Abstract
In the field of plant virology, the usage of reverse genetic systems has been reported for multiple purposes. One is understanding virus–host interaction by labelling viral cDNA clones with fluorescent protein genes to allow visual virus tracking throughout a plant, albeit this visualization depends on technical devices. Here we report the first construction of an infectious cDNA full‐length clone of beet mosaic virus (BtMV) that can be efficiently used for Agrobacterium‐mediated leaf inoculation with high infection rate in Beta vulgaris, being indistinguishable from the natural virus isolate regarding symptom development and vector transmission. Furthermore, the BtMV clone was tagged with the genes for the monomeric red fluorescent protein or the Beta vulgaris BvMYB1 transcription factor, which activates the betalain biosynthesis pathway. The heterologous expression of BvMYB1 results in activation of betalain biosynthesis genes in planta, allowing visualization of the systemic BtMV spread with the naked eye as red pigmentation emerging throughout beet leaves. In the case of BtMV, the BvMYB1 marker system is stable over multiple mechanical host passages, allows qualitative as well as quantitative virus detection and offers an excellent opportunity to label viruses in plants of the order Caryophyllales, allowing an in‐depth investigation of virus–host interactions on the whole plant level.
Keywords: beet mosaic virus, betalain biosynthesis, BvMYB1, Caryophyllales, infectious cDNA clone, virus tracking
The BvMYB1 transcription factor activates betalain biosynthesis in members of Caryophyllales and, by virus‐derived expression, allows visualization of virus spread by the naked eye.
1. INTRODUCTION
The infection of sugar beet with aphid‐transmissible yellowing viruses consisting of beet yellows virus (BYV, genus Closterovirus), beet mild yellowing virus (BMYV, genus Polerovirus), beet chlorosis virus (BChV, genus Polerovirus) and a mosaic disease caused by beet mosaic virus (BtMV, genus Potyvirus) is an increasing problem for European sugar beet growers. Today, aphid‐transmissible viruses have re‐emerged in Europe with widespread distribution in all sugar beet‐growing areas due to the ban of neonicotinoid seed coating in the European Union, resulting in a strong need for natural plant resistance and the development of biotests for fast and reliable identification and phenotypic characterization (Hauer et al., 2017; Hossain et al., 2021). The BtMV genome sequence with 9591 nucleotides (nt) in length was first published by Nemchinov et al. (2004), displaying a typical potyvirus genome organization with a single polyprotein translated and cleaved into 10 mature and one fusion protein by a virus‐encoded protease or autocatalytic processing (Nigam et al., 2019). BtMV causes mosaic‐like patterns on leaves and a stunted growth with an estimated yield loss of approximately 10%; however, it is highly probable that BtMV in mixed infection with other viruses of the yellowing virus complex increases disease severity in sugar beet and is potentially under‐represented in field monitoring activities due to the light mosaic deviating from the conspicuous yellowing caused by the other species (Wintermantel, 2005).
Virus‐induced symptoms in plants as well as yield losses do not always correlate with virus replication level and systemic tissue colonization, which is frequently inhomogeneous over time (Crespo et al., 2020; Kaweesi et al., 2014). This often requires extensive work for reliable and detailed evaluation of potential resistance/tolerance in breeding material relying on specialized personnel and cost‐intensive molecular analysis.
There was limited work done on virus–host interaction for the yellowing viruses in the sugar beet host, as only one reverse genetic system for BMYV has been reported being fully infectious (Klein et al., 2014). Other previously published cDNA clones of yellowing virus displayed low (Stephan & Maiss, 2006; Wetzel et al., 2018) or no infectivity (Hasan, 2004; Peremyslov et al., 1998) in beet using Agrobacterium‐mediated inoculation. The field of plant virology has benefitted from reporter genes integrated into the viral genome, enabling virus tracking throughout the plant for a better understanding of the viral lifecycle in plants (Oparka et al., 1996). Most of the tagged recombinant viruses rely on the use of green or red fluorescent proteins (Baulcombe et al., 1995; Cruz et al., 1996; Dietrich & Maiss, 2003) allowing virus tracking throughout the whole plant. Alternative plant‐derived fluorescent proteins made from light, oxygene, or voltage (LOV) domains have been described, such as the flavoprotein improved LOV (Chapman et al., 2008). All these reporter systems rely on technical equipment such as a UV light handlamp or fluorescence microscopes, which often cannot be used directly in a greenhouse. In addition to this, the need for technical equipment and suitable facilities limits the feasability of a high‐throughput resistance screen, and fluorescence microscopy does not allow the visualization on the whole plant level but only a rather small leaf section or tissue. A reporter system allowing visual virus tracking without the need for any technical devices was first reported by Bedoya et al. (2012). It was shown that the integration of the MYB‐related Rosea1 transcription factor gene derived from Antirrhinum majus into the genome of tobacco etch virus, tobacco mosaic virus and potato virus X resulted in an increased anthocyanin biosynthesis and subsequently clearly visible red pigmentation of Nicotiana tabacum or Nicotiana benthamiana leaves during viral replication and systemic colonization (Bedoya et al., 2012). Potato virus Y expressing Rosea1 initiates anthocyanin production in other plant species such as tomato and potato as well (Cordero et al., 2017). An alternative system used phytoene synthase (crtB) to initiate carotenoid biosynthesis and visualize virus spread (Majer et al., 2017). Still, every virus–host combination has to be tested for the suitability of this reporter system (Bedoya et al., 2012).
However, members of the order Caryophyllales such as beet, spinach, prickly pear or quinoa produce a different class of red pigments, the betalains (Polturak & Aharoni, 2018). Furthermore, no plant family is known to produce both betalains and anthocyanins in the same species (Stafford, 1994). Betalains possess pH stability in a wider range, which is thought to be beneficial mostly in the context of acidification for vacuoles of CAM and C4 plants (Jain & Gould, 2015). Betalains are mostly redundant in their cellular functions as an antioxidant compared to anthocyanins but it is hypothesized that the ability to synthesize betalains might have an evolutionary advantage under specific environmental conditions like high salinity (Jain & Gould, 2015; Timoneda et al., 2019).
Starting with tyrosine as a precursor from the shikimate pathway, L‐3,4‐dihydroxyphenylalanine (L‐DOPA) is formed by tyrosine hydroxylase activity of the cytochrome P450 CYP76AD1 and other CYP76AD‐family members (Polturak et al., 2016; Sunnadeniya et al., 2016). It was then shown that the DOPA 4,5‐dioxygenase (DODA) and CYP76AD1 are the catalysts for betalain biosynthesis resulting in the red/violet betalacyanins and yellow betaxanthins as a result of multiple spontaneous reactions (Hatlestad et al., 2012). Betalain synthesis is regulated differently compared to the anthocyanin biosynthesis. The beet Y locus, responsible for red‐fleshed beet and red leaf colour, encodes the BvMYB1 transcription factor responsible for the bottleneck for betalain pigmentation in beets. BvMYB1 is a plant R2R3‐MYB that contains R2 and R3 MYB domains and a C‐terminal activation domain; however, BvMYB1 does not interact with the basic helix–loop–helix (bHLH) of the heterologous MYB‐bHLH‐WD40 activation complex for anthocyanin biosynthesis, leaving BvMYB1 without an influence on pigmentation for anthocyanin‐producing plants (Hatlestad et al., 2015).
