Abstract
Purpose:
Cancer drug development is currently limited by a paradigm of preclinical evaluation that does not adequately recapitulate the complexity of the intact human tumor microenvironment (TME). To overcome this, we combined trackable intratumor microdosing (CIVO) with spatial biology readouts to directly assess drug effects in patient tumors in situ.
Experimental Design:
In a first-of-its-kind phase 0 clinical trial, we explored the effects of an investigational stage SUMOylation-activating enzyme (SAE) inhibitor, subasumstat (TAK-981) in 12 patients with head and neck carcinoma (HNC). Patients scheduled for tumor resection received percutaneous intratumor injections of subasumstat and vehicle control 1 to 4 days before surgery, resulting in spatially localized and graded regions of drug exposure (∼1,000–2,000 μm in diameter). Drug-exposed (n = 214) and unexposed regions (n = 140) were compared by GeoMx Digital Spatial Profiler, with evaluation at single-cell resolution in a subset of these by CosMx Spatial Molecular Imager.
Results:
Localized regions of subasumstat exposure revealed SUMO pathway inhibition, elevation of type I IFN response, and inhibition of cell cycle across all tumor samples. Single-cell analysis by CosMx demonstrated cell-cycle inhibition specific to the tumor epithelium, and IFN pathway induction commensurate with a TME shift from immune-suppressive to immune-permissive.
Conclusions:
Pairing CIVO with spatial profiling enabled detailed investigation of response to subasumstat across a diverse sampling of native and intact TME. We demonstrate that drug mechanism of action can be directly evaluated in a spatially precise manner in the most translationally relevant setting: an in situ human tumor.
Translational Relevance.
Early cancer drug development has been hampered by lack of predictive preclinical models of tumors on which to evaluate the activity of new anticancer agents. Central to this issue is the highly complex composition of the solid tumor microenvironment (TME) compounded by the uniqueness of every patient's cancer. This makes modeling outside of the cancer patient very challenging. To address this limitation, we paired trackable intratumor microdosing (CIVO) with two spatial profiling platforms, digital spatial profiler (GeoMx), and spatial molecular imaging (CosMx), to reveal the effects of an investigational SUMOylation inhibitor (subasumstat, TAK-981) on the intact, native TME of patients with head and neck cancer. The data generated are the first to show modulation of the immune TME by subasumstat and represent the first trial that uses this novel phase 0 trial approach to reveal detailed mechanistic effects of an investigational anticancer agent in situ.
Introduction
Tumor response to drug exposure is heavily influenced by the composition and activity state of the niche in which tumor cells grow, the tumor microenvironment (TME). The TME is complex and heterogeneous, consisting of myriad cell types, including tumor, immune, stromal, and vascular endothelial cells intertwined with noncellular components such as extracellular matrix (1, 2). This poses a significant challenge to preclinical evaluation and development of new investigational anticancer therapies (3–6). Despite numerous approaches, it has proven difficult to recapitulate the compositional complexity and interpatient tumor heterogeneity in laboratory models of cancer. Even in cases where patient diversity is captured, for example, using extensive collections of patient-derived xenografts (7–9), the contribution of the immune response to drug efficacy is often lacking due to growth of such models in immune-deficient hosts. As a result, investigational agents that perform well in preclinical models of cancer often fail to provide benefit to patients when evaluated clinically, making evaluation of antitumor drug efficacy dependent on clinical trials that are time- and resource-intensive and can expose patients to significant drug-induced adverse effects for agents that ultimately offer no clinical benefit (10–12). The lack of robust and meaningfully correlative preclinical to clinical models is a ubiquitous issue in drug development and is particularly challenging for new assets aiming to target nontumor components of the TME (5, 13).
Acknowledging this challenge, we have developed a novel platform for the clinical evaluation of investigational agents in the context of the authentic, native, and structurally intact TME of patients with human cancer. Specifically, we have combined trackable intratumoral microdosing of drugs (CIVO, Presage Biosciences) with high-plex spatial biology readouts in phase 0 trials in patients with surface-accessible tumors that are planned for surgical resection. The CIVO platform consists of a handheld injector that deposits a columnar array of drug, co-compounded with fluorescent microspheres (4 μm) to enable subsequent tracking of injection site locations and resolution of exact areas of drug deposition in resected tumor samples. Drug-induced responses are observed within the patient's tumor following surgical resection. These responses can be studied by processing the tumor for histological sectioning and conducting quantitative biomarker analysis at each site of drug deposition (14–16).
Until recently, assessment of drug response in intact tissue was restricted to low-plex IHC or ISH techniques, with higher content assays such as next-generation sequencing (NGS) only being applied to homogenized tissue. The emergence of high-plex spatial profiling platforms such as digital spatial profiling (GeoMx DSP, NanoString Technologies) and spatial molecular imaging (CosMx SMI, NanoString Technologies), provides the possibility for drug response readouts at the single-cell level in the preserved TME. Digital spatial profiler (DSP) uses NGS-based readouts from spatially defined regions in structurally intact tissues such as the TME (17), whereas SMI uses high-resolution imaging of approximately 1,000 genes, with single-cell and subcellular resolution (18). Critically, both methods can be used in formalin-fixed, paraffin-embedded (FFPE) sections that are readily generated from clinical material. Combined with the CIVO multiplexed microdosing platform, direct high-plex interrogation of the impact of drug exposure on tumor in situ at spatially defined regions of interest (ROI) is now possible. Importantly, analysis occurs without exposing patients to systemic drug and potential-associated toxicities. Trials combining these platforms provide early and precise insights into whether an investigational agent induces its intended pharmacodynamic (PD) mechanism of action, initiates tumor cell autonomous responses, or shifts the composition and activity state of the TME (15, 16).
We present a spatially precise, genomic, and single-cell analysis of the effect of an investigational agent, subasumstat (TAK-981), microdosed directly into tumors of head and neck squamous cell carcinoma (HNC) using the CIVO platform in a phase 0 clinical trial (NCT04065555). Subasumstat is an inhibitor of the small ubiquitin-like modifier (SUMO)–activating enzyme (SAE2). It is currently being evaluated in phase 1/2 clinical trials, namely NCT03648372, NCT04381650, NCT04074330, and NCT04776018. SAE2 is a component of the enzymatic cascade that facilitates the posttranslational modification of protein substrates with SUMO, thereby regulating their activity, subcellular localization, and stability (19–21). In addition to regulating cellular processes important for tumor cell proliferation and survival (22), SUMOylation has also been shown both in vitro and in syngeneic models of cancer to play a key role in regulating innate immune response (20, 23, 24). Our study provides both genomic and single-cell population evidence that SUMOylation initiates an innate immune response driven by type 1 IFN signaling, with concomitant drug-induced effects on multiple cell types, including changes to macrophages, cytotoxic T cells, and tumor cell activity, in the native and intact TME, up to 96 hours following drug exposure.
Materials and Methods
Study enrollment and CIVO procedure
Twelve patients (two females, 10 males, mean age 63.5 years) were enrolled in this IRB-approved, phase 0 clinical trial (PBI-TAK-01) at three centers in the United States (University of Washington, Seattle, WA; Portland VA/Oregon Health and Sciences University, Portland, OR; University of Michigan, Ann Arbor, MI). All subjects provided informed consent and were screened for eligibility. This study was conducted in accordance with the ethical guidelines outlined in the Declaration of Helsinki and the International Council on Harmonization guidelines on Good Clinical Practice. Key inclusion criteria included a pathologic diagnosis of HNC, and a surface-accessible tumor (depth up to approximately 3.0 cm) amenable to CIVO injection that was at least 2.0 cm in the shortest dimension and planned for surgical resection. The enrolled population included patients with metastatic disease (58.3%), local disease (25.0%), and local recurrent disease (15.7%). Of these, 83.8% were undergoing their first surgery for the study indication, none of whom received neoadjuvant therapy before tumor resection. Eligible patients underwent microdose injection with the CIVO device on an outpatient basis, approximately 1 to 4 days before planned surgery. The microdose selected was based on previously established FDA guidance that defines a microdose as less than 1/100th the recommended systemic dose of the agent under investigation. In the case of subasumstat, each microinjection delivered a maximum of 8.3 μL of a 1 mg/mL solution deposited by the CIVO device in a column extending 10 mm in length.
Following resection, the CIVO-injected portion of each patient's tumor was immediately gross-sectioned in a plane perpendicular to the microinjection drug columns. Tissue was then fixed and shipped to Presage Biosciences for analysis. Injection sites were visible through each stage of the process due to the presence of fluorescent tracking microspheres (FTM) in each injection column, enabling precise attribution of sites microdosed with subasumstat. The CIVO microdosing procedure was well-tolerated with no adverse effects, probably related to the injection or delivery of drug contents and only one case of mild, transient nausea and vomiting deemed possibly related to CIVO technology.
