Summary
Chemically modified mRNAs hold great potential for therapeutic applications in vivo. Currently, the base modification scheme largely preserves the canonical Watson-Crick base pairing, thus missing one mode of mRNA modulation by altering its secondary structure. Here we report the incorporation of base Z (2-aminoadenine) into mRNA to create Z-mRNA with improved translational capacity, decreased cytotoxicity, and drastically reduced immunogenicity compared to the unmodified mRNA in mammalian cells. In particular, the A-to-Z substitution renders modified mRNAs less immunogenic than the state-of-the-art base modification N1-methylpseudouridine (m1ψ) in mouse embryonic fibroblast cells. As a proof of concept, we developed a Z-mRNA-based vaccine against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Antigen-encoding Z-mRNA elicited substantial humoral and cellular immune responses in vivo in mice, albeit with relatively lower efficacy than the state-of-the-art m1ψ-mRNA. Z-mRNA expands the scope of mRNA base modifications toward noncanonical bases and could offer an advantageous platform for mRNA-based therapeutics where minimal immunogenicity is desired.
Subject areas: Biological sciences, Biochemistry, Medical biochemistry
Graphical abstract

Highlights
-
•
Replacing base A with noncanonical base Z to create Z-mRNA
-
•
Z-mRNA has higher translational capacity and lower immunogenicity than unmodified mRNA
-
•
RBD-encoding Z-mRNA elicited substantial immune response in mice
-
•
Z-mRNA expands the current scope of mRNA base modifications toward noncanonical bases
Biological sciences; Biochemistry; Medical biochemistry
Introduction
Chemically modified messenger RNA (mRNA) has emerged as a promising therapeutic agent over the past decade.1 Compared to unmodified mRNAs that are typically unstable and unfavorably immunogenic (i.e., adverse immune reactions such as inflammation activation), synthetic mRNAs containing modified bases such as pseudouridine (ψ) or N1-methylpseudouridine (m1ψ) demonstrate reduced immunogenicity and improved translational capacity.2,3 Importantly, mRNA vaccines for coronavirus disease 2019 (COVID-19) developed by both Pfizer-BioNTech and Moderna adopt the m1ψ modification to minimize the innate immune response to introduced mRNAs and to enhance antigen production.4,5 These mRNA-based vaccines have contributed enormously to combatting the global severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, validating this novel vaccine platform. Given the essential role modified bases play in the development of mRNA-based therapeutics,6 it is important to explore additional base modifications, potentially beyond the current modification scheme, that may further alleviate the unwanted side effects associated with high immunogenicity and improve the translational capacity of mRNA.
The current base modification scheme largely preserves the canonical Watson-Crick base pairing that is also involved in RNA secondary structure formation. Accumulating evidence has pointed out that mRNA secondary structure regulates protein expression, with GC-rich mRNAs overall exhibiting higher expression level than AU-rich ones.7,8,9 Moreover, studies have revealed that the enhanced translation of ψ/m1ψ-modified mRNA was associated with its improved stability and increased secondary structure.2,10 In fact, compared to base U, ψ could thermodynamically enhance its hydrogen bonding with A, thanks to its extra hydrogen bond donor (N1 imino proton).11,12
Inspired by these observations, we set out to explore if by replacing base A with base Z (2-aminoadenine), a noncanonical base that naturally forms three hydrogen bonds with base T and in theory with base U (Figure 1A),13,14 the resultant mRNA (named Z-mRNA) would entail distinct properties that are favorable for therapeutic applications. For instance, extra hydrogen bonds formed between Z:U pairs would, in theory, stabilize the secondary structure of Z-mRNA, which may in turn improve its expression. Furthermore, base Z originates from cyanophage13 and was also identified as an extraterrestrial nucleobase in carbonaceous meteorites.15 Considering the foreign nature of base Z to mammalian systems, we hypothesized the Z-mRNA may exhibit reduced immunogenicity as it might escape the innate immune surveillance when delivered into mammalian cells. This hypothesis was partially inspired by the fact that the Z-DNA genome of cyanophage confers an evolutionary advantage to escape the host defense system in bacteria (e.g., restriction enzyme digestion),13 a strategy adopted by nature to bypass the detection of foreign nucleic acids delivered into host cells.
Figure 1.
Overview and in vitro characterizations of Z-mRNA
(A) Comparison of base pairings between A:U, A:m1ψ, and Z:U.
(B) Schematic overview of mRNA synthesis, including in vitro transcription (IVT) followed by 5′ capping and 3′ tailing reactions.
(C) Denaturing gel electrophoresis of Z-mRNA synthesized from three different DNA templates, including GFP, mCherry, and RBD by two RNA polymerases (SP6 and T7), and a comparison of the Z-mRNA IVT yield by two RNA polymerases (N = 3). Error bars represent means ± SD (standard deviation). n.s. = not significant. Ladder: NEB ssRNA ladder. ∗: 500 bases. ∗∗: 1000 bases.
(D) IVT yield of mRNAs. N = 10 for unmodified (abbreviated as “Un”) and Z-mRNA. N = 8 for m1ψ-mRNA. Error bars represent means ± SEM (standard error of the mean). p value was calculated by two-tailed unpaired t test. n.s. = not significant. One-way ANOVA test of three groups: n.s.
(E) Denaturing gel electrophoresis of mRNAs prior to (top) or after 3′ polyA tailing (bottom). Un: unmodified mRNA. Z: Z-mRNA. m1ψ: m1ψ-mRNA. GFP/RBD mRNA (without the 3′ polyA tail): ∼960 bases. Ladder: NEB ssRNA ladder. ∗: 500 bases. ∗∗: 1000 bases.
(F) UV absorbance of unmodified and Z-mRNA over temperature (see STAR Methods for details). Dash line = 0.5. Representative image shown from two replicates.
(G) The median fluorescence intensity (MFI) of EGFP+ population in HCT116 cells transfected with Z-mRNAs under various capping and tailing configurations. Cells were analyzed 24 h after transfection. N = 3. Error bars represent means ± SD. p value was calculated by two-tailed unpaired t test, ∗∗: p < 0.01, ∗∗∗: p < 0.001. n.s. = not significant.
(H) Representative images showing the gating strategy used for flow cytometry analysis for four populations, including wild-type cells (no mRNA transfection, top left), cells that received Z-mRNA without cap or tail (top right), cells that received Z-mRNA with cap0 and a polyA tail (bottom left), and cells that received Z-mRNA with cap0 and a polyZ tail (bottom right). P5: gate for autofluorescence. P3: gate for EGFP+ cells. See also Figures S2 and S3.
However, to date it remains unknown if such a radical change in base pairing by the complete A-to-Z substitution in mRNA would adversely affect mRNA translation, a process that requires proper base pairing between the transfer RNA (tRNA) and mRNA for codon decoding.16 In this regard, it was previously reported that nonstandard codons such as ZGG and GZG did not significantly alter the eukaryotic translation process but preserved the unambiguous base pairing of Z with U during mRNA-tRNA interaction,17 suggesting the robustness of the decoding process. Nevertheless, the effect of global A-to-Z substitution in mRNA remains undetermined, and the characterization and application of Z-mRNA have not been reported before.
In this work, we demonstrate that, compared to unmodified mRNA, Z-mRNA exhibits improved translational capacity, decreased cytotoxicity, and minimal immunogenicity in mammalian cells. As a proof of concept for its therapeutic applications, we further developed an effective Z-mRNA-based COVID-19 vaccine that successfully elicited substantial humoral and cellular immune responses in vivo in mice. Z-mRNA leverages the robustness of the translation process and could shift the paradigm of current mRNA base modifications by bringing into play the noncanonical base that alters Watson-Crick base pairing for mRNA-based technologies.
