Abstract
Genomic stability in proliferating cells critically depends on telomere maintenance by telomerase reverse transcriptase. Here we report the development and proof-of-concept results of a single-molecule approach to monitor the catalytic activity of human telomerase in real time and with single-nucleotide resolution. Using zero-mode waveguides and multicolor FRET, we recorded the processive addition of multiple telomeric repeats to individual DNA primers. Unlike existing biophysical and biochemical tools, the novel approach enables the quantification of nucleotide-binding kinetics before nucleotide incorporation. Moreover, it provides a means to dissect the unique translocation dynamics that telomerase must undergo after synthesis of each hexameric DNA repeat. We observed an unexpectedly prolonged binding dwell time of dGTP in the enzyme active site at the start of each repeat synthesis cycle, suggesting that telomerase translocation is composed of multiple rate-contributing sub-steps that evade classical biochemical analysis.
Significance
Telomerase dysfunction or reactivation are hallmarks of stem-cell-failure diseases and most cancers. Structure-function studies have been hampered by the enzyme’s low natural abundance and its inherent conformational heterogeneity. The single-molecule method presented here benefits from low sample requirements and allows correlation of nucleotide addition with concurrent structural dynamics. A unique advancement of the method is its ability to detect the nucleotide-bound states of individual enzymes through real-time FRET, while maintaining near-physiological nucleotide concentrations. Existing telomerase assays depend on nucleotide incorporation and indirect DNA-product analysis. Paired with recent cryo-EM reconstructions of human and ciliate telomerases, our method opens avenues to address long-standing questions about telomerase translocation dynamics, DNA-product handling, and its interaction with telomere-associated proteins that are critical to human health.
Introduction
Telomere homeostasis is critical to tissue development and regeneration. The telomerase enzyme synthesizes the repetitive telomere DNA that, together with telomere-binding proteins, protects linear chromosome ends (1,2). Enzyme dysfunction causes premature aging diseases, whereas aberrantly upregulated telomerase activity confers replicative immortality to most cancers (3,4).
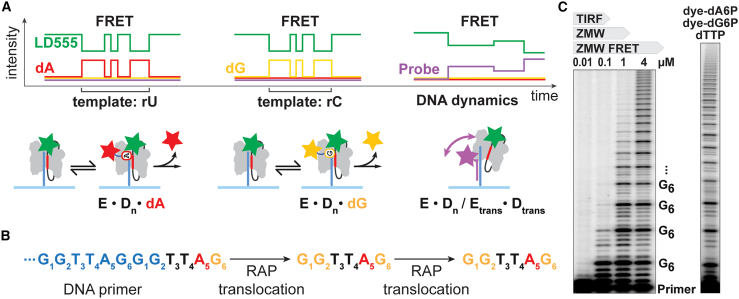
Telomerase is a ribonucleoprotein (RNP) reverse transcriptase (RT) that uses an integral RNA template to synthesize telomeric DNA repeats (Fig. 1 A). In humans, the telomerase RNA (TR) template sequence rC1-rC2-rA3-rA4-rU5-rC6 specifies the hexameric DNA-repeat sequence dG1-dG2-dT3-dT4-dA5-dG6 (1). The telomerase enzyme (E) is moderately processive and exhibits two modes of processivity (nucleotide-addition processivity (NAP) and repeat-addition processivity (RAP)). NAP describes the sequential binding and incorporation of up to six cognate nucleotides (N) into the DNA substrate (D), releasing pyrophosphate (PPi) upon nucleotide hydrolysis (Fig. 1 A; Eq. 1).
| (1) |
Figure 1.
The human telomerase catalytic cycle. (A) Schematic of the processive telomerase RNP reverse transcription reaction. Top, nucleotide addition occurs in six steps for a full telomeric DNA repeat. Binding and incorporation of the first dG1 nucleotide is shown in enhanced detail. RNA template positions are numbered in red. Below, processive repeat addition is enabled by DNA/RNA translocation steps that reset the telomerase enzyme. Two possible paths of DNA/RNA repositioning and hybrid formation are shown. (B) Gel-based analysis of the telomerase reaction extending a radiolabeled DNA primer (TTAGGG)3. Initial steps indicate the completion of the mid-repeat primer. Subsequent full repeats are numbered to the left of the gel. To see this figure in color, go online.
NAP is associated with single-nucleotide translocation steps of the TR template in the active site. However, for processive nucleotide addition to continue past the incorporation of dG6, the short integral RNA template must be repositioned to allow for subsequent dG1 binding, a process underlying RAP. RAP describes the addition of multiple repeats to a single DNA substrate and specifically refers to the successful transition from dG6 to dG1 incorporation, initiating a new round of nucleotide addition (Fig. 1 A, bottom) (5). RAP requires larger-scale translocation steps that include melting of the DNA/RNA duplex, resetting of the RNA template from rC6 to rC1, and formation of a new DNA/RNA hybrid in the telomerase active site. The structural rearrangements required to promote RAP may be described by the dynamic equilibrium (6) depicted in Eq. 2.
| (2) |
represents the totality of transient intermediate states during RAP translocation (Fig. 1 A, bottom). After melting of the RNA/DNA duplex, RAP relies on a tether between various telomerase “anchor sites” and the dynamic DNA product, reducing the probability of DNA dissociation (7,8). The RAP-translocation mechanism remains largely undetermined, and its analysis is challenged by the presence of dynamic and transient states sampled by telomerase and the DNA product.
Current methods to investigate telomerase activity include primer-extension assays that monitor the growing DNA chain (9). Using sequencing gels and radiography, ensemble telomerase reactions can be analyzed at defined endpoints and with single-nucleotide resolution (Dn vs. Dn+1) (Fig. 1 B) (10). The moderate degree of telomerase processivity becomes apparent in a characteristic gel-band pattern where each band represents extended DNA products. Notably, repeat synthesis by telomerase is slow, with an average time span of several minutes per individual repeat under our assay conditions (9). The RAP transitions comprise the slowest steps in the telomerase cycle, providing a proportionally larger time window for DNA dissociation to occur (11). The respective DNA products terminating in dG6 therefore dominate the telomerase product distribution (Fig. 1 B). The gel-band intensities enable global kinetic modeling of telomerase repeat synthesis (11); however, investigation of mechanistic sub-steps demand biophysical approaches with greater spatiotemporal resolution.
