Summary
Staying ahead of the arms race against rust and mildew diseases in cereal crops is essential to maintain and preserve food security. The methodological challenges associated with conventional resistance breeding are major bottlenecks for deploying resistance (R) genes in high‐yielding crop varieties. Advancements in our knowledge of plant genomes, structural mechanisms, innovations in bioinformatics, and improved plant transformation techniques have alleviated this bottleneck by permitting rapid gene isolation, functional studies, directed engineering of synthetic resistance and precise genome manipulation in elite crop cultivars. Most cloned cereal R genes encode canonical immune receptors which, on their own, are prone to being overcome through selection for resistance‐evading pathogenic strains. However, the increasingly large repertoire of cloned R genes permits multi‐gene stacking that, in principle, should provide longer‐lasting resistance. This review discusses how these genomics‐enabled developments are leading to new breeding and biotechnological opportunities to achieve durable rust and powdery mildew control in cereals.
Keywords: biotechnology, biotrophic pathogens, cereal rusts, genomics, powdery mildew, resistance
Introduction
Whole‐grain cereals within the Triticeae including wheat (Triticum sp.), barley (Hordeum vulgare), rye (Secale cereale) and triticale (× Triticosecale Wittmack) are rich sources of calories, essential vitamins, minerals and phytochemicals that both nourish and protect humans and animals from ailments such as heart attack and cancer (Aune et al., 2016). Of these, wheat is the most widely consumed globally, while barley is the world's fourth most important cereal, used primarily in malt production for alcoholic beverages and as grain feed for livestock and human food (Figure 1; Tables S1 and S2). Although cereal cultivation is widespread, global demand and the ability to reliably cultivate large quantities of cereal grain varies substantially between wheat and barley and within different regions of the world (Figure 1).
Figure 1.
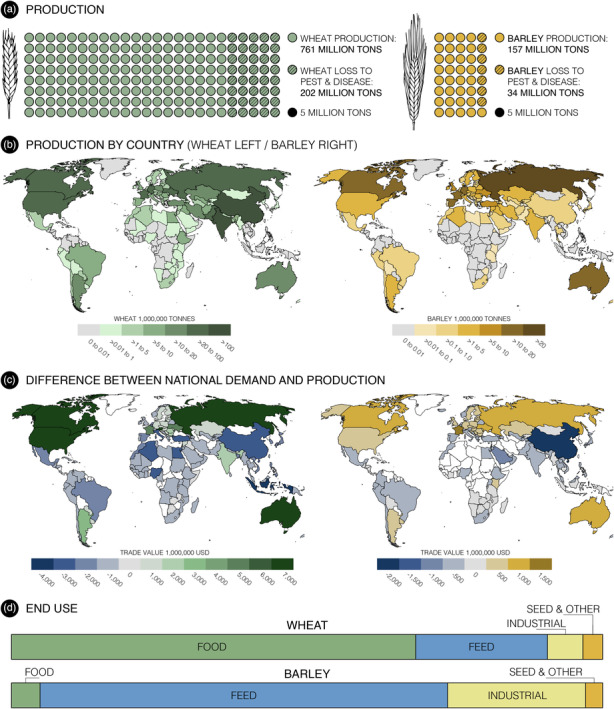
Global wheat and barley production relative to demand and the effect of pests and diseases on grain yield and end‐use data. (a) Estimates of yield loss to pests and diseases adapted from expert‐based assessments of crop health (Oerke and Dehne, 2004; Savary et al., 2019). (b) Worldwide wheat and barley production by country in 2017 (FAO stats accessed on 1 September 2022). (c) Trade deficit/surplus calculated as the value in US dollars between national demand and production for wheat and barley. (d) End‐use data for wheat and barley in 2020/21 as a proportion of whole production. https://www.igc.int/en/markets/marketinfo-sd.aspx (accessed on 1 September 2022). For the purpose of this map, the authors used production and trade data reported by the FAO and the World Bank. The authors remain neutral on issues of disputed regions and borders.
Wheat and barley are hosts for a myriad of pests and diseases that decrease projected yield and threaten crop production targets (Oerke and Dehne, 2004; Savary et al., 2019). For example, approximately 20% of global wheat production is lost to infection by pests and diseases (Figure 1a). Monoculture and climate change favour the emergence of new, highly virulent pathogen variants that reduce yield, posing a serious threat to global food security (Velásquez et al., 2018). Historically, foliar diseases of cereals have been controlled by (i) eradicating alternate hosts (Barnes et al., 2020), (ii) breeding resistant cvs. (Hafeez et al., 2021) and, more recently, (iii) spraying with systemic fungicides (Deising et al., 2008). Fungicides create selection pressure that encourages the emergence of fungicide‐resistant crop pathogen variants (Tucker et al., 2015) as well as environmentally ubiquitous yeast and Aspergillus variants which can transfer to humans to cause infections resistant to medical antifungals (Castelo‐Branco et al., 2021; Gow et al., 2022; Rhodes et al., 2022). Fungicides are also expensive, harmful to the environment and have been reported to deplete soil populations of mycorrhizal fungi, with negative consequences for crop mineral nutrition (Edlinger et al., 2022). Finally, exposure to fungicides has been linked to human cancers and other health disorders (Alhanti et al., 2022; Hutter et al., 2021; Lerro et al., 2021). Although genetic resistance has been used to control crop diseases, for example against powdery mildew in wheat and barley (Brown and Wulff, 2022), the durability of the resistance is often limited due to the ability of the pathogen to overcome major race‐specific resistance (R) genes (McDonald and Linde, 2002).
Fungal pathogens that cause disease in plants are categorized based on their feeding mechanisms. Biotrophic fungi colonize living plant cells and require carbon and nutrients provided by the host for growth and sporulation (Lorrain et al., 2019). Biotrophic fungal pathogens have a complex biochemical and structural relationship with their hosts (Dracatos et al., 2018). They contain specialized structures, called haustoria, that evaginate the host plasma membrane both creating a nutrient sink and suppressing host defences through the secretion of effector proteins (Lorrain et al., 2019). Often perceived to lack sophistication relative to their biotrophic counterparts, many necrotrophic fungal pathogens of cereals hijack host resistance responses deployed against biotrophs by targeting R genes and receptor kinases commonly associated with incompatible reactions in biotrophic interactions (Faris and Friesen, 2020; Hammond‐Kosack and Rudd, 2008). Therefore, rather than relying on haustoria, necrotrophic fungi tend to overpower their hosts with a cocktail of virulence factors and secreted molecules such as lytic enzymes (Koeck et al., 2011; Voegele and Mendgen, 2003). This often‐neglected perspective has important implications for the utility and pyramiding of R genes in breeding programmes and the development of gene stacks using biotechnological approaches.
The initial recognition of non‐self or pathogen‐associated molecular patterns (PAMPs) and subsequent mounting of downstream defence responses by plants is initiated at the cell surface by membrane‐associated pathogen recognition receptors (PRRs) and are collectively referred to as PAMP‐triggered immunity (PTI; Jones and Dangl, 2006). Adapted biotrophic pathogens have co‐evolved with their hosts and consequently produce effectors that enable them to evade PTI, colonize surrounding cells, and cause disease. The multi‐layered immune system from the host plant comprises large gene families encoding membrane‐bound and intracellular receptor proteins that recognize race‐specific pathogen effector molecules, leading to effector‐triggered immunity (ETI) (Dodds and Rathjen, 2010; Jones and Dangl, 2006). Recent studies in Arabidopsis have demonstrated that ETI and PTI potentiate each other (Ngou et al., 2021; Yuan et al., 2021) augmenting the importance of ETI gene‐based stacks. Our understanding of the genes involved in host‐biotrophic pathogen interactions is advanced, especially for membrane receptors that mount PTI and common R genes encoding nucleotide‐binding leucine‐rich repeat (NLR) proteins that trigger ETI. The products of other non‐canonical R genes underscore a level of mechanistic diversification that is less well understood (Sánchez‐Martín and Keller, 2021); however, in combination with NLRs, they are likely to provide a formidable barrier for pathogens to overcome (Brun et al., 2010).
New genomic approaches have enhanced the rate of gene isolation, improving our understanding of the molecular basis of host disease resistance and acquisition of virulence by the pathogen. In the case of biotrophic pathogens that infect cereals within the Triticeae, this molecular insight has been facilitated by an in‐depth knowledge of host–pathogen genetics for each of the respective pathosystems. Cereal rust (Puccinia spp.) and mildew (Blumeria spp.) pathogens are highly host‐specific, having co‐evolved with a narrow range of host species (Dracatos et al., 2018). Among Puccinia species, those that attack wheat are the most economically important (Savary et al., 2019). Stem rust, an epiphytotic disease caused by P. graminis f. sp. tritici (Pgt), can lead to crop failure during heavy epidemics, while stripe rust, caused by P. striiformis f. sp. tritici (Pst), and leaf rust, caused by P. triticina (Pt), cause losses of up to 50% on susceptible varieties. For the mildew fungus B. graminis, the pathogenic strains adapted to wheat (f. sp. tritici), or barley (f. sp. hordei) cause the most economic losses (Conner et al., 2003; Everts et al., 2001). In this review, we discuss recent developments in the molecular genetic analysis of wheat and barley interactions with rust‐ and powdery mildew‐causing fungi and explore opportunities to engineer durable host resistance.
Capturing functional diversity through genomics‐enhanced association genetics
The availability of reference genomes and the development of gene‐class‐specific capture arrays have undoubtedly enhanced R gene cloning efforts using map‐based cloning (Klymiuk et al., 2015; Walkowiak et al., 2020), mutational genomics (Sánchez‐Martín et al., 2016; Steuernagel et al., 2016) and genome‐wide association studies (GWAS) (Arora et al., 2019). Moreover, unbiased complexity reduction approaches, such as TACCA (targeted chromosome‐based cloning via long‐range assembly) or MutChromSeq, that are based on chromosome sorting and sequencing of wild‐type plants and chemically induced mutants have been applied to isolating genes involved in disease resistance (Dracatos et al., 2019; Sánchez‐Martín et al., 2016; Thind et al., 2017; Yu et al., 2022) and morphological characteristics (Ford et al., 2018) in cereals. A highly efficient approach combining PacBio Iso‐Seq of the wild‐type parental genotype with RNA sequencing of multiple mutants was used to clone the wheat leaf rust R gene Lr9 (Wang et al., 2023). This approach substantially decreased the cost and time of gene cloning, mitigating the necessity to sort chromosomes and is highly amenable for non‐reference wheat R gene donor lines. The necessity for genome complexity reduction methodology is decreasing as researchers capitalize more on the rapidly declining costs of DNA sequencing to generate reference genomes of resistant accessions (Athiyannan et al., 2022; Li et al., 2022; Yu et al., 2022) or sequence genetically diverse panels for GWAS (Gaurav et al., 2022).
