Abstract
Proteobacteria primarily utilize acyl-homoserine lactones (AHLs) as quorum-sensing signals for intra-/interspecies communication to control pathogen infections. Enzymatic degradation of AHL represents the major quorum-quenching mechanism that has been developed as a promising approach to prevent bacterial infections. Here we identified a novel quorum-quenching mechanism revealed by an effector of the type IVA secretion system (T4ASS) in bacterial interspecies competition. We found that the soil antifungal bacterium Lysobacter enzymogenes OH11 (OH11) could use T4ASS to deliver the effector protein Le1288 into the cytoplasm of another soil microbiome bacterium Pseudomonas fluorescens 2P24 (2P24). Le1288 did not degrade AHL, whereas its delivery to strain 2P24 significantly impaired AHL production through binding to the AHL synthase PcoI. Therefore, we defined Le1288 as LqqE1 (Lysobacter quorum-quenching effector 1). Formation of the LqqE1-PcoI complex enabled LqqE1 to block the ability of PcoI to recognize/bind S-adenosy-L-methionine, a substrate required for AHL synthesis. This LqqE1-triggered interspecies quorum-quenching in bacteria seemed to be of key ecological significance, as it conferred strain OH11 a better competitive advantage in killing strain 2P24 via cell-to-cell contact. This novel quorum-quenching also appeared to be adopted by other T4ASS-production bacteria. Our findings suggest a novel quorum-quenching that occurred naturally in bacterial interspecies interactions within the soil microbiome by effector translocation. Finally, we presented two case studies showing the application potential of LqqE1 to block AHL signaling in the human pathogen Pseudomonas aeruginosa and the plant pathogen Ralstonia solanacearum.
Subject terms: Microbial communities, Microbial ecology, Bacterial genetics
Introduction
Bacterial quorum sensing is a cell density-dependent intra-/inter-species communication mediated by small chemical signals known as auto-inducers [1–7]. These signals can be released by the bacteria themselves and accumulate in the local environment to concentrations required to regulate transcription of specific genes controlled by quorum-sensing [8–12]. Acyl-homoserine lactones (AHLs) produced by Proteobacteria are the most-studied quorum-sensing signals, which are converted by LuxI family AHL synthases from acyl–acyl carrier protein (acyl-ACP) and S-adenosy-L-methionine (SAM), two substrates necessary for AHL production [13–16]. This catalytic reaction proceeds in two steps: first, the acyl-SAM intermediate is formed by the transfer of the acyl group from the acyl-ACP to the amino group of the SAM, followed by lactonization of methionine moiety and release of methylthioadenosine [13, 17, 18]. As cell density increases, released AHLs accumulate correspondingly to a threshold concentration that can bind LuxR family transcription factors. AHL binding alters the DNA-binding affinity of LuxR, thereby fine-tuning the expression of downstream target genes [8].
Typically, AHL quorum sensing is required for full virulence of pathogenic bacteria [19, 20]. Consequently, bacteriologists have developed several effective strategies to interfere with quorum-sensing, a process known as quorum quenching, to prevent bacterial infection [21–24]. Three major canonical AHL quorum-quenching modes have been discovered and exploited for their potential as anti-infective agents [25–28]. These modes include, but not limited to, the design and utilization of inhibitors to block AHL synthesis, interference with AHL binding with LuxR-type receptors by AHL structural analogs, and degradation of AHL by bacterially-derived enzymes [26, 29–32]. The third was the first to be discovered, and several AHL-degrading enzymes were identified from multiple bacterial systems, including the well-characterized AiiA from Bacillus sp. 240B1, AttM from Agrobacterium tumefaciens A6, MomL from Muricauda olearia, and AiiD from Ralstonia sp. XJ12B [21, 33–35].
Lysobacter enzymogenes OH11 (referred to herein as OH11) is a plant-beneficial, antifungal bacterium originally isolated from the pepper rhizosphere [36–39]. This Proteobacteria did not produce AHL, due to the lack of AHL synthases [28]. Strain OH11 cannot kill Gram-negative bacteria by secreting antibacterial compounds. Instead, it uses a subset of the type IV secretion system (T4SS), termed type IVA secretion system (T4ASS), as an effective antibacterial weapon via cell-to-cell contact [40–42]. T4ASS is mostly employed for DNA delivery and is represented by the VirB/D4 system of Agrobacterium tumefaciens. T4ASS in pathogenic Xanthomonas citri and Stenotrophomonas maltophilia has a distinct function of delivering virulent effector proteins to susceptible competing cells, casing their cell lysis [41]. This distinct T4ASS is also employed by the plant-beneficial strain OH11 in establishing cell-to-cell contacts with ecologically relevant bacterial competitors, as recently mentioned in our laboratory [40]. Although type 4 effector proteins (T4Es) translocated by T4ASS have different domains and action modes, they all carry a conserved C-terminal domain known as the XVIPCD domain. These T4Es can bind VirD4, a T4ASS pathway-specific ATPase that specifically recognizes T4Es and hydrolyzes ATP to provide energy for effector translocation [41, 43]. The strain OH11 genome encodes 16 XVIPCD domain-containing proteins, three of which (Le0908, Le0989, and Le1288) are proved to trigger periplasmic toxicity in Escherichia coli upon arabinose-induced expressions [40]. Through cell-to-cell contact, strain OH11 appears to use T4ASS to deliver toxic T4Es into ecologically relevant competitors, such as soil microbiome members Pseudomonas fluorescens 2P24 (referred to herein as 2P24) to kill them and gain a competitive advantage [40].
During the study of strain OH11-derived T4Es, we were surprised to find that six randomly selected effectors (L4230, Le4232, Le0989, Le0908, Le1288, and Le3316) seemed to exhibit “avirulent” effect in the cytoplasm of E. coli when their gene expressions were induced [40]. This intriguing finding raises at least two possibilities: one is that these avirulent T4Es exhibit species-dependent toxicity, and this has been reported for certain effectors (i.e. TseL) of the type VI secretion system T6SS [44, 45]. Another is that these avirulent T4Es may use previously unknown functions when translocated or expressed in competing cells.
In this study, we focused on testing the second hypothesis by selecting the OH11-2P24 interaction model [46]. As an initial attempt, we aimed to find out which T4E genes of strain OH11 could be expressed in strain 2P24. To this end, we selected laboratory-available plasmids harboring 13 predicted T4E genes (Fig. 1A), and introduced them individually into 2P24 to express each T4E gene in the cytoplasm. We found that 2P24 could express these T4E genes and grow without any problem. We then asked if these T4Es could be expressed in strain 2P24, and what would be their roles? However, while multiple possibilities exist, we were more motivated to investigate whether these T4Es function in strain 2P24 to alter AHL production. This idea was inspired by our recent discovery that the genome of non-AHL-producing strain OH11 encodes several unique AHL-quenching proteins, including LqqP, Le4759, and Le0100 [28]. These proteins are defined as non-canonical AHL quenchers because they do not directly degrade AHL but block AHL synthesis by recognizing multiple AHL synthases through direct protein–protein interaction [27, 28]. We were therefore interested in understanding whether strain OH11 could naturally translocate one or more T4Es as interspecies AHL quenchers. This hypothesis was experimentally supported in this study, as we indeed found that Le1288 is one such functional T4E. Therefore, we defined Le1288 as LqqE1 (Lysobacter quorum-quenching effector 1) in the following contexts.
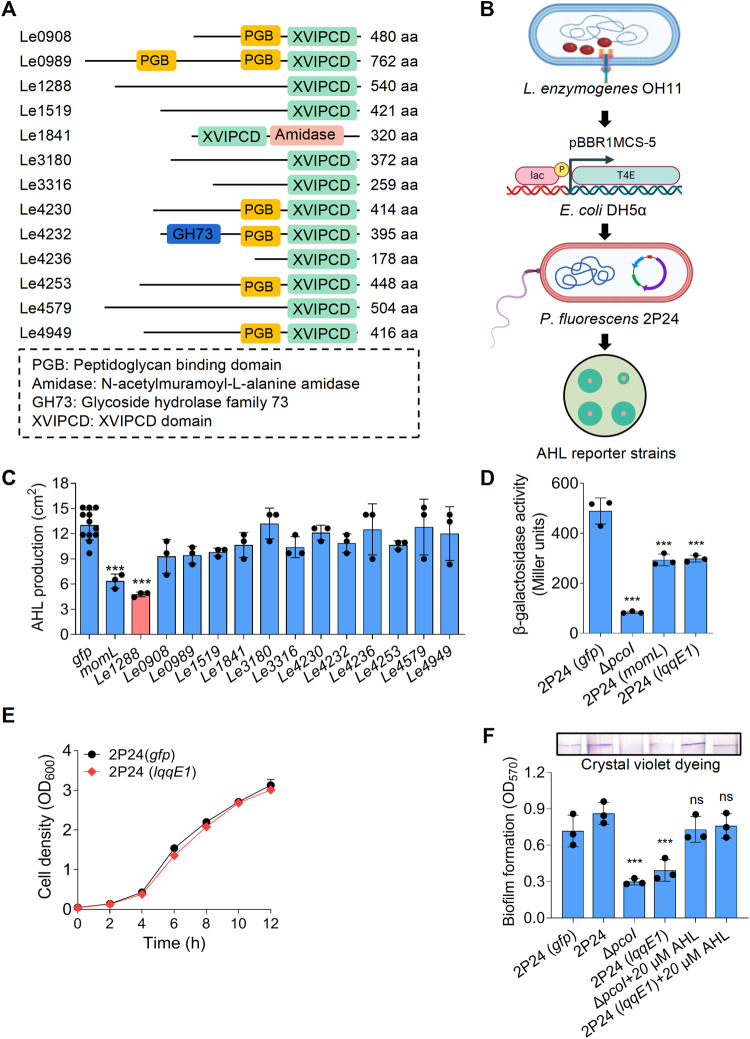
Fig. 1. Heterogenous expression of a predicted effector protein LqqE1 of the Lysobacter enzymogenes type IVA secretion system in Pseudomonas fluorescens impaired the AHL production.
A Schematic illustration of thirteen predicted type IVA secretion system effector (T4E) proteins encoded by L. enzymogenes OH11. B A procedure showing the steps of screening the L. enzymogenes T4E genes with capacities to block the AHL production in P. fluorescens 2P24 after expression by plasmid. The lqqE1 expression significantly blocked the AHL production in P. fluorescens 2P24 that is indicated by the AHL production zone (C) and beta-galactosidase activity (D) determined by an AHL biosensor JZA1. E The lqqE1 expression in P. fluorescens 2P24 did not affect the bacterial growth. F Pseudomonas fluorescens 2P24 expressing lqqE1 showed a significant decrease on biofilm formation that is a known AHL-controlled function. 2P24, wild type of P. fluorescens, served as a positive control; gfp, a negative control; momL encoding a known AHL-degrading enzyme was used as an additional positive control; ΔpcoI, a mutant of P. fluorescens 2P24 lacking the AHL synthase (PcoI) gene and served as an additional negative control. AHL production was calculated by measuring the area of the blue circle around the colony. Average data from three experiments was presented, ±SD. ***p < 0.0001, “ns” stands for not statistically significant, assessed by one-way ANOVA.