Results
Synthesis and characterization of Z-mRNA in vitro
To test our hypothesis, we first synthesized Z-mRNA via in vitro transcription (IVT) by replacing ATP with ZTP (2-Amino-ATP) in the reaction. The mRNA consists of a 5′ untranslated region (UTR), the Kozak sequence to enhance translation initiation, the coding sequence (CDS), and the 3′ UTR containing the bGH-plA termination signal (Figure 1B; Table S1). Both SP6 and T7 RNA polymerases could efficiently incorporate ZTP during IVT, yielding similar amounts of transcripts with correct size from all three templates tested (Figure 1C), including EGFP (enhanced green fluorescent protein), RBD (receptor binding domain of SARS-CoV-2), and mCherry fluorescent protein. When comparing Z-mRNA synthesis with unmodified or m1ψ-modified mRNA using the SP6 RNA polymerase, we observed no statistically significant difference in the IVT yield of three mRNAs, although the average yield of Z-mRNA was slightly lower than the other two (Figure 1D). When analyzed by the spectrophotometer, Z-mRNA exhibited lower spectral ratios (A260/280, A260/230) compared to the unmodified mRNA (Figure S1), which was in line with the typical spectral feature observed for the Z-DNA isolated from cyanophage S-2L.13
Interestingly, Z-mRNA appeared to have migrated faster in the denaturing gel compared to unmodified or m1ψ-modified GFP/RBD-encoding mRNA (Figure 1E, top), indicating it may encompass a more compact secondary structure. Similarly, when using UV spectroscopy to probe the folding energy of synthesized RNA,18 we found that Z-mRNA had a distinct absorbance profile at 260 nm with a higher melting temperature compared to its unmodified counterpart (Figure 1F). The distinct electrophoresis and spectroscopy profiles of Z-mRNA prompted us to further examine its biophysical property. Toward this goal, we tested the activity of 4 different nucleases (Table S2) on unmodified, Z- or m1ψ-mRNA in vitro as another indirect means of probing mRNA secondary structure. mRNAs were first subject to annealing to induce secondary structure formation before nuclease treatment (Figure S2A). Interestingly, we observed incomplete digestion of Z-mRNA by RNase If (Figure S2B, left), in contrast to the unmodified mRNA. Given RNase If selectively removes nucleotides in single-stranded RNAs,19 its incomplete digestion of Z-mRNA suggests an increased secondary structure of the modified mRNA with A-to-Z substitution. The remaining nucleases (RNase A, RNase R, S1 Nuclease) showed no obvious difference in substrate preference as all mRNAs were efficiently digested (Figure S2B, right), which was not unexpected considering these nucleases are less stringent on the secondary structure of an RNA substrate (Table S2). Taken together, these results strongly indicate that the A-to-Z substitution in mRNA increased and further stabilized the secondary structure of modified mRNA.
The 5′ cap and 3′ polyA tail are essential elements to regulate mRNA translation in eukaryotic cells.20 Hence, we systematically evaluated the effects of various combinations of 5′ capping and 3′ tailing on Z-mRNA expression using EGFP as a reporter (Figure S3A). We adopted the post-transcriptional capping and tailing route for two reasons: 1) co-transcriptional capping with cap analogs could reportedly reduce the yield of mRNA with modified bases and 2) given the “A-to-Z substitution” nature of Z-mRNA, co-transcriptional tailing using a DNA template with an encoded 3′ polyA tail would inevitably lead to Z-mRNA synthesized with a 3′ polyZ tail, of which the effect on mRNA translation was unknown.
We confirmed both the polyA tail and polyZ tail could be efficiently added to the mRNA transcript via a 3′ tailing reaction, with either ATP or ZTP provided (Figure 1E, bottom; Figure S3B). To evaluate the capping efficiency, we compared the expression level of EGFP-encoding Z-mRNA with or without a 5′ cap in HCT116 cells (human colorectal carcinoma cells). Our results showed that with no cap or tail, Z-mRNA expressed poorly in cells (Figures 1G, 1H, and S3C). By contrast, Z-mRNA under the “Cap0/1, polyA tail” configuration exhibited the highest expression level with >94% EGFP+ cells observed at 24 h after transfection (Figures 1G, 1H, and S3C), indicating the high efficiency of post-transcriptional capping and tailing methods. However, we noticed that a polyZ tail at the 3′ end adversely affected Z-mRNA expression compared to a polyA tail, thus necessitating the addition of the polyA tail to Z-mRNA in a post-transcription step (Figures 1G and S3C). Considering the polyA-binding protein is needed to initiate efficient mRNA translation in eukaryotic cells,21 A-to-Z substitutions in the 3′ tail likely reduced the binding affinity of polyA-binding protein to the mRNA 3′ tail, thus leading to decreased translational activity. For following studies, all mRNAs underwent the same 5′ capping (cap0) and 3′ tailing (polyA) reactions for efficient expression and direct comparison across groups.
Z-mRNA exhibited improved translational capacity in mammalian cells
To examine if the A-to-Z substitution would result in increased mRNA translation in mammalian cells, we delivered mRNA encoding EGFP or mCherry fluorescent protein into cultured cells via lipofectamine-based transfection and quantified mRNA expression by flow cytometry. Notably, in HCT116 cells Z-mRNA consistently outperformed its unmodified counterpart in EGFP expression over the monitored time course, by up to a 23.8-fold increase in MFI (median fluorescence intensity) at 24 h after transfection (Figures 2A, 2B, and S4A). When compared to m1ψ-mRNA as a control, Z-mRNA exhibited similar kinetics in translation with expression level peaked around 24 h after transfection (Figure 2A). However, its overall expression was moderately lower than that of the state-of-the-art m1ψ-mRNA by ∼ 40% in HCT116 (Figure 2A, right, quantified by the area under the curve).
Figure 2.
Translation of Z-mRNA in cultured cells
(A) EGFP expression in HCT116 cells as measured by the EGFP+ cells percentage (left) or MFI of the EGFP+ population (right) over time. MFI: median fluorescence intensity. N = 3. Error bars represent means ± SD.
(B) Representative fluorescent images of HCT116 cells transfected with unmodified or Z-mRNA encoding GFP. Images captured under the same exposure conditions. Scale bar: 50 μm.
(C) Normalized EGFP MFI in HEK293T cells (left) and in NIH3T3 cells (right). N = 3. Data collected at 24 h after transfection. Error bars represent means ± SD.
(D) Normalized mCherry MFI in HCT116 cells (left) and HEK293T cells (right). N = 3. Data collected at 24 h after transfection. Error bars represent means ± SD.
(E) Normalized EGFP MFI in primary BMDCs. N = 3. Data collected at 8 h after transfection. For normalization in (C-E), the absolute MFI of each group was divided by the MFI of EGFP+ or mCherry+ population transfected by unmodified mRNA. p value was calculated by two-tailed unpaired t test, ∗: p < 0.05, ∗∗: p < 0.01, ∗∗∗: p < 0.001, ∗∗∗∗: p < 0.0001. n.s. = not significant. Error bars represent means ± SD. In addition to the t test of neighboring two groups, we also performed the one-way ANOVA statistical analysis for all three groups shown in (C–E), and the results were summarized in Table S5. See also Figures S4–S6.
We further examined Z-mRNA expression in two other cell lines: HEK293T (immortalized human embryonic kidney cells) and NIH3T3 (immortalized mouse embryonic fibroblast [MEF] cells) and included mCherry (Table S3) as a second reporter. We selected cell lines of various origins to ensure that the results we observed would be translatable in different cell types. In particular, we included the HEK293 cell line also because it was used to evaluate the translational capacity of ψ- and m1ψ-modified mRNA in two seminal works reported previously.2,3 In all cases, Z-mRNA demonstrated superior translational capacity than the unmodified mRNA (Figures 2C, 2D, and S4B), with on average an 8.6-fold increase in the MFI of EGFP+/mCherry+ population. When compared to the state-of-the-art base modification m1ψ, Z-mRNA showed either comparable mCherry expression in HCT116 (Figure 2D, left; Figure S4B) or moderately lower expression in other cases by ∼ 48% on average (Figures 2C and 2D). The consistent increase in expression of Z-mRNAs encoding two different reporter proteins (EGFP and mCherry) also suggests that the enhanced translation of Z-mRNA is likely independent of the CDS of the protein.
We also tested if there was potential synergistic effect between base Z and other reported modifications, including ψ, m1ψ, and 5-methylcytidine (m5C).6 For this purpose, we synthesized mRNA with two base modifications (“Z + ψ”, “Z + m1ψ” and “Z + m5C”) and compared its expression to Z-mRNA (i.e., only A-to-Z substitution) in HCT116 cells using EGFP as the reporter. All four types of modified mRNAs resulted in high transfection efficiency, with >90% EGFP-positive cells observed at 24 h after transfection (Figure S5A) Interestingly, we observed drastically reduced EGFP expression when base Z was coupled with ψ, whereas adding a second modification such as m5C or m1ψ did not significantly affect mRNA expression compared to base Z modification alone (Figures S5B and S5C).
