Abstract
Cruciferous-rich diets, particularly broccoli, have been associated with reduced risk of developing cancers of various sites, cardiovascular disease and type-2 diabetes. Sulforaphane (SF), a sulfur-containing broccoli-derived metabolite, has been identified as the major bioactive compound mediating these health benefits. Sulforaphane is a potent dietary activator of the transcription factor Nuclear factor erythroid-like 2 (NRF2), the master regulator of antioxidant cell capacity responsible for inducing cytoprotective genes, but its role in glucose homeostasis remains unclear. In this study, we set to test the hypothesis that SF regulates glucose metabolism and ameliorates glucose overload and its resulting oxidative stress by inducing NRF2 in human hepatoma HepG2 cells. HepG2 cells were exposed to varying glucose concentrations: basal (5.5 mM) and high glucose (25 mM), in the presence of physiological concentrations of SF (10 μM). SF upregulated the expression of glutathione (GSH) biosynthetic genes and significantly increased levels of reduced GSH. Labelled glucose and glutamine experiments to measure metabolic fluxes identified that SF increased intracellular utilisation of glycine and glutamate by redirecting the latter away from the TCA cycle and increased the import of cysteine from the media, likely to support glutathione synthesis. Furthermore, SF altered pathways generating NADPH, the necessary cofactor for oxidoreductase reactions, namely pentose phosphate pathway and 1C-metabolism, leading to the redirection of glucose away from glycolysis and towards PPP and of methionine towards methylation substrates. Finally, transcriptomic and targeted metabolomics LC-MS analysis of NRF2-KD HepG2 cells generated using CRISPR-Cas9 genome editing revealed that the above metabolic effects are mediated through NRF2. These results suggest that the antioxidant properties of cruciferous diets are intricately connected to their metabolic benefits.
Keywords: Sulforaphane, NRF2, Glucose, Glutathione, One-carbon (1C) metabolism, Methionine, Pentose phosphate pathway, NADPH, CRISPR-Cas9
Graphical abstract
Highlights
-
•
High glucose blocks reduced glutathione production, whilst SF reverses the effects.
-
•
To support glutathione biosynthesis, SF results in glycine, cysteine and glutamate consumption.
-
•
In high glucose SF alters NADPH-producing pathways (Pentose Phosphate Pathway and 1C-metabolism) to meet glutathione demand.
-
•
CRISPR-Cas 9 reveals that the metabolic effects induced by SF are mediated by NRF2.
1. Introduction
The liver is the central metabolic organ in the body whose function is to govern energy metabolism. The liver comprises four distinct cell types; hepatocytes, hepatic stellate cells, Kupfer and sinusoidal epithelial cells; the predominant cell type is hepatocytes which account for approximately 80% of the liver's mass [1]. Hepatocytes are involved in various cellular and metabolic functions such as protein synthesis and storage, catabolism of amino acids, carbohydrate metabolism, cholesterol synthesis, bile acids/salts and phospholipids, and the detoxification of xenobiotics [2]. As a result, hepatocytes play a fundamental role in metabolic homeostasis and detoxification.
Diets rich in fats and sugars have been implicated in increased oxidative stress [3]. Oxidative stress is caused by an imbalance between the production and accumulation of reactive oxygen species (ROS) [4] in cells and tissues [5], leading to the activation of the transcription factor NF-ƙB, resulting in the expression of the following cytokines: tumour necrosis factor-alpha (TNFα) and interleukin 6 (IL6), thereby promoting inflammation, along with cellular senescence via cellular damage [4,6,7].
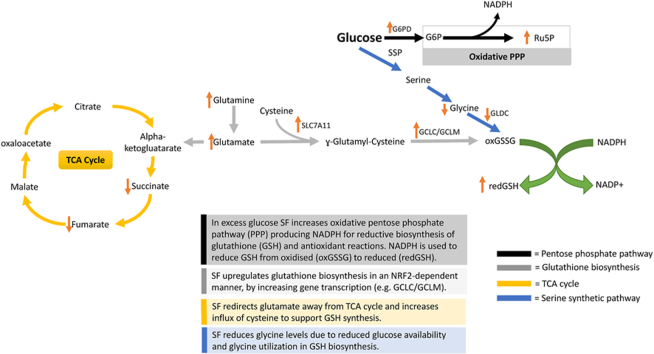
Broccoli belongs to the family of cruciferous vegetables or Brassicaceae (Cruciferae), which uniquely produce glucosinolates (GSLs) as a defence mechanism against pests and diseases [8]. Consumption of cruciferous vegetables, particularly broccoli, is associated with reduced cancer incidence and cardiovascular risk [8,9]. These benefits have been attributed to the breakdown products of GSLs, which in the case of broccoli is sulforaphane (SF), a highly bioactive dietary compound with chemopreventive, antioxidant and anti-inflammatory activities demonstrated in numerous in vitro and in vivo studies (ITCs) [10]. Although SF has demonstrable antioxidant effects, it cannot be considered a direct antioxidant, such as vitamin C; instead, it exerts its effect by strongly inducing an antioxidant response [10]. SF's gut formation leads it to be readily conjugated to glutathione, either passively or through the activity of glutathione S-transferases (GSTs) [11]. The glutathione conjugate is then exported to the systemic circulation, possibly via the multidrug resistance associated protein-1. The SF-glutathione conjugate is metabolised via the mercapturic acid pathway, which is finally excreted through the urine [12]. SF-GSH conjugate and other ITC thiol conjugates have been shown to inhibit phase I enzymes and induce phase II metabolism [12]. Indeed, SF is the most potent dietary activator of the nuclear factor erythroid 2-related factor transcription factor (NRF2) [13,14].
NRF2 is a master cellular regulator of oxidative stress, as it regulates the activity of several genes involved in scavenging free radicals [15,16]. Under normal conditions, NRF2 is inactive and bound to Kelch-like associated protein 1 (KEAP1) in the cytoplasm, ubiquitinated and sent for proteasomal degradation [17]. In the presence of electrophiles, SF interacts with sulfhydryl groups of cysteine residues of KEAP1, thereby releasing NRF2, where it binds to the sMaf protein, translocating to the nucleus and binding to the Antioxidant Response Element (ARE) present in the promoter of antioxidant response genes and inducing their transcription [18]. Among others, these include NADPH quinone oxidoreductase (NQO1), the glutathione biosynthetic genes (glutamate-cysteine ligase modifier and catalytic subunits; GCLM and GCLC) and thioredoxin reductase (TXNRD1) [15].
In addition to the antioxidant potential of broccoli diets, several human studies have recently reported SF as a potential modulator of metabolic health. Consumption of broccoli with increased GSL content reduced the levels of LDL cholesterol and attenuated genes involved in prostate cancer progression [[19], [20], [21]], and SF supplementation by itself improved glucose homeostasis in Type 2 diabetics [9]. However, the molecular mechanisms leading to such metabolic benefits have not been identified. Interestingly, the latest evidence now shows that NRF2 is involved in controlling the activity of several genes involved in central metabolism, such as glycolysis [22], TCA Cycle [23,24], Pentose Phosphate Pathway [25] and Lipid Metabolism [10,26]. This suggests for the first time that the metabolic benefits of broccoli-rich diets may also be mediated by the ability of SF to activate NRF2 potently.
In this study, we set to test the hypothesis that SF regulates glucose metabolism and ameliorates glucose overload and its resulting oxidative stress by inducing NRF2. We used a well-established hepatocellular carcinoma cell line, HepG2, as a liver model and exposed HepG2 to varying concentrations of glucose to induce oxidative stress ranging from basal (BG 5.5 mM) to high glucose (HG 25 mM). We measured the effects of dietary SF on whole-genome transcription and metabolism fluxes using a combination of metabolic phenotyping and stable isotope labelling experiments in wild-type and NRF2 knock-down cells generated by CRISPR-Cas9. The findings show that SF rewires central metabolism to support the antioxidant response in high glucose environments, demonstrating for the first time how the antioxidant properties of cruciferous diets are intricately connected to metabolic benefits.
2. Materials and methods
2.1. Preparation of sulforaphane
R, S-sulforaphane (LKT Laboratories #: S8044) was dissolved in 100% dimethyl sulfoxide (DMSO, Sigma #: D2650) to achieve a 5.5 M concentration a 100 mM master stock solution stored at −20 °C. 10 μL of the 100 mM stock solution was diluted to a 1 mM working solution. From the 1 mM solution, sulforaphane was diluted to final concentrations in the culture medium just before the addition to the cultures. The final concentration of DMSO in the culture was <0.01%, which is significantly lower than the 1% previously reported to have biochemical impact on sulfur metabolic pathways [27,28].
2.2. Cell culture
HepG2 cells, a human hepatocarcinoma cell line, were obtained from the American Type Culture Collection (ATCC, #: HB-8605). HepG2 was cultured in EMEM (Eagle's Minimum Essential Medium ATCC 30-2003) containing 10% fetal bovine serum (FBS) (Gibco #:10082147), 100 μg/ml-1 penicillin, and 100 μg/ml-1 streptomycin (Gibco #:15140122). Cells were maintained at 37 °C in a 5% CO2 environment.
2.3. RNA extraction and sequencing
The same RNA extraction protocol was used for the WT experiment under basal and high glucose, and the WT and NRF2-KD HepG2 cells were cultured in high glucose. Briefly, HepG2 cells were seeded on a 6-well plate (2 105 cells/well) and treated with 10 μM of SF or control (DMSO) in serum-free DMEM without glucose (Gibco #:11966025) supplemented with 1 mM sodium pyruvate (Gibco #:11360070), and 5.5 mM or 25 mM glucose (Gibco #: A2494001). RNA was extracted using the RNeasy Qiagen kit (QIAGEN #:74004) according to the manufacturer's instructions. RNA abundance and integrity were assessed using the Nanodrop ND-1000 spectrophotometer. RNA was sent to GeneWiz (Essex, United Kingdom). Both quality control and sequencing were carried out by GeneWiz through BioAnalyzer. Samples were then ribo depleted through the TruSeq RNA-Poly A (Illumina). Sequencing was performed on an Illumina NovaSeq 2x using 150-bp paired-end reads, generating 35 million reads/library.
The RNA-sequencing data analysis was divided into two steps: processing the raw RNA-sequencing data to estimate gene counts and then the comparative analysis of the transcriptomics data. Data processing followed the protocol for the "new Tuxedo" suite for short reads [29] and using the HPC environment managed by the Norwich Bioscience Institute's Computing infrastructure for Science, CiS [30]. Genes were aligned to the Human Reference Genome taken from Ensembl's release 97, GRCh38.p13" and used as reference the latest publication from Ensemble: https://doi.org/10.1093/nar/gkab1049.
Differential Expression analysis was performed using DESeq2 version 1.32.0 [25] using the raw gene counts with no replacement. Three to four biological replicates were used, and genes with less than ten counts were removed. For the statistical analysis, three samples were taken (as opposed to taking the average). Functional analysis was performed with the ranked list of expressed genes using the rank-rank hypergeometric overlap algorithm (RRHO) [31]. The RRHO algorithm compares two gene expression profiles as a ranked list, typically using the statistical results from the differential expression analysis. The algorithm uses the complete list of expressed genes instead of a subset of the top differentially expressed genes, increasing sensitivity to small but concordant changes. The motivation for using the whole gene expression profile is that groups with small but consistent changes tend to be discarded when taking only the top changing genes as representatives for genome-wide expression profiles. Considering all expressed genes allows for detecting enriched groups of related genes that would have been considered weakly differential on their own but with a concordant change in expression. The results of the differential expression test were represented as the list of expression changes ranked on the statistical significance of differential expression of basal glucose SF vs basal glucose control and high glucose SF vs high glucose control. The same genes were ranked using the (negative) -log10-transformed p-value multiplied by the sign of the log2-FC, where the sign denotes the change direction: positive for upregulated gene expression in samples and negative for downregulated expression in samples. The most significantly upregulated genes are at the top of the list, the most significantly downregulated genes are at the bottom, and those genes with small changes are in the middle.
