Abstract
Two consecutive nosocomial outbreaks of parainfluenza 3, in which 5 of 15 infected patients died, occurred in an adult bone marrow transplant unit. Parainfluenza 3 strain variation was assessed by reverse transcription-PCR sequencing of part of the parainfluenza 3 F gene, including the noncoding region, directly from clinical samples. Sequence data from the outbreaks were compared with those from 15 other parainfluenza 3 isolates circulating concurrently in the community; altogether, 13 strains which fell into three lineages were identified. Four immunosuppressed patients shed virus persistently for between 1 and 4 months without change in sequence. The first outbreak lasted 4 months and involved three parainfluenza 3 strains, and one persistently infected patient was implicated as the source of infection for three others. The second outbreak lasted for 1 month but involved only one strain. These data indicate that introduction of community parainfluenza 3 strains to the bone marrow transplant unit was followed by person-to-person transmission within the unit rather than reintroduction of virus from the community.
Parainfluenza viruses are well recognized as a cause of respiratory illness in children, ranging from mild upper respiratory tract symptoms to croup and pneumonia (5, 6). Almost all children encounter these viruses within the first few years after birth, but immunity is incomplete and reinfections occur throughout life. In the immunocompetent adult, a mild upper respiratory tract illness is the usual consequence of such reinfection, but lower respiratory tract disease in the immunocompromised adult is increasingly accepted as a cause of serious morbidity and mortality, especially in bone marrow transplant (BMT) patients (16, 22, 23). Of the parainfluenza viruses, type 3 (PIV3) seems to have the highest virulence, since PIV3-induced pneumonia has a mortality of about 40 to 50% in adult BMT patients (16, 23).
PIV3 infections in England and Wales are seasonal, occurring between May and September each year (11). Such community-wide PIV3 epidemics are known to be reflected in nosocomial outbreaks of PIV3 infection in pediatric wards (15), but so far there are no reports of nosocomial outbreaks in adults nor in immunosuppressed individuals. We now describe the molecular epidemiology of two consecutive outbreaks of PIV3 infection among adult patients in the BMT unit at the Hammersmith Hospital, London, United Kingdom. Respiratory samples from patients involved in the outbreaks were examined by sequence analysis for PIV3 strain variation, and the sequences were compared with the sequences of PIV3 strains concurrently circulating in the community in an attempt to clarify routes of nosocomial transmission and especially the role of shedding of PIV3 by immunosuppressed BMT patients. Such epidemiological information is essential for the planning of infection control measures to limit nosocomial spread of PIV3 infection in immunocompromised individuals.
(The results in this paper were given as oral presentations at meetings of the Society for General Microbiology/European Group for Rapid Viral Diagnosis, London, United Kingdom, January 1997, the European Group for Blood and Marrow Transplantation, Aix-les-Bains, France, March 1997, and the European Society for Clinical Virology, Bologna, Italy, September 1977.)
MATERIALS AND METHODS
Patient population.
Fifteen PIV3-infected immunosuppressed patients (P1 to P15) (13 BMT patients [3 autograft, 4 HLA-identical sibling donor, 1 HLA-mismatched related donor, and 5 volunteer unrelated donor (VUD) BMT patients] and 2 patients suffering hematological malignancies [mean age, 36.7 years; range, 17 to 64 years]) from the two consecutive outbreaks in the adult BMT unit at the Hammersmith Hospital were studied. The BMT patients, for whom fuller clinical details are reported elsewhere (12), became infected with PIV3 at a mean of 50 days (range, 7 to 153 days) after transplant, i.e., at a time when the patients were still severely immunosuppressed. Wards 1, 2, and 3 were involved but were geographically quite separate. The same medical staff visited all three wards, but nursing staff remained in their own ward. Ward 1 is the isolation ward for the hospital, and patients are each isolated in a single room with negative-pressure ventilation. Ward 2 is the main BMT ward. Here patients receive mainly allogeneic BMT and are nursed in, but not confined to, single rooms with positive-pressure ventilation. Ward 3 is a hematology ward consisting of single rooms and one four-bedded bay; in this ward, patients usually have hematological malignancies or have received an autologous BMT. The temporal relationship of the outbreaks to the annual community-wide epidemic in England and Wales is shown in Fig. 1. Since the incubation period for PIV3 is 2 to 3 days (21), we defined a community-acquired infection as one in which PIV3 was recovered from the patient within 4 days of admission to the hospital and a nosocomial infection as one in which PIV3 was recovered more than 4 days after admission.
