Abstract
Purpose:
Urinary comprehensive genomic profiling (uCGP) uses next-generation sequencing to identify mutations associated with urothelial carcinoma and has the potential to improve patient outcomes by noninvasively diagnosing disease, predicting grade and stage, and estimating recurrence risk.
Experimental Design:
This is a multicenter case–control study using banked urine specimens collected from patients undergoing initial diagnosis/hematuria workup or urothelial carcinoma surveillance. A total of 581 samples were analyzed by uCGP: 333 for disease classification and grading algorithm development, and 248 for blinded validation. uCGP testing was done using the UroAmp platform, which identifies five classes of mutation: single-nucleotide variants, copy-number variants, small insertion-deletions, copy-neutral loss of heterozygosity, and aneuploidy. UroAmp algorithms predicting urothelial carcinoma tumor presence, grade, and recurrence risk were compared with cytology, cystoscopy, and pathology.
Results:
uCGP algorithms had a validation sensitivity/specificity of 95%/90% for initial cancer diagnosis in patients with hematuria and demonstrated a negative predictive value (NPV) of 99%. A positive diagnostic likelihood ratio (DLR) of 9.2 and a negative DLR of 0.05 demonstrate the ability to risk-stratify patients presenting with hematuria. In surveillance patients, binary urothelial carcinoma classification demonstrated an NPV of 91%. uCGP recurrence-risk prediction significantly prognosticated future recurrence (hazard ratio, 6.2), whereas clinical risk factors did not. uCGP demonstrated positive predictive value (PPV) comparable with cytology (45% vs. 42%) with much higher sensitivity (79% vs. 25%). Finally, molecular grade predictions had a PPV of 88% and a specificity of 95%.
Conclusions:
uCGP enables noninvasive, accurate urothelial carcinoma diagnosis and risk stratification in both hematuria and urothelial carcinoma surveillance patients.
Translational Relevance.
The implementation of risk-stratified care in the management of urothelial carcinoma is a major challenge. New tools are needed for both primary diagnosis and detection of minimal residual disease (MRD) for recurrence risk prediction. Quantification of somatic genomic alterations in urine-derived DNA has notable promise to enhance risk stratification. In this study, we validated a urine-based test that performs urinary comprehensive genomic profiling (uCGP) to predict the presence and pathologic grade of urothelial carcinoma. In the initial diagnosis context, uCGP performance exceeds available urine biomarkers and cytology. In the surveillance context, uCGP significantly prognosticates future recurrence using a non-tumor informed (urine only) MRD paradigm that stratifies patients into high- and low-risk of recurrence. These findings demonstrate the unique utility of genomics to inform personalized risk stratification.
Introduction
In the United States, bladder cancer is the fourth most common cancer in men and fifth most common solid tumor overall (1). Though rare subtypes exist, most bladder cancers will be diagnosed as urothelial carcinoma (1). The most common symptom leading to initial diagnosis of urothelial carcinoma is hematuria (2). Guidelines recommend that hematuria severity and other clinical risk factors guide the use of transurethral white-light cystoscopy (WLC) to detect urothelial carcinoma (3). However, adherence to guidelines is poor and many patients at risk for urothelial carcinoma are never referred to urologists for evaluation (4, 5). High prevalence of hematuria in the adult population combined with low incidence of urothelial carcinoma and invasiveness of cystoscopy all contribute to under-evaluation (3, 6, 7). A noninvasive detection tool that identifies those most likely to have urothelial carcinoma could reduce the burden of evaluation, improving adherence to guidelines. For patients referred to the urologist, evaluation by WLC occurs only 35% of the time (8). A noninvasive diagnostic could help urologists reduce cystoscopy in patients with low likelihood of disease and prioritize patients with the highest risk.
WLC is also the standard of care for urothelial carcinoma surveillance following initial diagnosis and treatment. Under American Urological Association (AUA) guidelines, intermediate- and high-risk patients with urothelial carcinoma undergo post-treatment surveillance to visually assess the bladder for recurrence every 3–6 months for at least five years, and annually thereafter (9, 10). The invasiveness and frequency of surveillance is a challenge for many patients, who often have complicating comorbidities, resulting in a 40% adherence to recommended surveillance intensity (11). A noninvasive test that provides both diagnostic and prognostic information could, therefore, improve guideline compliance and support implementation of risk-adapted care.
Urine-based comprehensive genomic profiling (uCGP) using next-generation sequencing is well positioned to support these objectives for both initial diagnosis and surveillance, but challenges remain for clinical adoption. Although there is typically a clear distinction between tumor positive and negative in the initial diagnosis setting, tumor status in the urothelial carcinoma surveillance setting is often ambiguous. Because both muscle-invasive and non–muscle-invasive urothelial carcinoma recurs at a high rate (50%–70%; refs. 12, 13), minimal/molecular residual disease (MRD) following resection is expected to be common. However, whereas uCGP can detect mutations associated with these malignant cells before they become visually apparent, WLC may classify such patients as cancer negative until their tumor grows large enough for endoscopic visualization. In these cases, binary classification of cancer status becomes a matter of debate: Should classification align with WLC or with molecular disease presence and long-term recurrence outcomes? Prioritizing the detection of MRD maximizes test sensitivity but compromises specificity relative to cystoscopy and can present clinical actionability challenges for physicians. Favoring concordance with WLC allows for higher specificity but imposes the sensitivity ceiling of visualization by WLC (65%–71%; refs. 14, 15). Although detection of MRD for recurrence prediction has recognized utility in leukemia and some solid tumors (16–19), early implementation attempts for urothelial carcinoma surveillance have not achieved the performance necessary to be widely adopted.
In this study, we investigate the ability of the uCGP assay UroAmp to diagnose urothelial carcinoma, predict tumor grade and invasive potential, and stratify recurrence risk. UroAmp's development and technical performance were previously described in a matched tumor–urine correlation study with longitudinal monitoring of diverse clinical case studies (20). Here, we present blinded clinical validations of multiple diagnostic algorithms across diverse patient scenarios for detection and monitoring of bladder cancer. Test performance is compared with cystoscopy and cytology with histopathology confirmation and long-term outcomes.
Materials and Methods
Patients
We performed a blinded, case-controlled, multicenter validation study comprising consented patients recruited from 10 urology clinics in accordance with IRB-approved protocols and following the ethical guidelines of U.S. Common Rule. Subject informed consent was obtained verbally before participation, in accordance with the study's minimal risk designation. Validation cohort inclusion/exclusion criteria were defined in a prospective analysis plan before clinical data review and sample selection, and individual site PI's identified patients meeting inclusion criteria (Supplementary Table S1). Cases and controls were recruited for two clinical contexts: initial diagnosis of urothelial carcinoma and surveillance for urothelial carcinoma recurrence. Tumor-positive cases consisted of patients who underwent cystoscopy leading to a de novo urothelial carcinoma diagnosis (for the initial diagnosis context) or a recurrent urothelial carcinoma diagnosis (for the surveillance context); patients were eligible if they had pathologically confirmed urothelial carcinoma present in the urinary tract at the time of urine collection and the urine sample was collected before surgical resection of the lesion. For the initial diagnosis context, negative controls consisted of urology patients with urine samples collected at office visits for benign urologic conditions and were excluded if they had pathologic, cystoscopic, or imaging evidence of, or a known history of urothelial carcinoma; or pathologic, imaging, or laboratory evidence of current or prior prostate or renal cancer; or hematuria that was not evaluated to standard of care (cystoscopy and imaging of the upper tract). For the surveillance context, negative controls consisted of patients with prior definitive surgical treatment of urothelial carcinoma and negative cystoscopy within one month of urine collection, and a second negative cystoscopy 3–12 months following the collection. Tumor positive and surveillance-negative patients were excluded if they had prior history of pelvic radiation.
For validation, banked historical samples were provided to the sponsor (Convergent Genomics) in a blinded fashion, ensuring that the clinical status of samples was unknown by the sponsor laboratory or data analysis personnel. Before processing validation samples, the sponsor completed algorithm development and training, and all algorithms were “locked” with digital time stamps using the Git version control system. Disease classification predictions were made by the sponsor and returned to clinical site investigators for performance analysis. This work was performed in accordance with STARD/REMARK best practices for diagnostic accuracy studies (21, 22).