Here we present extensive genetic and biochemical data to show how LqqE1 quenched the AHL signaling system of strain 2P24, highlighting a novel quorum-quenching mechanism favoring the killing of strain OH11 against strain 2P24 in bacterial interspecies competition. We show that this natural T4E-triggered interspecies quorum quenching not only appear to be employed by various T4ASS-producting bacteria, but also has broad application potential in quenching AHL quorum-sensing in human and plant bacterial pathogens.
Materials and methods
Bacterial strains, plasmids, and growth conditions
Table S1 lists the strains and plasmids used in this study. Different E. coli strains were grown in Lysogeny broth (LB) medium at 37 °C with the addition of appropriate antibiotics kanamycin (Km), gentamicin (Gm), and tetracycline (Tc) for plasmid maintenance. Ralstonia solanacearum EP1 and its derivatives were cultured in liquid CTG medium (10.0 g of tryptone, 5.0 g of glucose, and 1.0 g of casamino acids per L) at 28 °C, L. enzymogenes OH11, Pseudomonas fluorescens 2P24 and their derivatives were cultured in liquid LB medium or 1/10 TSB at 28 °C, Pseudomonas aeruginosa PAO1 and their derivatives were cultured in liquid LB medium at 37 °C, and their growth was monitored by measuring the optical density at 600 nm (OD600) every 2 h. Each experiment was performed three times and the average OD600 of strains was used to generate the bacterial growth curve.
Genetic methods
In-frame deletions in L. enzymogenes OH11 were generated by homologous double-crossover recombination as previously described [47]. Briefly, two regions flanking the gene of interest were amplified by PCR and cloned into the suicide vector pEX18 (Table S1). Primers used for DNA amplification are listed in Table S2. The final construct was transformed into a wild-type strain by electroporation. Single crossover recombination was selected on LB agar supplemented with Km and Gm. Recombinant cultures were grown in LB without antibiotics for 4 h and then plated on LB agar containing 1/10 (wt/vol) sucrose and Km. PCR was performed using the primers listed in Table S2 to verify the deletion of in-frame genes caused by the double-crossover recombination.
Chromosomal complementation of mutants was performed by the double-crossover recombination approach [47] using the primers shown in Table S2. Briefly, the intact genes or those containing point mutations were amplified by PCR and cloned into the suicide vector pEX18-Gm. The resulting plasmids were transformed into wild type or gene deletion mutants via electroporation. Strains with double-crossover events were selected as described above.
Protein homolog identification
To identify homologous proteins of LqqE1, we ran local BLASTp using LqqE1 as a query to identify the corresponding homologs in selected bacterial genomes. When the E-value is lower than 10−5, similar protein was considered to exist, and the sequence identity with the corresponding homologous protein was usually higher than 25%.
Heterogeneous expression of AHL synthetase gene in E. coli
The coding regions of pcoI from P. fluorescens 2P24, rhlI from P. aeruginosa PAO1, and rasI from R. solanacearum EP1 were amplified by PCR using primers given in Table S2. The PCR products were cloned into the broad host vectors pBBR1-MCS5 and pVSP61 Restrictive using restriction enzyme digestion (Table S1). The plasmid was transformed into E. coli DH5α strain, and the integrity of the plasmid was determined by PCR and Western blotting.
AHL bioassay
AHL plate bioassays were performed as previously described [48]. An effective AHL bioassay strain, Agrobacterium tumefaciens JZA1, was used to determine AHL production [48]. The JZA1 strain was inoculated in 20 mL of LB liquid medium, and the final concentrations of antibiotics used were 30 μg/mL gentamicin, 30 μg/mL spectinomycin, and 12 μg/mL tetracycline, and cultured at 28 °C for 24 h. We then added 10 mL of JZA1 culture to 50 mL of solid LB medium at 50 °C and added 5-bromo-4-chloro-3-indolyl-d-galactoside (X-gal) to a final concentration of 40 mg/mL. After mixing, plates containing JZA1 were prepared, and 5 μL of the AHL samples were seeded on the surface of the plates. The prepared dishes were incubated at 28 °C for 24 h, and the size of blue circle was measured.
In previous work, the β-galactosidase method was used to quantify JZA1-based AHL [48]. Briefly, 20 mL liquid LB containing JZA1 and the antibiotics gentamicin (30 μg/mL), spectinomycin (30 μg/mL), and tetracycline (12 μg/mL) were shaken at 28 °C and 220 rpm for 24 h. 1 mL of JZA1 culture was added to 100 mL of fresh LB medium, followed by 1 mL of cell-free strain 2P24 and its derivative supernatant. The resulting culture was kept at 28 °C and shaken at 220 rpm until the OD600 reached 0.4–0.8, then 200 μL of the culture was added to 800 μL Z-Buffer (8.5 g/L Na2HPO4, 5.5 g/L NaH2PO4·H2O, 0.75 g KCl, 0.246 g/L MgCl2·7H2O, and 27 μL β-mercaptoethanol) in a 2 mL centrifuge tube supplemented with 10 μL SDS and 15 μL chloroform. After spinning for 20 seconds, 100 μL of the chromogenic substrate ONPG (2-Nitrophenyl β-D-galactopyranoside, Aladdin, Shanghai, China) at a concentration of 4 mg/mL was added and this time was recorded as T1. Once the reaction mixture turned yellow, 600 μL of 1 mM Na2CO3 was immediately added to terminate the reaction, and this time was recorded as T2. The solution was further centrifuged for 3 min before measuring the optical density of the supernatant at 420 nm (OD420). AHL activity was quantified as β-galactosidase activity/ (Miller units) and expressed by the formula 1000×OD420/OD600 (T2-T1)/0.2.
Reporter strain CV026 was used for the RhlI-dependent AHL bioassay of P. aeruginosa PAO1 according to the following procedure [49]. The CV026 strain was inoculated in 100 mL of LB liquid medium with a final concentration of kanamycin antibiotic at 30 μg/mL, and incubated at 28 °C for 24 h. Ten milliliters of CV026 culture was then added to 50 mL of solid LB medium at 50 °C. After mixing, 5 μL of AHL sample was seeded on the surface of the plates already containing CV026. The prepared plates were then incubated at 28 °C for 96 h, and the size of purple circle was measured.
Reporter strain YJL01 was used for the RasI-dependent AHL bioassay of R. solanacearum EP1 according to the following procedure [50]. After centrifuging the bacterial culture supernatants (20,000 × g, 30 min, 4 °C) to remove cells, the cells were extracted twice with an equal volume of acidified ethyl acetate (0.1 mL of glacial acetic acid/L). Ethyl acetate extracts was added to a 16-mL glass tube, and the ethyl acetate was evaporated under a gentle stream of N2 gas. The overnight culture of reporter strains YJL01 was then diluted to an OD600 of 0.05 and added to the tube containing signal solution to a final volume of 1.0 mL. Test tubes were then shaken and incubated at 30 °C for 16 h, and the fluorescence was measured as previously described [51].
Biofilm formation assays
Biofilm formation assays of P. fluorescens 2P24 was performed as previously described [52]. In summary, Pseudomonas fluorescens 2P24 and its derivative strains were grown in liquid LB until OD600 reached 1.0. Then 250 μL of bacterial culture was added to a 2-mL tube containing 250 μL of fresh LB and incubated at 28 °C for 48 h. Biofilm were washed 3 times with sterilized ultrapure water and then treated with 0.3% crystal violet (CV) for 15 min. Next, it was washed 3 times again with ultrapure water and resuspended in 95% ethanol (1 mL). Biomass was quantified at 570 nm with a spectrophotometer (Biotek, USA).
Identification of key amino residues critical to LqqE1 function by random mutagenesis using an E. coli mutant strain
Random mutagenesis assays were performed as previously described [53]. Briefly, lqqE1 was cloned into pBBR1-MCS5 and transformed into E. coli XL-1 Red competent cells. Since E. coli XL-1 Red strain lacks functional DNA repair pathway genes (mutS, mutD, and mutT), it is capable of introducing random mutations at a very high rate [53]. Approximately 40 ng of plasmid DNA was used for transformation, and the transformation plates were incubated at 37 °C for 24–30 h. All colonies scraped off the transformation plates using a sterile loop were inoculated into 100 mL LB broth containing appropriate antibiotics. After overnight cultivation at 37 °C, a mutagenized plasmid DNA library was prepared using the plasmid Miniprep kit (OmegaD6943, USA). The purified plasmid library was then fully transformed into an AHL-producing recombinant E. coli strain harboring heterogeneously expressed pcoI from strain 2P24, and AHL was finally detected by the AHL biosensor JZA1 to screen for strains that could not inhibit PcoI-dependent AHL production. The resulting strains were grown in LB, and plasmids within the resulting strains were extracted and sequenced to identify mutated residues, which were further confirmed by a site-specific mutagenesis approach previously descried in the laboratory [27, 28].
Protein structure prediction
The LqqE1 and PcoI structure predictions were performed using the https://www.uniprot.org/online site [54–56]. UniProt entries were found by using the peptide search function of the query peptide sequences. Additionally, AlphaFold was applied to generate a per-residue confidence score (pLDDT) between 0 and 100. Some regions with low pLDDT may be unstructured. We then mapped the amino acid residue positions of the LqqE1 and PcoI structure models using the structure tool of the online site. The Uniprot and AlphaFold accession numbers for LqqE1 and PcoI are A0A3N2RLI6 and Q6ITU1, respectively.
Bacterial two-hybrid assay
The BacterioMatch II two-hybrid (B2H) system (Agilent Technologies, USA) was used to detect protein–protein interactions [57]. In brief, the coding region of LqqE1 target protein was cloned into the pBT plasmid, and the coding region of PcoI, PcoR or VirD4 target proteins was cloned into the pTRG plasmid, which were transformed into the blue MRF Kan strain of E. coli. Positive controls included plasmids pBT-GacS and pTRG-GacS (Table S1), whilst negative controls included transformants with empty pTRG and pBT vectors. Selective medium was spotted onto each co-transformant and cultivated at 28 °C for two days. If the two proteins physically interact, transformed E. coli strain carrying both vectors should thrive on reference medium (+3AT+Strr) based on minimal medium (M9) supplemented with 5 mM 3-AT, 2 μg/mL Str, 12.5 μg/mL tetracycline, 34 μg/mL chloramphenicol, and 30 μg/mL kanamycin as described in previous work [57]. As previously mentioned, LB agar is a non-selective medium (-3AT-Strr) that includes 12.5 μg/mL tetracycline, 34 μg/mL chloramphenicol, and 30 μg/mL kanamycin [57]. This medium was designed to ensure successful transformation of both vectors into E. coli blue MRF Kan.