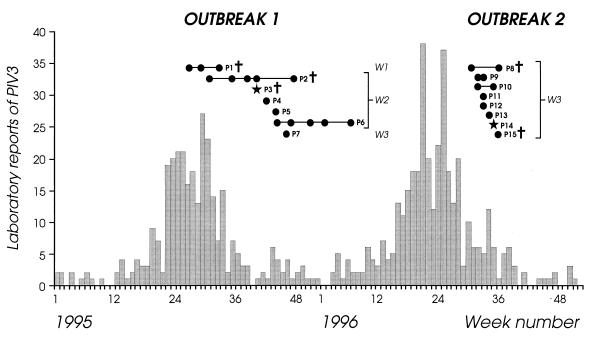
FIG. 1.
Timing of PIV3 isolates from patients in outbreaks 1 and 2 in relation to the prevalence of PIV3 in England and Wales. The data for the laboratory isolates are derived from reports made to the Public Health Laboratory Service Communicable Disease Surveillance Centre and are shown as a histogram. The timing of the outbreaks and the length of shedding of PIV3 by different patients (P1 to P15) are indicated. Symbols: •, clinical sample sequenced; ★, PIV3 detected, but insufficient material for sequence; †, patient died; W, patient’s ward.
The first outbreak (Fig. 1, outbreak 1) lasted for 4 months in 1995 (week 27 to week 46, in which the final case was detected) and involved seven BMT patients (P1 to P7, including two sibling donor [P3 and P7], one mismatched related donor [P2], and four VUD [P1 and P4 to P6] BMT patients), three of whom (P1 to P3) died. P1 to P6 met the definition for a nosocomial case, whereas P7 met the definition for a community-acquired case. The first patient, P1, was already isolated in ward 1 at week 27, at the start of his PIV3 infection; he remained symptomatic and shed virus for 6 weeks. At week 31, while P1 was still shedding virus, P2 in ward 2 became infected with PIV3, remained symptomatic, and shed virus for 17 weeks, i.e., until week 48. An additional four patients (P3 to P6) became infected with PIV3 between weeks 40 and 46, and P6 shed virus while remaining symptomatic for 16 weeks. All five patients (P2 to P6) in ward 2 had been nursed in single rooms, although ambulatory patients could meet together in the day room. No other patients became infected in ward 2 after week 44, at which time those patients who were still in ward 2 and shedding virus, namely, P2, P5, and P6, were transferred to the isolation ward (ward 1) and at the same time at which ward 2 was closed to admissions. The final patient to become infected (P7) was discharged from ward 2 15 days before a brief readmission to ward 3 at week 46 with a PIV3 upper respiratory tract infection.
The second outbreak (Fig. 1, outbreak 2) lasted for 1 month in 1996 (week 31 to week 36) and involved eight patients (P8 to P15), five of whom had been nursed in the four-bedded bay in ward 3. Six of the eight patients in this outbreak were BMT patients (three autograft [P12, P14, and P15], two sibling [P9 and P13], and one VUD [P8] BMT patients), and the remaining two had hematological malignancies (P10 and P12), acute myeloid leukemia and multiple myeloma, respectively. Two of the BMT patients (P8 and P15) died. The index case, P8, was admitted at week 31 from the community to the four-bedded bay in ward 3 with a PIV3 infection. He was transferred to ward 1 immediately after diagnosis of PIV3 infection but remained symptomatic and shed virus for 5 weeks until his death. An additional seven patients (P9 to P15) in ward 3 became infected in rapid succession over the next 5 weeks; P9, P10, P11, P12, P14, and P15 met the definition for nosocomial infection. The remaining patient, P13, was discharged from ward 3 the day after P8 was admitted but returned to the ward to visit three times, the last being 4 days prior to readmission in week 34 with PIV3 infection. Although all PIV3-infected patients were transferred to the isolation ward (ward 1) within 24 h of diagnosis or discharged, the outbreak did not stop until the closure of ward 3 in week 37.
