Visual Abstract
Key Words: ACMG classification, familial hypercholesterolemia, high-throughput characterization, LDLR
Highlights
-
•
Around 40% of LDLR variants are classified as VUS, posing as the biggest hurdle to reach a definitive FH diagnosis.
-
•
We optimized and validated a time- and cost-effective high-throughput cell-based assay to functionally profile rare LDLR variants.
-
•
This methodology is a valuable resource for systematic functional characterization of LDLR variants, with the potential to discriminate disruptive rare variants from silent ones, solving 1 of the major issues in definitive FH diagnosis.
Summary
Familial hypercholesterolemia (FH) is the most common inherited life-threatening disorder of lipid metabolism. Early diagnosis and treatment are the key to reduce the cumulative life-long cardiovascular burden of patients with FH. The high number of LDLR variants described as variants of unknown significance is the largest obstacle to achieve a definitive FH diagnosis. This study established a time- and cost-effective high-throughput cell-based assay to functionally profile LDLR variants, which allowed us to discriminate disruptive rare variants from silent ones. This work generated a valuable resource for systematic functional characterization of LDLR variants solving 1 of the major issues to achieve a definitive FH diagnosis.
Familial hypercholesterolemia (FH) (OMIM 143890) is a common autosomal semidominant disorder of lipid metabolism, with a heterozygous frequency around 1:250 in most populations, according to a recent meta-analysis.1 Clinically characterized by high concentration of plasma cholesterol, premature atherosclerosis, and coronary heart disease, FH is markedly underdiagnosed and undertreated in the general population, especially in children,2 in which early identification could lead to the implementation of important strategies for coronary heart disease prevention in adulthood. Currently, the genetic diagnosis is made by finding a causative variant in 1 of the 3 genes known to cause FH (LDLR, APOB, and PCSK9), with 90% of the cases traced to LDLR.3,4 The role of LDLR in cholesterol metabolism was first unveiled by Brown and Goldstein, namely its ability to bind and internalize low-density lipoprotein (LDL) particles being the key mechanism through which most cell types uptake cholesterol.5
As next-generation sequencing techniques became more widely used by diagnostic laboratories, the number of variants identified in genes associated with different disorders is continuously increasing, with FH being no exception.6 This new genomic era produces a huge quantity of data, which needs to be followed by rapid, low-cost, high-throughput, and validated functional assays.4 So far, >3,500 LDLR variants are listed in the ClinVar database (National Center for Biotechnology Information) with >2,300 unique LDLR variants detected in patients with FH, with about one-half of these being missense variants of unknown clinical significance.7 Despite the high number of variants, the majority lacks functional evidence to evaluate their pathogenicity, with <15% of these variants actually proven to affect LDLR function by functional studies, which is among the highest forms of evidence for a variant pathogenicity.8 LDLR variants are functionally divided into 5 categories: class I—no protein synthesis (null variants); class II—complete (IIa) or partial (IIb) retention of the protein at the endoplasmic reticulum; class III—defective binding; class IV—defective internalization or endocytosis; class V—defective recycling.9 Variants from classes II-V (except class IIa) are considered defective variants because the protein still retains some functionality. Variants from classes I and IIa produce no functional LDLR and therefore are considered null variants.
Early diagnosis and treatment are the key to reducing the cumulative life-long cardiovascular burden that these patients have.10 The likelihood of developing coronary artery disease is 3-fold higher in heterozygous of pathogenic variants compared with noncarriers for LDL plasma levels >190 mg/dL.11 Moreover, the type of variant and level of LDLR function impairment have been proven to be an important modulator of LDL cholesterol levels,11, 12, 13 and they can further influence the response of patients to statin therapy.14,15 These data highlight the relevance of correct classification of genetic variants according to their LDLR function for risk evaluation and treatment optimization, which poses as a unique opportunity for personalized treatment and disease management.
