Abstract
Risk assessment assays for chemically induced arrhythmia are critical, but significant limitations exist with current cardiotoxicity testing, including a focus on single select ion channels, the use of non-human species in vitro and in vivo, and limited direct physiological translation. To be predictive of actual adverse clinical arrhythmic risk, arrhythmia assessment models for chemicals and drugs should be fit-for-purpose and suited for evaluating compounds in which the mechanism of action may not be entirely known. Here, we describe methods for efficient and reliable screening for arrhythmogenic cardiotoxicity with a 3D human cardiac microtissue model using purified human-induced pluripotent stem cell (hiPSC)-derived cardiomyocytes and human cardiac fibroblasts. Applying optical mapping of voltage and calcium-sensitive dyes—an established approach to evaluate cardiac action potentials and calcium transients—to 3D heterotypic cardiac myocyte–fibroblast tissues allows for the generation and functional analysis of a large number of individual microtissues to provide greater throughput and high statistical power in analyses. Hundreds of microtissues in standard cell culture plates can be produced with low variability beat-to-beat, microtissue-to-microtissue, and across hiPSC-cardiomyocyte differentiation batches, reducing the number of microtissues required per condition for predictive outputs. The platform described here can be used as a sensitive, efficient, and predictive preclinical model validated for the purpose of assessing human pro-arrhythmic risk.
Keywords: Microtissues, Arrhythmia, Cardiac fibroblast, Early afterdepolarization, Action potential, Calcium transient, Optical mapping, Spheroids
1. Introduction
Cardiotoxicity is the occurrence of electrophysiological disturbance or muscle damage in the heart, and arrhythmia, which increases the risk for stroke, heart attack, heart failure, and sudden cardiac death, is the leading manifestation of chemical toxicity [1]. However, the causal relationship between specific chemicals and cardiotoxicity is not well understood. Cardiotoxic effects can be induced by industrial chemicals, environmental toxicants, and pharmaceutical drugs. Predictive assays for chemically induced arrhythmia are critical to protecting human cardiac safety, but significant limitations exist with current cardiotoxicity testing using existing in vitro models and animal models. Human ether-go-go (HERG) channel blockade and QTc prolongation have long been used as effective and selective “biomarkers” in compound screening to identify pro-arrhythmic risk that cause Torsades de Pointes (TdP) [2, 3]. It is now widely recognized this approach does not capture all drug-induced cardiac arrhythmia mechanisms [4–8]. In vitro cell-based models are often limited to two-dimensional monolayers and lack non-cardiomyocytes, which affect the arrhythmogenic phenotype [9]. Few 3D microtissue systems are used for thorough arrhythmia assessment, preferring to focus on cellular toxicity such as through live/dead, mitochondrial, and ER imaging [10] or contractile amplitude and kinetics from microtissue edge detection or force generation [11, 12]. 3D models aimed at detecting arrhythmia risk often have limited throughput [13, 14] and focus on metrics such as beating frequency [15] or conduction velocity [16]. Animal models are complex and have limited predictability of human biological responses due to species-specific differences in ion channel content and expression that effect depolarization and repolarization kinetics of cardiac action potentials (APs) and differential sensitivity to chemical agents [17, 18]. These methods describe an efficient and reliable screening for arrhythmogenic cardiotoxicity with a 3D human cardiac microtissue model using purified hiPSC-derived cardiomyocytes and human cardiac fibroblasts.
Our 3D heterotypic cardiac myocyte–fibroblast tissue platform for arrhythmia assessment described in this chapter allows for the generation and functional analysis of a large number of individual but consistent microtissues to provide greater throughput and high statistical power in analyses. Self-assembly in commercially available molds produces organotypic heterocellular interspersion and eliminates unnatural substrate or matrix stiffness. The 3D microtissue environment enables formation of many contact sites between cells due to nonplanar distribution of gap junctional proteins in hiPSC-CMs, facilitating electrical coupling between neighboring cells to increase fidelity of arrhythmia assessment compared to 2D monolayer culture. We incorporate human cardiac fibroblasts (hCFs) to enable heterotypic cell–cell interactions characteristic of the intact myocardium [19–21] and employ culturing techniques like metabolic selection to purify and mature the hiPSC-CMs [22]. In our experiments, we use 5% primary adult normal hCF content for stable, healthy cardiac electromechanical function based on our previous work [23], yet the platform allows for alterations in cell composition and ratios to mimic physiological and pathophysiological conditions. This platform can produce hundreds of microtissues in standard cell culture plates with low variability beat-to-beat, microtissue-to-microtissue, and across hiPSC-CM differentiation batches, reducing the number of microtissues required per condition for predictive outputs [24].