To assess variability in PIV3 strains circulating in the community at the same time as the two outbreaks, 15 individuals with PIV3 infections who were patients in four other London hospitals were selected as a source of control community PIV3 isolates. Eight cases (C1 to C8) occurred between May and July 1995 and served as controls for outbreak 1, and the other seven cases (C9 to C15) occurred between March and August 1996 and served as controls for outbreak 2.
Laboratory diagnosis of PIV3 infection.
At the Hammersmith Hospital, diagnosis of PIV3 infection was made by direct immunofluorescence of samples of sputum or bronchoalveolar lavage fluid by using fluorescein isothiocyanate-conjugated PIV3-specific monoclonal antibody (catalog no. K6104; Dako Diagnostics, Ely, Cambridgeshire, United Kingdom) and/or isolation of PIV3 from these samples in primary rhesus monkey kidney cells (European Cell Culture Collection, Porton, Salisbury, United Kingdom). The community control isolates of PIV3 were identified at the four other London hospitals from either nasopharyngeal aspirates or nasopharyngeal swabs by using similar methods.
Reverse transcription (RT)-PCR and sequence analysis.
Strain variation was assessed by sequence analysis of cDNA amplified by RT-PCR from a portion of the PIV3 F gene comprising part of the coding region and the proximal 5′ noncoding leader region. Original samples and isolates were stored at −70°C or in liquid nitrogen before investigation. Control material for the development of the RT-PCR and reference material for the sequencing reactions consisted of a laboratory-adapted PIV3 strain (Colindale/1/66) which was grown in primary monkey kidney and African green monkey (Vero) cells. Virus-infected cultures were harvested when an 80 to 100% cytopathic effect was observed (3 to 5 days postinoculation) by scraping cells into the tissue culture fluid to form a cell suspension which was then sonicated and stored at −70°C.
For the RT-PCR, RNA was extracted from 200 μl of patients’ samples or control PIV3-infected tissue culture fluid by using guanidinium thiocyanate (2). Viral RNA was eluted in 30 μl of water, and for cDNA synthesis, 22.2 μl of RNA was made up to a final volume of 40 μl containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 7.5 mM MgCl2, 1.5 mM each deoxynucleoside triphosphate, 25 ng of random primer (pdN)6 (Pharmacia, St. Albans, United Kingdom), 1.6 U of RNasin (Promega, Southampton, United Kingdom), and 200 U of Moloney murine leukemia virus transcriptase (Gibco BRL, Paisley, United Kingdom). The reaction mixture was incubated at room temperature for 10 min and then at 37°C for 45 min. Samples were then heated to 100°C for 5 min and cooled on ice. For the single-round PCR, 20 μl of cDNA product was made up to a final volume of 100 μl containing 2 mM MgCl2, 1.5 U of Taq polymerase (Gibco BRL), and 250 pmol each of two primers targeted to the 5′ noncoding and proximal coding region of the F gene of the prototype PIV3 strain Wash/47885/57 (9) (5′-GTCAATACCAACAACTATTAGC-3′ [genome sense, bases 38 to 60] and 5′-TAACCAATACACCTACATGC-3′ [mRNA sense, bases 296 to 316]). Each sample was overlaid with mineral oil and denatured at 95°C for 2 min, followed by 35 cycles of amplification in a DNA thermal cycler (Genetic Research Instrumentation, Essex, United Kingdom); each cycle consisted of denaturation at 94°C for 1 min, reannealing at 56°C for 1 min, and extension at 72°C for 1 min. Following the PCR, the reaction products were run on a 1.2% agarose gel (Seakem; Flowgen, Lichfield, Staffordshire, United Kingdom), and the amplicon of 237 bp was visualized by using ethidium bromide, excised, and purified on a silica matrix (Kristal; Stratech, Luton, Bedfordshire, United Kingdom). After purification, the nucleotide sequence was determined by using the Dye Deoxy Terminator Method (Applied Biosystems, Foster City, Calif.) and an Applied Biosystems 373A sequencer. All the samples were sequenced in both the forward and reverse orientations in duplicate. Neighbor-joining phylogenetic analysis (20) was performed following the alignment of the sequences of the 5′ noncoding and coding regions of the F gene of PIV3 by using the program Megalign version 1.03 (DNASTAR Inc., Madison, Wis.).
