Abstract
Background
Drought is most likely the most significant abiotic stress affecting wheat yield. The discovery of drought-tolerant genotypes is a promising strategy for dealing with the world’s rapidly diminishing water resources and growing population. A genome-wide association study (GWAS) was conducted on 298 Iranian bread wheat landraces and cultivars to investigate the genetic basis of yield, yield components, and drought tolerance indices in two cropping seasons (2018–2019 and 2019–2020) under rainfed and well-watered environments.
Results
A heatmap display of hierarchical clustering divided cultivars and landraces into four categories, with high-yielding and drought-tolerant genotypes clustering in the same group. The results of the principal component analysis (PCA) demonstrated that selecting genotypes based on the mean productivity (MP), geometric mean productivity (GMP), harmonic mean (HM), and stress tolerance index (STI) can help achieve high-yield genotypes in the environment. Genome B had the highest number of significant marker pairs in linkage disequilibrium (LD) for both landraces (427,017) and cultivars (370,359). Similar to cultivars, marker pairs on chromosome 4A represented the strongest LD (r2 = 0.32). However, the genomes D, A, and B have the highest LD, respectively. The single-locus mixed linear model (MLM) and multi-locus random-SNP-effect mixed linear model (mrMLM) identified 1711 and 1254 significant marker-trait association (MTAs) (-log10 P > 3) for all traits, respectively. A total of 874 common quantitative trait nucleotides (QTNs) were simultaneously discovered by both MLM and mrMLM methods. Gene ontology revealed that 11, 18, 6, and 11 MTAs were found in protein-coding regions (PCRs) for spike weight (SW), thousand kernel weight (TKW), grain number per spike (GN), and grain yield (GY), respectively.
Conclusion
The results identified rich regions of quantitative trait loci (QTL) on Ch. 4A and 5A suggest that these chromosomes are important for drought tolerance and could be used in wheat breeding programs. Furthermore, the findings indicated that landraces studied in Iranian bread wheat germplasm possess valuable alleles, that are responsive to water-limited conditions. This GWAS experiment is one of the few types of research conducted on drought tolerance that can be exploited in the genome-mediated development of novel varieties of wheat.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-023-04416-3.
Keywords: Drought tolerance indices, Genome-wide association study, MTAs, SNP markers, Wheat accessions
Background
Providing 25% of the total proteins and calories in the human diet and food supplies worldwide relies on wheat (Triticum aestivum L.) [1–3]. Wheat grain annual production and consumption reach 750 and 735 million tons, respectively [4]. Global climate change adversely affects wheat yield, raising concerns regarding food security in the future. The genetic dissection of agronomic traits that affects yield and stress tolerance is essential to improve wheat yield [5–7]. The Food and Agriculture Organization estimated that of the United Nations 13 million tons of wheat were produced in Iran in 2022, which is over 28% more compared to 10.1 million tons in 2021. The quarterly global Crop Prospects and Food Situation report’s forecast for 2023 production remains the same at 13 million tons. Drought stresses result in approximately over 50% loss in agricultural productivity [8]. Drought is the main concern for crop productivity in most wheat cultivation areas in Iran. In Iran, approximately 6 million hectares of arable lands have been assigned to wheat cultivation in 2020 [8]. Iran is located in a semi-dry region where end-season drought stress severely affects wheat growth [8].
Drought events adversely affect crop productivity by disrupting a wide range of biochemical/physiological functions on a global scale. Water deficit stress is the most severe environmental stress, limiting plant growth and development worldwide [2, 9]. Drought tolerance refers to the capability of plants to grow, develop, and produce a harvestable yield in the absence of water. [10, 11]. Screen and use of the available genetic resources of crop plants can help to alleviate the impact of climate change on agricultural productivity in arid and semi-arid regions [2, 12, 13]. Understanding how plants adapt to drought stress is crucial for developing new and improved methods to increase drought-tolerant plants [14].
Drought tolerance indices (DTIs) have been widely used for identifying compatible genotypes, by evaluating their performance under non-stressed and stressed environments [15, 16]. In the past, the tolerance index (TOL), geometric mean productivity index (GMP), stress tolerance index (STI), and stress susceptibility index (SSI) have all been employed for genotype selection [15]. The advancement of genetics and genomics has made it possible to decipher the genetic elements that control agronomic traits, as well as to determine their chromosomal locations and thus to identify QTLs in crops, including wheat [13, 17].
Wheat breeding programs need to use innovative technologies to explore the genetic basis of complicated quantitative traits [18]. The genome-wide association mapping (GWAS) technique is an efficient approach to dissect the genetic basis of complex traits by genotyping a large number of accessions with multiple single nucleotide polymorphisms (SNPs) and testing the association between SNPs and agronomic traits [19]. GWAS establishes a relationship between genotype and phenotype based on an assumption that linkage disequilibrium (LD) has developed within a population over several generations [20, 21]. The use of association mapping has been successful in evaluating several agronomic characteristics in plants/crops, including alfalfa [22], sorghum [23], maize [24], soybean [25], wheat [8], and rice [26]. A meta-analysis was conducted to identify the most stable QTLs for grain yield (GY) and grain quality traits in wheat [27]. The results revealed that 449 QTLs were successfully projected onto the genetic consensus map which condensed to 100 Meta-QTL (MQTLs) distributed on wheat chromosomes. The QTLs of thousand kernel weight (TKW) were frequently associated with QTLs for GY and grain protein content with co-localization occurring at 55% and 63%, respectively [27]. Meta-QTL analysis for drought tolerance was undertaken in bread wheat to identify consensus and robust MQTLs using 340 known QTLs from 11 earlier studies; accordingly, 13 MQTLs located on 6 chromosomes (1D, 3B, 5A, 6D, 7A, and 7D) were identified, with a maximum of 4 MQTLs on chromosome 5A. The in-silico expression analysis of these 228 cyclic glucans (CGs) allowed the identification of 14 important CGs, with +3 to −8 fold change in expression under drought (relative to normal conditions) in a tolerant cv. named TAM107 [28].
Our study used a collection of agronomic traits and drought tolerance indices between Iranian wheat varieties and landraces to identify significant SNP loci associated with drought-tolerance traits through a GWAS, using the MLM and mrMLM. The study further aimed to explore candidate genes for drought-resistance traits in bread wheat and to better understand the molecular mechanisms of the drought adaptation of bread wheat in order to facilitate the cultivation of drought-tolerant varieties.
Results
Phenotypic data summary
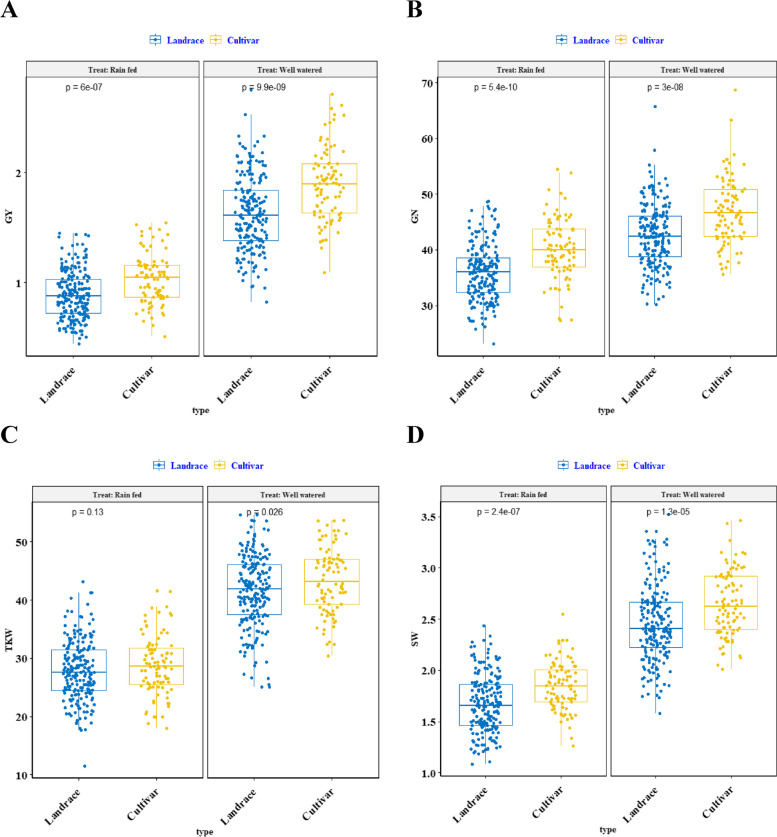
The box plots of wheat landraces and cultivars are displayed in Fig. 1. Compared to a normal situation, the mean of all traits decreased under stress in both cultivars and landraces. Based on the data distribution, there was considerable diversity in wheat accessions regarding agronomical properties. The variance was stronger in native populations. In both moisture conditions, the mean of all traits was lower in landraces than in cultivars.
Fig. 1.
Box-plot representation of the distribution for grain yield (A), grains number per spike (B), thousand kernel weight (C) and spike weight (D) for Iranian landraces and cultivars in the well-watered and rain-fed environments
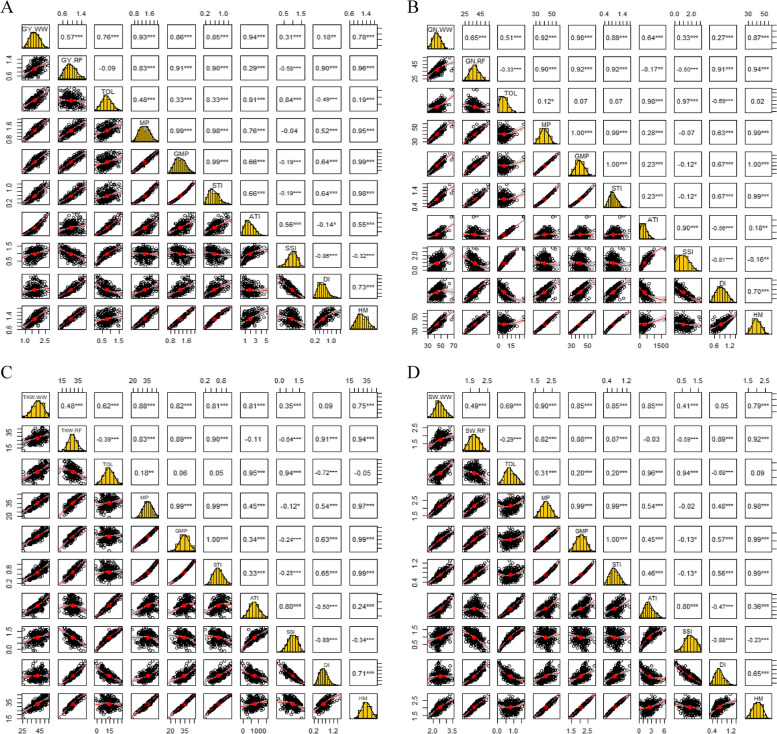
The traits GY, GN, TKW, and SW in the normal environment exhibited the highest significant, positive association with the drought tolerance index MP. However, in the rain-fed environment, the mentioned traits had the strongest significant, positive connection with the drought tolerance index HM (Fig. 2).
Fig. 2.
