Abstract
The spirochetes inhabiting the large intestines of humans and animals consist of a diverse group of related organisms. Intestinal spirochetosis caused by Serpulina pilosicoli is a newly recognized enteric disease of human beings and animals with potential public health significance. The purpose of this study was to determine the species identity of canine intestinal spirochetes by comparing 30 isolates obtained from dogs in Australia (n = 25) and the United States (n = 5) with reference strains representing Serpulina species and Brachyspira aalborgi, by phenotypic and genetically based typing methods. All of the canine isolates were indole negative and produced a weak β-hemolysis when cultured anaerobically on agar medium containing blood. Four isolates were identified as S. pilosicoli by 16S rRNA-specific PCR assays, rRNA gene restriction fragment length polymorphism or ribotyping, and multilocus enzyme electrophoresis. The remaining 26 isolates formed a cluster related to porcine Serpulina innocens as determined by multilocus enzyme electrophoresis but had a unique ribotype pattern. The data suggested the existence of a novel Serpulina species, provisionally designated “Serpulina canis,” colonizing the intestines of dogs.
Recent detailed phenotypic and genotypic characterizations of culturable intestinal spirochetes have led to the taxonomic classification of four new species within the genus Serpulina in addition to Serpulina hyodysenteriae, the cause of swine dysentery, and Serpulina innocens, a nonpathogenic commensal of the swine colon (11, 14, 18, 32). These include a pathogenic spirochete of chickens, Serpulina alvinipulli (34), and two spirochetes with unknown pathogenic potential, Serpulina intermedia, found in swine and poultry, and Serpulina murdochii, found in swine and rats (33). The fourth new species, Serpulina pilosicoli, is the etiologic agent of a colonic disease affecting human beings, nonhuman primates, dogs, pigs, and birds (1–6, 8, 9, 22, 23, 25–27, 28, 32, 37–41). The disease caused by S. pilosicoli, which is characterized by colonization of the surface and crypt mucin and intimate attachment of the spirochetes to the apical membrane of cecal and colonic enterocytes (2–7, 26–28, 36–38, 40, 43), has been referred to as intestinal spirochetosis (IS), cecal spirochetosis, colonic spirochetosis, colorectal spirochetosis, or rectal spirochetosis (1–6, 16, 19, 22, 23, 25–27, 28, 32, 35–43). Spirochetes structurally and phenotypically different from S. pilosicoli and called Brachyspira aalborgi also have been seen in human beings and rhesus macaques with IS (5, 13). Additionally, the cecal and colonic epithelium of North American opossums and laboratory guinea pigs can be colonized by spirochetes of unknown taxonomic classification but with morphological features resembling those of S. pilosicoli and B. aalborgi (4).
Spirochetes have been seen in the feces and colons of dogs for decades, but conflicting reports about their role in disease have made it difficult to determine their pathogenetic significance. Between the 1940s and 1970s, several investigators reported that spirochetes were more common in the feces of dogs with diarrhea, particularly puppies, than in those of healthy dogs (4, 12, 18, 44). Whereas some suggested that spirochetes may cause diarrhea, Leach and coworkers (21) found spirochetes within the colonic crypts of dogs without diarrhea. Based on this observation and the demonstration of large numbers of spirochetes in the feces of rats with osmotic diarrhea caused by the cathartic agent magnesium sulfate, it was suggested that spirochetes may be commensals that are mechanically dislodged from the crypts by diarrhea induced by other etiological factors. Harris and Kinyon (12) later cultured weakly β-hemolytic intestinal spirochetes (WBHIS) from the feces of a pup with mucohemorrhagic colitis, but Turek and Meyer (42) found similar organisms in the feces of apparently healthy mature dogs. On the basis of its ultrastructural morphology and its failure to cause disease in experimentally-inoculated dogs, pigs, and mice (10, 15, 17, 18), the canine isolate described by Kinyon and Harris (12) was considered a nonpathogenic commensal similar to porcine S. innocens. To this day, a causal relationship between large numbers of spirochetes and diarrhea continues to be reported (24), but the morphology and numbers of organisms are probably insufficient to establish a definitive role for spirochetes in enteric diseases of dogs.
Koch’s postulates for S. pilosicoli have been fulfilled by using gnotobiotic swine (28) and conventional swine (6, 36, 37, 40). Swine challenged with porcine or human S. pilosicoli develop diarrhea and reduced growth together with spirochetal attachment to the apical surfaces of colonic enterocytes (6, 36, 40). Similarly, crop inoculation of chicks with porcine and human S. pilosicoli results in spirochetal attachment to the cecal epithelium accompanied by invasion of the cecal wall (7, 26, 27, 38). Although the attachment of S. pilosicoli to the enterocytes is pathognomonic of IS (2–7, 26, 27, 36–38, 40), the mechanism of association appears different from that described previously for attaching and effacing gastroenteric bacterial pathogens (26).
Canine IS was first described in mature dogs with normal feces (43), but Duhamel and coworkers (3) later documented IS in a 3-month-old beagle pup with diarrhea that had a concurrent infection with Giardia spp. Spirochetes isolated from the beagle pup, designated 16242-94, and spirochetes isolated by Turek and Meyer (42), designated K9-12, were found to attach to cecal enterocytes of chicks by a mechanism similar to that described for porcine and human S. pilosicoli (26). These isolates also have been assigned to S. pilosicoli on the basis of phenotypic characteristics and the results of genetically based methods, including DNA-DNA reassociation and DNA fingerprinting by arbitrarily primed PCR (1) and flaA1 gene restriction fragment length polymorphism (RFLP) (9). Because the DNA homology between canine isolate 16242-94 and porcine S. pilosicoli P43/6/78T and human isolate SP16 was greater than 95% and because the DNA homology between 16242-94 and S. hyodysenteriae and S. innocens was less than 32%, it was concluded that S. pilosicoli colonizes dogs and causes IS (1).