ISH and IHC biomarker staining
ISH was completed using the RNAscope multiplex fluorescent reagent kit v2 (Advanced Cell Diagnostics). FFPE tumors were cut onto slides with a thickness of 4 μm. Slides were baked for 1 hour at 60°C, deparaffinized in Clear-Rite 3, and rehydrated via graded ethanol. Hydrogen peroxide was added for 10 minutes to quench endogenous peroxidase activity. Slides underwent a 15-minute target retrieval solution incubation at 100°C, followed by a 15-minute protease digestion at room temperature. The RNAscope ISH assays were completed with probes from Advanced Cell Diagnostics listed in Supplementary Table S1 and Opal Tyramide Signal Amplification (TSA) reagent detection (Akoya). The slides were counterstained with DAPI for 10 minutes and mounted with Prolong Gold mounting medium (Invitrogen).
A list of primary antibodies used is included in Supplementary Table S2. Sections were antigen retrieved and incubated with primary and secondary antibodies specified in Supplementary Table S3. An iterative staining approach was used whereby antibodies were detected with Opal TSA reagents, followed by antibody stripping, and re-staining with the next antibody. Sections were counterstained with DAPI.
Whole-slide imaging and image analysis
Immunofluorescent-stained slides were imaged using a Zeiss Axioscan 7 whole-slide imaging system. Image analysis to segment cells and quantify biomarkers was performed using custom tissue and object segmentation and biomarker-specific classification algorithms using the HALO Image Analysis Platform (Indica Labs). Microdose injection sites were identified via CIVO GLO microspheres co-injected through each needle within the CIVO device, and subasumstat injection sites were further identified by signal from the MIL113 antibody recognizing subasumstat-SUMO adducts. MIL113 staining was performed on adjacent slide sections as a proxy for subasumstat target engagement, and the percentage of MIL113-positive cells was scored for each ROI. ROI analyzed via GeoMx were mapped to closely adjacent immunofluorescent-stained slides. Using nuclear segmentation, cells were identified and classified and plotted as the percent of biomarker-positive or biomarker-negative cells within each ROI. Signal intensity from the tissue sections was classified as either negative, low, medium, or high for every mapped overlay of the corresponding GeoMx ROI.
GeoMx digital spatial profiling of CIVO microdosed tumors
Tissue sections were baked, deparaffinized, washed in PBS, and targets were retrieved for 20 minutes in 1× Tris-EDTA pH 9.0 buffer (Invitrogen No. 00–4956–58) at 100°C. Next, tissues were washed in PBS, incubated in 0.1 μg/mL proteinase K (Thermo Fisher Scientific, No. AM2546) in PBS for 15 minutes at 37°C and washed again in PBS. Protein targets were post-fixed in 10% neutral-buffered formalin for 5 minutes, followed by two 5-minute incubations in 12.25 mg/mL Tris base, 7.5 mg/mL Glycine stop buffer diluted in DEPC-treated water. Tissues were incubated overnight at 37°C with hybridization solution [Human Cancer Transcriptome Atlas (CTA) probe mix (NanoString No. 121400101), Buffer R (NanoString No. 121300313), and DEPC-treated water]. During incubation, slides were covered with HybriSlip hybridization covers (Grace BioLabs No. 714022). After incubation, HybriSlip covers were gently removed by soaking in 2× SSC buffer. Two 25-minute stringent washes were performed using 50% formamide in 2× SSC at 37°C. Tissues were washed for 5 minutes in 2× SSC and incubated in blocking buffer W (NanoString No. 121300313) for 30 minutes at room temperature in a humidity chamber. Primary antibody targeted against MIL113 was diluted in buffer W and incubated with the tissues for 1 hour followed by a 1-hour incubation with fluorescently labeled secondary antibody diluted in buffer W. After the slides were washed twice for 5 minutes in 5× SSC buffer, they were incubated for 1 hour in SYTO13 (50 nmol/L) and fluorescently conjugated antibody targeting PanCK in blocking buffer. Following staining, the sections were washed for 5 minutes in fresh 2× SSC. Following whole-slide imaging, multiple geometric ROI of 400 μm2 were placed in regions of subasumstat drug exposure according to MIL113 signal intensity and regions of vehicle or background. Indexing oligonucleotides were then cleaved and quantified via NGS on a NovaSeq 6000 Illumina sequencer as previously described (17).
NanoString GeoMx DSP analysis
Raw probe level counts from the NanoString DSP machine were subjected to QC and normalization as follows. ROI with deduplicated read counts of <10,000 were excluded. Count data were then normalized using TMM normalization (25) before performing limma analysis (26). The probe level data were used in the limma model to increase statistical power with the duplicate correlation function used to account for gene level replicates. For the definition of a common signature of subasumstat target engagement, linear models were fit to account for different patients, slides, injection sites, and logFC values extracted corresponding to the drug effect. Clustering was performed using ComplexHeatmap (27). To calculate signature scores (IFN signature, cell-cycle signature, etc.), the Gene Set Variation Analysis (GSVA) algorithm was used (28–30).
CosMx SMI of CIVO microdosed tumors
CosMx cyclic RNA readout was performed on tissue sections following the previously described Materials and Methods and protocol (18). Briefly, the tissue sections were dewaxed, rehydrated, and underwent target retrieval, protease digestion, and post-fixation. Hybridization chambers were applied to the slides and the probe set incubated on the tissue overnight. Unbound probe was removed through stringency washes. Nanostring flow cells were applied to the slides and loaded onto the SMI instrument. FOVs were selected that corresponded to the GeoMx ROI on parallel tissue sections, and cyclic RNA readouts were acquired. Following RNA readouts, the tissue sections were stained using a four-antibody cocktail stain of morphology markers (B2M, CD45, CD3, and PanCK) and imaged on the SMI instrument.
NanoString CosMx SMI analysis
NanoString data for patients 7 and 8 from the CosMx SMI machine were processed in Seurat (31) broadly following the vignette provided here (https://satijalab.org/seurat/articles/spatial_vignette_2.html). CosMx data consist of a sparse matrix of gene expression counts, a data frame of centroids (cell_ids and x, y coordinates), and a data frame of molecule pixel coordinates (gene_ids and x, y coordinates; ref. 18). The LoadNanoString function was used to load data in R, including coordinates for the cell boundaries provided by NanoString. Cells with <99 counts were excluded from downstream analysis: This step excluded 19,235 cells from patient 7 (28% of total), and 26,906 cells from patient 8 (37% of total), leaving a total of 47,417 and 46,075 cells for each patient, respectively. Data from the two patients were combined in a single seurat object, and gene counts were normalized using SCTransform (32) and PCA and UMAP performed with dimensions set to 20. Gene markers to distinguish UMAP clusters were identified using the FindAllMarkers function with min.pct = 0.25 and logfc.threshold = 0.25. Cell identities were then manually assigned to the clusters using the identified genes by reference to established marker sets. GSVA analysis for the CosMx data was performed using the enrichIt (33) function.
Although it is possible to measure gene expression at the single-cell level with the CosMx platform, and it is also possible to measure target engagement at the single-cell level using MIL113 staining, we were not able to perform these assays on the same serial sections because the CosMx protocol does not allow for the introduction of additional IHC/ISH reagents. Therefore, to evaluate the effect of subasumstat on localized regions of the TME, we performed MIL113 staining on sections immediately adjacent to the sections taken for CosMx analysis. Analytically, we then assigned an average MIL113 staining to each FOV and used this average to relate to the individual cell expression results.
All analyses for GeoMx DSP and CosMx SMI were performed in RStudio (version 2021.09.0) using R version 4.1.1.
Data availability
GeoMx gene expression data were deposited into the Gene Expression Omnibus database under the accession number GSE235475 and are available at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE235475. Additional data generated in this study are available upon reasonable request from the corresponding author.
