Abstract
Background
NVX-CoV2373 is an efficacious coronavirus disease 2019 (COVID-19) vaccine comprising full-length recombinant severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (rS) glycoprotein and Matrix-M adjuvant. Phase 2 of a randomized, placebo-controlled, phase 1/2 trial in healthy adults (18–84 years of age) previously reported good safety/tolerability and robust humoral immunogenicity.
Methods
Participants were randomized to placebo or 1 or 2 doses of 5-µg or 25-µg rS with 50 µg Matrix-M adjuvant 21 days apart. CD4+ T-cell responses to SARS-CoV-2 intact S or pooled peptide stimulation (with ancestral or variant S sequences) were measured via enzyme-linked immunosorbent spot assay and intracellular cytokine staining.
Results
A clearly discernable spike antigen-specific CD4+ T-cell response was induced after 1 dose, but markedly enhanced after 2 doses. Counts and fold increases in cells producing Th1 cytokines exceeded those secreting Th2 cytokines, although both phenotypes were clearly present. Interferon-γ responses to rS were detected in 93.5% of 2-dose 5-µg recipients. A polyfunctional CD4+ T-cell response was cross-reactive and of equivalent magnitude to all tested variants, including Omicron BA.1/BA.5.
Conclusions
NVX-CoV2373 elicits a moderately Th1-biased CD4+ T-cell response that is cross-reactive with ancestral and variant S proteins after 2 doses.
Clinical Trials Registration
Keywords: CD4+ T-cell response, COVID-19 vaccine, Matrix-M adjuvant, polyfunctional, variant cross-reactivity
Primary series of NVX-CoV2373 vaccine with Matrix-M adjuvant induces polyfunctional CD4+ T-cell responses of balanced to moderately Th1-biased phenotype. Essentially equivalent cytokine production by vaccinee cells can be stimulated by spike proteins of ancestral or multiple variant sequences.
The coronavirus disease 2019 (COVID-19) pandemic due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to produce disease waves driven by evolving variants. Vaccines based on multiple technologies, all targeting the SARS-CoV-2 spike (S) protein that mediates virus attachment to host cells, have shown moderate to high levels of efficacy [1–3] and good effectiveness against severe SARS-CoV-2 hospitalization and death in real-world data [4, 5]. Notably, booster doses with vaccines based on the ancestral S sequence continue to provide protection against severe disease [6], and this continues to be associated with improved variant-specific neutralization titers after these doses [7, 8].
The NVX-CoV2373 vaccine comprises 5 µg of full-length recombinant SARS-CoV-2 spike glycoprotein (rS) stabilized in prefusion conformation, with 50 µg of Matrix-M adjuvant [9, 10]. Matrix-M or similar saponin adjuvants provide antigen sparing in induction of antibody responses [11, 12], increase antibody affinity, expand the range of epitopes recognized [13], and broaden strain specificity [14, 15]. In addition, saponin adjuvants enhance polyfunctional CD4+ T-cell responses and may allow cross-presentation to induce CD8+ T-cell responses to exogenous protein antigens [9, 14–17].
In phase 1 of this phase 1/2 trial, 5 µg and 25 µg SARS-CoV-2 rS doses with 50 µg Matrix-M adjuvant were well tolerated in healthy adults 18–59 years old and induced immunoglobulin G (IgG) anti-S protein-binding antibodies and neutralizing antibodies [10], with CD4+ T-cell responses biased toward a type 1 T-helper (Th1) phenotype detected by the intracellular cytokine staining (ICCS) assay [10]. CD8+ T-cell responses in the phase 1 participants were not investigated, but other investigators detected antigen-specific CD8+ T cells using the activation-induced marker and ICCS assays [16, 18]. Other studies have found similar patterns of T-cell responses to SARS-CoV-2 infection and other vaccines [8].
Phase 2 of this trial investigated SARS-CoV-2 rS with Matrix-M adjuvant in healthy adults 18–84 years old. Both 5 µg and 25 µg SARS-CoV-2 rS doses with 50 µg Matrix-M adjuvant were again well tolerated and induced strong anti-S protein IgG and neutralizing antibody responses 2 weeks after the second dose [19]. The 5 µg SARS-CoV-2 rS with 50 µg Matrix-M formulation was subsequently studied in 2 large phase 3 clinical trials. In 15 187 adult participants (18–84 years of age) in the United Kingdom, 89.7% efficacy (95% confidence interval [CI], 80.2%–94.6%) against mild, moderate, or severe virologically confirmed COVID-19 was shown from 7 days after the second injection [20]. Extended follow-up (median of 4.5 months postvaccination) showed 82.7% efficacy (95% CI, 73.3%–88.8%) against all SARS-CoV-2 disease and 100% efficacy against severe disease [21]. In the second efficacy trial of 29 949 participants ≥18 years old in the United States and Mexico, NVX-CoV2373 demonstrated 90.4% efficacy (95% CI, 82.9%–94.6%) against the mild, moderate, or severe confirmed COVID-19 endpoint, including 92.6% against then-circulating variants of concern or interest [22].
