Abstract
Dissimilatory nitrate reduction to ammonia (DNRA) is a common biochemical process in the nitrogen cycle in natural and man-made habitats, but its significance in wastewater treatment plants is not well understood. Several ammonifying Trichlorobacter strains (former Geobacter) were previously enriched from activated sludge in nitrate-limited chemostats with acetate as electron (e) donor, demonstrating their presence in these systems. Here, we isolated and characterized the new species Trichlorobacter ammonificans strain G1 using a combination of low redox potential and copper-depleted conditions. This allowed purification of this DNRA organism from competing denitrifiers. T. ammonificans is an extremely specialized ammonifier, actively growing only with acetate as e-donor and carbon source and nitrate as e-acceptor, but H2 can be used as an additional e-donor. The genome of G1 does not encode the classical ammonifying modules NrfAH/NrfABCD. Instead, we identified a locus encoding a periplasmic nitrate reductase immediately followed by an octaheme cytochrome c that is conserved in many Geobacteraceae species. We purified this octaheme cytochrome c protein (TaNiR), which is a highly active dissimilatory ammonifying nitrite reductase loosely associated with the cytoplasmic membrane. It presumably interacts with two ferredoxin subunits (NapGH) that donate electrons from the menaquinol pool to the periplasmic nitrate reductase (NapAB) and TaNiR. Thus, the Nap-TaNiR complex represents a novel type of highly functional DNRA module. Our results indicate that DNRA catalyzed by octaheme nitrite reductases is a metabolic feature of many Geobacteraceae, representing important community members in various anaerobic systems, such as rice paddy soil and wastewater treatment facilities.
Subject terms: Water microbiology, Proteomics, Bacteriology
Introduction
There are two currently known anaerobic nitrate/nitrite respiratory processes (anammox is principally different and not considered here): denitrification and dissimilatory nitrate reduction to ammonia (DNRA). The two processes share the first step, where nitrate is reduced to nitrite, but further nitrite reduction steps either to N2 (denitrification) or ammonia (DNRA) are fundamentally different in their enzymology and ecological consequences. Nitrite reduction to N2 is a three-step process with at least one step depending on copper-containing reductases (the nitrous oxide reductase NosZ and in some denitrifiers also the copper-dependent nitrite reductase NirK), while its reduction to ammonia is a completely copper-independent, single-enzyme process [1–3]. The difference in final product of the two processes has large ecological implications: denitrification removes nitrogen from the system, with a negative impact on nitrogen-limited natural habitats, but desired in engineered nutrient-rich or anthropogenically eutrophicated systems. DNRA, by contrast, is an unwanted process in wastewater treatment but beneficial for instance in nitrogen (N)-limited soils.
Denitrification is studied much more intensively, because most denitrifiers are facultative anaerobes and easier to cultivate than DNRA organisms, which are obligate anaerobes except for the fermentative (enteric) gammaproteobacterial branch [1, 4]. Consequently, the enrichment and isolation in pure culture of ammonifiers using standard batch cultivation techniques is problematic and their targeted isolation is uncommon [5]. Often anaerobic microorganisms are isolated based on certain metabolic traits and it is discovered only later during standard physiological tests or from genomic context that they can ammonify [6, 7].
For a long time, it was obscure how these two nitrate respiration processes were competing for their electron (e) donors and acceptors. Early studies with anoxic marine and freshwater sediments demonstrated the Corg:N ratio as an important discriminator [8–11]. DNRA was favoured over denitrification at high C:N ratios, which describes settings where e-donors exceeded e-acceptors and the eight-electron reduction of nitrate to ammonium became more profitable then the five-electron reduction to N2 [6, 12, 13]. Contrastingly, denitrifiers are prevalent at low C:N ratios, i.e., under e-donor limited but nitrate excess conditions, and it was concluded from those experiments that DNRA organisms have higher affinity for nitrate. The outcome of the competition between distinct organisms with different types of nitrate respiration systems is thus determined by external environmental parameters optimal to maximize their energy and growth yield. One additional parameter favouring DNRA is the presence of sulfide, which inhibits N2O reductase catalyzing the last step of denitrification [14–17], although a prolonged sulfide exposure might promote proliferation of sulfide-oxidizing denitrifiers [18]. A combination of high organic load and low nitrate with low redox conditions due to the presence of reduced sulfur compounds (i.e., sulfide and FeS) is typical for eutrophic marine and freshwater sediments and particularly for activated sludge, thus potentially supporting proliferation of DNRA organisms.
The classical and well-investigated enzyme system responsible for DNRA includes two genetic variations: The NrfAH and NrfABCD systems, which both utilize the catalytic pentaheme cytochrome c subunit NrfA as the ammonifying nitrite reductase. NrfH, a homologue of NapC/CymA, is a membrane-bound tetraheme cytochrome c that serves as electron donor for NrfA. It acts as quinol dehydrogenase and is mostly present in obligately anaerobic ammonifying Epsilon- and Deltaproteobacteria. NrfBCD is fulfilling the analogous function mostly in facultatively anaerobic fermentative enteric Gammaproteobacteria [2, 3].
Recently, an octaheme cytochrome c nitrite reductase (ONR) present in members of the sulfur-oxidizing gammaproteobacterial genus Thioalkalivibrio has been shown to catalyze nitrite reduction to ammonium in vitro using low-potential electron donors [19, 20]. The enzyme is distantly related to the octaheme cytochrome c hydroxylamine dehydrogenase (HAO) family, has a five heme c core unit structurally highly similar to NrfA, and has multiple homologues in Bacteria, particularly in Geobacterales species. Despite an extremely high nitrite ammonifying activity in vitro, this enzyme is not functioning as the nitrite reductase in vivo in these aerobic bacteria. Furthermore, the extremely thermophilic crenarchaeon Igniococcus hospitalis encodes a membrane-associated ONR that is truly functioning as a nitrite reductase in vivo [21]. However, ammonia is not a product of this ONR and it likely has a role in NOx detoxification. Moreover, octaheme cytochromes (εHAO) acting in vivo as ammonifying nitrite reductases were described in several members of the Epsilonproteobacteria, including two Campylobacter species, Caminibacter mediatlanticus and Nautilia profundicola [22, 23]. Still, specific nitrite-reducing activity was at least 10-fold lower than of NrfA from Wolinella succinogenes, a member of the same phylogenetic group. Finally, a novel metabolism, sulfide-dependent DNRA, has recently been shown in two members of the Desulfobulbales, in the sulfur-disproportionating Desulfurivibrio alkaliphilus [24] and a freshwater "cable" bacterium Ca. Electronema [25]. In both, the classical NrfAH pentaheme nitrite reductase module is absent and, instead, octaheme cytochromes c that are members of the ONR (Desulfurivibrio) or εHAO clades (Ca. Electronema) represent the only putative ammonifying nitrite reductases.
In this study, we isolated Trichlorobacter ammonificans strain G1, an obligately anaerobic bacterium representing a novel species within the genus Trichlorobacter (formerly part of the genus Geobacter), which is a highly specialized acetate-oxidizing DNRA organism. We comprehensively describe its physiological, biochemical, and genomic properties. It is lacking the classical Nrf systems and instead encodes a DNRA module that includes a ammonifying ONR and the complete beta-subtype periplasmic nitrate reductase (NAP) in a single genetic locus. This ONR was the only known nitrite reductase expressed during ammonifying growth, and purification and biochemical characterisation demonstrated that this enzyme was highly active in vitro and, apparently, in vivo.
Material and methods
Isolation and batch cultivation
Trichlorobacter ammonificans strain G1 was isolated from a previously described nitrate-limited chemostat culture highly enriched in DNRA bacteria originally obtained from an activated sludge inoculum with acetate as e-donor and nitrate as e-acceptor at an electron molar ratio of 2.7 [13, 26]. For pure-culture isolation, this steady-state chemostat culture was serially ten-fold diluted up to (−10) in 12 ml serum bottles with 8 ml medium containing 5 mM acetate as e-donor and either 2 or 5 mM nitrate or 1–2 mM nitrite as e-acceptor. Before inoculation, bottles were made anoxic by several cycles of evacuation and subsequent argon flushing of the head space. The base medium contained 5 mM sodium acetate, 50 μM methionine, and 1 g l−1 of potassium phosphate buffer at pH 6.8. After sterilization, the base medium was supplemented with 1 ml l−1 of acidic trace metal solution [27], 0.5 mM MgSO4, 20 mg l−1 CaCl2, and variable concentrations of KNO3 from 1 M sterile stock solutions. To create copper-free conditions, CuCl2 was omitted from the trace element solution and the copper chelator tetrathiomolybdate was added to the medium at up to 0.2 mM. Sulfur reductant solutions (0.2 M cysteine hydrochloride, pH 6, and 0.1 M sodium sulfide) were prepared in boiled demineralized water cooled under a stream of argon, filter-sterilized into sterile serum bottles and kept under argon overpressure at room temperature. The sulfide solution was stable at least for 1 year, while cysteine needed to be replaced every 3–4 weeks. The same cultivation setup as described above using 12 ml serum bottles containing 70% liquid culture and 30% head space was used for pure culture batch experiments. In case of incubations with high initial nitrate concentrations (5–10 mM), 10% (v/v) CO2 was added into the gas phase in the early exponential growth phase to prevent the pH from raising above pH 8.