Here we report the construction of the first beet‐infecting BtMV cDNA full‐length clone and assess its biological properties, such as symptom development and vector transmission. Furthermore, the obtained BtMV clone was labelled by integration of monomeric red fluorescent protein (mRFP) or the BvMYB1 gene. Using the BvMYB1 as a reporter system in Beta vulgaris enables virus tracking with the naked eye in whole plants as red pigmentation was induced in B. vulgaris plants wherever the virus replicated. This makes it a useful molecular tool to study the virus biology, estimate viral loads and assess information about the disease severity within a leaf without the need for technical devices.
2. RESULTS
2.1. Generation of a BtMV cDNA clone for agroinoculation and infectivity testing in different host plant species
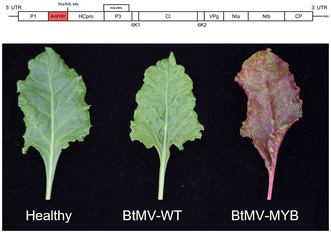
We obtained three different cDNA clones of BtMV from the cloning experiments (Figure 1). The first of the three clones (pDIVA‐BtMV1228) is identical to sequence of the BtMV wild‐type isolate (DSMZ PV‐1228) and was confirmed by Sanger sequencing. After successful cloning, different host plants from the order of Caryophyllales (B. vulgaris, n = 24) and Solanales (N. benthamiana, n = 20) were agroinoculated with the recombinant cDNA clone. The resulting recombinant virus was named BtMV1228. Each of the tested virus–host combinations resulted in 100% symptomatic plants at 7 days postinoculation (dpi) (Figures 2 and 3, and Table S1). First symptoms of BtMV1228 were observed starting at 5 dpi in emerging leaves of B. vulgaris plants and developed over the observation period of 4 weeks (Figure 2a–f). The start of symptom expression was comparable to the mechanically inoculated wild‐type BtMV isolate. Symptom severity at later time points did not show any discrimination between recombinant and wild‐type virus (Figure 2b–f and Table S1).
FIGURE 1.
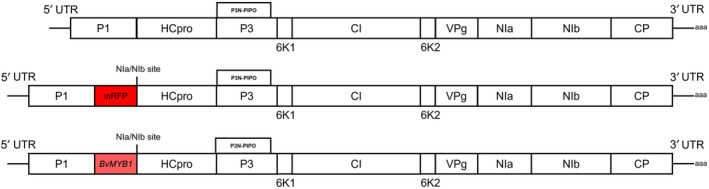
Schematic view of the BtMV genome organization for the constructed cDNA clones pDIVA‐BtMV1228, pDIVA‐BtMV‐mRFP and pDIVA‐BtMV‐MYB. The 5′ and 3′ untranslated regions (UTR) are indicated by straight lines. The poly(A) tail is indicated at the 3′ UTR. Genomic BtMV cistrons are shown as white boxes. Inserted mRFP and BvMYB1 cistrons are shown as red boxes; the additional NIa/NIb‐protease cleavage site is also indicated.
FIGURE 2.

BtMV symptom development in Beta vulgaris infected with the different recombinant viruses (BtMV1228, BtMV‐mRFP, BTMV‐MYB) compared to healthy plants, mock‐inoculated plants and mechanically BtMV DSMZ PV‐1228 (BtMV‐WT) infected plants. (a) 5 days postinoculation (dpi), (b) 7 dpi, (c) 10 dpi, (d) 14 dpi, (e) 21 dpi, (f) 28 dpi. (g) Side view of B. vulgaris plants 4 weeks postinoculation with the recombinant viruses in comparison to a healthy/mock‐inoculated plant. (a–f) Bar = 1 cm, (g) bar = 10 cm.
FIGURE 3.
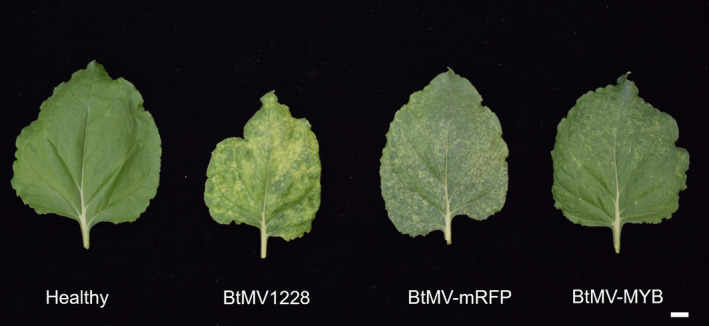
Leaf symptoms in Nicotiana benthamiana after inoculation with BtMV1228, BtMV‐mRFP and BtMV‐MYB recombinant viruses at 21 days postinoculation compared to a healthy plant. White bar = 1 cm.
2.2. Labelling of recombinant BtMV with mRFP or BvMYB1 and effect on infectivity
After BtMV1228 infectivity in B. vulgaris and N. benthamiana was shown, the reporter genes mRFP and BvMYB1 were integrated into the cDNA clone of pDIVA‐BtMV1228 without a stop codon, allowing translation of the viral polyprotein, resulting in the cDNA clones pDIVA‐BtMV‐mRFP and pDIVA‐BtMV‐MYB (Figure 1). The recombinant viruses were named BtMV‐mRFP and BtMV‐MYB, respectively. Here we have chosen an insertion between the P1 proteinase (P1) and the helper component proteinase (HC‐Pro), which has been used in previous works for potyvirus labelling (Beauchemin et al., 2005; Bedoya et al., 2012; Kelloniemi et al., 2008). By using the already existing autoproteolytic P1 cleavage site (RTMHY/SS) at the 5′ end of the insert and via a duplication of the NIa/NIb (EVVEQ/G) at the insert's 3′ end, proper translation and release of the heterologous proteins was ensured. Again, different host plants from the order of Caryophyllales (B. vulgaris, n = 24) and Solanales (N. benthamiana, n = 20) were agroinoculated with the recombinant cDNA clones pDIVA‐BtMV‐mRFP and pDIVA‐BtMV‐MYB. Independent of the tested host plant, the plants displayed less severe symptoms when infected by BtMV‐MYB and BtMV‐mRFP compared to BtMV1228 (Figures 2a–f and 3). The anthocyanin‐producing plant N. benthamiana did not accumulate red pigments at any time point after inoculation with BtMV‐MYB although virus symptoms were clearly visible (Figure 3). When B. vulgaris was infected by one of the recombinant labelled viruses, the plants displayed milder BtMV symptoms compared to a BtMV1228 infection; however, red pigments extensively accumulated over time in BtMV‐MYB infected plants (Figures 2 and 5) while BtMV‐mRFP infected plants displayed a mRFP fluorescence signal (Figure 4). Symptom development appeared to be slower and less severe in B. vulgaris plants inoculated with the labelled viruses as symptoms at 6 dpi were not as distinguishable as for the wild‐type viruses and fewer plants infected with BtMV‐MYB showed symptoms compared to early time points of BtMV1228 infection (Figure 2a and Table S1). However, at 7–8 dpi symptoms caused by each labelled clone increased, showing symptoms in all of the tested plants, being comparable in disease severity for BtMV‐MYB and BtMV‐mRFP (Figure 2b). In the case of BtMV‐mRFP a clear signal (607 nm) could be observed using a confocal laser‐scanning microscope correlating with the beginning of mosaic symptoms at 5 dpi (Figure 2a). Furthermore, BtMV‐MYB‐infected B. vulgaris plants displayed an accumulation of red pigments in oldest leaves first, which was particularly visible on the lower leaf surface by the naked eye starting at 8–10 dpi (Figure 5a,b) and could be confirmed photometrically to be caused by betalains, as the red pigments had a specific absorption peak at c.536 nm after chloroform extraction, which is typical for betalains (Khan & Giridhar, 2015). Old leaves displayed pigmentation that appeared to follow the veins and spread out from there into the surrounding tissue (Figure 5c); pigmentation in leaves that emerged after infection did not exclusively follow the veins as most leaves already displayed full viral symptoms and subsequently produced red pigments in a mosaic‐like pattern during emergence (Figure 5d,e). The beet root itself only displayed minor pigmentation except occasional red spots (Figure S1) and an inhomogeneous virus distribution throughout the plant was observed as well (Figures 2g and S2). To test for the stability of the inserted reporter genes into the genome of BtMV, five consecutive rounds of mechanical transmission of the recombinant viruses BtMV‐mRFP and BtMV‐MYB were carried out in B. vulgaris. Over the course of five passages, mRFP signal as well as red pigmentation was observed without a loss in signal intensity. Furthermore, all of the mentioned viruses were aphid transmissible, and the symptoms caused were indistinguishable to mechanical/agroinoculation (data not shown).
