Abstract
Sensory neurons in the dorsal root ganglia (DRG) convey somatosensory and metabolic cues to the central nervous system and release substances from stimulated terminal endings in peripheral organs. Sex-biased variations driven by the sex chromosome complement (XX and XY) have been implicated in the sensory-islet crosstalk. However, the molecular underpinnings of these male-female differences are not known. Here, we aim to characterize the molecular repertoire and the secretome profile of the lower thoracic spinal sensory neurons and to identify molecules with sex-biased insulin sensing- and/or insulin secretion-modulating activity that are encoded independently of circulating gonadal sex hormones. We used transcriptomics and proteomics to uncover differentially expressed genes and secreted molecules in lower thoracic T5-12 DRG sensory neurons derived from sexually immature three-week-old male and female C57BL/6J mice. Comparative transcriptome and proteome analyses revealed differential gene expression and protein secretion in DRG neurons in males and females. The transcriptome analysis identified, among others, higher insulin signaling/sensing capabilities in female DRG neurons; secretome screening uncovered several sex-specific candidate molecules with potential regulatory functions in pancreatic β cells. Together, these data suggest a putative role of sensory interoception of insulin in the DRG-islet crosstalk with implications in sensory feedback loops in the regulation of β-cell activity in a sex-biased manner. Finally, we provide a valuable resource of molecular and secretory targets that can be leveraged for understanding insulin interoception and insulin secretion and inform the development of novel studies/approaches to fathom the role of the sensory-islet axis in the regulation of energy balance in males and females.
Graphical Abstract:
Transcriptome and proteome analyses of DRG neurons in sexually immature three-week-old male and female mice revealed sex differences in cellular and molecular pathways relevant to the sensory neuron-islet crosstalk.
1. Introduction:
Sensory neurons in the dorsal root ganglia (DRG) arise from the neural crest cells and emerge from the intervertebral neural foramina to innervate the skin, bones, muscles and multiple internal organs. DRG neurons are pseudo-unipolar, with one axonal branch innervating peripheral tissues and another projecting to the spinal cord [1]. They comprise a variety of sensory subtypes that detect, transduce and respond to a myriad of physical, thermal and chemical stimuli [2]. Internal and external cues activate transduction receptors and ion channels on the sensory terminal endings and generate action potentials that propagate to the central nervous system (CNS) where signals are integrated, interpreted and relayed back to the target tissues via motor neurons [3]. Moreover, DRG neurons carry dual afferent-efferent functions, that is, in addition to their ability to convey to the CNS information arising from the tissues they innervate; these afferents also locally release neuromodulators such as calcitonin gene-related peptide (CGRP) and substance P (SP), in the target tissue [4]. DRG sensory neurons modulate multiple biological processes via the efferent pathway, including neurogenic inflammation [5], immunity [6], tissue repair and regeneration [7], and cell proliferation and function [8].
Although historically described for their roles in somatosensation and nociception [9], transient receptor potential vanilloid type 1 (TRPV1)-expressing neurons—an abundant DRG neuronal subtype—play critical roles in energy balance and lifespan [10]. These neurons innervate major metabolic tissues including the liver, adipose tissue and pancreas and express receptors for metabolic hormones such as insulin [11], leptin [12] and glucagon-like peptide (GLP-1) [13]. Surgical and chemical denervation of whole-body or pancreatic TRPV1+ neurons enhance insulin secretion and improves glucose clearance in male mice [14]. Worms and mice lacking TRPV1 exhibit extended lifespans and youthful metabolic profiles [15]. These beneficial outcomes are likely a direct consequence of CGRP downregulation in TRPV1 knockout mice [15]. Circulating levels of CGRP increase with age and treatment of old mice with CGRP receptor antagonist result in youthful metabolism due, in part, to alterations in insulin secretion [15]. Moreover, genetic ablation of CGRPα resulted in reduced lipid accumulation in adipose tissue and increased mitochondrial respiration from interscapular brown adipose tissue [16].
There are sex differences in the regulation of energy balance [17]; particularly at the level of pancreatic β-cell function [18]. Glucose-stimulated insulin secretion (GSIS) is higher in females—in mice [19], rats [20] and humans [21]. In healthy individuals, women exhibit superior GSIS compared to men [22–24]. The dysfunction of β-cell activity is also sex-biased. While obesity is more prevalent in women; type 2 diabetes is more prevalent in men [17]. Interestingly, while autoimmune conditions are known to affect more women than men, type 1 diabetes is exclusively more prevalent in men than women [25, 26]. Insulin sensitivity is different between men and women [27]. Sex differences are also evident in insulin sensitivity. Women tend generally to be more insulin sensitive than men even after adjustment for age and body mass index (BMI) [28]. Several tissues contribute to the overall sex-differential action of insulin, including adipose tissue [29] and hypothalamic neurons [30].
Sex differences in energy balance are largely attributed to the actions of sex steroid hormones, which can act directly via their receptors to modulate the activity of the CNS and various peripheral metabolic organs [25]. In pancreatic β cells, estrogens regulate insulin biosynthesis and enhance cell survival in metabolic stress conditions [31–33]; testosterone enhances GSIS in mouse and human islets in vitro [34, 35]; and both sex hormones regulate β-cell mass in vivo [36]. In addition to their direct action, we reported that male sex hormones regulate glucose clearance and GSIS via intricate gonadal-DRG-islet crosstalk [19]. Indeed, while chemical and surgical sensory denervation improved glucose excursion and insulin secretion in male mice, orchidectomy abrogated these effects. The action of male sex hormones in sensory modulation of GSIS is interpreted at least, in part, by their stimulatory action on the density of peri-islet of sensory CGRP+ in the adult male pancreas [19]. Beyond the action of gonadal sex hormones, sex differences may also arise as the sole consequence of the action of the sex chromosome complement (XX and XY) [17, 37]. Mounting evidence suggests that sex chromosomes play a role in glucose homeostasis, fatty liver, adiposity and feeding behavior—independently of the action of sex hormones [38]. We demonstrated that cell-autonomous sensory cues—potentially encoded by the sex chromosome complement—regulate GSIS differently in male and female mice [19].
Here, we paired genomics and proteomics to characterize the molecular repertoire and the secretome profile of lower thoracic DRG neurons harvested from sexually immature three-week-old male and female mice. Computational analysis uncovered differentially expressed genes in sensory neurons between sexes. Gene ontology revealed significant sex differences in epigenetic regulation, cell cycle, DNA replication, cell senescence, neuron projection and extension, and insulin signaling pathways. Several conventionally and unconventionally secreted proteins were differentially regulated in one sex compared to the other; some of these molecules have modulatory roles in insulin secretion. Collectively, we provide a valuable resource of molecular and secretory targets that can be leveraged for understanding sex differences in sensory modulation of energy balance via sensory-islet intercommunication.
2. Materials and Methods:
Animals:
All mice studied were on the C57BL/6J background (stock #000664, The Jackson Laboratory). Mice were housed in pathogen-free facilities and maintained on a 12-hour light/dark cycle in the Animal Care Facility at the Child Health Institute of New Jersey. All studies and protocols were approved by the Rutgers University Animal Care and Use Committee and were in accordance with the National Institutes of Health guidelines.