With this work our objective was to answer a fundamental challenge of contemporary genetics, namely distinguishing disruptive rare variants from benign ones. With this purpose, we established a time- and cost-effective high-throughput assay to functionally profile LDLR variants, which allows us to discriminate the biological effects and likely disease relevance of rare LDLR missense variants, contributing to an improved variant classification, and consequently to better diagnosis, management, and prognosis of the patient’s condition.
Methods
LDLR variant selection
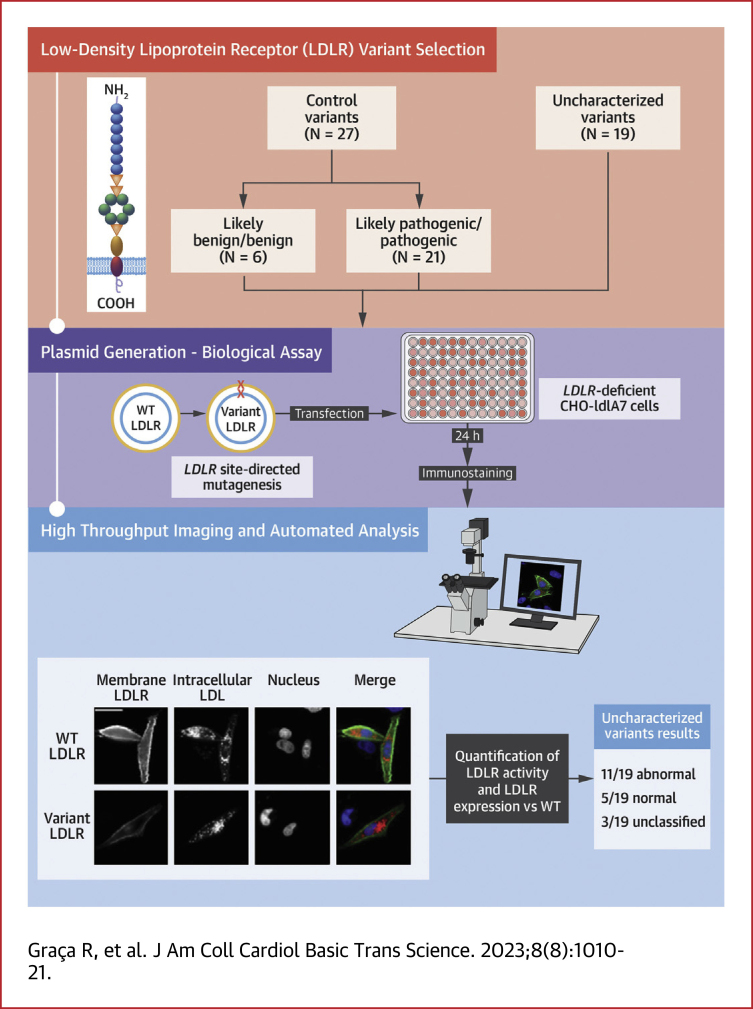
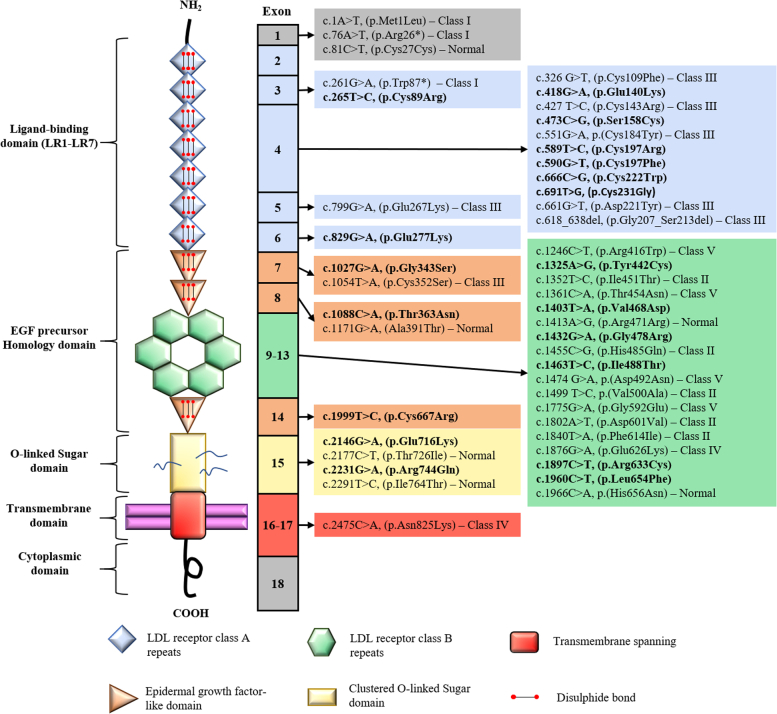
Two sets of variants were characterized in this work, adding up to 46 LDLR variants in total (Figure 1). The control set (n = 27) (Supplemental Table 1) consisted of previously functionally characterized variants (n = 22)16, 17, 18, 19 and a subset of LDLR variants reaching an American College of Medical Genetics (ACMG) classification of likely benign/benign (n = 3) and likely pathogenic/pathogenic (n = 2) without using the functional criteria, fulfilling the requirements for a validation study following the Clinical Genome Resource (ClinGen) guidelines.20 The variants of the control group were chosen to cover all 5 classes of LDLR variants, with 3 class I variants, 5 class II variants, 7 class III variants, 2 class IV variants, 4 class V, and 6 variants shown not to affect LDLR function. The test set included 19 uncharacterized LDLR variants, all identified in patients clinically diagnosed with FH (mean LDL cholesterol 252.2 ± 67.9 mg/dL), who were enrolled in the Portuguese FH study. Initiated in 1999, the Portuguese FH study follows the Simon Broome criteria: total cholesterol ≥260 mg/dL (6.7 mmol/L) or LDL cholesterol ≥155 mg/dL (4.0 mmol/L) for children under 16 years old or total cholesterol ≥290 mg/dL (7.5 mmol/L) or LDL cholesterol ≥190 mg/dL (4.9 mmol/L) for adults, as well as a family history of hypercholesterolemia or premature coronary heart disease.21,22 The study protocol and database have been approved by the National Institute of Health Ethics Committee and the National Data Protection Commission, respectively. All patients gave their informed consent to participate in the genetic study. Moreover, all uncharacterized variants are rare with a minor allele frequency <0.1% and were absent from a panel of 100 normolipidemic Portuguese control subjects.
Figure 1.
LDLR Variants Location
LDLR protein schematic showing domain and exon location of all characterized variants. Variants from the control set (n = 27) are listed with the correspondent variant class, whereas the uncharacterized variants, from the test set (n = 19), are in bold. EGF = epidermal growth factor; LDL = low-density lipoprotein; LR = LDL receptor class A repeats.
ACMG classification
All uncharacterized variants were classified according to the ClinGen FH Variant Curation Expert Panel (VCEP) LDLR specifications based on ACMG/Association for Molecular Pathology Variant Interpretation Guidelines,23 with and without integration of the functional data.
Site-directed mutagenesis
The mammalian expression vector pcDNA3 expressing human LDLR complementary DNA (NM_000527.4) was the template used to introduce all studied variants using NZYMutagenesis kit (NZYTech) according to the manufacturer’s instructions. Oligonucleotides used to generate the different LDLR variants carrying plasmids were all checked for single-nucleotide variations using SNPCheck (EMQN and Certus Technology). The presence of the correct variant introduced and integrity of the region were confirmed by direct Sanger sequencing.
Cell culture and transfection
LDLR-deficient Chinese hamster ovary cell line ldlA7 (CHO-ldlA7) cells (kindly provided by Dr Monty Krieger, Massachusetts Institute of Technology) were cultured in F-12 Nut mix (Gibco) medium supplemented with 10% (vol/vol) fetal bovine serum (Invitrogen), 100 units/mL penicillin, and 100 μg/mL streptomycin. For microscopy assays, cells were seeded in 24-well and 96-well glass bottom plates at a density of 2.5 × 104 cells/well and 2.5 × 103 cells/well, respectively. Twenty-four hours after seeding, cells were starved with Opti-MEM (Gibco) medium until transfection. Plasmids with wild-type or mutant LDLR variants were transfected using Lipofectamine 2000 reagent (Invitrogen) following manufacturer’s instructions. Microscopy assays were performed 24 hours after transfection.