Correlation coefficients between GYWW, GYRF and various drought tolerance indices (A), GNWW, GNRF and various drought tolerance indices (B), TKWWW, TKWRF and various drought tolerance indices (C) and SWWW, SWRF and various drought tolerance indices (D) for Iranian landraces and cultivars wheat in the well-watered and rain-fed environments. Abbreviations: GY, grain yield; GN, grains number per spike; TKW, thousand kernel weight; SW, spike weight; WW, well-watered; RF, rain-fed; TOL. tolerance index; MP, mean product; GMP, geometric mean product; STI, stress tolerance index; ATI, abiotic stress tolerance index; SSI, stress susceptibility index; DI, new drought resistance index; HM, harmonic mean
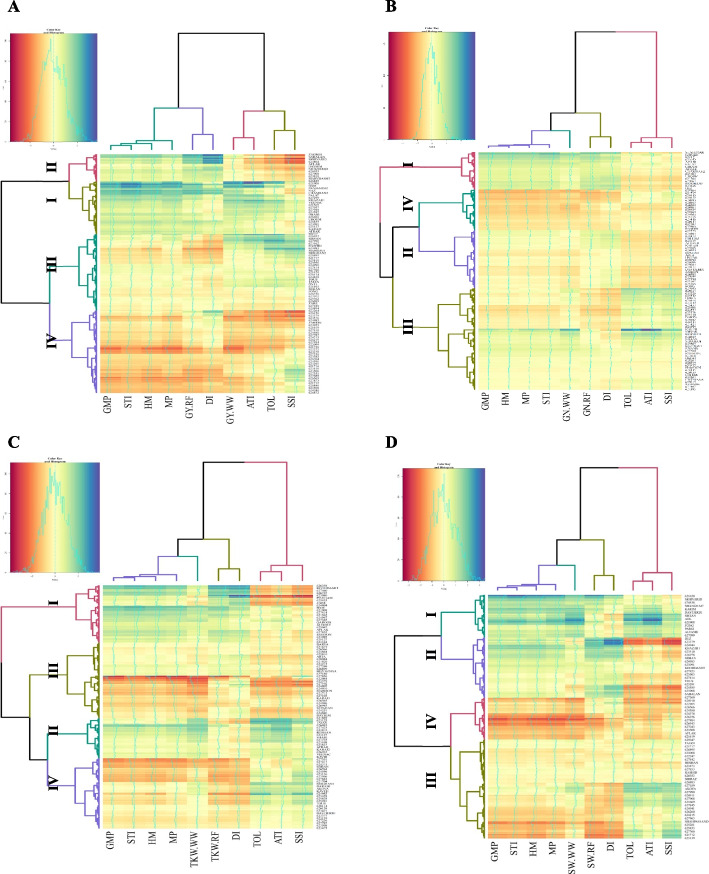
Clustering analysis
The heatmap was created using the average of agronomic features as well as a variety of drought tolerance indices (DTIs). Our purpose in grouping genotypes was to identify genotypes with common characteristics in terms of thousand kernel weight, grains number per spike, spike weight, and grain yield. Heatmap and clustering wheat accessions classified genotypes into four groups based on GYWW, GYRF, and DTIs (Fig. 3a). Group I contains 66 high-yielding genotypes (32 landraces and 34 cultivars). Further, Group II contains 34 genotypes with average-to-high yields (22 landraces and 12 cultivars), and Group III contains 179 genotypes with average-to-low yields (136 landraces and 43 cultivars). Finally, Group IV contains 19 low-yielding (18 landraces and 1 cultivar). Wheat genotypes were split into four categories using the GNWW, GNRF, and DTIs. Based on the results, 48 (27 cultivars and 21 landraces), 73 (20 cultivars and 53 landraces), 127 (38 cultivars and 89 landraces), and 50 (5 cultivars and 45 landraces), genotypes were found in the first, second, third, and fourth groups, respectively (Fig. 3b). Clustering and heatmap based on TKWWW, TKWRF, and DTIs were used to separate wheat accessions into four groups. The first and second groups consisted of 70 genotypes with a high TKW (22 cultivars and 48 landraces), and 49 genotypes with a medium-to-TKW (21 cultivars and 28 landraces), respectively. Moreover, the third and fourth groups contained 93 genotypes with a medium-to-low TKW (68 landraces and 25 cultivars), and 86 genotypes (22 cultivars and 64 landraces) with a low TKW (Fig. 3c), respectively. Wheat genotypes based on the SWWW, SWRF, and DTIs were classified into four groups; the I, II, III, and IV groups consisted of 53 (28 landraces and 25 cultivars), 74 (43 landraces and 31 cultivars), 121 (92 landraces and 29 cultivars), and 50 genotypes (45 landraces and 5 cultivars), respectively (Fig. 3d).
Fig. 3.
Hierarchical clustering and heatmap based on GYWW, GYRF and various drought tolerance indices (A), GNWW, GNRF and various drought tolerance indices (B), TKWWW, TKWRF and various drought tolerance indices (C) and SWWW, SWRF and various drought tolerance indices (D) for Iranian landraces and cultivars wheat in well-watered and rain-fed environments. Abbreviations: GY, grain yield; GN, grains number per spike; TKW, thousand kernel weight; SW, spike weight; WW, well-watered; RF, rain-fed; TOL. tolerance index; MP, mean product; GMP, geometric mean product; STI, stress tolerance index; ATI, abiotic stress tolerance index; SSI, stress susceptibility index; DI, new drought resistance index; HM, harmonic mean
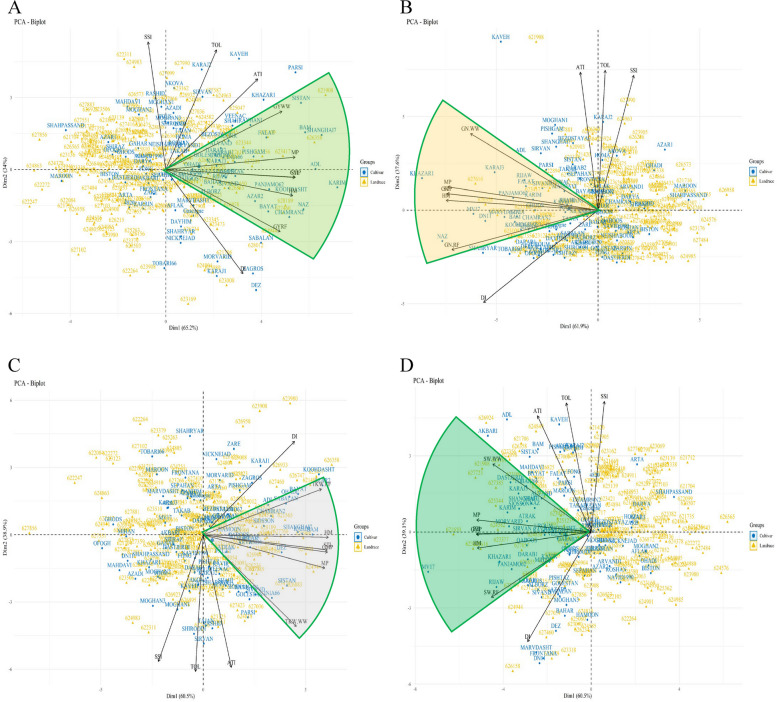
Principal component analysis (PCA) for drought tolerance indices
For the selection of superior genotypes based on the environment, the responses of all accessions were evaluated with a PCA. The results of PCA for DTIs (based on GY) showed that 65.2% and 34% of the variance were explained by the first and second components, respectively. The attributes GYWW, GYRF, MP, GMP, STI, and HM had direct associations with PC1. The PC2 had a positive correlation with the SSI, TOL, and ATI indices, whereas the DI index had a negative correlation with the PC2. Genotypes-based PCA revealed that several genotypes (623417, 627299, 628189, 627460, 626855, 622356, 623344, 623109, and 624944, BAM, ADL KARIM, AZARE2, CHAMRAN2, PISHGAM, and FALAT cultivars) had the highest GY in both rain-fed and well-watered environments (Fig. 4a). Nonetheless, selecting genotypes based on the MP, GMP, HM, and STI indices can help achieve high yield genotypes in the environment. The above explanations are also applicable to the other attributes of GN, TKW, and SW (Fig. 4b, c, d).
Fig. 4.
Principal component analysis of Iranian wheat germplasm exposed to well-watered irrigation and rain-fed environments using PC1 and 2. Biplot for GYWW, GYRF and various drought tolerance indices (A), Biplot for GNWW, GNRF, and various drought tolerance indices (B), Biplot for TKWWW, TKWRF, and various drought tolerance indices (C), and Biplot for SWWW, SWRF and various drought tolerance indices (D). Abbreviations: GY, grain yield; GN, grains number per spike; TKW, thousand kernel weight; SW, spike weight; WW, well-watered; RF, rain-fed; TOL. tolerance index; MP, mean product; GMP, geometric mean product; STI, stress tolerance index; ATI, abiotic stress tolerance index; SSI, stress susceptibility index; DI, new drought resistance index; HM, harmonic mean
Analysis of linked single-nucleotide polymorphisms (SNPs)
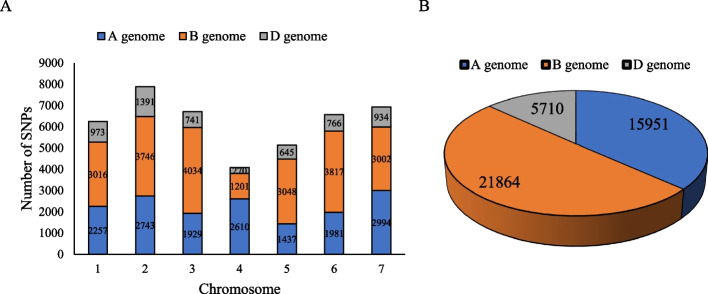
Among the 566,439,207 reads identified in eight Ion Proton runs, 458,363,607 (about 81%) were high-quality barcoded ones. Eventually, 133,039 unique SNPs were identified after removing duplicate reads. A total of 43,525 SNPs were detected across all 21 wheat chromosomes after imputation and discarding those with > 20% missing values, > 10% heterozygosity, and < 5% minor allele frequency. In addition, a set of 43,525 imputed SNPs was obtained using the W7984 reference genome, and these SNPs were used to estimate genetic diversity. Overall 15,951, 21,864, and 5,710 SNPs were mapped to the A, B, and D genomes, respectively, accounting 36.7%, 50.2%, and 13.1% of the total SNPs, respectively (Fig. 5). The highest and lowest numbers of SNPs were located on 3A (4034 SNPs) and 4D (270 SNPs), respectively.
Fig. 5.
Number of imputed SNPs used in different chromosomes of the wheat genomes (A), number of imputed SNPs used in wheat genomes (B)
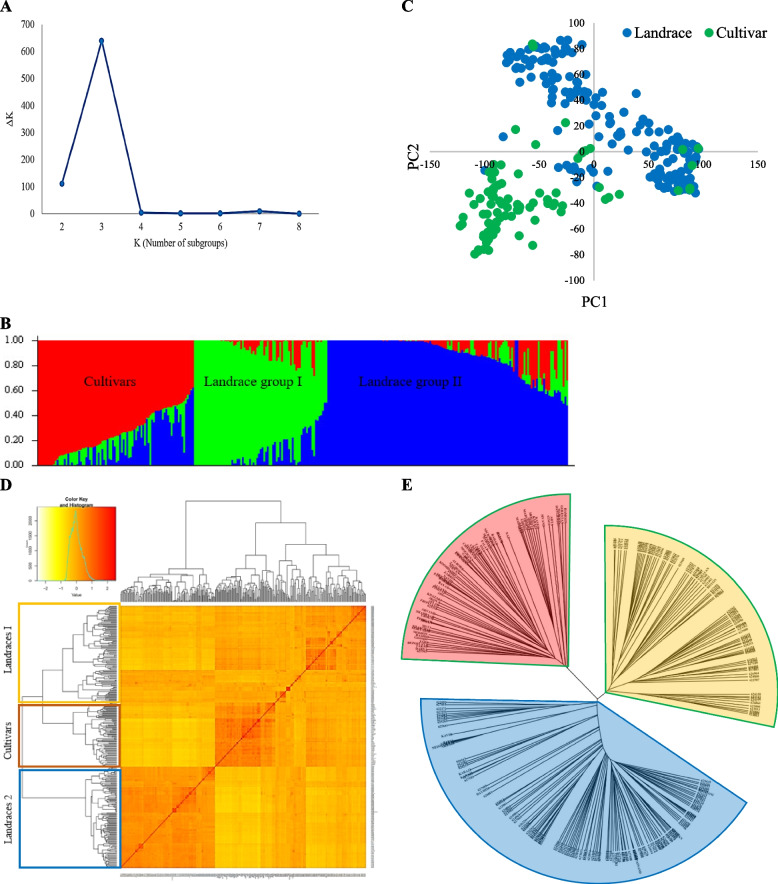
Population structure and kinship matrix
The number of clusters (K) and subpopulations (ΔK) was plotted against each other to determine the appropriate number of subpopulations. Three subpopulations were observed as the strongest ΔK value at K = 3. Three subpopulations of 298 accessions were created using the structure software, S-I, S-II, and S-III (Fig. 6a). S_I contained 113 genotypes (107 landraces and 6 cultivars). Furthermore, S_II encompasses 111 genotypes (97 landraces and 14 cultivars), and S_III consists of 74 genotypes (4 landraces and 70 cultivars) (Fig. 6b). According to a PCA based on molecular markers, the first and second components explain 16.9% and 6.3% of the total genetic variance between wheat accessions. This study could distinguish landraces from cultivars favorably. In the Iranian wheat landraces, a population structure was found with 30.5% genetic diversity, accounting for the first five eigenvalues. The selection effects in breeding programs are considered the reasons for such a genetic separation (Fig. 6c). Heatmap analysis using the kinship matrix for Iranian genotypes is illustrated in Fig. 6d. Clustering results identified two subgroups of native populations. Using imputed markers, we could also separate cultivars and landraces by utilizing the nearest neighbor clustering (Fig. 6e).
Fig. 6.