To examine if the increased expression of Z-mRNA was also true in primary cells, we performed the comparison in mouse bone marrow-derived dendritic cells (BMDCs). We observed comparable EGFP expression from Z-mRNA and m1ψ-mRNA in BMDCs, both of which were significantly higher than the unmodified mRNA (Figures 2E and S6A). We noticed that the absolute expression level of all mRNAs in BMDCs were much lower than in other cell lines (Figure S6B), and this difference was likely due to the hard-to-transfect nature of primary cells such as BMDCs. Taken together, our results demonstrate that Z-mRNA with a 3′ polyA tail could be successfully translated into functional proteins (e.g., EGFP and mCherry) in mammalian cells at a capacity greater than the unmodified mRNA. Compared to the state-of-the-art m1ψ-mRNA, Z-mRNA exhibited moderately lower or similar expression level depending on the cell line tested.
Z-mRNA demonstrated reduced cytotoxicity and immunogenicity in mammalian cells
While comparing the translational capacity of different mRNAs, we consistently noticed that cells transfected with Z-mRNA appeared less stressed and maintained healthier morphology (Figure S7). Indeed, cell viability assay revealed Z-mRNA as significantly less toxic to cells than unmodified mRNA in all three cell lines tested as well as in primary BMDCs (Figure 3A). Specifically, at 48 h following transfection, cells (HCT116, HEK293T, or NIH3T3) that received Z-mRNA or m1ψ-mRNA had similar viability that was significantly higher than cells transfected with unmodified mRNA (Figure 3A). Notably, in BMDCs we noticed m1ψ-mRNA exhibited cytotoxicity similar to the unmodified mRNA (Figure 3A, BMDCs), in line with a previous study.3 Interestingly, Z-mRNA was significantly less toxic in BMDCs compared to both unmodified mRNA and m1ψ-mRNA, suggesting a potential advantage of base Z modification over m1ψ.
Figure 3.
Cytotoxicity and immunogenicity of Z-mRNA in mammalian cells
(A) Viability of cells transfected with different mRNAs at 48 h after transfection, including HCT116, HEK293T, NIH3T3, and primary BMDCs. Lipo = lipofetamine MessengerMax only. Ctrl = no transfection. N = 3. Error bars represent means ± SD. p value was calculated by two-tailed unpaired t test, ∗: p < 0.05, ∗∗: p < 0.01, ∗∗∗: p < 0.001, ∗∗∗∗: p < 0.0001.
(B and C) Secretion of IFN-β (B) and CCL5 (C) by MEF cells following mRNAs transfection at indicated doses. N = 3. Error bars represent means ± SD. p value was calculated by two-tailed unpaired t test, ∗∗: p < 0.01, ∗∗∗: p < 0.001, ∗∗∗∗: p < 0.0001. n.s. = not significant. Values above each bar represent mean.
(D and E) Secretion of IFN-β (D) and CCL5 (E) by BMDCs following mRNAs transfection. N = 3. Cell culture supernatant was collected at 24 h after transfection for cytokine detection. Dashed line in (C–E) denotes the cytokine level detected from the no-mRNA control. N = 3. p value was calculated by two-tailed unpaired t test, ∗: p < 0.05, ∗∗: p < 0.01, ∗∗∗: p < 0.001, ∗∗∗∗: p < 0.0001. n.s. = not significant. Error bars represent means ± SD. In addition to the t test of neighboring two groups, we also performed the one-way ANOVA statistical analysis for all five groups in (A) and four groups in (D and E), and the results were summarized in Table S5. See also Figures S7–S9.
Considering the improved translational capacity and decreased cytotoxicity of modified mRNAs often correlate with the reduction of innate immune signaling activation,3,22,23 we sought to examine the immunogenicity of Z-mRNA in mammalian cells. Toward this goal, we first compared the induction of two cytokines in MEF cells by the enzyme-linked immunosorbent assay (ELISA): interferon beta (IFN-β), a major orchestrator of intracellular innate immune signaling,24 and CCL5/RANTES, a key cytokine significantly upregulated upon unmodified mRNA transfection.23 We chose MEF cells because they are commonly used for the functional characterization of innate immune signaling pathways,25 and they were also adopted to examine the immunogenicity of modified mRNAs in the original study by Karikó et al.2
We transfected EGFP-encoding mRNA into MEF cells using lipofectamine. As expected, at all three doses tested, unmodified mRNA induced the highest levels of IFN-β and CCL5 secretion (Figures 3B and 3C), confirming its highly immunogenic nature. In comparison, m1ψ-mRNA significantly reduced cytokines secretion in MEF cells, in line with its reported low immunogenicity.3 Strikingly, we observed significantly lower induction of IFN-β and CCL5 from MEF cells transfected with Z-mRNA compared to m1ψ-mRNA. Specifically, MEF IFN-β induction by Z-mRNA was on average ∼11-fold lower than m1ψ-mRNA (Figure 3B). In addition, CCL5 induction by Z-mRNA was statistically comparable to the background level (i.e., no mRNA transfected), whereas CCL5 induction by m1ψ-mRNA was ∼ 3-fold higher than the no-mRNA control (Figure 3C). Taken together, these results suggest incorporating base Z into mRNA could aid in the escape of innate immune surveillance against foreign mRNAs, and Z-mRNA outperformed m1ψ-mRNA with further reduced immunogenicity when assessed by IFN-β and CCL5 induction in MEF cells. We also confirmed that the minimal immunogenicity of Z-mRNA in MEF cells was not due to its low expression, as both Z- and m1ψ-mRNA gave significantly higher EGFP expression than unmodified mRNA in MEF cells (Figures S8A and S8B), consistent with what we observed in other cell types (Figure 2). To confirm the reduced cytokine induction by Z-mRNA in MEF cells was not caused by low cell viability, we determined the viability of MEF cells transfected with different mRNAs. Again, consistent with results obtained in other cell lines (Figure 3A), we found Z-mRNA exhibited significantly lower cytotoxicity in MEF cells than the unmodified mRNA (Figure S9).
To further characterize the immunogenicity of Z-mRNA in the context of adaptive immune system, we performed the comparison in mouse BMDCs. Consistent with results obtained in MEF cells, unmodified mRNA induced the highest levels of IFN-β and CCL5 secretion, which were significantly higher than the induction by both Z-mRNA and m1ψ-mRNA (Figures 3D and 3E). This difference was not caused by the potentially low expression of modified mRNAs in BMDCs, as both Z-mRNA and m1ψ-mRNA gave significantly higher expression than unmodified mRNA in BMDCs (Figure 2E). When comparing the immunogenicity of Z-mRNA against m1ψ-mRNA in BMDCs, we observed a relatively lower IFN-β induction by Z-mRNA (Figure 3D), although the difference was not considered as statistically significant (p = 0.12). Nonetheless, these results demonstrated that, in addition to adopting RNA modifications that are naturally present in mammalian systems, it is also possible to reduce the immunogenicity of synthetic mRNA by incorporating other types of base modifications such as base Z explored here.
Z-mRNA as an effective vaccine against SARS-CoV-2 in vivo in mice
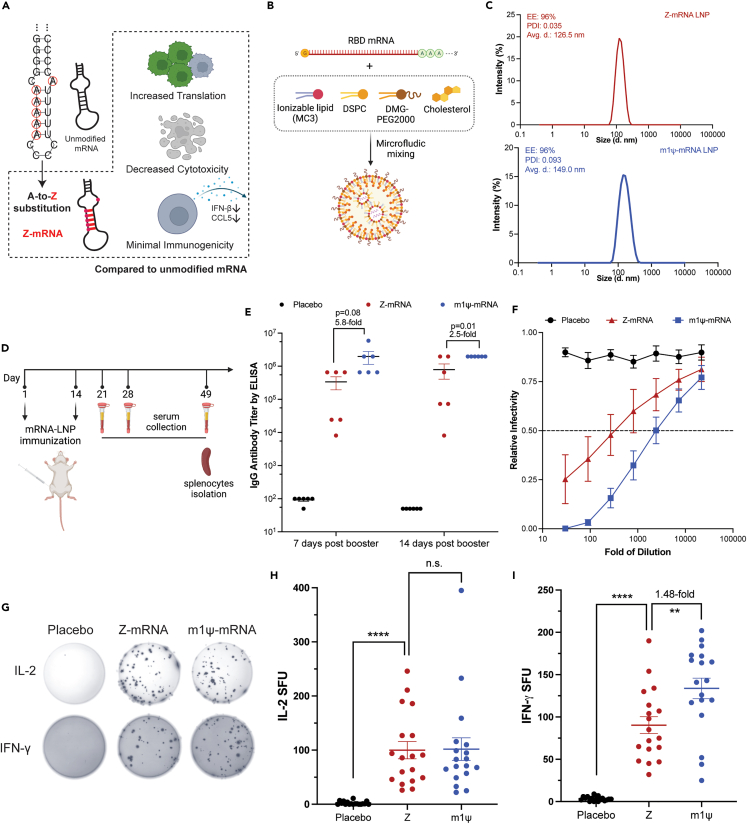
Compared to unmodified mRNA, the increased translational capacity, reduced cytotoxicity, and decreased immunogenicity of Z-mRNA poise it as a promising candidate for in vivo therapeutic applications (Figure 4A). As a proof of concept, we developed a Z-mRNA-based vaccine against SARS-CoV-2 and tested its efficacy in mice.