The output is an ES which is then often normalised to the size of the gene set and in correlations between gene sets and the expression dataset [32]. The sign of the enrichment score indicates whether the pathway is upregulated (positive) or down-regulated (negative). If a set is not enriched to a significant q value, the signal in the dataset is not enough to make any claims for that particular set, i.e., there is not enough evidence of changes in that set due to the treatment. Positive NES indicates gene set enrichment mostly from upregulated genes (located at the top of the ranked list); a negative ES indicates gene set enrichment mostly from down-regulated genes (located at the bottom of the ranked list). Raw data is stored in ArrayExpress under accessions E-MTAB-12851 and E-MTAB-12858.
2.4. Seahorse Analyser
HepG2 cells were seeded at a density of 5 × 104 cells/well in six wells of the eight-well Seahorse plate and grown for 48 h. For the OCR assay, the medium was replaced with Seahorse XF DMEM Medium, pH 7.4, supplemented with 10 mM d-Glucose, 1 mM pyruvate and 2 mM l-Glutamine, and cell cultures were allowed to equilibrate for 1 h at 37 °C in a no-CO2 incubator. For the ECAR assay, the medium was replaced with Seahorse XF DMEM Medium, pH 7.4, supplemented with 4 mM l-Glutamine. Seahorse assays were performed according to the manufacturer's instructions, as reported in Pasquale et al. [33]. At the end of the assay, protein from each well was obtained for both the OCR and ECAR assays was extracted to normalise for variation in cell number. Briefly, cells were lysed with 1X lysis buffer (Cell Signalling Technology, #: 9803S), and lysates were centrifuged at 17,000 g at 4 °C for 10 min. Protein concentrations were then quantified using the bicinchoninic protein assay (Sigma #: BCA1-1 KT).
2.5. Stable isotope tracers/gas chromatography coupled to mass spectrometry (GC-MS)
For the 13C6 glucose assay, HepG2 cells were seeded at a density of 2 × 105 cells/incubated for a further 24 h with DMEM (Sigma, #: D5030) supplemented with 30.5 mg of 13C6 glucose (Sigma, #: 389374) for the BG environment or 140 mg of 13C6 glucose for the HG environment and 12C5 glutamine (Roth, #: HN08.2), and sodium bicarbonate (Sigma, #: S5761) with 10 μM of SF or control (DMSO) without serum. For the 13C5 glutamine assay, HepG2 cells were treated as described for the 13C6 glucose assay, except using 12C6 glucose and 12 mg of 13C5 glutamine (Sigma, #: 605166). For the 1-2-13C6 glucose assay, HepG2 cells were treated as described for the 13C6 glucose assay except using 1-2-13C6 glucose tracer (Sigma, #: 453188) and 12C5 glutamine.
Metabolites were extracted and quantified as previously described [34]. GC-MS chromatograms were processed using the in-house developed software, MetaboliteDetector, v3.320200313 [35]. Mass isotopomer distributions were calculated according to the chemical formulas from Wegner et al. [36].
2.6. Glutathione, pyroglutamic acid and amino acid analysis through liquid chromatography coupled to mass spectrometry (LC-MS)
HepG2 cells were seeded on a 6-well plate (2 105 cells/well). Metabolites were extracted according to Labuschange and colleagues [37]. Amino acid separation was carried out according to the chromatographic method described by Prinsen and colleagues after injection of 1 μL of the sample [38]. Separation of oxidised/reduced glutathione was separated using Luna Omega Polar 1.6 μM Polar C18 (Thermofisher) column, and the temperature for both columns was set to 30 °C. For the chromatographic separation gradient, mobile phases were used. Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in methanol.
For separating all compounds, the mobile phase gradient was started from 1% B for 2 min, 2% B for 4.1 min, 10% B for 5 min, 20% B for 6 min. After 1 min the column was washed up with 90% B and re-equilibrated for 2 min by 1% B. The flow rate was 0.1 ml/min. The LC eluent flow was sprayed into the mass spectrometer interface without splitting. All ions were monitored using mass spectrometry in multiple reaction monitoring modes (MRM) in positive polarity with an electrospray ionisation (ESI) source. The source parameters were: [1] gas temperature of 200 °C with a gas flow of 16 l/min, [2] sheath gas temperature of 300 °C with a sheath gas flow of 11 l/min, and nebuliser pressure of 50 psi and capillary voltage of 3500 °C. 4 Quantification of metabolites was carried out as described by Perez-Moral [39].
2.7. CRISPR-Cas 9 transfection
Cell transfection in a 6-well plate was performed using Lipofectamine CRISPRMAX (Invitrogen #: CMAX00003). HepG2 cells were seeded at 2X105 cells per ml well and transfected with 12.5 μg of GeneArt platinum Cas9 nuclease and 144 μL of Cas9 plus reagent or 12.5 μg of GeneArt platinum Cas9 nuclease, 144 μL of Cas9 plus reagent, and 2.5 μg of sgRNA for 9 h as previously described [40]. The percentage of locus-specific indel formation was measured by GeneArt® Genomic Cleavage Detection Kit (GCD), according to Yu and colleagues [40]. Band intensities were quantitated using the ImageJ software.
2.8. NRF2 immunoblotting
Following 72 h post-transfection, the supernatant from the transfected cells was removed, washed once with 1X DPBS, and trypsinised. Edited cells were transferred to a T75 flask and were left to expand for 2-3 weeks, constantly changing the media every three days. Once cells reached 80% confluency, cells were passaged as previously described and were seeded onto 6 well plates. Cell lysates were extracted with cell lysis buffer containing protease and phosphates inhibitors (pH 6.8) on ice for 10 min. Denatured samples (30 μg) were separated as previously described [41].
2.9. Quantitative RT-PCR
RNA isolated from WT and NRF2-KD HepG2 and quantified. 4 ng of RNA was utilised, and NQO1, G6PD and TKT mRNA was assessed by real-time qPCR (StepOnePlus) and normalised to the housekeeping gene β-actin.
2.10. Statistical analysis
Statistical analysis was carried out using Graph Pad Prism version (9.2.0) Results are represented as means ± SD from three to five biological replicates. Significance was assessed using either a paired Student t-test or two-way ANOVA followed by the Benjamini Hochberg test were appropriate, with significance confirmed by p <0.05.
3. Results
3.1. SF induces antioxidant as well as central metabolism genes in different metabolic states in the HepG2 cells
We first assessed the effect of exposing HepG2 to two different glucose environments, basal (BG; 5.5 mM) and high (HG; 25 mM), representing HepG2 cells in different cellular metabolic states through RNA sequencing. As expected, exposure to excessive glucose concentration led to HepG2 cells upregulating gene sets involved in proliferation, such as calcium (NES = 1.64, q=0.049), Hedgehog (NES=1.79, q=0.005) and Wnt (NES= 1.65, q=0.042) signalling along with Focal adhesion (NES= 1.80, q=0.005 and basal cell carcinoma (NES= 1.75, q=0.009) (Fig. S1A), thereby increasing its cancer phenotype. There were 157 genes that were changed by high glucose (q <0.05, Supplementary Table 1).
We next assessed the effects of SF in modulating hepatic metabolism transcription by exposing HepG2 to 10 μM SF, representing a physiological concentration [42]. We identified that SF resulted in more enriched gene sets in the HG environment, 14, compared to 9 in the BG environment (Fig. S1B). SF treatment in the BG environment resulted in transcriptional changes to 1657 genes, of which 691 were upregulated and 966 were downregulated (q<0.05, Supplementary Table 2). In the HG environment, 536 were upregulated, and 714 were downregulated (q<0.05, Supplementary Table 3).In the HG environment, SF led to a positive enrichment of Cytokine-Cytokine receptor interaction genes (normalised enrichment score (NES) 1.75, q=0.023) and Toll-like receptor signalling pathway (NES 1.76, q=0.023). In both glucose environments, SF upregulated genes involved in Glutathione metabolism (NES 1.81 in BG, q=0.016 and 2.05 in HG, q=0.001) and Metabolism of Xenobiotics by P450 (NES 1.73 in BG, q=0.041 and 1.80 in HG, q=0.021), key NRF2 targets, and proteasome metabolism (NES 1.93 in BG, q=0.009 and 2.03 in HG, q <0.0001). SF downregulated glycine, serine and threonine metabolism in both environments (NES -1.76 in BG, q=0.027 and −1.74 in HG, q=0.047), and One Carbon Pool by Folate was upregulated by SF, albeit only in the HG environment (1.72 in HG, q=0.026), key pathways in one carbon (1C) metabolism (Fig. S1B).
As expected, antioxidant and Phase II metabolism genes directly regulated by NRF2 were upregulated to a greater extent in the HG vs BG. There was an extensive upregulation in the glutathione biosynthetic gene, the modifier (GCLM) and the catalytic subunit (GCLC) greater than 4-fold in both glucose environments (Table 1.). SF treatment also resulted in a 4-fold increase in TXNRD1 compared to a 2-fold increase in basal glucose. TXNRD1 maintains thioredoxin (TXN) in the reduced state by reducing cystine to cysteine in the presence of NADPH. SF treatment also resulted in a 2.8-fold increase in the high glucose environment in glutathione peroxidase (GPX2) compared to a 2-fold increase in basal glucose, whose functions is involved in reducing hydrogen peroxide through reduced glutathione. Finally, SF treatment resulted in a 5.4-fold increase in basal and high glucose in sulfiredoxin (SRXN1) involved in reducing the cysteine-sulfinic acid formed under exposure to oxidants [43].
Table 1.
Comparison of differentially expressed genes in response to SF in BG and HG involved in glutathione metabolism and the antioxidant response.
| Gene Symbol |
Ensembl ID |
Gene |
Basal Glucose |
High Glucose |
||
|---|---|---|---|---|---|---|
| Folda Change | q-valueb | Folda Change | Qvalueb | |||
| GCLC | ENSG00000001084 | Glutamate-cysteine ligase catalytic subunit | 4.2 | <0.001 | 4.4 | <0.001 |
| GCLM | ENSG00000023909 | Glutamate-cysteine ligase regulatory subunit | 6.0 | <0.001 | 5.0 | <0.001 |
| GLRX | ENSG00000173221 | Glutaredoxin-1 | 1.3 | 0.04 | 1.8 | <0.0001 |
| GPX2 | ENSG00000176153 | Glutathione Peroxidase 2 | 2.0 | 0.04 | 2.8 | 0.007 |
| GSR | ENSG00000104687 | Glutathione reductase | 2.3 | <0.001 | 2.3 | <0.001 |
| SLC7A11 | ENSG00000151012 | Cysteine/Glutamate transporter | 2.7 | 0.04 | 2.7 | 0.007 |
| SRXN1 | ENSG00000271303 | Sulfiredoxin 1 | 5.4 | <0.001 | 5.4 | <0.001 |
| TXN | ENSG00000136810 | Thioredoxin | 1.8 | <0.001 | 1.9 | 0.001 |
| TXNRD1 | ENSG00000198431 | Thioredoxin reductase 1 | 2.0 | <0.001 | 4.6 | 0.002 |
Fold change of SF treated cells compared to DMSO control.
q-values were determined through Benjamini-Hochberg p-value adjustment
3.2. SF results in increased reduced glutathione in the high but not basal glucose environment in HepG2 cells
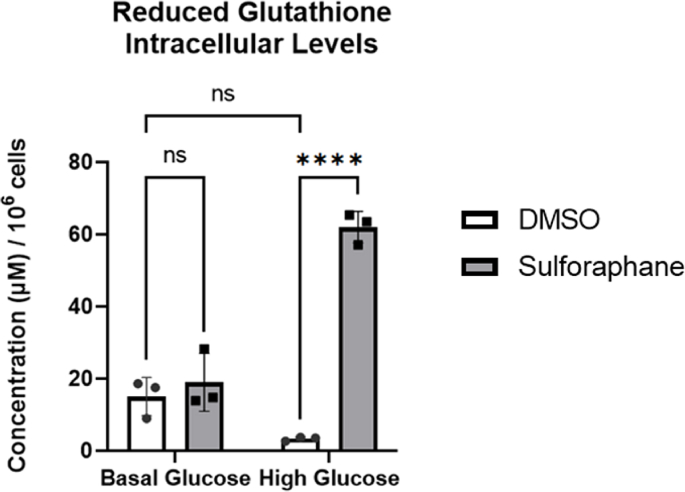
Glutathione is a potent antioxidant capable of preventing damage to cellular components caused by ROS, such as free radicals, peroxides and lipid peroxides [44]. We used LC-MS to assess reduced glutathione intracellular concentration levels, as the former indicates the cellular antioxidant capacity. Using LC-MS, we showed that HG impaired the cell's ability to accumulate reduced glutathione. In contrast, SF treatment resulted in a 20-fold increase from 3 μM to 61 μM (Fig. 1A). Interestingly, when cells are not metabolically stressed, such as at BG levels, SF did not affect intracellular concentration of reduced glutathione (BG control 15 μM compared to BG SF 18 μM) (Fig. 1A). We also identified that the intracellular concentration of oxidised glutathione was unaffected by either the glucose environment or SF treatment and was almost undetected (data not shown), suggesting that the increase in reduced glutathione might be linked to increased glutathione synthesis.
