Summary
The plant cell wall is the first line of defence against physical damage and pathogen attack. Wall‐associated kinase (WAK) has the ability to perceive the changes in the cell wall matrix and transform signals into the cytoplasm, being involved in plant development and the defence response. Downy mildew, caused by Hyaloperonospora brassicae, can result in a massive loss in Chinese cabbage (Brassica rapa L. ssp. pekinensis) production. Herein, we identified a candidate resistant WAK gene, BrWAK1, in a major resistant quantitative trait locus, using a double haploid population derived from resistant inbred line T12–19 and the susceptible line 91–112. The expression of BrWAK1 could be induced by salicylic acid and pathogen inoculation. Expression of BrWAK1 in 91–112 could significantly enhance resistance to the pathogen, while truncating BrWAK1 in T12–19 increased disease susceptibility. Variation in the extracellular galacturonan binding (GUB) domain of BrWAK1 was found to mainly confer resistance to downy mildew in T12–19. Moreover, BrWAK1 was proved to interact with BrBAK1 (brassinosteroid insensitive 1 associated kinase), resulting in the activation of the downstream mitogen‐activated protein kinase (MAPK) cascade to trigger the defence response. BrWAK1 is the first identified and thoroughly characterized WAK gene conferring disease resistance in Chinese cabbage, and the plant biomass is not significantly influenced by BrWAK1, which will greatly accelerate Chinese cabbage breeding for downy mildew resistance.
Keywords: Brassica rapa, QTL, Downy mildew, Wall‐associated kinase, BrWAK1
Introduction
Plants, with a sessile lifestyle, face the threats from a wide range of pathogens, which have prompted plants to evolve an immunity system to obtain protection from pathogen attack. As the first line of defence, pattern recognition receptors (PRRs) located on the cell membrane recognize pathogen‐associated molecular patterns (PAMPs) or damage‐associated molecular patterns (DAMPs), which is known as pattern‐triggered immunity (PTI) (Ngou et al., 2022). One of the most important PRRs is the receptor‐like protein kinase (RLK) superfamily, including somatic embryogenesis receptor kinase 3/brassinosteroid insensitive 1 associated kinase 1 (SERK3/BAK1), wall‐associated kinase 1 (WAK1), flagellin sensing 2 (FLS2) and elongation factor Tu (EF‐Tu) receptor (EFR) (Gómez‐Gómez and Boller, 2000; He et al., 1998; Heese et al., 2007; Zipfel et al., 2006). PTI could lead to an immune response that restricts pathogen colonization; however, the pathogens that escape from the first line of defence could secret virulence molecules, referred to as effectors, into host cells to trigger the secondary immune response, termed effector‐triggered immunity (ETI) (Liu et al., 2014). In the ETI response, intracellular nucleotide‐binding domain, leucine‐rich‐repeat containing receptors (NLRs) recognize and target the effectors to activate a rapid and robust defence response (Liu et al., 2014). Phytohormones, such as salicylic acid (SA), jasmonic acid (JA) and ethylene (ET), were also found to participate in the resistance response (Ding and Ding, 2020; Glazebrook et al., 2003; Jones and Dangl, 2006; Katagiri, 2004). The biosynthesis of SA could be enhanced by both PTI and ETI, and SA‐mediated systemic acquired resistance (SAR) could effective against biotrophic pathogens (Malamy et al., 1990; Métraux et al., 1990; Ton et al., 2002; Zeier, 2021).
Chinese cabbage (Brassica rapa L. ssp. pekinensis) is one of the most important vegetable crop plants in China. Brassica downy mildew is one of the most severe diseases of Chinese cabbage, which causes damage to nearly 90% of outer leaves in an epidemic year (Yu et al., 2009). The causal agent of Brassica downy mildew is the oomycetes of Hyaloperonospora parasitica var. Brassicae, which is a biotrophic pathogen with strict host specialization. Since the 1980s, researchers have been devoted to characterizing the resistance mechanism against H. Brassicae to breed downy mildew‐resistant Chinese cabbage. From the aspect of resistant loci mapping, a series of major quantitative trait loci (QTLs) for disease resistance have been identified, including BraDM on A08; BrRHP1 on A01; and six QTLs on A04, A06, and A08 (Kim et al., 2011; Li et al., 2011; Yu et al., 2009, 2016). This suggested that the genetic system controlling resistance was complicated, and is probably distinct in different varieties and developmental stages. Research into resistant phytohormones showed that the expression of major genes in the SA signalling pathways were induced after downy mildew invasion, which implied that SA plays a crucial role in downy mildew resistance (Chen et al., 2015; Gao et al., 2014). In transcriptomic analysis, immunity‐related genes, long non‐coding RNAs (lncRNAs), and target resistant genes have also been identified (Li et al., 2018; Zhang et al., 2021). Some candidate genes conferred resistance to downy mildew have been identified in Chinese cabbage (Zhang et al., 2018); however, no studies on the molecular mechanisms of resistant genes have appeared.
The WAK gene family is a unique subfamily of the RLK superfamily characterized by its epidermal growth factor (EGF)‐like domain in the extracellular domain, which also contains a galacturonan binding (GUB) domain N‐terminal to the EGF‐like repeats (Shiu and Bleecker, 2001). Through their extracellular domain and typical cytoplasmic Ser/Thr kinase domain, WAKs could directly connect the cell wall matrix and the cytoplasm (Anderson et al., 2001; He et al., 1996; Verica and He, 2002). The pectin and oligogalacturonides (OGs) that are disaggregated from the damaged cell wall after pathogen invasion could be perceived and translated it into cytoplasmic defence signals by WAKs (Brutus et al., 2010; Kohorn et al., 2009). Therefore, WAKs were found to function extensively in plant defence and developmental regulation. The function of Arabidopsis WAK1 (AtWAK1) was the first and the most studied. AtWAK1 could perceive OGs and its expression level could also be induced by OGs and SA to mediate resistance to Botrytis cinerea (Brutus et al., 2010; Denoux et al., 2008). In recent years, a series of WAK genes involved in defence response have been identified in rice, wheat, maize and other crops. In wheat, the TaWAK5 and TaWAK6 genes are involved in resistance to wheat sharp eyespot disease and leaf rust, respectively (Dmochowska‐Boguta et al., 2020; Yang et al., 2014). Wheat Stb6, a WAK‐like gene, was found to control gene‐for‐gene resistance to Zymoseptoria tritici (Saintenac et al., 2018). In rice, overexpression of OsWAK1 could enhance resistance to rice blast fungus (Li et al., 2009). OsWAK14/19/92 can regulate the resistance response to rice blast, and OsWAK91 is also involved in the production of H2O2 to enhance the expression of defence genes (Delteil et al., 2016). In maize, the ZmWAK gene is a key gene for resistance to head smut (Zuo et al., 2015). The variable splicing of the ZmWAK‐RLK1 gene increases susceptibility to big leaf spot disease (Hurni et al., 2015). In Chinese cabbage, a total 96 BrWAKs were identified, containing 11 BrWAKs that have close phylogenetic relationship with AtWAKs (Zhang et al., 2020). However, there has been no research on functions of BrWAKs in the defence response till now. In this study, we firstly identified three tandem duplicated BrWAK genes as candidate resistant genes in a major QTL region conferring the resistance to downy mildew in Chinese cabbage. Overexpression BrWAK1 significantly enhanced resistance to downy mildew and truncated BrWAK1 increased susceptibility. Furthermore, the functional variation and interaction proteins of BrWAK1 were investigated. These results revealed the molecular mechanism of a downy mildew resistance gene in Chinese cabbage, and will also lead to a deeper understanding of the involvement of the WAK gene family in defence responses.
Results
Fine mapping of Bra‐DM08
A major QTL, Bra‐DM08 (explaining 65.4% of the phenotypic variance), comprising 2.9 cM flanked by the markers PGM and K14‐1030, conferring downy mildew resistance in B. rapa, was previously identified in the doubled‐haploid (DH) population (DH100, 100 DH lines) derived from a cross between two Chinese cabbage inbred lines, 91–112 (highly susceptible, DI = 20.9) and T12–19 (highly resistant, DI = 83.8). An improved genetic map with high‐density molecular markers was also constructed (Yu et al., 2009; Figure 1a).
Figure 1.
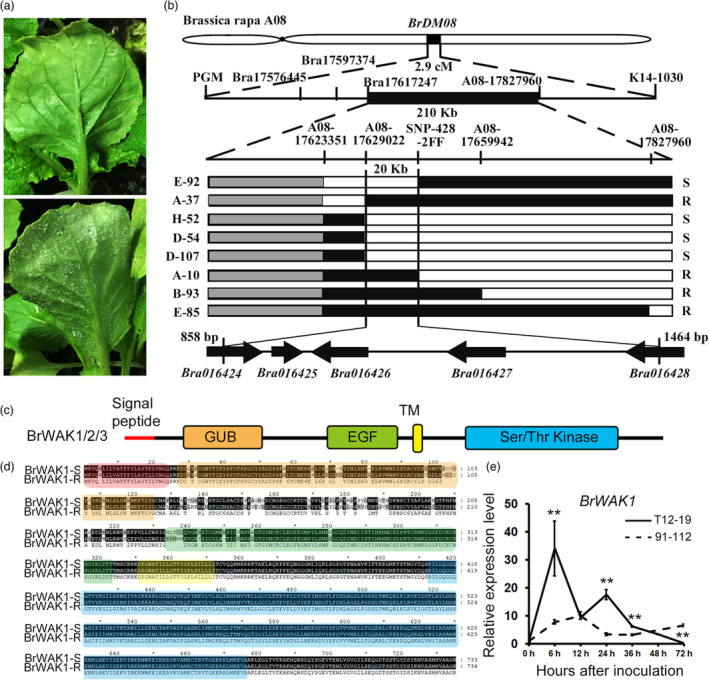
Fine mapping of the downy mildew resistant QTL Bra‐DM08. (a) Downy mildew symptoms in T12–19 (upper panel) and 91–112 (lower panel). (b) Fine mapping of Bra‐DM08. The vertical lines represent the sites of molecular markers. The grey, black, and white rectangle represent undetected, heterozygous, and homozygous sites from 91 to 112, respectively. The name of recombinant lines and phenotype are listed on the left and right, respectively. “R” means resistant and “S” means susceptible. The genes in Bra‐DM08 are shown by black arrows. (c) Schematic diagram of the conserved domains in BrWAK1/2/3. (d) The amino acid sequence alignment of BrWAK1 in T12–19 and 91–112. The sequences in the conserved domains are highlighted by different background colours, which are consistent with those in c. (e) The expression levels of BrWAK1 in T12–19 and 91–112 at 0, 6, 12, 24, 36, 48, and 72 h after inoculation. More than ten leaves were collected in each replicate. Expression of each gene at 0 h after inoculation in T12–19 or 91–112 was defined as 1.0. Values represent means ± SD (n = 3) from three biological replicates. Statistical significance between 0 h after inoculation in T12–19 or 91–112 was by a t‐test: **P < 0.01.
