Abstract
Two new species of Trechispora indigenous to southern China, T.laxa and T.tongdaoensis, are described and illustrated, and the first record of T.khokpasiensis in China is reported. Molecular phylogenetic analyses of the concatenated nuclear rDNA ITS1–5.8S–ITS2 and nuclear large subunit sequences supported the inclusion of the three species within the Trechispora clade, together with species formerly classified in Scytinopogon. The new species are similar in micromorphology to species of Trechispora (as traditionally circumscribed) but are distinguished by having coralloid basidiomata. A key to the known coralloid Trechispora species in China is provided.
Key words: Coral fungi, Phylogenetic analysis, Scytinopogon , Taxonomy
Introduction
The genus Trechispora P. Karst was established by Karsten (1890) with Trechisporaonusta P. Karst as the type species. Trechispora is the largest genus in the order Trechisporales (Larsson 2007), which is highly diverse in morphology: stipitate, clavarioid, or resupinate basidiomata (Meiras-Ottoni et al. 2021; Sommai et al. 2023); smooth, grandinioid, odontoid, hydnoid, or poroid hymenophores; short cylindric basidia, clamped, with 2 or 4 sterigmata on the basidia; and smooth or variously ornamented basidiospores. In addition, ampullate septa is an important character in Trechispora (Bernicchia and Gorjón 2010, Ordynets et al. 2015, Meiras-Ottoni et al. 2021, Liu et al. 2022). Calcium oxalate crystals usually accumulate on the mycelium or subhymenial hyphae, and the crystal morphology can be useful for species identification (Larsson 1994). Currently, approximately 90 species are accepted in Trechispora (Liu et al. 2022; Sommai et al. 2023). Consistent with previous studies (Birkebak et al. 2013), most Trechispora species are distributed in the tropics or subtropics (Chikowski et al. 2020). The placement of Trechispora in the order Trechisporales is supported by phylogenetic analyses of molecular data (Hibbett et al. 2007; Larsson 2007).
Scytinopogon Singer, erected by Singer (1945), has been assigned to several different families in the past: Clavariaceae Chevall (Corner 1970), Thelephoraceae Chevall (Donk 1964), and Gomphaceae (Maas Geesteranus 1962). The genus has also been suggested to be related to the Hydnodontaceae (Jülich 1981). Morphologically, Scytinopogon is characterized by clavarioid basidiomata with flattened and dense branches, which distinguish the species from Trechispora. However, phylogenetic analyses of molecular data indicate that Scytinopogon is nested within Trechispora (Hydnodontaceae), and no clear delimitation exists between the two genera. Because the name Trechispora has nomenclatural priority, Scytinopogon has been synonymized with Trechispora, thus rendering Trechispora a large, monophyletic genus (Meiras-Ottoni et al. 2021). To determine the correct name for Scytinopogon species, sequences and specimens for the type species were needed (Meiras-Ottoni et al. 2021). For this reason, to avoid misinterpretations in species delimitation within Trechispora, not all currently accepted Scytinopogon species have been transferred to Trechispora. Scytinopogoncryptomerioides W.R. Lin and P.H. Wang has recently been described from Taiwan (Lin et al. 2022).
During research on clavarioid fungi indigenous to southern China, two undescribed and one recently described coralloid Trechispora species were collected. Descriptions and illustrations of these three species are provided, and phylogenetic reconstructions based on nuclear rDNA ITS1–5.8S–ITS2 (ITS) and nuclear large subunit (LSU) sequences support the distinction of the new species and their placement in Trechispora.
Materials and methods
Specimen sources
Field work was conducted and specimens gathered by the authors from 2011 to 2022 in Hainan, Hunan, and Guangdong provinces, China. The habitat and morphological characters of fresh specimens were recorded in the field, including their dimensions and color. The fresh fruiting bodies were dried using heat or silica gel. The dried specimens were deposited in the Mycological Herbarium of Hunan Normal University (MHHNU), Changsha, China.
Morphological observation
Macroscopic characteristics were mainly derived from record sheets and photographs. The colors cited in the descriptions are based on those of Kornerup and Wanscher (1978) and Ridgway (1912). Dried fruiting body sections were placed in 3% KOH solution containing 1% Congo red solution. Microscopic characters were observed from a small portion of dried hymenial tissue using a light microscope to observe the basidiospores (100×), basidia, and hyphae. Scanning electron microscopy (SEM) was conducted with a TESCAN CLARA Xplore 30 operating at 2 keV. Forty basidiospores of each specimen were randomly selected for measurement. The spore size is expressed in the form (a–) b–c (– d), where a and d are the minimum and maximum dimensions of spores, respectively, and b and c encompass the majority of the spore dimensions. The abbreviation [n/m/p] refers to n spores measured from m basidiomata of p specimens. In addition, the Q value represents the length: width ratio of basidiospores, and the Qm value is the average Q ± standard deviation.
DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from dried specimens using the EZup Column Fungal Genomic DNA Extraction Kit (Sangon Biotech, Shanghai, China). A 20 mg sample of a dried specimen was ground to powder in liquid nitrogen in accordance with the manufacturer’s instructions. The primer pairs ITS4/ITS5 and LR5/LR0R were used to amplify the ITS and LSU regions, respectively (Vilgalys and Hester 1990; White et al. 1990; Gardes and Bruns 1993). The PCR amplification reactions were performed on an Eppendorf Mastercycler thermal cycler in a 25 µL volume containing 1 µL DNA, 2 µL primers, 9.5 µL ddH2O, and 12.5 µL 2× Es Taq Master Mix. The amplification procedure consisted of pre-denaturation at 94 °C for 4 min, then 32 cycles comprising denaturation at 94 °C for 40 s, annealing at 55 °C for 40 s, and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 8 min, and held at 4 °C (Liu et al. 2022). The PCR products were separated by electrophoresis on a 1% agarose gel. An ABI 3730 DNA Analyzer (PerkinElmer Inc., USA) was used to sequence the PCR products. The newly generated sequences (seven ITS and seven LSU) were deposited in GenBank (Table 1).
Table 1.