Mining germplasm collections for traits of interest and introducing them into crop breeding programmes is far from novel. Fundamental to this activity are the tens of thousands of crop accessions maintained in global gene banks along with passport data (a basic description of the accession such as accession name, genus, country of origin, acquisition date, etc.). However, errors in labelling can lead to duplications (Singh et al., 2019). Milner et al. (2019) generated molecular passport data by performing genotyping‐by‐sequencing (GBS) on the entire German ex situ gene bank of more than 23 000 accessions of wild and domesticated barley. This resource provided insight into the global population structure of domesticated barley and, through increased marker density and population size, greater analytical power for GWAS. Furthermore, phenotyping diverse subsets of the barley gene bank identified marker–trait associations underlying previously cloned genes controlling virus resistance and morphological traits (Milner et al., 2019). In a separate study, whole‐genome shotgun sequencing of 242 diverse Tausch's goatgrass (Aegilops tauschii) accessions revealed that the D genome of wheat was sourced from geographically distinct lineages, while GWAS identified and validated candidate genes underlying stem rust (SrTA1662) and powdery mildew (WTK4, [WHEAT TANDEM PROTEIN KINASE4]) R genes (Gaurav et al., 2022). These approaches facilitate rapid identification of previously uncharacterized alleles for resistance to biotrophic fungal pathogens in cereal crops.
Pan‐genomics reveal the extent of variation at R loci
Molecular passport data can be used to select representative genotypes suitable for sequencing and assembling chromosome‐scale reference genomes. Pan‐genome projects in bread wheat (Triticum aestivum) (Walkowiak et al., 2020) and barley (Jayakodi et al., 2020) have unravelled the extent of intraspecies structural variation on both micro and macro scales. The availability of a pan‐genome addresses the most common bottleneck in gene cloning projects, which is over‐reliance on gene order and representation in a single available reference genome. In the case of the barley pan‐genome, representative barley genotypes were selected from each of the major global gene pools based on the GBS data from 23 000 barley accessions reported in Milner et al. (2019). Further selection criteria were imposed to maximize geographical diversity including row type and growth habit. The utility of the barley and wheat pan‐genomes lies in being able to use the information from the most closely related genotype or directly from the sequenced genotype harbouring the trait of interest as a reference genome, thus limiting the number of single nucleotide polymorphisms (SNPs), presence/absence variations (PAVs) and inversions.
Gene duplication and diversifying selection have given rise to a clustered distribution of NLRs in plant genomes (Lee and Chae, 2020). Pan‐genome variants, such as PAVs, are especially relevant for NLRs and other types of R genes. Often, the NLR conditioning the resistance is absent or has reduced functionality in susceptible accessions, for example due to a premature stop codon (Sánchez‐Martín et al., 2016). A bread wheat pan‐genome study illustrated this point when examining NLR gene expansion in the genome assemblies of eight diverse elite wheat cvs., the Chinese Spring landrace and spelt wheat (Triticum spelta; Walkowiak et al., 2020). A de novo annotation of their NLR complements identified, on average, 2500 full‐length NLR loci per accession. Only 31%–34% of the NLR signatures were shared across all 10 genomes and the number of unique NLRs ranged from 22 to 192 per genome, suggesting that incorporating GM stacks of R genes should not be cause for concern. The total NLR complement of all 10 wheat lines consisted of 5905 (98% identity) to 7780 (100% identity) unique NLRs. Furthermore, the authors estimated that 90% of the NLR complement was reached at between eight (considering 95% sequence identity) and 11 wheat lines (considering 100% protein sequence identity). These findings highlight the size and complexity of the wheat immune receptor complement, the extent of PAVs between accessions, and the dynamic evolution of the plant immune system (Walkowiak et al., 2020).
The releases of the wheat and barley pan‐genomes have already been and will continue to be important for isolating non‐canonical R genes. For example, two recent studies utilized sequenced chromosome‐scale assemblies from the wheat and barley pan‐genomes to clone the leaf rust R genes Leaf rust 14a (Lr14a) (Kolodziej et al., 2021) and Resistance to Puccinia hordei 3 (Rph3) (Dinh et al., 2022) from wheat and barley, respectively. Most recently the resistance gene Rph7 was cloned using comparative sequence and expression analysis utilizing the updated barley pan‐genome (Chen et al., 2022). Lr14a, Rph3 and Rph7 encode an ankyrin membrane‐bound repeat protein, putative executor protein and NAC transcription factor, respectively, which are race‐specific non‐canonical resistance proteins. In the case of Rph3, identification of a large 80‐kb deletion in the resistant barley cv. Barke relative to the reference genome assembly of the susceptible cv. Morex delimited the candidate locus to two possible genes (Dinh et al., 2022). Similarly, cloning and characterization of R genes encoding kinase fusion proteins, such as Resistance to Puccinia graminis 1 (Rpg1) (Brueggeman et al., 2002), Yellow rust resistance 15 (Yr15) (Klymiuk et al., 2015), Lr9 (Wang et al., 2023), Stem rust resistance 43 (Sr43) (Yu et al., 2023), Sr60 (Chen et al., 2020), Sr62 (Yu et al., 2022), WTK4 (Gaurav et al., 2022), Powdery mildew resistance 4 (Pm4) (Sánchez‐Martín et al., 2021), Pm24 (Lu et al., 2020), Pm13 (Li et al., 2023), Pm57 (Liu et al., 2023) and Rwt4 (synonym of Resistance to Magnaporthe grisea 1; Rmg1) (Arora et al., 2023), along with the Kolodziej et al. (2021) and Dinh et al. (2022) studies suggest that as many as 35% of major, dominant, race‐specific R genes to biotrophic pathogens of wheat and barley do not encode canonical NLR immune receptors (Table S3). Many of these (18 out of 85, 20%) encode protein kinase proteins containing fusion domains. Despite the well‐documented involvement of protein kinases as resistance proteins across the plant kingdom, those including fusion domains have to date only been reported within the Triticeae (Fahima and Coaker, 2023). It has been postulated that the large and, in the case of wheat, polyploid genomes might potentiate this genomic innovation through de novo gene duplication and fusion events (Sánchez‐Martín and Keller, 2021).
Improvements to long‐read sequencing technologies continue to enhance our ability to generate ultra‐contiguous chromosome‐scale assemblies, thus further improving the efficacy of R gene identification. Athiyannan et al. (2022) combined PacBio high‐fidelity long reads, optical mapping and chromosome conformation capture (HiC) to generate an improved 14.7‐Gb chromosome‐scale assembly of the South African stripe rust‐resistant bread wheat cv. Kariega. These authors improved the N50 contig length previously reported for both short‐read and PacBio long‐read sequencing technologies by 150‐ to 600‐fold, to 30 Mb. The improved genome assembly permitted efficient isolation of the race‐specific R gene Yr27. Taken together, these wheat and barley pan‐genome studies highlight opportunities to accelerate the cloning and characterization of R genes. To maximize the potential of pan‐genomes and sequenced germplasm diversity panels, it will be critical to determine their R gene complements by performing detailed multi‐pathotype testing using diverse rust and mildew isolates with contrasting pathogenicity profiles. This comprehensive analysis will help determine the presence of previously identified genes, allowing promising accessions to be shortlisted for preliminary genetic analysis to map novel R loci.
NLR‐integrated domains reveal the diversity of pathogenicity targets in ETI
NLRs are immune receptor proteins that typically contain three domains. The central nucleotide‐binding domain is common to all NLRs. Most NLRs also have a C‐terminal leucine‐rich repeat (LRR) domain, which confers pathogen recognition specificity, and an N‐terminal interaction domain involved in signalling and subsequent cell death (Duxbury et al., 2020). In monocots (all cereal species), the N terminus typically contains a coiled coil (CC) domain. By contrast, dicots have either a CC domain or a toll interleukin receptor (TIR) domain. Recent NLR gene cloning and functional studies have unravelled a diverse array of atypical‐integrated domains (IDs) that act as decoys for pathogen effectors and trigger a specific immune reaction when interacting with the decoy (Marchal et al., 2022). Examples include zinc‐finger (ZF), BED, and WRKY domains of transcription factors, which are common targets for effector proteins as they play a crucial role in regulating signal transduction during basal plant immunity (Sarris et al., 2015). In some cases, NLR genes encode immune receptor proteins with ID combinations that are species‐specific or are common across different cereal and crop species. Map‐based cloning of the stripe rust R gene YrU1 from red wild einkorn wheat (Triticum urartu) revealed that it encodes a CC‐NBS‐LRR protein with an N‐terminal ankyrin‐repeat (ANK) domain and a C‐terminal WRKY domain (ANK‐NLR‐WRKY). Further empirical searches determined that this unusual NLR protein structure is only found in Triticum species (Wang et al., 2020).
The ZF‐BED domain was originally characterized in the fruit fly (Drosophila melanogaster) through mutagenesis studies and was subsequently shown to be essential for Xa1‐mediated resistance to bacterial blight in rice (Yoshimura et al., 1998). In bread wheat, two BED domain‐containing NLRs (Yr5 and Yr7), encoded by a complex NLR gene cluster with unique pathogen specificity, confer resistance to stripe rust (Marchal et al., 2018). The discovery that the barley leaf rust R gene Rph15 also encodes an NLR‐BED domain‐containing protein suggests that different rust pathogen species that are adapted to distinct hosts may have effectors targeting similar transcription factor domains (Chen et al., 2021). Functional studies in other plant species demonstrated that the binding of effectors with IDs can activate plant defence. The WRKY domain of RESISTANT TO RALSTONIA SOLANACEARUM 1 (RRS1), an NLR from Arabidopsis (Arabidopsis thaliana), is an integrated decoy that is targeted by two effector proteins from different pathogens whose binding activates RRS1‐mediated resistance (Sarris et al., 2015). Analogously, the NLR proteins RGA5 and Pik‐1 from rice contain heavy metal‐associated (HMA) domains that act as a decoy for effectors of the rice blast pathogen Magnaporthe oryzae (Guo et al., 2018). De la Concepcion et al. (2021) engineered a variant of Pik‐1 (Pik1h) with a single amino acid substitution in the HMA domain that broadens its recognition specificity to corresponding Avr‐Pik effectors. Increasing our understanding of the diversity of IDs within cereal NLR proteins will broaden our toolkit for engineering resistance to biotrophic pathogens such as rusts and mildews.
Diverse ‘gatekeepers’ of host specialization
Proteins encoded by structurally distinct R gene classes have key roles in forming host specificity barriers. A pertinent example is in wheat, where the two R genes Rwt3 and Rwt4, encoding an NLR and a tandem kinase, respectively, act as host specificity barriers against non‐Triticum blast pathotypes (Arora et al., 2023). In Hordeum spp., orthologous cell wall‐associated receptor kinase genes from cultivated (H. vulgare) and bulbous (H. bulbosum) barley conferred both partial and non‐host resistance to adapted and non‐adapted leaf rust pathogens, respectively (Wang et al., 2019). The partial resistance quantitative trait locus (QTL) Rphq2 (Resistance to P. hordei 2) from Dutch barley cv. Vada confers quantitative resistance to adapted strains of the leaf rust pathogen P. hordei and non‐host resistance to leaf rust strains adapted to bulbous barley. In parallel, Rph22 from H. bulbosum, which maps to the same physical position as Rphq2, confers partial resistance to P. bulbosii and non‐host resistance to P. hordei. Positional cloning and phenotypic validation of each orthologous kinase in a susceptible transgenic background determined that Rphq2 and Rph22 both conferred a stronger non‐host resistance response to heterologous leaf rust pathogens relative to the partial response to their respective adapted leaf rust pathogens (Wang et al., 2019). The authors subsequently hypothesized that leaf rust isolates adapted to cultivated (P. hordei) and bulbous (P. bulbosii) barley had co‐evolved with their respective hosts to mitigate perception by host receptors by lowering ligand recognition.