RESULTS
General analysis of PIV3 isolates from outbreaks and the community.
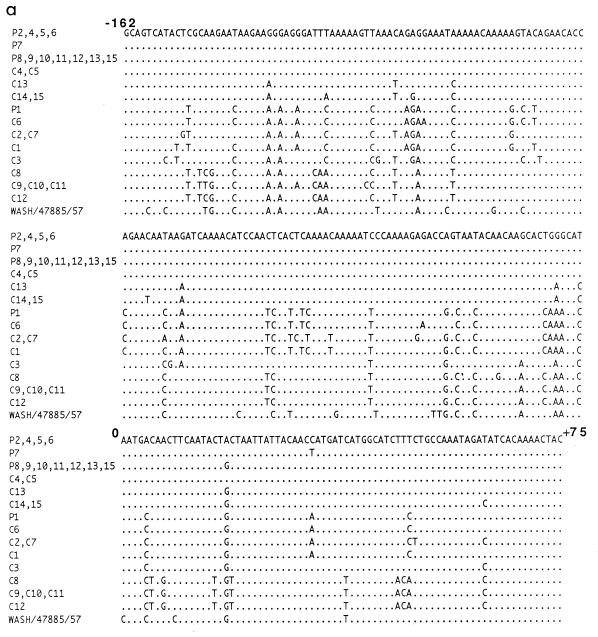
The PIV3 sequences obtained from the samples from patients were compared with those from control community PIV3 isolates circulating in London at the time of the hospital outbreaks, i.e., 1995 and 1996. Altogether, samples or viruses from 28 individuals were available for analysis. RT-PCR was performed on samples from 13 of the 15 patients from the two outbreaks (samples from P3 and P14 were insufficient for molecular analysis) and on all 15 of the control community isolates. In each case, the RT-PCR yielded a product of the expected 237 bp which spanned the 5′ noncoding leader and the proximal coding regions of the F gene (positions −162 to +75 with respect to the AUG start codon).
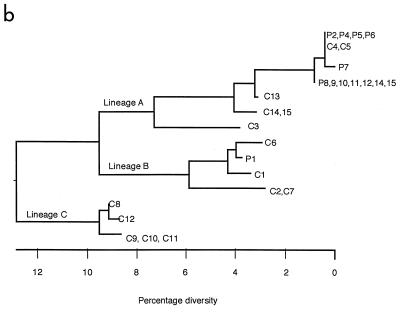
Sequence analysis of the RT-PCR products revealed that all sequences were colinear, with no insertions or deletions, and that there were no nonsense mutations within the 75-bp coding region (Fig. 2a). The sequences varied from the published F gene sequence of the prototype strain Wash/47885/57 (9) by 30 to 45 bases (13 to 19%) (Fig. 2a). Altogether, there were 13 different PIV3 sequences or strains which clustered into three lineages (lineages A, B, and C) (Fig. 2b). The maximum distance between the most divergent sequences was 16.9% (40 bp), and within lineages, the nucleotide variation was between 2 and 11% (5 to 26 bp). The clustering pattern of strains was maintained regardless of whether the coding region sequence was included in the sequence alignment. Nucleotide variation within the 5′ noncoding leader region was higher than that within the proximal coding region of the F gene (19.9% compared with 15.8%). There was a high degree of homology (88%) in the predominantly hydrophobic 25-amino-acid stretch of coding region. Within each lineage, amino acids were 100% homologous, but there were characteristic differences between lineages. Amino acid substitutions were noted at 3 of 25 positions, namely, at positions 2, 3, and 17, all of which lie in the signal sequence of the F protein. In comparison to lineage A, lineage B contained coding changes leading to amino acid substitutions at positions 2 and 17 whereas lineage C contained changes at all three positions.
