Abstract
The heat shock protein 90 (Hsp90) is a molecular chaperone, which plays a key role in eukaryotic protein homeostasis. Co-chaperones assist Hsp90 in client maturation and in regulating essential cellular processes such as cell survival, signal transduction, gene regulation, hormone signaling, and neurodegeneration. Aha1 (activator of Hsp90 ATPase) is a unique co-chaperone known to stimulate the ATP hydrolysis of Hsp90, but the mechanism of their interaction is still unclear. In this report, we show that one or two Aha1 molecules can bind to one Hsp90 dimer and that the binding stoichiometry affects Hsp90’s conformation, kinetics, ATPase activity, and stability. In particular, a coordination of two Aha1 molecules can be seen in stimulating the ATPase activity of Hsp90 and the unfolding of the middle domain, whereas the conformational equilibrium and kinetics are hardly affected by the stoichiometry of bound Aha1. Altogether, we show a regulation mechanism through the stoichiometry of Aha1 going far beyond a regulation of Hsp90’s conformation.
Significance
Many studies have shown that molecular chaperones, like the heat shock protein Hsp90, are regulated by co-chaperones. These studies were usually done at saturating co-chaperone conditions. As the functional form of Hsp90 is a dimer, it remains unclear if in these experiments two co-chaperones are bound per dimer or only one. Here, we directly control the binding stoichiometry and show that it has a significant effect using the co-chaperone Aha1. Most interestingly, the stoichiometry of Aha1 binding has different effects on different quantities like ATPase rate, conformational kinetics, or protein stability. Therefore, the stoichiometry can serve as an additional regulatory mechanism that has likely been underappreciated in the past.
Introduction
Hsp90 is a highly abundant protein in the cell and is conserved in all eukaryotes. The proteome is guarded by Hsp90 against heat and other stresses, and Hsp90 maintains protein homeostasis in eukaryotes (1). It enables correct folding, stabilization, and maturation of a diverse set of client proteins such as transcription factors, kinases, hormone receptors, tumor suppressors, and signal transducers (2). Co-chaperones are the key regulators that recruit client proteins to Hsp90 and enable it to regulate diverse cellular processes such as signal transduction, gene regulation, cell cycle control, cell survival, DNA repair, and neuronal signaling (2).
Hsp90 has a homodimeric structure, with each monomer consisting of an N-terminal (N) ATP binding domain, a middle (M) domain that is important for client binding, and a C-terminal (C) domain that facilitates Hsp90 dimerization (3,4). Hsp90 exists in a dynamic equilibrium between an open and a closed conformation. In the open conformation, it adopts a V-shaped structure where the N-terminal domain is only loosely connected to the middle domain (4,5,6). In the closed conformation, the N-terminal domains are dimerized and likely also more strongly associated with the M-domains (7,8). ATP binding and possibly also hydrolysis are critical for the function of Hsp90 (9); however, its opening and closing dynamics are only weakly associated with ATP hydrolysis (10). In the Hsp90 ATPase cycle, Hsp90 undergoes conformational changes while interacting with several co-chaperones leading to ATP hydrolysis and client maturation.
Some co-chaperones modulate the Hsp90 function by stimulating or reducing its ATPase activity, whereas others recruit a diverse set of client proteins to Hsp90 to fulfill crucial biological functions or coordinate the interaction with other chaperone systems like Hsp70 (1,11,12). Different co-chaperones bind to specific conformational states of Hsp90, and they may bind simultaneously, competitively, or sequentially (8). Hsp90’s co-chaperones can be classified into three functional groups: client recruiters (e.g., Hop, Cdc37, and Sgt1), remodeling co-chaperones (e.g., Aha1 and peptidyl-prolyl isomerases), and late-acting co-chaperones (e.g., Sba1/p23) (13). Hsp90 can bind to multiple co-chaperones during the client maturation process leading to multiprotein complex formation.
Aha1 (activator of Hsp90 ATPase) is the only known co-chaperone that significantly accelerates the ATPase activity of Hsp90 and by this enhances its function (14). It contributes to the activation of several client proteins such as protein kinases and steroid hormone receptors (8,14). It has been shown that Aha1, along with Hsp90, plays a critical role in the regulation of cystic fibrosis transmembrane conductance regulator (CFTR) quality control and folding. Interestingly, a reduced expression level of Aha1 coupled with a reduced ATPase activity of Hsp90 affect the folding of CFTR. The authors hypothesized that Aha1 acts as a “molecular referee” and regulates Hsp90-client interactions by modulating the client dwell time (15). In a recent study, it has also been shown that overexpression of Aha1 increases the production of aggregated tau in an Hsp90-dependent process, leading to cognitive defects in mouse models (16). These studies suggest that the effect of cellular Aha1 concentration, the ratio of Aha1:Hsp90, and the level of Hsp90’s ATPase activity play a vital role in client maturation.
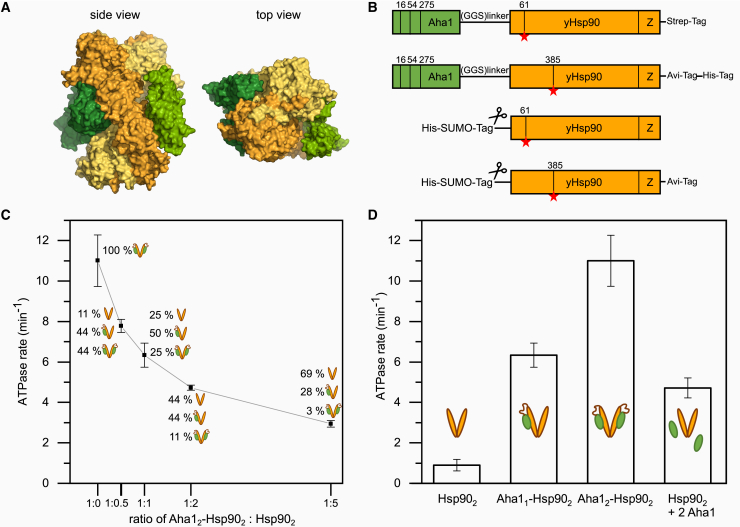
Aha1 consists of two domains: one N-terminal domain and one C-terminal domain, which are connected via a flexible linker. Aha1 interacts with Hsp90 in an antiparallel orientation where the N-terminal domain of Aha1 interacts with the middle domain of Hsp90, and Aha1’s C-terminal domain dynamically binds to the Hsp90 N-terminal domain (17,18). Aha1 binding shifts the equilibrium toward the closed conformational state of Hsp90 by facilitating the dimerization of its N-terminal domains and rearrangements in the N-terminal and middle domain (19,20,21). A few structural studies probing the Hsp90-Aha1 complex have been performed using techniques such as x-ray crystallography, NMR, and cryo-EM (17,19,22,23). However, the stoichiometry of Aha1 binding to Hsp90 remains elusive, indicative of one or two Aha1 molecules binding to an Hsp90 dimer (14,17,23,24). In this regard, a recent cryo-EM study of Aha1-Hsp90 complexes found both one and two molecules of Aha1 bound to the Hsp90 dimer (Fig. 1 A) (23).