Shared genetic architecture of R loci within and between closely related cereal genomes is often characterized by diverse pathogen specificity. The Mla (mildew resistance locus a) clade on the short arm of chromosome 1H in barley is exemplified by its pronounced allelic diversification (Jørgensen, 1994). Recent phylogenetic analysis of the wheat stem rust R proteins Sr33 and Sr50 showed them to be orthologs of barley Mla proteins based on amino acid similarity to Mla1, Mla6 and Mla9, and shared synteny between the group 1 chromosomes within the Triticeae (Dracatos et al., 2019). This observation suggests that, although evolving from a common ancestor, NLRs from the Mla clade have evolved distinct resistance specificities to different biotrophic fungi both within and between respective crop species. In some instances, however, NLRs can have dual‐pathogen recognition specificities conferring race‐specific resistance to adapted pathogens and resistance to pathogen variants adapted to closely related host species. For instance, a 522‐bp region of the sequence encoding the LRR domain within Mla8 determines dual specificity for recognizing barley powdery mildew and wheat stripe rust (Bettgenhaeuser et al., 2021). Mla8 represents a striking example of the shared genetic architecture within cereal genomes for resistance to both adapted and non‐adapted biotrophic pathogens of different genera, implicating this R gene in host species specificity to the wheat stripe rust pathogen. Barley is a host to adapted P. striiformis strains; however, Mla8 did not confer resistance to the P. striiformis f. sp. hordei isolates tested. These two studies emphasize the importance of further molecular characterization and understanding to be able to manipulate non‐host resistance traits in cereal crops for biotechnological applications. In parallel, understanding the molecular signatures of host specificity between biotrophic pathogen isolates specialized to closely related host cereal grasses (formae speciales) using pathogenomics may help predict future host jumps.
Utilizing cloned recognized effector genes to confirm function and predict NLR gene durability
Cloning the pathogen effectors corresponding to broadly effective individual race‐specific R genes plays an important role in determining R gene function. Compared to the 85 R genes cloned from Triticum and Hordeum spp. (including 49 NLRs), only 22 corresponding recognized effector genes encoding secreted pathogen effectors have been cloned (Tables S3 and S4). Effector discovery has lagged that of R gene cloning due to the challenges of working with obligate biotrophic fungal pathogens and due to the unexpected genetic complexity controlling avirulence in some cases, as attested by the recent discovery of two pathogen loci governing virulence to wheat resistance gene Pm1a (Hewitt et al., 2021; Kloppe et al., 2023). However, going forward, the large number of cloned R genes combined with recent progress in developing transient expression systems (Lorrain et al., 2019) and the development of gain‐of‐virulence mutant libraries towards targeted R genes, as recently applied to wheat rusts (Kangara et al., 2020; Upadhyaya et al., 2021), will facilitate effector biology studies by permitting rapid functional assays to study their specificity. Avr gene cloning studies could also be enhanced and/or more efficient through improved in silico rust effector predictor tools to reduce the number of candidate effectors for functional validation. For example, artificial intelligence‐guided computational structural genomics has led to more accurate and better prediction of candidate pathogen effector repertoires (Seong and Krasileva, 2021, 2023). Combining this with high‐throughput transient expression assays in protoplasts will likely increase the rate of Avr‐effector discovery (Arndell et al., 2023; Wilson et al., 2023).
Other major drivers include recent advancements in sequencing technologies, such as PacBio SMRT sequencing and chromosome conformation capture sequencing (Hi‐C), which have enabled haplo‐phasing of dikaryotic rust genomes to increase assembly quality (Duan et al., 2022; Schwessinger et al., 2018). Numerous recognized effector genes were previously cloned in the flax rust (Melampsora lini) experimental system; however, to date, only three Avr genes have been characterized from the wheat stem rust pathogen Pgt: AvrSr35 (Salcedo et al., 2017), AvrSr50 (Chen et al., 2017), and most recently AvrSr27 (Upadhyaya et al., 2021).
One mechanism contributing to the breakdown of rust resistance through loss of avirulence in pathogen populations is the proliferation and mobilization of transposable elements within rust pathogen genomes. For instance, transposon‐mediated disruption of AvrSr35 resulted in natural Sr35‐virulent Pgt isolates (Salcedo et al., 2017). Loss of avirulence can also occur via large‐scale chromosomal changes in pathogen genomes. For example, in the case of AvrSr50, a spontaneous deletion‐mutant was identified, which allowed the identification of AvrSr50 by comparative sequence analysis (Chen et al., 2017). The crystal structure of a natural AvrSr50 variant was recently solved, which showed structural similarity to cupin‐like proteins with a carbohydrate‐binding domain. Subsequently, site‐directed mutagenesis and transient expression assays were used to understand the potential mechanisms conferring the gain of virulence. AvrSr50 mutants escaped recognition by the stem rust resistance receptor Sr50 by means of DNA insertion or stop codon loss in AvrSr50, or via a single amino acid substitution of the surface‐exposed residue Q121 in the AvrSr50 protein (Ortiz et al., 2022). Furthermore, a recent study determined the key residues in the LRR domain of Sr50 responsible for AvrSr50 effector recognition and specificity by performing cell death assays in T. aestivum protoplasts and Nicotiana benthamiana leaves. These same authors engineered Sr33 from Tausch's goatgrass (Aegilops tauschii), which shares 80% similarity at the protein level with Sr50, to recognize AvrSr50 (Tamborski et al., 2022). These studies represent important steps towards understanding and monitoring rust pathogen evolution while providing avenues for R gene engineering in grasses.
The generation of haplo‐phased reference assemblies for cereal rust isolates has enabled several recent breakthroughs in cereal rust pathogenomics and effector biology (Duan et al., 2022; Henningsen et al., 2022). A genome assembly of the highly avirulent Pgt isolate 21‐0 led to the cloning of AvrSr27 (Upadhyaya et al., 2021). In the case of AvrSr27, virulence for Sr27 could be achieved experimentally or occur within the field via deletion mutations, copy number variation and expression level polymorphisms (Upadhyaya et al., 2021). More recently, a chromosome‐scale haplo‐phased reference assembly was generated for the founder isolate of the Ug99 wheat stem rust pathogen lineage (TTKSK) that enabled researchers to demonstrate that Ug99 arose due to somatic hybridization based on nuclei exchange between 21–0 and an uncharacterized Pgt isolate of African origin (Li et al., 2019). Further haplotype analysis of neighbouring AvrSr35 and AvrSr50 recognized effector genes revealed that both are heterozygous in the Ug99 founder lineage, highlighting the potential for single‐step mutations in the pathogen to overcome their corresponding cloned, effective stem rust R genes (Li et al., 2019). This information validates the utility of Avr gene cloning studies to forecast the durability of deployed NLR genes and confirm the function of stacked race‐specific R genes. For example, haplovariant mining analysis combined with functional characterization of natural variants of effector genes helps to understand gain‐of‐virulence mechanism and predict R gene durability and further guide R gene deployment strategies (Müller et al., 2022). Further information on the effectiveness of the different cereal rust and mildew R genes cloned to date is detailed in Tables S5–S9 and summarized in Figure 2 to inform potential future inclusion for pyramiding in gene stacks. The relative effectiveness of each cloned R gene is based on published phenotypic response data collected using diverse rust and mildew pathogen isolates from international cereal disease laboratories.
Figure 2.
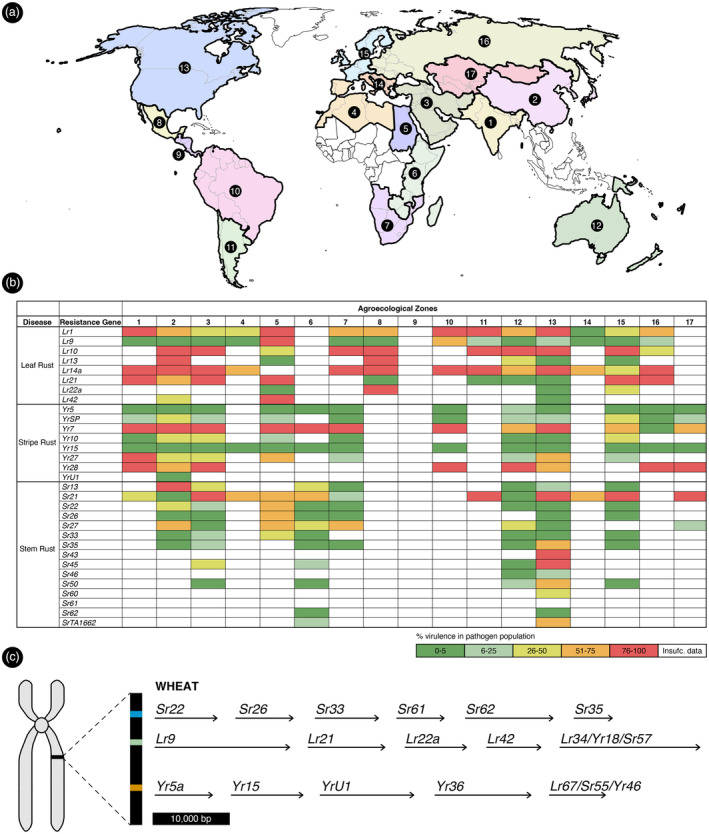
Efficacy of cloned resistance genes across agroecological zones. (a) Map of the world divided into 17 agroecological zones representing different target environments for wheat breeding. Adapted from Rajaram et al. (1994). (b) Efficacy of the 31 cloned wheat stem rust, stripe rust and leaf rust resistance (R) genes by agroecological zone. Colours from green to red represent virulence in the pathogen population expressed as a percentage of isolates tested against a given R gene that are virulent to this R gene. White indicates missing data (i.e. when less than five isolates from an agroecological zone have been tested against a given R gene). See Tables S5–S7 and S9 for further details. (c) Genes that can be combined to create polygene stacks in wheat against the three rust pathogens. The stacks were designed based on the most broad‐spectrum cloned R genes available for each respective disease in wheat across the 17 agroecological zone(s) (Table S9). The stacks have been conceptually targeted to the same chromosome location to facilitate breeding and to ensure co‐inheritance of the major dominant R genes and the multi‐disease R genes Lr34 and Lr67.