FIGURE 5.

Representative pictures of increasing betalain pigmentation as a result of BtMV‐MYB infection on the lower leaf side of Beta vulgaris ‘Albina Vereduna’ (a–e) in systemically infected tissue in comparison to a mock‐inoculated leaf (f). (a) First observable betalain pigmentation at 8 days postinoculation (dpi). Development of betalain pigmentation at 10 dpi (b), 14 dpi (c), 21 dpi (d) and 28 dpi (e). White bars = 1 cm.
FIGURE 4.
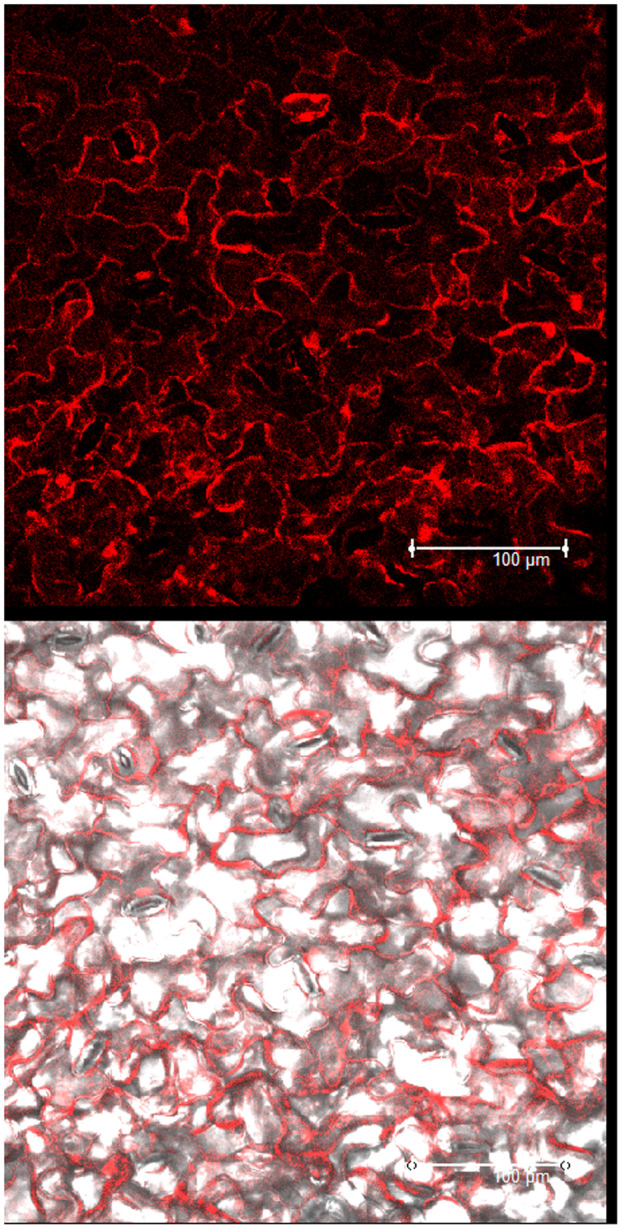
Red fluorescence recorded by confocal laser‐scanning microscopy in white beet epidermal cells from systemically BtMV‐mRFP‐infected leaf tissue displaying mRFP signal at 6 days postinoculation. White bar = 100 μm.
2.3. Quantification of viral load and betalain gene expression by reverse transcription‐quantitative PCR
The chloroform betalain extraction is a destructive method that does not allow betalain content and viral load from the same sample to be measured, therefore reverse transcription‐quantitative PCR (RT‐qPCR) was performed to analyse the influence of BtMV‐MYB on the mRNA expression of betalain biosynthesis genes BvMYB1, BvDODA and BvCYP76AD1 in comparison to BtMV1228 replication. Agrobacterium‐mediated inoculation was performed to allow better comparability of inoculum among treatments. White beets cv. Albina Vereduna were leaf infiltrated with Agrobacterium C58C1 harbouring no additional plasmid (mock control), pDIVA‐BtMV‐1228 or pDIVA‐BtMV‐MYB. Starting from 6 dpi, whole systemically infected leaves of the same age were sampled, homogenized and used for total RNA extraction on a weekly basis. BtMV was detectable by RT‐qPCR in all infected plants independent of the tested construct, while no amplification was observed in mock‐inoculated plants (data not shown). However, copy numbers of the viral genome at 6 dpi was 875 times higher in BtMV1228 than in BtMV‐MYB‐infected plants (Figure 6). Over the test period of 4 weeks, the difference in viral load between BtMV1228 and BtMV‐MYB continuously decreased significantly, resulting in a 22‐fold higher viral load for BtMV1228 compared to BtMV‐MYB at the end of the test period. This mainly resulted from a rising BtMV content in BtMV‐MYB‐infected leaves, and both viruses' accumulation increased significantly over time (Figure 6). The gene expression level of the betalain biosynthesis genes continuously increased during the experiment in plants infected with BtMV‐MYB compared to the steady gene expression in BtMV1228‐infected plants (Figure 7). After only 6 days the detected level of BvMYB1 mRNA was significantly up‐regulated in BtMV‐MYB‐ compared to BtMV1228‐infected plants, even though no symptoms were observed at this time point in BtMV‐MYB‐infected plants (Figure 7a). However, the two betalain biosynthesis genes BvDODA and BvCYP76AD1 only displayed a slightly increased expression in BtMV‐MYB‐infected plants at 6 dpi, which was not significantly increased compared to BtMV1228 (Figure 7a,b). Apart from that, BvDODA and BvCYP76AD1 expression significantly increased over time in BtMV‐MYB‐infected beets (Figure 7a,b), subsequently leading to an accumulation of red pigments being visible with the naked eye (Figure 5). With this, absolute viral content and relative betalain biosynthesis gene expression in BtMV‐MYB‐infected plants compared to BtMV1228‐infected plants were determined. To evaluate whether an increasing BtMV content in BtMV‐MYB‐infected plants correlates with an increased gene expression of BvMYB1, BvDODA and BvCYP76AD1, a linear regression model was used to prove a highly significant correlation (r 2 ≥ 0.95, p ≤ 0.001) between increasing BtMV content and increased gene expression for the betalain biosynthesis genes (Figure 8). The highest correlation (r 2 = 0.97, p ≤ 0.001) between BtMV content and gene expression was found for BvMYB1.
