Abstract
Efficient ovarian follicle development, maturation, and ovulation are critical for egg production performance. Previous research has underscored the importance of messenger RNAs (mRNAs) in regulating development and folliculogenesis in chicken ovarians. However, the molecular mechanism is not fully understood, especially in the late period of the laying cycle. In the present study, ovarian tissues from 80-week-old Hy-Line Brown layers (three with high and three with low rates of egg laying) were collected for transcriptome sequencing. A total of 306 differentially expressed genes (DEGs) were identified in this study, at a false discovery rate (FDR)-corrected P-value < 0.05 and a log2|fold change| (log2|FC|) ≥1.5. Among these DEGs, stanniocalcin 1 (STC1) was mainly related to cellular processes, single-organism processes, biological regulation, metabolic processes, developmental processes, and reproductive processes. Then, we further investigated the regulation of STC1 during chicken follicle development and found that STC1 inhibited the proliferation and stimulated the apoptosis of follicular granulosa cells (GCs), and decreased the expression of progesterone (P4) and estradiol (E2). Collectively, these results suggest that STC1 plays an important role in chicken follicle development by decreasing GC proliferation and steroidogenesis and stimulating GC apoptosis. This study contributes to the understanding of the reproductive biology of laying hens in the late period of the laying cycle and further lays a foundation for the improvement of egg production in poultry breeding.
Keywords: chicken, egg production, ovary, transcriptome sequencing, STC1
•Stanniocalcin 1 (STC1) expression is significantly lower in the ovaries of high-egg-laying chickens than in those of low-egg-laying chickens and can be used as an important indicator of the egg production performance of chickens.
•Stanniocalcin 1 (STC1) can significantly inhibit the proliferation and differentiation of chicken prefollicular granulosa cells, promote their apoptosis, inhibit the secretion of progesterone and estrogen, and inhibit the development of poultry follicles.
Introduction
Eggs are considered one of the most affordable sources of animal protein (Bain et al., 2016; Lesnierowski et al., 2018). As human demand for eggs increases, the number of layers are increasing rapidly. In poultry breeds, prolonging the egg-laying cycle is an important measure to improve egg production performance. The longer the laying cycle is, the greater the production and the higher the profit we obtain. However, in production practice, we found that some laying hens have a high-egg-laying rate, and some laying hens have a low-egg-laying rate in the late laying period (80 to 100 weeks). In addition to environmental and metabolic factors, egg production performance may be related to growth, development, ovulation, atresia, and regression in the chicken ovary follicle (Etches et al., 1983). Therefore, thoroughly understanding the process of follicular development and its mechanisms could aid in significantly improving egg production.
The exploration of the molecular mechanisms of ovarian follicle development has been a hot topic of research on poultry reproductive traits, and genetic differences are the determining factors affecting poultry reproduction. For decades, breeders have used traditional quantitative genetics and other technical tools to continuously select excellent reproductive traits with great success. However, reproductive traits are controlled by multiple genes with low heritability, and relying on traditional means of selection, the generation interval is long, the process is tedious, and new genetic progress occurs slowly. With the development of modern molecular biology, especially the application of molecular marker-assisted selection, high-throughput sequencing, and other molecular biology technologies, poultry breeders have been provided with new ideas and approaches.
Granulosa cells (GCs) proliferation and differentiation are an important part of follicular development, and the process of follicular development maturation must be accompanied by the proliferation and differentiation of GCs, which are mutually dependent. GCs proliferation, differentiation, and apoptosis determine follicular growth, selection, and atresia (Ghanem and Johnson, 2018). However, GCs synthesize the steroid hormones progesterone (P4) and estradiol (E2) under the stimulation of gonadotropins follicle-stimulating hormone (FSH) and luteinizing hormone (LH), respectively, to regulate follicular maturation and promote ovulation (Pierce and Parsons, 1981). During follicular growth and development, the coordination between GCs and oocytes is crucial for follicular development and oocyte maturation, and information exchange is complex. GCs provide 85% of nutrient metabolic requirements for oocytes mainly through gap junctions, regulate oocyte transcriptional activity, and induce post-translational modification of oocyte proteins (Jin et al., 2006). GCs secrete and express the growth factors insulin-like growth factor 1 (IGF1) and the local hormones follicle-stimulating hormone receptor (FSHR) in a paracrine manner to regulate follicular development and maturation, and activins (ACV), inhibin (INH), and follistatin (FST) expressed by GCs can regulate follicular differentiation and maturation with the help of FSH (Onagbesan et al., 2009; Liu et al., 2019b). In conclusion, GCs have a very important regulatory role in the developmental process of the follicle, and this role is closely related to the normal proliferation and differentiation of GCs and their secretory function.
Stanniocalcin (STC) is a homodimeric glycoprotein hormone that was first discovered in the corpuscles of Stannius of the teleost kidney endocrine gland (Yeung et al., 2012). It was found that STC in teleost tissue inhibited and promoted calcium uptake and phosphate reabsorption, respectively; therefore, STC was shown to act as an endocrine regulator, regulating the balance of calcium and phosphate (Lu et al., 1994; Schein et al., 2012). STC also play an important regulatory role in developmental and pathophysiological processes such as pregnancy, lactation, and organogenesis (Varghese et al., 1998; An et al., 2020). At present, two related STC genes, stanniocalcin 1 (STC1) and stanniocalcin 2 (STC2) have been identified in mammals (Joshi, 2020). Interestingly, the highest level of STC1 expression in mammals occurs in ovarian tissue, but no information is available on chicken species (Chang et al., 1996; Basini et al., 2010). Previous reports have shown that STC1 has different expression patterns during folliculogenesis, and STC1 has been proven to inhibit steroidogenesis (Baioni et al., 2009). Luo et al. (2004) showed that STC1 inhibited progesterone production in mouse ovarian GCs but had no effect on estrogen production, while studies in pigs showed that it could inhibit estrogen production (Baioni et al., 2009). However, even though STC1 plays an essential role in many cell processes, its function in chicken ovary folliculogenesis remains unclear. Therefore, the present study was undertaken to investigate the expression and function of STC1 in chicken ovaries.
In the present study, we adopted RNA sequencing (RNA-seq) technology to evaluate the predominant genes involved in the development of growing follicles and found that STC1 was differentially expressed in the two groups [Hy-Line Brown layers with high- and low-egg-laying rates (HH and HL)]. Then, the expression of STC1 in vitro was detected, and functional research was performed using a chicken follicular GCs model. In addition, STC1 was found to have a potential role in inhibiting follicular development in chickens.
Materials and Methods
Ethics statement
In this study, all of the animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Henan Agricultural University, Zhengzhou, China, and performed in strict accordance with the guidelines of the Animal Use Committee of the Chinese Ministry of Agriculture, Beijing, China (approval number: S20190196).