Figure 1.
ATPase rates of Aha1-Hsp90 fusion constructs. (A) Side view and top view for a combination of Hsp90 dimer (yellows) from PDB: 6XLF and the Aha1 (greens) positions from PDB: 6XLB (cryo-EM structures) (23). (B) Schematics of the protein constructs showing the design of the Aha1-Hsp90 fusion along with the linker and Hsp90 constructs used in this study. The scissors indicate the position of the tag, which is cleaved during purification. The mutation positions are indicated as vertical lines inside. The red star denotes the position of the dyes. (C) ATPase rates of 2 μM Aha12-Hsp902 and a series of monomer exchanges of 2 μM Aha12-Hsp902 with increasing concentrations of Hsp902 starting from 1 up to 10 μM. The experiment was performed twice (n = 2). The assay was performed at 37°C in 40 mM HEPES, 10 mM MgCl2, and 150 mM KCl (pH 7.5) buffer. The line is a guide to the eye. (D) ATPase rate comparison between Hsp902, Aha11-Hsp902, Aha12-Hsp902, and Hsp902 with freely added Aha1. The schematics show the Hsp90 dimer in yellow and the Aha1 in green. Error bars represent the standard deviation of the data set. The monomer exchange of a 1:1 mixture of Aha12-Hsp902 and Hsp902 is denoted as Aha11-Hsp902, and it follows a binomial distribution of one part Aha12-Hsp902, two parts Aha11-Hsp902, and one part Hsp902. Hsp902 with freely added Aha1 (2 μM each) shows a lower ATPase due to measurement conditions being below the KD. To see this figure in color, go online.
Here, we perform a combination of biochemical, fluorescence anisotropy, and single-molecule experiments to investigate the detailed interactions between yeast Aha1 and yeast (Saccharomyces cerevisiae) Hsp90 in vitro and demonstrate how different stoichiometries of Aha1 binding influence Hsp90’s thermodynamics, kinetics, and function. To obtain defined stoichiometries, we designed a fusion protein construct in which Aha1 and Hsp90 are connected via a flexible linker. Altogether, these results show that Hsp90’s ATPase, thermodynamics, kinetics, and stability are differently affected by Aha1’s binding stoichiometry to the Hsp90 dimer.
Materials and methods
Expression constructs
Hsp90 constructs contained the hsp82 gene from Saccharomyces cerevisiae, an N-terminal cleavable His6-SUMO-tag and a C-terminal coiled-coil domain ensuring stable dimer formation (9). The coiled-coiled motif is either followed by a Strep-tagII or an Avi-tag for in vivo biotinylation.
The basic Aha1-Hsp90 was purchased as synthetic gene construct (GeneScript, the Netherlands) in which all native cysteines of Aha1 (Saccharomyces cerevisiae, Uniprot: Q12449) were exchanged with alanines to avoid unspecific modification by fluorophores or DNA handles (C16A, C54A, C275A). The linker between the two proteins consists of in total 115 amino acids, including a TEV protease site with the following sequence: P-GGENLYFQS-(GGS)14-P(EAAAK)3-P-(GGS)15-P. The construct also carried the C-terminal coiled-coil motif followed by either a Strep-tagII, His6-tag, or Avi-His6-tag for in vivo biotinylation.
For FRET experiments, single cysteines for labeling with fluorescent dyes were introduced on Hsp90 at positions 61 and 385 (D61C, Q385C). For the optical tweezers (OT) experiments, a cysteine was introduced at the coiled-coil motif on position 6 (A6C) to covalently connect the Hsp90 monomers via disulfide bond and another at position 452 (D452C) to be able to apply force on the Hsp90 middle domain (MD).
All expression constructs were cloned into pET28b plasmid vectors.
Protein preparation
Expression and purification were performed in variations of established protocols as previously reported (23,24). In short, proteins were produced in E. Coli BL21Star (DE3) or BL21 (DE3) cod+ and purified depending on the affinity tag. For in vivo biotinylation on the Avi-tag, the plasmid pBirA (Avidity Nanomedicines, USA) was cotransformed, and the cell culture procedure was adapted according to Avidity’s in vivo biotinylation protocol.
His6-SUMO-tagged Hsp90 was isolated with affinity chromatography using a Ni-IMAC column (HisTrap HP, Cytiva, Germany), followed by SUMO cleavage with SenP protease and a second Ni-IMAC to separate the His-tag-free protein from uncleaved proteins and the His6-SUMO tag. The protein was then applied to an anion exchange column (HiTrap Q, Cytiva, Germany) and finally to a size exclusion chromatography (SEC) column (Superdex 200, Cytiva, Germany).
Strep-tagII and simple His6-tag preparations were performed with a two-step protocol, starting with affinity chromatography using either a Strep-Tactin column (Strep-Tactin Superflow, IBA Lifesciences, Germany) or an Ni-IMAC (HisTrap HP, Cytiva/Ni-NTA Agarose, Qiagen, Germany) followed by SEC (Superdex 200, Cytiva, Germany).
Sample preparation and fluorescent labeling for single-molecule FRET experiments
Fluorescent labels (Atto550-maleimide and Atto647N-maleimide) were purchased from Atto-tec (Siegen, Germany) and coupled to cysteines according to the provided protocol (https://www.atto-tec.com/images/ATTO/Procedures/Mal.pdf). D61C- and Q385C-constructs were labeled with the FRET donor and acceptor dyes Atto550 and Atto647N, respectively. For the Hsp902 measurements, the dye positions were swapped for two out of four measurements to rule out that different labeling schemes affect the results. For the Aha1-Hsp90 construct, the donor dye was on the D61C position and the acceptor dye on the Q385 position. We swapped the position of the Aha1, i.e., once on the Hsp90 with the D61C and once on the Hsp90 with Q385C, to rule out effects of the dye positions on Aha1 binding. Hsp90 heterodimers (Hsp90 labeled with donor dye + Hsp90 labeled with acceptor dye) were obtained by 1:1 mixing of Hsp90 homodimers in buffer containing 40 mM HEPES, 150 mM KCl, and 10 mM MgCl2 (pH 7.5) and incubating for 40 min at 42°C, 300 RPM. The sample was then centrifuged at 16,000 g and 4°C for 30 min to remove any aggregates.