For the resting cell experiment, 0.5 L batch culture was inoculated with 5 mL starting culture and grown with acetate and nitrate in the presence of cysteine and sulfide as reductants as described above until 10 mM nitrate were consumed after 12 days. Cells were collected by centrifugation, washed and resuspended in 20 mM potassium phosphate buffer, pH 7, at a final density of 0.15 mg cell protein ml−1. The anoxic incubations were done in 9 ml serum bottles with 3 ml cell suspension at 30 °C and metabolic products were analyzed after 14 and 40 h. Nitrate and nitrite were added at 5 mM, acetate at 10 mM, and reductants at 0.1 mM HS- and 0.2 mM cysteine.
Continuous culture operation
The pure culture of T. ammonificans G1 was cultivated in a glass bioreactor with a working volume of 2 l (Applikon) as described by van den Berg and coworkers [26], with the following adjustments. The reactor was autoclaved with 1 L of 9.16 mM phosphate buffer (pH 6.8) at 121 °C. The medium in the reactor was flushed with argon and after adding 1 L of the fully grown batch culture of strain G1, the flow of argon was maintained only through the headspace to avoid sulfide stripping. The reactor temperature was controlled at 30 °C. The redox potential was monitored using a platinum Redox probe (Ag/AgCl-based, Mettler Toledo, Tiel, The Netherlands). The pH was maintained at 7 by titration with 0.5 M HCl. Two separate media were fed to the reactor in equal amounts. Medium A contained varying amounts of NaNO3. Medium B contained 0.7 g l−1 KH2PO4 and K2HPO4 each and varying amounts of NaCH3COO × 3H2O. After autoclaving the following compounds were added to medium A: 1 mM MgSO4 × 7H2O, 1 ml l−1 10 % (w/v) CaCl2, and 2 ml l−1 Pfennig & Lippert trace element solution [27]. After flushing with argon for 15 min, 1 ml l−1 of 10% (w/v) neutralized anoxic filter-sterilized cysteine solution and 0.2 mM filter-sterilized Na2S were added to medium B. The media were connected to the reactor and, for volume compensation during pumping, an extra empty vessel was connected, which was continuously flushed with argon to provide an argon buffer for the flow into the media bottles. Both media were pumped at 23 ml h−1 into the reactor, with a total influent of 43 ± 4 ml h−1 resulting in a dilution rate of 0.023 h−1.
Analyses
The ONR of T. ammonificans G1 was purified from the soluble cell-free extract fraction and biochemically characterized. Furthermore, the genome of strain G1 was sequenced and annotated, and used for 16 S rRNA-based phylogenetic, phylogenomic, and comparative genomic analyses. A more detailed description is given in the Supplementary Methods. For proteomics, triplicate samples (technical replicates) were analyzed from a steady-state T. ammonificans G1 culture grown with acetate as e-donor and carbon source in a nitrate-limited chemostat (see above) at a dilution rate of 0.05 h−1. Growth in batch and chemostat cultures was monitored by OD660 measurements. Cell protein content in cultures was determined by the Lowry method [28]. Acetate, nitrate, nitrite, and ammonia were analyzed as described previously [13, 26]. N2O in the gas phase was detected by GC (Varian 2000, column Parapaq-Q, Ni-63 detector). Online gas measurements of O2, CO2, NO, and N2O during chemostat operation were performed using a gas analyzer (NGA 2000, Rosemount, Chanhassen, MN, U.S.A.). The flow of argon gas to the reactor was kept at 100 ml min−1 using a mass flow controller (Brooks Instrument, Ede, The Netherlands) to maintain sufficient flow through the gas analyzer (80 ml min−1). Sulfide was measured by the methylene blue method after fixation of the cell-free culture supernatant into 2% Zn-acetate [29]. The polar lipid fatty acid analysis of T. ammonificans G1 was performed by DSMZ (Braunschweig, Germany). For electron microscopy, washed cells were positively stained with 1% (w/v) uranyl acetate and examined with a transmission electron microscope (JEOL 100CX, Japan).
Results
Pure culture isolation
In a past study, a chemostat was inoculated with an activated sludge sample and provided with acetate as carbon and energy source and nitrate as sole electron acceptor, which enriched for a member of the Geobacteraceae performing DNRA [13, 26]. The high dominance of this organism over denitrifiers was achieved by gradually increasing the acetate to nitrate ratio (based on the electron molar ratio) from nitrate excess to nitrate-limited conditions [13].
Despite this high dominance, when we in this study inoculated batch cultures (liquid medium or soft agar dilutions) with this chemostat enrichment in the presence of 5 mM acetate and 2 mM nitrate or 1 mM nitrite, this resulted in the prevalence of betaproteobacterial denitrifiers that developed rapidly, with nitrate fully converted into gaseous products. Complete elimination of copper from the trace metal solution and, furthermore, addition of the specific copper chelator tetrathiomolybdate (up to 0.2 mM) resulted in copious nitrite formation in all positive dilutions (up to −9) and formation of up to 0.4 mM ammonium in the first dilution. It was suspected that the main cultivation bias towards denitrifiers was a much higher (starting) redox balance in the batch cultures (+50 mV) compared to the stable negative redox situation in the fully grown nitrate-limited chemostat culture (−350 mV). In the latter, the supernatant accumulated sulfide at 2–15 μM due to the presence of a background population of sulfate-reducing bacteria (Fig. S1). To verify the importance of the low redox potential conditions, cysteine (0.05–0.2 mM) was added over the dilution series. This resulted in ammonium formation up to 1 mM in dilutions up to (−6), but still did not fully eliminate growth of denitrifiers. Finally, use of cysteine (0.2 mM) together with sulfide (0.1 mM) plus complete copper elimination allowed domination of DNRA over denitrification up to the highest positive dilutions [(-8) to (-9)] and resulted in a morphologically homogenous ammonifying culture similar to the dominant morphotype in the chemostat. The strain was designated G1.
Pure culture characterization
The 16 S rRNA gene sequence analysis placed the isolate within the genus Trichlorobacter (Geobacterales) with a sequence identity of 97% to its closest related species, T. lovleyi. This, together with average nucleotides identities (ANI) below the species threshold of 95% to other cultured Geobacterales species (Table S1), indicated that the isolate represents a novel species, for which we propose the tentative name Trichlorobacter ammonificans G1. The phylogenetic affiliation with the genus Trichlorobacter was also confirmed by phylogenomic analysis [30] (Fig. 1, Fig. S2).
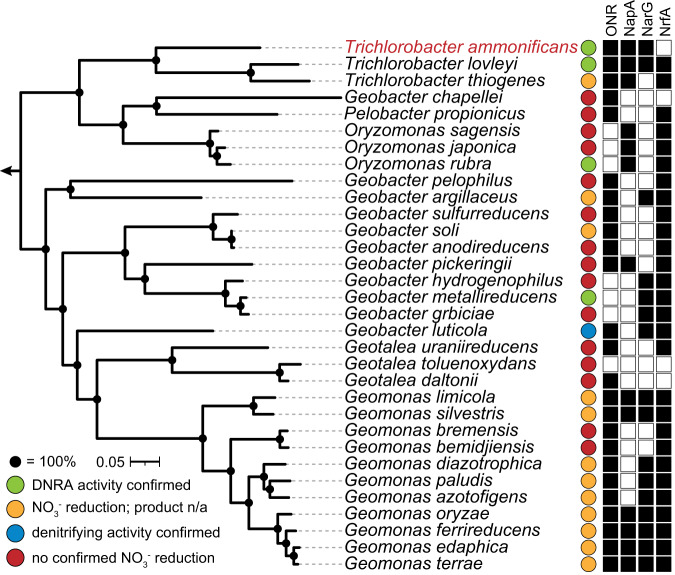
Fig. 1. Phylogenetic affiliation of T. ammonificans G1 and distribution of ONR compared to other N-transforming proteins in the order Geobacterales.
The maximum-likelihood tree is based on a concatenated alignment of 92 universal single-copy bacterial core genes. Two Desulfuromonas genomes served as outgroup. Bootstrap support values derived from 1000 ultrafast bootstrap iterations = 100% are represented by filled circles. The scale bar corresponds to 5% sequence divergence. The matrix on the right indicates presence and absence of ONR, NapA, NrfA and NarG (BLAST p ≥ 50% identity, ≥ 90% query coverage); white, no homologous protein; black, homologous protein to T. lovleyi. Coloured circles represent reductive nitrogen conversion potential; green, confirmed DNRA activity; yellow, confirmed nitrite reduction activity, but product not analyzed; blue, confirmed denitrification activity; red, no confirmed NO3- reduction. Except for T. ammonificans G1, the confirmed activity is based on previous studies. Accession numbers of included Geobacterales genomes are listed in Table S6. A phylogenomic tree including all Geobacterales genomes are shown in Fig. S2.