Recently, extensive cryo electron microscopy (cryo-EM) efforts have provided insight into the architecture of the ciliate and human telomerase RNP, including higher-resolution snapshots of the DNA-RNA hybrid in the active site in the presence of telomerase protein cofactors (12,13,14). Structural studies of telomerase are challenged by the conformational heterogeneity intrinsic to the enzyme; however, the static structures are invaluable to the design and interpretation of single-molecule approaches that aim to correlate structural heterogeneity with enzyme function. Recent single-molecule assays are able to probe the growing DNA product in real time ( vs. ), but mechanistic and kinetic insights into NAP and RAP cycles were obstructed by DNA structural dynamics (10,15,16). Here we present a FRET-based single-molecule assay that monitors telomerase NAP steps by means of fluorescence-dye-labeled nucleotide analogs. This approach allowed us to demarcate the RAP-translocation steps over multiple consecutive repeat cycles and simultaneously assess DNA conformational dynamics.
Materials and methods
RNA preparation
RNA in vitro transcription
In vitro transcription (IVT) DNA templates for hTR CR4/5 (hTR 239–328), hTR template/pseudoknot (hTR t/PK, 32–195 and 63–195), were generated by PCR fusing the T7 promotor to the respective DNA sequence of hTR (Table S1) (10). A total of 400 μL of PCR reactions were added directly to the IVT mix. IVTs were carried out using homemade T7 RNA polymerase in 5-mL reactions of 40 mM Tris-HCl, pH 8.0, 35 mM MgCl2, 0.01% (v/v) Triton X-100, 5 mM DTT, 1 mM spermidine, 2.5 mM each NTP, and 40 U RNasin Plus (Promega). The reactions were incubated overnight at 37°C followed by the addition of 10 U TURBO DNase (Thermo Fisher Scientific) and incubation for 15 min at 37°C. The RNA was enriched by isopropanol precipitation and purified by denaturing urea polyacrylamide gel electrophoresis (PAGE). The IVT mix of hTR t/PK 63–195 destined for splinted ligation reactions (see below) was modified to contain each NTP at 1 mM and 5 mM GMP to obtain 5′ monophosphate groups.
RNA dye labeling
To generate dye-labeled telomerase, the synthetic hTR template/pseudoknot (t/PK) fragment 32–62 was purchased from Dharmacon with an internal 5-aminohexylacrylamino-uridine (5-LC-N-U) at position U42 (Table S1) (17). The RNA was deprotected using provided deprotection buffer (Dharmacon) and following the manufacturer’s instructions. Deprotected RNA was desalted using G-25 MidiTrap gravity columns (GE Healthcare), followed by ethanol precipitation in the presence of 300 mM sodium acetate, pH 5.2. The pellet was resuspended in 100 μL of 0.1 M sodium bicarbonate and brought to a concentration of 200 nmol/mL. For dye coupling, 20 nmol of RNA were combined with an equal volume of anhydrous DMSO containing 100 nmol of LD555 (Lumidyne Technologies). The mix was incubated in the dark for 2 h at 37°C, desalted using gravity columns, and dried using a vacufuge (Eppendorf). RNA pellets were dissolved in 60 μL of 0.1 M triethylamine acetate (TEAA), pH 7.5, and dye-labeled RNA was purified by high-pressure liquid chromatography (HPLC) in 0.1 M TEAA to 60% acetonitrile gradient on a reversed phase C8 column (Agilent Technologies).
RNA ligation
Synthetic and dye-labeled hTR 32–62 was ligated to in vitro transcribed hTR t/PK 63–195 by splinted RNA ligation(17). A 200-μL reaction contained 800 pmol of LD555-hTR 32–62, 1600 pmol of hTR 63–195, and 1600 pmol of DNA splint (Table S1) in 0.5× T4 DNA ligase buffer (NEB). The mix was incubated for 5 min at 95°C and for a further 10 min at 30°C. Then 200 μL of ligation mix (1.5× T4 DNA ligase buffer, 8000 U T4 DNA ligase (NEB), 2 mM ATP, and 200 U RNasin Plus) was added to the reaction and incubated overnight at 30°C. Then 10 U of TURBO DNase were added and incubated for 15 min at 37°C. The RNA was then phenol-chloroform extracted, ethanol precipitated, and purified by urea PAGE.
Telomerase reconstitution
Human telomerase was reconstituted in rabbit reticulocyte lysate (RRL, Promega TnT Quick Coupled Transcription/Translation kit) following the manufacturers specifications (18). Specific to telomerase, 200 μL of TnT mix were supplemented with 5 μg of the plasmid pNFLAG-hTERT (19) and 1 μM in vitro transcribed (see above) and unlabeled hTR fragments (hTR template/pseudoknot and CR4/5). In reconstitutions of dye-labeled telomerase, the LD555 t/PK fragment (see above) was added at 0.1 μM final concentration owing to lower relative yields after RNA ligation and dye labeling. The reaction mix was incubated for 3 h at 30°C and quenched with 5 μL of 0.5 M EDTA, pH 8.0, for 30 min at room temperature. For immuno-purification of telomerase, beads from 50 μL of ANTI-FLAG M2-agarose bead slurry (Sigma-Aldrich) were washed three times by suspension in 750 μL of wash buffer (50 mM Tris-HCl, pH 8.3, 3 mM MgCl2, 2 mM DTT, 100 mM NaCl), centrifugation for 30 s at 2350 rcf, and removal of the supernatant. Beads were subsequently blocked twice for 15 min in blocking buffer (50 mM Tris-HCl, pH 8.3, 3 mM MgCl2, 2 mM DTT, 500 μg/mL BSA, 50 μg/mL glycogen, 100 μg/mL yeast tRNA) under end-over-end agitation at 4°C. Blocked beads were collected by centrifugation, resuspended in 200 μL of blocking buffer, and added to the telomerase reconstitution reaction in RRL. This binding step was left to proceed for 2 h at 4°C under end-over-end agitation. The beads were then pelleted and washed three times in wash buffer containing 300 mM NaCl, followed by three further wash steps in wash buffer containing 100 mM NaCl. Elution of telomerase from the beads was performed by adding 60 μL of elution buffer (50 mM Tris-HCl, pH 8.3, 3 mM MgCl2, 2 mM DTT, 750 μg/mL 3xFLAG peptide, 20% glycerol) to the beads and incubation for 1 h at 4°C under orbital agitation. The mix was then transferred to a Nanosep MF 0.45 μM centrifugal filter and the eluate was collected by centrifugation at 10,000 rcf for 1 min. Then 5-μL aliquots (3 μL in the case of dye-labeled telomerase) were prepared in Lo-bind tubes (Eppendorf), flash frozen in liquid nitrogen, and stored at −80°C until use.
32P-end labeling of DNA primers
DNA primer (50 pmol) was labeled with gamma-32P ATP and T4 polynucleotide kinase (NEB) in a 50-μL reaction in 1× PNK buffer (NEB). The mix was incubated for 1 h at 37°C followed by heat inactivation of T4 PNK at 65°C for 20 min. The primers were purified on Centrispin columns (Princeton Separations) and brought to a final concentration of 50 nM in nuclease-free water.