Figure 4.
Development and evaluation of Z-mRNA-based COVID-19 vaccine in vivo
(A) Schematic representation of the A-to-Z substitution in Z-mRNA and its advantages over unmodified synthetic mRNA.
(B) Synthesis of mRNA LNPs via microfluidic mixing. Ionizable lipid: D-Lin-MC3-DMA. DSPC: distearoylphosphatidylcholine. DMG-PEG2000: 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000.
(C) Characterization of Z- and m1ψ-mRNA LNPs. EE: encapsulation efficiency (top left). PDI: polydispersity index (top left). The size distribution was detected by dynamic light scattering and the average of three technical replicates was shown in the figure.
(D) Mouse vaccination scheme adopted in this study.
(E) SARS-CoV-2 RBD-specific IgG antibodies in sera determined by ELISA. Mice sera was collected 7 days or 14 days after second immunization and used for RBD-specific IgG antibody detection by ELISA. N = 6. Error bars represent means ± SEM. p value was calculated by two-tailed unpaired t test.
(F) Pseudovirus neutralization curve. Mice sera collected 14 days after second immunization was used for pseudovirus neutralization assay. N = 6. Error bars represent means ± SEM. Dashed line: 0.5.
(G) Representative well images of ELISPOT assays. Mouse splenocytes were isolated 35 days after second immunization for ELISPOT assay. Image here shows the spot forming units (based on IL-2 or IFN-γ secretion) from mouse splenocytes stimulated in vitro with the antigen RBD peptide-mix pool.
(H and I) ELISPOT assays of IL-2 (H) and IFN-γ (I) secretion. Error bars represent means ± SEM. N = 6 × 3 = 18 in (H) and (I) (three ELISPOT replicates per mouse splenocyte sample). p value was calculated by two-tailed unpaired t test, ∗: p < 0.05, ∗∗: p < 0.01, ∗∗∗: p < 0.001, ∗∗∗∗: p < 0.0001. n.s. = not significant. See also Figures S10–S12.
We selected the RBD (Table S3) of the SARS-CoV-2 spike protein as the antigen as previously reported26,27 and included m1ψ-mRNA to serve as a positive control. We first examined RBD-encoding mRNA expression in HEK293T cells. Consistent with the fluorescence reporter-based assay, we detected successful RBD expression by both western blot (Figure S10A) and ELISA (Figure S10B), with Z-mRNA showing significantly higher RBD expression compared to the unmodified mRNA. Furthermore, considering western blot and ELISA rely on specific antibody-antigen interactions for target detection, these results also indicate the protein translated from Z-mRNA (i.e., RBD) also maintained the proper conformation that is recognizable to its corresponding antibody.
To achieve efficient delivery and expression of mRNAs in vivo, we encapsulated synthesized mRNAs into lipid nanoparticles (LNPs) by microfluidic mixing (Figure 4B).28 We obtained ∼96% encapsulation efficiency (EE) for both Z- and m1ψ-mRNA LNPs as determined by the ribogreen-based encapsulation assay (Figure 4C, top left). The dynamic light scattering of LNPs revealed the average diameter of Z-mRNA and m1ψ-mRNA LNPs to be 126.5 nm and 149.0 nm, respectively (Figure 4C). The polydispersity index (PDI) of both LNPs was below 0.01 (Figure 4C, top left), suggesting the high quality of prepared LNPs.
To examine the efficacy of Z-mRNA LNPs as a COVID-19 vaccine in mice, we adopted the two-dose vaccination scheme with a two-week interval (Figure 4D). Three groups of BALB/c mice (n = 6) were immunized by the intramuscular (i.m.) route on day 1 and day 14 with either 15 μg Z-mRNA_RBD, or 15 μg m1ψ-mRNA_RBD, or with a placebo (phosphate-buffered saline only). The 15 μg dose was selected based on a relevant study that adopted the same mRNA dosage using RBD as the antigen.27 We collected serum samples from mice 7 days (day 21) and 14 days (day 28) after the second injection to evaluate the immune response. On both days we successfully detected RBD-specific immunoglobulin G (IgG) antibodies from Z-mRNA-immunized mice serum by ELISA with the mean antibody level > 105 (Figure 4E), demonstrating that Z-mRNA could be translated in vivo and elicit efficient antigen-specific humoral immune response. However, we observed that the level of RBD-specific IgG antibodies elicited by Z-mRNA was lower than that elicited by m1ψ-mRNA by ∼ 5.8-fold on day 21 (p = 0.08) and ∼2.5-fold on day 28 (p = 0.01), likely due to the lower expression level of RBD from Z-mRNA compared to m1ψ-mRNA (Figure S10B).
To evaluate anti-SARS-CoV-2 neutralizing antibodies in the serum, we performed the pseudovirus neutralization assay as previously described.29 Consistent with the ELISA-based quantification of RBD-specific IgG antibodies, we observed significant neutralization of pseudovirus following its incubation with serum from Z-mRNA- or m1ψ-mRNA-immunized mice (Figure 4F), with Z-mRNA having relatively lower NT50 (mean value 523.5) compared to m1ψ-mRNA (mean value 1858, p = 0.058) (Figure S10). Nonetheless, these results collectively demonstrate Z-mRNA-encapsulated LNPs can work as an effective vaccine to elicit humoral immune response in immunized mice, albeit with lower potency than the state-of-the-art m1ψ-mRNA.
We also performed the enzyme-linked immunospot (ELISPOT) assay to characterize cellular immune responses induced by the vaccine. We harvested spleens from immunized mice at 5 weeks (day 49) after the second injection (Figure 4D). Splenocytes were then isolated and stimulated in vitro with RBD peptide pools to evaluate cellular cytokines secretion. ELISPOT assays revealed that mice immunized with either Z-mRNA or m1ψ-mRNA produced T cells that could be activated by RBD to release cytokines interleukin-2 (IL-2) and IFN-γ, in contrast to the placebo group (Figure 4G). Specifically, splenocytes from Z-mRNA-immunized mice had a similar level of IL-2 secretion compared to that of m1ψ-mRNA-immunized mice (Figure 4H) and a slightly lower level of IFN-γ secretion (Figure 4I) following RBD stimulation. Collectively, these results demonstrate that alongside the humoral immune response detected from mouse serum, Z-mRNA also induced significant cellular immune response against the SARS-CoV-2 RBD antigen. In addition, we observed no significant difference in the body weight (Figure S12A, left), spleen weight (Figure S12A, middle), and spleen size (Figure S12A, right; Figure S12B) between Z-mRNA- and m1ψ-mRNA-immunized mice on day 49, suggesting no obvious toxicity of Z-mRNA in vivo within the time span of this study.
Discussion
In this work, we report a novel type of modified mRNA named Z-mRNA and demonstrated its in vivo application as an effective vaccine against SARS-CoV-2 in mice. To the best of our knowledge, Z-mRNA represents the first of its kind with the noncanonical base Z incorporated (Figure 1A) that alters Watson-Crick base pairing yet manifests favorable biological properties, including enhanced translation, decreased cytotoxicity, and minimal immunogenicity in mammalian cells compared to unmodified synthetic mRNA (Figure 4A).
