Abstract
Despite recent advances in molecularly targeted therapies and immunotherapies, the effective treatment of advanced-stage cancers remains a largely unmet clinical need. Identifying driver mechanisms of cancer aggressiveness can lay the groundwork for the development of breakthrough therapeutic strategies. Assembly factor for spindle microtubules (ASPM) was initially identified as a centrosomal protein that regulates neurogenesis and brain size. Mounting evidence has demonstrated the pleiotropic roles of ASPM in mitosis, cell-cycle progression, and DNA double-strand breaks (DSB) repair. Recently, the exon 18–preserved isoform 1 of ASPM has emerged as a critical regulator of cancer stemness and aggressiveness in various malignant tumor types. Here, we describe the domain compositions of ASPM and its transcript variants and overview their expression patterns and prognostic significance in cancers. A summary is provided of recent progress in the molecular elucidation of ASPM as a regulatory hub of development- and stemness-associated signaling pathways, such as the Wnt, Hedgehog, and Notch pathways, and of DNA DSB repair in cancer cells. The review emphasizes the potential utility of ASPM as a cancer-agnostic and pathway-informed prognostic biomarker and therapeutic target.
Introduction
Cancer cells in established tumors exploit and modify pathways and cellular programs, which benefit their growth, survival, spreading, and ability to cope with stresses they encounter during malignant evolution (1). Cancer progression, especially for advanced-stage events such as invasion and metastasis, may not rely on classical driver oncogenes but on the rewired oncogenic network and “cancer fitness genes” that are not responsible for initiating tumorigenesis (2). Cancer cells may become addicted to the genes and pathways essential for sustaining cancer progression and metastasis, whose targeting constitutes synthetic lethality with the underlying tumor genotype. Thus, identifying critical signaling nodes controlling cancer aggressiveness may unlock novel opportunities for the successful treatment of advanced-stage cancers.
Initially identified as a centrosomal protein that regulates neurogenesis and brain size (3), assembly factor for spindle microtubules (ASPM) has recently emerged as a regulatory hub of diverse oncogenic pathways critical for malignant progression. This review summarizes the expression pattern of ASPM and its isoforms in cancers, their binding partners, and the molecular mechanism of action (MOA) underlying their biological functions in cancers. We emphasize the role of the exon 18–preserved ASPM isoform 1 (ASPM-i1) in development-associated pathways, such as Wnt, Hedgehog (Hh), and Notch, and the cancer stem cell (CSC) phenotype. We consider ASPM as a cancer fitness gene and a hub connecting signaling modules critical for cancer progression (4). We propose the clinical utility of ASPM as a pathway-informed biomarker in human cancers and present feasible molecular targeting and synthetic lethal strategies to exploit the therapeutic opportunities associated with this novel oncoprotein.
ASPM in Neurogenesis and Germ Cell Development
Loss-of-function mutations of ASPM are the most common cause of autosomal recessive primary microcephaly in humans (5). The Drosophila ASPM orthologue, Asp, is involved in the organization of microtubules during spindle formation and cytokinesis (6, 7). The expression of mouse Aspm is restricted to embryonic days 11 to 17, which is substantially reduced by postnatal day 0 when neurogenesis is completed (5). Mutations in Asp arrest neuroblasts in metaphase, thus reducing central nervous system (CNS) development (8). Asp also regulates neuroepithelium morphogenesis by controlling the polarized distribution of myosin II along the apicobasal axis (9). Mammalian ASPM is expressed in the ventricular zone and outer subventricular of the embryonic forebrain, where it directs spindle alignment and mitotic orientation in dividing neural progenitor cells (NPC; refs. 5, 10–12). Loss of ASPM elicits premature differentiation and delamination of NPCs (11–14). ASPM controls microtubule disassembly at spindle poles by promoting the katanin-mediated severing of dynamic microtubules (15), spindle positioning by recruiting citron kinase (CITK), centriole biogenesis (16), ciliogenesis and the apical polarity complex formation (14), and passage through the G1 restriction point by enhancing the stability of cyclin E (13). In addition, although the mutations or deletion of ASPM in mice were not embryonically lethal, they caused a massive loss of germ cells, resulting in testicular hypoplasia and a severe decrease in the number of developing follicles and ovarian size (12, 17, 18). Notably, it has been shown that ASPM is also expressed in a wide variety of non-CNS fetal tissues during development, such as the liver, heart, lung, and kidney (3). Thus, it is still puzzling how the loss of ASPM leads to microcephaly, not primordial dwarfism, with relatively normal immune and digestive functions in humans (19).