DRG culture:
DRG neurons located at vertebrae levels T5-12 were prepared as previously described [39]. Briefly, lower thoracic DRG were harvested from three-week-old mice and dissociated in collagenase (5mg/ml) and dispase (1mg/ml) at 37°C for 70 minutes, and then cultured in 6-well plates coated with Poly-D-lysine (100μg/ml) and laminin (10μg/ml). DRG neurons were plated at a density of 5x104 cells/well in Neurobasal media containing B27 (200mM), NGF (50 ng/ml), GDNF (2ng/ml) and AraC (10μM). After three days of axonal outgrowth, DRG neurons were incubated in HEPES solution containing 75mM KCL for 60 minutes. Cells were lysed and total RNAs were extracted for RNA sequencing. Supernatants were collected for secretome analysis by LC-MS/MS.
RNA sequencing:
RNA samples were quantified using Qubit 2.0 Fluorometer (ThermoFisher Scientific, Waltham, MA, USA) and RNA integrity was checked with 4200 TapeStation (Agilent Technologies, Palo Alto, CA, USA). Samples were initially treated with TURBO DNase (Thermo Fisher Scientific, Waltham, MA, USA) to remove DNA contaminants. rRNA depletion sequencing library was prepared by using QIAGEN FastSelect rRNA HMR Kit (Qiagen, Hilden, Germany). RNA sequencing library preparation uses NEBNext Ultra II RNA Library Preparation Kit for Illumina by following the manufacturer’s recommendations (NEB, Ipswich, MA, USA). Briefly, enriched RNAs are fragmented for 15 minutes at 94 °C. First- and second-strand cDNA are subsequently synthesized. cDNA fragments are end-repaired and adenylated at 3’ends, and universal adapters are ligated to cDNA fragments, followed by index addition and library enrichment with limited cycle PCR. Sequencing libraries were validated using the Agilent Tapestation 4200 (Agilent Technologies, Palo Alto, CA, USA), and quantified using Qubit 2.0 Fluorometer (ThermoFisher Scientific, Waltham, MA, USA) as well as by quantitative PCR (KAPA Biosystems, Wilmington, MA, USA). The sequencing libraries were multiplexed and clustered onto a flowcell. After clustering, the flowcell was loaded onto the Illumina HiSeq instrument according to the manufacturer’s instructions. The samples were sequenced using a 2x150bp Paired-End (PE) configuration.
RNA sequencing data analysis:
Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using Illumina bcl2fastq 2.20 software. The RNA-seq data analyses were performed as previously described [40, 41]. Briefly, the FASTQ files were aligned to mouse mm10 genome using STAR 2.7.7a [42] with the following parameters: ‘–runThreadN 40 –outFilterMultimapNmax 1 –outFilterMismatchNmax 3 –outFilterScoreMinOverLread 0.25 –outFilterMatchNminOverLread 0.25’. A Perl script was used to calculate the total number of mapped reads and the number of reads that aligned to the exons of each gene. The table of raw read counts was normalized, and the top 1000 variable genes were selected to perform the principal component analysis by DESeq2 [43] and to perform the distance between variables by pheatmap. The edgeR [44] and the Genewise Negative Binomial Generalized Linear Models with Quasi-likelihood Tests (glmQLFTest) were used to compare the gene expression levels between the groups. The false discovery rate (FDR) < 0.05 was used as the cutoff to identify differentially expressed genes (DEGs). The functional enrichment analysis was performed using g:Profiler (version_e106_eg53_p16_65fcd97) and Gene set enrichment analysis (GSEA) software [45]. The list of DEGs were used as the input files for g:Profiler. Briefly, the Benjamini-Hochberg FDR with a threshold of 0.05 was used to determine the statistical significance and carried out to detect the Gene Ontology (GO), Reactome, and KEGG pathway. The −log10 FDR values were used to generate the bar plot for GO enrichment analysis. The TPM values from edgeR were used as input files for the GSEA analysis. Briefly, the msigdb.v2022.1.Mm.symbols.gmt was used as gene sets database and Mouse-Gene_Symbol_Remapping_MsigDB.v2022.1.Mm.Chip was used as the chip platform. The parameters of ‘1000 permutation’, ‘collapse’ and ‘gene_set as permutation type’ were selected to run GSEA. The scripts used in this study are available at GitHub repository (https://github.com/Jerry-Zhao/DRG2023).
Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR):
cDNA synthesis was carried out by using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Cat# 4368814) with total RNA samples. The primers used were listed in the supplemental Figure 1A. The cycling parameters for qRT-PCR amplification reactions were: AmpliTaq activation at 95°C for 10min, denaturation at 95°C for 15s, and annealing/extension at 60°C for 1min (40 cycles). The Ct value of each selected gene was normalized by the Ct value of internal reference (β-actin) and the relative expression values were calculated according to the 2−ΔΔCt method. Finally, to analyze the correlation, the individual 8 gene’s log2 fold change value was used to calculate the coefficient of determination (R2) and to evaluate the concordance between qRT-PCR results and RNA-seq data.
LC-MS/MS-based proteomics for secretome samples.
Secretome samples were prepared using a protocol based on S-Trap micro-columns (Protifi, NY) [46]. Briefly, 600 ul HEPES cultured media from each sample were reduced and alkylated by incubating with 5% SDS (Sigma-Aldrich) and 10 mm DTT (Thermo Scientific) for 30 min at 37 °C and with 40 mm iodoacetamide (Thermo Scientific) for another 30 min at room temperature, respectively. Then 27.5% aqueous phosphoric acid was added into the sample with a 1:10 ratio followed by adding S-Trap binding buffer (90% MeOH, 100 mm triethylammonium bicarbonate (TEAB)) with a 1:7 ratio. Translucent protein samples were transferred into S-Trap column and cleaned four times using 250 ul of S-Trap binding buffer per each washing. After cleaning, 1 µg of sequencing-grade trypsin (Promega) in 20ul 100 mM TEAB buffer was added to the top of the column. It was allowed to digest overnight at 37 °C. Finally, the peptides were eluted by 40ul 50 mM TEAB, 40ul 0.2% formic acid (FA) and 40 μL of 50% acetonitrile (ACN). The eluted peptides were pooled for LC-MS/MS Analysis. LC-MS/MS was performed as previously described [47]. Briefly, Briefly, the peptides were analyzed on a Q Exactive Plus mass spectrometer (Thermo Scientific) with a nano-electrospray ion source. The peptides were separated on 60 cm x 50 μm ID with Jupiter 3 μm C18 material (Phenomenex)) with an integrated PicoTip emitter (New Objective) using a 2 hr LC gradient. A data-dependent acquisition mode was employed to automatically trigger the precursor scan and the MS/MS scans. For the full scan, a resolution of 35,000, 3 × 106 AGC target, and a maximum ion trap time of 50 ms were used. Top-10 precursors were isolated for fragmentation under normalized collision energy of 32 with an exclusion time of 30 s. Injection time, resolution, and AGC target were configured as 300 ms, 17,500, and 2 × 105, respectively.