Both the batch and the chemostat ammonifying cultures of T. ammonificans G1 had an intensive pink colour typical for iron-reducing Geobacterales species (Fig. S3). In contrast to all known Trichlorobacter/Geobacter species with versatile anaerobic catabolic potentials [31, 32], strain G1 showed sustainable growth in batch culture only at a single combination of electron donor and acceptor, i.e., with acetate and nitrate (Fig. 2A). The nitrate-limited chemostat culture grown with acetate was also able to switch to nitrite-limited conditions. Nitrite can also be utilized in batch cultures pre-grown with limited amounts of nitrate, but when grown in batch, the culture never started to grow with nitrite, even at initial concentrations as low as 0.2 mM. Only when nitrite was added after full nitrate consumption, it was converted mostly to ammonia with N2O as a side product, both in batch and chemostat cultures (Fig. 2B). Tests with resting cells of T. ammonificans G1 pre-grown in batch culture with acetate and nitrate demonstrated the importance of low redox potential conditions for complete nitrite reduction to ammonia and showed that N2O formation increased under high redox conditions (Table S2). A separate test with acetate and N2O as electron acceptor showed that T. ammonificans G1 was unable to reduce N2O further to N2.
Fig. 2. Anaerobic growth dynamics of strain G1.
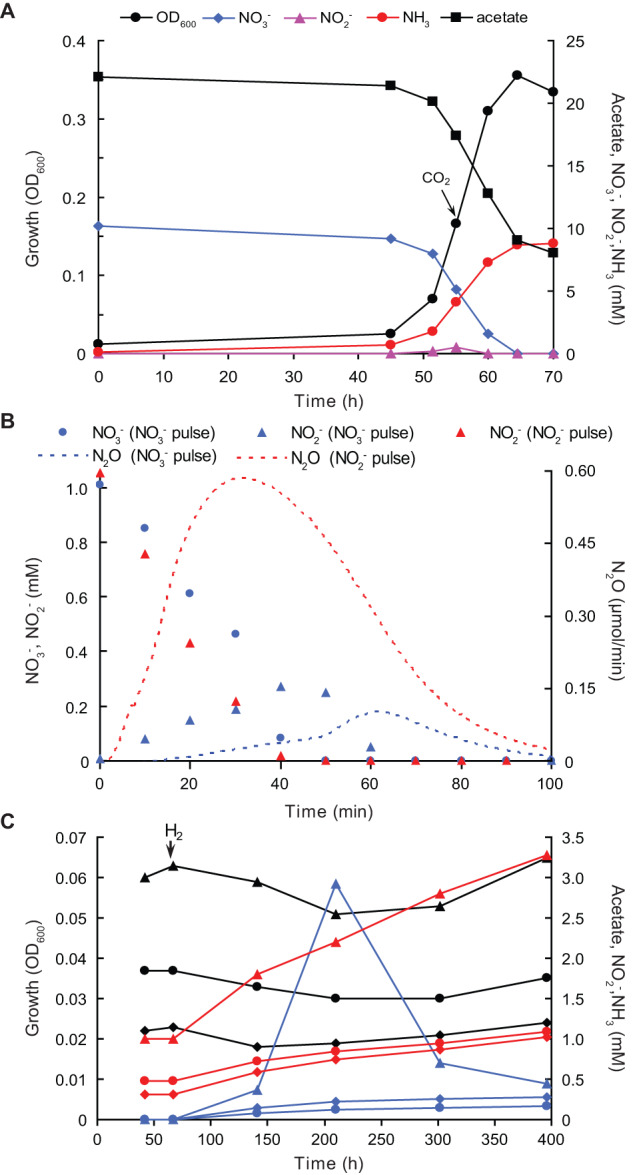
A Growth in nitrate-limited ammonifying batch culture in presence of 0.1 mM sulfide and 0.2 mM cysteine as reductants. B N2O formation from nitrate and nitrite (1 mM additions) pulse-added to the nitrate-limited chemostat culture. Total N2O emissions constituted 0.44% and 2.8% of the consumed nitrate and nitrite, respectively. C Use of H2 (20 ml culture in 32 ml flasks, 15 ml addition to the gas phase after 67 h incubation is indicated by arrow) as an e-donor during ammonification in an acetate-limited batch culture of strain G1 with 5 mM nitrate. Black lines, growth (OD600); blue lines, NO2- (mM); red lines, NH4+ (mM); diamonds, 0.5 mM acetate; circles, 1 mM acetate; triangles, 2 mM acetate. Results in panels (A) and (C) are mean values from two parallel incubations.
From the tested organic e-donors with 2 or 5 mM nitrate as e-acceptor, weak growth in batch cultures was observed only on succinate (10 mM), with nitrate being converted to nitrite only. Growth on 20 mM formate amended with 0.1 g l−1 yeast extract as the carbon source was negative. Testing H2 as a low-potential inorganic e-donor proved to be difficult, as T. ammonificans G1 cannot grow autotrophically, and acetate, which had to be added as carbon source since yeast extract was unable to replace it, also served as the preferred e-donor. However, using acetate-limited conditions allowed us to prove that G1 was able to utilize H2 as an additional e-donor for nitrate ammonification (Fig. 2C). This is also supported by the presence of three operons encoding periplasmic uptake [NiFe]-hydrogenases in the genome of T. ammonificans G1 (see below). In sharp contrast to the known Trichlorobacter species, G1 was unable to use any other e-acceptor with 5 mM acetate as e-donor, including different forms of Fe(III) and MnO2 (both at 20 mM), AQDS, S8, thiosulfate, selenate, arsenate, fumarate, acrylate, crotonate (all at 5 mM), and O2 (1% in the gas phase).
When cultivating T. ammonificans G1 in a chemostat under nitrate-limited conditions, we ensured low redox conditions by adding reductants to the reactor influent. This resulted in residual nitrate concentrations below detection limit (<20 μM). Under these conditions, the redox potential was in the range of −320 to −350 mV and the culture was stable, converting 87 ± 11% of the supplied nitrate to ammonia and incorporating 13 ± 2% of the N into biomass. No NO or N2O were detectable in the off-gas. The biomass yield was 12.1 ± 2.2 g VSS/mol acetate and 16.6 ± 2.3 VSS/mol NO3-. Nitrogen conversion rates and biomass yields were similar in steady state conditions without reductants in the influent. This is in sharp contrast to results from batch cultivation, where growth without reductants did not commence. In the nitrate-limited chemostat started with a dense batch culture, nitrate is immediately consumed to low µM concentrations, thus ensuring low redox potential conditions. Contrastingly, in batch culture the starting nitrate concentrations are in the mM range, hence requiring reduction of the initial redox potential. When the redox potential of the chemostat culture was increased by switching to acetate-limited conditions, nitrate and nitrite started to accumulate despite the addition of reductants and the cells aggregated, as was also observed in batch cultures. Ultimately, this led to culture collapse, affirming the oxygen-independent high redox sensitivity of T. ammonificans G1, accompanied with nitrite toxicity.
