Abstract
The purpose of this study was to evaluate the value of amide proton transfer-weighted (APTw) MRI radiomic features for the differentiation of tumor recurrence from treatment effect in malignant gliomas. Eighty-six patients who had suspected tumor recurrence after completion of chemoradiation or radiotherapy, and who had APTw-MRI data acquired at 3 T, were retrospectively analyzed. Using a fluid-attenuated inversion recovery (FLAIR) image-based mask, radiomics analysis was applied to the processed APTw and structural MR images. A chi-square automatic interaction detector decision tree was used for classification analysis. Models with and without APTw features were built using the same strategy. Tenfold cross-validation was applied to obtain the overall classification performance of each model. Sixty patients were confirmed as having tumor recurrence, and the remainder were confirmed as having treatment effect, at median time points of 190 and 171 days after therapy, respectively. There were 525 radiomic features extracted from each of the processed APTw and structural MR images. Based on these, the APTw-based model yielded the highest accuracy (86.0%) for the differentiation of tumor recurrence from treatment effect, compared with 74.4%, 76.7%, 83.7%, and 76.7% for T1w, T2w, FLAIR, and Gd-T1w, respectively. Model classification accuracy was 82.6% when using the combined structural MR images (T1w, T2w, FLAIR, Gd-T1w), and increased to 89.5% when using these structural plus APTw images. The corresponding sensitivity and specificity were 85.0% and 76.9% for the combination of structural MR images, and 85.0% and 100% after adding APTw image features. Adding APTw-based radiomic features increased MRI accuracy in the assessment of the treatment response in post-treatment malignant gliomas.
Keywords: glioblastoma, treatment effect, MRI, amide proton transfer-weighted imaging, radiomics
1 |. INTRODUCTION
Malignant gliomas are the most common and deadly primary brain cancers in adults worldwide. Even with the standard chemoradiation therapy following tumor resection, gliomas invariably recur.1 The MRI appearance of tumor recurrence is often indistinguishable from treatment-related changes, such as radiation necrosis2: both show T2-weighted (T2w) hyperintensity, fluid-attenuated inversion recovery (FLAIR) hyperintensity, and gadolinium (Gd) enhancement on T1-weighted (T1w) sequences. Notably, published data suggest that about 50% of patients have new or increased Gd enhancement on MRI within the first 1–3 months after completion of therapy.3,4 In this scenario, determining whether a patient shows tumor recurrence (requiring an alternative therapy) versus treatment effect (termed “pseudoprogression,” where the current therapy would be continued) remains pivotal in the post-chemoradiation management of patients with malignant gliomas. Unfortunately, current standard neuroimaging techniques have considerable limitations in differentiating these two pathologies,5,6 which prevents treatment adjustments in a timely manner. Therefore, there remains a need to develop novel and reliable approaches with which to precisely assess the treatment response in patients with gliomas.
There are many advanced functional and molecular imaging techniques (including MRI and positron emission tomography), as well as multiparametric imaging, to tackle this unmet clinical need.7–14 Multiparametric imaging could be promising, but has not been widely used due to its limitations, as discussed previously.10,13 Amide proton transfer-weighted (APTw) MRI is a novel molecular imaging approach that generates contrast based on endogenous mobile proteins.15 3D APTw-MRI allows fast, volumetric acquisition on 3 T clinical instruments,16–18 and consensus recommendations on this technique have been published recently.19 In the past several years, the technique has been validated extensively for the grading of tumors,20 accurate detection of recurrent malignant gliomas,21–24 prognostication,25–28 and identification of genetic markers.29–31 A recent radiographically guided stereotactic biopsy study showed that APTw signal intensities were significantly higher in regions that represented active tumor than in regions without histologic evidence of active tumor.32
Alternatively, radiomics analysis is capable of converting images into quantitative and objective features. Multiple radiomics studies have demonstrated that such quantitative measurements have potential in the diagnosis and subsequent management of patients with gliomas,33–42 including APTw-based radiomics studies for the molecular marker identification43 and differentiation of gliomas from metastases.44 In addition, researchers are beginning to develop computer-aided diagnostic models for radiomics analysis to assess the treatment response of gliomas.45–47 Here, we hypothesize that different treatment responses are reflected by different APTw image patterns that can be assessed quantitatively by radiomics analysis. If confirmed, radiomics analysis that includes APTw-MRI would add value to structural MRI sequences in the differentiation of treatment effect from tumor recurrence in malignant gliomas.
2 |. METHODS
2.1 |. Patients
This was a retrospective study of previously collected data. Some of the data in this study have been used previously21,32,48 and are being re-analyzed here. This retrospective study was approved by the institutional review board, and informed consent was waived. Patients with malignant gliomas treated between April 2010 and August 2017 were screened. Enrollment criteria were the following: 20 years old or more; pathologically proven primary malignant glioma (WHO Grades III and IV) that was treated with chemoradiation or radiotherapy alone; suspected tumor recurrence and a completed APTw imaging study after the completion of therapy, including T2w, FLAIR, T1w, and gadolinium-enhanced T1-weighted (Gd-T1w) sequences; integrated clinical diagnosis of tumor recurrence or treatment effect. Exclusion criteria included inferior image quality due to various factors.
For patients who underwent a clinically referred repeat surgery (biopsy only or biopsy followed by resection) within four weeks after APTw-MRI, the integrated clinical pathologic results were used to determine the final diagnosis of the lesions when APTw-MRI was performed. If clinical pathological reports were not available, the clinical MR images (before the initial surgery, as well as before and after chemoradiation or radiotherapy) and all other clinical reports in the electronic medical record system were reviewed to confirm the final diagnosis of the suspected lesions. The clinical follow-up MR images were evaluated according to the updated Response Assessment in Neuro-Oncology (RANO) criteria.5,6 Patients with image changes on conventional MRI but with complete response, partial response, and stable disease were grouped as “treatment effect,” and patients with progressive disease were grouped as “tumor recurrence.”
2.2 |. MRI data acquisition
MRI was performed on a 3 T MRI scanner (Achieva; Philips Medical Systems, Best, the Netherlands). A 3D, volumetric, APTw imaging sequence (saturation power = 2 μT; saturation time = 800 ms; repetition time (TR) = 3 s; echo time (TE) = 17 ms; field of view (FOV) = 212×186 mm2; 15 slices; slice thickness = 4.4 mm; matrix = 96 × 94, reconstructed to 256 × 256)49 was used. The total APTw scan time was 8 min 6 s. APTw images were calculated using the magnetization-transfer-ratio asymmetry at the 3.5 ppm offset from the water frequency. The water-saturation-shift-referencing method50 was used for B0 inhomogeneity corrections of APTw images.