LC-MS/MS Data Analysis:
MaxQuant software [48] was used for protein identification and quantification. The MS raw files were processed with MaxQuant (Version 1.6.2.10), and MS/MS spectra were searched by Andromeda search engine against the mouse UniProt database (fasta file dated July 14, 2020) with the following parameters: tryptic peptides with up to two missed cleavage sites; 10 ppm parent ion tolerance; 0.6 Da fragment ion mass tolerance; variable modification (methionine oxidation); fixed modification (cysteine carbamidomethyl). Search results were processed with MaxQuant and filtered with a false discovery rate (FDR) ≤ 1% at both protein and peptide levels. For label-free quantification (LFQ), the match between runs (MBR) function was activated with a matching window of 0.4 min and an alignment window of 20 min. Protein quantification was performed by using the LFQ function. The search result files “peptides.txt” and “proteinGroups.txt” were used for further data processing. Proteins detected in four or more samples were included in the downstream analysis. Protein level comparison was performed by the Student’s t-test, and the P-values were further corrected by multiple comparisons using the Benjamini-Hochberg method to obtain the FDRs. Proteins with an FDR < 0.1 were considered differentially expressed proteins. The functional enrichment analysis was performed using g:Profiler (version_e106_eg53_p16_65fcd97) and Gene set enrichment analysis (GSEA) software [45]. The differentially expressed proteins were converted into the gene ID and used as the input files for g:Profiler. Briefly, the Benjamini-Hochberg FDR with a threshold of 0.05 was used to determine the statistical significance and carried out to detect the GO, Reactome, and KEGG pathway. The −log10FDR values from edgeR were used as input files for the GSEA analysis. Briefly, the msigdb.v2022.1.Mm.symbols.gmt was used as gene sets database and Mouse-Gene_Symbol_Remapping_MsigDB.v2022.1.Mm.Chip was used as the chip platform. The parameters of ‘1000 permutation’, ‘Remap_Only’ and ‘Max_probe as collapsing mode for probe sets’ were selected to run GSEAPreranked.
3. Results:
Transcriptome and secretome profiling in male and female DRG neurons:
To gain insights into the transcriptome and secretome differences between male and female spinal sensory DRG neurons as they relate to insulin sensing and insulin secretion, we carried out RNA sequencing (RNA-Seq) and proteomics assays using lower thoracic DRG neurons known to project directly in the pancreas [49] and modulate islet β-cell activity [14, 19]. Briefly, DRG neurons located at vertebrae levels T5-12 were harvested from male and female C57BL/6J mice, dissociated and cultured as we previously described [19]. To identify sex-hormone-independent effects underlying sensory sex differences, we used neurons harvested from three-week-old—sexually immature—mice. After three days of culturing, the conditioned cell culture media DRG neurons were acutely incubated in serum-free depolarizing KCL (75 mM) solution to induce secretion for 60 minutes. After the incubation, the conditioned cell culture media was collected for secretome analysis, whereas DRG neurons were lysed and RNAs were extracted for the RNA-Seq experiment (Figure 1).
Figure 1: Schematic of the experimental plan.
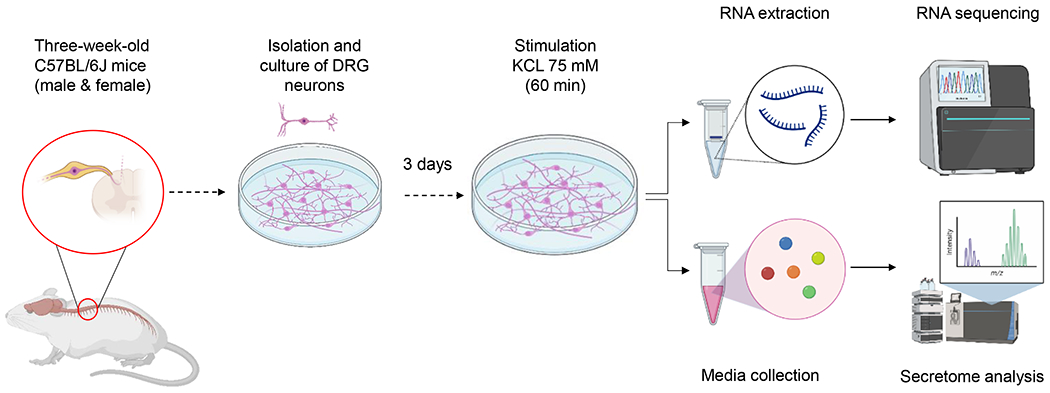
DRG neurons were harvested from three-week-old male and female C57BL/6J mice. After culture for three days, neurons were placed in serum-free HEPES media for one hour and then incubated with KCL 75 mM to enhance secretion for 1 hour. After stimulation, culture media was collected for secretome analysis and RNAs were extracted for sequencing.
Male and female DRG neurons exhibit distinct transcriptome profiles:
To determine the transcriptome differences between male and female DRG neurons, we carried out an RNA-seq experiment to unbiasedly assess gene transcription levels. We obtained 817 million high-quality RNA-Seq reads from sequencing five biological replicates of male and female DRG neurons (Supplemental Figure 1A). Pearson’s correlation analysis (Figure 2A) showed tight similarities among the biological replicates within each intragroup (Pearson’s correlation coefficient r > 0.99) and high intergroup dissimilarities (males versus females). To further assess the transcriptome profiles, we next performed a principal component analysis (PCA) and found that male and female datasets were segregated into two independent groups (Figure 2B), suggesting that male and female DRG neurons exhibit distinct transcriptome profiles. To identify genes that are differentially expressed between male and female DRG neurons, we quantified the expression levels of all mouse genes and compared them using edgeR method. As a result, we identified 403 genes that were differentially expressed (FDR < 0.05) between male and female DRG neurons, including 126 male-enriched genes and 277 female-enriched genes (Figure 2C and Supplemental Table 1). Furthermore, we found that these gene expression changes between male and female DRG neurons are consistent among the five biological replicates (Figure 2D). To verify the RNA-Seq results, we assessed the relative expression levels of a few genes using qRT-PCR and calculated the coefficient of determination R2 to assess the association between RNA-seq results and qRT-PCR results. This approach demonstrated a high correlation (R2 = 0.9864) between the RNA-seq and qRT-PCR datasets (Figure 2E and Supplemental Figure 1B–J), thus independently validating the RNA-seq results. Collectively, these results demonstrated that male and female DRG neurons exhibit distinct transcriptome profiles.
Figure 2: Male and female DRG neurons exhibit distinct transcriptome profiles.
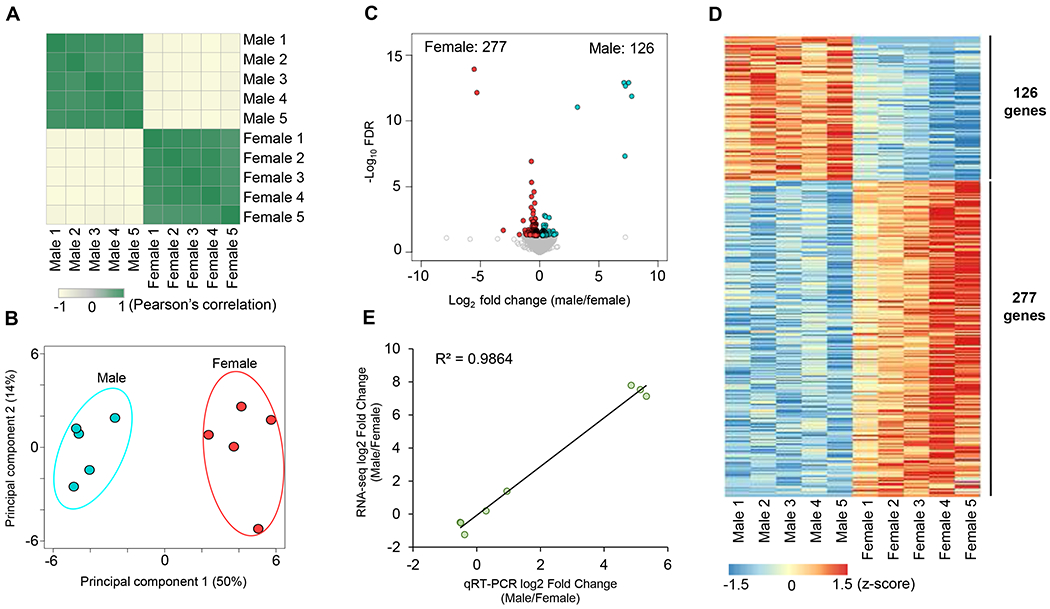
A. Heatmap of the Pearson’s correlation analysis of the gene expression profiles obtained by RNA sequencing of male and female DRG neurons. B. PCA plot of male and female DRG neurons RNA-Seq data. C. Volcano plots of male and female DRG neurons RNA-Seq data. The differentially expressed genes (FDR < 0.05) were highlighted in red for female-enriched genes and blue for male-enriched genes. D. Heatmap depicting differential gene relative expression in male versus female DRG neurons. E. Correlation of gene expression fold change (log2 fold change) between RNA-Seq (y-axis) and real-time qRT-PCR (x-axis) data.