Animal tissue sampling, RNA extraction, and sequencing
A total of 30 Hy-Line Brown layers from the Henan Innovative Engineering Research Center of Poultry Germplasm Resource were reared under the same environmental conditions with ad libitum access to water and food before sample collection. Based on similar reproductive traits, the population was divided into high-egg-laying rate (HH1, HH2, and HH3) and low-egg-laying rate (HL1, HL2, and HL3) groups, and ovarian tissues collected at 80 wks of age, and analyzed the egg production performance differences in the high- and low-egg-laying rates by SPSS 24.0 (IBM, Among, New York, USA). Fresh tissue samples were washed in Phosphate Buffered Saline (PBS) (Cat 70011044, Gibco, Fisher Scientific, Waltham, MA, USA) and immediately stored at −80 °C after quick freezing with liquid nitrogen. Total RNA was extracted using TRIzol reagent (Cat R401-01, Vazyme, Nanjing, China) following the manufacturer’s instructions, and RNA integrity was assessed by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Qualified total RNA was further purified by an RNA Clean XP Kit (Cat A63987, Beckman Coulter, Inc., Kraemer Boulevard Brea, CA, USA) and RNase-Free DNase Set (Cat 79254, QIAGEN, GmBH, Germany). The cDNA libraries were constructed by Shanghai Biotechnology Corp (Shanghai, China) and sequenced on an Illumina HiSeq 2500 ((Illumina, San Diego, CA, USA). The raw reads were filtered with Seqtk (https://github.com/lh3/seqtk), and the number of all clean reads for each gene in each sample was counted with HTSeq (version: 2.0.4) (Kim et al., 2015). The fragments per kilobase of exon model per million mapped reads (FPKM; Mortazavi et al., 2008) were calculated to estimate the expression level of genes in each sample using the Stringtie (version: 1.3.0) (Pertea et al., 2015, 2016). The raw transcriptome datasets for the Hy-Line Brown layers ovarian have been deposited in the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra) with accession number SRR6456830.
Differential expression and functional enrichment analyses
edgeR (Robinson et al., 2010) was used to perform differential gene analysis between samples and multiple hypothesis test correction. The threshold of the P-value was determined by controlling the false discovery rate (FDR; Deol et al., 2000). In addition, we calculated the differential expression multiple based on the FPKM value, that is, fold change (FC). In this study, the genes with P-value < 0.05 and log2|FC| ≥ 1.5 were identified as differentially expressed genes (DEGs). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed by clusterProfile, and P < 0.05 was considered as the threshold.
Plasmid construction and siRNA
The STC1 coding sequence was amplified from chicken follicle cDNA by PCR using gene-specific clone primers (Supplementary Table S1). The PCR product was cloned into the pcDNA3.1-EGFP vector (Cat V79020, Invitrogen, USA) within NheI and EcoRI sites. The successful STC1 overexpression vector was confirmed by agarose gel electrophoresis and DNA sequencing. siRNA specifically against chicken STC1 was obtained from RiboBio (Guangdong, China), and a nonspecific duplex was used as the control. The siRNAs used in this study are listed in Supplementary Table S2.
GC isolation culture and transfection
GCs were isolated and cultured from chicken follicles. First, the granulosa layers were dissected away from the pre-hierarchical follicles (6 to 8 mm) of 35-wk-old chicken, and then minced and homogenized in a centrifuge tube. To release single cells, the granulosa layers were digested with 0.25% trypsin (Cat 25200056, Gibco) for 10 min at 37 °C. After neutralization with complete medium, single cells were collected by centrifugation at 1800 × g. Subsequently, cell suspensions were seeded in cell culture medium containing DMEM/F12 (Cat SH30023.01B, HyClone, Logan, Utah, USA) supplemented with 2.5% fetal bovine serum (Cat 1099141C, Gibco) and 1% penicillin/streptomycin (Cat 15140122, Gibco). GCs were cultured at 37 °C in a 5% CO2 humidified atmosphere. Lipofectamine 2000 (Cat 11668-027, Invitrogen) was used for transient transfection. For specific procedures, refer to the reagent specification.
Cell proliferation assays
GCs were treated with STC1 overexpression vector and siRNA, and cell activity was detected at 12 h, 24 h, 36 h, and 48 h. First, 10 μL of Cell Counting Kit-8 solution (CCK-8) (Cat CK04, Dojindo, Kumamoto, Japan) was added into the blank, control, and treatment wells of the 96-well plate and incubated in the incubator for 2 h. Then, the absorbance at 450 nm was measured by Enzyme Labeler (Cat Synergy2, BioTek, Palo Alto, USA), and the GCs viability was calculated: (treatment wells−control wells)/(control wells−blank wells) × 100%.
A Cell-Light TM EdU DNA Cell Proliferation (EdU) (Cat C10310, RiboBio, Guangzhou, China) assay was performed on GCs treated with STC1 overexpression vector and siRNA. In the experiment, EdU medium was first added for incubation for 2 h and then fixed with paraformaldehyde for 30 min. Subsequently, glycine and Triton X-100 were added for incubation on the decolorization shaking bed. Finally, 1×Apollo staining reaction solution and DAPI were added for observation of cell proliferation under a fluorescence inverted microscope (Cat Eclipse Ts2, Nikon, Tokyo, Japan).
Cell cycle analysis
For analysis of the cell cycle of GCs, cells were collected and fixed in 75% ethanol overnight. After ethanol fixation, the cells were stained with propidium iodide using PI/RNase staining buffer (Cat 550825, BD Biosciences Pharmingen, San Diego, CA, USA) according to the manufacturer’s manual. Subsequently, flow cytometry (Cat Accuri C6, BD Biosciences Pharmingen) was used for detection, generally counting 2~3 × 104 cells, and the cell cycle was analyzed by Flow-Jo7.6 software (Stanford University, Stanford, CA, USA).
Cell apoptosis assay
Forty-eight hours after transfection, chicken GCs were harvested, and a cell-counting machine (Cat 6749, Corning, NY, USA) was used for the detection of apoptotic cells based on the principle of fluorescence-activated cell sorting (FACS). According to the indicated manufacturer’s protocol (Cat A211-01, Vazyme), the apoptosis rate was detected by flow cytometry (Cat Accuri C6, BD Biosciences Pharmingen) with the fluorescein isothiocyanate (FITC) and propidium iodide (PI) signals. The FlowJo v7.6 software (Stanford University, Stanford, CA, USA) was used to analyze the results.
Hormone analysis
Chicken GCs were cultured in a six-well plate with 2 mL of medium. After a 48 h of overexpression vector or siRNA treatment, the culture medium was assayed immediately or stored at −80 °C until assayed. The progesterone concentration in the conditioned medium was diluted to one-twentieth with saline and then measured using an enzyme-linked immunosorbent assay kit (ELISA) (Cat FY4369-B, Feiya Technology, Jiangsu, China) according to the manufacturer’s instructions. PG and E2 values were determined by extrapolating from a standard curve. Each sample was measured in triplicate.
Quantitative real-time PCR
Quantitative real-time PCR (qRT–PCR) was performed using a TB Green Premix Ex Taq II (Tli RNaseH Plus) Kit (Cat RR820B, TaKaRa, Dalian, China). The qRT–PCR gene-specific primers were designed using the NCBI online software (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and purchased from BiosSunya Co., Ltd (Zhengzhou, China), and all the qRT–PCR primers for these genes are described in Supplementary Table S3. The qRT–PCR amplification procedure was as follows: 95 °C for 3 min, 35 cycles of 95 °C for 12 s, 60 °C for 30 s, 72 °C for 30 s, and an extension for 10 min at 72 °C (n = 3). Quantification of the expression of all chosen mRNAs was conducted using the comparative CT (2−ΔΔCT) method with β-actin as an internal reference RNA (β-actin acts as an internal reference gene, and it is stably expressed in melanocytes; Livak and Schmittgen, 2001).
Statistical analysis
Statistical analysis was carried out by using Prism 8 software (GraphPad Software, Inc., USA). All results are presented as the means ± SEMs. The significance of the comparison between the two groups was assessed by one-sample t-test using SPSS 24.0 (IBM, Among, New York, USA). P < 0.05 was considered a significant difference.