The increased folding energy and electrophoresis mobility of Z-mRNA were in line with our prediction that Z-mRNA may encompass a more compact and stable secondary structure due to the extra hydrogen bonds introduced by base Z when paired with U. The increased secondary structure of Z-mRNA was likely associated with and could have contributed to its enhanced translational capacity in cells as previous studies suggested that the structure of mRNA regulates its expression.7,8,9,10 Moreover, Z-mRNA could be successfully translated into functional proteins in mammalian cells, indicating the Z:U base pairing was preserved during the decoding process, consistent with a previous report.17 Hence, the A-to-Z base substitution in Z-mRNA is, in theory, equivalent to an increased GC content of mRNA without CDS or codon alteration. Interestingly, the complete A-to-Z substitution in DNA drastically reduced the expression of encoded EGFP in HCT116 cells (Figure S13), suggesting a more stringent regulation on transcription than translation in mammalian cells. This observation was also consistent with a previous study reporting the inhibitory effect of base Z on transcription in human cells.30
The current base modification scheme for mRNA is largely restricted to naturally occurring RNA modifications such as ψ, m1ψ, N7-methylguanosine (m7G), N6-methyladenosine (m6A), and m5C.6 This strategy is based on the rationale that synthetic mRNA incorporated with natural eukaryotic RNA modifications may better mimic endogenous RNA to bypass the innate immune surveillance, thus reducing the immunogenicity of modified mRNA.1 Surprisingly, however, we observed minimal immunogenicity of Z-mRNA in mammalian cells, despite that base Z was originally discovered from the genomic DNA of cyanophage S-2L and its existence in eukaryotic cells has not been reported. In this regard, our work indicates that foreign nucleobases such as Z could also be harnessed to reduce the immunostimulatory effect of synthetic mRNA, thus expanding the scope of modified nucleosides for future exploration. Further research is needed to unravel the underlying mechanism of immunosuppression by Z-mRNA.
Interestingly, the base Z, commonly known as 2,6-diaminopurine or 2-aminoadenine, was once used as a drug to treat leukemia in humans thanks to its ability to arrest the proliferation of cancer cells.31 This fact raises certain safety concern over the usage of Z-mRNA in mammalian systems. In this respect, it is important to note that Z-mRNA and base Z are chemically distinct molecules. For a given Z-mRNA macromolecule, only corresponding A bases are replaced with Z bases, while the sugar backbone, phosphodiester bonds, and other nitrogenous bases remain unchanged. Hence, Z-mRNA may not exhibit antiproliferation activity in mammalian cells because the functional group (i.e., 2,6-diaminopurine) responsible for this activity is attached to the ribose via a covalent bond in the mRNA macromolecule. In line with this hypothesis, we did not observe adverse effect on cell growth in our cytotoxicity assay comparing Z-mRNA and unmodified mRNA in cultured cells. In fact, the 2,6-diaminopurine was used to treat leukemia because following administration this circulating compound could disrupt DNA replication in cancer cells.31 Z-mRNA, by contrast, consists of no free 2,6-diaminopurine. It is possible that following in vivo delivery, Z-mRNA could be digested by the host’s digestive system to generate free Z bases. However, the evidence for this possibility is lacking and future studies are needed to elucidate metabolic pathways involved in Z-mRNA digestion in vivo.
We anticipate the translational capacity of Z-mRNA could be further improved through systematic engineering efforts such as optimizing 5′ and 3′ UTRs.32 More importantly, the minimal immunogenicity of Z-mRNA in certain cell types (e.g., MEF cells) could make it a more attractive platform for in vivo therapeutic applications where minimal immunogenicity is desired, e.g., the treatment of autoimmune diseases.1
Limitations of the study
In this study we adopted the same UTR sequences (Table S1) for all reporter constructs tested. Considering UTRs play an important role in regulating mRNA translation, it would be of interest to test the compatibility of Z-mRNA with diverse UTRs. In addition, in our study we evaluated the immunogenicity of Z-mRNA in MEF cells and BMDCs. Although previous studies reported strong correlation between in vitro and in vivo measurements of mRNA immunogenicity,2,3,33 it would be beneficial to further determine the immunogenicity of Z-mRNA in vivo. Furthermore, although we did not observe obvious adverse effects in Z-mRNA-immunized mice, the duration of our in vivo study was relatively short (7 weeks) and the long-term safety profile of Z-mRNA warrants further investigation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SARS-CoV-2 RBD primary antibody | R&D Systems | Cat#MAB105401-SP |
| Secondary antibody anti-mouse IgG-HRP | Jackson Immunoresearch | Cat#115-036-003; AB_2338518 |
| Anti-mouse IgG-peroxidase rabbit antibody | Sigma-Aldrich | Cat#A9044; AB_258431 |
| Bacterial and virus strains | ||
| SARS-CoV-2 pseudovirus | This study | N/A |
| Biological samples | ||
| Splenocytes isolated from immunized mice | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| ZTP (2-Amino-ATP) | TriLink Biotechnologies | Cat#N-1001 |
| N1-Methylpseudouridine-5'-Triphosphate (m1ψTP) | TriLink Biotechnologies | Cat#N-1081 |
| SARS-CoV-2 Spike RBD recombinant protein | Sino Biological | Cat#40592-V08B |
| SARS-CoV-2 Spike RBD peptide pool | JPT | Cat#PM-WCPV-S-RBD-2 |
| Critical commercial assays | ||
| HiScribe SP6 RNA Synthesis Kit | NEB | Cat#E2070S |
| HiScribe T7 High Yield RNA Synthesis Kit | NEB | Cat#E2040S |
| Monarch RNA Clean-up Kit | NEB | Cat#T2040L |
| Lucigen Poly(A) Polymerase Tailing Kit | Lucigen | Cat#PAP5014H |
| Lipofectamine MessengerMAX | Thermo Fisher | Cat#LMRNA008 |
| CellTiter-Glo 2.0 | Promega | Cat#G9241 |
| Mouse IFN Beta ELISA Kit | PBL | Cat#42400-1 |
| Mouse CCL5/RANTES DuoSet ELISA kit | R&D systems | Cat#DY478-05 |
| Human SARS-CoV-2 RBD ELISA Kit | Life Technologies | Cat#NC2008933 |
| Mouse IL-2 ELISpot (plus) Kit | Mabtech | Cat#3441-4HPW-2 |
| Mouse IFN-γ ELISpot (basic) Kit | Mabtech | Cat#3321-2H |
| Experimental models: Cell lines | ||
| HCT116 | ATCC | Cat#CCL-247 |
| HEK293T | ATCC | Cat#CRL-3216 |
| NIH3T3 | ATCC | Cat#CRL-1658 |
| HEK293T-hACE2 | BEI Resources | Cat#NR-52511 |
| MEF | Gift from Dr. Huanyu Qiao (UIUC) | N/A |
| Experimental models: Organisms/strains | ||
| BALB/c mice | Jackson Laboratory | Cat#000651 |
| Recombinant DNA | ||
| pcDNA3.1_ spike_del19 | Addgene | Cat#155297 |
| pHAGE-CMV-Luc2-IRES-ZsGreen-W | Addgene | Cat#164432 |
| Software and algorithms | ||
| BD FACSDiva | BD Biosciences | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| GraphPad Prism | GraphPad | https://www.graphpad.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Huimin Zhao (zhao5@illinois.edu).
Materials availability
Plasmids generated in this study are available upon request.
Experimental model and subject details
Animals and ethics statement
BALB/c mice were purchased from the Jackson Laboratory and were housed in a temperature- and light cycle-controlled animal facility (Institute for Genomic Biology at UIUC). For vaccination, groups of 6-8 weeks old female BALB/c mice were immunized intramuscularly with LNP-Z-mRNA_RBD (15 μg, n = 6), LNP-m1ψ-mRNA_RBD (15 μg, n = 6), or Placebo (phosphate-buffered saline (PBS), only, n = 6) in 100 μL volume. All mice had access to food and water. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC), and were performed in accordance with the UIUC Division of Animal Resources (DAR) guidelines.