Pathway analyses of transcriptome profiles of male and female DRG neurons:
To identify the biological pathways in DRG neurons that predominate in one sex relative to the other, we performed Gene Ontology (GO) enrichment analysis and Gene Set Enrichment Analysis (GSEA). We found that the male- and female-selective gene expression patterns were enriched in distinct GO biological pathways (FDR < 0.05, Supplemental Tables 2 and 3). In the 126 genes enriched in male DRG neurons, we observed an upregulation of chromatin modification and epigenetic regulation of gene expression pathways, including chromatin modifying enzymes, histone deacetylases (HDACs), chromatin organization and remodeling, DNA packaging complex and nucleosomal DNA binding (Figure 3A). Male DRG neurons were also enriched in genes implicated in cell cycle (Figure 3A and Supplemental Figure 2F), DNA replication (Figure 3A and Supplemental Figure 2I), G2/M checkpoints (Figure 3A and Supplemental Figure 2J), E2F-mediated regulation of DNA replication and DNA helicase activity (Figure 3A). Another male-specific cluster of genes highlighted the predominance of cytokine signaling in the immune system and signaling by interleukins—particularly interleukin-7 signaling (Figure 3A and Supplemental Figure 2H). Remarkably, cell senescence and senescence-associated secretory phenotype (SASP) pathways were enhanced in male DRG neurons (Figure 3A). Lastly, GO enrichment analysis also showed enhanced signaling by nuclear receptors in male DRG neurons, including estrogen receptor (ESR)-mediated signaling and estrogen-dependent gene expression pathways (Figure 3A and Supplemental Figure 2C). The 277 genes enriched in female DRG neurons, on the other hand, were enriched in GO biological pathways associated with insulin action, such as response to insulin hormone, insulin signaling pathway, phosphatidylinositol 3-kinase signaling and forkhead box O (FOXO) signaling pathway (Figure 3B and Supplemental Figure 3E–I). The transmembrane receptor protein serine/threonine kinase signaling pathway was also preponderant in female sensory neurons (Figure 3B and Supplemental Figure 3B). Furthermore, several gene regulatory networks were linked to aspects of glucose homeostasis, positive regulation of metabolic processes, cellular carbohydrate metabolic process and response to ketone (Figure 3B and Supplemental Figure 3A). Among differentially regulated GO biological pathways between sexes, we found that extracellular matrix organization, extracellular matrix assembly and collagen-containing extracellular matrix were upregulated in female DRG neurons (Figure 3B). Lastly, our analysis revealed female-specific enrichment of biological pathways related to neuron projection development, axon guidance, positive regulation of dendrite extension, neuron projection extension and axon extension involved in axon guidance (Figure 3B and Supplemental Figure 3C). Notably, GSEA analysis results in male versus female DRG neurons (Supplemental Figures 2 and 3 and Supplemental Tables 4 and 5) showed similar trends compared to the GO enrichment analysis. Taken together, these analyses indicated the existence of male- and female-selective gene expression patterns that modulate distinct biological pathways in DRG neurons with the highlight of the insulin-sensing feature as an important female-enriched signaling pathway in the regulation of DRG-islet crosstalk.
Figure 3: Gene ontology enrichment analysis of male and female DRG neurons.
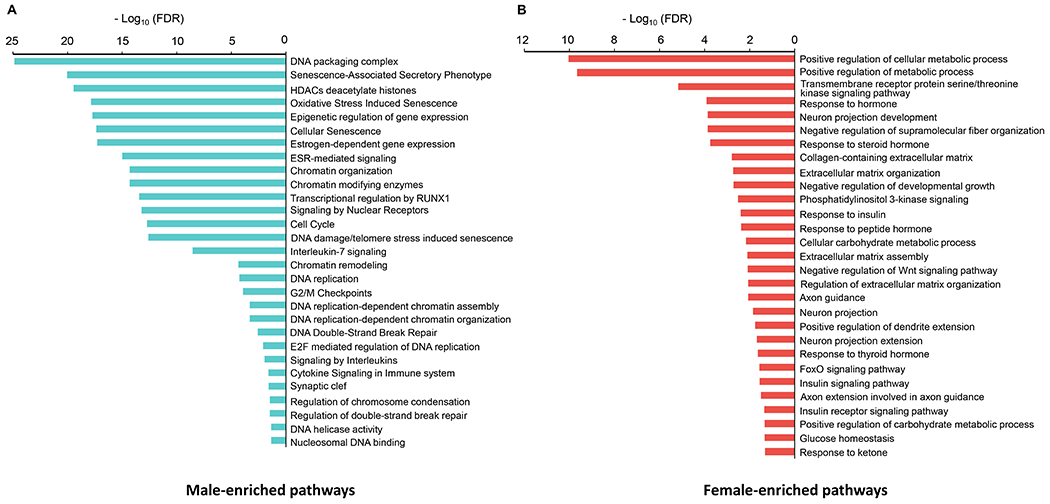
A. Gene ontology (GO) enrichment analysis in male DRG neurons, as compared to female DRG neurons. B. Gene ontology (GO) enrichment analysis in female DRG neurons, as compared to male DRG neurons. FDR < 0.05 was used as the cutoff criteria.
Male and female DRG neurons exhibit distinct secretome profiles:
To identify secretory proteins that are differentially regulated by male and female DRG neurons, we collected culture medium of male and female DRG neurons in depolarizing conditions (five biological replicates) and performed LC-MS/MS to characterize the secretome profiles (Figure 1). We detected 335 proteins in total (Supplemental Table 6). To determine the similarity of the secretome profiles, we carried out Pearson’s correlation analysis and showed a high correlation between pairs of the secretome of DRG neurons from the same-sex groups (Pearson’s correlation coefficient r > 0.99) and high divergence between opposite-sex groups (Figure 4A). The PCA analysis also showed a clear separation between male and female datasets (Figure 4B), which supports that male and female DRG neurons exhibit distinct secretome profiles. In addition, we compared the levels of the 335 secretory proteins between male and female datasets and identified 30 and 16 secretory proteins that were differentially enriched in male and female DRG secretomes, respectively (FDR < 0.1) (Figures 4C and 4D, Supplemental Table 6). Collectively, these results demonstrated that male and female DRG neurons exhibit distinct secretome profiles.