Sample preparation for optical tweezers experiments
To ensure that the majority of the homodimers are covalently linked with a disulfide bond in the coiled-coil motif, SEC elution fractions of highest purity were combined and incubated for 30 min at room temperature (21°C) with 10 mM TCEP for cysteine reduction. After removal of TCEP by Ni-IMAC (Ni-NTA Agarose, Qiagen, Germany), the protein was incubated at room temperature (21°C) for dimerization. The second cysteine at position 452 is considered less reactive to form disulfide bonds.
The protein constructs were then functionalized with single-stranded DNA oligonucleotides (Biomers, Germany) containing a maleimide group, which is reactive toward solvent-exposed cysteines, i.e., the cysteine at position 452 in each Hsp90 monomer. Free maleimide-DNA oligonucleotides and aggregated protein were removed by SEC. The DNA oligonucleotides are complementary to sticky ends of DNA handles, which were functionalized with either digoxigenin or biotin. DNA handles were made in-house using Lambda DNA as a template (New England Biolabs, USA) and custom-designed primers (Metabion, Germany) (25).
DNA sequences
DNA oligonucleotides containing a maleimide group
5′-GGCAGGGCTGACGTTCAACCAGACCAGCGAGTCG-maleimide-3′
Primers used to make DNA handles
5′-biotin-GGCGA(biotin-dT)CTGG(biotin-dT)CGTTGATTTG-3′
5′-digoxigenin-GGCGA(digoxigenin-dT)CTGG(digoxigenin-dT)CGTTGATTTG-3′
5′-CGACTCGCTGGTCTGGTTGAACGTCAGCCCTGCC(abasic site)CCTGCCCGGCTCTGGACAGG-3′
Single-molecule FRET measurements
Single-molecule FRET experiments were carried out in a custom-built prism-type TIRF setup using two lasers at wavelengths 532 nm (green, Coherent OBISTM LS) and 637 nm (red, Coherent OBISTM LX) (26). Measurements were conducted in a custom-built flow chamber, passivated with a mixture of methoxy-polyethylene glycol-silane (5000 Da, Rapp Polymere, Germany) and biotin-PEG-silane (3000 Da, Rapp Polymere, Germany) in an 80:3 ratio. The flow chamber was incubated with bovine serum albumin (BSA) (0.5 mg/mL in HEPES buffer) for 30 min before measurement. The flow chamber was further incubated with NeutrAvidin (0.25 mg/mL, Thermo Fisher Scientific, Germany) to enable the biotinylated protein to bind to the surface and then washed with HEPES buffer. Approximately 10 pM heterodimer was injected into the flow chamber with 10 min incubation and then washed with the buffer to discard the unbound protein. Measurements were performed in the presence of 2 mM ATP. The measurements were performed using alternating laser excitation of donor and acceptor dyes with an excitation time of 200 ms and a dark time of 50 ms. The fluorescence was collected by an oil immersion objective (100× magnification, Nikon) and detected by an EMCCD camera with 3 × 3 binning (iXon Ultra 897, Andor). The movies were recorded and saved as 16-bit TIFF stacks, which store the fluorescence intensities of the respective detection channels as successive frames (26).
For the selection of single-molecule time traces (donor fluorescence after donor excitation, acceptor fluorescence after acceptor excitation, acceptor fluorescence after donor excitation, i.e., FRET), an Igor Pro based in-house script (version 6.37, Wavemetrics, USA) was used. By searching for the brightest spots in five consecutive frames of the respective detection channel, the program identifies the positions of single molecules. For further analysis, trace selection criteria were flat plateaus and single bleaching steps, as well as anticorrelated behavior of donor and FRET intensity traces. The dynamic analysis of the selected traces was conducted using Single-Molecule Analysis of Complex Kinetic Sequences (SMACKS) (27), an IgorPro based script employing hidden Markov models.
ATPase assay
ATPase activity was determined using a regenerating ATPase assay (28) at 37°C. The ATP regenerating system contains a final concentration of 0.2 mM NADH Di-Na (Roche, CH), 2 mM phosphoenol pyruvate K-salt (PEP) (Bachem, CH), 2 U/mL pyruvate kinase (Roche, CH), 10 U/mL lactate dehydrogenase (Roche, CH) in HEPES buffer containing 40 mM HEPES, 150 mM KCl, 10 mM MgCl2 (pH 7.5). ATP hydrolysis, which is coupled to NADH oxidation, was calculated by monitoring the decrease in absorption at 340 nm. At the end of each measurement, 200 μM radicicol (Sigma, Germany) was added to measure the background signal. Radicicol background signal was subtracted from the protein signal for each measurement. The data was analyzed using linear fitting in Origin (OriginLAB, USA). Monomer exchanges in different ratios were performed as for the single-molecule FRET measurements. The statistical byproducts of homodimers are averaged out in the bulk ATPase assays, but they can be identified and disregarded in single-molecule experiments by their fluorescence or force fingerprints.
Optical tweezers experiments
For OT experiments, in-house made measurement chambers were passivated by incubation with 10 mg/mL BSA. Excess BSA was flushed out with buffer before sample loading. Measurements were performed in 40 mM HEPES/KOH, 20 mM KCl, 5 mM MgCl2 (pH 7.4) in presence of an oxygen scavenger system consisting of 1700 U/mL glucose catalase, 27 U/mL glucose oxidase, and 0.66% glucose to minimize photodamage caused by oxygen free radicals. Protein constructs functionalized with oligonucleotides were incubated with the DNA handles described above. DNA handles enable coupling of the protein construct to 1.29 μm or 1.28 μm diameter antidigoxigenin- or streptavidin-coated silica beads (Spherotech, USA). The DNA handle-protein construct was preincubated with antidigoxigenin beads. Using the two traps of the C-Trap dual beam OT instrument (Lumicks, Netherlands), one kind of each bead was trapped, and the trap stiffness was set to 0.25 pN/nm. In order to tether a single DNA handle-protein construct between the trapped beads, they were brought into contact with each other. Once a tether was formed, constant velocity experiments were performed at 20 nm/s with a wait time of 1 s between cycles. Data were collected at 78 kHz using Bluelake software (Lumicks, Netherlands).