Primer-extension assays
Bulk activity assays were performed using 5 μL of immuno-purified telomerase brought to a 15-μL reaction volume of 50 mM Tris-HCl, pH 8.3, 50 mM KCl, 3 mM MgCl2, and 2 mM DTT (6). 32P-end-labeled DNA primers were used at 50 nM and nucleotide concentrations were as indicated (ranging from 0.1 to 10 μM each of dATP, dTTP, and dGTP). Dye-dG6P and dye-dA6P were used at 3.5 and 8 μM, respectively, to be consistent with real-time sequencer (RS) assay conditions. dTTP was used at 10 μM when mixed with dye-dG6P and dye-dA6P. Reactions were incubated for 90 min at 30°C and quenched with 200 μL of TES (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.1% SDS). For kinetic analysis, reactions were chased after 10-min incubation with 20 μM unlabeled primer and samples quenched at 10-min intervals. DNA products were phenol-chloroform extracted and ethanol precipitated. DNA pellets were resuspended in formamide gel loading buffer (50 mM Tris base, 50 mM boric acid, 2 mM EDTA, 80% (v/v) formamide, 0.05% (w/v) each bromophenol blue and xylene cyanol) and resolved on a 12% denaturing urea PAGE gel. The gel was then dried and imaged using a storage phosphor screen and Typhoon scanner (GE Healthcare).
RS assay
Chip preparation and enzyme immobilization
RS chips (single-molecule real-time (SMRT) cell, PacBio) were washed three times each with 40 μL of TP50 buffer (50 mM Tris-HCl, pH 8.0, 50 mM KCl). Then 20 μL of NeutrAvidin was supplied to the chip and incubated for 5 min at room temperature. NeutrAvidin was removed, followed by three wash steps each with 40 μL of telomerase buffer (50 mM Tris-HCl, pH 8.3, 50 mM KCl, 3 mM MgCl2). Then 5 nM stocks of biotin-(TTAGGG)3-primer in TLi (50 mM Tris-HCl, pH 8.3, 50 mM LiCl) were heated to 95°C for 3 min to melt primer aggregates or G-rich structures. Primers were cooled on ice. Three micro-liters of immuno-purified LD555-telomerase was combined with 1 μL of 5 nM heat-treated primer and incubated for 20 min at 30°C. Then 16 μL of telomerase buffer was added to this mix at room temperature. The resulting 20-μL enzyme mix was supplied to an RS chip, sealed with parafilm, and incubated for 15 min at 30°C. The enzyme mix was removed, and the chip was washed three times each with telomerase buffer.
RS setup and chip imaging
The RSII (PacBio) was initialized from the user interface (20). The green laser (532 nm) was set to a laser power of 0.24 μW/μm2. A calibration measurement was previously conducted at the same laser power as a reference according to the manufacturer’s instructions. The video length was 30 min at a frame rate of 10 Hz. The chip clamp temperature (∼zero-mode waveguide (ZMW) temperature) was 30°C. A dispense protocol was selected for fluidic delivery of 20 μL of delivery solution (see below) to the chip at 140 s into the video. After initialization, enzyme-treated chips were supplied with telomerase imaging buffer (50 mM Tris-HCl, pH 8.3, 50 mM KCl, 3 mM MgCl2, 2.5% (v/v) N-formylmorpholine, 3.5 μM dye-dG6P, 8 μM dye-dATP, oxygen-scavenging system (2.5 mM protocatechuic acid (PCA), 2.5 mM TSY, and 2× protocatechuate-3,4-dioxygenase (PCD), purchased from Pacific Bioscience)) and the silica surface of the chip was cleaned with methanol. Chips were then mounted in the SMRT cell chamber of the RS. Telomerase imaging delivery solution (imaging buffer including 20 μM dTTP, and 40 nM DNA probe where indicated) was provided in a 96-well sample plate in the sample chamber of the RS and kept at 4°C until delivery. Data acquisition was started from the RS user interface (20).
Data analysis
Fluorescence trace extraction and data analysis were performed with a set of custom-written Matlab scripts (available upon request). Procedures adhered to the principal workflow described below. The RS generates approximately 150,000 four-color traces per ZMW experiment. To down-select data for ZMWs that, in principle, possessed active telomerase complexes, the raw data traces were computationally screened for fluorescence intensity anti-correlation between detection channels 1 (green) and 3 (red), indicating energy transfer between the respective fluorophores. The down-selected trace subset was then manually screened for photophysical artifacts or noise. The manually curated trace subset was used for two different analysis workflows as described below.
Semi-automated processing and idealized trace generation
Individual data traces were pre-processed for FRET state assignment using the previously published vbFRET software suite (21). Trace pre-processing was performed according to the after workflow: 1) crop the region of interest to remove photobleaching events or initial noise in the trace due to buffer exchange, 2) donor channel (Ch1) drift correction, 3) channel crosstalk correction, 4) baseline correction of acceptor channels (Ch2, Ch3, and/or Ch4), 5) calculation of effective FRET values for each channel and fine adjust of channel crosstalk correction due to optical variation between ZMWs, 6) visual removal of any artifactual (non-anticorrelated) signals from traces, 7) dynamic calculation of FRET threshold values to set unbound state to zero before automated state assignment, and 8) file export for each trace in vbFRET compatible format. Idealized traces were subsequently generated using the HMM module of vbFRET to fit a simple two-state model to each FRET trace. The output of vbFRET was then used to automatically extract on-time and off-time distributions for dG (Ch2) and dA (Ch3) binding events, which were fitted using the MEMLET analysis software (22). Detailed fit parameters are listed in Table S3. Figures were subsequently prepared using GraphPad Prism.
Manual assignment and kinetic analysis of telomerase phases in individual traces
Individual traces were viewed using a custom and interactive Matlab script (Fig. S11; all scripts are available upon request). The assignment script allows the user to highlight any suspected telomerase sub-step in a point-and-click approach: basically, the start (e.g., G6 intensity upward transition) and end (e.g., G6 intensity downward transition) of signal clusters were selected by mouse click, generating an idealized (noise-free) trace that displays all state transitions that were selected manually in the raw data window (Fig. S11, raw data window vs. idealized trace window). The script permitted state assignment in different colors and with different amplitudes to aid visual distinction of the states (Fig. S11, red states are A5, yellow states are G6, gray states are RAP and G1/G2 phases). The purpose of the idealized state trace was to demarcate the time points of state transitions for kinetic analysis. The time points of state transitions were extracted in Matlab and allowed calculation of both wait times in between signal clusters as well as lifetimes of signal clusters. Time distributions were fitted with exponential functions in Matlab MEMLET (22) and plotted in GraphPad Prism. Rate constants for single irreversible kinetic steps were calculated as the inverse of the mean of the lifetime population. Reported estimates of the SEM for each measured wait time were determined using the bootstrapping function within Matlab MEMLET (22,23,24). Although manual state assignment for kinetic analysis was performed using raw data traces (Fig. S11), we corrected traces for bleed-through and background for clarity in the figures (Fig. S8 A).