ASPM Isoforms, Domains, and Functions
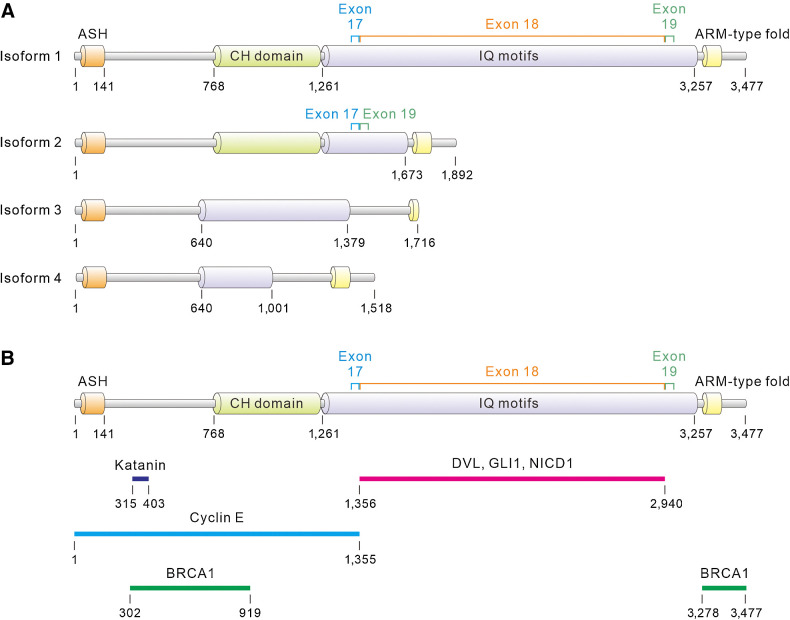
The human ASPM gene is located at chromosome 1q31 and contains a 10,434 base pair long-coding sequence with 28 exons (5). Several putative splicing variants of the ASPM transcripts may exist in human tissues (Fig. 1A; ref. 3). The largest variant, ASPM variant 1 (ASPM-v1), encodes ASPM-i1 (NCBI RefSeq: NP_060606.3) and has an estimated molecular weight (M.W.) of 409 kDa. The transcript variant 2 of ASPM (ASPM-v2) results from the alternative splicing between exons 17 and 19 and encodes a shorter isoform 2 (ASPM-i2; NCBI RefSeq: NP_001193775.1) with an estimated M.W. of 250 kDa. Compared with ASPM-i1, ASPM-i2 lacks the 67 isoleucine and glutamine (IQ) motifs encoded by exon 18 and thus has only 14 IQ motifs. The IQ motifs are 20 to 25 amino acids (a.a.) motifs with the core consensus sequence of IQXXXRGXXXR originally discovered in myosins (20, 21). The IQ motifs on ASPM vary from 14 to 38 a.a. in length and mediate the interaction with calmodulin (3). Intriguingly, the number of IQ motifs on ASPM undergoes rapid evolution and varies considerably among species (5). The central part of the IQ motif region (IQs 4–54) of ASPM displays a striking periodicity, predicting its possible organization into a higher-order repeat structure (3). The other putative ASPM isoforms, isoforms 3 and 4, lack the calponin-homology (CH) domain and some IQ motifs (9). The CH domain comprises approximately 100 a.a. and is involved in actin and microtubule binding (22, 23). The amino (N)-terminal region of ASPM contains the ASH (ASPM, SPD-2, Hydin) domain that has roles in the cilia, flagella, centrosome, and Golgi complex (9, 24). The carboxy (C)-terminal region of ASPM consists of HEAT/Armadillo-like repeats, which are repetitive amino acid sequences also found in β-catenin (9, 15).
Figure 1.
ASPM isoforms and their binding partners. A, The domain architectures of putative ASPM isoforms. ASH, ASPM, SPD-2, Hydin; CH, calponin-homology; ARM, armadillo; IQ, isoleucine and glutamine. The protein segments encoded by exons 17, 18, and 19 of ASPM are indicated with brackets. B, Schematics representing the protein segments or domains by which human ASPM interacts with its binding partners. The katanin-binding domain is predicted by aligning the sequences of human and murine ASPM according to ref. 15.
Fetal tissues predominantly express ASPM-v1 and ASPM-v2 (3). Cancer cells also mainly express these two ASPM isoforms (25–27). In pancreatic ductal adenocarcinoma (PDAC), gastric cancer, and small cell lung cancer (SCLC), ASPM-i1 is prominently overexpressed in cancer cells compared with normal cells, whereas the expression of ASPM-i2 remains unchanged or even decreases in cancer cells (25–27). Another interesting difference between ASPM-i1 and ASPM-i2 is their subcellular localizations in cancer cells. ASPM-i1 expression is localized to the cytoplasm, especially the cortical region, of cancer cells, whereas ASPM-i2 is predominantly nuclearly localized (25, 27). The N-terminal region (a.a. 1–640) of ASPM is shared by all isoforms and may mediate its functions in both normal and malignant cells. For instance, ASPM regulates microtubule disassembly at spindle poles by interacting with the microtubule-severing ATPase katanin via a small domain located within the N-terminal region of ASPM (15). The N-terminal region of ASPM binds to cyclin E to regulate its stability (Supplementary Fig. S1A and S1B), thereby controlling the restriction point progression in NPCs and cancer cells (13). Moreover, ASPM is recruited to the DNA double-strand break (DSB) through its N-terminal region to facilitate repair by homologous recombination (28). Recent studies uncovered that the exon 18–encoded segment of ASPM, which exists only in ASPM-i1, mediates its interaction with the critical regulatory nodes in development-associated signaling pathways, including Dishevelled (DVL) proteins in Wnt signaling, GLI family zinc finger 1 (GLI1) in Hh signaling, and NOTCH1 intracellular domain (NICD1) in Notch signaling (Fig. 1B; Supplementary Figs. S2A and S3; refs. 26, 27). The differences in the domain architectures and the binding partners of ASPM-i1 and ASPM-i2 may reflect the different cellular processes and functions that they mediate in normal and malignant cells.
ASPM Expression and Prognostic Significance in Cancers
Aside from fetal tissues, ASPM is expressed in some normal adult tissues, such as the testis, ovary, and spleen, and the stem/progenitor cells in gastric and colonic mucosa (25, 29–31). ASPM has been identified as an E2F transcription factor 1 (E2F1)-regulated gene expressed explicitly by the scattered stem/progenitor cells located in the isthmus zone of gastric oxyntic mucosa (30). ASPM is also expressed by the stem-cell-like cells in the crypt base of normal colonic mucosa (31). Perhaps related to its expression in stem/progenitor cells and fetal tissue, ASPM is widely overexpressed in malignant tissues, and its expression correlates with the poor prognosis for patients with various types of cancer (Supplementary Table S1; refs. 3, 30–54). Indeed, the interrogation into The Cancer Genome Atlas (TCGA) data sets revealed a marked increase in ASPM expression in most human solid tumors and leukemia (Supplementary Fig. S4). Several recent reports demonstrated that the expression of ASPM-i1 is specifically upregulated in cancer cells and has poor prognostic significance (Supplementary Table S1; refs. 25–27).