FIG. 2.
(a) Sequence analysis of community control PIV3 isolates and patient samples. Nucleotides are numbered with respect to the AUG start codon of the F gene (18). (b) Phylogenetic tree of the 237-bp region of the F gene showing relationships between outbreak strain sequences and control community PIV3 isolate sequences. The scale beneath the tree shows the percentage diversity between the sequences.
Analysis of PIV3 isolates from outbreak 1, in 1995.
Sequence analysis of the 237-bp amplicons from the 16 clinical samples from P1, -2, -4, -5, -6, and -7 in outbreak 1 (Fig. 1) yielded three different sequences (Fig. 2). All three samples taken from P1 over a 6-week period gave an identical sequence which fell into lineage B, whereas the sequences from samples from P2, -4, -5, -6, and -7 fell into lineage A, which differed by a maximum of 14.8% (35 bp). All 12 samples from P2, -4, -5, and -6 yielded an identical sequence; these samples were collected over a 7-month period during which P2 and P6 shed PIV3 for 17 and 16 weeks, respectively. The sequence obtained from the single sample from P7 differed from that of P2, -4, -5, and -6 by a single base in the coding region (Fig. 2a).
Analysis of control community PIV3 isolates from 1995.
The eight control community isolates C1 to C8 gave six different sequences which differed by a maximum of 15.6% (37 bp) and fell into lineages A, B, and C. Two sequences from C2 and C7 obtained from specimens taken on the same day in two separate hospitals were identical, as were two sequences from two young siblings (C4 and C5) infected at the same time (Fig. 2b, lineages B and A, respectively). The sequence from C4 and C5 was identical to the sequence amplified from P2, -4, -5, and -6 in outbreak 1. No PIV3 strain identical to that which infected P1 was found in any of the control individuals, although closely related strains differing by only 3 and 4 bases were detected in samples from C1 and -6, respectively.
Analysis of PIV3 isolates from outbreak 2, in 1996.
Sequence analysis of the 237-bp amplicons from 11 clinical samples from P8, -9, -10, -11, -12, -13, and -15 in outbreak 2 (Fig. 1) yielded the same sequence for all cases which fell into lineage A (Fig. 2b). This PIV3 strain was closely related to the two strains in lineage A which were identified in outbreak 1. Thus, it differed in the coding region by 1 base from the strain obtained from P2, -4, -5, and -6 and by 2 bases from the strain obtained from P7.
Analysis of control community PIV3 isolates from 1996.
The seven control community isolates from C9 to C15 gave four different sequences which differed by a maximum of 14.8% (35 bp) and fell into lineages A and C (Fig. 2b). C14 and C15 yielded a single strain with a sequence in lineage A and that came from an outbreak in a neonatal unit. Similarly C9, -10, and -11 yielded a single strain with a sequence in lineage C and that came from an outbreak in a different hospital. No PIV3 strain identical to that which infected the patients in outbreak 2 was found in any of the control individuals, although closely related strains differing by only 4 and 7 bases were detected in samples from C13 and from C14 and C15, respectively.