High-throughput microscopy screening
Biological assay
Culture medium was changed 24 hours after transfection start for Opti-MEM medium with 2.5 μg/mL DiI-LDL (Invitrogen), and internalization was stimulated for 3 hours at 37 °C. Cells were first briefly washed with imaging medium (Opti-MEM without phenol red, containing 30 mmol/L N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid and 0.5 g/L NaHCO3 [pH 7.4])/0.2% bovine serum albumin (Invitrogen) then noninternalized LDL was removed from plasma membrane with acidic wash (imaging medium pH 3.5) for 30 seconds. Cells were then fixed with 4% paraformaldehyde and counterstained for LDLR on plasma membrane (Mouse anti-human-LDLR IgG-C7 [Progen Biotechnik GmbH] and Alexa Fluor 488-conjugated chicken anti-mouse IgG [Invitrogen]) for nuclei (Hoechst [Thermo Scientific]) and cell outlines (Draq5 [Biostatus]).
Image acquisition and image data analysis
Images were acquired on a fully automated widefield Olympus IX81 microscope, using an UPlanApo 20 × 0.7 NA objective and ScanR software (version 2.1.0.15, Olympus Biosciences). For each 24-well plate and 96-well plate, images were taken with identical baseline settings from 100 and 36 different subpositions per well, respectively.
Quality control of experiments was visually performed using Image J24(National Institutes of Health) to exclude images not fulfilling predefined criteria (eg, out of focus or too high cell density). Image data analysis was performed with customized pipelines based on CellProfiler 2.2 software25 and in-house developed tool HTM Explorer.
Statistical methods
Cell-based functional profiling allowed us to distinguish functionally normal from functionally abnormal rare LDLR variants. Scatter plots depict LDLR activity (y-axis) and LDLR expression (x-axis), both in arbitrary units. LDLR activity and expression values were derived from the signal intensities of total DiI-LDL in endocytic subcellular compartments and from Alexa Fluor 488 signal, respectively. Each scatter plot illustrates 1 characterized variant (black dots) and the wild-type (red dots) from 1 experimental replica, where each dot represents a single cell.
Graph bars portray the mean value of LDLR expression and activity obtained from 4 experimental replicas. Error bars represent SD. For each LDLR variant the expression and activity was normalized against the wild type. Nominal values for each variant from the control set and test set were indexed in Supplemental Table 1 and Table 1, respectively.
Table 1.
ACMG/AMP Classification of the Test Set Variant With and Without Functional Data
| ClinVar ID | Number of Carriers (LDL-C [mg/dL])a | FS Results (%)b |
ACMG Classification Without FS | Criteria Met Without FS | ACMG Classification After FS Level 1c | ACMG Classification After FS Level 3d | ||
|---|---|---|---|---|---|---|---|---|
| Expression | Activity | |||||||
| c.265T>C, (p.Cys89Arg) | 251102 | 3 (260.7 ± 39.0) | 89/99 | 74/72 | Likely pathogenice | PM1, PM2, PM3, PP1, PP3, PP4, PS4_Supporting | Likely pathogenic | Likely pathogenic |
| c.418G>A, (p.Glu140Lys) | 251213 | 7 (218.2 ± 36.4) | 114/104 | 37/42 | Pathogenice | PP1_Strong, PM1, PM2, PS4_Supporting, PP3, PP4 | Pathogenic | Pathogenic |
| c.473C>G, (p.Ser158Cys) | 251245 | 1 (353) | 78/86 | 49/48 | VUS | PM1, PM2, PP4, BP4 | Likely pathogenic | Likely pathogenic |
| c.589T>C, (p.Cys197Arg) | 183091 | 9 (273.6 ± 56.5) | 99/101 | 27/37 | Likely pathogenic | PM1, PM2, PP3, PP4, PP1_Moderate, PS4_Supporting | Pathogenic | Likely pathogenic |
| c.590G>T, (p.Cys197Phe) | 251309 | 3 (290.5 ± 13.4) | 98/106 | 31/43 | Likely pathogenice | PM1, PM2, PP3, PP4, PP1_Moderate | Pathogenic | Likely pathogenic |
| c.666C>G, (p.Cys222Trp) | 251365 | 2 (196.5 ± 14.8) | 103/111 | 31/43 | Likely pathogenic | PM1, PM2, PP3, PP4 | Pathogenic | Likely pathogenic |
| c.691T>G, (p.Cys231Gly) | 251397 | 1 (289) | 94/108 | 29/35 | Likely pathogenic | PM1, PM2, PP3, PP4, PS4_Supporting | Pathogenic | Likely pathogenic |
| c.829G>A, (p.Glu277Lys) | 183097 | 6 (194.8 ± 52.8) | 94/107 | 86/79 | VUS | — | VUS | VUS |
| c.1027G>A, (p.Gly343Ser) | 183106 | 5 (205.1 ± 12.9) | 100/104 | 69/ 64 | Pathogenic | PP1_Strong, PM2, PS4_Moderate, PP3, PP4 | Pathogenic | Pathogenic |
| c.1088C>A, (p.Thr363Asn) | 251656 | 1 (202) | 90/107 | 95/98 | VUS | PM2, PP4, BP4 | Likely benign | Likely benign |
| c.1325A>G, (p.Tyr442Cys) | 251787 | 1 (224) | <10/<1 | <10/<10 | VUS | PM2, PP3, PP4, PS4_Supporting | Likely pathogenic | Likely pathogenic |
| c.1403T>A, (p.Val468Asp) | 251828 | 1 (143)f | 103/100 | 95/99 | VUSe | PM2, PP3, PP4 | VUS | VUS |
| c.1432G>A, (p.Gly478Arg) | 161277 | 9 (204.0 ± 69.5) | 33/43 | 68/66 | Pathogenice | PP1_Strong, PS4, PM2, PP3, PP4 | Pathogenic | Pathogenic |
| c.1463T>C, (p.Ile488Thr) | 251857 | 8 (224.4 ± 43.4) | 20/39 | 63/45 | Likely pathogenic | PM2, PP1, PP3, PP4, PS4_Supporting | Pathogenic | Likely pathogenic |
| c.1897C>T, (p.Arg633Cys) | 226379 | 2 (217.7 ± 47.5) | 95/99 | 99/107 | Likely pathogenic | PM2, PP1_Moderate, PS4_Moderate, PP3, PP4 | Likely pathogenic | Likely pathogenic |
| c.1960C>T, (p.Leu654Phe) | 252132 | 3 (233.7 ± 41.5) | 79/84 | 89/108 | VUS | PM2, PP4, PS4_Supporting | VUS | VUS |
| c.1999T>C, (p.Cys667Arg) | 252163 | 1 (273) | <10/<10 | <10/<10 | Likely pathogenic | PM1, PM2, PP3, PP4, PS4_Supporting | Pathogenic | Likely pathogenic |
| c.2146G>A, (p.Glu716Lys) | 252241 | 1 (299)g | 94/101 | 102/93 | VUS | PM2, PP4, BP4 | Likely benign | Likely benign |
| c.2231G>A, (p.Arg744Gln) | 68104 | 1 (253)h | 100/117 | 100/90 | Likely benigne | BP2, BP4, PS3_Supporting | Likely benign | Likely benign |
Percentages in bold represent values below the 70% cutoff and thus affect LDLR function.
