Abstract
We discuss pathological epigenetic events that are reversible (PEERs). A recent study by Poganik and colleagues showed that severe stress in mice and humans transiently elevates biological age of several tissues, and this transient age increase is reversible when the stress is removed. These studies suggest new strategies for reversing normal aging. However, it is important to note that developmental origin of health and disease studies have shown that developmental exposure to toxic chemicals such as lead causes permanent changes in neuron shape, connectivity and cellular hyperplasia of organs such as the heart and liver. In this review, the PEER hypothesis speculates that the hallmarks of aging and the hallmarks of developmental origin of health and disease intersect at PEERs.
Keywords: aging, developmental toxicity, DOHaD, epigenetic reprogramming, epigenomics, heterochronic parabiosis
Plain language summary
The main goal in aging research is to find treatments to reverse aging. There are nine hallmarks of aging which describe cellular mechanisms that change as we age. For example, one of the hallmarks of aging is cellular senescence, which means that cells stop dividing when they get old. In this review, we describe nine hallmarks of developmental origins of health and disease (DOHaD). DOHaD studies show that exposures of the mother during pregnancy to stress or toxic chemicals can alter the health of the child throughout the child's lifespan. We argue that six of the nine hallmarks of DOHaD overlap with the hallmarks of aging and are reversible by dietary restriction or by drugs such as rapamycin which affect nutrient signaling. Based on this finding, we have formulated a hypothesis that we call ‘pathological epigenetic events that are reversible’ that contain the six hallmarks that overlap between the hallmarks of aging and the hallmarks of DOHaD. With this unexpected connection between aging and DOHaD, we argue that findings in one field, such as drugs that reverse aging, can apply to treatments in the other field, such as ways to reverse the adverse effects of exposures during pregnancy.
Graphical abstract
The goal of much medical epigenetics is to find ways to reverse pathological epigenetic events. Many see in epigenetics the potential for a medical revolution. Richard C Francis (2012) [1].
Pathological epigenetic events (PEEs) are chemical exposures or other stressors that change the epigenome, usually during development in a dose-dependent manner. The word ‘Epigenetics’ in PEE refers to the broad sense of the word – including not only changes in DNA methylation, histones and noncoding RNAs, but also changes in mitochondrial function, loss of proteostasis (PROT), and the other hallmarks of aging and developmental origins of health and disease (DOHaD) discussed in this review. To avoid confusion, we will refer to changes in DNA methylation, histones and noncoding RNAs with the word ‘Epigenomics’. We will capitalize Epigenetics and Epigenomics to reinforce how the words are precisely used in this review.
The first use of the term ‘pathological epigenetic event’, to our knowledge, was in the above quote in a popular book by RC Francis titled, Epigenetics – How Environment Shapes our Genes [1]. PEE is a useful term because it concisely suggests an Epigenetic mechanism by which a transient stress, that is, a PEE, can have long-term adverse health outcomes. Some of the first, and most famous, studies showing transient stress having long-term consequences were by DJ Barker and colleagues in 1986, who studied pregnant women in impoverished areas of England and Wales who had poor nutrition during pregnancy. His hypothesis postulates that fetuses are molecularly and metabolically reprogrammed to adapt to the low nutrition conditions they experience and have a ‘thrifty phenotype’ during childhood and in later life in that they are efficient in using whatever low amounts of nutrition they can obtain [2]. However, when these thrifty babies enter the world and have a ready availability of highly processed foods, they are unable to metabolize the foods correctly and thereby have increased likelihood of developing deleterious diseases later in life such as obesity, Type 2 diabetes and hypertension-induced cardiovascular diseases. Barker's hypothesis was initially met with skepticism but was strongly supported by other studies over the past four decades. For example, in 1976, Rich-Edwards and colleagues studied over 70,000 nurses with birth weights in their medical records [3]. They showed that low birthweight children have an increased risk of developing cardiovascular diseases later in life.
The single-generation effects of malnutrition during pregnancy were extended to the next generation by Painter and colleagues in 2008 in the ‘Dutch famine birth cohort study’ [4]. Over 20,000 people died during the Dutch famine and thousands of women were subjected to famine conditions during all or part of their pregnancy. The Dutch famine was caused by a Nazi siege for several months at the end of World War II during 1944–1945. The babies born soon after the famine were frequently low body weight at birth, followed by compensatory overgrowth and increased likelihood of developing deleterious diseases such as obesity, Type 2 diabetes and cardiovascular diseases later in life [4]. Remarkably, the babies born from the women who experience famine during gestation (i.e., the F2) had reduced birth length (0.6 cm) and increased BMI (1.2 kg/m3). Furthermore, when the F2 grew up to adults, they had poor health 1.8-times more frequently than control F2 whose mothers did not endure famine conditions during pregnancy [4]. The Dutch famine study of 2008 stimulated numerous other multigenerational Epigenetic and Epigenomic studies in model organisms (e.g., plants [5]; Caenorhabditis elegans, reviewed in Frolows and Ashe [6]; Daphnia [7]; mouse [8–12]; and humans [13,14]). These studies suggest that diet or environmental chemical exposures or stress during early development can cause changes in the Epigenome (i.e., DNA methylation, miRNAs, piRNAs, etc.) that can be transmitted to future generations.
Barker's hypothesis, which originally dealt with malnutrition during pregnancy, is contained within the DOHaD hypothesis. The DOHaD hypothesis postulates that environmental exposures or stressors during sensitive windows of development, such as in the fetus or early childhood, can affect the health of the individual throughout life [15]. The first demonstration of a DOHaD-related early childhood exposure effect, to our knowledge, was in 1963 when Widdowson and McCance showed that, when rat pups were malnourished immediately after birth and during the 3 weeks of weaning, they grew slower over their lifetime [16]. However, malnourishment during weeks 9–12 had no long-term effect on growth [16]. This study was the first to demonstrate the main two tenets of the DOHaD hypothesis – environmental insults or stress early in life can have life-long consequences and the exposures must occur during a critical window of development to have maximal effects [15]. The effects of DOHaD are thought to be mediated by Epigenomics, with DNA methylation being the most studied mechanism, followed by small RNAs, modified histones and other mediators (reviewed in Felix and Cecil [17]).
In this review, we discuss our hypothesis that speculates that the hallmarks of aging and the hallmarks of DOHaD intersect at pathological epigenetic events that are reversible (PEERs). We developed the PEER hypothesis as a working model to explain many recent findings that gestational exposures to environmental chemicals or stress can accelerate Epigenetic clocks. For example, Pilsner and colleagues showed that sperm Epigenetic clocks correlate with adverse pregnancy outcomes in humans [18], and Oluwayiose and colleagues from the same laboratory showed that urinary phthalates correlate with advanced sperm Epigenomic aging [19]. Niemiec and colleagues showed using the Healthy Homes cohort that gestational exposure to perfluoroalkyl substances accelerates the Epigenomic age of DNA in umbilical cord blood [20]. Ladd-Acosta and colleagues showed using the Environmental Influences on Child Health Outcomes longitudinal cohort of over 50,000 children and mothers that pregnancy complications accelerate the Epigenomic gestational age of newborns [21]. There are many other studies along these lines, but these are beyond the scope of this review.
To support the PEER hypothesis, we describe: 1) Epigenetic clocks and the nine hallmarks of aging; 2) heterochronic parabiosis (HPB) experiments that alter the Epigenetic clocks of both the young and the old mouse; 3) the nine hallmarks of DOHaD and the overlap with six of the nine hallmarks of aging; 4) the six PEERS that intersect at the hallmarks of aging and DOHaD; and 5) the three hallmarks of DOHaD that are unlikely to be reversible. Finally, in the Conclusion and Future perspective sections, we discuss how the PEER hypothesis can help move aging and DOHaD research further along.