Altering the success of biotrophy utilizing naturally occurring recessive mutations
Although rare in nature, recessively inherited non‐lethal loss‐of‐function mutations can spontaneously occur within ‘susceptibility’ (S) genes that regulate and facilitate the biotrophic feeding habit of rust and mildew pathogens (van Schie and Takken, 2014). Due to the polyploid genome of bread wheat, spontaneous recessive loss‐of‐function mutations in S genes are either tolerated or their effect is masked by functionally redundant gene homeologs (Krasileva et al., 2017). On average, a greater proportion of rust and mildew R genes are recessive in barley (26%) relative to polyploid wheat (6.7%), likely because of redundancy between homoeologous genes in wheat (Uauy et al., 2017). Loss‐of‐function mutations in S genes derived from polyploid species, such as bread wheat, have to date conferred pleiotropic resistance to multiple biotrophic pathogen genera and species. However, these are often accompanied by deleterious yield penalty traits due to leaf tip necrosis (Rosewarne et al., 2006). The best examples of loss‐of‐function pleiotropic resistance are two cloned partial‐resistance genes, Lr34/Yr18/Sr57/Pm38 (Krattinger et al., 2009) and Lr67/Yr46/Sr55/Pm46/Ltn3 (Moore et al., 2015), from bread wheat that encode ABC and hexose transporters, respectively.
The most notable example in barley is mlo‐mediated resistance to the powdery mildew fungus that has remained effective for over 50 years after its introduction into agriculture (Kusch and Panstruga, 2017). mlo‐mediated resistance is derived from loss‐of‐function mutations in Mlo encoding a serpentine plasma membrane protein (Panstruga, 2005). The mechanism of resistance remains poorly understood, but mlo protein accumulates at powdery mildew microdomain infection sites that modulate cell actin cytoskeleton arrangement (Bhat et al., 2005). Several barley mlo alleles trigger a developmentally controlled defence mimic phenotype affecting yield; however, unlike the Lr34/Yr18/Sr57/Pm38, Lr46/Yr29/Sr58/Pm39 and Lr67/Yr46/Sr55/Pm46/Ltn3 pleiotropic partial resistance genes characterized in wheat, there is no evidence that any of the mlo alleles confer resistance to barley rust diseases. The adverse effects of mlo mutations, such as premature leaf senescence, are well documented and are consistent with Mlo having a broad role in delaying or preventing mesophyll cell death in pathogen‐challenged and non‐challenged leaves (Consonni et al., 2006; Piffanelli et al., 2002). However, Li et al. found that a 304‐kb deletion‐mutant (Tamlo‐R32) at the wheat Mlo B‐genome locus, originally edited to create mlo alleles in wheat, did not result in the growth defects observed in other wheat mlo mutants. The absence of the mlo‐related yield defects was not due to a deletion in the protein‐coding region of Mlo but rather to de‐repression of the neighbouring gene Tonoplast Monosaccharide Transporter 3 (TaTMT3B). TaTMT3B is normally silenced in wild‐type wheat, but the large adjacent deletion altered the local chromatin structure leading to its de‐repression (Li et al., 2022). These desirable effects were confirmed using elite wheat cvs. under field conditions in China, and in the model species Arabidopsis. The study by Li et al. (2022) provides a crucial gateway to improve the utilization of mlo‐induced resistance in cereals, other grasses, and, by extrapolation from the Arabidopsis results, in other vegetable and horticultural food crops susceptible to powdery mildew (Brown and Wulff, 2022; Spanu, 2022).
By contrast, a recent study in bread wheat suggested that a Ser/Thr kinase gene, Puccinia striiformis‐Induced Protein Kinase 1 (TaPsIPK1), acts as an S gene. The authors also cloned the Pst effector gene encoding PsSpg1, which targets and binds to TaPsIPK1, and showed that autophosphorylation and nuclear translocation of TaPsIPK1 is required for Pst virulence. The subsequent TaPsIPK1‐dependent phosphorylation of the transcription factor C‐REPEAT/DRE BINDING FACTOR 1d (TaCBF1d) regulated gene expression in the nucleus; however, disruption of all three TaPsIPK1 homoeologs led to a primed host immune response conferring broad‐spectrum resistance. A field assessment using geographically defined pathogen isolates and environmental conditions did not reveal any deleterious effects associated with disrupting TaPsIPK1 via gene editing. This study highlights the ability to target multiple classes of S genes by gene editing to improve resistance and enhance our understanding of host–pathogen interactions (Wang et al., 2022a,b).
Gene stacks utilizing mechanistic diversity
The central dogma of pyramiding R genes states that as the number of genes introgressed, transformed or bred into a cereal variety increase, the more challenging it will be for pathogen populations to evolve corresponding virulence to multiple resistance components. Pyramiding R genes using transgenic and cisgenic biotechnological approaches was successful for conditioning complete late blight (Phytophthora infestans) resistance in potato (Solanum tuberosum) (Song et al., 2003). The Lr34/Yr18/Sr57/Pm38 ABC transporter gene was transferred using transgenesis directly into several cereal crops including durum wheat (T. durum; Rinaldo et al., 2017), barley (Risk et al., 2013) and rice (Krattinger et al., 2016) with varying success. Severe deleterious effects, including senescence, were observed in diploid barley and rice transgenic lines expressing Lr34, whereas in the tetraploid durum wheat, Lr34‐induced seedling resistance to leaf rust, stripe rust and powdery mildew (Rinaldo et al., 2017). This finding suggests that the deleterious effects worsened at reduced ploidy levels, which may be a feature of genes with pleiotropic functions in their endogenous polyploid background. By contrast, the cloned wheat NLR genes Sr22, Sr33, Sr35 and Sr45 confer race‐specific resistance to Pgt when transformed into barley cv. Golden Promise without any noticeable agronomic effect (Hatta et al., 2021).
Taking advantage of technological developments in vector construction and the availability of a suite of cloned stem rust R genes, a stack comprising five distinct cloned R genes was transferred into bread wheat. The stack, referred to as the ‘Big Five’, comprises the cloned Lr67/Yr46/Sr55/Pm46 partial‐resistance gene and the all‐stage stem rust R genes Sr22, Sr35, Sr45 and Sr50 and has been rigorously assessed for effectiveness in response to diverse global races of Pgt. In the T1 progeny of three transgenic lines all five full‐length genes were inherited as a single locus confirming the co‐insertion of each gene. The R genes were transformed into the rust‐susceptible bread wheat cv. Fielder and, although conferring complete immunity to seven highly virulent global Pgt isolates, the transgenic seedlings were susceptible to leaf and stripe rust (Luo et al., 2021). This study highlights the effectiveness of pyramiding four highly effective race‐specific NLRs with an additional partial‐resistance gene to control stem rust. The technology also demonstrates the opportunity to introduce multiple genes and/or stacks of genes into the same cv. and design either disease‐specific or multi‐disease R gene stacks containing different gene combinations for durable disease control in wheat and barley.
Based on the available rust R genes cloned to date in wheat, Figure 2c depicts disease‐specific gene stacks that reflect a diverse mechanistic action and broad efficacy to the three rust pathogens of wheat. Both Lr34/Yr18/Sr57/Pm38 and Lr67/Yr46/Sr55/Pm46 were included in the leaf rust and stripe rust stacks, respectively. These genes provide incomplete yet broad‐spectrum resistance to powdery mildew, redressing the reduced efficacy of cloned mildew R genes across the different regions in the world (Tables S8 and S9). Moreover, Lr34/Yr18/Sr57/Pm38 and Lr67/Yr46/Sr55/Pm46 can potentiate race‐specific rust resistance genes (McCallum and Hiebert, 2022), thus likely improving the overall effectiveness of the stacks. For example, in the Swiss wheat cv. Forno, the recently cloned wheat leaf rust resistance gene Lr14a acts additively with the previously cloned pleiotropic APR gene Lr34/Yr18/Sr57/Pm38. Two additional QTLs, including the minor APR Lr75, constitute the leaf rust resistance complex in cv Forno, which has provided durable resistance for >30 years in Switzerland (Singla et al., 2017). Similarly, the race‐specific Lr13 gene is additive in the presence of Lr34/Yr18/Sr57/Pm38 and may contribute with other QTLs to the basis of durability in the cvs. Era, Alsen and Norm (Kloppers and Pretorius, 1997; Oelke and Kolmer, 2005). In the South African cv. Kariega, durable stripe rust resistance appears to be conditioned by the APR Lr34/Yr18/Sr57/Pm38, the recently cloned race‐specific resistance gene Yr27 and two additional QTLs (Athiyannan et al., 2022; Ezzahiri and Roelfs, 1989; McDonald et al., 2004; McIntosh et al., 1992). Cloning the remaining QTLs in cvs Forno and Kariega would allow the GM stacking of their leaf and stripe rust resistance complexes to test their durability in different genetic backgrounds and environments (Athiyannan et al., 2022).
Interestingly, Yr27 and Lr13 were found to be allelic, displaying 97% sequence identity, but conferring resistance to two different types of rusts (Athiyannan et al., 2022). It would be highly interesting to identify the amino acids that determine pathogen specificity and see whether this would allow engineering of a dual‐pathogen receptor. In a similar vein, the tandem kinases Rwt4 and Pm24 have been recently described to be alleles of the same gene but protecting against wheat blast and powdery mildew, respectively (Arora et al., 2023; Lu et al., 2020), whereas the leaf rust and powdery mildew R genes Lr9 (Wang et al., 2023) and Pm57 (Liu et al., 2023), encode tandem kinases fused to a von Willebrand factor A domain and share 88% amino acid homology (Liu et al., 2023). It can be hypothesized that Rwt4/Pm24 and Lr9/Pm57 are part of a three‐component (indirect) guardee system, where the pseudokinase in the tandem‐kinase configuration acts as sensor of pathogen effectors. This would be similar to the ZAR1 (NLR)—RKS1 (pseudokinase)—PBL2 (kinase) case, which confers resistance to Xanthomonas campestris (Breit‐McNally et al., 2022). When the pseudokinase RSK1 is ‘replaced’ by ZED1, another pseudokinase, there is resistance to Pseudomonas syringae (Wang et al., 2015). This is akin to the Pto and Fen kinases interacting with the NLR Prf to confer overlapping but differential ETI specificities in response to AvrPtoB (Mathieu et al., 2014). Therefore, the finding of the putative third component in Rwt4/Pm24 and Lr9/Pm57‐mediated resistance as well as their corresponding effectors may facilitate the future engineering of variants with broader‐spectrum resistance or indeed dual‐pathogen resistance.
Just like wheat breeders have combined distinct rust resistance genes in certain cvs. that have stood the test of time, barley breeders appear to have combined genes that provide near non‐host resistance to wheat stripe rust. This immunity is genetically simple being conditioned by three QTL, including Rps6 and Rps7 which encode NLRs (Bettgenhaeuser et al., 2021; Moscou and Dawson, 2017), and Rps8, a physically discrete digenic complex encoding an Exo70Fx and a receptor‐like kinase (Holden et al., 2021). Having obtained the molecular identity of these genes provides the exciting prospect of engineering this non‐host immunity into wheat.