Pull-down assays
Pull-down assays were performed as previously described [58]. Equal volumes of His-tagged protein and FLAG-tagged protein were mixed together in 1.5 mL of PBS buffer. The mixture was then supplemented with 50 μL of anti-His magnetic beads (Bimake, Shanghai, China), and incubated at 4 °C overnight. Magnetic beads were collected at 4 °C and washed 3 times with PBS containing 1% Triton X-100 to remove non-specifically bound proteins. Proteins captured on FLAG-beads were eluted by boiling with 4× SDS loading dye for 10 min, followed by SDS-PAGE and western blotting. Protein detection involved the use of FLAG-(M20008S) and His-(ab18184) specific antibodies (Abmart, Shanghai, China).
T4SS effector translocation assay
The effector translocation assay was performed as described previously [59]. The step-by-step procedures and schematic diagram of the experiment were shown in Fig. 2D and Fig. S2A, respectively. In brief, the respective L. enzymogenes donor strains-OH11/Cre-LqqE1, OH11/Cre, and ΔvirD4/Cre-LqqE1, and the recipient strain 2P24/loxP were grown together overnight in liquid LB at 28 °C, and then diluted 100-fold by fresh LB and cultured to OD 1.0 at 28 °C. Afterward, cells from each bacterial culture were collected by centrifugation (5000 rpm at 28 °C for 5 min) and resuspended with sterile water. The resulting culture of the recipient strain was mixed with each donor strain at a 10:1 ratio and inoculated on the LB plates at 28 °C for 16 h.
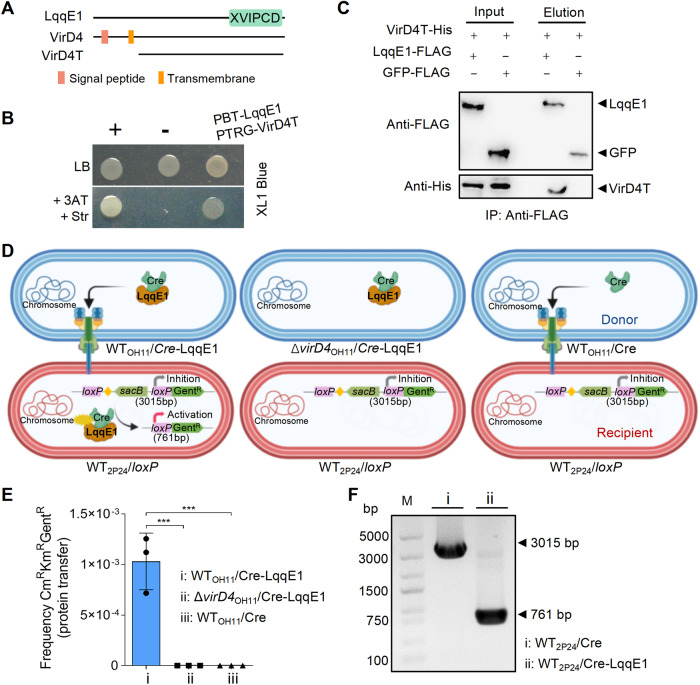
Fig. 2. LqqE1 is a T4E that could be translocated into P. fluorescens by L. enzymogenes using the T4ASS.
A Schematic maps of LqqE1 and VirD4, a T4ASS-pathway-specific ATPase. B Bacterial two-hybrid assays showing LqqE1 bound to VirD4. pTRG-VirD4 and pBT-LqqE1 were co-expressed in E. coli XL-1-blue. “+”, positive control with co-expression of plasmids pBT-GacS and pTRG-GacS; “-”, negative control with co-expression of empty pTRG and pBT vectors. LB, a non-selective condition; +3AT+Str, a selective condition. C Pull-down assays showing LqqE1-FLAG, but not the GFP-FLAG control, directly interacted with VirD4T-His. Immunoprecipitation (IP) was carried out using anti-FLAG antibody and Western blot was performed using anti-FLAG and anti-His antibodies. D Schematic diagram of the LqqE1 translocation assay. Lysobacter enzymogenes OH11 carrying the Cre-LqqE1 hybrid protein served as a donor strain resistant to kanamycin and gentamicin, while P. fluorescens 2P24 harboring pBBR-loxP as a recipient strain resistant to kanamycin and chloromycetin. No parent grew on selective medium containing kanamycin, gentamicin, and chloromycetin. The translocation of Cre-LqqE1 hybrid protein to the recipient strain results in the excision, through recombination at the loxP sites, of the DNA fragment that reconstitutes a functional loxP-GentR translational fusion. Only the recombined strain can survive on selective medium. Yellow diamond, transcriptional terminator; sacB, levansucrase; GentR, gentamycin acetyltransferase. E Transfer of Cre-LqqE1 hybrid protein from strain OH11 to strain 2P24. As described in Materials and Methods, protein transfer was performed by using either WTOH11 or ΔvirD4OH11 expressing the designated protein fusions as the donor strains and strain 2P24 expressing the pBBR-loxP as the recipient strains. The data show the efficiency of recombination (CmRKmRGentR/CmRKmR CFU recovered; recombined/total recipients). ***p < 0.001. The results are expressed as mean ± SD. F Analysis of Cre recombination by gel electrophoresis. The pBBR-loxP DNA fragment after Cre recombination can be amplified to 761-bp in size with M13F/M13R primers, and 3015-bp in size can be amplified without recombination. All experiments were repeated at least three times.
The mixtures were eluted separately and spotted on two different LB plates, one containing 50 μg/ml kanamycin and 100 μg/ml chloromycetin that can kill donor strains; the other containing 50 μg/ml kanamycin, 100 μg/ml chloromycetin, and 100 μg/ml gentamicin was used to select the Cre-recombined strain. The two plates were incubated in parallel at 28 °C for 48 h for calculating protein translocation efficiency.
Next, the strains surviving the selective medium containing 50 μg/ml kanamycin, 100 μg/ml chloromycetin, and 100 μg/ml gentamicin were amplified and cultured to extract plasmids and transformed into E. coli, which were spread on LB plate containing 25 μg/ml gentamicin. The grown colonies were amplified with M13F/M3R primers (Table S2) and the products were sent out for sanger sequencing.
Protein expression and purification
The PcoI proteins were expressed as His6-fusions and the LqqE1 protein was expressed as MBP-fusion protein and purified by affinity chromatography. The coding region of pcoI was cloned into plasmid pET30a (Table S1) using primers listed in Table S2. The coding region of LqqE1 was cloned into plasmid pMal-p2X (Table S1). The genes were then expressed in E. coli BL21(DE3) (Table S1). The His6-fusion proteins were purified from 400 mL E. coli BL21(DE3) carrying the pET30a or pMal-p2X plasmid derivatives using Ni-NTA resin (GE Healthcare, Shanghai, China) and amylose resin (New England Biolabs, Beijing, China). The strains were grown to OD600 of 0.4 at 37 °C, and then 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, Sigma, USA) was used to induce gene expression at 15 °C for 24 h to obtain PcoI and LqqE1. The concentration of purified proteins was determined by BCA protein assay kit (Sangon Biotech, Shanghai, China). Finally, protein purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Microscale thermophoresis assay (MST)
The protein–protein binding affinities were determined by MST using Monolith NT.115 as previously described [57]. In brief, the MBP-LqqE1 protein or MBP protein was labeled with the fluorescent dye RED-NHS (MO-L011) via lysine residues conjugation. A constant concentration (20 nM) of MBP-LqqE1 protein or MBP protein in MST buffer was titrated against PcoI-His protein (concentration range from 0.153 nM to 5 μM). MST premium-coated capillaries (Monolith NT.115 MO-K025) were used to load the samples into the MST instrument at 25 °C using medium MST power and 20% LED power. All experiments were conducted in triplicate. Data were analysed using NanoTemper Analysis v2.3 software (NanoTemper Technologies).
For LqqE1 and S-adenosy-L-methionine (SAM) competitive PcoI binding assay, the PcoI-His protein was labeled with the fluorescent dye RED-Tris-NTA (MO-L018). A constant concentration (100 nM) of PcoI-His protein in MST buffer was titrated against SAM (Solarbio, Beijing, China, S9990) (concentration range from 38.1 μM to 1.25 mM). The labeled PcoI-His protein was incubated with MBP-LqqE1 protein at room temperature for 10 min, and the same operation was performed for the control MBP protein. MST premium-coated capillaries (Monolith NT.115 MO-K025) were used to load the samples into the MST instrument at 25 °C using medium MST power and 90% LED power. All experiments were conducted in triplicate. Data were analyzed using NanoTemper Analysis v2.3 software (NanoTemper Technologies).
Pyocyanin assay
Determination of pyocyanin was performed as previously described [60]. In brief, the P. aeruginosa PAO1 strain was inoculated in LB medium at the proportion of 1% and cultured at 37 °C and 180 rpm for 24 h. The bacterial culture medium was centrifuged at 10,000 rpm for 3 min to remove bacteria, and then filtered into a new sterilized EP tube with a 0.22 μm sterile microporous filter. Five milliliters of the above supernatant and 3 mL of chloroform were mixed thoroughly, shook and extracted, followed by standing at room temperature for 5 min, and then centrifuged at 10,000 rpm for 10 min. One milliliter chloroform layer was mixed with 0.5 mL 0.2 M hydrochloric acid solution and stood evenly. After centrifugation at 10,000 rpm for 10 min, the supernatant was taken to determine the absorbance value at 520 nm.
Rhamnolipid assay
Determination of rhamnolipids was performed as previously described [61]. In brief, the P. aeruginosa PAO1 strain was inoculated in 1% M9 medium (Na2HPO4 6 g/L, KH2PO4 3 g/ L, NH4Cl 1 g/L, NaCl 0.5 g/ L, 1 mM MgSO4·7H2O, 0.1 mM CaCl2, and glucose 20 g/ L), cultured at 37 °C and 180 rpm for 24 h. The bacterial culture medium was centrifuged at 10,000 rpm for 12 min, and 1 mL of supernatant (pH adjusted to 2.0) was mixed with 1 mL of ethyl acetate and vortexed vigorously. After standing at room temperature for layer separation, the organic layer was transferred to a new sterile EP tube and dried in a vacuum pump, which was then dissolved in 500 μL sterile distilled water. One hundred microliters of this solution and 900 μL of 0.19% lichenol-concentrated H2SO4 solution (terbanol: 0.475 g, concentrated sulfuric acid: 139 mL, and distilled water: 111 mL) were fully mixed, then dipping in 80 °C water for 30 min, and cooled to room temperature. The absorbance of the above solutions at 421 nm were determined.
Swimming assay
The swimming assay of R. solanacearum EP1 was carried out as previously described [50]. In brief, 1 μL bacterial suspension (OD600 = 0.1) was inoculated in the center of plate with soft motility agar (10.0 g bacteriological peptone, 3.0 g bacteriological yeast extract, 3.0 g bacteriological agar per L). After inoculation, the plates were incubated at 28 °C for 72 h for the swimming assay. The diameter of the colony reflects the strength of swimming motility.