Details of the ITS and 28S rDNA sequences used for phylogenetic analyses. The sequences newly generated in this study are highlighted in bold, and all types marked with an asterisk.
| Taxon | Voucher | GenBank No. (ITS) | GenBank No. (28S) | Geographical origin | References |
|---|---|---|---|---|---|
| Trechisporaaraneosa | KHL8570 | AF347084 | AF347084 | Sweden | Larsson et al. (2004) |
| T.bambusicola | CLZhao3302 | MW544021 | MW520171 | China | Zhao et al. (2021) |
| T.bambusicola | He3381 | OM523405 | OM339227 | China | Liu et al. (2022) |
| T.chaibuxiensis | He5072 | OM523408 | OM339230 | China | Liu et al. (2022) |
| T.chaibuxiensis | LWZ2017081434 | OM523409 | OM339231 | China | Liu et al. (2022) |
| T.copiosa* | AMO422 | MN701013 | MN687971 | Brazil | Meiras-Ottoni et al. (2021) |
| T.copiosa | AMO427 | MN701015 | MN687973 | Brazil | Meiras-Ottoni et al. (2021) |
| T.copiosa | AMO453 | MN701018 | MN687975 | Brazil | Meiras-Ottoni et al. (2021) |
| T.confinis | KHL11064 | AF347081 | AF347081 | Sweden | Larsson et al. (2004) |
| T.confinis | LWZ2021092023b | OM523414 | OM339235 | China | Liu et al. (2022) |
| T.constricta | He5899 | OM523417 | OM339236 | China | Liu et al. (2022) |
| T.constricta | LWZ2021092430a | OM523418 | OM339237 | China | Liu et al. (2022) |
| T.caulocystidiata* | FLOR56314 | MK458772 | – | Brazil | Furtado et al. (2021) |
| T.crystallina | LWZ201707292 | OM523419 | OM339238 | China | Liu et al. (2022) |
| T.crystallina | LWZ 201710137 | OM523420 | OM339239 | Vietnam | Liu et al. (2022) |
| T.dimitiella | Dai21181 | OK298493 | OK298949 | China | Liu et al. (2022) |
| T.dimitiella* | Dai 21931 | OK298492 | OK298948 | China | Liu et al. (2022) |
| T.dealbata | FLOR56183 | MK458777 | – | Brazil | Furtado et al. (2021) |
| T.fimbriata | He 4873 | OM523424 | OM339243 | China | Liu et al. (2022) |
| T.fimbriata | He 6134 | OM523425 | OM339244 | China | Liu et al. (2022) |
| T.fissurata | He6190 | OM523427 | OM339245 | China | Liu et al. (2022) |
| T.fissurata | He6322 | OM523428 | OM339246 | China | Liu et al. (2022) |
| T.foetida* | FLOR56315 | MK458769 | – | Brazil | Furtado et al. (2021) |
| T.farinacea | KHL 8451 | AF347082 | AF347082 | Sweden | Larsson et al. (2004) |
| T.farinacea | KHL 8454 | AF347083 | AF347083 | – | Larsson (2001) |
| T.gelatinosa | AMO824 | MN701020 | MN687977 | Brazil | Meiras-Ottoni et al. (2021) |
| T.gelatinosa* | AMO1139 | MN701021 | MN687978 | Brazil | Meiras-Ottoni et al. (2021) |
| T.havencampii* | SFSUDED8300 | NR154418 | NG059993 | Africa | Desjardin and Perry (2015) |
| T.hymenocystis | KHL16444 | MT816397 | MT816397 | Norway | Meiras-Ottoni et al. (2021) |
| T.hymenocystis | KHL8795 | AF347090 | AF347090 | Sweden | Larsson et al. (2004) |
| T.khokpasiensis* | MMCR00009 | MZ687107 | MZ683197 | Thailand | Sommai S et al. (2023) |
| T.latehypha | He5848 | OM523446 | OM339262 | Sri Lanka | Liu et al. (2022) |
| T.latehypha | LWZ2017061116 | OM523447 | OM339263 | China | Liu et al. (2022) |
| T.longiramosa | HG140168 | OM523448 | OM339264 | China | Liu et al. (2022) |
| T.longiramosa | CH 19233 | OM523449 | – | China | Liu et al. (2022) |
| T.laxa | MHHNU10379 | OP959649 | OP954660 | China | This study |
| T.laxa* | MHHNU10714 | OP959650 | OP954661 | China | This study |
| T.malayana | Dai17876 | OM523452 | OM339265 | Singapore | Liu et al. (2022) |
| T.malayana | He4156 | OM523453 | OM339266 | Thailand | Liu et al. (2022) |
| T.minispora | AM170 | MK328885 | MK328894 | Mexico | Yuan et al. (2020) |
| T.minispora | AM176 | MK328886 | MK328895 | Mexico | Yuan et al. (2020) |
| T.mollusca | Dai 6191 | OM523455 | OM339269 | China | Liu et al. (2022) |
| T.mollusca | Dai 11085 | OM523457 | OM339270 | China | Liu et al. (2022) |
| T.nivea | LWZ201808043 | OM523461 | OM339273 | China | Liu et al. (2022) |
| T.nivea | MAFungi74044 | JX392832 | JX392833 | – | Telleria et al. (2013) |
| T.papillosa | AMO713 | MN701022 | MN687979 | Brazil | Meiras-Ottoni et al. (2021) |
| T.papillosa | AMO714 | – | MN687980 | Brazil | Meiras-Ottoni et al. (2021) |
| T.papillosa* | AMO795 | MN701023 | MN687981 | Brazil | Meiras-Ottoni et al. (2021) |
| T.pallescens | FLOR56184 | MK458767 | – | Brazil | A.N.M. Furtado et al. (2021) |
| T.pallescens | FLOR56188 | MK458774 | – | Brazil | A.N.M. Furtado et al. (2021) |
| T.aff.pallescens | RL 115 | MK328887 | MK328896 | Mexico | Unpublished |
| T.aff.pallescens | RL 132 | MK328889 | MK328898 | Mexico | Unpublished |
| T.aff.pallescens | RL 133 | MK328890 | MK328899 | Mexico | Unpublished |
| T.robusta | FLOR 56179 | MK458770 | – | Brazil | A.N.M. Furtado et al. (2021) |
| T.sanpapaoensis | MMCR00124.1 | MZ687109 | MZ683200 | Thailand | Sommai S et al. (2023) |
| T.saluangensis* | MMCR00260 | MZ687104 | MZ683201 | Thailand | Sommai S et al. (2023) |
| T.saluangensis | MMCR00261 | MZ687105 | MZ683202 | Thailand | Sommai S et al. (2023) |
| T.scabra | FLOR56189 | MK458773 | – | Brazil | A.N.M. Furtado et al. (2021) |
