Abstract
Congenital transmission of Trypanosoma cruzi is an important source of new Chagas infections worldwide. The mechanisms of congenital transmission remain poorly understood, but there is evidence that parasite factors are involved. Investigating changes in parasite strain diversity during transmission could provide insight into the parasite factors that influence the process. Here we use amplicon sequencing of a single copy T. cruzi gene to evaluate the diversity of infection in clinical samples from Chagas positive mothers and their infected infants. Several infants and mothers were infected with multiple parasite strains, mostly of the same TcV lineage, and parasite strain diversity was higher in infants than mothers. Two parasite haplotypes were detected exclusively in infant samples, while one haplotype was never found in infants. Together, these data suggest multiple parasites initiate a congenital infection and that parasite factors influence the probability of vertical transmission.
Keywords: amplicon sequencing, congenital chagas, Trypanosoma cruzi
Amplicon sequencing of congenital chagas infection reveals complex infection in infants, suggesting that congenital infection is the result of multiple inoculating parasite clones.
Trypanosoma cruzi is the causative agent of Chagas disease and is estimated to infect almost 6 million people worldwide [1]. Effective vector control has decreased the number of new infections, but congenital transmission has become an increasing concern, particularly in nonendemic areas. An estimated 22% of new Chagas cases occur via congenital transmission, and approximately 5% of mothers infected with T cruzi will transmit the parasite to their newborn [2–4]. The role that parasite genetics plays in transmission and congenital infection is poorly understood. By identifying parasite strains that are more likely to be vertically transmitted, researchers can uncover mechanisms underlying this growing source of new cases, which may lead to improved detection and treatment of congenital T cruzi infection.
Studies examining parasite diversity within T cruzi infections are often performed with a focus on T cruzi's 6 Discrete Typing Units (DTUs), TcI through TcVI, which are distinct genetic groups that segregate by genotyped markers. However, diversity has been observed within individual DTUs, both across isolates and within single infections [5–7]. Therefore, using DTUs alone likely underestimates parasite diversity, because several clones of the same DTU may possibly coinfect a patient [8–10]. Moreover, T cruzi has 2 hybrid DTUs, TcV and TcVI. Each of these are ancient hybrids of TcII and TcIII, with each homologous chromosome of the parasite thought to approximately match one of these ancestral parental haplotypes. In some parasite clones, there may be complex recombination events present between these parental haplotypes resulting in mosaic alleles [11]. Diversity arising from these events can only be identified by characterization of each individual haplotype. Whole-genome sequencing of clinical isolates can circumvent this problem and identify complex hybrids, but this is typically not feasible due to the low parasitemia during chronic infection. To ameliorate the problem of low parasitemia, some studies have targeted high copy number genes, such as the miniexon locus, to evaluate the complexity of T cruzi infection [12–14]. These genes frequently display variability, even within a single parasite strain, however, artificially raising the apparent number of parasite clones [5, 15].
In this study, we use amplicon sequencing of a single-copy gene, TcSC5D, to characterize the clonal diversity in clinical samples of Chagas-positive infants and mothers, including several twins. More importantly, this gene contains nucleotide polymorphisms that are distinct across several DTUs, allowing an additional rough DTU determination [16]. Our results reveal haplotypes that are present exclusively in infant or mother samples, indicating a possible relationship between parasite genotype and transmission. We also observed an increase in parasite diversity in infant samples relative to maternal samples, which suggests that congenital transmission may be the result of multiple colonizing parasites that infect the infant during pregnancy.
METHODS
Study Information
Mothers who tested positive for Chagas disease were recruited from Percy Boland Women's Hospital in Santa Cruz, Bolivia between the 2016 and 2018. Three hundred microliters of maternal venous blood and 300 µL of infant blood from heel puncture was taken at birth. For longitudinal timepoints at 1, 3, and 9 months, 300 µL of infant venous blood was taken. Mothers recruited to the study were given a survey regarding obstetrics and demographic characteristics. Analysis of these epidemiological characteristics are described elsewhere [17]. This collection protocol was approved by the ethics committee of the Bolivian Catholic University, international registration Federal Wide Assurance (FWA) 0017928 and PRISMA 00001219. Study analysis was given exempt status by the Institutional Review Board (IRB) at the University of North Carolina at Chapel Hill (IRB 19-3014).