Clinical patient monitoring
All urothelial carcinoma treatment decisions were made in accordance with clinical best practice guidelines for bladder cancer (10). Initial diagnosis of urothelial carcinoma required cystoscopic identification followed by pathologic confirmation. WLC or augmented endoscopy was permitted at the discretion of the treating physician. Urothelial carcinoma surveillance was performed in accordance with AUA risk stratification for non–muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC; refs. 23, 24). For example, surveillance of AUA high-risk patients with NMIBC was performed by cystoscopy and cytology at 3-month intervals for two years and 3- to 6-month intervals thereafter. Upper tract and abdominal/pelvic-imaging baselines were established and repeated every 1–2 years or as clinically indicated. Observed recurrences required pathology confirmation as prespecified in the study inclusion criteria.
uCGP
Voided urine specimens were collected before standard-of-care flexible WLC in clinic or rigid WLC at time of surgical resection. All urine voids (mid-stream, first-void, etc.) were considered. Urine samples provided by Ohio State University (OSU) and CURE clinical sites were collected at the site in Enhanced Preservation Media (20). All samples from these institutions were maintained at room temperature and shipped at ambient conditions by FedEx Clinical Pak, and samples were banked at –80°C within five days from collection. Samples provided by Massachusetts General Hospital (MGH) and Oregon Health and Science University (OHSU) were refrigerated immediately after collection and frozen at –80°C within four hours. Frozen samples were shipped to the sponsor laboratory on dry ice. Urine samples were de-identified of any protected health information at time of collection.
UroAmp (Convergent Genomics) testing methodology was performed as previously described (20). Briefly, purified urine DNA was fragmented by sonication (Covaris ME220, RRID:SCR_019818) before undergoing library preparation using the KAPA Hyper Prep Kit (Roche) protocol. xGen Dual Index adapters were synthesized by IDT, and a custom xGen Lockdown Probe panel (Integrated DNA Technology) was used for hybridization capture of DNA libraries. Target-enriched libraries were analyzed using a 2100 Bioanalyzer (RRID:SCR_018043) and diluted for sequencing. Libraries were loaded into a NextSeq 500/550 High Output Reagent Cartridge (v2, 300 cycles) and sequenced on a NextSeq 550 (RRID:SCR_016381).
UroAmp is a Clinical Laboratory Improvement Amendments–certified urine test developed to aid in the diagnosis of bladder cancer, predict risk of disease progression, and provide a longitudinal measure of genomic disease burden (GDB) in the bladder (20). Next-generation DNA sequencing is performed to measure approximately 250,000 genome locations at a high depth averaging approximately 10,000 molecules at each location, enabling highly sensitive detection of low-abundance single-nucleotide variants (SNV), insertion-deletions (INDEL), loss of heterozygosity (LOH), and copy-number variants (CNV) mutations. Simultaneously, the whole genome is sequenced at shallower depth (∼0.3×) to identify large-scale genomic alterations (aneuploidy). Mutation profiles serve as input features to disease and molecular-grade prediction algorithms. UroAmp risk algorithms are calculated independent of any clinical features.
Algorithm development
Three distinct algorithms are presented: An initial diagnosis test for hematuria evaluation, a surveillance test for urothelial carcinoma monitoring, and a molecular grade prediction test. Candidate samples for algorithm training underwent chart review to confirm tumor status and urothelial carcinoma history at time of collection, and samples were selected to balance positive and negative tumor status. For each sample, genomic features were extracted from UroAmp assay measurements. Selected samples were partitioned into distinct sets for initial diagnosis and surveillance algorithm training, which were then further divided into train and test splits.
Models with varying hyperparameters were trained using 10-fold cross validation. The models selected demonstrated good performance generalization between cross-validation results and the held-out test set. For initial diagnosis, several models achieved very high cross-validated sensitivity; thus, a boosted random forest model was selected that simultaneously optimized specificity while maintaining the high sensitivity appropriate for use in screening. For surveillance, a logistic regression model was chosen that optimized specificity and positive predictive value (PPV). Trained and locked algorithms were then used to make blinded predictions on validation cohorts.
The UroAmp molecular-grade prediction algorithm was developed to predict the presence of high-grade (HG) urothelial carcinoma. For grade training, the cohort was partitioned into a single train/test split. Genomic feature engineering and the optimization of a linear discriminant model was conducted in the training partition and validated on the test partition.
Statistical analysis
To ensure statistical confidence across the various clinical cohorts and define a study size, a prospective power analysis was performed, and 95% confidence intervals (CI) were calculated for each variable being tested [sensitivity, specificity, negative predictive value (NPV), and PPV]. Assuming an observed performance of 90%, cohort sample sizes of 30 and 50 samples provided CIs of (73.5%–97.9%) and (78.2%–96.7%), respectively. To minimize potential site selection bias between training and validation cohorts, samples from multiple sites are used in both cohorts. OHSU samples were used exclusively in training, whereas samples from MGH and OSU were used for independent validation. CURE samples were used in both training and blinded validations. To minimize selection bias between training and validation, CURE subjects were randomized according to their consent date.
The primary endpoints of the blinded validation study were UroAmp diagnostic performance and grade prediction. Secondary endpoints included: Likelihood ratios for initial diagnosis patients; recurrence-free survival (RFS) and hazard ratios (HR) between UroAmp recurrence-risk groups in WLC-negative surveillance patients; and odds ratios (OR) demonstrating the association between genomic features (e.g., disease burden, SNVs, CNVs, and INDELs) to tumor grade and invasion status.
UroAmp test performance was assessed by calculating sensitivity, specificity, PPV, and NPV. The Clopper–Pearson method was used to compute CIs for each performance metric, and receiver operating characteristic (ROC) curve analysis was used to assess overall classification accuracy. Time to recurrence was examined in survival analyses using the UroAmp-risk group as a predictor. Cox proportional hazard models were used to estimate the association between UroAmp features and RFS. Statistical significance for ORs was assessed using the Fischer's exact test. The P values were corrected for multiple tests using the Benjamini/Hochberg method with values below 0.05 considered significant.
The prevalence of urothelial carcinoma among UroAmp surveillance risk groups was compared with the prevalence of urothelial carcinoma among AUA surveillance guideline-risk groups. Pre- and post-test probabilities for urothelial carcinoma presence among AUA hematuria-risk categorizations of patients in our cohort was calculated as previously described (25) using the diagnostic likelihood ratio (DLR) determined from UroAmp test characteristics. DLR is defined in terms of the performance characteristics of the test, where positive DLR = Sensitivity/(1-Specificity) and negative DLR = (1-Sensitivity)/Specificity. DLRs relate pre-test odds to post-test odds, which then determine post-test probability (25). GDB is a percentile ranking of the sum of variant allele frequencies (VAF) for a urine specimen. To calculate GDB, the sum of VAFs was computed for all training samples to generate a representative empirical distribution. Each test sample was then ranked against that distribution to obtain its GDB percentile.
Data availability
Genome sequencing data used in this study have been deposited in the Sequence Read Archive (RRID:SCR_001370) at the National Center for Biotechnology Information (NCBI) and are available through BioProject ID PRJNA961222 (NCBI BioProject, RRID:SCR_004801).
Results
Characteristics of patients with urothelial carcinoma in the study cohorts
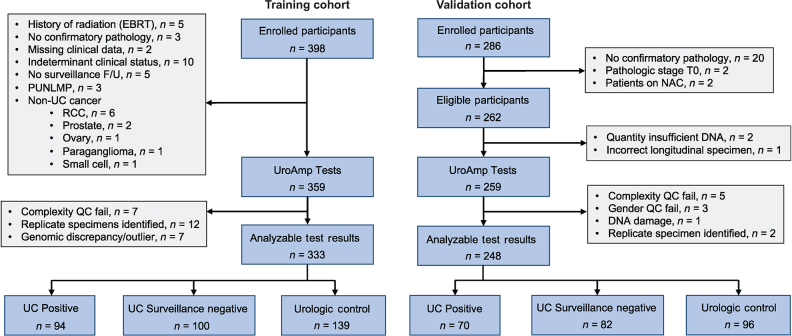
A total of 581 participant urine specimens were analyzed across all cohorts, with clinical and pathological characteristics detailed in Table 1. The training cohort consisted of 398 participants. Of these, 333 had analyzable test results, including pathology-confirmed diagnoses of de novo urothelial carcinoma (n = 56), pathology-confirmed recurrent urothelial carcinoma (n = 38), participants with a history of urothelial carcinoma with a negative surveillance cystoscopy (n = 100), and individuals without urothelial carcinoma but with other urologic morbidities, including hematuria (n = 139). The validation cohort included 286 participants, among which 248 had analyzable test results, consisting of 70 urothelial carcinoma positives (22 de novo tumors, 48 recurrent tumors), 82 urothelial carcinoma surveillance negatives, and 96 urology control subjects without evidence of urothelial carcinoma (Fig. 1). The UroAmp platform was used to determine urine-based comprehensive genomic profiles of the validation cohort (Supplementary Fig. S1A and S1B). Distribution of AUA risk categories for patients with hematuria (low, intermediate, and high) was consistent with those previously described (ref. 3; Supplementary Table S2), ensuring generalizability of results. Stage and grade distributions of tumor-positive cases were consistent with the natural distribution of disease at primary diagnosis and recurrence on surveillance (26, 27).