| T.sinensis | He3714 | OM523464 | OM339274 | China | Liu et al. (2022) |
| T.sinensis | He4314 | OM523465 | OM339275 | China | Liu et al. (2022) |
| T.stevensonii | MAFungi70645 | JX392843 | JX392844 | – | Telleria et al. (2013) |
| T.stevensonii | MAFungi70669 | JX392841 | JX392842 | – | Telleria et al. (2013) |
| T.subfissurata | LWZ2019061348 | OM523491 | – | China | Liu et al. (2022) |
| T.subfissurata | He3907 | OM523490 | OM339298 | China | Liu et al. (2022) |
| T.khokpasiensis | MHHNU07529 | ON897819 | ON898005 | China | This study |
| T.khokpasiensis | MHHNU10662 | ON897822 | ON898008 | China | This study |
| T.khokpasiensis | MHHNU10670 | ON897823 | ON898009 | China | This study |
| T.thailandica* | He4101 | OM523499 | OM339307 | Thailand | Liu et al. (2022) |
| T.thailandica | He 4112 | OM523500 | OM339308 | Thailand | Liu et al. (2022) |
| T.thelephora | URM85757 | – | MH280001 | Brazil | Chikowski et al. (2020) |
| T.thelephora | URM85758 | – | MH280002 | Brazil | Chikowski et al. (2020) |
| T.torrendii* | URM85886 | MK515148 | MH280004 | Brazil | Chikowski et al. (2020) |
| T.torrendii | URM85887 | – | MH280005 | Brazil | Chikowski et al. (2020) |
| T.tongdaoensis* | MHHNU11083 | OP959651 | OP954662 | China | This study |
| T.tongdaoensis | MHHNU11086 | OP959652 | OP954663 | China | This study |
| T.termitophila* | AMO396 | MN701025 | MN687983 | Brazil | Meiras-Ottoni et al. (2021) |
| T.termitophila | AMO893 | MN701026 | MN687984 | Brazil | Meiras-Ottoni et al. (2021) |
| T.termitophila | AMO1169 | MN701028 | MN687986 | Brazil | Meiras-Ottoni et al. (2021) |
| T.tropica | LWZ2017061314 | OM523502 | OM339310 | China | Liu et al. (2022) |
| T.tropica | LWZ2017061316 | OM523503 | OM339311 | China | Liu et al. (2022) |
| Scytinopogoncryptomerioides* | 0906RK10-23 | – | OK422242 | China | Lin et al. (2022) |
| Brevicelliciumatlanticum | LISU178566 9065IM | HE963773 | HE963774 | Portugal | Telleria et al. (2013) |
| B.olivascens | MAFungi23496 | HE963787 | HE963788 | Spain | Telleria et al. (2013) |
Alignment and phylogenetic analysis
The newly generated sequences were aligned with publicly available ITS and LSU sequences of Scytinopogon and Trechispora species from GenBank (see Table 1 for the accession numbers and sources of previously published sequences). Sequences for the corresponding regions from single accessions of Brevicelliciumolivascens K.H. Larss. and Hjortstam and Brevicelliciumatlanticum Melo, Tellería, M. Dueñas and M.P. Martín were used as the outgroup and included in the ITS+LSU sequence matrix. The ITS and LSU sequences were aligned using MAFFT v7.471 with default settings of gap openings and extension penalties (Katoh and Standley 2016). The final ITS+LSU dataset comprising 151 sequences and 1663 aligned positions (77 ITS and 74 LSU). It was assembled with SEQUENCEMATRIX v1.7.8 (Vaidya et al. 2011) and used for a multimarker phylogenetic analysis. A maximum likelihood (ML) analysis was conducted with RAxML v7.2.6 (Stamatakis et al. 2005, Stamatakis 2006) using the GTR+Gamma evolutionary model (Stamatakis et al. 2008). ML bootstrapping (BS) was performed with 1000 replicates. Bayesian inference (BI) was performed using MrBayes v3.2.7 (Ronquist and Huelsenbeck 2003); analyses were run for 2,000,000 generations using four Metropolis-coupled Monte Carlo Markov chains to calculate posterior probabilities (PP). FigTree 1.4.2 (Rambaut 2012) was used to visualize the tree files, which were edited using Adobe Photoshop CS6 (Adobe Systems Inc., USA).
Results
Phylogenetic analyses
The phylogeny derived from the ML analysis of the concatenated ITS+LSU dataset, with both PP and BS support values, is shown in Fig. 1. The BI phylogeny (not shown) was very similar in topology and branch support to the ML tree. The ML and BI analyses resolved that the three species collected from southern China each formed a monophyletic lineage within the genus Trechispora with high statistical support values. The species of Trechispora and Scytinopogon were intermixed, which was consistent with previous phylogenetic studies (Meiras-Ottoni et al. 2021; Chikowski et al. 2020). Trechisporalaxa and T.tongdaoensis were closely related to the species Trechisporahavencampii (D.E. Desjardin and B.A. Perry) Meiras-Ottoni and Gibertoni, Trechisporarobusta (Rick) S.L. Liu and L.W. Zhou, Trechisporafoetida (A.N.M. Furtado and M.A. Neves) S.L. Liu and L.W. Zhou, Trechisporalongiramosa S.L. Liu, G. He, L. Chen Shuang and L.W. Zhou, Trechisporasanpapaoensis Pinruan, Sommai and Khamsuntorn, and Trechisporatermitophila Meiras-Ottoni and Gibertoni. Trechisporakhokpasiensis Pinruan, Sommai and Khamsuntorn, Trechisporacopiosa Meiras-Ottoni and Gibertoni, and Scytinopogoncryptomerioides were grouped in a well-supported subclade (PP 0.99, BS 86%). These results strongly supported the phylogenetic distinction of T.laxa and T.tongdaoensis from other species within the Trechispora clade.