Results
Statistics of egg production rate
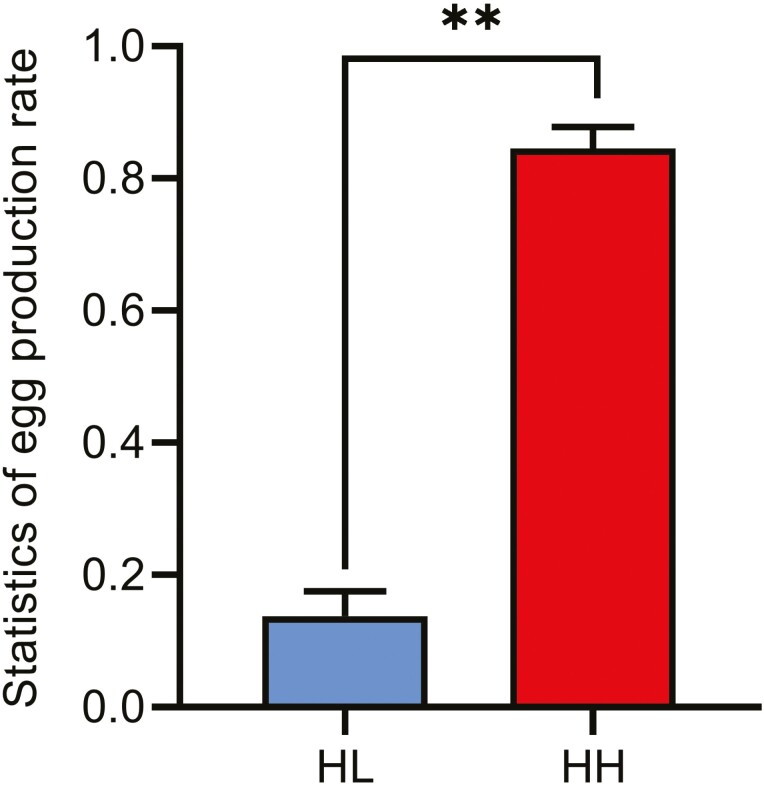
The average egg production rates at 80 weeks (mean ± SEM) were 85.11% ± 0.0212 and 15.60% ± 0.071 for HH and HL chickens, respectively. The egg production rate of HH chickens was significantly higher than that of HL chickens (P < 0.01, Figure 1), indicating that the two groups were suitable for identifying DEGs associated with reproductive traits.
Figure 1.
Comparison of the egg production rate of 80-wk-old HH and HL chickens. Data are presented as the mean ± SE (n = 15), and the error bar shows the standard deviation. **P < 0.01.
Identification and functional annotation of differentially expressed genes
In this study, we included six chicken ovarian cDNA libraries created by the Illumina HiSeq 2500 platform. After quality control, 147,492,401, 117,774,725, and 80,057,178 clean reads were generated for the three HHs, and 142,872,483 78,079,711, and 90,807,270 clean reads were generated for the three HLs. Approximately, 91% of the reads in each library were uniquely mapped to the GGA5 assembly of the chicken reference genome (Table 1).
Table 1.
Characteristics of the reads from six chicken ovarian transcriptomes
| Sample ID1 | Raw reads | Clean reads | Clean ratio2 (%) | All reads | Mapped reads | Mapped | Mapping ratio3 (%) |
|---|---|---|---|---|---|---|---|
| HH-1 | 154,392,790 | 147,492,401 | 95.53 | 143,663,524 | 131,113,327 | 129,937,309 | 91.26 |
| HH-2 | 123,416,200 | 117,774,725 | 95.43 | 114,535,538 | 104,276,259 | 103,432,922 | 91.04 |
| HH-3 | 83,774,408 | 80,057,178 | 95.56 | 78,055,678 | 71,071,119 | 70,446,136 | 91.05 |
| HL-1 | 149,512,210 | 142,872,483 | 95.56 | 139,337,904 | 127,556,400 | 126,481,832 | 91.54 |
| HL-2 | 81,256,850 | 78,079,711 | 96.09 | 76,637,518 | 69,923,560 | 69,366,288 | 91.24 |
| HL-3 | 94,542,336 | 90,807,270 | 96.05 | 88,805,590 | 82,822,947 | 82,080,840 | 93.26 |
1HH, 3 ovarian samples from high rates of egg production chickens; HL, 3 ovarian samples from low rates of egg production chickens.
2Clean ratio = clean reads/raw reads
3Mapping ratio = mapped reads/all reads.
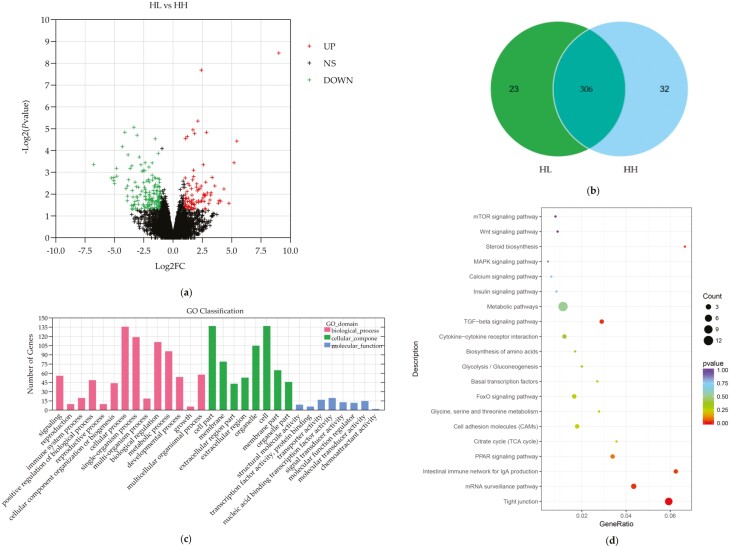
A total of 24,881 genes were identified in both groups, including 306 differentially expressed genes (P-value < 0.05 and log2|FC| ≥ 1.5) that were coexpressed in the high- and low-egg-laying rate groups, with 112 mRNAs being upregulated and 194 mRNAs being downregulated in HL compared to HH (Figure 2a and b).
Figure 2.
Differentially expressed genes between HH and HL chickens. (a) Volcano plot of differentially expressed genes between HL and HH chicken ovaries. (b) Venn diagram of coexpressed differentially expressed genes in HL and HH chicken ovaries. (c) GO enrichment of differentially expressed genes between HH and HL chicken ovaries. (d) KEGG pathway of differentially expressed genes between HH and HL chicken ovaries.
To explore the biological roles of the DEGs, we performed GO and KEGG enrichment analyses. A total of 175 differentially expressed genes were annotated to the biological processes, cellular components, and molecular functions of GO. Among them, in biological processes, differentially expressed genes were mainly involved in regulating cellular processes, bioregulation, metabolic processes, developmental processes, and reproduction processes; in cellular components, differentially expressed genes were associated with cells, organelles, cell membranes, and extracellular compartments; and in molecular functions, differentially expressed genes were associated with nucleic acid binding transcription factor activity, transporter protein activity, and molecular sensor activity (Figure 2c). Notably, STC1 is enriched in cell development, cell differentiation, tissue development, hormone activity, cell migration, and other biological processes (Supplementary Table S4).
In addition, the results of KEGG analysis revealed that DEGs were enriched mainly in metabolic pathways, tight junctions, and mRNA surveillance pathways. As shown in Figure 2d, the largest category was metabolic pathways, which had 12 annotated genes. Notably, specific enrichment of genes was observed for pathways involved in reproduction regulation (Supplementary Table S5), such as the FoxO signaling pathway, TGF–beta signaling pathway, MAPK-signaling pathway, PPAR-signaling pathway, and steroid biosynthesis. In addition, several genes are involved in multiple pathways, such as PCK1 (FoxO signaling pathway, glycolysis/gluconeogenesis, adipocytokine-signaling pathway, insulin signaling pathway), and FZD6 (mTOR-signaling pathway, Wnt-signaling pathway), which indicates that these genes can be taken as functional candidate genes associated with the difference in egg production.