Results and discussion
Concept
We developed a SMRT assay that directly visualizes telomerase-nucleotide complexes () and permits simultaneous detection of functional structural dynamics during the telomere synthesis reaction. The method overcomes several technical challenges in single-molecule studies, facilitating experiments that 1) increase the concentration of dye-coupled nucleotides to physiologically relevant concentrations, 2) differentiate enzyme-bound nucleotides from unspecific fluorescence signals, and 3) enable real-time identification of conformational dynamics that govern enzyme function (Fig. S1). The technique builds on the Pacific Biosciences SMRT-sequencing approach (Figs. 2 A and S1) (25). However, a key differentiating principle of our approach is the use of Förster resonance energy transfer (FRET) to measure the transfer of energy between dye-conjugated nucleotide analogs and an enzyme-linked fluorescent dye (Fig. 2 B).
Figure 2.
Principal components of the human telomerase ZMW-FRET approach. (A) Zero-mode waveguide array chips used for telomerase multicolor ZMW-FRET. (B) Schematic of human telomerase RT (TERT) and the telomerase RNA (TR, solid and dashed lines). LD555 at the U42 dye-labeling site is highlighted. The catalytic core is formed by TERT and essential domains of TR (solid lines). At right, the FRET acceptor components used in this study. To see this figure in color, go online.
In contrast to the direct excitation of fluorescent components across the visible light spectrum employed in SMRT sequencing, the use of FRET permits experiments to be conducted at higher, near-physiological, concentrations of acceptor-dye-conjugated nucleotide analogs (Figs. 3 and S1) (26). Of the three telomeric nucleotides (dTTP, dATP, dGTP), we employed dye-coupled dATP and dGTP analogs to serve as FRET acceptor molecules (Fig. S2).
Figure 3.
Concept of the telomerase ZMW-FRET approach. (A) Principal signals expected from multiplexed ZMW-FRET applied to real-time analysis of telomerase. Dye-coupled hexaphosphate nucleotide analogs dA6P (red) and dG6P (yellow) interact reversibly with telomerase (E). Eventual irreversible nucleotide incorporation into the DNA (D) releases the dye moiety and removes FRET. At right, a Cy5.5-labeled DNA probe (magenta) can bind to nascent telomere DNA and reports on DNA structural dynamics. Telomerase is immobilized via a telomeric DNA primer (B) that is extended repetitively guiding expectations and interpretation of telomerase ZMW-FRET measurements. (C) Telomerase primer-extension assays resolved on sequencing gels after 90-min reaction time (see section “materials and methods”). At left, telomerase requires micro-molar nucleotide concentrations. An approximated range of dye-coupled nucleotide analog concentrations accessible by different single-molecule methods is given above the gel. TIRF, total internal reflection fluorescence. At right, telomerase utilizes dye-dN6P at micro-molar concentrations as used in ZMW-FRET (3.5 μM dye-dG6P, 8 μM dye-dA6P, and 10 μM dTTP). To see this figure in color, go online.
We reasoned that the combination of unmodified dTTP with color-coded dATP/dGTP analogs will produce unambiguous FRET patterns during extension of a telomeric DNA primer by reporting on rU and rC template positions (Fig. 3 A and B). Also, the FRET patterns could be expected to delineate the start and endpoint of RAP-translocation steps in real time by reporting on the rC6 and rC1 template positions (G1G2TTA5G6—translocation—G1G2TTA5G6) (Fig. 3 B). As in SMRT sequencing, the phosphate-linked dye moieties are released upon nucleotide incorporation, removing the FRET signal, and yielding a natural DNA product that does not interfere with downstream processes, such as DNA structure formation or anchor-site interactions (25). In a multiplexed FRET approach, concurrent DNA structural dynamics could then be visualized in parallel using a spectrally distinct dye-labeled DNA probe bound to the nascent telomere DNA (Fig. 3 A). The rA3/4 template positions (dTTP) and RAP-translocation steps were expected to present as “dark” states in the absence of a FRET acceptor.
Approach and results
We employed this FRET-based approach to visualize complexes directly (i.e., states just before nucleotide incorporation) and in real time. A “self-healing” FRET-donor fluorescence dye (LD555) was coupled to the telomerase RNA (TR) subunit at residue U42 (Fig. 2 B) (6,27). Structural models suggest that this labeling site was within FRET range of the nucleotide-binding pocket as well as the DNA-product exit path, enabling the multiplexed FRET approach (Figs. 3 A and S3 A) (13,28). To facilitate site-specific dye labeling, the catalytic TERT protein (telomerase RT) was reconstituted with essential domains of telomerase RNA in vitro (Fig. 2 B). This core complex of the telomerase RNP showed comparable activity to the telomerase holoenzyme isolated from human cells, and a U42-coupled dye label did not interfere with DNA-repeat synthesis kinetics or processivity (Figs. S3 and S4) (10,13,28). Hexaphosphate-aminohexyl dATP and dGTP nucleotide analogs (dye-dN6P) were synthesized at PacBio and coupled to fluorescence dyes following established protocols (Fig. S2 B) (29). Importantly, telomerase incorporated both types of analogs in ensemble primer-extension assays with comparable activity to unmodified dNTP (Fig. 3 C).
To detect real-time telomerase activity using FRET, LD555-telomerase was incubated with a biotinylated (TTAGGG)3 DNA primer, which terminates mid-repeat after the dG2 position and forms stalled enzyme-DNA complexes in the absence of the subsequent nucleotide dTTP (30). Complexes were immobilized on a NeutrAvidin-coated surface inside individual ZMW wells using PacBio array chips (Figs. 2 and 3 A and B). The chips were then incubated with imaging mix containing dye-d(A/G)6P but deficient in dTTP to prevent initiation of telomerase catalysis.
For imaging, chips were mounted on a ZMW-compatible PacBio RSII. Previous work has demonstrated the utility and customization of the RS instrument for single-molecule fluorescence spectroscopy (Fig. S2 A) (20). Three- or four-color emission traces from each ZMW were recorded at 10-Hz detector frame rate (Figs. S5 and S6). To initiate telomerase primer extension, dTTP was added to the chip by automated pipetting after 2 min of data acquisition. The low background signal afforded by a combination of ZMW confinement with single green laser-line excitation at 532 nm allowed for dye-dN6P concentrations that support robust telomerase activity (3.5 μM dG6P, 8 μM dA6P, 10 μM dTTP; Fig. 3 C).