To be predictive of actual adverse clinical arrhythmic risk, arrhythmia models for chemicals and drugs should evaluate both trigger and substrate mechanisms for reentry. It is generally thought that cardiac arrhythmias start from premature ventricular contractions (PVCs) due to automaticity or triggered activity such as delayed (DADs) or early afterdepolarizations (EADs), that must occur in a localized tissue mass in order to propagate and form reentry to cause ventricular tachycardia and fibrillation [25]. Therefore, successful proarrhythmic drug screening should be able to detect changes in action potential shape associated with increased triggered activity of a small mass of tissue (not just single or a few cells), which may lead to arrhythmia initiation and propagation. In this 3D microtissue platform, visual detection is used with high temporal and spatial resolution for signal extraction and automated, unbiased quantification to define electrophysiological responses to chemicals. 3D microtissues are mounted on a temperature regulated chamber and loaded with reactive fluorescence dyes (e.g., voltage- or calcium-sensitive dyes), electrically stimulated with platinum electrodes to evoke action potentials and recorded with a high-speed camera [19]. Our analysis of cardiotoxicity as presented herein generates measurements of cardiac waveforms, including excitation and calcium, with the potential for expanding to include contraction. Measurements can include action potential excitability, stimulation delay time (ms), rise time (ms), action potential duration (APD) to 30%, 50%, 80%, 90%, and maximal relaxation (APD30, APD50, APD80, APD90, APDMxR, respectively), presence of EADs or DADs [24], or similar metrics from the calcium transient. Multiple doses of test compounds can be perfused to investigate changes in action potential or calcium transient metrics induced by the test compounds. Tissues can be exposed to compounds acutely (short-term exposure, typically minutes to hours) or chronically (long-term exposure, typically days). The platform described here can thus be used as a robust, predictive, and efficient preclinical model of human proarrhythmic risk.
2. Materials
2.1. Cardiomyocyte Differentiation
6-, 12-, or 24-well culture plates.
Matrigel.
Human-induced pluripotent stem cell line (hiPSC).
CDM3 basal media.
6 μM Chiron 99021.
5 μM IWP2.
RPMI 1640 medium with B27 supplement (RPMI/B27).
Lactate medium: DMEM without glucose, L-glutamine, phenol red, sodium pyruvate and sodium bicarbonate; 4 mM L-glutamine; 1× Non-Essential Amino Acids; 1× Glutamax; 4 mM lactate; pH 7.4.
Penicillin/streptomycin (p/s).
100 mm culture dish.
Human cardiac fibroblasts (hCFs).
hCF media: DMEM/F12; 10% fetal bovine serum (FBS); 1% p/s; 4 ng/mL basic fibroblast growth factor.
2.2. Fabrication of Hydrogels and 3D Culture
2% (weight/volume) ultrapure agarose in 1× phosphate-buffered saline.
Microwave or hot plate.
3D Petri Dish®.
Microtissue media: RPMI/B27; 10% FBS; 1% p/s.
Field culture stimulator, i.e., C-Pace EP (IonOptix).
2.3. Optical Mapping and Action Potential Analysis
Olympus Macroview system with filter set for di-4 ANEPPS.
Photometrics Evolve128+ EM-CCD camera (see Note 1).
Dual Automatic Temperature Controller TC-344B, Warner Instrument to maintain 35 ± 1 °C.
Motorized syringe pump.
Thorlabs motorized stage for automated repetitive recordings (see Note 2).
5 μM di-4-ANEPPS.
Thorlabs Solis LED light source.
Platinum electrode.
Ionoptix BioPacer field stimulator.
Tyrode solution of (in mM) 140 NaCl, 5.1 KCl, 1 MgCl2, 1 CaCl2, 0.33 NaH2PO4, 5 HEPES, and 7.5 glucose.
No. 1.5 glass coverslip.
3. Methods
3.1. Cardiomyocyte Differentiation and Cardiac Fibroblast Maintenance
Culture human-induced pluripotent stem cells (hiPSCs) in a high-density monolayer using CDM3 basal medium on plates coated with Matrigel.
Treat hiPSCs with 6 μM Chiron 99,021 (Tocris), a glycogen synthase kinase 3 (GSK3) inhibitor at day 1, followed by 5 μM IWP2 (Tocris), a chemical Wnt inhibitor at day 3. Cardiac phenotype, expressed by beating cells, is usually visible between days 8 and 12.