Epigentic clocks & the nine hallmarks of aging
The first Epigenomic clocks, and the entire field of biological clock research, were developed in 2013 by two laboratories. Horvath's group showed that DNA methylation in 353 CpG sites in over 8000 tissue samples correlate with the age of the individual, and tumor cells have accelerated aging up to 35 years [22]. Horvath concluded, “I propose that DNA methylation age measures the cumulative effect of an epigenetic maintenance system. This novel epigenetic clock can be used to address a host of questions in developmental biology, cancer and aging research” [22]. Also in 2013, Hannum and colleagues developed a similar Epigenomic clock that measures aging rate by looking at DNA methylation at CpG sites in the blood [23]. Using the convention discussed above, we refer to these first two clocks as Epigenomic clocks rather than the term used by the authors (Epigenetic clocks) because they were entirely based on DNA methylation patterns.
Since 2013, when the Horvath and Hannum clocks emerged, the biological clock field has exploded with thousands of publications. The Horvath and Hannum clocks, and similar clocks that solely measure DNA methylation levels at CpG sites, are called ‘first-generation Epigenomic clocks’. The CpG methylation levels from human tissues are measured primarily on Illumina human DNA methylation bead arrays, the most recent version called EPIC-2, which can measure DNA methylation levels at over 1 M CpG sites (Illumina, Inc., CA, USA). Arrays such as EPIC-2, and a recently developed mouse Illumina DNA methylation bead array [24], are often used to determine the methylation levels of specific CpG sites because they have greater accuracy, lower cost and greater lab-to-lab reproducibility than other techniques such as whole-genome bisulfite sequencing. Since the mouse bead array was only recently developed, the Horvath group developed in 2017 and 2018 reduced representation bisulfite sequencing-based assays to develop Epigenomic clocks for several mouse tissues [25,26]. Stubbs and colleagues also developed in 2017 a mouse first-generation Epigenomic clock based on 329 unique CpG sites [27].
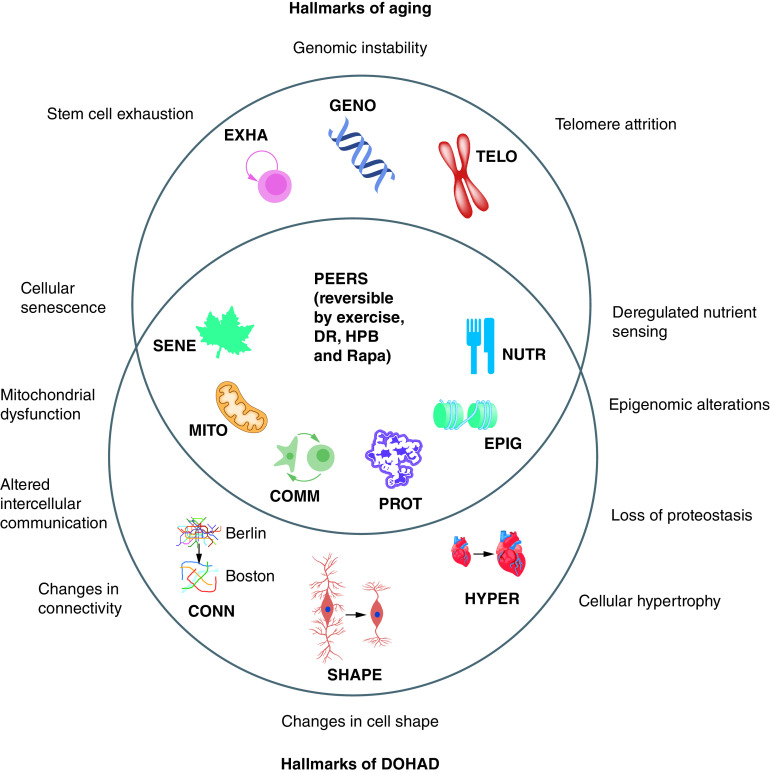
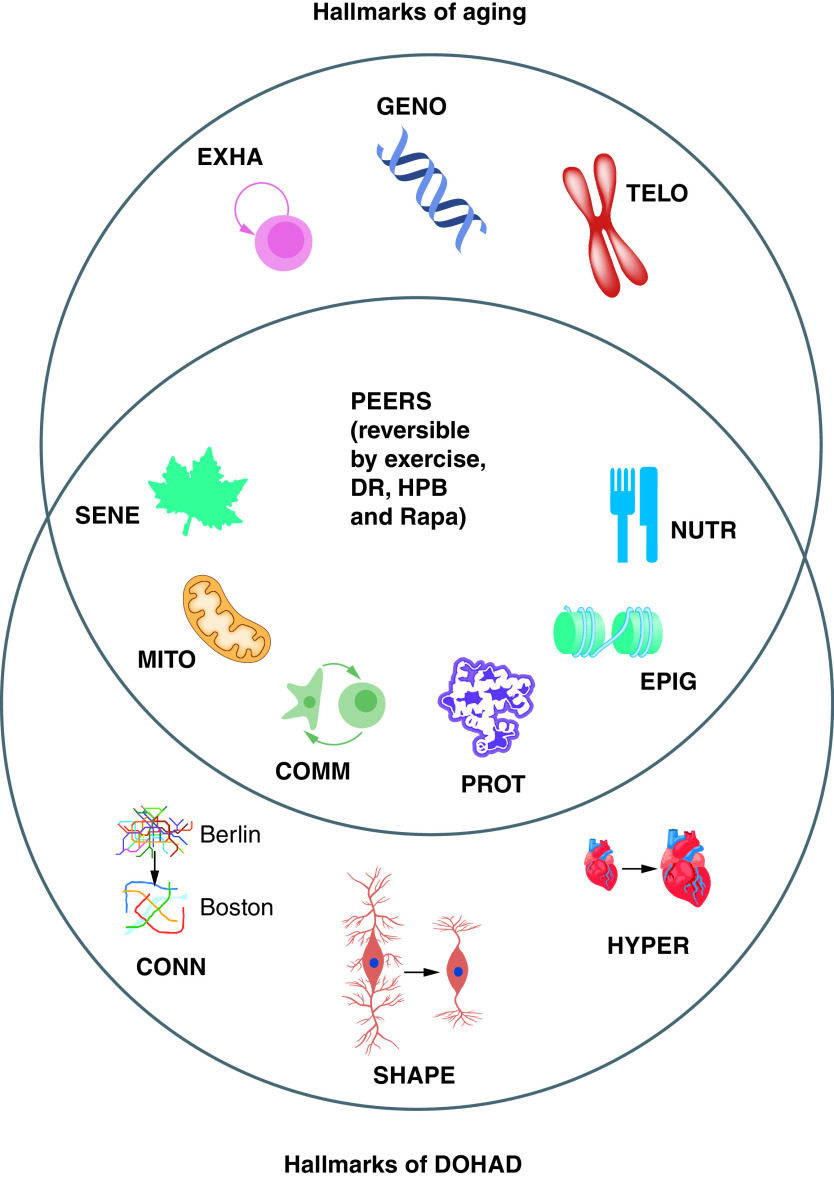
An influential 2013 review by Lopez-Otin and colleagues, with over 11,000 citations, discussed nine hallmarks of aging (Figure 1) [28]. The nine hallmarks of aging are: 1) cellular senescence (SENE – a leaf symbol), which is an indication of the limited number of cell divisions in differentiated cells (leaves die in the fall); 2) mitochondrial dysfunction (MITO – a mitochondrion symbol), which is an indication of mitochondrial integrity and polarity which decreases with aging; 3) intercellular communication (COMM – two-cells-interacting symbol), which indicates increased inflammation and altered cell-to-cell signaling in many organs; 4) PROT (a folded protein symbol), which indicates protein aggregates which increase in several organs during aging, primarily by a decrease in autophagy during aging; 5) Epigenomic alterations (EPIG – a histone on DNA symbol), such as DNA methylation changes, which are measured in Epigenomic and Epigenetic clocks; 6) deregulated nutrient sensing (a knife and fork symbol), which indicates altered metabolism during aging; 7) stem cell exhaustion (a symbol showing a stem cell self-renewal), which indicates that stem cells eventually lose their ability to self-renew as they age; 8) genomic instability (a DNA symbol), which indicates that cells have an increased rate of aneuploidy and mutations in aging cells; and 9) telomere attrition (a chromosome symbol), which indicates that telomeres shorten as a cell ages.
