therefore Emilin2 was used for Type I OMCs). Nevertheless, when we conducted a developmental trajectory analysis spanning from E13/E15 to P7, we observed an early divergence of type I OMCs from type II-IV OMCs (Figures S5C–S5F). This suggests an early bifurcation of the tympanic border cells (type I OMCs) from other cell types derived from OMCs. In summary, the spatial distribution of OMC subpopulations is distinct as early as E15 and consistent with OMC-derived cochlear structures in postnatal stages (Figure 2Q).
OMC subpopulations display distinct functional roles that correspond to their spatial localization
To better understand the functional roles of the OMC subpopulations, OMCs were extracted from the P7 dataset, unbiasedly clustered, and reidentified by their identified marker genes (Figures 3A and 3B). To identify genes more highly expressed in each OMC subpopulation, a differential analysis was performed using a non-parametric Wilcoxon rank-sum test (Table S4). A heatmap of the top 25 differentially expressed genes showcases the transcriptional distinctness of OMC subpopulations (Figure 3C). Performing the same differential analysis for E15 OMCs also showed unique gene expression profiles further indicating early OMC diversification (Figures S6A–S6B). Comparing the expression profiles of OMC subpopulations to the other three subpopulations (rest) identified upregulated genes in each subpopulation including 84 type I OMC genes, 88 type II OMC genes, 70 type III OMC genes, and 80 type IV OMC genes (log2 fold change cutoff = 0.6; Figure 3D). Interestingly, increasing the cluster resolution reveals type I, III, and IV OMC subpopulations can be further divided with unique marker genes indicating further diversification (Figure S7; Table S5).
Figure 3.
Identification of differentially expressed genes between P7 OMC subpopulations
(A) Type I-IV OMC subpopulations at P7, visualized in UMAP space. Spatial localization of OMC subpopulation also indicated.
(B) Marker gene expression for each OMC subpopulation visualized on P7 OMC subpopulation UMAP. Gene expression represented by gray-cyan-magenta color scale.
(C) Heatmap of the top 50 differentially expressed genes for each OMC subpopulation. Gene expression represented by gray-cyan-magenta color scale. Full differential expression shown in Table S4.
(D) Volcano plots representing log10 p value and log2 fold change of differentially expressed genes. Genes that display log2 fold change of 0.6 or higher are colored.
(E–H) Representative gene regulatory networks of the top 3 potential upstream regulators (transcription factors) for each OMC subpopulation, identified by RcisTarget.
To ascertain potential upstream regulators of each OMC subpopulation, we used RcisTarget to identify transcription factor binding motifs enriched in each list of genes differentially expressed in the OMC subpopulations.33 RcisTarget was used at P7 to identify genes directing distinct functional processes in each OMC subpopulation. This analysis identified 109, 49, 23, and 53 significant transcription factors for type I–IV OMC subpopulations, respectively (Table S6). Three candidate transcription factors were chosen for each mesenchyme subpopulation based on normalized enrichment score, uniqueness of motif to OMC subpopulation, and overall expression in the OMC subpopulation (Figures 3E–3H). Type III OMCs (modiolar/bone progenitors) were used to assess the accuracy of our upstream regulator analysis as transcription factors of bone development have been well characterized. Indeed, Runx2, Sp7, and Dlx6 have all been implicated in osteoblast differentiation (Figure 3G).34,35,36 We also identified type IV OMCs express the upstream regulators Sox8 and Sox9, both of which have been implicated in chondrogenesis.37,38 To identify transcription factors that may be critical for early diversification of OMCs, we completed the same analysis at E15 (Figures S6C–S6F; Table S6). Again, we looked at type III OMCs to validate our analyses and indeed we identified Zfp281 and Atf1, both of which have been shown to negatively regulate osteoclast differentiation (Figure S6E).39,40 Finally, we also identified Stat3 as a regulator of the type I OMCs, established to be necessary for hair cell differentiation.41
To assess the functional roles of each OMC subpopulation, gene ontology (GO) enrichment analysis was performed on the highly expressed genes identified each subpopulation (Figures 4A–4D; Table S7). Type I (basilar membrane) GO-terms signify roles in tissue, epithelium, and structural development suggesting a role of basilar membrane OMCs in the development of the sensory epithelium of the organ of Corti (Figure 4A). Interactions between OMCs and sensory epithelial have been shown to promote the development of cochlear hair cells.42,43 Interestingly, type I OMCs also express genes that play a role in nervous system development and neurogenesis. Consistently, OMCs have been shown to promote SGN survival and fasciculation.15,16 Indeed, SGN projections are established by P0, however, are not fully refined as synapse formation and SGN subtype refinement continue until adulthood.44 Type II (spiral limbus) GO-terms also indicate a role in structure, tissue, and sensory organ development (Figure 4B). The spiral limbus is an important structural component of the cochlea which helps form the interdental cells located on the top of the limbus, anchors the tectorial membrane that connects to the organ of Corti and is essential for sound-induced vibration of the basilar membrane.45 In addition, type II OMCs show functional roles of cell adhesion and cell migration, both thought to play key roles in auditory system development.46 Type III (modiolar bone) OMCs show GO-terms involved with bone ossification and development, which is ongoing at P7 (Figure 4C). Interestingly, patients with POU3F4 mutations exhibit hypoplasia of the modiolar bone which causes difficulties positioning a cochlear implant electrode, making it ineffective in some of these patients.47,48 Finally, type IV (lateral wall/spiral ligament) GO-terms include cartilage development, as well as functions related to angiogenesis, supporting the hypothesis that lateral wall fibrocytes play a role in the development of the blood-labyrinth barrier (Figure 4D).49,50
Figure 4.
OMC subpopulations display distinct functional roles that correspond to their spatial localization
(A–D) Biological gene ontology (GO) term analysis of each spatially distinct OM population visualized by -log10 p value. Expression of genes found in relevant GO-terms displayed by log2 fold change. Spatial localization of OMC subpopulations visualized by cochlear schematic.
(E) Dot plots of 5 protein categories: (1) transcription factors, (2) receptors, (3) signaling ligands, (4) extracellular matrix proteins (ECM-structural), and (5) potassium channels. Gene expression is represented by gray-cyan-magenta color scale. Percentage of cells within OM subpopulation expressing each gene is represented by dot size (25–100%).
To further elucidate the molecular signatures of each OMC subpopulation, we evaluated the differential expression of genes belonging to 5 different categories related to development and known OMC functions including: (1) transcription factors, (2) receptors, (3) signaling molecules, (4) extracellular matrix proteins (structural), and (5) potassium channels (Figure 4E). Strikingly, each OMC subpopulation displays enrichment of distinct sets of transcription factors, indicating diverse gene regulatory networks. OMC subpopulations also express unique signaling molecules and receptors, hinting at distinct signaling pathways. Finally, OMCs are known to play a role in the development and maintenance of the extracellular matrix and potassium recycling.51,52 Interestingly, each OMC subpopulation expresses different extracellular matrix genes essential for multicellular structures indicating spatially unique extracellular environments. Finally, type I and type IV OMCs express several unique potassium channels, supporting the hypothesis that the basilar membrane and lateral wall OMCs are recycling potassium ions and helping to maintain the endocochlear potential.53 Future subpopulation-specific analyses of OMCs will allow for better understanding of the diverse functional roles during cochlear development.
Cellular communication analysis reveal OMCs are the main contributors of outgoing signaling during cochlear development
We have shown that each P7 OMC subpopulation surrounds spatially distinct cochlear structures and expresses distinct signaling molecules and receptors. We, therefore, hypothesized that OMCs engage in robust signaling with surrounding cell types to facilitate region-specific cochlear development. To test this hypothesis, we applied CellChat, which uses a manually curated ligand-receptor interaction database, based on known protein structures and biological function, and our scRNA-seq gene expression data to reconstruct the cell-cell communications between cochlear cell types.54 CellChat was chosen for this analysis as it has been shown to have better performance than other cell-cell communication tools, due to integration of regulatory information, such as multimeric ligand receptor complexes.55 As our datasets were enriched for non-epithelial cells with only partial representation of epithelial cell types, we merged our datasets with data from epithelial enriched datasets generated at similar time points (Figures 5A and 6A).1
Figure 5.
Cell-cell communication analysis of the P7 cochlea
(A) Merged P7 epithelial enriched and mesenchymal enriched datasets, visualized in magenta and cyan, respectively, in UMAP space.
(B) P7 merged dataset visualized by cell type in UMAP space.
(C and D) Inferred outgoing/incoming signaling communication patterns of the P7 cochlea, which displays the cell types within each signaling pattern and the inferred signaling pathways. Thickness of flow indicates cell group or signaling pathway contribution to each pattern.
(E–F) Inferred outgoing/incoming signaling pathways of P7 OMCs. Top histogram displays amount each cell type contributes to the pattern. Histogram to the right displays the amount each signaling pathway contributes to the pattern. Relative strength represented by a gray-cyan-magenta color scale.
(G) Dot plot of relative incoming vs. outgoing interaction strength of all P7 cochlear cell types. Size of dot represents number of interactions of cell type. Dotted circle highlights mesenchyme.
DC: Deiters Cells, HeC: Hensen’s Cells; IHC: Inner Hair Cells, OHC: Outer Hair Cells, IPC: Inner Pillar Cells, LKO: Lateral Kölliker’s Organ, MKO: Medial Kölliker’s Organ, MLKO: Medial Lateral Kölliker’s Organ, OPC: Outer Pillar Cells, IPC: Inner Pillar Cells, IPhCs: Inner Phalangeal Cells.
Figure 6.
Cell-cell communication analysis of the E15/E16 cochlea
(A) Merged E16 epithelial enriched and E15 mesenchymal enriched datasets, visualized in magenta and cyan, respectively, in UMAP space.
(B) E15/E16 integrated dataset visualized by cell type in UMAP space.
(C and D) Inferred outgoing/incoming signaling communication patterns of the E15/E16 cochlea, which displays the cell types within each signaling pattern and the inferred signaling pathways. Thickness of flow indicates cell group or signaling pathway contribution to each pattern.