Figure 1.
Phylogenetic relationships of Trechispora species inferred from a concatenated ITS and LSU sequence dataset under the maximum likelihood optimality criterion. Bayesian posterior probabilities (PP) > 0.95 and bootstrap values (BS) >70% are reported at the nodes (PP/BS); “–” indicates that the support value was less than the respective threshold. The two newly described species and one newly recorded species from China are highlighted in bold.
Taxonomy
. Trechispora khokpasiensis
Pinruan, Sommai & Khamsuntorn
78E95334-3CBC-52E4-8ABC-2E7C6124D647
Figure 2.
Basidiomata of Trechisporakhokpasiensis (MHHNU7529). Scale bars: 1cm.
Figure 3.
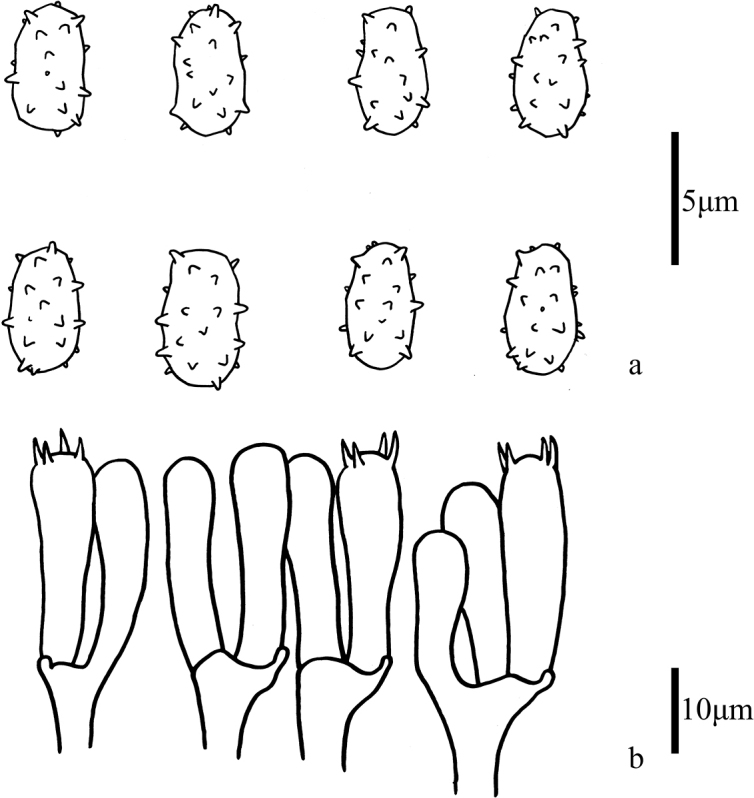
Microscopic features of Trechisporakhokpasiensis (MHHNU7529) a basidiospores b basidia.
Figure 4.
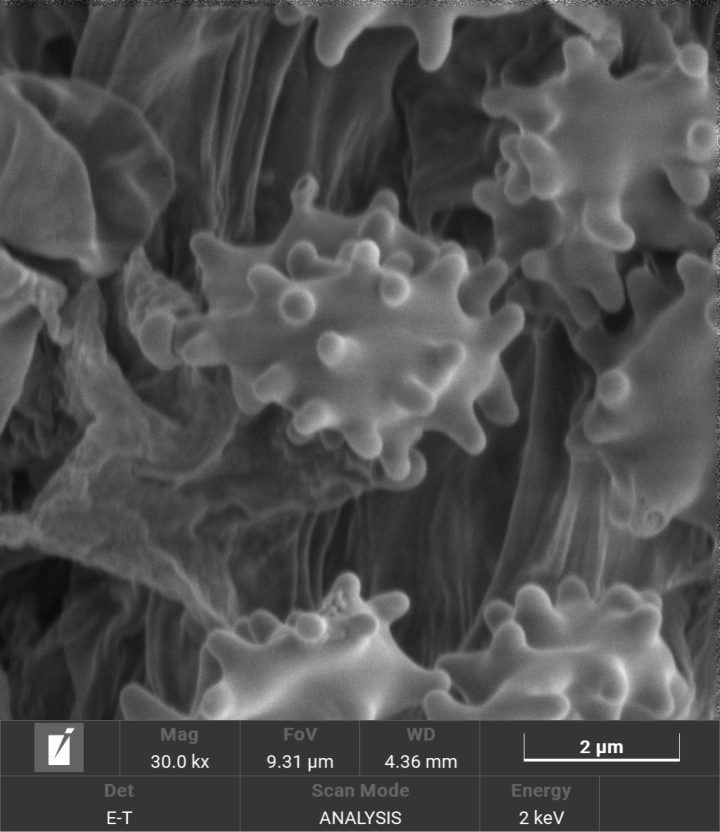
Scanning electron micrograph of basidiospores of Trechisporakhokpasiensis (MHHNU7529).
Basidiomata.
Clavarioid, scattered or fascicled, 25–30 mm tall, 15–36 mm broad, chalk white (1A1), slightly yellow (2A2) with age, apices white (1A1), yellowish white (3A4) when dry. Stipe single, short and flattened, 10–15 × 3–4 mm, white. Branches flattened or palmate, palmately branched from flattened stipe, dense, 4–7 mm wide, polychotomous below, dichotomous towards apices, internodes becoming gradually longer, branches 6–8 mm diam, divided 3–5 times, apices cristate or flattened, blunt, axils V-shaped. Flesh white to pale yellow, waxy. Taste and odor unrecorded.
Micromorphology.