FIGURE 6.
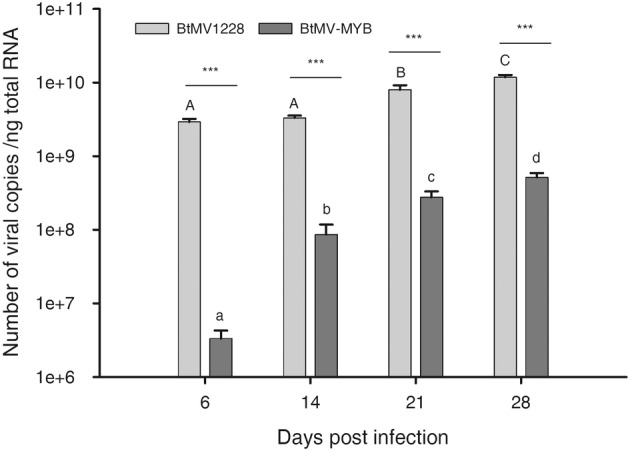
Quantification of wild‐type (BtMV1228) compared to recombinant (BtMV‐MYB) BtMV accumulation in systemically infected leaves of white beet cv. Albina Vereduna tested by reverse transcription‐quantitative PCR. Bars show means with standard deviation of number of copies per nanogram total RNA from five biological replicates. Different upper/lower case letters indicate significantly different virus contents between time points for each virus identified by two‐way analysis of variance with a post hoc Tukey test (p < 0.05, n = 5). Differences in virus accumulation between the tested viruses for each time point are indicated with asterisks (***p ≤ 0.001; n = 5).
FIGURE 7.
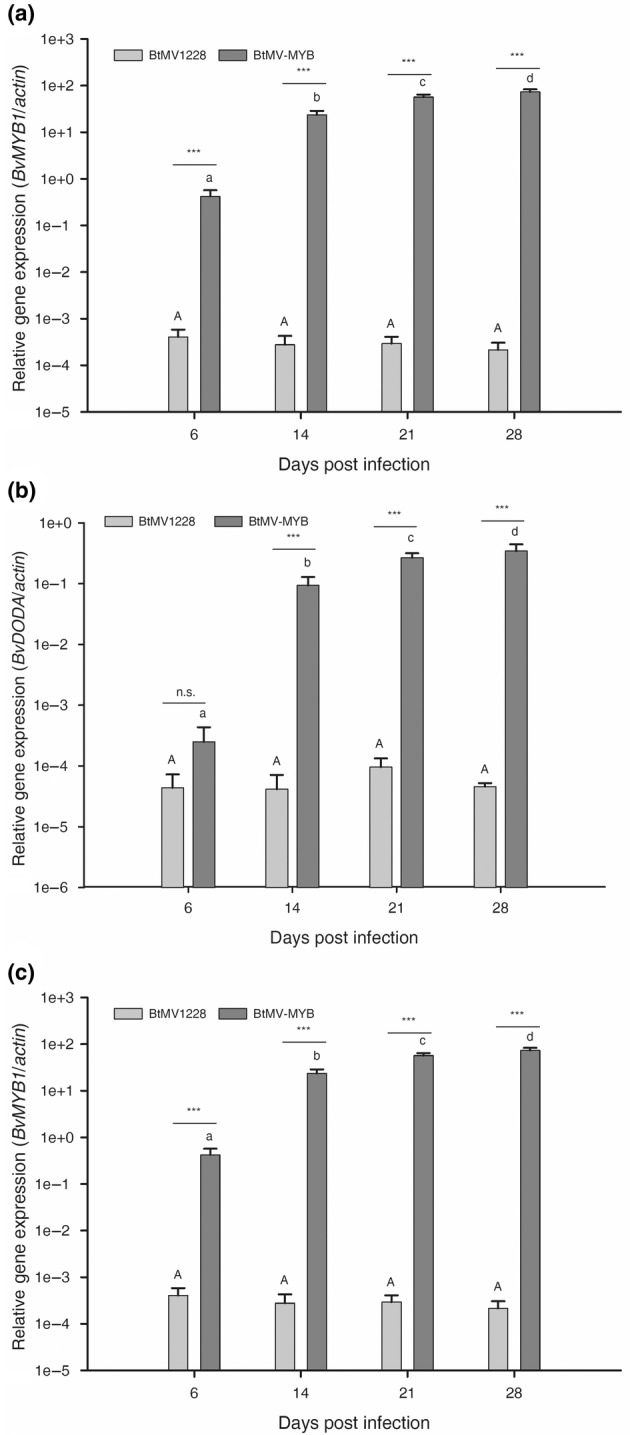
Relative gene expression of (a) BvMYB1, (b) BvCYP76AD1 and (c) BvDODA in Beta vulgaris ‘Albina Vereduna’ infected with BtMV‐MYB compared to BtMV1228 over time. Bars show means with standard deviation of relative gene expression compared to the internal control gene actin from five biological replicates. Different upper/lower case letters indicate significantly different gene expression over time for each virus identified by two‐way analysis of variance with a post hoc Tukey test (p < 0.05, n = 5). Differences in gene expression between the tested viruses for each time point are indicated with asterisks ( ***p ≤ 0.001; n.s., not significant; n = 5).
FIGURE 8.
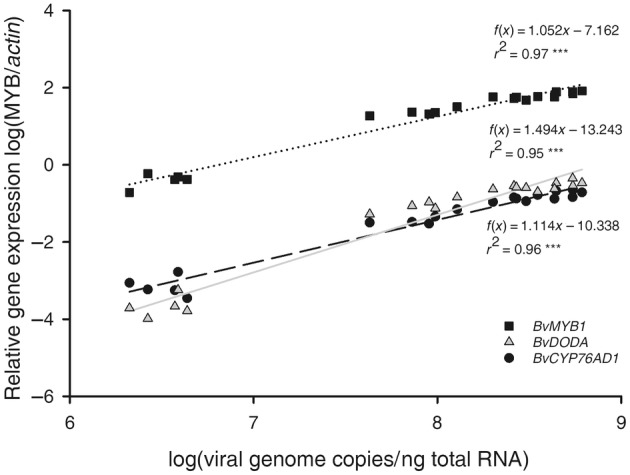
Correlation of log(relative gene expression compared to an internal control gene) for BvMYB1/BvDODA/BvCYP76AD1 in white beet cv. Albina Vereduna in relation to the BtMV‐MYB content shown as log(viral genome copies per ng total RNA) of 20 independent biological replicates. Linear regressions were performed, significant differences are indicated by ***p ≤ 0.001 (n = 20).
3. DISCUSSION
This study describes the successful construction and modification of the first recombinant BtMV full‐length clone with 100% infection rate in beets. No differences were observed regarding viral load, symptom development or transmissibility for BtMV1228 compared to a mechanically inoculated wild‐type virus isolate. Furthermore, we were able to effectively label the recombinant virus to allow virus tracking with the naked eye in systemically infected tissue and could show that viral load and betalain gene expression in beets were highly correlated after infection with the labelled recombinant BtMV‐MYB.