Verification of gene expression profiles
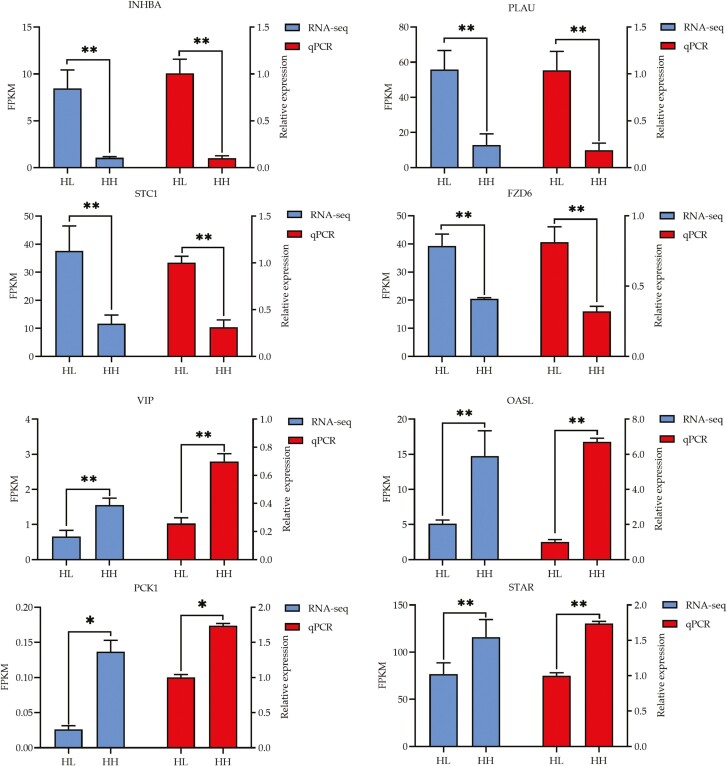
To verify the accuracy and reproducibility of the RNA-seq data, we selected 8 well-known candidate DEGs associated with egg reproduction, including INHBA, PLAU, STC1, FZD6, VIP, OASL, PCK1, and STAR, to conduct qRT–PCR. All eight mRNAs had similar expression patterns in comparison to the RNA-seq data (Figure 3), indicating that the RNA-seq data are reliable.
Figure 3.
Quantitative validation of transcriptome sequencing results.
Infection efficiency
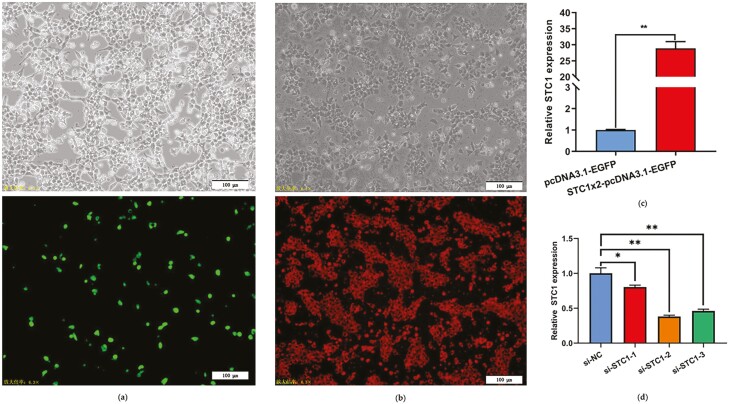
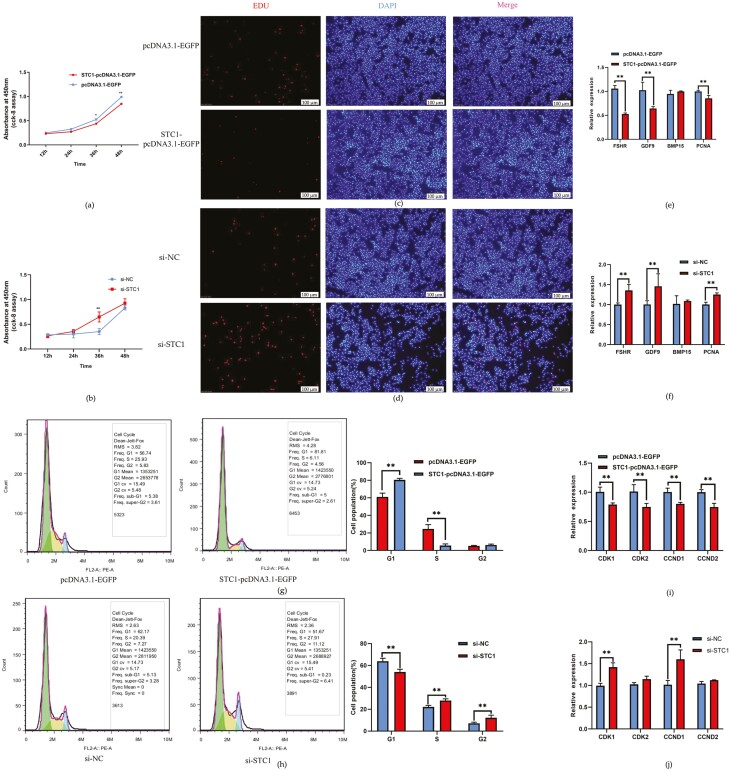
To study the function of STC1 in the chicken ovary, we constructed an STC1 overexpression vector and synthesized STC1-specific siRNA. The pcDNA3.1-EGFP-STC1 plasmid and STC1-specific siRNA were successfully transfected into GCs. EGFP and CY5 were detected by fluorescence microscopy in the transfected group (Figure 4a and b). qRT–PCR indicated a significant increase in STC1 abundance in the pcDNA3.1-EGFP-STC1 group (P < 0.01, Figure 4c). Among the three synthesized STC1-specific siRNAs, si-STC1-2 most significantly silenced STC1 expression in the chicken GCs (P < 0.01, Figure 4d), so we subsequently used this siRNA to conduct the following research.
Figure 4.
Detection of the transfection efficiency of STC1 in GCs. (a) GCs expressing green fluorescent protein (GFP) could be observed after STC1 overexpression. (b) GCs expressing red fluorescence (Cy5) could be observed after si-STC1 transfection. (c) STC1 mRNA expression levels in GCs after STC1 overexpression (n = 3). (d) STC1 mRNA expression levels in GCs after si-STC1 transfection (n = 3).
STC1 inhibits cell proliferation in chicken GCs
Because the normal proliferation and differentiation of GCs are crucial for follicular development, we tested the effects of STC1 on GC activity and proliferation. CCK-8 and the EdU assays demonstrated that STC1 significantly repressed GC viability, while its inhibition GC proliferation (P < 0.01, Figure 5a–d). We also detected the expression of marker genes for the proliferation and differentiation of GCs, and the results showed that the overexpression of STC1 in GCs can repress the expression of FSHR, GDF9, and PCNA (P < 0.01, Figure 5e), and that STC1 knockdown induced the expression of FSHR, GDF9, and PCNA (P < 0.01, Figure 5f).
Figure 5.
The inhibitory effect of STC1 on granulosa cell proliferation. (a) Results of the CCK-8 assay after STC1 overexpression (n = 3). (b) CCK-8 assay results after si-STC1 transfection (n = 3). (c) EdU assay results after STC1 overexpression. (d) EdU results after si-STC1 transfection. (e) The expression of cell proliferation-related genes after STC1 overexpression (n = 3). (f) The expression of cell proliferation-related genes after si-STC1 transfection (n = 3). (g) Results of flow cytometry after STC1 overexpression (n = 3). (h) Results of the flow cytometry cycle analysis after si-STC1 transfection (n = 3). (i) The expression of cell cycle-related genes after STC1 overexpression (n = 3). (j) The expression of cell cycle-related genes after si-STC1 transfection (n = 3).