Fig. 1.
SF increases reduced glutathione in HepG2 cells.
HepG2 cells were treated with 10 µM SF for 24 h in basal (5.5 mM) and high glucose (25 mM). After 24 h, reduced glutathione intracellular levels were quantified. DMSO BG vs SF BG p= 0.8, DMSO BG vs DMSO HG p= 0.13, and DMSO HG vs SF HG p<0.0001. Metabolites were normalised to the total number of cells. Values are mean ± SD. Results from in vitro experiments are representative of n= 3 biological replicates.
3.3. SF redirects glutamine towards glutathione biosynthesis in HepG2 cells
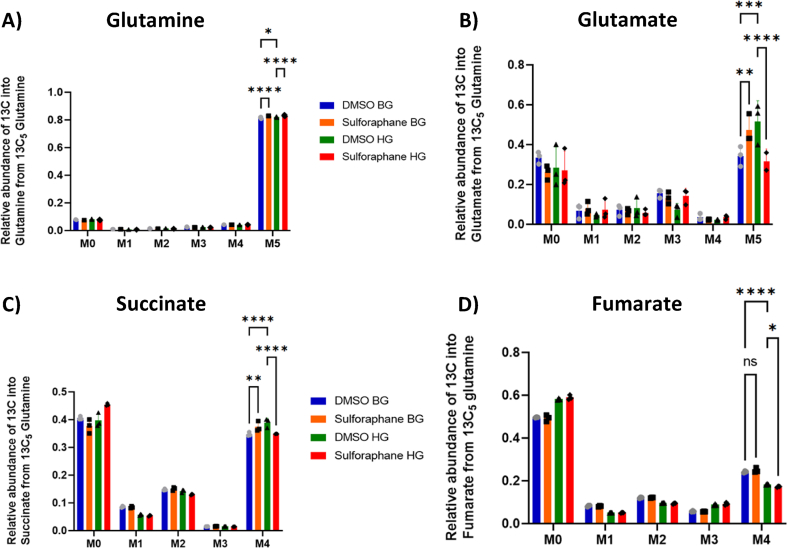
As reduced glutathione levels increased in the presence of SF, we next assessed the presence of glutamate, cysteine, and glycine, critical substrates required for glutathione biosynthesis. Glutamate is derived from glutamine and can act as an energy source by entering the TCA cycle [45]. Using LC-MS, we identified that SF significantly increased the intracellular concentration of glutamine in the HG environment from 27 μM to 46 μM (p<0.017 Fig. S2). In contrast, intracellular glutamate was significantly decreased, likely suggesting glutamate being consumed in downstream pathways (p<0.04, Fig. S2).
To test this and assess whether SF affected the glutamine flux towards glutathione, we cultured HepG2 in both glucose environments with a fully labelled glutamine tracer as the only glutamine source in the culture media. The glutamine tracer can be converted through glutaminase to M5 glutamate (all five carbon atoms labelled). Glutamate can then feed into the TCA cycle through glutamate dehydrogenase, resulting in M5 α-ketoglutarate (also referred to as 2-oxoglutarate), or be redirected towards other metabolic pathways. The abundance of glutamine, glutamate and the TCA metabolites succinate and fumarate were quantified using GC-MS. SF in both glucose environments increased glutamine flux in the cell M5 glutamine, followed by a decrease in M5 glutamate only in the high glucose environment (Fig. 2 A-B). The tracer also revealed a profound reduction in M4 succinate (four carbon atoms labelled due to releasing a carbon dioxide molecule during the reaction) and a decrease in M4 fumarate in the HG environment (Fig. 2 C-D).
Fig. 2.
SF affects glutamine metabolism, and TCA cycle intermediates in HepG2 cells.
HepG2 cells were treated with 10 μM SF for 24 h in the presence of the fully labelled glutamine tracer (13C5) in basal glucose (BG, 5.5 mM) or high glucose (HG, 25 mM). After 24 h, metabolites were extracted and quantified using GC-MS. (A) Labelling pattern of glutamine, (B) Labelling pattern of glutamate, (C) Labelling pattern of succinate, and (D) Labelling of fumarate. All values are expressed as mean ± SD. The x-axis M0 to M5 for glutamine and M0 to M4 for succinate and fumarate represent the mass isotopomer distribution (MID). This represents the incorporation of the isotope into the metabolite. M0 means that all the carbon atoms in the metabolite are from carbon-12, whereas M + n depicts that n carbon atoms are isotopes from the used tracer. The MID represents the relative abundances of M+0 to M + n isotopologues for each metabolite. Consequently, the sum of all fractions from M+0 to M + n is 100% or 1. M5 glutamine: DMSO BG vs SF BG q<0.0001, DMSO BG vs DMSO HG q=0.01, and DMSO HG vs DMSO SF q<0.0001. M5 glutamate: DMSO BG vs SF BG q=0.0021, DMSO BG vs DMSO HG q<0.0001, and DMSO HG vs SF HG q<0.0001. M4 succinate: DMSO BG vs SF BG q=0.0026, DMSO BG vs DMSO HG q<0.0001, and DMSO HG vs SF HG q<0.0001. M4 fumarate: DMSO BG vs SF BG q=0.14, DMSO BG vs DMSO HG q<0.0001, and DMSO HG vs SF HG q=0.02. Values are mean ± SD. Results from in vitro experiments are representative of n= 3 biological replicates.
Consistent with the substrate redirection, we showed that SF did not affect the mitochondrial function in BG as measured by the Seahorse Analyser. In contrast, SF-treated cells in HG demonstrated significantly (p=0.02) diminished maximal capacity of mitochondria activity (Fig. S2). Similarly, SF treated cells had reduced ATP production in the HG but not the BG environment (Fig. S2). Taken together, these results suggest that SF in the HG environment reduces glutamine flux into the tricarboxylic acid (TCA) cycle, leading to reduced capacity for maximal respiration, and instead redirects glutamine towards other metabolic pathways, potentially glutathione biosynthesis.
3.4. SF affects cysteine metabolism in HepG2 cells
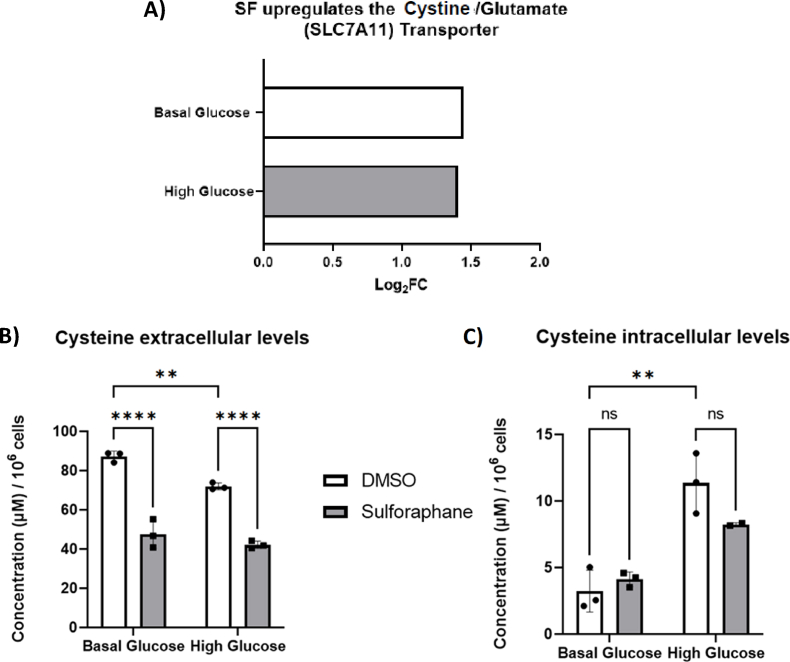
We next focused on assessing how SF affects cysteine metabolism. From the RNA sequencing analysis, SF treatment significantly increased the cystine/glutamate transporter expression (SLC7A11) expression by 2.5-fold in both glucose environments (Fig. 3A). SF treatment resulted in a significant cysteine reduction in the extracellular concentration (media) in both glucose environments (Fig. 3B), whereas levels of intracellular cysteine remained unchanged. Together, these results suggest an increased influx of cysteine which does not accumulate intracellularly but is likely utilised in subsequent pathways, namely glutathione biosynthesis and potentially through the backflow of the Transsulfuration pathway.
Fig. 3.
SF affects cysteine metabolism in HepG2 cells.
HepG2 cells were treated with 10 μM SF for 24 h in basal (5.5 mM) and high glucose (25 mM). After 24 h, metabolites were extracted and quantified using the LC-MS TripleQuad 6490 Agilent. (A) Whole transcriptome analysis on HepG2 looking at the cystine receptor q<0.05 corrected using the Benjamini Hochberg. (B) Cysteine extracellular levels: DMSO BG vs SF BG p<0.0001, DMSO BG vs DMSO HG p=0.077, and DMSO HG vs SF HG p<0.0001. (C) Cysteine intracellular levels: DMSO BG vs SF BG p=0.51, DMSO BG vs DMSO HG p=0.0014, and DMSO HG vs SF HG p=0.15. Metabolites were normalised to the number of cells. Values are mean ± SD. Results from in vitro experiments are representative of n= 3 biological replicates.
3.5. SF affects one carbon (1C) metabolism in HepG2 cells
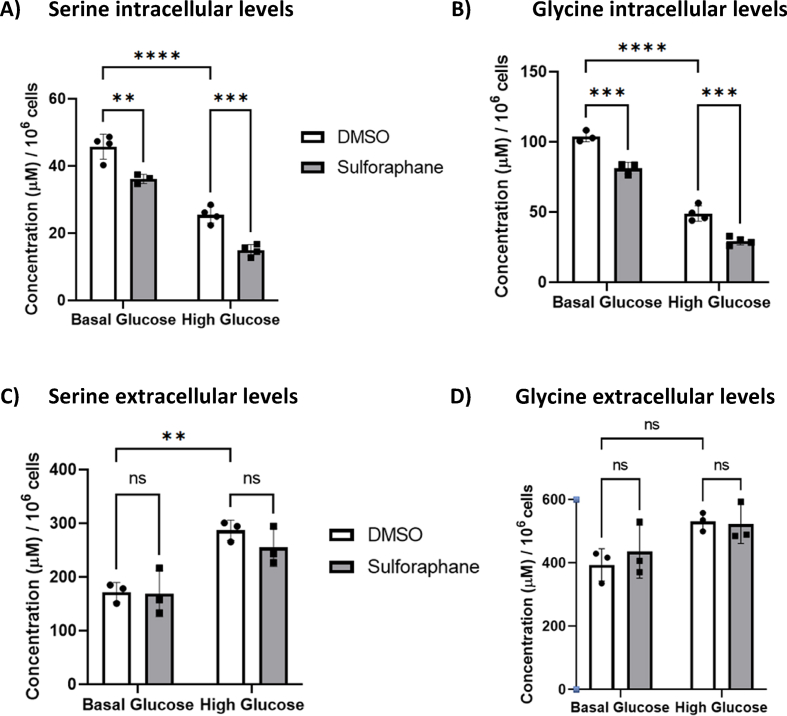
We next quantified glycine and serine and found that SF decreased both the serine and glycine intracellular pool (Fig. 4. A, B) without affecting their extracellular levels [46], suggesting that the observed decrease in serine and glycine could be from a reduction in serine biosynthesis and/or increased intracellular utilisation, likely as a substrate for glutathione biosynthesis.