Determination of subpopulations number in wheat genotypes based on ΔK values (A), A structure plot of the 298 wheat genotypes and landraces determined by K = 3 (B). Principle component analysis (PCA) for a total of 298 Iranian bread wheat accessions (C). Cluster analysis using Kinship matrix of imputed data for Iranian wheat accessions (D). The dendrogram of Neighbor-Joining clustering constructed using 43,525 SNPs and 298 Iranian wheat accessions (E)
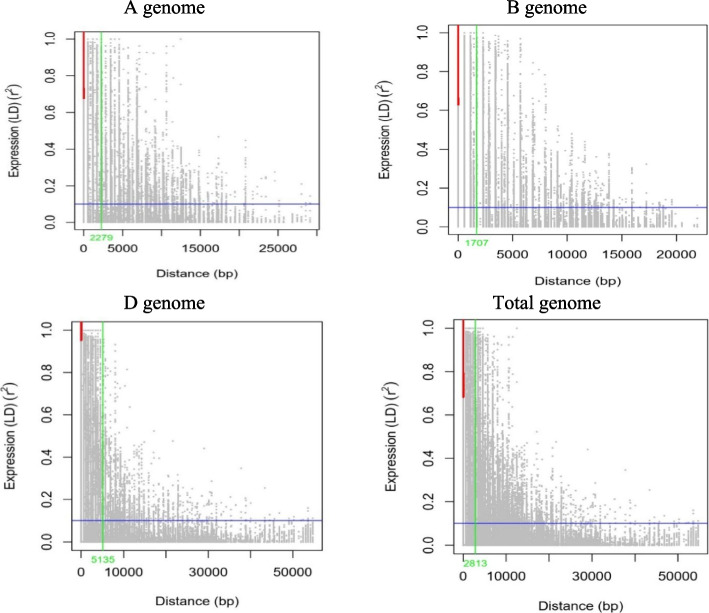
Linkage disequilibrium (LD)
The LD decreased with increased distances between SNPs and varied between and within chromosomes. There were 1,858,425 markers with r2 = 0.211 with varieties, out of which 700,991 (37.72%) had significant linkages at P < 0.001. The majority of significant marker pairs were located at a distance of < 10 cM, based on our observations. An analysis of landraces found 1,867,575 markers with r2 = 0.182, of which 847,725 (45.39%) displayed significant linkages at P < 0.001. Chromosome 4A marker pairs also demonstrated the strongest LD (r2 = 0.368). Most marker pairs with statistical significance were located at distances of < 10 cM. Genomes B and D had the most and least marker pairs (575,681 and 113,374), respectively (Fig. 7, Supplementary 1 Table 1).
Fig. 7.
Overview of the linkage disequilibrium (LD) within the whole association panel per genome using imputed SNPs
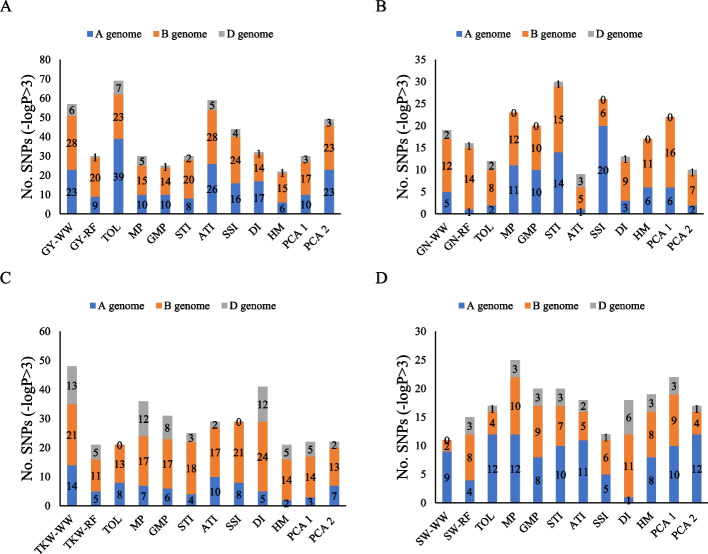
Single-nucleotide polymorphisms for agronomic traits and various drought tolerance indices
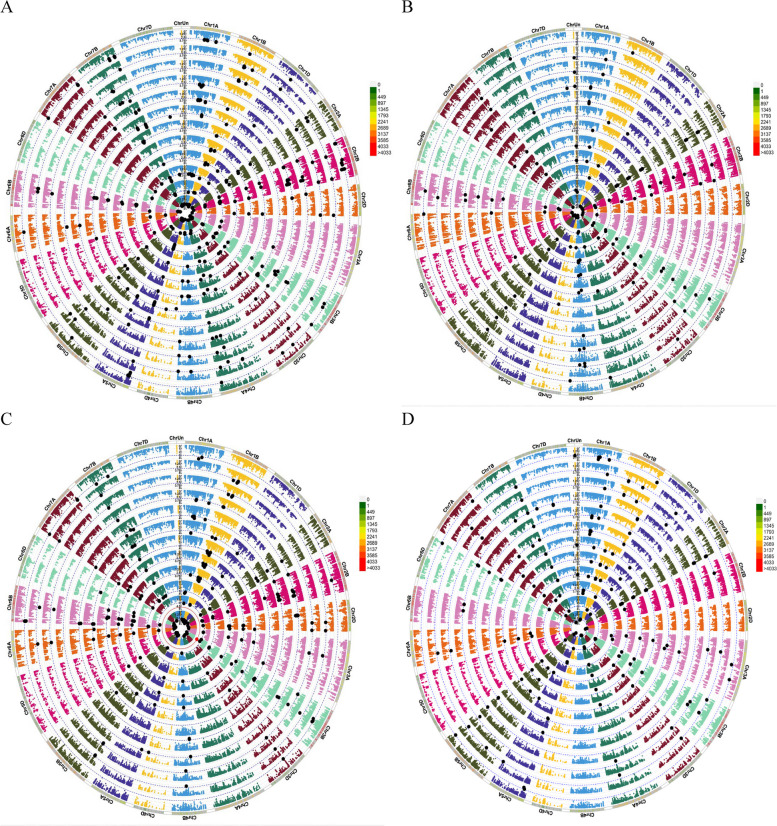
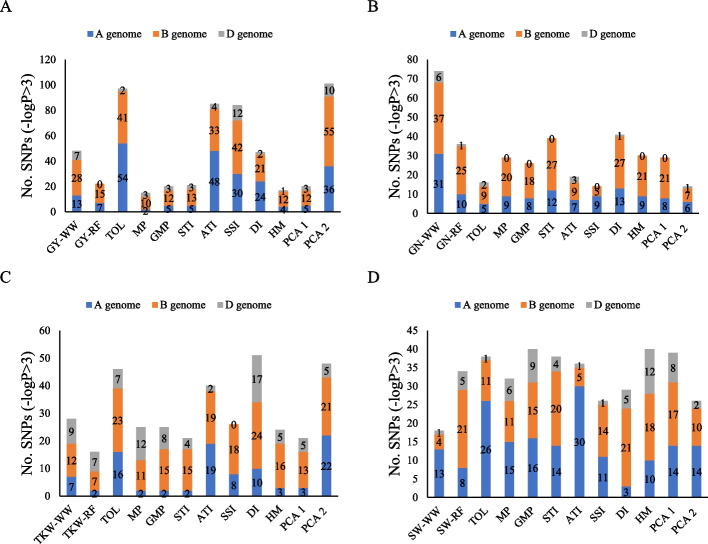
In total, 477 significant marker pairs (MTAs) were identified for GY and related stress tolerance indices [–log10 P > 3] by using MLM approaches. Of the total number of MTAs in the MLM method, 197, 241, and 39 MTAs were assigned to genomes A, B, and D, respectively. Using the MLM method, the number of significant MTAs for GYWW, GYRF, TOL, MP, GMP, STI, ATI, SSI, DI, HM, PCA1, and PCA2 traits were 57, 30, 69, 30, 25, 30, 59, 44, 32, 22, 30, and 49, respectively (Fig. 8a). The number of significant SNPs based on GN, TKW, SW, and stress tolerance indices associated with them were 217, 346, and 214, respectively. Drought tolerance indices for GN, TKW, and SW traits led to the discovery of 81, 79, and 102 significant SNPs for genome A, as well as 125, 200, and 83 significant SNPs for genome B. Additionally, 11, 67, and 29 significant SNPs were found for genome D, respectively. GN, TKW, SW, and stress tolerance indices associated with them led to the discovery of 81, 79, and 102 significant SNPs for genome A. In addition, for genomes B and D, 125, 200, and 83 as well as 11, 67, and 29 significant SNPs were found, respectively (Fig. 8b,c,d). SNPs with P values < 10–3 and < 10–5 (black and red) are highlighted in the Manhattan circle plot (Fig. 9).
Fig. 8.
GWAS results (MLM method) for GYWW, GYRF, various drought tolerance indices, PCA1 and PCA2 (A), GNWW, GNRF, various drought tolerance indices, PCA1 and PCA2 (B), TKWWW, TKWRF, various drought tolerance indices, PCA1 and PCA2 (C) and SWWW, SWRF, various drought tolerance indices, PCA1 and PCA2 (D) of Iranian landraces and cultivars wheat in well-watered and rain-fed environments. Abbreviations: GY, grain yield; GN, grains number per spike; TKW, thousand kernel weight; SW, spike weight; WW, well-watered; RF, rain-fed; TOL, tolerance index; MP, mean product; GMP, geometric mean product; STI, stress tolerance index; ATI, abiotic stress tolerance index; SSI, stress susceptibility index; DI, new drought resistance index; HM, harmonic mean; PCA, principal component analysis
Fig. 9.
Circular Manhattan plots (MLM method) to draw common regions associated with A = GYWW, GYRF, various drought tolerance indices, PCA1 and PCA2, B = GNWW, GNRF, various drought tolerance indices, PCA1 and PCA2, C = TKWWW, TKWRF, various drought tolerance indices, PCA1 and PCA2 and D = SWWW, SWRF, various drought tolerance indices, PCA1 and PCA2. Inner to outer circles represent average trait and breeding Values including YWW, YRF, TOL, MP, GMP, STI, ATI, SSI, DI, HM, PCA1 and PCA2, respectively. The chromosomes are plotted at the outmost circle where thin dotted blue and red lines indicate significant level at P value < 0.001 (− log10 (p) > 3) and < 0.00001 (− log10 (p) > 5), respectively. Green and red dots indicate genome-wide significantly associated SNPs at P value < 0.001 and < 0.00001 probability level, respectively. Scale between ChrUn and Chr1A indicates − log10 (p) values. Colored boxes outside on the top right side indicate SNP density across the genome where green to red indicates less dense to dense. Abbreviations: GY, grain yield; GN, grains number per spike; TKW, thousand kernel weight; SW, spike weight; WW, well-watered; RF, rain-fed; TOL, tolerance index; MP, mean product; GMP, geometric mean product; STI, stress tolerance index; ATI, abiotic stress tolerance index; SSI, stress susceptibility index; DI, new drought resistance index; HM, harmonic mean; PCA, principal component analysis
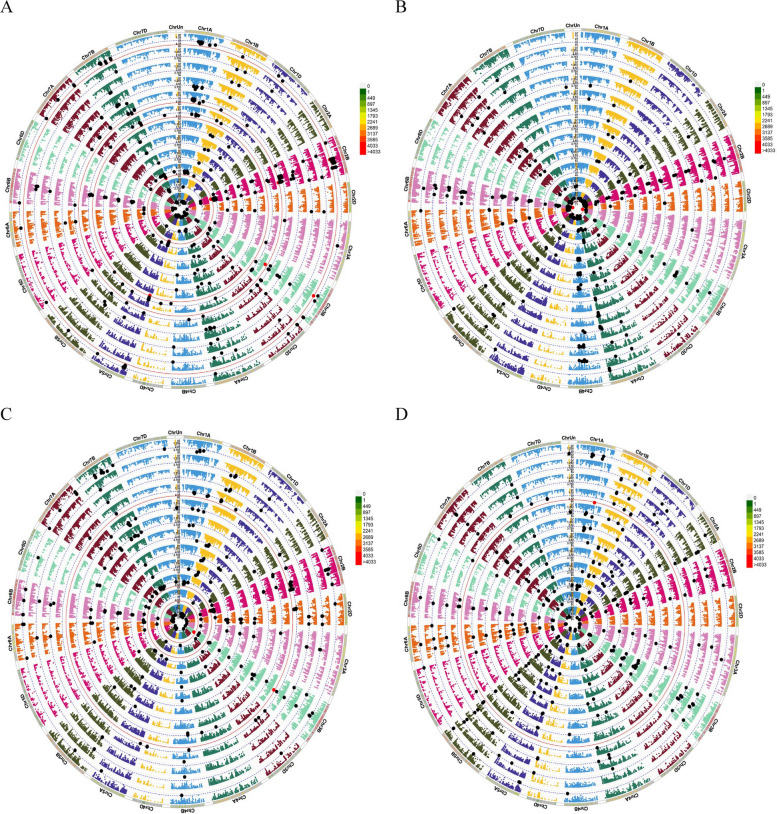
The mrMLM method discovered 233, 294, and 50 significant SNPs for GYWW, GYRF, DTIs, PCA1, and PCA2 in genomes A, B, and D, respectively. Further, GYWW, GYRF, TOL, MP, GMP, STI, ATI, SSI, DI, HM, PCA1, and PCA2 had 48, 22, 97, 15, 20, 21, 85, 84, 47, 17, 20, and 101 significant markers, respectively. The number of important SNPs based on GN, TKW, SW, and associated stress tolerance indices was 367, 371, and 396, respectively (Fig. 10). SNPs with P values < 10–3 and < 10–5 (black and red) are highlighted in the Manhattan circle plot (Fig. 11).
