Abstract
Microbial communities host many auxotrophs—organisms unable to synthesize one or more metabolites required for their growth. Auxotrophy is thought to confer an evolutionary advantage, yet auxotrophs must rely on other organisms that produce the metabolites they require. The mechanisms of metabolite provisioning by “producers” remain unknown. In particular, it is unclear how metabolites such as amino acids and cofactors, which are found inside the cell, are released by producers to become available to auxotrophs. Here, we explore metabolite secretion and cell lysis as two distinct possible mechanisms that result in the release of intracellular metabolites from producer cells. We measured the extent to which secretion or lysis of Escherichia coli and Bacteroides thetaiotaomicron amino acid producers can support the growth of engineered Escherichia coli amino acid auxotrophs. We found that cell-free supernatants and mechanically lysed cells provide minimal levels of amino acids to auxotrophs. In contrast, bacteriophage lysates of the same producer bacteria can support as many as 47 auxotroph cells per lysed producer cell. Each phage lysate released distinct levels of different amino acids, suggesting that in a microbial community the collective lysis of many different hosts by multiple phages could contribute to the availability of an array of intracellular metabolites for use by auxotrophs. Based on these results, we speculate that viral lysis could be a dominant mechanism of provisioning of intracellular metabolites that shapes microbial community structure.
Subject terms: Bacteria, Bacteriophages, Bacterial evolution, Bacterial secretion
Microbial communities are ubiquitous, contributing to global nutrient cycling, human health and agricultural productivity. Nutritional interdependence is a universal feature of microbial communities [1]. One form of nutritional interaction occurs between auxotrophs—organisms unable to produce a nutrient required for growth—and prototrophs or “producers”—organisms capable of synthesizing a nutrient required by others. Auxotrophy contributes to microbial community stability [2, 3], and many bacteria are predicted to be auxotrophic for one or more amino acids or cofactors [4, 5], likely because auxotrophy often confers a growth advantage when the required nutrient is available [5]. The prevalence of auxotrophy suggests that a consistent source of nutrients must be available over evolutionary timescales, yet it is unknown how metabolites that function intracellularly, such as amino acids and cofactors, become accessible to auxotrophs. We previously proposed that nutrient release resulting from secondary effects of evolved biological processes, such as secretion of excess metabolites or cell lysis, are possible mechanisms of nutrient provisioning [6]. Here, we experimentally test both mechanisms using two phylogenetically distinct producer species and engineered Escherichia coli amino acid auxotrophs.
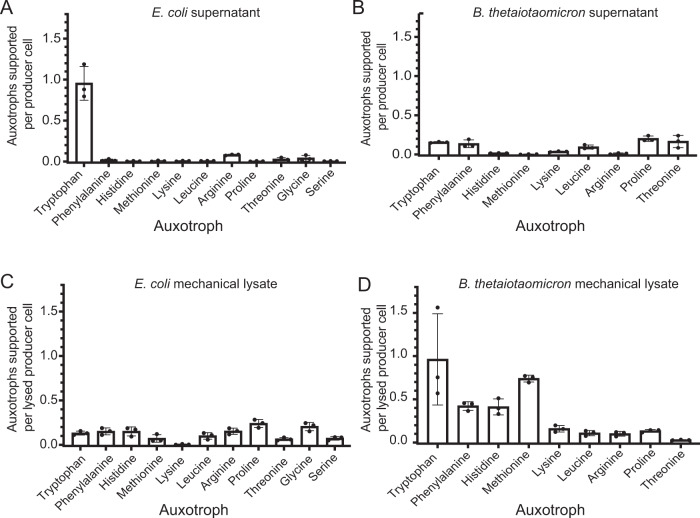
Amino acids and dipeptides have been found in exometabolomes of cultured microorganisms and soil biocrusts [7, 8]. Furthermore, certain pairs of amino acid auxotrophs can grow in coculture in the absence of amino acid supplementation, suggesting that bacteria may shed amino acids during growth [4, 9, 10]. To test whether amino acid secretion or leakage can be sufficient to support the growth of auxotrophs, we measured the extent to which cell-free supernatants from E. coli and Bacteroides thetaiotaomicron, both producers of all 20 proteogenic amino acids, could support the growth of up to 11 different E. coli amino acid auxotrophs in the absence of amino acid supplementation. Supernatants from both E. coli and B. thetaiotaomicron minimally supported most of the tested amino acid auxotrophs used in this study, with each producer cell supporting the growth of less than 0.2 auxotroph cells in all except the Trp auxotroph (Fig. 1A, B). Amino acid release was only slightly influenced by growth phase, as cell-free supernatants of producers harvested in stationary (Fig. 1) or exponential phase (Fig. S1) support only modest levels of auxotroph growth. Together, these results suggest that secretion or leakage may not be dominant forms of amino acid provisioning.
Fig. 1. Cell-free supernatants and mechanical lysates minimally support growth of amino acid auxotrophs.