Here we report on the CD4+ T-cell responses of participants in phase 2 of the phase 1/2 trial as well as cytokine responses to variant spike proteins after 2 doses of NVX-CoV2373.
MATERIALS AND METHODS
Trial Design and Oversight
The phase 2 component of the trial was conducted at 9 sites in Australia and 8 in the United States (Supplementary Table 1) in men and nonpregnant women 18–84 years old. The protocol was approved by the Alfred Hospital Human Research Ethics Committee (Melbourne, Australia) and Advarra Central Institutional Review Board (Columbia, Maryland), performed in accordance with the International Conference on Harmonization Good Clinical Practice guidelines and overseen by an independent safety monitoring committee. All participants provided written informed consent prior to enrollment. Details of the design, conduct, and analyses (including a diagram of participant enrollment and disposition through the principal period of sampling reflected here), safety, and antibody responses to ancestral antigens are provided in prior publications [19, 23] and in the Supplementary Methods and Supplementary Table 2. The trial is registered at ClinicalTrials.gov (NCT04368988).
Participants were randomized (1:1:1:1:1) to placebo or 1 of 4 vaccine groups, with stratification by study site and age (18–59 and 60–84 years). Each participant received, in a blinded manner, 2 intramuscular injections comprising placebo (group A) or NVX-CoV2373 in a 1-dose regimen (5 µg [group C] or 25 µg [group E] rS with Matrix-M adjuvant on day 0 followed by placebo on day 21 [−1 to +3 days]) or 2-dose regimen (5 µg [group B] or 25 µg [group D] rS with Matrix-M adjuvant on day 0 and day 21 [−1 to +3 days]). Blood samples for T-cell responses were drawn before treatment (day 0) and at 7 days after each injection of the primary 2-dose series.
The trial was designed by Novavax, Inc (Gaithersburg, Maryland), with funding support from the Coalition for Epidemic Preparedness Innovations. The authors assume responsibility for the accuracy and completeness of the data and analyses, as well as for the fidelity of the trial.
Immunogenicity Assessments
Heparinized blood was collected from participants at a subset of sites preselected for capacity to collect and ship samples under specified conditions. The samples were couriered overnight at room temperature to central laboratories in Australia (360Biolabs, Melbourne) or the United States (Cellular Technology Ltd [CTL], Shaker Heights, Ohio) for separation of peripheral blood mononuclear cells (PBMCs) by Ficoll density gradient centrifugation. PBMC fractions were frozen at −80°C and stored under vapor nitrogen until assay.
Enzyme-linked immunosorbent spot (ELISpot) assays were performed by CTL. ELISpot plates were coated with antibodies to human interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α), or interleukin (IL)–5. Stimulating antigens and PBMCs were added to wells in serum-free CTL-Test Medium and incubated for 48 hours at 37°C in a 7% carbon dioxide atmosphere. After incubation, cells were discarded, and the plates were washed. Spots representing cytokine-secreting cells were detected by incubation with enzyme-conjugated antibodies to IFN-γ, TNF-α, or IL-5 followed by chromogenic substrate, and spots were counted using an automated ImmunoSpot reader (from CTL).
ICCS assays were performed at Novavax on a subset of samples that were randomly selected from the overall ELISpot sample pool. Thawed PBMCs were cultured overnight, and viable counts were determined. PBMCs were then cultured for 6 hours at 37°C with stimulating antigens and positive and negative controls, with addition of BD GolgiPlug and BD GolgiStop (BD Biosciences, San Jose, California) for the last 4 hours. Cells were labeled with surface markers (CD3, CD4, and CD8; BD Biosciences) and LIVE/DEAD indicator dye (Invitrogen, Eugene, OR). Cells were fixed and permeabilized, and cytokines were detected with antibodies specific for IFN-γ, TNF-α, IL-2, IL-5, and IL-13 (BD Biosciences). Samples were processed using flow cytometers (Fortessa and Symphony A3) and analyzed using FlowJo software version 10 (Tree Star, Ashland, Oregon). Data shown were gated on the CD3+CD4+CD8− live CD4+ T-cell population. The gate of cytokine-positive cells was set up based on negative control cells that were treated with medium only.
More detailed descriptions of the immunogenicity methods are provided in the Supplementary Methods and Supplementary Figures 1 and 2.
Statistical Analysis
Analyses were conducted on the per-protocol immunogenicity PBMC analysis subset, which included participants in the per-protocol immunogenicity analysis population restricted to consenting participants at the subset of preselected sites where PBMCs were harvested. Results for both the ELISpot and ICCS assays were summarized for cells stimulated with N-terminal and C-terminal S peptide pools, for the combined counts for both peptide pools, and separately, for cells stimulated with intact rS protein. Counts below the lower limit of quantitation (LLOQ) were assigned values of LLOQ / 2. Responses were described by the geometric mean of counts per 1 million cells (GMCs) and geometric mean fold rise (GMFR), that is, the geometric mean of the within-participant fold rises from baseline to post-treatment assessments. For ELISpot data, where participant numbers were greater, the proportion of participants “responding” to a given stimulus was based on those attaining cytokine-producing cell counts ≥95th percentile of all baseline preimmunization values, which were collected early in the pandemic in a population 98.1% SARS-CoV-2 seronegative (Supplementary Table 3) at first sampling.