The overexpression of STC1 significantly decreased the numbers of S-phase and G2/M-phase cells and induced cell cycle arrest (P < 0.01, Figure 5g). Conversely, STC1 knockdown significantly increased S-phase cells relative to control group cells (P < 0.01, Figure 5h). In addition, STC1 overexpression reduced the expression of CDK1, CDK2, CCND1, CCND2 genes involved in the cell cycle pathway (P < 0.01, Figure 5i), and the siRNA of STC1 promoted the expression of CDK1 and CCND1 (P < 0.01, Figure 5j). Together, these data indicate that STC1 expression has a negative regulatory role in cell proliferation and decreases the cell cycle distribution of proliferating GCs.
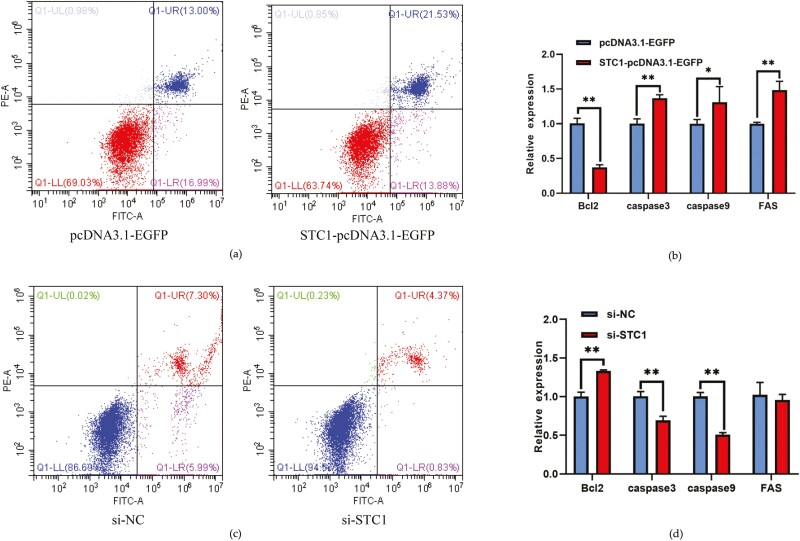
STC1 promotes cell apoptosis in chicken GCs
STC1 overexpression caused a significant upregulation in the apoptosis rate and proapoptotic gene caspase 3, caspase 9 mRNA levels in chicken GCs and a significant downregulation of antiapoptotic gene Bcl2 mRNA expression (P < 0.05, Figure 6a and b). Furthermore, the siRNA of STC1 revealed that the GC apoptosis rate and proapoptotic gene caspase 3 and caspase 9 mRNA levels were significantly inhibited and antiapoptotic gene Bcl2 mRNA level is significantly increased (P < 0.01, Figure 6c and d). These data revealed that STC1 promotes apoptosis in the chicken GCs.
Figure 6.
The promoting effect of STC1 on granulosa cell apoptosis. (a) Results of flow cytometry apoptosis after si-STC1 transfection. (b) The expression of cell apoptosis-related genes after STC1 overexpression (n = 3). (c) Results of flow cytometry apoptosis after si-STC1 transfection. (d) The expression of cell apoptosis-related genes after si-STC1 transfection (n = 3).
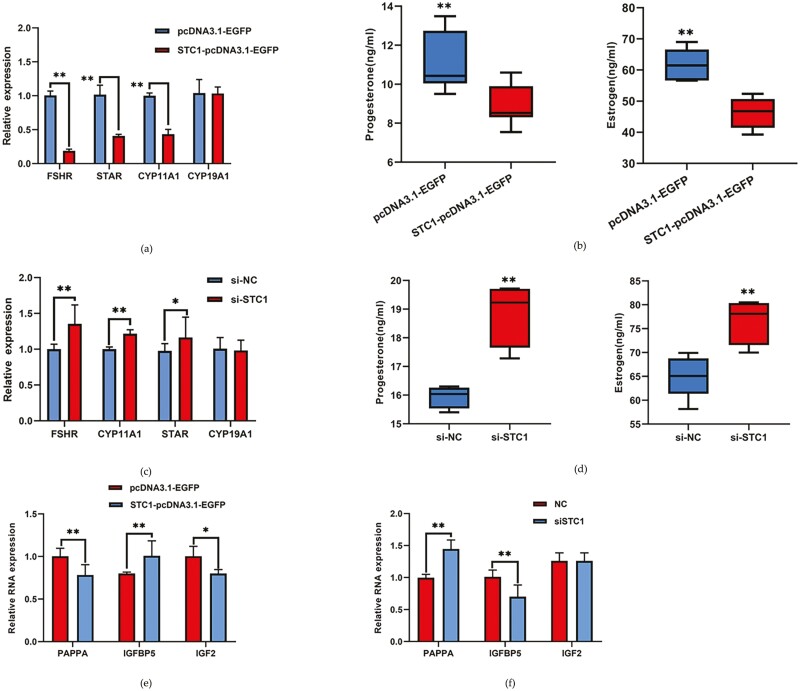
STC1 inhibits steroid hormone production in chicken GCs
The proliferation and apoptosis of GCs play an important role in follicular development. Another major function of GCs is to regulate the production and secretion of steroid hormones. Next, we tested the function of STC1 in the synthesis of steroids. Overexpression of STC1 repressed follicular development marker gene expression and the expression of genes related to the secretion of steroid hormones, such as FSHR, STAR, and CYP11A1 (P < 0.01, Figure 7a). ELISA results showed that overexpression of STC1 significantly reduced progesterone and estrogen production (P < 0.01, Figure 7b). However, the siRNA of STC1 promoted marker gene expression and the production and secretion of steroid hormones (P < 0.01, Figure 7c and d). To investigate the role of STC1 in the insulin-like growth factor pathway, we assessed the expression of PAPPA, IGFBP5, and IGF2 after overexpression and interference of STC1 in chicken GCs. The results showed that overexpression of STC1 could significantly downregulate the expression of PAPPA and IGF2 and significantly upregulate the expression of IGFBP5 (P < 0.05, Figure 7e). The siRNA of STC1 significantly upregulated PAPPA and downregulated IGFBP5 (P < 0.01), while IGF2 showed no significant change (Figure 7f).
Figure 7.
The inhibitory effect of STC1 on granulosa cell steroid hormone production. (a) The expression of steroid hormone secretion-related genes after STC1 overexpression (n = 3). (b) The secretion of steroid hormones after STC1 overexpression (n = 3). (c) The expression of steroid hormone secretion-related genes after si-STC1 transfection (n = 3). (d) The secretion of steroid hormones after si-STC1 transfection (n = 3). (e) The expression of insulin-like growth factor pathway-related genes after STC1overexpression (n = 3). (f) The expression of the insulin-like growth factor pathway-related genes after si-STC1 transfection (n = 3).
Discussion
The egg-laying rate of hens mainly depends on the normal development of follicles and ovulation in the ovary. The egg-laying process is mainly regulated by the hypothalamic–pituitary–ovarian axis, involving a series of sex steroid hormones and cytokines (Bronneberg et al., 2009; Brady et al., 2019). The development of follicles is orchestrated by numerous gene interactions and requires tight regulation of gene expression at both the transcriptional and post-transcriptional levels (Etches et al., 1983; Onagbesan et al., 2009). In recent years, an increasing number of studies have proven that high-throughput transcriptomic sequencing technology is efficient and widely used to generate whole transcriptome sequences and identify differentially expressed genes (Kang et al., 2009; Ozsolak and Milos, 2011; Terenina et al., 2017). Gaining insight into the gene expression patterns during ovarian follicle development by high-throughput sequencing could improve egg-laying performance.