Generative hyphae septate, clamped, interwoven, smooth, thin-walled, hyaline; tramal hyphae parallel arranged, 2–4 μm wide, smooth, thin-walled, hyaline. Subhymenial hyphae branched and wide, 3–8 μm; ampulliform septa present in the hyphae, 5−6 µm wide. Basidia: approximately 22–28 × 5.5–8 µm with four sterigmata 3–4.5 µm long, hyaline, subcylindrical to clavate, slight constriction, clamp connection in base. Cystidia absent. Basidiospores [40/4/3] 5–6 (–6.5) × 3–4 μm [Q = 1.33–1.72(1.83), Qm = 1.57 ± 0.16], ellipsoid, angular, finely verruculose or echinulate, hyaline, thin-walled, spines 0.5–1 μm long, apex slightly blunt; hilar appendage extremely small, obscured by spore ornamentation, inamyloid, contents usually uniguttulate.
Habit and distribution.
Solitary to caespitose, grows on humus in broadleaf forest or grows on soil; basidiomata generally occur in summer. Known from Thailand (Sommai et al. 2023), Laos and China.
Notes.
Trechisporakhokpasiensis is mainly characterized by chalk-white basidiomata and flattened branches. Trechisporapallescens (Bres.) Singer is easily mistaken for T.khokpasiensis in the field on account of its similar size, shape, and color. However, the two species occur in different habitats: T.khokpasiensis grows in the humus layer on soil without any plant root association. Trechisporachartacea (Pat.) Gibertoni also has flattened, narrowly spathulate branches, grayish white in age, axils U-shaped, arising from scarce white mycelia on the soil. However, T.khokpasiensis differs in that the axils are V-shaped, the basidiomata are pale yellow with age, and it grows on dead branches and leaves. Trechisporacaulocystidiata is distinguished from T.khokpasiensis by having subglobose basidiospores and possessing caulocystidia.
In the present phylogenetic analyses, Scytinopogoncryptomerioides was close to T.khokpasiensis, but the two species differ in that T.khokpasiensis has relatively smaller basidia (22–28 × 6–8 μm vs. 35–42 × 5.5–6 μm in S.cryptomerioides). Trechisporacopiosa has similar branches to T.khokpasiensis, but in T.copiosa the branches are moderately open and the basidia are primarily 2–4-spored.
. Trechispora laxa
P. Zhang & P.T. Deng sp. nov.
79637F69-608E-585D-AC85-834FC1A4715F
MycoBank No: 846842
Figure 5.
Basidiomata of Trechisporalaxa (MHHNU10714). Scale bars: 1 cm.
Figure 6.
Microscopic features of Trechisporalaxa (MHHNU10714) a basidiospores b basidia.
Figure 7.
Scanning electron micrograph of basidiospores of Trechisporalaxa (MHHNU10714).
Diagnosis.
Differs from Trechisporahavencampii by the loose branches and 4-spored basidia.
Type.
China, Hainan Province, Baoting County, Qixianling Hot Springs National Forest Park, 18°70′24″N, 109°69′35″E, 300 m asl, 31 July 2021, leg. P. Zhang (holotype MHHNU10714).
Etymology.
laxus (Latin), loose, referring to the loose branching.
Basidiomata.
Solitary or scattered, fleshy consistency, 45–55 mm tall, 30–35 mm broad, fresh color (8B6–7), apices white when young but turning grayish purple (14B6) with age, drying pale grayish beige (4C3). Stipe single, white (1A1), 10–15 mm tall. Branches polychotomous from the stipe, dichotomous towards apices, not flattened, loose, divided 3–5 times, terminal branches relatively short and with color transitions to lilac, apices pale purple or white, acute, axils U-shaped. Taste and odor not recorded.
Micromorphology.
Context with parallel arranged hyphae, 2–4 μm wide; generative hyphae clamped, smooth, thin-walled, hyaline, no calcium oxalate crystals. Subhymenial hyphae branched and wide, 3–8 μm; ampullate septa present at the base of the stipe, up to 6–8 μm wide. Basidia 20–26 × 7–9 µm with four sterigmata 4–5 µm long and a basal clamp connection, hyaline, subclavate, barrel-shaped. Cystidia absent. Basidiospores [40/3/2] 5–6 × 3–4 μm [Q = 1.25–1.71(1.83), Qm = 1.46 ± 0.16], ellipsoid, slightly irregular, inner side slightly concave, aculeate or finely verrucose, spines 1–1.5 μm long, apex not sharp but blunt; hilar appendage obscured by spore ornamentation; usually uniguttulate; hyaline, thin-walled, inamyloid.
Habit and distribution.
Solitary or scattered, grows in soil in broadleaf forest; basidiomata generally occur in summer. Known only from the type locality in China.
Notes.
The branches of T.laxa are scattered, not dense, and the apices are white or lilac gray with age. In the field, T.laxa and Trechisporahavencampii are similar because of their pale grayish brown coloration, but T.havencampii has dense branches, tips white and axils V-shaped, and 2-spored basidia with long sterigmata (5–9.5µm). Trechisporalongiramosa differs from T.laxa by having long terminal branches, densely branched and white to honey-yellow tips. Trechisporasanpapaoensis has smaller basidia (11–26 × 5.5–11.0 μm) and a grayish brown stipe. Trechisporatermitophila develops abundant basidiomata in active termite nests, but T.laxa generally grows in the soil of broadleaf forest. Trechisporafoetida has a reddish brown to deep brown, flattened stipe, and branching in one plane. In comparison, T.laxa is flesh colored, turning grayish purple in the terminal branches, the branches are not flattened, the branches are not white, and the stipe is non-flat. An additional pigmented species, Trechisporarobusta described from Brazil, is pale grayish with internodes irregular, branches flattened to subcylindrical, and inflated hyphae.
. Trechispora tongdaoensis
P. Zhang & P.T. Deng sp. nov.
DFDF06BA-CB3D-5783-B33D-EC0E19B42106
MycoBank No: 846849
Figure 8.
Basidiomata of Trechisporatongdaoensis (MHHNU11083). Scale bars: 1 cm.
Figure 9.
Microscopic features of Trechisporatongdaoensis (MHHNU11083) a Basidiospores b Basidia.
Figure 10.