Sample Processing and Amplicon Sequencing
Deoxyribonucleic acid (DNA) was extracted at the Infectious Diseases Research Laboratory of the Universidad Peruana Cayetano Heredia in Lima, Peru. Quantitative polymerase chain reaction (qPCR) was performed as previously described [18]. In this study, we do not estimate parasitemia and instead use the cycle threshold (CT) value as a proxy for number of parasites in the blood. Amplicon PCR, library preparation, and sequencing were performed at the University of North Carolina at Chapel Hill. Details of nested PCR and library preparation are described in Supplementary Methods. Raw sequencing data are available in National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number PRJNA891347.
Haplotype Calling
Demultiplexed reads were adapter-trimmed using CutAdapt [19]. After trimming, any read pairs less than 100 base pairs long were removed to eliminate adapter dimers and off-target amplicons from the pipeline. After adapter trimming, reads were aligned to a custom BLAST search against the target amplicon to remove nonspecific reads and orient amplicons in the same direction before calling haplotypes. Filtered reads were then run through DADA2 using a maximum expected error of 2 for both the R1 and R2 reads [20]. Any amplicon sequence variants that made up less than 0.01% of the total population of reads were removed from the analysis, and any sample that had fewer than 200 total reads after all filtering steps was removed from the analysis.
Two of the recovered haplotypes were identical, except for a small insertion at the end of the amplicon. Because it is possible that the shorter amplicon could be amplified in samples containing the longer amplicon, they were merged into 1 amplicon for the analysis. This amplicon sequence is available in the Supplemental Data as haplotype number 12, and it was merged with haplotype 0 for subsequent analysis.
The DTU typing was done by comparing called amplicons to previously published sequencing data targeting the same locus on reference DTUs. Sanger sequencing data were obtained from data previously published by Cosentino and Agüero [16], and whole-genome data were from TriTypDB. The SRA numbers for these sequences are provided in the Supplemental Methods.
Haplotype Diversity Analysis
Parasite diversity was measured using Shannon's diversity index using the Vegan R package [21]. Only haplotypes detected in both sample replicates were counted when analyzing the frequency of appearance of each haplotype (in Figure 1C), except in cases in which only 1 sample replicate passed filtering; in these cases, every detected haplotype was counted. All scripts used to process raw data, call haplotypes, and generate figures for analysis are available at https://github.com/MugnierLab/Hakim2022.
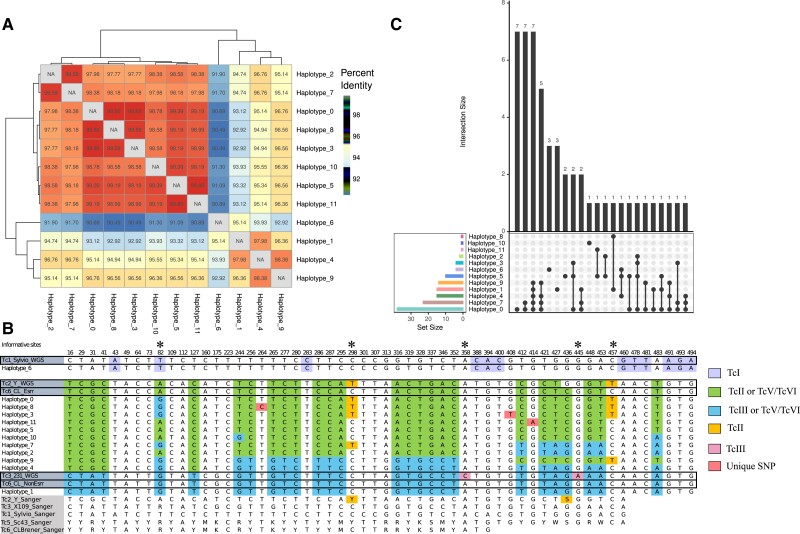
Figure 1.