Cancer cells within the same tumor exhibit substantial phenotypic and genetic heterogeneity. IHC studies revealed the considerable expressional heterogeneity of ASPM in solid human tumors (25, 27, 40, 44). For instance, only 5% of prostate cancer cells and 3.7% of hepatocellular carcinoma (HCC) cells display a high staining intensity of ASPM, respectively (40, 44). Similarly, only a small subset of cancer cells expresses a high level of ASPM-i1 in human PDAC (1.8%), gastric cancer (5.3%), and SCLC tissues (2.6%; refs. 25–27). Of note, ASPMhigh cancer cells were found to express a variety of stemness markers, such as active β-catenin and dishevelled 1 (DVL1) in HCC (44), LGR5 and SOX2 in gastric cancer (25), and GLI1 in SCLC (26), reflecting its role in the CSC phenotype as discussed below. Therefore, the overall expression level of ASPM or ASPM-i1 in bulk tumor tissues may not faithfully reflect its clinical and pathogenetic significance. An “ASPM positive score” has been used to enumerate cancer cells displaying varying expression levels of ASPM. The staining intensity of ASPM was quantified at the single-cell level, encompassing multiple tissue sections. The percentage of tumor cells exhibiting a moderate-to-high (≥2+) staining intensity of ASPM was considered the ASPM positive score. Thus, a high ASPM positive score (>18%) predicted a poor prognosis for patients with prostate cancer (40). Likewise, a high (≥1.5% in PDAC; ≥10% in gastric cancer and SCLC) ASPM-i1 positive score was associated with poor survival of patients with PDAC, gastric cancer, or SCLC (25–27).
The Roles of ASPM in the Malignant Phenotype and Stemness
Mounting data over recent years have indicated the critical role of ASPM in the malignant phenotypes of cancer cells. Lose-of-function studies reported that the genetic knockdown or knockout of ASPM would lead to reduced proliferation, migration, and invasion of cancer cells in various cancers (Supplementary Table S2; refs. 25–27, 33–37, 40, 43, 44, 47, 53, 55–59). Consistently, downregulating ASPM or ASPM-v1 expression markedly attenuated the growth of xenograft tumors in multiple cancers (26, 37, 40, 47, 55, 56, 59). ASPM deficiency also reduces the ability of cancer cells, such as colorectal cancer cells, to initiate distant metastasis (Supplementary Table S2; ref. 58).
Data accumulated over recent years indicate that ASPM is preferentially expressed by CSCs in various solid tumors, such as PDAC (37) and SCLC (26). In gastric cancer, the transcription factor Forkhead box M1 (FOXM1), a well-known regulator of stem cells and CSCs (60), was shown to enhance ASPM expression in CSCs (25). The elevated expression of ASPM in stem and progenitor cells and CSCs prompts speculation about its role in regulating cancer stemness (61–63). Indeed, ASPM contributes to the maintenance of CD44+CD133+ or CD44+CD24+ CSCs in PDAC (37, 64, 65), aldehyde dehydrogenase 1 (ALDH1)+ CSCs in prostate cancer (40), and Wnt-activityhighALDH1+EpCAM+ CSCs that mediate HCC progression (44). In gastric cancer, ASPM augments the Wnt pathway activity to maintain LGR5+ and ALDH1+ CSCs (25). ASPM-i1 supports CD133+ or UPAR+ CSCs by positively controling the Hh pathway in SCLC (26). Corroborating the role of ASPM in the CSC phenotype, its expression level positively correlates with CSC markers, such as ALDH1 (27), active β-catenin (25, 44), LGR5, SOX2, and FOXM1 in human cancer tissues (25, 60). Consistently, the expressions of representative stemness-associated genes, such as EZH2, KLF4, MYC, NANOG, POU5F1, SMO, and SOX2, are markedly downregulated in ASPM-deficient cancer cells (25, 40).
At the functional level, in vitro and in vivo studies have provided compelling evidence supporting the importance of ASPM in the CSC phenotypes. Downregulating ASPM or ASPM-v1 expression crippled the ability of cancer cells to form tumorspheres or the growth of xenograft tumors in immunodeficient mice (25, 26, 37, 40, 44). Of the two major ASPM isoforms, ASPM-i1 may play a more crucial role in regulating cancer stemness and CSCs due to its unique exon 18–preserved region and MOAs, as described below. The role of ASPM in regulating the small population of CSCs in cancer provides a plausible explanation for its cell-to-cell expressional heterogeneity as described above (25–27, 40, 44).
Regulation of ASPM Expression in Cancers
Despite the wealth of data showing the elevated expression of ASPM in malignant tissues, the molecular underpinnings of its overexpression in cancers remain incompletely understood. At least in glioblastoma, ASPM overexpression is not due to changes in the methylation level of its promoter or copy-number gain, and instead, its expression is mediated by the mutant EGFR (EGFRvIII) signaling (35). In glioma cells and CSCs in gastric cancer, the transcriptional factor FOXM1 regulates ASPM expression by directly binding to its promoter (25, 55). Interestingly, reports have also shown that FOXM1 promotes the nuclear translocation of β-catenin (66, 67), suggesting an interplay between FOXM1, ASPM, and β-catenin that reinforces the Wnt pathway activity (25).
The roles of ASPM in regulating chromosome stability, DNA DSB repair, and the CSC phenotype raise the possibility that its upregulated expression in cancer cells represents a cellular response to heightened DNA damage/replication stress and oxidative stress during malignant progression (1, 2). Additional screening strategies will be required to identify the upstream signaling pathways that induce ASPM expression in stressed cancer cells.