Scanning electron micrograph of basidiospores of Trechisporatongdaoensis (MHHNU11083).
Diagnosis.
Differs from Trechisporatermitophila by the white fruiting body and 4-spored basidia.
Type.
China, Hunan Province, Tongdao County, WanFo Mountain Nature Reserve, 26°32′54″N, 109°86′95″E, 523 m asl., 6 July 2022, leg. P. Zhang (holotype MHHNU11083).
Etymology.
tongdaoensis (Latin), referring to the currently known distribution of the species in Tongdao County, Hunan Province, China.
Basidiomata.
Clavarioid, gregarious to caespitose clusters, 60–90 mm tall, 30–45 mm broad, white (1A1), with pale yellow (1A3). Stipe single, 20–40 mm long, white (1A1), no change in color when dried. Branches dense, branching from the base, repeatedly dichotomous towards apices, divided 3–5 times, branches slender, 2–3 mm wide, internodes becoming gradually longer, terminal branches long and not flat, sometimes split at the tips, acute, axils V-shaped, terminal branches short, tips acute. Context pale yellow. Taste and odor not recorded.
Micromorphology.
Context hyphae compact, 3–5.5 μm wide, subparallel arranged, cylindric; generative hyphae with clamp connections but not at every septum, thin-walled, smooth, hyaline, no calcium oxalate crystals; ampullate septa present in the hyphae of the stipe, 7–8 µm wide. Basidia: approximately 18–28 × 6–8 µm with four sterigmata 3–5 µm long, hyaline, subclavate to cylindrical, with clamp connection in base. Cystidia absent. Basidiospores [40/3/2] 4–6(6.5) × 3–5.5 μm [Q = 1.25–1.57(1.83), Qm = 1.50 ± 0.11] ellipsoid, slightly angular, tuberculate or coarsely echinulate, spines 0.5–1 μm long, blunt; hilar appendage ambiguous by spore ornamentation, sometimes contents uniguttulate, inamyloid.
Habit and distribution.
Caespitose or gregarious on the soil of broadleaf forests; basidiomata generally occur in summer. Known only from the type locality in China.
Notes.
The fruiting body of T.tongdaoensis has a long stalk, 20–40 × 4–6 mm, the terminal branches are long, bifurcate, the tips are white, and the branches are not flattened in a plane. Trechisporafoetida differs from T.tongdaoensis by having reddish brown to deep brown basidiomata, and flattened branches and stipe. Trechisporacaulocystidiata has cystidia in the stipitipellis as caulocystidial hairs. This feature is obvious under a microscope, but we failed to observe this structure in T.tongdaoensis. In addition, the T.caulocystidiata stipe is relatively shorter (10–20 × 3–5 mm vs. 20–40 × 4–6 mm in T.tongdaoensis). Trechisporacopiosa differs from T.tongdaoensis in having acute or flattened branch tips and T.tongdaoensis has relatively smaller spores (4–6 (–6.5) × 3–5.5 μm vs. (5–) 5.5–6.5 (–7) × (3–) 3.5–4 (–4.5) μm in T.copiosa). Trechisporadealbata was classified in Ramariopsis (Donk) Corner based on the gelatinous context by Petersen (1984, 1988), whereas T.tongdaoensis lacks a gelatinous context and differs in having relatively smaller spores (4.0–4.5 × 2.5–3.5 µm vs. 4–6(6.5) × 3–5.5 μm in T.tongdaoensis). In the field, T.gelatinosa has a fleshy or gelatinous consistency, translucent when fresh, small basidia (12–26 × 5–6.5 μm), and small basidiospores (3–) 3.2–4.5 (–5) × (2–) 2.5–3.5 (–4) μm, which distinguishes the species from T.tongdaoensis. In the present phylogenetic analyses, T.tongdaoensis clustered with pigmented species.
Discussion
Trechispora has until recently been considered to encompass a variety of morphological characteristics and broad diversity in hymenophore structure ranging from smooth, grandinioid to odontioid, hydnoid, or poroid, but always resupinate basidiomata. Scytinopogon is characterized by having clavarioid basidiomata and flattened branches, and the basidiomata are mostly white in color with a tough texture. Over time, additional pigmented species of Scytinopogon have been described and some species have rounded instead of flattened branches. A relationship between the flattened coralloid Scytinopogon and the resupinate Trechispora was first suggested by Jülich (1981), who placed the genera in separate families of the order Trechisporales, Hydnodontales. Subsequently, Meiras-Ottoni et al. (2021) analyzed a number of sequences from specimens of each genus, including the type species of each genus, and concluded that Scytinopogon is a synonym of Trechispora. In phylogenetic reconstructions, several transitions between fruiting body types are indicated to have occurred (Meiras-Ottoni et al. 2021). In addition, the color of Trechispora species has changed many times over the course of evolution, from white to pigmented, but the evolutionary trend for the basidiomata is not understood. The transition from resupinate basidiomata to clavarioid basidiomata seems to represent an evolutionary trend. The effect of this transformation is presumably an increase to expand the surface area of the hymenophore. The driving force behind morphological differences in fruiting bodies seems likely to be associated with selection for efficient spore dispersal (Hibbett and Binder 2002). In micromorphology, variation in basidiospores (smooth or ornamented) and basidia (2 or 4 sterigmata) represents different evolutionary directions, but the effects of these differences remain unknown. Most species of Trechispora are distributed in the tropics or subtropics, including coralloid species. Indeed, the two new species described in this study were collected in the subtropics of southern China. However, the current distribution of stipitate and clavarioid species does not include temperate regions. To comprehend the diverse factors contributing to the evolution in Trechispora, a number of specific aspects should be considered: substrate, nutritional mode, and environmental conditions. The ITS region is highly variable among Trechispora species and cannot be aligned reliably within the genus (Meiras-Ottoni et al. 2021), such that other genetic markers (e.g., LSU, SSU, ef1, rpb1, and rpb2) should be used to for phylogenetic analyses in Trechispora.