Many haplotypes belonged to the same Discrete Typing Unit (DTU). (A) Heatmap showing percentage identity between haplotypes, clustered by similarity. (B) Table of all single-nucleotide polymorphisms (SNPs) across haplotypes found. The position of each polymorphism within the amplicon is noted at the top of each column. Whole-genome data (white background) of TcI, TcII, TcIII, and TcVI as well as Sanger sequencing data (gray background) for a portion of the amplicon from TcI, TcII, TcIII, TcV, and TcVI are included for reference. Bases are colored based on similarity to reference DTU data; only positions that are unique to a reference haplotype are colored. Asterixis denote informative SNP sites for DTU calls. (C) UpSet plot describing the frequency and co-occurrence of each haplotype and haplotype combination. Dots represent a haplotype or haplotype combination, with the frequency of that combination represented on the bar graph above. Colored bar graphs on the side represent the number of samples in which the haplotype was found.
RESULTS
Haplotype Sequences Were Largely Similar and Mostly Belonged to Hybrid Discrete Typing Unit TcV or TcVI
Amplicon sequencing targeting the TcSC5D gene was performed in duplicate on DNA extracted from peripheral blood from 75 total individuals. After eliminating samples with low-quality sequencing data, 44 participants were included in the analysis. These 44 individuals included 8 mother-infant sets (2 sets of twins, 6 singletons), 2 mothers without a matching infant sample, and 10 infants (including 1 set of twins) without a matching mother sample. All collected epidemiological data associated with these samples are available in Supplemental Table 1, alongside the total number of reads each sample replicate received. TcSC5D haplotypes were identified in each sample using DADA2 [20]. This tool, optimized for calling haplotypes from amplicons, corrects for both PCR and sequencing errors. Thus, haplotypes present in only 1 PCR replicate are mostly likely due to under sampling, rather than sequencing or PCR errors. Supporting this assumption, every haplotype except for 1 was detected in more than 1 infection. We therefore considered a haplotype detected in either PCR replicate to be present in that infection.
The 12 unique haplotype sequences identified in the study population showed between 90.9% and 99.8% sequence similarity (Figure 1A). Using reference whole-genome data as well as Sanger sequencing data at the same locus as a reference, we assigned each recovered haplotype a tentative DTU designation. The DTU TcVI is an ancestral hybrid of a TcIII and TcII strain, and both Esmeraldo (resembling the TcIII ancestor) and non-Esmeraldo (resembling the TcII ancestor) homologous chromosomes were included as reference haplotypes. Figure 1B shows a table of all single-nucleotide polymorphisms (SNPs) across the recovered haplotypes and reference sequences. Haplotype 6 shared 100% sequence identity to the TcI Sylvio strain, whereas all other haplotypes have some similarity to phased haplotypes of the hybrid CL Brener strain. Haplotypes 3, 8, and 11 had unique SNPs not found in the reference strains or in any other haplotype. Several haplotypes had stretches of similarity matching the TcII SNP pattern, before switching to match the TcIII pattern, and vice versa. Similarly, haplotypes 0, 3, and 8 have loci that suggest they may belong to TcII, but at site 82 all 3 have a nucleotide associated with TcIII. These patterns are consistent with recombination between homologous chromosomes in the hybrid strains. Although we cannot call these haplotypes’ DTU designation specifically, it is apparent that they are all hybrids, making them either TcV or TcVI.
We analyzed the combinations of haplotypes detected in each infection to determine whether certain haplotypes were more likely to co-occur or occur alone. Four haplotypes, haplotypes 0, 1, 5, and 6, were detected as the only clone within some infections (Figure 1C). This could indicate that these haplotypes represent individual parasite clones in which both homologous chromosomes contain the same sequence at the amplicon locus. It is unlikely that the same novel mutation in a conserved gene would occur in 2 independent infections. Thus, the fact that most haplotypes occur in multiple independent infections suggests that they are not novel mutations arising in infants, but instead they represent parasite clones that were transmitted to the infant from the mother. The analysis revealed 11 samples containing 3 or more haplotypes, which, because of T cruzi's diploid genome, likely indicates infection with at least 2 parasite clones. All of these samples were infant samples, with only 2 complex infections detected in the 14 maternal samples (Supplemental Figure 1).