Innovative approaches to improve transformation efficiency
The recalcitrance of crop plants to Agrobacterium (Agrobacterium tumefaciens)‐mediated transformation (ABMT) was, until recently, arguably the largest bottleneck limiting progress in plant biotechnology. This recalcitrance is largely due to the high frequency of host‐Agrobacterium incompatibility explained by strong plant‐induced defence responses and that Agrobacterium is predominantly adapted to infect dicot plants. The genetic basis of ABMT efficiency in plants is complex and strongly genotype‐dependent. Several QTLs have been mapped in Brassica oleracea (Cogan et al., 2004) and barley (Hisano et al., 2017) permitting a haplotype‐based assessment and selection for improved ABMT efficiency.
The options available to improve transformation efficiency are to isolate and manipulate the causal genes underlying ABMT, explore approaches to improve ABMT or devise alternative transformation systems. One approach to improve ABMT involves engineering Agrobacterium to express a type III secretion system and deliver the effectors AvrPto, AvrPtoB or HopA01 derived from the tomato (Solanum lycopersicum) pathogen Pseudomonas syringae to suppress plant defence. This technology increased ABMT efficiency by 250%–400% across the diverse plant species wheat, alfalfa (Medicago sativa) and switchgrass (Panicum virgatum; Raman et al., 2022). The system also permits direct delivery of a plant histone protein into somatic plant cells in a non‐transgenic manner to enhance ABMT efficiency. Enhancements in ABMT efficiency have also been demonstrated in monocot species using morphogenic regulation. Expression of a construct encoding a fusion protein combining wheat GROWTH‐REGULATING FACTOR 4 (GRF4) and its cofactor GRF‐INTERACTING FACTOR 1 (GIF1) substantially increased ABMT efficiency and regeneration speed in monocots and dicots (Debernardi et al., 2020). Overexpressing Baby boom and Wuschel improved ABMT in previously recalcitrant maize inbred lines and improved transformation frequencies in sorghum (Sorghum bicolor) immature embryos and in rice callus (Lowe et al., 2016). These morphogenic enhancers could, in principle, be delivered via an Agrobacterium strain expressing a type III secretion system to further improve ABMT efficiency in cereals. An alternative approach is to deliver clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR‐associated nuclease 9 (Cas9) ribonucleoproteins (RNPs) using biodegradable, polymeric nanocomplexes via endocytosis‐driven uptake into pollen grains (Gogoi et al., 2022). This approach was used to knock out both alleles of the cloned stem rust R gene Sr50 in the homozygous wheat genotype Gladius + Sr50. The ability to improve ABMT efficiency or provide an alternative to tissue culture has important implications for future biotechnology applications in cereal crops recalcitrant to transformation.
Future prospects
Major advances have been made in our understanding of the molecular basis of disease resistance in crops, facilitating informed gene editing opportunities. Rapid innovations, such as gene cassettes, gene editing in S genes and enhanced transformation efficiency, now more than ever offer the potential to improve crop protection outcomes. Effective R gene combinations should, ideally, prioritize mechanistic diversity to further enhance prospects for resistance durability. Naturally occurring loss‐of‐function mutations within specific S genes involved in fungal biotrophic/parasitic habit represent promising resistance mechanisms. S genes often encode nutrient transporters or enzymes that are well conserved at the amino acid level, especially between closely related crop species such as wheat and barley. Opportunities for gene editing, therefore, exploit comparative genomics between closely related crop species (i.e. wheat and barley); however, this approach may be less effective when editing S genes in wheat derived from more distant relatives such as rice.
While rarely durable individually, it is important to continue research on NLRs and their IDs that act as pathogen effector decoys because NLRs may confer durable resistance when used in polygene stacks. Although a large repertoire of wheat and barley rust and powdery mildew R genes have been cloned, providing the basis for engineering powerful gene stacks against these diseases in wheat and barley, our knowledge of the efficacy of many cloned genes is still incomplete (Figure 2; Tables S5–S9). To maximize the durability of resistance, it is important to incorporate multiple genes conferring resistance against each existing pathogen isolate. In parallel, it is also important to consider the potential effect of introducing R genes that have a dual function and serve as susceptibility genes to promoting infection of damaging necrotrophic fungal pathogens. Therefore, a more comprehensive overview of the spectrum of resistance provided by the cloned R genes against contemporary pathogen isolates from around the world is required.
Generating this knowledge using existing germplasm and pathogen resources is complicated by the fact that the cloned genes are typically in different backgrounds containing other effective R genes. These background genes can mask the resistance conferred by the gene of interest. For example, wheat cv. Fielder, which is typically used as a transformation line to confirm the function of cloned genes, likely contains stripe rust R genes Yr6 (Chen and Line, 1992) and Yr20 (Chen et al., 1995), and other catalogued Sr and Lr genes yet to be determined, thus masking the resistance conferred by any cloned rust R gene against isolates containing corresponding Avr genes. Today, researchers can take advantage of the low cost of DNA synthesis and the new protocols for cv.‐independent transformation to incorporate ‘all’ cloned wheat and barley rust and mildew R genes into universally susceptible cvs., that is those typically used as susceptible checks. The resulting true isogenic set could then be shared with laboratories around the world for deep phenotyping. This approach would provide the information required to exercise responsible gene stewardship by permitting the judicious design of stacks that minimize the risk of selecting resistance‐evading pathogen variants (Figure 3).
Figure 3.
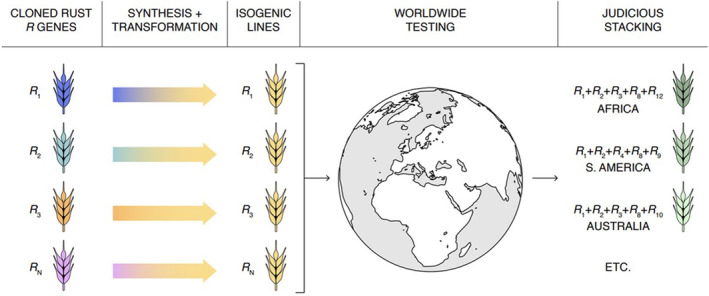
Creation and use of a wheat and barley isogenic library for determining the efficacy of cloned R genes. Cloned rust R genes are transformed into lines universally susceptible to either leaf rust (e.g. cv. Thatcher), stripe rust (e.g. cv. Avocet S), and stem rust (e.g. cv. Morocco). Genetically stable and publicly available homozygous isogenic lines can be shared and phenotyped around the world to determine the regional and worldwide efficacy of each gene to inform judicious stacking and deployment.
One important consideration for deploying transgene cassettes in wheat is the regulatory constraints and the socio‐political opposition toward genetic modification technologies in wheat‐growing regions around the world (Wulff and Dhugga, 2018). However, a genetically modified drought‐tolerant wheat line containing the Hb4 transcription factor gene from sunflower (Helianthus annuus) was deregulated in Argentina in August 2020 (González et al., 2019; https://investors.biocerescrops.com/news). The flour of this wheat was later approved in Brazil, United States, Colombia, Nigeria, Australia and New Zealand, and in March 2023 the cultivation of Hb4 wheat was approved in Brazil, thus heralding a new era for commercial biotech wheat. These developments will hopefully lower the barrier for introducing additional transgenic traits, such as for disease resistance.
Funding
This work was funded by the King Abdullah University of Science and Technology to BBHW, La Trobe University and the Alexander von Humboldt Foundation to PMD, and a China Scholarship Council fellowship to JL. JSM is recipient of the grant ‘Ramon y Cajal’ Fellowship RYC2021‐032699‐I funded by MCIN/AEI/10.13039/501100011033 and by the ‘European Union NextGenerationEU/PRTR’. JSM acknowledges the support of the Junta de Castilla y León through the projects ‘Escalera de Excelencia CLU‐2018‐04, and CL‐EI‐2021‐04 support to the internationalisation of AGRIENVIRONMENT ‐ Unidad Producción Agrícola y Medioambiente’ of the University of Salamanca, both co‐financed by the European Regional Development Fund (ERDF ‘Europe drives our growth’).
Conflict of interest
The authors declare they have no conflict of interest.
Author contributions
PMD and BBHW conceptualized the study and wrote the manuscript with contributions from JSM. JL performed background data consolidation for the rust efficacy tables and figures, whereas JSM collated background data for the powdery mildew efficacy table. All authors approved the final manuscript draft.
Supporting information
Table S1 Wheat and barley grain production and trade by country.
Table S2 Wheat and barley grain end use data.
Table S3 List of cloned wheat and barley R genes and their mode of inheritance (adapted from Uauy et al., 2017, and Yu et al., 2022).
Table S4 List of cloned Avr effector genes from wheat and barley pathogens (adapted from Hafeez et al., 2021, and Sánchez‐Martín and Keller, 2021).
Table S5 Efficacy of cloned wheat stem rust R genes.
Table S6 Efficacy of cloned wheat stripe rust R genes.
Table S7 Efficacy of cloned wheat leaf rust R genes.
Table S8 Efficacy of cloned wheat powdery mildew R genes.
Table S9 Efficacy summary for cloned wheat rust and powdery mildew R genes.
Acknowledgements
We thank Xianming Chen (Washington State University), Jim Kolmer, Bob McIntosh, Simon Krattinger and Erich‐Christian Oerke (University of Bonn) for helpful discussions and Emma Waller for the artwork.
[Correction added on 07 August 2023, after first online publication: The term ‘Lr34/Yr18/Sr57/Pm18’ has been changed to ‘Lr34/Yr18/Sr57/Pm38’ in all instances in this version.]