Genomic and proteomic analyses of acetate-driven DNRA
The genome of T. ammonificans G1 (Table S3) encodes key features shared with other members of Geobacterales including the metabolic machinery for transport, oxidation, and assimilation of acetate (Table S4, Fig. S4, Supplementary Text). The first step of both DNRA and denitrification is the reduction of nitrate to nitrite. This step can by catalyzed by the cytoplasmic (NAR) or the periplasmic (NAP) nitrate reductase complexes encoded in the T. ammonificans G1 genome and a few other Geobacterales species (Fig. 1). In the proteome of T. ammonificans G1 growing by acetate-driven DNRA in the nitrate-limited chemostat culture, NAP was the only nitrate reductase detected. The second step in DNRA is the dissimilatory reduction of nitrite to ammonia. While other described Geobacterales species employ the cytochrome c nitrite reductase NrfAH for this key step, genes encoding these proteins are missing in the T. ammonificans G1 genome. In contrast, it encodes an octaheme cytochrome c in a single gene cluster together with the complete beta-type NAP system (lacking the tetraheme c subunit NapC; Fig. 3), which includes the catalytic subunits NapAB, the cytoplasmic chaperone NapD, a putative NapF-like maturation factor and the two membrane-associated polyferredoxins NapGH that shuttle electrons from the menaquinol pool to NapAB [3]. Notably, a similar genomic region was recently identified in the freshwater cable bacterium Ca. Electronema sp. GS, and similar to T. ammonificans G1 it is hypothesized that the encoded proteins are involved in DNRA [25]. A striking difference, however, is the type of the octaheme cytochrome c protein present in these Desulfobacterales members, which forms a sister clade to the εHAO family. In contrast, the T. ammonificans G1 protein is related to the octaheme cytochrome c nitrite reductase (ONR) of Thioalkalivibrio (Fig. 4, Fig. S5) and contains also the CxxCK motif characteristic for NrfA and ONR [20, 33]. While NAP and putative ONR genes have been identified in several Geobacterales genomes (Fig. 1), this gene cluster is only conserved in few species. Still, as indicated by the wide distribution of octaheme cytochrome c proteins in Geobacterales, these enzymes might play a vital role in the N metabolism of its members. However, their exact function is poorly understood, mainly since octaheme cytochrome c proteins form a large enzyme group catalyzing the conversion of several nitrogen compounds, such as nitrite, hydroxylamine, and hydrazine. While other Geobacterales members possess the ONR in addition to a canonical NrfAH, ammonification capacity in T. ammonificans G1 is fully dependent on the nitrite-reducing activity of ONR. The importance of this NAP-ONR gene cluster is underlined by the identification of all encoded proteins in the proteome of T. ammonificans G1 cells from the nitrate-limited chemostat culture growing by acetate-driven DNRA (Fig. 4, Table S4). The highest level of expression within the NAP-ONR module was found for subunits NapA and ONR, catalyzing reduction of nitrate to nitrite and of nitrite to ammonia, respectively (Fig. 4C, Fig. S6). In addition to NAP and ONR, the hybrid cluster protein Hcp was also identified in the proteome of T. ammonificans G1, potentially detoxifying side products of the ONR such as nitric oxide [34–36]. Taken together, the high abundance of these proteins and the lack of NAR expressed under this growth condition further suggest NAP-ONR as the only functional DNRA module.
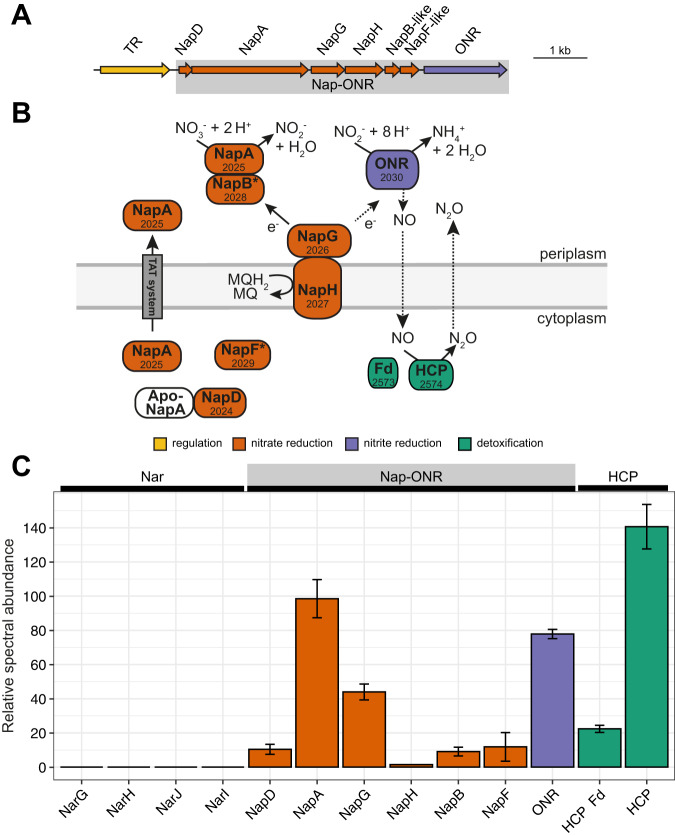
Fig. 3. Organization of the novel DNRA module in T. ammonificans G1 based on genomic and biochemical analyses.
A Schematic representation of the Nap-ONR gene locus. B Scheme of the Nap-ONR module reducing nitrate to ammonium: NapAB reduces nitrate to nitrite, ONR catalyzes the dissimilatory reduction of nitrite to ammonium, and the ferredoxins NapGH are involved in electron transport from the menaquinone pool. NO and N2O are minor side products. NO originates from partial nitrite reduction by ONR at high nitrite and suboptimal redox conditions, while N2O is produced from NO in a detoxification mechanism due to the activity of hydroxylamine reductase (HCP) and a ferredoxin (Fd). NapD is a maturation chaperone for NapA, a role also speculated for NapF. Gene identifiers are indicated as numbers in the proteins and asterisks indicate low sequence similarity to characterized proteins. C Expression level (relative spectral abundance) of the Nar operon, the Nap-ONR module proteins, and HCP in strain G1 cells grown in a nitrate-limited chemostat culture with acetate as e-donor and C-source. The proteomics data is summarized in Fig. S6 and Table S4. HCP, hydroxylamine reductase; Fd, ferredoxin; MQ, menaquinone; MQH2, menaquinol; Nap, periplasmic nitrate reductase; Nar, cytoplasmic nitrate reductase; ONR, octaheme nitrite reductase; TAT, twin-arginine translocation system; TR, transcriptional regulator.
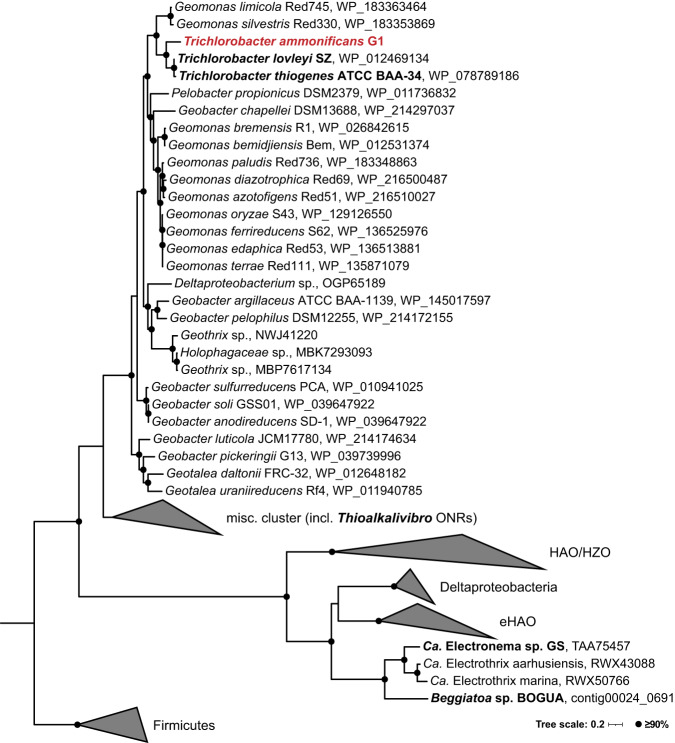
Fig. 4. Phylogeny of the octaheme cytochrome c proteins potentially involved in nitrogen redox conversions.
The maximum likelihood tree was calculated using iqtree with 1000 ultrafast bootstrap iterations. Proteins with confirmed nitrite ammonifying activity mentioned in the main text are shown in bold; the T. ammonificans G1 ONR in red. Bootstrap support values ≥90% are indicated by filled circles. NrfA proteins in Geobacterales members and Anaeromyxobacter spp. were used to root the tree. The scale bar corresponds to 20% sequence divergence. ONR, octaheme nitrite reductase; HAO, hydroxylamine dehydrogenase; HZO, hydrazine dehydrogenase; eHAO, εHAO. An extended phylogenetic tree of the octaheme cytochrome c oxidoreductases is shown in Fig. S5.
Properties of the ONR
Despite the bioinformatics-based prediction that the T. ammonificans G1 ONR (further referred to as TaNiR) is a periplasmic protein lacking transmembrane helices (TMH), it was mainly found in the cytoplasmic membrane fraction (Fig. S7A). However, after several types of membrane solubilization treatments (0.25 and 1 М NaCl; 0.05% dodecylmaltoside +0.25 M NaCl; 0.05% Triton Х-100 + 0.25 M NaCl), TaNiR was fully solubilized in 1 M NaCl (Fig. S7B) indicating that its membrane association was based on electrostatic interaction. This also explains the presence of nitrite reductase activity to varying extents in all cell fractions. Thus, TaNiR is a periplasmic protein with weak attachment to the external face of the cytoplasmic membrane. Such configuration places it apart from the classical DNRA module NrfAH, where the tetraheme subunit NfrH is an integral membrane protein. The TaNiR protein was purified to homogeneity (Fig. S7A) from the soluble fraction of cell homogenate using two-step anionic column chromatography followed by the size exclusion chromatography.