ACMG = American College of Medical Genetics; AMP = Association for Molecular Pathology; BP = benign supporting; FS = functional study; BS = benign strong; LDL-C = low-density lipoprotein cholesterol; PM = pathogenic moderate; PP = pathogenic supporting; PS = pathogenic strong; VUS = variant of uncertain significance.
Parenthetical values are mean ± SD.
Percentages for the 24-well and 96-well assays, on the left and right, respectively.
Classification with additional PS3 or BS3 point.
Classification with additional PS3_Supporting or BS3_Supporting point.
Classification performed by the ClinGen’s Familial Hypercholesterolemia Variant Curation Expert Panel.
LDL level achieved under combined therapy of statin and ezetimibe.
Individual also carrying variant NM_000527.5(LDLR): c.670G>A (p.Asp224Asn).
Individual also carrying variant NM_000384.3(APOB): c.10580G>A (p.Arg3527Gln).
Results
Microscope-based approach to functionally profile rare LDLR variants
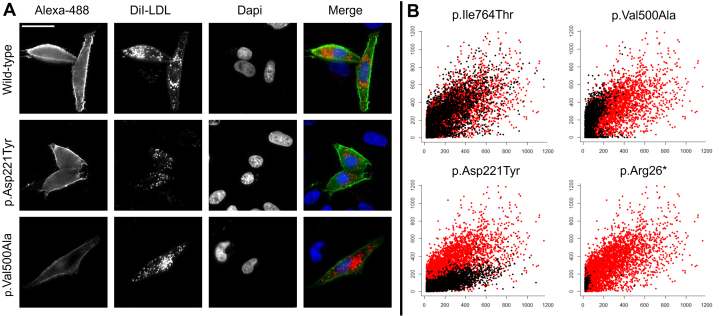
Aiming to functionally distinguish between variants in LDLR affecting (abnormal) and not affecting (normal) LDLR function, we established a quantitative and high-throughput in vitro assay to functionally profile LDLR variants. In our approach, we overexpressed wild-type or mutated LDLR in cultured cells without endogenous LDLR expression, bypassing an extra silencing step and avoiding interfering stimulus from basal expression. LDLR activity (quantified by the amount of LDL inside the cell) and LDLR expression at cell surface was quantified by multiparametric analysis from images acquired by high-content automated microscopy (Figure 2A).
Figure 2.
Image Acquisition
Cell-based functional profiling distinguishes functionally normal from functionally abnormal rare LDLR variants. (A) Images automatically acquired of Chinese hamster ovary (CHO)-ldlA7 cells transiently expressing wild-type LDLR, variant p.Asp221Tyr or variant p.Val500Ala. LDLR variants were characterized by monitoring cellular internalization of fluorescently labeled low-density lipoprotein (LDL) (DiI-LDL; red) and quantifying LDLR expression at cell surface (Alexa Fluor 488; green). Nuclei are labeled with 4′,6-diamidino-2-phenylindole (DAPI) (blue). All images were acquired on a fully automated widefield Olympus IX81 microscope, using an UPlanApo 20 × 0.7 NA objective and ScanR software (version 2.1.0.15, Olympus Biosciences). Bar = 25 μm. (B) Graphs depict relative signal intensities of total DiI-LDL in endocytic subcellular compartments (y-axis, in arbitrary units) plotted against LDLR expression (x-axis, in arbitrary units). Each individual scatter plot represents a LDLR variant (black dots) and the wild type (red dots), where each dot corresponds to a single cell. CHO-ldlA7 cells transiently expressing normal variant p.Ile764Thr (upper left), class II variant Val500Ala (upper right), class III variant p.Asp221Tyr (bottom left), and class I variant p.Arg26∗ (bottom right).
The range of functional phenotypes detected by this methodology can be divided in 4 variant groups, according to LDLR cell surface expression and LDLR activity (LDL binding plus uptake). An example of the results obtained can be seen in Figure 2A, which shows images automatically acquired of CHO-ldlA7 cells transiently expressing wild-type LDLR, variant p.Asp221Tyr or variant p.Val500Ala. Figure 2B shows an example of each group: 1) (upper left graph: p.Ile764Thr) variants with normal expression and activity, associated to silent variants that do not affect LDLR function and behave as the wild type; 2) (upper right graph: p.Val500Ala) variants with reduced expression and activity, associated to class IIb variants with partial ER retention or class V variants (defective recycling); 3) (bottom right graph: p.Arg26∗) variants showing no LDLR expression and activity, related to class I variants or class IIa variants with total ER retention (null variants); 4) (bottom left graph: p.Asp221Tyr) variants with normal expression and reduced activity, with reduced binding and/or reduced consequent internalization, related to class III (defective binding) or class IV (defective internalization) variants.17
Control set
Following the ACMG FH VCEP guidelines, the thresholds of >90% and <70% of wild-type activity were used to define variants with normal functionality and abnormal functionality, respectively.