RESULTS
Cellular Immunogenicity Population
Between 24 August 2020 and 25 September 2020, 1288 participants were randomized at 17 sites in Australia and the United States in the phase 2 component of the trial. A total of 1283 participants received the initial injection. A subset comprising consenting enrollees fulfilling the per-protocol criteria for immunogenicity at preselected sites whose PBMC specimens were received in good condition and without contamination (see Supplementary Material) totaled 155 participants and constituted the cellular immunogenicity subset. Treatment group sizes were 31, 32, 28, 33, and 31 for groups A, B, C, D, and E, respectively (Supplementary Table 3).
The median age was 56.0 years (range, 49.0–58.0 years), with 44.5% in the older (60–84 years) stratum (range, 39.4%–48.4%). Women comprised 47.7% (range, 36.4%–53.6%) of participants, and 83.2% of participants were White.
Immunogenicity Analysis
ELISpot
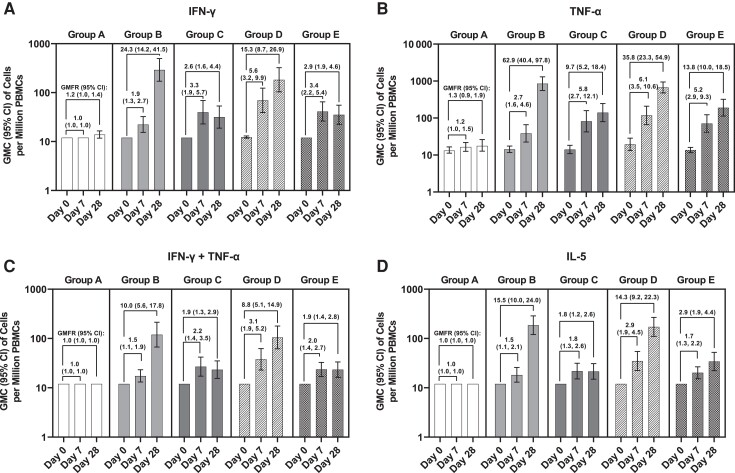
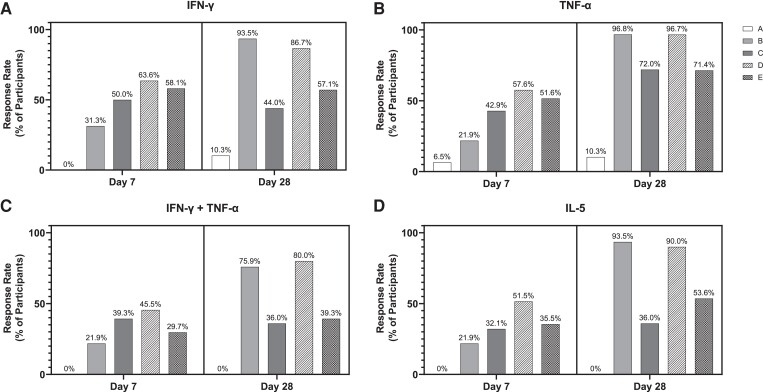
Figure 1 depicts counts of cytokine-secreting cells per 1 million PBMCs when stimulated with intact ancestral SARS-CoV-2 rS. Baseline (day 0) counts were low and near LLOQ, with minimal variation, for all cytokines assessed. In vaccine groups, counts of IFN-γ– or TNF-α–secreting cells (both indicative of a Th1 response) rose 1.9- to 5.6-fold (IFN-γ) or 2.7- to 6.1-fold (TNF-α) from baseline to day 7, whereas placebo recipients showed little change. Responses to 25 µg rS (group E) were slightly higher than those to 5 µg rS (group C) after 1 dose, albeit with overlapping 95% CIs (Figure 1). One week after the second dose, marked increases in IFN-γ–secreting cells were present in the 2-dose vaccine groups (group B: 24.3-fold, group D: 15.3-fold over baseline), while the 1-dose vaccine groups (groups C and E) declined modestly from day 7 (Figure 1). On day 28, 86.7% and 93.5% of 2-dose active vaccine recipients (groups D and B, respectively) had evidence of response versus 44.0% and 57.1% of 1-dose active vaccine recipients (groups C and E, respectively) (Figure 2). Placebo recipients showed 1.2-fold increases in IFN-γ–secreting cells, with 10.3% manifesting an apparent response (Figures 1 and 2). This small increase in placebo recipients might reflect assay variation near the LLOQ or exposure to circulating virus, at least in the United States.