DISCUSSION
In this study, we undertook the molecular investigation of two consecutive nosocomial outbreaks of PIV3 infection in a BMT unit with the aim of clarifying routes of viral transmission. PIV3 strain variation was assessed by analyzing the nucleotide sequence of a portion of the F gene, including the noncoding region. This region was chosen because it has a high degree of sequence diversity relative to the other sequenced parts of the PIV3 genome, including the HN (hemagglutinin-neuraminidase) gene (7) and the rest of the F gene (8, 18). The expected high variability of the chosen region was amply confirmed since sequence analysis of the outbreak and control community PIV3 isolates from the United Kingdom from 1995 and 1996 revealed 13 strains that differed as much from each other as they did from strains isolated between 1957 and 1987 outside Europe (8, 18). In the United Kingdom, a minimum of three different PIV3 lineages were circulating concurrently, each with a unique amino acid sequence in the predominantly hydrophobic signal sequence of the F protein. This sequence varied at a maximum of three amino acid positions, confirming the findings of Prinoski et al. (18). Cocirculation of four PIV3 lineages (isolates with distinct sequences) has also been described for a shorter period in a more limited geographical location, namely a 2-month outbreak of PIV3 in a children’s ward (15). The observed sequence divergence described here is not unexpected since other members of the paramyxovirus and orthomyxovirus groups, i.e., measles and respiratory syncytial viruses and influenza virus, also cocirculate as several separate evolutionary lineages (4, 10, 19).
With respect to the epidemiology of the two nosocomial outbreaks in the Hammersmith Hospital adult BMT unit, these might have resulted from multiple introductions of different strains from the community to the unit, presumably via staff or visitors, or alternatively from a single introduction from the community followed by person-to-person transmission within the unit. Considering the 1995 BMT outbreak in detail, there were three separate introductions, each of a different PIV3 strain to a different patient on three different wards. However, secondary cases were seen only in ward 2. Thus, 9 weeks after the start of the infection of P2, this patient was still on ward 2 shedding PIV3 when a further three inpatients became infected with the same strain. At this time, the community-wide PIV3 epidemic had ceased (Fig. 1), strengthening the conclusion that the chronically infected patient was the source of these subsequent infections. In the case of the 1996 BMT unit outbreak, a single PIV3 strain was introduced to ward 3 from the community by an infected patient, and an additional six patients became infected with this same strain. Although it was unclear whether the infection was acquired in hospital or the community for one of these six cases, overall the evidence strongly suggested person-to-person transmission on the ward. Such transmission of PIV3 has been demonstrated previously by Karron et al. (15), who investigated a PIV3 outbreak on a pediatric ward. However, our data additionally implicate prolonged shedding of virus by an immunosuppressed patient as a major factor in the first outbreak. In fact, symptoms together with virus shedding were observed for up to 4 months in several of the BMT patients despite treatment with ribavirin (12). Prolonged shedding of PIV3 is known to occur in immunosuppressed children (13) but has not previously been documented in adults.
As regards the extent of PIV3 strain diversity in the London community during 1995 and 1996, in two instances identical strains from different locations were identified, namely, (i) strains from the 1995 Hammersmith Hospital BMT outbreak and two siblings at another hospital and (ii) two community isolates from different hospitals. Overall, 13 strains were identified, including the two clusters of identical strains from the BMT unit outbreaks. However, strain variation was certainly underestimated since we inadvertently sampled three other clusters of identical strains, namely, strains from two neonatal unit outbreaks and strains from two siblings infected at the same time. Full assessment of PIV3 strain diversity will require a large prospective study in the same locality over several years.