The 5 control variants—p.Met1Leu, p.Arg26∗, p.Trp87∗, p.Ile451Thr, and p.Asp601Val—exhibiting a null phenotype (ie, lack of cell surface expression and protein activity) were easily identified both in the 24-well and 96-well assays (Figure 3A). The normal LDLR activity control variants—p.Cys27Cys, Ala391Thr, p.Arg471Arg, His656Asn, p.Thr726Ile, and p.Ile764Thr—were all correctly assigned in the 24-well and 96-well assays, with the exception of the synonymous variant, p.Cys27Cys, which in the 96-well assay had a activity value (85% ± 6%) slightly below the 90% threshold (Figure 3A). Class III variants, with affected binding—p.Cys109Phe, p.Cys143Arg, Cys184Tyr, p.Gly207_Ser213del, p.Asp221Tyr, p.Glu267Lys, and p.Cys352Ser—were all correctly identified in both assays, having normal expression but abnormal binding and internalization (Figure 3A). Variants p.Glu626Lys and p.Asn825Lys, both categorized as LDLR class IV variants, fell in the gray area between thresholds (70%, 90%) in the 96-well assay and p.Asn825Lys also in the 24-well assay, whereas the p.Glu626Lys was correctly identified as functionally abnormal in the 24-well format (Figure 3A). The remaining control variants included variants with partial expression at cell surface— p.His485Gln, p.Val500Ala, and p.Phe614Ile—or recycling defects—p.Arg416Trp, p.Thr454Asn, p.Asp492Asn, and p.Gly592Glu (Figure 3A). Of these, only variants p.Val500Ala and p.Gly592Glu were unanimously identified as functionally abnormal by 24-well and 96 well assays. Variants p.His485Gln, p.Asp492Asn, and p.Phe614Ile were equally identified as functionally abnormal, but exclusively in the 24-well assay, falling into the gray area for the 96-well assay by borderline values. The 2 remaining variants—p.Arg416Trp and p.Thr454Asn—both with recycling defects, exhibited expression and/or activity values between the 70% and 90% thresholds in the microscopy assays but not with the reference cytometry assay.
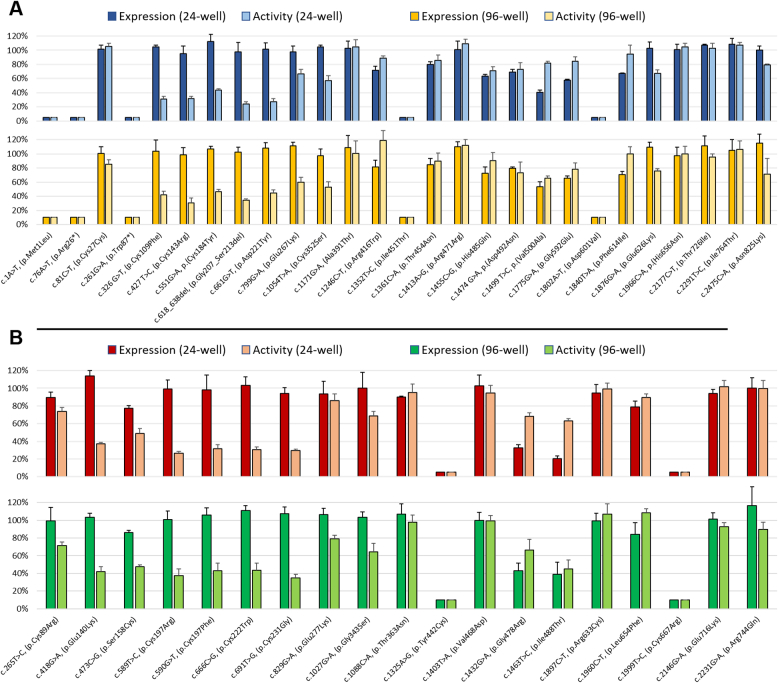
Figure 3.
LDLR Functional Profile
Functional profile of LDLR in CHO)-ldlA7 cells transfected with different LDLR variants. LDLR expression and activity of variants included in the control set (A) and test set (B). Graph bars represent the expression and overall activity of each sample normalized for wild-type LDLR, in both 24-well and 96-well assays. LDLR expression at cell surface was quantified by Alexa Fluor 488 signal, and LDLR activity was measured by total DiI-LDL signal in endocytic compartments. The values in bar graphs represent the mean of 4 experimental replicas. Error bars represent ± SD. Abbreviations as in Figures 1 and 2.
Test set
The above-mentioned thresholds for LDLR variants’ functional classification were applied to the test set variants. Accordingly, 11 LDLR variants exhibited abnormal function, 5 normal function, and 3 were inconclusive, in both 24-well and 96-well assays (Figure 3B, Table 1). Variants p.Glu140Lys, p.Ser158Cys, p.Cys197Arg, p.Cys197Phe, p.Cys222Trp, p.Cys231Gly, and p.Gly343Ser shared a similar phenotype with normal expression and reduced activity. Variants p.Tyr442Cys and p.Cys667Arg featured no expression or activity, whereas variants p.Gly478Arg and p.Ile488Thr had reduced expression and activity. Variants p.Thr363Asn, p.Val468Asp, p.Arg633Cys, p.Glu716Lys, and p.Arg744Gln were unanimously classified as normal variants in 24-well and 96-well assays. Variants p.Cys89Arg, p.Glu277Lys, and p.Leu654Phe had values for expression and/or activity between thresholds, [70%, 90%], in both assays (Figure 3B).
ACMG classification
All test variants (n = 19) were classified according to newly published ACMG FH VCEP LDLR specifications (Table 1).23 Prior to functional characterization, variants were classified as pathogenic/likely pathogenic (n = 11), variants of uncertain significance (VUSs) (n = 7), and likely benign (n = 1). After functional characterization, independently of considering the functional study level 1 (PS3) or level 3 (PS3_Supporting), the inclusion of functional data allowed us to reach a conclusive AMCG classification for 4 variants previously classified as VUSs. If level 1 was attributed to this assay, the classification of 6 variants would be upgraded from likely pathogenic to pathogenic.
Discussion
The approach for functional characterization of LDLR variants presented in this work was designed in compliance with all the specifications required to be classified as a level 1 functional study according to the ClinGen FH Expert Panel specifications to the ACMG/AMP variant interpretation guidelines.23 Briefly, the whole cycle of LDLR was studied in heterologous cells, without endogenous LDLR and transfected with mutant plasmid of interest. The described method was successfully validated with >2 variants classified as likely benign/benign, and 2 variants classified as likely pathogenic/pathogenic without functional evidence as requested by the functional analysis guideline.20
The functional characterization of LDLR variants has been performed over the last several decades by flow cytometry, a well-stablished and accepted level 1 functional study.23 Although this procedure gives an accurate and reliable characterization of variants, it is time-consuming and labor-intensive and does not allow a simple and rapid functional characterization of putative FH-causing variants, leaving many patients without a definite diagnosis. The work by Thormaehlen et al26 showed an approach to profile missense variants through systematic overexpression and complementation experiments in HeLa-Kyoto cells. However, the lack of control variants in these experiments as specified in ClinGen guidelines for the establishment and validation of functional assays20 and the use of a human cell line, with normal LDLR expression, prevents this approach from reaching a level 1 classification, and it leads to many inconclusive results. This methodology is classified by the ClinGen guidelines as level 3 functional study.