References
- Alhanti, B. , Van Wendel de Joode, B. , Soto Martinez, M. , Mora, A.M. , Gamboa, L.C. , Reich, B. , Lindh, C.H. et al. (2022) Environmental exposures contribute to respiratory and allergic symptoms among women living in the banana growing regions of Costa Rica. Occup. Environ. Med. 79, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndell, T. , Chen, J. , Sperschneider, J. , Upadhyaya, N.M. , Blundell, C. , Niesner, N. , Wang, A. et al. (2023) Pooled effector library screening in protoplasts rapidly identifies novel Avr genes. 10.1101/2023.04.28.538616v1.full.pdf [DOI] [PMC free article] [PubMed]
- Arora, S. , Steuernagel, B. , Gaurav, K. , Chandramohan, S. , Long, Y. , Matny, O. , Johnson, R. et al. (2019) Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotech. 37, 139–143. [DOI] [PubMed] [Google Scholar]
- Arora, S. , Steed, A. , Goddard, R. , Gaurav, K. , O'Hara, T. , Schoen, A. , Rawat, N. et al. (2023) A wheat kinase and immune receptor form host‐specificity barriers against the blast fungus. Nat. Plants, 9, 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athiyannan, N. , Abrouk, M. , Boshoff, W.H.P. , Cauet, S. , Rodde, N. , Kudrna, D. , Mohammed, N. et al. (2022) Long‐read genome sequencing of bread wheat facilitates disease resistance gene cloning . Nat. Genet. 54, 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aune, D. , Keum, N. , Giovannucci, E. , Fadnes, L.T. , Boffetta, P. , Greenwood, D.C. , Tonstad, S. et al. (2016) Whole grain consumption and risk of cardiovascular disease, cancer, and all cause and cause specific mortality: systematic review and dose‐response meta‐analysis of prospective studies. BMJ, 353, i2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, G. , Saunders, D.G. and Williamson, T. (2020) Banishing barberry: the history of Berberis vulgaris prevalence and wheat stem rust incidence across Britain. Plant Pathol. 69, 1193–1202. [Google Scholar]
- Bettgenhaeuser, J. , Hernández‐Pinzón, I. , Dawson, A.M. , Gardiner, M. , Green, P. , Taylor, J. , Smoker, M. et al. (2021) The barley immune receptor Mla recognizes multiple pathogens and contributes to host range dynamics. Nat. Commun. 12, 6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, R.A. , Miklis, M. , Schmelzer, E. , Schulze‐Lefert, P. and Panstruga, R. (2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl Acad. Sci. USA, 102, 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breit‐McNally, C. , Laflamme, B. , Singh, R.A. , Desveaux, D. and Guttman, D.S. (2022) ZAR1: Guardian of plant kinases. Front. Plant Sci. 13, 981684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.K.M. and Wulff, B.B.H. (2022) Diversifying the menu for crop powdery mildew resistance. Cell, 185, 761–763. [DOI] [PubMed] [Google Scholar]
- Brueggeman, R. , Rostoks, N. , Kudrna, D. , Kilian, A. , Han, F. , Chen, J. , Druka, A. et al. (2002) The barley stem rust‐resistance gene Rpg1 is a novel disease‐resistance gene with homology to receptor kinases. Proc. Natl Acad. Sci. USA, 99, 9328–9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun, H. , Chevre, A.M. , Fitt, B.D.L. , Powers, S. , Besnard, A.L. , Ermel, M. , Huteau, V. et al. (2010) Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus . New Phytol. 185, 285–299. [DOI] [PubMed] [Google Scholar]
- Castelo‐Branco, D.D. , Lockhart, S.R. , Chen, Y. , Santos, D.A. , Hagen, F. , Jane Hawkins, N. , Lavergne, R.A. et al. (2021) Collateral consequences of agricultural fungicides on pathogenic yeasts: a one health perspective to tackle azole resistance. Mycoses, 65, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. and Line, R.F. (1992) Inheritance of stripe rust resistance in wheat cultivars used to differentiate races of Puccinia striiformis in North America. Phytopathology, 82, 633–637. [Google Scholar]
- Chen, X. , Jones, S.S. and Line, R.F. (1995) Chromosomal location of genes for stripe rust resistance in spring wheat cultivars Compair, Fielder, Lee, and Lemhi and interactions of aneuploid wheats with races of Puccinia striiformis . Phytopathology, 85, 375–381. [Google Scholar]
- Chen, J. , Upadhyaya, N.M. , Ortiz, D. , Sperschneider, J. , Li, F. , Bouton, C. , Breen, S. et al. (2017) Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science, 358, 1607–1610. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Rouse, M.N. , Zhang, W. , Zhang, X. , Guo, Y. , Briggs, J. and Dubcovsky, J. (2020) Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol. 225, 948–959. [DOI] [PubMed] [Google Scholar]
- Chen, C. , Jost, M. , Clark, B. , Martin, M. , Matny, O. , Steffenson, B.J. , Franckowiak, J.D. et al. (2021) BED domain containing NLR from wild barley confers resistance to leaf rust. Plant Biotechnol. J. 19, 1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Jost, M. , Outram, M.A. , Friendship, D. , Periyannan, S. , Bartoš, J. , Holušová, K. et al. (2022) A pathogen‐induced putative NAC transcription factor mediates leaf rust resistance in barley. 10.1101/2022.11.30.518475v1 [DOI] [PMC free article] [PubMed]
- Cogan, N.O.I. , Newbury, J.H. , Oldacres, A.M. , Lynn, J.R. , Kearsey, M.J. , King, G.J. and Puddephat, I.J. (2004) Identification and characterization of QTL controlling Agrobacterium‐mediated transient and stable transformation of Brassica oleracea . Plant Biotechnol. J. 1, 59–69. [DOI] [PubMed] [Google Scholar]
- Conner, R.L. , Kuzyk, A.D. and Su, H. (2003) Impact of powdery mildew on the yield of soft white spring wheat cultivars. Can J. Plant Sci. 83, 725–728. [Google Scholar]
- Consonni, C. , Humphry, M.E. , Hartmann, H.A. , Livaja, M. , Durner, J. , Westphal, L. , Vogel, J. et al. (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38, 716–720. [DOI] [PubMed] [Google Scholar]
- De la Concepcion, J.C. , Maidment, J.H.R. , Longya, A. , Xiao, G. , Franceschetti, M. and Banfield, M.J. (2021) The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface. PLoS Pathog. 17, e1009368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi, J.M. , Tricoli, D.M. , Ercoli, M.F. , Hayta, S. , Ronald, P. , Palatnik, J.F. and Dubcovsky, J.A. (2020) GRF‐GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deising, H.B. , Reimann, S. and Pascholati, S.F. (2008) Mechanisms and significance of fungicide resistance. Br. J. Microbiol. 39, 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh, H.X. , Singh, D. , Gomez de la Cruz, D. , Hensel, G. , Kumlehn, J. , Mascher, M. , Stein, N. et al. (2022) The barley leaf rust resistance gene Rph3 encodes a predicted membrane protein and is induced upon infection by avirulent pathotypes of Puccinia hordei . Nat. Commun. 13, 2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant‐pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dracatos, P.M. , Haghdoust, R. , Singh, D. and Park, R.F. (2018) Exploring and exploiting the boundaries of host specificity using the cereal rust and mildew models. New Phytol. 218, 453–462. [DOI] [PubMed] [Google Scholar]
- Dracatos, P.M. , Bartoš, J. , Elmansour, H. , Singh, D. , Karafiátová, M. , Zhang, P. , Steuernagel, B. et al. (2019) The coiled‐coil NLR Rph1, confers leaf rust resistance in barley cultivar Sudan. Plant Physiol. 179, 1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, H. , Jones, A.W. , Hewitt, T. , Mackenzie, A. , Hu, Y. , Sharp, A. , Lewis, D. et al. (2022) Physical separation of haplotypes in dikaryons allows benchmarking of phasing accuracy in Nanopore and HiFi assemblies with Hi‐C data . Genome Biol. 23, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury, S. , Wang, C.I. , MacKenzie, J.L. , Tenthorey, X. , Zhang, S.U. , Huh, L. , Hu, L. et al. (2020) Induced proximity of a TIR signaling domain on a plant‐mammalian NLR chimera activates defense in plants. Proc. Natl Acad. Sci. USA, 117, 18832–18839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlinger, A. , Garland, G. , Hartman, K. , Banerjee, S. , Degrune, F. , García‐Palacios, P. , Hallin, S. et al. (2022) Agricultural management and pesticide use reduce the functioning of beneficial plant symbionts. Nat. Ecol. Evol. 6, 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts, K.L. , Leath, S. and Finney, P.L. (2001) Impact of powdery mildew and leaf rust on milling and baking quality of soft red winter wheat. Plant Dis. 85, 423–429. [DOI] [PubMed] [Google Scholar]
- Ezzahiri, B. and Roelfs, A.P. (1989) Inheritance and expression of adult plant resistance to leaf rust in era wheat. Plant Dis. 73, 549–551. [Google Scholar]
- Fahima, T. and Coaker, G. (2023) Pathogen perception and deception in plant immunity by kinase fusion proteins. Nat. Genet. 55, 908–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO . (2017) Data. http://www.fao.org/faostat/en/#data/QC
- Faris, J.D. and Friesen, T.L. (2020) Plant genes hijacked by necrotrophic fungal pathogens. Curr. Opin. Plant Biol. 56, 74–80. [DOI] [PubMed] [Google Scholar]
- Ford, B.A. , Foo, E. , Sharwood, R. , Karafiatova, M. , Vrána, J. , MacMillan, C. , Nichols, D.S. et al. (2018) Rht18 semi‐dwarfism in wheat is due to increased GA 2‐oxidaseA9 expression and reduced GA content. Plant Physiol. 177, 168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaurav, K. , Arora, S. , Silva, P. , Sánchez‐Martín, J. , Horsnell, R. , Gao, L. , Brar, G.S. et al. (2022) Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol. 40, 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogoi, N. , Kanwal, M. , Norman, M. , Downs, J. , Ahmad, N. , Mago, R. , Bariana, H. et al. (2022) Wheat pollen uptake of CRISPR/Cas9 RNP‐PDMAEMA nanoassemblies results in targeted loss of gene function in progeny. biorxiv . 10.1101/2022.06.02.494465v1.full.pdf [DOI]