The purpose of the present study was to determine the species identity of canine intestinal spirochetes by comparing 30 isolates obtained from dogs in Australia and the United States with reference strains of Serpulina spp. and B. aalborgi by phenotypic and genetically based typing methods. The identification of canine intestinal spirochetes will assist with the development of improved protocols for the diagnosis of enteric diseases of dogs.
MATERIALS AND METHODS
Spirochetes and growth conditions.
The sources, origins, clinical history, and phenotypic and genotypic characterization of reference and canine intestinal spirochetes investigated in this study are presented in Table 1. Pure cultures of spirochetes were propagated either on Trypticase soy agar containing 5% citrated sheep blood (TSAB) incubated at 42°C in the Gas Pak anaerobic system (BBL, Becton Dickinson Microbiology Systems, Cockeysville, Md.) for 7 to 14 days or in prereduced anaerobically sterilized Trypticase soy broth, as previously described (1, 20). Broth cultures were grown to late logarithmic phase (approximately 3 to 5 days; 108 cells per ml), while being stirred constantly at 37°C under a 10% hydrogen–10% carbon dioxide–80% nitrogen atmosphere.
TABLE 1.
Source, origins, clinical history, and phenotypic and genotypic characterization of reference and canine intestinal spirochetes investigated in this study
| Species and strain | Origin (animal/country or state/locale)a | Clinical historyb | Phenotypec | RFLP flaA1d | ET | Source/ reference(s)e |
|---|---|---|---|---|---|---|
| S. hyodysenteriae | ||||||
| B78T (ATCC 27164) | Pig/U.S.A./farm | 1 | I | I | 9 | 1/11,14,18 |
| A1 | Pig/U.K./farm | 1 | I | I | 13 | 1/11,14,22 |
| S. intermedia | ||||||
| PWS/AT (ATCC 51140) | Pig/U.K./farm | 2 | II | NDf | 34 | 2/33 |
| S. innocens | ||||||
| B256T (ATCC 29796) | Pig/U.S.A./farm | 3 | III | III | 38 | 2/8,14,18 |
| 4/71 | Pig/U.K./farm | 3 | III | III | 43 | 3/14,22,18 |
| “S. canis” | ||||||
| Dog B | Dog/W.A./shelter | 6 | ND | ND | D1 | 5/24 |
| Dog C14 | Dog/W.A./shelter | 6 | ND | ND | D2 | 5/24 |
| Dog C5 | Dog/W.A./shelter | 6 | ND | ND | D3 | 5/24 |
| Dog M2 | Dog/W.A./shelter | 6 | ND | ND | D4 | 5/24 |
| D97 | Dog/W.A./hosp. | 6 | ND | ND | D5 | 5/24 |
| D26 | Dog/W.A./Abor. comm. | 6 | ND | ND | D6 | 5/24 |
| D148 | Dog/W.A./hosp. | 6 | ND | ND | D7 | 5/24 |
| Dog T | Dog/W.A./shelter | 6 | ND | ND | D8 | 5/24 |
| WW20-2 | Dog/N.S.W./Abor. comm. | 6 | ND | ND | D8 | 5/24 |
| Dog A | Dog/W.A./shelter | 6 | ND | ND | D9 | 5/24 |
| Dog C1 | Dog/W.A./shelter | 6 | ND | ND | D9 | 5/24 |
| Dog C8 | Dog/W.A./shelter | 6 | ND | ND | D10 | 5/24 |
| D20-2 | Dog/W.A./Abor. comm. | 6 | ND | ND | D10 | 5/24 |
| WB38 | Dog/N.S.W./Abor. comm. | 6 | ND | ND | D10 | 5/24 |
| Dog A2 | Dog/W.A./shelter | 6 | ND | ND | D11 | 5/24 |
| D16 | Dog/W.A./Abor. comm. | 6 | ND | ND | D12 | 5/24 |
| D24 | Dog/W.A./Abor. comm. | 6 | ND | ND | D12 | 5/24 |
| D22 | Dog/W.A./hosp. | 6 | ND | ND | D13 | 5/24 |
| D30 | Dog/W.A./hosp. | 6 | ND | ND | D13 | 5/24 |
| D32 | Dog/W.A./hosp. | 6 | ND | ND | D13 | 5/24 |
| D15 | Dog/W.A./Abor. comm. | 6 | ND | ND | D14 | 5/24 |
| D27 | Dog/W.A./Abor. comm. | 6 | ND | ND | D14 | 5/24 |
| G108 | Dog/W.A./hosp. | 6 | ND | ND | D14 | 5/24 |
| Dog X | Dog/W.A./shelter | 6 | ND | ND | D15 | 5/24 |
| 14199-95 | Dog/U.S.A./hosp. | 5 | ND | ND | D16 | 4/1 |
| 24072-93B | Dog/U.S.A./hosp. | 4 | ND | ND | D17 | 4/1 |
| S. alvinipulli | ||||||
| C1T (ATCC 51933) | Chicken/U.S.A./farm | 7 | NAg | ND | C1 | 3/32,34 |
| S. murdochii | ||||||
| 56-150T (ATCC 51254) | Pig/Canada/farm | 2 | III | IV | 54 | 2/33 |
| S. pilosicoli | ||||||
| P43/6/78T (ATCC 51139) | Pig/U.K./farm | 8 | IV | V | 60 | 3/1,8,22,39 |
| SP16 (ATCC 49776) | Human/U.S.A./hosp. | 9 | IV | V | ND | 6/1,23 |
| Dog 17 | Dog/W.A./Abor. comm. | 2 | IV | V | D18 | 5/23 |
| K9-12 | Dog/U.S.A./shelter | 10 | IV | V | D19 | 7/1 |
| 24072-93A | Dog/U.S.A./hosp. | 4 | IV | ND | D20 | 4/1 |
| 16242-94 | Dog/U.S.A./res. col. | 11 | IV | V | D21 | 4/1 |
| B. aalborgi | ||||||
| 513AT (ATCC 43994) | Human/Denmark/hosp. | 12 | NA | VI | ND | 5/13,32 |
N.S.W., New South Wales, Australia; U.S.A., United States; U.K., United Kingdom; W. A., Western Australia; hosp., hospital; res. col., research colony; Abor. comm., Aboriginal community.