For the fine mapping of Bra‐DM08, the DH60 line (highly susceptible, DI = 85.2) and DH88 line (highly resistant, DI = 12.0) in the DH100 population, which are distinct in terms of Bra‐DM08, but have a similar genetic background (86.1% similarity), were selected as the parents to construct chromosomal segment substitution lines (CSSLs by multigenerational backcrossing to a susceptible DH line (DH60); Figure S1). We used DH60 as the backcrossing (BC) parent to develop BC1F1 (425 individuals), BC2F1 (386 individuals) and BC3F1 (489 individuals). The BC populations were also developed into DH populations, containing DH390 population (390 DH lines), DH327 and DH208 population (327 DH lines and 208 DH lines respectively). The BC populations, comprising 1300 lines, and more than 700 DH lines, were used for the fine mapping of the downy mildew resistant QTL. Finally, we narrowed Bra‐DM08 to about 20 kb region on Chromosomal A08, flanked with two single nucleotide polymorphism (SNP) markers, A08‐17629022 and SNP‐428‐2FF, using the recombinants generated from the backcrossing and DH populations derived from backcrossing populations (Figure 1b and Table S1).
Identification of candidate disease resistance genes
There are three intact genes, Bra016425, Bra016426 and Bra016427, and parts of two genes, Bra016424 and Bra016428, in Bra‐DM08 (Figure 1b and Table 1). Among these five genes, Bra016426, Bra016427 and Bra016428 encode proteins belonging to the WAK family and were named as BrWAK1, BrWAK2 and BrWAK3, respectively (Table 1). The WAK protein family comprises transmembrane kinase receptors containing GUB and EGF domains in their extracellular regions and are involved in plant resistance, stress response and development in other species (Figure 1c; Shiu and Bleecker, 2001). In the phylogenetic tree, BrWAK1, BrWAK2 and BrWAK3 showed a close relationship with AtWAK1–5 and their kinase domains were RD‐type kinase domains, the same as AtWAK1–5 (Figure S2a,b). Therefore, these three WAK genes were considered as the candidate resistance genes.
Table 1.
The annotated genes in Bra‐DM08
| Gene ID | Chr | Homologue in Arabidopsis | Annotation in Arabidopsis |
|---|---|---|---|
| Bra016424 | A08 | AT2G15480 | UDP‐glucosyl transferase 73B5; UDP‐glucosyltransferase |
| Bra016425 | A08 | AT1G76970 | VHS domain‐containing protein/GAT domain‐containing protein |
| Bra016426 | A08 | AT1G21230 | WAK5; WAK5 (WALL ASSOCIATED KINASE 5) |
| Bra016427 | A08 | AT1G21230 | WAK5; WAK5 (WALL ASSOCIATED KINASE 5) |
| Bra016428 | A08 | AT1G21250 | WAK1, PRO25; WAK1 (CELL WALL‐ASSOCIATED KINASE) |
To verify the sequences of the candidate genes, we constructed a BAC library of T12–19 and selected the clones containing Bra016426, Bra016427 and Bra016428. Through sequencing of BAC clones combined with cDNA amplification, the amino acid sequences of BrWAK1, BrWAK2 and BrWAK3 in T12–19 and 91–112 were obtained (Table S2). The distinct amino acid sequences of BrWAK1 between T12–19 and 91–112 were mainly distributed in the GUB domain, EGF domain and the intermediate region between them, while there are only two different amino acids for BrWAK2 (Figure 1d and Figure S3a). Similar to BrWAK1, most of differences in the amino acid sequences of BrWAK3 were in the GUB domain and the intermediate region (Figure S3b). During the H. Brassicae invasion, the expression levels of BrWAK1 and BrWAK2 increased significantly at 6 and 24 h after inoculation (HAI) in the resistant parent T12–19 compared with those in the susceptible parent 91–112, while BrWAK3 showed no obvious induction (Figure 1e and Figure S3c). We amplified the promoter sequences of T12–19 and 91–112 and analysed the distinct transcription elements. We found that the number of SA responsive element and TATA box were increased in T12–19 comparing with these in 91–112, which might cause the higher expression of BrWAK2 in T12–19 after inoculation (Figure S3d). The tissue‐specific expression pattern analysis by qRT–PCR showed that BrWAK1, BrWAK2 and BrWAK3 all have relatively consistent expression levels in cotyledon, shoot, stem, root, first and second leaf with sight higher expression levels in shoot and stem in 91–112 and T12–19 (Figure S3e).
To efficiently determine which BrWAK gene confers downy mildew resistance, the transient expression system mediated by agroinfiltration in Chinese cabbage cotyledons was performed. Three days after infiltration, BrWAK1, BrWAK2 and BrWAK3 proteins fused with GFP were detected in crude protein of cotyledons in 91–112 and the fluorescent signal located on the membrane could be also observed (Figures S4a,b and S5). Then, the pathogens were inoculated on the infiltrated cotyledons. Only those infiltrated with the resistant (R) genotype (sequence in T12‐19) BrWAK1‐R‐GFP showed a significant reduction in the percentage of susceptible cotyledons (reduced by ~40%) together with a comparatively similar gene expression level (Figure S4b–d). The cotyledons infiltrated with empty vector (GFP), the susceptible (S) genotype (sequence in 91–112) BrWAK1‐S‐GFP, and those without infiltration were used as control (Figure S4b). In T12–19, RNA interference (RNAi) experiments were also performed using the transient expression system. The expression level of BrWAK1, BrWAK2 and BrWAK3 in RNAi experiment were all remarkably decreased by a similar extent compared with those in the “No‐Injection” and “Vector” control groups, no matter whether they were inoculated or not (Figure S6a). However, only RNAi of BrWAK1 could significantly enhance the susceptibility of T12–19 (increased by ~20%) (Figure S6b,c). These results indicated that BrWAK1 probably plays a crucial role in resistance to downy mildew.
BrWAK1 confers resistance to downy mildew
To further validate the resistant function of BrWAK1, transgenic Chinese cabbage plants were constructed. Four overexpression lines of BrWAK1 (the genotype of T12–19) in 91–112, in which BrWAK1 was indeed upregulated at the mRNA and protein level, showed obviously disease resistance after inoculation compared with 91–112 and plants transformed with the empty vector (Figure 2a–c). The overexpression lines also showed a stronger resistance reaction, including producing more hydrogen peroxide and callose deposition (Figure 2d). Furthermore, two gRNAs for BrWAK1 in T12–19 were designed and a premature stop codon before the EGF domain was formed using cas9‐mediated gene editing system (Figure 3a,b). For off‐target detection, there are no gene has similar gRNA sequences with <5 bp mismatches using Cas‐OFFinder (http://www.rgenome.net/cas‐offinder/). The transgenic T12–19 seedlings containing truncated BrWAK1 showed significantly reduced resistance compared with T12–19 after inoculation (Figure 3c). DAB staining and callose deposition detection showed a weaker response to the pathogen in the gene edited T12–19 lines compared with that in the non‐edited BrWAK1 line (Figure 3d). Meanwhile, the down‐regulated expression of BrWAK1 in T12–19 also showed reduced resistance using virus induced gene silence (VIGS) system (Figure S7). These results showed that BrWAK1 has pivotal effort on defensing downy mildew in Chinese cabbage and it is the crucial resistant gene in Bra‐DM08, which means that it could mainly explain why T12–19 has higher resistance than 91–112.
Figure 2.
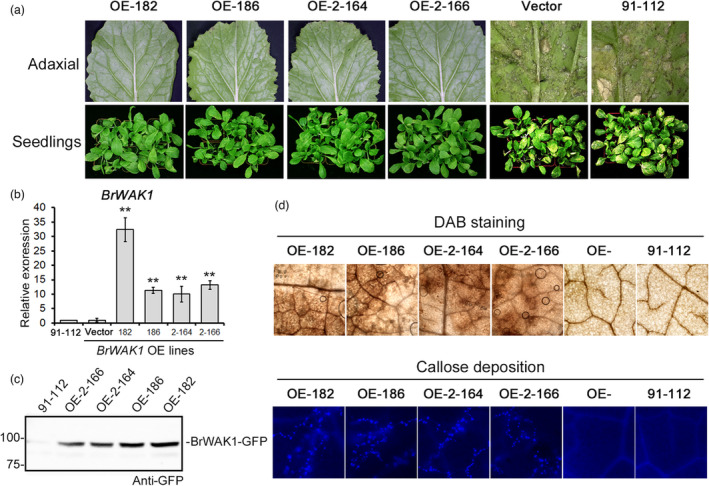
Overexpression of BrWAK1 in 91–112 could significantly enhance disease resistance. (a) Four‐week‐old seedlings of the T2 progeny of the BrWAK1 overexpression lines, OE‐182, OE‐186, OE‐2‐164 and OE‐2‐166 were inoculated. Approximately three‐quarters of the seedlings in each line showed obvious resistance to downy mildew. 91–112, the transgenic acceptor, and the lines transformed with the empty vector were used as controls. (b) The expression level of BrWAK1 in the overexpression lines. The expression of 91–112 was designated as “1”. Values represent means ± SD (n = 3) from three biological replicates. Statistical significance between 91 and 112 was by a t‐test: **P < 0.01. (c) The detection of GFP‐fused BrWAK1 in the overexpression lines by western blotting using anti‐GFP antibodies. (d) Images of DAB staining and callose deposition of the BrWAK1 overexpression lines at 24 HAI.
Figure 3.