In the present work, we merged these 2 approaches to get the best attributes of each one. We successfully combined the expedited characterization from high-throughput microscopy and the “clean” biological system (CHO-ldlA7 cells) used in the cytometry assays. In the first phase of the work, all LDLR variants were characterized in 24-well plates as a proof-of-concept of the proposed methodology. During the next phase, all variants were characterized in 96-well plates to scale up the assay to more high-throughput characterization methodology. If we take the time needed to characterize a 20-variant sample with the cytometry methodology used in recent works18,19 as a base of comparison, the 24-well and 96-well formats would take, respectively, one-third and one-sixth of the time to achieve the same output.
Our control set was chosen to be representative of the 5 LDLR variant classes and cover all main domains of the LDLR, namely, the ligand-binding domain, the epidermal growth factor precursor homology domain, the O-linked oligosaccharide-rich domain, and the transmembrane domain.27 With this variability of control variants and wide spectrum of functional phenotypes, we could ascertain the reliability and accuracy of this new approach in the functional profiling of the different classes of variants. For the phenotypes at the extremities of the spectrum, null and normal function variants, the assays had an excellent performance, correctly identifying all variants in these 2 groups, in both the 24-well and the 96-well plate assays. Likewise, all control variants with binding defects were identified as affecting the normal function of LDLR.
In the group of control variants with uptake defects and diminished cell surface expression (partial endoplasmic reticulum [ER] retention or recycling defects), it was not possible to reach a functional classification for 7 of 9 variants, in at least 1 of the formats used, because the levels of expression and/or activity fell within the gray zone of thresholds defined by ACMG guidelines for LDLR variant classification. For this subset of variants, the 24-well format had a better performance because it identified 6 of 9 defective variants. This type of hypomorphic variant is usually associated with a milder phenotype and so a milder reduction of LDLR activity, which creates difficulties in functional data interpretation. Indeed, 3 of these variants—p.Arg416Trp, p.Thr454Asn, and p.Asp492Asn—were previously characterized by cytometry as recycling defective variants, exhibiting expression, binding, and internalization levels around 60%.17,19 In a similar way, the control variants with uptake defects, p.Glu626Lys and p.Asn825Lys, were previously characterized by cytometry with internalization levels around 60%.17,19 Finally, control variants p.His485Gln and p.Phe614Ile, both falling in the gray area for functional classification in the 96-well assays, were functionally characterized as having expression, binding, and internalization levels near 50%.19 For this group of variants, it was also possible to observe that activity values exceeded the value of LDLR expression. Although in a smaller scale, this phenomenon was also observed in cytometry assays, but in this new assay this effect was possibly enhanced by specificities of the technique.
To overcome this apparent lack of specificity of this assay to accurately identify variants with expression and recycling defects, a conservative approach is recommended for now, and variants with results between 70% and 90% in the high-throughput assay should be reanalyzed by cytometry to fine tune the LDLR activity.
To improve the method’s sensitivity, in future work, we will fine-tune the assay to better identify variants with milder phenotypes. Here we are working with an overexpression system, which for these specific variants might hamper the best readout of the assay and lead to an overestimation of activity for class IIb and IV variants. To work out this situation, we could consider adjusting the transfection conditions. Another possibility of adjustment is the time of cell exposure to LDL. As described for the cytometry methodology, we stimulated LDL internalization for 3 hours at 37 ºC, and this long period of time can overstimulate these variants and result in an overestimation of their function. In subsequent works, the time can be shortened to 30-minute chase experiments.
In parallel to the characterization of the control set used to validate and fine-tune the pipeline, at the beginning of this project we characterized 19 rare LDLR variants without level 1 functional studies. In total, 11 variants displayed abnormal function, 5 normal function, and 3 remained unclassified and should therefore be reanalyzed by cytometry.
Variants p.Glu140Lys, p.Ser158Cys, p.Cys197Arg, p.Cys197Phe, p.Cys222Trp, p.Cys231Gly, and c.1027G>A, (p.Gly343Ser), share a similar functional profile with normal expression and reduced activity (Figure 3B). Six of these variants are in exon 4, which is the exon with the most described variants7 and most pathogenic variants confirmed by functional studies. This exon is a fundamental part of the ligand-binding domain and without surprise several variants in exon 4 have been functionally characterized as binding-defective variants.17, 18, 19,28 Altogether, the inability of these variants to internalize LDL and their location indicate that these point substitutions affect LDLR-binding ability and can be classified as class III LDLR variants. Variants p.Tyr442Cys and p.Cys667Arg have a near complete loss of LDLR expression at the cell surface and overall activity (Figure 3B). This functional behavior matches the profile of class IIa, with total ER retention and are therefore considered null variants. Besides, other variants in the same domain, epidermal growth factor precursor homology domain, have been descried with similar phenotypes.17, 18, 19 The last 2 functionally abnormal variants are p.Gly478Arg and p.Ile488Thr, both with reduced expression and activity (Figure 3B). This phenotype fits class IIb variants with partial ER retention and class V variants with defective recycling. However, considering the milder phenotype observed in class V variants,17,19 together with the behavior of the control variants with recycling defects, these 2 variants, with expression levels between 20% and 40%, are possibly partially retained at the ER. Experiments to characterize recycling variants18,19 can be performed for these variants to correctly attribute the functional class.
As previously mentioned, variants p.Thr363Asn, p.Val468Asp, p.Arg633Cys, p.Glu716Lys, and p.Arg744Gln showed cell surface expression and activity similar to wild-type LDLR. Thus, these variants are considered silent variants that do not affect LDLR function. Completing the test set, variants p.Cys89Arg, p.Glu277Lys, and p.Leu654Phe presented expression and/or activity values between the 70% and 90% thresholds in at least 1 of the performed assays. For this reason, these variants are neither classified as normal or abnormal functional LDLR variants. For these variants, the flow cytometry assay should be performed to see whether a more sensitive assay can attribute a final functional phenotype.