(E–F) Inferred outgoing/incoming signaling pathways of E15/E16 OMCs. Top histogram displays amount each cell type contributes to the pattern. Histogram to the right displays the amount each signaling pathways contributes to the pattern. Relative strength gray-cyan-magenta color scale.
(G) Relative incoming vs. outgoing interaction strength of all E15/E16 cochlear cell types. Size of dot represents number of interactions of cell type. Dotted circle highlights mesenchyme.
DC: Deiters Cells, HeC: Hensen’s Cells; IHC: Inner Hair Cells, OHC: Outer Hair Cells, IPC: Inner Pillar Cells, KO: Kölliker’s Organ, OPC: Outer Pillar Cells, IPC: Inner Pillar Cells, IPhCs: Inner Phalangeal Cells, Pro Epi/OMCs: progenitor epithelial/OMCs, Peri: Pericytes, IdC: Interdental Cells, IS: Inner Sulcus, LPro: Lateral Prosensory, MPro: Medial Prosensory, OS: Outer Sulcus, LER: Lesser Epithelial Ridge.
After merging the OMC-enriched and the epithelial-enriched P7 datasets, 24 clusters were identified and classified using previously described marker genes (Figure 5B).1 To understand how multiple groups of cells and their corresponding signaling pathways coordinate function, we utilized the pattern recognition method (non-negative matrix factorization) in CellChat. This allowed us to infer a set of communication patterns that groups cell types both via outgoing signaling (cell type is the sender of signal) or incoming signaling (cell type is the receiver of signal). CellChat inferred 6 groups of outgoing communication patterns from 56 significant signaling pathways (Figure 5C). Each signaling pattern encompassed distinct cellular populations including all OMCs and stria vascularis basal cells (pattern 1), ectoderm-derived epithelial cells (pattern 2), vascular/immune cells (pattern 3), neuronal and Hensen’s cells (pattern 4), inner hair cells (IHCs), satellite and marginal cells (pattern 5), and pericytes (pattern 6). The IHC pattern was interrogated to verify the accuracy of the predicted communication networks. Indeed, many of the signaling pathways identified by the IHC pattern have been reported to be necessary for normal IHC function including neuronal cell adhesion (NCAM), fibroblast growth factor (FGF), Tenascin (TENASCIN), and Claudins (CLDN).56,57,58,59 A heatmap of the significant outgoing pathways for the OMC pattern shows the relative contribution each cell type and signaling pathway has on the overall outgoing pattern (Figure 5E). Both global and subpopulation-specific signaling pathways were identified for OMCs. As a global signaling pathway, OMCs and basal cells are the main senders of PTN signaling, a neurotropic factor essential for normal auditory function.60 In contrast, fibronectin (FN1) signaling originates predominately by type I/basilar membrane OMCs; bone sialoprotein (BSP) is exclusively through type III/modiolar OMCs and chemokine (C-X-C motif) ligand (CXCL) is mainly by type II/spiral limbus OMCs (Figure S8A). Fibronectin, a large extracellular matrix protein, has been shown to be localized to the basilar membrane in both mice and humans and is thought to be essential to the mass and stiffness of the basilar membrane.61,62 BSP, also an extracellular matrix protein, is abundant in bone and has been shown to regulate osteoblast differentiation.63 Finally, CXCL12/CXCR4 signaling pathway is important for neurite extension and SGN survival.64
The P7 incoming signaling network shows similar patterns to the outgoing signaling network, including an OMC and stria vascularis basal cell pattern (Figure 5D). A heatmap of the significant incoming pathways of the OMC pattern includes migration inhibitory factory (MIF), FGF, and sonic hedgehog (SHH) signaling (Figure 5F). Both type I and IV OMCs receive signaling from MIF, suggesting spatially distinct roles of MIF signaling in postnatal cochlear development (Figure S8B). Indeed, MIF has been shown to be essential in the developing and mature cochlea, promoting neurite survival and outgrowth of SGNs while simultaneously preventing the apoptosis of the intermediate cells in the stria vascularis.65,66 The FGF signaling pathway plays diverse developmental roles in the cochlea, including signaling across the epithelial-mesenchymal boundaries.57 FGF signaling from the IHCs has been shown to regulate pillar cell development; however, FGF signaling to OMCs has yet to be explored beyond embryonic time points (Figure S8B).67 In addition, SHH signaling is a major incoming signaling to type II and III OMCs, with the majority coming from neuronal cell types (Figure S8B). Lack of SHH signaling from the auditory ganglion has been shown to cause cochlear duct shortening, and abnormalities in hair cell differentiation.68 Interestingly, OMC-specific transcription factors POU3F4 and TBX1 have been shown to be directly impacted by loss of SHH signaling, with Pou3f4-deficient mice exhibiting a shortened cochlear phenotype.14,69 Finally, to ascertain the overall contribution of each cell type to cochlear signaling, we visualized both the incoming and outgoing interaction strengths on a scatterplot (Figure 5G). This analysis, based on communication probability, indicates that OMCs contribute the most outgoing signaling during early postnatal cochlear development.
To identify if OMC subpopulations are secreting spatially distinct outgoing signaling molecules during embryonic development, we examined the cellular communication network of the E15/E16 cochlea. Twenty five clusters were identified in the merged embryonic dataset and classified using marker genes found here and previously described (Figure 6B).1 CellChat identified 64 significant outgoing pathways between five patterns. Much like the P7 pattern analysis, each pattern encompasses distinct cellular populations including an OMC pattern (pattern 2; Figure 6C). Interestingly, type III/modiolar OMCs are the main contributors of multiple signaling pathways including but not limited to EphrinB-Eph (EPHB), Semaphorin-5A (SEMA5), and CXCL (Figure 6E). These signaling pathways are mainly received by the surrounded SGNs (Figure S9A). Ephrin-B2/EphA4 interactions between SGNs and OMCs have been shown to promote SGN axon fasciculation.16 However, many other ephrin ligands and receptors are expressed by either OMCs or SGNs, indicating further ephrin-mediated signaling (Figure S9C). Additionally, Sema5B (expressed by immature hair cells) has been shown to interact with PlexinA1 (expressed by SGNs) to limit SGN terminal branching, however, the function of Sema5A, expressed by OMCs, is still unknown (Figure S9A).70 Furthermore, as stated earlier, CXCL12/CXCR4 signaling pathway is important for neurite extension (Figure S9A).64
In addition to the outgoing signaling cascades, we also examined the incoming signaling patterns of the embryonic cochlea elucidating 8 patterns with 49 significant pathways (Figure 6D). Again, much like the outgoing signaling patterns, patterns encompassed similar cell populations including an OMC pattern (pattern 1). OMC incoming signaling pathways include but are not limited to transforming growth factor β (TGFb), neurexin (NRXN), and platelet-derived growth factor (PDGF) signaling (Figure 6F). OMCs are the main receivers of TGFb signaling pathway which is essential for otic capsule formation, cochlear tonotopic organization, and spiral ganglion formation.71,72,73 Also, OMCs and glia are receivers of NRXN signals from neurons, which have been shown to play a role in synaptic transmission.74 Finally, OMCs are the receivers of PDGF signaling, which may play a role in the proliferation of embryonic cochlear hair cells (Figure S9B).75 Finally, much like the P7 OMCs, E15 OMCs contribute the most to outgoing signaling in late embryonic cochlear development (Figure 6G). These data suggest that OMCs are the largest influencer of cochlear cell type development at both E15 and P7.
Utilizing gEAR, a cloud-based data resource, for the sharing, visualization, and analysis of OMC-centric data
To allow biologists with a diverse range of informatic expertise meaningful access to these data, we created an OMC-focused page on the gene Expression Analysis Resource portal (UMgEAR.org/mesenchyme; Figure 7).31 This innovative OMC profile allows users full access to explore all datasets presented in this publication (Figure 7A). Additional user-friendly informatic tools allow for further investigation of these data, including newly generated graphics to spatially display expression levels in color gradients, comparative gene expression tools, and a fully functional scRNA-seq workbench (Figures 7B–7D).
Figure 7.
OMC-centric gEAR profile for visualization, sharing and analyses
(A) Overview of the OMC-centric gEAR profile including the E15/E16 whole cochlea, E15 mesenchymal, P7 whole cochlea, and P7 mesenchymal datasets. Gene displayed is Pou3f4.
(B) Examples of the custom graphical displays for both the E15 and P7 mesenchymal datasets. Gene displayed is Car3. Gene expression represented by gray to magenta color scale.
(C) gEAR compare gene expression tool. Allows for users to compare expression across any two conditions within a single dataset, here showing differentially expressed genes between type III and type IV OMCs.
(D) The single-cell workbench allows user to perform analyses from raw data (“de novo”) or use a curated stored analysis to explore marker genes and compare gene expression across clusters. Shown here are marker genes for each P7 OMC subpopulation in both dot plot and violin form. Additionally, comparative analysis of type I and type IE OMC subpopulations are shown via split-violin plots.