Table 1.
Clinical demographics of training and validation cohorts.
| Initial diagnosis | Surveillance | |||||||
|---|---|---|---|---|---|---|---|---|
| Cohort | Train | Validation | Train | Validation | ||||
| Cancer Status | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive |
| No. subjects | 139 | 56 | 96 | 22 | 100 | 38 | 82 | 48 |
| Median age (IQR) | 62 (31) | 75 (16) | 65 (24) | 70 (17) | 72 (14) | 77 (13) | 72 (13) | 69 (13) |
| Sex (%) | ||||||||
| Male/female | 55/45 | 71/29 | 65/35 | 73/27 | 74/26 | 82/18 | 77/23 | 71/29 |
| Race (%) | ||||||||
| White | 58 | 55 | 60 | 59 | 75 | 55 | 78 | 79 |
| Black or African American | 12 | 14 | 11 | 9 | 7 | 8 | 7 | 15 |
| Asian | 16 | 9 | 18 | 9 | 14 | 19 | 7 | 4 |
| American Indian or Alaska Native | 1 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| Hispanic or Latino | 2 | 0 | 1 | 0 | 1 | 0 | 4 | 0 |
| other | 11 | 20 | 9 | 23 | 2 | 18 | 4 | 2 |
| Smoking History (%) | ||||||||
| Current | 10 | 7 | 5 | 14 | 7 | 3 | 15 | 13 |
| Former | 18 | 14 | 26 | 45 | 36 | 47 | 52 | 50 |
| Never | 62 | 20 | 69 | 41 | 55 | 32 | 32 | 38 |
| Unknown | 10 | 59 | 0 | 0 | 2 | 18 | 1 | 0 |
| Urologic comorbidities (%)a | ||||||||
| UTI | 17 | 0 | 10 | 5 | 13 | 3 | 1 | 4 |
| LUTS | 21 | 11 | 21 | 18 | 27 | 8 | 16 | 2 |
| BPH | 16 | 9 | 27 | 5 | 41 | 24 | 24 | 8 |
| Kidney stone history (%) | ||||||||
| Current | 10 | 0 | 5 | 5 | 2 | 3 | 1 | 0 |
| Past | 16 | 4 | 11 | 0 | 5 | 3 | 1 | 2 |
| Urine chemistry/microscopy (%)a | ||||||||
| Hematuria | 50 | 23 | 39 | 50 | 31 | 24 | 2 | 10 |
| Leukocytes | 35 | 13 | 15 | 5 | 25 | 16 | 1 | 6 |
| Grade (%)b | ||||||||
| LG | — | 25 | — | 36 | 45 | 47 | 49 | 65 |
| HG | — | 75 | — | 64 | 32 | 45 | 51 | 35 |
| Unknown | — | — | 23 | 8 | 0 | 0 | ||
| Stage (%)b | ||||||||
| Cis | — | 5 | — | 0 | 6 | 5 | 7 | 11 |
| Ta | — | 38 | — | 68 | 51 | 58 | 71 | 79 |
| T1 | — | 25 | — | 14 | 15 | 8 | 17 | 8 |
| T2 | — | 29 | — | 18 | 3 | 11 | 3 | 2 |
| Unknown | — | 4 | — | 0 | 25 | 18 | 2 | 0 |
aAs reported at the time of sample collection, >1+ urine microscopy or >3 RBC/hpf or any gross.
bGrade and stage of surveillance negatives indicate the pathology of the primary tumor.
Figure 1.
Standards for the reporting of diagnostic accuracy studies (STARD) diagram detailing training and validation specimen usage in this study.
Urothelial carcinoma initial diagnosis clinical performance
The UroAmp disease classification algorithm for initial diagnosis of urothelial carcinoma was trained to a sensitivity of 98% and specificity of 96%. In the blinded validation cohort, the initial diagnosis algorithm demonstrated a sensitivity of 95% and specificity of 90% among de novo tumor cases and urology negative controls (Table 2; Supplementary Table S3). Among these patients, UroAmp identified the most worrisome tumors (HG and muscle-invasive tumors) with 100% sensitivity. NMIBC was identified with a sensitivity of 94%, and low-grade (LG) disease was identified with a sensitivity of 87%. The overall NPV for a de novo tumor was 99% in patients with hematuria.
Table 2.
Initial diagnosis disease classification performance.
| Initial diagnosis performance | ||
|---|---|---|
| Train | Validation | |
| Sensitivity (%; Sample No. Train/Validation) | ||
| Overall (56/22) | 98 (90–100) | 95 (77–100) |
| High grade (42/14) | 100 (92–100) | 100 (77–100) |
| Low grade (14/8) | 93 (66–100) | 87 (47–100) |
| NMIBC (38/18) | 97 (86–100) | 94 (73–100) |
| MIBC (16/4) | 100 (79–100) | 100 (40–100) |
| Upper tract UC (7/2) | 100 (59–100) | 100 (16–100) |
| Specificity (%; Sample No. Train/Validation) | ||
| All urology controls (139/96) | 96 (91–98) | 90 (82–95) |
| Controls with UTI (23/10) | 100 (85–100) | 90 (56–100) |
| Controls with hematuria (71/37) | 96 (88–99) | 87 (71–96) |
| Controls with BPH (23/26) | 96 (77–100) | 85 (65–96) |
| Controls with Leukocytes (48/14) | 98 (89–100) | 86 (57–98) |
| Controls with LUTS (29/20) | 93 (77–99) | 80 (56–94) |
| Positive predictive value (%) | ||
| Hematuria/initial diagnosis (10% prevalence) | 72 | 51 |
| Negative predictive value (%) | ||
| Hematuria/initial diagnosis (10% prevalence) | 100 | 99 |
Note: Clinical demographic subsets, including non-muscle invasive bladder cancer (NMIBC), muscle invasive bladder cancer (MIBC), urologic tract infection (UTI), benign prostatic hyperplasia (BPH), and lower urologic tract symptoms (LUTS). Values in parenthesis denote 95% confidence intervals.
To investigate how UroAmp may support screening of patients with hematuria, we performed a DLR analysis considering levels of disease prevalence across AUA hematuria-risk categories. UroAmp demonstrated a positive DLR of 9.2 and a negative DLR of 0.05 (Table 3), numbers that compare favorably with FDA-approved and other available biomarker approaches (Supplementary Table S4; ref. 25). Critically, in AUA low- and intermediate-risk microhematuria, where the natural prevalence of urothelial carcinoma is 0.5%–1%, respectively, a negative UroAmp test result decreased the likelihood of urothelial carcinoma to 0.03%–0.05%.
Table 3.
Initial diagnosis disease classification performance.
| Initial diagnosis likelihood ratios | |||||
|---|---|---|---|---|---|
| AUA-risk group | Pre-test UC prob. | Positive DLR | Post-test UC prob. UroAmp+ | Negative DLR | Post-test UC prob. UroAmp− |
| Low | 0.5% | 9.2 (5.1–16.6) | 4.4% | 0.05 (0.01–0.34) | 0.03% |
| Intermediate | 1.0% | 8.5% | 0.05% | ||
| High (microhematuria) | 3.0% | 22.1% | 0.16% | ||
| High (gross hematuria) | 10.0% | 50.5% | 0.56% | ||
Note: Pre- and post-test probability of urothelial carcinoma given UroAmp diagnostic likelihood ratios (DLR) for a positive or negative test. Pre-test probability of urothelial carcinoma based on Woldu et. al. 2021 (25). Values in parenthesis denote 95% confidence intervals; AUA, American Urological Association.