Data analysis was done using custom-written software in Igor Pro (Wavemetrics, USA). For fitting the stretching of the DNA handles, force extension traces were fitted with an extensible worm-like chain (eWLC) with an expected DNA contour length of 370 nm, a persistence length of 20 nm, and a Hookean contribution of 400 pN. In order to determine contour length gains of the protein constructs resulting from either protein dissociation or unfolding, a WLC with a fixed persistence length of 0.7 nm was used for fitting. The first WLC was fixed to the contour length estimated for unstructured regions of the protein. In a next step, measured contour length gains were assigned to structural elements of the probed protein. In order to determine the theoretical contour length gain of a structural element (e.g., one domain) the number of involved amino acids was multiplied with the average length per amino acid of 0.365 nm (29). From this length, the initial distance between the first and the last amino acid of the structural element was subtracted. For this purpose, structural information about the protein was used.
Results and discussion
Construction of Aha1-linker-Hsp90 fusion protein
In order to obtain defined stoichiometries between Aha1 and Hsp90, we first designed an Aha1-linker-Hsp90 fusion construct (Fig. 1 B). The purified Aha1-Hsp90 fusion contains two Aha1 molecules per Hsp90 dimer (Aha12-Hsp902) where each Aha1 molecule is linked to one monomer of Hsp90. To obtain an Aha11-Hsp902 heterodimer in which only one Aha1 is interacting with the Hsp90 dimer, we performed a 1:1 monomer exchange of Aha12-Hsp902 dimer fusion and Hsp90 dimer. The motivation for using fusion constructs, on the one hand, is to define the stoichiometries and, on the other hand, to overcome the mutual low affinities, which are difficult for single-molecule experiments, because they have to be conducted at concentrations far below the dissociation constant, KD (Fig. S1). Based on the structural report of Aha1’s C-terminal domain binding to the Hsp90 N-terminal domain, the two proteins are fused between Aha1’s C-terminus and Hsp90’s N-terminus with a long and flexible linker peptide. The flexible linker peptide consists of 115 amino acids (P-GGENLYFQS-(GGS)14-P(EAAAK)3-P-(GGS)15-P), containing random coil-forming GGS repeats, alpha helices forming EAAAK repeats (representing stiff rods) (30,31), and prolines in between, which collectively constitutes a random coil-stiff rod-random coil structure (32,33). In this study, we used an Hsp90 construct with an artificial C-terminal coiled-coil zipper motif to stabilize the dimeric form and to prevent dissociation, which has been previously reported and used (Fig. 1 B). For the purpose of site-specific labeling and to perform smFRET measurements, three native cysteines in Aha1 have been mutated to alanines (C16A, C54A, C275A), and artificial cysteines have been introduced in Hsp90 either in amino acid position D61C or Q385C (Fig. 1 B). For all our single-molecule and bulk biochemical assays, we used the same protein constructs.
Effect of different stoichiometric Aha1 binding on Hsp90’s ATPase function
ATPase activity is important for the function of Hsp90. It has been reported that Aha1 stimulates Hsp90’s ATPase activity about 12-fold (14). We tested the ATPase activity of the Aha12-Hsp902-fusion protein to monitor the effect of 1 versus 2 Aha1 molecules binding to the Hsp90 dimer by performing ATPase assays using an ATP regenerating system. We performed a series of monomer exchanges mixing Aha12-Hsp902 and Hsp90 dimer in different ratios and measured the ATPase activities of all the monomer exchanged products. The ATPase catalytic rate of Aha12-Hsp902 was found to be ∼11 per min, as shown in Fig. 1 C and D. These figures also include the ATPase rates of all different monomer exchange products. We found that the Aha12-Hsp902 has the highest ATPase activity, followed by the Aha11-Hsp902 heterodimer (∼6 per min), whereas the dimeric Hsp90 alone has the lowest ATPase activity (∼1 per min). The 11-fold increase in ATPase activity we found for the Hsp90 construct with two Aha1 molecules bound is consistent with the literature values, demonstrating that our construct is functional. We showed that free Aha1 and covalently linked Aha1 behave similarly when introducing mutations or in low-salt conditions (Fig. S2). In addition, our results show that two Aha1 molecules are needed to fully stimulate the ATPase activity of Hsp90. In summary, different stoichiometries of Aha1 stimulate the ATPase activity of Hsp90 to different extents.
Effect of different stoichiometric Aha1 binding on Hsp90’s conformation equilibrium
N-terminal open and closed conformations are important for binding and processing different co-chaperones and clients. The ratio between the two conformations therefore likely defines the amount of binding of certain co-chaperones and clients. In a TIRF microscope (Fig. 2 A), the effect of Aha1-binding to Hsp90’s N-terminal conformations was monitored in real time using smFRET. In order to observe FRET, we labeled Hsp90 monomers at positions 61 and 385 with a donor and acceptor fluorophore. In vivo biotinylation of the C-terminal end of one Hsp90 monomer enabled its surface immobilization on a polyethylene glycol passivated microscope slide using biotin-neutravidin coupling. SmFRET measurements were performed for the Hsp90-homodimer, as well as for the Aha11-and Aha12-Hsp902 heterodimers. Fig. 2 B shows examples of such smFRET traces for those experiments, respectively, in presence of saturating ATP concentrations (2 mM). SmFRET traces show anticorrelated behavior between donor and acceptor signal, which indicates conformational transitions between the open (low FRET) and closed (high FRET) conformational states of Hsp90 (FRET efficiency: blue trace, acceptor fluorescence after donor excitation: red trace, donor fluorescence after donor excitation: green trace). Fig. 2 C shows the FRET efficiencies of all smFRET traces and therefore represents the thermodynamic equilibrium for the distribution of open and closed states. For the Aha11-Hsp902 heterodimer, the population of the closed state increased from (41 ± 4) % to (58 ± 4) % compared with Hsp90 (Table S1); this is independent of the Hsp90 cysteine mutant the first Aha1 is attached to (see Fig. S3). For the Aha12-Hsp902 fusion, the population of the open and closed conformation is similar to the one of the Aha11-Hsp902 heterodimer (Table S1). Although one Aha1 clearly induces the closed conformational state in Hsp90, surprisingly, a second Aha1 does not further increase the amount of closed dimers. In contrast, the ATPase strongly depends on Aha1’s binding stoichiometry. Therefore, the acceleration of Hsp90’s ATPase is not directly linked to promoting the closed state.
Figure 2.