One of the factors involved in viral strain diversity is the natural evolution rate of individual viruses. The rate of nucleotide substitution in the relatively variable region of the PIV3 genome under investigation in this study seems intrinsically low, being about 1 to 2 bases per year, assuming a rate of change of 5 × 10−3 to 7 × 10−3 bases per annum (18). This was confirmed in our study, since the 237-bp region sequenced did not change in the two BMT patients who shed virus for 4 months. The fact that these individuals were immunosuppressed probably did not influence the rate of nucleotide substitution since the noncoding region and signal sequence of the F gene are unlikely to be subject to selective immune pressure. In the context of this intrinsically low rate of change, it is interesting to note that, with the exception of a strain from one patient, the outbreaks at the Hammersmith Hospital involved very closely related strains from the same lineage. Thus, the strain infecting patients on ward 2 in 1995 differed by just 1 base from that which infected the patient admitted to ward 3 later in that year and by a different base from that reintroduced from the community to ward 3 in 1996, suggesting the natural evolution of one PIV3 strain persisting locally. It is certainly possible that the 1995 PIV3 strain persisted in the London community until 1996, since persistent asymptomatic shedding of parainfluenza viruses has been described previously in normal individuals (17) and might account for overwintering of the virus.
Finally, considering the implications of our study for hospital infection control policies, prolonged shedding of PIV3 by immunosuppressed patients can clearly be a major factor in nosocomial person-to-person transmission of virus. Although exact modes of transmission are not easily identified in a retrospective study, transmission of virus by secretions on hands (1) and fomites (3), as well as via large droplets, are the most probable routes, by analogy with respiratory syncytial virus (14). Infection control measures involving strict hand washing and wearing of gloves and aprons were rigorously adhered to in both outbreaks, and decontamination of surfaces was instituted. However, both outbreaks 1 and 2 were terminated only by closure of the relevant wards together with transfer of infected patients to the isolation ward. The protracted time course of the outbreaks described here and the high mortality associated with PIV3 infection in BMT patients suggest that stringent infection control measures should be implemented as soon as a single case is identified in a ward, particularly if the patient is immunosuppressed and other such patients are nearby.
ACKNOWLEDGMENTS
We thank the diagnostic virology departments of St. Mary’s, St. Thomas’, Queen Charlotte’s, and St. George’s Hospitals for the provision of PIV3 isolates. We also thank Dan Dedman of the respiratory section of the Communicable Disease Surveillance Centre for provision of the aggregated laboratory reports of PIV3 in England and Wales in 1995 and 1996.
This study was supported by the Public Health Laboratory Service, London, United Kingdom, and the Royal Postgraduate Medical School, London, United Kingdom.
REFERENCES
- 1.Ansari S A, Springthorpe V S, Sattar S A, Rivard S, Rahman M. Potential role of hands in the spread of respiratory virus infections: studies with human parainfluenza 3 and rhinovirus 14. J Clin Microbiol. 1991;29:2115–2119. doi: 10.1128/jcm.29.10.2115-2119.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boom R, Sol C J, Salimans M M, Jansen C L, Wertheim van Dillen P M, Van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:493–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady M T, Evans J, Cuartas J. Survival and disinfection of parainfluenza viruses on environmental surfaces. Am J Infect Control. 1990;18:18–23. doi: 10.1016/0196-6553(90)90206-8. [DOI] [PubMed] [Google Scholar]
- 4.Cane P A, Mathews D A, Pringle C R. Analysis of relatedness of subgroup A respiratory syncytial viruses isolated worldwide. Virus Res. 1992;25:15–22. doi: 10.1016/0168-1702(92)90096-r. [DOI] [PubMed] [Google Scholar]
- 5.Chanock R M, Bell J A, Parrott R H. Natural history of parainfluenza infection. Perspect Virol. 1961;2:126–138. [Google Scholar]
- 6.Clarke S K R. Parainfluenza virus infections. Postgrad Med J. 1973;49:792–797. doi: 10.1136/pgmj.49.577.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coelingh K V W, Winter C C, Murphy B R. Nucleotide and deduced amino acid sequences of hemagglutinin-neuraminidase genes of human type 3 parainfluenza viruses from 1957–1983. Virology. 1988;163:137–143. doi: 10.1016/0042-6822(88)90402-3. [DOI] [PubMed] [Google Scholar]
- 8.Coelingh K V W, Winter C C. Naturally occurring human parainfluenza 3 viruses exhibit divergence in amino acid sequence of their protein neutralization epitopes and cleavage sites. J Virol. 1990;64:1329–1334. doi: 10.1128/jvi.64.3.1329-1334.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Côté M-J, Storey D G, Yong Kang C, Dimock K. Nucleotide sequence of the coding and flanking regions of the human parainfluenza virus type 3 fusion glycoprotein gene. J Gen Virol. 1987;68:1003–1010. doi: 10.1099/0022-1317-68-4-1003. [DOI] [PubMed] [Google Scholar]
- 10.Cox N J, Bender C A. Molecular epidemiology of influenza viruses. Semin Virol. 1995;6:359–370. [Google Scholar]
- 11.Easton A J, Eglin R P. Epidemiology of parainfluenza 3 in England and Wales over a 10 year period. Epidemiol Infect. 1989;102:531–535. doi: 10.1017/s0950268800030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elizaga, J., E. Olavarria, K. M. Murphy, N. J. Philpott, J. F. Apperley, J. M. Goldman, and K. N. Ward. Unpublished data.