Results
Trackable, spatially defined regions of subasumstat exposure exhibit dose-dependent target engagement and reduction of SUMOylation following intratumor microdosing with CIVO
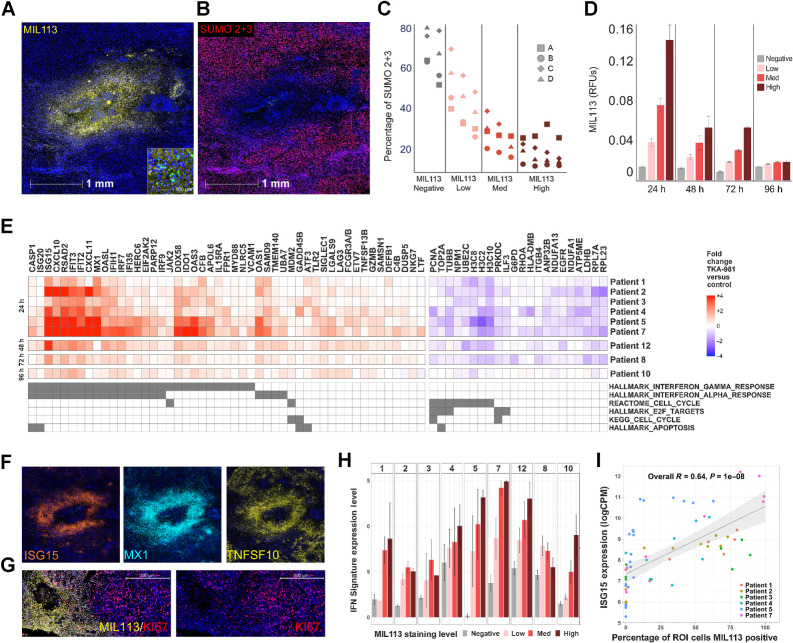
To directly assess the impact of drug target engagement within the complexity of the intact patient TME, tumors of 12 patients with accessible HNC lesions were microinjected with subasumstat using CIVO technology as previously described (ref. 16; Supplementary Table S4). Tumors were surgically resected 24 to 96 hours after microdosing with CIVO. Of these, nine tumors exhibited trackable sites of drug microinjection [denoted by clearly visible fluorescent signal from the co-microinjected fluorescent-tracking microspheres (FTM)] and were of sufficient tissue quality to be selected for further analysis by DSP, SMI, and standard IHC or ISH (Fig. 1). Of the three tumors that were not evaluated, two exhibited severe tissue degradation between surgery and pathology, and one exhibited high-quality injection sites; but these were confined to the tumor capsule, not malignant tissue. Accurate tracking of subasumstat distribution within the TME was aided by IHC staining with an antibody (MIL113) developed to provide a target engagement biomarker that specifically recognizes subasumstat-SUMO adducts formed by mechanism-based inhibition of SAE (refs. 20, 21; Fig. 2A). Consistent with the anticipated mechanism of action, localized regions of adduct (MIL113 staining) exhibited concomitant inhibition of SUMOylation as shown by a decrease in staining for SUMO2/3-conjugated proteins (Fig. 2B). Moreover, as observed in previous studies (14), CIVO microinjection resulted in graded distribution of drug, from the highest exposure closest to the epicenter of the injection site (marked by CIVO GLO fluorescent microspheres) to lower and ultimately no drug exposure with increasing radial distance outward from the site of drug microinjection (Supplementary Fig. S1). The observed gradient of target engagement was directly related to inhibition of SUMOylation, as detected by an inverse relationship between MIL113 and SUMO 2/3 staining (Fig. 2C). Target engagement was time-dependent with MIL113 staining intensity strongest at 24 hours, observable above background through 72 hours, and low by 96 hours following microinjection (Fig. 2D). With target engagement and pathway inhibition established, we evaluated the biological impact of SUMOylation inhibition on the native TME.
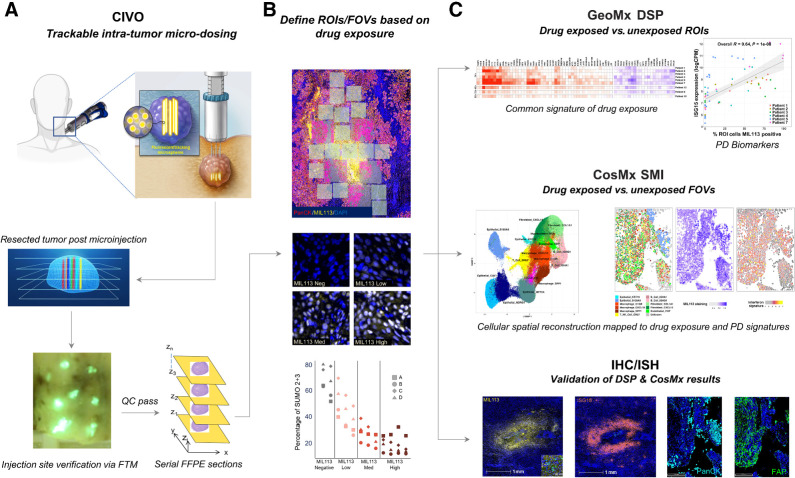
Figure 1.
CIVO + GeoMx + CosMx Workflow. A, Drugs are delivered intratumorally in a columnar fashion to spatially defined positions in head and neck tumors using the CIVO microinjector. The position of each site of drug injection is identified and tracked from the initial injection procedure, through surgical resection, and ultimately spatial profiling by co-injection of fluorescent-tracking microspheres (FTM). Following incubation for 4 to 96 hours in the living intact tumor, the tumor is surgically resected and cross-sectioned perpendicular to the injection column plane. Tumors exhibiting distinct injection sites, as visualized by the presence of the FTM (fluorescent green in appearance) and verified as located in tumor tissue are processed as formalin-fixed, paraffin-embedded (FFPE) sections for downstream analysis via spatial profiling technologies and conventional IHC/ISH assays. B, Regions from FFPE sections containing trackable sites of drug target engagement are analyzed using GeoMx Digital Spatial Profiling RNA assays. Identification of localized regions of target engagement was enhanced by staining with the MIL113 antibody that specifically recognizes formation of subasumstat-SUMO adducts (yellow) and verified for inhibition of the SUMO pathway using an antibody specific for SUMO 2+3. To establish drug-induced responses across a compositionally diverse TME, multiple regions of interest (ROI) are placed at sites within and outside of graded areas of MIL113 staining (from high to low surrounding the FTM) for transcriptional profiling using probe sets such as the Cancer Transcriptome Atlas (1,800+ gene targets). C, Differential gene expression analysis was performed to determine drug-specific gene expression signatures across the patient tumor sample set. Similarly, regions within CIVO microdosed tumors can be analyzed using spatial molecular imaging such as Nanostring's CosMx technology. Gene expression at the cellular level is determined through iterative ISH and imaging for up to 1,000 gene targets. Segmented cells are assigned to cell types and cell type–specific expression levels can be compared at the cellular level within the spatial context of the intact TME. Drug-induced changes in transcript expression, detected by either DSP or SMI, are then validated at each spatially defined position of drug exposure within the TME by conventional IHC or ISH.
Figure 2.
Microinjection of subasumstat with CIVO induces exposure-dependent inhibition of SUMOylation, activation of an IFN response, and reduction of cell-cycle–associated gene expression across multiple tumors. A, A representative single CIVO site of subsumstat microinjection. MIL113 staining (yellow) tracks regions of target engagement (subasumstat-SUMO adduct formation) around the site of injection and demonstrates spatially localized distribution of drugs; scale bar, 1,000 μm. The original epicenter of the injection site is identified by the presence of fluorescent tracking microspheres (CIVO GLO green; inset; scale bar, 50 μm). B, The same site of localized subsumstat exposure as shown in (A) evaluated by IHC with an antibody specific for SUMO 2+3 (red) shows reduction in cells staining positive for this proximal biomarker of SUMOylation pathway activity. C, Quantification of the percentage of cells staining positive for SUMO2+3 shows an inverse relationship between level of subasumstat target engagement (MIL113 staining) and SUMOylation (%SUMO 2+3 positive). D, MIL113 staining intensity for four binned categories (negative, low, medium, and high) for tumors processed at different timepoints (24, 48, 72, and 96 hours). Error bars represent the standard error of the mean. E, Heat map of genes identified as responsive to subasumstat across nine individual tumors resected at different timepoints (24, 48, 72, and 96 hours). Linear models were fit to the data to account for different patients, slides, and injection sites and fold-change values extracted corresponding to the drug effect—subasumstat negative (negative) versus subasumstat-exposed (high, med, and low). Genes were selected if they showed a >1.5-fold change in expression between drug exposed and nonexposed ROI at Padj < 0.05 across eight of the nine patients (note patient 10 was excluded from this analysis because of weak signal at the late timepoint of 96 hours), with a consistent direction of change across all nine patients. The genes are annotated for membership of pathways identified by enrichment analysis using MSigDB (Supplementary Table S6A and S6B). F, Visualization of subasumstat-induced elevation of specific IFN response genes with ISH probes specific for ISG15 (orange), MX1 (cyan), and TNFSF10 (yellow). Note that these images are taken from the same region and the same scale as that shown for A and B. G, Visualization of subasumstat inhibition of cell cycle by dual IHC with MIL113 (yellow) and KI67 (red). Left and right panels are the same except the right panel has the channel for MIL113 staining removed to show the effect on KI67 staining; scale bar, 500 μm. H, The IFN signature score (level of signature expression) was determined for regions of high (n = 61), medium (n = 71), low (n = 55), or no MIL113 staining (n = 125). Signature scores are plotted by patient, and the level of subasumstat drug target engagement was determined by MIL113 stain intensity. Error bars represent the standard error of the mean. I, For each ROI analyzed via DSP, the relative expression of the highly IFN-responsive gene ISG15 is plotted as a function of percentage of cells that are MIL113-positive within the ROI. A linear model fit is shown with associated Pearson correlation, and P value as well as 95% confidence intervals (shaded). ISG15 expression levels are highly correlated with the percentage of cells exposed to subasumstat.