Harvest and replate hiPSC-CM to new culture plates coated with Matrigel in RPMI 1640 medium with B27 supplement.
Deprive these cells of media change for 4 days and then feed with lactate medium for 4 days, refreshing medium every 2 days. After lactate purification, feed cells with RPMI +B27 + 1% penicillin/streptomycin until harvested for microtissues assembly.
Maintain commercially available human cardiac fibroblasts in hCF media (see Note 3).
Pass cells at a 1:4 ratio and incorporate into cardiac microtissues at cell passages P2-P4.
3.2. Fabrication of Hydrogels and 3D Culture
Heat 2% agarose with microwave or hot plate to boiling.
Pipet molten 2% agarose into 3D Petri dish mold (i.e., molds for 24-well plates with 800-μm-diameter rounded pegs).
Cool to room temperature (~5 min), separate agarose gel from the mold, and transfer to 24-well plate.
Equilibrate in 1 mL microtissue media for 24 h.
Harvest hiPSC-CM and hCF. Prepare cell seeding solution of 400,000–800,000 hiPSCs with 5% hCFs in 100 μL of microtissue media (per well) (see Note 4).
Remove equilibration media from molds including media in the center cell seeding chamber. Pipet 100 μL of cell seeding solution into the cell seeding chamber of each mold (Fig. 1a).
Allow cells to settle into recesses for 15–30 min. Add media to the well, outside of the 3D Petri dish to cover (1 mL for 24-well plate, 2–3 mL for 12-well plate).
Culture for 6–8 days with 1 Hz, 10.0 V, 4.0 ms duration bipolar pulse train field stimulation. (Fig. 1b, c, see Note 5).
Fig. 1.
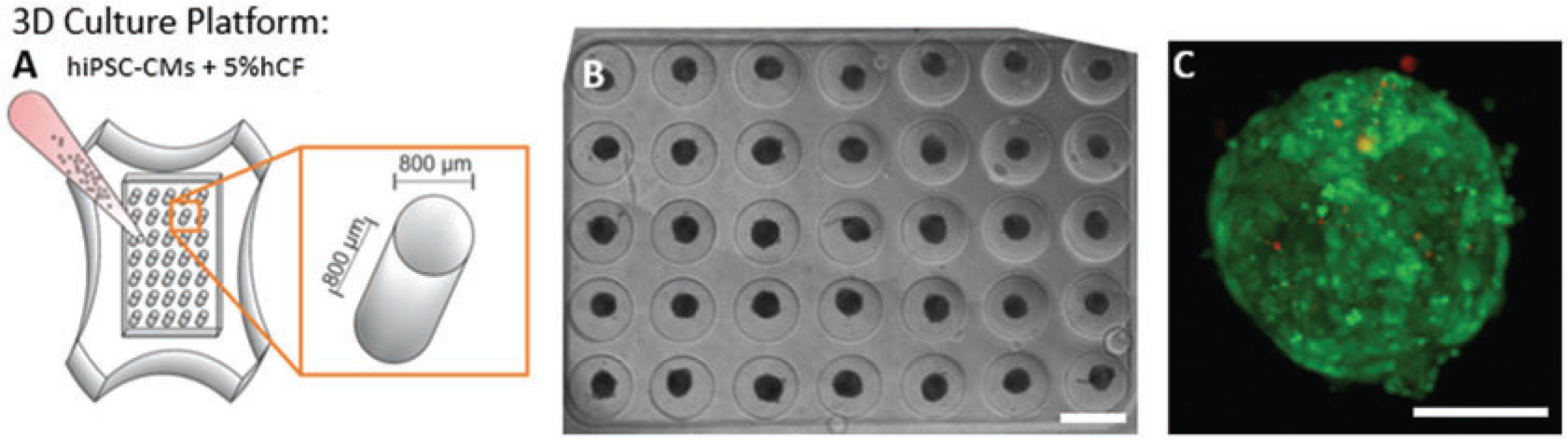
Formation of cardiac microtissues. (a) Schematic of three-dimensional (3D) cardiac microtissue generation shows non-adhesive agarose gels with cylindrical recesses with hemispherical bottoms that guide self-assembly. (b) Phase contrast image shows consistent spherical microtissue formation after 5 days of 3D culture in all 35 microwells. Cardiac microtissues were cultured for 6–8 days with 1 Hz pacing. Scale bar, 800 μm. (c) Compressed confocal z-stack image shows a representative cardiac tissue with hiPSC-CM (green) and hCF (red) stained with CellTracker dyes with a low number of interspersed hCFs. Scale bar, 100 μm
3.3. Optical Mapping and Automated Action Potential Analysis
Setup for optical mapping is shown in Fig. 2a.