Urothelial carcinoma surveillance clinical performance
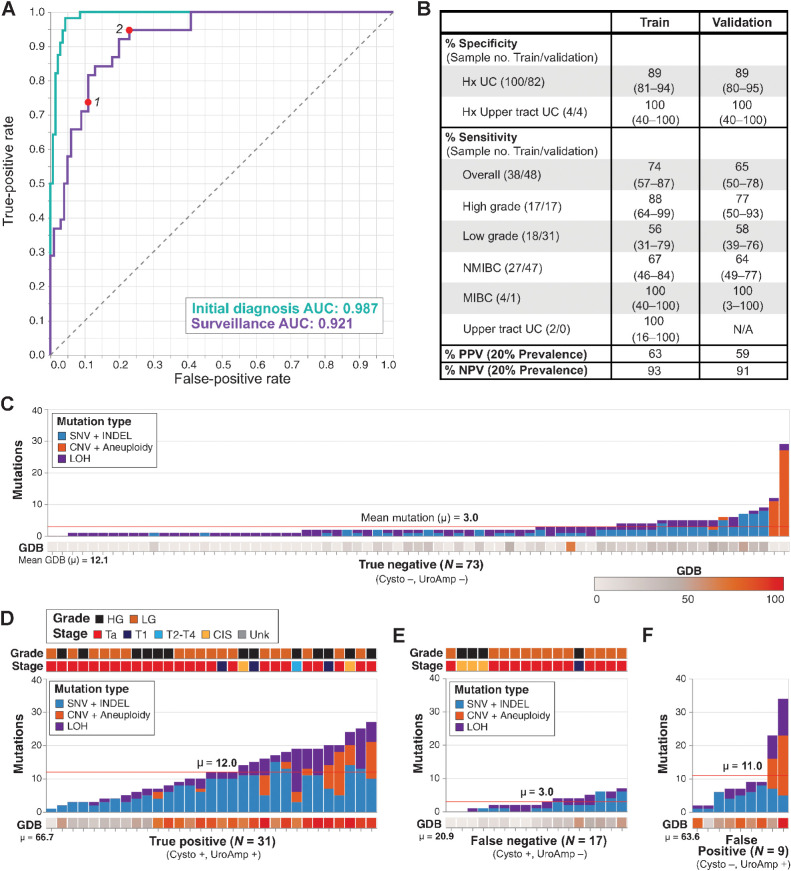
The UroAmp disease classification algorithm for urothelial carcinoma surveillance was assessed among surveillance-positive (recurrent tumor) cases and surveillance-negative controls. Consistent with the distinct clinical demographics of patients receiving an initial diagnosis of urothelial carcinoma versus those undergoing surveillance for urothelial carcinoma recurrence, a larger trade-off between sensitivity and specificity was observed during surveillance algorithm training. ROC analysis yielded an AUC of 0.921, as compared with the initial diagnosis value of 0.987 (Fig. 2A). To optimize concordance with WLC, we selected a surveillance model that prioritized specificity and PPV. In training, this model produced a sensitivity of 74%, a specificity of 89%, and a prevalence-adjusted PPV of 63% (Fig. 2B). The ROC curve suggests that adjusting the classification threshold to prioritize sensitivity over specificity could produce a model with a sensitivity as high as 95% while still maintaining 76% specificity in the training cohort (Fig. 2A, ROC point 2).
Figure 2.
Binary classification of urothelial carcinoma surveillance patients. A, ROC curves for the training of initial diagnosis (n = 195) and surveillance (n = 138) cohorts. Points 1 and 2 denote performance for the specificity-prioritized and sensitivity-prioritized models, respectively. B, Disease classification performance for the surveillance cohort. Clinical demographic subsets include non-muscle invasive bladder cancer (NMIBC) and muscle invasive bladder cancer (MIBC). Values in parenthesis denote 95% confidence intervals. C–F, Mutation number, type, and genomic disease burden (GDB) for patients in the surveillance validation cohort. Stage and grade are indicated for the original primary tumor for (C) True Negative and (F) False Positive or the recurrent tumors in (D) True Positive and (E) False Negative. Mean mutation count and mean GDB are indicated for each group.
With the selected high-specificity model, in the blinded validation cohort of surveillance patients, HG disease was detected with 76% sensitivity, whereas sensitivity for LG disease was 58%, and the overall prevalence-adjusted NPV and PPV were 91% and 59%, respectively. The single MIBC surveillance case was correctly detected (Fig. 2B). Overall, surveillance cases were classified with a specificity of 89%, demonstrating generalizability of the high-specificity performance observed in the training cohort, and a sensitivity of 65%, consistent with the performance ceiling imposed by WLC as the reference standard.
To better understand performance limitations in the surveillance cohort, we compared the mutation profiles of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). As expected, TP and TN patients largely demonstrated distinct mutation profiles, with TPs accumulating more mutations (mean of 12 vs. 3) and much higher GDBs (mean of 67 vs. 12) than TNs (Fig. 2C and 2D). Outliers—TNs with high signal and TPs with low signal—were found in each group. To assess limitations to sensitivity, we evaluated FN patients. Among these FNs, some patients had low signal, aligning with the majority of TNs. We also saw samples with modest GDBs and mutation counts; these samples look similar to both some TNs and some TPs (Fig. 2E). Finally, to explore specificity, we reviewed the nine FP patients. We saw substantial diversity in mutation signal from these samples but, on average, these patients had mutation and GDB values indistinguishable from TPs (Fig. 2F). Of note, two FP patients with a history of HG primary tumors (stage Ta and T1) had extremely high mutation signals, including the strongest overall mutation signal in the entire study cohort. Though WLC negative, the strong mutation signals present in the urine of these samples was consistent with the presence of minimal residual disease (MRD), leading us to consider whether such mutations are predictive of future recurrence.
RFS by uCGP-predicted risk
We next asked whether uCGP-detected MRD can be used to risk-stratify patients to predict likelihood of future recurrence. In related clinical scenarios, liquid biopsy MRD mutations have been shown to serve as predictive markers for future recurrence and therapy response (18, 19, 28, 29). To assess the prognostic potential of uCGP-identified mutations, we implemented a two-class recurrence risk model—low and high risk—defined by the binary surveillance algorithm output and the presence of HG-associated mutations (Supplementary Table S5).
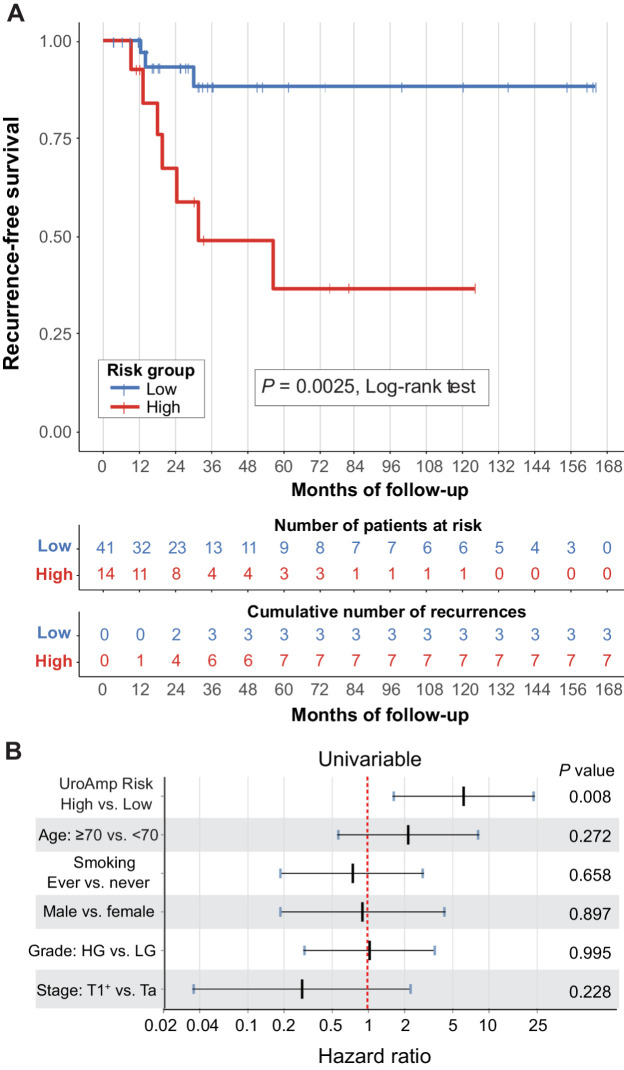
In an analysis of individuals in the validation group with at least three months of follow-up, UroAmp recurrence risk was assigned to patients undergoing urothelial carcinoma surveillance who were negative by WLC at time of collection (n = 55). The stage and grade distribution of the primary tumor at diagnosis was Ta (76%), CIS (2%), T1 (16%), T2 (2%), T3 (2%), Tx (2%), with concomitant CIS in 9%, and 49/51% LG/HG (Supplementary Table S6). Median follow-up after specimen collection was 30 months. UroAmp-predicted high-recurrence–risk subjects (n = 14) had a Kaplan–Meier recurrence rate estimate of 63% at five years, whereas low-recurrence–risk subjects (n = 41) had a recurrence rate estimate of 12%, resulting in a statistically significant difference in RFS (P = 0.0025, log-rank test, Fig. 3A). In the low-recurrence–risk group, no recurrences were observed within the first 12 months of surveillance. A sub-analysis by key clinical demographics (primary tumor grade and intravesical therapy history) supports generalizability of the recurrence prediction (Supplementary Fig. S2A–S2D). Univariable Cox proportional hazards regression demonstrated that UroAmp risk was a significant predictor of recurrence (HR, 6.2), whereas no clinical features showed significant predictive value (Fig. 3B; Supplementary Table S7).