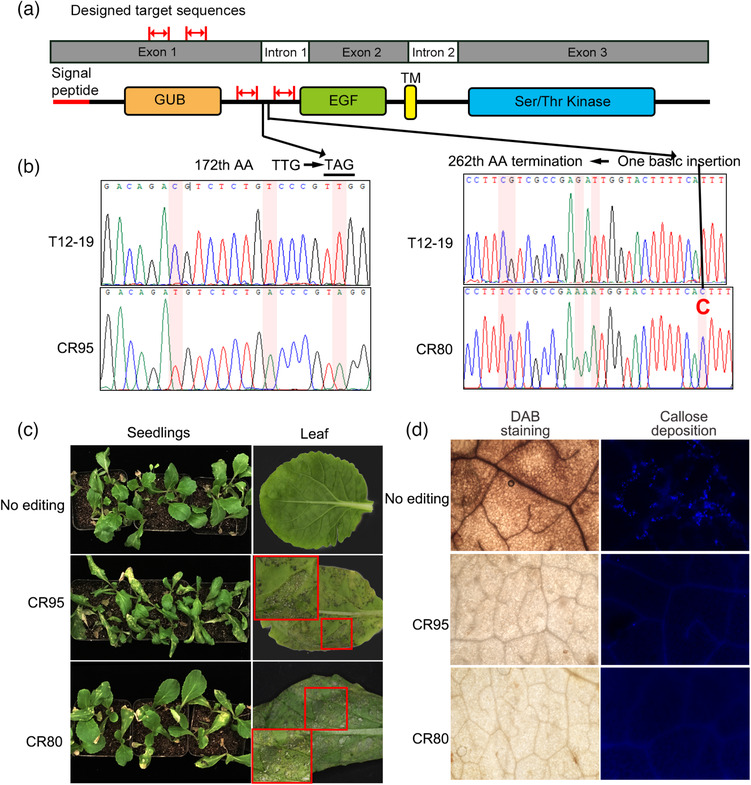
Truncated BrWAK1 by cas9‐mediated gene editing in T12–19 could weaken the resistance ability. (a) The schematic diagram of the gRNA (red arrows) location on the genome (upper panel) and conserved protein domains (lower panel) of BrWAK1. (b) Sanger‐sequencing peak map of edited sites. The mutated sites are highlighted in red. The sequences of T12–19 are on the top and those of BrWAK1 editing lines CR95 and CR80 are below it. (c) The disease symptom of BrWAK1 editing lines. Lines with non‐edited BrWAK1 were used as controls. The image in small red rectangle is magnified in the large red rectangle. (d) Images of DAB staining and callose deposition of the BrWAK1 editing lines at 24 HAI.
Resistant function of BrWAK1 depends on variation in the GUB domain
The expression level of BrWAK1 was noticeably upregulated after H. Brassicae invasion. Therefore, to clarify the regulation mechanism of BrWAK1, sequence analysis of BrWAK1's promoter in T12–19 and 91–112 was performed. The promoter sequence in T12–19 (R‐promoter) has one more TCA‐element (SA responsive element) and a 5′ UTR Py‐rich stretch cis‐transcription element (confers high‐level transcription) compared with that in 91–112 (S‐promoter) (Figure 4a). SA is an important phytohormone involved in the response to disease resistance and it could greatly increase the transcriptional activity of the R‐promoter at 3 h and 6 h compared with that of the S‐promoter (Figure 4b). Likewise, the expression level of BrWAK1 was substantially induced from 6 h to 24 h after SA treatment in T12–19, while it showed no obvious change in 91–112 (Figure 4c). The contents of SA were significantly increased at 72 and 120 HAI in T12–19 and the BrWAK1 overexpression line, while those of JA exhibited no induction and decline in 91–112 and the transgenic plants without BrWAK1 overexpression (Figure 4d,e). Therefore, the content of SA was up‐regulated, which could induce the transcription of BrWAK1 to trigger a downstream defence response.
Figure 4.
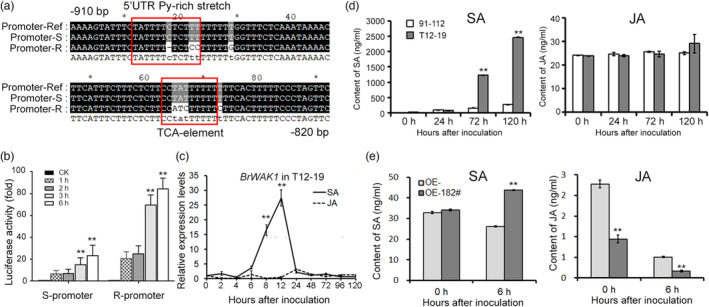
SA could accumulate and induce the expression of BrWAK1 after inoculation. (a) Part of the BrWAK1 promoter sequence in T12–19 with distinct cis‐transcription element from that in 91–112. (b) The activity of the BrWAK1 promoter in 91–112 (S‐promoter) and in T12–19 (R‐promoter) after SA treatment in the double‐LUC reporter assay in Chinese cabbage protoplast cells. Values represent means ± SD (n = 3) from three biological replicates. Statistical significance between CK sample (without SA treatment) was by a t‐test: **P < 0.01. (c) The expression of BrWAK1 in T12–19 at 0, 2, 4, 6, 8, 12, 24, 48, 72, 96, and 120 h after 1.5 mm SA and 1 mm JA treatment. More than five leaves were collected in each replicate. Expression of BrWAK1 at 0 h in SA and JA treatment (CK) was defined as 1.0. Values represent means ± SD (n = 3) from three biological replicates. Statistical significance between CK was by a t‐test: **P < 0.01. (d) The contents of SA and JA in 91–112 and T12–19 at 0, 24, 72, and 120 HAI. More than five leaves were collected in each replicate. Values represent means ± SD (n = 3) from three biological replicates. Statistical significances between 91 and 112 samples were by a t‐test: **P < 0.01. (e) The contents of SA and JA in the BrWAK1 overexpression line (OE‐182) and the transgenic line without BrWAK1 overexpression (OE‐) at 0, 24, 72, and 120 HAI. More than five leaves were collected in each replicate. Values represent means ± SD (n = 3) from three biological replicates. Statistical significances between 91 and 112 samples were by a t‐test: **P < 0.01.
In addition to the distinction in expression level of BrWAK1 between T12–19 and 91–112, there are also sequence variations in their coding regions. To investigate whether the sequence variation or different expression level of BrWAK1 caused its different function in response to the pathogen between T12–19 and 91–112, we constructed a vector containing BrWAK1‐S (the genotype of 91–112) driven by the R‐promoter (~1.5 kb) (Figure 5a). In the transient expression system, overexpression of BrWAK1‐S could not enhance the resistance of 91–112 (~80% susceptible cotyledons) (Figure 5b,c). Furthermore, we designed and constructed chimeric receptors to clarify which variations conferred the resistance function of BrWAK1. The EGF domain and GUB domain (including the intermediate region between GUB and EGF domain) from T12–19 were embedded in BrWAK1‐S, which were designated as EGF R ‐BrWAK1 and GUB R ‐BrWAK1, respectively (Figure 5a). In cotyledons of 91–112, the overexpression of GUB R ‐BrWAK1 could substantially reduce the percentage of susceptible cotyledons (~60%), while EGF R ‐BrWAK1 could not (Figure 5b–d). Under OG treatment, the transgenic Arabidopsis lines in which BrWAK1‐R and GUB R ‐BrWAK1 were overexpressed exhibited resistant reactions, including hydrogen peroxide accumulation and callose deposition, while those in which BrWAK1‐S and EGF R ‐BrWAK1 were overexpressed could not (Figure 5e). In summary, BrWAK1‐R and GUBR‐BrWAK1 could perceive extracellular OGs to activate resistance responses, and the sequence variations, especially the variations in the GUB domain, play a more dominant role in downy mildew resistance in Chinese cabbage than the promoter variations.
Figure 5.
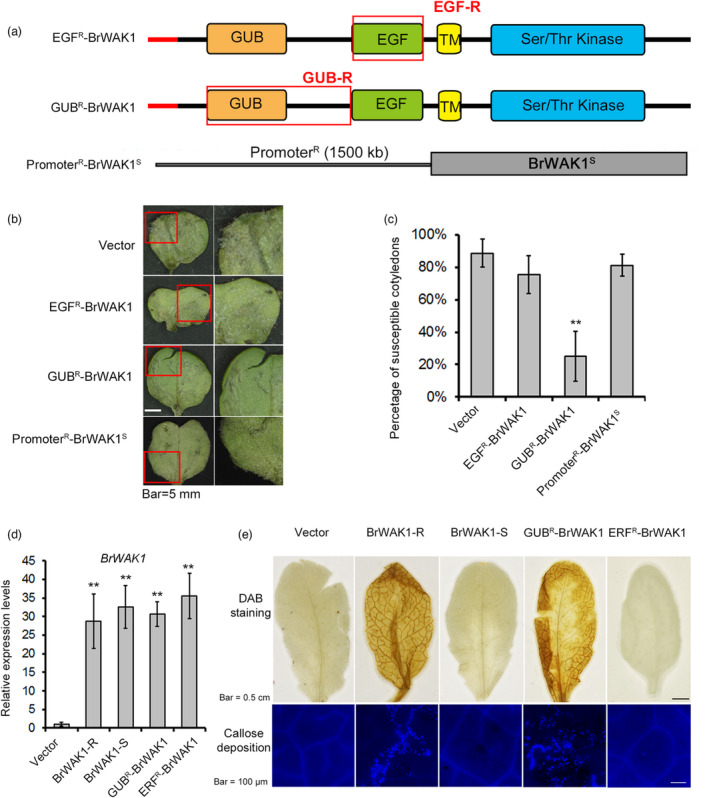
The sequence variation in the GUB domain could confer BrWAK1 disease resistance and ability to perceive extracellular signals. (a) Schematic image of the construction of the vectors containing chimeric receptors (EGRR‐BrWAK1 and GUBR‐BrWAK1) and BrWAK1‐S driven by the R‐promoter (PromoterR‐BrWAK1S). “R” means the sequences from T12‐19 and “S” stands for those from 91 to 112. (b) The disease symptoms of the cotyledons infiltrated with chimeric vectors (EGRR‐BrWAK1 and GUBR‐BrWAK1) and PromoterR‐BrWAK1S after inoculation. The images in red rectangles are enlarged in the right panels. Cotyledons infiltrated with empty vector (pSuper1300) were used as controls. (c) The percentage of susceptible infiltrated cotyledons after inoculation. More than fifty leaves were observed after inoculation in each replicate. Values represent means ± SD (n = 3) from three biological replicates. Statistical significance between transgenic lines with empty vector (Vector) was by a t‐test: **P < 0.01. (d) The expression levels of BrWAK1 in infiltrated cotyledons after inoculation. Expression of BrWAK1 in Vector was defined as 1.0. Values represent means ± SD (n = 3) from three biological replicates. Statistical significance between Vector was by a t‐test: **P < 0.01. (e) The images of DAB staining and callose deposition of Arabidopsis seedlings transformed with BrWAK1‐S, BrWAK1‐R, EGRR‐BrWAK1 and GUBR‐BrWAK1 after 200 μg/mL of OG treatment.