Fig. 10.
GWAS results (mrMLM method) for GYWW, GYRF, various drought tolerance indices, PCA1 and PCA2 (A), GNWW, GNRF, various drought tolerance indices, PCA1 and PCA2 (B), TKWWW, TKWRF, various drought tolerance indices, PCA1 and PCA2 (C) and SWWW, SWRF, various drought tolerance indices, PCA1 and PCA2 (D) of Iranian landraces and cultivars wheat in well-watered and rain-fed environments. Abbreviations: GY, grain yield; GN, grains number per spike; TKW, thousand kernel weight; SW, spike weight; WW, well-watered; RF, rain-fed; TOL, tolerance index; MP, mean product; GMP, geometric mean product; STI, stress tolerance index; ATI, abiotic stress tolerance index; SSI, stress susceptibility index; DI, new drought resistance index; HM, harmonic mean; PCA, principal component analysis
Fig. 11.
Circular Manhattan plots (mrMLM method) to draw common regions associated with A = GYWW, GYRF, various drought tolerance indices, PCA1 and PCA2, B = GNWW, GNRF, various drought tolerance indices, PCA1 and PCA2, C = TKWWW, TKWRF, various drought tolerance indices, PCA1 and PCA2 and D = SWWW, SWRF, various drought tolerance indices, PCA1 and PCA2. Inner to outer circles represent average trait and breeding Values including YWW, YRF, TOL, MP, GMP, STI, ATI, SSI, DI, HM, PCA1 and PCA2, respectively. The chromosomes are plotted at the outmost circle where thin dotted blue and red lines indicate significant level at P value < 0.001 (− log10 (p) > 3) and < 0.00001 (− log10 (p) > 5), respectively. Green and red dots indicate genome-wide significantly associated SNPs at P value < 0.001 and < 0.00001 probability level, respectively. Scale between ChrUn and Chr1A indicates − log10 (p) values. Colored boxes outside on the top right side indicate SNP density across the genome where green to red indicates less dense to dense. Abbreviations: GY, grain yield; GN, grains number per spike; TKW, thousand kernel weight; SW, spike weight; WW, well-watered; RF, rain-fed; TOL, tolerance index; MP, mean product; GMP, geometric mean product; STI, stress tolerance index; ATI, abiotic stress tolerance index; SSI, stress susceptibility index; DI, new drought resistance index; HM, harmonic mean; PCA, principal component analysis
Using 43,525 SNPs and a significant value of –log10 (P > 5), MLM and mrMLM models identified a total of 67 and 24 MTAs for yield attributes and DTIs, respectively. There were a total of 26, 36, and 5 MTAs based on MLM, as well as 4, 20, and zero MTAs based on mrMLM, respectively, for genomes A, B, and D (Table 1).
Table 1.
A summary of marker-trait associations for yield traits of Iranian wheat accessions
| Genome | GY | GN | TKW | SW | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mrMLM | MLM | mrMLM | MLM | mrMLM | MLM | mrMLM | MLM | |||||||||
| Significant | –log10 P > 3 | –log10 P > 5 | –log10 P > 3 | –log10 P > 5 | –log10 P > 3 | –log10 P > 5 | –log10 P > 3 | –log10 P > 5 | –log10 P > 3 | –log10 P > 5 | –log10 P > 3 | –log10 P > 5 | –log10 P > 3 | –log10 P > 5 | –log10 P > 3 | –log10 P > 5 |
| MTA | 577 | 11 | 477 | 30 | 367 | 8 | 217 | 2 | 371 | 3 | 346 | 16 | 396 | 2 | 214 | 19 |
| Genome A | 233 | 2 | 197 | 13 | 127 | 1 | 81 | 0 | 96 | 0 | 79 | 0 | 174 | 1 | 102 | 13 |
| Genome B | 294 | 9 | 241 | 17 | 226 | 7 | 125 | 2 | 194 | 3 | 200 | 11 | 167 | 1 | 83 | 6 |
| Genome D | 50 | 0 | 39 | 0 | 14 | 0 | 11 | 0 | 81 | 0 | 67 | 5 | 55 | 0 | 29 | 0 |
Abbreviations: GY Grain yield, GN Grains number per spike, TKW Thousand kernel weight, SW Spike weight
Gene ontology
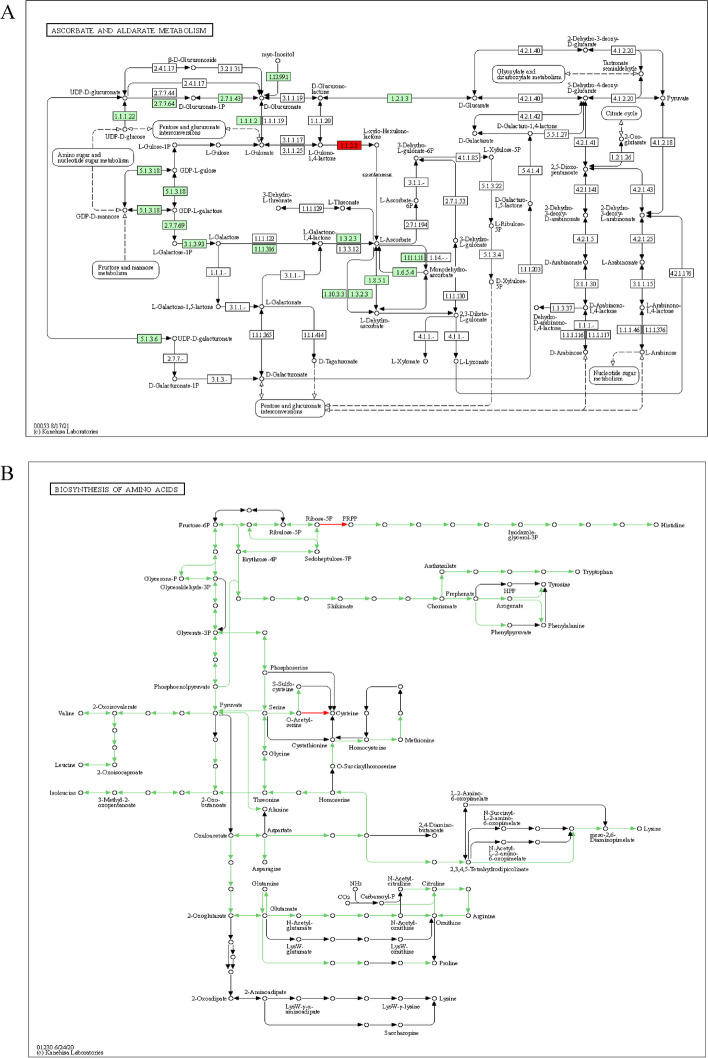
The markers with the highest significance (P value < 0.0001) and pleiotropic impact were investigated thoroughly. Based on GO for DTIs for GN, TKW, and SW traits, 11, 6, 18, and 11 markers containing overlapping genes were identified, which are contained in important molecular and biological processes. Several biological and molecular processes were attributed to some of the discovered MTAs, including defense response, glycolytic process, lipid biosynthetic process, lipid metabolic process, fatty acid biosynthetic process, and response to wounding. The other processes were carbohydrate metabolic process, protein binding, ATP binding, nucleic acid binding, DNA binding, zinc ion binding, oxidoreductase activity, sulfotransferase activity, lipid binding, RNA binding and DNA binding (Table 2). Different pathways were found through rice reference genomes, including ascorbate and aldarate metabolism (Fig. 12a), biosynthesis of amino acids (Fig. 12b), oxidative phosphorylation (Supplementary 1 Fig. 1), fatty acid elongation (Supplementary 1 Fig. 2), and metabolic pathways (Supplementary 1 Fig. 3).
Table 2.
Description of expected MTAs using imputed SNPs for tolerance indices of Iranian wheat accessions
| No | Trait | SNP | Sequence | Trait- Index | Chromosome | Position (bp) | Gene ID | ological process | Cellular component | Molecular process |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | GN | rs18828 | TGCAGCATAACCTCAAGGGTGTTGGACACTTAATTCACCTAAGGTACCTTGGTCTATCAGGTAC | ATI | 1D | 12,505 | TraesCS1A02G026400 | defense response | - | ADP binding |
| 2 | rs65348 | TGCAGTTTTCCGATCGGATATGTCAGCGGCGTCGAGGACCATGCATGGATCGTTTAAAGGTGAT | SSI, DI | 1A | 44,512 | TraesCS1A02G148300 | - | - | protein binding | |
| 3 | rs5402 | TGCAGAGACTCCTTAGTCTTATTCAGCACATTTGGAGCCTTGGAAATAGCAATACCCACCTCAG | TOL, ATI, PCA2 | 1B | 26,163 | TraesCS1B02G047700 | regulation of transcription, DNA-templated | - | double-stranded DNA binding | |
| 4 | rs5109 | TGCAGAGAAAAAGAGATGTATCATTCAGAGGCCAAACAAAAGTGAAGAGACAAATTTGATCACG | GN,MP, GMP,STI | 2A | 59,228 | TraesCS2A02G251800 | protein phosphorylation, signal transduction, response to nutrient, response to water deprivation, potassium ion import across plasma membrane, stomatal movement, phosphorylation, intracellular signal transduction | nucleus, cytosol, plasma membrane, plastid | nucleotide binding, protein kinase activity, protein serine/threonine kinase activity, ATP binding, kinase activity, transferase activity, protein serine kinase activity, protein threonine kinase activity | |
| 5 | rs30934 | TGCAGCGCGTGGGCATCGGCCACGTTCTAAATGTCACTGCCATGGTCGCTGCCGCACTGGTGGA | GNWW | 4A | 125,389 | TraesCS4A02G440300 | transmembrane transport | membrane | transmembrane transporter activity | |
| 6 | rs56604 | TGCAGTAGAAACCACCACTAGATGCAGTTATTCTAGTGAAACATCCGAGATCGGAAGAGCGGGA | STI | 5D | 7959 | TraesCS5A02G014400 | glycolytic process | phosphopyruvate hydratase complex | phosphopyruvate hydratase complex, phosphopyruvate hydratase activity | |
| 7 | TKW | rs804 | TGCAGAAATACACTCCTAATTTTACATGAACGCTACTCATATGGATTTATCCACACGACGCTAC | TKWRF, SSI,DI | 1A | 52,471 | TraesCS1A02G312800 | - | - | - |
| 8 | rs165 | TGCAGAAAACTAAGATGCAGTTATTTTCTGAGTGATGCGTACTGTTAATTCGATCACAGGAGTT | DI | 1B | 27,300 | TraesCS1B02G049300 | DNA repair, protein ADP-ribosylation, response to oxidative stress, response to abscisic acid | nucleus, integral component of membrane | DNA binding, NAD + ADP-ribosyltransferase activity, zinc ion binding, NAD binding | |
| 9 | rs15519 | TGCAGCACTCTGCAAGAAAAACGTCAAAGTAAGAACCACCTACCCACATCTGCTCCAATTCAAA | TKWWW | 1D | 47,767 | TraesCS1D02G147800 | lipid biosynthetic process | integral component of membrane | iron ion binding, oxidoreductase activity | |
| 10 | rs2355 | TGCAGAAGTGCAGATAATCAGACAAGCCAGCGAAACCCTAAGCCCTCGACTAATCTGTTCGATC | SSI, DI | 2B | 0 | TraesCS2D02G011200 | positive regulation of transcription, DNA-templated | - | transcription coactivator activity, chromatin DNA binding | |
| 11 | rs16199 | TGCAGCAGCAACCACCACCATGGAAAGAGAGAGACAGAGACGGTGAGCTCCTCTGGACAGCGAG | DI | 2D | 28,183 | TraesCS2D02G082700 | lipid metabolic process | - | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water | |
| 12 | rs51900 | TGCAGGGTGGGGGCGGAGAAAAAGGAGGAGGGGCGGCCGAGATCGGAAGAGCGGGATCACCGAC | TKWRF, DI | 2D | 28,183 | TraesCS2D02G082900 | vesicle-mediated transport | plasma membrane, integral component of membrane | protein binding | |
| 13 | rs59777 | TGCAGTCTTTCAGAAGTGCAGATGTAAACGTATTGCTATATCAGTGGTTTGAACTACATGGTAA | TKWWW, TOL, ATI | 2D | 58,883 | TraesCS2D02G152500 | protein ubiquitination, positive regulation of protein catabolic process | - | protein binding | |