Cell lines
HCT116 (#CCL-247), HEK293T (#CRL-3216) and NIH3T3 (#CRL-1658) cells were purchased from ATCC. MEF cells were provided by Prof. Huanyu Qiao at College of Agricultural, Consumer and Environmental Sciences at UIUC. HEK293T-hACE2 (#NR-52511) were purchased from BEI Resources. HCT116 cells were cultured in McCoy’s 5A medium supplemented with 10% fetal bovine serum (FBS). HEK293T, NIH3T3, MEF and HEK293T-hACE2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS. Murine DCs were generated from bone marrow (BM) cells of femurs and tibia of 4-12 weeks old. BMDC cells were cultured in RPMI 1640 (Thermo Fisher #11875093) media containing 100 U/mL penicillin/streptomycin, 10% fetal calf serum, 50 μM 2-mercaptoethanol and 20 ng/mL muGM-CSF (Thermo Fisher #BMS325). BMDC Cells were maintained with fresh media every 2 days and used on day 8. All cells were maintained at 37°C in a 5% CO2 incubator in our laboratory.
Method details
mRNA synthesis by in vitro transcription (IVT)
DNA templates for IVT were amplified by PCR using the Q5 High Fidelity DNA Polymerase from New England Biolabs (NEB) (NEB #M0491S). Primers for PCR can be found in Table S4. PCR product was checked by gel electrophoresis for specificity and then purified using the Zymo DNA Clean-up and Concentration kit (Zymo #D4033). DNA was eluted with nuclease-free water and stored at -20°C until use. IVT reactions were set up with the HiScribe™ SP6 RNA Synthesis Kit (NEB #E2070S) or HiScribe™ T7 High Yield RNA Synthesis Kit (NEB #E2040S). Reactions were prepared in a volume of 25 μL following manufacturer's protocol. To generate Z-mRNA or m1ψ-mRNA by IVT, 1.25 μL of 100 mM ZTP (TriLink Biotechnologies N-1001, 2-Amino-ATP) or 1.25 μL of 100 mM m1ψTP (TriLink Biotechnologies N-1081) was used to replace ATP or UTP, respectively. Reactions were incubated at 37°C for 2-4 hours. After incubation, 25 μL of nuclease-free water was added to each reaction along with 2 μL of DNase I (NEB #M0303A), and the reaction was incubated at 37°C for 15 minutes. Reactions were then purified using the Monarch RNA Clean-up Kit (NEB #T2040L) following manufacturer’s protocol. mRNA was eluted with nuclease free water and the concentration was determined by the Thermo Scientific NanoDrop 2000/2000c. mRNAs were stored at -80°C until use.
5’ capping and 3’ polyA tailing
The mRNA from IVT was 5’ capped using the Vaccinia Capping System (NEB #M2080S) following manufacturer's protocol to add a 5’ cap0 or cap1. 25 μg of mRNA was diluted in nuclease-free water and heated at 65°C for 5 minutes followed by cooling on ice for 5 minutes. After cooling, capping reactions were set up in 50 μL volumes per manufacturer’s protocol with the addition of 1.25 μL RNase Inhibitor (NEB #M0314L). Capping reactions were incubated at 37°C for 30 minutes and then purified using the Monarch RNA Clean-up Kit (NEB #T2040L). PolyA tailing of 5’ capped mRNA was performed using the Lucigen Poly(A) Polymerase Tailing Kit (Lucigen #PAP5014H). Reactions were set up in 50 μL per manufacturer’s protocol with 12.5 μg of capped mRNA and 1.25 μL of RNase Inhibitor (NEB #M03014L). PolyZ tailing was set up in the same way but replacing ATP with ZTP in the reaction. For the tailing of RBD mRNA used for vaccine, we increased the mRNA amount from 12.5 μg to 25 μg per reaction to conserve reagent. Reactions were incubated at 37°C for 30 minutes and then purified using the Monarch RNA Clean-up Kit (NEB #T2040L). Concentrations of capped and tailed mRNAs were determined using the ThermoScientific NanoDrop 2000/2000c. mRNAs were stored at -80°C until use.
UV spectroscopy
RNA samples were heat denatured at 90°C for 5 minutes in 900 μl of water, snap cooled on ice for 10 minutes before adding 100 μl of 10X buffer (100 mM sodium cacodylate (pH = 7), 5 mM MgCl2, 1 M KCl) and incubating at room temperature for 30 minutes. UV absorbance was collected by heating RNA from 20°C to 85 °C at 1°C/min using Cary UV-Vis spectrometer. Readings were taken at 260 nm every 30 seconds. The RNA transcript used for UV spectroscopy analysis had ∼ 1600 bases with ∼19% base A content.
In vitro RNase digestion
Capped and polyA tailed mRNAs were used for all digestion reactions. 2 μg mRNAs were first heated in nuclease-free water at 95°C for 2 minutes, and then placed on the bench and cooled down to room temperature over the course of 30 minutes. Control reactions (without nuclease treatment) with 2 μg mRNA in nuclease-free water (total volume 10 μL) were kept at room temperature. Nucleases were then added with matching buffers (if any) as detailed below. RNase If reactions were prepared in 10 μL with indicated amount of mCherry mRNAs. Each reaction contained 0.2 μl of RNase If (NEB #M0243S), 1 μL of NEBuffer 3 (NEB #B7003S). Reactions were incubated at 37°C for 10 minutes followed by heat inactivation at 70°C for 20 minutes. For S1 Nuclease reactions, S1 Nuclease (ThermoFisher #EN032) was diluted 10-fold in S1 Nuclease Dilution Buffer. Reactions were prepared with 2 μg of EGFP mRNA, 1 μL of diluted S1 Nuclease and 3 μL of 10× Buffer in a total volume of 30 μL. Reactions were incubated at room temperature for 30 minutes. RNase A reactions were set up in 10 μL (350 mM NaCl) with 2 μg of EGFP mRNA and 0.5 μL RNase A (NEB #T3018L). Reactions were incubated at 37°C for 30 minutes. RNase R reactions were set up in 10 μL with 2 μg of EGFP mRNA, 1 μL 10× RNase R Buffer (Lucigen #SS000769-D1) and 1.5 μL RNase R (Lucigen #E0111-20D1). Reactions were incubated at 37°C for 30 minutes. mRNAs after digestion were analyzed in pre-stained denaturing gels (1% agarose). Gels were prepared in 1× MOPS buffer with ∼7% formaldehyde (final concentration). Electrophoresis was performed in 1× MOPS buffer at 4 V/cm.
mRNA translation in cultured cells
For fluorescence reporter-based assays, around 100,000 HCT116 cells, 150,000 HEK293T cells, 150,000 NIH3T3 or 100,000 BMDCs cells were plated per well in 24-well plate at ∼12 hrs before transfection. For transfection per well, 2 μL lipofectamine MessengerMAX (Thermo Fisher #LMRNA008) was added in 25 μL Opti-MEM (Thermo Fisher #31985062) and incubated at room temperature for 10 minutes. Following that, 500 ng mRNA was added in 25 μL Opti-MEM and mixed with lipofectamine MessengerMAX. The mixed solution was incubated at room temperature for 5 minutes and then added to each well. Cell culture media was refreshed at 24 hrs (except for BMDCs) after transfection if the sample would be analyzed after the 24-hr time point. For RBD expression in HEK293T cells, around 30,000 cells were plated per well in 96-well plate before transfection. Transfection was performed as described above but with reduced reagent volume (0.3 μL lipofectamine MessengerMAX in 5 μL Opti-MEM followed by 300 ng mRNA in 5 μL Opti-MEM). Cell culture supernatant was collected at 36 hrs after transfection for ELISA without refreshing culture media at 24 hrs.
Flow cytometry
Cells were trypsinized and resuspended in ∼ 500 μL PBS for analysis with either BD LSR II (for EGFP) or BD FACS ARIA II (for mCherry) flow cytometer. At least 10,000 events were recorded for each sample. Representative images of gating strategies can be found in Figures 1, S5 and S6. Data were analyzed using the BD FACSDiva software.
Widefield fluorescence microscopy
HCT116 cells were fixed at 24 hrs after transfection with EGFP-encoding mRNAs. The fluorescence images were collected using the Zeiss Axiovert 200M widefield fluorescence microscopy.