Molecular Mechanisms Underlying the Oncogenic Function of ASPM
Canonical Wnt signaling
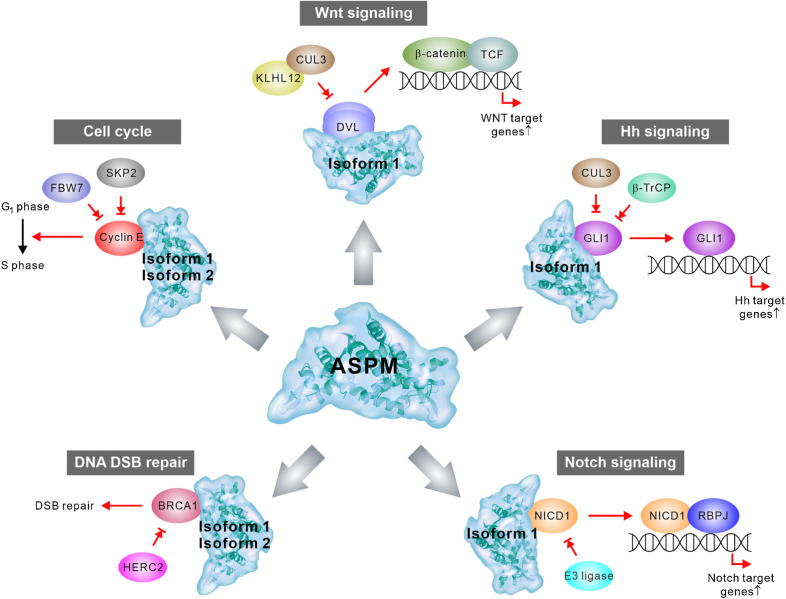
The earliest evidence suggesting ASPM as a positive Wnt regulator was reported by a genome-wide small-interfering RNA (siRNA) screening conducted in colorectal cancer cells (68). In a mouse teratocarcinoma cell line, the knockdown of ASPM expression reduced the Wnt reporter activity (69). The critical role of ASPM in Wnt signaling was affirmed in subsequent studies (25, 36, 37, 40, 43, 44, 54, 56). Mechanistically, ASPM interacts with and stabilizes the upstream Wnt-pathway regulator, dishevelled 2 (DVL2), to facilitate the relay of Wnt signals to β-catenin in PDAC cells. Another DVL isoform, dishevelled 3 (DVL3), is involved in the ASPM-regulated Wnt pathway activity in prostate cancer cells (40). ASPM regulates different DVL proteins among different types of cancers, such as DVL1 in HCC cells (44) and DVL3 in gastric cancer cells (25). ASPM stabilizes DVL proteins by competing with its specific E3 ubiquitin-ligase Cullin 3 and the adaptor protein Kelch-like 12 (25, 44, 70). Notably, because ASPM-i1 interacts with DVL through its distinct exon 18–encoded segment (Fig. 1B; Supplementary Fig. S3; ref. 26), the DVL proteins were found to interact with ASPM-i1 but not ASPM-i2 (Supplementary Fig. S2A; refs. 26, 27).
Hh and Notch signaling
Recently, a pathway reporter screening has extended the repertoires of ASPM-regulated pathways to other development- and stemness-associated signaling pathways, notably the Hh and Notch pathways (26). Like the MOA by which ASPM-i1 regulates the stability of DVL proteins, it stabilizes the Hh pathway transcriptional factor GLI1 by inhibiting its binding with the E3-ubiquitin ligase β-TrCP and Cullin 3, thus preventing it from proteasomal degradation. Interestingly, ASPM-i1 also enhances the transcription of the membrane regulator Smoothened (SMO) in Hh signaling indirectly through augmenting the Wnt−DVL3−β-catenin axis, representing an intersection between Wnt and Hh signaling pathways in cancers (26). Interestingly, ASPM-v1 deficiency also significantly reduced the Notch pathway activity in SCLC cells (26). Indeed, our follow-on molecular studies showed that ASPM-i1 contributes to the cellular responsiveness to Notch ligands by regulating the expression of NICD1 at the protein level (Supplementary Fig. S5A, S5B and S5C), potentially by inhibiting its specific E3-ubiquitin ligases (Fig. 2; ref. 71). Like the interaction between ASPM-i1 and DVL, ASPM-i1 interacts with GLI1 and NICD1 specifically through its distinct exon 18–encoded segment (Fig. 1B; Supplementary Fig. S3).
Figure 2.
Model depicting the pleiotropic pro-oncogenic functions of ASPM. ASPM isoform 1 competes with specific E3 ligases to bind to DVL, GLI1, and NICD1 to prevent their ubiquitination and proteasome-dependent degradation, thereby procuring the Wnt, Hh, and Notch signaling activities in cancer cells, especially CSCs. Both ASPM isoforms 1 and 2 bind to and stabilize cyclin E and BRCA1 to promote cell-cycle progression and DNA DSB repair. The protein structure of Saquinus labiatus ASPM (predicted by AlphaFold; https://alphafold.ebi.ac.uk/), which has a high sequence similarity with the human paralog, is used in the diagram.
DNA damage repair and chromosome stability
ASPM plays a pleiotropic role in mediating DNA damage repair and chromosome stability of normal and cancer cells (28, 72, 73). Downregulating ASPM expression inhibited DNA DSB repair and enhanced the sensitivity of cancer cells and fibroblasts to ionizing radiation (IR) exposure and DNA-damaging agents (72). The role of ASPM in DNA DSB repair has been ascribed to nonhomologous end-joining (NHEJ) through the DNA-PK-dependent pathway, but the supportive evidence was indirect (72). Another study has more directly pointed to the specific role of ASPM in BRCA1-dependent DNA DSB repair by homologous recombination in 293T and HeLa cells (28). ASPM interacts with and stabilizes BRCA1 by interfering with its binding with the E3-ubiquitin ligase HERC2, thereby maintaining the chromosome stability of cancer cells and promoting their survival following X-ray irradiation. Downregulating ASPM expression increased the sensitivity of cancer cells to IR exposure and exhibited synthetic lethality with the PARP inhibitor olaparib, potentially yielding a therapeutic opportunity. Notably, the ability of ASPM to regulate BRCA1-dependent DNA DSB repair is unlikely ASPM-i1-specific as ASPM interacts with BRCA1 through the N-terminal region and the extreme C-terminus, which is shared by ASPM-i1 and ASPM-i2 (28). In addition, ASPM was recently reported to bind to the stalled replication forks during replication stress in a FAD17-dependent manner. The fork-enriched ASPM promotes RAD9 and TopBP1 loading to chromatin, facilitating ATR-CHK1 activation to promote fort restart. ASPM also antagonizes MRE11 loading to protect nascent DNA from degradation to maintain chromosome stability (73).
Mitotic progression
As described above, ASPM and the products of Drosophila asp and mouse Aspm bind to the microtubule minus ends during cell division and contribute to aster formation, spindle pole focusing and orientation, chromosome segregation, and cytokinesis (15, 16, 74). ASPM regulates mitotic processes in various types of cells, including neuroblasts, epithelial cells, phagocytes, and human cancer cell lines, likely representing its conserved and essential function in cells. Molecular studies demonstrated that ASPM interacts with the microtubule-severing protein katanin through its N-terminal region shared by ASPM-i1 and ASPM-i2 (Fig. 1B; Supplementary Fig. S2B; ref. 15), reinforcing the notion that the effects of ASPM in cellular mitotic processes are not restricted to stem/progenitor cells or CSCs. A recent comprehensive review provides details of the role of ASPM and its orthologs in mitotic processes (74). Whether or how the mitosis-regulatory function of ASPM contributes to its pro-oncogenic and pro-stemness properties remains unclear and requires further elucidation.