In recent years genetic testing has emerged as a key part of FH diagnosis. Accordingly, several international entities have officially recommended genetic testing for FH.2,10 To meet this end, a correct and definitive variant classification is essential. In this work, the addition of new functional data made it possible to determine 4 of 7 variants of uncertain significance to reach a clear classification (Table 1) that are now classified as likely pathogenic (p.Ser158Cys and p.Tyr442Cys) or likely benign (p.Thr363Asn and p.Glu716Lys). Currently, around 50% of the described LDLR missense variants are classified as VUSs,8 for which functional data can provide substantial help to upgrade its status to an actionable classification.8 If the proportion of solved VUSs in our test set (57%) is maintained in the remaining LDLR VUSs, it would be a major step for variant classification, with one-half of these variants reaching a clear classification and allowing a definitive diagnosis of FH. This percentage could be even higher if case level data as informative cosegregation in 4-5 relatives could be obtained for these variants because all remaining VUSs would be upgraded to likely pathogenic with just this additional information.
Supporting the accuracy of this methodology, we verified that for 10 of the 11 test variants (91%) with a definitive classification (prior to functional evidence), either likely pathogenic/pathogenic or likely benign/benign, the functional characterization data were concordant with the variants’ classification. The only variant that did not show this straight correlation was variant c.265T>C, (p.Cys89Arg) with values for LDLR activity of 74% ± 5% and 72% ± 4% in the 24-well assays and 96-well assays, respectively.
As a new methodology for functional characterization, it is expected that the functional study level attributed will raise discussion. Although our approach was designed to meet all requirements to be considered a level 1 functional study, the FH VCEP guidelines categorize high-throughput assays as level 3 functional studies.23 However, the guidelines also mention that after validation of any new assay as described in the ClinGen guidelines,23 the assay can be upgraded to level 2 or even to level 1, depending on the assay specificities. Hopefully the new evidence provided here for this methodology will be taken into consideration when this guideline is revised, allowing this new assay to reach a level 1 functional assay.
Study limitations
It is important to note that our methodology did not misclassify any of the control variants, it was simply unable to determine, according to the currently defined cutoffs, whether some variants affect or do not affect LDLR normal function. Moreover, independently of the attributed level to the study, in our test set, functional data had the power to clarify 57% of the tested VUSs. These hallmarks make our approach an excellent screening method to quickly and accurately differentiate abnormal and normal LDLR variants, decreasing the number of variants that need to be characterized by the more time-consuming and laborious reference method. It thus can be expected that the implementation of this methodology will allow in the future several VUSs to reach an improved classification, either reaching an actionable classification (likely pathogenic or pathogenic) or highlighting the need to search for another cause for the unexplained hypercholesterolemia (likely benign and benign variants).
Conclusions
With this work we established a new high-throughput cell-based strategy to characterize the biological effects and likely disease relevance of rare LDLR variants. As with all methodologies, this new approach has its strengths and weaknesses. Although at this time the data output does not allow us to unequivocally distinguish class IIb and class V variants or class III and class IV variants, it can rapidly and firmly discriminate silent variants that do not affect LDLR function from variants that strongly affect LDLR function. With the results obtained it was possible for us to validate the cutoffs for this type of high-throughput assay, allowing the assay to be used to functionally profile LDLR variants. Moreover, further work will be necessary to fine-tune the microscopy-based approach to improve the assays’ specificity for more finicky variants with milder phenotypes (those in the gray area). Altogether, our work generates a valuable resource for systematic functional characterization of LDLR variants, with the potential to quickly functionally profile a great part of LDLR VUSs, solving a major issue in definitive FH diagnosis, which is the clarification of the effect on the LDLR function of VUSs, improving its classification.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Genetic testing plays a major role in clinical evaluation and management of patients with FH and their families; however, in many cases, the genetic analysis fails to identify a disease-causing variant. This is in part caused by difficulties in classifying newly detected rare genetic variants as well as VUSs. Currently, the high number of LDLR variants described as VUSs is the biggest obstacle to achieve a definitive FH diagnosis. Here, we present a time- and cost-effective alternative to the current methodology used to functionally characterize rare LDLR variants. Our control set of variants showed that no variant was misclassified, thereby validating the assay. Additionally, 19 rare LDLR variants found worldwide were functionally characterized with this assay: 11 displayed abnormal function, 5 normal function, and 3 remain unclassified. This assay will help several patients worldwide to achieve a definite FH diagnosis allowing for treatment optimization and improvement in prognosis.
TRANSLATIONAL OUTLOOK: The high-throughput cell-based methodology described in this work, if used correctly, is a valuable resource for systematic functional characterization of LDLR variants. The elucidation of the effect on the protein level for many VUSs will lead to an improvement in the classification of such variants and, most importantly, in the diagnosis of patients with FH.
Funding Support and Author Disclosures
Mr Graça has received PhD fellowship funding from the Science and Technology Foundation (PD/BD/131427/2017) and a Christian Boulin Fellowship. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors would also like to thank Dr Monty Krieger for kindly providing LDLR-deficient CHO-ldlA7 cells, Joana Chora for her expertise in variant classification, and the team of the Advanced Light Microscopy Facility at the European Molecular Biology Laboratory for support in image acquisition.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For a supplemental table, please see the online version of this paper.