1, swine dysentery; 2, unknown age with diarrhea; 3, healthy; 4, 4-month-old poodle littermate pups with a chronic mucoid diarrhea containing flecks of blood; 5, mature German shepherd that died suddenly of an unknown cause with normal stools; 6, not available; 7, chicken with typhilitis; 8, grower pig with IS and diarrhea; 9, human immunodeficiency virus-positive adult homosexual male with diarrhea; 9, mature dog with IS and normal stools; 11, 3-month-old beagle pup with diarrhea and wasting caused by IS and giardiasis; 12, adult with IS and unknown immune and enteric status.
Phenotype as described by Fellström and coworkers (8).
RFLP flaA1, RFLP of Serpulina sp. flaA1 gene (9).
1, J. M. Kinyon, College of Veterinary Medicine, Iowa State University, Ames; 2, American Type Culture Collection, Manassas, Va.; 3, T. B. Stanton, National Animal Disease Center, Ames, Iowa; 4, G. E. Duhamel; 5, D. J. Hampson; 6, R. M. Smibert, Virginia Polytechnic Institute, Blacksburg, Va.; 7, M. J. Wannemuehler, College of Veterinary Medicine, Iowa State University.
ND, not determined.
NA, not applicable.
Phenotypic characterization.
The intensity and pattern of hemolysis of the canine spirochetes were compared with those of type strains of intestinal spirochetes that produce either a strong β-hemolysis characteristic of S. hyodysenteriae B78T, a weak β-hemolysis as seen with S. innocens B256T and S. pilosicoli P43/6/78T, or a very weak β-hemolysis indicative of B. aalborgi. Each isolate was streaked onto TSAB and incubated at 42°C in the Gas Pak anaerobic system (BBL, Becton Dickinson Microbiology Systems) for up to 7 days, as previously described (1). The production of indole, a characteristic of S. hyodysenteriae and S. intermedia, was determined from broth cultures of each canine isolate as previously described (1, 22).
PCR.
Total DNA was obtained from reference and canine intestinal spirochetes by a previously described method (27). Amplification of a 16S rRNA gene sequence specific for S. pilosicoli was done by previously described PCR assays (27, 29). Briefly, a 21-base forward primer extending from base position 65 of the porcine S. pilosicoli P43/6/78T 16S rRNA gene (GenBank accession no. U14927; National Center for Biotechnology Information, Bethesda, Md.) with the nucleotide sequence 5′-AGAGGAAAGTTTTTTCGCTTC-3′ and either a 22-base universal eubacterial 16S rRNA gene sequencing primer (′1492r′) with the nucleotide sequence 5′-TACGGCTACCTTGTTACGACTT-3′ (29) or a 20-base Serpulina sp. conserved 16S rRNA gene reverse primer at position 506 with the sequence 5′-TCCGCCTACTCACCCTTTAC-3′ (27) were used. Negative controls consisting of samples with DNA from S. hyodysenteriae, S. innocens, or B. aalborgi or with no template DNA were included in all assays. The DNA was amplified with a thermocycler (Perkin-Elmer Cetus, Norwalk, Conn.) in a total volume of 50 μl containing 6 mM MgCl2; 1× PCR buffer; 0.4 mM concentrations of dATP, dTTP, dGTP, and dCTP (Perkin-Elmer Cetus); 100 pmol of primers; and 1.5 U of Taq DNA polymerase (Perkin-Elmer Cetus) in sterile filtered autoclaved water. Initial denaturing was for 10 min at 94°C, followed by 35 cycles (45 s at 55°C, 45 s at 72°C, and 90 s at 94°C). The amplified products were visualized in 2% agarose gels run at 3 V/cm after being stained with ethidium bromide.
MEE.