- González, F.G. , Capella, M. , Ribichich, K.F. , Curín, F. , Giacomelli, J.I. , Ayala, F. , Watson, G. et al. (2019) Field‐grown transgenic wheat expressing the sunflower gene HaHB4 significantly outyields the wild type. J. Exp. Bot. 70, 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow, N.A.R. , Johnson, C. , Berman, J. , Coste, A.T. , Cuomo, C.A. , Perlin, D.S. , Bicanic, T. et al. (2022) The importance of antimicrobial resistance in medical mycology. Nat. Commun. 13, 5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L. , Cesari, S. , de Guillen, K. , Chalvon, V. , Mammri, L. , Ma, M. , Meusnier, I. et al. (2018) Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces. Proc. Natl Acad. Sci. USA, 115, 11637–11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafeez, A.N. , Arora, S. , Ghosh, S. , Gilbert, D. , Bowden, R.L. and Wulff, B.B.H. (2021) Creation and judicious application of a wheat resistance gene atlas. Mol. Plant, 14, 1053–1070. [DOI] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Rudd, J.J. (2008) Plant resistance signalling hijacked by a necrotrophic fungal pathogen. Plant Signal. Behav. 3, 993–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta, M.A.M. , Arora, S. , Ghosh, S. , Matny, O. , Smedley, M.A. , Yu, G. , Chakraborty, S. et al. (2021) The wheat Sr22, Sr33, Sr35 and Sr45 genes confer resistance against stem rust in barley. Plant Biotechnol. J. 19, 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningsen, E.C. , Hewitt, T. , Dugyala, S. , Nazareno, E.S. , Gilbert, E. , Li, F. , Kianian, S.F. et al. (2022) A chromosome‐level, fully phased genome assembly of the oat crown rust fungus Puccinia coronata f. sp. avenae: a resource to enable comparative genomics in the cereal rusts. bioRxiv . 12, jkac149. 10.1101/2022.01.26.477636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt, T. , Müller, M.C. , Molnár, I. , Mascher, M. , Holušová, K. , Šimková, H. , Kunz, L. et al. (2021) A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis . New Phytol. 229, 2812–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano, H. , Meints, B. , Moscou, M.J. , Cistue, L. , Echávarri, B. , Sato, K. and Hayes, P.M. (2017) Selection of transformation‐efficient barley genotypes based on TFA (transformation amenability) haplotype and higher resolution mapping of the TFA loci. Plant Cell Rep. 36, 611–620. [DOI] [PubMed] [Google Scholar]
- Holden, S. , Bergum, M. , Green, P. , Bettgenhaeuser, J. , Hernández‐Pinzón, I. , Thind, A. , Clare, S. et al. (2021) A lineage‐specific Exo70 is required for receptor kinase–mediated immunity in barley. Sci. Adv. 8, eabn7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter, H.P. , Poteser, M. , Lemmerer, K. , Wallner, P. , Kundi, M. , Moshammer, H. and Weitensfelder, L. (2021) Health symptoms related to pesticide use in farmers and laborers of ecological and conventional banana plantations in Ecuador. Int. J. Environ. Res. Public Health, 18, 1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakodi, M. , Padmarasu, S. , Haberer, G. , Bonthala, V.S. , Gundlach, H. , Monat, C. , Lux, T. et al. (2020) The barley pan‐genome reveals the hidden legacy of mutation breeding. Nature, 588, 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jørgensen, J.H. (1994) Genetics of powdery mildew resistance in barley. Critic. Rev. Plant Sci. 13, 97–119. [Google Scholar]
- Kangara, N. , Kurowski, T.J. , Radhakrishnan, G.V. , Ghosh, S. , Cook, N.M. , Yu, G. , Arora, S. et al. (2020) Mutagenesis of Puccinia graminis f. sp. tritici and selection of gain‐of‐virulence mutants. Front. Plant Sci. 11, 570180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloppe, T. , Whetten, R.B. , Kim, S.B. , Powell, O.R. , Lück, S. , Douchkov, D. , Whetten, R.W. et al. (2023) Two pathogen loci determine Blumeria graminis f. sp. tritici virulence to wheat resistance gene Pm1a . New Phytol. 238, 1546–1561. [DOI] [PubMed] [Google Scholar]
- Kloppers, F.J. and Pretorius, Z.A. (1997) Effects of combinations amongst genes Lr13, Lr34 and Lr37 on components of resistance in wheat to leaf rust. Plant Pathol. 46, 737–750. [Google Scholar]
- Klymiuk, V. , Yaniv, E. , Huang, L. , Raats, D. , Fatiukha, A. , Chen, S. , Feng, L. et al. (2015) Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase‐pseudokinase family. Nat. Commun. 9, 3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeck, M. , Hardham, A.R. and Dodds, P.N. (2011) The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cellul. Microbiol. 13, 1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej, M.C. , Singla, J. , Sánchez‐Martín, J. , Zbinden, H. , Šimková, H. , Karafiátová, M. , Doležel, J. et al. (2021) A membrane‐bound ankyrin repeat protein confers race‐specific leaf rust disease resistance in wheat. Nat. Commun. 12, 956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva, K.V. , Vasquez‐Gross, H.A. , Howell, T. , Bailey, P. , Paraiso, F. , Clissold, L. , Simmonds, J. et al. (2017) Uncovering hidden variation in polyploid wheat. Proc. Natl Acad. Sci. USA, 114, E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger, S.G. , Lagudah, E.S. , Spielmeyer, W. , Singh, R.P. , Huerta‐Espino, J. , McFadden, H. , Bossolini, E. et al. (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science, 323, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Krattinger, S.G. , Sucher, J. , Selter, L.L. , Chauhan, H. , Zhou, B. , Tang, M. , Upadhyaya, N.M. et al. (2016) The wheat durable, multi‐pathogen resistance gene Lr34 confers partial blast resistance in rice. Plant Biotechnol. J. 14, 1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, S. and Panstruga, R. (2017) mlo‐Based resistance: an apparently universal “weapon” to defeat powdery mildew disease. Mol Plant Microbe Interact. 30, 179–189. [DOI] [PubMed] [Google Scholar]
- Lee, R.R.Q. and Chae, E. (2020) Variation patterns of NLR clusters in Arabidopsis thaliana genomes. Plant Commun. 1, 100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerro, C.C. , Beane Freeman, L.E. , DellaValle, C.T. , Andreotti, G. , Hofmann, J.N. , Koutros, S. , Parks, C.G. et al. (2021) Pesticide exposure and incident thyroid cancer among male pesticide applicators in agricultural health study. Environ. Int. 146, 106187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Upadhyaya, N.M. , Sperschneider, J. , Matny, O. , Nguyen‐Phuc, H. , Mago, R. , Raley, C. et al. (2019) Emergence of the Ug99 lineage of the wheat stem rust pathogen through somatic hybridisation. Nat. Commun. 10, 5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Lin, D. , Zhang, Y. , Deng, M. , Chen, Y. , Lv, B. , Li, B. et al. (2022) Genome‐edited powdery mildew resistance in wheat without growth penalties. Nature, 602, 455–460. [DOI] [PubMed] [Google Scholar]
- Li, H. , Men, W. , Ma, C. , Liu, Q. , Dong, Z. , Tian, X. , Wang, C. et al. (2023) Wheat powdery mildew resistance gene Pm13 from Aegilops longissima encodes a unique mixed lineage kinase domain‐like protein. https://assets.researchsquare.com/files/rs-2844171/v1_covered_24e431bb-50c8-48d9-9e0c-04a15dea19f8.pdf?c=1682655036 [DOI] [PMC free article] [PubMed]
- Liu, W. , Zhao, Y. , Dong, Z. , Miao, J. , Liu, Q. , Ma, C. , Tian, X. et al. (2023) Pm57 from Aegilops searsii encodes a novel tandem kinase protein conferring powdery mildew resistance in bread wheat. Research Square . 10.21203/rs.3.rs-2844708/v1 [DOI] [PMC free article] [PubMed]
- Lorrain, C. , Gonçalves Dos Santos, K.C. , Germain, H. , Hecker, A. and Duplessis, S. (2019) Advances in understanding obligate biotrophy in rust fungi. New Phytol. 2, 1190–1206. [DOI] [PubMed] [Google Scholar]
- Lowe, K. , Wu, E. , Wang, N. , Hoerster, G. , Hastings, C. , Cho, M.J. , Scelonge, C. et al. (2016) Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell, 28, 1998–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P. , Guo, L. , Wang, Z. , Li, B. , Li, J. , Li, Y. , Qiu, D. et al. (2020) A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 11, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M. , Xie, L. , Chakraborty, S. , Wang, A. , Matny, O. , Jugovich, M. , Kolmer, J.A. et al. (2021) A five‐transgene cassette confers broad‐spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotech. 39, 561–566. [DOI] [PubMed] [Google Scholar]
- Marchal, C. , Zhang, J. , Zhang, P. , Fenwick, P. , Steuernagel, B. , Adamski, N.M. , Boyd, L. et al. (2018) BED‐domain‐containing immune receptors confer diverse resistance spectra to yellow rust. Nat. Plants, 4, 662–668. [DOI] [PubMed] [Google Scholar]
- Marchal, C. , Michalopoulou, V.A. , Zou, Z. , Cevik, V. and Sarris, P.F. (2022) Show me your ID: NLR immune receptors with integrated domains in plants. Essays Biochem. 66, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, J. , Schwizer, S. and Martin, G.B. (2014) Pto kinase binds two domains of AvrPtoB and Its proximity to the effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog. 10, e1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum, B.D. and Hiebert, C.W. (2022) Interactions between Lr67 or Lr34 and other leaf rust resistance genes in wheat (Triticum aestivum). Front. Plant Sci. 13, 871970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, B.A. and Linde, C. (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. [DOI] [PubMed] [Google Scholar]
- McDonald, D. , McIntosh, R. , Wellings, C. , Singh, R.P. and Nelson, J.C. (2004) Cytogenetical studies in wheat XIX. Location and linkage studies on gene Yr27 for resistance to stripe (yellow) rust. Euphytica, 136, 239–248. [Google Scholar]
- McIntosh, R.A. , Wellings, C.R. and Park, R.F. (1992) Wheat Rusts: An Atlas of Resistance Genes. Victoria, Australia: CSIRO Publishing. [Google Scholar]
- Milner, S.G. , Jost, M. , Taketa, S. , Mazón, E.R. , Himmelbach, A. , Oppermann, M. , Weise, S. et al. (2019) Genebank genomics highlights the diversity of a global barley collection. Nat. Genet. 51, 319–326. [DOI] [PubMed] [Google Scholar]
- Moore, J.W. , Herrera‐Foessel, S. , Lan, C. , Schnippenkoetter, W. , Ayliffe, M. , Huerta‐Espino, J. , Lillemo, M. et al. (2015) A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. [DOI] [PubMed] [Google Scholar]
- Moscou, M.J. and Dawson, A.M. (2017) Wheat stripe rust resistance genes and methods of use. https://patents.google.com/patent/WO2017079286A1/en
- Müller, M.C. , Kunz, L. , Schudel, S. , Lawson, A.W. , Kammerecker, S. , Isaksson, J. , Wyler, M. et al. (2022) Ancient variation of the AvrPm17 gene in powdery mildew limits the effectiveness of the introgressed rye Pm17 resistance gene in wheat. Proc. Natl Acad. Sci. USA, 119, e2108808119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou, B.P.M. , Ahn, H.K. , Ding, P. and Jones, J.D.G. (2021) Mutual potentiation of plant immunity by cell‐surface and intracellular receptors. Nature, 592, 110–115. [DOI] [PubMed] [Google Scholar]
- Oelke, L.M. and Kolmer, J.A. (2005) Genetics of leaf rust resistance in spring wheat cultivars Alsen and Norm. Phytopathology, 95, 773–778. [DOI] [PubMed] [Google Scholar]
- Oerke, E.‐C. and Dehne, H.‐W. (2004) Safeguarding production‐losses in major crops and the role of crop protection. Crop Protect. 23, 275–285. [Google Scholar]
- Ortiz, D. , Chen, J. , Outram, M.A. , Saur, I.M.L. , Upadhyaya, N.M. , Mago, R. , Ericsson, D.J. et al. (2022) The stem rust effector protein AvrSr50 escapes Sr50 recognition by a substitution in a single surface‐exposed residue. New Phytol. 234, 592–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga, R. (2005) Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 33, 389–392. [DOI] [PubMed] [Google Scholar]
- Piffanelli, P. , Zhou, F.S. , Casais, C. , Orme, J. , Jarosch, B. , Schaffrath, U. , Collins, N.C. et al. (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 129, 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram, S.M. , van Ginkel, M. and Fischer, R.A. (1994) CIMMYT's wheat breeding mega‐environments. 8th International Wheat Genetics Symposium, July 1994, Beijing, China.