Figure 3.
uCGP predicted recurrence risk. Urothelial carcinoma surveillance patients with negative cystoscopy and long-term follow-up with outcomes were analyzed for recurrence risk (n = 55, validation cohort). A, Kaplan–Meier curves for recurrence-free survival by UroAmp-predicted risk. Significance: P = 0.0025 (Log-rank test). B, Univariable Cox proportional-hazard regression analysis of UroAmp recurrence-risk groups and clinical-risk factors. For stage, T1+ indicates the grouping of patients with T1, T2, and T3 disease.
uCGP versus urine cytology
Validation specimens with available urine cytology results (n = 66) were investigated to compare cytology to UroAmp classification. Cytology returned atypical findings in 32% of samples, whereas UroAmp provided definitive findings in all samples. We evaluated performance characteristics of cytology under three scenarios where atypical findings were either omitted, deemed negative, or deemed positive. When atypical findings were omitted, cytology correctly identified 50% of HG urothelial carcinomas and 13% of LG urothelial carcinomas with a specificity of 94% (Supplementary Table S8). When atypical findings were interpreted as negative, cytology correctly identified 29% of HG urothelial carcinomas and 8% of LG urothelial carcinomas with a specificity of 96%. When interpreted as positive, 71% of HG urothelial carcinomas and 42% of LG urothelial carcinomas were correctly identified whereas specificity was 66%. In contrast, UroAmp correctly identified 100% of HG urothelial carcinomas and 67% of LG urothelial carcinomas with a specificity of 83%. Among samples with an atypical cytology result, UroAmp correctly identified 100% of HG urothelial carcinomas and 75% of LG urothelial carcinomas with a 71% specificity.
Molecular grade and stage prediction
The grade algorithm was optimized for specificity. Among disease positive training subjects, it performed with a PPV of 94% and specificity of 94%. In the blinded validation cohort, molecular prediction of HG achieved a PPV of 88%, a specificity of 95%, and an OR of 15.2 (P < 0.0001; Table 4).
Table 4.
uCGP molecular-grade prediction performance.
| Molecular-grade prediction performance | ||
|---|---|---|
| Train | Validation | |
| Percentage of PPV (for high-grade) | 94 (80–99) | 88 (62–98) |
| PPV [%] (for high-grade) and Specificity [%] | 94 (79–99) | 95 (83–99) |
| Odds ratio | 17.8 (3.9–81.4) | 15.2 (3.1–74.5) |
| Total samples (HG/LG) | 91 (59/32) | 70 (31/39) |
Note: Percentage of PPV is UroAmp-predicted HG that is confirmed by HG pathology. Specificity is the percent of pathologically LG samples not classified as HG by UroAmp. Odds ratio is the fold-increase in odds that a tumor is pathologically HG when predicted UroAmp HG. Values in parenthesis denote 95% confidence intervals.
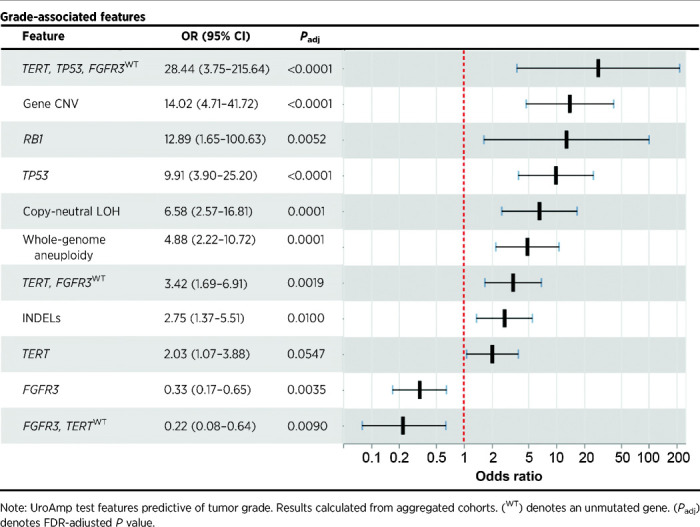
In the aggregated set of tumor-positive subjects (train and validation, n = 164), UroAmp identified genomic features associated with grade and stage (Table 5; Supplementary Table S5). The presence of urinary mutations in both TP53 and TERT in combination with wild-type (WT) FGFR3 was indicative of invasion and was the strongest indicator of HG disease, with an OR of 28.4 (P < 0.0001). In general, mutations in TP53 were strong indicators of muscle invasion, with TP53 mutations enriched in MIBC compared the NMIBC (OR, 7.4; P = 0.0004) and in HG T1–4 disease compared the HG Ta/CIS disease (OR, 6.0; P = 0.0025). Gene-level CNV and whole-genome aneuploidy were all enriched in HG and muscle-invasive disease, and CDKN1A and RB1 mutations were found almost exclusively in HG lesions. Among CIS lesions, mutations in TP53, as well as TERT mutations combined with WT FGFR3, were enriched compared with Ta tumors (Supplementary Table S5). Finally, the strongest predictor of LG disease was mutation in FGFR3 (OR, 0.3; P = 0.0035), with FGFR3 mutation combined with WT TERT being observed exclusively in NMIBC.
Table 5.
uCGP molecular-grade prediction features.
Discussion
This study constitutes the first blinded clinical validation of the UroAmp cancer diagnostic platform. Compared with published reports of currently available tests, we report superior performance metrics for the detection of urothelial carcinoma during initial diagnosis, with higher combined sensitivity and specificity and improved DLRs. This performance is paired with high-PPV grade prediction. In the urothelial carcinoma surveillance setting, UroAmp significantly prognosticated future recurrence.
Initial diagnosis
Clinical evidence makes it clear that better tools are needed to accurately risk-stratify patients with hematuria. Despite recommendations, patients with hematuria continue to be under-referred for evaluation, and urologists continue to underuse cystoscopy in intermediate- and high-risk patients (4, 5, 8). As a result, the prevalence of muscle-invasive urothelial carcinoma has been stable for the last 30 years (27), despite significant advances in other types of cancer (1). Clearly, new approaches to early detection are needed to improve compliance with guidelines among primary care physicians and change the paradigm of diagnosis.
We know that detecting urothelial carcinoma in patients with microhematuria correlates with a lower stage of disease compared with those with gross hematuria (30). Therefore, increasing evaluation of microhematuria is likely to improve outcomes overall. However, due to the high prevalence of microhematuria, referring every case for cystoscopic evaluation would overwhelm urology practices. Instead, patient referral should be strategic and use of cystoscopy for evaluation should be selective. Clinical-risk factors, such as smoking history and hematuria severity, are helpful, and risk calculators developed to assign bladder cancer likelihood to patients with hematuria have demonstrated impressive performance (3, 31, 32). But the adoption of these calculators has been poor, as the necessary clinical risk factors are not routinely charted or accurately collected (33). Although these calculators are helpful at identifying extremes (very low-risk or very high-risk), they still rely on population averages rather than measuring individual biologic risk. UroAmp's high sensitivity and specificity (95%/90%) in initial diagnosis means it could be used to triage patients with microhematuria, reducing the burden of evaluation while identifying those most likely to have urothelial carcinoma. Furthermore, because UroAmp does not use clinical demographics or risk factors, it can complement clinical-risk stratification systems, as demonstrated in Table 3.
