Abstract
Background
The potent immunosuppressive properties of sialic acid-binding immunoglobulin-like lectin-9 (Siglec-9) on myeloid cells and lymphocytes provide a strong rationale for serving as a therapeutic target. However, the expression profile and critical role of Siglec-9 in high-grade serous ovarian cancer (HGSC) remain obscure. This study aimed to elucidate the prognostic significance of Siglec-9 expression and its predictive value for immunotherapy in HGSC.
Methods
Study enrolled two cohorts, consisting of 120 tumor microarray specimens of HGSC for immunohistochemistry (IHC) and 40 fresh tumor specimens for flow cytometry (FCM). Expression profile of Siglec-9 in immune cells was analyzed by both bioinformatics analysis and FCM. Role of Siglec-9 was studied to identify that Siglec-9+TAMs linked with an immunosuppressive phenotype by IHC and FCM, and block Siglec-9 was sensitive to immunotherapy by ex vivo and in vitro assays.
Results
Siglec-9 is predominantly expressed on tumor-associated macrophages (TAMs). High Siglec-9+TAMs were associated with inferior overall survival (OS). Both tumor-conditioned medium (TCM) and tumor ascites induced enrichment of Siglec-9+TAMs with protumorigenic phenotypes. Siglec-9+TAMs were associated with immunosuppressive tumor microenvironment (TME) characterized by exhausted CD8+T cells and increased immune checkpoint expression. Blockade of Siglec-9 suppressed phosphorylation of the inhibitory phosphatase SHP-1 and repolarized TAMs to antitumorigenic phenotype and retrieved cytotoxic activity of CD8+T cells in vitro and ex vivo. Responders toward antiprogrammed death receptor-1 (anti-PD-1) therapy present more Siglec-9+TAMs than non-responders. Furthermore, blockade Siglec-9 synergized with anti-PD-1 antibody to enhance the cytotoxic activity of CD8+T cells in tissues with higher Siglec-9+TAMs.
Conclusions
Siglec-9+TAMs may serve as an independent prognostic of poor survival but a predictive biomarker for anti-PD-1/antiprogrammed death ligand-1 immunotherapy in HGSC. In addition, the potential of immunosuppressive Siglec-9+TAMs as a therapeutic target is worth further exploration.
Keywords: Immunotherapy, Tumor Microenvironment, Tumor Escape
WHAT IS ALREADY KNOWN ON THIS TOPIC
Sialic acid-binding immunoglobulin-like lectin-9 (Siglec-9) is a novel target for immunotherapy by counteracting immune-suppressive signaling from tumor cells overexpressing glycans containing sialic acids, but its expression pattern and prognostic potential have not been investigated in high-grade serous ovarian cancer (HGSC).
WHAT THIS STUDY ADDS
The study demonstrated that Siglec-9 is predominantly expressed on tumor-associated macrophages (TAMs) with an immune-suppressive phenotype and is associated with the adverse prognosis of patients with HGSC. A context with higher Siglec-9+TAMs had significant prognostic value in HGSC patients receiving programmed death-1 (PD-1) blockaded therapy. Therefore, the monoclonal antibody (mAbs) targeting Siglec-9 repolarize TAMs from protumor to antitumor phenotypes and enhance response to anti-PD-1 in ex vivo.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our study unravels Siglec-9+TAMs as a novel biomarker for predicting HGSC patients who would benefit from anti-PD-1/programmed death ligand-1 therapy and as a promising target for cancer immunotherapy that could be exploited to enhance antitumor immunity and improve immunotherapy management in HGSC patients.
Introduction
Ovarian cancer ranks as the fifth leading cause of female cancer-related death.1 2 High-grade serous ovarian cancer (HGSC) is recognized as the most widespread and deadly subtype of ovarian cancer.3 Patients who have been diagnosed with early-stage disease (International Federation of Gynecology and Obstetrics, FIGO stage I and II) are often effectively treated with surgery and chemotherapy containing platinum and are more prone to recover.4 Most patients with advanced-stage HGSC (International Federation of Gynecology and Obstetrics (FIGO) stage III and IV) rarely achieve a cure through treatment. Although antiangiogenic agents5 and PARP inhibitor therapy6 7 are combined, most patients with advanced disease suffer a relapse within 24 months of undergoing their initial chemotherapy treatment.8 Recent clinical trials have demonstrated the potential of immunotherapies in treating HGSC, such as immune checkpoint blockade (ICB) targeting the programmed death receptor-1 (PD-1)/programmed death ligand-1 (PD-L1) axis. While some patients may benefit greatly from second-line therapy with ICB due to an increase in T and B cell infiltration that heats the tumor microenvironment (TME), others may not respond to treatment at all.9–11 Thus, there is an urge to explore more effective combination therapies, such as combining ICB with chemotherapy or targeted therapies, than ICB alone in certain patients.
Recent research has shown that tumor cells also educate myeloid cells to evade the immune system,12 besides directly suppressing T cell immune responses through binding immune checkpoints. Tumor-associated macrophages (TAMs), which are a significant type of myeloid cells within the TME, play a role in regulating the immune response to tumors.13 The ability of macrophages to adapt to changing conditions within the microenvironment is a key characteristic known as plasticity, which encompasses both immunostimulatory and immunosuppressive subsets.14 15 Hence, a noteworthy treatment strategy involves the reprogramming of immunosuppressive TAMs, which is viewed as a complementary or synergistic approach to be used in conjunction with existing backbone regimens.
Sialic acid sugars present on the surface of cancer cells have been identified as immune modulators, contributing to both the immunosuppressive microenvironment and the ability of tumors to evade the immune system.16 Sialic acid binding immunoglobulin-like lectin 9 (Siglec-9) belongs to the sialic acid binding immunoglobulin-like lectins (Siglecs) family, which showed high affinity for α2,3 and α2,6 binding sialoglycans.17 Functions of Siglec-9 on myeloid cells, natural killer (NK) cells and T cells have been reported recently.17–19 Siglec-9 expression has been shown to skew macrophage polarization to a protumorigenic phenotype with upregulation of PD-L1 in macrophages20 and contribute to forming an immunosuppressive environment.21 Blockade and ablating of Siglecs-9 may potentially enhance the therapeutic effects of immune checkpoint inhibitors in animal models.22–25 Additionally, high expression of Siglec-9 promotes monocytes differentiating toward macrophages and pancreatic cancer tumorigenesis by inducing immune tolerance.26 Currently, there is no research available on the impact of Siglec-9 on tumor immunity in HGSC, nor has its potential as a prognostic biomarker for prediction and as an adjuvant therapy to enhance ICB response has been investigated.
Here, we discovered that Siglec-9, highly expressed in both primary tumors and ascites, could be an independent prognostic factor for predicting poor prognosis in HGSC. Furthermore, Siglec-9 is expressed predominantly on TAMs in HGSC, which displays an immunosuppressive phenotype along with dysfunction of T cells. Siglec-9 neutralizing antibody could reverse the immunosuppressive effects of TAMs and restore the antitumor immune response of T cells. It is noteworthy that the combination of Siglec-9 and PD-1 blockades can significantly improve the antitumor activity compared with PD-1 blockade alone. Siglec-9 is expected to emerge as a new target for immunotherapy and a prognostic biomarker for predicting the response of HGSC patients.
Materials and methods
HGSC patient tissue
Specimens from 120 patients with primary carcinoma of HGSC (online supplemental table S1) who had undergone surgery between 2013 and 2015 were obtained from the Obstetrics and Gynecology Hospital of Fudan University with approval. All patients were followed up until April 2019. None of the patients had an autoimmune disorder or a history of prior cancer. None of the patients was treated with chemotherapy, radiation, or any other antitumor medicines before tumor resection. Overall survival (OS) was calculated from the date of surgery to the date of death or last follow-up. Fresh tumor tissue samples were obtained from 40 patients with HGSC (online supplemental table S1) during surgery at the Department of Gynecology of our hospital, including 24 HGSC tissues used for ex vivo stimulation and immunofluorescence. For immunohistochemical and immunofluorescence studies, once the surgical specimens were obtained, all specimens were snap-frozen immediately in liquid nitrogen. The clinical characteristics of all tissue samples from HGSC patients were summarized in online supplemental table S1. Written consent was obtained from all the donors. The detailed procedure of specimen processing was provided in online supplemental methods.
jitc-2023-007099supp001.pdf (245.8KB, pdf)
jitc-2023-007099supp012.pdf (49.6KB, pdf)
jitc-2023-007099supp013.pdf (43.8KB, pdf)
Monocyte isolation and macrophage stimulation
Buffy coats were obtained from healthy donors (Shanghai, China). Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation with a leukocyte separation solution (HISTOPAQUE-1077; Sigma-Aldrich), to later purify CD14+ monocytes using MACS CD14 MicroBeads (Miltenyi). To compare the phenotype with cytokine-induced macrophages, monocytes were stimulated with 100 ng/mL macrophage colony-stimulating factor (M-CSF) or 100 ng/mL granulocyte macrophage colony-stimulating factor (GM-CSF) for 3 days. The polarization of M-CSF-induced macrophages was performed by incubation with 20 ng/mL IL-4, 20 ng/mL IL-13, or 20 ng/mL IFN-γ and 10 ng/mL lipopolysaccharide (LPS). The phenotype was studied by flow cytometry. To stimulate macrophages with tumor cell-derived conditioned medium (TCM) or ascites, we cultured PBMC-derived macrophages with the complete medium containing 50% TCM or ascites (volume). For Siglec blocking experiments, macrophages were preincubated with neutralizing antibodies for Siglec-9 (R&D Systems), with a final concentration of 5 µg/mL. The phenotype was studied by flow cytometry.