| 14 | rs16023 | TGCAGCAGAGGTGGTTTGGAGGTTTGGTGGCGGCAGGATTCCCCTCCCGCGGGCGGCTCGGCTC | TKWRF, DI | 3B | 56,892 | TraesCS3B02G373500 | auxin-activated signaling pathway, transmembrane transport, intracellular auxin transport | membrane, integral component of membrane | - | |
| 15 | rs60485 | TGCAGTGCAGAAGGCGGCGTCCATCTGGTCCATGGCCTTGAGCTCGCCG | SSI | 4B | 34,449 | TraesCS4B02G027000 | - | - | - | |
| 16 | rs23471 | TGCAGCCCCATGGCTGGCCACTGCCCCGCCGACGCCACCTGCGGGTTTGGAGACGCCACCACGC | TKWWW | 5A | 58,225 | TraesCS5A02G431100 | - | - | RNA binding | |
| 17 | rs31132 | TGCAGCGCTCTCGGCGGTGACTCGTCGTCGCTCGGTGGCATCACCATCAACAAGACACGCGCGC | TKWWW | 5A | 58,225 | TraesCS5A02G433100 | lipid transport | - | lipid binding | |
| 18 | rs44444 | TGCAGGAGCTGAGCAACGAGGCCACAGCCGCCGCAGAAAAGGAGTCCCTGAACGGCACACTGGC | TKWWW, MP, STI | 6B | 92,187 | TraesCS6A02G402500 | ubiquitin-dependent protein catabolic process, protein deubiquitination | intracellular anatomical structure | thiol-dependent deubiquitinase | |
| 19 | rs44444 | TGCAGACACTAGTATCATTGGAAGCACAGGATGAGTCCGTTAGACAGTTGGGGGAGCTGAGGCA | K2STI | 4A | 61,015 | TraesCS6B02G151700 | - | - | ADP binding | |
| 20 | rs37640 | TGCAGCTCGTCATCACCGCTCGCCCGCCCGCGTGGATGCAGAAGTGCTCGAACGCCGTGCGGAA | PCA1,TKWRF, MP, GMP, STI, HM | 6A | 55,893 | TraesCS6D02G274600 | fatty acid biosynthetic process | membrane | acyltransferase activity, acyltransferase activity, transferring groups other than amino-acyl groups | |
| 21 | rs9536 | TGCAGATGGGGCGTTTCACGACGCTAGGCTATTGAGTGGAACTGGCAGTTTAGGGCTAAGCAGT | PCA2,TOL, SSI | 7A | 0 | TraesCS7A02G020300 | - | - | ||
| 22 | rs49562 | TGCAGGGACAGAGCCACCATGCATGCCATTCTGCTGCACTGGGTCAGGCTCCACGTCCCCAACA | TKWWW | 7B | 45,510 | TraesCS7B02G057400 | - | membrane, integral component of membrane | ionotropic glutamate receptor activity, ligand-gated ion channel activity | |
| 23 | rs30571 | TGCAGCGCGAGGCGGCGGAAGGCCCTCTTGACCTCGCCCCTGGAGGCCCCCGAGATCGGAAGAG | PCA2, TOL, SSI | 7B | 94,660 | TraesCS7B02G415000 | - | - | - | |
| 24 | rs36375 | TGCAGCTCCAGGTACTGGACGATGTCGTGAGGGTTGCGGCGGCGGAGCGGGTGGTCAAGGCCAC | PCA2,TOL, ATI | 7A | 135,625 | TraesCS7B02G498400 | - | - | sulfotransferase activity | |
| 25 | SW | rs49008 | TGCAGGCTCTGGAAACTTCCCAGTGCTTCCCTCCATGGAACTCATGTTCTTCAATGAACTATTG | SWRF,MP, GMP, HM,PCA1 | 1B | 21,612 | TraesCS1B02G033900 | response to wounding | - | serine-type endopeptidase inhibitor activity |
| 26 | rs57106 | TGCAGTAGTATTAACTGAAGCCAAAGTCGTTTTTGTTGTTGTTGTGATGACAGGGGCAATGAGG | MP, SWWW,GMP, STI, HM,PCA1 | 2A | 92,517 | TraesCS2A02G570400 | - | - | - | |
| 27 | rs2179 | TGCAGAAGGCATCTCAATTCAAGGCCCAGCCGATGTTACGGCGAGCTCCAATGATAAAGCCTAT | TOL, ATI, SSI,PCA2 | 3A | 75,276 | TraesCS3A02G405600 | - | integral component of membrane | - | |
| 28 | rs64448 | TGCAGTTGTAATCTTCCATGGAATCCCAACAAGTTTAGAGCGTGTCGATTCGTGGTAGATGGAT | SWRF, MP, GMP, STI, HM,PCA1 | 3B | 56,892 | TraesCS3B02G373400 | - | membrane, integral component of membrane | monooxygenase activity, iron ion binding, oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, heme binding | |
| 29 | rs61637 | TGCAGTGGGCGCACGGATCCCTCAACGCGCTCTCCTGGGGCCTCCTGCTCCCCGTGGGCGCGGC | STI | 4A | 111,120 | TraesCS4B02G265500 | - | - | - | |
| 30 | rs39277 | TGCAGCTGCTGCTGGAGCAGCGGGTGCATCGCCGTCGCCGCCGAGATCGGAAGAGCGGGATCAC | SWRF | 4A | 111,179 | TraesCS4D02G312700 | - | - | - | |
| 31 | rs55954 | TGCAGTAATCTACTAGATACGGAGTACTATGGAAGTGAGGGAGCAGCCGCGCGCGCTTTTAATG | TOL, ATI, PCA2 | 5A | 9093 | TraesCS5A02G303400 | - | - | oxidoreductase activity | |
| 32 | rs36801 | TGCAGCTCCGTGAGGCACACCGCGCACACGTCGTCGTCGCCGCCCGCGCCGCCGTCGTGGGCGC | ATI | 6D | 117,663 | TraesCS6D02G389000 | - | integral component of membrane | - | |
| 33 | rs63819 | TGCAGTTGACATGGTGGTTTTGTTCAAGGCACGGGCTCAAGAATCCAAAGAAACAAAATTTATT | DI | 6D | 117,663 | TraesCS6D02G389600 | - | integral component of membrane | aspartic-type endopeptidase activity | |
| 34 | rs63548 | TGCAGTTCGTCGTGGCGATGTTCGCCATGGACACGTGGCAATACTTCGTGCACAGGTATCTCCA | SW, TOL | 7B | 50,057 | TraesCS7B02G092300 | lipid biosynthetic process | integral component of membrane | iron ion binding, oxidoreductase activity | |
| 35 | rs40986 | TGCAGCTTCGGTTGGCCAGAGCCACACAACTCCACAAGTGAGCACATGCTAGCTGATGCATCCA | SWRF, DI | 7B | 51,193 | TraesCS7B02G200200 | - | - | - | |
| 36 | GY | rs7126 | TGCAGAGTCAGGCGGTAGGAAAAGGAGCGAAGGGGAACGGGGAACTCAGATCTGACGGGCAGCG | TOL | 1A | 46,787 | TraesCS1B02G273100 | regulation of transcription, DNA-templated | - | nucleic acid binding, DNA binding, zinc ion binding |
| 37 | rs6689 | TGCAGAGGCCTGTTGACGTTGTGGTACAAGCCCGAGATCGGAAGAGCGGGATCACCGACTGCCC | SSI, PCA2 | 2B | 7969 | TraesCS2B02G029300 | - | - | - | |
| 38 | rs15903 | TGCAGCAGAGAATAATAGATGGAGGGAGGGGTGGTGCAAGTATAGCACCCGAGATCGGAAGAGC | GYRF, GMP, STI, HM,PCA1 | 2B | 34,160 | TraesCS2B02G084300 | - | - | ADP binding | |
| 39 | rs7932 | TGCAGATATTTATCGCCCAAGAGCAAAGATGCTTGACCAGGATTTGGATTGCGGACCGAGATCG | TOL, SSI, DI, PCA2 | 2B | 86,479 | TraesCS2B02G552400 | transcription, DNA-templated | - | DNA binding, DNA-directed 5ʹ-3ʹ RNA polymerase activity, ribonucleoside binding | |
| 40 | rs2997 | TGCAGAATTGACAGATGCATCAAAATTGGTAGCCGCTGAAGCTAACAATGCTCATGTTGATGTT | TOL | 3A | 77,549 | TraesCS3A02G417600 | regulation of growth | - | ||
| 41 | rs63419 | TGCAGTTCGAGCGCCGATGGTGCCTCTTGTTGTGTTGTGTCCCCCCTCGCCATGTGTTGTCCAT | GYRF,TOL | 4A | 61,015 | TraesCS4A02G156500 | cation transport, calcium ion transport, transmembrane transport | integral component of membrane | cation transmembrane transporter activity, calcium:proton antiporter activity | |
| 42 | rs51991 | TGCAGGGTTCGCTCGTCGACGTCAACCCTTTGGAAGCGCAGCTCGAGCGCGGCATCCTTCTGGA | GYRF, DI | 4A | 129,369 | TraesCS4A02G437000 | - | ATP binding, ATPase | ||
| 43 | rs42463 | TGCAGGAACTCCGCTCTGTCCTCGTCCTCGTCGTCGCTGTCGGCCTTGCTCGAGGTGCCGAGAT | TOL, ATI, PCA2 | 5A | 9093 | TraesCS5A02G295400 | carbohydrate metabolic process | - | hydrolase activity, hydrolyzing O-glycosyl compounds | |
| 44 | rs55429 | TGCAGGTTTCAATTACGGAGGGAAAAACTCCAAGAAACTTATTGTTAGCAAGACGAAGTGACTG | GYWW | 5A | 109,694 | TraesCS5A02G542100 | - | - | protein binding | |
| 45 | rs8046 | TGCAGATCACTATGCTTGTATGGTGGATCTCTATGGCCGAGCTGGACTTATTGAAGAAGCACAC | ATI | 6B | 58,062 | TraesCS6A02G338900 | - | - | protein binding | |
| 46 | rs24860 | TGCAGCCGAGCTAATGGTGGTAGGGGTGGATGCTTTTTGTGAGCGACTGAGCGATGAGGAATGA | TOL | 6B | 1137 | TraesCS6D02G008200 | aromatic compound biosynthetic process, methylation | - | methyltransferase activity, O-methyltransferase activity, S-adenosylmethionine-dependent methyltransferase activity, transferase activity, protein dimerization activity |
Abbreviations: GY Grain yield, GN Grains number per spike, TKW Thousand kernel weight, SW Spike weight, TOL Tolerance index, MP Mean product, GMP Geometric mean product, STI Stress tolerance index, ATI Abiotic stress tolerance index, SSI Stress susceptibility index, DI New drought resistance index, HM Harmonic mean
Fig. 12.
The KEGG pathway of ascorbate and aldarate metabolism (A) and pathway of biosynthesis of amino acids (B). The authors declare all that permissions were obtained for the appropriate copyright KEGG image depicted
Discussion
It is possible to examine how GY is affected under normal and drought conditions by using DTIs to select optimal genotypes. DTIs STI, GMP, MP, and HM are key indices for screening high-yielding genotypes in different moisture conditions [29, 30]. The previous study also demonstrated that STI is a useful parameter for distinguishing high-yield genotypes growing in both high-yielding and drought-tolerant environments [31–33]. The MP, GMP, STI, and HM parameters were related to GY. According to Ravari et al. [34] YSI, GMP, STI, and HM parameters were all well associated with the dependent variable for stress.
PCA and heatmap analyses have already been confirmed to be effective in identifying drought-tolerant and high-yield genotypes in tomato [35], chickpea [36], and switchgrass [37]. The MP, GMP, STI, and HM indices are effective for finding DT genotypes suitable for planting in a water-limited region according to the cluster analysis. Based on the PCA of Iranian cultivars and landraces, the first component could be linked to GYWW, GYRF, MP, STI, and GMP, and cultivars with high yields and DT could be identified as having this component. A similar finding was reported by Farshadfar et al. [38] for DTIs. According to our PCA results, GMP and HM were the most appropriate indices for screening in local varieties.