Figure 4: Male and female DRG neurons exhibit distinct secretome profiles.
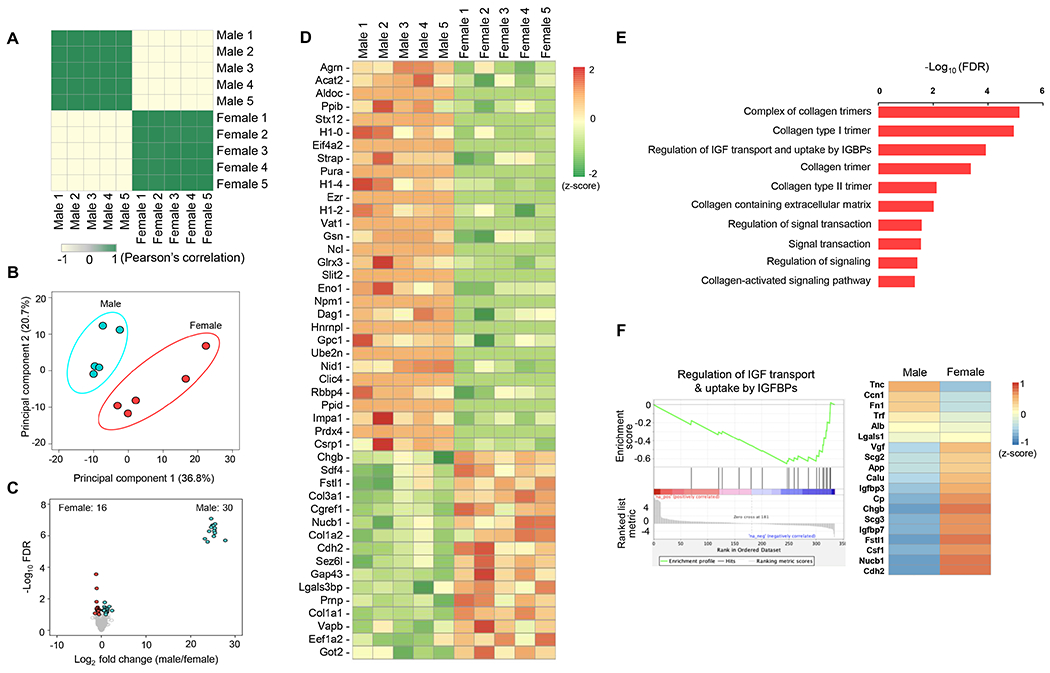
A. Heatmap of the Pearson’s correlation analysis of secretome profiles captured via proteomics of male and female DRG neurons. B. PCA plot showing the secretome data corresponding to male and female DRG neurons (five biological replicates). C. Volcano Plots of differentially secreted proteins (FDR < 0.1) in male and female DRG neurons. D. Heatmap depicting differentially secreted proteins in male versus female DRG neurons. E. Gene ontology (GO) enrichment analysis in proteomics of female DRG neurons, as compared to male DRG neurons. FDR < 0.1 was used as the cutoff criteria. F. Gene set enrichment analysis (GSEA) plot of the biological pathway of regulation for IGF transport and uptake by IGFBPs and heatmap for proteins in this pathway in male versus female DRG neurons.
Pathway analyses of secretome profiles of male and female DRG neurons:
To identify the biological pathways in the secretome of DRG neurons that are differentially expressed between males and females, we performed GO enrichment analysis and GSEA. We found that the male- and female-enriched secretory proteins were enriched in distinct GO biological pathways (FDR < 0.1, Supplemental Tables 7 and 8). Notably, GO enrichment analysis results were consistent between secretome and transcriptome datasets. In the 16 secretory proteins enriched in female DRG neurons, we observed upregulation of collagen and extracellular matrix pathways, including a collagen trimer, a complex of collagen trimers, collagen type I and III trimer, collagen-containing extracellular matrix, and collagen-activated signaling pathway (Figure 4E). The secretome of female DRG neurons was also enriched in signal transduction and regulation of signaling and signaling transduction GO terms (Figure 4E). Lastly, GO enrichment analysis revealed the regulation of insulin-like growth factor (IGF) transport and uptake by insulin-like growth factor binding proteins (IGFBPs) pathways (Figure 4E). GSEA analysis results further confirmed differential regulation of IGF transport and uptake by the IGFBP pathway in the male and female secretomes (Figure 4F and Supplemental Table 9). Interestingly, among the 19 secretory proteins involved in this pathway, the levels of cadherin 2 (Cdh2), nucleobindin 1 (Nucb1), follistatin 1 (Fstl1) and chromogranin B (Chgb) were significantly higher in the secretome of female DRG neurons compared to that of male DRG neurons (FDR < 0.1). Surprisingly, the 30 secretory proteins enriched in male DRG neurons were enriched in GO terms associated with chromatin modification and DNA, such as chromatin organization, remodeling, assembly, DNA binding, and DNA replication (Supplemental Table 8) consistent with the transcriptomic signature of male DRG neurons. GSEA analysis results also showed that DNA binding pathways were enriched in male DRG neurons (Supplemental Table 10), hence mirroring the GO enrichment analysis. Finally, secretome screening uncovered several proteins with known roles in the modulation of insulin synthesis and/or secretion (Figure 4D and Supplemental Table 6), including peroxiredoxin-4 (Prdx-4), seizure 6-like protein (Sez6l), Agrin, Slit guidance ligand 2 (Slit-2), collagen 1 alpha 1 (Col1a1), collagen 1 alpha 2 (Col1a2) and collagen 3 alpha 1 (Col3a1). Taken together, these analyses revealed male- and female-specific secretory proteins that modulate distinct biological pathways, including those with reported roles in the modulation of pancreatic β-cell function.
4. Discussion:
Understanding the biological differences between men and women requires the identification of the cellular and molecular mechanisms that directly or indirectly cause sex differences. In the central nervous system, it has long been hypothesized that gonadal hormones set permanently the sex differences during early development “organizational effects” and maintain them throughout adulthood via interaction with their respective sex hormone receptors “activational effects” [17]. However, the sex chromosome complement has also been implicated as an important contributor to sex differences [37, 50]—particularly in aspects of energy homeostasis, including the regulation of body weight, cholesterol and liver triglycerides [51, 52]. We have recently reported that sensory neuromodulation of islet β-cell function is sex-biased [14] and that sensory neuron-derived cell-autonomous factors regulate insulin secretion independently of the action of gonadal sex hormones [19]. Here, we report the transcriptome and secretome profiles of lower thoracic DRG neurons derived from sexually immature male and female mice. We found that DRG neurons exhibit sex differences in the gene expression profile and secreted proteins, independent of circulating sex hormones. Furthermore, GO and GSEA analyses revealed that male and female spinal sensory neurons display differences in biological pathways associated—among others—with insulin sensing, chromatin remodeling, cell senescence, axon extension, and insulin secretion.