Figure 1.
Cytokine-secreting cells after recombinant severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike glycoprotein (rS) stimulation, assessed by enzyme-linked immunosorbent spot assay (ELISpot). Cells secreting interferon gamma (IFN-γ; A), tumor necrosis factor alpha (TNF-α; B), IFN-γ and TNF-α (C), or interleukin 5 (IL-5; D) after SARS-CoV-2 rS stimulation were detected by ELISpot. Geometric mean count (GMC) of cells per million peripheral blood mononuclear cells (PBMCs) secreting the requisite cytokine is plotted with 95% confidence intervals (CIs). Geometric mean fold rise (GMFR) in cytokine-secreting cells relative to day 0 (preimmunization) is indicated above the bars along with 95% CIs. Group A: placebo (day 0/7, n = 31; day 28, n = 29). Group B: 2-dose 5 µg rS + 50 µg Matrix-M adjuvant (n = 32, n = 31). Group C: 1-dose 5 µg rS + 50 µg Matrix-M adjuvant (n = 28, n = 25). Group D: 2-dose 25 µg rS + 50 µg Matrix-M adjuvant (n = 33, n = 30). Group E: 1-dose 25 µg rS + 50 µg Matrix-M adjuvant (n = 31, n = 28).
Figure 2.
Response rate of participants producing cytokines after recombinant severe acute respiratory syndrome coronavirus 2 spike glycoprotein (rS) stimulation, assessed by enzyme-linked immunosorbent spot assay (ELISpot). The response rate, defined as the proportion of participants in each treatment group at that time point with a count that exceeds the 95th percentile of pooled day 0 counts, was determined for each group, assessed by ELISpot. Response rate is plotted, shown as a percentage, for interferon gamma (IFN-γ; A), tumor necrosis factor alpha (TNF-α; B), IFN-γ and TNF-α (C), or interleukin 5 (IL-5; D). Group A: placebo (day 0/7, n = 31; day 28, n = 29). Group B: 2-dose 5 µg rS + 50 µg Matrix-M adjuvant (n = 32, n = 31). Group C: 1-dose 5 µg rS + 50 µg Matrix-M adjuvant (n = 28, n = 25). Group D: 2-dose 25 µg rS + 50 µg Matrix-M adjuvant (n = 33, n = 30). Group E: 1-dose 25 µg rS + 50 µg Matrix-M adjuvant (n = 31, n = 28).
Patterns of TNF-α–secreting cell responses elicited by rS protein stimulation were generally similar to IFN-γ (Figure 1), although the magnitudes of increase at day 28 in the 2-dose active vaccine groups were greater (GMC = 850.9/million [62.9-fold rise in group B] and 671.1/million [35.8-fold rise in group D]). In contrast to IFN-γ–secreting cell counts, TNF-α–secreting cells continued to increase modestly at day 28 in the 1-dose vaccine groups. Counts of PBMCs secreting both IFN-γ and TNF-α, and thus manifesting a polyfunctional response to rS, showed patterns similar to IFN-γ alone, with 36% to 80% of participants who were given active vaccine showing a response producing both cytokines upon stimulation (vs none in the placebo group) (Figure 2).
Counts of PBMCs secreting the type 2 T-helper (Th2) phenotype cytokine IL-5 showed no response above LLOQ at the baseline visit and remained unchanged in placebo recipients (Figure 1). Participants receiving the first dose of vaccine had modest increases in IL-5 at day 7 (1.5- and 1.8-fold in the 5-µg dose groups, B and C, respectively; 2.9- and 1.7-fold in the 25-µg groups, D and E, respectively). Participants receiving a second dose of vaccine had further increases in IL-5 at day 28 (15.5-fold in the 5-µg dose group B; 14.3-fold in the 25-µg dose group D); IL-5 response rates in groups B and D were 93.5% and 90.0%, respectively (Figure 2).
Supplementary Tables 4–6 provide ELISpot data for stimulations using overlapping peptide pools representing the N-terminal or C-terminal sequences of the S protein and summed data reflecting peptides spanning the full S protein. These results demonstrate the same trends noted with the intact protein, with GMCs/million PBMCs of IFN-γ–secreting cells, cells secreting both IFN-γ and TNF-α, and cells secreting IL-5 being similar in the pooled peptide analysis (Supplementary Table 6) and the intact rS analysis (Figure 1). The principal difference was increased numbers of TNF-α–secreting cells at baseline when stimulated with both the N-terminal and C-terminal peptide pools. Despite strong increases in absolute GMCs of TNF-α–secreting cells, especially at day 28 in the 2-dose vaccine groups, GMFRs were limited by the high baseline counts. Notably, absolute GMCs and GMFRs of IL-5–producing cells remained less than those of IFN-γ–secreting cells for all assays with peptide stimulation.