Historically, urothelial carcinoma tests have positioned themselves as either “rule-out” (identify low-risk patients) or “rule-in” (identify high-risk patients). Our results suggest that uCGP has the potential to act as both. UroAmp has a high prevalence-adjusted NPV (99%) and a low negative DLR (0.05), which means AUA low-risk patients with hematuria who test negative by UroAmp have only a 0.03% chance of being urothelial carcinoma positive. This noninvasive rule-out test would dramatically ease the burden of evaluation and focus WLC on the patients most at risk. Existing noninvasive WLC adjuncts, such as cytology, UroVysion FISH, and NMP22, struggle to effectively stratify patients with microhematuria. The much higher negative DLRs of these tests (0.35–0.42; ref. 25) mean that a negative result only reduces a patient's chance of having cancer from 0.5% (pre-test) to approximately 0.2% (post-test). UroAmp thus has the potential to provide an order-of-magnitude improvement at ruling out urothelial carcinoma compared with these legacy tests.
Rule-in tests rely on their high prevalence-adjusted PPV and positive DLR to identify patients who are urothelial carcinoma positive. This is particularly true of cytology, which can return the diagnostic result of “atypical”—lying somewhere between positive and negative—and trades sensitivity in favor of optimizing specificity. Cytology's high specificity affords it a robust positive DLR (7.67). UroAmp, which also has high prevalence-adjusted PPV (51%) and a positive DLR (9.2) that exceeds even cytology, can similarly help identify urothelial carcinoma–positive patients, but does so without compromised sensitivity. Moreover, UroAmp gives only clear positive or negative results for initial diagnosis and has demonstrated the ability to correctly stratify patients with “atypical” results by cytology. Thus, uCGP can identify those most likely to have urothelial carcinoma (effective rule-in) in addition to reducing the burden of evaluation (effective rule-out).
Recurrence surveillance
Compared with initial diagnosis, tumor detection in the surveillance setting provides multiple additional challenges. One cubic centimeter of urothelial carcinoma tumor consists of approximately one billion cells (34). This means that patients who develop a WLC-visible recurrence will have visually occult malignant cells (potentially many millions) present during negative cystoscopies preceding eventual clinical detection. Molecular testing can detect the mutations arising from these malignant cells before they achieve the critical mass necessary to become visually apparent. Furthermore, roughly 60% of patients will experience recurrence during the first five years of surveillance (12, 13). Hence, a urine marker with predictive power is more likely to correlate with long-term outcomes than current cystoscopy status.
Genomic insights provide opportunities for enhanced clinical management, but their application to binary (positive/negative) disease classification continues to be constrained by the limited sensitivity of WLC, and specificity is similarly hindered by the absence of long-term follow-up. In meta-analyses comparing WLC with mapping biopsy or photodynamic enhanced cystoscopy, WLC had an estimated sensitivity of 65%–71% and specificity of approximately 81% in bladder cancer surveillance (14, 15, 35). Confounding factors, such as operator thoroughness, equipment, and condition of the bladder, all lead to a high degree of ambiguity in recurrence diagnosis. These sources of inconsistency may not correlate with any particular genomic profile, confounding a quantitative algorithm's ability to learn a tumor-associated pattern. We are thus faced with the challenge of developing a quantitative diagnostic tool using a qualitative and mercurial reference standard. As prior work has shown, urologists are approximately six times more likely to find recurrent tumors by WLC if they are given positive biomarker test results before cystoscopy (36).
The tradeoff of concordance with WLC is illustrated in the surveillance classification ROC curve (Fig. 2A). Point 2 on the curve shows that UroAmp might attain much higher sensitivity (∼95%) with the compromise of modest specificity (∼76%). To achieve this, a sample would be called positive based on lower levels of mutations. This would result in more FN samples (Fig. 2E) being correctly called positive (improved sensitivity) but would also lead to TN samples with similar profiles (Fig. 2C) being incorrectly called positive (lower specificity). To limit such apparent false negatives, we chose a model represented by point 1 on the ROC curve, giving high specificity and a sensitivity that approximates that of WLC.
Improving this performance requires an alternative reference standard to WLC and the study of long-term recurrence outcomes. Our study of recurrence among negative cystoscopy surveillance patients identifies a high rate of recurrence among subjects with a UroAmp algorithm-positive prediction. When clinical-risk factors of age, smoking history, gender, grade, and stage were assessed in a univariable model, they did not achieve significance in prediction of future recurrence, suggesting that UroAmp MRD recurrence prediction has significant potential to enable the delivery of personalized and risk-stratified care.
Molecular grading
The predictive capability of uCGP comes with the challenge of how to best manage patients whose disease is months from being visibly apparent. Watchful waiting may be appropriate for low-risk disease whereas treatment escalation may be required for occult but high-risk disease. Urine-based biomarker tests, which have similarly struggled with positive test results in cystoscopy-negative patients, have adopted “anticipatory positives” for false-positive test results. Biomarker anticipatory positives, whether true or not (37), have proven to have limited clinical utility in part due to tests relying on generic indicators of malignancy that cannot distinguish one tumor from another or predict severity. In contrast, urine cytology, which can also yield anticipatory positives, maintains its value because it informs potential cancer severity with tumor grading. Similarly, MRD detection with uCGP assigns a molecular fingerprint to a cancer based on the tumor's unique constellation of mutant alleles, allowing for uCGP to not only distinguish new tumors from recurrent tumors, but also to calculate risk based on a mutation profile associated with stage and grade. Here, UroAmp provides a molecular grade assessment with 88% PPV (UroAmp HG predictions are confirmed by pathology 88% of the time) and characterizes molecular features associated with increased risk of invasion (Supplementary Table S5). Within our training and validation cohorts, multiple instances of disease progression were observed, including the case of an initial Ta LG tumor that recurred as T4a HG. UroAmp identified this patient as high-risk 18 months ahead of diagnosis, noting their elevated risk for HG recurrence. Intensification of WLC, BLC, or mapping biopsies may have been warranted if UroAmp had been clinically available. Moving forward, a positive UroAmp test may prompt consideration of additional diagnostic investigations or altered therapeutic strategies, enabling personalized care.
Comprehensive genomic profiling
The comprehensive nature of the genomic profile provides much more information than traditional urinary biomarkers. uCGP, as validated here, measures diverse mutation types, including SNVs, INDELs, gene level CNV, copy-number neutral LOH, and whole-genome chromosomal aneuploidy. In addition, it detects somatic variants at low allele frequencies along with germline variants. These diverse mutation types are interpreted with annotations from The Cancer Genomic Atlas Project, AACR Project GENIE, and large population-level studies such as The Genome Aggregation Database (38–40). The breadth of measurement provides a genomic fingerprint of each tumor that provides insight into its etiology. For example, tumors with high numbers of INDELs are more likely to be driven by defects in DNA mismatch repair genes (41), whereas tumors with high aneuploidy are likely to have errors in chromosome segregation during cell division (42). Tumors may also be driven primarily by mutation of a single gene, such as a case with high level ERBB2 (HER2) amplification.
Such genomic profiles also have the potential to identify molecular subtypes of cancer and predict response to FDA-approved therapies. For example, detection of FGFR3 mutations may indicate therapy with FGFR3 inhibitors, whereas mutations in genes such as ERBB2 or ATM could identify candidates for clinical trials and drugs approved in other indications (e.g., trastuzumab or olaparib). By leveraging the broad and diverse measurements of a genomic profile for diagnosis and prognosis in cancer, pattern recognition algorithms can learn to prioritize certain genes above others and, with sufficient data, could discover complex interactions of genes in tumor development. For example, in Supplementary Fig. S1 we observe chromatin-modifying enzymes like KMT2D and KMT2C are commonly mutated in tumor-positive patients but also constitute the most frequent mutations in cystoscopy-negative patients. In contrast, an oncogene like FGRF3 is one of the most frequently mutated genes in tumor-positive patients but is almost never seen in cystoscopy-negative patients. Ultimately, these algorithms may identify previously unknown tumor sub-types and their correlation to prognosis or response to therapies.
Limitations
Study limitations include likely interobserver variation in pathological grading due to absence of centralized pathology review (43, 44). Clinical sites received inclusion criteria to collect patients with hematuria, but the histories of hematuria, urine microscopy, and smoking pack-years were not uniformly charted in the EHR, limiting our ability to define AUA hematuria-risk categories in some subjects (2). Characterization of clinical demographics was limited by variable clinical documentation of benign diseases. Patients with upper tract urothelial carcinoma (UTUC) were included in the study, but their sample size was too small to enable UTUC-specific analyses. Likewise, certain urothelial carcinoma stages (CIS, T3/T4) are not well represented, and we are underpowered to assess performance in these clinically meaningful subtypes. In the surveillance monitoring cohort, long-term surveillance times were variable: Recurrence events were observed for some patients beyond 36 months, whereas many patients were lost to follow-up before 36 months. Finally, identification of patients at high-risk for disease progression would have great utility to guide risk-stratified care, but the current cohort is underpowered to identify these rare cases.