Subasumstat induces dose-dependent IFN response signatures across multiple heterogeneous microdosed HNC tumors
To investigate the effects of subasumstat, we compared expression profiles derived from drug-exposed and unexposed regions of the TME by DSP. MIL113 staining was used as a guide for ROI placement on slide sections, and a total of 354 ROI representing a wide sampling of diverse TME across nine patients with HNC microdosed with subasumstat was analyzed for differential gene expression (Supplementary Table S5) using the NanoString CTA probe set. Differential gene expression analysis focused on common signatures of response to subasumstat that could be observed across TME regions of diverse cellular composition. Common signatures were defined as genes that showed at least a 1.5-fold up- or downregulation (adjusted P value <0.01) upon comparison of subasumstat-exposed versus control in all patients (we excluded the 96-hour sample, patient 10, from this signature selection procedure due to low signal at this late timepoint). The resulting common signature comprised 49 upregulated genes and 22 downregulated genes (Fig. 2E). Gene enrichment analysis using the BROAD Molecular Signatures Database (MSigDB; refs. 34, 35) revealed that the upregulated genes are highly enriched in curated IFN response pathways, both type 1 and 2, whereas the downregulated genes map to pathways involved in cell-cycle checkpoint control (Fig. 2E; Supplementary Table S6A and S6B). Of note, this common signature was observed in all patients at 24 to 72 hours following intratumoral injection with subasumstat, and even in the one tumor sample resected at 96 hours (patient 10) albeit at reduced levels (Fig. 2E). To confirm the fidelity of the DSP-derived signatures, IFNβ, select IFN response genes, and the well-established biomarker for cell proliferation, KI67, were evaluated by ISH and IHC, respectively. Although it is notable that we did not detect expression of IFNA1 or IFNB1 in the common signature (due to low expression in some samples), elevation of IFNβ transcript was detected by both DSP and ISH in a subset of tumor samples following localized exposure to subasumstat (Supplementary Figs. S2 and S3). Co-hybridization with cell type–specific in situ probes demonstrated IFNβ elevation in multiple immune cell populations, including dendritic cells, macrophages, and T cells (Supplementary Fig. S3). Consistent with robust elevation of IFN signatures highlighted by DSP, marked elevation of representative IFN response genes, including ISG15, MX1, and TNFSF10, was observed corresponding closely to spatially defined regions of inhibited SUMOylation (Fig. 2F). Staining for IFN response genes involved both the TME and the tumor epithelium (Supplementary Fig. S4). Similarly, decreased KI67 was also observed tightly correlating to regions of subasumstat target engagement (Fig. 2G). These data represent the first demonstration of drug-induced cellular responses and shifts in the activity state of the TME induced by inhibition of SUMOylation in patient tumor samples.
In addition to capturing drug-exposed versus unexposed comparisons, we further explored the relationship between target engagement and gene expression changes by leveraging the quantitative analysis of MIL113 staining across patient samples. As noted above, MIL113 staining intensity was used to classify tumor ROI as having negative, low, medium, or high subasumstat target engagement that corresponded with reduction in SUMO2/3 conjugates as anticipated (Fig. 2C). With some tumor-specific effects noted, expression of both the IFN and cell-cycle signatures exhibited clear drug dose responsiveness across patient samples (Fig. 2H; Supplementary Fig. S5). In addition, striking concordance between subasumstat target engagement and elevation of single-gene transcripts such as the IFN response gene ISG15 was observed (Fig. 2I; Supplementary Figs. S2 and S6). This highlights the utility of our approach to reveal sensitive and specific PD biomarkers of TME responsiveness to subasumstat that have potential for future use in conventional clinical evaluations of subasumstat dosed intravenously in patients with solid tumors.
Cell type–specific responses to subasumstat revealed by SMI
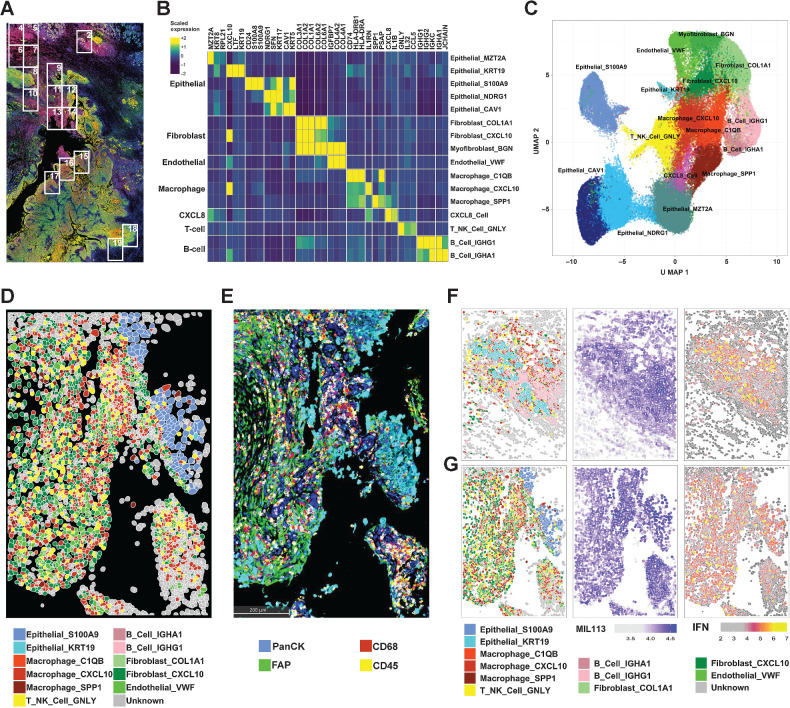
We next sought to better define mechanistic effects of subasumstat at the level of individual cell subtypes within the TME. To obtain single-cell resolution on subasumstat-induced gene expression changes while preserving spatial information within the tumor, we leveraged the CosMx SMI platform. We selected slide sections from patients 7 (24 hours) and 8 (72 hours) and recovered data from multiple drug-exposed and unexposed fields of view (FOV): 17 FOV for patient 7 and 20 FOV for patient 8 (Fig. 3A; Supplementary Fig. S7). Expression data for 91,873 cells (45,799 for patient 7 and 46,074 for patient 8) were retained after processing and filtering (see Materials and Methods). Major cell type categories were assigned to cell clusters identified by UMAP clustering based on specific genes from established lineage marker gene sets (ref. 36; Fig. 3B and C; Supplementary Figs. S8 and S9). In total, 16 molecularly distinct cell subtypes were defined. As anticipated, malignant epithelial cell populations tend to be specific to the tumor of origin, whereas stromal and immune components are shared across samples (Supplementary Figs. S8B and S8C, S9, and S10). Next, a spatial map of the TME consisting of each genetically defined cell type was reconstructed for patients 7 and 8. Remarkably, cells that cluster together in mathematical space, based on common patterns of gene expression (UMAP projection) are also found spatially adjacent in the reconstructed section (Fig. 3D and E; Supplementary Fig. S9C and S9D). We validated the epithelial, immune cell, and stromal cell assignments derived from CosMx by IHC (Fig. 3E; Supplementary Fig. S9C and S9D).
Figure 3.
CosMx Spatial Imaging identifies subasumstat-responsive cells in CIVO microinjected tumors. A, Formalin-fixed, paraffin-embedded sections of tumor from patient 7 stained with DAPI, panCK, and MIL113 adjacent to the section used in the CosMx analysis. Boxes represent fields of view (FOV) collected. B, Differentially expressed genes used to identify cell types in UMAP clusters. Genes were identified for clusters from UMAP projection using the FindAllMarkers function in Seurat using parameters only.pos = TRUE, min.pct = 0.25, and logfc.threshold = 0.25. The top three genes for each cluster are shown (see Supplementary Fig. S8A for all genes and cells). Expression is the average for each cell type of the scaled gene level data across tumors 7 and 8. On the basis of the identity of the genes for each cluster a cell type was established by reference to established marker sets. C, UMAP projection of 16 cell types identified from clustering of data from patients 7 and 8. UMAP clustering was performed in Seurat using a standard workflow following data normalization and PCA. Cells (n = 93,491) are colored according to the cell identities from (B). D, Spatial reconstruction of patient 7: FOV 14. Cell identities are based on UMAP cluster (C). E, IHC staining of adjacent section to (D) showing distribution of immune cells (CD45), macrophages (CD68), epithelial cells (PanCK), and fibroblasts (FAP). F, Patient 7: FOV 2 representing adjacent normal tissue region containing salivary gland exposed to subasumstat showing reconstruction of cell types, MIL113 staining intensity, and IFN signature expression in respective panels. Note that the MIL113 staining data are from an adjacent section. G, Patient 7: FOV 14 representing a region of tumor cells exposed to subasumstat showing reconstruction of cell types, MIL113 staining intensity, and IFN signature expression in respective panels. Note that the MIL113 staining data are from an adjacent section.