Figure 1. . The PEER hypothesis: the hallmarks of aging and the hallmarks of DOHAD overlap at PEERs.
The PEERs have the potential for recovery by interventions by exercise, DR, HPB and pro-longevity drugs such as Rapa.
COMM: Intercellular communication; CONN: Change in connectivity; DOHAD: Developmental origins of adult disease; DR: Dietary restriction; EPIG: Epigenomic alteration; EXHA: Stem cell exhaustion; GENO: Genomic instability; HPB: Heterochronic parabiosis; HYPER: Change in hypertrophy; MITO: Mitochondrial dysfunction; NUTR: Deregulated nutrient sensing; PEER: Pathological Epigenetic event that is reversible; PROT: Loss of proteostasis; Rapa: Rapamycin; SENE: Cellular senescence; SHAPE: Change in cell shape; TELO: Telomere attrition.
Newer clocks that take into account phenotypes and other hallmarks of aging, such as the PhenoAge clock developed by Levine and colleagues in 2018 [29], and the GrimAge clock developed by Lu and colleagues in 2019 [30], are called ‘second-generation Epigenetic clocks’ (here, we use the word ‘Epigenetic’ for second-generation clocks since more than DNA methylation is involved). To measure the pace of aging in a longitudinal aging cohort, Belsky and colleagues in 2022 developed the DunedinPACE second-generation Epigenetic clock using data from the Dunedin, New Zealand, Study, a 1972–1973 birth cohort of 1037 subjects that tracked within-individual decline in 19 indicators of organ-system integrity [31]. The 19 indicators measured at four time points spanning two decades are: 1) cardiovascular; 2) metabolic; 3) renal; 4) hepatic; 5) immune; 6) dental; 7) pulmonary; and 12 measures of physical and mental fitness: 8) balance; 9) gait speed; 10) steps in place; 11) chair stands; 12) grip strength; 13) motor coordination; 14) physical limitations; 15) perceptual reasoning; 16) working memory; 17) processing speed; 18) self-rated health and 19) facial aging. The 19 indicators of organ system integrity in the DunedinPACE clock are not direct measures of the hallmarks of aging, but rather are surrogate measures that reflect them.
HPB alters the epigenetic clocks of both the young & old mouse
How sad it is! I shall grow old, and horrible, and dreadful. But this picture will remain always young. It will never be older than this particular day of June… If it were only the other way! If it were I who was to be always young, and the picture that was to grow old! For that – for that – I would give everything! Yes, there is nothing in the whole world I would not give! I would give my soul for that! Oscar Wilde, The Picture of Dorian Gray (1891) [32].
I tried to kill him for the strengthening my vital powers by the assimilation with my own body of his life through the medium of his blood – relying, of course, upon the scriptural phrase, for the blood is the life. Bram Stoker, Dracula (1899) [33].
Reversing the effects of aging has been a theme in both classic and modern fiction. Stories of the ‘Fountain of Youth’ trace back to Herodotus in the 5th century, BC [34]. The Fountain of Youth is a magical spring that restores the youth of anyone who drinks or bathes in its waters. In the 16th century, Juan Ponce de Leon was supposedly charged by the King of Spain to find the Fountain of Youth in the mythical land of Bimini in the Caribbean [35]. In the late 19th century, age reversal shifted to other means. For example, in the classic novel The Picture of Dorian Gray, by Oscar Wilde in 1891 [32], youth is bestowed to the title character by a magical picture, which unfortunately shows his true age when he looks into it. The second of the above quotes is from the classic novel Dracula, by Bram Stoker in 1899 [33]. In Dracula, he reveals the secret to longevity for vampires is for them to drink their victim's blood.
From the literary concept that youthful blood can reverse the effects of aging came the idea of HPB experiments starting in the 1950s. In HPB experiments, the circulatory system of a young mouse is fused to that of an old genetically identical mouse by surgically joining the two animals together through a flap of skin (early references cited in Conboy et al. [36]). In 2005, HPB reemerged as an important model by Conboy and colleagues who focused on phenotypes such as brain function, tissue regeneration and stem cell characteristics [36]. While they confirmed the rejuvenating effects of the old mouse's stem cells, they also noted that the young mouse became “pale, shriveled and anemic” [36]. As an analogy, the old mouse is Dorian Gray who maintains his youth but at the expense of his picture, that is, the young mouse, that becomes old and debilitated [32]. The phenotypic effects of HPB were followed up by further studies on its effects in the brain [37,38], heart [39] and bone in both the old and young parabiotes [40,41]. The HPB studies consistently demonstrate robust rejuvenating effects on the old mouse, while at the same time severe age-promoting effects on the young mouse.
In a recent paper, Poganik and colleagues used modern molecular techniques, such as second-generation Epigenetic clocks, to study the effects of HPB on Epigenetic aging, gene expression and metabolomics [42]. Their HPB experiments consisted of surgically connecting the circulatory systems of a young and an old mouse for 3 months, separating them and allowing them to recover for 2 months (Figure 2) [42]. These HPB experiments stand out from other HPB experiments (over 100 publications so far – almost half since 2020) in that the mice were surgically separated and allowed to recover, which is not typically done in HPB experiments. What they demonstrated was that the young mouse's Epigenetic clocks in brain, adipose, liver, heart and kidney were accelerated after 3 months of HPB, but mostly completely restored to their correct time after only 2 months of recovery [42]. The recovery in these tissues was demonstrated by using multiple second-generation Epigenetic clocks [42], which take into account both the biological age and phenotypic characteristics. No effect was observed using traditional first-generation epigenetic clocks, suggesting that phenotype needs to be taken into account [42]. In the next section, we discuss the nine hallmarks of DOHaD and their overlap with the hallmarks of aging.
Figure 2. . Heterochromatic parabiosis model for stress-induced aging.