Several standard structural MR images were acquired for reference, including T2w (dual-echo TSE sequence; TR = 4 s; TE = 80 ms; FOV = 212 × 190 mm2; 60 slices; thickness = 2.2 mm; matrix = 192 × 179; reconstructed to 512 × 512), FLAIR (TR = 11 s; TE = 120 ms; inversion recovery time = 2.8 s; FOV = 212 × 189 mm2; 60 slices; thickness = 2.2 mm; matrix = 256 × 192; reconstructed to 512 × 512), and T1w and Gd-T1w (three-dimensional magnetization-prepared rapid gradient echo sequence; TR = 3 s; TE = 3.7 ms; inversion recovery time = 843 ms; FOV = 212 × 172 × 165 mm3; matrix = 212 × 212 × 150; reconstructed to 512 × 512 × 150). The Gd-T1w imaging (0.2 mL/kg body weight; Magnevist; Berlex, Wayne, New Jersey) was the last sequence acquired.
2.3 |. Data preprocessing
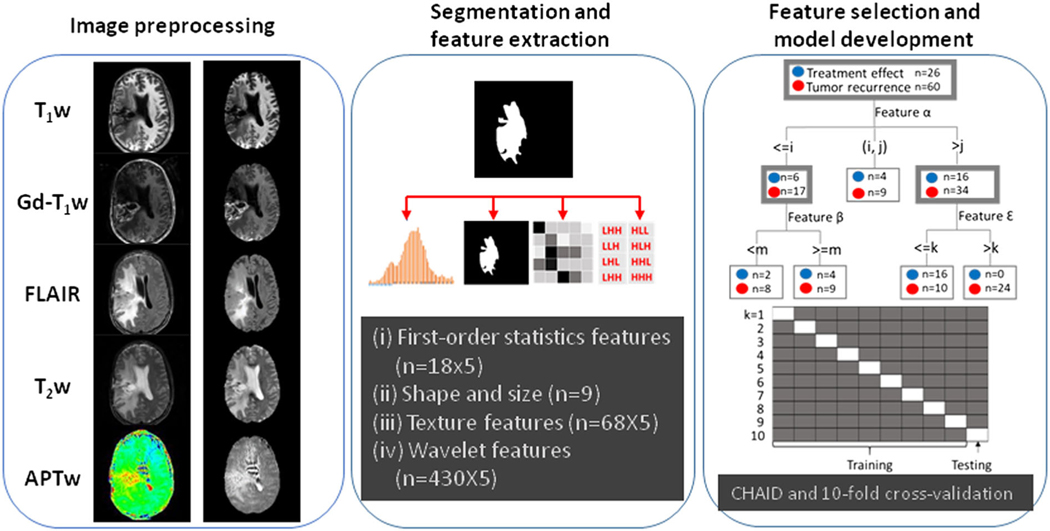
The data processing workflow is presented in Figure 1. Data preprocessing, including co-registration, skull-stripping, N4-bias field-correction, and MRI scale standardization, were performed sequentially.51,52 All MRI volumes (T1w, T2w, FLAIR, Gd-T1w, and APTw images) were re-sampled and co-registered to the saturated images at 3.5 ppm (with the same spatial resolution as the APTw images) with rigid-body registration using SimpleITK.53 An atlas-based skull-stripping was performed after adopting Simple Skull Stripping (S3).54 To preserve the visual details that might easily be neglected by MRI standardization, we adopted and optimized a proposed method52 to the window and level for image display. Notably, based on our experience during the image preprocessing, MRI scale standardization was not performed on APTw-MRI to avoid missing the distinguishing radiographic patterns on APTw-MRI. Each patient had 15 slices with five MR sequences.
FIGURE 1.
Overall workflow and pipeline of the project. First, data preprocessing, including co-registration, skull-stripping, N4-bias field correction, and MRI standardization, were performed sequentially. Second, the lesion masks on T2w MR images were identified and annotated. Features were extracted using PyRadiomics, and features with high redundancy were removed using a statistical strategy. Finally, the top features were selected and used to construct CHAID models.
2.4 |. Lesion segmentation, feature extraction, and annotation
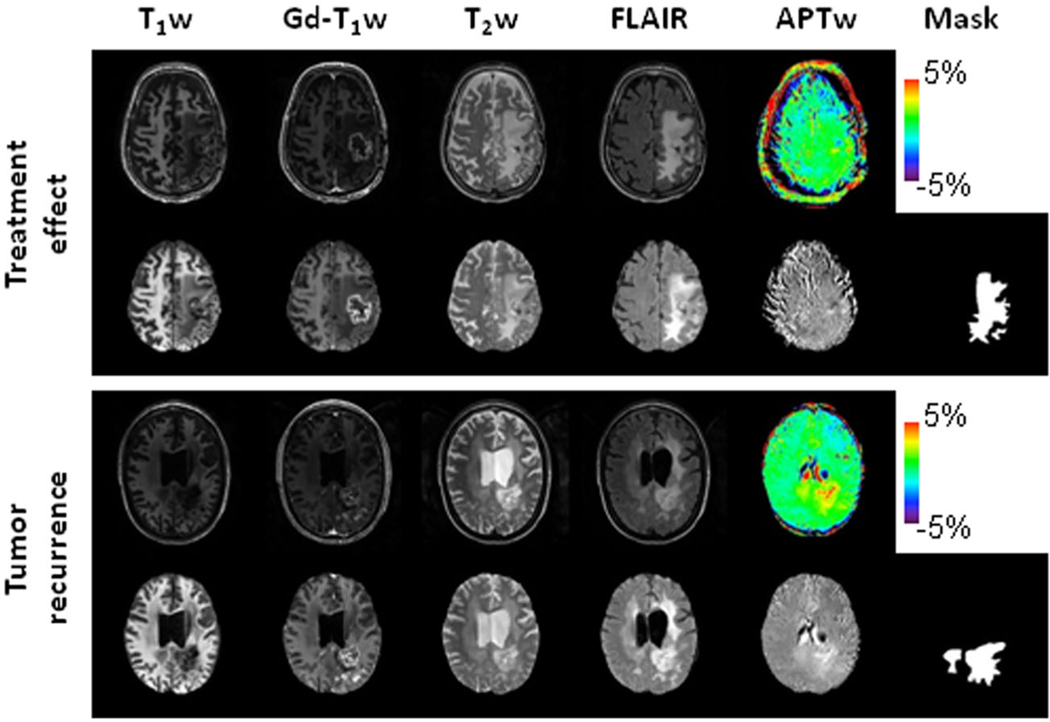
Manual annotation was performed by two readers independently to segment the regions with abnormal FLAIR signal intensities on the co-registered FLAIR images (including Gd-enhancing regions, edema, necrosis, and resection cavities) to generate masks using ITK-SNAP (Version 3.8.0). Then, one of the 15 slices with the largest Gd-enhancing region for each subject was selected for feature extraction. All radiomic features (except for shape) were calculated on original and derived images and extracted from the tumor regions of interest (ROIs) using a customized PyRadiomics program.55–57 Each MRI sequence yielded 525 radiomic features. Since the same mask was shared across the preprocessed images derived from five MRI sequences, the values of size- and shape-based features (n = 9) were the same in each of the five sequences. Thus, a total of 2589 features were extracted for each subject. For all the features, a bin width of 32 was empirically chosen for radiomics analysis. Figure 2 shows structural and APTw MR images after preprocessing for two glioblastoma patients, one with tumor recurrence and one with treatment effect.