A high level of variation was uncovered in the studied traits for Iranian wheat accessions, suggesting the potential of the GWAS technique for exploring QTLs. A strong correlation between yield traits can be justified by indirect or direct contributions from other traits [39]. Regarding the wheat genome, the genetic areas responsible for such yield traits can be similar [40]. Mwadzingeni et al. [41], for example, discovered that a single locus influences numerous wheat traits such as plant height, spike length, and, grains per spike, all of which are connected frequently [42]. However, some loci affect only one crop attribute [41].
The frequency of the linked SNPs was higher in genome B than that of the other genomes. Because chromosome B is smaller than chromosome A, it appears that there is a clear association between chromosome size and SNP density [43, 44]. The increased frequency of SNP in the B genome was the result of evolutionary processes. Alipour et al. [45] and Mourad et al. [46] also reported this inference. Three separate subpopulations of wheat accessions were identified. Considering that wheat accessions have different pedigrees, this issue is expectable. There may be some relationships among accessions when common parents or origins exist in their pedigrees [40, 47].
Genomes D, A, and B have the highest LD, respectively. The strongest LD was recorded between marker pairs on chromosome 4A [48]. LD differences between genomes and accessions indicate the effects of breeding schedules in addition to evolutionary processes. In wheat Pakistan/China collections, Liu et al. [26] found that the distance of LD decay in native populations is less than that in cultivated varieties.
The number of GO-based on GY, GN, TKW, and SW were 11, 6, 18, and 11 markers containing overlapping genes were identified. Although only those connections with P < 0.0001 were considered significant, the remaining MTAs may be useful in improving wheat drought tolerance. These connections can be found in genomic areas that influence agronomic traits. Given that yield is a highly complex genetic trait with low heritability, MTAs for yield appeared significant at a higher P value. Most of the identified markers were located on chromosomes 4A, 5A, 7B, 1A, 1B, 6B, and 2B based on the studied traits and their related tolerance indices. A number of MTAs/QTLs have been found for GY in wheat chr. 7A [49–52], 7B [49, 52, 53], 3D [49], 3A [49, 52–54], 5B [49, 52, 55], 1B [49, 53, 56], and 2B [49, 53, 54, 56, 57]. MTAs/QTLs for TKW have been found in previous reports on chr. 7B [52], 7D [50], 5B [58], 3A [57, 58], 3B [50], 2D [56], 2A [50], 2B [50, 52, 57], and 1A [52, 56–58]. Therefore, MTAs on chr. 5A, 1B, 6B, and 1D are novel for TKW. As a result, MTA on chr. 4A and 5A have never been documented, and it is novel for wheat output.
The results of our study are in line with those of some studies made on the bread wheat of Iran, including Salarpour et al. [59], Salarpour et al. [8], Sobhanian et al. [60], Tahmasebi et al. [61], and Heidari et al. [62]. Tahmasebi et al. [61], stated that the most important QTLs for the thousand-grain weight, and GY were detected on chromosomes 1B, 1D-a, and 7D-b. In another study, Heydari et al. [62] reported that the major QTL located at the Hair–Xpsp2999 interval on chromosome 1A controlled the expression of grains/spike (R2 = 12.9% in 2004 and 22.4% in 2005), grain weight/spike (R2 = 21.4% in 2004 and 15.8% in 2005), and spike number (R2 = 15.6% in 2004 and 5.4% in 2005). The QTL for GY located on chromosomes 6A, 6B, and 6D totally accounted for 27.2% and 31.7% of the total variation in this trait in 2004 and 2005, respectively.
The flanking sequences of 43,525 SNPs were compared to RefSeq v2.0 and aligned accordingly. Surprisingly, the majority of marker pairs were found in the protein-coding areas, which regulate transcription. Other factors contributing to drought tolerance include DNA binding, transcription factor activity, and transmembrane transport. Some pathways were discovered using the rice reference genome, including ascorbate and aldarate metabolism, oxidative phosphorylation, biosynthesis of amino acids, fatty acid elongation, and metabolic pathways. In the metabolism of ascorbate and aldarate sucrose synthase, and sucrose-phosphate synthase are both genes that are involved in a metabolic pathway that is associated with DS tolerance [63]. Drought stimulates energy-intensive activities such as osmolyte production and oxidative phosphorylation, as well as increased respiratory rates [64, 65]. In oats [66] and wheat [67], fatty acid synthesis is beneficial in combating DS. The amino acid pathway is one of the amino acids produced by proline. The amino acid proline has been related to a number of osmoprotective properties that scavenge reactive oxygen species [68–71]. Under DS, drought-tolerant genotypes gained proline content faster and in higher proportions than sensitive equivalents, emphasizing its importance in drought-tolerance breeding. It has been discovered that genes that control proline content have cumulative effects [72, 73].
Conclusion
MTAs are key elements for detecting genomic areas linked to agronomic traits in wheat under drought stress. The identified markers might be used to clone and fine map underlying genes, as well as perform gene introgression and marker-based selection in wheat under normal and drought conditions. The discovery of QTL-rich regions on Ch. 4A and 5A supports the theory that this chromosome is important for drought tolerance and should be utilized for wheat breeding. Furthermore, a large number of SNP correlations were discovered at the genome level on the B genome, which has been related to drought resistance. Further, the use of association mapping based on several drought tolerance indices can be highly effective in finding the most essential markers for drought tolerance as well as discovering linked gene networks.
Materials and methods
Experimental site
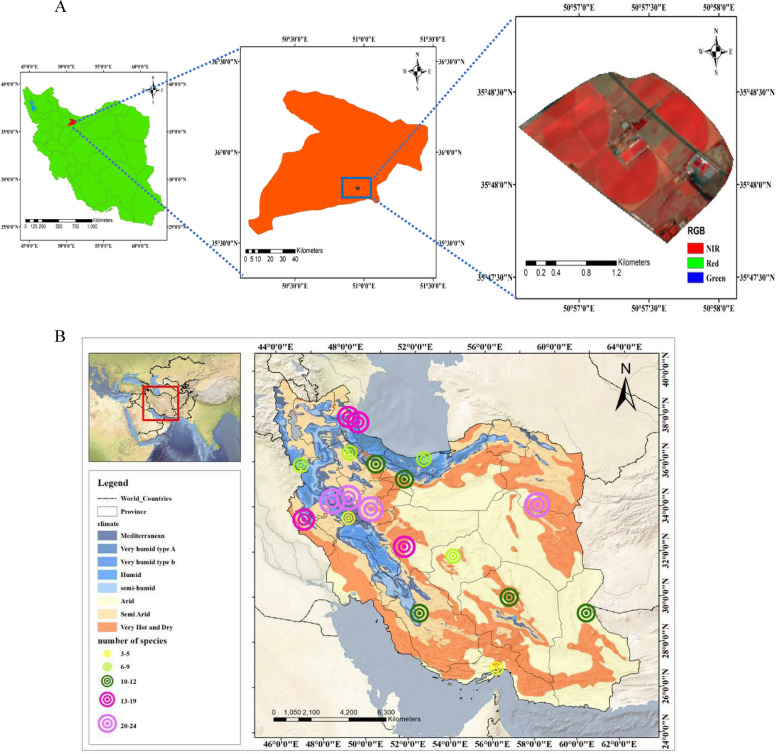
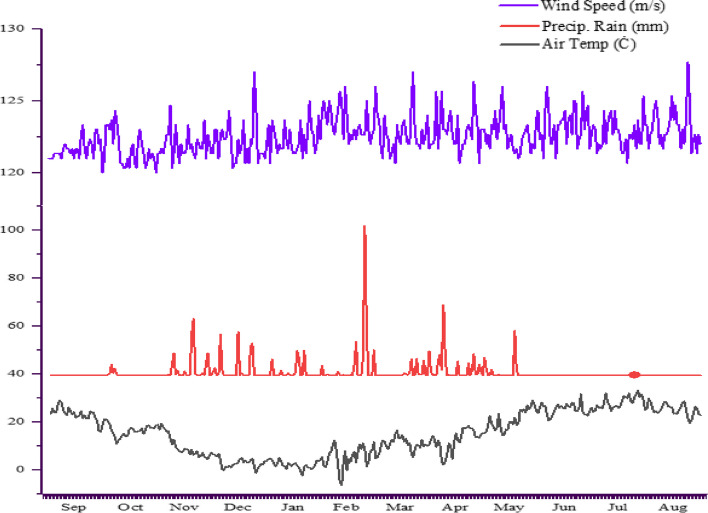
The research was conducted at the Agricultural and Natural Resources Campus of Tehran University. Figure 13 illustrates the geographical location of the study area (35°48′59′′N, 51°58′48′′E, 1321 m elevation) and the geographic distribution of Iranian wheat landraces collected between 1931 and 1968 years. Figure 14 shows the climatic characteristics of this field (Supplementary 1 Table 2). It covers approximately 246 ha and its main crops are barley, corn, wheat, and alfalfa. The climate in this region is dry and warm. A majority of the soil is clay and silt with an average annual temperature and precipitation of 22 °C and 248 mm.
Fig. 13.
The geographical location of the study area (a) and the geographic distribution of Iranian wheat landraces collected between 1931 and 1968 years (b)
Fig. 14.
Climatic data in the studied environments
Plant materials and experimental design
This study followed 298 wheat genotypes collected from various regions and climates of Iran (Supplementary 1 Table 3) in alpha lattice design with two repeats during two crop seasons (2018–19 and 2019–20) under rain-fed (drought) and well-watered (normal) conditions. The plots consisted of four rows (1*1 m2) spaced 50 cm. The plant density was 300 plants/m2, and the sowing and harvesting dates in both years were November 1 and June 30. The threshold for irrigation implementation was determined based on 40 mm evaporation from an evaporation pan for well-watered crops. We used a crop coefficient of determination (KC) as well as a reference crop evapotranspiration equation, ET0 = Epan*Kpan, where Epan is the evaporation depth below the pan surface (40 mm), and Kpan denotes the pan coefficient (0.8) for each month, to determine evapotranspiration (ETC = KC * ET0) [2, 74]. In this study, the irrigation time was calculated by dividing the applied water for 1400 m2 (the cultivation area for 298 genotypes in two repeats) by the water discharge (10.8 m3/h). Water requirement (m3/ha) was estimated by multiplying the depth of ET0 (mm) by 10. Wheat is grown under the rain-fed regime and only receives rainfall as a source of water. Table 1 presents the patterns of rainfall during the cropping seasons. After physiological maturity, GY per plant was measured by isolating 20 plants and pounding their spikes, then weighing their seeds, which were weighed, followed by calculating the yield of a single plant. The traits measured in this study were GY (g per plant), spike weight (SW, gr), GN (per spike), and TKW (gr). The calculations of the DTIs were based on the trait yield for normal (YP) and stress (YS) conditions, and the total average trait (GY, GN, TKW, and SW) for normal () and stress () environments with the formulas listed in Table 3. Samples of plants are provided by the Gene Bank of Agronomy and Plant Breeding Group and these samples are available at USDA with USDA PI number (Supplementary 1 Table 3), respectively. The authors declare that all study complies with relevant institutional, national, and international guidelines and legislation for plant ethics in the methods section. The authors declare that all that permissions or licenses were obtained to collect the wheat plant.
Table 3.
Drought tolerance indices used for investigation of Iranian wheat germplasm
Genotyping-by-sequencing and imputation
In accordance with Alipour et al. [45], GBS libraries for Iranian wheat genotypes were established and sequenced. As reads were trimmed to 64bp and categorized into tags, SNPs were detected based on internal alignments, allowing for up to 3 bp of mismatch. The GBS pipeline was called Universal Network-Enabled Analysis Kit SNPs and discarded reads with a low-quality score (< 15). The imputation was performed with BEAGLE v3.3.2 and the w7984 reference genome [48]. Finally, SNPs with heterozygotes of < 10% and minor allele frequency greater than > 5% were considered for further analysis.
Population structure and kinship matrix
STRUCTURE (version 2.3.4) was used to analyze the population structure of the landraces and cultivars of Iranian wheat [75]. This study employed a 30,000-step simulation phase, along with an admixture model, covering K = 1 to 10. The most likely number of sub-populations in this study was estimated by using ΔK. For association studies, Q-matrix was utilized as a structural matrix. Based on pairwise distance matrices counted in TASSEL [76], a neighbor-joining tree was formed and visualized using Archaeopteryx to explore the relationships between the cultivars and landraces of Iranian wheat.