Assay methods
The details methods of flow cytometry and immunohistochemistry were also listed in the online supplemental methods. The experimental procedures of in vitro and ex vivo treatment assay with human specimens were described as online supporting information. All antibodies used in immunohistochemistry were listed in online supplemental table S2 and used in flow cytometry in online supplemental table S3.
jitc-2023-007099supp014.pdf (44.1KB, pdf)
Transcriptomics analysis
Transcriptomic, mutation, and clinical data of TCGA-OV were downloaded from the GDC Data Portal (https://portal.gdc.cancer.gov/) in December 2019. The endpoint OS from the TCGA Pan-Cancer Clinical Data Resource (TCGA-OV)27 was used to analyze patients’ clinical outcomes. The package limma28 was used for the analysis of the differential gene expression, with false discovery rate (FDR) correction for multiple comparisons. Gene set enrichment analysis (GSEA) on a single-sample basis using the GSVA package.29 Ecotype discovery was performed with EcoTyper.30 Cell ecotypes were discovered in the TCGA bulk RNA-seq dataset according to steps in the analysis tutorial of Bulk Expression Data (https://ecotyper.stanford.edu/carcinoma/). The detailed procedure of single-cell RNA sequencing data analysis was provided in online supplemental methods.
Statistics analysis
All statistical analysis was performed in R (V.4.1.3). Unpaired two-sided Wilcoxon rank-sum tests were used for pair-wise comparisons, and the Kruskal-Wallis rank sum test was used for comparisons between more than two groups, followed by Dunn’s test for multiple comparisons. Correlation between variables was performed using Spearman’s rank correlation, and CIs were calculated according to the method of Bonett and Wright. Statistical significance was accepted for p<0.05. Minimal p value method provided by X-tile (V.3.6.1, Yale University) was used to automatically find the cut-off for the ‘H score’ of Siglec-9 ligand and the number of Siglec-9+TAMs in Fudan cohort, which was 78 score and 1 cell/high-powered field (HPF) (online supplemental figure S1A,B), respectively. For differential expression calculations in single-cell gene expression data and bulk RNA-seq data, the R package limma fits gene-wise linear models and implements empirical Bayes moderated t-statistics to determine statistical significance. For all differential expression and gene set testing analyses, p values were corrected for multiple testing using the Benjamini-Hochberg method. The cut-off value of Siglec-9+TAMs signature score was 6.94 calculated by minimum log-rank p value. Survival of two groups of patients with different expression statuses using cut-offs determined by the X-Tile software (V.3.6.1) was compared by the Kaplan-Meier method and log-rank test.
jitc-2023-007099supp002.pdf (720.3KB, pdf)
Result
Subgroup high Siglec-9+TAMs infiltration associated with poor survival in HGSC
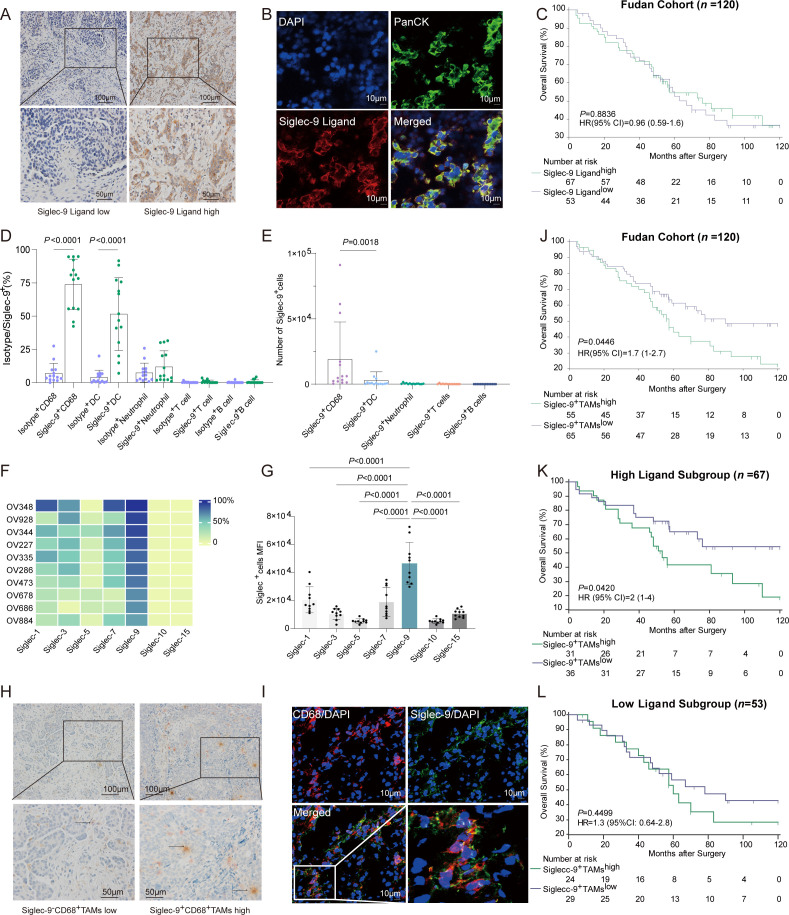
Given that sialylated structures can serve as ligands for Siglec receptors, we next evaluated whether HGSC could be recognized by a human Siglec-9 Fc chimera. Biotinylated human Siglec-9 Fc chimeras were used to detect the expression of Siglec-9 ligand (figure 1A–B). No correlation was observed between the Siglec-9 ligand, Siglec-9+TAMs and FIGO stages (online supplemental figure S1C,D). Grouping of HGSC patients based on Siglec-9 ligand did not show any correlation with overall survival (figure 1C). SKOV3 and HO-8190 cell lines had a significant level of Siglec-9 ligand expression compared with the A2780 cell line (online supplemental figure S1E). Treatment with neuraminidase was found to decrease the expression of the Siglec-9 ligand (online supplemental figure S1F).31
Figure 1.
Siglec-9+TAMs interacting with Siglec-9 ligand expressing on tumor cells associated with poor overall survival in HGSC. (A) Representative immunohistochemistry images show HGSC specimens with low (left) and high (right) infiltration of the Siglec-9 ligand. Scale bars: 100 mm. (B) Immunofluorescence staining of Siglec-9 ligand (red), pan CK (green) and DAPI (blue). Siglec-9 ligand coexpressing with pan CK was shown in HGSC. Scale bars: 10 mm. (C) Kaplan-Meier curves of overall survival (OS) stratified by expression of Siglec-9 ligand in Fudan cohort (n=120) using the log-rank test. Percentages of Siglec-9 among immune cells from HGSC tumor infiltrating lymphocytes (TILs). (D) Relative numbers of Siglec-9+immune cells from HGSC TILs. Kruskal–Wallis test. (E) Absolute cell counts of Siglec-9+immune cells from HGSC TILs. Kruskal–Wallis test. (F) Expression of Siglec receptors on macrophages in primary tumor tissues was detected by flow cytometry. (G) Flow cytometry analysis of mean fluorescence intensity (MFI) of Siglec in primary tumor tissues from HGSC. (H) Representative immunohistochemistry images (left) of double staining for Siglec-9 (red) and CD68 (brown). HGSC tissues were colabeled with Sigelc-9 and macrophage marker CD68. Black arrowheads indicate Siglec-9+CD68+cells. Scale bars: 100 and 50 µm for top and bottom panels, respectively. (I) Immunofluorescence staining of CD68 (red), Siglec-9 (green) and DAPI (blue). Siglec-9 coexpressing with CD68 was shown in HGSC. Scale bars: 10 mm. (J–L) Kaplan-Meier curves of OS stratified by infiltration of Siglec-9+TAMs (J) in Fudan cohort (n=120) as well as for high Siglec-9 ligand (K) and low Siglec-9 ligand (L) proportion in patients with HGSC tumors using the log-rank test. HGSC, high-grade serous ovarian cancer; Siglec-9, sialic acid-binding immunoglobulin-like lectin 9; TAMs, tumor-associated macrophages.