FIGURE 2.
Structural and APTw MR images before and after image preprocessing, as well as masks for two post-treatment patients. Top case, a glioblastoma patient (male, 65 years) with treatment effect. The lesion showed a homogeneous isointensity to minimal hyperintensity on the APTw image. Bottom case, an anaplastic astrocytoma patient (female 42 years) with tumor recurrence. The lesion showed a strong hyperintensity on the APTw image. In this case, the cerebrospinal fluid in ventricles presented some artifact (APTw hyperintensity). Notably, the masks covered only the tumor and tumor-associated vasogenic edema. To focus on the direct effects related to the tumor, the masks did not include the periventricular edema in the bilateral frontal lobes, which were detached from the original location (bottom case).
2.5 |. Statistical analysis
All the values of the features were normalized across patients by Z-score transformation.58 The intraclass correlation coefficient (ICC) analysis was used to assess inter-observer reproducibility of radiomic features extracted from two sets of independently drawn masks. ICC values lower than 0.5 indicated poor reliability.59 The feature selection was based on the univariate analysis (to identify features with a significant difference between treatment effect and tumor recurrence, P < 0.05) and on the Pearson correlation analysis (between the features). Features with higher P values and with correlation coefficients |r| > 0.85 (in addition to ICC < 0.5) were removed to avoid the curse of dimensionality.60 Then, we used Bonferroni-corrected, 10-fold, cross-validated decision-tree learning (chi-square automatic interaction detector algorithm or CHAID) for the multi-variate classification analysis.61,62 CHAID, as a predictive model with the important advantage of visual presentation for interpretation, applies a chi-squared test (χ2) to determine the splitting condition. This algorithm is used to prevent data overfitting and to inhibit further decision-tree splitting. To reduce overfitting and to ensure the clarity and interpretability of the results, the maximum tree depth was set to three levels. The minimal case number was set to 10 for parent nodes and to three for child nodes. The parameters used in the CHAID model were experimentally set up as follows: alpha for splitting = 0.05; alpha for merging = 0.05; maximum iterations for convergence = 100. Epsilon = 0.001 determined the convergence criterion. The diagnostic performance was evaluated by the receiver operating characteristic (ROC) analysis. The DeLong test was implemented to compare differences in the area under the curve (AUC) between two ROC curves.63 The accuracy, sensitivity, and specificity of the classifier were also calculated. The same statistical strategy was applied to feature sets extracted from images of (i) all five MR sequences (T1w, T2w, FLAIR, Gd-T1w, APTw), (ii) each of the five MR sequences, (iii) all MR sequences except APTw, and (iv) all MR sequences except Gd-T1w. All analyses were performed using SPSS 26.0 (IBM SPSS, Chicogo, Illinois) and MATLAB R2021a (MathWorks, Natick, Massachusetts). P values less than 0.05 were considered statistically significant.
3 |. RESULTS
3.1 |. Patient characteristics
A total of 116 patients with treated malignant gliomas were screened for this study. After 30 patients were excluded (not high-grade glioma, treated with tumor resection only, no Gd-T1w images or inferior image quality), 86 patients (age 23–78 years old; 33 females and 53 males; 26 grade III and 60 grade IV; see Table 1) were enrolled. The APTw-MRI study was performed at a median time point of 185 days post-chemoradiation or post-radiotherapy (range, 18 days to 3655 days). Sixty patients were confirmed as cases of tumor recurrence, and the remaining 26 patients were confirmed as cases of treatment effect, based on biopsy (24 cases) or integrated clinical diagnosis. There were no significantly different demographic characteristics between the groups of tumor recurrence and treatment effect.
TABLE 1.
Demographic characteristics of subjects
| Treatment effect | Tumor recurrence | P value | |
|---|---|---|---|
| Subject number (%) | 26 (30.2%) | 60 (69.8%) | |
| Age (mean ± SD; year) | 55.6 ± 13.1 | 50.7 ± 12.4 | 0.323 |
| Gender (n) Male |
14 | 39 | 0.345 |
| Female | 12 | 21 | |
| Time after (chemo)radiation completion (median, range; day) | 171, 28–1746 | 190, 18–3655 | 0.502 |
| WHO grade (n) III |
4 | 22 | 0.073 |
| IV | 22 | 38 | |
| Surgery (n) Gross total resection |
15 | 31 | 0.607 |
| Other surgical procedures | 11 | 29 | |
| 2nd line therapy (n) With |
7 | 16 | 0.980 |
| Without | 19 | 44 | |
| Pathological type (n) Glioblastoma |
21 | 36 | 0.073 |
| Anaplastic oligodendroglioma | 1 | 4 | |
| Anaplastic astrocytoma | 3 | 20 | |
| Other | 1 gliosarcoma | 0 | |
| Diagnostic standards (n) Integrated clinical diagnosis |
19 | 43 | 0.893 |
| Pathological report | 7 | 17 |
Independent t test, Pearson’s chi-squared test and Kruskal–Wallis test were applied for numerical data or categorical data separately.
3.2 |. Diagnostic performance
For T1w, T2w, FLAIR, Gd-T1w, or APTw MR images, 34, 61, 47, 18, or 176 radiomic features with a significant difference between treatment effect and tumor recurrence were identified by the univariate analysis, respectively. Among them, 16, 20, 23, 12, or 30 features were further selected, respectively, after the reduction (jrj > 0.85 and ICC < 0.5 removed).
The CHAID models achieved accuracies of 89.5% with the combined radiomic features extracted from all five MRI sequences (Model I), of 86.0%, 74.4%, 76.7%, 83.7%, and 76.7% with the features from APTw, T1w, T2w, FLAIR, and Gd-T1w, respectively (Models IIA-E), of 82.6% with the combined features from T1w, T2w, FLAIR, and Gd-T1w (Model III), and of 90.7% with the combined features from T1w, T2w, FLAIR, and APTw (Model IV). In terms of sensitivity to identify tumor recurrence in patients with positive test results, Model IIE was highest, at 100%. For specificity to diagnose treatment effect by a negative test result, Model I was highest, at 100%. The use of APTw features (Model IIA) reached a more accurate assessment of the tumor response compared with any other single MR sequence (Models IIB–E). Moreover, the APTw-based model achieved an accuracy and specificity higher than the model that combined four structural MR methods (Model III). Notably, when combining T1w, T2w, FLAIR, and APTw without gadolinium administration, Model IV yielded much higher accuracy (90.7% versus 82.6%) and specificity (96.2% versus 76.9%) than, and a comparable sensitivity (88.3% versus 85.0%) to, that achieved when combining T1w, T2w, FLAIR, and Gd-T1w with gadolinium administration (Model III). Based on the comparison of ROC curves, AUCs for Models I versus III and for Models III versus IV were significantly different (P = 0.031 and 0.006, respectively). Accuracy, sensitivity, specificity, and AUC data are summarized in Table 2 for the CHAID algorithm.