Conclusion
The findings presented here provide strong evidence for the utility of uCGP to diagnose urothelial carcinoma among patients with hematuria, identify risk of HG cancer, and detect MRD to predict future recurrence risk in patients under surveillance for urothelial carcinoma. UroAmp-risk assessment is calculated independent of any clinical features, providing novel information for physicians to integrate with clinical-risk factors. Considering its performance in multiple areas of urothelial carcinoma diagnosis and monitoring, uCGP shows great promise to enhance delivery of risk-stratified care. Ongoing studies are expected to support the generalizability of these findings.
Supplementary Material
Supplemental Tables 1-8
Supplementary Figure 1. Genomic profiles of the validation cohort.
Supplemental Figure 2. uCGP Predicted Recurrence Risk by Subgroup.
Acknowledgments
The authors thank Gbolahan Fatuga and Isabella Tse for review of clinical data and Bob Schmidt for statistical support. We would like to acknowledge all of the patients who participated in this study and the many CURE Biobank clinical site staff who facilitated collection of samples and data. We would like to acknowledge the American Association for Cancer Research and its financial and material support in the development of the AACR Project GENIE registry as well as members of the consortium for their commitment to data sharing. Interpretations are the responsibility of the study authors. Finally, we would like to acknowledge the TCGA Research Network. Research reported in this publication was supported by the National Cancer Institute of the National Institute of Health under Award Number R44CA200174. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. This award supported sample procurement, materials and supplies, computer services, and personnel (T.G. Levin, T.M. Koppie, M. Garcia-Ransom, and K.G. Phillips). All other costs were self-funded by Convergent Genomics. Convergent maintains a financial conflict of interest (FCOI) policy in accordance with 42 CFR Part 50 and 45 CRF Part 94 to protect the integrity and objectivity of all research activities.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
K. Salari reports grants from Convergent Genomics during the conduct of the study as well as grants from Urology Care Foundation and Prostate Cancer Foundation, and personal fees from OrigiMed outside the submitted work. D. Sundi reports other support from Janssen and Arquer outside the submitted work. A. Pienkny reports personal fees from Research support from Convergent Genomics during the conduct of the study. C.K. Lee reports other support from Convergent outside the submitted work. G. Barme reports other support from Convergent Genomics during the conduct of the study. R. Kahn reports personal fees from Golden Gate Urology during the conduct of the study. K.G. Philips reports grants from NCI during the conduct of the study; other support from Convergent Genomics outside the submitted work; as well as reports a patent for Diagnostic assay for urine monitoring of bladder cancer pending and a patent for Methods and systems for urine-based detection of urologic conditions pending. V.M. Caruso reports grants from National Cancer Institute during the conduct of the study as well as other support from Convergent Genomics outside the submitted work. M. Goudarzi reports grants from the National Cancer Institute of the National Institute of Health during the conduct of the study and reports employment and shareholder of Convergent Genomics. P.S. Lentz reports grants from the National Cancer Institute of the National Institute of Health during the conduct of the study and reports employment and shareholder of Convergent Genomics. M.E. Evans-Holm reports grants from the National Cancer Institute of the National Institute of Health during the conduct of the study and being an employee and shareholder of Convergent Genomics. A.R. MacBride reports grants from The National Cancer Institute of the National Institute of Health during the conduct of the study and reports employment and shareholder of Convergent Genomics. D.S. Fischer reports grants from National Cancer Institute of the National Institute of Health during the conduct of the study and reports employment and shareholder of Convergent Genomics. I.J. Haddadzadeh reports grants and other support from The National Cancer Institute of the National Institute of Health during the conduct of the study and reports employment and shareholder of Convergent Genomics. B.C. Mazzarella reports other support from the National Cancer Institute of the National Institute of Health during the conduct of the study; other support from Janssen, Pfizer, Invitae, MDx Health, Candel Therapeutics, as well as other support from Myovant outside the submitted work and reports employment and shareholder of Convergent Genomics. J.W. Gray reports other support from Convergent Geomics during the conduct of the study. T.M. Koppie reports personal fees from Convergent Genomics during the conduct of the study and is a founder and shareholder in Convergent Genomics. V.T. Bicocca reports grants from the National Cancer Institute of the National Institute of Health during the conduct of the study and reports employment and shareholder of Convergent Genomics. T.G. Levin reports grants from the National Cancer Institute of the National Institute of Health during the conduct of the study; as well as reports a patent for Diagnostic Assay for Urine Monitoring of Bladder Cancer pending; and reports employment and shareholder of Convergent Genomics. Y. Lotan reports personal fees from convergent genomics during the conduct of the study and personal fees from pacific edge, Nonagen, and Nucleix outside the submitted work. A.S. Feldman reports grants from Convergent Genomics during the conduct of the study; and stock ownership in Scentient, Inc. and Vessi Medical as well as Urogen Pharma consulting fees. No disclosures were reported by the other authors.
Authors' Contributions
K. Salari: Conceptualization, resources, data curation, formal analysis, supervision, investigation, methodology, writing–review and editing. D. Sundi: Conceptualization, data curation, formal analysis, investigation, methodology, writing–review and editing. J.J. Lee: Resources, data curation, formal analysis, investigation. S. Wu: Resources, data curation. C.-L. Wu: Resources, supervision. G. DiFiore: Data curation. Q.R. Yan: Data curation. A. Pienkny: Data curation. C.K. Lee: Data curation. D. Oberlin: Data curation. G. Barme: Data curation. J. Piser: Data curation. R. Kahn: Data curation. E. Collins: Data curation. K.G. Phillips: Data curation, software, formal analysis, funding acquisition, methodology, writing–original draft. V.M. Caruso: Data curation, software, formal analysis, visualization, methodology, writing–review and editing. M. Goudarzi: Data curation, software, formal analysis, visualization. M. Garcia-Ransom: Data curation, funding acquisition, project administration. P.S. Lentz: Investigation. M.E. Evans-Holm: Investigation, project administration. A.R. MacBride: Data curation, software, project administration. D.S. Fischer: Data curation, software. I.J. Haddadzadeh: Funding acquisition, project administration. B.C. Mazzarella: Conceptualization, data curation, investigation, methodology, writing–review and editing. J.W. Gray: Data curation. T.M. Koppie: Conceptualization, data curation, funding acquisition, investigation, methodology. V.T. Bicocca: Formal analysis, funding acquisition, investigation, visualization, writing–original draft. T.G. Levin: Conceptualization, formal analysis, funding acquisition, investigation, methodology, writing–review and editing. Y. Lotan: Formal analysis, methodology, writing–review and editing. A.S. Feldman: Conceptualization, resources, formal analysis, supervision, methodology, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7–33. [DOI] [PubMed] [Google Scholar]
- 2. Barocas DA, Boorjian SA, Alvarez RD, Downs TM, Gross CP, Hamilton BD, et al. Microhematuria: AUA/SUFU guideline. J Urol 2020;204:778–86. [DOI] [PubMed] [Google Scholar]
- 3. Woldu SL, Ng CK, Loo RK, Slezak JM, Jacobsen SJ, Tan WS, et al. Evaluation of the new American urological association guidelines risk classification for hematuria. J Urol 2021;205:1387–93. [DOI] [PubMed] [Google Scholar]
- 4. Elias K, Svatek RS, Gupta S, Ho R, Lotan Y. High-risk patients with hematuria are not evaluated according to guideline recommendations. Cancer 2010;116:2954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buteau A, Seideman CA, Svatek RS, Youssef RF, Chakrabarti G, Reed G, et al. What is evaluation of hematuria by primary care physicians? Use of electronic medical records to assess practice patterns with intermediate follow-up. Urol Oncol 2014;32:128–34. [DOI] [PubMed] [Google Scholar]