BrWAK1 can interact with BrBAK1 to activate MAPK cascades
BAK1 and SOBIR1 are known as the crucial disease resistant receptor proteins, which interact with other receptor proteins to activate the downstream disease resistance response (Heese et al., 2007; Liebrand et al., 2013; van der Burgh et al., 2019). Therefore, BAK1 and SOBIR1 were preliminarily assessed as the candidate interaction proteins with BrWAK1. According to the homology ratio and phylogenetic relationship with AtBAK1 and AtSOBIR1, Bra034562 and Bra022870 were identified as BrBAK1 and BrSOBIR1, respectively (Figure S8). In the firefly luciferase complementation (FLC) system and bimolecular fluorescence complementation (BiFC) system, strong interaction signals were observed between BrWAK1‐R and/or GUBR‐BrWAK1 and BrBAK1, but not BrSOBIR1 (Figure 6a,b). There were no or very weak interactions between BrWAK1 and BrWAK1, which implied that BrWAK1 could not form a dimer in vivo (Figure 6a). BrWAK‐S and EGFR‐BrWAK1 also exhibited weak interaction with BrBAK1 in the BiFC system (Figure S9). The interaction between BrWAK1‐R and BrBAK1 was further validated using a CO–IP assay (Figure 6c). Thus, BrWAK1 could interact with BrBAK1 and the sequence variations, especially those in GUB domain, would probably weaken the interaction.
Figure 6.
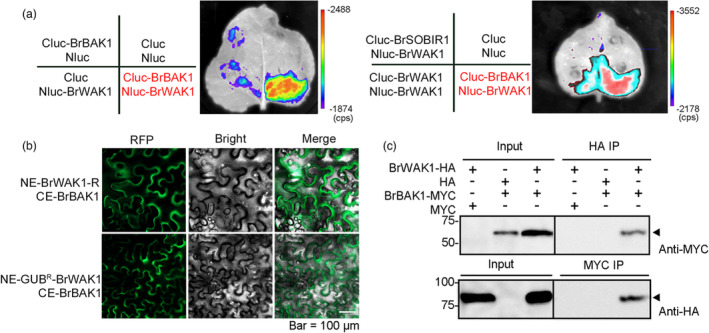
BrWAK1‐R could interact with BrBAK1. (a) The interaction of BrWAK1 and BrBAK1 in the firefly luciferase complementation system. (b) The interaction of BrWAK1‐R and BrBAK1, GUBR‐BrWAK1, and BrBAK1 in tobacco (Nicotiana benthamiana) leaves in the BiFC assay. (c) Co‐IP assay of BrWAK1‐R and BrBAK1 in tobacco (Nicotiana benthamiana) leaves.
After the interaction of BrWAK1 and BrBAK1, BAK1 could trigger the downstream MAPK cascade to activate resistance reactions at 2 HAI in T12–19 and BrWAK1 overexpression lines (Figure 7a). In 91–112, less phosphorylated MAPK was detected, which indicated that BrWAK1‐R could induce a stronger resistance response mediated by the interaction with BrBAK1. However, BrWAK1 with the kinase activity site mutated could not enhance disease resistance of 91–112, which implied that the kinase activity of BrWAK1 is crucial for its resistant function (Figure 7b,c). Camalexin, an indole phytoalexin, is the product of resistance signalling, and was found to accumulate in T12–19 and the BrWAK1 overexpression lines at 12 HAI (Figure 7d). The expression levels of the key genes involved in camalexin biosynthesis and signalling, including MAPK6, WRKY33 and CYP79β2, were likewise upregulated in T12–19 and the BrWAK1 overexpression lines (Figure 7d and Table S3). In addition, after inoculation, transcriptome analysis of the BrWAK1 overexpression (OE) and gene editing lines showed that the upregulated and downregulated genes in the OE and gene editing lines were mostly enriched into the GO items involved in the defence response and protein phosphorylation pathway, such as protein amino acid phosphorylation, regulation of innate immune response, regulation of hydrogen peroxide metabolic process, salicylic acid metabolic process, protein kinase cascade and plant‐type hypersensitive response (Figure S10). Meanwhile, in the RNAseq data, the expression levels of crucial genes involved the resistance response, including BAK1, SOBIR1, MAPK4 and WRKY51, were increased in the OE and T12–19 lines and reduced in 91–112 and the BrWAK1 gene editing lines (Figure S11 and Table S3). In a word, BrWAK1 could interact with BrBAK1 to activate downstream resistance signalling, including the MAPK cascade, camalexin biosynthesis, hydrogen peroxide metabolism and phytohormone signalling.
Figure 7.
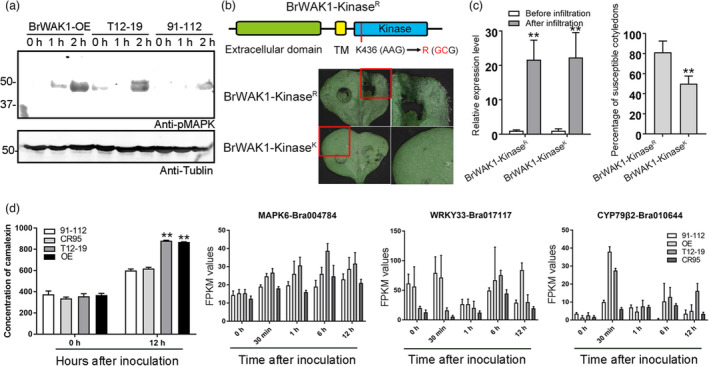
BrWAK1 could interact with BrBAK1 to activate downstream signalling. (a) MAPK phosphorylation in the BrWAK1 overexpression lines, T12–19, and 91–112 at 0, 1, and 2 HAI. The tubulin protein in each sample was used as the loading control. (b) The schematic diagram of kinase activity site mutated BrWAK1 (K436R, BrWAK1‐KinaseR) and the disease symptoms of cotyledons infiltrated with BrWAK1‐KinaseR and BrWAK1 without mutation (BrWAK1‐KinaseK) after inoculation. The images in red rectangles are enlarged in the right panels. (c) The expression levels of BrWAK1‐Kinase R and BrWAK1‐Kinase K in injected cotyledons before and after inoculation and the susceptible percentage of infiltrated cotyledons after inoculation. Expression of BrWAK1 in samples before infiltration in each sample were defined as 1.0. Values represent means ± SD (n = 3) from three biological replicates. Statistical significances between samples before infiltration were by a t‐test: **P < 0.01. More than fifty leaves were observed after inoculation in each replicate. Values represent means ± SD (n = 3) from three biological replicates. Statistical significance between transgenic lines with kinase activity site mutated BrWAK1 (BrWAK1‐KinaseR) was by a t‐test: **P < 0.01. (d) The contents of camalexin and the expression of crucial genes involved in camalexin biosynthesis and signalling, including MAPK6, WRKY33, and CYP79β2, in 91–112, T12–19, the BrWAK1 overexpression line and the gene editing line (CR95) after inoculation. More than five leaves were collected in each replicate. Values represent means ± SD (n = 3) from three biological replicates. Statistical significances between the camalexin concentrations of 91–112 at 0 HAI were by a t‐test: **P < 0.01.
Gain‐ and loss‐function of BrWAK1 do not significantly influence plant biomass
To characterize the function of BrWAK1 in plant development, the phenotypes of the BrWAK1 overexpression lines (OE‐182 and OE‐186) and the BrWAK1 truncated lines (CR95 and CR80) were observed during the whole growth period. Similar plant morphology was observed between 91–112 and OE‐186, and T12–19 and CR95 (Figure 8a). Compared with 91–112, the BrWAK1 overexpression lines exhibited slight larger leaves in terms of both leaf length and width, while they were similar in terms of plant height and yield per plant (Figure 8b). There was no significant difference in leaf length, leaf width, plant height and yield per plant between T12–19 and the BrWAK1 truncated lines (Figure 8b). Therefore, the overexpression and gene editing of BrWAK1 could only change the disease resistance ability, with no obvious developmental changes except for a slightly larger leaf in the overexpression lines (Figure 8). These results implied that BrWAK1 will helpful for further resistance breeding efforts to control downy mildew in Chinese cabbage.
Figure 8.
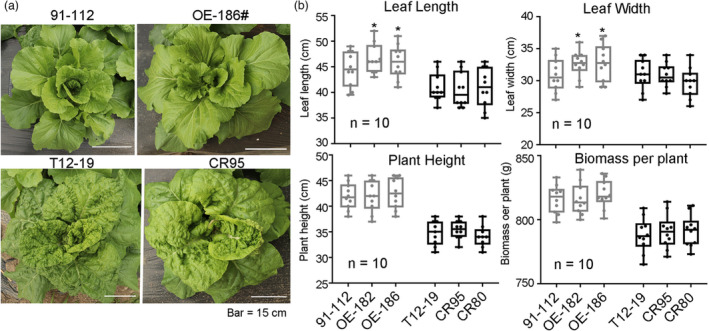
The phenotype of BrWAK1 overexpression and gene editing lines during the whole growth period. (a) The rosette stage of 91–112, T12‐19, the BrWAK1 overexpression line (OE‐186), and the gene editing line (CR95). (b) The leaf length, leaf width, plant height, and biomass per plant of 91–112, T12–19, the BrWAK1 overexpression lines (OE‐186 and OE‐182), and the gene editing lines (CR95 and CR80). A total of 5–10 seedlings in each line were measured. Values represent means ± SD (n = 10). Statistical significances between 91112 or T12–19 were by a t‐test: *P < 0.05.