Introducing the BvMYB1 transcription factor into the genome of BtMV resulted in an activation of the beet endogenous betalain biosynthesis pathway. Due to its nuclear functional localization, it is anticipated that the transcription factor BvMYB1 is not only properly and functionally released from the polyprotein during viral translation and subsequent replication but also transported into the nucleus. Throughout the viral infection cycle, heterologous expression of a protein with a size of 25.4 kDa (mRFP) or 26.3 kDa (BvMYB1) was possible while retaining their function in planta. However, the integration of additional 675 nt (BvMYB1) into the genome of BtMV had a negative influence on viral replication and speed of virus spread throughout the plant, especially during early infection compared to BtMV1228, similar to what was observed by Bedoya et al. (2012). For BtMV‐MYB infection, similarities in the estimated delay of approximately 2 days between first mild symptoms and subsequent pigmentation were observed and verified by the gene expression experiments performed, limiting the application for such a cDNA clone in specific experiments especially investigating viral cell‐to‐cell movement. At 6 dpi BvMYB1 expression was already significantly up‐regulated compared to BtMV1228, while BvDODA and BvCYP76AD1 expression just started to increase (Figure 7). This indicates that even though the virus is present and replicating, betalain biosynthesis needs additional time after infection in planta, as the BvMYB1 must be released from the polyprotein, localized to the nucleus, the transcription of the other biosynthesis genes started, and pigments must accumulate in such a high amount that they are visible by naked eye without the need for technical devices. The 2‐day delay in pigmentation also displays one of the disadvantages of the BtMV‐MYB clone compared to fluorescence‐labelled ones. While fluorescently labelled clones, depending on the fluorophore, supply a real‐time picture, a BvMYB1‐labelled clone is only indirectly labelled, as the virus is already present in green tissue before betalains accumulate to a visually detectable amount. On the other hand, the reporter can be used without any artificial irradiation and offers a potential way to investigate the virus spread on the whole plant level, especially in non‐model plants such as B. vulgaris. As betalains are stored in the vacuole (Jain & Gould, 2015), movement tracking through cells without or with small vacuoles, such as vascular tissue, can be limited. Besides the limitation inside the plant, the viral genome organization is an additional factor that has to be considered, as the genome size of the recombinant virus negatively influences viral replication and efficacy of fluorescence labels (Chapman et al., 2008). Additionally, partial RNA silencing due to the presence of the BvMYB1 cistron cannot be excluded either, as small quantities of intracellular BvMYB1 transcript can be found in white beet without being infected with BtMV‐MYB. Similar to virus‐induced gene silencing (VIGS), this could lead to a lack of pigmentation in some cells (Lu et al., 2003). Nonetheless, besides these limitations BtMV‐MYB showed similar properties comparable to the wild‐type virus regarding in planta localization, symptom development, host range and transmission. Strikingly the virus distribution was not homogenous throughout the plant, which has implications for representative sampling and foreshadowing the advantage of such a cDNA clone to get a better understanding of virus–host interactions. In addition to that, betalain biosynthesis genes showing such a strong correlation to the BtMV content is an indicator for the suitability of this clone to be used in noninvasive high‐throughput screens, correlating an image‐determined betalain content to the viral load. Studies determining the anthocyanin content in plant material from pictures (e.g., apples) have already been published (Grimm et al., 2022). Such a quantification algorithm combined with a fast‐phenotyping apparatus through sensor technology could be used to reduce the need for experts to evaluate, for example, disease severity, making the selection procedure of tolerant plant material less error prone (Mahlein, 2016). Even without the use of such technical equipment, the more distinguishable symptoms caused by BtMV‐MYB would facilitate the evaluation of disease severity for an expert. Still, a decrease or loss of betalain pigmentation for a tested genotype does not directly imply a resistance, as the R‐locus, which is responsible for red hypocotyl colour, mapped to be BvCYP76AD1 and is a prerequisite to express betalains (Hatlestad et al., 2012). With these limitations, the use of BtMV‐MYB for a breeding programme does need additional future research; however, it is the first proof of principle that the betalain biosynthesis pathway in general can be manipulated by a plant virus, as already shown for anthocyanins (Bedoya et al., 2012) and carotenoids (Majer et al., 2017). We expect that every virus–host combination needs careful evaluation for the suitability of such a reporter gene before being used in scaled‐up experiments.
4. EXPERIMENTAL PROCEDURES
4.1. Virus source and cloning strategy for BtMV cDNA full‐length clone construction
Total RNA was extracted from B. vulgaris systemically infected with BtMV (DSMZ PV‐1228, GenBank accession no. MT815987) using the NucleoSpin RNA plant mini kit (Machery‐Nagel). Four different cDNAs were synthesized with RevertAid H minus reverse transcriptase (Themo Fisher Scientific) covering the virus genome, using the primers C1 for cDNA1, C2 for cDNA2, C3 for cDNA3 and C4 for cDNA4. All primers used for cloning purposes are listed in Table S1. Using C4, a poly‐A‐tail of 22 nt was added to the genomic BtMV sequence, increasing the genome size up to 9613 nt. These cDNAs were processed by Phusion high‐fidelity DNA polymerase (Thermo Fisher Scientific) to a total of six overlapping PCR products covering the entire BtMV genome. The PCR products were separated by gel electrophoresis and purified by NucleoSpin gel and PCR clean‐up kit (Macherey‐Nagel). All subsequent cloning steps were performed by introducing PCR products into a small binary vector pDIVA (accession no. KX665539) downstream of the 2 × 35S promoter from cauliflower mosaic virus and upstream of a hepatitis delta virus ribozyme and the nopaline synthase terminator using the standard protocol for Gibson isothermal assembly (Gibson et al., 2009). Using PCR primers, appropriate overlaps to the PCR fragments were introduced, resulting in five different subclones that were used as templates to assemble the full‐length cDNA clone. Chemically competent Escherichia coli NM522 cells were transformed with the plasmids (Inoue et al., 1990). After plasmid purification (Birnboim & Doly, 1979), positive clones were identified by restriction enzyme digest and complete sequencing of the viral insert. Plasmid preparations of six to eight positive clones were mixed for population‐based cloning. For each subclone assembly the pDIVA vector backbone was amplified in two overlapping PCR products using primers PDIVA1 + PDIVA2 and PDIVA3 + PDIVA4, which were subsequently combined with partial BtMV genome PCR fragments. Starting with BtMV cDNA1, pDIVA‐BtMVsub1 was produced using a PCR product corresponding to BtMV nt 1–2370 with the primers M1 and M2. pDIVA‐BtMVsub2 using cDNA2 consisted of two BtMV PCR products amplified by M3 + C1 and M4 + M5, respectively. These PCR products correspond to the nt 2344–3077 and 3058–4704, respectively. Using primers M6 + C3 and cDNA3, pDIVA‐BtMVsub3 was generated corresponding to the nt 4679–6762 of the BtMV genome. The last subclone pDIVA‐BtMVsub4 consisted again of two BtMV PCR products amplified by primers M7 + M8 and M9 + C4 using cDNA4, which amplified the genomic BtMV nt 6736–8708 and 8684–9614, respectively. The correct sequence of all subclones was confirmed by Sanger sequencing. Amplifying not only PCR products from the BtMV cDNAs, but additional vector sequences, the pDIVA‐BtMVsub34 was generated from pDIVA‐BtMVsub3 using primers V1 + PDIVA4 and pDIVA‐BtMVsub4 using primers V2 + PDIVA3. After the subcloning steps were performed, and three final PCR products including genomic BtMV sequence and pDIVA sequence were generated from the subclones pDIVA‐BtMVsub1 pDIVA‐BtMVsub2 pDIVA‐BtMVsub34 using primers M1 + M10, M11 + C3 and M12 + pDIVA1, respectively, to assemble the final clone pDIVA‐BtMV1228. Correct assembly was confirmed by complete Sanger sequencing of the final product.