Single-molecule FRET measurements of Hsp90 and Aha1. (A) Schematics of the smFRET TIRF setup showing an immobilized, labeled Aha12-Hsp90 dimer within the evanescent field. Fluorescence signal is separated by wavelengths using dichroic mirrors.
For a Figure360 author presentation of Figure 2, see https://doi.org/10.1016/j.bpj.2023.07.020. (B) Single-molecule FRET time traces in presence of 2 mM ATP for Hsp90, Aha11-Hsp902, and Aha12-Hsp902. SmFRET trajectories show conformational dynamics in real time. Green trace represents donor fluorescence signal after donor excitation (DemDex), red trace represents acceptor fluorescence signal after donor excitation (AemDex), black traces represent acceptor fluorescence after acceptor excitation (AemAex), and blue trace represents calculated FRET efficiency (right axis). (C) FRET efficiency histogram (normalized) obtained from the smFRET traces of Hsp902 (orange line), Aha11-Hsp902 (light blue line), and Aha12-Hsp902 (dark blue line) shows the population of open and closed states. (D) Quantitative rate constants of Hsp902 (orange), Aha11-Hsp902 (light blue), and Aha12-Hsp902 (dark blue) using a four-state model for Hsp90’s conformational dynamics. (E) Kinetic models represented as graphs of the Markov chain. Conformational kinetics are depicted by four states: 0 and 1 (open states) and 2 and 3 (closed states). The assay was performed at room temperature in 40 mM HEPES, 5 mM MgCl2, and 20 mM KCl (pH 7.5) buffer condition in presence of 2 mM ATP (Hsp90: 896 traces from four independent experiments; Aha11-Hsp902: 1422 traces from four independent experiments; Aha12-Hsp902: 441 traces from two independent experiments). To see this figure in color, go online.
Effect of different stoichiometric Aha1 binding on Hsp90’s conformational kinetics
As surprisingly the ATPase rate was affected by the stoichiometry of Aha1 whereas the thermodynamic equilibrium hardly was, we next tested to what extent the conformational kinetics were impacted by Aha1’s stoichiometry. Hence, we resolved the kinetic rates of opening and closing conformational dynamics after analyzing the smFRET traces using the SMACKS software (27). Hsp90’s conformation cycle can be described as a four-state model using SMACKS. This model comprises two open conformational states (0 and 1) and two closed conformational states (2 and 3), where 0 and 3 are the long-lived states, and 1 and 2 are the short-lived states (27,34). The states 0 and 1, as well as 2 and 3, cannot be separated by FRET efficiency but differ kinetically. Fig. 2 D and E shows the full kinetic analysis and reveals different effects on different transitions. Most of the transitions are hardly affected by Aha1, only showing slight changes upon co-chaperone addition. For the transitions 01 and 10, the addition of one Aha1 leads to a slight rate reduction, whereas the addition of a second Aha1 seems to have an opposing effect when compared with Hsp90 alone. In contrast, the rate 21 is slightly increased in the presence of a single Aha1, whereas the second Aha1 cancels this weak effect. Altogether, the full kinetic analysis shows that there are effects of Aha1’s stoichiometry on the transition rates, but neither in an additive nor in an independent way. This is in line with the findings above, namely that the conformational equilibrium and the acceleration of Hsp90’s ATPase are affected in different ways by Aha1’s stoichiometry.
Effect of different stoichiometric Aha1 binding on Hsp90’s unfolding force
Finally, we tested if the stability of the Hsp90 is affected by Aha1 binding and its stoichiometry. A previous study could reproducibly observe contour length changes when unfolding Hsp90 harboring force application points at amino acid position 452 in the MD. By comparing the contour length changes to the crystal structure of Hsp90, these were assigned to C-terminal domain (CTD) dissociation, CTD unfolding, and the unfolding of the part of the middle domain below the force application point, MDpart (amino acids 453–527) (35). In our study, OT experiments were done with Aha12-Hsp902,452C having the same force application points as Hsp902,452C (Fig. 3 C and D). Data from three molecules of Aha12-Hsp902,452C were collected from constant velocity experiments done at 20 nm/s. This resulted in 38 analyzed unfolding traces in total (as each molecule was unfolded and refolded several times). The Aha12-Hsp90452C construct showed additional unfolding events of reproducible contour length gains when compared with the Hsp902,452C construct (example in Fig. 3 C and D). These additional contour length gains, 14.8 ± 2.3, 35.2 ± 3.0, 44.5 ± 3.1, 52.8 ± 2.1, and 64.6 ± 2.2 nm, were observed repeatedly in different traces.
Figure 3.
Optical tweezers measurements of Hsp90 with Aha1. (A) Schematic illustration of one monomer of the Aha12-Hsp902,452C construct with assigned amino acid positions at the start and end of each structural element. (B) The optical tweezers (OT) experimental setup and the interaction of Aha1 domains with each other and Hsp90apo based on PDB: 6XLB (23). Aha12-Hsp902,452C is tethered between two trapped silica beads using long DNA handles. Displayed are the Aha1 NTD (green), Aha1 CTD (green), one Hsp90 monomer in gray, and the domains of the other Hsp90 monomer in color: Hsp90 NTD (blue), Hsp90 linker (purple), Hsp90 MD (orange), and Hsp90 CTD (yellow). Introduced cysteine mutations are indicated by yellow circles. (C) Example unfolding trace (black) of Hsp90452C in constant velocity (500 nm/s) OT experiment. The trace shows first dissociation of the Hsp90 CTDs (Cdiss) and then unfolding of both Hsp90 CTDs (C) and finally unfolding of both Hsp90 part MDs (Mpart). (D) Example unfolding trace (black) of Aha12-Hsp902,452C in constant velocity (20 nm/s) OT experiments. After a first small unfolding event (see main text for details), the trace shows first dissociation of the Hsp90 CTDs (Cdiss) and then unfolding of both Hsp90 CTDs (C), followed by two additional contour length gains likely resulting from the interaction with Aha1 (A1 and A2, red arrows) and finally unfolding of a Hsp90 part MD (Mpart). Dissociation and unfolding forces are smaller than in Fig. 3C due to the lower pulling velocity. Data were analyzed using a down-sampling factor of 4. Data were boxcar average filtered using a width of 21 data points. Worm-like chain model fits are displayed by dotted lines. (E) Stoichiometry histogram of Aha1 binding to Hsp90 in Aha12-Hsp902,452C during repeated cycles of constant velocity OT experiments. Assignments of binding of either 0, 1, 1–2, or 2 Aha1 were done based on the observation of reproducible, additional contour length gains (A1, A2) observed for Aha12-Hsp902,452C in comparison to Hsp90452C. To see this figure in color, go online.