- Raman, V. , Rojas, C.M. , Vasudevan, B. , Dunning, K. , Kolape, J. , Oh, S. , Yun, J. et al. (2022) Agrobacterium expressing a type III secretion system delivers Pseudomonas effectors into plant cells to enhance transformation. Nat. Commun. 13, 2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, J. , Abdolrasouli, A. , Dunne, K. , Sewell, T.R. , Zhang, Y. , Ballard, E. , Brackin, A.P. et al. (2022) Population genomics confirms acquisition of drug‐resistant Aspergillus fumigatus infection by humans from the environment. Nat. Microbiol. 7, 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo, A. , Gilbert, B. , Boni, R. , Krattinger, S.G. , Singh, D. , Park, R.F. , Lagudah, E. et al. (2017) The Lr34 adult plant rust resistance gene provides seedling resistance in durum wheat without senescence. Plant Biotechnol. J. 15, 894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risk, J.M. , Selter, L.L. , Chauhan, H. , Krattinger, S.G. , Kumlehn, J. , Hensel, G. , Viccars, L.A. et al. (2013) The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnol. J. 11, 847–854. [DOI] [PubMed] [Google Scholar]
- Rosewarne, G.M. , Singh, R.P. , Huerta‐Espino, J. , William, H.M. , Bouchet, S. , Cloutier, S. , McFadden, H. et al. (2006) Leaf tip necrosis, molecular markers and beta1‐proteasome subunits associated with the slow rusting resistance genes Lr46/Yr29 . Theor. Appl. Genet. 112, 500–508. [DOI] [PubMed] [Google Scholar]
- Salcedo, A. , Rutter, W. , Wang, S. , Akhunova, A. , Bolus, S. , Chao, S. , Anderson, N. et al. (2017) Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99 . Science, 358, 1604–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Martín, J. and Keller, B. (2021) NLR immune receptors and diverse types of non‐NLR proteins control race‐specific resistance in Triticeae. Curr. Opin. in Plant Biol. 62, 102053. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Martín, J. , Steuernagel, B. , Ghosh, S. , Herren, G. , Hurni, S. , Adamski, N. , Vrána, J. et al. (2016) Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Martín, J. , Widrig, V. , Herren, G. , Wicker, T. , Zbinden, H. , Gronnier, J. , Spörri, L. et al. (2021) Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants, 7, 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris, P.F. , Duxbury, Z. , Huh, S.U. , Ma, Y. , Segonzac, C. , Sklenar, J. , Derbyshire, P. et al. (2015) A plant immune receptor detects pathogen effectors that target WRKY Transcription factors. Cell, 161, 1089–1100. [DOI] [PubMed] [Google Scholar]
- Savary, S. , Willocquet, L. , Pethybridge, S.J. , Esker, P. , McRoberts, N. and Nelson, N. (2019) The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 430, 439. [DOI] [PubMed] [Google Scholar]
- van Schie, C.C. and Takken, F.L. (2014) Susceptibility genes 101: how to be a good host. Ann. Rev. Phytopathol. 52, 551–581. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B. , Sperschneider, J. , Cuddy, W.S. , Garnica, D.P. , Miller, M.E. , Taylor, J.M. , Dodds, P.N. et al. (2018) A near‐complete haplotype‐phased genome of the dikaryotic wheat stripe rust fungus Puccinia striiformis f. sp. tritici reveals high interhaplotype diversity. mBio, 9, e02275‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong, K. and Krasileva, K.V. (2021) Computational structural genomics unravels common folds and novel families in the secretome of fungal phytopathogen Magnaporthe oryzae . Mol. Plant Microbe Interact. 34, 1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong, K. and Krasileva, K.V. (2023) Prediction of effector protein structures from fungal phytopathogens enables evolutionary analyses. Nat. Microbiol. 8, 174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N. , Wu, S. , Raupp, W.J. , Sehgal, S. , Arora, S. , Tiwari, V. , Vikram, P. et al. (2019) Efficient curation of genebanks using next generation sequencing reveals substantial duplication of germplasm accessions. Sci. Rep. 9, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla, J. , Lüthi, L. , Wicker, T. , Bansal, U. , Krattinger, S.G. and Keller, B. (2017) Characterization of Lr75: a partial, broad‐spectrum leaf rust resistance gene in wheat. Theor. Appl. Genet. 130, 1–12. [DOI] [PubMed] [Google Scholar]
- Song, J. , Bradeen, J.M. , Naess, S.K. , Raasch, J.A. , Wielgus, S.M. , Haberlach, G.T. , Liu, J. et al. (2003) Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl Acad. Sci. USA, 100, 9128–9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu, P.D. (2022) Slicing the cost of bread. Nat. Plants, 8, 200–201. [DOI] [PubMed] [Google Scholar]
- Steuernagel, B. , Periyannan, S.K. , Hernández‐Pinzón, I. , Witek, K. , Rouse, M.N. , Yu, G. , Hatta, A. et al. (2016) Rapid cloning of disease‐resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655. [DOI] [PubMed] [Google Scholar]
- Tamborski, J. , Seong, K. , Liu, F. , Staskawicz, B. and Krasileva, K.V. (2022) Engineering of Sr33 and Sr50 plant immune receptors to alter recognition specificity and autoactivity. Bioarchives . 10.1101/2022.03.05.483131 [DOI] [PMC free article] [PubMed]
- Thind, A.K. , Wicker, T. , Simkova, H. , Fossati, D. , Moullet, O. , Brabant, C. , Vrana, J. et al. (2017) Rapid cloning of genes in hexaploid wheat using cultivar‐specific long‐range chromosome assembly. Nat. Biotechnol. 35, 793–796. [DOI] [PubMed] [Google Scholar]
- Tucker, M.A. , Moffat, C.S. , Ellwood, S.R. , Tan, K.C. , Jayasena, K. and Oliver, R.P. (2015) Development of genetic SSR markers in Blumeria graminis f. sp. hordei and application to isolates from Australia. Plant Pathol. 64, 337–343. [Google Scholar]
- Uauy, C. , Wulff, B.B.H. and Dubcovsky, J. (2017) Combining traditional mutagenesis with new high‐throughput sequencing and genome editing to reveal hidden variation in polyploid wheat. Ann. Rev. Genet. 51, 1435–1454. [DOI] [PubMed] [Google Scholar]
- Upadhyaya, N.M. , Mago, R. , Panwar, V. , Hewitt, T. , Luo, M. , Chen, J. , Sperschneider, J. et al. (2021) Genomics accelerated isolation of a new stem rust avirulence gene‐wheat resistance gene pair. Nat. Plants, 7, 1220–1228. [DOI] [PubMed] [Google Scholar]
- Velásquez, A.C. , Castroverde, C.D.M. and He, S.Y. (2018) Plant‐pathogen warfare under changing climate conditions. Curr. Biol. 28, R619–R634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele, R.T. and Mendgen, K. (2003) Rust haustoria: nutrient uptake and beyond. New Phytol. 159, 93–100. [DOI] [PubMed] [Google Scholar]
- Walkowiak, S. , Gao, L. , Monat, C. , Haberer, G. , Kassa, M.T. , Brinton, J. , Ramirez‐Gonzalez, R.H. et al. (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature, 588, 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Roux, B. , Feng, F. , Guy, E. , Li, L. , Li, N. , Zhang, X. et al. (2015) The decoy substrate of a pathogen effector and a pseudokinase specify pathogen‐induced modified‐self recognition and immunity in plants. Cell. Host Microbe, 18, 285–295. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Subedi, S. , de Vries, H. , Doornenbal, P. , Vels, A. , Hensel, G. , Kumlehn, J. et al. (2019) Orthologous receptor kinases quantitatively affect the host status of barley to leaf rust fungi. Nat. Plants, 5, 1129–1135. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Zou, S. , Li, Y. , Lin, F. and Tang, D. (2020) An ankyrin‐repeat and WRKY‐domain‐containing immune receptor confers stripe rust resistance in wheat. Nat. Commun. 11, 1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Abrouk, M. , Gourdoupis, S. , Koo, D. , Karafiátová, M. , Molnár, I. , Doležel, J. et al. (2022a) An unusual tandem kinase fusion protein confers leaf rust resistance in wheat. Nat. Genet. 55, 914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Tang, C.L. , Fan, X. , He, M.Y. , Gan, P.F. , Zhang, S. , Hu, Z.Y. et al. (2022b) Inactivation of a wheat protein kinase gene confers broad‐spectrum resistance to rust fungi. Cell, 185, 2961–2974. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Abrouk, M. , Gourdoupis, S. , Koo, D.H. , Karafiátová, M. , Molnár, I. , Holušová, K. et al. (2023) An unusual tandem kinase fusion protein confers leaf rust resistance in wheat. Nat. Genet. 55, 914–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, S. , Dagvadorj, B. , Tam, R. , Murphy, L. , Heng, N. and Crean, E. (2023) Multiplexed effector screening for recognition by endogenous resistance genes using positive defense reporters in wheat protoplasts. 10.1101/2023.04.30.538885v1 [DOI] [PubMed]
- Wulff, B.B.H. and Dhugga, K.S. (2018) Wheat‐the cereal abandoned by GM. Science, 361, 451–452. [DOI] [PubMed] [Google Scholar]
- Yoshimura, S. , Yamanouchi, U. , Katayose, Y. , Toki, S. , Wang, Z.X. , Kono, I. , Kurata, N. et al. (1998) Expression of Xa1, a bacterial blight‐resistance gene in rice, is induced by bacterial inoculation. Proc. Natl Acad. Sci. USA, 95, 1663–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Matny, O. , Champouret, N. , Steuernagel, B. , Moscou, M.J. , Hernández‐Pinzón, I. , Green, P. et al. (2022) Aegilops sharonensis genome‐assisted identification of stem rust resistance gene Sr62 . Nat. Commun. 13, 1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Matny, O. , Gourdoupis, S. , Rayapuram, N. , Aljedaani, F.R. , Wang, Y.L. , Nürnberger, T. et al. (2023) The wheat stem rust resistance gene Sr43 encodes an unusual protein kinase. Nat. Genet. 55, 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M. , Jiang, Z. , Bi, G. , Nomura, K. , Liu, M. , Wang, Y. , Cai, B. et al. (2021) Pattern‐recognition receptors are required for NLR‐mediated plant immunity. Nature, 592, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Wheat and barley grain production and trade by country.
Table S2 Wheat and barley grain end use data.
Table S3 List of cloned wheat and barley R genes and their mode of inheritance (adapted from Uauy et al., 2017, and Yu et al., 2022).
Table S4 List of cloned Avr effector genes from wheat and barley pathogens (adapted from Hafeez et al., 2021, and Sánchez‐Martín and Keller, 2021).
Table S5 Efficacy of cloned wheat stem rust R genes.
Table S6 Efficacy of cloned wheat stripe rust R genes.
Table S7 Efficacy of cloned wheat leaf rust R genes.
Table S8 Efficacy of cloned wheat powdery mildew R genes.
Table S9 Efficacy summary for cloned wheat rust and powdery mildew R genes.