Cell viability assay
Around 100,000 HEK293T/HCT116/NIH3T3 cells were plated per well in 24-well plate or around 30,000 BMDCs were plated per well in 96-well plate ∼ 24 hrs before transfection. Transfections were performed as described above. Cell culture media was refreshed at 24 hrs after transfection (except for BMDCs). 48 hrs after transfection, for transfection in 24-well plate, culture media was carefully aspirated away and 150∼200 μl PBS was added per well followed by equal volume of the CellTiter-Glo 2.0 reagent (Promega #G9241). For BMDCs transfection in 96-well plate, 100 μl CellTiter-Glo 2.0 reagent was directly added to each well. The reagents were mixed for 2 minutes on an orbital shaker to induce cell lysis and then the plate was incubated at room temperature for 10 minutes before measurement. Luminescence signal was detected with the SpectraMax M5 plate reader (Molecular Devices) with an integration time of 1000 ms. The absolute luminescence signal of each well was then normalized to the control group (no mRNA added) to obtain the relative cell viability.
Immunogenicity assay
Around 50,000 MEF cells (at low passage) or 30,000 BMDCs were plated per well in a 96-well plate at ∼ 24 hrs prior to transfection. Transfections were performed with lipofectamine MessengerMAX as described above with the following recipes per well for varying doses: 0.3 μL lipofectamine MessengerMAX in 5 μL Opti-MEM followed by 300 ng mRNA in 5 μL Opti-MEM, or 0.9 μL lipofectamine MessengerMAX in 15 μL Opti-MEM followed by 900 ng mRNA in 15 μL Opti-MEM, or 1.5 μL lipofectamine MessengerMAX in 25 μL Opti-MEM followed by 1500 ng mRNA in 25 μL Opti-MEM. 24 hrs after transfection, cell culture supernatant was collected for cytokine quantification. IFN-β was measured using the Mouse IFN Beta ELISA Kit (PBL #42400-1) per manufacturer’s protocol, without diluting the supernatant. CCL5/RANTES was measured using the Mouse CCL5/RANTES DuoSet ELISA kit (R&D systems #DY478-05) per manufacturer’s protocol, with samples being diluted by 8-fold (cells transfected with mRNAs) or 4-fold (lipo-only control). Absorbance at 450 nm was measured with the TECAN infinite M1000 Pro plate reader with the reference wavelength set at 570 nm.
Detection of RBD by Western blot
Cell culture supernatants from HEK293T cells were collected ∼36 hrs after transfection to detect RBD protein by Western blotting. For image shown in Figure S10A, 15 μL samples were combined with 10X loading buffer and separated in 12% SDS-PAGE gel (Biorad #4561046). Proteins were then transferred to a PVDF membrane (Biorad #1704156). The membrane was blocked with 5% non-fat milk in water for 1 hr. RBD protein was detected with the primary antibody (diluted in 5% non-fat milk, R&D systems #MAB105401-SP) after incubation at 4C overnight, followed by the treatment with secondary antibody goat anti-mouse IgG-HRP (Jackson Immunoresearch #115-036-003) at room temperature for 25 minutes. The membrane was developed by adding the Clarity Western ECL Substrate (BioRad #1705060S) and imaged using the Amersham ImageQuant studio.
Quantification of RBD expression by ELISA
Cell culture supernatant was collected at 38 hrs after transfection and was diluted by 2000-fold for ELISA. ELISA was performed using the Life Technologies Human SARS-CoV-2 RBD ELISA Kit (#NC2008933) following manufacturer’s protocol. Absorbance at 450 nm was measured with the TECAN infinite M1000 Pro plate reader with the reference wavelength set at 630 nm.
Lipid-nanoparticle encapsulation of mRNA
mRNAs were diluted in 50 mM citrate buffer (pH 3.0) as the aqueous phase. Lipids were prepared by dissolving MC3-lipid, DSPC, cholesterol, PEG2000-DMG in ethanol at molar ratios of 50:10:38.5:1.5 (total concentration 12.5 mM). The lipids mixture was then mixed with the aqueous phase (mRNA) using the NanoAssemblr Benchtop (Precision #NIT0046) in compatible microfluidic cartridges. The formulated mRNA-LNP solution was first dialyzed against 1X PBS (pH ∼7) at 4°C overnight, and then concentrated by ultrafiltration using the Amicon Ultra Centrifugal Filter Unit (Millipore). The encapsulation efficiency and concentration of mRNAs were determined by the QuantiFluor RNA system (Promega #E3310). The size of mRNA-LNP particles was determined by dynamic light scattering (DLS) using samples diluted in PBS. DLS was performed using the Malvern Zetasizer (Malvern).
Animal experiments
For mouse vaccination, groups of 6-8 weeks old female BALB/c mice were immunized intramuscularly with LNP-Z-mRNA_RBD (15 μg, n = 6), LNP-m1ψ-mRNA_RBD (15 μg, n = 6), or Placebo (phosphate-buffered saline (PBS), only, n = 6) in 100 μL volume using sterile BD U-100 insulin syringes. The sera of immunized mice were collected at indicated time points in Figure 4D. At 5 weeks after second immunization, mice were sacrificed and splenocytes were isolated to detect cellular immune responses by ELISPOT as described below.
Quantification of RBD-specific IgG antibodies from serum by ELISA
ELISA plates (Thermo Fisher #3455) were coated overnight at 4°C with 2 μg/mL of SARS-CoV-2 Spike RBD (Sino Biological #40592-V08B) in bicarbonate buffer (Sigma-Aldrich #88975) and blocked by 1% BSA in PBS with 0.1% Tween 20 (PBST) for 1 hr at room temperature. Serum samples were 3-fold serially diluted (in blocking buffer), starting at 1:100, and loaded to each well (100 μL). Following incubation at 37°C for 60 minutes, plates were washed 3 times with 1× PBS. Then 100 μL of diluted anti-mouse IgG-peroxidase rabbit antibody (Sigma-Aldrich #A9044, 1:10,000 dilution) was added per well and incubated at 37°C for 30 minutes. Plates were washed 3 times following incubation, and TMB substrates were added (100 μL/well) and incubated for 15-20 minutes at room temperature. Then, the ELISA stop buffer was added and the absorbance (450/630 nm) was measured with the TECAN infinite M1000 Pro plate reader. The endpoint IgG titers were defined as the highest dilution fold of sera to yield an absorbance greater than 2.1-fold of the background values (without serum but the secondary antibody was added) as previously described.27 Antibody titer below the limit of detection was determined as half the limit of detection.
Pseudovirus packaging and neutralization assay
To package SARS-CoV-2 pseudovirus, plasmids of 0.4 μg pcDNA3.1_ spike_del19 (Addgene #155297), 3.6 μg dR8 plasmid and 4 μg pHAGE-CMV-Luc2-IRES-ZsGreen-W (Addgene #164432) were transfected into HEK293T cells (100 mm plate) with FuGENE HD (Promega #E2311). ∼60 hrs after transfection, the supernatant containing pseudoviral particles was harvested, centrifuged, and filtered through a 0.45 μm sterilized membrane. Aliquots were stored at -80°C until use. The pseudovirus neutralization assay including pseudovirus titration was performed as previously described in details.29 We used the Bright-Glo luciferase assay system (Promega #E2610) to measure the infectivity of pseudovirus. Luminescence was measured with the SpectraMax M5 plate reader (Molecular Devices) with an integration time of 1000 ms. The neutralizing activity of serum (neutralizing antibody titer NT50) was calculated by fitting the relative infectivity data using the asymmetrical (five parameter) regression model in Prism (Version 9.4.0) with the bottom baseline fixed to zero.
ELISPOT assay
ELISPOT assays were performed to detect antigen-specific T lymphocyte responses. Spleens were removed from immunized BALB/c mice at 5 weeks post second injection and splenocytes were isolated. ELISPOT assays were performed with Mabtech Mouse IL-2 ELISpot (plus) Kit (#3441-4HPW-2) and Mabtech Mouse IFN-γ ELISpot (basic) Kit (#3321-2H) following manufacturer’s protocols. The viability of isolated mouse splenocytes was determined by the BIO-RAD TC20 automated cell counter and ∼ 1 × 105 live cells per well were added to the plate. A pool with 53 peptides consisting of 15 mers with 11 amino acids overlap spanning the RBD of the SARS-CoV-2 spike protein (JPT, #PM-WCPV-S-RBD-2) was added as the antigen stimulant (1 μg/mL). Phytohemagglutinin (10 μg/mL) was added as a positive control. Cells incubated without stimulation were included as a negative control. ELISPOT plates were developed per manufacturer’s protocol. Plates were analyzed by a third-party service provider (ZellNet Consulting) for ELISPOT plate reading.