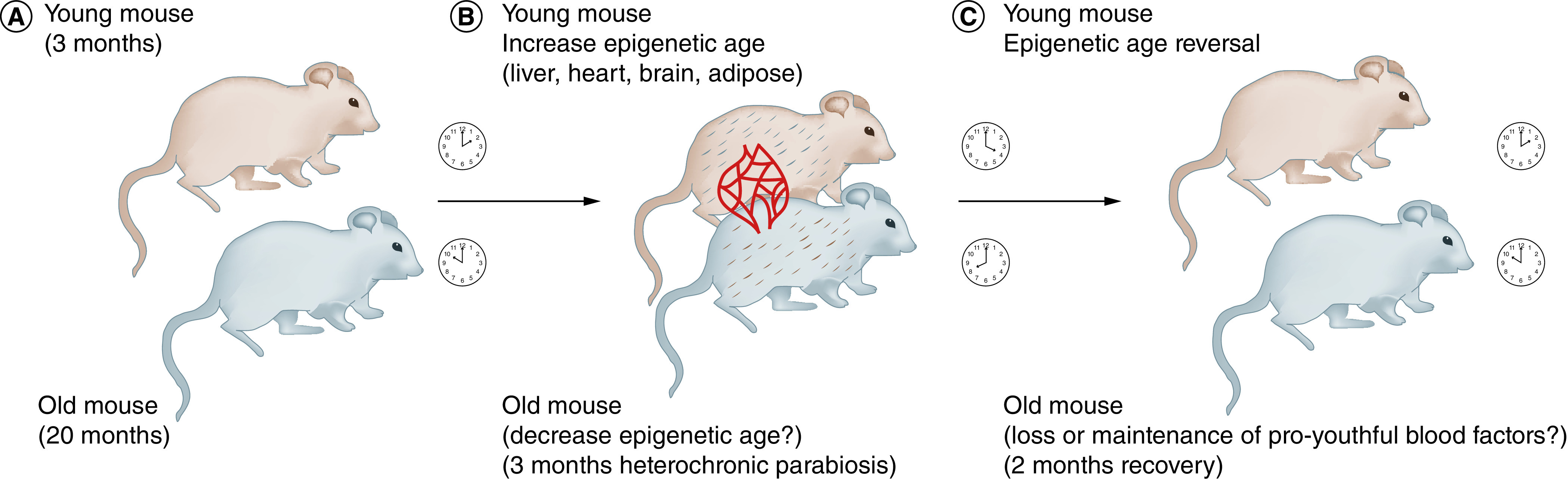
(A) In the Poganik and colleagues' paper [42], a young mouse (3 months) is surgically joined to an old mouse (20 months). The clocks represent the Epigenetic age – 2 PM for the young mouse and 10 PM for the old mouse. (B) The young mouse is joined to the old mouse for 3 months by a skin flap and shared vasculature (red lines). The clocks show that the Epigenetic age is increased by 2 h for the young mouse (gray hairs indicate aging acceleration) and decreased by 2 h for the old mouse (brown hairs indicate aging reversal). (C) The mice are separated and allowed 2 months of recovery. The young mouse demonstrates age reversal in several organs (clock moves back to 2 PM and gray hairs are gone) and the old mouse ages normally (brown hairs revert back to gray and clock moves back to 10 PM).
The nine hallmarks of DOHaD & the overlap with six of the nine hallmarks of aging
The nine hallmarks of DOHaD, which have not been previously elucidated, appear to overlap with at least six of the hallmarks of aging (Figure 1). The rationale for including them as hallmarks of DOHaD is discussed below.
The six hallmarks of aging that overlap with the hallmarks of DOHaD are outlined in the following sections.
Cellular senescence
Cells become senescent because they have a limited number of cell divisions. In 2022, Ryan and colleagues showed that advanced age on a second-generation Epigenetic clock trained on leptin predicted a significantly earlier gestational age at delivery (beta = -0.15; p = 0.009) [43]. This suggests that SENE might be a hallmark of DOHaD because leptin is able to induce cell cycle arrest and senescence by activating the p53–p21 pathway and inhibiting Sirt1 [44].
Mitochondrial dysfunction
Mitochondrial integrity and polarity decreases with age. In a 2021 review, Oke and colleagues discuss the role of oxidative stress, MITO, endoplasmic reticulum stress and inflammation in the pathogenesis of metabolic disease in interuterine growth-restricted (IUGR) offspring [45]. IUGR can lead to low birth weight, which, as discussed above, Barker and many others have shown to be a DOHaD hallmark [46].
Intercellular communication
Aging is associated with altered COMM, such as increased inflammation in several organs. Several studies have shown that increased inflammation during pregnancy can have adverse pregnancy outcomes, such as IUGR and low birth weight [47–50]. COMM is therefore a likely DOHaD hallmark.
Loss of proteostasis
Protein aggregates, an example of PROT, increase in several organs during aging, such as amyloid-β plaques in aging brains of people with Alzheimer's disease. In a 2020 study, Klein and colleagues found that maternal exercise promotes changes in the rat offspring's cerebellum that are still evident in young adult life [46]. They conclude that these changes may contribute to a protective phenotype against amyloid-β-induced neurotoxicity in young adult male rat offspring.
Epigenomic alterations
EPIG changes induced by toxic chemicals and stress during gestation were discussed already in this review (e.g., Niemiec et al. [20]).
Deregulated nutrient sensing
As discussed earlier, Barker and colleagues showed that maternal malnutrition during pregnancy can lead to several adverse outcomes later in life that reflect NUTR [2]. Also, several studies have shown that maternal obesity can increase the risk of IUGR and low birth weight, which can have a DOHaD effect [51–54].
The three hallmarks of aging that do not appear to overlap with the hallmarks of DOHaD are described below.
Stem cell exhaustion. In older individuals, stem cells eventually lose their ability to self-renew, which is a characteristic of EXHA. We could not find evidence that this would be a hallmark of DOHaD.
Genomic instability. Cells have an increased rate of aneuploidy in aging cells, which is a characteristic of GENO. This would not likely occur in early development unless the stress was genotoxic. While some chemical toxicants are genotoxic, such as some of the chemicals in cigarette smoke (e.g., benzo(a)pyrene), we nevertheless decided to not include this as a hallmark of DOHaD.
Telomere attrition. Telomeres shorten as a cell ages. This would probably not occur during gestation or early development and is therefore probably not a hallmark of DOHaD.
The six PEERs intersect at the hallmarks of aging & DOHaD
When we looked at the overlaps between the nine hallmarks of aging and the nine hallmarks of DOHaD we noticed that they apparently overlap at six PEERs (Figure 1). These PEERs have been shown to be reversible by interventions such as HPB [55], exercise [56], dietary restriction [57], young bone marrow transplants [58–60], and longevity drugs such as rapamycin [61] and metformin [62]. The three hallmarks of aging that have limited interventions are telomere shortening, stem cell exhaustion and genome instability (Figure 1).
Figure 3. . Hallmarks of developmental origin of health and disease that would be difficult or impossible to reverse.
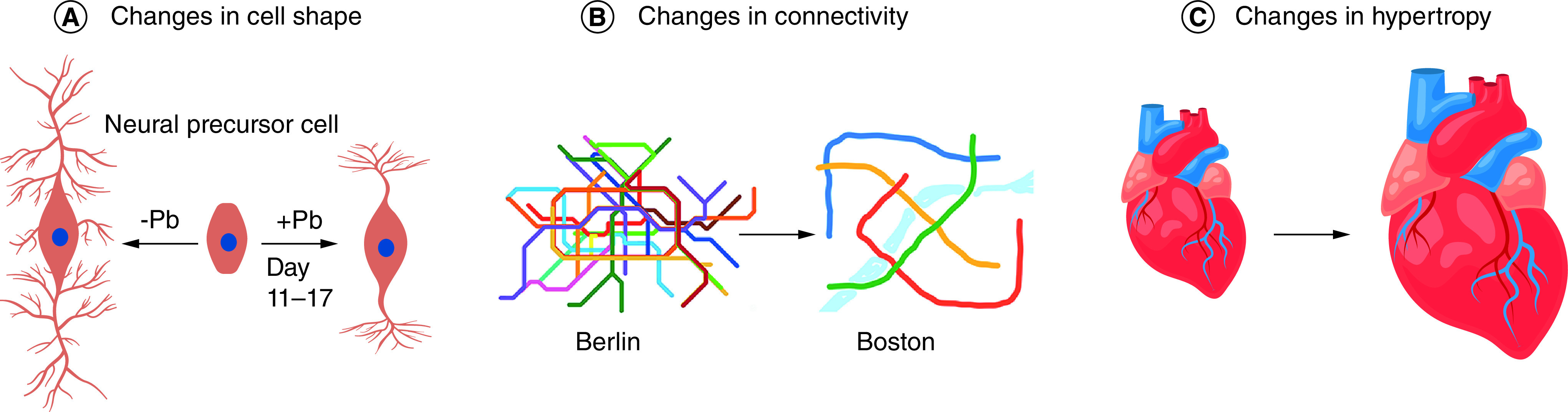
(A) Changes in cell shape. Left, human neural precursor cell (center) is differentiated in culture for 17 days by Senut and colleagues [78] and shows a stereotypical pattern of neurite outgrowth. Right, after exposure to Pb for days 11–17 of differentiation, the neurites are shorter and there is less branching. (B) Changes in connectivity. Shown are the subway maps of Berlin and Boston, with Berlin having much more connectivity. Subways are a metaphor for neuronal connectivity that would be reduced by Pb exposure, for instance. (C) Changes in hypertrophy. Shown is a normal heart (left) and one greatly increased by cellular hypotrophy of cardiomyocytes. Such hypertrophy of the heart can be caused by developmental exposure to Pb, cadmium or arsenic [79], while hypertrophy of the liver can be caused by exposure to some perfluoralkyl substances [80,81].