Discussion
Understanding how cochlear cell types influence cochlear development provides insights into the molecular mechanisms required for normal auditory function. Here, we provide a comprehensive characterization of the gene expression profiles of OMCs, identifying molecularly and functionally distinct OMC subtypes and elucidating OMC signaling cascades that may be critical for cochlear development. Interestingly, our data suggest OMCs are distinct during embryonic development, indicating either early diversification from a common progenitor or disparate embryonic origins. Early diversification is likely, as all OMCs express several transcription factors (e.g., Pou3f4, Tbx18) but further research is necessary to uncover transcriptional cascades necessary for this diversification. Interestingly, type I basilar membrane, type III modiolar, and type IV lateral wall OMCs show further diversification at P7 (Figure S7). In the case of type I OMCs, this diversification could reflect tonotopic differences accounting for the basilar membrane stiffness gradient, the main basis for cochlear frequency tuning.76 Indeed, both Nov (extracellular matrix protein) and Tnc (glycoprotein) mark type I OMC subpopulations, have been localized to the basilar membrane and have been shown to exhibit increasing base-to-apex tonotopic expression.73 In contrast, both type III and type IV OMC further diversification seems to reflect refined spatial populations. Previous studies spatially localized Car8 and Panx3, marking cluster 3B and 3C, respectively, to different parts of the modiolar bone.77,78 Type IV OMC subpopulations most likely echo the 5 distinct fibrocyte populations found in the mature lateral wall, however, mature fibrocyte marker genes are unable to fully confirm this hypothesis (Figure S7).51
Basal cells of the stria vascularis and cochlear pericytes both express OMC-specific transcription factors while not representing OMCs and were therefore excluded from our OMC analyses. Specifically, OMC-derived basal cells at P7 still express low levels of Pou3f4, Tbx18, and Tbx1 and cluster closely with OMCs.79 Pericytes, contractile cells on capillaries, are both Pou3f4 and Tbx18 positive, confirmed with a separate P7 cochlear dataset (Figure S1D).80 Pericytes derive from the same embryonic germ layers as OMCs, therefore possibly originate from the same progenitor cells and are known to be heterogeneous from organ to organ.81,82,83,84 The blood labyrinthine barrier in the stria vascularis, which is essential for maintaining the endocochlear potential, often referred to as the “cochlear battery” or driving force for hearing function, is established in part by cochlear pericytes.85,86 Interestingly, both Pou3f4 and Tbx18-deficient mice exhibit a complete disruption in endocochlear potential thought to be caused by disruption of outgoing signaling from the surrounding fibrocytes. However, this disruption in endocochlear potential could also be caused by defects in OMC-like pericytes and their signaling to and from the surrounding vascular.13,87 Indeed, we identified several signaling pathways between OMC-like pericytes and vascular cells including angiopoietin/Tie signaling known to be critical for vascular stability and Notch signaling shown to be necessary for pericyte survival (Figures S8C and S8D).88,89 Determining the mechanism by which Pou3f4 contributes to generating and maintaining the endocochlear potential will require conditional deletion of Pou3f4 in type IV lateral wall OMCs as well as OMC-like pericytes.
The maturation of the cochlea requires precise temporal and spatial signaling patterns necessary for morphogenetic events and cell fate decisions.90 Our data show that OMCs are the main contributors to outgoing signaling in the developing cochlea. This outgoing signaling of OMCs consists of both a coordinated global response (i.e., universal OMC pathways) as well as spatially distinct communication patterns unique to each one of the OMC subpopulations. During embryonic development, OMCs are known to play a role in axonal guidance and SGN survival and our data suggest that the adjacent type III modiolar OMCs facilitate many of the previously identified SGN signaling pathways including EPHB, SEMA5, and CXCL. However, our analysis also identified the growth and differentiation factor (GDF) pathway, mainly Gdf10 (ligand) as expressed by type III OMCs and Acvr1b/2a (receptors) by SGNs (Figure S9C). Gdf10 is known to facilitate axonal sprouting and more recently the GDF10-ACVR1b/ACVR2a pathway was identified to be a factor in early enteric nervous system development.91,92 Interestingly, another enteric nervous system pathway, endothelin (EDN), was identified with Edn3 (ligand) expressed by type III OMCs and Ednrb (receptor) expressed by glial cells (Figure S9C). Both EDN3 and EDNRB are known human deafness genes and have been shown to inhibit both neuronal and glial differentiation.93,94 In addition to neuronal development, we also identified angiopoietin-like (ANGPTL) and vascular endothelial growth factor (VEGF), known to play a role in angiogenesis, in the modiolar OMCs (Figure S9D). Even though most cochlear vasculature networks are found in the lateral wall (∼80%), the modiolus/spiral lamina also contains vascularity (∼19–24%) which is critical for normal neuronal activity.95,96,97
In addition to furthering our cell type-specific understanding of cochlear development, this study is pivotal to developing mesenchyme-containing inner ear organoids which are instructive for the identification of new regenerative drugs and therapeutics for hearing loss. Early in otic vesicle development, the combination of numerous signaling pathways from within the cochlear epithelium itself and the surrounding cell types, including the OMCs, enables otic progenitor cells to commit to cochlear cell fates.90 Our data suggest that OMCs are heavily involved in cell-cell signaling, most likely influencing the development of numerous cell types during cochlear development. This is supported by the fact that explanted otocysts to ectopic locations do not develop normally without some OMC tissue present.43,98 However, current in vitro organoid cochlear models appear to contain a small subset of cochlear cell types, with the focus being on neurosensory cell types.99,100,101,102 Interestingly, the hair cells generated from these in vitro studies are often described as hair cell-like cells and predominantly vestibular in character.103,104 Cochlear-specific mesenchymal cells, currently not known to be present in these cultures, could be one of the missing variables needed for more mature cochlear organoids including terminal differentiated cochlear hair cells. Indeed, recent publications highlight the importance of organ-specific mesenchyme in organoid development.105,106,107,108,109 Also, OMCs not only influence the differentiation of developing hair cells, but potentially could be a source of regeneration, as tympanic border cells are self-renewing in a WNT-dependent manner, and able to trans-differentiate into supporting cell-like and hair cell-like cells.110 This further indicates that characterization of the type I basilar membrane OMC gene regulatory network may lead to the discovery of novel genes necessary for a hair cell regeneration pathway.
In summary, we have shown that OMC diversification occurs shortly after the formation of the otocyst with further refinement until the onset of hearing, well before terminal differentiation. Our data also suggest OMCs are the main contributors of outgoing signaling during cochlear development, showcasing their importance in influencing surrounding cochlear cell types. Without cochlear OMCs and their later terminally differentiated cell types, normal auditory function would not be feasible highlighting the importance of tissue-specific mesenchymal cells in cochlear development.
Limitations of this study
This study has several limitations. Firstly, due to the high-cost nature of scRNA-seq, this study has limited biological replicates for each scRNA-seq dataset. Secondly, without more developmental time points, we were unable to resolve the full developmental trajectory of each OMC subpopulation. Thirdly, our data suggest regulators of each OMC subpopulation; however, further multiomic analyses are needed to identify transcription factors necessary for OMC diversity. Finally, the cell-cell commination pathways presented in this study are purely based on scRNA-seq gene expression and currently lack a spatial component and experimental validation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken polyclonal anti-POU3F4 | Coate et al., 2012,16 Aves Labs | RRID: AB_2814704 |
| Rat monoclonal anti-CD326 (Ep-CAM) | Biolegend | Cat# 118213, RRID:AB_1134105 |
| Rabbit polyclonal anti-MYO6 | Proteus Biosciences | Cat# 25–6791, RRID:AB_10013626 |
| Mouse monoclonal anti-KCNQ1 | Santa Cruz Biotechnology | Cat# sc-365186, RRID:AB_10707688 |
| Mouse polyclonal anti-TUJ1 | Biolegend | Cat# MMS-435P, RRID: AB_2313773 |
| Rabbit polyclonal anti-EMILIN2 | Amma et al., 200322 | RRID:AB_2941943 |
| Rabbit monoclonal anti-TGFBI | Abcam | Cat# ab170874, RRID:AB_2895231 |
| Rabbit monoclonal anti-RUNX2 | Cell Signaling Technology | Cat# 12556, RRID:AB_2732805 |
| Rabbit polyclonal anti-CAR3 | Proteintech | Cat# 15197-1-AP, RRID:AB_2243603 |
| Critical commercial assays | ||
| Chromium single cell 3′reagent kit V3 | 10x Genomics | Cat# PN-1000075 |
| Deposited data | ||
| E15 and P7 raw cochlear scRNA-seq datasets | This paper | GEO: GSE217727 |
| E15 and P7 analyzed cochlear scRNA-seq datasets | This paper | UMgEAR.org/mesenchyme |
| E16 and P7 cochlear epithelial scRNA-seq datasets | Kolla et al., 20201 | GEO: GSE137299 |
| Experimental models: Organisms/strains | ||
| Mouse: Crl:CD1(ICR) | Charles River | RRID:IMSR_CRL:022 |
| Mouse: C57BL/6J Pou3f4tm1Cren/y | Phippard et al., 199914 | RRID:MGI:3046172 |
| Oligonucleotides | ||
| Primer: Pou3f4 Forward Primer: CACTCTGATGAA GAGACTCCAAC | This paper | N/A |
| Primer: Pou3f4 Reverse Primer: CACCGTGTGCGA ATAAACCTC | This paper | N/A |
| Software and algorithms | ||
| ImageJ (FIJI) | Schindelin et al., 2012111 | RRID:SCR_002285 |
| R Project for Statistical Computing | http://www.r-project.org/ | RRID:SCR_001905 |
| Cell Ranger | 10x genomics112,113 | RRID:SCR_017344 |
| Seurat (R package) | https://satijalab.org/seurat/114,115 | RRID:SCR_016341 |
| IpathwayGuide | Advaita Bio116 | N/A |
| RcisTarget (R package) | Aibar et al., 201733 | N/A |
| EnhancedVolcano (R package) | https://bioconductor.org/packages/EnhancedVolcano | RRID:SCR_018931 |
| Monocle3 (R package) | https://cole-trapnell-lab.github.io/monocle3117,118,119 | RRID:SCR_018685 |
| gene Expression Analysis Resource (gEAR) | https://umgear.org31 | RRID:SCR_017467 |
| Biorender | http://biorender.com | RRID:SCR_018361 |
| CellChat (R package) | https://github.com/sqjin/CellChat54 | RRID:SCR_021946 |
| Other | ||
| Chromium Controller | 10x Genomics | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Ronna Hertzano (ronna.hertzano@nih.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Mouse models
All procedures involving animals were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and have been approved by the Institutional Animal Care and Use Committee at the University of Maryland, Baltimore (protocol numbers 0721005 and 0918005). Mice were housed in temperature and humidity-controlled facilities with a 12-hour-light/dark cycle and with ad libitum access to food and water.
Only wildtype male pups with a C57BL/6J background, generated from a cross between Pou3f4+/− females and Pou3f4y/- males were used in the generation of both E15 and P7 scRNA-seq datasets.14 Male and female C57BL/6J pups were used for immunohistochemistry experiments (Veterinary Resources, University of Maryland, Baltimore). Time-pregnant CD-1(ICR) females were purchased from Charles River to generate pups at P2, P5 and P7 and inner ears were extracted for immunohistochemistry (strain code: 022).