TABLE 2.
Average diagnostic performance of 10-fold cross-validation on different CHAID models for tumor recurrence prediction
| Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC (%) | |
|---|---|---|---|---|
| (I) All | 89.5 (81.1–95.1) | 85.0 (73.4–92.9) | 100 (86.8–100) | 92.5 (84.8–97.1) |
| (IIA) APTw | 86.0 (76.7–89.9) | 70.6 (58.7–75.4) | 96.2 (88.4–99.3) | 87.8 (79.0–93.9) |
| (IIB) T1w | 74.4 (63.9–83.2) | 96.7 (88.5–99.6) | 23.1 (9.0–43.7) | 59.9 (48.7–70.3) |
| (IIC) T2w | 76.7 (66.3–83.6) | 58.3 (45.9–66.6) | 90.0 (81–95.9) | 77.9 (67.6–86.1) |
| (IID) FLAIR | 83.7 (74.2–90.8) | 88.3 (77.4–95.2) | 73.1 (52.2–88.4) | 80.7 (70.8–88.4) |
| (IIE) Gd-T1w | 76.7 (70.3–76.7) | 100 (53.5–100) | 75.0 (71.5–75) | 61.5 (50.4–71.8) |
| (III) T1w, T2w, FLAIR, Gd-T1w | 82.6 (72.9–89.9) | 85.0 (73.4–92.9) | 76.9 (56.4–91) | 81.0 (71.1–88.6) |
| (IV) T1w, T2w, FLAIR, APTw | 90.7 (82.5–95.9) | 88.3 (77.4–95.2) | 96.2 (80.4–99.9) | 92.2 (84.4–96.9) |
Note: Data in parentheses are 95% confidence intervals. Based on the comparison of ROC curves, AUCs between I and III and between III and IV were significantly different (P = 0.031 and 0.006, respectively); AUCs between IIA and IIB and between IIA and IIE were also significantly different (P < 0.0001); AUCs between IIA and IIC were marginally significantly different (P = 0.061).
3.3 |. Independent predictors
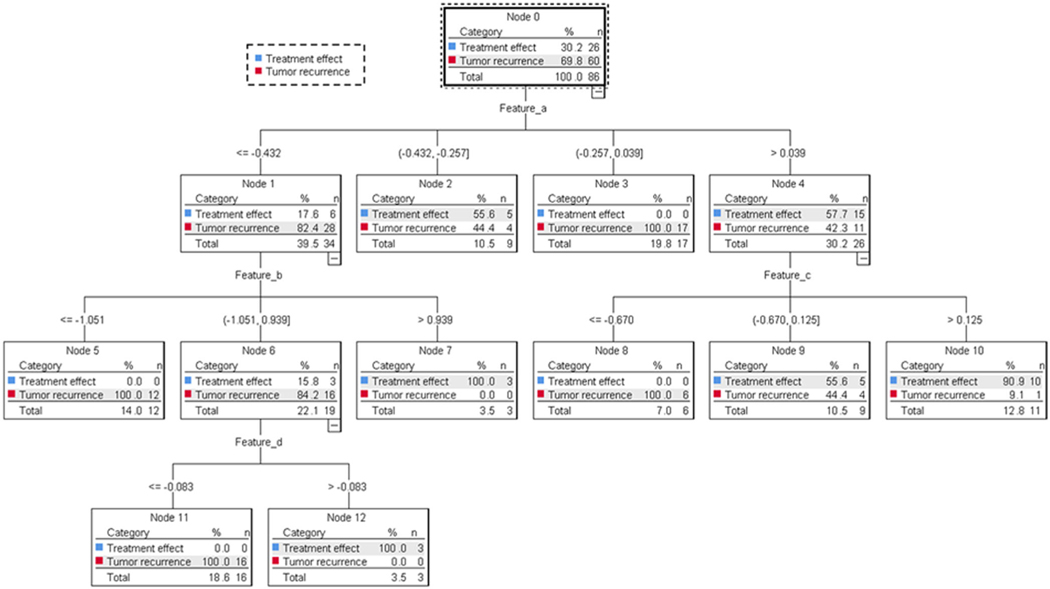
Classification was achieved using the CHAID decision tree based on the heuristic parameters. Based on the combined data from APTw and structural MRI (Model I), CHAID decision tree analysis (Figure 3) revealed that two features (b and d; see Table 3) extracted from APTw, one (a) from FLAIR, and one (c) from Gd-T1w were the most significant predictors of tumor recurrence. The details of CHAID decision tree analyses based on the data from APTw (Model IIA) and all structural MRI sequences (Model III) are shown in Supplementary Figures S1 and S2.
FIGURE 3.
CHAID decision tree analysis based on the data from APTw and structural MR images (Model I). Four features (a–d) were identified as independent predictors with which to build a CHAID decision tree with three levels of tree depth and nine terminal nodes.
TABLE 3.
The influential radiomic features identified by different CHAID models
| Connected node |
|||||||
|---|---|---|---|---|---|---|---|
| Model | Tree depth | Basic feature name | Filter | Feature class | MRI sequence | ICC value | Feature label |
| (I) All | 1 | Robust mean absolute deviation | LoG (5 mm) | first order | FLAIR | 0.915 | a |
| 2 | Informal measure of correlation 2 | LoG (1 mm) | GLCM | APTw | 0.976 | b | |
| 2 | Informal measure of correlation 2 | LoG (1 mm) | GLCM | Gd-T1w | 0.959 | c | |
| 3 | Large dependence low gray level emphasis | wavelet (H) | GLDM | APTw | 0.683 | d | |
| (IIA) APTw | 1 | Informal measure of correlation 2 | LoG (1 mm) | GLCM | APTw | 0.976 | b |
| 2 | Minor axis length | original | first order | mask | 0.893 | e | |
| 2 | Long run low gray level emphasis | original | GLRLM | APTw | 0.932 | f | |
| (IIB) T1w | 1 | Minor axis length | original | first order | mask | 0.893 | e |
| (IIC) T2w | 1 | Minor axis length | original | first order | mask | 0.893 | e |
| 2 | Small area emphasis | LoG (5 mm) | GLSZM | T2w | 0.906 | g | |
| (IID) FLAIR | 1 | Robust mean absolute deviation | LoG (5 mm) | first order | FLAIR | 0.915 | a |
| 2 | Minor axis length | original | first order | mask | 0.893 | e | |
| (IIE) Gd-T1w | 1 | Informal measure of correlation 2 | LoG (1 mm) | GLCM | Gd-T1w | 0.959 | c |
| 2 | Gray level non-uniformity | original | GLSZM | Gd-T1w | 0.955 | h | |
| (III) T1w, T2w, FLAIR, | 1 | Robust mean absolute deviation | LoG (5 mm) | first order | FLAIR | 0.915 | a |
| Gd-T1w | 2 | Informal measure of correlation 1 | LoG (1 mm) | GLCM | Gd-T1w | 0.931 | i |
| 2 | Informal measure of correlation 2 | LoG (1 mm) | GLCM | Gd-T1w | 0.959 | c | |
| (IV) T1w, T2w, FLAIR, | 1 | Robust mean absolute deviation | LoG (5 mm) | first order | FLAIR | 0.915 | a |
| APTw | 2 | Informal measure of correlation 2 | LoG (1 mm) | GLCM | APTw | 0.976 | b |
| 2 | Minor axis length | original | first order | mask | 0.893 | e | |
| 3 | Large dependence low gray level emphasis | wavelet (H) | GLDM | APTw | 0.683 | d | |
Abbreviations: GLCM, gray level co-occurrence matrix; GLDM, gGray level dependence matrix; GLSZM, gray level size zone matrix; GLRLM, gray level run length matrix; LoG, Laplacian of Gaussian.