We also performed separate experiments in the presence of Cy5.5-labeled DNA oligonucleotides that are complementary to 2.5 telomeric repeats, serving to probe DNA synthesis and conformational changes during real-time telomerase activity (Figs. 2 B and 3 A) (10,12). We have previously shown that individual DNA probes anneal to the nascent telomeric DNA product within FRET range of a U42-coupled donor dye (10). Importantly, the binding of DNA probe to the DNA product depended on telomerase activity and extrusion of nascent DNA from telomerase (10).
We obtained two datasets of 876 and 2009 ZMW traces (plus and minus DNA probe, respectively) that contained discernible FRET events between LD555-telomerase and dye-dG6P (, yellow) or dye-dA6P (, red) (Figs. 4 and S5–S7). In this way, bona fide binding events between telomerase and the dye-conjugated dNTPs can be distinguished from occasional “sticking” events, wherein, e.g., a dye-conjugated dG6P transiently interacts with the ZMW surface, accessible to direct and unintended green laser excitation.
Figure 4.
Representative ZMW-FRET traces showing dye-labeled dNTP analog-binding events to telomerase. (A) A representative donor (LD555, Ch1) dye intensity trace (top, green), acceptor-dye-labeled dG analog (middle, Ch2 yellow), and estimated Ch1-Ch2 FRET trajectory (bottom, blue) with idealized smFRET trace fit in vbFRET shown in red (see section “materials and methods” for details). Times spent in the dG-bound state (tbound) and times spent in the nucleotide-unbound state (tunbound) are indicated on the top of the plot. (B) For the same trajectory shown in (A), the same donor (LD555, Ch1) dye intensity trace (top, green) is shown, along with an acceptor-dye-labeled dA analog (middle, Ch3 red) and estimated Ch1-Ch3 FRET trajectory (bottom, blue) with idealized smFRET trace fit in vbFRET shown in red. Times spent in the dA-bound state (tbound) and times spent in the nucleotide-unbound state (tunbound) are indicated on the top of the plot. To see this figure in color, go online.
Raw data were corrected for drift and channel crosstalk, and acceptor channel dark periods were set to zero to calculate an approximate FRET value for each donor-acceptor pair. We note the FRET values in our experiments are estimates since the true baseline of the donor channel could not accurately be determined for all traces analyzed. Nevertheless, the estimated FRET trajectories for each channel provide a useful signal for generating idealized state assignments using the previously published vbFRET software (21) (Fig. 4). Recent systematic evaluations of several state transition detection algorithms show that each has benefits and pitfalls, with the vbFRET algorithm performing well under standard camera-based smFRET experimental conditions (31,32). When appropriate, automated single-molecule FRET analysis programs were employed to extract unbiased values for the lifetime distributions of high (bound) and low (unbound) FRET states. However, in certain contexts (see below), we elected to use an analysis approach that employs manual state assignment to reveal the full information content of the data. For example, currently available procedures (31) cannot take advantage of the information specific to telomerase that enables distinction between dG incorporation pre- and post-translocation events. Furthermore, each repeat cycle has the potential for local modulation (10), so averaging across multiple repeats would lose this important detail.
Estimated FRET trajectories derived from 219 analyzed molecules were fitted to a two-state hidden Markov model in vbFRET, and the output was used to extract lifetime distributions of the telomerase bound and unbound states for dG6P or dA6P (Figs. 4 and 5). Interestingly, although the distribution of bound lifetimes for the complex was well fitted to a single-exponential function, the distribution of bound lifetimes for the complex displayed kinetic heterogeneity that was better fitted by a double-exponential function (compare Figs. 5 A and B). This unexpected result suggested the presence of at least two resolvable kinetically distinct complexes observed in our experiments. Similarly, the telomerase unbound lifetime distributions for both the dG6P (Fig. 5 C) and dA6P (Fig. 5 D) showed substantial kinetic heterogeneity and were better fitted by a double-exponential model when compared to a single-exponential fit. Interestingly, this result is consistent with a recent report of kinetic heterogeneity in RNA-DNA and RNA-RNA hybrid duplex formation as measured by smFRET (33). In the context of the telomerase system studied here, our results suggest the nucleotide-unbound states measured in our experiments encompass multiple distinct kinetic steps during telomerase catalysis, including the translocation steps necessary for RAP (Fig. 1).
Figure 5.
Lifetime histograms for nucleotide analog bound and unbound states. Cumulative lifetime histograms for telomerase in the indicated nucleotide analog bound or unbound states as depicted in Fig. 4A and B. (A) Cumulative lifetime histogram of telomerase-dG-bound complexes fit with a single (red) or double (green) exponential function (n = 2198). (B) Cumulative lifetime histogram of telomerase-dA-bound complexes fit with a single (red) exponential function (n = 1958). (C) Cumulative lifetime histogram of telomerase-dG-unbound states (time between binding events) fit with a single (red) or double (green) exponential function (n = 1999). (D) Cumulative lifetime histogram of telomerase-dA-unbound states (time between binding events) fit with a single (red) or double (green) exponential function (n = 1958). All fits were performed in MEMLET (22) and dashed lines in all plots represent the 95% confidence interval for the fit determined by MEMLET’s bootstrapping function. Detailed fit parameters are listed in Table S3. To see this figure in color, go online.
Having used semi-automated analysis to establish the presence of kinetic heterogeneity in our data, we next employed a manual analysis workflow to correlate the specific sequence of FRET signals with specific stages of telomerase catalysis, an approach that leverages a priori knowledge of the telomeric sequence. Non-cognate sampling events with lifetimes on the micro-second timescale are too short to be resolved in the RS measurement (100 ms per frame) and will contribute to the average background fluorescence (Fig. S8 A). However, a feature of our method is the sensitivity to the reversible nature of cognate nucleotide association and dissociation that may occur before nucleotide incorporation () (Figs. 3 A and S8 B). A given template position thereby often manifested as a cluster of FRET signals, rather than a single FRET peak, similar to what was previously described in ZMW analysis of HIV RT using a SMRT-sequencing-related approach (34).