Siglec-9 expression has been observed on various cell types, including myeloid cells, NK cells, and certain subsets of T cells.19 Gating strategies (online supplemental figure S2A–C) were used to investigate the pattern of Siglec-9 expression in primary tumors, and results revealed that macrophages had the highest expression of Siglec-9, while T cells, B cells, and dendritic cells (DCs) did not exhibit any significant expression of Siglec-9 (figure 1D–E). Siglec-9 had the highest expression level on macrophages compared with other members of the Siglec family in HGSC specimens (figure 1F–G). Double-stain immunohistochemistry and immunofluorescence further identified the presence of CD68+macrophages expressing Siglec-9 in primary HGSC (figure 1H–I). Therefore, Siglec-9 expression was predominantly observed on TAMs in HGSC.
jitc-2023-007099supp003.pdf (465.9KB, pdf)
To verify the association between Siglec-9+TAMs and survival outcomes, a high level of infiltration of Siglec-9+TAMs was associated with a significant decrease in overall survival rates (p=0.0446, figure 1J), which was confirmed in patients with high Siglec-9 ligand (p=0.0420, figure 1K), but not in those with low Siglec-9 ligand (p=0.4499, figure 1L). Furthermore, Siglec-9+TAMshigh was an independent prognostic marker for OS adjusted for age, FIGO staging, lymph node invasion, PD-L1, and CD163 (online supplemental table S4), which was confirmed in Siglec-9 ligandhigh subgroup (online supplemental table S5). Taken together, a high infiltration of Siglec-9+TAMs was linked to inferior survival outcomes in HGSC, and this association was especially significant in patients with high expression levels of the Siglec-9 ligand.
jitc-2023-007099supp015.pdf (41.2KB, pdf)
jitc-2023-007099supp016.pdf (43KB, pdf)
Delineation of the protumoral phenotype of Siglec-9+TAMs in HGSC
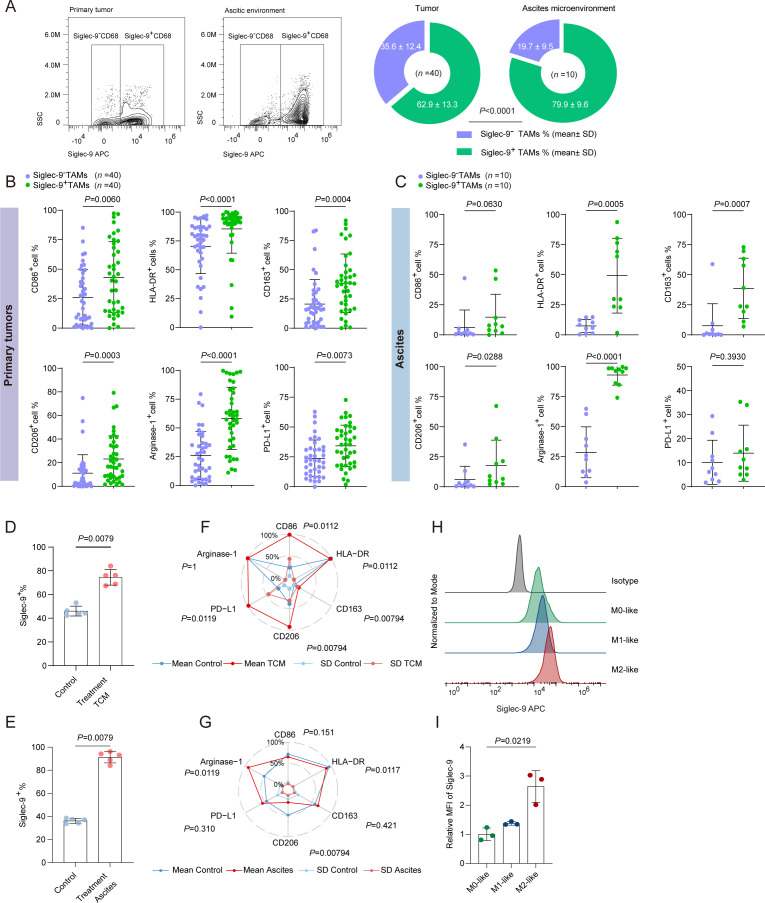
To illustrate the variation of Siglec-9+TAMs between primary and ascitic environments, a higher percentage of Siglec-9+TAMs was observed in ascites than in primary tumors (79.9% vs 62.9%) (figure 2A). Specifically, Siglec-9+TAMs decreased the expression of antitumor markers (like CD86 and HLA-DR) in primary tumors (figure 2B, online supplemental figure S3A,B) and increased the expression of protumor markers (like CD163, CD206, and arginase-1) in both primary and ascitic environment than Siglec-9−TAMs subgroup (figure 2B–C, online supplemental figure S3A–D). Under both TCM and ascitic treatment, an increase of Siglec-9+TAMs was observed (figure 2D–E). Siglec-9+TAMs upregulated the expression of proinflammatory markers (like CD86 and HLA-DR) and immunosuppressive factors (like CD163, CD206 and PD-L1) compared with Siglec-9−TAMs in the TCM treatment group (figure 2F, online supplemental figure S3E,F). Meanwhile, Siglec-9+TAMs showed relatively low expression of CD86 and HLA-DR in the ascites treatment group (figure 2G and online supplemental figure S3G,H). To validate the polarization status of Siglec-9+TAMs, the expression of Siglec-9 increased significantly in M2-like macrophages compared with the M1-like macrophages (figure 2H–I). To summarize, Siglec-9+TAMs possessed protumor traits with plasticity expressing proinflammatory markers.
Figure 2.
Protumoral phenotype of Siglec-9+TAMs presented in HGSC. (A) Flow cytometry gating and quantification of Siglec-9+ or Siglec-9− cells in total CD68+ (macrophages) cells in tumor tissues and corresponding ascites fluid samples. Significance was assessed by χ2 test. (B and C) Flow cytometry analysis of M1-like (CD86 and HLA-DR) and M2-like (CD163, CD206, arginase-1, and PD-L1) populations in TIL in Siglec-9+TAMs compared with Siglec-9−TAMs in primary tumor tissues (B) and ascites fluid samples (C) from HGSC. Cells were pregated on CD45 and CD68. (D and E) Statistical analysis of Siglec-9+TAMs in human peripheral blood monocyte (PBMC)-derived macrophages treated with tumor cell conditioned medium (TCM) (D) or ascites (E) for 48 hours. (F and G) Radar chart of flow cytometric data demonstrating surface coexpression of Siglec-9 with CD86, HLA-DR, CD163, CD206, arginase-1 and PD-L1 on human PBMC-derived macrophages from HGSC TILs treated with TCM (F) or ascites (G) for 48 hours. Blue and red lines: mean; light blue and light red lines: SD. (H and I) The expression of Siglec-9 in the different phenotypes of human PBMC-derived macrophages (lipopolysaccharide (LPS) plus IFN-γ induced M1 macrophages, IL-4 plus IL-13-induced M2 macrophages) was detected by flow cytometry (H) and quantified by the ratio of the relative mean fluorescence intensity (MFI), which was calculated by: relative MFI=MFI sample/MFI isotype (n = 3) (I). Plot showed the mean±SD. Significance was assessed by Mann-Whitney U test. HGSC, high-grade serous ovarian cancer; PD-L1, programmed death ligand-1; Siglec-9, sialic acid-binding immunoglobulin-like lectin 9; TAMs, tumor-associated macrophages.