Discussion
During cool and humid days, H. Brassicae can invade leaves of Chinese cabbage. The pathogens develop and reproduce in leaves and eventually the sporangium and sporangia grow out of epidermal cells to spread spores to infect other leaves. The resistant line T12–19 could trigger PTI immune reactions to inhibit the invasion from epidermal cells. In this study, BrWAK1 was cloned for quantitative resistance to downy mildew in T12–19. The resistant parent T12–19 used in this article is introduced from Japan. It was cultivated in recent decades and it was not a common variety in Chinese cabbage breeding. We performed haplotype analysis in more than 200 natural varieties and we found that this resistant variation only exists in T12–19. This indicated that the resistant locus in T12–19 is a rare variation, but not a domesticated locus. BrWAK1 has a comparatively higher expression in leaf than in other tissues and its expression could be markedly induced after pathogen inoculation. Therefore, the spatiotemporal expression pattern of BrWAK1 showed that BrWAK1 could encounter the pathogen at the proper time and location. BrWAK1 could interact with BrBAK1 to activate the MAPK phosphorylation cascade and downstream signalling, including biosynthesis of resistant phytohormones and camalexin. Whether the intracellular kinase activity of BrWAK1 could phosphorylate other proteins to activate resistance signalling in addition to BAK1 is also worthy of investigation. A yeast library of the leaf in T12–19 after inoculation was constructed and a candidate resistant protein was successfully identified using the intracellular part of BrWAK1 as the bait (data unpublished). These results helped us to hypothesize the disease resistance mechanism of BrWAK1 (Figure 9). Intriguingly, we found that resistant phytohormone SA could upregulate the expression of BrWAK1, which implied that there is probably positive feedback between SA and BrWAK1 for further expansion of the resistance signals (Figure 9). However, the factors involved in regulating this positive feedback, e.g., the factors that could terminate resistance signalling transduction, are unknown and are valuable for further investigation. In addition, BrWAK1 is a receptor‐like kinase protein and is located on the cell membrane to participate PTI immune reactions in response to downy mildew; however, the resistant genes participating in ETI immune reactions response to downy mildew in Chinese cabbage should also be explored.
Figure 9.
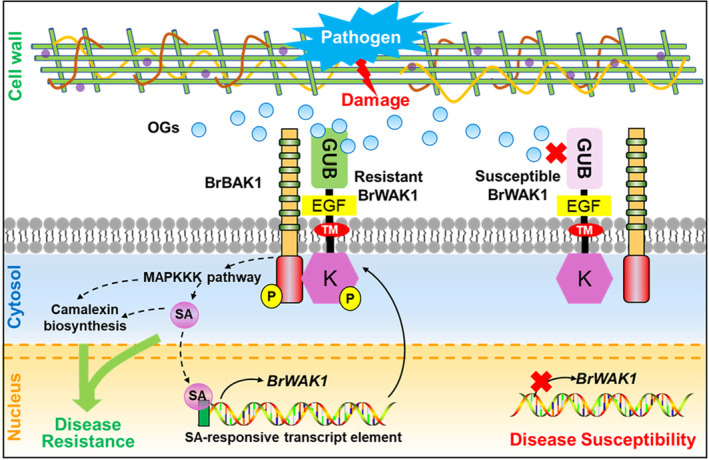
Schematic diagram of the BrWAK1 working model in downy mildew resistant and susceptible lines. OGs released from the damaged cell wall caused by pathogen could be perceived by resistant genotype of BrWAK1 (Resistant BrWAK1). Then, it could trigger downstream resistance responses, such as MAPK cascade, SA and camalexin biosynthesis through the interaction with BrBAK1. Increased SA could bind SA‐responsive transcript element in the promoter of Resistant BrWAK1 to enhance the transcription of BrWAK1, which could continuously perceive signals caused by pathogen invasion. However, the susceptible genotype of BrWAK1 (Susceptible BrWAK1) could not perceive OGs and could not activate downstream resistance responses.
The WAK gene family participates extensively in cell shape determination, leaf development, metal ion stress and the disease resistance response (He et al., 1998; Kohorn et al., 2006). In contrast to most transmembrane receptor proteins, the WAK family are associated with the cell wall and can perceive signals from the cell wall and then transduce them into the cytoplasm. Consequently, the WAK family could play an important role in cell adaptation to various circumstances and intercellular communication during plant development. Overexpression of BrWAK1 in 91–112 made the leaves slightly larger than those in 91–112, which implied that BrWAK1 probably has extra functions in leaf development. Besides, the expression level of BrWAK1 could be induced under high‐humidity condition, which was facilitate for H. Brassicae infection (Zhang et al., 2020). It implied that higher expression level of BrWAK1 under high‐humidity condition in T12–19 might also contribute to disease resistance. Likewise, under other abiotic stress treatments, T12–19 and the BrWAK1 overexpression lines showed stronger stress tolerance than 91–112 (data unpublished). All these results implied that BrWAK1 might be the crucial node in the crosstalk of multi‐stress responses, which would help to improve our understanding of stress crosstalk mediated by BrWAK1 in Chinese cabbage. As a disease resistant gene, its overexpression could enhance plant resistance typically along with the loss of biomass. Fortunately, BrWAK1 overexpression and gene edited lines do not have significant changes in biomass, which is probably owing to its functions in developmental regulation. It will also greatly assist with disease resistance breeding of Chinese cabbage. In this study, the identification and characterization of BrWAK1 in downy mildew resistance not only paves the way to clarify the molecular mechanism of disease resistance, but also promotes further molecular associated breeding efforts for downy mildew resistance in Chinese cabbage.
Materials and methods
Plant material
The mapping population used in this study was derived from a cross between T12–19, a downy mildew highly resistant line, and 91–112, a highly susceptible line. 91–112 is a Chinese cabbage inbred line (B. rapa ssp. pekinensis) that was self‐fertilized for nine generations. T12–19 is a double haploid (DH) line derived from a cross between two Chinese cabbage inbred lines, Orange Queen and DaBaiKou. The DH population (DH100) derived from across between 91–112 and T12–19 was used for preliminary mapping of the major resistant QTL and a 2.9 cM region flanked by the random amplification of polymorphic DNA (RAPD) marker K14‐1030 and isozyme marker PGM were identified on Chromosome A08 (Yu et al., 2009). For fine mapping, chromosomal segment substitution lines (CSSLs) were constructed. We identified DH60, the susceptible DH line from the DH100 population, as the backcrossing (BC) parent to develop BC1F1, BC2F1 and BC3F1. The BC populations were also developed into DH populations. The BC populations, comprising 1300 lines, and more than 700 DH lines, were used for the fine mapping of the downy mildew resistant QTL.
Evaluation of downy mildew resistance phenotype
Two‐week‐old seedlings, growing at 25 °C under a 16 h light/8 h dark cycle, were used for downy mildew inoculation. The acquisition of pathogen isolates and the method of maintaining the inoculum are described by Yu et al. (2016). For inoculation, about 2 × 105 spores/μL of the H. brassicae isolate were collected as a conidial suspension and then sprayed on the abaxial side of leaves (Yu et al., 2009). After inoculation, the seedlings need high‐humidity treatment for 3–4 days with the first 24 h in the dark for pathogen invasion. Three inoculation replicates were conducted, with ten plants per replicate (n = 30). The phenotypic data were then collected, and disease indices (DIs) for each line were calculated as described in Yu et al. (2009).
Phylogenetic analysis
The phylogetic trees of the WAKs/WAKLs, BAK1 and SOBIR1 (suppressor of BIR1 1) from Brassica rapa and Arabidopsis were constructed using MEGA 7.0 with the Neighbour‐Joining method (Kumar et al., 2016). The amino acid sequences were obtained from The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/) and Brassicaceae Database (BRAD, http://brassicadb.cn/).
Gene sequence analysis and vector construction
The reference genomic DNA and coding sequences of BrWAK1 (Bra016426), BrWAK2 (Bra016427), BrWAK3 (Bra016428), BrBAK1 (Bra034562) and BrSOBIR1 (Bra022670) were derived from BRAD. The upstream reference sequence (~1.5 kb) of BrBAK1 was also obtained from BRAD. Based on the reference sequences, primers were designed for amplification in T12–19 and 91–112. The amplified coding sequences of BrWAK1 were sequenced and constructed into vector pMDC43 (Invitrogen, Waltham, MA) for transgenic overexpression. The coding sequences of BrWAK1, BrWAK1 K436R , BrWAK2 and BrWAK3 were cloned into the vector pSuper1300 for transient expression as a C‐terminal green fluorescent protein (GFP) tag fusion protein (Gong et al., 2002). The vectors for RNAi were constructed as described in Zhang et al. (2018). The specific fragments of BrWAK1/2/3 were synthesized (BGI, Beijing, China). For cas9‐mediated gene editing of BrWAK1, the guide RNA (gRNA) sequences of BrWAK1 were identified and evaluated using CRISPRscan (https://www.crisprscan.org/?page=sequence) and Cas‐OFFinder (http://www.rgenome.net/cas‐offinder/), respectively. Using vector pCBC–DT1T2 as the template, two AtU6 promoter‐gRNA‐AtU6 terminator cassettes were induced into plasmid pKSE401 (Xing et al., 2014). To detect protein interactions, the full length coding sequences of BrWAK1, BrBAK1 and BrSOBIR1 were cloned into vectors pSPYNE, pSPYCE (Waadt et al., 2008), pCAMBIA‐NLuc and pCAMBIA‐CLuc (Chen et al., 2008) for the Bimolecular Fluorescence Complementation (BiFC) and Firefly Luciferase Complementation (FLC) systems using homologous recombination technology. For chimeric receptor construction, the sequences of chimeric BrWAK1 containing GUBT12–19(1–244)‐EGF91–112(234–330)‐TM‐Kinase or GUB91‐112(1–233)‐EGFT12–19(245–331)‐TM‐Kinase were synthesized (BGI) and constructed into vector pSuper1300. The sequence containing Promoter91–112::BrWAK1T12–19 was also synthesized (BGI) and constructed into vector pSuper1300 using the BamHI and KpnI sites (the Super35S promoter was removed). All the primers used are listed in Table S4.