The multilocus enzyme electrophoresis (MEE) analyses were done by the procedures described previously for porcine and human intestinal spirochetes (22, 23, 25). Broth cultures in late log phase were harvested by centrifugation (10,000 × g; 20 min), washed in phosphate-buffered saline (pH 7.2), resuspended in 0.5 ml of sterile distilled water, and lysed by three 30-s cycles of sonication at 50 W (Labsonic 1510 sonicator). Supernatants containing the constitutive enzymes were obtained by centrifugation (20,000 × g; 20 min) and stored at −70°C until needed. The electrophoretic mobilities of 15 constitutive enzymes were determined in 11.4% horizontal starch gels (30). The enzymes examined were acid phosphatase, alcohol dehydrogenase, adenylate kinase, alkaline phosphatase, esterase, fructose-1,6-diphosphatase, glucose phosphate isomerase, guanine deaminase, glutamate dehydrogenase, hexokinase, mannose phosphate isomerase, nucleoside phosphorylase, l-leucylglycylglycine peptidase, phosphoglucomutase, and superoxide dismutase. Each enzyme was localized by the addition of a suitable substrate under appropriate buffer conditions, as described previously (22). Differences in the electrophoretic mobility of a given enzyme for different isolates indicated different alleles at the corresponding structural gene locus. Isolates were characterized by their alleles at each enzyme locus, with isolates having the same alleles at all 15 loci belonging to the same electrophoretic type (ET). The genetic distance between ETs was calculated as the proportion of loci at which different alleles occurred. A matrix of these distances was prepared, and as previously described (25), these distances were compared and clustered by the unweighted pair group method of arithmetic averages strategy. The genetic relatedness between isolates was depicted as a phenogram by combining the results of this study with previous results for porcine intestinal spirochetes and S. alvinipulli C1T (22, 23, 32, 34).
Ribotyping.
Total DNA was obtained from reference spirochetes and a subset of canine intestinal spirochetes by a previously described method (27). Restriction fragments of the genes encoding rRNA were obtained by digestion of chromosomal DNA with Sau3AI followed by separation by electrophoresis in an 0.8% agarose gel and transfer to a nylon membrane (Amersham, Arlington Heights, Ill.), as previously described (14). The rRNA from S. hyodysenteriae isolate A1 was obtained with an RNA isolation kit (RNA Track; Biotecx Laboratories Inc., Houston, Tex.) and labeled with photobiotin (PHOTOPROBE Biotin; Vector Laboratories, Burlingame, Calif.) by following the manufacturer’s recommended procedure. Hybridization of the rRNA probe was performed as previously described (14), and the rRNA banding pattern of each isolate was visualized after incubation with streptavidin-alkaline phosphatase and nitroblue tetrazolium (BluGene; Gibco-BRL, Life Technologies, Gaithersburg, Md.).
RESULTS
Phenotypic characterization.
All of the canine spirochetes grew within 7 days as a thin film that gave a ground-glass appearance to the surface of the agar medium and produced discrete weak β-hemolysis without a zone of enhanced hemolysis or ring phenomenon when growing in areas where the agar had been stabbed. B. aalborgi produced very weak β-hemolysis without a ring phenomenon when grown anaerobically at 37°C for 14 to 21 days but did not grow at 42°C. None of the canine spirochetes produced indole.
PCR.
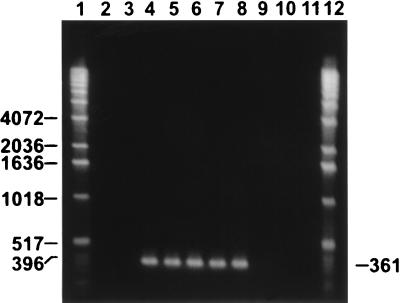
Amplification of chromosomal DNA from each isolate with primers for the S. pilosicoli 16S rRNA gene sequence by PCR methods yielded specific products with porcine S. pilosicoli P43/6/78T and canine isolates K9-12, 16242-94, 24072-93A, and Dog 17 but not with S. hyodysenteriae, S. innocens, B. aalborgi, the remaining canine isolates, and a sample without template DNA (Fig. 1 and Table 1).
FIG. 1.
Ethidium bromide-stained 2% agarose gel of PCR-amplified 361-bp products obtained with primers specific for an S. pilosicoli 16S rRNA gene sequence (27). Lanes: 1 and 12, molecular size standard (1-kb DNA ladder; Gibco-BRL); 2, porcine S. hyodysenteriae B78T; 3, porcine S. innocens B256T; 4, porcine S. pilosicoli P43/6/78T; 5, canine isolate K9-12; 6, canine isolate 16242-94; 7, canine isolate 24072-93A; 8, canine isolate Dog 17; 9, canine isolate 24072-93B; 10, canine isolate 14199-95; 11, no template DNA (negative control).
MEE.
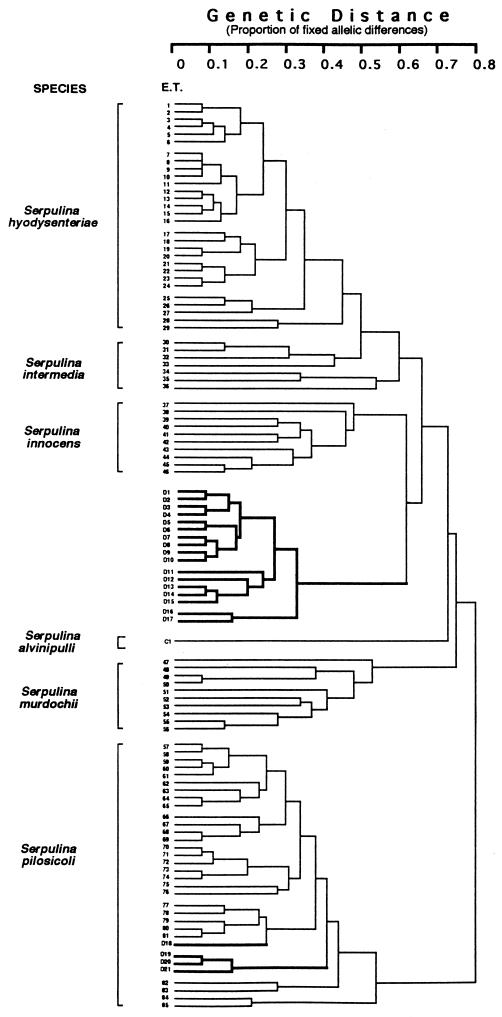
A phenogram of genetic distance expressed as percentage of fixed allelic differences among 85 ETs of porcine intestinal spirochetes and S. alvinipulli indicated that the canine isolates clustered into two distinct groups. Twenty-six of the canine isolates clustered within a new division of the phenogram at ETs D1 to D17 (Fig. 2 and Table 1), which formed a tight cluster of ETs spanning a genetic distance of 0.32. Three ETs each contained three isolates, three contained two isolates, and the other ETs were represented by single isolates (Table 1). Three subclusters could be distinguished at a genetic distance of 0.28. There was no obvious clustering of isolates based on their geographic origins, except that the two isolates from the United States formed a third subcluster (ETs D16 and D17), separated from the Australian isolates at a genetic distance of 0.32. The new cluster was clearly separated from other species groupings, with a genetic distance of 0.6 separating it from its nearest neighboring group, S. innocens (Fig. 2).