Pb: Lead.
Evidence that the six overlapping hallmarks of aging and DOHaD are PEERs is as follows.
Cellular senescence
Senescent cell removal by drugs has longevity-promoting effects in mice [63,64]. Also, in 2020, Yousefzadeh and colleagues showed that HPB regulates the extent of SENE in multiple tissues [65].
Mitochondrial dysfunction
In 2020, Kiss and colleagues showed that HPB promotes a transcriptional footprint of mitochondrial protection and attenuation of oxidative stress [94]. Similary, in 2019, Morrison and colleagues showed that HPB partially reverses aging-induced metabolic changes and oxidant stress in mouse red blood cells [95].
Intercellular communication
In 2019, Yousefzadeh and colleagues showed that HPB-derived aged blood impairs COMM of hippocampal neural precursor activity and activates microglia via brain endothelial cell VCAM1 [65]. Also, in a detailed single-cell RNA sequencing analysis of both the young and old brain in a HPB experiment, Ximerakis and colleagues showed, using the software CellChat [66], that the old HPB mouse has fewer connections than the old control mouse, and that many of the connections in the old mouse were related to inflammation signaling [55].
Loss of proteostasis
In 2018, Liu and colleagues showed that young plasma reverses age-dependent alterations in liver function through the restoration of autophagy [67].
Epigenomic alterations
Several HPB studies have shown that Epigenetic clock reversal can occur in the old parabiont (reviewed in Rando and Jones [68]). Also, in the HPB study discussed above, the clock acceleration in the young parabiont is reversed 2 months after surgical separation [42].
Deregulated nutrient sensing
Reversal of deregulated nutrient sensing is evident by longevity drugs such as rapamycin [61] and metformin [62].
The three hallmarks of DOHaD that are unlikely to be reversible
Many PEEs, especially those that affect brain, skeletal or other organ development in the fetus or young childhood, are probably irreversible. Such irreversible PEEs are the initiating event that could mechanistically explain some of the permanent consequences of developmental exposure. In Figure 3, we argue that three of the nine hallmarks of DOHaD are unlikely to be reversible. These are: 1) changes in cell shape, represented by neurons with many and few neurites (Figure 3A); 2) changes in connectivity, represented by subway maps of Berlin and Boston, with Berlin having a much more connected subway (Figure 3B); and 3) changes in hypertrophy, represented by a small and large heart (Figure 3C). In this section, we describe how these three hallmarks arise after developmental exposures, with a focus on lead (Pb), which can cause all three of these irreversible hallmarks (Figure 3).
Changes in cell shape
In studies with Pb-exposed human neural precursor cells in culture, Senut and colleagues showed that exposure to 1.9 uM Pb for days 11–19 (1 week) in neuron-differentiation media caused the neurite length to decrease [78]. In addition, exposure to even lower concentrations of Pb (0.4 uM) during days 11–19 caused a decrease in branching of the neurites in a dose–response manner that increases linearly with Pb dose (Figure 3) [78]. Such effects of Pb on neuron shape and size might explain the decreased brain volume neural connectivity defects observed in adults with childhood Pb exposure [74]. Similar exposures in vivo have shown that Pb in both human and rodents can permanently change the shape and connectivity of neurons [76,77]. Based on these and numerous other studies, we postulate that Pb exposure during early brain development would be an example of a PEE that would be very difficult if not impossible to reverse. Reversing such physical changes in a cell or organ would require removing the old and improperly shaped cells and replacing them with new cells, which is likely prohibitively difficult.
Changes in connectivity
Pb is a common environmental exposure that affects development of many organs but is especially damaging to the brain [69,70,71]. The CDC level of concern for Pb is currently 5 µg/dl (0.24 uM) but the CDC has also stated that there is “no safe level of lead” [72]. The PEE induced by Pb might be acute, such as a toddler only once eating Pb-containing paint or one-time sucking on a Pb-containing toy. A PEE might also be chronic, such as continuous Pb exposure for many months or years in a young child whose housing is not immediately remediated for Pb paint. The degree of recovery to a PEE depends on the duration of exposure and the plasticity of the organ or system that is altered. In the case of Pb, chronic exposure during a sensitive period during fetal or early childhood would likely cause permanent and irreparable structural damage to the neurons, especially if the exposure continues until after synaptogenesis and neurodevelopment is completed. For example, childhood Pb exposure correlates with cognitive deficits and a decrease in gray matter and brain volume later in life [73,74,75]. Similar results have been found in the mouse model with maternal Pb exposure and Pb mixtures with other chemicals [76,77]. Once the brain is hardwired and the connections are made, it would be difficult if not impossible to reverse.
Changes in hypertrophy
Hypertrophy of the heart can be caused by developmental exposure to Pb, cadmium or arsenic (reviewed in Lamas et al. [79]). Hypertrophy of the liver can be caused by exposure to some perfluoralkyl substances [80,81]. What is the mechanism by which exposures to toxicants such as Pb cause hypertrophy of the heart and change the shape and connectivity of neurons? Svoboda and colleagues have shown in 2019 and 2020 that developmental exposure to Pb in mice can change the heart epigenome [82,83]. Similarly, several groups have shown that both the long- and short-term effects of Pb exposure are likely mediated by changes in DNA methylation in neurons. In human epidemiological studies, Montrose and colleagues showed changes in DNA methylation in blood that correlates with Pb exposure [84]. Similarly, Sen and colleagues showed that exposure to Pb over 5 µg/dl correlates with changes in blood DNA methylation levels in humans [85]. In another study, Sen and colleagues showed that Pb exposure in human umbilical cord blood over 5 µg/dl causes changes in both DNA methylation and hydroxymethylation (5mC and 5hmC) levels at dozens of genes [86]. In a third study, Sen and colleagues showed that grandmaternal exposure to Pb correlates with changes in DNA methylation in the newborn blood of the grandchildren [14]. While further mechanistic studies are needed, these and other studies suggest that many of the long-term effects of Pb exposure, as well as multigenerational Epigenetic effects of Pb exposure, are mediated at least in part by changes in DNA methylation, piRNAs and other small RNAs that are heritable in the sperm and eggs, and the information transmitted is not erased in the embryo [13,87–89]. While EPIG changes are reversible, once an organ such as a heart or liver undergoes hypertrophy, it would be difficult if not impossible to reverse this phenotype other than by transplanting a healthy organ. In the next section, we discuss PEEs that, unlike Pb exposure during early development, have recently been shown to be reversible.