Intracellular Cytokine Staining
Induction of T-cell responses of Th1 and Th2 phenotypes was also assessed at days 0 and 28 by ICCS using cells from a randomly selected subset of participants (n = 75), which included only 3 placebo recipients. These data reflect counts of cytokine-positive cells per million CD4+ T cells, gated on CD3+CD4+CD8– live cell populations (Supplementary Figure 3, Supplementary Table 7).
At day 0, the GMCs of CD4+ T cells responding to intact rS ranged from 12.5 to 19.1 per million cells for IFN-γ, 66.1 to 128.7 for TNF-α, and 27.0 to 54.2 for IL-2, with overlapping 95% CIs for the various treatment groups (Supplementary Figure 3A–C).
One week following the second vaccination (day 28), geometric mean IFN-γ–secreting cells were markedly increased in the 2-dose groups (282.3 and 422.9 per million cells for groups B and D, with 17.3 and 25.5 GMFRs, respectively). The 1-dose vaccine groups had lesser responses, with 1.9 and 3.3 GMFRs in groups C and E, respectively, while the placebo recipients showed no change in GMC (Supplementary Figure 3A).
Similarly, GMCs of TNF-α–secreting cells were increased at day 28 in the 2-dose vaccine groups (4452.3 and 3956.5 per million cells for groups B and D, with 37.0 and 50.6 GMFRs, respectively). The 1-dose groups again demonstrated lesser responses, with 10.0 and 8.2 GMFRs in groups C and E, respectively (Supplementary Figure 3B). Likewise, day 28 GMCs of IL-2–secreting cells were increased sharply in the 2-dose vaccine groups, attaining 3366.6 and 1826.4 per million cells for groups B and D, with 65.4 and 51.6 GMFRs, respectively. Again, 1-dose groups demonstrated lesser responses, with GMFRs similar to those for TNF-α (Supplementary Figure 3C).
Again, in ICCS, CD4+ T cells producing Th1 cytokines in response to intact rS (Supplementary Figure 3D and 3E) tended to demonstrate a polyfunctional phenotype. For example, cells producing all 3 Th1 cytokines in response to rS at the day 28 visit accounted for 63.5% to 90.1% of IFN-γ–producing cells in the 2-dose vaccine groups (B and D) and 53.1% to 78.3% in the 1-dose vaccine groups (C and E).
Supplementary Table 7 demonstrates similar trends in Th1 cytokine induction detected by ICCS when stimulation was performed with pooled peptides, although background day 0 GMCs for IL-2–producing cells were markedly higher, thus reducing the GMFRs for that cytokine.
One week following second vaccination, geometric mean counts of CD4+ T cells producing IL-5 in response to intact rS were increased by 2.5-fold (group B) and 2.0-fold (group D) in the 2-dose active vaccine groups (128.6 and 96.3 per million cells for groups B and D, respectively), with lesser-fold rises in the 1-dose 5 µg (group C; GMFR 1.4) and 25 µg (group E; GMFR 1.1) groups (Supplementary Figure 3F). Increases in IL-13–producing cells were more dynamic, with 7.9-fold and 2.2-fold increases in group B and group D, respectively (Supplementary Figure 3G). Patterns observed with pooled peptide stimulation (Supplementary Table 7) were again similar. The amplitude of Th2 responses was notably less in ICCS data as compared with ELISpot (compare Figures 1 and 2 and Supplementary Figure 3H, Supplementary Tables 6 and 7).
T-Cell Responses to Variant Spike Sequences
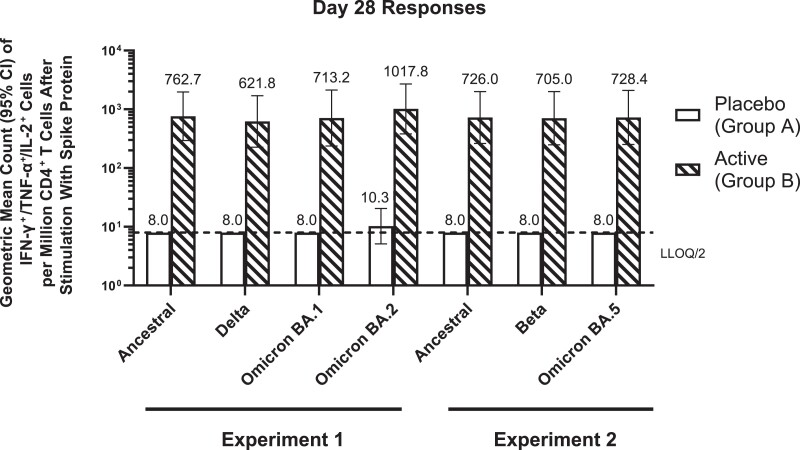
NVX-CoV2373, which contains rS of the ancestral sequence, is able to elicit neutralizing antibody responses to variant viruses after completion of a 2-dose primary series and a third (booster) vaccine dose [23]. To assess whether Th1 phenotype CD4+ T-cell responses induced by NVX-CoV2373 also showed cross-reactivity, we stimulated day 28 PBMCs from a randomly selected subset of 6 placebo (group A) and 18 NVX-CoV2373 (group B) recipients with rS proteins of variants including Beta, Delta, and Omicron BA.1, BA.2, and BA.5. As shown in Figure 3, polyfunctional CD4+ T cells expressing all 3 Th1 cytokines tested (IFN-γ, IL-2, and TNF-α) increased by almost 2 orders of magnitude (78- to 99-fold) in the immunized participants relative to placebo recipients regardless of the variant S sequence used for stimulation; 94.4%–100% of actively immunized participants responded to the variant proteins, with polyfunctional Th1 responses greater than in any placebo recipient (Figure 3, Supplementary Table 8).