Parasite Strain Diversity Is Higher in Infants Than in Mothers
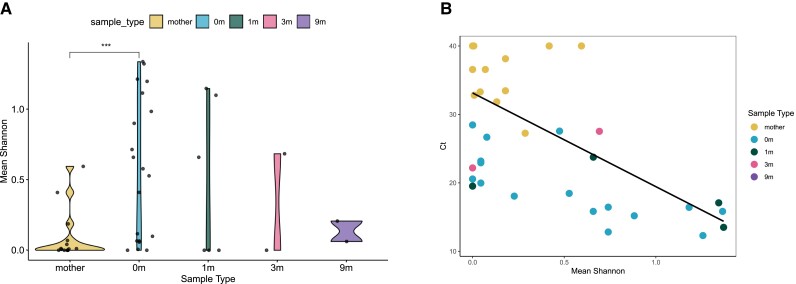
Given the presence of multiple parasite clones in many samples, we assessed the changes in parasite diversity that occur during congenital transmission. Under many modes of infectious transmission, a bottleneck occurs, and the colonized site has less strain diversity than the source. However, we find that maternal samples are less diverse, as measured by Shannon's diversity, than samples of infants at birth (Figure 2A). Shannon's diversity is more robust to sampling error than comparing the number of detected haplotypes, because it considers the proportion at which each haplotype is found [22]. Diversity appears to gradually decrease as infants get older, although this effect is not statistically significant. A potential explanation for this observation could be that the low parasitemia in the maternal blood sample causes undersampling of the true maternal diversity of circulating parasites. We found that the parasitemia of each patient as measured by qPCR was correlated to the Shannon's diversity for each sample (Spearman's rho = −0.62, P = 2.3e-4) (Figure 2B). However, because there were no chronically infected maternal samples of a high enough parasitemia to directly compare to the acutely infected infant samples, and because there may be a causative link between blood parasite load and parasite diversity, it is impossible to eliminate the role that sampling may play in the reduced parasite diversity found in mother's blood.
Figure 2.
Haplotype diversity in mothers is lower than in infants at birth. (A) Mean Shannon's diversity between sample replicates for each sample type: 0 m is infant's sample at birth, 1 m = 1 month of age, 3 m = 3 months, 9 m = 9 months; Wilcoxon's rank-sum test, P = .0024. (B) Association between parasitemia measured by cycle threshold (CT) and average Shannon's diversity for each replicate, with undetermined samples set at CT = 40. Spearman's correlation was done excluding samples with undetermined CT. Mother's samples: Spearman's rho = −0.14, P = .752. 0 m samples: Spearman's rho = −0.82, P = 1.16e-4. Overall: Spearman's rho = −0.62, P = 2.3e-4.
Several Haplotypes Were Exclusive to the Mother or the Infant
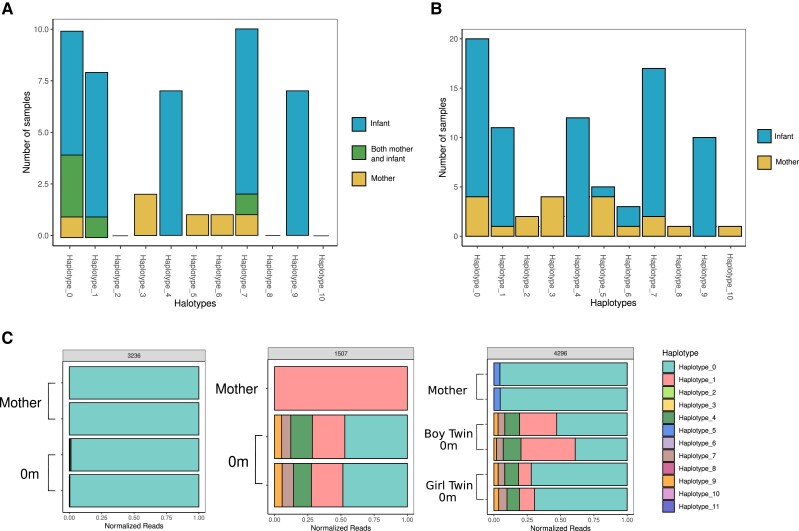
Parasite genetics may play a role in the probability of congenital transmission of T cruzi. To address this possibility, we searched for haplotypes that were more or less likely to be transmitted from the mother to her infant. Haplotypes found in a mother, but not her infant, could represent clones that are less likely to be congenitally transmitted. For this analysis, we counted the number of times a haplotype was found in a mother, her paired infant, or in both the mother and her infant (Figure 3A). Haplotypes 0, 1, and 7 appeared in both samples of some mother-infant pairs, indicating that these haplotypes were likely to be transmitted and detected in maternal blood. Haplotypes 3, 5, and 6 were only found in mothers and not paired infants. Haplotypes 4 and 9 were only ever found in the infant samples of mother-infant pairs. To further explore the effect of haplotype on transmission, we also analyzed haplotype presence in samples without considering the mother-infant pairs (Figure 3B). Among these samples, haplotypes 5 and 6 were no longer exclusive to the mother and were found in some infant samples, whereas haplotype 3 remained exclusive to mothers. It is interesting to note that haplotypes 4 and 9 remain exclusive to infant samples. This indicates that these haplotypes are highly likely to be transmitted, and that there may be additional biological mechanisms resulting in the lack of these haplotype's detection in the maternal blood at time of birth.