Set temperature control to 35 °C, and set constant perfusion flow (0.1–1 mL/min).
Transfer 3D Petri dish to temperature regulated chamber on motorized stage (Fig. 2b).
Place stimulation electrode, perfusion, and suction tubing in solution with 3D Petri dish. Two leads of platinum electrode should be 1 or 2 cm apart for easy calculation of stimulation strength in V/cm.
To reduce water vibration noise, float a glass coverslip on top of the solution in the well.
Load microtissues with voltage-sensitive di-4-ANEPPS in Tyrode solution for 10 min at 35 °C for measurements of membrane potential (Vm) and wash out with Tyrode solution for 10 min.
Set up the stimulation protocol using 2 ms biphasic pulse with 10–15 V/cm strength, 2 s interval (0.5 Hz).
Adjust field of view and focus of the image. The locations of the four corners of the mold are registered from the data acquisition software to automate sequential recording of action potentials from all 35 microtissues.
Acquire fluorescence images at 979 frames/s using Photometrics Evolve +128 EM-CCD camera (2 × 2 binning to 64 × 64 pixels, 18.7 × 18.7-μm2 resolution, 1.2 × 1.2-mm2 field of view). Record four microtissues simultaneously per scan at this magnification (see Note 6). Record action potentials from all of the microtissues in a single mold (35 microtissues per mold). Filter fluorescence images using nonlinear bilateral filter (spatial filter: 5 × 5 window, temporal filter: 21-point window) to preserve AP upstrokes from blurring (Fig. 2c).
Check how many microtissues show action potentials evoked by electrical stimulation. For quality control, ensure more than 70% of microtissues show action potentials under 2 s pacing (> 25/35 microtissues, typically excitability of 95% is an indication of a good preparation).
After baseline recording of all microtissues in this mold, change perfusion solution to include drug or chemical of interest. Expose for desired time frame (i.e., 20-min exposure) to E4031, a high-risk hERG channel blocker, or other compounds of interest.
Repeat data acquisition (step 8) and perfusion (step 10) for each concentration of compound to be tested. Total imaging time should be no more than about 1 h to minimize effects of dye toxicity.
The baseline drifting caused by water level fluctuation or photobleaching of dye can be subtracted using asymmetric least square method [26].
Process the fluorescence signal using fast Fourier transform (FFT) to construct an FFT image of the amplitude. Apply Otsu’s thresholding and then segment the image to average the fluorescence signals from the regions containing each microtissue to generate an AP signal for each microtissue. Use the first and second derivatives of the AP to quantify parameters (Fig. 3a–g).
Quantify proarrhythmic risk metrics including excitability, stimulation time delay between stimulation pulse and peak AP upstroke (stimulation delay time, stimΔ), rise time of AP upstroke, AP duration (APD) to 30%, 50%, and 80% repolarization (APD30, APD50, APD80), AP duration to end of maximum repolarization rate (APDMxR, determined with d2F/Dt2max), APD triangulation (APDtri = APDMxR –APD50), and the presence of EADs (% of microtissues showing EAD) (Fig. 3h, i). Changes in these metrics can provide insights into alteration in ion channel kinetics as demonstrated in Table 1.
Statistical analysis can compare data for AP recordings from the same mold before and after drug treatment with paired t-tests or for AP recordings from the different molds with unpaired t-tests. Variability between microtissues, molds, or batches of hiPSC-CMs can be quantified.
Fig. 2.