Fig. 4.
SF affects serine and glycine levels in HepG2 cells.
HepG2 cells were treated with 10 μM SF for 24 h in basal (5.5 mM) and high glucose (25 mM). After 24 h, metabolites were extracted and quantified using the LC-MS TripleQuad 6490 Agilent. (A) Serine intracellular levels: DMSO BG vs SF BG p=0.0024, DMSO BG vs DMSO HG p<0.0001, and DMSO HG vs SF HG p=0.0006. (B) Glycine intracellular levels: DMSO BG vs SF BG p=0.0003, DMSO BG vs DMSO HG p<0.0001, and DMSO HG vs SF HG p=0.0003. (C) Serine extracellular levels: DMSO BG vs SF BG p=0.99, DMSO BG vs DMSO HG p=0.0075, and DMSO HG vs SF HG p=0.6. (D) Glycine extracellular levels: DMSO BG vs SF BG p= 0.82, DMSO BG vs DMSO HG p= 0.08, and DMSO HG vs SF HG p=0.99. Values are mean ± SD. Results from in vitro experiments are representative of n= 4 biological replicates.
SF also downregulated the first and the rate-limiting step in serine biosynthesis, phosphoglycerate dehydrogenase (PHGDH), in both glucose environments by about 1.5 fold. In addition, SF downregulated the expression of two genes involved in the catabolism of glycine: glycine decarboxylase (GLDC) by 1.4 and 1.3 fold in the BG and HG environment, respectively, which catalyses glycine degradation to H-protein-S-aminomethyldihydrolipoyllysine and carbon dioxide (Table 2).
Table 2.
Comparison of differentially expressed genes in response to SF in BG and HG involved in 1C Metabolism, Pentose Phosphate Pathway and Glycolysis.
| Gene Symbol |
Ensembl ID |
Gene |
Basal Glucose |
High Glucose |
||
|---|---|---|---|---|---|---|
| Folda Change | q-valueb | Folda Change | Qvalueb | |||
| PHGDH | ENSG00000092621 | Phosphoglycerate dehydrogenase | −1.7 | 0.005 | −1.6 | 0.01 |
| GLDC | ENSG00000178445 | Glycine decarboxylase | −1.4 | 0.005 | −1.3 | 0.01 |
| GNMT | ENSG00000124713 | Glycine-N-methyltransferase | −7.0 | 0.001 | 3.0 | 0.03 |
| ALDH1L1 | ENSG00000144908 | 10-formyltetrahydrofolate dehydrogenase | 1.9 | <0.001 | 2.3 | <0.001 |
| MTHFD1L | ENSG00000120254 | monofunctional C1-tetrahydrofolate | 1.3 | 0.01 | 1.3 | 0.04 |
| BHMT2 | ENSG00000132840 | betaine--homocysteine S-methyltransferase 2 | 3.6 | <0.001 | 3.2 | <0.001 |
| G6PD | ENSG00000160211 | Glucose-6-Phosphate Dehydrogenase | 1.2 | 0.3 | 1.6 | 0.01 |
| TKT | ENSG00000163931 | Transketolase | 1.5 | 0.02 | 1.7 | 0.005 |
| TALDO1 | ENSG00000177156 | Transaldolase 1 | 2.0 | <0.001 | 2.2 | <0.001 |
| ENO3 | ENSG00000108515 | Enolase | −2.3 | <0.001 | −1.7 | 0.01 |
| PGM1 | ENSG00000079739 | Phosphoglucomutase-1 | −1.5 | 0.004 | −1.4 | 0.01 |
| GCKR | ENSG00000084734 | Glucose kinase regulatory protein | 2.4 | <0.001 | 2.4 | <0.001 |
Fold change of SF treated cells compared to DMSO control.
q-values were determined through Benjamini-Hochberg p-value adjustment
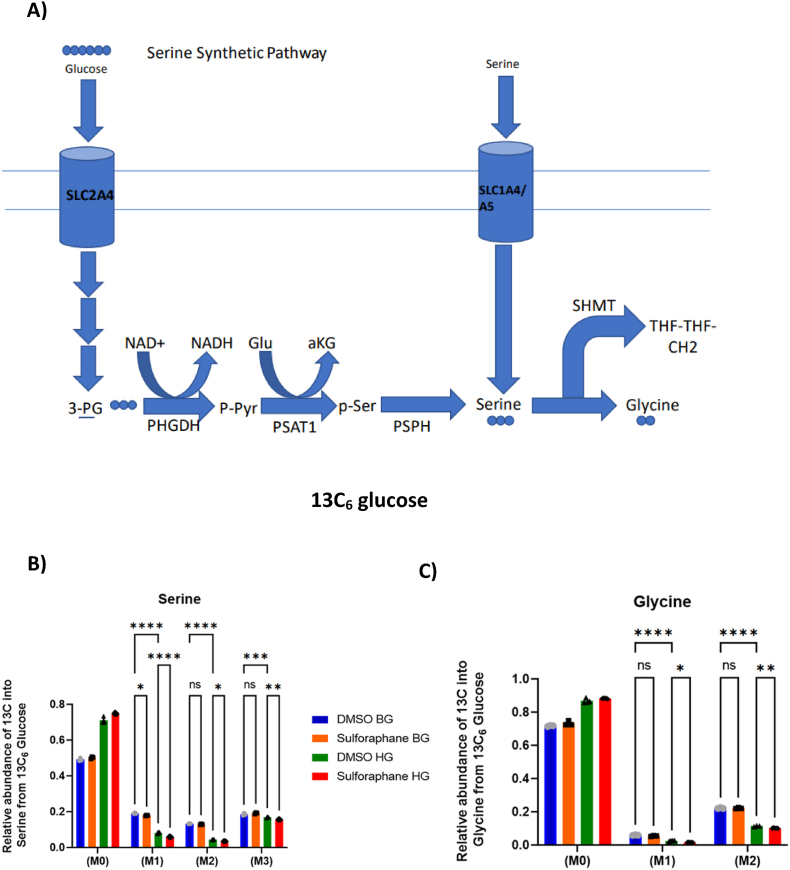
To better understand how serine and glycine are utilised intracellularly in the presence of SF, we cultured HepG2 in both glucose environments with a fully labelled glucose tracer [U13C6] as the only glucose source in the culture media. This tracer is catabolised via the glycolytic intermediate 3-phosphoglycerate to M3 serine and, subsequently, to M2 glycine through the combined action of the serine synthetic pathway (SSP) and serine hydroxymethyltransferase (SHTM1/2). A summary of how serine is derived from glucose through the serine synthetic pathway is shown in (Fig. 5A).
Fig. 5.
SF affects serine and glycine metabolism in HepG2 cells.
(A) Summary of serine biosynthesis from glucose through the serine synthetic pathway. HepG2 cells were treated with 10 μM SF for 24 h in basal glucose (5.5 mM) or high glucose (25 mM). After 24 h, metabolites were extracted and quantified using GC-MS. (B) Serine labelling from the 13C6glucose tracer. M3 serine BG DMSO vs BG SF q=0.20, BG DMSO vs HG DMSO q=0.0007, HG DMSO vs HG SF q=0.0078. M2 Serine BG DMSO vs BG SF q=0.42, BG DMSO vs HG DMSO q<0.0001, HG DMSO vs HG SF q=0.02. M1 serine BG DMSO vs BG SF q=0.01, BG DMSO vs HG DMSO q<0.0001, HG DMSO vs HG SF q<0.0001. (C) Glycine labelling from the 13C6glucose tracer. M2 Glycine BG DMSO vs BG SF q=0.93, BG DMSO vs HG DMSO q<0.001 HG DMSO vs HG SF q=0.0093, M1 Glycine BG DMSO vs BF SF q =0.21, BG DMSO vs HG DMSO q<0.0001, HG DMSO vs HG SF q=0.03. Values are mean ± SD. Results from in vitro experiments are representative of n=5 biological replicates.
SF had no effect in labelled serine in BG except for M1 serine but led to a significant reduction across M1, M2 and M3 serine in HG, indicating a decreased serine biosynthesis from glucose (M3) and reduced activity of SHMT (M1, M2), which is further confirmed in reduced fractions of M1 and M2 glycine isotopologues (Fig. 5B and C).
3.6. SF affects the methionine cycle in HepG2 cells
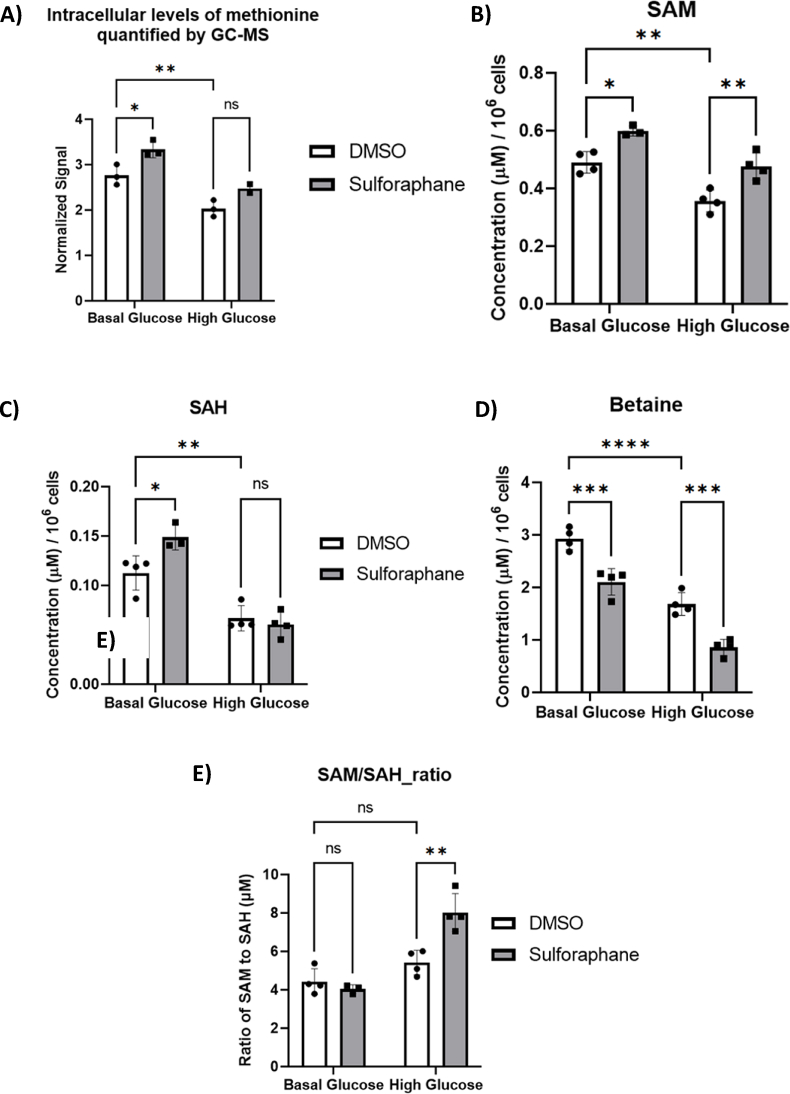
We next measured methionine cycle metabolites and found that firstly SF increased the methionine pool in BG but not significantly in HG (Fig. 6A). These methionine levels were also correlated with increased SAM levels, the first step in the methionine cycle (Fig. 6B). SAM is used as a methyl donor for methylation reactions in the cell, yielding SAH as a byproduct, which was not changed in response to SF in the HG environment (Fig. 6C). Finally, we identified that in the HG environment, SF significantly increased the SAM/SAH ratio (Fig. 6E).