Figure 3.
Certain haplotypes are found only in mothers or only in infants. (A) Among families with paired infant-mother samples, the number of times a haplotype was found only in the mother sample, only in the newborn sample, or in both the mother and the newborn sample. (B) Number of times a haplotype was found in any mother or any newborn, regardless of family, including samples without mother-infant pairs. (C) Representative plots of relative haplotype abundance in each sample grouped by family. Read count was normalized to total reads in each sample.
By examining individual families, we observed diverse transmission patterns (Figure 3C, Supplemental Figures 2 and 3). Family 1507 shows the maternal detection of haplotype 1, whereas within the infant sample we detect haplotype 1 in addition to several other haplotypes, consistent with our previous observation of increased diversity in infant samples relative to mother's samples. Transmission within family 4296 is perhaps the most informative. In this case, 2 haplotypes are detected in the maternal sample, whereas a larger number of haplotypes are detected in each of the 2 fraternal twins. It is notable that the haplotype distribution in each twin is almost identical. Because these were fraternal twins of discordant sex, with 2 separate placentas, this finding supports the hypothesis that transmission is not the result of a single inoculating parasite.
DISCUSSION
These data describe the changes in parasite diversity that exist during the process of T cruzi congenital transmission and suggest a role for parasite genetics in the likelihood of transmission. By directly sequencing blood samples from patients and targeting a single-copy gene by amplicon sequencing, we were able to identify complex infections in mother and infant samples in an endemic setting and determine changes in parasite strain diversity that occur during the process of transmission. Although our findings are somewhat limited by small sample sizes, our approach allows a direct assessment of parasite diversity beyond simple DTU classification and reveals new features of congenital transmission of T cruzi that warrant further investigation.
The method used in this paper allows us to avoid overestimating complexity of infection for a sample. In this study, we assume the single locus gene occurs twice per parasite, with a unique allele on each homologous chromosome, thereby potentially underestimating the parasite diversity. This assumption of exclusive diploidy may not always hold; karyotypic instability has been found in T cruzi, and gene duplication commonly occurs [23, 24]. However, duplicated genes often encode surface genes involved in immune evasion, and the conserved metabolic gene targeted in this work is not thought to be expressed on the surface and is likely not under selective pressure. An additional strength of this work is that parasites were sequenced directly from patient samples and did not undergo expansion and potential strain selection in culture. Thus, we avoid the possibility of selecting for clones that are better adapted to culture at the expense of the true parasite diversity. This study demonstrates the feasibility of this approach for characterizing parasite diversity across congenital infection.
The majority of the haplotypes detected in our samples were hybrid DTUs, likely TcV considering other strain typing work from the same setting [2, 25, 10]. In this study, we observe potential ancestral recombination events between the parental TcII and TcIII alleles in our recovered haplotypes. This suggests additional diversity exists in the hybrid strains beyond what has been sequenced in the reference CL Brener strain. Within these hybrid DTU types, we observed several haplotypes that occurred exclusively in mother or infant samples. This suggests that there may be genetic factors within DTUs that influence parasite transmission and underscores the fact that current DTU designations are insufficient to appropriately assess the diversity of parasite strains in a single infection. Investigation into specific virulence factors, either by transcriptomic or genomic analysis, rather than DTU typing, is likely to be required to uncover the factors that influence transmission. An analysis including mothers infected with a more diverse set of parasite DTUs and mothers who did not transmit to their infant may shed light on the degree to which DTU alone can influence the probability of transmission.