jitc-2023-007099supp004.pdf (774.6KB, pdf)
Protumoral phenotype of Siglec-9+TAMs was confirmed in scRNA
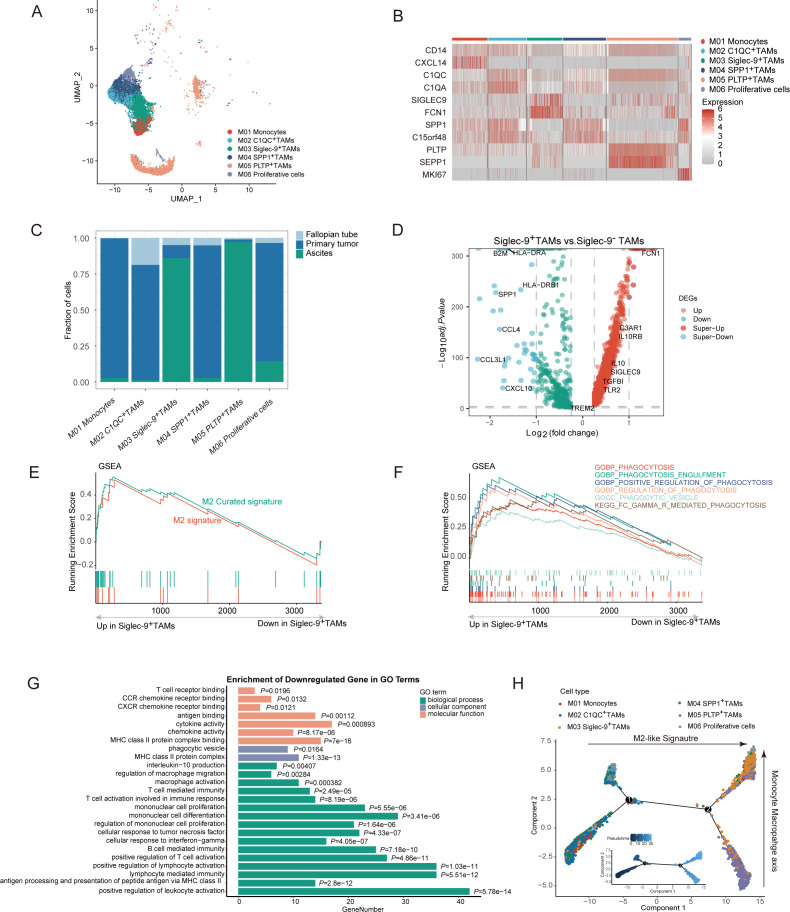
To further characterize the functional phenotype of Siglec-9+TAMs, we extracted myeloid cells identified from the full dataset based on marker expression and subclustered them including 4 scRNA-sequencing datasets(online supplemental figure S4A–C, online supplemental table S6), identifying one cluster of monocytes (classical CD14 monocytes), and five clusters of TAMs, which expressed a broad range of immunomodulatory genes (figure 3A–B).32–34 Most Siglec-9+TAMs were derived from ascites than primary tumors, accounting for more than 75% (figure 3C). Genes associated with the M2 phenotype, including CD163, IL10, and TGFB1, were upregulated in Siglec-9+TAMs (online supplemental figure S4D). Among these differentially expressed genes (DEGs), Siglec-9+TAMs expressed genes involved in the positive regulation of immune system processes including B and T cell chemoattractants (CXCL10, CCL3L1, and CCL4), antigen processing and presentation (B2M, HLA-DRA, and HLA-DRB1), and the marker of TAMs (SPP1) (figure 3D). Siglec-9+TAMs were enriched in two M2-like macrophage pathways (figure 3E) and five activated phagocytosis pathways (figure 3F, online supplemental figure S4E) and also were fueled by fatty acid uptake and oxidation in the mitochondria, identical to metabolic changes in M2 macrophage35 (online supplemental figure S4F). It was worth mentioning that phagocytosis markers in Siglec-9+TAMs, including significant expression of genes linked to FCN1, RACK1, IGLC2, and IGKC in the process by which immune cells engulf and digest foreign particles such as pathogens (online supplemental figure S4E). The immune signaling pathways were associated with Siglec-9+TAMs, such as positive regulation of leukocyte activation, antigen processing and presentation, cellular response to IFN-γ, and cellular response to tumor necrosis factor (figure 3G). An increase in IL-10 production by not only cancer-associated fibroblasts and DCs but also Siglec-9+TAMs, which were regulated by the IL-10 produced by themselves36(figure 3D and online supplemental figure S4G). STAT3 and JUN were critical transcription factors and regulatory molecules involved in the IL-10 related signaling pathway (online supplemental figure S4H), which contributed to the development of a protumorigenic M2-like phenotype. Thus, the Siglec-9+TAMs were identified as a protumoral phenotype.
Figure 3.
Phenotype of Siglec-9+TAMs by scRNA-seq in HGSC. (A and B) Characterization and quantification of the myeloid cell population found in the scRNA-seq analysis of HGSC tumors. (C) Heatmap highlighting the markers characterizing each myeloid population. (D) Volcano plots of differentially expressed genes between tumors with Siglec-9+TAMs and Siglec-9− TAMs in HGSC (adjusted p<0.05, |log2 fold change|>0.25). P value adjustment is performed using bonferroni correction. (E and F) Gene Set Enrichment Analysis (GSEA) plots of M2-like signature (E) and phagocytic pathways (F) were shown in Siglec-9+TAMs compared with Siglec-9− TAMs. The signature was defined by genes with significant expression changes. (G) Barplot showing gene ontology (GO) analysis of the downregulated DEGs between Siglec-9−TAMs and Siglec-9+TAMs. Orange refers to enriched pathways in molecular function; blue refers to enriched pathways in cellular components, and green refers to enriched pathways in biological processes. P, multiple testing-corrected p values were determined using the Benjamini-Hochberg method. (H) Analysis of the differentiation of monocytes toward macrophages using the R package monocle2. Pseudotime trajectory analysis of selected myeloid cells (monocytes cells, C1QC+TAMs, SiGlec-9+TAMs, SPP1+TAMs, PLTP+TAMs and proliferative cells; n=13 249) with high variable genes. Each dot represents one single cell, colored according to its cluster label. The inlet plot showed each cell with a pseudotime score from dark blue to yellow, indicating early and terminal states, respectively. For myeloid cell clusters, 13 249 cells were randomly selected for the analysis. HGSC, high-grade serous ovarian cancer; Siglec-9, sialic acid-binding immunoglobulin-like lectin 9; TAMs, tumor-associated macrophages
jitc-2023-007099supp005.pdf (2.1MB, pdf)
jitc-2023-007099supp017.pdf (32.6KB, pdf)
To gain a deeper understanding of the transition from monocytes to macrophages, the developmental trajectory was found to have a branched structure, with the Siglec-9+TAMs and PLTP+TAMs clusters located at the opposite end of the monocytes. C1QC+TAMs, SPP1+TAMs, and proliferative cells were located in between, indicating their intermediate functional states (figure 3H, online supplemental figure S4E). Hence, Siglec-9+TAMs represented terminally differentiated macrophages with protumoral characteristics.
To investigate the potential mechanism between CD8+T cells and Siglec-9+TAMs in scRNA analysis, 22 out of 63 signaling pathways were highly active in both primary and ascitic microenvironments, which included nine pathways involved in inflammatory and immune responses, such as CXCL, GALACTIN, COMPLEMENT, MIF, integrin, and glycoprotein (black in online supplemental figure S4I). Chemokines (CCL5-CCR1), adhesive connection (ITGAL-ICAM1, ANXA1-FPR1), and immune regulation (HAVCR2-LGALS9, MIF-(CD74+CD44) were potential interaction pathways between CD8+T cells and Siglec-9+TAMs in the ascitic microenvironment (online supplemental figure S4J). Consequently, Siglec-9+TAMs, which are a type of macrophage with protumoral properties and have reached the final stage of macrophage development, can inhibit the immune response of CD8+T cells through both direct and indirect contact.
Identification of the immunosuppressive microenvironment in specimens of high Siglec-9+TAMs
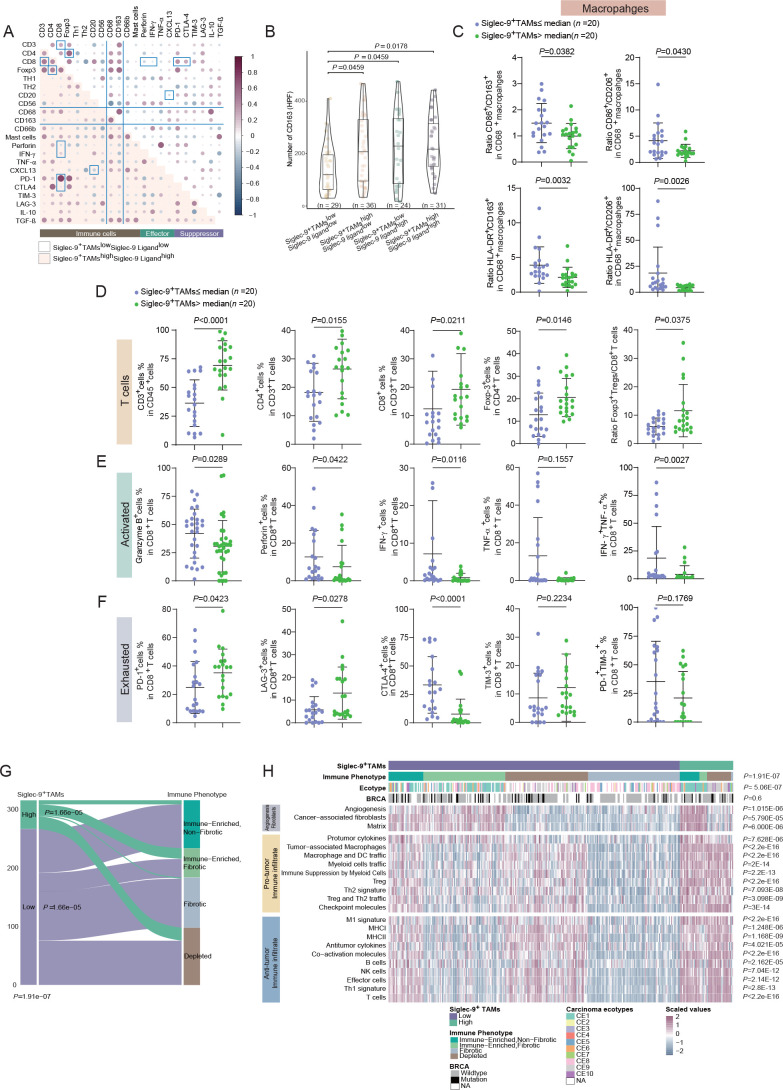
To examine the distinct features of immune contexture concerning the expression of Siglec-9 and its ligand, a positive correlation between CD8+T cells and CD4+T cells, as well as CD163+macrophages and Th2 in Siglec-9+TAMshighSiglec-9 ligandhigh subgroup (figure 4A). Conversely, CD163+macrophages exhibited a negative correlation with tumor necrosis factor-α (TNF-α), while both CD68+macrophages and CD163+macrophages were positively associated with CD66b+neutrophils, mast cells, and transforming growth factor-β (TGF-β) in the Siglec-9+TAMshighSiglec-9 ligandhigh subgroup (figure 4A). CD163+cells mostly infiltrate in high Siglec-9+TAMs despite the different levels of Siglec-9 ligand (figure 4B). Findings in the TCGA cohort supported the results in Fudan cohort, which annotated Siglec-9+TAMs manifested protumoral phenotype and had no association with FIGO stages (online supplemental figure S5A–C). Furthermore, high infiltration of Siglec-9+TAMs was found to be a predictor of poor prognosis (p=0.0005, online supplemental figure S5D). A higher M2/M1 ratio was observed in Siglec-9+TAMshigh subgroup rather than the Siglec-9+TAMslow subgroup (figure 4C). CD163 expression did not show correlation between high and low Siglec-9+TAMs subgroups (online supplemental figure S5E) and the combination of CD163 and Siglec-9+TAMs was not a prognostic indicator for OS (online supplemental figure S5F).