The purified protein was identified as the product of the putative ONR gene (GEAMG1_2030). The protein consists of 545 amino acids (aa), including a Sec/SP1-type signal peptide of 28 aa, seven heme c-binding CXXCH motifs, and a single CXXCK motif characteristic of the ammonifying penta- and octaheme enzymes NrfA and ONR [37]. The mature protein consists of 517 аa with a theoretical pI of 9.08. Direct heme c analysis after pyridine extraction confirmed the presence of eight hemes per protein (7.4 ± 0.5). The calculated mass of the mature form containing eight hemes c is 63.3 kDa, corresponding well to the results of SDS-PAGE (Fig. S7A). Closely related homologues of TaNiR are encoded in genomes of many Geobacterales species and some other bacterial taxa (Fig. 4, Fig. S5). The closest predicted homologues to TaNiR (sequence coverage ≥95% and identity ≥75%) are the ammonia-forming ONRs of Trichlorobacter lovleyi (WP_012469134.1) and Trichlorobacter thiogenes (WP_078789186.1). Structurally and functionally characterized homologues of TaNiR with sequence coverages ≥94% and identities ≤50% are ONRs from the haloalkaliphilic sulfur-oxidizing gammaproteobacteria Thioalkalivibrio nitratireducens (WP_015257125.1) and Thioalkalivibrio paradoxus (WP_006746849.1; Fig. S8). It has been shown previously that the primary structure of ONR proteins from the members of Geobacterales include all key loci essential for the catalytic properties proven for the Thioalkalivibrio ONR, including the catalytic heme c coordinated by CXXCK, the active site residues His, Arg, Tyr, and Cys characteristic of ONR, and the residues responsible for binding a structurally important Ca2+ ion located in the vicinity of the catalytic heme [19, 20]. All those structural features are also present in TaNiR (Fig. S8), indicating that this protein is a typical ammonifying octaheme cytochrome c nitrite reductase.
Spectral characteristics of reduced and oxidized forms of TaNiR are very similar to those of characterized ONRs [20, 38], including an absorbance maximum of the oxidized form at 408 nm and of the reduced form at 422 nm (γ-band), 525 nm (β-band) and 554 nm (α-band; Fig. 5A). TaNiR is present in solution (рН 8.0, 150 mM NaCl) as a mixture of hexa- (20%), tri- (70%) and monomers (10%; Fig. S9). The trimer becomes the only form upon increasing NaCl concentrations to 1 M, in contrast to the Thioalkalivibrio ONR that is stabilized in the hexamer form at high salt conditions. Potentiometric titration of TaNiR hemes (monitored at А554) showed that it is redox-active in a broad range from −350 to +50 mV (Fig. 5B). The reductive and oxidative curves are superimposable, indicating that under these experimental conditions the protein can reversibly cycle between the fully reduced and fully oxidized states. These broad redox windows reflect the contribution of multiple hemes with overlapping redox potentials to the activity of ONR. This redox potential window of TaNiR is slightly more positive than the Thioalkalivibrio ONR (−450 to +50 mV).
Fig. 5. Functional properties of purified T. ammonificans G1 ONR (TaNiR).
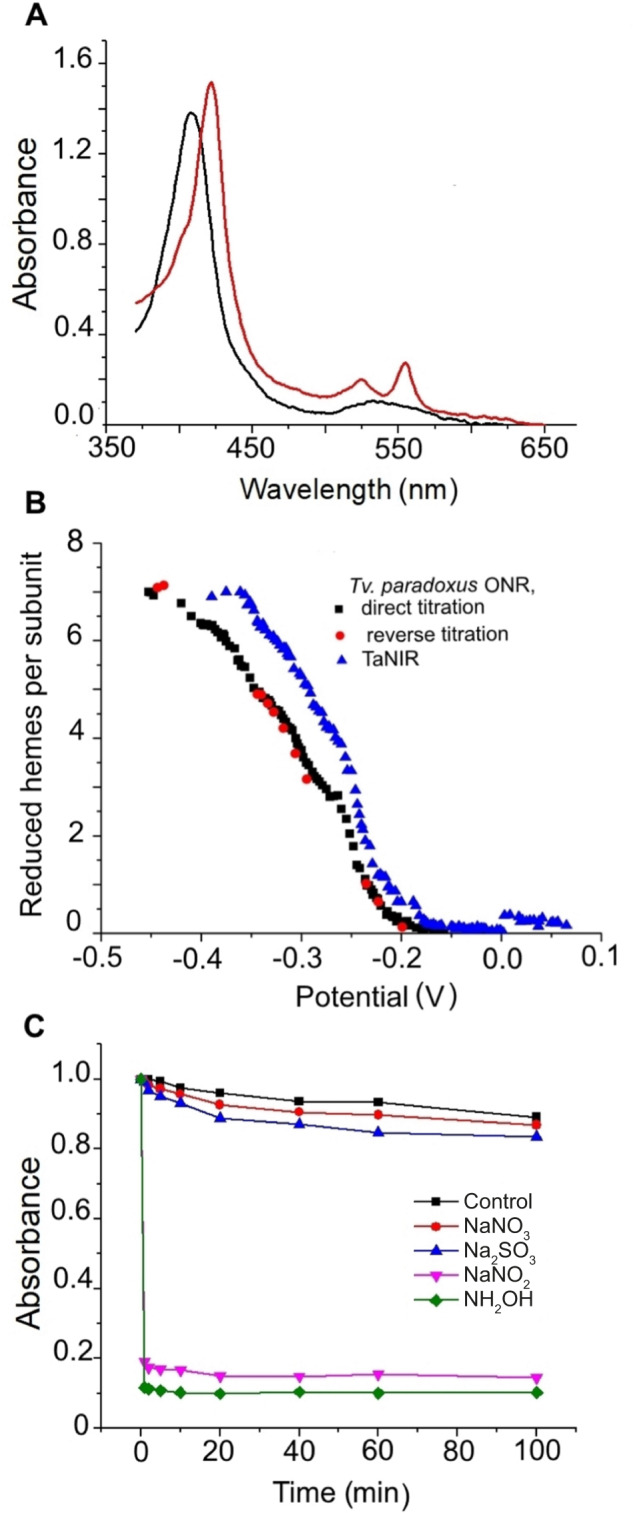
A Visible spectra of oxidized and reduced forms of TaNiR. B Potentiometric titration of TaNiR and its homologue Tv. paradoxus ONR. C Screening of the TaNiR catalytic activities by oxidation of reduced TaNiR with potential substrates. 1.9 µM of EuCl2-reduced TaNiR was incubated in 50 mM HEPES (pH 7.5) with 1.4 mM of the potential substrates NaNO3, NaNO2, NH2OH, or Na2SO3. In the control experiment the corresponding volume of buffer was added.
Oxidative titration of dithionite-reduced TaNiR showed that nitrite and hydroxylamine oxidize the reduced enzyme rapidly and completely (Fig. 5C, Fig. S10). Nitrate and sulfite did not oxidize the reduced enzyme and, therefore, are not its substrates. These results were also confirmed by in-gel zymography (Fig. S11). MALDI-TOF-MS analysis of the mixed proteins present in sixteen gel strips revealed presence of TaNiR protein in the samples 1–7 and 12–16, in agreement with the nitrite reductase activity distribution in cellular fractions. The presence of TaNiR in soluble protein fractions of T. ammonificans G1 with different charges and masses may indicate that it either forms complexes with those proteins or that it is present in small fragments of the cytoplasmic membrane. However, the MALDI-TOF-MS analysis of the protein bands coeluted with TaNiR did not show a presence of NapGH or other functional proteins involved in DNRA, except for sample 1 where the catalytic subunit NapA was identified.
Reductive titration of oxidized TaNiR with 10 mM hydroxylamine did not result in protein reduction. Obviously, TaNiR cannot function as hydroxylamine dehydrogenase. The nitrite and hydroxylamine-reducing activities of TaNiR at standard conditions with 1 mM nitrite or 10 mM hydroxylamine were 1500 and 250 µmol (min mg)-1, respectively. Like NrfA and other ONR, TaNiR catalyzes the six-electron reduction of nitrite to ammonia. Hydroxylamine was not detected during nitrite reduction.
The pH optimum of the nitrite reduction reaction was 7.5 (Fig. S12). The kinetic characteristics of nitrite and hydroxylamine reduction by TaNiR at optimal pH are summarized in Table 1 and Fig. S13. The nitrite-reducing activity of TaNiR is comparable with those observed for other ONR and pentaheme NrfA containing the lysin-coordinated heme c (CXXCK) in the active center, but is much higher than in other octaheme cytochrome c groups, including the tetrathionate reductase from Shewanella oneidensis MR-1 [39] and the εHAO mentioned above. Members of these groups are considered as alternatives for NrfA and ONR for the DNRA process [22], although, obviously, their ammonifying nitrite reduction is an unspecific side reaction. Apart from the reaction rate, the substrate affinity for nitrite in TaNiR is higher than in εHAO, which also confirms that the ONR family members are more specialized in nitrite ammonification. Finally, this is further corroborated by the difference in ratio of the specific affinity constants kcat/Km for nitrite and hydroxylamine reduction, showing that nitrite is the preferred substrate of TaNiR.
Table 1.