Genome-wide association study
Both MLM (mixed linear models) and mrMLM (multi-locus random-SNP-effect MLM) were applied to provide an unbiased estimation of marker effects. Using the MLM approach, it was possible to accurately associate marker traits between accessions and various MLM models for controlling both population structure (Q) and diffused associations (K) between accessions with the GAPIT package. In RStudio, GWAS was performed with the MLM and mrMLM using the GAPIT package [77]. The MLM approach considers accessions to be a random effect, the relevance of each is defined by a kinship matrix. The cluster analysis was conducted using kinship matrix elements as likeness measures, and the clusters were visualized by the unweighted double group approach with arithmetic mean (UPGMA) using a heat map. Moreover, –log10 (P) > 3 and –log10 (P) > 5 thresholds were used for statistically significant MTAs [78]. Confidence intervals for each chromosome were determined using LD decay [79]. A Manhattan plot was obtained by applying the CMplot package to explore associations between genotypes and phenotypes [80].
Annotation of genes
The sequences surrounding all significantly associated SNPs were obtained by aligning them with IWGSC RefSeq v2.0 of the wheat 90 K SNP database used for Gramene (http://www.gramene.org/) gene annotation assessments. The identification of putative candidate genes was evaluated according to two parameters: a) being located in the vicinity of the peak marker, and b) having known functions and involvement in the studied traits in wheat (http://ensembl.gramene.Org; https://wheaturgi.versailles.inra.fr/SeqRepository/Annotations). Moreover, the significant SNPs were utilized in the enrichment analysis of gene ontology via KOBAS version 2.0 for testing in the KEGG. Finally, gene pathways were identified through the rice reference genome) Finally, gene pathways were identified through the rice reference genome ([80–82]; www.kegg.jp/kegg/kegg1.html).
Statistical analysis
Descriptive statistics and correlation coefficients were obtained by R 4.1 using the ggplot2, dplyr, ggpubr, and psych packages to reveal the distribution of wheat traits. An analysis of heatmaps was performed in RStudio to classify wheat genotypes. Eventually, the evaluation and dispersion of wheat traits and genotypes across the biplot diagram were analyzed using PCA and the factoextra packages in RStudio.
Supplementary Information
Additional file 1: Supplementary Table 1. A summary of LD observed among marker pairs and the number of significant marker pairs per genome and chromosome. Supplementary Table 2. Climatic data in the studied environments. Supplementary Table 3. Overview on the landraces and cultivars of Iranian wheat studied. Supplementary Fig 1. The KEGG pathway of oxidative phosphorylation. Supplementary Fig 2. The KEGG pathway of fatty acid elongation. Supplementary Fig 3. The KEGG pathway of metabolic pathways.
Acknowledgements
Not applicable.
Abbreviations
- DS
Drought stress
- GWAS
Genome-Wide Association Study
- MTAs
Marker-trait associations
- LD
Linkage disequilibrium
- TKW
Thousand kernel weight
- GN
Grains number per spike
- SW
Spike weight
- GY
Grain yield
- WW
Well-watered
- RF
Rain-fed
- TOL
Tolerance index
- MP
Mean productivity
- GMP
Geometric mean productivity
- STI
Stress tolerance index
- ATI
Abiotic stress tolerance index
- SSI
Stress susceptibility index
- DI
New drought resistance index
- HM
Harmonic mean
- GBS
Genotyping-by-sequencing
- MAF
Minor allele frequencies
- PCA
Principal component analysis
Authors’ contributions
M.R.B proposed the idea and helped to provide the plant materials, E.R, K.A and S.J performed field trials, analyzed the data, and wrote a draft version of the manuscript, H.A proposed the idea and helped in the genomic data analysis, M.E.M and V.M were involved in designing and conducting the experiment. All authors contributed to revising and editing the manuscript. All authors have read and approved the final manuscript.
Funding
This research did not receive any specific funding.
Availability of data and materials
The datasets generated and analyzed during the current study are available in Supplementary 2.
Declarations
Ethics approval and consent to participate
Samples of plants are provided from the Gene Bank of Agronomy and Plant Breeding Group and these samples are available at USDA with USDA PI number (Supplementary 1 Table 3), respectively. The authors declare that all study complies with relevant institutional, national, and international guidelines and legislation for plant ethics in the methods section. The authors declare that all that permissions or licenses were obtained to collect the wheat plant.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Reza Bihamta, Email: mrghanad@ut.ac.ir.
Hadi Alipour, Email: ha.alipour@urmia.ac.ir.
References
- 1.Alipour H, Abdi H. Interactive effects of vernalization and photoperiod loci on phenological traits and grain yield and differentiation of Iranian wheat landraces and cultivars. J Plant Growth Regul. 2021;40(5):2105–2114. [Google Scholar]
- 2.Rabieyan E, Bihamta MR, Moghaddam ME, Mohammadi V, Alipour H. Imaging-based screening of wheat seed characteristics towards distinguishing drought-responsive Iranian landraces and cultivars. Crop Pasture Sci. 2022;73(4):337–355. [Google Scholar]
- 3.Rabieyan E, Bihamta MR, Moghaddam ME, Mohammadi V, Alipour H. Morpho-colorimetric seed traits for the discrimination, classification and prediction of yield in wheat genotypes under rainfed and well-watered conditions. Crop Pasture Sci. 2022;74(4):294–311. [Google Scholar]
- 4.Abid M, Ali S, Qi LK, et al. Physiological and biochemical changes during drought and recovery periods at tillering and jointing stages in wheat (Triticum aestivum L.) Sci Rep. 2018;8:4615. doi: 10.1038/s41598-018-21441-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alipour H, Rabieyan E. NGS-based multiplex assay of trait-linked molecular markers revealed the genetic diversity of Iranian bread wheat landraces and cultivars. Crop Pasture Sci. 2021;72(3):173–182. [Google Scholar]
- 6.Qaseem MF, Qureshi R, Shaheen H. Effects of pre-anthesis drought, heat and their combination on the growth, yield and physiology of diverse wheat (Triticum aestivum L.) genotypes varying in sensitivity to heat and drought stress. Sci Rep. 2019;9:6955. doi: 10.1038/s41598-019-43477-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ilyas M, Nisar M, Khan N, Hazrat A, Khan AH, Hayat K, Fahad S, Khan A, Ullah A. Drought tolerance strategies in plants: a mechanistic approach. J Plant Growth Regul. 2021;40(3):926–944. [Google Scholar]
- 8.Salarpour M, Pakniyat H, Abdolshahi R, Heidari B, Razi H, Afzali R. Mapping QTL for agronomic and root traits in the Kukri/RAC875 wheat (Triticum aestivum L.) population under drought stress conditions. Euphytica. 2020;216:1–19. [Google Scholar]
- 9.Batool M, El-Badri AM, Hassan MU, Haiyun Y, Chunyun W, Zhenkun Y, Jie K, Wang B, Zhou G. Drought stress in Brassica napus: effects, tolerance mechanisms, and management strategies. J Plant Growth Regul. 2022;17:1–25. [Google Scholar]
- 10.Zhao T, Dai A. The magnitude and causes of global drought changes in the twenty-first century under a low–moderate emissions scenario. J Clim. 2015;28(11):4490–4512. [Google Scholar]
- 11.Abou-Elwafa SF. Association mapping for yield and yield-contributing traits in barley under drought conditions with genome-based SSR markers. C R Biol. 2016;339:153–162. doi: 10.1016/j.crvi.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds M, Langridge P. Physiological breeding. Curr Opin Plant Biol. 2016;31:162–171. doi: 10.1016/j.pbi.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Abou-Elwafa SF. Association mapping for drought tolerance in barley at the reproductive stage. C R Biol. 2016;339(2):51–59. doi: 10.1016/j.crvi.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Rizhsky L, Liang H, Mittler R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2022;130(3):1143–1151. doi: 10.1104/pp.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mursalova J, Akparov Z, Ojaghi J, Eldarov M, Belen S, Gummadov N, Morgounov A. Evaluation of drought tolerance of winter bread wheat genotypes underdrip irrigation and rain-fed conditions. Turk Tarim ve Orman Derg/Turk J Agric For. 2015;39(5):817–824. [Google Scholar]
- 16.Abdolshahi R, Nazari M, Safarian A, Sadathossini TS, Salarpour M, Amiri H. Integrated selection criteria for drought tolerance in wheat (Triticum aestivum L.) breeding programs using discriminant analysis. Field Crops Res. 2015;174:20–29. [Google Scholar]
- 17.Mathew I, Shimelis H, Shayanowako AIT, Laing M, Chaplot V. Genome-wide association study of drought tolerance and biomass allocation in wheat. PLoS One. 2019;14(12):e0225383. doi: 10.1371/journal.pone.0225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kehel Z, Sanchez-Garcia M, El Baouchi A, Aberkane H, Tsivelikas A, Charles C, Amri A. Predictive characterization for seed morphometric traits for genebank accessions using genomic selection. Front Ecol Evol. 2020;8:1–32. [Google Scholar]
- 19.Alemu A, Feyissa T, Tuberosa R, Maccaferri M, Sciara G, Letta T, Abeyo B. Genome-wide association mapping for grain shape and color traits in Ethiopian durum wheat (Triticum turgidum ssp. durum) Crop J. 2020;8(5):757–768. [Google Scholar]
- 20.Huang C, Nie X, Shen C, You C, Li W, Zhao W, Lin Z. Population structure and genetic basis of the agronomic traits of upland cotton in China revealed by a genome-wide association study using high-density SNPs. Plant Biotechnol J. 2017;15(11):1374–1386. doi: 10.1111/pbi.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Wang Y, Wang X, Deng D. Integrated meta-QTL and genome-wide association study analyses reveal candidate genes for maize yield. J Plant Growth Regul. 2020;39(1):229–238. [Google Scholar]
- 22.Yu LX, Zheng P, Zhang T, Rodringuez J, Main D. Genotyping-by-sequencing-based genome-wide association studies on Verticillium wilt resistance in autotetraploid alfalfa (Medicago sativa L.) Mol Plant Pathol. 2017;18:187–194. doi: 10.1111/mpp.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mace ES, Tai S, Gilding EK, Li Y, Prentis PJ, Bian L, Wang J. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat Commun. 2013;4:1–9. doi: 10.1038/ncomms3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Wang H, Liu S, Ferjani A, Li J, Yan J, Qin F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet. 2016;48:1233–1241. doi: 10.1038/ng.3636. [DOI] [PubMed] [Google Scholar]
- 25.Hwang EY, Song Q, Jia G, Specht JE, Hyten DL, Costa J, Cregan PB. A genome-wide association study of seed protein and oil content in soybean. BMC Genomics. 2014;15:1–12. doi: 10.1186/1471-2164-15-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Rasheed A, He Z, Imtiaz M, Arif A, Mahmood T, Xia X. Genome-wide variation patterns between landraces and cultivars uncover divergent selection during modern wheat breeding. Theor Appl Genet. 2019;132(9):2509–2523. doi: 10.1007/s00122-019-03367-4. [DOI] [PubMed] [Google Scholar]
- 27.Shariatipour N, Heidari B, Tahmasebi A, Richards C. Comparative genomic analysis of quantitative trait loci associated with micronutrient contents, grain quality, and agronomic traits in wheat (Triticum aestivum L.). Front Plant Sci. 2021;12:709817. 10.3389/fpls.2021.709817. [DOI] [PMC free article] [PubMed]
- 28.Kumar A, Saripalli G, Jan I, Kumar K, Sharma PK, Balyan HS, Gupta PK. Meta-QTL analysis and identification of candidate genes for drought tolerance in bread wheat (Triticum aestivum L.) Physiol Mol Biol Plants. 2020;26:1713–25. doi: 10.1007/s12298-020-00847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mardeh ASS, Ahmadi A, Poustini K, Mohammadi V. Evaluation of drought resistance indices under various environmental conditions. Field Crops Res. 2006;98:222–229. [Google Scholar]
- 30.Mohammadi R. Efficiency of yield-based drought tolerance indices to identify tolerant genotypes in durum wheat. Euphytica. 2016;211(1):71–89. [Google Scholar]
- 31.Rizza F, Badeck FW, Cattivelli L, Lidestri O, Di Fonzo N, Stanca AM. Use of a water stress index to identify barley genotypes adapted to rainfed and irrigated conditions. Crop Sci. 2004;44(6):2127–2137. [Google Scholar]