In China, previous studies have reported one new coralloid species of Trechsipora (T.longiramosa; Liu et al. 2022) and three species of Scytinopogon (S.cryptomerioides, S.echinosporus (Berk. and Broome) Corner and S.pallescens; Zhang and Yang 2003, Lin et al. 2022). The present phylogenetic analysis confirmed that S.cryptomerioides was firmly nested within Trechispora. Together with the previous studies mentioned here, this finding highlights that further exploration for coralloid species of Trechispora is needed. The present study expands our understanding of clavarioid species of Trechispora by providing descriptions and illustrations for two new species. The findings enrich our knowledge of the distribution of coralloid Trechispora species in China and the overall diversity of Trechispora.
Key to coralloid Trechispora species in China
| 1 | Basidiomata pure white to pale yellow | 2 |
| – | Basidiomata grayish brown to pale purple | 4 |
| 2 | Basidiomata with no flattened branches | T . tongdaoensis |
| – | Basidiomata with flattened branches | 3 |
| 3 | Basidiomata only grow in soil | T.pallescens |
| – | Basidiomata grow in the humus layer on soil | T . khokpasiensis |
| 4 | Basidiomata with dense branches and long terminal branches | T.longiramosa |
| – | Basidiomata with loose branches | T . laxa |
Supplementary Material
Acknowledgements
We thank Robert McKenzie, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the English text of a draft of this manuscript.
Citation
Deng P-T, Yan J, Liu X-F, He Z-M, Lin Y, Lu M-X, Zhang P (2023) Three coralloid species of the genus Trechispora (Trechisporales, Basidiomycota) in China: two newly discovered taxa and one reported for the first time. MycoKeys 99: 153–170. https://doi.org/10.3897/mycokeys.99.109375
Funding Statement
This study was financially supported by Key Research and Development Program of Hunan Province (No. 2020SK2103)
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This study was financially supported by the Key Research and Development Program of Hunan Province (No. 2020SK2103)
Author contributions
Conceptualization: Ping Zhang; methodology: Peng-Tao Deng and Jun Yan; performing the experiment: Peng-Tao Deng, Xiang-Fen Liu; resources: Ping Zhang, Peng-Tao Deng, Jun Yan, Ming-Xin Lu and Yuan Lin; writing – original draft preparation: Peng-Tao Deng; writing – review and editing: Ping Zhang and Zheng-Mi He; supervision: Ping Zhang; project administration: Ping Zhang; funding acquisition: Ping Zhang. All authors have read and agreed to the published version of the manuscript.
Author ORCIDs
Peng-Tao Deng https://orcid.org/0000-0002-8755-7965
Jun Yan https://orcid.org/0000-0002-2832-8046
Zheng-Mi He https://orcid.org/0000-0001-8754-3427
Ping Zhang https://orcid.org/0000-0002-8751-704X
Data availability
The sequence data generated in this study are deposited in NCBI GenBank.
Supplementary materials
Multiple sequence alignment
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Peng-Tao Deng, Jun Yan, Xiang-Fen Liu, Zheng-Mi He, Yuan Lin, Ming-Xin Lu, Ping Zhang
Data type
fasta file
References
- Bernicchia A, Gorjón SP. (2010) Fungi Europaei 12. Corticiaceae s.l. Edizioni Candusso, Italia.
- Birkebak JM, Mayor JR, Ryberg KM, Matheny PB. (2013) A systematic, morphological and ecological overview of the Clavariaceae (Agaricales). Mycologia 105(4): 896–911. 10.3852/12-070 [DOI] [PubMed] [Google Scholar]
- Chikowski RS, Larsson KH, Gibertoni TB. (2020) Taxonomic novelties in Trechispora (Trechisporales, Basidiomycota) from Brazil. Mycological Progress 19(12): 1403–1414. 10.1007/s11557-020-01635-y [DOI] [Google Scholar]
- Corner EJH. (1970) Supplement to “A monograph of Clavaria and allied genera”. Beihefte zur Nova Hedwigia 33: 87–92. [Google Scholar]
- Desjardin DE, Perry BA. (2015) A new species of Scytinopogon from the island of Príncipe, Republic of São Tomé and Príncipe, West Africa. Mycosphere: Journal of Fungal Biology 6(4): 433–440. 10.5943/mycosphere/6/4/5 [DOI] [Google Scholar]
- Donk MA. (1964) A conspectus of the families of Aphyllophoralles. Persoonia 3: 199–324. [Google Scholar]
- Furtado ANM, Daniëls PP, Reck MA, Neves MA. (2021) Scytinopogoncaulocystidiatus and S.foetidus spp. nov., and five other species recorded from Brazil. Mycotaxon 136(1): 107–130. 10.5248/136.107 [DOI] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology 2(2): 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M. (2002) Evolution of complex fruiting-body morphologies in homobasidiomycetes. Proceedings of the Royal Society B, Biological Sciences 269(1504): 1963–1969. 10.1098/rspb.2002.2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Lumbsch T, Lutzoni F, Matheny PB, Mclaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde K, Ironside JE, Kõljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miądlikowska J, Miller A, Moncalvo JM, Mozley Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüßler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiß M, White MM, Winka K, Yao YJ, Zhang N. (2007) A higher-level phylogenetic classification of the Fungi. Mycological Research 111(5): 509–547. 10.1016/j.mycres.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Jülich W. (1981) Higher taxa of Basidiomycetes. Bibliotheca Mycologica 85: 1–485. [Google Scholar]
- Karsten PA. (1890) Fragmenta mycologica XXIX. Hedwigia 29: 147–149. [Google Scholar]
- Katoh K, Standley DM. (2016) A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics (Oxford, England) 32(13): 1933–1942. 10.1093/bioinformatics/btw108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. (1978) Methuen handbook of colour, 3rd edn. Methuen, London.