Kinetic parameters of nitrite and hydroxylamine reduction by octaheme and pentaheme cytochrome c proteins with nitrite ammonifying activity.
| Source | Nitrite reduction | Hydroxylamine reduction | ||||
|---|---|---|---|---|---|---|
| kcat,* s−1 | Km, mM | kcat/Km, M−1s−1 | kcat,* s−1 | Km, mM | kcat/Km, M−1s−1 | |
| Octaheme NiR (ONR) | ||||||
| T. ammonificans | 2113 ± 235 (1500) | 0.22 ± 0.06 | 9.6 × 106 | 1201 ± 135 (250) | 33.5 ± 7.2 | 35.8 × 103 |
| Tv. paradoxus [20] | 4160 ± 320 | 0.10 ± 0.02 | 4.2 × 107 | 2950 ± 440 | 230 ± 50 | 12.8 × 103 |
| Tv. nitratireducens [20] | 3100 ± 300 | 0.18 ± 0.05 | 1.7 × 107 | 45 | ||
| Pentaheme NiR (NrfA) | ||||||
| E. coli [50] | 770 | 0.028 | 3.5 × 107 | 2380 | 30 | 7.9 × 104 |
| W. succinogenes [22] | (1066 ± 27) | 159 ± 6 | ||||
| Shewanella oneidensis [51] | 824 ± 33 | 0.023 ± 0.004 | 3.5 × 107 | 2380 ± 160 | 8.3 ± 2.4 | 2.8 × 105 |
| Octaheme tetrathionate reductase (OTR) | ||||||
| Shewanella oneidensis [39] | 2.8 ± 0.4 | 0.005 ± 0.001 | 5.6 × 105 | 849 ± 7 | 2.2 ± 0.4 | 3.85 × 105 |
| εHAO [22] | ||||||
| Campylobacter fetus | (181 ± 3) | 0.89 ± 0.0 | (149 ± 17) | 1.9 ± 0.1 | ||
| Campylobacter curvus | (27 ± 5) | 2. 7 ± 0.3 | (68 ± 3) | 6.6 ± 0.6 | ||
| Caminibacter mediatlanticus | (1.0 ± 0.02) | 1.5 ± 0.5 | (158 ± 2) | 6.0 ± 0.5 | ||
| Nautilia profundicola | 1.2 ± 0.1) | (29 ± 1) | ||||
*kcat was calculated as Vmax/[E]; Specific activity in µmol × min−1 mg−1 is indicated in brackets.
Discussion
In this study, we report on the isolation of the ammonifying Trichlorobacter ammonificans strain G1 from an enrichment culture originally derived from an activated sludge sample. T. ammonificans G1 only grew by nitrate-dependent acetate oxidation under low redox conditions and, unlike most well-characterized DNRA organisms, encodes a DNRA module consisting of an ammonifying ONR apparently forming a complex with the NAP system. This shows that DNRA-catalyzing systems are structurally and functionally more diverse than previously assumed, possibly resulting in underestimation of DNRA potential in the environment when relying on molecular tools using functional marker genes.
Ecological implications
We isolated the ammonifying T. ammonificans G1 from a chemostat enrichment culture where it highly dominated over denitrifiers. However, the difficulties faced during isolation, and the way this bacterium was finally obtained in pure culture indicated that the competition between the two dissimilatory nitrate respiratory processes is not captured well with existing theory on the ratio between electron donor and acceptor as main driver [1–3, 6, 8, 12, 40–44]. Our isolation attempts clearly showed that under conditions where the electron acceptor (nitrate or nitrite) is present at high concentrations additional factors, such as the redox potential and the availability of copper, start to play a dominant role. These three factors appear independent from each other, but in environmental reality they might be tightly interlinked. A combination of high concentrations of reduced electron donor and iron, low nitrate and copper, and low redox potential is found, for example, in sulfidic sediments and flooded soils, which is exactly where high DNRA activity has consistently been observed [42, 43]. The enzyme systems catalyzing DNRA depend on two heavy metals: molybdenum for nitrate reduction and iron for nitrite ammonification, which both are present in sulfidic zones in the form of metal sulfides [45]. Contrastingly, several enzymes in the denitrification pathway (NirK and NosZ) require copper, which is less available in sulfidic sediments, since CuS has a much lower solubility and is more difficult to solubilize than the more abundant amorphous FeS [46]. Thus, we hypothesize that there is a niche segregation between DNRA and denitrification, with the former dominating in reduced nitrate and copper-depleted zones rich in organics, sulfide, and reduced iron, and the latter in more oxidized conditions with lower electron donor but higher copper and NOx availability. This also coincides with the fact that most dedicated DNRA organisms are obligate anaerobes, in contrast to the mostly facultatively anaerobic denitrifiers. Thus, T. ammonificans G1 can be considered as an outstanding example of a highly specialized, low redox potential-dependent DNRA organism. The cultivation-independent evidence for heavy involvement of Geobacterales bacteria in acetate-dependent ammonification in paddy soils with low redox potential [47] corroborates this assumption and shifts the paradigm regarding the members of this environmentally important group of anaerobes as mostly metal-reducers. Nonetheless, still more research on these specialized ammonifiers is necessary to validate our findings in other environmental settings.
Genomic and enzymology analyses evidence a novel DNRA module
Over the last years, our understanding of the DNRA machinery increased considerably, mainly through the description of two types of octaheme cytochrome c proteins, namely ONR and εHAO. There is ample evidence supporting this function for the εHAO based on physiological data (ability for DNRA and genomic analysis), with the most recent organisms including the marine orange mat-forming Beggiatoa [48, 49], several Epsilonproteobacteria [22, 23], cable bacteria from the Desulfobulbia [25], and Limisalsivibrio acetivorans from the Geovibrionaceae/Deferribacteres [49] (see Supplementary text). Contrastingly, the only available pure culture with ONR as best candidate to perform the observed DNRA phylotype was the sulfur-disproportionating Desulfurivibrio alkaliphilus from the Desulfobulbia [25]. Moreover, experimental verification of the proposed functions of these modules is still lacking.
The ONR described here is only the second enzyme after the one of Desulfurivibrio alkaliphilus [24] with a demonstrated role of dissimilatory ammonifying nitrite reductase in vivo. In contrast to Desulfurivibrio, however, the T. ammonificans G1 ONR gene is co-located with the NAP operon, lending strong support that they are forming a new DNRA module where both NapAB and ONR obtain electrons from the membrane-bound menaquinol-oxidizing ferredoxins NapGH. Formation of such a complex might explain the observed inability of strain G1 to initiate growth with nitrite as electron acceptor, implying transcriptional control of the complete module by nitrate, similar to recent observations for the Deferribacteraceae DNRA bacterium Limisalsivibrio acetivorans [49].
Our enzymological characterization of the TaNiR provides first in vivo evidence of function for this type of ONR. Unlike for the two previously described ONR proteins from aerobic sulfur-oxidizing Thioalkalivibrio species [20, 38] where the actual function is still unknown, the TaNiR represents a valid counterpart for the more widely known pentaheme NrfA. Although T. ammonificans G1 is the only species within the Geobacteraceae confirmed to catalyze DNRA using the NAP-ONR system, most members of this family contain either NrfA, ONR, or both systems (Fig. 1 and Supplementary Fig. S5). However, the final product of nitrate reduction was not analyzed for most of these species, thus making it impossible to confirm their DNRA phenotype. Furthermore, some of these species lack known nitrate reductases, but as in most studies nitrate was tested as electron acceptor, it remains to be determined whether these organisms might be able to reduce nitrite to ammonium using either one of these nitrite reductase systems. Thus, our strain is not the only member of the Geobacteraceae possessing ONR, and ONR-catalyzed DNRA might be a common metabolic feature of this particular group of obligate anaerobes that represent important members of the microbial community in various systems, like, for instance, rice paddy soil [47] and wastewater treatment facilities [26].
In conclusion, the pure culture isolation of T. ammonificans G1 showed that it is a highly specialised ammonifying bacterium requiring low redox conditions and copper limitation to outcompete denitrifiers, as for instance prevalent in sulfidic sediments and flooded soils. This demonstrates that the competition between denitrification and DNRA is influenced by factors other than the electron donor to acceptor ratio only. Furthermore, purification and characterisation of the ONR enzyme of T. ammonificans G1 has for the first time proven that this type of octaheme cytochrome c protein functions as true dissimilatory nitrite reductase. Lastly, genomic and proteomic analyses suggest an association with the NAP complex, which implies that DNRA-catalyzing systems are structurally and functionally more versatile than previously assumed. Together, this indicates that non-NrfA type DNRA is more widespread than assumed, possibly resulting in underestimation of DNRA potential in the environment.
Description of Trichlorobacter ammonificans sp. nov
am.mo.ni'fi.cans. N.L. neut. n. ammonium, ammonia; L. part. suff. -ficans, making (from L. v. facere, to make); N.L. masc. part. adj. ammonificans, ammonifying.