Note:Filter “original” indicates no filter applied. For filter “LoG”, the value in brackets indicates the filter width used for the Gaussian kernel. For filter “wavelet”, the label in brackets indicates the filter (H, high-pass filter; L, low-pass filter) applied.
One to four radiomic features were identified as independent predictors of tumor recurrence for each CHAID model (Table 3). Of the nine independent predictors (a–i) identified to build eight CHAID models, one feature (e) was extracted from a mask, one (g) was extracted from T2w images, one (a) was extracted from FLAIR images, three (c, h, and i) were extracted from Gd-T1w images, and three (b, d, and f) were extracted from APTw images. The four features (a–d) identified as independent predictors in Model I were repeatedly selected for different models (Models IIA, IID, IIE, III, and IV). The most frequently repeated feature was feature e, which was used in Models I, IIA, IIB, IIC, ID, and IV.
4 |. DISCUSSION
In this work, we developed and tested a classifying framework that integrated structural and APTw-MRI radiomic features for the non-invasive diagnosis of tumor recurrence versus treatment effect in malignant gliomas. The majority of radiomics studies—with variable degrees of success—have been restricted to structural MRI data. There are only a few published MRI-based radiomics studies that have used advanced MRI data to explore the prediction of treatment responsiveness or survival for patients with post-treatment malignant gliomas.64–66 These studies show various diagnostic performances. Two of them reported accuracies for responsiveness prediction of 86.7% and 75–84%, respectively.65,66 APTw-MRI generates contrast based on endogenous cellular proteins and peptides in tissue without contrast agent administration.67,68 The APTw signal intensity is closely correlated with cellularity and proliferation, and therefore has great potential as a valuable biomarker for the assessment of viable malignancy. Recently, two initial APTw-based radiomics studies have been conducted to identify a molecular marker in newly diagnosed gliomas43 and to differentiate gliomas from metastases.44 More attempts to determine the feasibility and efficacy of implementing APT-based radiomic features for response assessment are needed.
We used PyRadiomics to extract 525 radiomic features from each of the processed APTw and structural MR images. Among several published papers using MRI for the assessment of brain tumors, the numbers of radiomic features for each ROI per MRI sequence varied widely from 117 to 1731.69,70 In our study, due to the smaller APTw volume coverage, any features related to 3D images were not included (see limitation (iii) below). As an exploratory study of the application of APTw image-based radiomics in post-treatment glioma patients, features were extracted only from derived images using two classic filters (Laplacian of Gaussian with three sigma levels and wavelet with two levels). Fewer filter approaches were implemented to derive images for feature extraction, since unsatisfactory reproducibility has been previously found for the features extracted from images derived with a filter.71
In our study, we applied the CHAID decision tree algorithm to build the prediction model. Compared with the “black-box” supervised classifiers, such as the support vector machine, naive Bayes, and the artificial neural network, the decision tree is the most interpretable classification algorithm in machine learning.62,72,73 The CHAID has been previously applied in the medical field because its prediction rules are easy to interpret in clinical settings. The joint model (III), built from the most frequently applied four structural MRI sequences (T1w, T2w, FLAIR, and Gd-T1w), was presented with three independent predictors (FLAIR and Gd-T1w image-based features) to yield an accuracy of 82.6%. After adding features extracted from APTw-MRI, forming Model I, the accuracy was increased to 89.5%. Notably, the single APTw-based CHAID model using three independent predictors yielded diagnostic performances with a 95% CI accuracy of 76.7–89.9%, while adding APTw-based features to routine structural MR images improved the specificity from 86.8 to 100%. Given the greatly reduced scan time and no need for contrast agents, the single APTw-based model (IIA) is highly promising, compared with the joint models (I and III). In terms of sensitivity, the ability of a model to correctly identify patients with tumor recurrence, Model IIE, consisting of two Gd-T1w features, achieved the highest sensitivity, 100%, among all of the radiomic models.
The radiomic features extracted from APTw images yielded a diagnostic performance comparable to those in several previous APTw studies performed at 3 T.21–24,32 These previous APTw MRI studies used ROIs from solid tumor areas that covered Gd-enhancing areas (often avoiding liquefactive necrosis, intratumoral vessels, and hemorrhage). In contrast, our radiomics study used masks that covered the whole abnormality on FLAIR images as ROIs from which to extract possible distinguishing features. This APTw radiomics approach does not require professional skills or intensive labor; thus, it provides a potential automatic method with which to analyze brain tumors that display highly sophisticated radiographic patterns.
Nine radiomic features were selected in the presented models, with five features (a–e) used repeatedly. Feature a (see Table 3), repeatedly presented in Models I, IID, III, and IV, was in the first order feature category from FLAIR. Feature a was defined as the mean distance of all intensity values from the mean intensity, calculated on a subset of the image array with gray levels in between or equal to the 10th and 90th percentiles after Gaussian (LoG) (σ = 5 mm) filtering.55 Features b and c were the same feature, after Gaussian (LoG) (σ = 1 mm) filtering, from APTw and Gd-T1w MRI, respectively. Feature d was extracted from APTw-MRI with a high-pass wavelet filter. This measured the joint distribution of large dependence with lower gray-level values. Interestingly, feature e, extracted from a mask, was the most frequently repeated feature. It yielded the second-largest axis length of the ROI-enclosing ellipsoid, which was performed using the physical coordinates of the voxel (pixel) centers that defined the ROI. When minor axis length serves as an independent predictor at the first tree depth in the decision tree, lesions with masks of longer second-largest axis length are prone to tumor recurrence. However, the potential underlying mechanism needs to be further validated. Features c and h extracted from Gd-T1w yielded the highest detection sensitivity for tumor recurrence (100%). Feature h measured the variability of gray-level intensity values in the image.