Fig. 6.
SF affects methionine metabolism in HepG2 cells.
HepG2 cells were treated with 10 μM SF for 24 h in basal glucose (5.5 mM) or high glucose (25 mM). After 24 h, metabolites were extracted and quantified using GC-MS and LC-MS. (A) Methionine: BG DMSO vs BG SF p=0.031 and HG DMSO vs HG SF p=0.13. (B) S-Adenosyl methionine (SAM): BG DMSO vs BG SF p= 0.0094, BG DMSO vs HG DMSO p=0.0012, and HG DMSO vs HG SF p=0.0019. (C) S-Adenosyl homocysteine (SAH): BG DMSO vs BG SF p= 0.028, BG DMSO vs HG DMSO p= 0.037, and HG DMSO vs HG SF p=0.92. (D) Betaine: BG DMSO vs BG SF p<0.0001, BG DMSO vs HG DMSO p<0.0001, and HG DMSO vs HG SF p=0.0006. (E) SAM to SAH ratio: BG DMSO vs BG SF p=0.891, BG DMSO vs HG DMSO p=0.261, and HG DMSO vs HG SF p=0.0016. Values are mean ± SD. Results from in vitro experiments are representative of n= 4 biological replicates.
We next sought to identify the source of methyl groups to support the increased methionine levels in response to SF. 5-methyl tetrahydrofolate [47] is the primary methyl donor for the reaction that remethylates homocysteine into methionine and is produced in the folate cycle via the transfer of the serine methyl group to THF. Using the [U13C6] glucose tracer, we identified that most of the intracellular methionine was unlabelled (data not shown), suggesting 5-methyl THF is not the methyl donor and the methyl group from serine is transferred to THF for purine rather than methionine biosynthesis. Betaine is an alternative methyl donor for methionine, an essential cofactor for converting homocysteine back to methionine in the methionine cycle. Interestingly, SF decreased betaine (Fig. 6D), making it a likely methyl source for the production of methionine in response to SF. In support of this, there was a prominent transcriptional upregulation in the BHMT2 betaine homocysteine methyl transferase 2, by 3.6 and 3.2 fold in the BG and HG environment, respectively, a gene that transfers methyl groups from betaine to homocysteine to regenerate methionine (Table 2.).
3.7. SF redirects glucose towards the pentose phosphate pathway in the high but not basal glucose in HepG2 cells
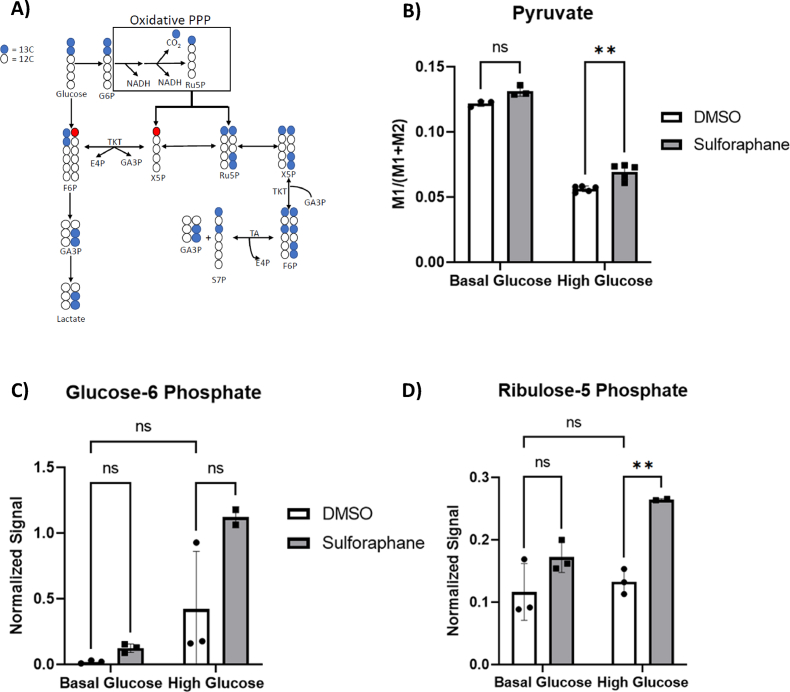
We next sought to test whether the reduction in glucose flux through the serine synthesis pathway was due to the glucose being redirected towards the PPP [24] and used a [1,213C2] glucose tracer to test this. The glucose catabolism through the oxidative PPP leads to M1 ribulose-5-phosphate (Ru5P) accumulation, which can finally be converted to M1 pyruvate and M1 lactate via the reductive PPP and LDH activity. On the other hand, glycolysis generates M2 lactate (Fig. 7A). The fraction of M1 lactate in relation to the sum of M1 and M2 lactate and pyruvate indicates oxidative PPP activity. Notably, SF increased oxidative PPP activity in HG but not BG, identified by a significant increase in M1 pyruvate (Fig. 7B). In addition, in the HG environment, SF treatment led to a significant increase in Ru5P, the end product of the oxidative phase of the PPP (p<0.01) whilst the increase of glucose-6-phosphate was insignificant (G6P, p=0.06) (Fig. 7C and D). In addition, HepG2 cells treated with SF demonstrated reduced glycolytic activity measured by the glycolysis stress test using the Seahorse XPF Analyser (Fig. S3). The data suggest a reduction in glucose flux and likely redirection in the PPP pathway.
Fig. 7.
SF affects glycolysis and PPP in HepG2 cells.
(A) Summary of how the 1-2-13C2 glucose tracer is metabolised to assess glycolysis and PPP activity. Following the phosphorylation of glucose through hexokinase to generate glucose-6 phosphate (G6P), the labelled G6P molecule can continue glycolysis, producing M2 lactate and pyruvate, or if it is metabolised through the oxidative phase of PPP to generate M1 ribulose 5-phosphate (Ru5P), the end product of the oxidative branch of the PPP. Blue circles represent the labelled 13C carbon, whilst red circles represent the labelled 13C carbon derived from the non-oxidative branch of the PPP. (B) HepG2 cells were treated with 10 μM SF for 24 h in basal glucose (5.5 mM) or high glucose (25 mM). After 24 h, metabolites were extracted and quantified using GC-MS. Pyruvate labelling through the 1–2 13C6tracer: BG DMSO vs BG SF p =0.06, BG DMSO vs HG DMSO p<0.0001, HG DMSO vs HG SF p=0.0014. (C) Glucose-6 phosphate quantification from the untargeted metabolomics: BG DMSO vs BG SF p =0.95, BG DMSO vs HG DMSO p=0.25, HG DMSO vs HG SF p=0.06. (D) Ribulose-5 phosphate quantification from the untargeted metabolomics: BG DMSO vs BG SF p =0.19, BG DMSO vs HG DMSO p=0.90, HG DMSO vs HG SF p=0.0077. Values are mean ± SD. Results from in vitro experiments are representative of n= 3 biological replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Consistent with this observation, SF significantly upregulated the expression of the rate-limiting enzyme glucose-6-phosphate dehydrogenase (G6PD) in the PPP in the HG environment only by 1.6 fold, and upregulated expression of transketolase (TKT) and transaldolase (TALDO), both involved in the non-oxidative branch of the PPP, 1.7-fold and 2.2-fold in HG cells (Table 2.).
We also showed that SF downregulated key glycolytic genes enolase 3 (ENO3, BG DMSO vs BG SF q= 0.0006, HG DMSO vs HG SF q=0.01) and phosphoglucomutase-1 (PGM1, BG DMSO vs BG SF q=0.003, HG DMSO vs HG SF q=0.02) and upregulated expression of glucose kinase regulatory protein (GCKR), involved in binding and controlling the function of glucokinase, the first gene in glycolysis. GCKR phosphorylates glucose and allows it to be either broken down through glycolysis or redirected towards the PPP (Table 2.).
3.8. CRISPR-Cas9 reveals that NRF2 mediates the metabolic effects of SF
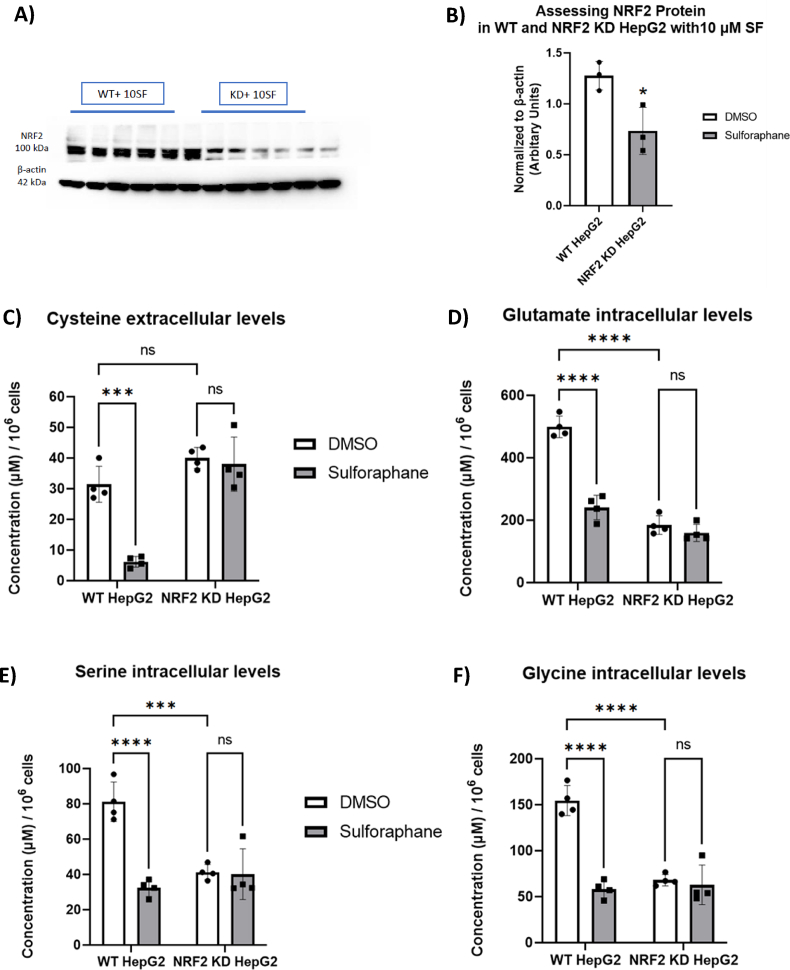
We utilised the genome editing technique CRISPR-Cas9 to develop a novel hepatic cell line with substantially impaired NRF2 activity. We first assessed the efficiency of NRF2 editing by stimulating WT and NRF2-KD HepG2 cells with 10 μM SF. There was a 60% reduction in NRF2 protein; notably, the lower band of the NRF2 was deleted entirely, which suggests that the editing resulted in a truncated NRF2 protein (Fig. 8A and B).
Fig. 8.
SF effects on amino acids related to glutathione metabolism are altered in NRF2 KD HepG2 cells. (A) Western blot analysis of NRF2 protein in NRF2-KD HepG2 treated with 10 μM SF overnight. sVisualised protein lysates. (B) Band Intensity was quantified using Fiji, and NRF2 was normalised to beta-actin. p=0.03. (C) Cysteine extracellular levels WT DMSO vs WT SF p= 0.0002, WT DMSO vs NRF2-KD DMSO p=0.19, and NRF2-KD DMSO vs NRF2-KD SF p=0.95. (75) Glutamate intracellular levels WT DMSO vs WT SF p<0.0001, WT DMSO vs NRF2-KD DMSO p<0.0001, and NRF2-KD DMSO vs NRF2-KD SF p=0.70.(E) Serine intracellular levels, WT DMSO vs WT SF p<0.0001, WT DMSO vs NRF2-KD DMSO p=0.0004, and NRF2-KD DMSO vs NRF2-KD SF p=0.99. (F) Glycine intracellular levels WT DMSO vs WT SF p<0.0001, WT DMSO vs NRF2-KD DMSO p<0.0001, and NRF2-KD DMSO vs NRF2-KD SF p=0.95. Values are mean ± SD. Results from in vitro experiments are representative of n= 4 biological replicates.