We believe that the additional contour length gains result from the interaction of Aha1 with Hsp90, because they have not been observed for the Hsp902,452C construct. Aha1 has been found to have interaction interfaces with both Hsp90 monomers in nucleotide-free (apo) state (Fig. 3 B). More precisely, Aha1 bridges the two Hsp90 monomers (23). Those interaction interfaces can be found above and below position 452 in the Hsp90MD, where the force is applied. The additionally observed contour length changes in presence of Aha1 may result either from dissociation of Aha1 from Hsp90 or from unfolding of structural elements of Aha1. We cannot only observe these additional contour lengths in experiments with the fusion construct but also with soluble Aha1 (Fig. S4 A). The Aha1 N-terminal domain (NTD) and Aha1 CTD were shown to directly interact with one another in a cryo-electron microscopy (cryo-EM) structure of Hsp90 apo with two bound Aha1 molecules (PDB: 6XLB) (23). The observed contour length gain of 14.8 ± 2.3 nm matches well with the length of the unstructured region between the Aha1NTD and Aha1CTD. Therefore, the contour length gain of 14.8 ± 2.3 nm likely results from pulling apart and therefore undocking these two domains. Subtracting this length from the two observed additional contour length gains (52.8 ± 2.1 and 64.6 ± 2.2 nm) results in length changes that are similar to the remaining repeatedly observed contour length gains of 35.2 ± 3.0 and 44.5 ± 3.1 nm, respectively.
Altogether, these findings support the following assignment of the additional unfolding events in presence of Aha1: the contour length gain of 14.8 ± 2.3 nm to pulling apart the two Aha1 domains, the contour length gains of 35.2 ± 3.0 and 52.8 ± 2.1 nm to interaction of Hsp90 with one of the two Aha1 domains, and the contour length gains of 44.5 ± 3.1 and 64.6 ± 2.2 nm to interaction of Hsp90 with the other Aha1 domain.
Finally, a first event of 31.1 ± 5.8 nm was observed in 14 unfolding traces before the dissociation and unfolding of the Hsp90 CTDs and unfolding of the MDpart domains, which was not observed for Hsp90 alone. An additional event of similar stability and contour length was also observed for a standard Hsp90 construct when Aha1 was added in solution (data not shown). At this point, we cannot assign this event to a specific unfolding, but we speculate that Aha1 stabilizes a more compact conformation of Hsp90, which is extended at a very low force of 6.9 ± 1.3 pN. The distance between the force application points at amino acid position 452, in the Hsp90 MD is 5.6 nm in a cryo-EM structure of Hsp90 apo with two bound Aha1 (PDB: 6XLB) (23). In the open state of Hsp90, the distance between those residues was determined to be 7.9 nm in single-molecule FRET experiments (36), corresponding to an increase of (7.9 nm – 5.6 nm = ) 2.3 nm. Upon the application of force, the stretching of an unstructured region connecting the Hsp90 MD and CTD (37) and the loop region around the force application point gives an extra 5.5 nm extension per Hsp90 monomer. The total expected increase in length from the closed state of Hsp90 to the open state with the C-terminal dimerization interface still intact is therefore approximated as (5.5 nm 2 + 2.3 nm = ) 13.3 nm. This approximation is less than half of the observed contour length gain. However, we consider this approximation as a lower limit because the open state of Hsp90 measured in OT experiments under force application is likely to be more extended than the open state of Hsp90 observed in FRET experiments without applied force.
In order to quantify the effect of the stoichiometry of Aha1 and Hsp90, we counted how often different contour length gains occurred. The results are shown in the histogram in Fig. 3 E. When one contour length gain assigned to the interaction with one Aha1 domain and one contour length gain assigned to the interaction with the other Aha1 domain were observed in an unfolding trace, this was counted as “1–2” observed bound Aha1, because it is not clear whether these contour length gains result from the interaction of Hsp90 with one or two Aha1 molecules. Fig. 3 E depicts the result of this assignment, which shows that we have different binding stoichiometries and allows us to investigate their effect on Hsp90’s stability. Therefore, in the following, we focus on the unfolding of the Hsp90 MD.
OT experiments previously done with Hsp90452C resulted in one population of Hsp90 MDpart unfolding events (35). These data have been collected at a velocity of 500 nm/s. Data collected with Hsp90452C at the same velocity of 20 nm/s, as chosen for experiments with Aha12-Hsp902, 452C, also showed a single population of Hsp90 MDpart unfolding events with a measured unfolding force of 22.4 ± 2.2 pN (see Fig. S4 B; Table S2). We decided to show the data collected at 500 nm/s in Fig. 3 C, because there the same dimerization strategy was used as for the Aha1 construct. For the 20 nm/s traces, we used a different dimerization strategy (YBBR-based), which affects some length gains as expected.
Interestingly, an additional population of Hsp90 MDpart unfolding events was observed when experiments were done with the Aha12-Hsp902,452C construct. Besides Hsp90 MDpart unfolding events at 23.0 ± 2.5 pN, which is in good agreement with the MDpart unfolding force observed for Hsp90452C, a population of MDpart unfolding events at lower force of 10.9 ± 2.7 pN was observed (Table S2). A cryo-EM structure of apo-Hsp90 with bound Aha1 (PDB: 6XLB) (23) reveals that the Hsp90 MD is rotated 8° relative to the Hsp90 CTD in comparison to the fully open state of apo HtpG crystal structure (PDB: 2IOQ). This change in MD-CTD interface induced by binding of Aha 1 could be a possible explanation for the observation of Hsp90 MDpart unfolding events at reduced force in presence of Aha1. Unfolding traces were observed with both of the two Hsp90 MDpart unfolding at low force as well as traces in which one of the MDpart unfolds at regular force, whereas the second MDpart unfolds at low force. Hsp90 MDpart unfolding events of reduced force were not detected in any of the traces with no Aha1-related events. In traces with observed Aha1-related events, at least one Hsp90 MDpart unfolded at reduced force. In only four of the 19 traces assigned to observation of two bound Aha1, one of the Hsp90 MDpart unfolding events happened at regular force. However, none of these Hsp90 MDpart unfolding events at regular force were followed by any of the events assigned to interaction with Aha1; i.e., in these cases the interaction with Aha1 likely was already disrupted. This suggests that one Aha1 reduces the stability of one Hsp90 MDpart, and two Aha1 can reduce the stability of both Hsp90 MDparts. Overall, 46 of the 61 observed MDpart unfolding events happened at reduced force translating into a probability for observing Hsp90 MDpart unfolding events at low force in Aha12-Hsp902,452C of 0.75.