Our transcriptomics-based screening highlighted the presence of insulin signaling machinery (e.g., insulin receptors) in lower thoracic DRG neurons. This finding strengthens the previously reported data suggesting the expression of insulin receptors in DRG sensory neurons directly projecting in the pancreas [53] and strongly suggests a role for insulin and insulin-like growth factor 1 (IGF-1) as key interoceptive molecules that bidirectionally links the CNS to the endocrine pancreas via thoracic DRG neurons [54]. Interestingly, GO and GSEA analyses demonstrated that the insulin signaling pathway is enhanced in DRG neurons harvested from female vs. male mice. The higher insulin sensitivity in female sensory neurons is consistent with the well-known sex difference in insulin action [29]. Why do female spinal sensory neurons exhibit enhanced insulin sensitivity? A teleological explanation may lie in the requirement to compensate for the lesser abundant sensory innervation of the female pancreas. Indeed, while the peri-islet sensory innervation is reduced in female mice as compared to their male counterparts [19], the enhanced insulin signaling/sensing may constitute a sensory advantage to capture subtle variations in insulin release from the poorly innervated islets. An alternative explanation is that improved insulin sensitivity would enhance sensory neurite growth and branching within pancreatic islets [11] as a compensatory strategy to counter the peri-islet sensory hypo-innervation observed in females vs. males who a higher number of islet-projecting DRG neurons [19]. Consistent with this hypothesis, GO and GSEA analyses indicated enrichment of cluster genes associated with neuron projection/extension and axonal guidance in DRG cell cultures derived from female mice (Figure 3B and Supplemental Figure 3C). Interestingly, the concept “more neurons in males; more processes in females” has been also reported in the cerebral cortex [55]. It is intriguing to identify gene clusters and GO terms associated with cell cycle, cell proliferation and DNA synthesis pathways in the DRG neural preparation. While spinal sensory neurons become postmitotic after differentiation during embryonic life [56], other non-neuronal cell types composing the DRG have proliferative capabilities, including satellite glial cells (SGCs) [57] and macrophages [58]. Comparative GO analysis showed an upregulation of cell cycle gene sets in male versus female DRG neural cultures that is consistent with recent studies [58, 59]. The increase in self-renewal of macrophages and SCGs within the male DRG underscores the importance of continuous self-renewal of these cells to maintain/support the highly abundant peri-islet sensory neurons in male mice [19]. The enhanced macrophage proliferation is also consistent with the heightened inflammatory response in the male DRG and points to interleukin-7 signaling (Supplemental Figure 2H) as an important pathway in the expansion of DRG resident immune cells, including macrophages [60]. Another important sex-biased/male-enriched biological feature identified in DRG neurons is cellular senescence/senescence-associated secretory phenotype (SASP). In light of the recent studies suggesting the propensity of islet β cells in males (vs. females) to cellular senescence [61] and the reduction of β-cell function during aging in men but not women [62], our pathway analyses suggest that the pancreas-projecting thoracic DRG neurons act within the islet β-cell niche and contribute to islet β-cell dysfunction in males. The reduction of the peri-islet sensory innervation density [19] and the DRG-expressed SASP in females (vs. males) palliates the deleterious inflammatory milieu generated by the senescent DRG fibers, thus resulting in superior GSIS in females [19–21]. Finally, our study revealed sex-differential/female-enriched expression of pathways associated with collagen-containing extracellular matrix (ECM). Interestingly, collagen production has been associated with insulin secretion and collagen supplementation in transplantation settings has been shown to improve β-cell engraftment and attenuate hypoxia which ultimately enhance β-cell response to glucose [63]. Moreover, studies have demonstrated that islet ECM is altered/remodeled in T1D in a fashion that renders islets more permissive to immune cell infiltration and impairment of β-cell function. The higher collagen expression and secretion from the pancreas-innervating thoracic DRG neurons in females (vs. males) is consistent with the recently evoked concept of β-cell resilience in response to islet stress in female mice [64]. We did not observe major congruence in pathways identified by transcriptome and secretome analyses. This is expected as the secretome does not necessarily reflect the proteome signature but rather the pool of molecules packaged in the secretory granules ready to be released in the extracellular media under stimulatory conditions such as occurring experimentally in response to KCL 75 mM (Figure 1). Secretome screening unraveled several sex-differentially secreted molecules with previously described modulatory roles in pancreatic β-cell activity, including Prdx-4, Sez6l, Agrin, Slit-2, Col1a, Col1a2 and Col3a1. Prdx-4 was reported to improve insulin synthesis and glucose-induced insulin secretion in vitro [65]; Sez6l was recently identified as a putative target for Bace1/2; plasma membrane proteases known to regulate β-cell function [66]; post-translational modification-specific proteomics identified Agrin signaling pathway to be relevant in GSIS [67]; Slit-2 increases the frequency of glucose-induced calcium oscillations and potentiates insulin secretion [68] and several collagen molecules have been reported to have stimulatory effect on insulin secretion [69–71]. Finally, secretome screening highlighted the enrichment of IGFBPs in female thoracic DRG culture media preparations which suggests the ability of females to exert a tighter control of IGF action within the islet niche. Together, our genomics and proteomics analyses unraveled sex differences in multiple converging cellular pathways relevant to the bidirectional crosstalk between DRG neurons and pancreatic β cells.
We are cognizant that the sex-differential expression and secretion of the DRG molecules might not be due, exclusively, to the action of the sex chromosomes (XX or XY). While our experimental design rules out the occurrence of activational effects (occurring in adulthood) of gonadal sex hormones in three-week-old mice, the organizational testosterone surge taking place during the neonatal period might be playing a role [72, 73] as previously described for the hypothalamic POMC neurons [74]. It is possible that perinatal testosterone shapes the genetic and secretome profiles in the male DRG through epigenetic mechanisms. Nevertheless, it is relevant to mention that this has been largely described in the CNS [75] and it is currently unknown whether similar masculinization effects of the neonatal testosterone surge take place in the peripheral nervous system. Moreover, while the perinatal testosterone surge is regarded as a significant contributor to the masculinization of the CNS, recent insights from disorders of sexual development underscored a role for the Y chromosome genes and X-chromosome dosage in the moderation of the biological processes linked to the sexual differentiation of the mammalian brain [76, 77]. Additional studies are warranted to separate the effects of testosterone surge from those of the sex chromosomes and to determine whether these male-female differences in the DRG transcriptome and secretome have sex-specific functional consequences on pancreatic β-cell activity.
5. Conclusion:
Our study unveiled sex differences in the transcriptome and secretome profiles of the lower thoracic DRG sensory neurons, including those projecting directly into the endocrine pancreas. These differences appear prior to puberty and likely arise from a combination of perinatal testosterone surge and sex-chromosome effects. The transcriptome analysis revealed higher insulin-sensing capabilities in female (vs. male) DRG neurons. The secretome screening uncovered several sex-specific candidate molecules with potential regulatory functions in pancreatic β cells. Together, these data suggest a putative role of sensory interoception of insulin in the DRG-islet crosstalk with potential implications of sensory feedback loops in the sex-biased regulation of β-cell activity. The characterization of these cellular and molecular pathways will be relevant in the context of strategies aimed at developing sex-based therapeutic and/or preventive measures for men and women with compromised β-cell function.
Supplementary Material
Acknowledgements:
We are grateful for the HIRN and NIH funding to conduct this study.
Funding information:
This research was supported by the NIDDK-supported Human Islet Research Network (HIRN, RRID:SCR_014393; https://hirnetwork.org; UC4 DK104162; to A.E.), R01 DK122167 (to A.E.), U01 DK124020 (to W.J.Q.) and NINDS R15 NS130456 (to J.Y.Z.). The Child Health Institute of New Jersey was supported by the Robert Wood Johnson Foundation (grant #74260).
Footnotes
Disclosures: The authors declare no competing interests.