We first looked at downstream NRF2 target genes, known to have an ARE sequence, including NQO1, an essential Phase II gene, and two genes in the PPP, G6PD and TKT, under the HG environment, as that is where we observed SF increasing reduced glutathione and interfering with 1C metabolism. SF treatment in NRF2-KD cells attenuated NQO1 induction (1.5-fold increase in NRF2-KD cells vs 5-fold upregulation in WT; (Fig. S4B), and failed to induce expression of G6PD and TKT in NRF2-KD, compared to a 1.5 and 2-fold increase in WT, respectively (Figs. S4C–D).
In contrast to the NRF2 wild-type cells, SF did not affect extracellular cysteine or the intracellular glutamate, serine and glycine levels in NRF2-KD cells (Fig. 8 C–F). Interestingly, untreated NRF2-KD cells had significantly lower serine, glycine and glutamate levels, suggesting that NRF2 plays a crucial role in regulating baseline serine/glycine/glutamate levels.
We also identified that for betaine but not methionine, the NRF2-KD cells had higher levels than WT cells, suggesting that the regulation of betaine is independent of NRF2. SF treatment for the WT cells significantly reduced methionine and betaine, but the effect was abolished in the NRF2-KD cells (Figs. S4E–F).
As expected, SF affected transcription increasingly over time in WT cells, as seen by the separation of the control and SF-treated samples (Fig. S4G). However, this was abolished when NRF2-KD cells were exposed to SF (Fig. S4H), suggesting that NRF2 mediates the overwhelming majority of gene induction by SF.
NRF2 editing affected the ability of SF to induce genes involved in Glutathione Metabolism (NES WT 1.8, q=0.008 vs 1.1 NRF2 KD, q=0.57), Metabolism of Xenobiotics by Cytochrome P450 (NES WT 1.83, q=0.006 vs 1.2 NRF2-KD, q=0.53) and PPP (NES WT 1.73, q=0.004 vs 0.72 NRF2-KD, q=0.91) (Table 3, Fig. S4I). Glycine, Serine and Threonine metabolism pathway was enriched despite NRF2 status, although to a lesser extent in NRF2KD cells (NES WT -1.79, q=0.03 vs −1.63 NRF2-KD, q=0.07) (Fig. S4I).
Table 3.
Comparison of differentially expressed genes in response to SF in the WT and NRF2-KD HepG2 cell lines involved in the antioxidant response, PPP, glycolysis, glycine serine and threonine metabolism.
| Gene Symbol |
Ensembl ID |
Gene |
Wild type |
NRF2KD |
||
|---|---|---|---|---|---|---|
| Folda Change | q-valueb | Folda Change | Qvalueb | |||
| GCLC | ENSG00000001084 | Glutamate-cysteine ligase catalytic subunit | 3.2 | <0.001 | 1.3 | <0.001 |
| GCLM | ENSG00000023909 | Glutamate-cysteine ligase regulatory subunit | 4.1 | <0.001 | 1.4 | <0.001 |
| SLC7A11 | ENSG00000151012 | Cysteine/Glutamate transporter | 3.3 | <0.001 | 1.6 | <0.001 |
| GPX2 | ENSG00000176153 | Glutathione Peroxidase 2 | 2.0 | <0.001 | 1.0 | 0.92 |
| GSR | ENSG00000104687 | Glutathione Reductase | 2.2 | <0.001 | 1.2 | <0.001 |
| TXN | ENSG00000136810 | Thioredoxin | 2.0 | <0.001 | 1.0 | 0.99 |
| TXNRD1 | ENSG00000198431 | Thioredoxin reductase 1 | 4.3 | <0.001 | 1.4 | <0.001 |
| G6PD | ENSG00000160211 | Glucose-6-Phosphate Dehydrogenase | 1.5 | <0.001 | 1.0 | 0.60 |
| PGD | ENSG00000142657 | Phosphogluconate dehydrogenase | 1.5 | <0.001 | 1.0 | 0.94 |
| RPE | ENSG00000197713 | ribulose-5-phosphate-3-epimerase | 1.2 | <0.001 | 1.0 | 0.01 |
| TALDO1 | ENSG00000177156 | Transaldolase 1 | 2.2 | <0.001 | 1.1 | <0.001 |
| TKT | ENSG00000163931 | Transketolase | 1.7 | <0.001 | 1.1 | 0.41 |
| GPI | ENSG00000105220 | Glucose Phosphate Isomerase | 1.3 | <0.001 | 1.0 | <0.001 |
| PFKP | ENSG00000067057 | Phospho Fructo Kinase platelet | 1.7 | <0.001 | 1.1 | 0.41 |
| ALDOA | ENSG00000149925 | Aldolase A | 1.5 | <0.001 | 1.1 | 0.10 |
| PRPS1 | ENSG00000147224 | Phosphoribosyl pyrophosphate synthase 1 | 1.3 | <0.001 | 1.1 | 0.21 |
| PHGDH | ENSG00000092621 | Phosphoglycerate Dehydrogenase | −1.2 | <0.001 | −1.0 | <0.001 |
| PSPH | ENSG00000146733 | Phosphoserine Phosphatase | −1.4 | <0.001 | −1.0 | <0.001 |
| GLDC | ENSG00000178445 | Glycine Decarboxylase | −1.3 | <0.001 | −1.1 | <0.001 |
| GNMT | ENSG00000124713 | Glycine-N-methyl transferase | −2.9 | <0.001 | −2 | <0.001 |
| ALDH1L1 | ENSG00000144908 | Aldehyde Dehydrogenase family 1 member L1 | 2.3 | <0.001 | 1.0 | 0.60 |
| MTHFD1L | ENSG00000120254 | Methylenetetrahydrofolate dehydrogenase | 1.3 | <0.001 | 1.1 | 0.02 |
| BHMT2 | ENSG00000132840 | betaine--homocysteine S-methyltransferase 2 | 2.3 | <0.001 | 1.3 | <0.001 |
Fold change of SF treated cells compared to DMSO control.
q-values were determined through Benjamini-Hochberg p-value adjustment
4. Discussion
In this study, we set out to assess for the first time the effect and the mechanisms by which SF regulates metabolism that potentially underpins the observed health benefits of consuming SF-rich diets, which include protection from type-2 diabetes, cancer progression and cardiovascular disease. SF, compared to other phytochemicals such as quercetin (found in apples, berries, onion, tea and red wine), curcumin (found in turmeric), beta-carotene (present in carrots and sweet potatoes) and resveratrol (red wine), is the most potent activator of the key cellular antioxidant transcriptional regulator, NRF2 [14]. To date, the mechanistic focus has been the ability of SF to elicit a systemic antioxidant response through NRF2, but the potential mechanism by which SF may regulate metabolism is less clear. In addition, although NRF2 has recently been implicated in regulating genes involved in metabolic pathways, it is not yet clear whether the putative SF effects on metabolic pathways are mediated solely via NRF2. To assess these, we evaluated the effect of physiological concentrations of SF on HepG2 cells cultured in two distinct glucose environments to represent a liver not subject to oxidative stress and a high-glucose induced insulin resistant liver under oxidative stress. We first identified that dietary activation of NRF2 by SF is still maintained during excess glucose [48] and showed for the first time that SF is indeed a metabolic regulator and specifically interferes with the metabolism of glutamine and cysteine along with serine and glycine in 1C metabolism, interconnected with its ability to increase antioxidant cellular capacity. In doing so, SF resolves glucose imbalances, increases antioxidant capacity, and supports methylation by increasing the methyl pool. Using CRISPR-Cas9 genome editing, we found that these effects were entirely mediated by the ability of SF to induce NRF2.
When hepatocytes are cultured in excessive glucose ranging from 25 to 100 mM, oxidative stress is enhanced through increased expression of TNF-α and IL-6. Moreover, the excess glucose load can result in the oxidation of thiol groups on cysteine residues and blocking insulin signalling [[49], [50], [51]]. To counterbalance the increased oxidative stress, we show that SF treatment in the HG but not the BG environment results in a profound increase in reduced glutathione and induction in the antioxidant response genes (Fig. 1A). It is highly plausible that dietary compounds could have minimal effects in normal conditions but only fully mobilise their protective mechanisms when the cells are in metabolic dysregulation. An increase in GSH synthesis is primarily controlled by the cellular level of the amino acid cysteine, the availability of which is the rate-limiting step [44] but also requires glutamate and glycine. To meet glutathione demands, we showed SF upregulated the activity of the cystine transporter (SLC7A11), thereby increasing cysteine flux (Fig. 3A). Although SF increased glutamine levels, the resulting glutamate did not enter the TCA cycle, as seen by the observed reduced levels of the TCA intermediates, succinate and fumarate. (Fig. 2). The reduced glutamate levels in response to SF suggest glutamate was subsequently bound to cysteine to produce ɣ-Glutamyl-Cysteine, a metabolic precursor of GSH. The final GSH amino acid, glycine, was also reduced by SF (Fig. 4), partly due to increased utilisation towards GSH biosynthesis.
The increased glutathione-mediated antioxidant capacity of cells following SF treatment requires the availability of NADPH. NADPH is the primary reducing agent in the body, and many oxidoreduction reactions, including reducing oxidised Glutathione and thioredoxin, occur through the oxidation of NADPH to NADP+ [52]. PPP is the major source of NADPH in the cell, and the increase in the glucose flux away from glycolysis to the PPP, as well as the transcriptional activation of PPP genes, such as G6PD, PGD, TKT, and TALDO, which are NRF2 targets observed by SF only under HG conditions, will lead to an increase in NADPH pool that supports antioxidant reactions [25]. In addition, previous findings have shown that serine catabolism through SHMT1-MTHFD1-ALDH1L1 generates two NADPH per serine, supporting hepatic lipogenesis [53]. Such metabolic and transcriptional response to SF will allow the cells to maximise NADPH production for the continuous regeneration of GSH, which is needed to suppress the inflammatory cytokines and ROS produced by the high glucose environment [49]. In addition, other NADPH-consuming pathways, such as lipid biosynthesis, are also reduced, as described previously [26,54], and also seen here as the downregulation of genes involved in the biosynthesis of unsaturated fatty acids (Fig. S4F).
The availability of glucose likely drives the increased PPP capacity seen here. Glucose can feed into glycolysis, the PPP, but also serves as a one-carbon donor for serine and glycine biosynthesis in 1C metabolism. In high-glucose environments, SF redirected glucose away from glycolysis, as seen by a marked reduction of glycolytic activity (Fig. S3) and downregulation of glycolytic genes (Table 2), and towards the oxidative phase of the PPP. In the presence of SF, excess glucose was promptly converted to glucose-6-phosphate (Fig. 7), driven by the increased transcription of the NRF2-dependent rate-limiting glucose-6-phosphate dehydrogenase (G6PD) enzyme, which led to increased ribulose-5-phosphate (Ru5P) levels (Fig. 7). The NADPH generated in the process is necessary for reductive biosynthesis of glutathione and antioxidant reactions.
The redirection of glucose flux from glycolysis towards the PPP may also partly explain the established chemopreventive activity of SF [55]. Reduced glucose flux implies a glucose reduction through the SSP, ultimately decreasing serine and glycine biosynthesis [56]. Many cancer cells depend highly on serine and glycine for proliferation [37,57]. Tumours that have a loss of p53 become addicted to serine, and serine starvation has been shown to decrease the growth of such tumours considerably [51]. In addition, SSP genes such as PHGDH and SHTMT are often upregulated in lung, breast and melanoma cancer cell lines [58,59]. The reduction of the total intracellular serine by SF was due to decreased serine biosynthesis, as evidenced by a down-regulation of PHGDH, the rate-limiting biosynthetic gene, PSPH and a reduction in glucose flux (Table 3). Such reduction of the intracellular pool of serine and glycine seen here may reflect fewer methyl units entering the folate cycle, potentially resulting in reduced proliferation, which suggests a novel mechanism for the chemopreventive effects of SF.