This study did have some limitations. (i) Our study was limited by a relatively small and retrospective sample. In terms of generalizability, the use of an independent dataset from an outside institution for validation is needed to produce robust results. However, the radiographic patterns on APTw images are heavily affected by the MRI scanning parameters. Unfortunately, the APTw scanning protocol had not been standardized on brain tumors until the consensus recommendations were released very recently in 2022.19 (ii) Our data were collected from patients with suspected tumor recurrence during a wide time range of 18 days to 10 years after the completion of therapy. The timing of MRI scans can also affect the distinction between recurrence and treatment changes. Accurate treatment response assessment in malignant gliomas remains a clinical challenge, and even biopsy yields variable results due to the intra-tumoral heterogeneity of treatment response. (iii) The feature extraction was performed on one of the 15 slices that showed the largest Gd-enhancing area for each subject. To reduce the scanning time, the APTw MRI protocol used in the previous prospective study covered only 66 mm in the z direction (4.4 mm × 15 slices). APTw volumes covered the whole region with the FLAIR abnormality only in a small number of cases. Volumetric radiomic features are largely determined by the 3D masks (ROIs) where the features are extracted, as well as by the mutual relationship between voxels within the same 3D masks. APTw-FLAIR mismatched 3D masks (where APTw slides did not cover FLAIR-based ROIs) may lead to incorrect radiomic measurements; moreover, the coarse resolution of APTw images limited the use of volumetric feature extraction. Thus, caution should be taken when comparing the diagnostic performance with other radiomics studies that used the entire tumor volume, as well as pulse-sequence-specific ROI masks (such as a Gd-enhancing lesion mask) and solid tumor ROI masks (such as those that remove intra-tumoral necrosis and vessels). (iv) MGMT methylation or IDH mutation strongly influence patient prognosis. The lack of data for these prognostic pathologic markers in this retrospective study profoundly impeded our effort to perform a comprehensive assessment of treatment. (v) As discussed in the recent consensus paper,19 the B1 variations in the brain at 3 T are typically within ±10% and can be up to ±30% in some regions, such as in the infratentorial region and in the superior part of the brain,74 which may affect the APTw signal. A body coil and parallel transmit were used in this study, so the related effect may not be sufficient to substantially affect APTw contrast within most brain slices. However, this could be an issue only in the infratentorial and superior brain regions, and further assessments are needed. Further, it is known that the APTw signal consists of APT and a few other contributions, such as the downfield nuclear Overhauser effect, spillover, and magnetization transfer contrast dilution. Possible implications for radiomics analyses from these technical factors need to be assessed in the future.
5 |. CONCLUSION
Our results in this study indicate that an APTw radiomic feature-based CHAID model can diagnose tumor response in malignant gliomas better than one with individual structural MRI-based features to a certain extent. Moreover, adding APTw features may boost the accuracy of the responsiveness assessment model using only features extracted from traditional MRI. This study showed preliminary data from CHAID decision-tree modeling using APTw-based radiomic features. Further multicenter and large-scale studies should be performed to improve and verify the model, as well as the potential for application in the clinical management of malignant gliomas.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institutes of Health (R37CA248077, R01CA228188, and P41EB015909). The authors thank Ms Lindsay Blair for patient coordination and Ms Mary McAllister for editorial assistance.
Funding information
National Institutes of Health, Grant/Award Numbers: P41EB015909, R01CA228188, R37CA248077
Abbreviations:
- APTw
amide proton transfer weighted
- AUC
area under the curve
- CHAID
chi-square automatic interaction detector
- FLAIR
fluid-attenuated inversion recovery
- FOV
field of view
- Gd-T1w
gadolinium enhanced T1 weighted
- ICC
intraclass correlation coefficient
- RANO
Response Assessment in Neuro-Oncology
- ROC
receiver operating characteristic
- ROI
region of interest
- T1w
T1 weighted
- T2w
T2 weighted
- T E
echo time
- T R
repetition time
Footnotes
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10): 987–996. doi: 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 2.Kumar AJ, Leeds NE, Fuller GN, et al. Malignant gliomas: MR imaging spectrum of radiation therapy- and chemotherapy-induced necrosis of the brain after treatment. Radiology. 2000;217(2):377–384. doi: 10.1148/radiology.217.2.r00nv36377 [DOI] [PubMed] [Google Scholar]
- 3.Dunbar E, Blakeley J, Ye XB, et al. A systemic review of the magnetic resonance imaging features immediately postoperative and one-month postradiation and concurrent temozolomide. Neuro-Oncology. 2008;10:892–893. [Google Scholar]
- 4.Sanghera P, Perry J, Sahgal A, et al. Pseudoprogression following chemoradiotherapy for glioblastoma multiforme. Can J Neurol Sci. 2010;37(1):36–42. doi: 10.1017/S0317167100009628 [DOI] [PubMed] [Google Scholar]
- 5.Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: Response Assessment in Neuro-Oncology Working Group. J Clin Oncol. 2010;28(11):1963–1972. doi: 10.1200/JCO.2009.26.3541 [DOI] [PubMed] [Google Scholar]
- 6.Eisele SC, Wen PY, Lee EQ. Assessment of brain tumor response: RANO and its offspring. Curr Treat Options Oncol. 2016;17:35. [DOI] [PubMed] [Google Scholar]
- 7.Prager AJ, Martinez N, Beal K, Omuro A, Zhang Z, Young RJ. Diffusion and perfusion MRI to differentiate treatment-related changes including pseudoprogression from recurrent tumors in high-grade gliomas with histopathologic evidence. Am J Neuroradiol. 2015;36:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazda T, Bulik M, Pospisil P, et al. Advanced MRI increases the diagnostic accuracy of recurrent glioblastoma: single institution thresholds and validation of MR spectroscopy and diffusion weighted MR imaging. NeuroImage Clin. 2016;11:316–321. doi: 10.1016/j.nicl.2016.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Dijken BRJ, van Laar PJ, Holtman GA, van der Hoorn A. Diagnostic accuracy of magnetic resonance imaging techniques for treatment response evaluation in patients with high-grade glioma, a systematic review and meta-analysis. Eur Radiol. 2017;27:4129–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suh CH, Kim HS, Jung SC, Choi CG, Kim SJ. Multiparametric MRI as a potential surrogate endpoint for decision-making in early treatment response following concurrent chemoradiotherapy in patients with newly diagnosed glioblastoma: a systematic review and meta-analysis. Eur Radiol. 2018;28: 2628–2638. [DOI] [PubMed] [Google Scholar]