Conclusion
In this review, we discussed the hallmarks of aging and DOHaD and discovered that their intersection are PEERs. The most promising hallmarks in both aging and DOHaD that have the greatest potential for reversibility are EPIGs because much is known about the mechanisms of DNA and histone modifications during development and disease. Other reversible hallmarks in both aging and DOHaD are PROT and MITO, which both can be reversed by decreasing oxidative stress. Altered COMM of the immune system can be reversed by decreasing inflammation. However, it is important to keep in mind that there are three DOHaD hallmarks that are not reversible – altered cell shape, connectivity and hypertrophy. PEEs during development that permanently alter the shape or connectivity of neurons or the size of organs would be difficult, if not impossible to reverse.
Future perspective
The PEER hypothesis might help explain some of the paradoxes in the aging and DOHaD fields. For example, PEEs that altered cell shape, connectivity and hypertrophy might help explain the growth hormone (GH)/IGF-1 paradox [90]. Overexpression or supplementation with either protein has youth-restoring effects in old mice and humans (reviewed in Bickel et al. [91]). However, the paradox is that mice with deficiencies in either protein during early development have greatly increased longevity. In fact, the Methuselah prize for the longest-lived mouse has both a GH and an IGF-1 deficiency [92].
One possibility to explain this paradox, as suggested by Sun and colleagues, is that GH/IGF-1 mutant mice have early developmental origins for their longevity phenotype [93]. In other words, the GH/IGF-1 paradox might be explained if the longevity of the GH/IGF2 deficient mice reflects the irreversible aspect of the DOHaD hypothesis, such as the connectivity of the neurons involved in GH/IGF-1 signaling (Figure 3B). Conversely, the longevity of the adult supplemented GH/IGF-1 mice might reflect a reversible aspect of the DOHaD hypothesis, a PEER, such as Epigenomic changes that alter the signaling strength of GH/IGF-1 in neurons (Figure 1, middle).
Finally, the PEER hypothesis speculates that treatments that reverse aging that are identified in HPB experiments might also reverse some of the phenotypes caused by DOHaD exposures. In this manner, findings in aging research could potentially apply to DOHaD, and vice versa. Further experiments are needed to determine the degree of cross-fertilization between the DOHaD and aging fields.
Executive summary.
Epigentic clocks & the nine hallmarks of aging
The first section discusses Epigenetic clocks and the nine hallmarks of aging. The nine hallmarks of aging are: 1) cellular senescence (SENE – a leaf symbol), which is an indication of the limited number of cell divisions in differentiated cells; 2) mitochondrial dysfunction (MITO – a mitochondrion symbol), which is an indication of mitochondrial integrity and polarity which decreases with aging; 3) intercellular communication (COMM – two-cells-interacting symbol), which indicates increased inflammation in many organs; 4) loss of proteostasis (PROT – a folded protein symbol), which indicates protein aggregates which increase in several organs during aging, primarily by a decrease in autophagy during aging; 5) Epigenomic alterations (EPIG – a histone on DNA symbol), which are measured in first-generation Epigenomic clocks; 6) deregulated nutrient sensing (NUTR – a knife and fork symbol), which indicates altered metabolism during aging; 7) stem cell exhaustion (a symbol showing a stem cell self-renewal), which indicates that stem cells eventually lose their ability to self-renew; 8) genomic instability (a DNA symbol), which indicates that cells have an increased rate of aneuploidy in aging cells; and 9) telomere attrition (a chromosome symbol), which indicates that telomeres shorten as a cell ages.
Heterochronic parabiosis alters the epigenetic clocks of both the young & old mouse
The second section discusses heterochronic parabiosis experiments that alter the Epigenetic clocks of both the young and the old mouse. The young mouse has aging acceleration and the old mouse has aging reversal. These are reversed and restored to normal when the mice are separated for 2 months.
The nine hallmarks of developmental origin of health and disease & the overlap with six of the nine hallmarks of aging
The third section discusses the nine hallmarks of developmental origin of health and disease (DOHaD) and argues that they overlap with six of the nine hallmarks of aging; the overlapping hallmarks of DOHaD and aging are SENE, MITO, COMM, PROT, EPIG and NUTR.
The six pathological epigenetic events that are reversible intersect at the hallmarks of aging & DOHaD
The fourth section discusses the six pathological epigenetic events that are reversible (PEERs). The PEER hypothesis argues that the six PEERs intersect at the hallmarks of aging and DOHaD. The six PEERs are SENE, MITO, COMM, PROT, EPIG and NUTR.
The three hallmarks of DOHaD that are unlikely to be reversible
The fifth section discusses the new three hallmarks of DOHaD that are unlikely to reversible. They are cell connectivity (CONN – represented by Berlin and Boston subway maps), cell shape (SHAPE – represented by neurons with many and few neurites) and hyperplasty (HYPER – represented by large and small human hearts).
Future perspective
Finally, in the Future perspective section we discuss how the PEER hypothesis can help move aging and DOHaD research further along. We argue that findings and treatments in the aging field that reverse some of the hallmarks of aging will also advance the DOHaD field and reverse some of the hallmarks of DOHaD, and vice versa.
Footnotes
Author contributions
DMR wrote the first draft of the paper and AS and DAR helped edit subsequent drafts and suggested new ideas to include in the final version.
Financial & competing interests disclosure
This work was supported by NIH grant nos. UH3 OD023285, P42 ES030991 and P30 ES020957 to the first author and was deposited in the NIHMS system/PMC. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Francis RC. Epigenetics: How Environment Shapes our Genes (1st Edition). WW Norton, NY, USA: (2012). [Google Scholar]; • This popular book coined the term ‘pathological epigenetic events’.
- 2.Barker DJ, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet 1(8489), 1077–1081 (1986). [DOI] [PubMed] [Google Scholar]; • This is the classic paper by Barker and Osmond.
- 3.Rich-Edwards JW, Stampfer MJ, Manson JE et al. Birth weight and risk of cardiovascular disease in a cohort of women followed up since 1976. BMJ 315(7105), 396–400 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Painter RC, Osmond C, Gluckman P, Hanson M, Phillips DI, Roseboom TJ. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG 115(10), 1243–1249 (2008). [DOI] [PubMed] [Google Scholar]; • This is the classic Dutch famine study.