Method details
Genotyping
Genotyping of Pou3f4 alleles was performed by PCR with the following primers: Pou3f4-FP 5′ CAC TCT GAT GAA GAG ACT CCA AC 3’; Pou3f4-RP 5′ CAC CGT GTG CGA ATA AAC CTC 3’. Amplification of the Pou3f4 alleles results in a wild-type band of 510bp. The primers for genotyping the Pou3f4 alleles are located within the Pou3f4 gene, resulting in no amplification in Pou3f4 null mice. Genotyping for E15 pups was performed simultaneously with cochlear preparations before 10x Genomic single-cell capture. Genotyping for P7 pups was performed the day before cochlear preparations/10x Genomic single-cell capture and pups were identified via tattoo.
Cochlear dissections, single cell dissociations, FACS, and 10x genomics capture
For both E15 and P7 scRNA-seq datasets, cochlear dissections were performed by extracting the inner ear from the mice followed by extracting the cochlear duct from the overlying bone/cartilage. To ensure our analysis encompassed all OMCs in an unbiased manner, the entirety of the cochlear duct was utilized, including the modiolus and lateral wall. Micro-dissected cochlear tissue from 2 mice was pooled for each dataset and dissociation was performed by incubating cochleae in Thermolysin (Sigma) for 20 min at 37°C followed by two 5-min incubations with papain (Worthington Biochemical) at 37°C with mechanical disruption by titration after each incubation. After addition of 20% ovomucoid protease inhibitor (Worthington Biochemical), the cell suspension was passed through a 20μm filter (Falcon) to remove large debris. To ensure high-quality samples, fluorescence-activated cell sorting (FACS) was performed using a BD FACSAria™ II Cell Sorter (BD Biosciences), at the University of Maryland School of Medicine (UMSOM) Flow Cytometry Core. Strict forward and side scatter (FSC and SSC) gates were used to remove confounding debris via size and granularity, respectively. Before the 10x Genomics single cell capture occurred, an aliquot of each sample was used to assess quality and concentration of cells using a TC20 Automated Cell Counter (Bio-Rad). A target capture of 10,000 cells per sample was chosen. Samples were processed by the Genomics Resource Center (GRC) at the UMSOM Institute for Genome Sciences (IGS) for single-cell micro-fluidic capture via the GemCode platform with v3 chemistry (10x Genomics). Time from euthanasia to 10x Genomics single cell capture was ∼3 hours.
Library preparation and sequencing
After droplet-based single-cell capture via the 10x Genomics platform, single-cell gene expression libraries were generated using the Chromium single cell 3′ reagent kit v3 (10x Genomics). To assess library quality, traces were analyzed traces using the LapChip GC Touch HT. Sequencing was performed on an Illumina NovaSeq 6000 at a target depth of 100,000 reads per cell with paired-end 75 bp reads. All steps for library preparation and sequencing were performed by the GRC at UMSOM IGS.
Immunohistochemistry
Inner ears from E15, P2, P7 or P14 mice were harvested and placed into 10% buffered formalin (Sigma) overnight rotating at 4°C. An additional decalcification incubation for P7 and P14 mice was performed overnight at 4°C with 0.5M EDTA (Quality Biological). All inner ears for this study were paraffin-embedded (Leica), sectioned at 7μm (Leica RM2135 microtome), mounted on positively charged glass slides (Superfrost Plus; Thermo Fisher Scientific), and dried in a HybEZ Oven (Advanced Cell Diagnostics) at 37°C overnight. Sectioned cochleae were deparaffined with Xylenes (Sigma Aldrich), dehydrated by ethanol washes (100%, 95%, 80% and 70%) and rehydrated with deionized water. Prior to staining, antigen retrieval was performed with a 10mM citric acid solution (0.05% Tween, pH 6.0) incubated at 95°C for 25 min followed by a 1-h blocking incubation at room temperature with 10% fetal bovine serum (Gemini Bio-Products) and 1% bovine serum albumin (Millipore Sigma) in PBS with 0.2% Tween 20 (MP Biomedicals). All sections were labeled with anti-Pou3f4 (1:10,000, Coate et al., 2012), DAPI (1:2500, Thermo Fisher) and one of the following: anti-TUJ1 (1:500, BioLegend), anti-CD326 (1:50, BioLegend), anti-MYO6 (1:1000, Proteus Biosciences), anti-KCNQ1 (1:50, Santa Cruz Biotechnology), anti-EMILIN2 (1:1000, Amma et al., 2003), anti-TGFBI (1:1000, Abcam), anti-RUNX2 (1:1000, Cell Signaling Technology), or anti-CAR3 (1:1000, Proteintech).16 All secondary antibodies were used at 1:1000 and include: goat anti-chicken IgY Alexa Fluor 488 (Thermo Fisher Scientific), donkey anti-rabbit IgG Alexa Fluor 548 (Thermo Fisher Scientific), donkey anti-mouse IgG Alexa Fluor 488 (Thermo Fisher Scientific), and chicken anti-rat IgG Alexa Fluor 488 (Thermo Fisher Scientific). Primary antibodies were incubated overnight at 4°C and secondary antibodies, including 300nM of DAPI, were incubated for 1-h at room temperature. Coverslips were mounted onto slides with ProLong gold (Invitrogen) and sections were visualized using the Nikon E600 fluorescence microscope connected to an Infinity3-6UR microscope camera (Lumenera).
Quantification and statistical analysis
Data preprocessing and initial analyses of both E15 and P7 scRNA-seq datasets
Illumina BCL files were processed via Cell Ranger v6.1.2 pipeline, with reads aligned to the GRCm38 mm10 genome assembly. Cell Ranger uses STAR for alignment and manufacturer’s software for all other steps.112,113 Seurat v3 was used for processing of the Cell Ranger output.114,115 Firstly, quality control was performed removing cells with less than 200 genes, genes expressed in less than 3 cells and cells expressing more than 30% mitochondrial genes. After quality control, 7,703 P7 and 6,018 E15 cochlear cells were included in the final analyses. E15 cochlear cells come from two separate scRNA-seq datasets, integrated via Seurat’s anchoring methodology.114 Next, the expression data were log transformed, normalized, and scaled for sequencing depth. Linear dimensionality reduction was performed via principal component analysis (PCA), and unbiased graph-based clustering was performed using the top 15–20 principal components. Non-linear dimensionality reduction was performed via the UMAP algorithm for visualization and cluster identification was performed manually by identification of known marker genes (Figures S1B and S3A).120 OMC clusters were identified based on the expression of canonical markers (Pou3f4, Tbx18, Tbx1 and Pou3f3, Figure S1C). To find marker genes for each OMC subpopulation, differential expression analysis was performed via a non-parametric Wilcoxon rank-sum test (Seurat’s “FindAllMarkers” function; Tables S1 and S2).
Sub-clustering analyses of OMCs at E15 and P7
To fully investigate the differences between OMC subpopulations, all OMCs were separated into new datasets for both E15 and P7. Dimensionality reduction via PCA and UMAP was repeated. OMCs were subclustered by an unbiased graph-based method and OMC subpopulations were identified via previously identified marker genes at P7. The resolution of the unbiased clustering at P7 was increased to identify additional OMC subpopulations that could represent tonotopic differences (type I) or further refined spatial distribution (type III/IV; Figure S7; Table S5). To help identify E15 OMC subpopulations, E15 and P7 OMC datasets were integrated via Seurat’s anchoring methodology to allow for direct comparison (Figure S5B). Differential expression analysis was performed at both E15 and P7 as per above (Tables S3 and S4). The Bioconductor R-package EnhancedVolcano was used to generate differential expression volcano plots of E15 and P7 OMCs with a fold change cutoff of 0.6 (https://github.com/kevinblighe/EnhancedVolcano). P7 differential gene lists were then used for GO-term enrichment analysis (biological processes) by Advaita Bio’s iPathwayGuide (Table S7).116 RcisTarget was used for motif analysis, using its gene-motif rankings for mouse genes which is based on sequences 500bp upstream of the transcriptional start sites.33 visNetwork was used for visualization of the gene regulatory networks of the top 3 potential upstream regulators for each OMC subpopulation (https://datastorm-open.github.io/visNetwork/). Monocle3 was used for the developmental trajectory analysis, using the raw count matrix from the E13, E15, and P7 integrated OMC dataset.117,118,119 Gene modules were calculated using the find_gene_modules command using differentially expressed genes across pseudotime with a q-value cutoff of <0.05.
Cell-cell communication network analysis of the E15/16 and P7 cochlea
To have the most robust datasets possible, our E15 and P7 mesenchyme-enriched datasets were merged with previously published E16 and P7 epithelial-enriched datasets via Seurat’s merge function resulting in combined raw count matrices.1 After merging, the expression data were log transformed, normalized, scaled for sequencing depth, dimensionality reduced and clustered as performed above. Resulting cell clusters were identified with already known marker genes previously described.1 Ligand-receptor expression and cell interactions were ascertained by using the R-package CellChat v1.1.3 (http://www.cellchat.org/).54 The analyses were performed according to the author’s most updated vignette (https://github.com/sqjin/CellChat). Briefly, the normalized counts for each cell type were imported from Seurat into a CellChat object, preprocessed to find over-expressed ligand-receptor interactions and biological significance was inferred by assigning a probability value to each interaction (default CellChat method trimean was used) and performing a permutation test. Finally, CellChat employs a non-negative matrix factorization pattern recognition method to identify global communication patterns. The number of patterns used was representative of when both Cophenetic and Silhouette values begin to drop suddenly as per suggested by CellChat authors. CellChat provides various functionality for visualization, which were used here, including: river (alluvial) plots for global communication patterns, heatmaps for signaling patterns and centrality scores, scatterplots for interaction strength, bubble plots for significant ligand-receptor pairs from one cell group to another, and histograms for visualization of relative contributions of each ligand-receptor pair. Interactions with a p value of <0.05 were considered significant.