4.2. Integration of reporter genes into pDIVA‐BtMV1228
The pDIVA‐BtMV1228 clone was PCR‐amplified in two overlapping fragments using the primers PDIVA4 + M14 and M15 + PDIVA6. With the primer M15, a duplication of the NIa/NIb‐cleavage site (EVVEQ/G) was introduced 5′ to the HC‐Pro coding sequence. The coding sequence without the stop codon of B. vulgaris BvMYB1 (accession no. JF432080.1) was RT‐PCR amplified from total RNA extracts of red beet cv. Bulls Blood (Pieterpikzonen B.V.), which was extracted by NucleoSpin RNA plant mini kit (Machery‐Nagel). The cDNA was synthesized with RevertAid H minus reverse transcriptase (Thermo Fisher Scientific) and random hexamer primers followed by a Phusion High‐Fidelity PCR (Thermo Fisher Scientific) with primers BMYB1 + BMYB2. The monomeric red fluorescent protein (mRFP) coding sequence was amplified from plasmid pDIVA‐BNYVV‐P2‐mRFP (Laufer, Mohammad, Christ, et al., 2018) using primers BRFP1 + BRFP2. Both primer pairs added appropriate overlaps to the inserts, allowing an integration of the reporter between P1 and HC‐Pro coding sequence in pDIVA‐BtMV1228 using Gibson assembly (Table S2).
4.3. Plant inoculation with pDIVA‐BtMV variants
White beet B. vulgaris var. conditiva ‘Albina Vereduna’ (Pieterpikzonen B.V.), and N. benthamiana were grown under controlled greenhouse conditions (24°C/14 h light, 18°C/10 h dark photoperiod). Agrobacterium tumefaciens C58C1 cells were transformed with the plasmid constructs using electroporation at 1440 V. A. tumefaciens C58C1 containing the different plasmids pDIVA‐BtMV1228, pDIVA‐BtMV‐MYB or pDIVA‐BtMV‐mRFP were grown overnight at 28°C. OD600 was adjusted to 0.5 (N. benthamiana) or 1.5 (B. vulgaris), respectively, in inoculation buffer (10 mM MES, pH 5.6, 10 mM MgCl2, 150 μM acetosyringone). After 3 h of incubation at room temperature, beet cotyledons were inoculated with a needleless syringe (agroinoculation) 14 days after emergence, while first true leaves of N. benthamiana were inoculated 21 days after emergence (Laufer, Mohammad, Maiss, et al., 2018). Virus symptom development was evaluated daily over 6 weeks.
4.4. Betalain extraction
To verify that produced red pigments in BtMV‐MYB‐infected plants were betalains, 100 mg of red pigmented leaf tissue was harvested and homogenized in liquid nitrogen. Using a chloroform–methanol–water mixture (2:1:1) red pigments were separated from green leaf tissue after centrifugation for 10 min at 21,000 g. Red pigments were taken from the aqueous phase and extinction at 476 nm (betaxanthins) and 536 nm (betacyanins) was measured photometrically and compared to a red beet extract (Chang et al., 2021).
4.5. Fluorescence microscopy
To investigate the recombinant BtMV‐mRFP, infiltration patches, as well as systemically infected leaves of beet and N. benthamiana, were evaluated for a distinguishable signal at 600–610 nm. The mRFP fluorescence was visualized with the TCS‐SP5 confocal laser‐scanning microscope (Leica Microsystems). Excitation/emission wavelengths for mRFP were 584/607 nm. All confocal images were processed with LAS‐AF software v. 2.6.3.8173 (Leica Microsystems).
4.6. BtMV transmission procedures
Leaves of infected and symptomatic plants were ground 1:10 (wt/vol) in 10 mM phosphate buffer (pH 7.3). Infectious plant sap was mechanically inoculated together with Celite 535 on 14‐day‐old seedlings of beet and evaluated for symptoms and presence of reporter.
For analysis of BtMV aphid transmissibility, nonviruliferous aphids (Myzus persicae) were placed for 24 h on beet plants infected with each recombinant BtMV variant for virus acquisition. Ten aphids were then brush transferred to healthy plants and left for an additional day on these plants before being treated with a systemic insecticide (Teppeki; Belchim Crop Protection). Plants were evaluated for BtMV symptoms over a period of 4 weeks and the presence of reporter gene signal, if applicable.
4.7. Virus and plant mRNA quantification with RT‐qPCR
Total RNA was extracted from virus‐infected white beet cv. Albina Vereduna on a weekly basis with the GeneJET Plant RNA purification kit (Thermo Fisher Scientific). All subsequent RT‐qPCRs were carried out using the CFX96 Real time system (Bio‐Rad). cDNA synthesis was performed with 1 μg of total RNA and 5 μM oligo(dT)18 primer. All primers used for gene expression analysis can be found in Table S3.
The RT‐qPCR for quantification of BvMYB1, BvDODA and BvCYP76AD1 transcripts was performed with specific primers and iTaq Universal SYBR Green Supermix (Bio‐Rad) as described in Hatlestad et al. (2015) using actin as a housekeeping gene.
Viral load in BtMV‐infected plants was quantified by one‐step RT‐qPCR using 100 ng of total RNA. For cDNA synthesis, Superscript IV reverse transcriptase (Thermo Fisher Scientific) was used. In addition to that 2× Maxima Probe qPCR mix (Thermo Fisher Scientific) with 400 nM primers BtMVqs1, BtMVqas1 and the Fam‐BHQ1‐labelled probe BtMVq‐p1 were added to the reaction mix. A calibration curve to determine absolute viral genome copies for each recombinant virus was produced using serial dilution of the corresponding purified full‐length clone plasmid of known concentration. Beet cytochrome oxidase was used as a housekeeping gene to confirm integrity of samples with 400 nM primers COX‐F, COX‐R and the Fam‐BHQ1‐labelled probe COX‐P‐FAM as described (Mahillon et al., 2022). Cycler conditions were 3 min at 95°C, 10 min at 52°C and 10 min at 95°C for cDNA synthesis, followed by 40 cycles 15 s at 95°C and 60 s at 60°C. Gene expression data was analysed using the ΔC t method (Schmittgen & Livak, 2008).
4.8. Statistical analysis
Relative gene expression was calculated by the ΔC t method, values have been log‐transformed before being used for statistical analysis using SigmaPlot v. 14.5 (Systat Software). Mean values are displayed with standard deviation (SD). Mean values of expression data between different viruses and time points were tested for differences by a two‐way analysis of variance with a subsequent post hoc Tukey test (p ≤ 0.05). Linear regressions were performed, and significant differences were determined and indicated by *p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001.