FIG. 2.
Phenogram of genetic distances (expressed as percentages of fixed allelic differences) among ETs of 86 intestinal spirochetes from swine, a chicken, and dogs, clustered by the unweighted pair group method of arithmetic averages strategy. Isolates in ETs 1 to 29 belong to S. hyodysenteriae, those in ETs 30 to 36 belong to S. intermedia, those in ETs 37 to 46 belong to S. innocens, those in ETs 47 to 56 belong to S. murdochii, and those in ETs 57 to 85 belong to S. pilosicoli. Canine isolates are indicated by ETs with the prefix D and are outlined in the phenogram in boldface. They are contained in ETs D1 to D17, the proposed new species “S. canis,” and ETs D18 to D21 of S. pilosicoli. The chicken S. alvinipulli C1T is located between ET 46 and ET 47 in a branch separate from that of “S. canis.”
The remaining four canine isolates were located in four ETs, designated D18 to D21, together with the porcine isolates constituting ETs 57 to 85 of S. pilosicoli. The three canine isolates from the United States were closely related, whereas Australian isolate Dog 17 in ET D18 was separated from these by a genetic distance of 0.4.
Ribotyping.
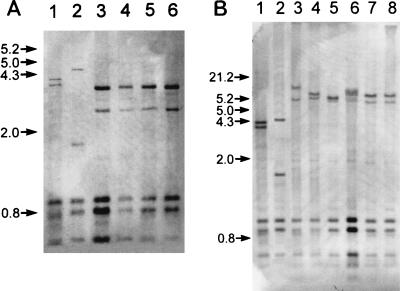
Hybridization of photobiotin-labeled rRNA of S. hyodysenteriae to purified chromosomal DNA of intestinal spirochetes digested with Sau3AI revealed distinct banding patterns that segregated the canine isolates into two groups, with one group having a pattern that was distinct from previously described ribotypes of Serpulina species (14). Canine isolates 24072-93B, 14199-95, D27, and D148 had patterns that were identical to each other and different from that of S. innocens (Fig. 3A). Canine isolates Dog 17, 16242-94, K9-12, and 24072-93A had ribotypes similar to those of porcine S. pilosicoli P43/6/78T and human S. pilosicoli SP16 (Fig. 3B).
FIG. 3.
Hybridization of photobiotin-labeled rRNA of S. hyodysenteriae porcine isolate A1 to purified chromosomal DNA of reference and canine intestinal spirochetes. Purified chromosomal DNA was digested with Sau3AI (Gibco-BRL), electrophoresed in an 0.8% agarose gel, and transferred to a nylon membrane. Panel A lanes: 1, porcine S. hyodysenteriae B78T; 2, porcine S. innocens B256T; 3, canine isolate 24072-93B; 4, canine isolate 14199-95; 5, canine isolate D27; 6, canine isolate D148. Panel B lanes: 1, S. hyodysenteriae B78T; 2, porcine S. innocens 4/71; 3, porcine S. pilosicoli P43/6/78T; 4, human S. pilosicoli SP16; 5, canine isolate Dog 17; 6, canine isolate 16242-94; 7, canine isolate K9-12; 8, canine isolate 24072-93A. Numbers along the left side of each panel are kilobase size markers.
DISCUSSION
Several schemes have been proposed for the identification of intestinal spirochetes. The most widely accepted methods are based on MEE, either alone (22, 23, 25) or in combination with 16S rRNA gene sequencing (32) or RFLP-PCR assay of rDNA encoding the 16S rRNA (34). With these methods, group I is defined as S. hyodysenteriae, whereas groups II, III, V, and VI consist of S. intermedia, S. innocens, S. murdochii, and S. pilosicoli, respectively. The chicken S. alvinipulli C1T forms group IV, and the human B. aalborgi forms group VII. There has been good agreement between Serpulina species identification by MEE and other proposed schemes that are based on either biochemical analysis combined with 16S rRNA sequencing (8) or RFLP of the flaA1 gene (9). Consequently, it was not surprising to find a good correlation between (i) the results of phenotypic characterization and each of the genetically based typing methods and (ii) those of previous characterizations of canine spirochetes by phenotypic methods as well as DNA-DNA reassociation and arbitrarily primed PCR fingerprint analysis (1) and RFLP of the flaA1 gene (9). However, by MEE we found that the 26 canine indole-negative WBHIS isolates that were non-S. pilosicoli by 16S rRNA gene-specific PCR assays formed a new cluster that was clearly separated from all other Serpulina spp., with genetic differences from these species exceeding some genetic distances known to exist between the recognized species. Similarly, these isolates had the same ribotype pattern, which was distinct from those of the other species. These results indicated that this new group may represent a new species, provisionally designated “Serpulina canis.” Further studies, including DNA-DNA reassociation studies, are now required to confirm the new species designation.