RNA extraction and qRT–PCR analysis
Total RNA was extracted from leaves and the reverse transcribed into cDNA using a plant RNeasy kit (TIANGEN, Beijing, China) and PrimeScript™ RT reagent Kit (Takara, Shiga, Japan), respectively. The SYBR Green I Master Mix (Roche, Basel, Switzerland) was used for the quantitative real‐time PCR (qPCR) reactions. The relative expression levels of genes were quantified using the Light Cycler 480 II system (Roche). The reaction conditions were as follows: an initial denaturation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s (denaturation), 60 °C for 30 s (annealing) and 72 °C for 45 s (extension). PCR amplification was followed by heating for 1 min at 60–95 °C for melting curve analysis. Each sample reaction was performed with three replicates using 5 μL of Master Mix, 0.25 μm of each primer, 1 μL of diluted cDNA and DNase‐free water to a final volume of 10 μL. The Chinese cabbage GADPH gene (encoding glyceraldehyde‐3‐phosphate dehydrogenase) was used as internal control (Qi et al., 2010).
Agroinfiltration
Agroinfiltration for gene transient expression and subsequent infection experiments were performed according to a previous description (Zhang et al., 2018). Three days after infiltration, GFP fluorescence was observed under a confocal microscope (Zeiss LSM700, Tokyo, Japan). Seven days after inoculation, the disease‐resistant phenotype of the infiltrated cotyledons was investigated.
Generation of transgenic plants
Chinese cabbage transfromation was performed according to the method described by Su et al. (2021). The plasmids pMDC43‐BrWAK1 and pSK401‐gRNABrWAK1 were transformed into Agrobacterium tumefaciens strain LBA4404, which was cultured to OD600 = 2.0 in Luria‐Bertani broth with and without 50 mg/L kanamycin successively. The cotyledon petioles of Chinese cabbage were inoculated with Agrobacterium cells diluted in Murashige & Skoog (MS) medium (pH 5.7) for 5 s and then transferred to cocultivation medium (MS medium supplemented with 500 mg/L 2‐(4‐morpholino)ethanesulfonic acid (MES), 4 mg/L 6‐benzylaminopurine (BAP), 0.1 mg/L 1‐naphthalene acetic acid (NAA), and 2.75% Phytagel (P8169, Sigma, St. Louis, MO), pH 5.7) under low‐light condition at 25 °C for 2 days. The transformation positive explants were then selected under 25 mg/L kanamycin and identified by PCR and qRT–PCR. In the BrWAK1 editing transgenic lines, the target sequences were amplified and cloned into vector pMD™ using the 18‐T Vector Cloning Kit (Takara, Dalian, China) for sequencing. More than 20 clones from each sample were sequenced and then sequence analysis was performed.
Agrobacterium with chimeric BrWAK1 vectors were transformed into Arabidopsis using the flower‐dipping method (Clough and Bent, 1998). Transgenic plants were selected on MS‐agar plates with 50 mg/L kanamycin. To test the expression of the transgenes, the fully expanded leaves from 4‐week‐old soil‐grown plants were taken, quickly frozen in liquid nitrogen, and stored at −80 °C until use. Homozygous transgenic lines of the T3 generation of were used for the phenotype analysis.
SA and JA treatment
SA and JA treatments were performed according to a previous description (Zhang et al., 2018). SA at 1.5 mm (Sigma‐Aldrich, Lot V900072) and 1 mm methyl‐(Me)‐JA (Sigma‐Aldrich, Lot 392 707) dissolved in water were sprayed on the abaxial side of the leaves of T12–19 and 91–112 plants. Seedlings sprayed with water were designated as the controls. The ratio of gene expression under phytohormone treatment to that under water treatment was considered as the gene induction level by the phytohormone.
Measurement of Camalexin, SA, and JA
The leaves of T12–19, 91–112, and the BrWAK1 overexpression lines were collected after downy mildew inoculation. Leaves of five seedlings were mixed to form three biological repeats. SA and JA were measured using liquid chromatography mass spectrometry (LCMS) (Fan‐Xing Biological Technology Beijing Co., Ltd, Beijing, China). Twelve hours after inoculation, the leaves of T12–19, 91–112, the BrWAK1 gene edited lines and the overexpression lines were sampled. The seedlings under high‐humity conditions without inoculation were used as controls. Leaves of 5–10 seedlings were mixed into three biological repeats. Camalexin was measured using an HITACHI F4500 spectrofluorometer (315 nm excitation and 385 nm emission; Hitachi, Tokyo, Japan), and the concentration of camalexin was determined by comparison with a camalexin standard curve (Fan‐Xing Biological Technology‐ Beijing Co., Ltd).
Bimolecular fluorescence complementation
The Agrobacterium strains GV3101 containing the BiFC vectors pSCYCE and pSCYNE (Waadt et al., 2008) were infiltrated into leaves of 1‐ or 1.5‐month‐old tobacco (Nicotiana benthamiana) plants as described previously (Waadt and Kudla, 2008). Fluorescence signals were observed 3–5 days later using a fluorescence microscope (Zeiss, Axio Imager M2). Images were analysed using Image ZEN software (Zeiss).
Luciferase (LUC) activity measurement
The Agrobacterium strains GV3101 carrying pCambia‐NLuc or pCambia‐CLuc were co‐infiltrated into the leaves of tobacco (Nicotiana benthamiana). Two or three days after infiltration, the leaves were collected and one millimolar luciferin was sprayed onto the leaves. The leaves were kept in dark for 6 min and placed into a low‐light cooled charge‐coupled device (CCD) imaging apparatus (NightSHADE LB985 with Indigo software; Berthold Technologies GmbH & Co. KG, Bad Wildbad, Germany) to capture the LUC images. An exposure time of 2 min was used and the relative LUC activity was measured as described previously (Chen et al., 2008). Relative LUC activity was equivalent to luminescence intensity/0.2 mm2 leaf area. At least three independent experiments were performed for each assay.
Transient expression in Chinese cabbage protoplasts
For promoter activity analysis, the double‐LUC reporter assay vector containing a firefly LUC gene driven by the 1.5 kb promoter sequences of BrWAK1 in T12–19 or 91–112 and an internal control Renilla LUC (REN) driven by the 35S promoter, which was modified based on the pGreenII 0800‐LUC reporter vector (Hellens et al., 2005). The constructed effector and reporter plasmids were co‐transformed into Chinese cabbage protoplasts (Su et al., 2019). The co‐transfected protoplasts were incubated for 16 h at 25 °C before harvest. For SA treatment, 50 μm SA was added for 1, 2, 3, and 6 h and then protoplasts were lysed in 100 mL of lysis buffer (E4550; Promega, Madison, WI). After centrifugation, the supernatant was used for LUC and REN LUC activities analysis using a dual‐LUC assay kit (Promega) on a Tecan M1000 reader (TECAN, Männedorf, Switzerland). The results were calculated using the ratio of LUC to REN, which defined the relative promoter activity. At least three biological repeats were assayed for each construct.
Virus‐induced gene silencing (VIGS)
The recombinant plasmid pTY‐S (the empty pTY‐S plasmid was provided by Professor Jinghua Yang, Horticulture department, Zhejiang university) containing specific palindromic 80‐nt oligonucleotides of BrWAK1 was extracted in large quantities for virus infiltration. The method of infiltration was performed as described by Zhang et al. (2021). First, silicon carbide powder was sprinkled onto the leaf surface of four‐fully expanded‐leaf seedlings. After gentle friction, 8 μL of purified pTY‐S recombinant plasmid (300 ng/μL) was applied evenly onto the leaf for 2–5 min, and then the leaves were washed with water. The infiltrated seedlings were placed under dark conditions for 16 h. Three or four infiltrations were performed over 2 weeks. Two weeks after infiltration, the seedlings were inoculated with H. brassicae and the phenotypes were observed and the gene expression levels measured.
Co‐immunoprecipitation and MAPK phosphorylation
Whole proteins were extracted from infiltrated N. benthamiana leaves using 2 × extraction buffer (50 mm Tris–HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, pH 8.0, 0.1% Triton‐X‐100, 0.1% Nonidet P‐40, 5 mm DTT, 20 μm MG132 and 1 × protease inhibitor cocktail tablets). For co‐immunoprecipitation, the mixture was centrifuged at 13,000 g for 20 min and anti‐HA tag agarose (MBL, Woburn, MA) or anti‐Myc agarose (Santa Cruz Biotechnology, Santa Cruz, CA) was added to the supernatant for incubation at 4 °C for 2–4 h. After incubation, the agarose beads were washed for three to five times using wash buffer (50 mm Tris–HCl, pH 7.5, 150 mm NaCl, and 0.1% NP‐40) and then SDS sample buffer was added before boiling 10 min for immunoblot analysis. For immunoblot analysis, proteins were separated by SDS–PAGE in 12% acrylamide gels and electroblotted to polyvinylidene fluoride membrane (Millipore, Billerica, MA) at 360 V for 60 min. After reaction with primary and secondary antibodies, the immunoreactive protein bands were detected using ECL Prime Western Blotting Detection Reagent (GE Healthcare, Chicago, IL). Antibodies and the dilutions used in these experiments were as follows: anti‐HA antibody (Labcorp Drug Development (formerly Covance), Princeton, NJ; lot MMS‐101R, 1 : 1000), anti‐Myc antibody (Sigma‐Aldrich; M4439, 1 : 5000), anti‐GFP antibody (Abmart, Shanghai, China; M2004, 1 : 2000), and goat anti‐mouse horseradish peroxidase‐conjugated antibody (BPI, Beijing, China; lot AbP71003‐D‐HRP, 1 : 10 000).
After inoculation, the leaves from at least three 4‐week‐old seedlings were mixed into one sample at 0, 1, and 2 h for phosphorylated MAPK detection. Proteins were extracted using lysis buffer (50 mm Tris–HCl, pH 7.5, 0.25% sodium deoxycholate, 15 mm EGTA, 100 mm NaCl, 10 μg/mL Aprotenin, 10 μg/mL Leupeptin, 1 mm PMSF, 1 mm NaF, 1 mm Na3VO4, 10 mm β‐glycerophosphate, 0.1% NP‐40 and 1 × protease inhibitor cocktail). Total protein levels were quantified using the Bradford assay. Total protein was also normalized by immunoblotting with tubulin antibodies (Cell Signalling Technology, Danvers, MA, #2144, 1 : 1000). Phospho‐p44/42 MAPK (Erk1/2) (Thr202/Tyr204) rabbit monoclonal antibodies (Cell Signalling Technology, #4370L, 1 : 2000) were used for immunoblotting by overnight incubation at 4 °C.