To further evaluate if Aha1 binding results in the occurrence of Hsp90 MDpart unfolding at low force in a binding stoichiometry-dependent way, the binding probability of Aha1 was compared with the population of all observed Hsp90 MDpart unfolding events at low force. The effective concentration of Aha1 can be estimated by the following (38):
| (1) |
The “volume of sphere” is the volume allowed by the linker, which attaches Aha1 to each Hsp90 monomer. Based on the use of the flexible linker of 115 amino acids, i.e., about 42 nm to connect Aha1 to Hsp90, and the close proximity of Hsp90 monomers in dimeric Hsp90, two Aha1 molecules were estimated to be in the spherical volume of 3.1 nm3. This results in The binding probability (39) of Aha1 to Hsp90 was then calculated using this effective concentration and a KD of 7.8 μM, determined by fluorescence anisotropy (Fig. S1):
| (2) |
We consider this as a lower limit, as it is more likely that Aha1 is in the center of the sphere than in the periphery due to the entropic elasticity of the linker, which would result in a higher effective concentration. Therefore, this is in good agreement with the probability for observing Hsp90 MDpart unfolding at low force of 0.75 (see above). Altogether, these experiments support that Aha1 binding to Hsp90 reduces the Hsp90 MDpart unfolding force of either one or both Hsp90 MDs in a binding stoichiometry-dependent way.
Conclusion
Many previous studies have shown that the molecular chaperone and heat shock protein Hsp90 is regulated by co-chaperones and nucleotides. These studies were usually done at saturating co-chaperone or nucleotide conditions. As the functional form of Hsp90 is a dimer, it remains unclear if in these experiments two co-chaperones/nucleotides are bound per dimer or only one. Here, we showed that the binding stoichiometry has a significant effect using the co-chaperone Aha1. Therefore, the stoichiometry can serve as an additional regulatory mechanism that has likely been underappreciated in the past. Most interestingly, the stoichiometry of Aha1 binding has different effects on different quantities. In some cases, binding of a second Aha1 amplifies the effect of the first bound Aha1, whereas in other cases, it cancels the effect of the first bound Aha1 or has no effect at all. The ATPase or the reduction of the Hsp90MDpart unfolding force, for example, is amplified by a second bound Aha1, whereas changes in some rate constants are reversed by the second bound Aha1, and the thermodynamics of the conformational landscape is not changed at all.
Our results show that the binding stoichiometry of co-chaperones to Hsp90 has significant effects on many quantities. Therefore, we anticipate that tuning the concentration of co-chaperones in cells (i.e., their binding stoichiometry) is an important regulation mechanism for the many Hsp90-related processes.
Author contributions
T.H., T.M., and K.T. designed the research; T.M., L.-M.S., J.S., and L.V. performed the measurements; T.M., B.H., and L.-M.S. made the protein constructs. T.M., L.-M.S., J.S., and L.V. analyzed the data after consultation with T.H. and K.T.; all authors wrote the manuscript and have given approval to the final version of the manuscript.
Acknowledgments
This work was supported by the European Research Council (grant agreement No. 681891) and the Deutsche Forschungsgemeinschaft (DFG) under Germany’s Excellence Strategy (CIBSS EXC-2189 Project ID 390939984) and the SFB1381 programme (Project ID 403222702). We want to thank Marianne Birkle and Michael Witt for their help with protein production and Martin Jetzlaff for his support with ATPase assays.
Declaration of interests
The authors declare no competing interests.
Editor: Doug Barrick.
Footnotes
Tanumoy Mondol, Laura-Marie Silbermann, Julia Schimpf, and Leonie Vollmar contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.07.020.
Contributor Information
Katarzyna (Kasia) Tych, Email: k.m.tych@rug.nl.
Thorsten Hugel, Email: thorsten.hugel@pc.uni-freiburg.de.
Supporting material
References
- 1.Taipale M., Jarosz D.F., Lindquist S. HSP90 at the Hub of Protein Homeostasis: Emerging Mechanistic Insights. Nat. Rev. Mol. Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 2.Biebl M.M., Buchner J. Structure, Function, and Regulation of the Hsp90 Machinery. Cold Spring Harbor Perspect. Biol. 2019;11:a034017. doi: 10.1101/cshperspect.a034017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali M.M.U., Roe S.M., et al. Pearl L.H. Crystal Structure of an Hsp90–Nucleotide–P23/Sba1 Closed Chaperone Complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiau A.K., Harris S.F., et al. Agard D.A. Structural Analysis of E. Coli Hsp90 Reveals Dramatic Nucleotide-Dependent Conformational Rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 5.Prodromou C., Panaretou B., et al. Pearl L.H. The ATPase Cycle of Hsp90 Drives a Molecular ‘Clamp’ via Transient Dimerization of the N-Terminal Domains. EMBO J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jahn M., Rehn A., et al. Hugel T. The Charged Linker of the Molecular Chaperone Hsp90 Modulates Domain Contacts and Biological Function. Proc. Natl. Acad. Sci. USA. 2014;111:17881–17886. doi: 10.1073/pnas.1414073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer P., Prodromou C., et al. Pearl L.H. Structural and Functional Analysis of the Middle Segment of Hsp90: Implications for ATP Hydrolysis and Client Protein and Cochaperone Interactions. Mol. Cell. 2003;11:647–658. doi: 10.1016/S1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 8.Schopf F.H., Biebl M.M., Buchner J. The HSP90 Chaperone Machinery. Nat. Rev. Mol. Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 9.Panaretou B., Prodromou C., et al. Pearl L.H. ATP Binding and Hydrolysis Are Essential to the Function of the Hsp90 Molecular Chaperone Invivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mickler M., Hessling M., et al. Hugel T. The Large Conformational Changes of Hsp90 Are Only Weakly Coupled to ATP Hydrolysis. Nat. Struct. Mol. Biol. 2009;16:281–286. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- 11.Mayer M.P., Le Breton L. Hsp90: Breaking the Symmetry. Mol. Cell. 2015;58:8–20. doi: 10.1016/j.molcel.