The present study identified a total of 24,881 mRNAs in chicken ovaries using the Illumina sequencing method, including 306 differentially expressed mRNAs, some of which may participate in ovarian follicular development, maturation, and ovulation. GO enrichment analysis revealed nine differentially expressed genes associated with reproduction during biological processes, including two known genes, inhibin beta A (INHBA) and zona pellucida glycoprotein 3 (ZP3). KEGG analysis showed that the differentially expressed genes were significantly enriched in reproduction regulation-related pathways, including the FoxO signaling pathway, TGF-β signaling pathway, MAPK signaling pathway, PPAR signaling pathway, and steroid biosynthesis. INHBA is a subunit of inhibin that belongs to the transforming growth factor beta (TGF-beta) superfamily, which affects follicular development by regulating the production of inhibin (Burger et al., 1988; Bao et al., 2021). INH is a macromolecular glycoprotein hormone secreted by testicular support cells in males and ovarian GCs in females, and studies have shown that INH indirectly regulates follicular growth and development by inhibiting the synthesis and secretion of FSH (Jimenez-Krassel et al., 2003). Our transcriptome analysis showed that the expression of the INHBA gene was significantly increased in the ovaries of low-egg-laying Hy-Line Brown layers compared to high-egg-laying Hy-Line Brown layers. Therefore, it is hypothesized that the upregulation of INHBA in the ovaries of low-yielding layers may suppress the effect of FSH on reproductive activity. The ZP3 gene is a member of the zona pellucida glycoprotein family, and mutations in the ZP3 gene cause abnormal oocyte and follicle development in humans, resulting in female infertility (Li et al., 2022; Zhang et al., 2022). It has been shown that chicken ZP3 is synthesized in follicular GCs, and its encoded protein serves as a sperm receptor (Waclawek et al., 1998; Nishio et al., 2018). There are relatively few studies on the specific mechanism of action of the ZP3 gene in poultry.
We found that STC1 was significantly enriched in cell development, cell differentiation, tissue development, hormone activity, cell migration, and other biological processes. STC1 is a glycoprotein hormone involved in mineral homeostasis that was originally found in fish in 1839 (Wagner and Dimattia, 2006; Bishop et al., 2021), whereas mammalian STC1 shows a diverse tissue expression pattern, with the ovary exhibiting the highest level (Varghese et al., 2002; Yoshiko and Aubin, 2004). In recent years, studies have indicated that STC1 plays a key role in cell proliferation and apoptosis (Dai et al., 2016; Pan et al., 2017). However, the exact role and molecular regulatory mechanisms of STC1 in the follicular development of the hen ovary remain unclear. In the current study, we found that STC1 inhibited GC proliferation and decreased the mRNA expression levels of FSHR, GDF9, PCNA, CDK1, CDK2, CCND1, and CCND2, demonstrating that STC1 may suppress follicle selection, growth, and maturation by hindering GC proliferation and differentiation during the follicular development of the hen ovary. These data attested that the STC1 gene exerts an inhibitory effect on the development of the follicles.
Poultry reproduction is strictly regulated by sex steroid hormones (Johnson et al., 2002). Sex steroid hormones, such as progesterone and estradiol, are involved in the regulation of ovulation, gonadal differentiation, and sexual and nesting behaviors in birds through interactions with their intracellular receptors (Ghanem and Johnson, 2019; Liu et al., 2019b). In some GC research, STC1 has been proven to dampen gonadotropin stimulation of GC differentiation by paracrine regulation and inhibit progesterone secretion (Luo et al., 2004; Baioni et al., 2011). It is tempting to speculate that STC1 may have a pivotal role in the development of follicles. In the current study, overexpression of STC1 in chicken GCs inhibited the expression of FSHR, STAR, and CYP11A1. Conversely, the siRNA of STC1 in chicken GCs increased the expression of FSHR, STAR, and CYP11A1. As expected, ELISA detection also showed decreased progesterone and estradiol accumulation in the STC1 overexpressing group, and opposite results were obtained with siRNA of STC1. These results suggest that STC1 acts as a negative regulator of follicular growth and development in the hen ovary. Several studies (Argente et al., 2017) have shown that when STC1 acts on follicles, the IGF signaling pathway is inhibited. Recently, the stanniocalcin (STC1 and STC2) was identified as a novel inhibitor of pregnancy-associated plasma protein A (PAPPA) (Jepsen et al., 2015, 2016; Kløverpris et al., 2015), which is a key regulator of IGF bioactivity, by releasing IGFs from their corresponding IGF-binding proteins (IGFBPs; Spitschak and Hoeflich, 2018; Liu et al., 2019a). Thus, STC1 may regulate folliculogenesis via PAPPA in the chicken ovary. Similarly, our results showed that STC1 inhibited the expression of PAPPA and promoted the expression of IGFBPs, suggesting that there may be some interactions between STC1 and PAPPA that regulate follicular development. Moreover, further studies focusing on the function of the chicken STC1-PAPPA-IGF axis in the ovary are needed.
Conclusion
In conclusion, our results demonstrate a novel role and mechanism of STC1 in regulating GC proliferation, apoptosis, and steroid secretion (P4 and E2) in chicken GCs. We first verified that STC1 inhibited the expression of STAR and CYP11A1, and suppressed the expression of PAPPA through the IGF pathway in chickens. These findings show that STC1 plays a critical role in follicle development and maturation by maintaining steroid hormone homeostasis.
Supplementary Material
Acknowledgments
This study was supported by the Key Research Project of the Shennong Laboratory (SN01-2022-05) and the China Agriculture Research System (CARS-40-K04), National Natural Science Foundation of China (32072710 and 32102540).
Glossary
Abbreviations:
- ACV
activins
- DEG
differentially expressed genes
- E2
estradiol
- ELISA
enzyme-linked immunosorbent assay
- FC
fold-change
- FDR
false discovery rate
- FPKM
fragments per kilobase million
- FSH
follicle-stimulating hormone
- FSHR
follicle-stimulating hormone receptor
- FST
follistatin
- GCs
follicular granulosa cells
- GO
gene ontology
- HH
Hy-Line Brown layers with high-egg-laying rates
- HL
Hy-Line Brown layers with low-egg-laying rates
- IGF1
insulin-like growth factor 1
- INH
inhibin
- INHBA
inhibin beta A
- KEGG
kyoto encyclopedia of genes and genomes
- LH
luteinizing hormone
- log2|FC|
log2|fold change|
- mRNA
messenger RNA
- P4
progesterone
- PBS
phosphate buffered saline
- PG
prostaglandin
- qRT-PCR
quantitative real-time PCR
- RNA-seq
RNA sequencing
- STC
stanniocalcin
- STC1
stanniocalcin 1
- STC2
stanniocalcin 2
- ZP3
zona pellucida glycoprotein 3
Contributor Information
Junwei Sun, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Pengwei Zhang, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Dongxue Wang, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Shuaipeng Zhu, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Xiangfei Ma, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Zhenwei Du, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Jiechang Zhang, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Shuangyuan Yang, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Hetian Huang, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Ruirui Jiang, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Yadong Tian, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Wenting Li, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; The Shennong Laboratory, Zhengzhou 450002, China.
Xiangtao Kang, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; The Shennong Laboratory, Zhengzhou 450002, China.
Fengbin Yan, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Guirong Sun, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; The Shennong Laboratory, Zhengzhou 450002, China.
Donghua Li, College of Animal Science and Technology, Henan Agricultural University, Zhengzhou 450046, China; Henan Key Laboratory for Innovation and Utilization of Chicken Germplasm Resources, Zhengzhou 450046, China.