Cells are comma-shaped, 1.5–4 × 0.5 μm, motile with a single polar flagellum, and tend to form semicircles and aggregates in old cultures. The culture has a distinctive pink colour, concentrated cell biomass is red. Colony formation was not observed. The dominant polar lipid fatty acids included unsaturated 16:1ω9c (differentiating the novel isolate from the closely related Trichlorobacter species) and two saturated fatty acids commonly present in the known Trichlorobacter species, 14:0 and 16:0 (Table S5). The organism is an obligate anaerobe with a very restricted metabolism limited to acetate-dependent dissimilatory nitrate/nitrite ammonification. H2 can be used as an additional (to acetate) electron donor. Weak growth was also observed with succinate, with only partial nitrate reduction to nitrite. The substrates tested negative with nitrate as acceptor included formate, ethylene glycol, glycine, ethanol, propionate, propanol, lactate, pyruvate, butyrate, butanol, malate, fumarate, valerate, yeast extract, and peptone. The electron acceptors tested negative with acetate as donor included different forms of Fe(III), AQDS, MnO2, arsenate and arsenite, selenate and selenite, fumarate, S8, and S2O32−. The organism is an alkalitolerant neutrophile, growing optimally at pH 6.8–7.2 but tolerating pH values up to 8.5. The optimal growth temperature is 28–30 °C. The genomic G + C content is 61.3%. The type strain is G1 (=DSM 105480, =UNIQEM 10005) isolated from activated sludge.
Supplementary information
Acknowledgements
The authors thank Ben Abbas for technical support, Geert Cremers for assistance in genome assembly and closure, and Berhard Schink for nomenclatural advise. DYS and MCMvL were supported by the Gravitation Program of the Dutch Ministry of Education, Culture and Science (SIAM grant 024.002.002); TVT, NID, AYS and VOP by the Russian Science Foundation (grant 23-74-30004); HK and SL by the Netherlands Organization for Scientific Research (grants VI.Veni.192.086 and 016.Vidi.189.050, respectively). In addition, Russian authors also had support from the Russian Ministry of Science and Higher Education. The LABGeM (CEA/Genoscope & CNRS UMR8030), the France Génomique and French Bioinformatics Institute national infrastructures (funded as part of Investissement d'Avenir program managed by Agence Nationale pour la Recherche, contracts ANR-10-INBS-09 and ANR-11-INBS-0013) are acknowledged for support within the MicroScope annotation platform.
Author contributions
DYS, TVT, GJK, and MCMvL conceived the study and designed the research. GJK, VOP, MCMvL, and SL supervised the project. DYS, TVT, EMvdB, RSH, NID, and AYS performed experiments and data analysis. HK, MP, and SL performed proteomic and bioinformatic analyses. DYS, HK, and SL wrote the manuscript with input from all authors. All authors discussed results and commented on the manuscript.
Data availability
The complete and annotated genome of Trichlorobacter ammonificans G1 is deposited at the European Nucleotide Archive under project PRJEB49551. The mass spectrometry proteomics raw data have been deposited in the ProteomeXchange consortium database with the dataset identifier PXD031212.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dimitry Y. Sorokin, Email: d.sorokin@tudelft.nl
Sebastian Lücker, Email: s.luecker@science.ru.nl.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01473-2.
References
- 1.Kraft B, Strous M, Tegetmeyer HE. Microbial nitrate respiration - genes, enzymes and environmental distribution. J Biotechnol. 2011;155:104–17. doi: 10.1016/j.jbiotec.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 2.Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nat Rev Microbiol. 2018;16:263–76. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 3.Simon J, Klotz MG. Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. BBA Bioenerg. 2013;1827:114–35. doi: 10.1016/j.bbabio.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Welsh A, Chee-Sanford JC, Connor LM, Loffler FE, Sanford RA. Refined NrfA phylogeny improves PCR-based nrfA gene detection. Appl Environ Microbiol. 2014;80:2110–9. doi: 10.1128/AEM.03443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrendt A, de Beer D, Stief P. Vertical activity distribution of dissimilatory nitrate reduction in coastal marine sediments. Biogeosciences. 2013;10:7509–23. doi: 10.5194/bg-10-7509-2013. [DOI] [Google Scholar]
- 6.Yoon S, Cruz-Garcia C, Sanford R, Ritalahti KM, Loffler FE. Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3-/NO2- reduction pathways in Shewanella loihica strain PV-4. ISME J. 2015;9:1093–104. doi: 10.1038/ismej.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorokin DY, Foti M, Tindall BJ, Muyzer G. Desulfurispirillum alkaliphilum gen. nov sp nov., a novel obligately anaerobic sulfur- and dissimilatory nitrate-reducing bacterium from a full-scale sulfide-removing bioreactor. Extremophiles. 2007;11:363–70. doi: 10.1007/s00792-006-0048-8. [DOI] [PubMed] [Google Scholar]
- 8.Cole JA, Brown CM. Nitrite reduction to ammonia by fermentative bacteria: a short circuit in the biological nitrogen cycle. FEMS Microbiol Lett. 1980;7:65–72. doi: 10.1111/j.1574-6941.1980.tb01578.x. [DOI] [Google Scholar]
- 9.King D, Nedwell DB. The influence of nitrate concentration upon the end-products of nitrate dissimilation by bacteria in anaerobic salt-marsh sediment. FEMS Microbiol Ecol. 1985;31:23–28. doi: 10.1111/j.1574-6968.1985.tb01127.x. [DOI] [Google Scholar]
- 10.Sørensen J. Capacity for denitrification and reduction of nitrate to ammonia in a coastal marine sediment. Appl Environ Micro. 1978;35:301–5. doi: 10.1128/aem.35.2.301-305.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiedje JM, Sexstone AJ, Myrold DD, Robinson JA. Denitrification−ecological niches, competition and survival. A Leeuw J Micro. 1982;48:569–83. doi: 10.1007/BF00399542. [DOI] [PubMed] [Google Scholar]
- 12.Kraft B, Tegetmeyer HE, Sharma R, Klotz MG, Ferdelman TG, Hettich RL, et al. The environmental controls that govern the end product of bacterial nitrate respiration. Science. 2014;345:676–9. doi: 10.1126/science.1254070. [DOI] [PubMed] [Google Scholar]
- 13.van den Berg EM, Boleij M, Kuenen JG, Kleerebezem R, van Loosdrecht MCM. DNRA and denitrification coexist over a broad range of acetate/N-NO3- ratios, in a chemostat enrichment culture. Front Microbiol. 2016;7:1842. doi: 10.3389/fmicb.2016.01842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An SM, Gardner WS. Dissimilatory nitrate reduction to ammonium (DNRA) as a nitrogen link, versus denitrification as a sink in a shallow estuary (Laguna Madre/Baffin Bay, Texas) Mar Ecol Prog Ser. 2002;237:41–50. doi: 10.3354/meps237041. [DOI] [Google Scholar]
- 15.Brunet RC, GarciaGil LJ. Sulfide-induced dissimilatory nitrate reduction to ammonia in anaerobic freshwater sediments. FEMS Microbiol Ecol. 1996;21:131–8. doi: 10.1111/j.1574-6941.1996.tb00340.x. [DOI] [Google Scholar]
- 16.Caffrey JM, Bonaglia S, Conley DJ. Short exposure to oxygen and sulfide alter nitrification, denitrification, and DNRA activity in seasonally hypoxic estuarine sediments. FEMS Microbiol Lett. 2019;366:fny288. doi: 10.1093/femsle/fny288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy AE, Bulseco AN, Ackerman R, Vineis JH, Bowen JL. Sulphide addition favours respiratory ammonification (DNRA) over complete denitrification and alters the active microbial community in salt marsh sediments. Environ Microbiol. 2020;22:2124–39. doi: 10.1111/1462-2920.14969. [DOI] [PubMed] [Google Scholar]
- 18.Pang YM, Wang JL, Li SJ, Ji GD. Long-term sulfide input enhances chemoautotrophic denitrification rather than DNRA in freshwater lake sediments. Environ Pollut. 2021;270:116201. doi: 10.1016/j.envpol.2020.116201. [DOI] [PubMed] [Google Scholar]
- 19.Popinako A, Antonov M, Tikhonov A, Tikhonova T, Popov V. Structural adaptations of octaheme nitrite reductases from haloalkaliphilic Thioalkalivibrio bacteria to alkaline pH and high salinity. Plos One. 2017;12:e0177392. doi: 10.1371/journal.pone.0177392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tikhonova T, Tikhonov A, Trofimov A, Polyakov K, Boyko K, Cherkashin E, et al. Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species. FEBS J. 2012;279:4052–61. doi: 10.1111/j.1742-4658.2012.08811.x. [DOI] [PubMed] [Google Scholar]