We next evaluated the impact of subasumstat on distinct cell populations identified within the reconstructed spatial map of the TME from patients 7 and 8 starting with IFN response. An IFN signature from the CosMx probe set was derived from the GeoMx common gene signature. As expected, regions of subasumstat target engagement defined by MIL113 staining from tissue sections directly adjacent to those analyzed by CosMx showed strong correspondence with the spatial expression pattern of IFN pathway activation (Fig. 3F and G; Supplementary Fig. S11). We identified evidence of strong IFN pathway activation in distinct cell populations located within normal and malignant regions of the sections from patient 7, with less activation in patient 8 consistent with the DSP results (Fig. 2E and H; Supplementary Fig. S13C). For instance, pathological examination of a section from patient 7 had noted a region of normal salivary gland tissue, and our analysis clearly identified a unique epithelial cell population (Epithelial_KRT19) that mapped to this structure (Fig. 3F; Supplementary Fig. S8). A robust subasumstat-activated IFN response was observed from these epithelial salivary gland cells (Epithelial_KRT19), associated with a profound B-cell inflammatory response, including the recruitment of significant quantities of IgA B cells (B_Cell_IGHA; Fig. 3F). This finding is consistent with a tissue-specific immune response to IFN pathway activation that likely represents a specific and important protective mechanism at mucosal surfaces. Furthermore, consistent with previous studies performed in vitro and in mouse models of cancer (20, 22, 24) strong activation of IFN signaling in the TME was observed across multiple immune cell types, including T cells, stromal fibroblasts, and macrophages as well as in tumor cells (Fig. 3G; Supplementary Fig. S14). Therefore, we next evaluated how induction of IFN signaling by subasumstat impacts the biological activity of specific cell types within the intact TME.
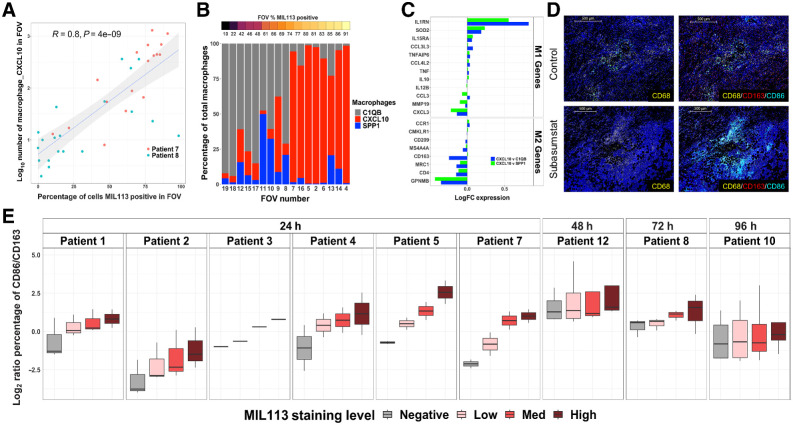
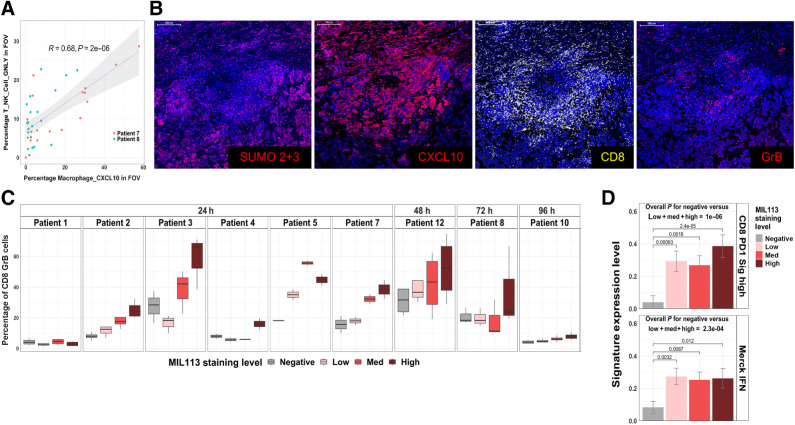
Further analysis by SMI revealed strong evidence that subasumstat induced significant changes to the activity state of specific immune and stromal cell populations within the TME that included tumor-associated macrophages (TAM), fibroblasts, and T cells. Cluster analysis revealed three subsets of macrophages and four sets of fibroblasts within the TME, each of which were named on the basis of a uniquely expressed transcript that was subsequently used to identify the subpopulation. For instance, we defined the three main macrophage subtypes as Macrophage_CXCL10, Macrophage_SPP1, and Macrophage_C1QB. The chemokine CXCL10 (IP-10) was one of the most robustly elevated transcripts identified as part of the common signature of response to subasumstat (Fig. 2E), and elevated expression has been previously shown to coincide with subasumstat-induced IFN1 response (20). Although several different cell types express CXCL10, we observed the highest expression in macrophages and fibroblasts. This expression showed a strong correlation between localized regions of subasumstat target engagement and the presence of the Macrophage_CXCL10 subpopulation within the TME (Fig. 4A; Supplementary Figs. S12–S14). A similar phenomenon was observed with a subpopulation of fibroblasts: Fibroblast_CXCL10 (Supplementary Figs. S12–S14). The increase in these populations appears to occur at the expense of macrophages and fibroblasts that do not express CXCL10 (Fig. 4B; Supplementary Figs. S12, S14, and S15). For example, in patient 7, a clear subasumstat-dependent increase in CXCL10-expressing cells appears to come at the expense of loss of C1QB-expressing cells, whereas in patient 8, the SPP1 population appears to be replaced by Macrophage_CXCL10 (Fig. 4B; Supplementary Fig. S15). Interestingly, this appears to be consistent with a conversion of the subasumstat-exposed TME from an immune suppressive to a more active state. Comparison of the expression profile of Macrophage_CXCL10 to either Macrophage_C1QB or Macrophage_SPP1 showed an upregulation of genes associated with M1 macrophages and a downregulation of genes associated with M2 macrophages (36), indicating that one effect of subasumstat is to shift the polarization state of macrophages within the TME (Fig. 4C). To further confirm this effect, we performed IHC with antibodies specific for CD68 (a general macrophage marker), CD163 (a marker of M2 polarization), and CD86 (a marker of M1 polarization) and observed elevation of dual positive CD86/CD68 cells in localized regions specifically exposed to subasumstat (Fig. 4D). Importantly, we observed this shift in the ratio of expression of CD86 over CD163 occurring in the CD68 population across all patient samples examined 24 hours after CIVO-mediated intratumor delivery of subasumstat (Fig. 4E). Taken together with induction of IFN signaling and increased expression of proinflammatory chemokines such as CXCL10, the observed skewing of macrophage polarization from M2 to M1 indicates that inhibition of SUMOylation induces a shift in the activity state of the TME from immune suppressive to immune active, commonly referred to as “cold-to-hot” (29, 37, 38). This phenomenon was not limited to macrophage polarization. As further evidence to this effect, we detected a strong correlation between the percentage of cytotoxic cells (T_NK_Cell_GNLY likely encompassing both T and NK cells expressing granulolysin) and the percentage of Macrophage_CXCL10 (R = 0.68; P = 2e−06) within the reconstructed TME (Fig. 5A). A similar relationship was observed between MIL-113-positive cells and the percentage of T_NK_Cell GNLY within the FOV evaluated by SMI (Supplementary Fig. S12). This was further observed by IHC and ISH biomarker analysis that demonstrated that localized inhibition of SUMOylation coincided with an overt increase in CXCL10, enrichment of CD8-positive T cells, and an elevation of granzyme B (Fig. 5B). Again, with some patient-specific differences noted, elevated cell activation was not limited to patient samples evaluated by SMI but trended with subasumstat target engagement across most patient tumor samples in our study (Fig. 5C). Consistent with subasumstat induced T-cell activation, further evidence was obtained by reference back to the DSP data in which we detected subasumstat-dependent increases in two published signatures of T-cell activation: “CD8 PD1 Sig high” and “Merck IFN” (refs. 29, 30; Fig. 5D; Supplementary Fig. S16). Evaluation of the cellular composition of multiple nondrug-exposed regions collected from each tumor in our study set, representing pre-existing TME conditions before the introduction of subasumstat, suggested that intrinsic differences of immune cell populations, specifically the ratio of CD8 to CD68-positive cells significantly influence immune response to subasumstat (Supplementary Fig. S17).
Figure 4.
Subasumstat induces an increase in proinflammatory CXCL10-expressing macrophages in the TME that exhibit a shift in polarization state from M2 to M1. A, Correlation of number of Macrophage_CXCL10 cells (log10) to subasumstat drug target engagement measured by the percentage of cells MIL113-positive in each FOV. A linear model fit is shown with associated Pearson correlation, P value, as well as 95% confidence intervals (shaded). Note that the MIL113-staining data are from an adjacent section. B, The percentage of each of three macrophage populations in individual FOVS from patient 7. FOVs are ordered from left to right by increasing MIL113 positivity. Note that the MIL113 staining data are from an adjacent section. C, Differential expression of M1 and M2 genes in comparison of Macrophage CXCL10 with either Macrophage_C1QB or Macrophage_SPP1. Data are from both patients 7 and 8. D, Immunofluorescent staining for macrophages (CD68) and mIF staining for CD68/CD163/CD86 at regions of subasumstat drug target engagement (TAK-981) and regions of no subasumstat drug target engagement (control) from patient 7. CIVO GLO tracking microspheres shown in green; scale bar, 500 μm. E, M1/M2 ratio for each patient sample shown as log2 of the ratio of the percentage of CD86+ cells to the percentage of CD163 cells. Box plot represents the distribution and the median value of multiple tumor sections taken from different tumor regions within each patient and analyzed for response to subasumstat target engagement. Note that only one section from patient 3 was available for this analysis.