- 11.Chiang GC, Kovanlikaya I, Choi C, Ramakrishna R, Magge R, Shungu DC. Magnetic resonance spectroscopy, positron emission tomography and radiogenomics—relevance to glioma. Front Neurol. 2018;9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barajas RF, Hamilton BE, Schwartz D, et al. Combined iron oxide nanoparticle ferumoxytol and gadolinium contrast enhanced MRI define glioblastoma pseudoprogression. Neuro-Oncology. 2019;21(4):517–526. doi: 10.1093/neuonc/noy160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.le Fèvre C, Constans JM, Chambrelant I, et al. Pseudoprogression versus true progression in glioblastoma patients: a multiapproach literature review. Part 2—Radiological features and metric markers. Crit Rev Oncol Hematol. 2021;159:103230. doi: 10.1016/j.critrevonc.2021.103230 [DOI] [PubMed] [Google Scholar]
- 14.Martin-Noguerol T, Mohan S, Santos-Armentia E, Cabrera-Zubizarreta A, Luna A. Advanced MRI assessment of non-enhancing peritumoral signal abnormality in brain lesions. Eur J Radiol. 2021;143:109900. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Heo H-Y, Knutsson L, van Zijl PCM, Jiang S. APT-weighted MRI: techniques, current neuro applications, and challenging issues. J Magn Reson Imaging. 2019;50:347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, Jones CK, van Zijl PCM, Barker PB, Zhou J. Fast 3D chemical exchange saturation transfer (CEST) imaging of the human brain. Magn Reson Med. 2010;64:638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X, Wen Z, Zhang G, et al. Three-dimensional turbo-spin-echo amide proton transfer MR imaging at 3-Tesla and its application to high-grade human brain tumors. Mol Imaging Biol. 2013;15(1):114–122. doi: 10.1007/s11307-012-0563-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Togao O, Keupp J, Hiwatashi A, et al. Amide proton transfer imaging of brain tumors using a self-corrected 3D fast spin-echo Dixon method: comparison with separate B0 correction. Magn Reson Med. 2017;77(6):2272–2279. doi: 10.1002/mrm.26322 [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Zaiss M, Knutsson L, et al. Review and consensus recommendations on clinical APT-weighted imaging approaches at 3T: application to brain tumors. Magn Reson Med. 2022;88:546–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sotirios B, Demetriou E, Topriceanu CC, Zakrzewska Z. The role of APT imaging in gliomas grading: a systematic review and meta-analysis. Eur J Radiol. 2020;133:109353. [DOI] [PubMed] [Google Scholar]
- 21.Ma B, Blakeley JO, Hong X, et al. Applying amide proton transfer-weighted MRI to distinguish pseudoprogression from true progression in malignant gliomas. J Magn Reson Imaging. 2016;44(2):456–462. doi: 10.1002/jmri.25159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park KJ, Kim HS, Park JE, Shim WH, Kim SJ, Smith SA. Added value of amide proton transfer imaging to conventional and perfusion MR imaging for evaluating the treatment response of newly diagnosed glioblastoma. Eur Radiol. 2016;26:4390–4403. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Li C, Chen Y, et al. Diagnostic performance of multiparametric MRI in the evaluation of treatment response in glioma patients at 3T. J Magn Reson Imaging. 2020;51(4):1154–1161. doi: 10.1002/jmri.26900 [DOI] [PubMed] [Google Scholar]
- 24.Park YW, Ahn SS, Kim EH, et al. Differentiation of recurrent diffuse glioma from treatment-induced change using amide proton transfer imaging: incremental value to diffusion and perfusion parameters. Neuroradiology. 2021;63:363–372. doi: 10.1007/s00234-020-02542-5 [DOI] [PubMed] [Google Scholar]
- 25.Mehrabian H, Myrehaug S, Soliman H, Sahgal A, Stanisz GJ. Evaluation of glioblastoma response to therapy with chemical exchange saturation transfer. Int J Radiat Oncol Biol Phys. 2018;101:713–723. [DOI] [PubMed] [Google Scholar]
- 26.Paech D, Dreher C, Regnery S, et al. Relaxation-compensated amide proton transfer (APT) MRI signal intensity is associated with survival and progression in high-grade glioma patients. Eur Radiol. 2019;29(9):4957–4967. doi: 10.1007/s00330-019-06066-2 [DOI] [PubMed] [Google Scholar]
- 27.Joo B, Han K, Ahn SS, et al. Amide proton transfer imaging might predict survival and IDH mutation status in high-grade glioma. Eur Radiol. 2019; 29(12):6643–6652. doi: 10.1007/s00330-019-06203-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meissner JE, Korzowski A, Regnery S, et al. Early response assessment of glioma patients to definitive chemoradiotherapy using chemical exchange saturation transfer imaging at 7 T. J Magn Reson Imaging. 2019;50(4):1268–1277. doi: 10.1002/jmri.26702 [DOI] [PubMed] [Google Scholar]
- 29.Jiang S, Zou T, Eberhart CG, et al. Predicting IDH mutation status in grade II gliomas using amide proton transfer-weighted (APTw) MRI. Magn Reson Med. 2017;78(3):1100–1109. doi: 10.1002/mrm.26820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paech D, Windschuh J, Oberhollenzer J, et al. Assessing the predictability of IDH mutation and MGMT methylation status in glioma patients using relaxation-compensated multi-pool CEST MRI at 7.0 Tesla. Neuro-Oncology. 2018;20(12):1661–1671. doi: 10.1093/neuonc/noy073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang S, Wen Z, Ahn SS, et al. Applications of chemical exchange saturation transfer magnetic resonance imaging in identifying genetic markers in gliomas. NMR Biomed. 2022;e4731. doi: 10.1002/nbm.4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang S, Eberhart CG, Lim M, et al. Identifying recurrent malignant glioma after treatment using amide proton transfer-weighted MR imaging: a validation study with image-guided stereotactic biopsy. Clin Cancer Res. 2019;25(2):552–561. doi: 10.1158/1078-0432.CCR-18-1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu CF, Hsu FT, Hsieh KL, et al. Machine learning-based radiomics for molecular subtyping of gliomas. Clin Cancer Res. 2018;24:4429–4436. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Li Y, Qian Z, et al. A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. NeuroImage Clin. 2018;20:1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arita H, Kinoshita M, Kawaguchi A, et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas. Sci Rep. 2018;8(1):11773. doi: 10.1038/s41598-018-30273-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Yang J, Ho A, et al. A predictive model for distinguishing radiation necrosis from tumour progression after gamma knife radiosurgery based on radiomic features from MR images. Eur Radiol. 2018;28(6):2255–2263. doi: 10.1007/s00330-017-5154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park YW, Choi YS, Ahn SS, Chang JH, Kim SH, Lee SK. Radiomics MRI phenotyping with machine learning to predict the grade of lower-grade gliomas: a study focused on nonenhancing tumors. Korean J Radiol. 2019;20:1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nie D, Lu J, Zhang H, et al. Multi-channel 3D deep feature learning for survival time prediction of brain tumor patients using multi-modal neuroimages. Sci Rep. 2019;9(1):1103. doi: 10.1038/s41598-018-37387-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei J, Yang G, Hao X, et al. A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication. Eur Radiol. 2019;29(2):877–888. doi: 10.1007/s00330-018-5575-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K, Qiao Z, Zhao X, et al. Individualized discrimination of tumor recurrence from radiation necrosis in glioma patients using an integrated radiomics-based model. Eur J Nucl Med Mol Imaging. 2020;47(6):1400–1411. doi: 10.1007/s00259-019-04604-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chougule T, Gupta RK, Saini J, et al. Radiomics signature for temporal evolution and recurrence patterns of glioblastoma using multimodal magnetic resonance imaging. NMR Biomed. 2022;35(3):e4647 doi: 10.1002/nbm.4647 [DOI] [PubMed] [Google Scholar]