Bacterial competition assays
For fluorescent microscopy, plasmid pYC12 carrying mCherry gene driven by the plasmid constitutive promoter (Ptac) was introduced into the killer strain - L. enzymogenes OH11. Similarly, plasmid pBBR1-MCS5 with a constitutively expressed gfp gene was transformed into the competing strains - P. fluorescens 2P24; further, plasmid pBBR1-MCS5 with a constitutively expressed lqqE1 gene and pUCP26 with a constitutively expressed gfp gene were co-transformed into the P. fluorescens 2P24 competing strain. All strains were cultured to OD600 1.0 at 28 °C, then centrifuged and resuspended with sterile water. After mixing in different proportions, 5 μL mixed bacterial solution were taken for culture on 1/10 TSB agar dishes, and observed by fluorescence microscope (Nikon SMZ25, Nikon, Japan) after culture for 24 h. After that, the number of recovered CFU of competing cells was determined and the mean log10 CFU calculated. All the experiments were carried out three times and three replicates were used for each treatment.
Results
LqqE1 was a predicted L. enzymogenes T4E acting as an interspecies AHL-quencher
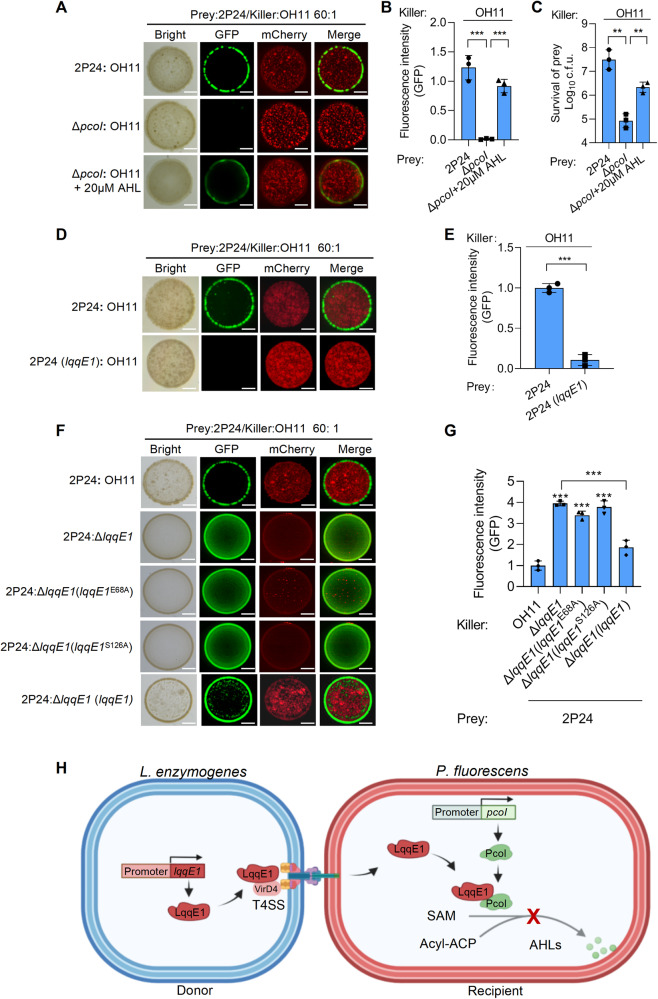
To test whether strain OH11 could inject any T4E into bacterial competitors to achieve AHL quorum quenching, we chose as a working model the laboratory-available strain 2P24, a soil-borne, AHL-producing antifungal bacterium with a similar niche with strain OH11 [46]. The genome of strain OH11 encodes a total of 16 T4E genes, 13 of which were cloned into pBBR1-MCS5 in the laboratory as previously descried [40]. These 13 available recombinant plasmids were introduced into the cytoplasm of strain 2P24 (Fig. 1A, B) to mimic the expression of these T4E genes in strain 2P24 after natural translocation. As previously descried, we detected AHL by the AHL biosensor JZA1 [48] and indeed found a T4E called Le1288 (LqqE1), whose gene expression in strain 2P24 significantly reduced the amount of AHL, while the remaining 12 T4E genes did not (Fig. 1C and Fig. S1A). The role of LqqE1 in blocking AHL production in strain 2P24 was further validated by quantifying the amount of AHL revealed by β-galactosidase activity (Fig. 1D). momL was used as a positive control because it encodes a known AHL-degrading enzyme [33] and was efficient in reducing AHL levels in strain 2P24 (Fig. 1D). Expression of lqqE1 in strain 2P24 was found not to impair bacterial growth under AHL assay conditions compared to a gfp control (Fig. 1E). To functionally support above findings, we further tested whether lqqE1 expression in strain 2P24 affected biofilm formation, a known AHL-controlled function [52]. As expected, lqqE1-expressing strain 2P24 significantly decreased biofilm biomass compared to the gfp-expressing control strain 2P24 (Fig. 1F). This phenotype was consistent with the 2P24-derived AHL-deficient mutant ΔpcoI, which contains an in-frame deletion of the AHL synthase pcoI gene (Fig. 1F). The biofilm defects of ΔpcoI or strain 2P24 were rescued to a level similar to the gfp-expressing control strain 2P24 (Fig. 1F) when cultured in medium supplemented with commercial AHL standard [N-3-oxo-octanoyl-L-homoserine lactone, 3-oxo-C8-HSL] in the physiological range (20 μM). Together, these results revealed that LqqE1, an OH11-drived T4E, might act as an interspecies AHL-quencher.
Considering that strain OH11 was originally isolated from the pepper rhizosphere, we aimed to understand whether LqqE1-triggered interspecies quorum quenching also occurred in the pepper rhizosphere microbiome. To test this, we searched the available Pseudomonas library in laboratory and selected an AHL-producing Pseudomonas A8, an unpublished strain isolated from the pepper rhizosphere that shared the same ecological niche as strain OH11. Strain A8 was further identified as Pseudomonas chlororaphis (Fig. S1B). We found that expression of lqqE1 in P. chlororaphis A8 also significantly reduced AHL amount (Fig. S1C), providing additional evidence to support the conclusion that LqqE1 might act as a natural interspecies AHL-quencher. In the following study, considering that we have set up a well-established system for studying the OH11-2P24 interspecies interaction, we further focused on this system to reveal the underlying mechanism by which LqqE1 achieved AHL quorum quenching in strain 2P24.
LqqE1 is a bone fide T4E that was naturally translocated to strain 2P24
To address the question of whether LqqE1 is a bone fide T4E, we first investigated whether LqqE1 binds to VirD4, as this protein–protein interaction is a prominent feature of T4ASS [41, 43, 62]. Both bacterial two-hybrid (B2H) and pull-down assays indicated that LqqE1 did interact directly with VirD4 (Fig. 2A–C), providing evidence that LqqE1 is a T4E. Next, we tested whether LqqE1 could naturally be translocated from strain OH11 to strain 2P24 using T4ASS. To achieve this, we modified the genetic Cre/loxP system to monitor LqqE1 translocation through cell-to-cell contact between strain OH11 and strain 2P24 [59, 63]. We then constructed three plasmids including pBBR-loxP, pBBR-Cre, and pBBR-Cre-LqqE1, and transformed the pBBR-loxP plasmid into wild-type strain 2P24 to generate the recipient strain 2P24/loxP. We also generated three donor strains, including WTOH11/Cre-LqqE1, ΔvirD4OH11/Cre-LqqE1, and WTOH11/Cre. We expected that the recombinant Cre-LqqE1 vector could be translocated to strain 2P24, and the fused Cre could cleave the loxP element to confer gentamicin resistance, whereas Cre-LqqE1 in ΔvirD4 and Cre in WTOH11 did not (Fig. 2D). We first mixed the three donor strains individually with the recipient strain at a ratio of 1:10 and inoculated these mixed cultures on LB plates. One day after inoculation, we spread the mixed cultures onto selective LB plates containing 100 μg/ml chloramphenicol, 50 µg/mL kanamycin, and 50 µg/mL gentamicin to select for strain 2P24 derivatives with translocated LqqE1. We made this choice because once Cre-LqqE1 was translocated to the recipient strain 2P24/loxP, the loxP element (3015 bp) could be specifically cleaved to generate a short fragment loxP-GentR (761 bp), which would result in gentamicin resistance. With this strategy, we found that only the WTOH11/Cre-LqqE1 and 2P24/loxP pair grew on selective LB plates (Fig. 2E), whereas the T4ASS-deficient mutant ΔvirD4/Cre-LqqE1 and 2P24/loxP pair or negative control, the WTOH11/Cre and 2P24/loxP pair, failed to do so (Fig. 2E). These results indicated that Cre-LqqE1 could naturally be translocated to the cytoplasm of strain 2P24 through strain OH11 using T4ASS. To further verify this, we carried out PCR to test the size of the loxP-GentR fragment. As expected, we found that strain 2P24 carrying the loxP-GentR fragment generated a 761-bp product, while the control plasmid generated a 3015-bp product (negative control) (Fig. 2F). DNA sequencing also proved that the loxP element was correctly cleaved into the loxP-GentR fragment (Fig. S2A, B). Overall, we thought that LqqE1 is a bone fide T4E that was naturally injected into strain 2P24 by strain OH11.
Identification of key amino acid residues required for LqqE1 function
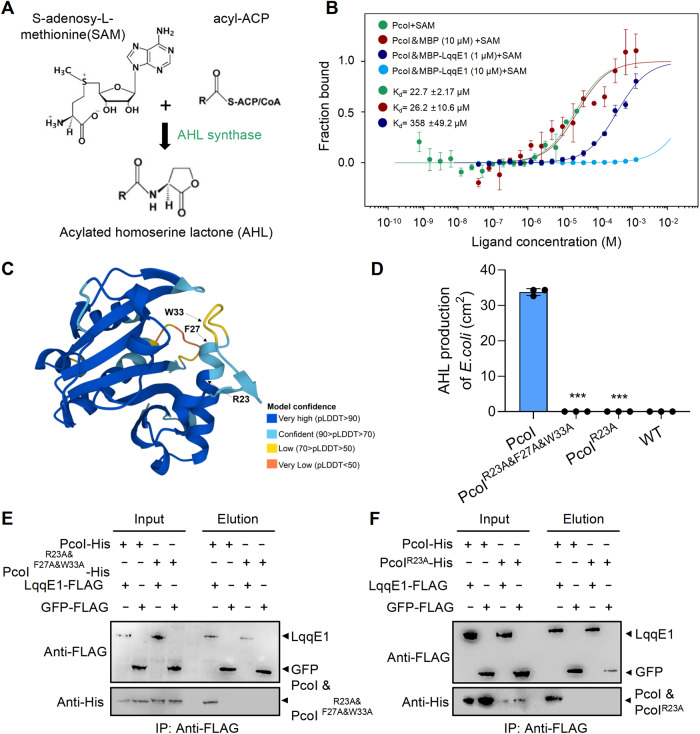
To double validate the role of LqqE1 in quenching the AHL system and facilitate the identification of key amino acid residues responsible for LqqE1 function, we switched our working model from strain 2P24 to an AHL-producing recombinant E. coli strain harboring a heterogeneously expressed pcoI gene from strain 2P24 (Fig. 3A). We found that expression of lqqE1 in this recombinant E. coli also significantly impaired the PcoI-dependent AHL production (Fig. 3A, B). This AHL inhibition effect was similar to that of the AHL-degrading enzyme MomL (Fig. 3B and Fig. S3A). Through BLASTP searches, we found that LqqE1 was defined as a hypothetical protein in most cases. To identify the amino acid residues required for LqqE1 function, we first searched for LqqE1 homologous, which led to the finding that LqqE1 was conserved in many bacterial species (Fig. 3C). Notably, many representative T4ASS-producing bacteria were found to encode LqqE1 homologs (Fig. 3C). We selected the laboratory-available strains (marked with an asterisk) in Fig. 3C as experimental material to test whether their genome-encoded LppE1 homologs have an AHL-quenching effect. Using AHL-producing recombinant E. coli (Fig. 3A), we found three LqqE1 homologs, GLE4749 from L. enzymogenes C3, Lg4270 from L. gummosus OH17, and XAC0574 from X. citri 306 were functionally active. Individual expression of GLE4749, Lg4270 or XAC0574 genes significantly suppressed the amounts of AHL in recombinant E. coli, similar to LqqE1 (Fig. 3D and Fig. S3B).