- 5.(. Yadav NS, Titov V, Ayemere I, Byeon B, Ilnytskyy Y, Kovalchuk I. Multigenerational exposure to heat stress induces phenotypic resilience, and genetic and epigenetic variations in Arabidopsis thaliana offspring. Front. Plant. Sci. 13, 2022) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(. Frolows N, Ashe A. Small RNAs and chromatin in the multigenerational epigenetic landscape of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 376 1826), (2021) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis LA, Kissane S, Hoffman E et al. Multigenerational exposures of daphnia magna to pristine and aged silver nanoparticles: epigenetic changes and phenotypical ageing related effects. Small 16(21), e2000301 (2020). [DOI] [PubMed] [Google Scholar]
- 8.(. Singh NP, Yang X, Bam M, Nagarkatti M, Nagarkatti P. 2,3,7,8-Tetrachlorodibenzo-p-dioxin induces multigenerational alterations in the expression of microRNA in the thymus through epigenetic modifications. PNAS Nexus 2 1), (2023) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raad G, Serra F, Martin L et al. Paternal multigenerational exposure to an obesogenic diet drives epigenetic predisposition to metabolic diseases in mice. Elife 10, e61736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buck JM, Sanders KN, Wageman CR, Knopik VS, Stitzel JA, O'Neill HC. Developmental nicotine exposure precipitates multigenerational maternal transmission of nicotine preference and ADHD-like behavioral, rhythmometric, neuropharmacological, and epigenetic anomalies in adolescent mice. Neuropharmacology 149, 66–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xin F, Susiarjo M, Bartolomei MS. Multigenerational and transgenerational effects of endocrine disrupting chemicals: a role for altered epigenetic regulation? Semin. Cell Dev. Biol. 43, 66–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leslie FM. Multigenerational epigenetic effects of nicotine on lung function. BMC Med. 11, 27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton NO, Greer EL. Multigenerational epigenetic inheritance: transmitting information across generations. Semin. Cell Dev. Biol. 127, 121–132 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sen A, Heredia N, Senut MC et al. Multigenerational epigenetic inheritance in humans: DNA methylation changes associated with maternal exposure to lead can be transmitted to the grandchildren. Sci. Rep. 5, (2015) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillman MW. Developmental origins of health and disease. N. Engl. J. Med. 353(17), 1848–1850 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Widdowson EM, McCance RA. The effect of finite periods of undernutrition at different ages on the composition and subsequent development of the rat. Proc. R. Soc. Lond. B. Biol. Sci. 158, 329–342 (1963). [DOI] [PubMed] [Google Scholar]
- 17.Felix JF, Cecil CAM. Population DNA methylation studies in the developmental origins of health and disease (DOHaD) framework. J. Dev. Orig. Health Dis. 10(3), 306–313 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Pilsner JR, Saddiki H, Whitcomb BW et al. Sperm epigenetic clock associates with pregnancy outcomes in the general population. Hum. Reprod. 37(7), 1581–1593 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(. Oluwayiose OA, Houle E, Wu H et al. Urinary phthalate metabolites and their mixtures are associated with advanced sperm epigenetic aging in a general population. Environ. Res. 214 Pt 4), (2022) (Online). [DOI] [PubMed] [Google Scholar]
- 20.(. Niemiec SS, Kechris K, Pattee J et al. Prenatal exposures to per- and polyfluoroalkyl substances and epigenetic aging in umbilical cord blood: the Healthy Start study. Environ. Res. 231 Pt 2), (2023) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladd-Acosta C, Vang E, Barrett ES et al. Analysis of pregnancy complications and epigenetic gestational age of newborns. JAMA Netw. Open 6(2), e230672 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 14 10), (2013) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper and Hannum et al. started the epigenomic clock field.
- 23.Hannum G, Guinney J, Zhao L et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49(2), 359–367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper and Horvath stated the epigenetic clock field in 2013.
- 24.(. Perez-Correa JF, Tharmapalan V, Geiger H, Wagner W. Epigenetic clocks for mice based on age-associated regions that are conserved between mouse strains and human. Front. Cell. Dev. Biol. 10, 2022) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meer MV, Podolskiy DI, Tyshkovskiy A, Gladyshev VN. A whole lifespan mouse multi-tissue DNA methylation clock. Elife 7, e40675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petkovich DA, Podolskiy DI, Lobanov AV, Lee SG, Miller RA, Gladyshev VN. Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metab. 25(4), 954–960.e956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stubbs TM, Bonder MJ, Stark AK et al. Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 18(1), 68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell 153(6), 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine ME, Lu AT, Quach A et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 10(4), 573–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu AT, Quach A, Wilson JG et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 11(2), 303–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belsky DW, Caspi A, Corcoran DL et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife 11, e73420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilde O. The Picture of Dorian Gray. Ward, Lock, London, UK: (1891). [Google Scholar]
- 33.Stoker B. Dracula. Doubleday and McClure Co., NY, USA: (1899). [Google Scholar]
- 34.Johnson WA. The Essential Herodotus: Translation, Introduction, and Annotations. Oxford University Press, NY, USA: (2017). [Google Scholar]
- 35.Slavicek LC. Juan Ponce de Leon. Chelsea House, PA, USA: (2003). [Google Scholar]
- 36.Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433(7027), 760–764 (2005). [DOI] [PubMed] [Google Scholar]; • This paper rejuvenated the heterochronic parabiosis (HPB) field from the earlier studies done in the 1950s.
- 37.Ruckh JM, Zhao JW, Shadrach JL et al. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell 10(1), 96–103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villeda SA, Luo J, Mosher KI et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477(7362), 90–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loffredo FS, Steinhauser ML, Jay SM et al. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 153(4), 828–839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baht GS, Silkstone D, Vi L et al. Exposure to a youthful circulaton rejuvenates bone repair through modulation of beta-catenin. Nat. Commun. 6, 7131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vi L, Baht GS, Soderblom EJ et al. Macrophage cells secrete factors including LRP1 that orchestrate the rejuvenation of bone repair in mice. Nat. Commun. 9(1), 5191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poganik JR, Zhang B, Baht GS et al. Biological age is increased by stress and restored upon recovery. Cell Metab. 35(5), 807–820.e805 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• These are the HPB experiments discussed in Figure 2.
- 43.Ryan CP, Rege RJ, Lee NR et al. Maternal epigenetic clocks measured during pregnancy do not predict gestational age at delivery or offspring birth outcomes: a replication study in metropolitan Cebu, Philippines. Clin. Epigenetics 14(1), 78 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abella V, Scotece M, Conde J et al. Leptin in the interplay of inflammation, metabolism and immune system disorders. Nat. Rev. Rheumatol. 13(2), 100–109 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Oke SL, Hardy DB. The role of cellular stress in intrauterine growth restriction and postnatal dysmetabolism. Int. J. Mol. Sci. 22(13), 6986 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein CP, Hoppe JB, Saccomori AB et al. Protective effect of maternal exercise against amyloid-beta neurotoxicity in the male rat offspring's cerebellum. J. Dev. Orig. Health Dis. 11(5), 521–532 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Lodge-Tulloch NA, Toews AJ, Atallah A, Cotechini T, Girard S, Graham CH. Cross-generational impact of innate immune memory following pregnancy complications. Cells 11(23), 3935 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kondo R, Ozawa R, Satomi T et al. Severe maternal stress alters placental function, resulting in adipose tissue and liver dysfunction in offspring of mice. Mol. Cell. Endocrinol. 560, (2023) (Online). [DOI] [PubMed] [Google Scholar]
- 49.Sanches APV, de Oliveira JL, Ferreira MS et al. Obesity phenotype induced by high-fat diet leads to maternal-fetal constraint, placental inefficiency, and fetal growth restriction in mice. J. Nutr. Biochem. 104, (2022) (Online). [DOI] [PubMed] [Google Scholar]
- 50.Goldstein JA, Gallagher K, Beck C, Kumar R, Gernand AD. Maternal-fetal inflammation in the placenta and the developmental origins of health and disease. Front. Immunol. 11, (2020) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheidl TB, Brightwell AL, Easson SH, Thompson JA. Maternal obesity and programming of metabolic syndrome in the offspring: searching for mechanisms in the adipocyte progenitor pool. BMC Med. 21(1), 50 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell AR, Glastras SJ. Maternal weight management to prevent the developmental programming of MAFLD in offspring of obese mothers. Nutrients 15(9), 2155 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.(. Nash MJ, Dobrinskikh E, Soderborg TK et al. Maternal diet alters long-term innate immune cell memory in fetal and juvenile hematopoietic stem and progenitor cells in nonhuman primate offspring. Cell Rep. 42 4), (2023) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Huo Y, Zhang J, Xu D, Bai F, Gui Y. Association between high-fat diet during pregnancy and heart weight of the offspring: a multivariate and mediation analysis. Nutrients 14(20), 4237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ximerakis M, Holton KM, Giadone RM et al. Heterochronic parabiosis reprograms the mouse brain transcriptome by shifting aging signatures in multiple cell types. Nat. Aging 3(3), 327–345 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper shows how HPB experiments can reverse some of the hallmarks of aging.