- Larsson KH. (1994) Poroid species in Trechispora and the use of calcium oxalate crystals for species identification. Mycological Research 98(10): 1153–1172. 10.1016/S0953-7562(09)80200-1 [DOI] [Google Scholar]
- Larsson KH. (2001) The position of Poria mucida inferred from nuclear ribosomal DNA sequences. Harvard Papers in Botany 6: 131–138. [Google Scholar]
- Larsson KH. (2007) Rethinking the classification of corticioid fungi. Mycological Research 111(9): 1040–1063. 10.1016/j.mycres.2007.08.001 [DOI] [PubMed] [Google Scholar]
- Larsson KH, Larsson E, Kõljalg U. (2004) High phylogenetic diversity among corticioid homobasidiomycetes. Mycological Research 108(9): 983–1002. 10.1017/S0953756204000851 [DOI] [PubMed] [Google Scholar]
- Lin WR, Wang PH, Hsieh SY. (2022) Scytinopogoncryptomerioides (Hydnodontaceae), a new species from Taiwan. Phytotaxa 552(1): 78. 10.11646/phytotaxa.552.1.6 [DOI] [Google Scholar]
- Liu WH, Yan J, Deng PT, Qin WQ, Zhang P. (2022) Two new species of Phaeoclavulina (Gomphaceae, Gomphales) from Hunan Province, China. Phytotaxa 561(1): 3. 10.11646/phytotaxa.561.1.3 [DOI] [Google Scholar]
- Liu SL, He SH, Wang XW, May Tw, He G, Chen SL, Zhou LW. (2022) Trechisporales emended with a segregation of Sistotremastrales ord. nov. (Basidiomycota). Mycosphere 13(1): 862–954. 10.5943/mycosphere/13/1/11 [DOI] [Google Scholar]
- Maas Geesteranus RA. (1962) Hyphal structures in Hydnums. Persoonia 2(3): 377–405. [Google Scholar]
- Meiras-Ottoni A, Larsson KH, Gibertoni TB. (2021) Additions to Trechispora and the status of Scytinopogon (Trechisporales, Basidiomycota). Mycological Progress 20(2): 203–222. 10.1007/s11557-021-01667-y [DOI] [Google Scholar]
- Ordynets A, Larsson Kh, Langer E. (2015) Two new Trechispora species from La Réunion Island. Mycological Progress 14(11): 1–11. 10.1007/s11557-015-1133-0 [DOI] [Google Scholar]
- Petersen RH. (1984) Type studies in the clavarioid fungi. VIII. Persoonia 12(3): 225–237. [Google Scholar]
- Petersen RH. (1988) Notes on clavarioid fungi. XXII. Three interesting South American collections. Mycologia 80(4): 571–576. 10.1080/00275514.1988.12025581 [DOI] [Google Scholar]
- Rambaut A. (2012) FigTree v1.4. University of Edinburgh, Edinburgh, UK.
- Ridgway R. (1912) Color standards and color nomenclature. Published by the author, Washington. 10.5962/bhl.title.144788 [DOI]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Singer (1945) New genera of fungi II. Lloydia 8: 139–144. [Google Scholar]
- Sommai S, Pinruan U, Khamsuntorn P, Lueangjaroenkit P, Somrithipol S, Luangsaard J. (2023) Three new species of Trechispora from Northern and Northeastern Thailand. Mycological Progress 22(6): 42. 10.1007/s11557-023-01886-5 [DOI] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: Maximum likelihoodbased phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England) 22(21): 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. (2005) RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics (Oxford, England) 21(4): 456–463. 10.1093/bioinformatics/bti191 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the raxml web servers. Systematic Biology 57(5): 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Telleria MT, Melo I, Dueñas M, Larsson KH, Paz Martín MP. (2013) Molecular analyses confirm Brevicellicium in Trechisporales. IMA Fungus 4(1): 21–28. 10.5598/imafungus.2013.04.01.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) Sequencematrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27(2): 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols, a guide to methods and applications. Academic Press, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Yuan HS, Lu X, Dai YC, Hyde KD, Kan YH, Kušan I, He SH, Liu NG, Sarma VV, Zhao CL, Cui BK, Yousaf N, Sun G, Liu SY, Wu F, Lin CG, Dayarathne MC, Gibertoni TB, Conceição LB, Garibay-Orijel R, Villegas-Ríos M, Salas-Lizana R, Wei TZ, Qiu JZ, Yu ZF, Phookamsak R, Zeng M, Paloi S, Bao DF, Abeywickrama PD, Wei DP, Yang J, Manawasinghe IS, Harishchandra D, Brahmanage RS, de Silva NI, Tennakoon DS, Karunarathna A, Gafforov Y, Pem D, Zhang SN, de Azevedo SALCM, Bezerra JDP, Dima B, Acharya K, Alvarez-Manjarrez J, Bahkali AH, Bhatt VK, Brandrud TE, Bulgakov TS, Camporesi E, Cao T, Chen YX, Chen Y, Devadatha B, Elgorban AM, Fan LF, Du X, Gao L, Gonçalves CM, Gusmão LFP, Huanraluek N, Jadan M, Jayawardena RS, Khalid AN, Langer E, Lima DX, Lima-Júnior NC, Lira CRS, Liu JKJ, Liu S, Lumyong S, Luo ZL, Matočec N, Niranjan M, Oliveira-Filho JRC, Papp V, Pérez-Pazos E, Phillips AJL, Qiu PL, Ren Y, Ruiz RFC, Semwal KC, Soop K, de Souza CAF, Souza-Motta CM, Sun LH, Xie ML, Yao YJ, Zhao Q, Zhou LW. (2020) Fungal diversity notes 1277–1386: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 104(1): 1–266. 10.1007/s13225-020-00461-7 [DOI] [Google Scholar]
- Zhang P, Yang ZL. (2003) Scytinopogon, a genus of clavarioid basidiomycetes new to China. Mycosystema 22(4): 663–665. 10.13346/j.mycosyste ma.2003.04.027 [DOI] [Google Scholar]
- Zhao W, Zhao CL. (2021) The phylogenetic relationship revealed three new wood-inhabiting fungal species from genus Trechispora. Frontiers in Microbiology 12: 650195. 10.3389/fmicb.2021.650195 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Peng-Tao Deng, Jun Yan, Xiang-Fen Liu, Zheng-Mi He, Yuan Lin, Ming-Xin Lu, Ping Zhang
Data type
fasta file
Data Availability Statement
The sequence data generated in this study are deposited in NCBI GenBank.