- 32.Drikvand R, Doosty B, Hosseinpour T. Response of rainfed wheat genotypes to drought stress using drought tolerance indices. J Agric Sci. 2012;4(7):126–131. [Google Scholar]
- 33.El-Hendawy SE, Hassan WM, Al-Suhaibani NA, Schmidhalter U. Spectral assessment of drought tolerance indices and grain yield in advanced spring wheat lines grown under full and limited water irrigation. Agric Water Manag. 2017;182:1–12. [Google Scholar]
- 34.Ravari SZ, Dehghani H, Naghavi H. Assessment of salinity indices to identify Iranian wheat varieties using an artificial neural network. Ann Appl Biol. 2016;168(2):185–194. [Google Scholar]
- 35.Aghaie P, Tafreshi SAH, Ebrahimi MA, Haerinasab M. Tolerance evaluation and clustering of fourteen tomato cultivars grown under mild and severe drought conditions. Sci Hortic. 2018;232:1–12. [Google Scholar]
- 36.Jha UC, Basu P, Shil S, Singh NP. Evaluation of drought tolerance selection indices in chickpea genotypes. Int J Stress Manag. 2016;7(6):1244–1248. [Google Scholar]
- 37.Liu Y, Zhang X, Tran H, Shan L, Kim J, Childs K, Zhao B. Assessment of drought tolerance of 49 switchgrass (Panicum virgatum) genotypes using physiological and morphological parameters. Biotechnol Biofuels. 2015;8(1):1–18. doi: 10.1186/s13068-015-0342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farshadfar E, Mohammadi R, Farshadfar M, Dabiri S. Relationships and repeatability of drought tolerance indices in wheat-rye disomic addition lines. Aust J Crop Sci. 2013;7(1):130–138. [Google Scholar]
- 39.Dholakia B, Ammiraju J, Singh H, Lagu M, RoÈder M, Rao V. Molecular marker analysis of kernel size and shape in bread wheat. Plant Breed. 2003;122(5):392–395. [Google Scholar]
- 40.Rabbi SMHA, Kumar A, Mohajeri Naraghi S, Simsek S, Sapkota S, Solanki S, Alamri MS, Elias EM, Kianian S, Missaoui A, Mergoum M. Genome-wide association mapping for yield and related traits under drought stressed and non-stressed environments in wheat. Front Genet. 2021;12:649988. doi: 10.3389/fgene.2021.649988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mwadzingeni L, Shimelis H, Rees DJG, Tsilo TJ. Genome-wide association analysis of agronomic traits in wheat under drought-stressed and non-stressed conditions. PLoS One. 2017;12(2):e0171692. doi: 10.1371/journal.pone.0171692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kashif M, Khaliq I. Heritability, correlation and path coefficient analysis for some metric traits in wheat. Int J Agric Biol. 2004;6(1):138–142. [Google Scholar]
- 43.Rahimi Y, Bihamta MR, Taleei A, Alipour H, Ingvarsson PK. Genome-wide association study of agronomic traits in bread wheat reveals novel putative alleles for future breeding programs. BMC Plant Biol. 2019;19:1–19. doi: 10.1186/s12870-019-2165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berkman PJ, Visendi P, Lee HC, Stiller J, Manoli S, Lorenc MT, Edwards D. Dispersion and domestication shaped the genome of bread wheat. Plant Biotechnol J. 2013;11:564–571. doi: 10.1111/pbi.12044. [DOI] [PubMed] [Google Scholar]
- 45.Alipour H, Bihamta MR, Mohammadi V, Peyghambari SA, Bai G, Zhang G. Genotyping-by-sequencing (GBS) revealed molecular genetic diversity of Iranian wheat landraces and cultivars. Front Plant Sci. 2017;8:1293. doi: 10.3389/fpls.2017.01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mourad AMI, Belamkar V, Baenziger PS. Molecular genetic analysis of spring wheat core collection using genetic diversity, population structure, and linkage disequilibrium. BMC Genomics. 2020;21(1):1–12. doi: 10.1186/s12864-020-06835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhatta M, Morgounov A, Belamkar V, Baenziger PS. Genome-wide association study reveals novel genomic regions for grain yield and yield-related traits in drought-stressed synthetic hexaploid wheat. Int J Mol Sci. 2018;19(10):3011. doi: 10.3390/ijms19103011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alipour H, Bai G, Zhang G, Bihamta MR, Mohammadi V, Peyghambari SA. Imputation accuracy of wheat genotyping-by-sequencing (GBS) data using barley and wheat genome references. PLoS One. 2019;14(1):e0208614. doi: 10.1371/journal.pone.0208614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bordes J, Goudemand E, Duchalais L, Chevarin L, Oury FX, Heumez E, Lapierre A, Perretant MR, Rolland B, Beghin D, et al. Genome-wide association mapping of three important traits using bread wheat elite breeding populations. Mol Breed. 2014;33:755–768. doi: 10.1007/s11032-013-0004-0. [DOI] [Google Scholar]
- 50.Sukumaran S, Lopes M, Dreisigacker S, Reynolds M. Genetic analysis of multi-environmental spring wheat trials identify genomic regions for locus-specific trade-offs for grain weight and grain number. Theor Appl Genet. 2018;131:985–998. doi: 10.1007/s00122-017-3037-7. [DOI] [PubMed] [Google Scholar]
- 51.Kumar N, Kulwal PL, Balyan HS, Gupta PK. QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Mol Breed. 2007;19:163–177. doi: 10.1007/s11032-006-9056-8. [DOI] [Google Scholar]
- 52.Neumann K, Kobiljski B, Denčić S, Varshney R, Börner A. Genome-wide association mapping: a case study in bread wheat (Triticum aestivum L.) Mol Breed. 2011;27(1):37–58. doi: 10.1007/s11032-010-9411-7. [DOI] [Google Scholar]
- 53.Sehgal D, Autrique E, Singh R, Ellis M, Singh S, Dreisigacker S. Identification of genomic regions for grain yield and yield stability and their epistatic interactions. Sci Rep. 2017;7(1):1–12. doi: 10.1038/srep41578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffstetter A, Cabrera A, Sneller C. Identifying quantitative trait loci for economic traits in an elite soft red winter wheat population. Crop Sci. 2016;56(2):547–558. doi: 10.2135/cropsci2015.06.0332. [DOI] [Google Scholar]
- 55.Edae EA, Byrne PF, Haley SD, Lopes MS, Reynolds MP. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor Appl Genet. 2014;127:791–807. doi: 10.1007/s00122-013-2257-8. [DOI] [PubMed] [Google Scholar]
- 56.Ogbonnaya FC, Rasheed A, Okechukwu EC, Jighly A, Makdis F, Wuletaw T, Hagras A, Uguru MI, Agbo CU. Genome-wide association study for agronomic and physiological traits in spring wheat evaluated in a range of heat prone environments. Theor Appl Genet. 2017;130:1819–1835. doi: 10.1007/s11032-006-9056-8. [DOI] [PubMed] [Google Scholar]
- 57.Lozada DN, Mason RE, Babar MA, Carver BF, Guedira GB, Merrill K, Arguello MN, Acuna A, Vieira L, Holder A, et al. Association mapping reveals loci associated with multiple traits that affect grain yield and adaptation in soft winter wheat. Euphytica. 2017;213(9):1–15. doi: 10.1007/s10681-017-2005-2. [DOI] [Google Scholar]
- 58.Sun C, Zhang F, Yan X, Zhang X, Dong Z, Cui D, Chen F. Genome-wide association study for 13 agronomic traits reveals the distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China. Plant Biotechnol J. 2017;15:953–969. doi: 10.1111/pbi.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salarpour M, Abdolshahi R, Pakniyat H, Heidari B, Aminizadeh S. Mapping quantitative trait loci for drought tolerance/susceptibility indices and estimation of breeding values of doubled haploid lines in wheat (Triticum aestivum) Crop Pasture Sci. 2021;72(7):500–513. [Google Scholar]
- 60.Sobhaninan N, Heidari B, Tahmasebi S, Dadkhodaie A, McIntyre CL. Response of quantitative and physiological traits to drought stress in the SeriM82/Babax wheat population. Euphytica. 2019;215:1–5. [Google Scholar]
- 61.Tahmasebi S, Heidari B, Pakniyat H, McIntyre CL. Mapping QTLs associated with agronomic and physiological traits under terminal drought and heat stress conditions in wheat (Triticum aestivum L.) Genome. 2016;60(1):26–45. doi: 10.1139/gen-2016-0017. [DOI] [PubMed] [Google Scholar]
- 62.Heidari B, Sayed-Tabatabaei BE, Saeidi G, Kearsey M, Suenaga K. Mapping QTL for grain yield, yield components, and spike features in a doubled haploid population of bread wheat. Genome. 2011;54(6):517–527. doi: 10.1139/g11-017. [DOI] [PubMed] [Google Scholar]
- 63.Wu KC, Wei LP, Huang CM, Wei YW, Cao HQ, Xu L, Li YR. Transcriptome reveals differentially expressed genes in Saccharum spontaneum GX83-10 leaf under drought stress. Sugar Tech. 2018;20(6):756–764. [Google Scholar]
- 64.Lambers H, Ribas-Carbó M. From cell to ecosystem. loan/open shelves. Plant Respir. 2005:1–15. 10.1007/1001-4020-3589-1006.
- 65.Florez-Sarasa ID, Bouma TJ, Medrano H, Azcon-Bieto J, Ribas-Carbo M. Contribution of the cytochrome and alternative pathways to grown respiration and maintenance respiration in Arabidopsis thaliana. Physiol Plant. 2010;129:140–151. doi: 10.1111/j.1399-3054.2006.00796.x. [DOI] [Google Scholar]
- 66.Sánchez-Martín J, Canales FJ, Tweed JK, Lee MR, Rubiales D. Fatty acid profle changes during gradual soil water depletion in oats suggests a role for jasmonates in coping with drought. Front Plant Sci. 2018;9:1–17. doi: 10.3389/fpls.2018.01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alipour H, Abdi H, Rahimi Y, Bihamta MR. Dissection of the genetic basis of genotype-by-environment interactions for grain yield and main agronomic traits in Iranian bread wheat landraces and cultivars. Sci Rep. 2021;11(1):1–17. doi: 10.1038/s41598-021-96576-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marek Ž, Jana R, Katarína O, Brestiè M. Osmotic adjustment in winter wheat varieties and its importance as a mechanism of drought tolerance. Cereal Res Commun. 2009;37:569–572. doi: 10.1556/CRC.37.2009.Suppl.4. [DOI] [Google Scholar]
- 69.Zadehbagheri M, Azarpanah A, Javanmardi S. Proline metabolite transport an efficient approach in corn yield improvement as response to drought conditions. Nature. 2014;566:76–485. doi: 10.5829/idosi.aejaes.2014.14.05.12328. [DOI] [Google Scholar]
- 70.Hayat S, Hayat Q, Alyemeni MN, Wani AS, Pichtel J, Ahmad A. Role of proline under changing environments: a review. Plant Signal Behav. 2012;7:1456–1466. doi: 10.4161/psb.21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carvalho K, de Campos MKF, Domingues DS, Pereira LFP, Vieira LGE. The accumulation of endogenous proline induces changes in gene expression of several antioxidant enzymes in leaves of transgenic Swingle citrumelo. Mol Biol Rep. 2013;40:3269–3279. doi: 10.1007/s11033-012-2402-5. [DOI] [PubMed] [Google Scholar]
- 72.Maleki A, Jalal S, Shekari F. Inheritance of proline content in bread wheat (Triticum aestivum L.) under rainfed conditions. J Food Agric Environ. 2010;8:155–157. [Google Scholar]
- 73.Mwadzingeni L, Shimelis H, Tesfay S, Tsilo TJ. Screening of bread wheat genotypes for drought tolerance using phenotypic and proline analyses. Front Plant Sci. 2016;7:1276. doi: 10.3389/fpls.2016.01276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kang S, Gu B, Du T, Zhang J. Crop coefficient and ratio of transpiration to evapotranspiration of winter wheat and maize in a semi-humid region. Agric Water Manag. 2003;59(3):239–254. [Google Scholar]
- 75.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 77.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Zhang Z. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- 78.Rabieyan E, Bihamta MR, Moghaddam ME, Mohammadi V, Alipour H. Genome-wide association mapping for wheat morphometric seed traits in Iranian landraces and cultivars under rain-fed and well-watered conditions. Sci Rep. 2022;12(1):1–21. doi: 10.1038/s41598-022-22607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rabieyan E, Bihamta MR, Moghaddam ME, Mohammadi V, Alipour H. Genome-wide association mapping and genomic prediction for pre-harvest sprouting resistance, low α-amylase and seed color in Iranian bread wheat. BMC Plant Biol. 2022;22(1):1–23. doi: 10.1186/s12870-022-03628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. A summary of LD observed among marker pairs and the number of significant marker pairs per genome and chromosome. Supplementary Table 2. Climatic data in the studied environments. Supplementary Table 3. Overview on the landraces and cultivars of Iranian wheat studied. Supplementary Fig 1. The KEGG pathway of oxidative phosphorylation. Supplementary Fig 2. The KEGG pathway of fatty acid elongation. Supplementary Fig 3. The KEGG pathway of metabolic pathways.
Data Availability Statement
The datasets generated and analyzed during the current study are available in Supplementary 2.