Figure 5.
Presence of CXCL10 macrophages correlates with an increase in localized recruitment of cytotoxic immune cells. A, Correlation of the percentage of T, NK, and granulolysin (GNLY)-positive cells in CosMx FOVs to the percentage of Macrophage_CXCL10 cells. A linear model fit is shown with associated Pearson correlation, and P value as well as 95% confidence intervals (shaded). Note that the MIL113 staining data are from an adjacent section. B, Immunofluorescent staining for SUMO 2+3, CXCL10, CD8, and granzyme B at site of subasumstat drug target engagement in patient 7; scale bar, 500 μm. C, Relationship between activated T cells and subasumstat target engagement across patient samples is plotted as the percentage of CD8+GrB+ cells. Box plot represents the distribution and the median value of multiple tumor sections taken from different tumor regions within each patient and analyzed for response to subasumstat target engagement. D, Expression of two signatures of T-cell activation; “CD8 PD1 Sig high” (45) and “Merck IFN” (44) grouped by MIL113 grade. Signature values were calculated by GSVA and grouped by MIL113-staining levels from DSP data for 354 ROI from nine patients. Error bars represent standard error of the mean for grouped data. P values for CD8 PD1 signature high: low = 0.00083, med = 0.0016, high = 2.4×10−5. P values for Merck IFN signature: low = 0.0032, med = 0.0087, high = 0.012. The overall P value represents a t test comparing the drug-unexposed (negative) to drug-exposed (low, med, high) ROI.
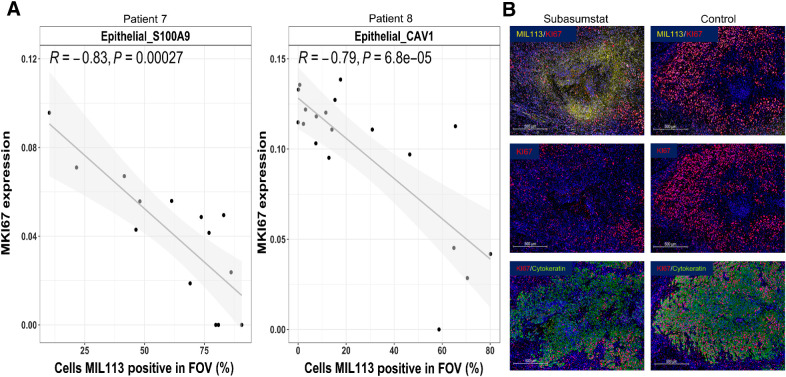
Finally, we used SMI to explore the decrease in cell-cycle signature, observed by DSP in subasumstat-exposed regions of the TME, at the cellular level. Although the available CosMx gene set represents very few of the genes in the cell-cycle response signature, we were able to identify changes to MKI67, a well-established marker of cell proliferation, associated with subasumstat-target engagement. Most notably, we observed a strong negative correlation of MKI67 expression with MIL113 positivity that appears to be restricted solely to the tumor cell components of the TME (Epithelial_S100A9 in patient 7 and Epithelial_CAV1 in patient 8; Fig. 6A; Supplementary Fig. S18). This tumor cell–specific effect was confirmed by IHC (Fig. 6B). This is consistent with known effects of both SUMO inhibition and IFN1 signaling on cell-cycle progression (22). Furthermore, in addition to cell-cycle inhibition, we detected elevation of cleaved caspase 3 (CC3)–positive cells proximal to the subasumstat injection sites (noted by the presence of the FTM). It is notable that increased apoptosis was coincident with enrichment of activated T cells dual positive for CD8 and granzyme B (GrB) infiltrating into CK-rich regions of tumor epithelium compared with nondrug-exposed controls (Supplementary Fig. S19). This is also consistent with previous observations that antitumor effects of subasumstat are dependent upon antitumor immune cell activity (20).
Figure 6.
Subasumstat target engagement is specifically associated with a reduction in markers of cell-cycle progression in malignant epithelial cells from patients 7 and 8. A, Correlation of MKI67 expression to drug target engagement measured by the percentage of cells MIL113-positive in Epithelial_S100A9 cells (patient 7) and Epithelial_CAV1 cells (patient 8), respectively. Linear models were fit and are shown with associated Pearson correlation, P value, as well as 95% confidence intervals (shaded). B, Immunofluorescent staining for subasumstat-SUMO adducts (MIL113, yellow), cell proliferation marker KI67 (red), and tumor cell marker (cytokeratin, green) showing the reduction in proliferating tumor epithelium in regions of subasumstat drug target engagement compared with a directly adjacent uninjected region, no drug control; scale bar, 500 μm.
Discussion
This study addresses a long-standing challenge in early cancer drug development: the lack of available platforms to generate reliable mechanistic understanding of drug activity in intact tumors (39). Using CIVO technology as the delivery platform and high content spatial biology technologies as the readout, we show dose-responsive molecular effects at cellular resolution in tumors in situ and elaborate the specific cellular and molecular effects of a novel investigational oncology agent under active clinical development. Rarely is it possible to reveal this level of mechanistic insight so early in clinical development and crucially, in an actual patient population for whom the drug is being developed.
Within the limits and controls of this exploratory phase 0 clinical trial, we observed that subasumstat-mediated inhibition of SUMOylation induces specific cellular changes in both the malignant cells of the tumor and the associated TME. Mechanistically, these effects partition into a reduction of the expression of genes associated with cell-cycle progression, which appears to be largely confined to the malignant epithelial cells and a pronounced upregulation of genes associated with IFN responses that affect multiple components of the TME, including malignant epithelial, immune, and stromal cells. It is unclear whether the cell-cycle effects are mediated directly via inhibition of SUMOylation in malignant cells themselves or indirectly through the effects on the TME, although mechanisms have been described for both, and IFN-induced cell-cycle arrest in tumor cells has been well documented (20, 21, 40). In contrast, the effects on the TME are consistent with a direct downstream effect of subasumstat on IFN1 expression and stimulation of IFN1 signaling. One key effect of subasumstat on the TME, observed across patient samples in our study, was conversion of macrophages from immune suppressive (M2) to inflammatory (M1).This was initially detected by SMI comparing MIL113-negative to MIL113-positive regions of the TME that revealed a shift from macrophage subtypes expressing C1Q or SPP1, biomarkers associated with M2 polarized macrophages, to a subtype expressing CXCL10, an M1-associated marker, and then confirmed by IHC. This phenotypic shift is consistent with the observed elevation of IFN signatures across patient samples, the previous established role of IFNγ as a potent inducer of M1 polarization (41, 42), and emerging data showing that type 1 IFN signaling also lead to a similar shift in macrophage activity (24, 43–45). Importantly, the study by Nakamura and colleagues (24) demonstrated that this shift occurs via direct effect of subasumstat on IFN1 activation, M1 polarization, and stimulation of phagocytic activity in cultured human monocyte–derived macrophages. The data presented here demonstrate that the direct effects of subasumstat can be observed in the context of the intact TME, where TAMs are situated, and likely actively promote, a complex immune-suppressive environment. This is further confirmed and expanded by the finding that in the precise locations of drug exposure within the same tumor samples where we observed M1 polarization, we also observed recruitment of CXCR3-expressing cells, including cytotoxic CD8 T and NK cells to regions of SUMOylation inhibition, characteristic of a shift in the TME from “cold-to-hot” (46, 47).
Although exposure of tumor tissue to subasumstat leads to observable changes in the TME that render the tumor more amenable to anti-neoplastic activity, our study was limited to the phase 0 space, which is inherently constrained to evaluating local PD response and does not expand to informing or directing patient care. Ideally, combining our approach with systemic validation of outcomes when investigational assets and approved agents are dosed both intratumorally and systemically will provide the most salient application of our technology, especially when combined with high-plex analytic tools. Identifying responder signatures across an array of biomarkers by DSP and SMI provides a theoretical mechanism of truly personalized medicine, unlike any other platform to date. Ultimately, for both phase 0 translational investigation and for personalized medicine applications of the approach described here, a well-designed trial demonstrating concordance between the localized responses induced by CIVO and responses induced by systemically delivered drug will be important. In this regard, encouraging data have emerged from ongoing phase 1/2 clinical evaluations of subasumstat that show that intravenous administration results in dose-dependent elevation of the same biomarkers highlighted in this phase 0 investigation, including CXCL10 and CD86 in peripheral blood sample, concordant with our findings in the TME (48, 49).