- 56.Brett JO, Arjona M, Ikeda M et al. Exercise rejuvenates quiescent skeletal muscle stem cells in old mice through restoration of Cyclin D1. Nat. Metab. 2(4), 307–317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brandhorst S, Choi IY, Wei M et al. A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab. 22(1), 86–99 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inoue A, Piao L, Yue X et al. Young bone marrow transplantation prevents aging-related muscle atrophy in a senescence-accelerated mouse prone 10 model. J. Cachexia Sarcopenia Muscle 13(6), 3078–3090 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kovina MV, Karnaukhov AV, Krasheninnikov ME et al. Extension of maximal lifespan and high bone marrow chimerism after nonmyeloablative syngeneic transplantation of bone marrow from young to old mice. Front. Genet. 10, 310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das MM, Godoy M, Chen S et al. Young bone marrow transplantation preserves learning and memory in old mice. Commun. Biol. 2, 73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arriola Apelo SI, Pumper CP, Baar EL, Cummings NE, Lamming DW. Intermittent administration of rapamycin extends the life span of female C57BL/6J mice. J. Gerontol. A Biol. Sci. Med. Sci. 71(7), 876–881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin-Montalvo A, Mercken EM, Mitchell SJ et al. Metformin improves healthspan and lifespan in mice. Nat. Commun. 4, 2192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miura Y, Endo K, Komori K, Sekiya I. Clearance of senescent cells with ABT-263 improves biological functions of synovial mesenchymal stem cells from osteoarthritis patients. Stem Cell Res. Ther. 13(1), 222 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang J, Wang Y, Shao L et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22(1), 78–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yousefzadeh MJ, Wilkinson JE, Hughes B et al. Heterochronic parabiosis regulates the extent of cellular senescence in multiple tissues. Geroscience 42(3), 951–961 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jin S, Guerrero-Juarez CF, Zhang L et al. Inference and analysis of cell–cell communication using CellChat. Nat. Commun. 12(1), 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu A, Guo E, Yang J et al. Young plasma reverses age-dependent alterations in hepatic function through the restoration of autophagy. Aging Cell 17(1), e12708 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rando TA, Jones DL. Regeneration, rejuvenation, and replacement: turning back the clock on tissue aging. Cold Spring Harb. Perspect. Biol. 13(9), 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanders T, Liu Y, Buchner V, Tchounwou PB. Neurotoxic effects and biomarkers of lead exposure: a review. Rev. Environ. Health 24(1), 15–45 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White LD, Cory-Slechta DA, Gilbert ME et al. New and evolving concepts in the neurotoxicology of lead. Toxicol. Appl. Pharmacol. 225(1), 1–27 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Bellinger DC. The protean toxicities of lead: new chapters in a familiar story. Int. J. Environ. Res. Public Health 8(7), 2593–2628 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuehn BM. Panel advises tougher limits on lead exposure. JAMA 307(5), 445 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Brubaker CJ, Schmithorst VJ, Haynes EN et al. Altered myelination and axonal integrity in adults with childhood lead exposure: a diffusion tensor imaging study. Neurotoxicology 30(6), 867–875 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cecil KM, Brubaker CJ, Adler CM et al. Decreased brain volume in adults with childhood lead exposure. PLoS Med. 5(5), e112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stokes L, Letz R, Gerr F et al. Neurotoxicity in young adults 20 years after childhood exposure to lead: the Bunker Hill experience. Occup. Environ. Med. 55(8), 507–516 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi J, Kim YS, Kim MH, Kim HJ, Yoon BE. Maternal lead exposure induces sex-dependent cerebellar glial alterations and repetitive behaviors. Front. Cell. Neurosci. 16, (2022) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y, Liu S, Xu H et al. Maternal exposure to low dose BDE209 and Pb mixture induced neurobehavioral anomalies in C57BL/6 male offspring. Toxicology 418, 70–80 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Senut MC, Sen A, Cingolani P, Shaik A, Land SJ, Ruden DM. Lead exposure disrupts global DNA methylation in human embryonic stem cells and alters their neuronal differentiation. Toxicol. Sci. 139(1), 142–161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper describes the experiments in Figure 3.
- 79.Lamas GA, Bhatnagar A, Jones MR et al. Contaminant metals as cardiovascular risk factors: a scientific statement from the American Heart Association. J. Am. Heart Assoc. 12(13), e029852 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chappell GA, Thompson CM, Wolf JC, Cullen JM, Klaunig JE, Haws LC. Assessment of the mode of action underlying the effects of GenX in mouse liver and implications for assessing human health risks. Toxicol. Pathol. 48(3), 494–508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crebelli R, Caiola S, Conti L et al. Can sustained exposure to PFAS trigger a genotoxic response? A comprehensive genotoxicity assessment in mice after subacute oral administration of PFOA and PFBA. Regul. Toxicol. Pharmacol. 106, 169–177 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Svoboda LK, Wang K, Goodrich JM et al. Perinatal lead exposure promotes sex-specific epigenetic programming of disease-relevant pathways in mouse heart. Toxics 11(1), 85 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Svoboda LK, Wang K, Jones TR, Colacino JA, Sartor MA, Dolinoy DC. Sex-specific alterations in cardiac DNA methylation in adult mice by perinatal lead exposure. Int. J. Environ. Res. Public Health 18(2) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Montrose L, Goodrich JM, Morishita M et al. Neonatal lead (Pb) exposure and DNA methylation profiles in dried bloodspots. Int. J. Environ. Res. Public Health 17(18), 6775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sen A, Heredia N, Senut MC et al. Early life lead exposure causes gender-specific changes in the DNA methylation profile of DNA extracted from dried blood spots. Epigenomics 7(3), 379–393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sen A, Cingolani P, Senut MC et al. Lead exposure induces changes in 5-hydroxymethylcytosine clusters in CpG islands in human embryonic stem cells and umbilical cord blood. Epigenetics 10(7), 607–621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeid D, Gould TJ. Chronic nicotine exposure alters sperm small RNA content in C57BL/6J mouse model. Dev. Psychobiol. 65(2), e22367 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yokota S, Takeda K, Oshio S. Spatiotemporal small non-coding RNAs expressed in the germline as an early biomarker of testicular toxicity and transgenerational effects caused by prenatal exposure to nanosized particles. Front. Toxicol. 3, (2021) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sarkies P. Molecular mechanisms of epigenetic inheritance: possible evolutionary implications. Semin. Cell Dev. Biol. 97, 106–115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sonntag WE, Csiszar A, Decabo R, Ferrucci L, Ungvari Z. Diverse roles of growth hormone and insulin-like growth factor-1 in mammalian aging: progress and controversies. J. Gerontol. A Biol. Sci. Med. Sci. 67(6), 587–598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bickel MA, Csik B, Gulej R, Ungvari A, Nyul-Toth A, Conley SM. Cell non-autonomous regulation of cerebrovascular aging processes by the somatotropic axis. Front. Endocrinol. (Lausanne) 14, (2023) (Online). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Duran-Ortiz S, List EO, Ikeno Y et al. Growth hormone receptor gene disruption in mature-adult mice improves male insulin sensitivity and extends female lifespan. Aging Cell 20(12), e13506 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun LY, Fang Y, Patki A et al. Longevity is impacted by growth hormone action during early postnatal period. Elife 6, e24059 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kiss T, Tarantini S, Csipo T et al. Circulating anti-geronic factors from heterochonic parabionts promote vascular rejuvenation in aged mice: transcriptional footprint of mitochondrial protection, attenuation of oxidative stress, and rescue of endothelial function by young blood. GeroScience 42, 727–748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morrison EJ, Champagne DP, Dzieciatkowska M et al. Parabiosis incompletely reverses aging-induced metabolic changes and oxidant stress in mouse red blood cells. Nutrients 11(6), 1337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]