Based upon the known sequence of the TR template, a successful telomerase single-molecule assay was expected to yield an initial “dark” period before dTTP delivery and during extension of the (TTAGGG)3 primer by dT3 and dT4 (Figs. 3 B and 6 A and B). Completion of the initial repeat by dA5 and dG6 was expected to present as onset of FRET in the red and yellow channels, respectively (Figs. 3 B and 6 A and B). These initial patterns were observed in ∼90% of all traces. Subsequent FRET signals were expected to adhere to full telomeric repeat cycles, separated by a dark phase that accounted for the telomerase translocation dynamics independent of the presence of nucleotides. In a total of 817 representative traces across both datasets, we identified 1134 repeat patterns that extended beyond the initial dA5/dG6 signals and indicated processive telomerase repeat addition (i.e., RAP). Analysis of each repeated pattern confirmed the presence of distinct phases that we assigned to known sub-steps in the telomerase reaction cycle: 1) a dark RAP-translocation phase, 2) G1/G2-FRET clusters, 3) a dark T3/T4-phase, and 4) an A5/G6-FRET cluster concluding the repeat (Figs. 6 B and S9 A). FRET signals for the G1/G2 pair are treated as a combined cluster in absence of differentiating information.
Figure 6.
Real-time ZMW analysis of human telomerase. (A) Four-color trace from a single telomerase molecule. Detection channels are shown separately for clarity. Dashed lines separate discernible repeat patterns. (B) At left, cropped gel lane visualizes early primer extension and informs expectations for ZMW sequencing. Sequence and numbers indicate telomeric DNA repeats and the respective template position for each nucleotide. The RAP-translocation steps occur after completion of each respective repeat. At right, example of a repetitive FRET pattern consistent with a telomeric DNA repeat. Repeat sub-steps corresponding to nucleotide positions or the RAP-translocation steps are indicated. The mean duration of each sub-step is given below; nRAP-translocation = 219, nG1G2 = 373, nT3T4 = 153, nA5 = 577, nG6 = 576. Errors are SE of the mean. (C) A5/G6 motifs enlarged from trace in (A). dG6 incorporations as repeat-end signatures are indicated (dashed lines) and yield repeat wait times for kinetic analysis (Δt), cf. Fig.S9. Below, mean wait times for individual repeats derived from two RS datasets. Errors are SE of the mean. (D) Mean wait times for individual repeats derived from ZMW and telomerase gel assay. Error bars indicate the standard deviation across duplicate ZMW experiments. The rate constants are consistent with values derived from gel-based telomerase ensemble assays (blue data points) (10). To see this figure in color, go online.
To validate the real-time measurement of processive telomere repeat synthesis, we extracted repeat wait times between individual G6 incorporation (Figs. 6 C and S9 B). The obtained values from both independent datasets were consistent with previously reported corresponding rate constants derived from kinetic modeling of telomerase ensemble assays (10). Thus, ensemble biochemical and ZMW-based single-molecule telomerase assays yield similar kinetic parameters, further supporting the validity of the ZMW-FRET assay (Fig. 6 D) (10). Consistent with prior biochemical analysis of ensemble telomerase activity, the dark RAP-translocation phase was the slowest sub-step within an individual repeat cycle with a mean duration of 165 ± 8 s (Fig. 6 B). An unexpected feature of our data was that dG6 incorporation occurred relatively soon after dG6 binding (5 ± 0.2 s), whereas the G1/G2-FRET clusters exhibited a substantially increased mean duration (76 ± 5 s; Figs. 4 B, S7, and S9 A). The prolonged G1/G2 clusters are unlikely to be explained by the simple combination of two successive dG-incorporation events and may suggest the existence of a long-lived state of the enzyme that results in slow dG-incorporation kinetics at the beginning of each telomeric DNA repeat (Fig. S10). This result is consistent with the global analysis of dG binding kinetics that suggested the existence of multiple, kinetically distinct, complexes (Fig. 5 A). Although the precise structural basis of this observation remains to be elucidated, it is plausible that additional translocation-related events occur after template repositioning and dG1 binding. These may include re-formation of the RNA/DNA hybrid, re-priming of the DNA 3′ end, and active-site closure (Figs. 1 A and S10) (8).
In the presence of Cy5.5-DNA probe, 605 of 2009 traces displayed FRET between LD555-telomerase and the Cy5.5-labeled probe (magenta) bound to the nascent telomere DNA product (Figs. 6 A, 7, and S6). This observation represented a direct measurement of product DNA dynamics resolved concurrently with nucleotide-addition cycles. Interestingly, we identified repeated instances of a Cy5.5-FRET increase during the RAP-translocation phase and preceding the onset of G1/G2 clusters. We hypothesize the DNA product undergoes positional rearrangements during RAP translocation and thereby becomes accessible for the Cy5.5-DNA probe to bind (Figs. 1 A and 7). Once bound to the DNA product, the DNA probes could be expected to report on DNA-product movements during subsequent repeat-addition cycles and their RAP-translocation steps (Fig. 7). For example, although the onset of G1/G2 clusters suggested successful translocation of the RNA template in the active site, future experiments are needed to assign translocation-related Cy5.5-FRET events to DNA-product dynamics before, during, or after formation of the new DNA/RNA duplex (Figs. 1 A and 7). However, the ability to resolve individual nucleotide-bound enzymes and product DNA dynamics at specific stages of telomerase repeat synthesis provides a powerful illustration of the information-rich data that can be obtained by combining SMRT sequencing with single-molecule FRET in the study of telomerase.
Figure 7.
Multiplexed ZMW telomerase assay. At left, schematic of the experimental setup inside individual ZMWs. At right, telomerase DNA-product dynamics are visualized in real time through binding of an acceptor-labeled DNA probe (top, detection channels 1 + 4, arrows highlight selected events). The time axis is broken to enlarge regions of interest; cf. Fig. S5. Concurrent nucleotide addition by the same telomerase enzyme (detection channels 2 + 3) is shown below. To see this figure in color, go online.
Discussion
The ZMW-FRET approach presented here tracks individual telomerase enzymes in real time through multiplexed and quantitative single-molecule analysis. We were thus able to monitor the cyclic nature of telomere repeat synthesis, differentiating the association, incorporation, and dynamics of nucleotides and the DNA substrate. The method enabled the determination of wait-time distributions for the synthesis of individual telomeric repeats as well as for kinetic sub-steps, such as the DNA/RNA translocation pause. Notably, the average repeat synthesis wait times were consistent with gel-based kinetic analysis of telomerase repeat synthesis, supporting the validity of ZMW-FRET in studying the mechanism of telomerase.