The canine S. pilosicoli isolate Dog 17, which was isolated from a diarrheal sample, has an ET that is closely related to that of S. pilosicoli isolated from Aboriginal children with diarrhea living in the same community (23). Dogs can be colonized by S. hyodysenteriae after consuming dysenteric feces from pigs affected with the disease (31) or following oral inoculation with colonic material from experimentally infected pigs (10). Thus, it can be argued that because the dog infected with Dog 17 was living in close contact with affected children, it was a passive carrier that consumed human feces contaminated with S. pilosicoli and that the diarrhea was from an unrelated cause. Although the number of isolates in the present study is small, S. pilosicoli also was found in intestinal specimens obtained from dogs without a history of exposure to contaminated feces and living in midwestern urban communities of the United States. On the basis of this finding, dogs likely serve as a natural reservoir for S. pilosicoli and possibly transmit the spirochete to other animals and human beings. This is supported by previous observations made by Koopman and coworkers (19), indicating that WBHIS isolated from five dogs with diarrhea, nine human immunodeficiency virus-positive men with intestinal disorders, and a woman with gastrointestinal complaints in The Netherlands had identical RFLP of 16S rRNA, flaA, and hemolysin genes. More recently, Trott and coworkers (41) found S. pilosicoli strains isolated from dogs with pulsed field gel electrophoresis patterns similar to those of S. pilosicoli strains isolated from human beings in villages in Papua New Guinea.
Results from the present study extend previous findings and confirm that the large intestines of dogs, like those of humans and other animals including birds, are colonized by more than one species of spirochete. The isolation of S. pilosicoli from intestinal specimens correlated with lesions of IS in the two dogs in which morphologic examination of the colon was done. This finding is consistent with previous information concerning the association of S. pilosicoli with IS in other animals (2–6, 26, 27, 36, 38, 40). Conversely, the absence of attachment of “S. canis” isolate 24072-93B to cecal enterocytes in the chick infection model (26) suggests that spirochetes in this group are nonpathogenic commensals of the dog colon. In fact, most of the “S. canis” isolates were obtained from healthy dogs, further suggesting that they are likely commensals. Passive shedding of these spirochetes during diarrheal disease from other causes may be one explanation for previous conflicting reports about the role of spirochetes in diarrheal disease of dogs.
We concluded that both S. pilosicoli and “S. canis” can be isolated from intestinal specimens from dogs living on two continents, including a community that is far remote from Western civilization. This finding provides a basis for future epidemiological studies aimed at determining the role of intestinal spirochetes either as primary etiologic agents of enteric disease or as concurrent causes of diarrhea and wasting in dogs. It is proposed that initial identification by veterinary diagnostic laboratories which isolate spirochetes from canine intestinal specimens begin with determinations of the intensity and pattern of hemolysis. A possible scheme for characterization of weakly β-hemolytic isolates would involve amplification of an S. pilosicoli 16S rRNA gene-specific sequence by PCR. A positive PCR result would suggest a role for S. pilosicoli in the disease, whereas a negative result may be interpreted to indicate passive shedding of “S. canis” and the need to search for an alternative cause of diarrhea.
In humans, S. pilosicoli has been identified in intestinal specimens obtained from immunocompetent hosts, mostly children with diarrhea in developing countries, and human immunodeficiency virus-positive immunocompromised adults with chronic diarrhea and wasting in developed countries (1, 16, 19, 23, 41). It is hypothesized that IS occurs as a subclinical disease in mature immunocompetent hosts, whereas clinical signs of diarrhea may be indicative of either massive infection, which occurs in a poor hygienic environment, or compromised intestinal function because of concurrent etiological factors. Because S. pilosicoli was identified in a broad host range affected with IS, there is a possibility that this spirochete is zoonotic and has public health significance.
ACKNOWLEDGMENTS
We thank J. M. Kinyon, T. B. Stanton, M. J. Wannemuehler, and R. M. Smibert for providing strains of intestinal spirochetes.
This work was supported by funds provided by the United States Department of Agriculture; Regional Research Project NC-62; Enteric Diseases of Swine and Cattle: Prevention, Control, and Food Safety, and by Murdoch University.
Footnotes
Paper no. 12157 of the Agriculture Research Division, Institute for Agriculture and Natural Resources, University of Nebraska—Lincoln.