Figure 4.
Identification of the immunosuppressive microenvironment with Siglec-9+TAMs infiltration. (A) A Spearman correlation dot plot between immune cells, effectors and suppressors as proportions out of all cells, divided by Siglec-9+TAMs infiltration and Siglec-9 ligand expression (n=31 Siglec-9+TAMshighSiglec-9 ligandhigh subgroup, n=29 Siglec-9+TAMslowSiglec-9 ligandlow subgroup). Dots in white background and light red background represent correlations in low-infiltrating Siglec-9+TAMs and low-expressing Siglec-9 ligand tumors, respectively. Dots shaded purple represent positive correlations, while dots shaded blue represent negative correlations. Black lines separate the immune, effectors, and suppressor columns and rows. Correlations with a p value <0.05 were shown. (B) Violin plot showing portions of CD163 expression stratified by Siglec-9+TAMs and Siglec-9 ligand in HGSC. In the box plots inside violin plots, the black horizontal lines represented the sample means, the boxes extend from the first to third quartile, and the whiskers indicated values at 1.5 times the IQR. Data were analyzed by Kruskal-Wallis test. (C) Ratio of M1-like to M2-like TAMs in Siglec-9+TAMs high/low infiltration group. (D) The frequency of T cells in Siglec-9+TAMs high/low infiltration group. (E) Expression of effector molecules on CD8+T cells in Siglec-9+TAMs high/low infiltration group. (F) Expression of coinhibitory receptors on CD8+T cells stratified by infiltration of Siglec-9+TAMs. Data were analyzed by Mann-Whitney U test and presented as mean and SD. (G) Sankey diagram showing the percentages of Siglec-9+TAMs across immune phenotypes among HGSC patients and vice versa. The χ2 test was used to compare groups. (H) Unsupervised hierarchical clustering from TCGA cohorts showing relative enrichment of indicated 22 functional gene expression signatures (rows). Data were patient scaled and immune population z-scored for visualization. Each column represents one patient (n=316). Data were analyzed by Kruskal-Wallis test. HGSC, high-grade serous ovarian cancer; Siglec-9, sialic acid-binding immunoglobulin-like lectin 9; TAMs, tumor-associated macrophages.
jitc-2023-007099supp006.pdf (604.5KB, pdf)
To investigate the impact of Siglec-9 and its ligand on the function of T cells, an increase of CD4+ and CD8+T cells was observed in the Siglec-9+TAMshigh subgroup (figure 4D), while the cytotoxic ability of CD8+T was remarkably attenuated with downregulation of effector molecules containing TNF-α, IFN-γ, granzyme B (GZMB) and perforin than Siglec-9+TAMslow subgroup (figure 4E). Meanwhile, the multifunctional CD8+T cell response in the Siglec-9+TAMshigh subgroup was impaired, as measured by the coexpression of TNF-α and IFN-γ (figure 4E, online supplemental figure S5G). Conversely, inhibitory receptor expression analysis revealed that CD8+T cells in Siglec-9+TAMshigh subgroup upregulated the expression of PD-1 and TIM-3 and downregulated CTLA-4 expression (figure 4A and F, online supplemental figure S5H) compared with Siglec-9+TAMslow subgroup. CD8+T cells predicted a favorable prognosis in patients with FIGO stage III/IV, which was not found in Siglec-9+TAMshigh subgroup (online supplemental figure S5I–K). An increase of Foxp3+CD4+T cells proportion and Tregs/CD8+T cells ratio was observed in the Siglec-9+TAMshigh subgroup compared with Siglec-9+TAMslow subgroup (figure 4D). Taken together, Siglec-9+TAMs showed immunosuppressive phenotype and were closely associated with T cell immune suppression in HGSC.
To confirm the correlation between immune phenotype and Siglec-9+TAMs, an increase in tumors defined as depleted (D, 28%–45%) and immune enriched and non-fibrotic (ie, 12%–31%), and a drastic decrease in tumors defined as immune enriched and fibrotic (IE/F, 28% to 17%) and fibrotic (F, 32% to 7%) from high Siglec-9+TAMs to low Siglec-9+TAMs tumors (figure 4G).30 High Siglec-9+TAMs subgroup increased accumulation of CE1, CE9, and CE10, along with a higher infiltration of M2-like macrophages (figure 4H). As illustrated, a high Siglec-9+TAMs signature score was associated with higher levels of both protumor immune infiltration (TAMs, immune suppression by myeloid cells, Tregs, Th2 signature, and checkpoint molecules) and matrix signature (angiogenesis and cancer-associated fibroblasts)37 (figure 4H). Two typical immune effector cells, CD8+T cells, and NK cells were more abundant in the Siglec-9+TAMshigh subgroup (figure 4H). Collectively, Siglec-9+TAMs displayed characteristics that had both immunosuppressive and immunoreactive effects.
Siglec-9 blockade shifts macrophages to an antitumoral phenotype and restores the antitumor activity of CD8+T cells
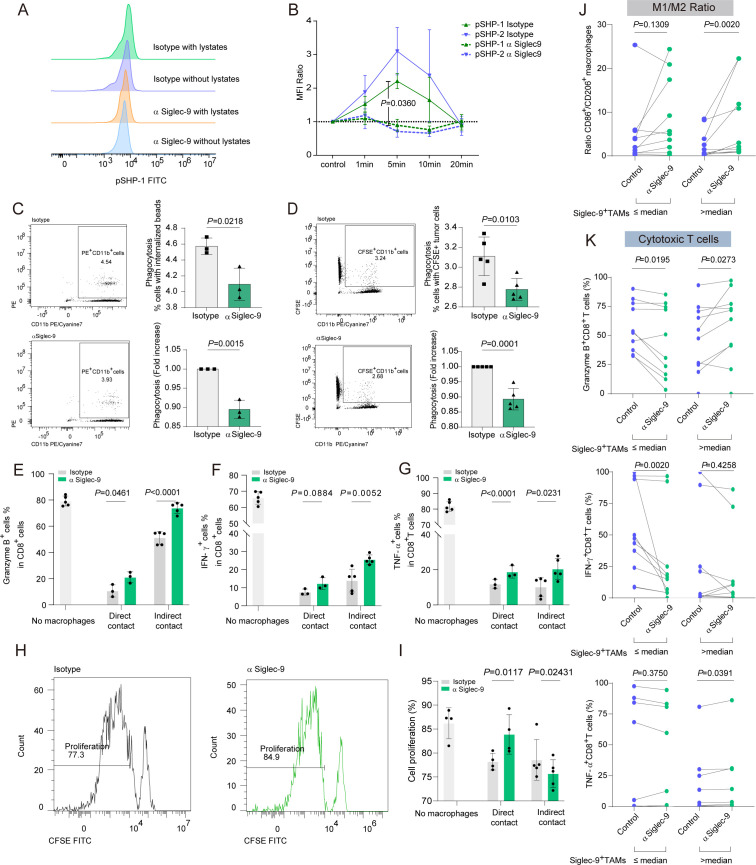
To confirm that immunosuppressive signals were being transmitted through tumor lysate-pulsed Siglec-9+TAMs, rapid phosphorylation of SHP-1 within 5 min was observed,17 19 38 which was suppressed by the Siglec-9 blockade (figure 5A,B). By blocking Siglec-9, Siglec-9+TAMs reduced phagocytosis under coculture with non-specific phagocytosis of latex beads and CFSE-labeled cells than the isotype-treated group (figure 5C,D). Consequently, anti-Siglec-9 reprogrammed TAMs toward an immunogenic M1-like phenotype.
Figure 5.