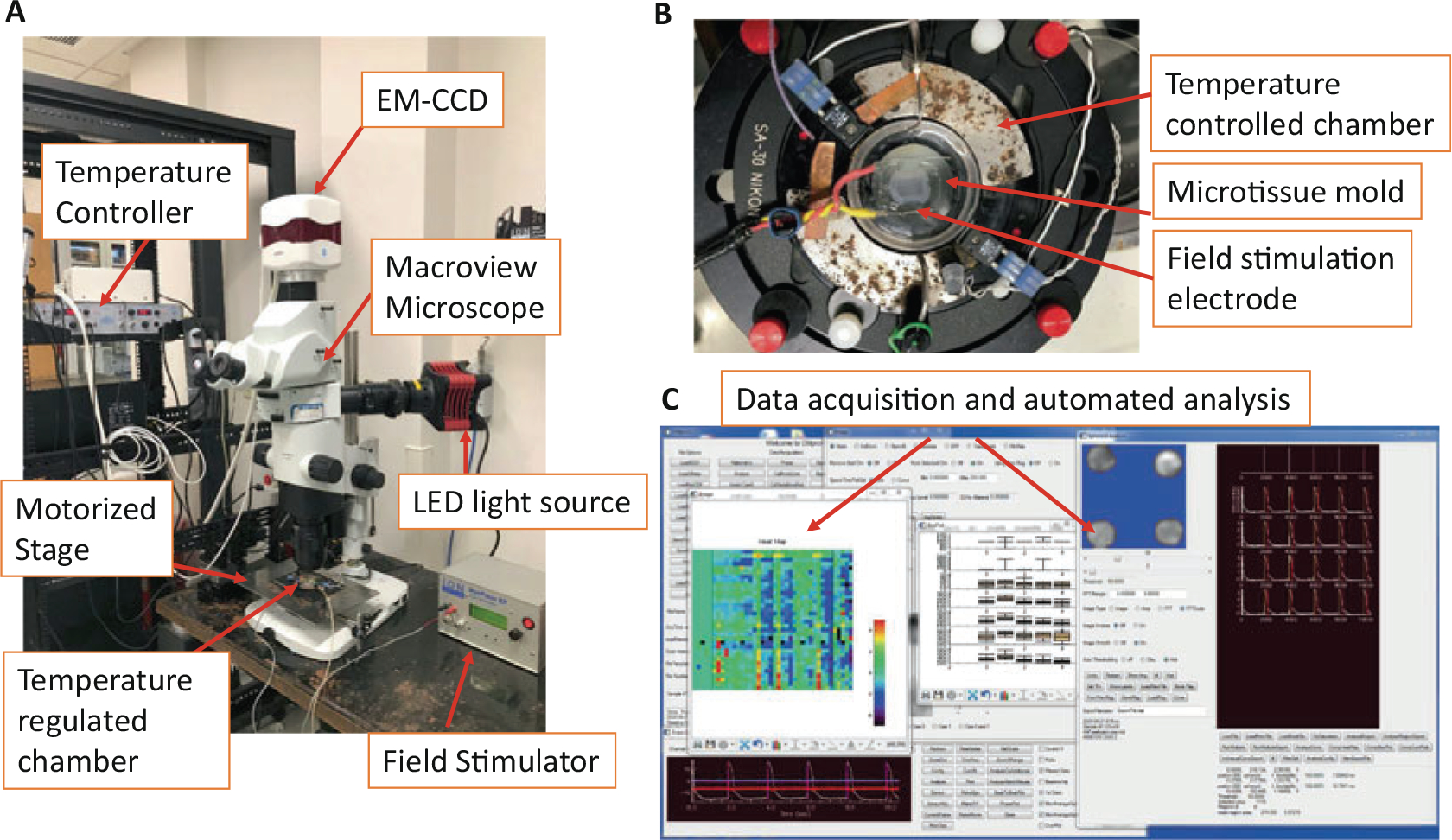
Optical mapping acquisition and analysis. (a) Microscope and stimulation setup to acquire fluorescence images at 979 frames/s in 1.2 × 1.2-mm2 field of view. (b) Stimulation electrode setup during optical mapping image acquisition. Two linear platinum electrodes (2 cm apart) were used to field-stimulate (20 V, 4 ms biphasic stimulation pulse with 10 V/cm amplitude) all of the microtissues simultaneously (adapted from Kofron et al. 2021, Supplemental Figures). (c) Example of data acquisition and custom-automated analysis user interface
Fig. 3.