- 6. Tan WS, Feber A, Sarpong R, Khetrapal P, Rodney S, Jalil R, et al. Who should be investigated for haematuria? Results of a contemporary prospective observational study of 3,556 patients. Eur Urol 2018;74:10–4. [DOI] [PubMed] [Google Scholar]
- 7. Ingelfinger JR. Hematuria in adults. N Engl J Med 2021;385:153–63. [DOI] [PubMed] [Google Scholar]
- 8. David SA, Patil D, Alemozaffar M, Issa MM, Master VA, Filson CP. Urologist use of cystoscopy for patients presenting with hematuria in the United States. Urology 2017;100:20–6. [DOI] [PubMed] [Google Scholar]
- 9. Woldu SL, Bagrodia A, Lotan Y. Guideline of guidelines: non–muscle-invasive bladder cancer. BJU Int 2017;119:371–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flaig TW, Spiess PE, Agarwal N, Bangs R, Boorjian SA, Buyyounouski MK, et al. Bladder Cancer, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2020;18:329–54. [DOI] [PubMed] [Google Scholar]
- 11. Tobert CM, Nepple KG, McDowell BD, Charlton ME, Mott SL, Gruca TS, et al. Compliance with American urological association guidelines for nonmuscle invasive bladder cancer remains poor: assessing factors associated with noncompliance and survival in a rural state. Urology 2019;132:150–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cumberbatch MGK, Foerster B, Catto JWF, Kamat AM, Kassouf W, Jubber I, et al. Repeat transurethral resection in non–muscle-invasive bladder cancer: a systematic review. Eur Urol 2018;73:925–33. [DOI] [PubMed] [Google Scholar]
- 13. Jeong SH, Han JH, Jeong CW, Kim HH, Kwak C, Yuk HD, et al. Clinical determinants of recurrence in pTa bladder cancer following transurethral resection of bladder tumor. BMC Cancer 2022;22:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mowatt G, N'Dow J, Vale L, Nabi G, Boachie C, Cook JA, et al. Photodynamic diagnosis of bladder cancer compared with white light cystoscopy: systematic review and meta-analysis. Int J Technol Assess Health Care 2011;27:3–10. [DOI] [PubMed] [Google Scholar]
- 15. Russo GI, Sholklapper TN, Cocci A, Broggi G, Caltabiano R, Smith AB, et al. Performance of Narrow Band Imaging (NBI) and Photodynamic Diagnosis (PDD) fluorescence imaging compared to White Light Cystoscopy (WLC) in detecting non–muscle-invasive bladder cancer: a systematic review and lesion-level diagnostic meta-analysis. Cancers 2021;13:4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hughes T, Morgan G, Martiat P, Goldman J. Detection of residual leukemia after bone marrow transplant for chronic myeloid leukemia: role of polymerase chain reaction in predicting relapse. Blood 1991;77:874–8. [PubMed] [Google Scholar]
- 17. Cavé H, van der Werff ten Bosch J, Suciu S, Guidal C, Waterkeyn C, Otten J, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia. N Engl J Med 1998;339:591–8. [DOI] [PubMed] [Google Scholar]
- 18. Faham M, Zheng J, Moorhead M, Carlton VEH, Stow P, Coustan-Smith E, et al. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood 2012;120:5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bicocca VT, Phillips KG, Fischer DS, Caruso VM, Goudarzi M, Garcia-Ransom M, et al. Urinary comprehensive genomic profiling correlates urothelial carcinoma mutations with clinical risk and efficacy of intervention. J Clin Med 2022;11:5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015;351:h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK). J Natl Cancer Inst 2005;97:1180–4. [DOI] [PubMed] [Google Scholar]
- 23. Chang SS, Boorjian SA, Chou R, Clark PE, Daneshmand S, Konety BR, et al. Diagnosis and treatment of non–muscle-invasive bladder cancer: AUA/SUO guideline. J Urol 2016;196:1021–9. [DOI] [PubMed] [Google Scholar]
- 24. Chang SS, Bochner BH, Chou R, Dreicer R, Kamat AM, Lerner SP, et al. Treatment of non-metastatic muscle-invasive bladder cancer: AUA/ASCO/ASTRO/SUO guideline. J Urol 2017;198:552–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woldu SL, Souter L, Boorjian SA, Barocas DA, Lotan Y. Urinary-based tumor markers enhance microhematuria risk stratification according to baseline bladder cancer prevalence. Urol Oncol 2021;39:787. [DOI] [PubMed] [Google Scholar]
- 26. Nielsen ME, Smith AB, Meyer AM, Kuo TM, Tyree S, Kim WY, et al. Trends in stage-specific incidence rates for urothelial carcinoma of the bladder in the United States: 1988 to 2006. Cancer 2014;120:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Surveillance Epidemiology and End Results (SEER) Program. SEER*Stat Database: incidence - SEER Research Plus Data, 8 Registries, Nov 2021 Sub (1975–2019) - Linked To County Attributes - Total U.S., 1969–2020 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2022, based on the November 2021 submission. 2022.
- 28. Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res 2019;25:4255–63. [DOI] [PubMed] [Google Scholar]
- 29. Parikh AR, van Seventer EE, Siravegna G, Hartwig AV, Jaimovich A, He Y, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin Cancer Res 2021;27:5586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramirez D, Gupta A, Canter D, Harrow B, Dobbs RW, Kucherov V, et al. Microscopic haematuria at time of diagnosis is associated with lower disease stage in patients with newly diagnosed bladder cancer. BJU Int 2016;117:783–6. [DOI] [PubMed] [Google Scholar]
- 31. Khadhouri S, Gallagher KM, MacKenzie KR, Shah TT, Gao C, Moore S, et al. The IDENTIFY study: the investigation and detection of urological neoplasia in patients referred with suspected urinary tract cancer—a multicentre observational study. BJU Int 2021;128:440–50. [DOI] [PubMed] [Google Scholar]
- 32. Khadhouri S, Gallagher KM, MacKenzie KR, Shah TT, Gao C, Moore S, et al. Developing a diagnostic multivariable prediction model for urinary tract cancer in patients referred with haematuria: results from the IDENTIFY collaborative study. Eur Urol Focus 2022;8:1673–82. [DOI] [PubMed] [Google Scholar]
- 33. Kukhareva PV, Caverly TJ, Li H, Katki HA, Cheung LC, Reese TJ, et al. Inaccuracies in electronic health records smoking data and a potential approach to address resulting underestimation in determining lung cancer screening eligibility. J Am Med Inform Assoc 2022;29:779–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Avanzini S, Kurtz DM, Chabon JJ, Moding EJ, Hori SS, Gambhir SS, et al. A mathematical model of ctDNA shedding predicts tumor detection size. Sci Adv 2020;6:eabc4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng C, Lv Y, Zhong Q, Wang R, Jiang Q. Narrow band imaging diagnosis of bladder cancer: systematic review and meta-analysis. BJU Int 2012;110:E680–7. [DOI] [PubMed] [Google Scholar]
- 36. van der Aa MNM, Steyerberg EW, Bangma C, van Rhijn BWG, Zwarthoff EC, van der Kwast TH. Cystoscopy revisited as the gold standard for detecting bladder cancer recurrence: diagnostic review bias in the randomized, prospective CEFUB trial. J Urol 2010;183:76–80. [DOI] [PubMed] [Google Scholar]
- 37. Gopalakrishna A, Fantony JJ, Longo TA, Owusu R, Foo WC, Dash R, et al. Anticipatory positive urine tests for bladder cancer. Ann Surg Oncol 2017;24:1747–53. [DOI] [PubMed] [Google Scholar]
- 38. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sweeney SM, Cerami E, Baras A, Pugh TJ, Schultz N, Stricker T, et al. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov 2017;7:818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao H, Thienpont B, Yesilyurt BT, Moisse M, Reumers J, Coenegrachts L, et al. Mismatch repair deficiency endows tumors with a unique mutation signature and sensitivity to DNA double-strand breaks. Elife 2014;3:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer 2001;1:109–17. [DOI] [PubMed] [Google Scholar]
- 43. Sharkey FE, Sarosdy MF. The significance of central pathology review in clinical studies of transitional cell carcinoma in situ. J Urol 1997;157:68–71. [PubMed] [Google Scholar]
- 44. Coblentz TR, Mills SE, Theodorescu D. Impact of second opinion pathology in the definitive management of patients with bladder carcinoma. Cancer 2001;91:1284–90. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Tables 1-8
Supplementary Figure 1. Genomic profiles of the validation cohort.
Supplemental Figure 2. uCGP Predicted Recurrence Risk by Subgroup.
Data Availability Statement
Genome sequencing data used in this study have been deposited in the Sequence Read Archive (RRID:SCR_001370) at the National Center for Biotechnology Information (NCBI) and are available through BioProject ID PRJNA961222 (NCBI BioProject, RRID:SCR_004801).