CONFLICT OF INTEREST STATEMENT
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
Figure S1 (a) Horizontal cross‐section for comparison of betalain pigmentation in healthy beets of white beet cv. Albina Vereduna compared to BtMV‐MYB‐infected beets 28 days postinoculation. (b) Vertical cross‐section of the same BtMV‐MYB‐infected beet. White bars = 1 cm.
Figure S2 Top view on Beta vulgaris plants systemically infected with the recombinant viruses (a) BtMV 1228 (b) BtMV‐mRFP and (c) BtMV‐MYB 4 weeks postinoculation in comparison to a mock‐inoculated plant. (d) Side view of BtMV‐MYB‐infected B. vulgaris plants 6 weeks postinoculation in comparison to a mock‐inoculated plant. (a–c) Bar = 10 cm, (d) bar = 1 cm.
Table S1 Symptom development of BtMV infection in Beta vulgaris after mechanical inoculation of BtMV DSMZ PV‐1228 (BtMV‐WT) or agroinoculation of the recombinant viruses BtMV‐1228/MYB/‐mRFP until all plants displayed clear systemic BtMV symptoms. Shown are numbers of B. vulgaris plants (n = 24) displaying BtMV symptoms from 4 to 8 days postinoculation. Mock‐inoculated plants (n = 12) were infiltrated with Agrobacterium tumefaciens C58C1 harbouring no additional plasmid, while healthy plants (n = 12) were untreated.
Table S2 Primers used for generation of pDIVA‐BtMV1228, pDIVA‐BtMV‐mRFP and pDIVA‐BtMV‐MYB. To distinguish between vector and viral sequence, the vector sequences are italicized.
Table S3 Reverse transcription‐quantitative PCR primers used in this study to confirm and quantify BtMV accumulation and evaluate gene expression of the betalain biosynthesis pathway in Beta vulgaris.
ACKNOWLEDGEMENTS
The authors would like to thank all staff of the Institute of Sugar Beet Research, Leibniz University Hannover and Leibniz Institute DSMZ for their technical support on laboratory work.
Rollwage, L. , Maiss, E. , Menzel, W. , Hossain, R. & Varrelmann, M. (2023) Beet mosaic virus expression of a betalain transcription factor allows visual virus tracking in Beta vulgaris . Molecular Plant Pathology, 24, 1319–1329. Available from: 10.1111/mpp.13372
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Baulcombe, D.C. , Chapman, S. & Santa Cruz, S. (1995) Jellyfish green fluorescent protein as a reporter for virus infections. The Plant Journal, 7, 1045–1053. [DOI] [PubMed] [Google Scholar]
- Beauchemin, C. , Bougie, V. & Laliberté, J.‐F. (2005) Simultaneous production of two foreign proteins from a polyvirus‐based vector. Virus Research, 112, 1–8. [DOI] [PubMed] [Google Scholar]
- Bedoya, L.C. , Martínez, F. , Orzáez, D. & Daròs, J.‐A. (2012) Visual tracking of plant virus infection and movement using a reporter MYB transcription factor that activates anthocyanin biosynthesis. Plant Physiology, 158, 1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim, H.C. & Doly, J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Research, 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.‐C. , Chiu, Y.‐C. , Tsao, N.‐W. , Chou, Y.‐L. , Tan, C.‐M. , Chiang, Y.‐H. et al. (2021) Elucidation of the core betalain biosynthesis pathway in Amaranthus tricolor . Scientific Reports, 11, 6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, S. , Faulkner, C. , Kaiserli, E. , Garcia‐Mata, C. , Savenkov, E.I. , Roberts, A.G. et al. (2008) The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proceedings of the National Academy of Sciences of the United States of America, 105, 20038–20043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero, T. , Mohamed, M.A. , López‐Moya, J.‐J. & Daròs, J.‐A. (2017) A recombinant potato virus Y infectious clone tagged with the Rosea1 visual marker (PVY‐Ros1) facilitates the analysis of viral infectivity and allows the production of large amounts of anthocyanins in plants. Frontiers in Microbiology, 8, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo, O. , Robles, C. , Ruiz, L. & Janssen, D. (2020) Antagonism of cucumber green mottle mosaic virus against tomato leaf curl New Delhi virus in zucchini and cucumber. Annals of Applied Biology, 176, 147–157. [Google Scholar]
- Cruz, S.S. , Chapman, S. , Roberts, A.G. , Roberts, I.M. , Prior, D.A. & Oparka, K.J. (1996) Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proceedings of the National Academy of Sciences of the United States of America, 93, 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, C. & Maiss, E. (2003) Fluorescent labelling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. Journal of General Virology, 84, 2871–2876. [DOI] [PubMed] [Google Scholar]
- Gibson, D.G. , Young, L. , Chuang, R.‐Y. , Venter, J.C. , Hutchison, C.A. & Smith, H.O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods, 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Grimm, E. , Kuhnke, F. , Gajdt, A. , Ostermann, J. & Knoche, M. (2022) Accurate quantification of anthocyanin in red flesh apples using digital photography and image analysis. Horticulturae, 8, 145. [Google Scholar]
- Hasan, H. (2004) Generation of an infectious beet mosaic virus (BtMV) full‐length clone based on the complete nucleotide sequence of a German isolate. Doctoral dissertation. Hannover: Leibniz Universität Hannover. [Google Scholar]
- Hatlestad, G.J. , Akhavan, N.A. , Sunnadeniya, R.M. , Elam, L. , Cargile, S. , Hembd, A. et al. (2015) The beet Y locus encodes an anthocyanin MYB‐like protein that activates the betalain red pigment pathway. Nature Genetics, 47, 92–96. [DOI] [PubMed] [Google Scholar]
- Hatlestad, G.J. , Sunnadeniya, R.M. , Akhavan, N.A. , Gonzalez, A. , Goldman, I.L. , McGrath, J.M. et al. (2012) The beet R locus encodes a new cytochrome P450 required for red betalain production. Nature Genetics, 44, 816–820. [DOI] [PubMed] [Google Scholar]
- Hauer, M. , Hansen, A.L. , Manderyck, B. , Olsson, Å. , Raaijmakers, E. , Hanse, B. et al. (2017) Neonicotinoids in sugar beet cultivation in central and northern Europe: efficacy and environmental impact of neonicotinoid seed treatments and alternative measures. Crop Protection, 93, 132–142. [Google Scholar]
- Hossain, R. , Menzel, W. , Lachmann, C. & Varrelmann, M. (2021) New insights into virus yellows distribution in Europe and effects of beet yellows virus, beet mild yellowing virus, and beet chlorosis virus on sugar beet yield following field inoculation. Plant Pathology, 70, 584–593. [Google Scholar]
- Inoue, H. , Nojima, H. & Okayama, H. (1990) High efficiency transformation of Escherichia coli with plasmids. Gene, 96, 23–28. [DOI] [PubMed] [Google Scholar]
- Jain, G. & Gould, K.S. (2015) Are betalain pigments the functional homologues of anthocyanins in plants? Environmental and Experimental Botany, 119, 48–53. [Google Scholar]