- 21.Parey K, Fielding AJ, Sorgel M, Rachel R, Huber H, Ziegler C, et al. In meso crystal structure of a novel membrane-associated octaheme cytochrome c from the Crenarchaeon Ignicoccus hospitalis. FEBS J. 2016;283:3807–20. doi: 10.1111/febs.13870. [DOI] [PubMed] [Google Scholar]
- 22.Haase D, Hermann B, Einsle O, Simon J. Epsilonproteobacterial hydroxylamine oxidoreductase (epsilon Hao): characterization of a ‘missing link’ in the multihaem cytochrome c family. Mol Microbiol. 2017;105:127–38. doi: 10.1111/mmi.13690. [DOI] [PubMed] [Google Scholar]
- 23.Hanson TE, Campbell BJ, Kalis KM, Campbell MA, Klotz MG. Nitrate ammonification by Nautilia profundicola AmH: experimental evidence consistent with a free hydroxylamine intermediate. Front Microbiol. 2013;4:180. doi: 10.3389/fmicb.2013.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorup C, Schramm A, Findlay AJ, Finster KW, Schreiber L. Disguised as a sulfate reducer: growth of the deltaproteobacterium Desulfurivibrio alkaliphilus by sulfide oxidation with nitrate. Mbio. 2017;8:e00671–17. doi: 10.1128/mBio.00671-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marzocchi U, Thorup C, Dam AS, Schramm A, Risgaard-Petersen N. Dissimilatory nitrate reduction by a freshwater cable bacterium. ISME J. 2022;16:50–57. doi: 10.1038/s41396-021-01048-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Berg EM, van Dongen U, Abbas B, van Loosdrecht MCM. Enrichment of DNRA bacteria in a continuous culture. ISME J. 2015;9:2153–61. doi: 10.1038/ismej.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfennig N, Lippert KD. Über das vtamin B12-Bedürfnis phototropher Schwefelbakterien. Arch Microbiol. 1966;55:245–56. [Google Scholar]
- 28.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–75. doi: 10.1016/S0021-9258(19)52451-6. [DOI] [PubMed] [Google Scholar]
- 29.Trüper HG, Schlegel HG. Sulphur Metabolism in Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium Okenii. Antonie Leeuwenhoek. 1964;30:225–38. doi: 10.1007/BF02046728. [DOI] [PubMed] [Google Scholar]
- 30.Na SI, Kim YO, Yoon SH, Ha SM, Baek I, Chun J. UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol. 2018;56:280–5. doi: 10.1007/s12275-018-8014-6. [DOI] [PubMed] [Google Scholar]
- 31.Reguera, G & K Kashefi. The electrifying physiology of Geobacter bacteria, 30 years on. Adv Microb Physiol. 2019;74:1–96. [DOI] [PubMed]
- 32.Röling, WFM. The Family Geobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E & Thompson F (eds). The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. Berlin, Heidelberg: Springer; 2014. pp 157–72.
- 33.Doyle R, Marritt SJ, Gwyer JD, Lowe TG, Tikhonova TV, Popov VO, et al. Contrasting catalytic profiles of multiheme nitrite reductases containing CxxCK heme-binding motifs. J Biol Inorg Chem. 2013;18:655–67. doi: 10.1007/s00775-013-1011-7. [DOI] [PubMed] [Google Scholar]
- 34.Cole JA. Anaerobic bacterial response to nitric oxide stress: widespread misconceptions and physiologically relevant responses. Mol Microbiol. 2021;116:29–40. doi: 10.1111/mmi.14713. [DOI] [PubMed] [Google Scholar]
- 35.van Wonderen JH, Burlat B, Richardson DJ, Cheesman MR, Butt JN. The nitric oxide reductase activity of cytochrome c nitrite reductase from Escherichia coli. J Biol Chem. 2008;283:9587–94. doi: 10.1074/jbc.M709090200. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Vine CE, Balasiny BK, Rizk J, Bradley CL, Tinajero-Trejo M, et al. The roles of the hybrid cluster protein, Hcp and its reductase, Hcr, in high affinity nitric oxide reduction that protects anaerobic cultures of Escherichia coli against nitrosative stress. Mol Microbiol. 2016;100:877–92. doi: 10.1111/mmi.13356. [DOI] [PubMed] [Google Scholar]
- 37.Klotz MG, Schmid MC, Strous M, Op den Camp HJM, Jetten MSM, Hooper AB. Evolution of an octahaem cytochrome c protein family that is key to aerobic and anaerobic ammonia oxidation by bacteria. Environ Microbiol. 2008;10:3150–63. doi: 10.1111/j.1462-2920.2008.01733.x. [DOI] [PubMed] [Google Scholar]
- 38.Tikhonova TV, Slutsky A, Antipov AN, Boyko KM, Polyakov KM, Sorokin DY, et al. Molecular and catalytic properties of a novel cytochrome c nitrite reductase from nitrate-reducing haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio nitratireducens. BBA Proteins Proteom. 2006;1764:715–23. doi: 10.1016/j.bbapap.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 39.Atkinson SJ, Mowat CG, Reid GA, Chapman SK. An octaheme c-type cytochrome from Shewanella oneidensis can reduce nitrite and hydroxylamine. FEBS Lett. 2007;581:3805–8. doi: 10.1016/j.febslet.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Akunna JC, Bizeau C, Moletta R. Nitrate reduction by anaerobic sludge using glucose at various nitrate concentrations - ammonification, denitrification and methanogenic activities. Environ Technol. 1994;15:41–49. doi: 10.1080/09593339409385402. [DOI] [Google Scholar]
- 41.Behrendt A, Tarre S, Beliavski M, Green M, Klatt J, de Beer D, et al. Effect of high electron donor supply on dissimilatory nitrate reduction pathways in a bioreactor for nitrate removal. Bioresour Technol. 2014;171:291–7. doi: 10.1016/j.biortech.2014.08.073. [DOI] [PubMed] [Google Scholar]
- 42.Buresh RJ, Patrick WH. Nitrate reduction to ammonium and organic nitrogen in an estuarine sediment. Soil Biol Biochem. 1981;13:279–83. doi: 10.1016/0038-0717(81)90063-8. [DOI] [Google Scholar]
- 43.Matheson FE, Nguyen ML, Cooper AB, Burt TP, Bull DC. Fate of 15N-nitrate in unplanted, planted and harvested riparian wetland soil microcosms. Ecol Eng. 2002;19:249–64. doi: 10.1016/S0925-8574(02)00093-9. [DOI] [Google Scholar]
- 44.Rütting T, Boeckx P, Müller C, Klemedtsson L. Assessment of the importance of dissimilatory nitrate reduction to ammonium for the terrestrial nitrogen cycle. Biogeosciences. 2011;8:1779–91. doi: 10.5194/bg-8-1779-2011. [DOI] [Google Scholar]
- 45.Dahl TW, Chappaz A, Hoek J, McKenzie CJ, Svane S, Canfield DE. Evidence of molybdenum association with particulate organic matter under sulfidic conditions. Geobiology. 2017;15:311–23. doi: 10.1111/gbi.12220. [DOI] [PubMed] [Google Scholar]
- 46.Rickard D. The solubility of FeS. Geochim Cosmochim Acta. 2006;70:5779–89. doi: 10.1016/j.gca.2006.02.029. [DOI] [Google Scholar]
- 47.Masuda Y, Itoh H, Shiratori Y, Isobe K, Otsuka S, Senoo K. Predominant but previously-overlooked prokaryotic drivers of reductive nitrogen transformation in paddy soils, revealed by metatranscriptomics. Microbes Environ. 2017;32:180–3. doi: 10.1264/jsme2.ME16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buckley A, MacGregor B, Teske A. Identification, expression and activity of candidate nitrite reductases from orange Beggiatoaceae, Guaymas Basin. Front Microbiol. 2019;10:644. doi: 10.3389/fmicb.2019.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spring S, Rohde M, Bunk B, Spröer C, Will SE, Neumann-Schaal M. New insights into the energy metabolism and taxonomy of Deferribacteres revealed by the characterization of a new isolate from a hypersaline microbial mat. Environ Microbiol. 2022;24:2543–75. doi: 10.1111/1462-2920.15999. [DOI] [PubMed] [Google Scholar]
- 50.Corker H, Poole RK. Nitric oxide formation by Escherichia coli - dependence on nitrite reductase, the NO-sensing regulator FNR, and flavohemoglobin Hmp. J Biol Chem. 2003;278:31584–92. doi: 10.1074/jbc.M303282200. [DOI] [PubMed] [Google Scholar]
- 51.Youngblut M, Judd ET, Srajer V, Sayyed B, Goelzer T, Elliott SJ, et al. Laue crystal structure of Shewanella oneidensis cytochrome c nitrite reductase from a high-yield expression system. J Biol Inorg Chem. 2012;17:647–62. doi: 10.1007/s00775-012-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete and annotated genome of Trichlorobacter ammonificans G1 is deposited at the European Nucleotide Archive under project PRJEB49551. The mass spectrometry proteomics raw data have been deposited in the ProteomeXchange consortium database with the dataset identifier PXD031212.