Author Contributions
Junwei Sun, Pengwei Zhang, Dongxue Wang: methodology and writing-original draft. Shuaipeng Zhu, Xiangfei Ma, Zhenwei Du: investigation and software. Jiechang Zhang: formal analysis. Ruirui Jiang: validation and data curation. Shuangyuan Yang and Yadong Tian: methodology and software. Wenting Li: investigation and data curation. Xiangtao Kang and Guirong Sun: resources, supervision, and project administration. Hetian Huang and Fengbin Yan: writing-review and editing. Donghua Li: conceptualization, writing-review, and editing.
Conflict of Interest Statement
All authors disclosed no relevant relationships.
Literature Cited
- An, X., Ma H., Liu Y., Li F., Song Y., Li G., Bai Y., and Cao B... 2020. Effects of miR-101-3p on goat granulosa cells in vitro and ovarian development in vivo via STC1. J. Anim. Sci. Biotechnol. 11:102. doi: 10.1186/s40104-020-00506-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argente, J., Chowen J. A., Pérez-Jurado L. A., Frystyk J., and Oxvig C... 2017. One level up: abnormal proteolytic regulation of IGF activity plays a role in human pathophysiology. EMBO Mol. Med. 9:1338–1345. doi: 10.15252/emmm.201707950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain, M. M., Nys Y., and Dunn I. C. J. B... 2016. Increasing persistency in lay and stabilising egg quality in longer laying cycles. What are the challenges? Br Poult Sci. 57:330–338. doi: 10.1080/00071668.2016.1161727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baioni, L., Basini G., Bussolati S., and Grasselli F... 2009. Stanniocalcin 1 is a potential physiological modulator of steroidogenesis in the swine ovarian follicle. Vet. Res. Commun. 33:73–76. doi: 10.1007/s11259-009-9252-1 [DOI] [PubMed] [Google Scholar]
- Baioni, L., Basini G., Bussolati S., and Grasselli F... 2011. Stanniocalcin 1 affects redox status of swine granulosa cells. Regul. Pept. 168:45–49. doi: 10.1016/j.regpep.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Bao, Y., Yao X., Li X., Ei-Samahy M. A., Yang H., Liang Y., Liu Z., and Wang F... 2021. INHBA transfection regulates proliferation, apoptosis and hormone synthesis in sheep granulosa cells. Theriogenology. 175:111–122. doi: 10.1016/j.theriogenology.2021.09.004 [DOI] [PubMed] [Google Scholar]
- Basini, G., Baioni L., Bussolati S., Grolli S., Kramer L. H., Wagner G. F., and Grasselli F... 2010. Expression and localization of stanniocalcin 1 in swine ovary. Gen. Comp. Endocrinol. 166:404–408. doi: 10.1016/j.ygcen.2009.12.013 [DOI] [PubMed] [Google Scholar]
- Bishop, A., Cartwright J. E., and Whitley G. S... 2021. Stanniocalcin-1 in the female reproductive system and pregnancy. Hum. Reprod. Update. 27:1098–1114. doi: 10.1093/humupd/dmab028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, K., Porter T. E., Liu H. C., and Long J. A... 2019. Characterization of gene expression in the hypothalamo-pituitary-gonadal axis during the preovulatory surge in the turkey hen. Poult. Sci. 98:7041–7049. doi: 10.3382/ps/pez437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronneberg, R. G., Vernooij J. C., Stegeman J. A., and Taverne M. A... 2009. Follicle dynamics and its relation with plasma concentrations of progesterone, luteinizing hormone and estradiol during the egg-laying cycle in ostriches. Reprod. Domest. Anim. 44:705–713. doi: 10.1111/j.1439-0531.2007.01057.x [DOI] [PubMed] [Google Scholar]
- Burger, H. G., Igarashi M., Baird D. T., Bardin W., Chappel S., de Jong F., Demoulin A., de Kretser D., Findlay J., Forage R.,. et al. 1988. Inhibin: definition and nomenclature, including related substances. Clin. Endocrinol. 28:448–449. doi: 10.1111/j.1365-2265.1988.tb03678.x [DOI] [PubMed] [Google Scholar]
- Chang, A. C., Dunham M. A., Jeffrey K. J., and Reddel R. R... 1996. Molecular cloning and characterization of mouse stanniocalcin cDNA. Mol. Cell. Endocrinol. 124:185–187. doi: 10.1016/s0303-7207(96)03929-9 [DOI] [PubMed] [Google Scholar]
- Dai, D., Wang Q., Li X., Liu J., Ma X., and Xu W... 2016. Klotho inhibits human follicular thyroid cancer cell growth and promotes apoptosis through regulation of the expression of stanniocalcin-1. Oncol. Rep. 35:552–558. doi: 10.3892/or.2015.4358 [DOI] [PubMed] [Google Scholar]
- Deol, H. K., Varghese R., Wagner G. F., and Dimattia G. E... 2000. Dynamic regulation of mouse ovarian stanniocalcin expression during gestation and lactation. Endocrinology. 141:3412–3421. doi: 10.1210/endo.141.9.7658 [DOI] [PubMed] [Google Scholar]
- Etches, R. J., MacGregor H. E., Morris T. F., and Williams J. B... 1983. Follicular growth and maturation in the domestic hen (Gallus domesticus). J. Reprod. Fertil. 67:351–358. doi: 10.1530/jrf.0.0670351 [DOI] [PubMed] [Google Scholar]
- Ghanem, K., and Johnson A. L... 2018. Follicle dynamics and granulosa cell differentiation in the turkey hen ovary. Poult. Sci. 97:3755–3761. doi: 10.3382/ps/pey224 [DOI] [PubMed] [Google Scholar]
- Ghanem, K., and Johnson A. L... 2019. Relationship between cyclic follicle recruitment and ovulation in the hen ovary. Poult. Sci. 98:3014–3021. doi: 10.3382/ps/pez100 [DOI] [PubMed] [Google Scholar]
- Jepsen, M. R., Kløverpris S., Bøtkjær J. A., Wissing M. L., Andersen C. Y., and Oxvig C... 2016. The proteolytic activity of pregnancy-associated plasma protein-A is potentially regulated by stanniocalcin-1 and -2 during human ovarian follicle development. Hum. Reprod. 31:866–874. doi: 10.1093/humrep/dew013 [DOI] [PubMed] [Google Scholar]
- Jepsen, M. R., Kløverpris S., Mikkelsen J. H., Pedersen J. H., Füchtbauer E. M., Laursen L. S., and Oxvig C... 2015. Stanniocalcin-2 inhibits mammalian growth by proteolytic inhibition of the insulin-like growth factor axis. J. Biol. Chem. 290:3430–3439. doi: 10.1074/jbc.M114.611665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Krassel, F., Winn M. E., Burns D., Ireland J. L., and Ireland J. J... 2003. Evidence for a negative intrafollicular role for inhibin in regulation of estradiol production by granulosa cells. Endocrinology. 144:1876–1886. doi: 10.1210/en.2002-221077. [DOI] [PubMed] [Google Scholar]
- Jin, Y., Zhang C., Lin X., and Zeng W... 2006. Prostaglandin involvement in follicle-stimulating hormone-induced proliferation of granulosa cells from chicken prehierarchical follicles. Prostaglandins Other Lipid Mediat. 81:45–54. doi: 10.1016/j.prostaglandins.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Johnson, A. L., Solovieva E. V., and Bridgham J. T... 2002. Relationship between steroidogenic acute regulatory protein expression and progesterone production in hen granulosa cells during follicle development. Biol. Reprod. 67:1313–1320. doi: 10.1095/biolreprod67.4.1313 [DOI] [PubMed] [Google Scholar]