Z-containing DNA synthesis
Z-containing DNA was prepared by PCR with Q5 polymerase (NEB) and by replacing dATP with dZTP (TriLink BioTechnologies N1046) in the reaction. PCR products were analyzed by gel electrophoresis and purified by standard column-based purification method. DNA was stored at -20°C until further use.
Quantification and statistical analysis
All statistical details of experiments can be found in figures and corresponding figure legends. All statistical analyses were performed using GraphPad Prism 9.
Acknowledgments
This work was supported by the Steve L. Miller Endowed Chair fund and partly by the U.S. National Institutes of Health (U54DK107965, R01GM143723) (H.Z.). We thank Dr. Teresa Anne Martin (UIUC) for administrative help with IACUC and IBC protocols. We thank Prof. Cecilia Leal (UIUC, MatSE) for sharing the NanoAssemblr Benchtop equipment. The dynamic light scattering experiment was performed at the Materials Research Laboratory (UIUC). We thank Jamie Leann Ludwig (UIUC, Division of Animal Resources) for training with mouse experiments. We thank Dr. Sandra Kay McMasters for providing cell culture media (UIUC, SCS Cell Media Facility). We thank Dr. Gaurav Sahay and Dr. Milan Gautam (Oregon State University) for helpful suggestions on LNPs preparation. We thank Dr. Marcin Krzysztof Wozniak and Kate Janssen (UIUC, Cytometry and Microscopy to Omics Facility) for assistance with flow cytometry. We thank Dr. Austin Joseph Cyphersmith for help with widefield microscopy. We thank Dr. Chunshuai Huang for help with ChemDraw, Dr. Hengqian Ren and Guanhua Xun for helpful discussions. We thank Dr. Huanyu Qiao (UIUC, College of Veterinary Medicine) for providing the MEF cell line. We thank Dr. Hua Wang and Joonsu Han (UIUC, MatSE) for providing BMDCs. Certain figures in this manuscript were created using BioRender.
Author contributions
H.Z. and M.Z. conceived the project and designed the experiments. M.Z. and M.E.E. conducted the in vitro and cellular experiments. M.Z. performed LNPs encapsulation with help from L.Z. for LNPs characterization. M.Z. and N.S. carried out mouse experiments. M.Z. and H.Z. wrote the paper with inputs from all authors.
Declaration of interests
A provisional patent application has been filed based on this study.
Published: August 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107739.
Supplemental information
Data and code availability
-
•
Data supporting findings of this study are included in this paper. All other relevant data are available from the lead contact upon reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 2.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 4.Nance K.D., Meier J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021;7:748–756. doi: 10.1021/acscentsci.1c00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhary N., Weissman D., Whitehead K.A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021;20:817–838. doi: 10.1038/s41573-021-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao M., Zhang Q., Feng X.-H., Liu J. Synthetic modified messenger RNA for therapeutic applications. Acta Biomater. 2021;131:1–15. doi: 10.1016/j.actbio.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kudla G., Lipinski L., Caffin F., Helwak A., Zylicz M. High Guanine and Cytosine Content Increases mRNA Levels in Mammalian Cells. PLoS Biol. 2006;4:e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courel M., Clément Y., Bossevain C., Foretek D., Vidal Cruchez O., Yi Z., Bénard M., Benassy M.-N., Kress M., Vindry C., et al. GC content shapes mRNA storage and decay in human cells. Elife. 2019;8:e49708. doi: 10.7554/eLife.49708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mordstein C., Savisaar R., Young R.S., Bazile J., Talmane L., Luft J., Liss M., Taylor M.S., Hurst L.D., Kudla G. Codon Usage and Splicing Jointly Influence mRNA Localization. Cell Syst. 2020;10:351–362.e8. doi: 10.1016/j.cels.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mauger D.M., Cabral B.J., Presnyak V., Su S.V., Reid D.W., Goodman B., Link K., Khatwani N., Reynders J., Moore M.J., McFadyen I.J. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. USA. 2019;116:24075–24083. doi: 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kierzek E., Malgowska M., Lisowiec J., Turner D.H., Gdaniec Z., Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014;42:3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X., Ma S., Yi C. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr. Opin. Chem. Biol. 2016;33:108–116. doi: 10.1016/j.cbpa.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Kirnos M.D., Khudyakov I.Y., Alexandrushkina N.I., Vanyushin B.F. 2-Aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature. 1977;270:369–370. doi: 10.1038/270369a0. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y., Xu X., Wei Y., Cheng Y., Guo Y., Khudyakov I., Liu F., He P., Song Z., Li Z., et al. A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science. 2021;372:512–516. doi: 10.1126/science.abe4882. [DOI] [PubMed] [Google Scholar]
- 15.Callahan M.P., Smith K.E., Cleaves H.J., Ruzicka J., Stern J.C., Glavin D.P., House C.H., Dworkin J.P. Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases. Proc. Natl. Acad. Sci. USA. 2011;108:13995–13998. doi: 10.1073/pnas.1106493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranjan N., Leidel S.A. The epitranscriptome in translation regulation: mRNA and tRNA modifications as the two sides of the same coin? FEBS Lett. 2019;593:1483–1493. doi: 10.1002/1873-3468.13491. [DOI] [PubMed] [Google Scholar]
- 17.Hoernes T.P., Faserl K., Juen M.A., Kremser J., Gasser C., Fuchs E., Shi X., Siewert A., Lindner H., Kreutz C., et al. Translation of non-standard codon nucleotides reveals minimal requirements for codon-anticodon interactions. Nat. Commun. 2018;9:4865. doi: 10.1038/s41467-018-07321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan Y., Qu K., Ouyang Z., Kertesz M., Li J., Tibshirani R., Makino D.L., Nutter R.C., Segal E., Chang H.Y. Genome-wide measurement of RNA folding energies. Mol. Cell. 2012;48:169–181. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grünberg S., Coxam B., Chen T.-H., Dai N., Saleh L., Corrêa I.R., Nichols N.M., Yigit E. E. coli RNase I exhibits a strong Ca2+-dependent inherent double-stranded RNase activity. Nucleic Acids Res. 2021;49:5265–5277. doi: 10.1093/nar/gkab284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 21.Wells S.E., Hillner P.E., Vale R.D., Sachs A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 22.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Andries O., De Filette M., De Smedt S.C., Demeester J., Van Poucke M., Peelman L., Sanders N.N. Innate immune response and programmed cell death following carrier-mediated delivery of unmodified mRNA to respiratory cells. J. Control. Release. 2013;167:157–166. doi: 10.1016/j.jconrel.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 24.McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y.S., Lei Y.L. Generation and Culture of Mouse Embryonic Fibroblasts. Methods Mol. Biol. 2019;1960:85–91. doi: 10.1007/978-1-4939-9167-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tai W., Zhang X., Drelich A., Shi J., Hsu J.C., Luchsinger L., Hillyer C.D., Tseng C.-T.K., Jiang S., Du L. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 2020;30:932–935. doi: 10.1038/s41422-020-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Q., Ji K., Tian S., Wang F., Huang B., Tong Z., Tan S., Hao J., Wang Q., Tan W., et al. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat. Commun. 2021;12:776. doi: 10.1038/s41467-021-21037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan Y., You C., Park J., Kim H.S., Guo S., Schärer O.D., Wang Y. Transcriptional Perturbations of 2,6-Diaminopurine and 2-Aminopurine. ACS Chem. Biol. 2022;17:1672–1676. doi: 10.1021/acschembio.2c00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weckbecker G., Cory J.G. Metabolic activation of 2,6-diaminopurine and 2,6-diaminopurine-2′-deoxyriboside to antitumor agents. Adv. Enzyme Regul. 1989;28:125–144. doi: 10.1016/0065-2571(89)90068-X. [DOI] [PubMed] [Google Scholar]
- 32.Castillo-Hair S.M., Seelig G. Machine Learning for Designing Next-Generation mRNA Therapeutics. Acc. Chem. Res. 2022;55:24–34. doi: 10.1021/acs.accounts.1c00621. [DOI] [PubMed] [Google Scholar]
- 33.Krienke C., Kolb L., Diken E., Streuber M., Kirchhoff S., Bukur T., Akilli-Öztürk Ö., Kranz L.M., Berger H., Petschenka J., et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science. 2021;371:145–153. doi: 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data supporting findings of this study are included in this paper. All other relevant data are available from the lead contact upon reasonable request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.