- 13.Fishaut M, Tubergen M D, McIntosh K. Cellular response to respiratory viruses with particular reference to children with disorders of cell-mediated immunity. J Paediatr. 1980;96:179–186. doi: 10.1016/s0022-3476(80)80799-2. [DOI] [PubMed] [Google Scholar]
- 14.Hall C B. The nosocomial spread of respiratory syncytial virus infections. Annu Rev Med. 1983;34:311–319. doi: 10.1146/annurev.me.34.020183.001523. [DOI] [PubMed] [Google Scholar]
- 15.Karron R A, O’Brien K L, Froehlich J L, Brown V A. Molecular epidemiology of a parainfluenza type 3 outbreak on a paediatric ward. J Infect Dis. 1993;167:1441–1445. doi: 10.1093/infdis/167.6.1441. [DOI] [PubMed] [Google Scholar]
- 16.Lewis V A, Champlin R, Englund J, Couch R, Goodrich J M, Rolston K, Przepiorka D, Mirza N Q, Yousuf H M, Luna M, Bodey G P, Whimbey E. Respiratory disease due to parainfluenza virus in adult bone marrow transplant recipients. Clin Infect Dis. 1996;23:1033–1037. doi: 10.1093/clinids/23.5.1033. [DOI] [PubMed] [Google Scholar]
- 17.Muchmore H G, Parkinson A J, Humphries J E, Scott E N, McIntosh D A, Scott L V, Cooney M K, Miles J A. Persistent parainfluenza virus shedding during isolation at the South Pole. Nature. 1981;289:187–189. doi: 10.1038/289187a0. [DOI] [PubMed] [Google Scholar]
- 18.Prinoski K, Côté M-J, Yong Kang C, Dimock K. Evolution of the fusion protein gene of human parainfluenza virus 3. Virus Res. 1991;22:55–69. doi: 10.1016/0168-1702(92)90089-r. [DOI] [PubMed] [Google Scholar]
- 19.Rota P A, Rota J S, Bellini W J. Molecular epidemiology of measles virus. Semin Virol. 1995;6:379–386. [Google Scholar]
- 20.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Tyrrell D A J, Bynoe M L, Birkum Petersen K, Sutton R N P, Pereira M S. Inoculation of human volunteers with parainfluenza viruses types 1 and 3 (HA2 and HA1) Br Med J. 1959;2:909–911. doi: 10.1136/bmj.2.5157.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wendt C H, Weisdorf D J, Jordan M C, Balfour H H, Hertz M I. Parainfluenza virus respiratory infection after bone marrow transplantation. N Engl J Med. 1992;326:921–926. doi: 10.1056/NEJM199204023261404. [DOI] [PubMed] [Google Scholar]
- 23.Whimbey E, Vartivarian S E, Champlin R E, Elting L S, Luna M, Bodey G P. Parainfluenza virus infection in adult bone marrow transplant recipients. Eur J Clin Microbiol. 1993;12:699–701. doi: 10.1007/BF02009383. [DOI] [PubMed] [Google Scholar]