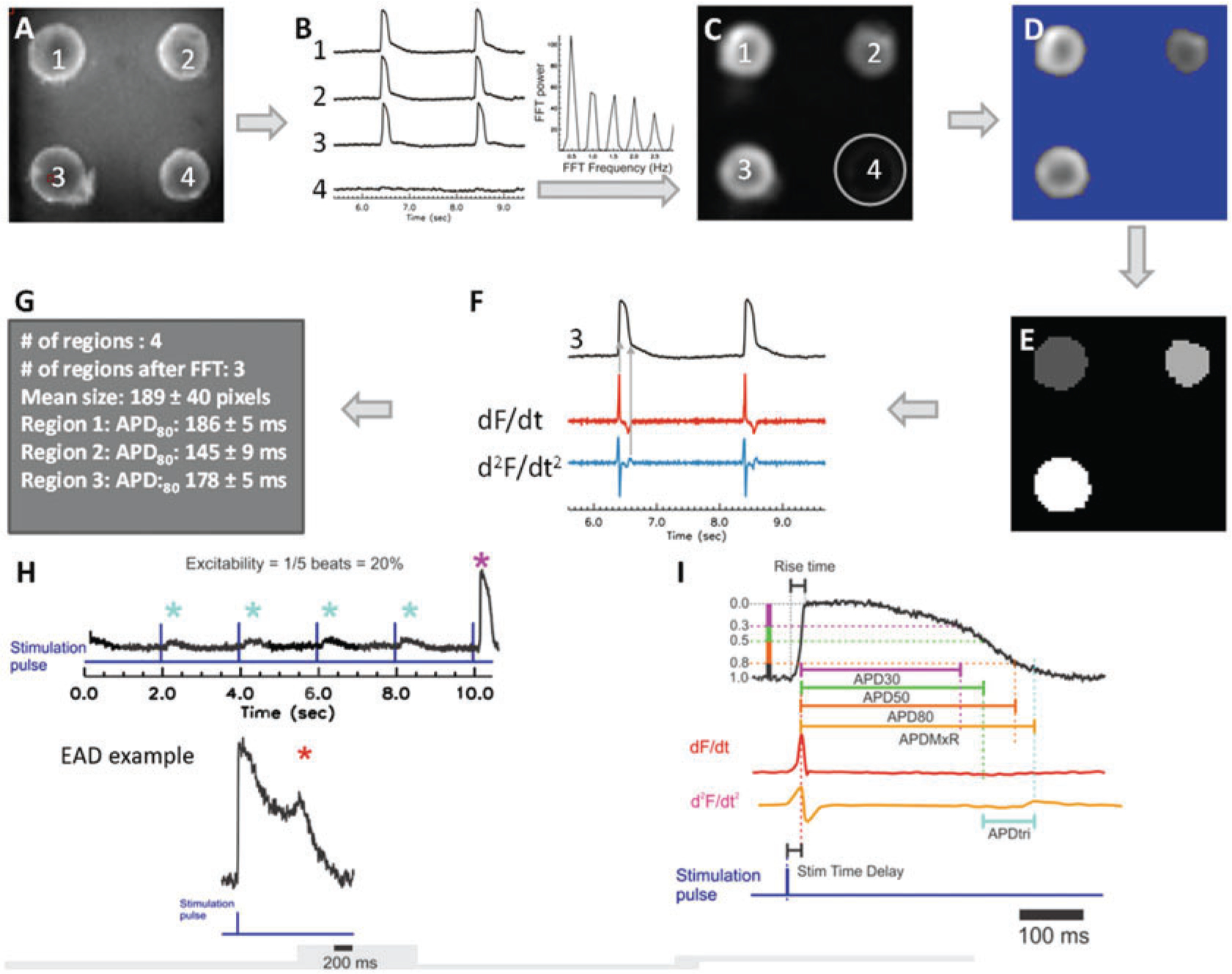
Automated analysis pipeline and definition of metrics. (a) A grayscale snapshot of fluorescence from microtissues at 3.2× magnification during optical mapping shows individual microtissues are numbered 1–4. (b) Sample membrane voltage (Vm) traces recorded from the corresponding microtissues in panel a. Note that electrical stimulation did not evoke APs in microtissue #4 (where the fluorescence trace shows a flat line). Fast Fourier transform (FFT) was used to distinguish responsive microtissues from non-responsive microtissues lacking APs. (c) FFTmax image shows that the microtissue (#4) without APs is automatically removed in FFTmax (circle). (d) Automated thresholding using Otsu’s thresholding to remove background and speed analyses. (e) Blob coloring algorithm to detect individual microtissues. Signals within the same microtissue are averaged to acquire high signal-to-noise ratio and fidelity of data analysis. (f) Automated analysis (of sample trace #3, black) uses the first derivative (dF/dt, red trace) for detecting AP upstroke to calculate AP duration to a defined recovery amplitude (see Fig. 1g) or uses the maximum peak of the second derivative (d2F/dt2, blue trace) for assessing the end of maximum repolarization rate to calculate AP duration APDMxR. APDMxR is useful when motion artifacts or other interference on fluorescence recordings such as fluctuation in water level is suspected to elevate fluorescence level during repolarization or reproducible APD80 measurement with low variation is not as reliable. (g) Sample output shows APD80 statistics from the three excitable individual microtissues. Schematics of the AP metrics that were defined (with units) as: (h) “excitability” (%) measured from the percentage of captured APs during 10 s duration of recording with 2 s pacing cycle length (top) and occurrence of “early afterdepolarization” (EAD) reported as (%) of microtissues showing EADs (bottom, *) and (i) “stimulation time delay” (ms; stim delay) between stimulation pulse and evoked AP upstroke (dF/dtmax), “rise time” (ms) of AP, “AP duration” (ms) to 30%, 50%, and 80% repolarization (APD30, APD50, APD80), “APD to maximum repolarization” (ms; APDMxR) defined as time between AP upstroke and the end of rapid repolarization marked by d2F/dt2max, “APD triangulation” (ms; APDtri) defined as APDMxR–APD50. Adapted from Kofron et al., 2021, Fig. 1 and Supplemental Figures
Table 1.