Figure 3.
Stimulation of polyfunctional type 1 T-helper (Th1) CD4+ T-cell responses by variant spike proteins in peripheral blood mononuclear cells (PBMCs) of individuals treated with 2 doses of placebo or NVX-CoV2373. PBMCs from a convenience sample of 6 placebo 2-dose and 18 NVX-CoV2373 2-dose recipients at day 28 were stimulated for 6 hours with intact recombinant spike proteins of ancestral, Delta, Beta, and Omicron BA.1, BA.2, and BA.5 sequences. CD4+ T cells producing all 3 Th1 cytokines (interferon gamma [IFN-γ], tumor necrosis factor alpha [TNF-α], and interleukin 2 [IL-2]) were quantified via intracellular cytokine staining assay. Geometric mean counts of triple Th1 cytokine-positive CD4+ T cells per million CD4+ T cells (with 95% confidence intervals [CIs]) are shown. Two different experiments were run, with the first testing against ancestral, Delta, Omicron BA.1, and Omicron BA.2 strains, and the second testing against ancestral, Beta, and Omicron BA.5 strains.
DISCUSSION
Initial data based on small numbers of samples from NVX-CoV2373 recipients in phase 1 [10] indicated a robust CD4+ T-cell response of predominantly Th1 phenotype to the ancestral sequence rS contained in NVX-CoV2373, complementing anti-S IgG and neutralizing antibodies; induction of strong IFN-γ T-cell responses to spike protein peptides was also noted in the efficacy trial conducted in the United Kingdom [21]. Current data from a sampling of participants in the phase 2 component of the trial confirm several points. In ELISpot data with intact protein stimulation (Figure 1), a balanced to moderately Th1-dominant CD4+ T-cell response was underway at 7 days after the first dose but was markedly enhanced after 2 doses. The higher antigen dose level (25 µg) demonstrated a marginal advantage after 1 dose but became comparable to the 5-µg dose after the second immunization. Cells responding with production of IL-5 (a Th2 cytokine) were clearly present, but of somewhat lower numbers than concurrent Th1 cytokine expression (Figure 1). Induction of balanced or Th1-biased responses, even when administration of the antigen alone tends to evoke Th2 responses, has been a feature of Matrix-M adjuvant in animal models [9, 17] and is apparent here. During early SARS-CoV-2 vaccine development, concerns were raised that Th2-predominant responses might lead to vaccine-enhanced disease [24]. While vaccine-enhanced disease with SARS-CoV-2 has not materialized, to our knowledge, the induction of pathogen-specific, balanced or Th1-biased polyfunctional T-cell responses, as seen here, is characteristic of convalescence from SARS-CoV-2. Moreover, the polyfunctional Th1 response is both theoretically desirable for the control of intracellular pathogens and correlated with protection against other respiratory viruses such as influenza [25, 26]. ELISpot data were also developed using peptide pools representing the N-terminal (Supplementary Table 4) and C-terminal (Supplementary Table 5) portions of the ancestral spike protein and peptides representing the entire sequence (Supplementary Table 6). Aside from slightly lower amplitudes of response with the C-terminal peptides, the patterns with regard to dose, dose number, and moderate Th1 predominance were similar to those measured in assays using intact S protein.
To measure a more complex cytokine expression profile at a single cell–based level, we also examined cytokine production upon antigen stimulation within the CD4+ T-cell population by ICCS. Again, a strong CD4+ T-cell response was observed after immunization, detected using either intact rS or peptide pool stimulation and featuring a predominance of cells producing Th1 cytokines. Moreover, these antigen-specific CD4+ T cells were predominantly polyfunctional as measured by double and triple Th1 cytokine expression (Figure 2, Supplementary Table 7). Interestingly, while the counts of IFN-γ–producing cells in day 28 samples were of similar magnitude using the ELISpot and ICCS detection methods, postimmunization antigen-specific IL-5–producing cell counts were considerably lower in the ICCS data and associated with a lower GMFR of Th2 responses. This may reflect the durations of stimulation in the 2 methods, with production of Th2 cytokines previously reported to be activated more slowly, thus favoring detection via the longer antigen exposure of the ELISpot methodology [27].