3,3′‐Diaminobenzidine (DAB) and callose staining
For OG treatment, four‐week‐old Arabidopsis seedlings were used for water or OG (200 μg/mL) infiltration using a needleless syringe. For H. brassicae treatment, Chinese cabbage seedlings with four fully expanded leaves were inoculated. DAB staining was used to detect the accumulation of H2O2 (Thordal‐Christensen et al., 1997). Leaves after inoculation or OG treatment were dipped in freshly prepared 100 μg/mL DAB solution, containing 1 mg/mL of 3,3′‐diaminobenzidine‐HCl, pH 5.0 for 12 h in the dark. Stained leaves were placed in boiling ethanol for 10 min to remove chlorophyll and then transferred to ethanol for 2 h at room temperature prior to photography (Orozco‐Cardenas and Ryan, 1999). For callose staining, the leaves were dehydrated with 100% ethanol and fixed in an acetic acid: ethanol (1:3) solution for 2 h. Sequentially, they were incubated for 15 min in 75% ethanol, 50% ethanol, and phosphate buffer (150 mm, pH 8.0), and then stained in dyeing solution (0.01% (w/v) aniline blue in 150 mm phosphate buffer, pH 8.0) for 1 h at 25 °C. After staining, callose was detected using UV epifluorescence under microscope (Zeiss, Axio Imager Z2). Each sample contained about five leaves from at least three independent plants.
RNAseq analysis
mRNAs were isolated and cDNA libraries were constructed by the Beijing Genomics Institute (BGI, Shenzhen, China). Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's protocols. Strand‐specific sequencing libraries were constructed according to previously described protocols (Borodina et al., 2011). The sequencing libraries were then subjected to high‐throughput sequencing on an Illumina HiSeq Xten paired‐end 150 (PE150) instrument (Illumina, San Diego, CA). Clean reads were obtained by removing reads with adapters, reads with poly‐N sequences, and low‐quality reads. The clean reads were mapped to the B. rapa reference genome (v1.5) using TOPHAT (Pollier et al., 2013). The alignment was assembled and the transcripts were quantified using StringTie (Pertea et al., 2015) and then the assembled transcripts were annotated using the Cufflinks package (Trapnell et al., 2010). The fragments per kilobase of exon per million mapped fragments (FPKM) values of coding mRNAs were calculated using Cuffdiff (v2.1.1) software (Trapnell et al., 2010). For all pairwise comparisons, the expression levels of mRNAs with corrected P‐values <0.05 and absolute fold‐change values >2.0 were considered to be differentially expressed (Liang et al., 2017). The gene ontology (GO) analysis in this study was performed using agriGO (Tian et al., 2017). The GO terms of biological processes (P) (P < 0.01) were selected for further analysis.
Statistical analysis
A t‐test was performed using SPSS software to determine the statistical significance of differences at P < 0.01 and used for the following comparisons: gene relative expression analysis, GO analysis, phytohormone content comparisons, the percentage of susceptible cotyledons comparison, luciferase activity comparison and developmental phenotypes of transgenic plants.
Conflict of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
Y.S.C., Z.B. and Z.F.L. designed experiments. Z.B., Y.S.C., S.T.B, X.X.Y., L.P.R., W.W.H., Z.X.Y., Z.D.S. and Y.Y.J. carried out the experiment. Z.B., S.T.B., Y.S.C. and Z.F.L. wrote the article. All the authors discussed the results and commented on the article.
Supporting information
Figure S1 The schematic diagram of the construction of the fine mapping population.
Figure S2 The phylogenetic analysis and sequence alignment between BrWAK1/23 and AtWAKs/WAKLs.
Figure S3 The amino acid sequence variation and expression level after inoculation of BrWAK2 and BrWAK3 in T12–19 and 91–112.
Figure S4 Overexpression of BrWAK1/2/3 using the agroinfiltration‐mediated transient expression system in the Chinese cabbage cotyledons.
Figure S5 The subcellular location of BrWAK1‐GFP in infiltrated Chinese cabbage cotyledons.
Figure S6 RNAi of BrWAK1/2/3 using an agroinfiltration‑mediated transient expression system in Chinese cabbage cotyledons.
Figure S7 The gene silencing of BrWAK1 in the VIGS system reduced the disease resistance of T12–19.
Figure S8 The phylogenic analysis of AtBAK1 (a) and AtSOBIR1 (b) with their homologues in Chinese cabbage using amino acid sequences.
Figure S9 The interaction of BrWAK1‐S and BrBAK1, EGFR‐BrWAK1, and BrBAK1 in the BiFC assay.
Figure S10 GO analysis of upregulated genes in the BrWAK1 overexpression lines (a) and downregulated genes in the BrWAK1 editing lines (b) after inoculation.
Figure S11 The RPKM values of marker genes involved in disease resistance in the BrWAK1 overexpression lines (a) and BrWAK1 editing lines (b) in the RNAseq data.
Table S1 The genotypes of molecular markers among Bra‐DM08 in recombinants.
Table S2 The sequences of BAC clone containing part of Bra‐DM08.
Table S3 The FPKM values of disease resistant‐related genes in RNAseq data.
Table S4 The sequences of Primers used in this study.
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2022YFF1003001), Beijing Joint Research Program for Germplasm Innovation and New Variety Breeding (G20220628003‐01), the Natural Science Foundation of China (31872126, 32072567), the Outstanding Scientists Training program of BAAFS (JKZX201906), and the earmarked fund for China Agriculture Research System (CARS‐23‐A‐05). [Corrections added on 17 July 2023, after first online publication: Names of funding projects in the Acknowledgements section have been corrected in this version.]
Contributor Information
Fenglan Zhang, Email: zhangfenglan@nercv.org.
Shuancang Yu, Email: yushuancang@nercv.org.
References
- Anderson, C.M. , Wagner, T.A. , Perret, M. , He, Z.‐H. , He, D. and Kohorn, B.D. (2001) WAKs: cell wall‐associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol. Biol. 47, 197–206. [PubMed] [Google Scholar]
- Borodina, T. , Adjaye, J. and Sultan, M. (2011) Chapter five – a strand‐specific library preparation protocol for RNA sequencing. In Methods in Enzymology ( Jameson, D. , Verma, M. and Westerhoff, H.V. , eds), pp. 79–98. Cambridge, MA: Academic Press. [DOI] [PubMed] [Google Scholar]
- Brutus, A. , Sicilia, F. , Macone, A. , Cervone, F. and De Lorenzo, G. (2010) A domain swap approach reveals a role of the plant wall‐associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA, 107, 9452–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burgh, A.M. , Postma, J. , Robatzek, S. and Joosten, M.H.A.J. (2019) Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol. 20, 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Zou, Y. , Shang, Y. , Lin, H. , Wang, Y. , Cai, R. , Tang, X. et al. (2008) Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol. 146, 323–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.F. , Qing‐Hua, M.A. , Jin‐Hua, M.U. and Wang, B.C. (2015) PRs genes expression in Chinese cabbage induction by Peronospora parasitica . J. Jilin Agric. Sci. 40, 60–64. [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium ‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Delteil, A. , Gobbato, E. , Cayrol, B. , Estevan, J. , Michel‐Romiti, C. , Dievart, A. , Kroj, T. et al. (2016) Several wall‐associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 16, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoux, C. , Galletti, R. , Mammarella, N. , Gopalan, S. , Werck, D. , De Lorenzo, G. , Ferrari, S. et al. (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant, 1, 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, P. and Ding, Y. (2020) Stories of salicylic acid: a plant defense hormone. Trends Plant Sci. 25, 549–565. [DOI] [PubMed] [Google Scholar]
- Dmochowska‐Boguta, M. , Kloc, Y. , Zielezinski, A. , Werecki, P. , Nadolska‐Orczyk, A. , Karlowski, W.M. and Orczyk, W. (2020) TaWAK6 encoding wall‐associated kinase is involved in wheat resistance to leaf rust similar to adult plant resistance. PLoS One, 15, e0227713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, T. , Yu, S. , Zhang, F. , Chen, X. , Yu, Y. , Zhang, D. , Zhao, X. et al. (2014) Expression analysis of major genes involved in signaling pathways during infection of Chinese cabbage with Hyaloperonospora brassicae . Sci. Hortic. 167, 27–35. [Google Scholar]
- Glazebrook, J. , Chen, W. , Estes, B. , Chang, H.‐S. , Nawrath, C. , Métraux, J.‐P. , Zhu, T. et al. (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217–228. [DOI] [PubMed] [Google Scholar]
- Gómez‐Gómez, L. and Boller, T. (2000) FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gong, Z. , Lee, H. , Xiong, L. , Jagendorf, A. , Stevenson, B. and Zhu, J.‐K. (2002) RNA helicase‐like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. 99, 11507–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z.‐H. , Fujiki, M. and Kohorn, B.D. (1996) A cell wall‐associated, receptor‐like protein kinase. J. Biol. Chem. 271, 19789–19793. [DOI] [PubMed] [Google Scholar]
- He, Z.‐H. , He, D. and Kohorn, B.D. (1998) Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 14, 55–63. [DOI] [PubMed] [Google Scholar]
- Heese, A. , Hann, D.R. , Gimenez‐Ibanez, S. , Jones, A.M.E. , He, K. , Li, J. , Schroeder, J.I. et al. (2007) The receptor‐like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA, 104, 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P. , Allan, A.C. , Friel, E.N. , Bolitho, K. , Grafton, K. , Templeton, M.D. , Karunairetnam, S. et al. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods, 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurni, S. , Scheuermann, D. , Krattinger, S.G. , Kessel, B. , Wicker, T. , Herren, G. , Fitze, M.N. et al. (2015) The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall‐associated receptor‐like kinase. Proc. Natl. Acad. Sci. USA, 112, 8780–8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Katagiri, F. (2004) A global view of defense gene expression regulation – a highly interconnected signaling network. Curr. Opin. Plant Biol. 7, 506–511. [DOI] [PubMed] [Google Scholar]