Quantitative metrics and physiological targets of the AP
| Metric (units) | Ion channels involved | Ion currents |
|---|---|---|
|
| ||
| Excitability (%) and stimulation time delay | Na channels (Nav1.5, 1.1, others) | INa |
| Inward rectifier K+ channel (Kir2.1) | Ik1 | |
| Rise time (ms) | Na channel (Nav1.5) | INa |
| APD30 (ms) | Late Na channels (Nav1.1, others) | INa,late |
| Ca channels (Cav1.2) | ICaL | |
| K channels (Kv4.2/4.3, Kv1.4) | Ito | |
| APD50 (ms) | Late Na channels (Nav1.1, others) | INa,late ICaL Ito, IKr, IKs |
| Ca channels (Cav1.2) | ||
| K channels (hERG, KvLQT, Kv4.2/4.3, Kv1.4) | ||
| APD80, APDMxR (ms) | K channels (hERG, KvLQT, Kir2.1) | IKr, IKs, IK1 |
| APDtri (ms) = APDMxR-APD50 | Late Na channels (Nav1.1, others) | INa,late |
| Ca channels (Cav1.2) | ICaL | |
| K channels (hERG, KvLQT, Kv4.2/4.3, Kv1.4, Kir2.1) | Ito, IKr, IKs, IK1 | |
| EADs (count/AP) | K channels (hERG, KvLQT) | IKr, Iks |
| Late Na channel (Nav1.5) | INa,late | |
4. Notes
Other systems and cameras could be suitable. The Photometrics Evolve128+ EM-CCD camera records from 0.3 × 0.3 mm2 to 3 × 3 mm2 field of view with a sampling rate of 979 frames per sec.
In our setup, the temperature-controlled chamber is mounted on a custom-built motorized stage for automated repetitive recordings. The microscope and the perfusion system are mounted on the isolation table to eliminate possible vibration noise.
hCFs were maintained in 10 cm dishes and split 1:4 when 90% confluent.
Seeding density may need to be optimized with different phenotypes of cardiac myocytes and fibroblasts. This seeding density yielded reproducible, synchronously beating tissues that could be easily imaged.
Stimulation protocol and/or culture time can be adjusted for different cell types and investigations, but this protocol and timeline yields reproducible microtissues with AP similar to in vivo.
The data acquisition, stimulation protocol, and motorized stage are incorporated in a custom-built software (e.g., written in Interactive Data Language (IDL, Harris Geospatial Solutions) and C routines). The spatial and temporal filtering using bilateral filter, baseline adjustment using asymmetric least square algorithm, and FFT of fluorescence signals are calculated in a multicore graphics card (Nvidia Quadro P4000) to shorten calculation time.