Cell-cycle progression
Another important molecular function of ASPM is regulating the passage of cells through the G1 restriction point in the cell cycle, which has been shown in NPCs and cancer cells (13, 36, 55, 57, 75). Mechanistically, ASPM binds to cyclin E through its N-terminal region to stabilize cyclin E by inhibiting its binding to the E3-ubiquitin ligases FBXW7 and SKP2 (Figs. 1B and 2; ref. 13). Although both ASPM-i1 and ASPM-i2 bind to cyclin E (Supplementary Fig. S2B), ASPM-i2 has been shown to play a relatively more important in cell-cycle regulation (27). However, because cyclin D is a major Wnt target gene, ASPM-i1 may also regulate the cell-cycle progression of cancer cells, especially CSCs, indirectly by augmenting the Wnt pathway activity (44). In line with this possibility, ASPM expression has been shown to contribute to the expression of cyclin D1 and cyclin-dependent kinase 4 (CDK4) in lung cancer cells (46). Whether or how the molecular function of ASPM, especially ASPM-i1, in cell-cycle regulation contributes to the self-renewal of CSCs and their stemness properties still awaits further investigation.
Kif11
ASPM has been recently reported to stabilize the microtubule motor protein KIF11, a potential oncoprotein, by inhibiting its protein ubiquitination and degradation in anaplastic thyroid cancer cells (59). The ASPM-regulated KIF11 expression may be functionally important, as the overexpression of KIF11 could partially rescue the ASPM-deficient phenotype in cancer cells.
The MOA of ASPM-mediated protein stabilization
The ability of ASPM to compete with various E3-ubiquitin ligases to inhibit the proteasome-dependent degradation of its binding partners represents a recurring and unifying MOA behind its oncogenic function (Fig. 2). Indeed, ubiquitin-dependent proteolysis is known to regulate various biological processes (76). The ability of ASPM to compete with different E3-ubiquitin ligases for substrate binding may endow it with the ability to control multiple oncogenic pathways, presumably in a context- and spatial-dependent manner. It is also noteworthy that ASPM interacts with its binding partners either through the isoform-shared N-terminal region (BRCA1 and cyclin E) or the ASPM-i1–specific exon 18–encoded segment (DVL, GLI1, and NICD1; Fig. 1B). Therefore, the protein-stabilizing function of ASPM may not rely on distinct domains but uncharacterized higher-order protein conformations that still await further investigation.
Concluding Remarks
During the past two decades, ASPM and its orthologs have been extensively studied for their roles in microtubule organization, mitotic spindle alignment, and neurogenesis (3, 5–8, 12). Although many clinical correlative studies have reported ASPM overexpression and its prognostic significance in human cancers, the molecular mechanisms underlying its oncogenic properties have not been rigorously studied until the identification of its role in Wnt signaling and the CSC phenotype (25, 37, 40). Emerging evidence suggests that ASPM may be the regulatory hub of multiple development- and stemness-associated signaling pathways beyond Wnt signaling (Fig. 2; refs. 25–27). From the systems biology perspective, ASPM may meet the definition of the conceptual “date hub,” which serves as the node connecting multiple modules to maintain the integrity of the interactome inside a cell (4). The pleiotropic pro-oncogenic functions of ASPM provide a plausible explanation for its ubiquitous overexpression in cancers (3). Of note, although the current review emphasizes the role of ASPM-i1 in regulating development- and stemness-associated pathways, ASPM-i2 may also participate in malignant progression as it governs the essential functions of cancer cells, including cell-cycle progression and DNA damage repair (13, 28).
The prognostic importance of ASPM in various cancers opens a window of opportunity for further developing a pathway-informed and “cancer-agnostic” prognostic biomarker. Potential assays that can be used to detect and measure ASPM or ASPM-i1 expression in cancer tissues include IHC staining, in situ hybridization, and proximal ligation assays. It is crucial to consider the cell-to-cell variation in ASPM expression while developing these assays. Similar issues have been resolved in other biomarkers exhibiting a heterogeneous expression in cancer, such as EGFR and programmed death ligand 1 (PDL1). The development of clinically useful ASPM-related biomarkers can be facilitated by whole-tissue single-cell profiling using high-resolution digital pathology platforms assisted with automated image analysis. On the other hand, given that overexpression or mutations in genes can lead to alteration in the tumor microenvironment, including the composition of immune cells (77), it would also be interesting to study the potential link between ASPM and the tumor microenvironment. In support of this possibility, a recent report demonstrated that ASPM upregulation in cancer cells might contribute to the infiltration of immune cells to tumors, such as B and T cells (78).
Cancer fitness genes have been implicated in late-stage tumor progression as they provide survival advantages to tumor cells (2). One distinguishing feature of cancer fitness genes from classical oncogenes is that they do not have the transforming ability per se and are not essential for normal tissue cells. We consider ASPM, specifically ASPM-i1, a candidate cancer fitness gene since (i) its expression is low or negligible in normal tissues, (ii) it is expressed by CSCs to promote tumor stemness, and (iii) the mutations or genetic knockout of ASPM are not embryonically lethal and does not affect the development of normal organs or tissues in mice or higher mammals (12, 17). Complementing these findings, ASPM is not classified as an essential gene of cells according to the Cancer Dependency Map (https://depmap.org/portal/; ref. 79). Conceptually, cancer fitness genes may serve as more attractive and feasible therapeutic targets than driver oncogenes as they play cardinal roles in late-stage cancer progression and metastasis and are associated with a larger therapeutic window (2). Substantiating this notion, a regulatory hub in the signaling networks and the interactome of cancer cells, such as ASPM, can represent a targetable vulnerability in cancers (4). We envision the therapeutics capable of inhibiting ASPM-i1 expression or disabling its interaction with DVL, GLI1, NICD1, and/or other binding partners may have clinical utility in treating advanced-stage cancers. Because the selective silencing of ASPM-v1 can be achieved by targeting exon 18 of the ASPM gene (25–27), the genetic targeting of ASPM-v1 using antisense oligonucleotide, siRNA, or gene-editing strategies may become promising cancer therapeutics that have minimal effects on the essential functions of normal cells. Such a strategy may also unlock an unprecedented opportunity to simultaneously block the Wnt, Hh, and Notch pathways in cancer cells and possibly enhances the tumoral responsiveness to cytotoxic agents in light of the role of cancer stemness in treatment resistance (80). Moreover, therapeutics disrupting the interaction of ASPM with BRCA1 may also provide a new avenue for improving the antitumor efficacy of radiation, DNA-damaging agents, and PARP inhibitors in cancers. In this approach, destabilizing BRCA1 by inhibiting ASPM may constitute synthetic lethality to broaden the clinical indication of PARP inhibitors (81). Whether these potential ASPM-targeting strategies can be successfully developed to become breakthrough cancer therapies remains to be seen in the coming decade.