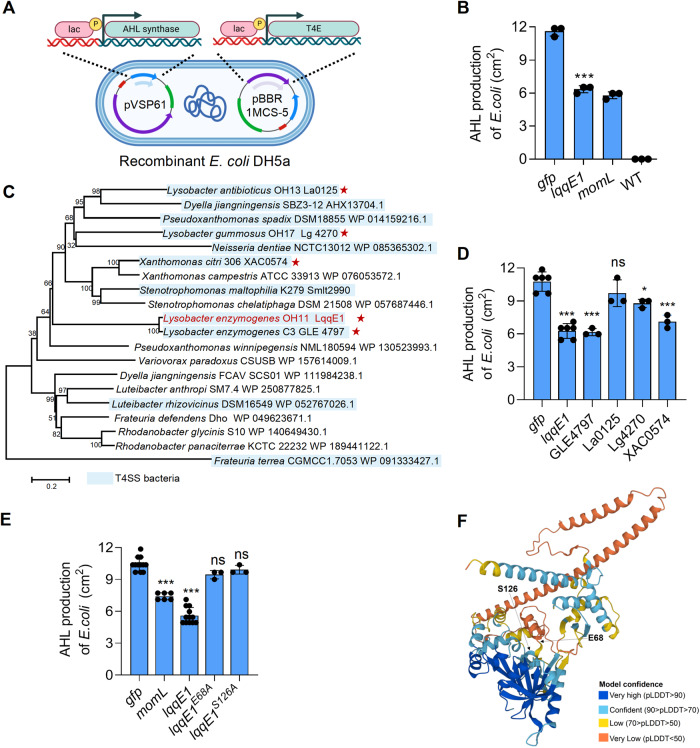
Fig. 3. Identification of key amino residues responsible for LqqE1 functioning as a quorum-quenching protein.
A A schematic map showing how to co-express the lqqE1 gene with AHL synthase genes by plasmids in E. coli. B Co-expression of the lqqE1 gene with the AHL synthase PcoI gene of P. fluorescens 2P24 in E. coli significantly reduced AHL production. Both the wild-type E. coli and the plasmid harboring gfp were used as negative controls; MomL, a known AHL-degrading enzyme, served as a positive control. C The distribution of the LqqE1 in diverse bacterial genomes. The gene name/NCBI accession number of the LqqE1 homologs was listed after the bacterial species. T4ASS-producing bacteria were highlighted by light blue. Red asterisk indicated these LqqE1 homologs were selected for testing their AHL-quenching activity. D Three selected LqqE1 homologs (GLE4749 from L. enzymogenes C3, Lg4270 from L. gummosus OH17, and XAC0574 from Xanthomonas citri 306) showed an AHL-quenching activity when their genes were co-expressed with the PcoI gene in E. coli, according to the protocol shown in (A). E Co-expression of the pcoI gene with lqqE1 variant genes in E. coli showed that two residues (E68, S126) are required for LqqE1 to execute AHL-quenching activity. lqqE1 variant genes were generated by site-directed mutagenesis, where each residue was substituted by alanine (A). gfp, a negative control; momL, a positive control. F A LqqE1 structural modeling predicted by AlphaFold Protein Structure Database, and two key functions residue (E68, S126) shared in LqqE1 were shown in the predicted structure. In all panels, average data from three experiments was presented, ±SD. ***p < 0.0001, *p < 0.05, “ns” stands for not statistically significant, assessed by one-way ANOVA.
To identify key amino residues required for LqqE1 function, we introduced mutations into the pBBR-lqqE1 plasmid by using the E. coli mutator strain XL-1 Red [53], which was described in details in the Materials and Methods section. After screening, we found two PcoI-harboring E. coli colonies generated AHL amounts similar to the gfp control. Sequencing of the DNAs of these two plasmids revealed mutations at E68 and S126 in LqqE1. Subsequently, we replaced E68 and S126 with alanine (A), and each LqqE1 point mutation variant was expressed in AHL-producing recombinant E. coli harboring PcoI. We found that these two amino acid residues, E68 and S126, were essential for the AHL-quenching function of LqqE1 (Fig. 3E and Fig. S3C). As a control, point substitution of Q48, S494, and Q526 by alanine did not affect the LqqE1-triggered quenching function of AHL (Fig. S3D). Meanwhile, Western blotting revealed that each substitution did not affect the abundance of lqqE1 expression in E. coli (Fig. S3E). Furthermore, we mapped the positions of these two crucial functional residues (E68 and S126) to the predicted LqqE1 structural model (Fig. 3F) in the AlphaFold Protein Structure Database. On top of these, sequence alignment revealed that E68 and S126 were conserved in LqqE1 and its three functionally active homologs (GLE4749, Lg4270, and XAC0574) (Fig. S3F). These results supported a role for LqqE1 as an interspecies AHL-quencher, which appeared to be encoded by multiple T4ASS producing bacteria.
LqqE1 targets multiple AHL synthases through direct physical interactions
How does LqqE1 function in strain 2P24? To address this question, we first tested whether LqqE1 is an AHL-degrading enzyme. We ruled out this possibility because we found that LqqE1 was unable to degrade two commercial AHLs: 3-oxo-C10-HSL [N-(3-oxodecanoyl)-L-homoserine lactone] and C8-HSL (N-octanoyl-L-homoserine lactone) (Fig. S4A, B). We then tested whether LqqE1 functions by directly binding to PcoI. By bacterial two-hybrid (B2H), we indeed found that LqqE1 interacted with PcoI, but not PcoR, the PcoI-paired AHL-binding transcription factor (Fig. 4A). Further pull-down assays confirmed the LqqE1-PcoI physical interaction, where LqqE1-FLAG could bind PcoI-His, but not PcoR-His (Fig. 4B, C). To quantify LqqE1-PcoI binding affinity, we further applied the microscale thermophoresis (MST) method and our results showed that MBP-LqqE1 could bind PcoI-His with moderate to strong affinity (Kd, 330 nM; Fig. 4D).
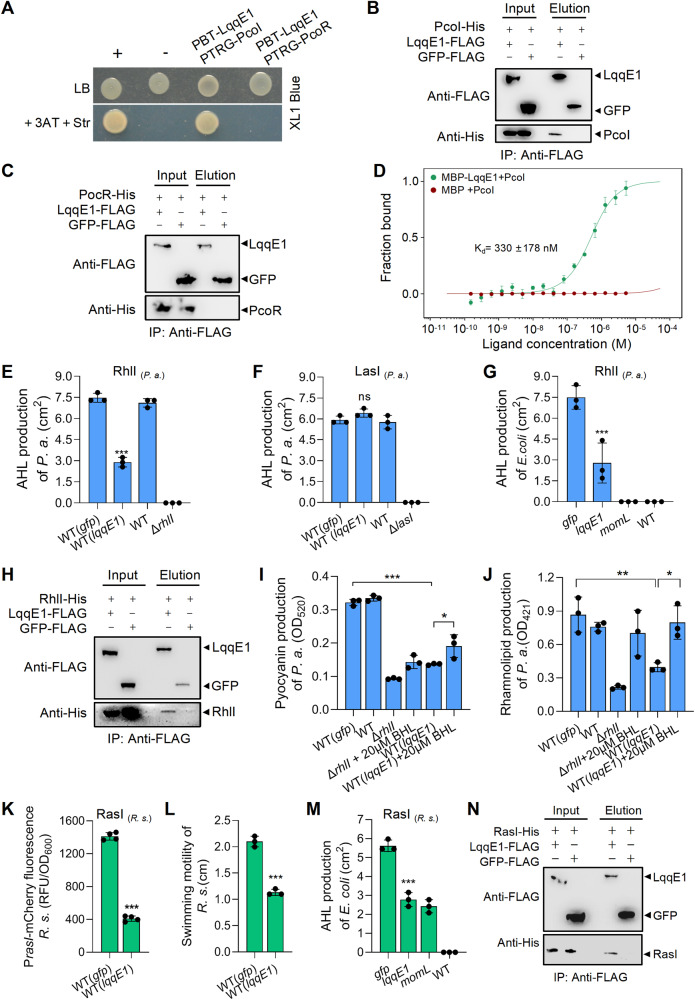
Fig. 4. LqqE1 targets multiple AHL synthases via physical interactions.
A Bacterial two-hybrid assay showing the direct binding between LqqE1 and PcoI but not PcoR. PcoI, the AHL synthase of P. fluorescens 2P24; PcoR, a neighbor of PcoI, is a LuxR-type receptor protein of AHL. Pull-down assay confirmed the direct binding of LqqE1-FLAG with PcoI-His (B) but not PcoR-His (C). Immunoprecipitation (IP) was carried out using anti-FLAG antibody and western blot was performed using anti-FLAG and anti-His antibodies. D Microscale thermophoresis (MST) showed MBP-LqqE1 bound to PcoI-His with moderately strong affinity (Kd, 330 nM). The lqqE1 expression in Pseudomonas aeruginosa PAO1 remarkably inhibited the RhlI-dependent (E), but not the LasI-dependent (F) AHL production. Both RhlI and LasI are AHL synthases of P. aeruginosa, and they are responsible for generating butyl-homoserine lactone (C4-HSL) and N-(3-oxododecanoyl)-L-homoserine lactone (3-oxo-C12-HSL), respectively. G Co-expression of the lqqE1 gene with the AHL synthase rhlI gene of P. aeruginosa PAO1 in E. coli significantly reduced AHL production. P. s. stands for P. aeruginosa PAO1. H Pull-down assay showing the LqqE1-FLAG bound to RhlI-His. The lqqE1 expression in P. aeruginosa PAO1 remarkably impaired the production of pyrazocyanin (I) and rhamnoolipids (J), two RhlI-controlled functions. K The lqqE1 expression in Ralstonia solanacearum EP1 inhibited the RasI-dependent AHL production. RasI is the major AHL synthase encoded by R. solanacearum EP1. R. s. stands for R. solanacearum EP1. L The lqqE1 expression in R. solanacearum EP1 impaired the RasI-controlled swimming motility. M Co-expression of the lqqE1 gene with the EP1 AHL synthase rasI gene in E. coli reduced AHL production. N Pull-down assay showing the LqqE1-FLAG bound to RasI-His. In all panels, average data from three experiments was presented, ±SD. ***p < 0.0001, **p < 0.01, *p < 0.05, “ns” stands for not statistically significant. Assessed by one-way ANOVA.