References
- 1.Virani SS et al. (2020) Heart disease and stroke statistics-2020 update: a report from the american heart association. Circulation 141: e139–e596. 10.1161/CIR.0000000000000757 [DOI] [PubMed] [Google Scholar]
- 2.Haraguchi Y, Ohtsuki A, Oka T, Shimizu T (2015) Electrophysiological analysis of mammalian cells expressing hERG using automated 384-well-patch-clamp. BMC Pharmacol Toxicol 16:39. 10.1186/s40360-015-0042-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wacker S, Noskov SY (2018) Performance of machine learning algorithms for qualitative and quantitative prediction drug blockade of hERG1 channel. Comput Toxicol 6:55–63. 10.1016/j.comtox.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferdinandy P et al. (2019) Definition of hidden drug cardiotoxicity: paradigm change in cardiac safety testing and its clinical implications. Eur Heart J 40:1771–1777. 10.1093/eurheartj/ehy365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alinejad S, Kazemi T, Zamani N, Hoffman RS, Mehrpour O (2015) A systematic review of the cardiotoxicity of methadone. EXCLI J 14: 577–600. 10.17179/excli2015-553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heranval A et al. (2016) Drugs with potential cardiac adverse effects: retrospective study in a large cohort of parkinsonian patients. Rev Neurol (Paris) 172:318–323. 10.1016/j.neurol.2015.11.007 [DOI] [PubMed] [Google Scholar]
- 7.Sun C, Brice JA, Clark RF (2018) Brugada-type pattern on electrocardiogram associated with high-dose loperamide abuse. J Emerg Med 54:484–486. 10.1016/j.jemermed.2017.12.040 [DOI] [PubMed] [Google Scholar]
- 8.Ramalho D, Freitas J (2018) Drug-induced life-threatening arrhythmias and sudden cardiac death: a clinical perspective of long QT, short QT and Brugada syndromes. Rev Port Cardiol 37:435–446. 10.1016/j.repc.2017.07.010 [DOI] [PubMed] [Google Scholar]
- 9.Kurokawa YK, George SC (2016) Tissue engineering the cardiac microenvironment: Multi-cellular microphysiological systems for drug screening. Adv Drug Deliv Rev 96:225–233. 10.1016/j.addr.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Archer CR et al. (2018) Characterization and validation of a human 3D cardiac microtissue for the assessment of changes in cardiac pathology. Sci Rep 8:10160. 10.1038/s41598-018-28393-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beauchamp P et al. (2020) 3D co-culture of hiPSC-derived cardiomyocytes with cardiac fibroblasts improves tissue-like features of cardiac spheroids. Front Mol Biosci 7:14. 10.3389/fmolb.2020.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feric NT et al. (2019) Engineered cardiac tissues generated in the biowire II: a platform for human-based drug discovery. Toxicol Sci 172(1):89–97. 10.1093/toxsci/kfz168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beauchamp P et al. (2015) Development and characterization of a scaffold-free 3D spheroid model of induced pluripotent stem cell-derived human cardiomyocytes. Tissue Eng Part C Methods 21:852–861. 10.1089/ten.TEC.2014.0376 [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y et al. (2019) A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 176:913–927. e918. 10.1016/j.cell.2018.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergstrom G, Christoffersson J, Schwanke K, Zweigerdt R, Mandenius CF (2015) Stem cell derived in vivo-like human cardiac bodies in a microfluidic device for toxicity testing by beating frequency imaging. Lab Chip 15:3242–3249. 10.1039/c5lc00449g [DOI] [PubMed] [Google Scholar]
- 16.Petersen AP et al. (2018) Microenvironmental modulation of calcium wave propagation velocity in engineered cardiac tissues. Cell Mol Bioeng 11:337–352. 10.1007/s12195-018-0522-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanner MR, Beeton C (2018) Differences in ion channel phenotype and function between humans and animal models. Front Biosci (Landmark Ed) 23:43–64. 10.2741/4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bracken MB (2009) Why animal studies are often poor predictors of human reactions to exposure. J R Soc Med 102:120–122. 10.1258/jrsm.2008.08k033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kofron CM et al. (2017) Gq-activated fibroblasts induce cardiomyocyte action potential prolongation and automaticity in a three-dimensional microtissue environment. Am J Physiol Heart Circ Physiol 313:H810–H827. 10.1152/ajpheart.00181.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kofron CM, Mende U (2017) In vitro models of the cardiac microenvironment to study myocyte and non-myocyte crosstalk: bioinspired approaches beyond the polystyrene dish. J Physiol 595:3891–3905. 10.1113/JP273100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuppinger C (2019) 3D cardiac cell culture: a critical review of current technologies and applications. Front Cardiovasc Med 6:87. 10.3389/fcvm.2019.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tohyama S et al. (2013) Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 12:127–137. 10.1016/j.stem.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 23.Rupert CE, Kim TY, Choi BR, Coulombe KLK (2020) Human cardiac fibroblast number and activation state modulate electromechanical function of hiPSC-cardiomyocytes in engineered myocardium. Stem Cells Int 2020: 9363809. 10.1155/2020/9363809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kofron CM et al. (2021) A predictive in vitro risk assessment platform for pro-arrhythmic toxicity using human 3D cardiac microtissues. Sci Rep 11(1):10228. 10.1038/s41598-021-89478-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss JN, Garfinkel A, Karagueuzian HS, Chen PS, Qu Z (2010) Early afterdepolarizations and cardiac arrhythmias. Heart Rhythm 7:1891–1899. 10.1016/j.hrthm.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng J et al. (2010) Asymmetric least squares for multiple spectra baseline correction. Anal Chim Acta 683:63–68. 10.1016/j.aca.2010.08.033 [DOI] [PubMed] [Google Scholar]