The number of auxotroph cells supported per producer cell was calculated for auxotrophs grown in defined medium containing (A, B) cell-free supernatants and (C, D) French pressed lysates of (A, C) E. coli and (B, D) B. thetaiotaomicron. The auxotrophs are ordered from left to right on the x-axis based on the estimated biosynthetic cost of producing one molecule of the amino acid [4]. The genotype of each auxotroph is listed in Supplementary Table 1. In all except one case (Met), a single gene in the biosynthetic pathway is deleted [4]. Bars represent the mean of three replicates; error bars represent standard deviation.
We explored the possibility that cell lysis can release amino acids at sufficient levels to support auxotrophs. Lysis occurs naturally in bacteria following exposure to toxins or as part of developmental genetic programs such as sporulation in Bacillus subtilis and fruiting body formation in Myxcococcus xanthus [11–13]. A common trigger of microbial lysis is viral infection, as viruses are abundant and ubiquitous across many environments [14]. In marine environments, viruses lyse an estimated 20% of microbial biomass per day and are major drivers of microbial turnover and elemental cycling, a process known as the viral shunt [15, 16]. Further, in a laboratory coculture, addition of a lytic phage can lead to increased growth yield of a partner strain [17, 18].
To determine whether the levels of intracellular amino acids liberated via lysis of a producer could be sufficient to support the growth of amino acid auxotrophs, we tested the ability of the 11 amino acid auxotrophs to grow in media supplemented with mechanically lysed E. coli or B. thetaiotaomicron cells. Lysates of both producers contain a higher level of bioavailable amino acids than their respective supernatants, though less than one auxotroph cell was supported by each lysed producer cell (Fig. 1C, D). Comparison of lysates of E. coli and B. thetaiotaomicron suggests that these bacteria contain different levels of several amino acids, which likely reflects their distinct metabolic characteristics. The strains that were most supported by mechanical lysates of B. thetaiotaomicron were those auxotrophic for the more biosynthetically costly amino acids (Fig. 1D) (in all graphs, the auxotrophs are ordered based on their estimated biosynthetic cost [4]). Yet, with less than one auxotroph cell supported for each lysed producer cell, the ratio of producer to auxotroph cells in a community would need to be high.
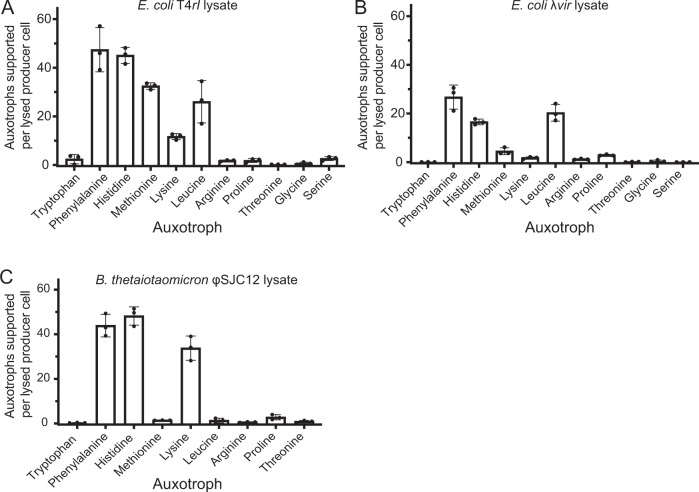
During infection, viral energy demands are met by reprograming their host’s metabolism by overexpressing translational genes and elevating levels of intracellular metabolites [19, 20]. We therefore tested whether producers lysed by a phage could support auxotroph growth to a greater extent than mechanically lysed cells. Indeed, E. coli T4rI phage lysates, filtered to remove phage particles, supported the growth of all 11 auxotrophs at levels higher than supernatants and mechanically lysed E. coli, with nine of the 11 auxotrophs at a level exceeding one auxotroph supported by one producer (Fig. 2A). Five auxotrophs were supported at levels ranging from 11 to 47 auxotrophs per producer, suggesting that producers and auxotrophs can coexist at a low ratio (Fig. 2A). Similarly, E. coli λvir lysates provided high levels of certain amino acids, supporting as many as 27 auxotrophs per producer cell (Fig. 2B). Upon observing that the two E. coli phage lysates showed differences in the levels of bioavailable amino acids, we compared these results with nine auxotrophs treated with lysates of B. thetaiotaomicron infected with the lytic phage ФSJC12 and found they supported auxotroph growth to a higher degree than B. thetaiotaomicron supernatants or mechanical lysates with, in three cases, over 30 auxotroph cells supported per producer cell (Fig. 2C). Together, these results demonstrate that phage lysis contributes a higher level of most amino acids compared to mechanical lysis or secretion; different phage infections and host cells vary in the levels of bioavailable amino acids released upon lysis; and in general, biosynthetically costly amino acids are released at higher levels than less costly amino acids. Though these results are limited to two producer bacteria and three phages in artificial conditions, they suggest that infection of diverse bacteria with different phages could support a variety of auxotrophs in microbial ecosystems. We postulate that phage lysis may be a dominant mechanism of intracellular nutrient release, potentially shaping the nutritional environment and contributing to the evolution of auxotrophy.
Fig. 2. Phage lysates robustly support growth of certain amino acid auxotrophs.