Neutralizing antibodies against pseudoviruses bearing more recent SARS-CoV-2 variant spike proteins were previously shown to be substantially lower than levels against the ancestral strain in participants who received a 2-dose primary series of NVX-CoV2373 [23]. However, neutralizing titers against Beta, Delta, and Omicron variants rose markedly after a third (booster) dose of NVX-CoV2373, the latter into the range previously associated with good clinical efficacy against the ancestral strain [23]. Here, we demonstrate that polyfunctional CD4+ T-cell responses stimulated by Beta, Delta, and Omicron BA.1, BA.2, and BA.5 sequence rS proteins were essentially indistinguishable from the ancestral rS responses after only 2 doses of NVX-CoV2373 (Figure 3). This pattern of primary series–induced cross-reactivity in the CD4+ T-cell compartment, regardless of neutralizing antibody responses, has been reported for messenger RNA and adenovirus-vectored vaccines [8], has been shown in animal models with ancestral and Beta variant spike nanoparticle vaccines with Matrix-M adjuvant in animals [28], and is now demonstrated here for NVX-CoV2373 in humans. Cross-reactivity to evolving variants at the CD4+ T-cell level may help limit the severity of COVID-19 due to these viruses directly (as has been suggested in the case of preexisting cross-reactive IFN-γ–producing cytotoxic CD4+ T cells limiting illness due to influenza challenge in persons seronegative for the challenge virus [26], and potentially also provide help for the progressive acquisition of neutralizing responses to variants evoked by boosting with the prototypical ancestral vaccine, as demonstrated for third and fourth doses of NVX-CoV2373 bearing the ancestral spike sequence [23, 29]. These data thus support the persisting clinical utility of NVX-CoV2373 in the face of evolving variants.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Louis Fries, Clinical Development.
Neil Formica, Clinical Development.
Raburn M Mallory, Clinical Development.
Haixia Zhou, Discovery.
Joyce S Plested, Clinical Immunology, Novavax, Inc, Gaithersburg, Maryland.
Raj Kalkeri, Clinical Immunology, Novavax, Inc, Gaithersburg, Maryland.
Ioana Moldovan, Cellular Technology Ltd, Shaker Heights, Ohio.
Nita Patel, Discovery.
Gary Albert, Medical Writing.
Michelle Robinson, Clinical Operations.
Iksung Cho, Biostatistics, Novavax, Inc, Gaithersburg, Maryland.
Gordon Chau, Biostatistics, Novavax, Inc, Gaithersburg, Maryland.
Filip Dubovsky, Clinical Development.
Gregory M Glenn, Discovery.
for the 2019nCoV-101 Study Group:
Mark Adams, Mark Arya, Eugene Athan, Ira Berger, Paul Bradley, Richard Glover, II, Paul Griffin, Joshua Kim, Scott Kitchener, Terry Klein, Amber Leah, Charlotte Lemech, Jason Lickliter, Mary Beth Manning, Fiona Napier-Flood, Paul Nugent, Susan Thackwray, and Mark Turner
Notes
Acknowledgments. We thank all study participants who volunteered for this study. We also thank Katia Alves for her role in the development of the manuscript. Editorial assistance on the preparation of this manuscript was provided by Kelly Cameron, PhD, and Rebecca Harris, PhD, of Ashfield MedComms (New York), an Inizio company, supported by Novavax, Inc. We gratefully acknowledge the contributions of the 2019nCoV-101 Study Group: Mark Adams, Mark Arya, Eugene Athan, Ira Berger, Paul Bradley, Richard Glover II, Paul Griffin, Joshua Kim, Scott Kitchener, Terry Klein, Amber Leah, Charlotte Lemech, Jason Lickliter, Mary Beth Manning, Fiona Napier-Flood, Paul Nugent, Susan Thackwray, and Mark Turner.
Data sharing. All relevant data are included in the article or supplement.
Financial support. This work was supported by Novavax, Inc., and by the Coalition for Epidemic Preparedness Innovations (CEPI).