Telomerase is a slow enzyme and, within the 30-min measurement window on the PacBio RSII, we rarely observed more than five consecutive repeats, as can be expected for stochastic events with a time constant of around 5 min. While gel-based assays allow for the study of longer DNA products (11), the finite measurement time in ZMW-FRET will bias the distributions of later repeats toward shorter wait times. Previous studies demonstrated that the length of the nascent DNA product influences telomerase activity (10); longer DNA products can fold into DNA structures that modulate telomerase kinetics, and telomerase processivity after the first repeat addition was lower than subsequent repeats (35), possibly because the short DNA primers form weaker interactions with the anchor sites of telomerase. Future ZMW-FRET experiments can employ varying lengths of the starting primer to investigate the influence of short and long DNA products on telomerase function in real time, thereby avoiding a wait-time bias that arises after five or more repeats.
In addition to the observation of multiple consecutive repeats, our proof-of-concept data also revealed differential binding and incorporation kinetics of dGTP at the end and beginning of individual telomeric repeats (dG6 versus dG1, respectively). This finding is a specific feature of the ZMW-FRET approach in that it can detect nucleotide-bound states of the enzyme in real time. Due to cognate (and non-cognate) nucleotide sampling of the enzyme active site, the identity of template positions might be encoded in signal clusters rather than single pulses (34). ZMW measurements of fast polymerase systems, which require higher frame rates, may visualize a larger contribution of non-cognate nucleotide sampling. The slow reaction speed of telomerase enables lower frame rates, which reduce the impact of this limitation. Adding to existing models for telomerase translocation mechanisms, we hypothesize that the prolonged G1/G2 clusters present a cognate dGTP-bound state after successful relocation of the TR template. The unique telomerase RAP translocation may therefore include rate-contributing rearrangements of the TERT-protein or DNA 3′ end that are uncoupled from TR-template positioning but are required to complete dG1 incorporation. The processivity-stimulating cofactors protection of telomeres 1 (POT1) and telomere protection protein 1 (TPP1) are known to engage the TERT protein as well as the telomerase DNA product to speed up the telomerase reaction (35,36). The ZMW-FRET approach is uniquely suited to test the hypothesis that POT1-TPP1 will enhance DNA-translocation efficiency, thereby reducing the duration of the G1/G2 clusters and increasing telomerase RAP. Our proof-of-concept experiments include a dye-labeled DNA probe that can bind to the nascent DNA product of actively extending telomerase in a manner analogous to POT1-TPP1. More generally, we expect the method described herein will find future application in telomere and telomerase research, enabling direct correlation of structural states observed in static cryo-EM structures with dynamic sub-steps required for telomere length maintenance.
Author contributions
J.H. and M.D.S. conceived the ZMW telomerase assay method. J.H. performed RS experiments and acquired ZMW-FRET data. J.C. provided training and advised on RS experimentation. J.C. and L.I.J. performed preliminary ZMW telomerase experiments. J.C., C.P.L., and J.W. contributed analytic tools and provided advice on data analysis. J.H. analyzed the data with input from C.R.B., M.D.S., and J.D.P. C.R.B. advised on kinetic analysis and interpretation. J.H. and L.I.J. reconstituted enzymes and performed gel-based assays. M.B. contributed dye-labeled enzyme reagents, conducted bulk telomerase kinetics assays, and analyzed ZMW data. J.H. interpreted the data, designed graphics, and wrote the manuscript with input from C.R.B., J.D.P., and M.D.S. All authors provided comments on the manuscript.
Acknowledgments
We are grateful to Lubomir Sebo and Jonas Korlach of Pacific Biosciences for synthesizing and providing dye-coupled nucleotide analogs for this study. We thank Carol Greider for discussion of the results. J.H. was supported by a Swiss National Science Foundation (SNSF) Early Postdoc Mobility fellowship (P2EZP3_181605). J.C. was supported by a Stanford Bio-x Fellowship and is a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation (DRG-#2403-20). C.P.L. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-#2321-18), and J.W. was supported by a postdoctoral scholarship from the Knut and Alice Wallenberg Foundation (KAW 2015.0406). This work was supported by National Institutes of Health grants F99CA212439 (to L.I.J.) and R01GM095850 (to M.D.S.), as well as grants GM51266, GM011378, and AI150464 to J.D.P.
Declaration of interests
The authors declare no competing interests.
Editor: Gijs Wuite.
Footnotes
Jendrik Hentschel’s present address is Alida Biosciences, 11535 Sorrento Valley Road, San Diego, California.
Junhong Choi’s present address is Department of Genome Sciences, University of Washington, Seattle, Washington.
Linnea I. Jansson’s present address is Department of Chemistry and Biochemistry, University of Colorado Boulder, Colorado.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.07.019.
Supporting material
References
- 1.Blackburn E.H., Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harbor Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003558. https://www.ncbi.nlm.nih.gov/pubmed/20660025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. https://www.ncbi.nlm.nih.gov/pubmed/16166375 [DOI] [PubMed] [Google Scholar]
- 3.Aubert G., Lansdorp P.M. Telomeres and aging. Physiol. Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. https://www.ncbi.nlm.nih.gov/pubmed/18391173 [DOI] [PubMed] [Google Scholar]
- 4.Shay J.W., Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. https://www.ncbi.nlm.nih.gov/pubmed/9282118 [DOI] [PubMed] [Google Scholar]
- 5.Greider C.W. Telomerase is processive. Mol. Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. https://www.ncbi.nlm.nih.gov/pubmed/1875940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parks J.W., Stone M.D. Coordinated DNA dynamics during the human telomerase catalytic cycle. Nat. Commun. 2014;5:4146. doi: 10.1038/ncomms5146. https://www.ncbi.nlm.nih.gov/pubmed/24923681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lue N.F. A physical and functional constituent of telomerase anchor site. J. Biol. Chem. 2005;280:26586–26591. doi: 10.1074/jbc.M503028200. https://www.ncbi.nlm.nih.gov/pubmed/15905172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu R.A., Upton H.E., et al. Collins K. Telomerase Mechanism of Telomere Synthesis. Annu. Rev. Biochem. 2017;86:439–460. doi: 10.1146/annurev-biochem-061516-045019. https://www.ncbi.nlm.nih.gov/pubmed/28141967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greider C.W., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. https://www.ncbi.nlm.nih.gov/pubmed/3907856 [DOI] [PubMed] [Google Scholar]