- Kaweesi, T. , Kawuki, R. , Kyaligonza, V. , Baguma, Y. , Tusiime, G. & Ferguson, M.E. (2014) Field evaluation of selected cassava genotypes for cassava brown streak disease based on symptom expression and virus load. Virology Journal, 11, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelloniemi, J. , Mäkinen, K. & Valkonen, J.P.T. (2008) Three heterologous proteins simultaneously expressed from a chimeric potyvirus: infectivity, stability and the correlation of genome and virion lengths. Virus Research, 135, 282–291. [DOI] [PubMed] [Google Scholar]
- Khan, M.I. & Giridhar, P. (2015) Plant betalains: chemistry and biochemistry. Phytochemistry, 117, 267–295. [DOI] [PubMed] [Google Scholar]
- Klein, E. , Brault, V. , Klein, D. , Weyens, G. , Lefèbvre, M. , Ziegler‐Graff, V. et al. (2014) Divergence of host range and biological properties between natural isolate and full‐length infectious cDNA clone of the beet mild yellowing virus 2ITB. Molecular Plant Pathology, 15, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer, M. , Mohammad, H. , Christ, D.S. , Riedel, D. , Maiss, E. , Varrelmann, M. et al. (2018) Fluorescent labelling of beet necrotic yellow vein virus and beet soil‐borne mosaic virus for co‐ and superinfection experiments in Nicotiana benthamiana . Journal of General Virology, 99, 1321–1330. [DOI] [PubMed] [Google Scholar]
- Laufer, M. , Mohammad, H. , Maiss, E. , Richert‐Pöggeler, K. , Dall'Ara, M. , Ratti, C. et al. (2018) Biological properties of beet soil‐borne mosaic virus and beet necrotic yellow vein virus cDNA clones produced by isothermal in vitro recombination: insights for reassortant appearance. Virology, 518, 25–33. [DOI] [PubMed] [Google Scholar]
- Lu, R. , Martin‐Hernandez, A.M. , Peart, J.R. , Malcuit, I. & Baulcombe, D.C. (2003) Virus‐induced gene silencing in plants. Methods, 30, 296–303. [DOI] [PubMed] [Google Scholar]
- Mahillon, M. , Groux, R. , Bussereau, F. , Brodard, J. , Debonneville, C. , Demal, S. et al. (2022) Virus yellows and syndrome “basses richesses” in western Switzerland: a dramatic 2020 season calls for urgent control measures. Pathogens, 11, 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlein, A.‐K. (2016) Plant disease detection by imaging sensors – parallels and specific demands for precision agriculture and plant phenotyping. Plant Disease, 100, 241–251. [DOI] [PubMed] [Google Scholar]
- Majer, E. , Llorente, B. , Rodríguez‐Concepción, M. & Daròs, J.‐A. (2017) Rewiring carotenoid biosynthesis in plants using a viral vector. Scientific Reports, 7, 41645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemchinov, L.G. , Hammond, J. , Jordan, R. & Hammond, R.W. (2004) The complete nucleotide sequence, genome organization, and specific detection of beet mosaic virus. Archives of Virology, 149, 1201–1214. [DOI] [PubMed] [Google Scholar]
- Nigam, D. , LaTourrette, K. , Souza, P.F.N. & Garcia‐Ruiz, H. (2019) Genome‐wide variation in potyviruses. Frontiers in Plant Science, 10, 1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J. , Boevink, P. & Santa Cruz, S. (1996) Studying the movement of plant viruses using green fluorescent protein. Trends in Plant Science, 1, 412–418. [Google Scholar]
- Peremyslov, V.V. , Hagiwara, Y. & Dolja, V.V. (1998) Genes required for replication of the 15.5‐kilobase RNA genome of a plant closterovirus. Journal of Virology, 72, 5870–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polturak, G. & Aharoni, A. (2018) “La Vie en Rose”: biosynthesis, sources, and applications of betalain pigments. Molecular Plant, 11, 7–22. [DOI] [PubMed] [Google Scholar]
- Polturak, G. , Breitel, D. , Grossman, N. , Sarrion‐Perdigones, A. , Weithorn, E. , Pliner, M. et al. (2016) Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytologist, 210, 269–283. [DOI] [PubMed] [Google Scholar]
- Schmittgen, T.D. & Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative C(T) method. Nature Protocols, 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Stafford, H.A. (1994) Anthocyanins and betalains: evolution of the mutually exclusive pathways. Plant Science, 101, 91–98. [Google Scholar]
- Stephan, D. & Maiss, E. (2006) Biological properties of beet mild yellowing virus derived from a full‐length cDNA clone. Journal of General Virology, 87, 445–449. [DOI] [PubMed] [Google Scholar]
- Sunnadeniya, R. , Bean, A. , Brown, M. , Akhavan, N. , Hatlestad, G. , Gonzalez, A. et al. (2016) Tyrosine hydroxylation in betalain pigment biosynthesis is performed by cytochrome P450 enzymes in beets (Beta vulgaris). PLoS One, 11, e0149417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timoneda, A. , Feng, T. , Sheehan, H. , Walker‐Hale, N. , Pucker, B. , Lopez‐Nieves, S. et al. (2019) The evolution of betalain biosynthesis in Caryophyllales . New Phytologist, 224, 71–85. [DOI] [PubMed] [Google Scholar]
- Wetzel, V. , Brault, V. & Varrelmann, M. (2018) Production of a beet chlorosis virus full‐length cDNA clone by means of Gibson assembly and analysis of biological properties. Journal of General Virology, 99, 1522–1527. [DOI] [PubMed] [Google Scholar]
- Wintermantel, W.M. (2005) Co‐infection of beet mosaic virus with beet yellowing viruses leads to increased symptom expression on sugar beet. Plant Disease, 89, 325–331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 (a) Horizontal cross‐section for comparison of betalain pigmentation in healthy beets of white beet cv. Albina Vereduna compared to BtMV‐MYB‐infected beets 28 days postinoculation. (b) Vertical cross‐section of the same BtMV‐MYB‐infected beet. White bars = 1 cm.
Figure S2 Top view on Beta vulgaris plants systemically infected with the recombinant viruses (a) BtMV 1228 (b) BtMV‐mRFP and (c) BtMV‐MYB 4 weeks postinoculation in comparison to a mock‐inoculated plant. (d) Side view of BtMV‐MYB‐infected B. vulgaris plants 6 weeks postinoculation in comparison to a mock‐inoculated plant. (a–c) Bar = 10 cm, (d) bar = 1 cm.
Table S1 Symptom development of BtMV infection in Beta vulgaris after mechanical inoculation of BtMV DSMZ PV‐1228 (BtMV‐WT) or agroinoculation of the recombinant viruses BtMV‐1228/MYB/‐mRFP until all plants displayed clear systemic BtMV symptoms. Shown are numbers of B. vulgaris plants (n = 24) displaying BtMV symptoms from 4 to 8 days postinoculation. Mock‐inoculated plants (n = 12) were infiltrated with Agrobacterium tumefaciens C58C1 harbouring no additional plasmid, while healthy plants (n = 12) were untreated.
Table S2 Primers used for generation of pDIVA‐BtMV1228, pDIVA‐BtMV‐mRFP and pDIVA‐BtMV‐MYB. To distinguish between vector and viral sequence, the vector sequences are italicized.
Table S3 Reverse transcription‐quantitative PCR primers used in this study to confirm and quantify BtMV accumulation and evaluate gene expression of the betalain biosynthesis pathway in Beta vulgaris.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