References
- 1. Fadlyana E, Rusmil K, Tarigan R, et al. . A phase III, observer-blind, randomized, placebo-controlled study of the efficacy, safety, and immunogenicity of SARS-CoV-2 inactivated vaccine in healthy adults aged 18–59 years: an interim analysis in Indonesia. Vaccine 2021; 39:6520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Polack FP, Thomas SJ, Kitchin N, et al. . Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 2020; 383:2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Voysey M, Clemens SAC, Madhi SA, et al. . Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021; 397:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jara A, Undurraga EA, González C, et al. . Effectiveness of an inactivated SARS-CoV-2 vaccine in Chile. N Engl J Med 2021; 385:875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Q, Qin C, Liu M, Liu J. Effectiveness and safety of SARS-CoV-2 vaccine in real-world studies: a systematic review and meta-analysis. Infect Dis Poverty 2021; 10:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kirsebom FCM, Andrews N, Stowe J, et al. . COVID-19 vaccine effectiveness against the Omicron (BA.2) variant in England. Lancet Infect Dis 2022; 22:931–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garcia-Beltran WF, St Denis KJ, Hoelzemer A, et al. . mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 2022; 185:457–66.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. GeurtsvanKessel CH, Geers D, Schmitz KS, et al. . Divergent SARS-CoV-2 Omicron-reactive T and B cell responses in COVID-19 vaccine recipients. Sci Immunol 2022; 7:eabo2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bengtsson KL, Song H, Stertman L, et al. . Matrix-M adjuvant enhances antibody, cellular and protective immune responses of a Zaire Ebola/Makona virus glycoprotein (GP) nanoparticle vaccine in mice. Vaccine 2016; 34:1927–35. [DOI] [PubMed] [Google Scholar]
- 10. Keech C, Albert G, Cho I, et al. . Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N Engl J Med 2020; 383:2320–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fries L, Cho I, Krähling V, et al. . Randomized, blinded, dose-ranging trial of an Ebola virus glycoprotein nanoparticle vaccine with Matrix-M adjuvant in healthy adults. J Infect Dis 2020; 222:572–82. [DOI] [PubMed] [Google Scholar]
- 12. Fries LF, Smith GE, Glenn GM. A recombinant viruslike particle influenza A (H7N9) vaccine. N Engl J Med 2013; 369:2564–6. [DOI] [PubMed] [Google Scholar]
- 13. Chung KY, Coyle EM, Jani D, et al. . ISCOMATRIX adjuvant promotes epitope spreading and antibody affinity maturation of influenza A H7N9 virus like particle vaccine that correlate with virus neutralization in humans. Vaccine 2015; 33:3953–62. [DOI] [PubMed] [Google Scholar]
- 14. Shinde V, Cai R, Plested J, et al. . Induction of cross-reactive hemagglutination inhibiting antibody and polyfunctional CD4+ T-cell responses by a recombinant Matrix-M–adjuvanted hemagglutinin nanoparticle influenza vaccine. Clin Infec Dis 2021; 73:e4278–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith G, Liu Y, Flyer D, et al. . Novel hemagglutinin nanoparticle influenza vaccine with Matrix-M adjuvant induces hemagglutination inhibition, neutralizing, and protective responses in ferrets against homologous and drifted A(H3N2) subtypes. Vaccine 2017; 35:5366–72. [DOI] [PubMed] [Google Scholar]
- 16. Rydyznski Moderbacher C, Kim C, Mateus J, et al. . NVX-CoV2373 vaccination induces functional SARS-CoV-2–specific CD4+ and CD8+ T cell responses. J Clin Invest 2022; 132:e160898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tian JH, Patel N, Haupt R, et al. . SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat Comm 2021; 12:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Z, Mateus J, Coelho CH, et al. . Humoral and cellular immune memory to four COVID-19 vaccines. Cell 2022; 185:2434–51.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Formica N, Mallory R, Albert G, et al. . Different dose regimens of a SARS-CoV-2 recombinant spike protein vaccine (NVX-CoV2373) in younger and older adults: a phase 2 randomized placebo-controlled trial. PLoS Medicine 2021; 18:e1003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heath PT, Galiza EP, Baxter DN, et al. . Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N Engl J Med 2021; 385:1172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heath PT, Galiza EP, Baxter DN, et al. . Safety and efficacy of the NVX-CoV2373 coronavirus disease 2019 vaccine at completion of the placebo-controlled phase of a randomized controlled trial. Clin Infect Dis 2023; 76:398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dunkle LM, Kotloff KL, Gay CL, et al. . Efficacy and safety of NVX-CoV2373 in adults in the United States and Mexico. N Engl J Med 2022; 386:531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mallory RM, Formica N, Pfeiffer S, et al. . Safety and immunogenicity following a homologous booster dose of a SARS-CoV-2 recombinant spike protein vaccine (NVX-CoV2373): a secondary analysis of a randomised, placebo-controlled, phase 2 trial. Lancet Infect Dis 2022; 22:1565–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gartlan C, Tipton T, Salguero FJ, Sattentau Q, Gorringe A, Carroll MW. Vaccine-associated enhanced disease and pathogenic human coronaviruses. Front Immunol 2022; 13:882972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen KW, Linderman SL, Moodie Z, et al. . Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med 2021; 2:100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilkinson TM, Li CKF, Chui CSC, et al. . Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med 2012; 18:274–80. [DOI] [PubMed] [Google Scholar]
- 27. Duechting A, Przybyla A, Kuerten S, Lehmann PV. Delayed activation kinetics of Th2- and Th17 cells compared to Th1 cells. Cells 2017; 6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Logue J, Johnson RM, Patel N, et al. . Immunogenicity and protection of a variant nanoparticle vaccine that confers broad neutralization against SARS-CoV-2 variants. Nat Commun 2023; 14:1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alves K, Plested JS, Galbiati S, et al. . Immunogenicity of a fourth homologous dose of NVX-CoV2373. N Engl J Med 2023; 388:857–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.