Previously SF has been shown to act as an epigenetic modulator, mainly through downregulating DNA methyltransferases and acting as an HDAC inhibitor [60,61]. SF treatment on LnCap prostate cancer cells decreased the expression of specific DNA methyltransferases such as DNMT1 and DNMT3b and lowered the methylation in the promoter region of the cyclin D2 gene [62]. In addition, HepG2 cells treated with concentrations of SF ranging from 2 to 32 μM resulted in SF inhibiting histone deacetylases (HDACs), resulting in a change in the methylation status in the promoter region of several oncogenic transcription factors, ultimately resulting in induced cell death through apoptosis and cell cycle arrest, thereby further highlighting its chemopreventive action [63]. In this study, we also demonstrate that SF directly interferes with 1C metabolism and the methionine cycle, thereby regulating the methylation capacity of the cell (Fig. 6).
The two critical components of 1C metabolism are folate and methionine; both substrates provide methyl groups to the methyl donor SAM, which acts as a substrate for numerous methyl transferase reactions and is critical for the maintenance and adaptation of the epigenome [[64], [65], [66]]. During the process, SAH is produced, a potent inhibitor of a wide range of methyltransferases that can be hydrolysed to homocysteine and converted back to methionine to complete the methionine cycle. Here, we report that SF affected 1C metabolism and the methionine cycle by affecting the methionine and SAM levels in the cells in the HG environment. Notably, the SAM: SAH ratio levels were significantly higher in SF-treated cells under excess glucose (Fig. 6). The ratio of SAM: SAH is frequently used as an indicator of cellular methylation capacity, whereby an increased ratio predicts enhanced cellular methylation potential [67]. The increase in SAM is consistent with the observation that SF downregulates glycine-N-methyltransferase (GNMT), which catalyses the synthesis of N-methyl glycine (sarcosine) from glycine using S-Adenosyl methionine (SAM) as the methyl donor, thereby reducing SAM levels. The conversion of SAH back to methionine can occur through the transmethylation reaction of either betaine or vitamin B12 through methionine synthase. The utilisation of betaine, which is part of a one-carbon metabolism via the methionine cycle, occurs mainly in the mitochondria of liver and kidney cells. In this reaction, BHMT catalyses the addition of a methyl group from betaine to homocysteine to form methionine, forming dimethylglycine (DM). In the case of SF, we suspect the decrease in betaine is due to the remethylation of homocysteine. These reactions are essential in animals because they conserve methionine and detoxify homocysteine, which is a cause of cardiovascular disease [68].
Through CRISPR/Cas9 genome editing, we were also able to demonstrate that the metabolic shifts observed were directly linked to the ability of SF to activate NRF2, as cells with significantly impaired NRF2 levels could not affect metabolic fluxes or induce transcriptional changes to facilitate these.
There are emerging reports of the metabolic health benefits observed in SF-rich diets. Such diets have been shown to reduce Hb1Ac and improve glucose homeostasis in Type 2 diabetics [9] and significantly decrease serum insulin concentration and homeostatic model assessment of insulin resistance (HOMA-IR) [69,70]. Insulin signalling is reported to activate NRF2 [71], and insulin and NRF2 activation trigger cellular glucose uptake. However, some contradictory observations remain at the interface of NRF2-dependent cellular detoxification and glucose metabolism. For example, a recent mouse study showed that non-canonical or constituent NRF2 activation increases carbohydrate flux through the polyol pathway, resulting in a pro-diabetic shift in glucose homeostasis [72]. However, constitutive activation of NRF2 has previously been shown to have adverse metabolic effects, resulting in continuous serine and glycine biosynthesis through the upregulation of serine biosynthetic genes, allowing cancer cells to proliferate and metastasise [73]. This apparent contradiction may be explained by the different nature of NRF2 activation, whereby dietary activation of NRF2 by SF leads to transient NRF2 activation as opposed to the permanent constitutive supraphysiological NRF2 activation occurring in genetic models, which is associated with deleterious effects [74].
5. Conclusion
To conclude, we have shown that the antioxidant capacity of SF is directly linked to its ability to act as a metabolic regulator. In the liver, SF rewires central metabolic fluxes, namely glutamine and glucose, as well as 1C metabolism and methionine cycle to support the production of reduced glutathione and the antioxidant reactions that affect cellular redox status, and, in the process, resolving the metabolic stress as a result of excess glucose in an NRF2-dependent manner. In this way, we demonstrate for the first time how SF coordinates the regulation of antioxidant capacity, glucose metabolism, and the epigenetics landscape, which underpin SF's health benefits, particularly around Type 2 diabetes and cancer prevention.
Author contributions
MHT and FB conceptualised the study. FB performed all the experiments and undertook all the statistical data analyses. AM, TL, and KH analysed the isotope data. SS assisted FB with the LC-MS analysis of methionine, its intermediates and amino acids. PTR developed the pipeline to analyse the RNAseq data and assisted FB with the RNAseq analysis. TK and RFM provided scientific insights. FB and MHT wrote the manuscript, with contributions from all authors. All authors reviewed the manuscript before submission.
Declaration of competing interest
MHT and RFM are inventors of patents concerned with the development and use of high glucoraphanin broccoli.
Acknowledgements
This work was supported by the UKRI Biotechnology and Biological Sciences Research Council (BBSRC) Norwich Research Park Biosciences Doctoral Training Partnership grant number BB/M011216/1, the BBSRC Institute Strategic Programme (ISP) Food Innovation and Health BB/R012512/1 and its constituent project BBS/E/F/000PR10347 (Theme 4, Regulation of Metabolic Homeostasis). MHT was additionally supported by the BBSRC Core Capability Grant BB/CCG2260/1 and its constituent project BBS/E/QU/23NB0006 (Food & Nutrition National Bioscience Research Infrastructure). T.K. was supported by the Earlham Institute (Norwich, UK) in partnership with the Quadram Institute (Norwich, UK) and strategically supported by the UKRI BBSRC UK grants (BB/J004529/1, BB/P016774/1, and BB/CSP17270/1). T.K. was also funded by a BBSRC ISP grant for Gut Microbes and Health BB/R012490/1 and its constituent projects, BBS/E/F/000PR10353 and BBS/E/F/000PR10355.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102878.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
we have submitted our whole genome transcriptome data in ArrayExpress database and shared the accession IDs in the manuscript
References
- 1.Ding C., Li Y., Guo F., Jiang Y., Ying W., Li D., et al. A cell-type-resolved liver proteome. Mol. Cell. Proteomics. 2016;15(10):3190–3202. doi: 10.1074/mcp.M116.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rui L. Energy metabolism in the liver. Compr. Physiol. 2014;4(1):177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bojková B., Kurhaluk N., Winklewski P.J. In: Cancer. second ed. Preedy V.R., Patel V.B., editors. Academic Press; San Diego: 2021. Chapter 11 - the interconnection of high-fat diets, oxidative stress, the heart, and carcinogenesis; pp. 111–120. [Google Scholar]
- 4.Elmarakby A.A., Sullivan J.C. Relationship between oxidative stress and inflammatory cytokines in diabetic nephropathy. Cardiovasc Ther. 2012;30(1):49–59. doi: 10.1111/j.1755-5922.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 5.Pizzino G., Irrera N., Cucinotta M., Pallio G., Mannino F., Arcoraci V., et al. Oxidative stress: harms and benefits for human health. Oxid. Med. Cell. Longev. 2017;2017 doi: 10.1155/2017/8416763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman I., Gilmour P.S., Jimenez L.A., MacNee W. Oxidative stress and TNF-alpha induce histone acetylation and NF-kappaB/AP-1 activation in alveolar epithelial cells: potential mechanism in gene transcription in lung inflammation. Mol. Cell. Biochem. 2002;234–235(1–2):239–248. [PubMed] [Google Scholar]
- 7.MacKellar J., Cushman S.W., Periwal V. Waves of adipose tissue growth in the genetically obese Zucker fatty rat. PLoS One. 2010;5(1) doi: 10.1371/journal.pone.0008197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Traka M., Mithen R. Glucosinolates, isothiocyanates and human health. Phytochemistry Rev. 2008;8(1):269–282. [Google Scholar]
- 9.Axelsson A.S., Tubbs E., Mecham B., Chacko S., Nenonen H.A., Tang Y., et al. Sulforaphane reduces hepatic glucose production and improves glucose control in patients with type 2 diabetes. Sci. Transl. Med. 2017;9(394) doi: 10.1126/scitranslmed.aah4477. [DOI] [PubMed] [Google Scholar]
- 10.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39(4):199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Kolm R.H., Mannervik B., Talalay P. Reversible conjugation of isothiocyanates with glutathione catalyzed by human glutathione transferases. Biochem. Biophys. Res. Commun. 1995;206(2):748–755. doi: 10.1006/bbrc.1995.1106. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y. Role of glutathione in the accumulation of anticarcinogenic isothiocyanates and their glutathione conjugates by murine hepatoma cells. Carcinogenesis. 2000;21(6):1175–1182. [PubMed] [Google Scholar]
- 13.Thimmulappa R.K., Mai K.H., Srisuma S., Kensler T.W., Yamamoto M., Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62(18):5196–5203. [PubMed] [Google Scholar]
- 14.Lee Y.J., Lee S.H. Sulforaphane induces antioxidative and antiproliferative responses by generating reactive oxygen species in human bronchial epithelial BEAS-2B cells. J. Kor. Med. Sci. 2011;26(11):1474–1482. doi: 10.3346/jkms.2011.26.11.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins L.G., Kelleher M.O., Eggleston I.M., Itoh K., Yamamoto M., Hayes J.D. Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Toxicol. Appl. Pharmacol. 2009;237(3):267–280. doi: 10.1016/j.taap.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., et al. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell Biol. 2009;29(2):493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tebay L.E., Robertson H., Durant S.T., Vitale S.R., Penning T.M., Dinkova-Kostova A.T., et al. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med. 2015;88(Pt B):108–146. doi: 10.1016/j.freeradbiomed.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armah C.N., Derdemezis C., Traka M.H., Dainty J.R., Doleman J.F., Saha S., et al. Diet rich in high glucoraphanin broccoli reduces plasma LDL cholesterol: evidence from randomised controlled trials. Mol. Nutr. Food Res. 2015;59(5):918–926. doi: 10.1002/mnfr.201400863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Traka M.H., Melchini A., Coode-Bate J., Al Kadhi O., Saha S., Defernez M., et al. Transcriptional changes in prostate of men on active surveillance after a 12-mo glucoraphanin-rich broccoli intervention-results from the Effect of Sulforaphane on prostate CAncer PrEvention (ESCAPE) randomized controlled trial. Am. J. Clin. Nutr. 2019;109(4):1133–1144. doi: 10.1093/ajcn/nqz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traka M., Gasper A.V., Melchini A., Bacon J.R., Needs P.W., Frost V., et al. Broccoli consumption interacts with GSTM1 to perturb oncogenic signalling pathways in the prostate. PLoS One. 2008;3(7) doi: 10.1371/journal.pone.0002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bollong M.J., Lee G., Coukos J.S., Yun H., Zambaldo C., Chang J.W., et al. A metabolite-derived protein modification integrates glycolysis with KEAP1–NRF2 signalling. Nature. 2018;562(7728):600–604. doi: 10.1038/s41586-018-0622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmstrom K.M., Baird L., Zhang Y., Hargreaves I., Chalasani A., Land J.M., et al. Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biology open. 2013;2(8):761–770. doi: 10.1242/bio.20134853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Athale J., Ulrich A., MacGarvey N.C., Bartz R.R., Welty-Wolf K.E., Suliman H.B., et al. Nrf2 promotes alveolar mitochondrial biogenesis and resolution of lung injury in Staphylococcus aureus pneumonia in mice. Free Radic. Biol. Med. 2012;53(8):1584–1594. doi: 10.1016/j.freeradbiomed.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22(1):66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Nagata N., Xu L., Kohno S., Ushida Y., Aoki Y., Umeda R., et al. Glucoraphanin ameliorates obesity and insulin resistance through adipose tissue browning and reduction of metabolic endotoxemia in mice. Diabetes. 2017;66(5):1222–1236. doi: 10.2337/db16-0662. [DOI] [PubMed] [Google Scholar]