These findings revealed that LqqE1 directly bound to PcoI to prevent AHL production. We were then interested in understanding whether this mode of action is unique to PcoI or shared by other AHL synthases. To address this, we first selected the human pathogen Pseudomonas aeruginosa PAO1 carrying two AHL synthases, RhlI and LasI [64], as a test. We found that expression of lqqE1 in P. aeruginosa PAO1 significantly reduced the amount of RhlI-mediated butyl-homoserine lactone (C4-HSL) but not LasI-mediated N-(3-oxododecanoyl)-L-homoserine lactone (3-oxo-C12-HSL) (Fig. 4E, F and Fig. S5A, B). Using a similar protocol as described in Fig. 3A, co-expression of lqqE1 with the rhlI genes by plasmids in E. coli DH5a significantly attenuated the ability of RhlI to produce AHL (Fig. 4G, and Fig. S5C). This suggested that RhlI might be recognized by LqqE1 through direct binding, which we indeed confirmed using pull-down assays in which LqqE1-FLAG bound to RhlI-His (Fig. 4H). Consistently, lqqE1 expression in P. aeruginosa PAO1 not only significantly reduced the production of RhlI-dependent AHL, but also impaired RhlI-controlled functions such as pyocyanin and rhamnolipid production [65]. Furthermore, supplementation with C4-HSL could partially rescue the defects in pyocyanin and rhamnoolipid production in lqqE1-expressing P. aeruginosa PAO1 (Fig. 4I, J).
Ralstonia solanacearum EP1 is an AHL-producing phytopathogen that causes bacterial wilt in eggplants [50, 66], and RasI is the major AHL synthase encoded in this strain. Previously, RasI was shown to promote motility essential for infection by this strain. We thus tested whether lqqE1 expression in R. solanacearum EP1 impaired AHL production and swimming motility, as confirmed by our results shown in Fig. 4K, L and Fig. S5D. Furthermore, significant inhibition of AHL production was also observed by co-expression of lqqE1 and rasI genes in E. coli DH5a (Fig. 4M, Fig. S5E). Pull-down assays also revealed direct binding between LqqE1-FLAG and RasI-His (Fig. 4N). These results collectively revealed that LqqE1 was able to bind multiple AHL synthases to quench their AHL quorum sensing.
Binding to LqqE1 blocked recognition of substrates required for AHL synthesis by the AHL synthase PcoI
How does LqqE1 achieve quorum quenching by binding to AHL synthases? To address this question, we focused on representative LqqE1-PcoI pairs. We first tested whether lqqE1 expression could reduce PcoI abundance. To facilitate this study, we further selected AHL-producing recombinant E. coli (Fig. 3A), in which LqqE1 was functional. We collected recombinant E. coli cells co-expressing LqqE1-FLAG and PcoI-His at OD600 of 1.0 and 1.5, and detected LqqE1 and PcoI protein levels by western blot using anti-FLAG or anti-His antibody. We found that lqqE1 expression did not seem to impair PcoI protein abundance at two selected cell densities (Fig. S6A).
Considering that the synthesis of AHL by synthases requires two substrates, acyl–acyl carrier protein (acyl-ACP) and S-adenosy-L-methionine (SAM) [13, 14] (Fig. 5A), we investigated whether LqqE1 binding would interfere with the recognition of acyl-ACP and/or SAM by PcoI. According to a previous report [52], we artificially synthesized C6-ACP and C8-ACP as a preliminary attempt (Fig. S6B), but found that their co-culture with PcoI in vitro failed to produce AHL for unknown reasons (Fig. S6C, D). We also tried using C6-oxo-ACP and C8-oxo-ACP as alternatives, but they were difficult to synthesize or obtain commercially. Therefore, we dropped such tests. Alternatively, we purchased another common AHL substrate, SAM, for further study.
Fig. 5. Binding LqqE1 blocks PcoI to recognize S-adenosy-L-methionine, a substrate required for AHL synthesis.
A Schematic illustration of AHL synthesis by AHL synthase using two substrates - acyl–acyl carrier protein (acyl-ACP) and S-adenosy-L-methionine (SAM). B Microscale thermophoresis (MST) showed PcoI bound to SAM. LqqE1 supplement impaired the PcoI-SAM binding affinity in a dose-dependent manner. C A PcoI structural modeling predicted by AlphaFold Protein Structure Database. Three amino residues (R23, F27, and W33) potentially required for the PcoI-SAM binding were predicted according to an earlier study and highlighted in the predicted structural model. D Contribution of the predicted amino residues to the PcoI-dependent AHL production in E. coli. Expression of the pcoI variant genes in E. coli were generated by site-directed mutagenesis, where each conserved residue was substituted by alanine (A) individually or in combination. Pull-down assays showing both the PcoIR23A&F27A&W33A-His (E) and PcoIR23A-His (F) did not interact with LqqE1-FLAG. In B and D, average data from three experiments was presented, ±SD. ***p < 0.0001, assessed by one-way ANOVA.
By MST, we did observe direct binding of PcoI-His to SAM (Kd, 22 μM, Fig. 5B), consistent with the fact that AHL synthase directly recognizes its substrate. But under similar MST conditions, LqqE1 failed to bind SAM (Fig. S6E). Next, we added different concentrations of purified MBP-LqqE1 to the MST solution containing PcoI-His and SAM to test whether LqqE1 could alter PcoI-SAM binding. Our MST results clearly showed that LqqE1 did impair PcoI-SAM binding affinity in a dose-dependent manner. Addition of 1 μM LqqE1 decreased the affinity of PcoI-SAM binding from an initial 22 μM to 358 μM, while addition of 10 μM LqqE1 completely inhibited PcoI-SAM binding (Fig. 5B). These lines of evidences suggested that LqqE1 binding attenuated PcoI-SAM binding affinity, thereby quenching AHL quorum sensing in strain 2P24.
To further validate the finding that binding of LqqE1 attenuates the binding affinity of PcoI to SAM, we carried out additional assays. Based on an earlier report, we predicted three PcoI amino acid residues (R23, F27, and W33) that maybe crucial for its recognition of SAM [67]. Through multiple sequence alignment, we found that these three amino acid residues were highly conserved among different AHL synthases (Fig. S7A). For better visualization, we also mapped the positions of these three residues to the predicted PcoI structure model (Fig. 5C) by AlphaFold Protein Structure Database. To facilitate residue-to-function studies, we first generated a PcoI variant by simultaneously substituting all three residues with alanine (A). The resulting PcoI -variant (PcoIR23A&F27A&W33A) and the native pcoI gene were separately expressed in E. coli BL21. Using the AHL assay, we found that native PcoI efficiently generated AHL, but not PcoIR23A&F27A&W33A variant (Fig. 5D and Fig. S7B). Guided by this finding, we further randomly generated an R23A mutant, and found that expression of this pcoI variant (PcoIR23A) gene in E. coli BL21 also failed to produce AHL (Fig. 5D and Fig. S7B). We then performed pull-down assays in which neither PcoIR23A&F27A&W33A-His nor PcoIR23A-His interacted with LqqE1-FLAG (Fig. 5E, F). These results raised a possibility that LqqE1 might interact with PcoI through these three key residues individually or in combination, thereby interfering with PcoI-SAM recognition.
LqqE1-triggered quorum quenching conferred a competitive advantage to L. enzymogenes during co-culture with P. fluorescens
What is the ecological implication of interspecies quorum quenching triggered by strain OH11 “designer” LqqE1? We investigated whether it conferred an advantage to strain OH11 in bacterial interspecies competition, as we previously showed that strain 2P24 could be killed by strain OH11 using T4ASS during co-culture at a 1:1 ratio in agar plates [40]. In support, we expressed a strain OH11-drived T4E gene (Le0908) in the periplasm of strain 2P24, because this gene was previously shown to induce virulence in the periplasm of E. coli [40]. Indeed, we found that artificial expression of Le0908 in the periplasm of strain 2P24 inhibited its growth (Fig. S7C), suggesting strain OH11 could deliver toxic T4E effectors into strain 2P24 to kill this competitor. Based on these points, we further tested the above hypothesis. We co-cultured mCherry-labeled strain OH11 and GFP-labeled strain 2P24 on 1/10 TSB agar plates at a ratio of 1: 60 after several rounds of pre-assays. Using such a resulting ratio, rather than the common 1:1 ratio, we could better reflect the involvement of LqqE1 in the killing process against strain 2P24 (see below). After one day of co-culture, we found that the AHL-deficient mutant ΔpcoI was completely killed by the strain OH11 compared to the wild-type strain 2P24, as no GFP signal could be observed in the mixed colonies using fluorescent microscopy (Fig. 6A, B). Addition of 20 μM of commercial AHL [N-3-oxo-octanoyl-L-homoserine lactone, 3-oxo-C8-HSL] to OH11-ΔpcoI co-culture restored the GFP signal intensity to that of wild-type strain 2P24 (Fig. 6A, B). Similar results were obtained by quantifying the numbers of viable cells of strain 2P24 and ΔpcoI in their respective mixed colonies (Fig. 6C).
Fig. 6. The LqqE1-triggered quorum quenching confers L. enzymogenes competitive advantages during co-culture with P. fluorescens.
A Fluorescent images showing the mCherry-labeled L. enzymogenes kills the AHL-deficient P. fluorescens labeled with GFP more efficiently when both species were co-cultured on agar plates with 1:60 rations. B Quantification of the GFP signal intensity in mixed colonies corresponding to (A). C Living cell numbers of P. fluorescens in mixed colonies corresponding to the panel A. Fluorescent images showing the lqqE1 expression P. fluorescens favored the killing efficacy by L. enzymogenes (D) during their co-culture on agar plants, and this is confirmed by quantifying the GFP signal intensity (E) in mixed colonies. Fluorescent images showing lqqE1 in-frame deletion in L. enzymogenes (ΔlqqE1) weakened the killing against P. fluorescens 2P24 (F) during their co-culture on agar plants. The chromosomal complement of lqqE1 at the ΔlqqE1 background restored this killing effect, while chromosomal complementation of lqqE1E68A and lqqE1S126A genes with mutations that affect its AHL-quenching function did not. G Quantification of the GFP signal intensity in mixed colonies corresponding to the panel F. In all panels, average data from three experiments was presented, ±SD. ***p < 0.0001, **p < 0.01, assessed by one-way ANOVA. H A proposed model showing the LqqE1-triggered interspecies quorum quenching during the L. enzymogenes-P. fluorescens competition occurring in soil microbiome. Via cell-to-cell contact, L. enzymogenes using T4ASS to kill the competitor, P. fluorescens. To protect the survival of the community members, P. fluorescens forms biofilms via producing the quorum-sensing signal AHL. As a counterattack, L. enzymogenes delivers a T4E protein LqqE1 into the cytoplasm of P. fluorescens. Upon the delivery, LqqE1 directly binds to the AHL synthase PcoI to interfere its recognition with SAM, one of the substrate required for AHL synthesis, by which the P. fluorescens quorum sensing is naturally quenched. Such an interspecies quorum quenching appears to prevent P. fluorescens to form community/biofilms, where the biofilms may act as a physical barrier to prevent the penetration of T4SS and/or delivery of toxic effector proteins from OH11, such as Le0908 presented in this study, enabling L. enzymogenes using T4ASS to more efficiently kill the free-living members of P. fluorescens, thereby gaining a competitive advantage.