Supplementary Material
This file contains Supplementary Figures 1-5, Supplementary Tables 1-2, and Supplementary Materials and Methods.
Acknowledgments
This work was supported by the National Science and Technology Council (NSTC), Taiwan (MOST 108–2314-B-038–105 and MOST 111–2314-B-038–075 to KKT), and the TMU Research Center of Cancer Translational Medicine from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
K.K. Tsai reports grants from Ministry of Science and Technology, Taiwan, and Ministry of Education, Taiwan, during the conduct of the study. K.K. Tsai also has a patent for U.S. provisional patent application pending to Taipei Medical University. No disclosures were reported by the other authors.
References
- 1. Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 2009;136:823–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shen M, Kang Y. Cancer fitness genes: emerging therapeutic targets for metastasis. Trends Cancer 2023;9:69–82. [DOI] [PubMed] [Google Scholar]
- 3. Kouprina N, Pavlicek A, Collins NK, Nakano M, Noskov VN, Ohzeki J, et al. The microcephaly ASPM gene is expressed in proliferating tissues and encodes for a mitotic spindle protein. Hum Mol Genet 2005;14:2155–65. [DOI] [PubMed] [Google Scholar]
- 4. Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 2004;430:88–93. [DOI] [PubMed] [Google Scholar]
- 5. Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet 2002;32:316–20. [DOI] [PubMed] [Google Scholar]
- 6. Wakefield JG, Bonaccorsi S, Gatti M. The drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J Cell Biol 2001;153:637–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. do Carmo Avides M, Tavares A, Glover DM. Polo kinase and Asp are needed to promote the mitotic organizing activity of centrosomes. Nat Cell Biol 2001;3:421–4. [DOI] [PubMed] [Google Scholar]
- 8. Gonzalez C, Saunders RD, Casal J, Molina I, Carmena M, Ripoll P, et al. Mutations at the asp locus of drosophila lead to multiple free centrosomes in syncytial embryos, but restrict centrosome duplication in larval neuroblasts. J Cell Sci 1990;96 (Pt 4):605–16. [DOI] [PubMed] [Google Scholar]
- 9. Rujano MA, Sanchez-Pulido L, Pennetier C, le Dez G, Basto R. The microcephaly protein Asp regulates neuroepithelium morphogenesis by controlling the spatial distribution of myosin II. Nat Cell Biol 2013;15:1294–306. [DOI] [PubMed] [Google Scholar]
- 10. van der Voet M, Berends CW, Perreault A, Nguyen-Ngoc T, Gonczy P, Vidal M, et al. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat Cell Biol 2009;11:269–77. [DOI] [PubMed] [Google Scholar]
- 11. Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci U S A 2006;103:10438–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson MB, Sun X, Kodani A, Borges-Monroy R, Girskis KM, Ryu SC, et al. Aspm knockout ferret reveals an evolutionary mechanism governing cerebral cortical size. Nature 2018;556:370–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Capecchi MR, Pozner A. ASPM regulates symmetric stem cell division by tuning cyclin E ubiquitination. Nat Commun 2015;6:8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jayaraman D, Kodani A, Gonzalez DM, Mancias JD, Mochida GH, Vagnoni C, et al. Microcephaly proteins Wdr62 and Aspm define a mother centriole complex regulating centriole biogenesis, apical complex, and cell fate. Neuron 2016;92:813–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang K, Rezabkova L, Hua S, Liu Q, Capitani G, Altelaar AFM, et al. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat Cell Biol 2017;19:480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gai M, Bianchi FT, Vagnoni C, Verni F, Bonaccorsi S, Pasquero S, et al. ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules. Embo Rep 2016;17:1396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pulvers JN, Bryk J, Fish JL, Wilsch-Brauninger M, Arai Y, Schreier D, et al. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci U S A 2010;107:16595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mori M, Tando S, Ogi H, Tonosaki M, Yaoi T, Fujimori A, et al. Loss of abnormal spindle-like, microcephaly-associated (Aspm) disrupts female folliculogenesis in mice during maturation and aging. Reprod Biol 2022;22:100673. [DOI] [PubMed] [Google Scholar]
- 19. Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends Genet 2009;25:501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin SR, Bayley PM. Regulatory implications of a novel mode of interaction of calmodulin with a double IQ-motif target sequence from murine dilute myosin V. Protein Sci 2002;11:2909–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bahler M, Rhoads A. Calmodulin signaling via the IQ motif. FEBS Lett 2002;513:107–13. [DOI] [PubMed] [Google Scholar]
- 22. Gimona M, Djinovic-Carugo K, Kranewitter WJ, Winder SJ. Functional plasticity of CH domains. FEBS Lett 2002;513:98–106. [DOI] [PubMed] [Google Scholar]
- 23. Ito A, Goshima G. Microcephaly protein Asp focuses the minus ends of spindle microtubules at the pole and within the spindle. J Cell Biol 2015;211:999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ponting CP. A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics 2006;22:1031–5. [DOI] [PubMed] [Google Scholar]
- 25. Hsu CC, Liao WY, Chang KY, Chan TS, Huang PJ, Chiang CT, et al. A multi-mode Wnt- and stemness-regulatory module dictated by FOXM1 and ASPM isoform I in gastric cancer. Gastric Cancer 2021;24:624–39. [DOI] [PubMed] [Google Scholar]
- 26. Cheng LH, Hsu CC, Tsai HW, Liao WY, Yang PM, Liao TY, et al. ASPM activates Hedgehog and Wnt signaling to promote small cell lung cancer stemness and progression. Cancer Res 2023;83:830–844. [DOI] [PubMed] [Google Scholar]