- Joshi, A. D. 2020. New insights into physiological and pathophysiological functions of stanniocalcin 2. Front. Endocrinol. 11:172. doi: 10.3389/fendo.2020.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B., Guo J. R., Yang H. M., Zhou R. J., Liu J. X., Li S. Z., and Dong C. Y... 2009. Differential expression profiling of ovarian genes in prelaying and laying geese. Poult. Sci. 88:1975–1983. doi: 10.3382/ps.2008-00519 [DOI] [PubMed] [Google Scholar]
- Kim, D., Langmead B., and Salzberg S. L... 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12:357–360. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kløverpris, S., Mikkelsen J. H., Pedersen J. H., Jepsen M. R., Laursen L. S., Petersen S. V., and Oxvig C... 2015. Stanniocalcin-1 potently inhibits the proteolytic activity of the metalloproteinase pregnancy-associated plasma protein-A. J. Biol. Chem. 290:21915–21924. doi: 10.1074/jbc.M115.650143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnierowski, G., and Stangierski J... 2018. What’s new in chicken egg research and technology for human health promotion? - a review. Trends Food Sci. Technol. 71:46–51.doi:10.1016/j.tifs.2017.10.022. [Google Scholar]
- Li, W., Li Q., Xu X., Wang C., Hu K., and Xu J... 2022. Novel mutations in TUBB8 and ZP3 cause human oocyte maturation arrest and female infertility. Eur. J. Obstet. Gynecol. Reprod. Biol. 279:132–139. doi: 10.1016/j.ejogrb.2022.10.017 [DOI] [PubMed] [Google Scholar]
- Liu, C., Li S., Noer P. R.,Kjaer-Sorensen K., Ke C., Oxvig C., and Duan C... 2019a. The metalloproteinase Papp-aa functions as a molecular switch linking IGF signaling to adaptive epithelial growth. bioRxiv 792978. doi: 10.1101/792978. [DOI] [Google Scholar]
- Liu, Y. X., Zhang Y., Li Y. Y., Liu X. M., Wang X. X., Zhang C. L., Hao C. F., and Deng S. L... 2019b. Regulation of follicular development and differentiation by intra-ovarian factors and endocrine hormones. Front. Biosci. 24:983–993. doi: 10.2741/4763 [DOI] [PubMed] [Google Scholar]
- Livak, K. J., and Schmittgen T. D... 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu, M., Wagner G. F., and Renfro J. L... 1994. Stanniocalcin stimulates phosphate reabsorption by flounder renal proximal tubule in primary culture. Am. J. Physiol. 267:R1356–R1362. doi: 10.1152/ajpregu.1994.267.5.R1356 [DOI] [PubMed] [Google Scholar]
- Luo, C. W., Kawamura K., Klein C., and Hsueh A. J... 2004. Paracrine regulation of ovarian granulosa cell differentiation by stanniocalcin (STC) 1: mediation through specific STC1 receptors. Mol. Endocrinol. 18:2085–2096. doi: 10.1210/me.2004-0066 [DOI] [PubMed] [Google Scholar]
- Mortazavi, A., Williams B. A., McCue K., Schaeffer L., and Wold B... 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 5:621–628. doi: 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Nishio, S., Okumura H., and Matsuda T... 2018. Egg-coat and zona pellucida proteins of chicken as a typical species of Aves. Curr. Top. Dev. Biol. 130:307–329. doi: 10.1016/bs.ctdb.2018.02.008 [DOI] [PubMed] [Google Scholar]
- Onagbesan, O., Bruggeman V., and Decuypere E... 2009. Intra-ovarian growth factors regulating ovarian function in avian species: a review. Anim. Reprod. Sci. 111:121–140. doi: 10.1016/j.anireprosci.2008.09.017 [DOI] [PubMed] [Google Scholar]
- Ozsolak, F., and Milos P. M... 2011. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12:87–98. doi: 10.1038/nrg2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., Jiang B., Liu J., Ding J., Li Y., Sun R., Peng L., Qin C., Fang S., and Li G... 2017. STC1 promotes cell apoptosis via NF-κB phospho-P65 Ser536 in cervical cancer cells. Oncotarget. 8:46249–46261. doi: 10.18632/oncotarget.17641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M., Kim D., Pertea G. M., Leek J. T., and Salzberg S. L... 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11:1650–1667. doi: 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., and Salzberg S. L... 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33:290–295. doi: 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce, J. G., and Parsons T. F... 1981. Glycoprotein hormones: structure and function. Annu. Rev. Biochem. 50:465–495. doi: 10.1146/annurev.bi.50.070181.002341 [DOI] [PubMed] [Google Scholar]
- Robinson, M. D., McCarthy D. J., and Smyth G. K... 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein, V., Cardoso J. C., Pinto P. I., Anjos L., Silva N., Power D. M., and Canário A. V... 2012. Four stanniocalcin genes in teleost fish: structure, phylogenetic analysis, tissue distribution and expression during hypercalcemic challenge. Gen. Comp. Endocrinol. 175:344–356. doi: 10.1016/j.ygcen.2011.11.033 [DOI] [PubMed] [Google Scholar]
- Spitschak, M., and Hoeflich A... 2018. Potential functions of IGFBP-2 for ovarian folliculogenesis and steroidogenesis. Front. Endocrinol. 9:119. doi: 10.3389/fendo.2018.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenina, E., Fabre S., Bonnet A., Monniaux D., Robert-Granié C., SanCristobal M., Sarry J., Vignoles F., Gondret F., Monget P.,. et al. 2017. Differentially expressed genes and gene networks involved in pig ovarian follicular atresia. Physiol. Genomics. 49:67–80. doi: 10.1152/physiolgenomics.00069.2016 [DOI] [PubMed] [Google Scholar]
- Varghese, R., Gagliardi A. D., Bialek P. E., Yee S. P., Wagner G. F., and Dimattia G. E... 2002. Overexpression of human stanniocalcin affects growth and reproduction in transgenic mice. Endocrinology. 143:868–876. doi: 10.1210/endo.143.3.8671 [DOI] [PubMed] [Google Scholar]
- Varghese, R., Wong C. K., Deol H., Wagner G. F., and DiMattia G. E... 1998. Comparative analysis of mammalian stanniocalcin genes. Endocrinology. 139:4714–4725. doi: 10.1210/endo.139.11.6313 [DOI] [PubMed] [Google Scholar]
- Waclawek, M., Foisner R., Nimpf J., and Schneider W. J... 1998. The chicken homologue of zona pellucida protein-3 is synthesized by granulosa cells. Biol. Reprod. 59:1230–1239. doi: 10.1095/biolreprod59.5.1230 [DOI] [PubMed] [Google Scholar]
- Wagner, G. F., and Dimattia G. E... 2006. The stanniocalcin family of proteins. J. Exp. Zool. A: Comp. Exp. Biol. 305:769–780. doi: 10.1002/jez.a.313 [DOI] [PubMed] [Google Scholar]
- Yeung, B. H., Law A. Y., and Wong C. K... 2012. Evolution and roles of stanniocalcin. Mol. Cell. Endocrinol. 349:272–280. doi: 10.1016/j.mce.2011.11.007 [DOI] [PubMed] [Google Scholar]
- Yoshiko, Y., and Aubin J. E... 2004. Stanniocalcin 1 as a pleiotropic factor in mammals. Peptides. 25:1663–1669. doi: 10.1016/j.peptides.2004.04.015 [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Guo Q., Jia L., Zhou C., He S., Fang C., Zhang M., Sun P., Zeng Z., Wang M.,. et al. 2022. A novel gene mutation in ZP3 loop region identified in patients with empty follicle syndrome. Hum. Mutat. 43:180–188. doi: 10.1002/humu.24297 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.