Considering that quantification of GFP signal intensity was consistent with numbers of isolated viable cells of 2P24/ΔpcoI in mixed colonies, we focused on the former approach to facilitate the following assays. By observing and quantifying the GFP signal intensity, we found that the expression of lqqE1 in strain 2P24 indeed conferred stronger killing effect of strain OH11 compared to the control (Fig. 6D, E). Notably, more strain 2P24 cells were detected in mixed colonies compared to wild-type strain OH11 co-cultured with an in-frame lqqE1 deletion mutant (Fig. 6F, G). We also complemented ΔlqqE1 with native lqqE1 and its variant genes (lqqE1E68A and lqqE1S126A) with mutations that affect its AHL-quenching function by a gene knock-in approaches via double-crossover recombination. We found that chromosomal complementation of native lqqE1 restored killing against strain 2P24, while chromosomal complementation of the lqqE1E68A and lqqE1S126A genes did not restore this function (Fig. 6F, G). Collectively, these results suggested that LqqE1-triggered quorum quenching conferred an advantage on L. enzymogenes in competition with P. fluorescens.
Discussions
There are two main classes of T4SS: one is T4ASS, and the other is T4BSS represented by the Dot/Icm system, which is mainly used for intracellular pathogens such as Legionella pneumophila to inject effector proteins into eukaryotic hosts to target various immune signaling pathways, promoting bacterial infections [68, 69]. Unlike T4BSS, T4ASS is generally believed to translocate toxic effector proteins into bacterial competitors to induce cell death [41, 42]. Contrary to this common view, our present study revealed a unique function of T4E by discovering LqqE1 as an ecologically important quorum-quencher protein (Fig. 6H). We proposed that in the soil microbiome, L. enzymogenes and P. fluorescens competed for limited nutrients. Through cell-to-cell contact, L. enzymogenes used T4ASS to kill P. fluorescens competitors. In this case, quorum sensing of strain 2P24 might enhance protection against killing activity conferred by T4SS of strain OH11. As a counterattack strategy, L. enzymogenes delivered LqqE1 into the cytoplasm of P. fluorescence to form a protein complex with the AHL synthase PcoI, thereby preventing PcoI from recognizing/binding to S-adenosy-L-methionine (SAM, one of the substrates required for AHL synthesis) to naturally quench P. fluorescence AHL system. The lessening of quorum-sensing might lead to the increased sensitivity of strain 2P24. This quorum-sensing-mediated protection appeared to be associated with quorum-sensing-controlled phenotypes, such as biofilm formation, where biofilms can act as a physical barrier to prevent the penetration of T4SS and/or delivery of toxic effector proteins from OH11, such as Le0908 presented in this study (Fig. S7C). This immune protein-independent protection mechanism has been reported in T6SS-mediated bacterial interspecies interactions [70]. Therefore, LqqE1-triggered interspecies quorum quenching conferred a competitive advantage on L. enzymogenes by preventing P. fluorescens from forming biofilms to enhance its killing efficiency against free-living members of P. fluorescens (Fig. 6H). In this way, LqqE1 served as the first T4E to link interspecies quorum quenching to bacterial competition. This suggested that T4ASS could translocate functionally diverse effectors into competitors to manipulate various cellular processes of the “host” beyond common growth inhibition.
How does LqqE1 achieve AHL quorum quenching? Earlier studies found that AHL-quencher proteins achieve this by two main mechanisms.: one is represented by AHL-degrading enzymes, which quench AHL signaling by directly degrading AHL [21], and the other is exemplified by non-canonical AHL-quencher proteins, which could not degrade AHL but block AHL synthesis by binding to AHL synthases, as described in our laboratory [27, 28]. LqqE1 seemed to employ a mechanism similar to non-canonical AHL quenchers, but they worked quite differently. We have shown that non-canonical AHL-quencher proteins from strain OH11 (i.e. LqqP and Le4759) can bind PcoI to reduce the abundance of its free form [27, 28], whereas LqqE1 did not act in this manner. Alternatively, PcoI binding allowed LqqE1 to block the PcoI-SAM interaction, thereby quenching the AHL system. The mechanism of this LqqE1-triggered AHL quorum-quenching was previously unknown. Yet, this novel mechanism appeared to be common in T4ASS-producing bacteria as well, because we demonstrated experimentally that at least three LqqE1 homologs had similar AHL-quenching effects to LqqE1. Therefore, T4E effector-triggered quorum quenching might be a common phenomenon in bacterial interspecies interactions.
How did strain OH11 evolve an LqqE1-triggered interspecies AHL-quenching “weapon” without affecting itself? While many Proteobacteria employ the AHL for quorum sensing, almost all Lysobacter species do not, as their genomes do not have an AHL synthase homolog [28, 71]. This “deficiency” might serve as an intrinsic, evolutionarily acquired “advantage” to ensure that LqqE1 targeted a competitor’s AHL synthases rather than Lysobacter itself. Lysobacter members produce DSF (diffusible signal factor) as an alternative quorum-sensing signal, which is chemically distinct from AHLs [71]. Therefore, one might question whether LqqE1 is also involved in DSF signaling, which is an issue we are now testing in the laboratory.
Our findings also provided a feasible approach to extend the scope of biocontrol spectrum of strain OH11 from agriculture to medical treatment. Strain OH11 has traditionally been considered an antifungal agent because it produces and secretes a broad-spectrum antifungal metabolite called heat-stable antifungal factor (HSAF) [38, 39]. Due to its diffusible properties, HSAF can be considered a “long-range” weapon whose action does not require cell-to-cell contact between the OH11 strain and the soil-borne fungal pathogens. Although the OH11 strain does not produce compounds against Gram-negative bacteria, we also extended its biocontrol spectrum to phytopathogens, as we recently discovered that T4ASS acts as a “short-range” antibacterial weapon via cell-to-cell contact [40]. Here, we further included strain OH11 as a promising alternative microbial resource to protect humans from P. aeruginosa infection, as we found that expression of lqqE1 in P. aeruginosa could directly target RhlI to significantly impair its production of AHL levels known to control virulence [64]. We believed that efficient delivery of LqqE1 into P. aeruginosa via T4ASS of strain OH11 or via cell-free nanotechnology would be a potentially feasible approach.
In conclusion, this study demonstrates that naturally occurring T4E-mediated quorum quenching is not only ecologically important in promoting bacterial interspecies competition, but also has broad application potential in preventing bacterial infection by quenching AHL signaling.
Supplementary information
Supplementary Materials for Quorum quenching by a type IVA secretion system effector
Acknowledgements
This study was funded by the Natural Key Research and Development Program (2022YFD1400200 to GQ), followed by the National Natural Science Foundation of China (U22A20486, 32072470, and 31872016 to GQ), Science and technology project of Shanxi Branch of China National Tobacco Corporation (KJ-2022-04), the Fundamental Research Funds for the Central Universities (KJJQ202001, KYT201805, and KYTZ201403 to GQ), and the Jiangsu University advantage discipline construction project (80900246 to XS).
Author contributions
GQ and JL conceived the project and designed experiments. JL carried out experiments. JL, ZL, DX, DS, LW, L Liao, and XS analyzed data and prepared figures and tables. JL, GQ, and XS wrote the manuscript. GQ, XS, L Lin, PL, LZ, and HW revised the manuscript. All authors read and approved the final manuscript.
Data availability
The sequence data from the present study have been submitted to the NCBI GenBank under the following accession numbers: MW052467 (Le0908), MW052468 (Le0989), MW052469 (Le1288), MW052471 (Le1519), MW052472 (Le1841), MW052473 (Le3180), MW052474 (Le3316), MW052488 (Le4230), MW052489 (Le4232), MW052491 (Le4236), MW052493 (Le4253), MW052494 (Le4579), MW052495 (Le4949), OQ192195 (La0125), OQ192196 (Lg4270), OQ179943 (16s rRNA), OQ192197 (GyrB), OQ192199 (RpoB), and OQ192198 (RpoD).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01457-2.