Appendix
References
- 1.Beheshti S.O., Madsen C.M., Varbo A., Nordestgaard B.G. Worldwide prevalence of familial hypercholesterolemia: meta-analyses of 11 million subjects. J Am Coll Cardiol. 2020;75(20):2553–2566. doi: 10.1016/j.jacc.2020.03.057. [DOI] [PubMed] [Google Scholar]
- 2.Nordestgaard B.G., Chapman M.J., Humphries S.E., et al. European Atherosclerosis Society Consensus Panel Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: Guidance for clinicians to prevent coronary heart disease. Eur. Heart J. 2013;34(45):3478–3490. doi: 10.1093/eurheartj/eht273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourbon M., Alves A.C., Sijbrands E.J. Low-density lipoprotein receptor mutational analysis in diagnosis of familial hypercholesterolemia. Curr Opin Lipidol. 2017;28(2):120–129. doi: 10.1097/MOL.0000000000000404. [DOI] [PubMed] [Google Scholar]
- 4.Berberich A.J., Hegele R.A. The complex molecular genetics of familial hypercholesterolaemia. Nat Rev Cardiol. 2019;16(1):9–20. doi: 10.1038/s41569-018-0052-6. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein J.L., Brown M.S. The LDL receptor. Arterioscler Thromb Vasc Biol. 2010;29(4):431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutting G.R. Annotating DNA variants is the next major goal for human genetics. Am J Hum Genet. 2014;94(1):5–10. doi: 10.1016/j.ajhg.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iacocca M.A., Chora J.R., Carrié A., et al. ClinGen FH Variant Curation Expert Panel ClinVar database of global familial hypercholesterolemia-associated DNA variants. Hum Mutat. 2018;39(11):1631–1640. doi: 10.1002/humu.23634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chora J.R., Medeiros A.M., Alves A.C., Bourbon M. Analysis of publicly available LDLR, APOB, and PCSK9 variants associated with familial hypercholesterolemia: Application of ACMG guidelines and implications for familial hypercholesterolemia diagnosis. Genet Med. 2018;20(6):591–598. doi: 10.1038/gim.2017.151. [DOI] [PubMed] [Google Scholar]
- 9.Hobbs H.H., Brown M.S., Goldstein J.L. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1(6):445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 10.Sturm A.C., Knowles J.W., Gidding S.S., et al. Convened by the Familial Hypercholesterolemia Foundation. Clinical genetic testing for familial hypercholesterolemia: JACC Scientific Expert Panel. J Am Coll Cardiol. 2018;72(6):662–680. doi: 10.1016/j.jacc.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 11.Khera A.V., Won H.-H., Peloso G.M., et al. Diagnostic yield of sequencing familial hypercholesterolemia genes in severe hypercholesterolemia. J Am Coll Cardiol. 2016;67(22):2578–2589. doi: 10.1016/j.jacc.2016.03.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourbon M., Alves A.C., Alonso, et al. Mutational analysis and genotype-phenotype relation in familial hypercholesterolemia: the SAFEHEART registry. Atherosclerosis. 2017;262:8–13. doi: 10.1016/j.atherosclerosis.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Abul-Husn N.S., Manickam K., Jones L.K., et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science. 2016;354(6319) doi: 10.1126/science.aaf7000. [DOI] [PubMed] [Google Scholar]
- 14.Santos P.C.J.L., Morgan A.C., Jannes C.E., et al. Presence and type of low density lipoprotein receptor (LDLR) mutation influences the lipid profile and response to lipid-lowering therapy in Brazilian patients with heterozygous familial hypercholesterolemia. Atherosclerosis. 2014;233(1):206–210. doi: 10.1016/j.atherosclerosis.2013.12.028. [DOI] [PubMed] [Google Scholar]
- 15.Perez De Isla L., Alonso R., Watts G.F., et al. SAFEHEART Investigators Attainment of LDL-cholesterol treatment goals in patients with familial hypercholesterolemia: 5-year SAFEHEART registry follow-up. J Am Coll Cardiol. 2016;67(11):1278–1285. doi: 10.1016/j.jacc.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Graça R., Fernandes R., Alves A.C., Menezes J., Bourbon M. Characterization of two variants at Met 1 of the human LDLR gene encoding the same amino acid but causing different functional phenotypes. Biomedicines. 2021;9(9):1219. doi: 10.3390/biomedicines9091219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etxebarria A., Benito-Vicente A., Palacios L., et al. Functional characterization and classification of frequent low-density lipoprotein receptor variants. Hum Mutat. 2015;36(1):129–141. doi: 10.1002/humu.22721. [DOI] [PubMed] [Google Scholar]
- 18.Alves A.C., Azevedo S., Benito-Vicente A., et al. LDLR variants functional characterization: contribution to variant classification. Atherosclerosis. 2021;329:14–21. doi: 10.1016/j.atherosclerosis.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Graça R., Alves A.C., Zimon M., Pepperkok R., Bourbon M. Functional profiling of LDLR variants: important evidence for variant classification. J Clin Lipidol. 2022;16(4):516–524. doi: 10.1016/j.jacl.2022.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Brnich S.E., Tayoun A.N.A., Couch F.J., et al. Clinical Genome Resource Sequence Variant Interpretation Working Group. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019;12(1):3. doi: 10.1186/s13073-019-0690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scientific Steering Committee on behalf of the Simon Broome Register Group Risk of fatal coronary heart disease in familial hypercholesterolaemia. Br Med J. 1991;303(6807):893–896. doi: 10.1136/bmj.303.6807.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourbon M., Rato Q. Estudo Português de hipercolesterolemia familiar: Apresentação do estudo e resultados preliminares [Portuguese Familial Hypercholesterolemia Study: presentation of the study and preliminary results] Rev Port Cardiol. 2006;25(11):999–1013. [PubMed] [Google Scholar]
- 23.Chora J.R., Iacocca M.A., Tichý L., et al. ClinGen Familial Hypercholesterolemia Expert Panel The Clinical Genome Resource (ClinGen) Familial Hypercholesterolemia Variant Curation Expert Panel consensus guidelines for LDLR variant classification. Genet Med. 2022;24(2):293–306. doi: 10.1016/j.gim.2021.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Rueden C.T., Schindelin J., Hiner M.C., et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18(1):529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenter A.E., Jones Tr, Lamprecht M.R., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thormaehlen A.S., Schuberth C., Won H.-H., et al. Systematic cell-based phenotyping of missense alleles empowers rare variant association studies: a case for LDLR and myocardial infarction. PLoS Genet. 2015;11(2) doi: 10.1371/journal.pgen.1004855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeon H., Blacklow S.C. Structure and physiologic function of the low-density lipoprotein receptor. Annu Rev Biochem. 2005;74:535–562. doi: 10.1146/annurev.biochem.74.082803.133354. [DOI] [PubMed] [Google Scholar]
- 28.Etxebarria A., Benito-Vicente, Stef M., et al. Activity-associated effect of LDL receptor missense variants located in the cysteine-rich repeats. Atherosclerosis. 2015;238:304–312. doi: 10.1016/j.atherosclerosis.2014.12.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.