- 27.Nikolaou N., Green C.J., Gunn P.J., Hodson L., Tomlinson J.W. Optimizing human hepatocyte models for metabolic phenotype and function: effects of treatment with dimethyl sulfoxide (DMSO) Phys. Rep. 2016;4(21) doi: 10.14814/phy2.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verheijen M., Lienhard M., Schrooders Y., Clayton O., Nudischer R., Boerno S., et al. DMSO induces drastic changes in human cellular processes and epigenetic landscape in vitro. Sci. Rep. 2019;9(1):4641. doi: 10.1038/s41598-019-40660-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plaisier S.B., Taschereau R., Wong J.A., Graeber T.G. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38(17):e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasquale V., Ducci G., Campioni G., Ventrici A., Assalini C., Busti S., et al. Profiling and targeting of energy and redox metabolism in grade 2 bladder cancer cells with different invasiveness properties. Cells. 2020;9(12):2669. doi: 10.3390/cells9122669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isaacs-Ten A., Moreno-Gonzalez M., Bone C., Martens A., Bernuzzi F., Ludwig T., et al. Metabolic regulation of macrophages by SIRT1 determines activation during cholestatic liver disease in mice. Cellular and Molecular Gastroenterology and Hepatology. 2022;13(4):1019–1039. doi: 10.1016/j.jcmgh.2021.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiller K., Hangebrauk J., Jäger C., Spura J., Schreiber K., Schomburg D. MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal. Chem. 2009;81(9):3429–3439. doi: 10.1021/ac802689c. [DOI] [PubMed] [Google Scholar]
- 36.Wegner A., Weindl D., Jäger C., Sapcariu S.C., Dong X., Stephanopoulos G., et al. Fragment formula calculator (FFC): determination of chemical formulas for fragment ions in mass spectrometric data. Anal. Chem. 2014;86(4):2221–2228. doi: 10.1021/ac403879d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labuschagne C.F., van den Broek N.J., Mackay G.M., Vousden K.H., Maddocks O.D. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7(4):1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 38.Prinsen H., Schiebergen-Bronkhorst B.G.M., Roeleveld M.W., Jans J.J.M., de Sain-van der Velden M.G.M., Visser G., et al. Rapid quantification of underivatized amino acids in plasma by hydrophilic interaction liquid chromatography (HILIC) coupled with tandem mass-spectrometry. J. Inherit. Metab. Dis. 2016;39(5):651–660. doi: 10.1007/s10545-016-9935-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Moral N., Saha S., Pinto A.M., Bajka B.H., Edwards C.H. In vitro protein bioaccessibility and human serum amino acid responses to white bread enriched with intact plant cells. Food Chem. 2023;404 doi: 10.1016/j.foodchem.2022.134538. [DOI] [PubMed] [Google Scholar]
- 40.Yu X., Liang X., Xie H., Kumar S., Ravinder N., Potter J., et al. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol. Lett. 2016;38:919–929. doi: 10.1007/s10529-016-2064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kemmerer Z.A., Ader N.R., Mulroy S.S., Eggler A.L. Comparison of human Nrf2 antibodies: a tale of two proteins. Toxicol. Lett. 2015;238(2):83–89. doi: 10.1016/j.toxlet.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sivapalan T., Melchini A., Saha S., Needs P.W., Traka M.H., Tapp H., et al. Bioavailability of glucoraphanin and sulforaphane from high-glucoraphanin broccoli. Mol. Nutr. Food Res. 2018;62(18) doi: 10.1002/mnfr.201700911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niso-Santano M., González-Polo R.A., Bravo-San Pedro J.M., Gómez-Sánchez R., Lastres-Becker I., Ortiz-Ortiz M.A., et al. Activation of apoptosis signal-regulating kinase 1 is a key factor in paraquat-induced cell death: modulation by the Nrf2/Trx axis. Free Radic. Biol. Med. 2010;48(10):1370–1381. doi: 10.1016/j.freeradbiomed.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 44.Pizzorno J. Glutathione! Integr Med (Encinitas) 2014;13(1):8–12. [PMC free article] [PubMed] [Google Scholar]
- 45.Zielke H.R., Zielke C.L., Ozand P.T. Glutamine: a major energy source for cultured mammalian cells. Fed. Proc. 1984;43(1):121–125. [PubMed] [Google Scholar]
- 46.Fell D., Steele R.D. Effect of methionine on in vivo histidine metabolism in rats. J. Nutr. 1983;113(4):860–866. doi: 10.1093/jn/113.4.860. [DOI] [PubMed] [Google Scholar]
- 47.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46(D1) doi: 10.1093/nar/gkx1132. D649-d55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heiss E.H., Schachner D., Zimmermann K., Dirsch V.M. Glucose availability is a decisive factor for Nrf2-mediated gene expression. Redox Biol. 2013;1:359–365. doi: 10.1016/j.redox.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panahi G., Pasalar P., Zare M., Rizzuto R., Meshkani R. High glucose induces inflammatory responses in HepG2 cells via the oxidative stress-mediated activation of NF-κB, and MAPK pathways in HepG2 cells. Arch. Physiol. Biochem. 2018;124(5):468–474. doi: 10.1080/13813455.2018.1427764. [DOI] [PubMed] [Google Scholar]
- 50.Chen J.-Y., Chou H.-C., Chen Y.-H., Chan H.-L. High glucose-induced proteome alterations in hepatocytes and its possible relevance to diabetic liver disease. J. Nutr. Biochem. 2013;24(11):1889–1910. doi: 10.1016/j.jnutbio.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 51.Nakajima K., Yamauchi K., Shigematsu S., Ikeo S., Komatsu M., Aizawa T., et al. Selective attenuation of metabolic branch of insulin receptor down-signaling by high glucose in a hepatoma cell line, HepG2 cells. J. Biol. Chem. 2000;275(27):20880–20886. doi: 10.1074/jbc.M905410199. [DOI] [PubMed] [Google Scholar]
- 52.Ju H.-Q., Lin J.-F., Tian T., Xie D., Xu R.-H. NADPH homeostasis in cancer: functions, mechanisms and therapeutic implications. Signal Transduct. Targeted Ther. 2020;5(1):231. doi: 10.1038/s41392-020-00326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z., TeSlaa T., Xu X., Zeng X., Yang L., Xing G., et al. Serine catabolism generates liver NADPH and supports hepatic lipogenesis. Nat. Metab. 2021;3(12):1608–1620. doi: 10.1038/s42255-021-00487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X., Dai M., Lao F., Chen F., Hu X., Liu Y., et al. Effect of glucoraphanin from broccoli seeds on lipid levels and gut microbiota in high-fat diet-fed mice. J. Funct.Foods. 2020;68 [Google Scholar]
- 55.Nandini D.B., Rao R.S., Deepak B.S., Reddy P.B. Sulforaphane in broccoli: the green chemoprevention!! Role in cancer prevention and therapy. J. Oral Maxillofac. Pathol. : JOMFP. 2020;24(2):405. doi: 10.4103/jomfp.JOMFP_126_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang M., Vousden K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer. 2016;16(10):650–662. doi: 10.1038/nrc.2016.81. [DOI] [PubMed] [Google Scholar]
- 57.Locasale J.W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Locasale J.W., Grassian A.R., Melman T., Lyssiotis C.A., Mattaini K.R., Bass A.J., et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011;43(9):869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaufman-Szymczyk A., Majewski G., Lubecka-Pietruszewska K., Fabianowska-Majewska K. The role of sulforaphane in epigenetic mechanisms, including interdependence between histone modification and DNA methylation. Int. J. Mol. Sci. 2015;16(12):29732–29743. doi: 10.3390/ijms161226195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pop S., Enciu A.M., Tarcomnicu I., Gille E., Tanase C. Phytochemicals in cancer prevention: modulating epigenetic alterations of DNA methylation. Phytochemistry Rev. 2019;18(4):1005–1024. [Google Scholar]
- 62.Su X., Jiang X., Meng L., Dong X., Shen Y., Xin Y. Anticancer activity of sulforaphane: the epigenetic mechanisms and the Nrf2 signaling pathway. Oxid. Med. Cell. Longev. 2018;2018 doi: 10.1155/2018/5438179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dos Santos P., Machado A.R.T., De Grandis R.A., Ribeiro D.L., Tuttis K., Morselli M., et al. Transcriptome and DNA methylation changes modulated by sulforaphane induce cell cycle arrest, apoptosis, DNA damage, and suppression of proliferation in human liver cancer cells. Food Chem. Toxicol. 2020;136 doi: 10.1016/j.fct.2019.111047. [DOI] [PubMed] [Google Scholar]
- 64.Kalhan S.C., Marczewski S.E. Methionine, homocysteine, one carbon metabolism and fetal growth. Rev. Endocr. Metab. Disord. 2012;13(2):109–119. doi: 10.1007/s11154-012-9215-7. [DOI] [PubMed] [Google Scholar]
- 65.Chorley B.N., Campbell M.R., Wang X., Karaca M., Sambandan D., Bangura F., et al. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40(15):7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirotsu Y., Katsuoka F., Funayama R., Nagashima T., Nishida Y., Nakayama K., et al. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012;40(20):10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caudill M.A., Wang J.C., Melnyk S., Pogribny I.P., Jernigan S., Collins M.D., et al. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine β-synthase heterozygous mice. J. Nutr. 2001;131(11):2811–2818. doi: 10.1093/jn/131.11.2811. [DOI] [PubMed] [Google Scholar]
- 68.Zhao G., He F., Wu C., Li P., Li N., Deng J., et al. Betaine in inflammation: mechanistic aspects and applications. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bahadoran Z., Mirmiran P., Hosseinpanah F., Hedayati M., Hosseinpour-Niazi S., Azizi F. Broccoli sprouts reduce oxidative stress in type 2 diabetes: a randomized double-blind clinical trial. Eur. J. Clin. Nutr. 2011;65(8):972–977. doi: 10.1038/ejcn.2011.59. [DOI] [PubMed] [Google Scholar]
- 70.Bahadoran Z., Tohidi M., Nazeri P., Mehran M., Azizi F., Mirmiran P. Effect of broccoli sprouts on insulin resistance in type 2 diabetic patients: a randomized double-blind clinical trial. Int. J. Food Sci. Nutr. 2012;63(7):767–771. doi: 10.3109/09637486.2012.665043. [DOI] [PubMed] [Google Scholar]
- 71.Wang X., Wu H., Chen H., Liu R., Liu J., Zhang T., et al. Does insulin bolster antioxidant defenses via the extracellular signal-regulated kinases-protein kinase B-nuclear factor erythroid 2 p45-related factor 2 pathway? Antioxidants Redox Signal. 2012;16(10):1061–1070. doi: 10.1089/ars.2011.4460. [DOI] [PubMed] [Google Scholar]
- 72.Liu P., Dodson M., Li H., Schmidlin C.J., Shakya A., Wei Y., et al. Non-canonical NRF2 activation promotes a pro-diabetic shift in hepatic glucose metabolism. Mol. Metabol. 2021;51 doi: 10.1016/j.molmet.2021.101243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DeNicola G.M., Chen P.-H., Mullarky E., Sudderth J.A., Hu Z., Wu D., et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 2015;47(12):1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tebay L.E., Robertson H., Durant S.T., Vitale S.R., Penning T.M., Dinkova-Kostova A.T., et al. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med. 2015;88:108–146. doi: 10.1016/j.freeradbiomed.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
we have submitted our whole genome transcriptome data in ArrayExpress database and shared the accession IDs in the manuscript