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 12.Kravats A.N., Hoskins J.R., et al. Wickner S. Functional and Physical Interaction between Yeast Hsp90 and Hsp70. Proc. Natl. Acad. Sci. USA. 2018;115:E2210–E2219. doi: 10.1073/pnas.1719969115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahasrabudhe P., Rohrberg J., et al. Buchner J. The Plasticity of the Hsp90 Co-Chaperone System. Mol. Cell. 2017;67:947–961.e5. doi: 10.1016/j.molcel.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Panaretou B., Siligardi G., et al. Prodromou C. Activation of the ATPase Activity of Hsp90 by the Stress-Regulated Cochaperone Aha1. Mol. Cell. 2002;10:1307–1318. doi: 10.1016/S1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 15.Wang X., Venable J., et al. Balch W.E. Hsp90 Cochaperone Aha1 Downregulation Rescues Misfolding of CFTR in Cystic Fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 16.Shelton L.B., Baker J.D., et al. Dickey C.A. Hsp90 Activator Aha1 Drives Production of Pathological Tau Aggregates. Proc. Natl. Acad. Sci. USA. 2017;114:9707–9712. doi: 10.1073/pnas.1707039114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Retzlaff M., Hagn F., et al. Buchner J. Asymmetric Activation of the Hsp90 Dimer by Its Cochaperone Aha1. Mol. Cell. 2010;37:344–354. doi: 10.1016/j.molcel.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Koulov A.V., LaPointe P., et al. Balch W.E. Biological and Structural Basis for Aha1 Regulation of Hsp90 ATPase Activity in Maintaining Proteostasis in the Human Disease Cystic Fibrosis. MBoC. 2010;21:871–884. doi: 10.1091/mbc.e09-12-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer P., Prodromou C., et al. Pearl L.H. Structural Basis for Recruitment of the ATPase Activator Aha1 to the Hsp90 Chaperone Machinery. EMBO J. 2004;23:1402–1410. doi: 10.1038/sj.emboj.7600141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulze A., Beliu G., et al. Neuweiler H. Cooperation of Local Motions in the Hsp90 Molecular Chaperone ATPase Mechanism. Nat. Chem. Biol. 2016;12:628–635. doi: 10.1038/nchembio.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J., Richter K., et al. Buchner J. Integration of the Accelerator Aha1 in the Hsp90 Co-Chaperone Cycle. Nat. Struct. Mol. Biol. 2013;20:326–331. doi: 10.1038/nsmb.2502. [DOI] [PubMed] [Google Scholar]
- 22.Oroz J., Blair L.J., Zweckstetter M. Dynamic Aha1 Co-chaperone Binding to Human Hsp90. Protein Sci. 2019;28:1545–1551. doi: 10.1002/pro.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., Sun M., et al. Agard D.A. Cryo-EM Structures Reveal a Multistep Mechanism of Hsp90 Activation by Co-Chaperone Aha1. Biophysics. 2020 doi: 10.1101/2020.06.30.180695. Preprint at. [DOI] [Google Scholar]
- 24.Siligardi G., Hu B., et al. Prodromou C. Co-Chaperone Regulation of Conformational Switching in the Hsp90 ATPase Cycle. J. Biol. Chem. 2004;279:51989–51998. doi: 10.1074/jbc.M410562200. [DOI] [PubMed] [Google Scholar]
- 25.Tych K., Rief M. In: Gennerich A., editor. Vol. 2478. Methods in Molecular Biology; Springer US; 2022. Using Single-Molecule Optical Tweezers to Study the Conformational Cycle of the Hsp90 Molecular Chaperone; pp. 401–425. (Optical Tweezers). [DOI] [PubMed] [Google Scholar]
- 26.Götz M., Wortmann P., et al. Hugel T. Using Three-Color Single-Molecule FRET to Study the Correlation of Protein Interactions. JoVE. 2018;131 doi: 10.3791/56896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid S., Götz M., Hugel T. Single-Molecule Analysis beyond Dwell Times: Demonstration and Assessment in and out of Equilibrium. Biophys. J. 2016;111:1375–1384. doi: 10.1016/j.bpj.2016.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura J.K., Gellert M. Characterization of the ATP Binding Site on Escherichia Coli DNA Gyrase. Affinity Labeling of Lys-103 and Lys-110 of the B Subunit by Pyridoxal 5’-Diphospho-5’-Adenosine. J. Biol. Chem. 1990;265:21342–21349. [PubMed] [Google Scholar]
- 29.Tych K., Žoldák G. In: Kister A.E., editor. Vol. 1958. Methods in Molecular Biology; Springer New York; 2019. Stable Substructures in Proteins and How to Find Them Using Single-Molecule Force Spectroscopy; pp. 263–282. (Protein Supersecondary Structures). [DOI] [PubMed] [Google Scholar]
- 30.Evers T.H., van Dongen E.M.W.M., et al. Merkx M. Quantitative Understanding of the Energy Transfer between Fluorescent Proteins Connected via Flexible Peptide Linkers. Biochemistry. 2006;45:13183–13192. doi: 10.1021/bi061288t. [DOI] [PubMed] [Google Scholar]
- 31.Arai R., Ueda H., et al. Nagamune T. Design of the Linkers Which Effectively Separate Domains of a Bifunctional Fusion Protein. Protein Eng. 2001;14:529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- 32.Williamson M.P. The Structure and Function of Proline-Rich Regions in Proteins. Biochem. J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wortmann P. Technische Universität München; 2017. The Dynamics of the Hsp90 Machine. [Google Scholar]
- 34.Schmid S., Hugel T. Controlling Protein Function by Fine-Tuning Conformational Flexibility. Elife. 2020;9 doi: 10.7554/eLife.57180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tych K.M., Jahn M., et al. Rief M. Nucleotide-Dependent Dimer Association and Dissociation of the Chaperone Hsp90. J. Phys. Chem. B. 2018;122:11373–11380. doi: 10.1021/acs.jpcb.8b07301. [DOI] [PubMed] [Google Scholar]
- 36.Hellenkamp B., Wortmann P., et al. Hugel T. Multidomain Structure and Correlated Dynamics Determined by Self-Consistent FRET Networks. Nat. Methods. 2017;14:174–180. doi: 10.1038/nmeth.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jahn M., Tych K., et al. Rief M. Folding and Domain Interactions of Three Orthologs of Hsp90 Studied by Single-Molecule Force Spectroscopy. Structure. 2018;26:96–105.e4. doi: 10.1016/j.str.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 38.Grison M. Technische Universität München; 2017. Single-Molecule Cohesion and Adhesion in Muscle Cells. [Google Scholar]
- 39.Schnellbächer N.D. Particle-Based Computer Simulations of Biological Reaction-Diffusion Systems. 2017. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.