- 10.Jansson L.I., Hentschel J., et al. Stone M.D. Telomere DNA G-quadruplex folding within actively extending human telomerase. Proc. Natl. Acad. Sci. USA. 2019;116:9350–9359. doi: 10.1073/pnas.1814777116. https://www.ncbi.nlm.nih.gov/pubmed/31019071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagshaw C.R., Hentschel J., Stone M.D. The Processivity of Telomerase: Insights from Kinetic Simulations and Analyses. Molecules. 2021;26 doi: 10.3390/molecules26247532. https://www.ncbi.nlm.nih.gov/pubmed/34946615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang J., Wang Y., et al. Feigon J. Structure of Telomerase with Telomeric DNA. Cell. 2018;173:1179–1190.e13. doi: 10.1016/j.cell.2018.04.038. https://www.ncbi.nlm.nih.gov/pubmed/29775593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghanim G.E., Fountain A.J., et al. Nguyen T.H.D. Structure of human telomerase holoenzyme with bound telomeric DNA. Nature. 2021;593:449–453. doi: 10.1038/s41586-021-03415-4. https://www.ncbi.nlm.nih.gov/pubmed/33883742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekne Z., Ghanim G.E., et al. Nguyen T.H.D. Structural basis of human telomerase recruitment by TPP1-POT1. Science. 2022;375:1173–1176. doi: 10.1126/science.abn6840. https://www.ncbi.nlm.nih.gov/pubmed/35201900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang H., Opresko P., Myong S. Single-molecule real-time detection of telomerase extension activity. Sci. Rep. 2014;4:6391. doi: 10.1038/srep06391. https://www.ncbi.nlm.nih.gov/pubmed/25263700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patrick E.M., Slivka J.D., et al. Schmidt J.C. Observation of processive telomerase catalysis using high-resolution optical tweezers. Nat. Chem. Biol. 2020;16:801–809. doi: 10.1038/s41589-020-0478-0. https://www.ncbi.nlm.nih.gov/pubmed/32066968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akiyama B.M., Stone M.D. Assembly of complex RNAs by splinted ligation. Methods Enzymol. 2009;469:27–46. doi: 10.1016/S0076-6879(09)69002-9. https://www.ncbi.nlm.nih.gov/pubmed/20946783 [DOI] [PubMed] [Google Scholar]
- 18.Weinrich S.L., Pruzan R., et al. Morin G.B. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. https://www.ncbi.nlm.nih.gov/pubmed/9398860 [DOI] [PubMed] [Google Scholar]
- 19.Drosopoulos W.C., Direnzo R., Prasad V.R. Human telomerase RNA template sequence is a determinant of telomere repeat extension rate. J. Biol. Chem. 2005;280:32801–32810. doi: 10.1074/jbc.M506319200. https://www.ncbi.nlm.nih.gov/pubmed/16061476 [DOI] [PubMed] [Google Scholar]
- 20.Chen J., Dalal R.V., et al. Puglisi J.D. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc. Natl. Acad. Sci. USA. 2014;111:664–669. doi: 10.1073/pnas.1315735111. https://www.ncbi.nlm.nih.gov/pubmed/24379388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bronson J.E., Fei J., et al. Wiggins C.H. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 2009;97:3196–3205. doi: 10.1016/j.bpj.2009.09.031. https://www.ncbi.nlm.nih.gov/pubmed/20006957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woody M.S., Lewis J.H., et al. Ostap E.M. MEMLET: An Easy-to-Use Tool for Data Fitting and Model Comparison Using Maximum-Likelihood Estimation. Biophys. J. 2016;111:273–282. doi: 10.1016/j.bpj.2016.06.019. https://www.ncbi.nlm.nih.gov/pubmed/27463130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efron B.a.T., R. Society for Industrial and Applied Mathematics; 1982. The Jackknife, the Bootstrap and Other Resampling Plans. [Google Scholar]
- 24.Numerical Recipes in C++: The Art of Scientific Computing. Cambridge University Press; W.H. Press; 2002. [Google Scholar]
- 25.Eid J., Fehr A., et al. Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. https://www.ncbi.nlm.nih.gov/pubmed/19023044 [DOI] [PubMed] [Google Scholar]
- 26.Goldschen-Ohm M.P., White D.S., et al. Goldsmith R.H. Observing Single-Molecule Dynamics at Millimolar Concentrations. Angew Chem. Int. Ed. Engl. 2017;56:2399–2402. doi: 10.1002/anie.201612050. https://www.ncbi.nlm.nih.gov/pubmed/28116856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Q., Jockusch S., et al. Blanchard S.C. Electronic tuning of self-healing fluorophores for live-cell and single-molecule imaging. Chem. Sci. 2017;8:755–762. doi: 10.1039/C6SC02976K. https://www.ncbi.nlm.nih.gov/pubmed/28377799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen T.H.D., Tam J., et al. Collins K. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature. 2018;557:190–195. doi: 10.1038/s41586-018-0062-x. https://www.ncbi.nlm.nih.gov/pubmed/29695869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korlach J., Bibillo A., et al. Turner S.W. Long, processive enzymatic DNA synthesis using 100% dye-labeled terminal phosphate-linked nucleotides. Nucleosides, Nucleotides Nucleic Acids. 2008;27:1072–1083. doi: 10.1080/15257770802260741. https://www.ncbi.nlm.nih.gov/pubmed/18711669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallweber G., Gryaznov S., et al. Pruzan R. Interaction of human telomerase with its primer substrate. Biochemistry. 2003;42:589–600. doi: 10.1021/bi026914a. https://www.ncbi.nlm.nih.gov/pubmed/12525188 [DOI] [PubMed] [Google Scholar]
- 31.Götz M., Barth A., et al. Schmid S. A blind benchmark of analysis tools to infer kinetic rate constants from single-molecule FRET trajectories. Nat. Commun. 2022;13:5402. doi: 10.1038/s41467-022-33023-3. https://www.ncbi.nlm.nih.gov/pubmed/36104339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadzic M.C.A.S., Börner R., et al. Sigel R.K.O. Reliable State Identification and State Transition Detection in Fluorescence Intensity-Based Single-Molecule Forster Resonance Energy-Transfer Data. J. Phys. Chem. B. 2018;122:6134–6147. doi: 10.1021/acs.jpcb.7b12483. https://www.ncbi.nlm.nih.gov/pubmed/29737844 [DOI] [PubMed] [Google Scholar]
- 33.Steffen F.D., Khier M., et al. Sigel R.K.O. Metal ions and sugar puckering balance single-molecule kinetic heterogeneity in RNA and DNA tertiary contacts. Nat. Commun. 2020;11:104. doi: 10.1038/s41467-019-13683-4. https://www.ncbi.nlm.nih.gov/pubmed/31913262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilfan I.D., Tsai Y.C., et al. Korlach J. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription. J. Nanobiotechnol. 2013;11:8. doi: 10.1186/1477-3155-11-8. https://www.ncbi.nlm.nih.gov/pubmed/23552456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latrick C.M., Cech T.R. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. https://www.ncbi.nlm.nih.gov/pubmed/20094033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F., Podell E.R., et al. Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. https://www.ncbi.nlm.nih.gov/pubmed/17237768 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.