Acknowledgments
We would like to thank Benjamin Shuster and Dr. Kathleen Gwilliam (National Institute on Deafness and Other Communication Disorder) for their technical assistance in generation of embryonic scRNA-seq datasets. We would also like to thank Dr. Douglas Forrest (National Institute of Diabetes and Digestive and Kidney Diseases) for providing the EMILIN2 antibody. We would like to thank Dr. Lisa D. Sadzewicz and Luke J. Tallon (Genomics Resource Center, Institute for Genome Sciences) for their technical assistance in library preparation, sequencing, and quality control analyses of all scRNA-seq datasets presented in this study. Finally, we would like to thank Dr. Paige Brooks (Georgtown University), Dr. Mansa Gurjar (Georgetown University), and Dr. Ran Elkon (Tal Aviv University) for their scientific discussions. This research was supported by the NIDCD/NIH RO1DC13817 (R.H.), RO1DC016595 (T.M.C), F31DC019513 (K.P.R) and the NIDCD/NIH Intramural Research Program to R.H (DC000094-01). This work was also supported by The United States – Israel Binational Science Foundation (2017218; Drs. Ronna Hertzano and Ran Elkon).
Author contributions
K.P.R., T.M.C., and R.H. oversaw project conceptualization and methodology. K.P.R. O.Z., R.H., G.M., and B.M. performed experiments. K.P.R. analyzed data. K.P.R. and Y.S. generated gEAR visualizations. T.M.C., R.H., and K.P.R. acquired funding. K.P.R. wrote the manuscript with help from T.M.C. and R.H. All authors contributed comments.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107769.
Supplemental information
(A–O) Markers identified via differential analysis of each cluster against all other clusters via non-parametric Wilcoxon rank sum test. Each row represents identified marker gene ranked by p-value. Columns from left to right: p-value, log-scale fold change, fraction of cells expressing marker gene in selected cluster, fraction of cells expressing marker gene in all other clusters, adjusted p-value, cluster, and gene name.
(A–I) Markers identified via differential analysis of each cluster against all other clusters via non-parametric Wilcoxon rank sum test. Each row represents identified marker gene ranked by p-value. Columns from left to right: p-value, log-scale fold change, fraction of cells expressing marker gene in selected cluster, fraction of cells expressing marker gene in all other clusters, adjusted p-value, cluster, and gene name.
(A–F) Markers identified via differential analysis of each cluster against all other clusters via non-parametric Wilcoxon rank sum test. Each row represents identified marker gene ranked by p-value. Columns from left to right: p-value, log-scale fold change, fraction of cells expressing marker gene in selected cluster, fraction of cells expressing marker gene in all other clusters, adjusted p-value, cluster, and gene name.
(A–D) Markers identified via differential analysis of each cluster against all other clusters via non-parametric Wilcoxon rank sum test. Each row represents identified marker gene ranked by p-value. Columns from left to right: p-value, log-scale fold change, fraction of cells expressing marker gene in selected cluster, fraction of cells expressing marker gene in all other clusters, adjusted p-value, cluster, and gene name.
(A–O) Markers identified via differential analysis of each cluster against all other clusters via non-parametric Wilcoxon rank sum test. Each row represents identified marker gene ranked by p-value. Columns from left to right: p-value, log-scale fold change, fraction of cells expressing marker gene in selected cluster, fraction of cells expressing marker gene in all other clusters, adjusted p-value, cluster, and gene name.
(A and B) Rcistarget analysis identifying potential upstream regulators of each OMC subpopulation at P7 and E15. Each row represents an identified potential upstream motif, ranked by normalized enrichment score (NES). Columns from left to right: rank: rank of motif, logo: URL to motif’s genomic sequence, geneSet: OMC subpopulation, motif: ID of the motif, NES: normalized enrichment score of the motif in the geneSet, AUC: area under the curve (used to calculate the NES), TF_highConf: transcription factors annotated to the motif according to confidence annotation (two asterisks), TF_lowConf: transcription factors annotated to the motif according to confidence annotation (one asterisk), enrichedGenes: genes that are highly ranked for the given motif, nErnGenes: number of genes highly ranked, rankAtMax: ranking at the maximum enrichment, used to determine the number of enriched genes.
(A–D) Biological gene ontology analysis for P7 OMC subpopulations. Each row represents identified biological GO-term ranked by p-value. Columns from left to right: GO-term ID, GO-term name, number of DE genes in GO-term, number of overall genes in GO-term, and p-value.
Data and code availability
All data supporting the findings of this study are available within the article itself, supplemental information or available upon reasonable request. All scRNA-seq datasets generated for this manuscript have been deposited at GEO (GEO: GSE217727) and are publicly available as of the date of publication. This paper does not generate or report any original code. Any reasonable request for additional information required to reanalyze the data reported in this manuscript is available from the lead contact, Dr. Ronna Hertzano.
References
- 1.Kolla L., Kelly M.C., Mann Z.F., Anaya-Rocha A., Ellis K., Lemons A., Palermo A.T., So K.S., Mays J.C., Orvis J., et al. Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun. 2020;11:2389. doi: 10.1038/S41467-020-16113-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shrestha B.R., Chia C., Wu L., Kujawa S.G., Liberman M.C., Goodrich L.V. Sensory Neuron Diversity in the Inner Ear Is Shaped by Activity. Cell. 2018;174:1229–1246.e17. doi: 10.1016/J.CELL.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun S., Babola T., Pregernig G., So K.S., Nguyen M., Su S.S.M., Palermo A.T., Bergles D.E., Burns J.C., Müller U. Hair Cell Mechanotransduction Regulates Spontaneous Activity and Spiral Ganglion Subtype Specification in the Auditory System. Cell. 2018;174:1247–1263.e15. doi: 10.1016/J.CELL.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu S., Olszewski R., Taukulis I., Wei Z., Martin D., Morell R.J., Hoa M. Characterization of rare spindle and root cell transcriptional profiles in the stria vascularis of the adult mouse cochlea. Sci. Rep. 2020;10:18100–18115. doi: 10.1038/s41598-020-75238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korrapati S., Taukulis I., Olszewski R., Pyle M., Gu S., Singh R., Griffiths C., Martin D., Boger E., Morell R.J., Hoa M. Single Cell and Single Nucleus RNA-Seq Reveal Cellular Heterogeneity and Homeostatic Regulatory Networks in Adult Mouse Stria Vascularis. Front. Mol. Neurosci. 2019;12:316. doi: 10.3389/FNMOL.2019.00316/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menendez L., Trecek T., Gopalakrishnan S., Tao L., Markowitz A.L., Yu H.V., Wang X., Llamas J., Huang C., Lee J., et al. Generation of inner ear hair cells by direct lineage conversion of primary somatic cells. Elife. 2020;9 doi: 10.7554/eLife.55249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petitpré C., Faure L., Uhl P., Fontanet P., Filova I., Pavlinkova G., Adameyko I., Hadjab S., Lallemend F. Single-cell RNA-sequencing analysis of the developing mouse inner ear identifies molecular logic of auditory neuron diversification. Nat. Commun. 2022;13:3878. doi: 10.1038/s41467-022-31580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chessum L., Matern M.S., Kelly M.C., Johnson S.L., Ogawa Y., Milon B., McMurray M., Driver E.C., Parker A., Song Y., et al. 2018. Helios Is a Key Transcriptional Regulator of Outer Hair Cell Maturation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-Añoveros J., Clancy J.C., Foo C.Z., García-Gómez I., Zhou Y., Homma K., Cheatham M.A., Duggan A. Tbx2 is a master regulator of inner versus outer hair cell differentiation. Nature. 2022;298:605. doi: 10.1038/s41586-022-04668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi Z., Li X., Ren M., Gu Y., Zhu T., Li S., Wang G., Sun S., Sun Y., Liu Z. Development and transdifferentiation into inner hair cells require Tbx2. Natl. Sci. Rev. 2022;9:nwac156. doi: 10.1093/NSR/NWAC156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trainor P.A., Tam P.P. Cranial paraxial mesoderm and neural crest cells of the mouse embryo: co-distribution in the craniofacial mesenchyme but distinct segregation in branchial arches. Development. 1995;121:2569–2582. doi: 10.1242/DEV.121.8.2569. [DOI] [PubMed] [Google Scholar]
- 12.Braunstein E.M., Crenshaw E.B., Morrow B.E., Adams J.C. Cooperative Function of Tbx1 and Brn4 in the Periotic Mesenchyme is Necessary for Cochlea Formation. J. Assoc. Res. Otolaryngol. 2008;9:33–43. doi: 10.1007/S10162-008-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trowe M.O., Maier H., Schweizer M., Kispert A. Deafness in mice lacking the T-box transcription factor Tbx18 in otic fibrocytes. Development. 2008;135:1725–1734. doi: 10.1242/DEV.014043. [DOI] [PubMed] [Google Scholar]