- Kim, S. , Song, Y.H. , Lee, J.‐Y. , Choi, S.R. , Dhandapani, V. , Jang, C.S. , Lim, Y.P. et al. (2011) Identification of the BrRHP1 locus that confers resistance to downy mildew in Chinese cabbage (Brassica rapa ssp. pekinensis) and development of linked molecular markers. Theor. Appl. Genet. 123, 1183–1192. [DOI] [PubMed] [Google Scholar]
- Kohorn, B.D. , Kobayashi, M. , Johansen, S. , Riese, J. , Huang, L.‐F. , Koch, K. , Fu, S. et al. (2006) An Arabidopsis cell wall‐associated kinase required for invertase activity and cell growth. Plant J. 46, 307–316. [DOI] [PubMed] [Google Scholar]
- Kohorn, B.D. , Johansen, S. , Shishido, A. , Todorova, T. , Martinez, R. , Defeo, E. and Obregon, P. (2009) Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 60, 974–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Zhou, S.‐Y. , Zhao, W.‐S. , Su, S.‐C. and Peng, Y.‐L. (2009) A novel wall‐associated receptor‐like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 69, 337–346. [DOI] [PubMed] [Google Scholar]
- Li, H. , Yu, S.‐C. , Zhang, F.‐L. , Yu, Y.‐J. , Zhao, X.‐Y. , Zhang, D.‐S. and Zhao, X. (2011) Development of molecular markers linked to the resistant QTL for downy mildew in Brassica rapa L. ssp. pekinensis . Yi Chuan, 33, 1271–1278. [DOI] [PubMed] [Google Scholar]
- Li, J. , Ding, Q. , Wang, F. , Li, H. , Zhang, Y. , Liu, L. , Jiao, Z. et al. (2018) Genome‐wide gene expression profiles in response to downy mildew in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Eur. J. Plant Pathol. 151, 861–873. [Google Scholar]
- Liang, R. , Han, B. , Li, Q. , Yuan, Y. , Li, J. and Sun, D. (2017) Using RNA sequencing to identify putative competing endogenous RNAs (ceRNAs) potentially regulating fat metabolism in bovine liver. Sci. Rep. 7, 6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T.W.H. , van den Berg, G.C.M. , Zhang, Z. , Smit, P. , Cordewener, J.H.G. , America, A.H.P. , Sklenar, J. et al. (2013) Receptor‐like kinase SOBIR1/EVR interacts with receptor‐like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA, 110, 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Liu, J. , Triplett, L. , Leach, J.E. and Wang, G.‐L. (2014) Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52, 213–241. [DOI] [PubMed] [Google Scholar]
- Malamy, J. , Carr, J.P. , Klessig, D.F. and Raskin, I. (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science, 250, 1002–1004. [DOI] [PubMed] [Google Scholar]
- Métraux, J.P. , Signer, H. , Ryals, J. , Ward, E. , Wyss‐Benz, M. , Gaudin, J. , Raschdorf, K. et al. (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science, 250, 1004–1006. [DOI] [PubMed] [Google Scholar]
- Ngou, B.P.M. , Jones, J.D.G. and Ding, P. (2022) Plant immune networks. Trends Plant Sci. 27, 255–273. [DOI] [PubMed] [Google Scholar]
- Orozco‐Cardenas, M. and Ryan, C.A. (1999) Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. 96, 6553–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M. , Pertea, G.M. , Antonescu, C.M. , Chang, T.‐C. , Mendell, J.T. and Salzberg, S.L. (2015) StringTie enables improved reconstruction of a transcriptome from RNA‐seq reads. Nat. Biotechnol. 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollier, J. , Rombauts, S. and Goossens, A. (2013) Analysis of RNA‐Seq data with tophat and cufflinks for genome‐wide expression analysis of jasmonate‐treated plants and plant cultures. In Jasmonate Signaling: Methods and Protocols ( Goossens, A. and Pauwels, L. , eds), pp. 305–315. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Qi, J. , Yu, S. , Zhang, F. , Shen, X. , Zhao, X. , Yu, Y. and Zhang, D. (2010) Reference gene selection for real‐time quantitative polymerase chain reaction of mRNA transcript levels in chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Mol. Biol. Rep. 28, 597–604. [Google Scholar]
- Saintenac, C. , Lee, W.‐S. , Cambon, F. , Rudd, J.J. , King, R.C. , Marande, W. , Powers, S.J. et al. (2018) Wheat receptor‐kinase‐like protein Stb6 controls gene‐for‐gene resistance to fungal pathogen Zymoseptoria tritici . Nat. Genet. 50, 368–374. [DOI] [PubMed] [Google Scholar]
- Shiu, S.‐H. and Bleecker, A.B. (2001) Receptor‐like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. 98, 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, T. , Li, P. , Wang, H. , Wang, W. , Zhao, X. , Yu, Y. , Zhang, D. et al. (2019) Natural variation in a calreticulin gene causes reduced resistance to Ca2+ deficiency‐induced tipburn in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Cell Environ. 42, 3044–3060. [DOI] [PubMed] [Google Scholar]
- Su, T. , Wang, W. , Li, P. , Xin, X. , Yu, Y. , Zhao, X. , Zhang, D. et al. (2021) Natural variations of BrHISN2 provide a genetic basis for growth‐flavour trade‐off in different Brassica rapa subspecies. New Phytol. 231, 2186–2199. [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z. , Wei, Y. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Tian, T. , Liu, Y. , Yan, H. , You, Q. , Yi, X. , Du, Z. , Xu, W. et al. (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, J. , Van Pelt, J.A. , Van Loon, L.C. and Pieterse, C.M.J. (2002) Differential effectiveness of salicylate‐dependent and jasmonate/ethylene‐dependent induced resistance in Arabidopsis. Mol. Plant Microbe Interact. 15, 27–34. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Williams, B.A. , Pertea, G. , Mortazavi, A. , Kwan, G. , van Baren, M.J. , Salzberg, S.L. et al. (2010) Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica, J.A. and He, Z.‐H. (2002) The cell wall‐associated kinase (WAK) and WAK‐like kinase gene family. Plant Physiol. 129, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt, R. and Kudla, J. (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protocols, 2008, pdb.prot4995. [DOI] [PubMed] [Google Scholar]
- Waadt, R. , Schmidt, L.K. , Lohse, M. , Hashimoto, K. , Bock, R. and Kudla, J. (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56, 505–516. [DOI] [PubMed] [Google Scholar]
- Xing, H.‐L. , Dong, L. , Wang, Z.‐P. , Zhang, H.‐Y. , Han, C.‐Y. , Liu, B. , Wang, X.‐C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K. , Qi, L. and Zhang, Z. (2014) Isolation and characterization of a novel wall‐associated kinase gene TaWAK5 in wheat (Triticum aestivum). Crop J. 2, 255–266. [Google Scholar]
- Yu, S. , Zhang, F. , Yu, R. , Zou, Y. , Qi, J. , Zhao, X. , Yu, Y. et al. (2009) Genetic mapping and localization of a major QTL for seedling resistance to downy mildew in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Breed. 23, 573–590. [Google Scholar]
- Yu, S. , Su, T. , Zhi, S. , Zhang, F. , Wang, W. , Zhang, D. , Zhao, X. et al. (2016) Construction of a sequence‐based bin map and mapping of QTLs for downy mildew resistance at four developmental stages in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Mol. Breed. 36, 44. [Google Scholar]
- Zeier, J. (2021) Metabolic regulation of systemic acquired resistance. Curr. Opin. Plant Biol. 62, 102050. [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Li, P. , Su, T. , Li, P. , Xin, X. , Wang, W. , Zhao, X. et al. (2018) BrRLP48, encoding a receptor‐like protein, involved in downy mildew resistance in Brassica rapa . Front. Plant Sci. 9, 1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Li, P. , Su, T. , Li, P. , Xin, X. , Wang, W. , Zhao, X. et al. (2020) Comprehensive analysis of wall‐associated kinase genes and their expression under abiotic and biotic stress in Chinese cabbage (Brassica rapa ssp. pekinensis). J. Plant Growth Regul. 39, 72–86. [Google Scholar]
- Zhang, B. , Su, T. , Li, P. , Xin, X. , Cao, Y. , Wang, W. , Zhao, X. et al. (2021) Identification of long noncoding RNAs involved in resistance to downy mildew in Chinese cabbage. Hortic. Res. 8, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D.G. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]
- Zuo, W. , Chao, Q. , Zhang, N. , Ye, J. , Tan, G. , Li, B. , Xing, Y. et al. (2015) A maize wall‐associated kinase confers quantitative resistance to head smut. Nat. Genet. 47, 151–157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The schematic diagram of the construction of the fine mapping population.
Figure S2 The phylogenetic analysis and sequence alignment between BrWAK1/23 and AtWAKs/WAKLs.
Figure S3 The amino acid sequence variation and expression level after inoculation of BrWAK2 and BrWAK3 in T12–19 and 91–112.
Figure S4 Overexpression of BrWAK1/2/3 using the agroinfiltration‐mediated transient expression system in the Chinese cabbage cotyledons.
Figure S5 The subcellular location of BrWAK1‐GFP in infiltrated Chinese cabbage cotyledons.
Figure S6 RNAi of BrWAK1/2/3 using an agroinfiltration‑mediated transient expression system in Chinese cabbage cotyledons.
Figure S7 The gene silencing of BrWAK1 in the VIGS system reduced the disease resistance of T12–19.
Figure S8 The phylogenic analysis of AtBAK1 (a) and AtSOBIR1 (b) with their homologues in Chinese cabbage using amino acid sequences.
Figure S9 The interaction of BrWAK1‐S and BrBAK1, EGFR‐BrWAK1, and BrBAK1 in the BiFC assay.
Figure S10 GO analysis of upregulated genes in the BrWAK1 overexpression lines (a) and downregulated genes in the BrWAK1 editing lines (b) after inoculation.
Figure S11 The RPKM values of marker genes involved in disease resistance in the BrWAK1 overexpression lines (a) and BrWAK1 editing lines (b) in the RNAseq data.
Table S1 The genotypes of molecular markers among Bra‐DM08 in recombinants.
Table S2 The sequences of BAC clone containing part of Bra‐DM08.
Table S3 The FPKM values of disease resistant‐related genes in RNAseq data.
Table S4 The sequences of Primers used in this study.