- 27. Hsu CC, Liao WY, Chan TS, Chen WY, Lee CT, Shan YS, et al. The differential distributions of ASPM isoforms and their roles in Wnt signaling, cell cycle progression, and pancreatic cancer prognosis. J Pathol 2019;249:498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu S, Wu X, Wang P, Cao SL, Peng B, Xu X. ASPM promotes homologous recombination-mediated DNA repair by safeguarding BRCA1 stability. iScience 2021;24:102534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luers GH, Michels M, Schwaab U, Franz T. Murine calmodulin binding protein 1 (Calmbp1): tissue-specific expression during development and in adult tissues. Mech Dev 2002;118:229–32. [DOI] [PubMed] [Google Scholar]
- 30. Vange P, Bruland T, Beisvag V, Erlandsen SE, Flatberg A, Doseth B, et al. Genome-wide analysis of the oxyntic proliferative isthmus zone reveals ASPM as a possible gastric stem/progenitor cell marker over-expressed in cancer. J Pathol 2015;237:447–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. An X, Huang Y, Zhao P. Expression of ASPM in colonic adenocarcinoma and its clinicopathologic significance. Int J Clin Exp Pathol 2017;10:8968–73. [PMC free article] [PubMed] [Google Scholar]
- 32. Bruning-Richardson A, Bond J, Alsiary R, Richardson J, Cairns DA, McCormack L, et al. ASPM and microcephalin expression in epithelial ovarian cancer correlates with tumour grade and survival. Br J Cancer 2011;104:1602–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu Y, You Y, Chen L, Liu Y, Liu Y, Lou W, et al. Abnormal spindle-like microcephaly-associated protein promotes proliferation by regulating cell cycle in epithelial ovarian cancer. Gland Surg 2022;11:687–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bikeye SN, Colin C, Marie Y, Vampouille R, Ravassard P, Rousseau A, et al. ASPM-associated stem cell proliferation is involved in malignant progression of gliomas and constitutes an attractive therapeutic target. Cancer Cell Int 2010;10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A 2006;103:17402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen X, Huang L, Yang Y, Chen S, Sun J, Ma C, et al. ASPM promotes glioblastoma growth by regulating G1 restriction point progression and Wnt-beta-catenin signaling. Aging (Albany NY) 2020;12:224–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang WY, Hsu CC, Wang TY, Li CR, Hou YC, Chu JM, et al. A gene expression signature of epithelial tubulogenesis and a role for ASPM in pancreatic tumor progression. Gastroenterology 2013;145:1110–20. [DOI] [PubMed] [Google Scholar]
- 38. Tian X, Wang N. Upregulation of ASPM, BUB1B and SPDL1 in tumor tissues predicts poor survival in patients with pancreatic ductal adenocarcinoma. Oncol Lett 2020;19:3307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xie JJ, Zhuo YJ, Zheng Y, Mo RJ, Liu ZZ, Li BW, et al. High expression of ASPM correlates with tumor progression and predicts poor outcome in patients with prostate cancer. Int Urol Nephrol 2017;49:817–23. [DOI] [PubMed] [Google Scholar]
- 40. Pai VC, Hsu CC, Chan TS, Liao WY, Chuu CP, Chen WY, et al. ASPM promotes prostate cancer stemness and progression by augmenting Wnt-Dvl-3-beta-catenin signaling. Oncogene 2019;38:1340–53. [DOI] [PubMed] [Google Scholar]
- 41. Lin SY, Pan HW, Liu SH, Jeng YM, Hu FC, Peng SY, et al. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res 2008;14:4814–20. [DOI] [PubMed] [Google Scholar]
- 42. Wang F, Chang Y, Li J, Wang H, Zhou R, Qi J, et al. Strong correlation between ASPM gene expression and HCV cirrhosis progression identified by co-expression analysis. Dig Liver Dis 2017;49:70–6. [DOI] [PubMed] [Google Scholar]
- 43. Wu B, Hu C, Kong L. ASPM combined with KIF11 promotes the malignant progression of hepatocellular carcinoma via the Wnt/beta-catenin signaling pathway. Exp Ther Med 2021;22:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liao WY, Hsu CC, Chan TS, Yen CJ, Chen WY, Pan HW, et al. Dishevelled 1-regulated superpotent cancer stem cells mediate wnt heterogeneity and tumor progression in hepatocellular carcinoma. Stem Cell Reports 2020;14:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu W, Xu J, Wang Z, Jiang Y. Weighted gene correlation network analysis identifies specific functional modules and genes in esophageal cancer. J Oncol 2021;2021:8223263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yuan YJ, Sun Y, Gao R, Yin ZZ, Yuan ZY, Xu LM. Abnormal spindle-like microcephaly-associated protein (ASPM) contributes to the progression of lung squamous cell carcinoma (LSCC) by regulating CDK4. J Cancer 2020;11:5413–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang J, Liang J, Li H, Han J, Jiang J, Li Y, et al. Oncogenic role of abnormal spindle-like microcephaly-associated protein in lung adenocarcinoma. Int J Oncol 2021;58:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu Z, Zhang Q, Luh F, Jin B, Liu X. Overexpression of the ASPM gene is associated with aggressiveness and poor outcome in bladder cancer. Oncol Lett 2019;17:1865–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saleh AA, Gohar SF, Hemida AS, Elgharbawy M, Soliman SE. Evaluation of ASPM and TEF gene expressions as potential biomarkers for bladder cancer. Biochem Genet 2020;58:490–507. [DOI] [PubMed] [Google Scholar]
- 50. Tang J, Lu M, Cui Q, Zhang D, Kong D, Liao X, et al. Overexpression of ASPM, CDC20, and TTK confer a poorer prognosis in breast cancer identified by gene co-expression network analysis. Front Oncol 2019;9:310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang C, Zhang B, Meng D, Ge C. Comprehensive analysis of DNA methylation and gene expression profiles in cholangiocarcinoma. Cancer Cell Int 2019;19:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu J, He Z, Zhu Y, Jiang C, Deng Y, Wei B. ASPM predicts poor clinical outcome and promotes tumorigenesis for diffuse large B-cell lymphoma. Curr Cancer Drug Targets 2021;21:80–9. [DOI] [PubMed] [Google Scholar]