- 14.Phippard D., Lu L., Lee D., Saunders J.C., Crenshaw E.B. Targeted Mutagenesis of the POU-Domain GeneBrn4/Pou3f4 Causes Developmental Defects in the Inner Ear. J. Neurosci. 1999;19:5980–5989. doi: 10.1523/JNEUROSCI.19-14-05980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks P.M., Rose K.P., MacRae M.L., Rangoussis K.M., Gurjar M., Hertzano R., Coate T.M. Pou3f4-Expressing Otic Mesenchyme Cells Promote Spiral Ganglion Neuron Survival in the Postnatal Mouse Cochlea. J. Comp. Neurol. 2020;528:1967–1985. doi: 10.1002/CNE.24867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coate T.M., Raft S., Zhao X., Ryan A.K., Crenshaw E.B., Kelley M.W. Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway. Neuron. 2012;73:49–63. doi: 10.1016/J.NEURON.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parzefall T., Shivatzki S., Lenz D.R., Rathkolb B., Ushakov K., Karfunkel D., Shapira Y., Wolf M., Mohr M., Wolf E., et al. 2013. Cytoplasmic Mislocalization of POU3F4 Due to Novel Mutations Leads to Deafness in Humans and Mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peeleman N., Verdoodt D., Ponsaerts P., Van Rompaey V. On the Role of Fibrocytes and the Extracellular Matrix in the Physiology and Pathophysiology of the Spiral Ligament. Front. Neurol. 2020;11:1240. doi: 10.3389/FNEUR.2020.580639/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song M.H., Choi S.Y., Wu L., Oh S.K., Lee H.K., Lee D.J., Shim D.B., Choi J.Y., Kim U.K., Bok J. Pou3f4 deficiency causes defects in otic fibrocytes and stria vascularis by different mechanisms. Biochem. Biophys. Res. Commun. 2011;404:528–533. doi: 10.1016/J.BBRC.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Mutai H., Nagashima R., Sugitani Y., Noda T., Fujii M., Matsunaga T. Expression of Pou3f3/Brn-1 and its genomic methylation in developing auditory epithelium. Dev. Neurobiol. 2009;69:913–930. doi: 10.1002/DNEU.20746. [DOI] [PubMed] [Google Scholar]
- 21.Russell I.J., Lukashkina V.A., Cho Y.W., Levic S., Lukashkin A.N., Ng L., Forrest D., Forrest D. Emilin 2 promotes the mechanical gradient of the cochlear basilar membrane and resolution of frequencies in sound. Sci. Adv. 2020;6:eaba2634. doi: 10.1126/SCIADV.ABA2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amma L.L., Goodyear R., Faris J.S., Jones I., Ng L., Richardson G., Forrest D. An emilin family extracellular matrix protein identified in the cochlear basilar membrane. Mol. Cell. Neurosci. 2003;23:460–472. doi: 10.1016/S1044-7431(03)00075-7. [DOI] [PubMed] [Google Scholar]
- 23.Kim J.E., Kim S.J., Lee B.H., Park R.W., Kim K.S., Kim I.S. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, betaig-h3. J. Biol. Chem. 2000;275:30907–30915. doi: 10.1074/JBC.M002752200. [DOI] [PubMed] [Google Scholar]
- 24.Chang J.L., Brauer D.S., Johnson J., Chen C.G., Akil O., Balooch G., Humphrey M.B., Chin E.N., Porter A.E., Butcher K., et al. Tissue-specific calibration of extracellular matrix material properties by transforming growth factor-β and Runx2 in bone is required for hearing. EMBO Rep. 2010;11:765–771. doi: 10.1038/EMBOR.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otto F., Kanegane H., Mundlos S. Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum. Mutat. 2002;19:209–216. doi: 10.1002/HUMU.10043. [DOI] [PubMed] [Google Scholar]
- 26.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.H., Inada M., et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/S0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 27.Adeva-Andany M.M., Fernández-Fernández C., Sánchez-Bello R., Donapetry-García C., Martínez-Rodríguez J. The role of carbonic anhydrase in the pathogenesis of vascular calcification in humans. Atherosclerosis. 2015;241:183–191. doi: 10.1016/J.ATHEROSCLEROSIS.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Murray S.A., Morgan J.L., Kane C., Sharma Y., Heffner C.S., Lake J., Donahue L.R. Mouse gestation length is genetically determined. PLoS One. 2010;5:e12418. doi: 10.1371/JOURNAL.PONE.0012418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma T., Van Tine B.A., Wei Y., Garrett M.D., Nelson D., Adams P.D., Wang J., Qin J., Chow L.T., Harper J.W. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunkin S.M., Ng L., Lau C., Dolbeare T., Gilbert T.L., Thompson C.L., Hawrylycz M., Dang C. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013;41:D996–D1008. doi: 10.1093/NAR/GKS1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orvis J., Gottfried B., Kancherla J., Adkins R.S., Song Y., Dror A.A., Olley D., Rose K., Chrysostomou E., Kelly M.C., et al. gEAR: Gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. Nat. Methods. 2021;18:843–844. doi: 10.1038/S41592-021-01200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y., Wang L., Zhu T., Wu B., Wang G., Luo Z., Li C., Wei W., Liu Z. Single-cell transcriptomic landscapes of the otic neuronal lineage at multiple early embryonic ages. Cell Rep. 2022;38 doi: 10.1016/J.CELREP.2022.110542. [DOI] [PubMed] [Google Scholar]
- 33.Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komori T. Regulation of osteoblast differentiation by Runx2. Adv. Exp. Med. Biol. 2010;658:43–49. doi: 10.1007/978-1-4419-1050-9_5. [DOI] [PubMed] [Google Scholar]
- 35.Hojo H., Ohba S., He X., Lai L.P., McMahon A.P. Sp7/Osterix Is Restricted to Bone-Forming Vertebrates where It Acts as a Dlx Co-factor in Osteoblast Specification The Sp7 Sp family variant correlates with the emergence of bone-forming vertebrates Accession Numbers GSE76187. Dev. Cell. 2016;37:238–253. doi: 10.1016/j.devcel.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levi G., Gitton Y. Dlx genes and the maintenance of bone homeostasis and skeletal integrity. Cell Death Differ. 2014;21:1345–1346. doi: 10.1038/cdd.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song H., Park K.H. Regulation and function of SOX9 during cartilage development and regeneration. Semin. Cancer Biol. 2020;67:12–23. doi: 10.1016/J.SEMCANCER.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Herlofsen S.R., Høiby T., Cacchiarelli D., Zhang X., Mikkelsen T.S., Brinchmann J.E. Brief report: importance of SOX8 for in vitro chondrogenic differentiation of human mesenchymal stromal cells. Stem Cell. 2014;32:1629–1635. doi: 10.1002/STEM.1642. [DOI] [PubMed] [Google Scholar]
- 39.Seo K.W., Roh K.H., Bhandari D.R., Park S.B., Lee S.K., Kang K.S. ZNF281 Knockdown Induced Osteogenic Differentiation of Human Multipotent Stem Cells In Vivo and In Vitro. Cell Transplant. 2013;22:29–40. doi: 10.3727/096368912X654948. [DOI] [PubMed] [Google Scholar]
- 40.Liu L.L., Meng J., Ma H.Y., Cao H., Liu W.J., Zhong J.H., Jia P., Liu W.Y. ATF1/miR-214-5p/ITGA7 axis promotes osteoclastogenesis to alter OVX-induced bone absorption. Anim. Biotechnol. 2022;28:1–10. doi: 10.1186/S10020-022-00476-7/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q., Quan Y., Wang N., Xie C., Ji Z., He H., Chai R., Li H., Yin S., Chin Y.E., et al. Inactivation of STAT3 Signaling Impairs Hair Cell Differentiation in the Developing Mouse Cochlea. Stem Cell Rep. 2017;9:231–246. doi: 10.1016/J.STEMCR.2017.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J., Zuo N., Wu C., Chen P., Ma J., Wang C., Li W., Liu S. Role of the periotic mesenchyme in the development of sensory cells in early mouse cochlea. J. Otolaryngol. 2020;15:138–143. doi: 10.1016/J.JOTO.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montcouquiol M., Kelley M.W. Planar and vertical signals control cellular differentiation and patterning in the mammalian cochlea. J. Neurosci. 2003;23:9469–9478. doi: 10.1523/JNEUROSCI.23-28-09469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liberman L.D., Liberman M.C. Postnatal maturation of auditory-nerve heterogeneity, as seen in spatial gradients of synapse morphology in the inner hair cell area. Hear. Res. 2016;339:12–22. doi: 10.1016/J.HEARES.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson G.P., Lukashkin A.N., Russell I.J. The tectorial membrane: one slice of a complex cochlear sandwich. Curr. Opin. Otolaryngol. Head Neck Surg. 2008;16:458–464. doi: 10.1097/MOO.0B013E32830E20C4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelley M.W. Determination and commitment of mechanosensory hair cells. Sci. World J. 2002;2:1079–1094. doi: 10.1100/TSW.2002.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee H.K., Lee S.H., Lee K.Y., Lim E.J., Choi S.Y., Park R.K., Kim U.K. Novel POU3F4 mutations and clinical features of DFN3 patients with cochlear implants. Clin. Genet. 2009;75:572–575. doi: 10.1111/J.1399-0004.2009.01181.X. [DOI] [PubMed] [Google Scholar]
- 48.Tekin A.M., Matulic M., Wuyts W., Assadi M.Z., Mertens G., Rompaey V.v., Li Y., Heyning P.v.d., Topsakal V. A New Pathogenic Variant in POU3F4 Causing Deafness Due to an Incomplete Partition of the Cochlea Paved the Way for Innovative Surgery. Genes. 2021;12 doi: 10.3390/GENES12050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai M., Shi X. Fibro-vascular coupling in the control of cochlear blood flow. PLoS One. 2011;6:e20652. doi: 10.1371/JOURNAL.PONE.0020652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carraro M., Harrison R.V. Degeneration of stria vascularis in age-related hearing loss; a corrosion cast study in a mouse model. Acta Otolaryngol. 2016;136:385–390. doi: 10.3109/00016489.2015.1123291. [DOI] [PubMed] [Google Scholar]
- 51.Furness D.N. Forgotten Fibrocytes: A Neglected, Supporting Cell Type of the Cochlea With the Potential to be an Alternative Therapeutic Target in Hearing Loss. Front. Cell. Neurosci. 2019;13:532. doi: 10.3389/FNCEL.2019.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerchman E., Hilfer S.R., Brown J.W. Involvement of extracellular matrix in the formation of the inner ear. Dev. Dynam. 1995;202:421–432. doi: 10.1002/AJA.1002020411. [DOI] [PubMed] [Google Scholar]