Although parasite genetics may influence the probability of transmission, we found complex infections observed in both mother and infant samples. Surprisingly, we observed an increase in parasite diversity after transmission, with many haplotypes recovered in the infant that were undetectable in maternal blood. This finding is corroborated in a similar study, in which Lewellyn et al [26] found novel strain types in infants compared to their paired mothers using a diverse low copy number gene. In that study, it was unclear whether the observed “strain types” represented individual parasite clones or diversity that was generated during infection, because the locus analyzed was a variant surface protein that is likely to diversify during the course of infection. Because our approach analyzes a single-copy metabolic gene and most haplotypes were identified independently in multiple infections, we are able to more confidently assume that detected haplotypes were not the result of diversification during infection.
The increased diversity in the infant samples may be explained in several ways. Perhaps most intriguing is the possibility that some parasite clones prefer the placental environment, remaining undetectable in the maternal blood. If these T cruzi clones are sequestered in the placenta, they might not circulate at detectable levels in the mother's blood. In a previous study, researchers performed kDNA PCR on mothers who tested positive for Chagas disease and found multiple patients with negative bloodstream but positive placental PCR, and in 2 cases they found placental minicircle fragments that were not detectable in the bloodstream of the same patient, which suggests placental tropism for specific parasite clones [27]. Placental tropism of specific T cruzi strains has also been observed in mouse models of infection [28, 29].
An additional explanation is that parasite transmission occurs throughout pregnancy. If parasite clones cross the placenta during multiple waves of infection, the composition in the circulation of the infant would reflect the set of parasite clones present in the mother throughout pregnancy, even if these clones are absent from the mother's circulation at the time of delivery. It is possible that waves of parasitemia lead to the expansion of different parasite clones at different time points in the maternal blood during pregnancy, and that clones found in the maternal blood at birth may not represent the true diversity of parasite clones infecting the mother.
Although compelling biological explanations for this observed increase in diversity exist, it remains possible that this observation is simply a matter of sampling. The lower parasitemia of chronically infected mothers means that fewer parasites are sampled from mothers for sequencing, potentially biasing diversity estimates. The correlation between parasitemia and Shannon's diversity means we cannot preclude this possibility.
More importantly, however, the observation of complex infections in infants is not affected by these issues. If infants are infected with multiple clones, regardless of each clone's presence in the mother, this is most likely the result of several parasites successfully colonizing the infant. Little is known about the dynamics of congenital transmission of T cruzi, but the relatively low rate of congenital transmission (estimated at 5%) might suggest that it occurs as a result of a single parasite occasionally breaching the placental barrier. Contrary to this model, our data suggest that congenital infection is the result of several parasites infecting the infant. The parasite profile from fraternal twin samples, distinct from the mother but identical between siblings, supports this model of transmission. Each of the 2 placentas were infected with the same set of clones, probably during multiple independent infection events during their gestation. The temporal dynamics of this process remain unclear. Parasites may cross the placenta at certain times during infection, which is in line with previous reports showing that parasitemia during the third trimester is most predictive of congenital infection [27]. Alternatively, parasite clones might cross the placenta during multiple waves of infection. Why transmission of multiple parasites occurs in some pregnancies, whereas most others result in no infection, will be a central question going forward. The fact that multiple parasites cross the placenta suggests that host factors, including the integrity of the placental barrier and the potency of the maternal immune response, likely influence the probability of transmission.
CONCLUSIONS
In this study, we define the changes in parasite diversity that occur during congenital transmission and raise interesting questions about the mechanism of the process. We find infant samples with increased parasite diversity compared with maternal samples, which, together with haplotype data from the fraternal twins in this study, support a model in which multiple parasites colonize the infant during pregnancy. Moreover, we detected 2 haplotypes unique to newborn samples that were not detected in maternal peripheral blood, which together with previously published data suggest a link between parasite genetics and transmission probability [9]. Understanding the mechanisms influencing transmission of T cruzi may help inform better diagnostics and lead to more effective treatment, limiting the global burden of Chagas disease.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Jill M C Hakim, Department of Molecular Microbiology and Immunology, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA.
Andreea Waltmann, Institute for Global Health and Infectious Disease, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Freddy Tinajeros, Asociación Benéfica PRISMA, Lima, Peru.