References
- 1.Papenfort K, Bassler BL. Quorum sensing signal–response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016;14:576–88. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng Y, Wu JE, Tao F, Zhang L. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem Rev. 2011;111:160–73. doi: 10.1021/cr100354f. [DOI] [PubMed] [Google Scholar]
- 3.Galloway WRJD, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem Rev. 2011;111:28–67. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]
- 4.Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551:313–20. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 6.González JE, Marketon MM. Quorum sensing in nitrogen-fixing Rhizobia. Microbiol Mol Biol Rev. 2003;67:574–92. doi: 10.1128/MMBR.67.4.574-592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Federle MJ, Bassler BL. Interspecies communication in bacteria. J Clin Investig. 2003;112:1291–9. doi: 10.1172/JCI20195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–46. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–75. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–81. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Murphy PJ, Kerr A, Tate ME. Agrobacterium conjugation and gene regulation by N-acyl-L-homoserine lactones. Nature. 1993;362:446–8. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 12.de Kievit TR, Iglewski BH. Bacterial quorum sensing in pathogenic relationships. Infect Immun. 2000;68:4839–49. doi: 10.1128/IAI.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsek MR, Val DL, Hanzelka BL, Cronan JJ, Greenberg EP. Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci USA. 1999;96:4360–5. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaefer AL, Val DL, Hanzelka BL, Cronan JE, Greenberg EP. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong S, Frane ND, Christensen QH, Greenberg EP, Nagarajan R, Nair SK. Molecular basis for the substrate specificity of quorum signal synthases. Proc Natl Acad Sci USA. 2017;114:9092–7. doi: 10.1073/pnas.1705400114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moré MI, Finger LD, Stryker JL, Fuqua C, Eberhard A, Winans SC. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–8. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 17.Watson WT, Minogue TD, Val DL, von Bodman SB, Churchill MEA. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol Cell. 2002;9:685–94. doi: 10.1016/S1097-2765(02)00480-X. [DOI] [PubMed] [Google Scholar]
- 18.Raychaudhuri A, Tullock A, Tipton PA. Reactivity and reaction order in acyl homoserine lactone formation by Pseudomonas aeruginosa RhlI. Biochemistry. 2008;47:2893–8. doi: 10.1021/bi702009n. [DOI] [PubMed] [Google Scholar]
- 19.Pearson JP, Feldman M, Iglewski BH, Prince AA. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect Immun. 2000;68:4331–4. doi: 10.1128/IAI.68.7.4331-4334.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pirhonen M, Flego D, Heikinheimo R, Palva ET. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. The EMBO Journal. 1993;12:2467–76. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Zhang L, Wang L, Zhang H, Xu J, Dong Y. Quenching quorum-sensingdependent bacterial infectionby an N-acyl homoserine lactonase. Nature. 2001;411:813–7. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 22.Sikdar R, Elias M. Quorum quenching enzymes and their effects on virulence, biofilm, and microbiomes: a review of recent advances. Expert Rev Anti Infect Ther. 2020;18:1221–33. doi: 10.1080/14787210.2020.1794815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murugayah SA, Gerth ML. Engineering quorum quenching enzymes: progress and perspectives. Biochem Soc Trans. 2019;47:793–800. doi: 10.1042/BST20180165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fetzner S. Quorum quenching enzymes. J Biotechnol. 2015;201:2–14. doi: 10.1016/j.jbiotec.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Grandclément C, Tannières M, Moréra S, Dessaux Y, Faure D. Quorum quenching: role in nature and applied developments. Fems Microbiol Rev. 2016;40:86–116. doi: 10.1093/femsre/fuv038. [DOI] [PubMed] [Google Scholar]
- 26.Lade H, Paul D, Kweon JH. Quorum quenching mediated approaches for control of membrane biofouling. Int J Biol Sci. 2014;10:550–65. doi: 10.7150/ijbs.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao J, Shen D, Lin L, Chen H, Jin Y, Chou S, et al. Bacterial quorum sensing quenching activity of Lysobacter leucyl aminopeptidase acts by interacting with autoinducer synthase. Comput Struct Biotechnol J. 2021;19:6179–90. doi: 10.1016/j.csbj.2021.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao J, Li Z, Xiong D, Shen D, Wang L, Shao X, et al. A novel and efficient platform for discovering noncanonical quorum-quenching proteins. Microbiol Spectr. 2023;11:e343722. doi: 10.1128/spectrum.03437-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong Y, Xu J, Li X, Zhang L. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci USA. 2000;97:3526–31. doi: 10.1073/pnas.97.7.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong Y, Gusti AR, Zhang Q, Xu J, Zhang L. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus Species. Appl Environ Microbiol. 2002;68:1754–9. doi: 10.1128/AEM.68.4.1754-1759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reina JC, Romero M, Salto R, Cámara M, Llamas I. AhaP, A quorum quenching acylase from Psychrobacter sp. M9-54-1 that attenuates Pseudomonas aeruginosa and Vibrio coralliilyticus virulence. Mar Drugs. 2021;19:16. doi: 10.3390/md19010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh BN, Prateeksha, Upreti DK, Singh BR, Defoirdt T, Gupta VK, et al. Bactericidal, quorum quenching and anti-biofilm nanofactories: a new niche for nanotechnologists. Crit Rev Biotechnol. 2017;37:525–40. doi: 10.1080/07388551.2016.1199010. [DOI] [PubMed] [Google Scholar]
- 33.Tang K, Su Y, Brackman G, Cui F, Zhang Y, Shi X, et al. MomL, a Novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Appl Environ Microbiol. 2015;81:774–82. doi: 10.1128/AEM.02805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin YH, Xu JL, Hu J, Wang LH, Ong SL, Leadbetter JR, et al. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol Microbiol. 2003;47:849–60. doi: 10.1046/j.1365-2958.2003.03351.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang HB, Wang LH, Zhang LH. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2002;99:4638–43. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen P, Cook FD. Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int J Syst Bacteriol. 1978;28:367–93. doi: 10.1099/00207713-28-3-367. [DOI] [Google Scholar]
- 37.Puopolo G, Tomada S, Pertot I. The impact of the omics era on the knowledge and use of Lysobacter species to control phytopathogenic micro-organisms. J Appl Microbiol. 2018;124:15–27. doi: 10.1111/jam.13607. [DOI] [PubMed] [Google Scholar]
- 38.Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, et al. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc. 2011;133:643–5. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, et al. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother. 2006;51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen X, Wang B, Yang N, Zhang L, Shen D, Wu H, et al. Lysobacter enzymogenes antagonizes soilborne bacteria using the typeIV secretion system. Environ Microbiol. 2021;23:4673–88. doi: 10.1111/1462-2920.15662. [DOI] [PubMed] [Google Scholar]
- 41.Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, et al. Bacterial killing via a type IV secretion system. Nat Commun. 2015;6:6453. doi: 10.1038/ncomms7453. [DOI] [PubMed] [Google Scholar]
- 42.Bayer-Santos E, Cenens W, Matsuyama BY, Oka GU, Di Sessa G, Mininel IDV, et al. The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. Plos Pathog. 2019;15:e1007651. doi: 10.1371/journal.ppat.1007651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alegria MC, Souza DP, Andrade MO, Docena C, Khater L, Ramos CHI, et al. Identification of new protein-protein interactions involving the products of the chromosome-and plasmid-encoded Type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv. citri. J Bacteriol. 2005;187:2315–25. doi: 10.1128/JB.187.7.2315-2325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12:137–48. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci USA. 2013;110:2623–8. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao G, Yin D, Chen S, Xia F, Yang J, Li Q, et al. Effect of biocontrol agent Pseudomonas fluorescens 2P24 on soil fungal community in cucumber rhizosphere using T-RFLP and DGGE. Plos One. 2012;7:e31806. doi: 10.1371/journal.pone.0031806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian G, Wang Y, Qian D, Fan J, Hu B, Liu F. Selection of available suicide vectors for gene mutagenesis using chiA (a chitinase encoding gene) as a new reporter and primary functional analysis of chiA in Lysobacter enzymogenes strain OH11. World J Microbiol Biotechnol. 2012;28:549–57. doi: 10.1007/s11274-011-0846-8. [DOI] [PubMed] [Google Scholar]
- 48.Zhu J, Chai Y, Zhong Z, Li S, Winans SC. Agrobacterium bioassay strain for ultrasensitive detection of N-acyl homoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl Environ Microbiol. 2003;69:6949–53. doi: 10.1128/AEM.69.11.6949-6953.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acyl homoserine lactones. Microbiology. 1997;143:3703–11. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 50.Yan J, Li P, Wang X, Zhu M, Shi H, Yu G, et al. RasI/R Quorum sensing system controls the virulence of Ralstonia solanacearum Strain EP1. Appl Environ Microbiol. 2022;88:e0032522. doi: 10.1128/aem.00325-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao L, Schaefer AL, Coutinho BG, Brown PJB, Greenberg EP. An aryl-homoserine lactone quorum-sensing signal produced by a dimorphic prosthecate bacterium. Proc Natl Acad Sci USA. 2018;115:7587–92. doi: 10.1073/pnas.1808351115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei H, Zhang L. Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Van Leeuwenhoek. 2006;89:267–80. doi: 10.1007/s10482-005-9028-8. [DOI] [PubMed] [Google Scholar]
- 53.Muteeb G, Sen R. Random mutagenesis using a mutator strain. In: Braman J, editor. In vitro mutagenesis protocols. 3rd ed., vol. 634. Totowa, NJ: Humana Press; 2010. p. 411–9. [DOI] [PubMed]
- 54.Coudert E, Gehant S, de Castro E, Pozzato M, Baratin D, Neto T, et al. Annotation of biologically relevant ligands in UniProtKB using ChEBI. Bioinformatics. 2022;39:btac793. doi: 10.1093/bioinformatics/btac793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bateman A, Martin M, Orchard S, Magrane M, Agivetova R, Ahmad S, et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callaway E. ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures. Nature. 2020;588:203–4. doi: 10.1038/d41586-020-03348-4. [DOI] [PubMed] [Google Scholar]
- 57.Xu G, Han S, Huo C, Chin K, Chou S, Gomelsky M, et al. Signaling specificity in the c-di-GMP-dependent network regulating antibiotic synthesis in Lysobacter. Nucleic Acids Res. 2018;46:9276–88. doi: 10.1093/nar/gky803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang M, Ren S, Shen D, Yang N, Wang B, Han S, et al. An intrinsic mechanism for coordinated production of the contact-dependent and contact-independent weapon systems in a soil bacterium. Plos Pathog. 2020;16:e1008967. doi: 10.1371/journal.ppat.1008967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo Z, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841–6. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee DJ, Jo AR, Jang MC, Nam J, Choi HJ, Choi G, et al. Analysis of two quorum sensing-deficient isolates of Pseudomonas aeruginosa. Microb Pathog. 2018;119:162–9. doi: 10.1016/j.micpath.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 61.Koch AK, Käppeli O, Fiechter A, Reiser J. Hydrocarbon assimilation and biosurfactant production in Pseudomonas aeruginosa mutants. J Bacteriol. 1991;173:4212–9. doi: 10.1128/jb.173.13.4212-4219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Redzej A, Ukleja M, Connery S, Trokter M, Felisberto Rodrigues C, Cryar A, et al. Structure of a VirD4 coupling protein bound to a VirB typeIV secretion machinery. EMBO J. 2017;36:3080–95. doi: 10.15252/embj.201796629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hersch SJ, Lam L, Dong TG. Engineered Type Six secretion systems deliver active exogenous effectors and Cre recombinase. mBio. 2021;12:e0111521. doi: 10.1128/mBio.01115-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell. 2015;6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brint JM, Ohman DE. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–63. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aoun N, Tauleigne L, Lonjon F, Deslandes L, Vailleau F, Roux F, et al. Quantitative disease resistance under elevated temperature: genetic basis of new resistance mechanisms to Ralstonia solanacearum. Front Plant Sci. 2017;8:1387. doi: 10.3389/fpls.2017.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gould TA, Schweizer HP, Churchill MEA. Structure of the Pseudomonas aeruginosa acyl-homoserinelactone synthase LasI. Mol Microbiol. 2004;53:1135–46. doi: 10.1111/j.1365-2958.2004.04211.x. [DOI] [PubMed] [Google Scholar]
- 68.Grohmann E, Christie PJ, Waksman G, Backert S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol. 2018;107:455–71. doi: 10.1111/mmi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voth DE, Broederdorf LJ, Graham JG. Bacterial Type IV secretion systems: versatile virulence machines. Future Microbiol. 2012;7:241–57. doi: 10.2217/fmb.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hersch SJ, Manera K, Dong TG. Defending against the type six secretion system: beyond immunity genes. Cell Rep. 2020;33:108259. doi: 10.1016/j.celrep.2020.108259. [DOI] [PubMed] [Google Scholar]
- 71.Han Y, Wang Y, Tombosa S, Wright S, Huffman J, Yuen G, et al. Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl Microbiol Biotechnol. 2015;99:801–11. doi: 10.1007/s00253-014-6120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials for Quorum quenching by a type IVA secretion system effector
Data Availability Statement
The sequence data from the present study have been submitted to the NCBI GenBank under the following accession numbers: MW052467 (Le0908), MW052468 (Le0989), MW052469 (Le1288), MW052471 (Le1519), MW052472 (Le1841), MW052473 (Le3180), MW052474 (Le3316), MW052488 (Le4230), MW052489 (Le4232), MW052491 (Le4236), MW052493 (Le4253), MW052494 (Le4579), MW052495 (Le4949), OQ192195 (La0125), OQ192196 (Lg4270), OQ179943 (16s rRNA), OQ192197 (GyrB), OQ192199 (RpoB), and OQ192198 (RpoD).