- 53.Zdebik A.A., Wangemann P., Jentsch T.J. Potassium ion movement in the inner ear: insights from genetic disease and mouse models. Physiology. 2009;24:307–316. doi: 10.1152/PHYSIOL.00018.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., Myung P., Plikus M.V., Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/S41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dimitrov D., Türei D., Garrido-Rodriguez M., Burmedi P.L., Nagai J.S., Boys C., Ramirez Flores R.O., Kim H., Szalai B., Costa I.G., et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat. Commun. 2022;13:1–13. doi: 10.1038/s41467-022-30755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harley R.J., Murdy J.P., Wang Z., Kelly M.C., Ropp T.J.F., Park S.H., Maness P.F., Manis P.B., Coate T.M. Neuronal Cell Adhesion Molecule (NrCAM) is Expressed by Sensory Cells in the Cochlea and is Necessary for Proper Cochlear Innervation and Sensory Domain Patterning During Development. Dev. Dynam. 2018;247:934–950. doi: 10.1002/DVDY.24629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebeid M., Huh S.H. FGF signaling: diverse roles during cochlear development. BMB Rep. 2017;50:487–495. doi: 10.5483/BMBREP.2017.50.10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitlon D.S., Zhang X., Kusakabe M. Tenascin-C in the cochlea of the developing mouse. J. Comp. Neurol. 1999;406:361–374. doi: 10.1002/(sici)1096-9861(19990412)406:3<361::aid-cne5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 59.Ben-Yosef T., Belyantseva I.A., Saunders T.L., Hughes E.D., Kawamoto K., Van Itallie C.M., Beyer L.A., Halsey K., Gardner D.J., Wilcox E.R., et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum. Mol. Genet. 2003;12:2049–2061. doi: 10.1093/HMG/DDG210. [DOI] [PubMed] [Google Scholar]
- 60.Bertram S., Roll L., Reinhard J., Groß K., Dazert S., Faissner A., Volkenstein S. Pleiotrophin increases neurite length and number of spiral ganglion neurons in vitro. Exp. Brain Res. 2019;237:2983–2993. doi: 10.1007/S00221-019-05644-6. [DOI] [PubMed] [Google Scholar]
- 61.Liu W., Atturo F., Aldaya R., Santi P., Cureoglu S., Obwegeser S., Glueckert R., Pfaller K., Schrott-Fischer A., Rask-Andersen H. Macromolecular organization and fine structure of the human basilar membrane - RELEVANCE for cochlear implantation. Cell Tissue Res. 2015;360:245–262. doi: 10.1007/S00441-014-2098-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keithley E.M., Ryan A.F., Woolf N.K. Fibronectin-like immunoreactivity of the basilar membrane of young and aged rats. J. Comp. Neurol. 1993;327:612–617. doi: 10.1002/CNE.903270411. [DOI] [PubMed] [Google Scholar]
- 63.Gordon J.A.R., Hunter G.K., Goldberg H.A. Activation of the mitogen-activated protein kinase pathway by bone sialoprotein regulates osteoblast differentiation. Cells Tissues Organs. 2009;189:138–143. doi: 10.1159/000151728. [DOI] [PubMed] [Google Scholar]
- 64.Zhang W., Sun J.Z., Han Y., Chen J., Liu H., Wang Y., Yue B., Chen Y. CXCL12/CXCR4 signaling pathway regulates cochlear development in neonatal mice. Mol. Med. Rep. 2016;13:4357–4364. doi: 10.3892/MMR.2016.5085. [DOI] [PubMed] [Google Scholar]
- 65.Bank L.M., Bianchi L.M., Ebisu F., Lerman-Sinkoff D., Smiley E.C., Shen Y.C., Ramamurthy P., Thompson D.L., Roth T.M., Beck C.R., et al. Macrophage migration inhibitory factor acts as a neurotrophin in the developing inner ear. Development (Camb.) 2012;139:4666–4674. doi: 10.1242/DEV.066647/-/DC1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang W., Zheng J., Meng J., Neng L., Chen X., Qin Z. Macrophage migration inhibitory factor mediates viability and apoptosis of PVM/Ms through PI3K/Akt pathway. Neuroscience. 2017;360:220–229. doi: 10.1016/J.NEUROSCIENCE.2017.06.063. [DOI] [PubMed] [Google Scholar]
- 67.Mueller K.L., Jacques B.E., Kelley M.W. Fibroblast growth factor signaling regulates pillar cell development in the organ of corti. J. Neurosci. 2002;22:9368–9377. doi: 10.1523/JNEUROSCI.22-21-09368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bok J., Zenczak C., Hwang C.H., Wu D.K. Auditory ganglion source of Sonic hedgehog regulates timing of cell cycle exit and differentiation of mammalian cochlear hair cells. Proc. Natl. Acad. Sci. USA. 2013;110:13869–13874. doi: 10.1073/PNAS.1222341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riccomagno M.M., Martinu L., Mulheisen M., Wu D.K., Epstein D.J. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002;16:2365–2378. doi: 10.1101/GAD.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jung J.S., Zhang K.D., Wang Z., McMurray M., Tkaczuk A., Ogawa Y., Hertzano R., Coate T.M. Semaphorin-5B Controls Spiral Ganglion Neuron Branch Refinement during Development. J. Neurosci. 2019;39:6425–6438. doi: 10.1523/JNEUROSCI.0113-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frenz D.A., Galinovic-Schwartz V., Liu W., Flanders K.C., Van de Water T.R. Transforming growth factor beta 1 is an epithelial-derived signal peptide that influences otic capsule formation. Dev. Biol. 1992;153:324–336. doi: 10.1016/0012-1606(92)90117-Y. [DOI] [PubMed] [Google Scholar]
- 72.Marzella P.L., Gillespie L.N., Clark G.M., Bartlett P.F., Kilpatrick T.J. The neurotrophins act synergistically with LIF and members of the TGF-beta superfamily to promote the survival of spiral ganglia neurons in vitro. Hear. Res. 1999;138:73–80. doi: 10.1016/S0378-5955(99)00152-5. [DOI] [PubMed] [Google Scholar]
- 73.Son E.J., Wu L., Yoon H., Kim S., Choi J.Y., Bok J. Developmental gene expression profiling along the tonotopic axis of the mouse cochlea. PLoS One. 2012;7:e40735. doi: 10.1371/JOURNAL.PONE.0040735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang W., Rohlmann A., Sargsyan V., Aramuni G., Hammer R.E., Südhof T.C., Missler M. Extracellular domains of alpha-neurexins participate in regulating synaptic transmission by selectively affecting N- and P/Q-type Ca2+ channels. J. Neurosci. 2005;25:4330–4342. doi: 10.1523/JNEUROSCI.0497-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee Y.W., Ozeki M., Juhn S.K., Lin J. Expression of platelet-derived growth factor in the developing cochlea of rats. Acta Otolaryngol. 2004;124:558–562. doi: 10.1080/00016480410016577. [DOI] [PubMed] [Google Scholar]
- 76.Robles L., Ruggero M.A. Mechanics of the mammalian cochlea. Physiol. Rev. 2001;81:1305–1352. doi: 10.1152/PHYSREV.2001.81.3.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu L., Sagong B., Choi J.Y., Kim U.K., Bok J. A systematic survey of carbonic anhydrase mRNA expression during mammalian inner ear development. Dev. Dynam. 2013;242:269–280. doi: 10.1002/DVDY.23917. [DOI] [PubMed] [Google Scholar]
- 78.Zhao H.B. Expression and function of pannexins in the inner ear and hearing. BMC Cell Biol. 2016;17(Suppl 1):16. doi: 10.1186/S12860-016-0095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trowe M.O., Maier H., Petry M., Schweizer M., Schuster-Gossler K., Kispert A. Impaired stria vascularis integrity upon loss of E-cadherin in basal cells. Dev. Biol. 2011;359:95–107. doi: 10.1016/J.YDBIO.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 80.Kalra G., Milon B., Casella A.M., Herb B.R., Humphries E., Song Y., Rose K.P., Hertzano R., Ament S.A. Biological insights from multi-omic analysis of 31 genomic risk loci for adult hearing difficulty. PLoS Genet. 2020;16:e1009025. doi: 10.1371/JOURNAL.PGEN.1009025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sims D.E. Diversity within pericytes. Clin. Exp. Pharmacol. Physiol. 2000;27:842–846. doi: 10.1046/J.1440-1681.2000.03343.X. [DOI] [PubMed] [Google Scholar]
- 82.Reyahi A., Nik A.M., Ghiami M., Gritli-Linde A., Pontén F., Johansson B.R., Carlsson P. Foxf2 Is Required for Brain Pericyte Differentiation and Development and Maintenance of the Blood-Brain Barrier. Dev. Cell. 2015;34:19–32. doi: 10.1016/J.DEVCEL.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 83.Korn J., Christ B., Kurz H. Neuroectodermal origin of brain pericytes and vascular smooth muscle cells. J. Comp. Neurol. 2002;442:78–88. doi: 10.1002/CNE.1423. [DOI] [PubMed] [Google Scholar]
- 84.Etchevers H.C., Vincent C., Le Douarin N.M., Couly G.F. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. 2001;128:1059–1068. doi: 10.1242/DEV.128.7.1059. [DOI] [PubMed] [Google Scholar]
- 85.Shi X., Han W., Yamamoto H., Tang W., Lin X., Xiu R., Trune D.R., Nuttall A.L. The cochlear pericytes. Microcirculation. 2008;15:515–529. doi: 10.1080/10739680802047445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Neng L., Zhang W., Hassan A., Zemla M., Kachelmeier A., Fridberger A., Auer M., Shi X. Isolation and culture of endothelial cells, pericytes and perivascular resident macrophage-like melanocytes from the young mouse ear. Nat. Protoc. 2013;8:709–720. doi: 10.1038/NPROT.2013.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Minowa O., Ikeda K., Sugitani Y., Oshima T., Nakai S., Katori Y., Suzuki M., Furukawa M., Kawase T., Zheng Y., et al. Altered cochlear fibrocytes in a mouse model of DFN3 nonsyndromic deafness. Science. 1999;285:1408–1411. doi: 10.1126/SCIENCE.285.5432.1408. [DOI] [PubMed] [Google Scholar]
- 88.Nadeem T., Bogue W., Bigit B., Cuervo H. Deficiency of Notch signaling in pericytes results in arteriovenous malformations. JCI Insight. 2020;5 doi: 10.1172/JCI.INSIGHT.125940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fuxe J., Tabruyn S., Colton K., Zaid H., Adams A., Baluk P., Lashnits E., Morisada T., Le T., O’Brien S., et al. Pericyte requirement for anti-leak action of angiopoietin-1 and vascular remodeling in sustained inflammation. Am. J. Pathol. 2011;178:2897–2909. doi: 10.1016/J.AJPATH.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Driver E.C., Kelley M.W. Development of the cochlea. Development. 2020;147 doi: 10.1242/DEV.162263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li S., Nie E.H., Yin Y., Benowitz L.I., Tung S., Vinters H.V., Bahjat F.R., Stenzel-Poore M.P., Kawaguchi R., Coppola G., Carmichael S.T. GDF10 is a signal for axonal sprouting and functional recovery after stroke. Nat. Neurosci. 2015;18:1737–1745. doi: 10.1038/nn.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Memic F., Knoflach V., Morarach K., Sadler R., Laranjeira C., Hjerling-Leffler J., Sundström E., Pachnis V., Marklund U. Transcription and Signaling Regulators in Developing Neuronal Subtypes of Mouse and Human Enteric Nervous System. Gastroenterology. 2018;154:624–636. doi: 10.1053/J.GASTRO.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watanabe Y., Stanchina L., Lecerf L., Gacem N., Conidi A., Baral V., Pingault V., Huylebroeck D., Bondurand N. Differentiation of Mouse Enteric Nervous System Progenitor Cells Is Controlled by Endothelin 3 and Requires Regulation of Ednrb by SOX10 and ZEB2. Gastroenterology. 2017;152:1139–1150.e4. doi: 10.1053/J.GASTRO.2016.12.034. [DOI] [PubMed] [Google Scholar]