Oksana Kharabora, Institute for Global Health and Infectious Disease, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Edith Málaga Machaca, Asociación Benéfica PRISMA, Lima, Peru; Infectious Diseases Research Laboratory, Department of Cellular and Molecular Sciences, Universidad Peruana Cayetano Heredia, Lima, Perú.
Maritza Calderon, Infectious Diseases Research Laboratory, Department of Cellular and Molecular Sciences, Universidad Peruana Cayetano Heredia, Lima, Perú.
María del Carmen Menduiña, Hospital Percy Boland Rodríguez, Ministerio de Salud Bolivia, Santa Cruz, Bolivia.
Jeremy Wang, University of North Carolina, Chapel Hill School of Medicine, Chapel Hill, North Carolina, USA.
Daniel Rueda, Facultad de Ciencias, Universidad Nacional de Ingeniería, Lima, Perú.
Mirko Zimic, Infectious Diseases Research Laboratory, Department of Cellular and Molecular Sciences, Universidad Peruana Cayetano Heredia, Lima, Perú.
Manuela Verástegui, Infectious Diseases Research Laboratory, Department of Cellular and Molecular Sciences, Universidad Peruana Cayetano Heredia, Lima, Perú.
Jonathan J Juliano, Institute for Global Health and Infectious Disease, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA; Division of Infectious Diseases, Department of Medicine, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, North Carolina, USA.
Robert H Gilman, Department of International Health, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA.
Monica R Mugnier, Department of Molecular Microbiology and Immunology, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland, USA.
Natalie M Bowman, Division of Infectious Diseases, Department of Medicine, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, North Carolina, USA.
Notes
Financial support. This work was supported by the National Institutes of Health (Grant Numbers R01AI151295, K23AI113197, R01AI136722 [to NMB, RHG, MRM, and JMCH], K24AI134990 [to JJJ], and TL1TR002491 [to AW]), the Doris Duke Charitable Foundation (Grant Number 2015213 [to NMB]), and the Sherrilyn and Ken Fisher Center for Environmental and Infectious Disease (Award Number 001MUG2022 [to MRM]).
References
- 1. Chagas disease in Latin America: an epidemiological update based on 2010 estimates. Wkly Epidemiol Rec 2015; 90:33–43. [PubMed] [Google Scholar]
- 2. Bern C, Verastegui M, Gilman RH, et al. Congenital Trypanosoma cruzi transmission in Santa Cruz, Bolivia. Clin Infect Dis 2009; 49:1667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Howard EJ, Xiong X, Carlier Y, Sosa-Estani S, Buekens P. Frequency of the congenital transmission of Trypanosoma cruzi: a systematic review and meta-analysis. BJOG 2014; 121:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Messenger LA, Gilman RH, Verastegui M, et al. Toward improving early diagnosis of congenital Chagas disease in an endemic setting. Clin Infect Dis 2017; 65:268–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cura CI, Mejía-Jaramillo AM, Duffy T, et al. Trypanosoma cruzi I genotypes in different geographical regions and transmission cycles based on a microsatellite motif of the intergenic spacer of spliced-leader genes. Int J Parasitol 2010; 40:1599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Llewellyn MS, Rivett-Carnac JB, Fitzpatrick S, et al. Extraordinary Trypanosoma cruzi diversity within single mammalian reservoir hosts implies a mechanism of diversifying selection. Int J Parasitol 2011; 41:609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reis-Cunha JL, Baptista RP, Rodrigues-Luiz GF, et al. Whole genome sequencing of Trypanosoma cruzi field isolates reveals extensive genomic variability and complex aneuploidy patterns within TcII DTU. BMC Genomics 2018; 19:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rendell VR, Gilman RH, Valencia E, et al. Trypanosoma cruzi-infected pregnant women without vector exposure have higher parasitemia levels: implications for congenital transmission risk. PLoS One 2015; 10:e0119527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herrera C, Truyens C, Dumonteil E, et al. Phylogenetic analysis of Trypanosoma cruzi from pregnant women and newborns from Argentina, Honduras, and Mexico suggests an association of parasite haplotypes with congenital transmission of the parasite. J Mol Diagn 2019; 21:1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bowman NM, Balasubramanian S, Gilman RH, et al. Deep sequencing to detect diversity of Trypanosoma cruzi infection in patients coinfected with human immunodeficiency virus and Chagas disease. J Infect Dis 2022; 225:243–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Westenberger SJ, Barnabé C, Campbell DA, Sturm NR. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 2005; 171:527–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burgos JM, Altcheh J, Bisio M, et al. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int J Parasitol 2007; 37:1319–27. [DOI] [PubMed] [Google Scholar]