Siglec-9 blockade decreases the number of intratumoral M2 macrophages and increases CD8+T effectors. (A) Representative histograms of SHP-1 phosphorylation staining in human peripheral blood mononuclear cells (PBMCs)-derived Siglec-9+TAMs evaluated by flow cytometry. Statistical significance was determined by unpaired two-tailed Mann-Whitney U-test. (B) Analysis of flow cytometry demonstrating SHP-1 or SHP-2 phosphorylation in Siglec-9+TAMs from healthy donors treated with α Siglec-9 or isotype control (both 5 µg/mL) for 1 or 5, 10, and 20 min compared with time-matched controls (ratio). Plots showed the mean±SEM. Significance was assessed by Mann-Whitney U test. (C–D) Representative flow cytometry plots and flow cytometry based quantification of phagocytosis depicting the phagocytosis of (C) carboxylate-modified red fluorescence latex beads and (D) HO-8190 cells treated with α Siglec-9, compared with the IgG control. (E–G) Intracellular cytokine production of (E) granzyme B or (F) IFN-γ or (G) TNF-α by healthy donor CD8+T cells cocultured with TCM-educated macrophages in direct or indirect contact on engagement by α Siglec-9 (5 µg/mL) (n=3–5, t-test). (H and I) T-cell proliferation in the presence of TCM-educated macrophages. Histograms showed the percentage of proliferating T cells cocultured (1:2) with macrophages treated by α Siglec-9 or isotype IgG (H). Bar graph showed the percentage of proliferating T cells cocultured with macrophages in direct or indirect contact (n=3–5) (I). (J and K) HGSC single cell suspension was incubated with control or Siglec-9-neutralizing antibody and subjected to flow cytometric analysis to determine the expression of the ratio of M1-like to M2-like TAMs (J), effector molecules (GZMB, IFN-γ, and TNF-α) on CD8+T cells (K) stratified by infiltration of Siglec-9+TAMs infiltration. Two-sided Wilcoxon rank-sum test was used for pairwise comparisons. CFSE, carboxyfluorescein succinimidyl ester; GZMB, granzyme B; HGSC, high-grade serous ovarian cancer; Siglec-9, sialic acid-binding immunoglobulin-like lectin 9; TCM, tumor-conditioned medium; TNF-α, tumor necrosis factor-α.
To investigate the impact of blocking Siglec-9 on macrophages and its effect on T cell function, CD8+T cells increased levels of cytotoxic markers, including IFN-γ, GZMB, and TNF-α, as well as enhanced proliferation in anti-Siglec-9 compared with isotype group, which was observed both in direct and indirect cell–cell contact conditions (figure 5E–I, online supplemental figure S6A–C). Ex vivo results supported the in vitro findings that blocking Siglec-9 in the Siglec-9+TAMshigh subgroup could upregulate the expression of the M1/2 ratio (CD86/CD206 ratio, figure 5J). Additionally, it upregulated the expression of proinflammatory markers (CD86 and HLA-DR) (online supplemental figure S6D) and downregulated the expression of CD163, CD206, PD-L1, and arginase-1 in the Siglec-9+TAMshigh subgroup with Siglec-9 blockade (online supplemental figure S6E, online supplemental figure S7A). Furthermore, blocking Siglec-9 in the Siglec-9+TAMshigh subgroup led to an expansion in the expression of effector cytokines, such as GZMB and TNF-α, in CD8+T cells (figure 5K, online supplemental figure S7B). Targeting Siglec-9 by mAb not only reversed the protumorigenic phenotype of macrophages but also enhanced CD8+T cell immune response through both direct and indirect cell–cell contact.
jitc-2023-007099supp007.pdf (306.8KB, pdf)
jitc-2023-007099supp008.pdf (641.5KB, pdf)
Combining PD-1 blockade with targeting Siglec-9 improved the CD8+T activity in tumors with high-infiltrating Siglec-9+TAMs
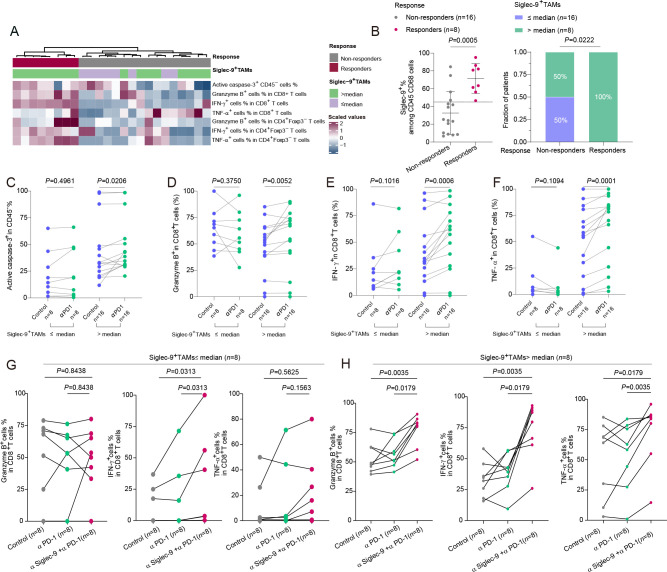
To recognize the potential biomarker of Siglec-9+TAMs and targeted therapy augmenting ICB response by Siglec-9 blockade,10 patients were classified into two groups, responders (n=8) and non-responders (n=16) to PD-1 blockade treatment, based on changes observed in the level of cytotoxic T cells and tumor apoptosis (figure 6A). More Siglec-9+TAMs accumulated in responders towards anti-PD-1 therapy (figure 6B). Targeting PD-1 led to the induction of apoptotic tumor cells evidenced by upregulation of active caspase-3 (figure 6C) and an increase in the cytotoxicity of CD8+T cells demonstrated by upregulation of GZMB, IFN-γ, and TNF-α (figure 6D–F) in Siglec-9+TAMshigh subgroup.
Figure 6.
Intratumoural CD8+T cells were functionally restored following PD-1 blockade in high-infiltrating Siglec-9+TAMs patients. (A) Hierarchical clustering of active caspase-3 among CD45− cells and cytotoxic markers (GZMB, IFN-γ, and TNF-α) of both CD8+T and CD4+Foxp3− T cells to identify the response of PD-1 blockade between Siglec-9+TAMs low and high subgroups (n=24). Expression values were scaled. Samples were clustered using complete linkage and Euclidean distance. (B) Relationship between response to PD-1 blockade and Siglec-9+TAMs infiltration in patients (n=24). Flow cytometric analysis by Mann-Whitney U test (left) and χ2 test (right) was shown. (C–F) The effect of pembrolizumab on the expression of active caspase-3 among CD45− cells (C) and cytotoxic markers (GZMB, IFN-γ, and TNF-α) of CD8+T cells (D–F) in Siglec-9+TAMs cells high/low infiltration group from HGSC tissue samples (n=24). Two-sided Wilcoxon rank-sum test was used for pairwise comparisons. (G and H) The frequency of cytotoxic markers after isotype, Siglec-9-neutralizing antibody, PD-1 blockade, and Siglec-9-neutralizing antibody combined with PD-1 blockade treatment in HGSC tumor tissues between high and low Siglec-9+TAMs subgroups in Fudan cohort (n=16). GZMB, granzyme B; HGSC, high-grade serous ovarian cancer; PD-1, programmed death receptor-1; Siglec-9, sialic acid-binding immunoglobulin-like lectin 9; TAMs, tumor-associated macrophages; TNF-α, tumor necrosis factor-α.
To examine the effect of dual blockade of PD-1 and Siglec-9 on the cytotoxic function of CD8+T cells than the PD-1 blockade, no significant changes of GZMB or TNF-α on CD8+T cells were found in Siglec-9+TAMslow subgroup (figure 6G), whereas prominent increases in GZMB, IFN-γ, and TNF-α on CD8+T cells in Siglec-9+TAMshigh subgroup (figure 6H). Furthermore, a significant correlation between Siglec-9+TAMs and various immune-responsive factors such as DNA damage immune response, immune checkpoint, tertiary lymphoid structure, and IFN-γ (online supplemental figure S8A), as well as good prognosis signatures that were responsive to ICB in patients with HGSC (online supplemental figure S8B). Generally, high infiltration of Siglec-9+TAMs predicted a better response toward PD-1 blockade than low infiltration and targeting Siglec-9 enhanced the therapeutic effects of PD-1 blockade in HGSC.
jitc-2023-007099supp009.pdf (412.8KB, pdf)
Characterization of Siglec-9+TAMs infiltration and PD-L1 expression across survival outcomes and therapeutic implication
To categorize patients based on their PD-L1 expression and Siglec-9+TAMs infiltration in survival analysis, patients with higher levels of Siglec-9+TAMs infiltration and increased PD-L1 expression had poorer outcomes (online supplemental figure S9A). Patients in the Siglec-9+TAMslowPD-L1pos subgroup revealed a survival advantage compared with other subgroups (online supplemental figure S9B). A positive correlation between the signature of Siglec-9+TAMs and the expression of CD274 in the TCGA cohort was proved (online supplemental figure S9C). DNA damage immune response, ATR/BRCA pathway, VEGF-VEGFR pathway and immune-related pathways were associated with Siglec-9+TAMslowPD-L1pos and Siglec-9+TAMshighPD-L1pos subgroups, which indicated favorable clinical decisions (online supplemental figure S9D).
jitc-2023-007099supp010.pdf (850.2KB, pdf)
Discussion
Enhancing our understanding of the mechanisms through which HGSC evades the immune system will aid in the development of new strategies to increase the effectiveness of ICB therapy. Our study revealed that macrophages in HGSC expressed Siglec-9 to a significant extent, while the levels of Siglec-1 and Siglec-7 were relatively low. Siglec-9+TAMs correlated with adverse prognosis, which showed a protumor phenotype and promoted immune evasion. Inhibiting Siglec-9 could transform macrophages into a more proinflammatory phenotype and restore the functional ability of T cells based on in vitro and ex vivo experiments. Siglec-9+TAMs might be served as a prognostic biomarker in HGSC. Targeting Siglec-9 expression on TAMs may be a possible strategy to improve the effectiveness of immune checkpoint blockade therapy.