Data availability statement:
RNA-seq data that supports the findings of this study are deposited in NCBI Gene Expression Omnibus under GSE228189. The reviewer may access the data at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE228189 using accession token cpmtigocjhcdzqx. All data will be made publicly available after final acceptance of the manuscript for publication. All the computational scripts that were used to analyze the data generated in this study are available at the GitHub repository (https://github.com/Jerry-Zhao/DRG2023).
References:
- 1.Nascimento AI, Mar FM, and Sousa MM, The intriguing nature of dorsal root ganglion neurons: Linking structure with polarity and function. Prog Neurobiol, 2018. 168: p. 86–103. [DOI] [PubMed] [Google Scholar]
- 2.Zheng Y, et al. Deep Sequencing of Somatosensory Neurons Reveals Molecular Determinants of Intrinsic Physiological Properties. Neuron, 2019. 103(4): p. 598–616 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Julius D, TRP channels and pain. Annu Rev Cell Dev Biol, 2013. 29: p. 355–84. [DOI] [PubMed] [Google Scholar]
- 4.Chiu IM, von Hehn CA, and Woolf CJ, Neurogenic inflammation and the peripheral nervous system in host defense and immunopathology. Nat Neurosci, 2012. 15(8): p. 1063–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan B, et al. Dorsal Root Ganglion Field Stimulation Prevents Inflammation and Joint Damage in a Rat Model of Rheumatoid Arthritis. Neuromodulation, 2018. 21(3): p. 247–253. [DOI] [PubMed] [Google Scholar]
- 6.Chiu IM, et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature, 2013. 501(7465): p. 52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabiller L, et al. Pain sensing neurons promote tissue regeneration in adult mice. NPJ Regen Med, 2021. 6(1): p. 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang PX, et al. Dorsal root ganglion neurons promote proliferation and osteogenic differentiation of bone marrow mesenchymal stem cells. Neural Regen Res, 2015. 10(1): p. 119–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caterina MJ, et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature, 1997. 389(6653): p. 816–24. [DOI] [PubMed] [Google Scholar]
- 10.Riera CE and Dillin A, Emerging Role of Sensory Perception in Aging and Metabolism. Trends Endocrinol Metab, 2016. 27(5): p. 294–303. [DOI] [PubMed] [Google Scholar]
- 11.Lazar BA, et al. Insulin Confers Differing Effects on Neurite Outgrowth in Separate Populations of Cultured Dorsal Root Ganglion Neurons: The Role of the Insulin Receptor. Front Neurosci, 2018. 12: p. 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang KT, et al. Data on the expression of leptin and leptin receptor in the dorsal root ganglion and spinal cord after preganglionic cervical root avulsion. Data Brief, 2017. 15: p. 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anand U, et al. Glucagon-like peptide 1 receptor (GLP-1R) expression by nerve fibres in inflammatory bowel disease and functional effects in cultured neurons. PLoS One, 2018. 13(5): p. e0198024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bou Karam J, et al. TRPV1 neurons regulate beta-cell function in a sex-dependent manner. Mol Metab, 2018. 18: p. 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riera CE, et al. TRPV1 pain receptors regulate longevity and metabolism by neuropeptide signaling. Cell, 2014. 157(5): p. 1023–36. [DOI] [PubMed] [Google Scholar]
- 16.Makwana K, et al. Sensory neurons expressing calcitonin gene-related peptide alpha regulate adaptive thermogenesis and diet-induced obesity. Mol Metab, 2021. 45: p. 101161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mauvais-Jarvis F, Arnold AP, and Reue K, A Guide for the Design of Pre-clinical Studies on Sex Differences in Metabolism. Cell Metab, 2017. 25(6): p. 1216–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gannon M, et al. Sex differences underlying pancreatic islet biology and its dysfunction. Mol Metab, 2018. 15: p. 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEwan S, et al. Deconstructing the origins of sexual dimorphism in sensory modulation of pancreatic beta cells. Mol Metab, 2021. 53: p. 101260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T, et al. Sex effect on insulin secretion and mitochondrial function in pancreatic beta cells of elderly Wistar rats. Endocr Res, 2016. 41(3): p. 167–79. [DOI] [PubMed] [Google Scholar]
- 21.Hall E, et al. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol, 2014. 15(12): p. 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu A, Dube S, and Basu R, Men Are from Mars, Women Are from Venus: Sex Differences in Insulin Action and Secretion. Adv Exp Med Biol, 2017. 1043: p. 53–64. [DOI] [PubMed] [Google Scholar]
- 23.Horie I, et al. Sex differences in insulin and glucagon responses for glucose homeostasis in young healthy Japanese adults. J Diabetes Investig, 2018. 9(6): p. 1283–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basu R, et al. Effects of age and sex on postprandial glucose metabolism: differences in glucose turnover, insulin secretion, insulin action, and hepatic insulin extraction. Diabetes, 2006. 55(7): p. 2001–14. [DOI] [PubMed] [Google Scholar]
- 25.Mauvais-Jarvis F, Sex differences in metabolic homeostasis, diabetes, and obesity. Biol Sex Differ, 2015. 6: p. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauvais-Jarvis F, Gender differences in glucose homeostasis and diabetes. Physiol Behav, 2018. 187: p. 20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tramunt B, et al. Sex differences in metabolic regulation and diabetes susceptibility. Diabetologia, 2020. 63(3): p. 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kautzky-Willer A, et al. Influence of increasing BMI on insulin sensitivity and secretion in normotolerant men and women of a wide age span. Obesity (Silver Spring), 2012. 20(10): p. 1966–73. [DOI] [PubMed] [Google Scholar]
- 29.Macotela Y, et al. Sex and depot differences in adipocyte insulin sensitivity and glucose metabolism. Diabetes, 2009. 58(4): p. 803–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu J, et al. Estradiol Protects Proopiomelanocortin Neurons Against Insulin Resistance. Endocrinology, 2018. 159(2): p. 647–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonso-Magdalena P, et al. Pancreatic insulin content regulation by the estrogen receptor ER alpha. PLoS One, 2008. 3(4): p. e2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Contreras JL, et al. 17beta-Estradiol protects isolated human pancreatic islets against proinflammatory cytokine-induced cell death: molecular mechanisms and islet functionality. Transplantation, 2002. 74(9): p. 1252–9. [DOI] [PubMed] [Google Scholar]
- 33.Wong WP, et al. Extranuclear estrogen receptor-alpha stimulates NeuroD1 binding to the insulin promoter and favors insulin synthesis. Proc Natl Acad Sci U S A, 2010. 107(29): p. 13057–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro G, et al. Androgen excess in pancreatic beta cells and neurons predisposes female mice to type 2 diabetes. JCI Insight, 2018. 3(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro G, et al. Extranuclear Actions of the Androgen Receptor Enhance Glucose-Stimulated Insulin Secretion in the Male. Cell Metab, 2016. 23(5): p. 837–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mauvais-Jarvis F, Role of Sex Steroids in beta Cell Function, Growth, and Survival. Trends Endocrinol Metab, 2016. 27(12): p. 844–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Vries GJ, et al. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J Neurosci, 2002. 22(20): p. 9005–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Link JC, et al. Metabolic impact of sex chromosomes. Adipocyte, 2013. 2(2): p. 74–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huebner EA, et al. Diltiazem Promotes Regenerative Axon Growth. Mol Neurobiol, 2019. 56(6): p. 3948–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon S and Zhao YT, Spatial, temporal and cell-type-specific expression profiles of genes encoding heparan sulfate biosynthesis enzymes and proteoglycan core proteins. Glycobiology, 2021. 31(10): p. 1308–1318. [DOI] [PubMed] [Google Scholar]