REFERENCES
- 1.Duhamel G E, Muniappa N, Mathiesen M R, Johnson J L, Toth J, Elder R O, Doster A R. Certain canine weakly β-hemolytic intestinal spirochetes are phenotypically and genotypically related to spirochetes associated with human and porcine intestinal spirochetosis. J Clin Microbiol. 1995;33:2212–2215. doi: 10.1128/jcm.33.8.2212-2215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duhamel G E, Muniappa N, Gardner I, Anderson M A, Blanchard P C, DeBey B M, Mathiesen M R, Walker R L. Porcine colonic spirochetosis: a diarrheal disease associated with a newly recognized species of intestinal spirochaetes. Pig J. 1995;35:101–110. [Google Scholar]
- 3.Duhamel G E, Hunsaker B D, Matheisen M R, Moxley R A. Intestinal spirochetosis and giardiasis in a beagle pup with diarrhea. Vet Pathol. 1996;33:360–362. doi: 10.1177/030098589603300318. [DOI] [PubMed] [Google Scholar]
- 4.Duhamel G E. Intestinal spirochaetosis in non-production animals. In: Hampson D J, Stanton T B, editors. Intestinal spirochaetosis in domestic animals and humans. Wallingford, England: CAB International; 1997. pp. 301–320. [Google Scholar]
- 5.Duhamel, G. E., R. O. Elder, N. Muniappa, M. R. Mathiesen, V. J. Wong, and R. P. Tarara. 1997. Colonic spirochetal infections of nonhuman primates associated with Brachyspira aalborgi, Serpulina pilosicoli, and unclassified flagellated bacteria. Clin. Infect. Dis. 25(Suppl. 2):186–188. [DOI] [PubMed]
- 6.Duhamel G E. Colonic spirochetosis caused by Serpulina pilosicoli. Large Anim Pract. 1998;19:14–22. [Google Scholar]
- 7.Dwars R M, Davelaar F G, Smit H F. Infection of broiler chicks (Gallus domesticus) with human intestinal spirochetes. Avian Pathol. 1992;21:559–568. doi: 10.1080/03079459208418877. [DOI] [PubMed] [Google Scholar]
- 8.Fellström C, Pettersson B, Thomson J, Gunnarsson A, Persson M, Johansson K. Identification of Serpulina species associated with porcine colitis by biochemical analysis and PCR. J Clin Microbiol. 1997;35:462–467. doi: 10.1128/jcm.35.2.462-467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher L N, Mathiesen M R, Duhamel G E. Restriction fragment length polymorphism of the periplasmic flagellar flaA1 gene of Serpulina species. Clin Diagn Lab Immunol. 1997;4:681–686. doi: 10.1128/cdli.4.6.681-686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glock R D, Kinyon J M, Harris D L. Proceedings of the 5th International Pig Veterinary Society Congress. 1978. Transmission of Treponema hyodysenteriae by canine and avian vectors; p. KB 63. [Google Scholar]
- 11.Hampson D J, Atyeo R F, Combs B G. Swine dysentery. In: Hampson D J, Stanton T B, editors. Intestinal spirochaetosis in domestic animals and humans. Wallingford, England: CAB International; 1997. pp. 175–209. [Google Scholar]
- 12.Harris D L, Kinyon J M. Significance of anaerobic spirochetes in the intestines of animals. Am J Clin Nutr. 1974;27:1297–1304. doi: 10.1093/ajcn/27.11.1297. [DOI] [PubMed] [Google Scholar]
- 13.Hovind-Hougen K, Birch-Andersen A, Henrik-Nielsen R, Orholm M, Pedersen J O, Teglbjærg P S, Thaysen E H. Intestinal spirochetosis: morphological characterization and cultivation of the spirochete Brachyspira aalborgi gen. nov., sp. nov. J Clin Microbiol. 1982;16:1127–1136. doi: 10.1128/jcm.16.6.1127-1136.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen N S, Casey T A, Stanton T B. Characterization of S. hyodysenteriae and related intestinal spirochetes by ribosomal RNA gene restriction patterns. FEMS Microbiol Lett. 1992;93:235–242. doi: 10.1016/0378-1097(92)90468-4. [DOI] [PubMed] [Google Scholar]
- 15.Joens L A, Glock R D, Kinyon J M. Differentiation of Treponema hyodysenteriae from T. innocens by enteropathogenicity testing in the CF1 mouse. Vet Rec. 1980;107:527–529. [PubMed] [Google Scholar]
- 16.Jones M J, Miller J M, George W L. Microbiological and biochemical characterization of spirochetes isolated from the feces of homosexual males. J Clin Microbiol. 1986;24:1071–1074. doi: 10.1128/jcm.24.6.1071-1074.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinyon J M, Harris D L, Glock R D. Enteropathogenicity of various isolates of Treponema hyodysenteriae. Infect Immun. 1977;15:638–646. doi: 10.1128/iai.15.2.638-646.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinyon J M, Harris D L. Treponema innocens, a new species of intestinal bacteria, and emended description of the type strain of Treponema hyodysenteriae Harris et. al. Int J Syst Bacteriol. 1979;29:102–109. [Google Scholar]
- 19.Koopman M B H, Käsbohrer A, Bechman G, van der Zeijst B A M, Kusters J G. Genetic similarities of intestinal spirochetes from humans and various animal species. J Clin Microbiol. 1993;31:711–716. doi: 10.1128/jcm.31.3.711-716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkle R A, Harris D L, Kinyon J M. Autoclaved liquid medium for propagation of Treponema hyodysenteriae. J Clin Microbiol. 1986;24:669–671. doi: 10.1128/jcm.24.4.669-671.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leach W D, Lee A, Stubbs R P. Localization of bacteria in the gastrointestinal tract: a possible explanation of intestinal spirochaetosis. Infect Immun. 1973;7:961–972. doi: 10.1128/iai.7.6.961-972.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J I, Hampson D J, Lymbery A J, Harders S J. The porcine intestinal spirochaetes: identification of new genetic groups. Vet Microbiol. 1993;34:273–285. doi: 10.1016/0378-1135(93)90017-2. [DOI] [PubMed] [Google Scholar]