The number of auxotroph cells supported per producer cell was calculated for auxotrophs grown in defined medium containing (A) E. coli T4rI phage lysate, (B) E. coli λvir lysate and (C) B. thetaiotaomicron ϕSJC12 lysate. The amino acids are ordered as described in Fig. 1. Bars represent the mean of three replicates; error bars represent standard deviation.
Supplementary information
Acknowledgements
We thank Richard Calendar (in memoriam) and Andrew Hryckowian for providing phage stocks. We are grateful to members of the Taga lab for helpful discussions, the UC Berkeley Department of Statistics consulting service for advice on statistical analysis, and Janani Hariharan, Alexa Nicolas, and Zachary Hallberg for critical reading of the manuscript. This work was funded by an NSF doctoral fellowship to GJP and NIH grant R35GM139633 to MET.
Author contributions
GJP and MET conceived the study and designed the experiments. GJP performed the experiments and analyzed the data. MET secured funding and supervised the study. All authors contributed to manuscript writing.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01452-7.
References
- 1.Seth EC, Taga ME. Nutrient cross-feeding in the microbial world. Front Microbiol. 2014;5:350. doi: 10.3389/fmicb.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson WM, Alexander H, Bier RL, Miller DR, Muscarella ME, Pitz KJ, et al. Auxotrophic interactions: a stabilizing attribute of aquatic microbial communities? FEMS Microbiol Ecol. 2020;96:fiaa115. doi: 10.1093/femsec/fiaa115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pande S, Merker H, Bohl K, Reichelt M, Schuster S, de Figueiredo LF, et al. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J. 2014;8:953–62. doi: 10.1038/ismej.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA. 2014;111:E2149–56. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Souza G, Waschina S, Pande S, Bohl K, Kaleta C, Kost C. Less is more: selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution. 2014;68:2559–70. doi: 10.1111/evo.12468. [DOI] [PubMed] [Google Scholar]
- 6.Gude S, Pherribo GJ, Taga ME. Emergence of metabolite provisioning as a by-product of evolved biological functions. mSystems. 2020;5:e00259–20. doi: 10.1128/mSystems.00259-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baran R, Brodie EL, Mayberry-Lewis J, Hummel E, Da Rocha UN, Chakraborty R, et al. Exometabolite niche partitioning among sympatric soil bacteria. Nat Commun. 2015;6:8289. doi: 10.1038/ncomms9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paczia N, Nilgen A, Lehmann T, Gatgens J, Wiechert W, Noack S. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Micro Cell Fact. 2012;11:122. doi: 10.1186/1475-2859-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammarlund SP, Gedeon T, Carlson RP, Harcombe WR. Limitation by a shared mutualist promotes coexistence of multiple competing partners. Nat Commun. 2021;12:619. doi: 10.1038/s41467-021-20922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giri S, Oña L, Waschina S, Shitut S, Yousif G, Kaleta C, et al. Metabolic dissimilarity determines the establishment of cross-feeding interactions in bacteria. Curr Biol. 2021;31:5547–57.e6. doi: 10.1016/j.cub.2021.10.019. [DOI] [PubMed] [Google Scholar]
- 11.Lopez D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol. 2009;74:609–18. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Gestel J, Vlamakis H, Kolter R. Division of labor in biofilms: the ecology of cell differentiation. Microbiol Spectr. 2015;3:MB-0002–2014. doi: 10.1128/microbiolspec.MB-0002-2014. [DOI] [PubMed] [Google Scholar]
- 13.Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–35. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokol NW, Slessarev E, Marschmann GL, Nicolas A, Blazewicz SJ, Brodie EL, et al. Life and death in the soil microbiome: how ecological processes influence biogeochemistry. Nat Rev Microbiol. 2022;20:415–30. doi: 10.1038/s41579-022-00695-z. [DOI] [PubMed] [Google Scholar]
- 15.Suttle CA. Marine viruses–major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 16.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–8. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 17.Fazzino L, Anisman J, Chacon JM, Heineman RH, Harcombe WR. Lytic bacteriophage have diverse indirect effects in a synthetic cross-feeding community. ISME J. 2020;14:123–34. doi: 10.1038/s41396-019-0511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma C, Ma A, Gong G, Liu Z, Wu Z, Guo B, et al. Cracking Streptococcus thermophilus to stimulate the growth of the probiotic Lactobacillus casei in co-culture. Int J Food Microbiol. 2015;210:42–6. doi: 10.1016/j.ijfoodmicro.2015.04.034. [DOI] [PubMed] [Google Scholar]
- 19.Ankrah NY, May AL, Middleton JL, Jones DR, Hadden MK, Gooding JR, et al. Phage infection of an environmentally relevant marine bacterium alters host metabolism and lysate composition. ISME J. 2014;8:1089–100. doi: 10.1038/ismej.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard-Varona C, Lindback MM, Bastien GE, Solonenko N, Zayed AA, Jang H, et al. Phage-specific metabolic reprogramming of virocells. ISME J. 2020;14:881–95. doi: 10.1038/s41396-019-0580-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.