Previous studies showed the plasticity and diversity of TAMs can clarify their varying roles in the tumor microenvironment.14 39 Various Siglec receptors have been shown to affect macrophage differentiation and transformation toward an M2 immunosuppressive, protumor phenotype.26 35 40 In line with our observation, Siglec-9+TAMs represented immunosuppressive macrophage populations demonstrating high plasticity and differentiated from monocytes and progressed to a terminally differentiated state. Given that inhibitors of both Siglec-7 and Siglec-9 are in preclinical development,38 we further found that TAMs expressed high levels of Siglec-9 on TAMs in HGSC, while Siglec-7 and Siglec-1 were expressed at relatively low levels.19 26 Siglec-9+TAMs were found to be correlated with worse patient outcomes and poorer prognosis in HGSC as well as in other cancers like glioblastoma.24 Siglec-9+TAMshigh subgroup showed immune evasion24 25 mechanisms with increased infiltration of CD8+T cells into the tumor, which involved downregulation of effector molecules and upregulation of inhibitory receptors of the CD8+T cells.24 25
Numerous therapeutic strategies targeting TAMs have been studied, which include depleting, reprogramming TAMs, and preventing TAMs recruitment.14 Identical to the recent studies, Siglec receptors may be considered novel immune checkpoints, which can not only reverse the immunosuppression of M2-like TAMs but also restore the effector function of T cells using in vitro assays and an established ex vivo model.40 Furthermore, we observed Siglec-9+TAMshigh subgroup predicted a better response toward PD-1 blockade by upscaling the cytotoxic T cells.24 25 41 42 Congruously, the combined intervention of Siglec-9 and PD-1/PD-L1 could be highly effective in activating CD8 T cells against tumors, indicating Siglec-9+TAMs could be a prominent target in treatment.
Evidence has proved that mutations could drive immunoediting mutational signatures in HGSC.43 44 Siglec-9 expression has been shown to skew macrophage polarization to a protumorigenic phenotype and increase PD-L1 expression in macrophages.20 The Siglec-9+TAMs and PD-L1 panel may guide the identification of potential therapeutic targets (online supplemental figures S9E and S10A–D). Tumors of the Siglec-9+TAMslowPD-L1neg subtype could benefit from BET or Notch inhibitors, whereas the Siglec-9+TAMslowPD-L1pos subtype might be susceptible to Nrf2 pathway inhibitors. The Siglec-9+TAMshighPD-L1neg subtype could be sensitive to Siglec-9-targeted therapy and WNT or HIPPO inhibitor. The Siglec-9+TAMshighPD-L1pos subtype could benefit from Siglec-9-targeted therapy, ICB combined with TGF-β, or JAK-STAT inhibitor. Accumulatively, the combined intervention of Siglec-9+TAMs and PD-1/PD-L1 was a potential target to improve the effects of assorted types of targeted therapies.
jitc-2023-007099supp011.pdf (538.5KB, pdf)
The limitations of this study are the following. Evidence showed the interaction of Siglec-9 and the aberrantly glycosylated MUC1 and MUC16 (CA125) on tumor cells modulates the TME.20 45 In our study, Siglec-9+TAMs upregulated PD-L1 and IL-10, and inhibiting Siglec-9 downregulated PD-L1 and IL-10, which was consistent with previous studies.46 However, the specific mechanisms by which the tumor cell microenvironment or ascites fluid might lead to the expression of Siglec-9 on macrophages remain unknown. Furthermore, CCR1 negative TAMs ignited CD8+T cell-mediated antitumor immunity,47 and CCL5 in cancer modulated the stability of PD-L1 promoting immune escape.48 Although the actual interaction between CD8+T cells and Siglec-9+TAMs has not been fully explored, the impact of CD8+T cells on Siglec-9+TAMs through direct (LGALS9-HAVCR2) and indirect (CCL5-CCR1) contact has been proposed. In addition, a randomized trial on a larger scale in the future will involve multiple centers and offer more robust evidence to validate the role of Siglec-9 as a prognostic biomarker and prominent target prospectively.
Overall, this study has revealed a distinct subset of TAMs in HGSC that express Siglec-9, exhibit a protumoral phenotype, and facilitate immune evasion. Further investigation into the potential of Siglec-9+TAMs as both a prognostic biomarker and therapeutic target, particularly in combination with ICB, is a promising avenue for future research in the treatment of HGSC.
Acknowledgments
We would like to thank Dr Jing Zhao (Department of Pathology, Obstetrics and Gynecology Hospital of Fudan University, Shanghai, China) for her excellent pathological technology help and Dr Hongbo Zhao (Bank of Tumor Resources, Obstetrics and Gynecology Hospital of Fudan University, Shanghai, China) for her help.
Footnotes
YW and MH contributed equally.
Contributors: YW and MH for the acquisition of data, analysis and interpretation of data, statistical analysis, and drafting of the manuscript; CZ, KC, GZ and MY for technical and material support; YH, WJ, and HL for study concept and design, analysis and interpretation of data, drafting of the manuscript, obtained funding, and study supervision. YH, WJ, and HL are responsible for the overall content as guarantors. All authors contributed to the revision of the manuscript.
Funding: This study was funded by grants from the National Natural Science Foundation of China grant 82072881, 82273205, and 82203665. Natural Science Foundation of Shanghai grant 23ZR1408300, Shanghai Sailing Program grant 21YF1403900, and Science and Technology Commission of Shanghai Municipality grant 22Y31900502. All these study sponsors have no roles in the study design, in the collection, analysis, and interpretation of data.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Not applicable.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Clinical Research Ethics Committee of the Obstetrics and Gynecology Hospital of Fudan University with approval (Kyy2016-49, Kyy2017-27). Participants gave informed consent to participate in the study before taking part.