- 53. Lin P, Liang LY, Dong YZ, Ren ZP, Zhao HJ, Li GS. Identification of abnormal spindle microtubule assembly as a promising therapeutic target for osteosarcoma. Orthop Surg 2020;12:1963–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang T, Chi Z, Liu G, Hong X, Cao S, Cheng K, et al. Screening ANLN and ASPM as bladder urothelial carcinoma-related biomarkers based on weighted gene co-expression network analysis. Front Genet 2023;14:1107625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zeng WJ, Cheng Q, Wen ZP, Wang JY, Chen YH, Zhao J, et al. Aberrant ASPM expression mediated by transcriptional regulation of FoxM1 promotes the progression of gliomas. J Cell Mol Med 2020;24:9613–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang H, Yang X, Zhu L, Li Z, Zuo P, Wang P, et al. ASPM promotes hepatocellular carcinoma progression by activating Wnt/beta-catenin signaling through antagonizing autophagy-mediated Dvl2 degradation. FEBS Open Bio 2021;11:2784–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xia C, Xu X, Ding Y, Yu C, Qiao J, Liu P. Abnormal spindle-like microcephaly-associated protein enhances cell invasion through Wnt/beta-catenin-dependent regulation of epithelial-mesenchymal transition in non-small cell lung cancer cells. J Thorac Dis 2021;13:2460–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang L, Hu XD, Li SY, Liang XY, Ren L, Lv SX. ASPM facilitates colorectal cancer cells migration and invasion by enhancing beta-catenin expression and nuclear translocation. Kaohsiung J Med Sci 2022;38:129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fang Q, Li Q, Qi Y, Pan Z, Feng T, Xin W. ASPM promotes migration and invasion of anaplastic thyroid carcinoma by stabilizing KIF11. Cell Biol Int 2023;47:1209–21. [DOI] [PubMed] [Google Scholar]
- 60. Sher G, Masoodi T, Patil K, Akhtar S, Kuttikrishnan S, Ahmad A, et al. Dysregulated FOXM1 signaling in the regulation of cancer stem cells. Semin Cancer Biol 2022;86(Pt 3):107–21. [DOI] [PubMed] [Google Scholar]
- 61. Hermann PC, Huber SL, Heeschen C. Metastatic cancer stem cells: a new target for anti-cancer therapy? Cell Cycle 2008;7:188–93. [DOI] [PubMed] [Google Scholar]
- 62. Santamaria-Martinez A, Huelsken J. The niche under siege: novel targets for metastasis therapy. J Intern Med 2013;274:127–36. [DOI] [PubMed] [Google Scholar]
- 63. Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 2008;8:755–68. [DOI] [PubMed] [Google Scholar]
- 64. Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007;1:313–23. [DOI] [PubMed] [Google Scholar]
- 65. Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res 2007;67:1030–7. [DOI] [PubMed] [Google Scholar]
- 66. Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell 2011;20:427–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sheng Y, Yu C, Liu Y, Hu C, Ma R, Lu X, et al. FOXM1 regulates leukemia stem cell quiescence and survival in MLL-rearranged AML. Nat Commun 2020;11:928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Major MB, Roberts BS, Berndt JD, Marine S, Anastas J, Chung N, et al. New regulators of Wnt/beta-catenin signaling revealed by integrative molecular screening. Sci Signal 2008;1:ra12. [DOI] [PubMed] [Google Scholar]
- 69. Buchman JJ, Durak O, Tsai LH. ASPM regulates Wnt signaling pathway activity in the developing brain. Genes Dev 2011;25:1909–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jin X, Jeon HM, Jin X, Kim EJ, Yin J, Jeon HY, et al. The ID1-CULLIN3 axis regulates intracellular SHH and WNT signaling in glioblastoma stem cells. Cell Rep 2016;16:1629–41. [DOI] [PubMed] [Google Scholar]
- 71. Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem 2001;276:34371–8. [DOI] [PubMed] [Google Scholar]
- 72. Kato TA, Okayasu R, Jeggo PA, Fujimori A. ASPM influences DNA double-strand break repair and represents a potential target for radiotherapy. Int J Radiat Biol 2011;87:1189–95. [DOI] [PubMed] [Google Scholar]
- 73. Wu X, Xu S, Wang P, Wang ZQ, Chen H, Xu X, et al. ASPM promotes ATR-CHK1 activation and stabilizes stalled replication forks in response to replication stress. Proc Natl Acad Sci U S A 2022;119:e2203783119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Razuvaeva AV, Graziadio L, Palumbo V, Pavlova GA, Popova JV, Pindyurin AV, et al. The multiple mitotic roles of the ASPM orthologous proteins: insight into the etiology of ASPM-dependent microcephaly. Cells 2023;12:922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang Q, Qi M, Chen Y, Tian S, Liao F, Dong W. ASPM is a novel candidate gene associated with colorectal cancer cell growth. DNA Cell Biol 2021;40:921–35. [DOI] [PubMed] [Google Scholar]
- 76. Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998;67:425–79. [DOI] [PubMed] [Google Scholar]
- 77. Dhainaut M, Rose SA, Akturk G, Wroblewska A, Nielsen SR, Park ES, et al. Spatial CRISPR genomics identifies regulators of the tumor microenvironment. Cell 2022;185:1223–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Deng T, Liu Y, Zhuang J, Tang Y, Huo Q. ASPM is a prognostic biomarker and correlates with immune infiltration in kidney renal clear cell carcinoma and liver hepatocellular carcinoma. Front Oncol 2022;12:632042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell 2017;170:564–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dias MP, Moser SC, Ganesan S, Jonkers J. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat Rev Clin Oncol 2021;18:773–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Figures 1-5, Supplementary Tables 1-2, and Supplementary Materials and Methods.