- 13. Villanueva-Lizama L, Teh-Poot C, Majeau A, Herrera C, Dumonteil E. Molecular genotyping of Trypanosoma cruzi by next-generation sequencing of the mini-exon gene reveals infections with multiple parasite discrete typing units in Chagasic patients from Yucatan, Mexico. J Infect Dis 2019; 219:1980–8. [DOI] [PubMed] [Google Scholar]
- 14. Murillo-Solano C, Ramos-Ligonio A, López-Monteon A, et al. Diversity of Trypanosoma cruzi parasites infecting Triatoma dimidiata in Central Veracruz, Mexico, and their one health ecological interactions. Infect Genet Evol 2021; 95:105050. [DOI] [PubMed] [Google Scholar]
- 15. Maiguashca Sánchez J, Sueto SOB, Schwabl P, Grijalva MJ, Llewellyn MS, Costales JA. Remarkable genetic diversity of Trypanosoma cruzi and Trypanosoma rangeli in two localities of southern Ecuador identified via deep sequencing of mini-exon gene amplicons. Parasit Vectors 2020; 13:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cosentino RO, Agüero F. A simple strain typing assay for Trypanosoma cruzi: discrimination of Major evolutionary lineages from a single amplification product. PLoS Negl Trop Dis 2012; 6:e1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klein MD, Tinajeros F, del Carmen Menduiña M, et al. Risk factors for maternal Chagas disease and vertical transmission in a Bolivian Hospital. Clin Infect Dis 2021; 73:e2450–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Piron M, Fisa R, Casamitjana N, et al. Development of a real-time PCR assay for Trypanosoma cruzi detection in blood samples. Acta Trop 2007; 103:195–200. [DOI] [PubMed] [Google Scholar]
- 19. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011; 17:10–2. [Google Scholar]
- 20. Callahan B, McMurdie P, Rosen M, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13:581–83. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Helene Wagner JO. Vegan: community ecology package. Available at: http://CRAN.R-project.org/package=vegan. Accessed 10 March 2022.
- 22. He Y, Zhou BJ, Deng GH, Jiang XT, Zhang H, Zhou HW. Comparison of microbial diversity determined with the same variable tag sequence extracted from two different PCR amplicons. BMC Microbiol 2013; 13:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Talavera-López C, Messenger LA, Lewis MD, et al. Repeat-driven generation of antigenic diversity in a major human pathogen, Trypanosoma cruzi. Front Cell Infect Microbiol 2021; 11:614665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matos GM, Lewis MD, Talavera-López C, et al. Microevolution of Trypanosoma cruzi reveals hybridization and clonal mechanisms driving rapid genome diversification. eLife 2022; 11:e75237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanchez L, Messenger LA, Bhattacharyya T, et al. Congenital Chagas disease in Santa Cruz department, Bolivia, is dominated by Trypanosoma cruzi lineage V. Trans R Soc Trop Med Hyg 2022; 116:80–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Llewellyn MS, Messenger LA, Luquetti AO, et al. Deep sequencing of the Trypanosoma cruzi GP63 surface proteases reveals diversity and diversifying selection among chronic and congenital Chagas disease patients. PLoS Negl Trop Dis 2015; 9:e0003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bisio M, Seidenstein ME, Burgos JM, et al. Urbanization of congenital transmission of Trypanosoma cruzi: prospective polymerase chain reaction study in pregnancy. Trans R Soc Trop Med Hyg 2011; 105:543–9. [DOI] [PubMed] [Google Scholar]
- 28. Andrade SG. The influence of the strain of Trypanosoma cruzi in placental infections in mice. Trans R Soc Trop Med Hyg 1982; 76:123–8. [DOI] [PubMed] [Google Scholar]
- 29. Juiz NA, Solana ME, Acevedo GR, et al. Different genotypes of Trypanosoma cruzi produce distinctive placental environment genetic response in chronic experimental infection. PLoS Negl Trop Dis 2017; 11:e0005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.