- 94.Bondurand N., Natarajan D., Barlow A., Thapar N., Pachnis V. Maintenance of mammalian enteric nervous system progenitors by SOX10 and endothelin 3 signalling. Development. 2006;133:2075–2086. doi: 10.1242/DEV.02375. [DOI] [PubMed] [Google Scholar]
- 95.Nakashima T., Suzuki T., Iwagaki T., Hibi T. Effects of anterior inferior cerebellar artery occlusion on cochlear blood flow – a comparison between laser-Doppler and microsphere methods. Hear. Res. 2001;162:85–90. doi: 10.1016/S0378-5955(01)00372-0. [DOI] [PubMed] [Google Scholar]
- 96.Gyo K. Vol. 3. 2013. Experimental Study of Transient Cochlear Ischemia as a Cause of Sudden Deafness; pp. 1–15.http://www.wjgnet.com/ [DOI] [Google Scholar]
- 97.Angelborg C., Axelsson A., Larsen H.C. Regional Blood Flow in the Rabbit Cochlea. Arch. Otolaryngol. 1984;110:297–300. doi: 10.1001/ARCHOTOL.1984.00800310021004. [DOI] [PubMed] [Google Scholar]
- 98.Swanson G.J., Howard M., Lewis J. Epithelial autonomy in the development of the inner ear of a bird embryo. Dev. Biol. 1990;137:243–257. doi: 10.1016/0012-1606(90)90251-D. [DOI] [PubMed] [Google Scholar]
- 99.Lahlou H., Nivet E., Lopez-Juarez A., Fontbonne A., Assou S., Zine A. Enriched Differentiation of Human Otic Sensory Progenitor Cells Derived From Induced Pluripotent Stem Cells. Front. Mol. Neurosci. 2018;11:452. doi: 10.3389/FNMOL.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gunewardene N., Bergen N.V., Crombie D., Needham K., Dottori M., Nayagam B.A. Directing human induced pluripotent stem cells into a neurosensory lineage for auditory neuron replacement. Biores. Open Access. 2014;3:162–175. doi: 10.1089/BIORES.2014.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ohnishi H., Skerleva D., Kitajiri S.I., Sakamoto T., Yamamoto N., Ito J., Nakagawa T. Limited hair cell induction from human induced pluripotent stem cells using a simple stepwise method. Neurosci. Lett. 2015;599:49–54. doi: 10.1016/J.NEULET.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 102.Mattei C., Lim R., Drury H., Nasr B., Li Z., Tadros M.A., D’Abaco G.M., Stok K.S., Nayagam B.A., Dottori M. Generation of Vestibular Tissue-Like Organoids From Human Pluripotent Stem Cells Using the Rotary Cell Culture System. Front. Cell Dev. Biol. 2019;7:25. doi: 10.3389/FCELL.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ronaghi M., Nasr M., Ealy M., Durruthy-Durruthy R., Waldhaus J., Diaz G.H., Joubert L.M., Oshima K., Heller S. Inner ear hair cell-like cells from human embryonic stem cells. Stem Cell. Dev. 2014;23:1275–1284. doi: 10.1089/SCD.2014.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koehler K.R., Nie J., Longworth-Mills E., Liu X.P., Lee J., Holt J.R., Hashino E. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol. 2017;35:583–589. doi: 10.1038/NBT.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van der Valk W.H., Steinhart M.R., Zhang J., Koehler K.R. Building inner ears: recent advances and future challenges for in vitro organoid systems. Cell Death Differ. 2020;28:24–34. doi: 10.1038/s41418-020-00678-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee J., Rabbani C.C., Gao H., Steinhart M.R., Woodruff B.M., Pflum Z.E., Kim A., Heller S., Liu Y., Shipchandler T.Z., Koehler K.R. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature. 2020;582:399–404. doi: 10.1038/S41586-020-2352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Taguchi A., Nishinakamura R. Higher-Order Kidney Organogenesis from Pluripotent Stem Cells. Cell Stem Cell. 2017;21:730–746.e6. doi: 10.1016/J.STEM.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 108.Sharma A., Sances S., Workman M.J., Svendsen C.N. Multi-lineage Human iPSC-Derived Platforms for Disease Modeling and Drug Discovery. Cell Stem Cell. 2020;26:309–329. doi: 10.1016/J.STEM.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Han L., Chaturvedi P., Kishimoto K., Koike H., Nasr T., Iwasawa K., Giesbrecht K., Witcher P.C., Eicher A., Haines L., et al. Single cell transcriptomics identifies a signaling network coordinating endoderm and mesoderm diversification during foregut organogenesis. Nat. Commun. 2020;11:4158. doi: 10.1038/S41467-020-17968-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jan T.A., Chai R., Sayyid Z.N., van Amerongen R., Xia A., Wang T., Sinkkonen S.T., Zeng Y.A., Levin J.R., Heller S., et al. Tympanic border cells are Wnt-responsive and can act as progenitors for postnatal mouse cochlear cells. Development (Camb.) 2013;140:1196–1206. doi: 10.1242/DEV.087528/-/DC1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/BIOINFORMATICS/BTS635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/J.CELL.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Draghici S., Khatri P., Tarca A.L., Amin K., Done A., Voichita C., Georgescu C., Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/GR.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/NBT.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McInnes L., Healy J., Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arxiv. 2018 doi: 10.48550/arXiv.1802.03426. Preprint at. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–O) Markers identified via differential analysis of each cluster against all other clusters via non-parametric Wilcoxon rank sum test. Each row represents identified marker gene ranked by p-value. Columns from left to right: p-value, log-scale fold change, fraction of cells expressing marker gene in selected cluster, fraction of cells expressing marker gene in all other clusters, adjusted p-value, cluster, and gene name.
(A–I) Markers identified via differential analysis of each cluster against all other clusters via non-parametric Wilcoxon rank sum test. Each row represents identified marker gene ranked by p-value. Columns from left to right: p-value, log-scale fold change, fraction of cells expressing marker gene in selected cluster, fraction of cells expressing marker gene in all other clusters, adjusted p-value, cluster, and gene name.
(A–F) Markers identified via differential analysis of each cluster against all other clusters via non-parametric Wilcoxon rank sum test. Each row represents identified marker gene ranked by p-value. Columns from left to right: p-value, log-scale fold change, fraction of cells expressing marker gene in selected cluster, fraction of cells expressing marker gene in all other clusters, adjusted p-value, cluster, and gene name.
(A–D) Markers identified via differential analysis of each cluster against all other clusters via non-parametric Wilcoxon rank sum test. Each row represents identified marker gene ranked by p-value. Columns from left to right: p-value, log-scale fold change, fraction of cells expressing marker gene in selected cluster, fraction of cells expressing marker gene in all other clusters, adjusted p-value, cluster, and gene name.
(A–O) Markers identified via differential analysis of each cluster against all other clusters via non-parametric Wilcoxon rank sum test. Each row represents identified marker gene ranked by p-value. Columns from left to right: p-value, log-scale fold change, fraction of cells expressing marker gene in selected cluster, fraction of cells expressing marker gene in all other clusters, adjusted p-value, cluster, and gene name.
(A and B) Rcistarget analysis identifying potential upstream regulators of each OMC subpopulation at P7 and E15. Each row represents an identified potential upstream motif, ranked by normalized enrichment score (NES). Columns from left to right: rank: rank of motif, logo: URL to motif’s genomic sequence, geneSet: OMC subpopulation, motif: ID of the motif, NES: normalized enrichment score of the motif in the geneSet, AUC: area under the curve (used to calculate the NES), TF_highConf: transcription factors annotated to the motif according to confidence annotation (two asterisks), TF_lowConf: transcription factors annotated to the motif according to confidence annotation (one asterisk), enrichedGenes: genes that are highly ranked for the given motif, nErnGenes: number of genes highly ranked, rankAtMax: ranking at the maximum enrichment, used to determine the number of enriched genes.
(A–D) Biological gene ontology analysis for P7 OMC subpopulations. Each row represents identified biological GO-term ranked by p-value. Columns from left to right: GO-term ID, GO-term name, number of DE genes in GO-term, number of overall genes in GO-term, and p-value.
Data Availability Statement
All data supporting the findings of this study are available within the article itself, supplemental information or available upon reasonable request. All scRNA-seq datasets generated for this manuscript have been deposited at GEO (GEO: GSE217727) and are publicly available as of the date of publication. This paper does not generate or report any original code. Any reasonable request for additional information required to reanalyze the data reported in this manuscript is available from the lead contact, Dr. Ronna Hertzano.