- 41.Moon S and Zhao YT, Convergent biological pathways underlying the Kallmann syndrome-linked genes Hs6st1 and Fgfr1. Hum Mol Genet, 2022. 31(24): p. 4207–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 2013. 29(1): p. 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, and Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014. 15(12): p. 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson MD, McCarthy DJ, and Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010. 26(1): p. 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A, 2005. 102(43): p. 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.HaileMariam M, et al. S-Trap, an Ultrafast Sample-Preparation Approach for Shotgun Proteomics. J Proteome Res, 2018. 17(9): p. 2917–2924. [DOI] [PubMed] [Google Scholar]
- 47.Duan J, et al. Quantitative Profiling of Protein S-Glutathionylation Reveals Redox-Dependent Regulation of Macrophage Function during Nanoparticle-Induced Oxidative Stress. ACS Nano, 2016. 10(1): p. 524–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cox J and Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol, 2008. 26(12): p. 1367–72. [DOI] [PubMed] [Google Scholar]
- 49.Fasanella KE, et al. Distribution and neurochemical identification of pancreatic afferents in the mouse. J Comp Neurol, 2008. 509(1): p. 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arnold AP, et al. Sex Hormones and Sex Chromosomes Cause Sex Differences in the Development of Cardiovascular Diseases. Arterioscler Thromb Vasc Biol, 2017. 37(5): p. 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X, et al. The number of x chromosomes causes sex differences in adiposity in mice. PLoS Genet, 2012. 8(5): p. e1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Link JC, et al. Increased high-density lipoprotein cholesterol levels in mice with XX versus XY sex chromosomes. Arterioscler Thromb Vasc Biol, 2015. 35(8): p. 1778–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lazar BA, et al. The Insulin Receptor Is Colocalized With the TRPV1 Nociceptive Ion Channel and Neuropeptides in Pancreatic Spinal and Vagal Primary Sensory Neurons. Pancreas, 2018. 47(1): p. 110–115. [DOI] [PubMed] [Google Scholar]
- 54.Chen WG, et al. The Emerging Science of Interoception: Sensing, Integrating, Interpreting, and Regulating Signals within the Self. Trends Neurosci, 2021. 44(1): p. 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rabinowicz T, et al. Gender differences in the human cerebral cortex: more neurons in males; more processes in females. J Child Neurol, 1999. 14(2): p. 98–107. [DOI] [PubMed] [Google Scholar]
- 56.Landy MA, Goyal M, and Lai HC, Nociceptor subtypes are born continuously over DRG development. Dev Biol, 2021. 479: p. 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, et al. Chronic pain induces nociceptive neurogenesis in dorsal root ganglia from Sox2-positive satellite cells. Glia, 2019. 67(6): p. 1062–1075. [DOI] [PubMed] [Google Scholar]
- 58.Yu X, et al. Dorsal root ganglion macrophages contribute to both the initiation and persistence of neuropathic pain. Nat Commun, 2020. 11(1): p. 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stephens KE, et al. Sex differences in gene regulation in the dorsal root ganglion after nerve injury. BMC Genomics, 2019. 20(1): p. 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen D, et al. Interleukin-7 Biology and Its Effects on Immune Cells: Mediator of Generation, Differentiation, Survival, and Homeostasis. Front Immunol, 2021. 12: p. 747324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker EM, et al. Sex-biased islet beta cell dysfunction is caused by the MODY MAFA S64F variant by inducing premature aging and senescence in males. Cell Rep, 2021. 37(2): p. 109813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drigo R.A.e., Aging of human endocrine pancreatic cell types is heterogeneous and sex-specific. bioRxiv, 2019. [Google Scholar]
- 63.Zbinden A, et al. Collagen and Endothelial Cell Coculture Improves beta-Cell Functionality and Rescues Pancreatic Extracellular Matrix. Tissue Eng Part A, 2021. 27(13–14): p. 977–991. [DOI] [PubMed] [Google Scholar]
- 64.Brownrigg GP, et al. Sex differences in islet stress responses support female beta cell resilience. Mol Metab, 2023. 69: p. 101678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mehmeti I, et al. Peroxiredoxin 4 improves insulin biosynthesis and glucose-induced insulin secretion in insulin-secreting INS-1E cells. J Biol Chem, 2014. 289(39): p. 26904–26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stutzer I, et al. Systematic proteomic analysis identifies beta-site amyloid precursor protein cleaving enzyme 2 and 1 (BACE2 and BACE1) substrates in pancreatic beta-cells. J Biol Chem, 2013. 288(15): p. 10536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang T, et al. Characterization of the Molecular Mechanisms Underlying Glucose Stimulated Insulin Secretion from Isolated Pancreatic beta-cells Using Post-translational Modification Specific Proteomics (PTMomics). Mol Cell Proteomics, 2018. 17(1): p. 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang YH, et al. Intraislet SLIT-ROBO signaling is required for beta-cell survival and potentiates insulin secretion. Proc Natl Acad Sci U S A, 2013. 110(41): p. 16480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu Y, et al. Collagen type I enhances cell growth and insulin biosynthesis in rat pancreatic cells. J Mol Endocrinol, 2021. 67(3): p. 135–148. [DOI] [PubMed] [Google Scholar]
- 70.Duner P, et al. Adhesion G Protein-Coupled Receptor G1 (ADGRG1/GPR56) and Pancreatic beta-Cell Function. J Clin Endocrinol Metab, 2016. 101(12): p. 4637–4645. [DOI] [PubMed] [Google Scholar]
- 71.Huang G, et al. alpha3(V) collagen is critical for glucose homeostasis in mice due to effects in pancreatic islets and peripheral tissues. J Clin Invest, 2011. 121(2): p. 769–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corbier P, Edwards DA, and Roffi J, The neonatal testosterone surge: a comparative study. Arch Int Physiol Biochim Biophys, 1992. 100(2): p. 127–31. [DOI] [PubMed] [Google Scholar]
- 73.Weisz J and Ward IL, Plasma testosterone and progesterone titers of pregnant rats, their male and female fetuses, and neonatal offspring. Endocrinology, 1980. 106(1): p. 306–16. [DOI] [PubMed] [Google Scholar]
- 74.Nohara K, et al. Early-life exposure to testosterone programs the hypothalamic melanocortin system. Endocrinology, 2011. 152(4): p. 1661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murray EK, et al. Epigenetic control of sexual differentiation of the bed nucleus of the stria terminalis. Endocrinology, 2009. 150(9): p. 4241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bonthuis PJ, Cox KH, and Rissman EF, X-chromosome dosage affects male sexual behavior. Horm Behav, 2012. 61(4): p. 565–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Savic I, et al. Role of testosterone and Y chromosome genes for the masculinization of the human brain. Hum Brain Mapp, 2017. 38(4): p. 1801–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data that supports the findings of this study are deposited in NCBI Gene Expression Omnibus under GSE228189. The reviewer may access the data at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE228189 using accession token cpmtigocjhcdzqx. All data will be made publicly available after final acceptance of the manuscript for publication. All the computational scripts that were used to analyze the data generated in this study are available at the GitHub repository (https://github.com/Jerry-Zhao/DRG2023).