- 42.Chawla S, Bukhari S, Afridi OM, et al. Metabolic and physiologic magnetic resonance imaging in distinguishing true progression from pseudoprogression in patients with glioblastoma. NMR Biomed. 2022;35(7):e4719. doi: 10.1002/nbm.4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han Y, Wang W, Yang Y, et al. Amide proton transfer imaging in predicting isocitrate dehydrogenase 1 mutation status of Grade II/III gliomas based on support vector machine. Front Neurosci. 2020;14:144. doi: 10.3389/fnins.2020.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sartoretti E, Sartoretti T, Wyss M, et al. Amide proton transfer weighted (APTw) imaging based radiomics allows for the differentiation of gliomas from metastases. Sci Rep. 2021;11(1):5506. doi: 10.1038/s41598-021-85168-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su C, Jiang J, Zhang S, et al. Radiomics based on multicontrast MRI can precisely differentiate among glioma subtypes and predict tumour-proliferative behaviour. Eur Radiol. 2019;29(4):1986–1996. doi: 10.1007/s00330-018-5704-8 [DOI] [PubMed] [Google Scholar]
- 46.Li J, Liu S, Qin Y, Zhang Y, Wang N, Liu H. High-order radiomics features based on T2 FLAIR MRI predict multiple glioma immunohistochemical features: a more precise and personalized gliomas management. PLoS ONE. 2020;15:e0227703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park JE, Kim HS, Kim D, et al. A systematic review reporting quality of radiomics research in neuro-oncology: toward clinical utility and quality improvement using high-dimensional imaging features. BMC Cancer. 2020;20(1):29. doi: 10.1186/s12885-019-6504-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo P, Wang P, Yasarla R, Zhou J, Patel VM, Jiang S. Anatomic and molecular MR image synthesis using confidence guided CNNs. IEEE Trans Med Imaging. 2020;40(10):2832–2844. doi: 10.1109/TMI.2020.3046460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J, Zhu H, Lim M, et al. Three-dimensional amide proton transfer MR imaging of gliomas: Initial experience and comparison with gadolinium enhancement. J Magn Reson Imaging. 2013;38(5):1119–1128. doi: 10.1002/jmri.24067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim M, Gillen J, Landman BA, Zhou J, van Zijl PCM. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn Reson Med. 2009;61:1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans Med Imaging. 2000;19:143–150. [DOI] [PubMed] [Google Scholar]
- 53.Lowekamp BC, Chen DT, Ibáñez L, Blezek D. The design of SimpleITK. Front Neuroinform. 2013;7:45. doi: 10.3389/fninf.2013.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lipkova J, Angelikopoulos P, Wu S, et al. Personalized radiotherapy design for glioblastoma: integrating mathematical tumor models, multimodal scans, and Bayesian inference. IEEE Trans Med Imaging. 2019;38(8):1875–1884. doi: 10.1109/TMI.2019.2902044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.PyRadiomics. https://pyradiomics.readthedocs.io
- 56.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21): e104–e107. doi: 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizzo S, Botta F, Raimondi S, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2(1):36. doi: 10.1186/s41747-018-0068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Avanzo M, Wei LS, Stancanello J, et al. Machine and deep learning methods for radiomics. Med Phys. 2020;47(5):E185–E202. doi: 10.1002/mp.13678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Jong EEC, Sanders KJC, Deist TM, et al. Can radiomics help to predict skeletal muscle response to chemotherapy in stage IV non-small cell lung cancer? Eur J Cancer. 2019;120:107–113. doi: 10.1016/j.ejca.2019.07.023 [DOI] [PubMed] [Google Scholar]
- 61.Kass GV. An exploratory technique for investigating large quantities of categorical data. Appl Stat. 1980;29(2):119. doi: 10.2307/2986296 [DOI] [Google Scholar]
- 62.Baltzer PA, Dietzel M, Kaiser WA. A simple and robust classification tree for differentiation between benign and malignant lesions in MR-mammography. Eur Radiol. 2013;23:2051–2060. [DOI] [PubMed] [Google Scholar]
- 63.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 64.Ion-Margineanu A, Van Cauter S, Sima DM, et al. Classifying glioblastoma multiforme follow-up progressive vs. responsive forms using multi-parametric MRI features. Front Neurosci. 2016;10:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qian X, Tan H, Zhang J, Zhao W, Chan MD, Zhou X. Stratification of pseudoprogression and true progression of glioblastoma multiform based on longitudinal diffusion tensor imaging without segmentation. Med Phys. 2016;43(11):5889–5902. doi: 10.1118/1.4963812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akbari H, Rathore S, Bakas S, et al. Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma. Cancer. 2020;126(11):2625–2636. doi: 10.1002/cncr.32790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou J, Payen J, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat Med. 2003;9:1085–1090. [DOI] [PubMed] [Google Scholar]
- 68.Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PCM. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med. 2003;50: 1120–1126. [DOI] [PubMed] [Google Scholar]
- 69.Cho H, Lee S, Kim J, Park H. Classification of the glioma grading using radiomics analysis. PeerJ. 2018;6:e5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li G, Li L, Li Y, et al. An MRI radiomics approach to predict survival and tumour-infiltrating macrophages in gliomas. Brain. 2022;145(3):1151–1161. doi: 10.1093/brain/awab340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Depeursinge A, Andrearczyk V, Whybra P, et al. Standardised convolutional filtering for radiomics. arXiv. 2021:2006.05470v05475. [Google Scholar]
- 72.Safavian SR, Landgrebe D. A survey of decision tree classifier methodology. IEEE Trans Syst Man Cybern. 1991;21:660–674. [Google Scholar]
- 73.Wu MX, Zhong XL, Peng QZ, et al. Prediction of molecular subtypes of breast cancer using BI-RADS features based on a “white box” machine learning approach in a multi-modal imaging setting. Eur J Radiol. 2019;114:175–184. doi: 10.1016/j.ejrad.2019.03.015 [DOI] [PubMed] [Google Scholar]
- 74.Mueller S, Stirnberg R, Akbey S, et al. Whole brain snapshot CEST at 3T using 3D-EPI: aiming for speed, volume, and homogeneity. Magn Reson Med. 2020;84(5):2469–2483. doi: 10.1002/mrm.28298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.