That said, these outcomes have the potential to enable a level of precision and efficiency in the bridge between preclinical and clinical drug development heretofore unattainable. We are able to directly test whether a proposed mechanism of action of a novel agent does, or does not, produce its intended effect in the most applicable context: an in situ human tumor with its functioning TME. This has the potential to accelerate development by discriminating between novel drug candidates that produce on-target biological responses and those that do not. Whereas the results presented here for subasumstat were consistent with preclinical studies, this does not always hold true for other agents. For example, in contrast with preclinical results, we previously showed that the anti-PDGF receptor antagonizing antibody olaratumab induced no effect on CIVO-mediated delivery to localized tumor regions in patients with soft tissue sarcoma (16). Ultimately, olaratumab also showed no benefit in phase III trials (50).
It is also worth noting that the CIVO platform is multiplexed, which allows for microdosing of up to eight different drugs, or combinations of drugs. As such, it can be used to inform a development strategy that includes combination dosing. Especially in the context of a development asset that is intended to complement or potentiate the effect of an existing drug (or drug class), identifying which drug combinations exhibit the most promising antitumor responses in patients before initiation of late-stage validation trials is essential.
We also raise the importance of interpatient heterogeneity in interpreting drug performance. As with all clinical trials, sample size is critical, and although the paradigm in which phase 0 microdosing studies is conducted is limited in patient sample size (∼12), and inherently exploratory, the combination of CIVO and spatial biology affords the opportunity to investigate drug response across a diverse range of TME even in a single tumor. Here, 354 ROI representing many distinct TME were evaluated by DSP across nine patients. So, although patient sample size is low, sampling of TME diversity is sufficient to evaluate drug-induced changes in the TME. Although a drug with pleotropic effects such as subasumstat lends itself to exploration of a wide variety of biological responses, we focused on robust responses that were observed in common across intratumor and interpatient samples. Our goal was to demonstrate that if responses could be detected in common across a wide range of TME samples, those observed effects may be applicable to broader disease, including metastatic lesions in a given patient. Although it will ultimately be interesting to evaluate patient-specific responses using the approach presented here, this was outside the scope of the current article. Even within the context of responses observed in common across patient samples, it is evident that there is significant heterogeneity that must be acknowledged. In the DSP data, we see a clear partition between strong IFN responders like patients 5 and 7, and weak responders like patients 1 and 3 (Fig. 2C and D). The CosMx data show that the TME are significantly different between patients 8 and 7, with a much larger population of SPP1-expressing macrophages in the former (Supplementary Fig. S9). Given the association of SPP1 macrophages with an immunosuppressive environment (51), it is tempting to speculate that this influences the stronger overall response of patient 7 compared with patient 8. A more robust sample size would help drive statistical assertions of outcomes.
Ultimately, and in the context of a precision medicine approach, including an ever-growing panoply of therapeutic interventions, a future can be envisaged in which multiplex microdosing of tumors allows direct relevant testing of treatment options before committing to systemic drug delivery, thereby accelerating the path to the most effective therapy for the individual patient. In the interim, rigorous and safe evaluation of candidate assets for on- or off-target mechanistic validation in a truly robust setting provides a critical stage gate early in the drug development pipeline.
Supplementary Material
ISH Probe Information
Primary Antibody Information
Secondary Antibody Information
Patient Demographics
GeoMx ROI Information
Enrichment analysis for genes upregulated by Subasumstat
Enrichment analysis for genes Downregulated by Subasumstat
Subasumstat Target Engagement Grading by MIL113 Staining
Expression of IFNB1 and ISG15 as a function of subasumstat exposure.
Subasumstatinduces IFNβ in immune cells.
Subasumstat induces IFN response genes in CIVO micro-injected tumors
Subasumstat dependent decrease in cell cycle signature observed across the tumor sample set by DSP
Subasumstat induces interferon-response genes in a dose responsive fashion
FOV placement for SMI Analysis for Patients 7 and 8 showing variation between sampled regions of the tumor
UMAP Cluster Analysis for Patients 7 and 8
Spatial Reconstruction of Patient 7 and Patient 8
Percentages of Cells identified by UMAP analysis from Tumors 7 and 8
Interferon Signature Expression closely matches MIL113 staining
Correlation of Percentage of cell types in TME to Percent of Cells MIL113 Positive
TME cell spatial distributions in Patient 7 and Patient 8
Subasumstat induces changes in the TME associated with Interferon Signature expression
Percentage of each of three macrophage populations in individual FOVS from Patient 8
Percentage of each of three macrophage populations in individual FOVS from Patient 8
Subasumstat-induced Interferon signature expression level is predicted by the CD8/CD68 ratio of the baseline tumor
Correlation of MKI67 gene expression levels to Percent MIL113 Positivity for Tumors 7 and 8
Subasumstat initiates cell death and cytotoxic T cell activity
Acknowledgments
This study was funded by Takeda Pharmaceutical Company and Presage Biosciences, Inc. The authors would like to thank Kate Gillespie and Bre Mills for their expert technical assistance in running GeoMx and IHC experiments for this study. We would also like to acknowledge the many contributions from the Presage clinical team in conducting the clinical trial and the clinical teams and pathologists at all patient enrolling sites.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
J.M.J. Derry reports other support from Takeda during the conduct of the study, employment with Presage Biosciences, and holds stock options in the company. C. Burns reports other support from Takeda during the conduct of the study and reports employment and stock options with Presage Biosciences. J.P. Frazier reports other support from Takeda Pharmaceuticals during the conduct of the study, other support from Presage Biosciences outside the submitted work, and holds a patent for CIVO injection technology. E. Beirne reports other support from Takeda during the conduct of the study and other support from Presage Biosciences, Inc. outside the submitted work. M. Grenley reports personal fees from Takeda Pharmaceuticals during the conduct of the study, personal fees from Presage Biosciences outside the submitted work, and holds a patent for 10478157B2 issued. C.C. DuFort reports other support from Takeda Pharmaceuticals during the conduct of the study and other support from Presage Biosciences outside the submitted work. E. Killingbeck reports other support from Presage Biosciences, Inc. and personal fees from NanoString Technologies during the conduct of the study. M. Leon reports other support from NanoString Technologies outside the submitted work. C. Williams reports employment with NanoString Technologies and holds NanoString stock/stock options. M. Gregory reports personal fees from NanoString Technologies during the conduct of the study and reports a patent for US2022/02823113 pending, US10415080 issued, US11473142 issued, a patent for US11549139 issued, and WO2021257795A1 pending. D. Huszar reports other support from Takeda Pharmaceuticals outside the submitted work and holds a patent for PCT/IB 2022/060413 pending to Takeda Pharmaceuticals. A. Berger reports full-time employment and is a stockholder of Takeda Development Center Americas, Inc. R.A. Klinghoffer reports other support from Takeda Pharmaceutical Company during the conduct of the study, other support from Takeda Pharmaceutical Company, Merck and Company, Bristol Myers Squibb, and Pure Biologics outside the submitted work; reports employment with Presage Biosciences and is a shareholder in the company; and reports stock options in Presage. No disclosures were reported by the other authors.