References
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics. CA Cancer J Clin 2022;72:7–33. 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 2.Beltra J-C, Manne S, Abdel-Hakeem MS, et al. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 2020;52:825–41. 10.1016/j.immuni.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labidi-Galy SI, Papp E, Hallberg D, et al. High grade serous ovarian carcinomas originate in the Fallopian tube. Nat Commun 2017;8:1093. 10.1038/s41467-017-00962-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collinson F, Qian W, Fossati R, et al. Optimal treatment of early-stage ovarian cancer. Ann Oncol 2014;25:1165–71. 10.1093/annonc/mdu116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman RL, Brady MF, Herzog TJ, et al. Bevacizumab and paclitaxel-carboplatin chemotherapy and secondary cytoreduction in recurrent, platinum-sensitive ovarian cancer (NRG oncology/gynecologic oncology group study GOG-0213): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 2017;18:779–91. 10.1016/S1470-2045(17)30279-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray-Coquard I, Pautier P, Pignata S, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 2019;381:2416–28. 10.1056/NEJMoa1911361 [DOI] [PubMed] [Google Scholar]
- 7.González-Martín A, Pothuri B, Vergote I, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2019;381:2391–402. 10.1056/NEJMoa1910962 [DOI] [PubMed] [Google Scholar]
- 8.Cheng Z, Mirza H, Ennis DP, et al. The genomic landscape of early-stage ovarian high-grade serous carcinoma. Clin Cancer Res 2022;28:2911–22. 10.1158/1078-0432.CCR-21-1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaulin N, Chilakapati SR, Zsiros E. Turning cold into hot: combination of pembrolizumab with bevacizumab and oral metronomic cyclophosphamide increases immune cell migration into the tumor microenvironment in responding patients with recurrent ovarian cancer (090). Gynecologic Oncology 2022;166:S61. 10.1016/S0090-8258(22)01316-6 [DOI] [Google Scholar]
- 10.Konstantinopoulos PA, Cannistra SA. Immune checkpoint inhibitors in ovarian cancer: can we bridge the gap between Imagynation and reality J Clin Oncol 2021;39:1833–8. 10.1200/JCO.21.00571 [DOI] [PubMed] [Google Scholar]
- 11.Porter RL, Matulonis UA. Checkpoint blockade: not yet NINJA status in ovarian cancer. J Clin Oncol 2021;39:3651–5. 10.1200/JCO.21.01886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim TK, Vandsemb EN, Herbst RS, et al. Adaptive immune resistance at the tumour site: mechanisms and therapeutic opportunities. Nat Rev Drug Discov 2022;21:529–40. 10.1038/s41573-022-00493-5 [DOI] [PubMed] [Google Scholar]
- 13.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19:369–82. 10.1038/s41577-019-0127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov 2018;17:887–904. 10.1038/nrd.2018.169 [DOI] [PubMed] [Google Scholar]
- 15.Christofides A, Strauss L, Yeo A, et al. The complex role of tumor-infiltrating macrophages. Nat Immunol 2022;23:1148–56. 10.1038/s41590-022-01267-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Büll C, Boltje TJ, Balneger N, et al. Sialic acid blockade suppresses tumor growth by enhancing T-cell-mediated tumor immunity. Cancer Res 2018;78:3574–88. 10.1158/0008-5472.CAN-17-3376 [DOI] [PubMed] [Google Scholar]
- 17.RodrÍguez E, Schetters STT, van Kooyk Y. The tumour Glyco-code as a novel immune checkpoint for immunotherapy. Nat Rev Immunol 2018;18:204–11. 10.1038/nri.2018.3 [DOI] [PubMed] [Google Scholar]
- 18.Rodrigues Mantuano N, Natoli M, Zippelius A, et al. Tumor-associated carbohydrates and immunomodulatory lectins as targets for cancer immunotherapy. J Immunother Cancer 2020;8:e001222. 10.1136/jitc-2020-001222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith BAH, Bertozzi CR. Author correction: the clinical impact of glycobiology: targeting selectins, Siglecs and mammalian glycans. Nat Rev Drug Discov 2021;20:244. 10.1038/s41573-021-00160-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beatson R, Tajadura-Ortega V, Achkova D, et al. The Mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat Immunol 2016;17:1273–81. 10.1038/ni.3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van de Wall S, Santegoets KCM, van Houtum EJH, et al. Sialoglycans and Siglecs can shape the tumor immune microenvironment. Trends Immunol 2020;41:274–85. 10.1016/j.it.2020.02.001 [DOI] [PubMed] [Google Scholar]
- 22.Ibarlucea-Benitez I, Weitzenfeld P, Smith P, et al. Siglecs-7/9 function as inhibitory immune checkpoints in vivo and can be targeted to enhance therapeutic antitumor immunity. Proc Natl Acad Sci USA 2021;118:26. 10.1073/pnas.2107424118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nalle S, Lam H, Leung L, et al. 875 Al009, a fusion protein and multi-Siglec inhibitor, repolarizes suppressive myeloid cells and potentiates anti-cancer effects. J Immunother Cancer 2021;9:A917. 10.1136/jitc-2021-SITC2021.875 [DOI] [Google Scholar]
- 24.Stanczak MA, Rodrigues Mantuano N, Kirchhammer N, et al. Targeting cancer glycosylation repolarizes tumor-associated macrophages allowing effective immune checkpoint blockade. Sci Transl Med 2022;14:eabj1270. 10.1126/scitranslmed.abj1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmassmann P, Roux J, Buck A, et al. Targeting the Siglec–Sialic acid axis promotes antitumor immune responses in preclinical models of glioblastoma. Sci Transl Med 2023;15:705. 10.1126/scitranslmed.adf5302 [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez E, Boelaars K, Brown K, et al. Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat Commun 2021;12:1270. 10.1038/s41467-021-21550-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA Pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018;173:400–16. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 2013;14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luca BA, Steen CB, Matusiak M, et al. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell 2021;184:5482–96. 10.1016/j.cell.2021.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jandus C, Boligan KF, Chijioke O, et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J Clin Invest 2014;124:1810–20. 10.1172/JCI65899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geistlinger L, Oh S, Ramos M, et al. Multiomic analysis of subtype evolution and heterogeneity in high-grade serous ovarian carcinoma. Cancer Res 2020;80:4335–45. 10.1158/0008-5472.CAN-20-0521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinh HQ, Lin X, Abbasi F, et al. Single-cell transcriptomics identifies gene expression networks driving differentiation and tumorigenesis in the human Fallopian tube. Cell Rep 2021;35:108978. 10.1016/j.celrep.2021.108978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izar B, Tirosh I, Stover EH, et al. A single-cell landscape of high-grade serous ovarian cancer. Nat Med 2020;26:1271–9. 10.1038/s41591-020-0926-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez MA, Lu DR, Yousefi M, et al. Phagocytosis increases an oxidative metabolic and immune suppressive signature in tumor macrophages. J Exp Med 2023;220:e20221472. 10.1084/jem.20221472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ando M, Tu W, Nishijima K-I, et al. Siglec-9 enhances IL-10 production in macrophages via tyrosine-based motifs. Biochem Biophys Res Commun 2008;369:878–83. 10.1016/j.bbrc.2008.02.111 [DOI] [PubMed] [Google Scholar]
- 37.Bagaev A, Kotlov N, Nomie K, et al. Conserved Pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 2021;39:845–65. 10.1016/j.ccell.2021.04.014 [DOI] [PubMed] [Google Scholar]
- 38.Läubli H, Nalle SC, Maslyar D. Targeting the Siglec–Sialic acid immune axis in cancer: current and future approaches. Cancer Immunol Res 2022;10:1423–32. 10.1158/2326-6066.CIR-22-0366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pittet MJ, Michielin O, Migliorini D. Author correction: clinical relevance of tumour-associated macrophages. Nat Rev Clin Oncol 2022;19:424. 10.1038/s41571-022-00632-2 [DOI] [PubMed] [Google Scholar]
- 40.Stanczak MA, Läubli H. Siglec receptors as new immune checkpoints in cancer. Mol Aspects Med 2023;90:101112. 10.1016/j.mam.2022.101112 [DOI] [PubMed] [Google Scholar]
- 41.Läubli H, Pearce OMT, Schwarz F, et al. Engagement of myelomonocytic Siglecs by tumor-associated ligands modulates the innate immune response to cancer. Proc Natl Acad Sci U S A 2014;111:14211–6. 10.1073/pnas.1409580111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mei Y, Wang X, Zhang J, et al. Siglec-9 acts as an immune-checkpoint molecule on macrophages in glioblastoma, restricting T-cell priming and immunotherapy response. Nat Cancer 2023;2023:1–19. 10.1038/s43018-023-00598-9 [DOI] [PubMed] [Google Scholar]
- 43.Vázquez-García I, Uhlitz F, Ceglia N, et al. Ovarian cancer mutational processes drive site-specific immune evasion. Nature 2022;612:778–86. 10.1038/s41586-022-05496-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garsed DW, Pandey A, Fereday S, et al. The genomic and immune landscape of long-term survivors of high-grade serous ovarian cancer. Nat Genet 2022;54:1853–64. 10.1038/s41588-022-01230-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belisle JA, Horibata S, Jennifer GAA, et al. Identification of Siglec-9 as the receptor for Muc16 on human NK cells, B cells, and monocytes. Mol Cancer 2010;9:118. 10.1186/1476-4598-9-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shoji T, Higuchi H, Nishijima K-I, et al. Effects of Siglec on the expression of IL-10 in the macrophage cell line Raw264. Cytotechnology 2015;67:633–9. 10.1007/s10616-014-9717-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Brown KL, Yan W, et al. Abstract P053: ablation of CCR1 relieves immunosuppression in pancreatic cancer. Cancer Immunol Res 2022;10:P053. 10.1158/2326-6074.TUMIMM21-P053 [DOI] [Google Scholar]
- 48.Liu C, Yao Z, Wang J, et al. Macrophage-derived CCL5 facilitates immune escape of colorectal cancer cells via the p65/STAT3-CSN5-PD-L1 pathway. Cell Death Differ 2020;27:1765–81. 10.1038/s41418-019-0460-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007099supp001.pdf (245.8KB, pdf)
jitc-2023-007099supp012.pdf (49.6KB, pdf)
jitc-2023-007099supp013.pdf (43.8KB, pdf)
jitc-2023-007099supp014.pdf (44.1KB, pdf)
jitc-2023-007099supp002.pdf (720.3KB, pdf)
jitc-2023-007099supp003.pdf (465.9KB, pdf)
jitc-2023-007099supp015.pdf (41.2KB, pdf)
jitc-2023-007099supp016.pdf (43KB, pdf)
jitc-2023-007099supp004.pdf (774.6KB, pdf)
jitc-2023-007099supp005.pdf (2.1MB, pdf)
jitc-2023-007099supp017.pdf (32.6KB, pdf)
jitc-2023-007099supp006.pdf (604.5KB, pdf)
jitc-2023-007099supp007.pdf (306.8KB, pdf)
jitc-2023-007099supp008.pdf (641.5KB, pdf)
jitc-2023-007099supp009.pdf (412.8KB, pdf)
jitc-2023-007099supp010.pdf (850.2KB, pdf)
jitc-2023-007099supp011.pdf (538.5KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Not applicable.