- 23.Lee J I, Hampson D J. Genetic characterisation of intestinal spirochaetes and their association with disease. J Med Microbiol. 1994;40:365–371. doi: 10.1099/00222615-40-5-365. [DOI] [PubMed] [Google Scholar]
- 24.Lee J I, Hampson D J. The prevalence of intestinal spirochaetes in dogs. Aust Vet J. 1996;74:25–26. doi: 10.1111/j.1751-0813.1996.tb07574.x. [DOI] [PubMed] [Google Scholar]
- 25.Lymbery A J, Hampson D J, Hopkins R M, Combs B, Mhoma J R I. Multilocus enzyme electrophoresis for identification and typing of Treponema hyodysenteriae and related spirochaetes. Vet Microbiol. 1990;22:89–99. doi: 10.1016/0378-1135(90)90127-h. [DOI] [PubMed] [Google Scholar]
- 26.Muniappa N, Duhamel G E, Mathiesen M R, Bargar T W. Light microscopic and ultrastructural changes in the ceca of chicks inoculated with human and canine Serpulina pilosicoli. Vet Pathol. 1996;33:542–550. doi: 10.1177/030098589603300509. [DOI] [PubMed] [Google Scholar]
- 27.Muniappa N, Mathiesen M R, Duhamel G E. Laboratory identification and enteropathogenicity testing of Serpulina pilosicoli associated with porcine colonic spirochetosis. J Vet Diagn Invest. 1997;9:165–171. doi: 10.1177/104063879700900210. [DOI] [PubMed] [Google Scholar]
- 28.Neef N A, Lysons R J, Trott D J, Hampson D J, Jones P W, Morgan J H. Pathogenicity of porcine intestinal spirochetes in gnotobiotic pigs. Infect Immun. 1994;62:2395–2403. doi: 10.1128/iai.62.6.2395-2403.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park N Y, Chung C Y, McLaren A J, Atyeo R F, Hampson D J. Polymerase chain reaction for identification of human and porcine spirochaetes recovered from cases of intestinal spirochaetosis. FEMS Microbiol Lett. 1995;125:225–230. doi: 10.1111/j.1574-6968.1995.tb07362.x. [DOI] [PubMed] [Google Scholar]
- 30.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Songer J G, Glock R D, Schwartz K J, Harris D L. Isolation of Treponema hyodysenteriae from sources other than swine. J Am Vet Med Assoc. 1978;172:464–466. [PubMed] [Google Scholar]
- 32.Stanton T B, Trott D J, Lee J I, McLaren A J, Hampson D J, Paster B J, Jensen N S. Differentiation of intestinal spirochaetes by multilocus enzyme electrophoresis analysis and 16S rRNA sequence comparisons. FEMS Microbiol Lett. 1996;136:181–186. doi: 10.1111/j.1574-6968.1996.tb08046.x. [DOI] [PubMed] [Google Scholar]
- 33.Stanton T B, Fournié-Amazouz E, Postic D, Trott D J, Grimont P A D, Baranton G, Hampson D J, Saint Girons I. Recognition of two new species of intestinal spirochetes: Serpulina intermedia sp. nov. and Serpulina murdochii sp. nov. Int J Syst Bacteriol. 1997;47:1007–1012. doi: 10.1099/00207713-47-4-1007. [DOI] [PubMed] [Google Scholar]
- 34.Stanton, T. B., D. Postic, and N. S. Jensen. Serpulina alvinipulli, sp. nov., a new Serpulina species enteropathogenic for chickens. Submitted for publication. [DOI] [PubMed]
- 35.Surawicz C M. Colorectal spirochetosis. In: Surawicz C M, Owen R, editors. Gastrointestinal and hepatic infections. Philadelphia, Pa: The W. B. Saunders Co.; 1995. pp. 287–293. [Google Scholar]
- 36.Taylor D J, Simmons J R, Laird H M. Production of diarrhoea and dysentery in pigs by feeding pure cultures of a spirochaete differing from Treponema hyodysenteriae. Vet Rec. 1980;106:326–332. doi: 10.1136/vr.106.15.326. [DOI] [PubMed] [Google Scholar]
- 37.Thomson J R, Smith W J, Murray B P, McOrist S. Pathogenicity of three strains of Serpulina pilosicoli in pigs with a naturally acquired intestinal flora. Infect Immun. 1997;65:3693–3700. doi: 10.1128/iai.65.9.3693-3700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trott D J, McLaren A J, Hampson D J. Pathogenicity of human and porcine intestinal spirochetes in one-day-old specific-pathogen-free chicks: an animal model of intestinal spirochetosis. Infect Immun. 1995;63:3705–3710. doi: 10.1128/iai.63.9.3705-3710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trott D J, Stanton T B, Jensen N S, Duhamel G E, Johnson J L, Hampson D J. Serpulina pilosicoli sp. nov, the agent of porcine intestinal spirochetosis. Int J Syst Bacteriol. 1996;46:206–215. doi: 10.1099/00207713-46-1-206. [DOI] [PubMed] [Google Scholar]
- 40.Trott D J, Huxtable C R, Hampson D J. Experimental infection of newly weaned pigs with human and porcine strains of Serpulina pilosicoli. Infect Immun. 1996;64:4648–4654. doi: 10.1128/iai.64.11.4648-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trott, D. J., A. S. J. Mikosza, B. G. Combs, S. L. Oxberry, and D. J. Hampson. Population genetic analysis of Serpulina pilosicoli and its molecular epidemiology in villages in the eastern highlands of Papua New Guinea. Submitted for publication. [DOI] [PubMed]
- 42.Turek J J, Meyer R C. Studies on canine intestinal spirochete. I. Its isolation, cultivation, and ultrastructure. Can J Comp Med. 1977;41:332–337. [PMC free article] [PubMed] [Google Scholar]
- 43.Turek J J, Meyer R C. Studies on a canine intestinal spirochete: scanning electron microscopy of canine colonic mucosa. Infect Immun. 1978;20:853–855. doi: 10.1128/iai.20.3.853-855.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zymet C L. Canine spirochetosis and its association with diarrhea. Vet Med Small Anim Clin. 1969;64:885–887. [PubMed] [Google Scholar]