Authors' Contributions
J.M.J. Derry: Conceptualization, data curation, software, formal analysis, supervision, methodology, writing–original draft, writing–review and editing. C. Burns: Data curation, formal analysis, writing–review and editing. J.P. Frazier: Conceptualization, resources, supervision, methodology, writing–review and editing. E. Beirne: Formal analysis, supervision, methodology, writing–review and editing. M. Grenley: Formal analysis. C.C. DuFort: Writing–review and editing. E. Killingbeck: Formal analysis. M. Leon: Formal analysis. C. Williams: Formal analysis. M. Gregory: Formal analysis. J. Houlton: Investigation. D. Clayburgh: Investigation. P. Swiecicki: Investigation. D. Huszar: Conceptualization, supervision, investigation, writing–review and editing. A. Berger: Conceptualization, supervision, investigation, writing–review and editing. R.A. Klinghoffer: Conceptualization, resources, data curation, supervision, writing–original draft, writing–review and editing.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 2. Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013;14:1014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cox TR. The matrix in cancer. Nat Rev Cancer 2021;21:217–38. [DOI] [PubMed] [Google Scholar]
- 4. Bejarano L, Jordāo MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov 2021;11:933–59. [DOI] [PubMed] [Google Scholar]
- 5. Xiao Y, Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther 2021;221:107753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Son B, Lee S, Youn H, Kim E, Kim W, Youn B. The role of tumor microenvironment in therapeutic resistance. Oncotarget 2017;8:3933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 2015;21:1318–25. [DOI] [PubMed] [Google Scholar]
- 8. Letai A, Bhola P, Welm AL. Functional precision oncology: testing tumors with drugs to identify vulnerabilities and novel combinations. Cancer Cell 2022;40:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chuprin J, Buettner H, Seedhom MO, Greiner DL, Keck JG, Ishikawa F, et al. Humanized mouse models for immuno-oncology research. Nat Rev Clin Oncol 2023;20:192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pound P, Ritskes-Hoitinga M. Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail. J Transl Med 2018;16:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greek R, Hansen LA. Questions regarding the predictive value of one evolved complex adaptive system for a second: exemplified by the SOD1 mouse. Prog Biophys Mol Biol 2013;113:231–53. [DOI] [PubMed] [Google Scholar]
- 12. Wong CH, Siah KW, Lo AW. Estimation of clinical trial success rates and related parameters. Biostatistics 2019;20:273–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seyhan AA. Lost in translation: the valley of death across preclinical and clinical divide—identification of problems and overcoming obstacles. Transl Med Commun 2019;4:18. [Google Scholar]
- 14. Klinghoffer RA, Bahrami SB, Hatton BA, Frazier JP, Moreno-Gonzalez A, Strand AD, et al. A technology platform to assess multiple cancer agents simultaneously within a patient's tumor. Sci Transl Med 2015;7:284ra58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frazier JP, Bertout JA, Kerwin WS, Moreno-Gonzalez A, Casalini JR, Grenley MO, et al. Multidrug analyses in patients distinguish efficacious cancer agents based on both tumor cell killing and immunomodulation. Cancer Res 2017;77:2869–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gundle KR, Deutsch GB, Goodman HJ, Pollack SM, Thompson MJ, Davis JL, et al. Multiplexed evaluation of microdosed antineoplastic agents in situ in the tumor microenvironment of patients with soft tissue sarcoma. Clin Cancer Res 2020;26:3958–68. [DOI] [PubMed] [Google Scholar]
- 17. Zollinger DR, Lingle SE, Sorg K, Beechem JM, Merritt CR. GeoMx RNA assay: high multiplex, digital, spatial analysis of RNA in FFPE tissue. Methods Mol Biol 2020;2148:331–45. [DOI] [PubMed] [Google Scholar]
- 18. He S, Bhatt R, Brown C, Brown EA, Buhr DL, Chantranuvatana K, et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol 2022;40:1794–806. [DOI] [PubMed] [Google Scholar]
- 19. Seeler J-S, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer 2017;17:184–97. [DOI] [PubMed] [Google Scholar]
- 20. Lightcap ES, Yu P, Grossman S, Song K, Khattar M, Xega K, et al. A small-molecule SUMOylation inhibitor activates antitumor immune responses and potentiates immune therapies in preclinical models. Sci Transl Med 2021;13:eaba7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Langston SP, Grossman S, England D, Afroze R, Bence N, Bowman D, et al. Discovery of TAK-981, a first-in-class inhibitor of SUMO-activating enzyme for the treatment of cancer. J Med Chem 2021;64:2501–20. [DOI] [PubMed] [Google Scholar]
- 22. He X, Riceberg J, Soucy T, Koenig E, Minissale J, Gallery M, et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat Chem Biol 2017;13:1164–71. [DOI] [PubMed] [Google Scholar]
- 23. Decque A, Joffre O, Magalhaes JG, Cossec J-C, Blecher-Gonen R, Lapaquette P, et al. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat Immunol 2016;17:140–9. [DOI] [PubMed] [Google Scholar]
- 24. Nakamura A, Grossman S, Song K, Xega K, Zhang Y, Cvet D, et al. The SUMOylation inhibitor subasumstat potentiates rituximab activity by IFN1-dependent macrophage and NK cell stimulation. Blood 2022;139:2770–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gu Z, Eils R, Schlesner M. Complex heat maps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 28. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFNγ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bassez A, Vos H, Van Dyck L, Floris G, Arijs I, Desmedt C, et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med 2021;27:820–32. [DOI] [PubMed] [Google Scholar]
- 31. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choudhary S, Satija R. Comparison and evaluation of statistical error models for scRNA-seq. Genome Biol 2022;23:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Borcherding N, Vishwakarma A, Voigt AP, Bellizzi A, Kaplan J, Nepple K, et al. Mapping the immune environment in clear cell renal carcinoma by single-cell genomics. Commun Biol 2021;4:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zaitsev A, Chelushkin M, Dyikanov D, Cheremushkin I, Shpak B, Nomie K, et al. Precise reconstruction of the TME using bulk RNA-seq and a machine learning algorithm trained on artificial transcriptomes. Cancer Cell 2022;40:879–94. [DOI] [PubMed] [Google Scholar]
- 37. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960–4. [DOI] [PubMed] [Google Scholar]
- 38. Emens LA, Silverstein SC, Khleif S, Marincola FM, Galon J. Toward integrative cancer immunotherapy: targeting the tumor microenvironment. J Transl Med 2012;10:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shih H-P, Zhang X, Aronov AM. Drug discovery effectiveness from the standpoint of therapeutic mechanisms and indications. Nat Rev Drug Discov 2018;17:19–33. [DOI] [PubMed] [Google Scholar]
- 40. Sangfelt O. Mechanisms of interferon-induced cell-cycle arrest. Front Biosci 2000;5:d479. [DOI] [PubMed] [Google Scholar]
- 41. Zhu L, Zhao Q, Yang T, Ding W, Zhao Y. Cellular metabolism and macrophage functional polarization. Int Rev Immunol 2015;34:82–100. [DOI] [PubMed] [Google Scholar]
- 42. Ivashkiv LB. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease, and cancer immunotherapy. Nat Rev Immunol 2018;18:545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ming-Chin Lee K, Achuthan AA, De Souza DP, Lupancu TJ, Binger KJ, Lee MKS, et al. Type I interferon antagonism of the JMJD3–IRF4 pathway modulates macrophage activation and polarization. Cell Rep 2022;39:110719. [DOI] [PubMed] [Google Scholar]
- 44. Liao J, Zeng DN, Li JZ, Hua QM, Huang CX, Xu J, et al. Type I IFNs repolarized a CD169+ macrophage population with antitumor potentials in hepatocellular carcinoma. Mol Ther 2022;30:632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol 2015;33:445–74. [DOI] [PubMed] [Google Scholar]
- 46. House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res 2020;26:487–504. [DOI] [PubMed] [Google Scholar]
- 47. Chow MT, Ozga AJ, Servis RL, Frederick DT, Lo JA, Fisher DE, et al. Intratumoral activity of the CXCR3 chemokine system Is required for the efficacy of anti-PD-1 therapy. Immunity 2019;50:1498–1512.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dudek A, Juric D, Dowlati A, Vaishampayan U, Assad H, Rodón J, et al. 476 First-in-human phase 1/2 study of the first-in-class SUMO-activating enzyme inhibitor TAK-981 in patients with advanced or metastatic solid tumors or relapsed/refractory lymphoma: phase 1 results. J Immunother 2021:9;A505–6. Available from:https://jitc.bmj.com/content/9/Suppl_2/A505 [Google Scholar]
- 49. Saggu G, Stroopinsky D, Dudek AZ, Olszanski AJ, Juric D, Dowlati A, et al. Subasumstat, a first-in-class inhibitor of SUMO-activating enzyme, demonstrates dose-dependent target engagement and SUMOylation inhibition, leading to rapid activation of innate and adaptive immune responses in the dose escalation portion of a phase 1/2 clinical study. Eur J Cancer 2022:174;S125–6. Available from:http://www.ejcancer.com/article/S0959804922011340/fulltext [Google Scholar]
- 50. Schulman K. Olaratumab for STS disappoints in phase III. Cancer Discov 2019;9:312–3. [DOI] [PubMed] [Google Scholar]
- 51. Shurin MR. Osteopontin controls immunosuppression in the tumor microenvironment. J Clin Invest 2018;128:5209–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ISH Probe Information
Primary Antibody Information
Secondary Antibody Information
Patient Demographics
GeoMx ROI Information
Enrichment analysis for genes upregulated by Subasumstat
Enrichment analysis for genes Downregulated by Subasumstat
Subasumstat Target Engagement Grading by MIL113 Staining
Expression of IFNB1 and ISG15 as a function of subasumstat exposure.
Subasumstatinduces IFNβ in immune cells.
Subasumstat induces IFN response genes in CIVO micro-injected tumors
Subasumstat dependent decrease in cell cycle signature observed across the tumor sample set by DSP
Subasumstat induces interferon-response genes in a dose responsive fashion
FOV placement for SMI Analysis for Patients 7 and 8 showing variation between sampled regions of the tumor
UMAP Cluster Analysis for Patients 7 and 8
Spatial Reconstruction of Patient 7 and Patient 8
Percentages of Cells identified by UMAP analysis from Tumors 7 and 8
Interferon Signature Expression closely matches MIL113 staining
Correlation of Percentage of cell types in TME to Percent of Cells MIL113 Positive
TME cell spatial distributions in Patient 7 and Patient 8
Subasumstat induces changes in the TME associated with Interferon Signature expression
Percentage of each of three macrophage populations in individual FOVS from Patient 8
Percentage of each of three macrophage populations in individual FOVS from Patient 8
Subasumstat-induced Interferon signature expression level is predicted by the CD8/CD68 ratio of the baseline tumor
Correlation of MKI67 gene expression levels to Percent MIL113 Positivity for Tumors 7 and 8
Subasumstat initiates cell death and cytotoxic T cell activity
Data Availability Statement
GeoMx gene expression data were deposited into the Gene Expression Omnibus database under the accession number GSE235475 and are available at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE235475. Additional data generated in this study are available upon reasonable request from the corresponding author.