Abstract
Assessing the severity of eczema in clinical research requires face-to-face skin examination by trained staff. Such approaches are resource-intensive for participants and staff, challenging during pandemics, and prone to inter- and intra-observer variation. Computer vision algorithms have been proposed to automate the assessment of eczema severity using digital camera images. However, they often require human intervention to detect eczema lesions and cannot automatically assess eczema severity from real-world images in an end-to-end pipeline. We developed a model to detect eczema lesions from images using data augmentation and pixel-level segmentation of eczema lesions on 1,345 images provided by dermatologists. We evaluated the quality of the obtained segmentation compared with that of the clinicians, the robustness to varying imaging conditions encountered in real-life images, such as lighting, focus, and blur, and the performance of downstream severity prediction when using the detected eczema lesions. The quality and robustness of eczema lesion detection increased by approximately 25% and 40%, respectively, compared with that of our previous eczema detection model. The performance of the downstream severity prediction remained unchanged. Use of skin segmentation as an alternative to eczema segmentation that requires specialist labeling showed the performance on par with when eczema segmentation is used.
Introduction
Atopic dermatitis (AD) (synonym with eczema and atopic eczema) is the most common chronic inflammatory skin disease affecting 15–30% of children and 2–10% of adults worldwide (Langan et al., 2020). Assessment of AD severity can include symptoms such as itching, disease signs such as excoriation, or other aspects of the disease such as control and QOL. Simple global methods or short symptom and QOL questionnaires can be used in clinical practice (Leshem et al., 2020).
However, objective assessments of AD severity are increasingly used to assess eligibility for systemic medicines that might be needed for severe disease (National Institute for Health and Care Excellence, 2018). In research studies such as clinical trials, objective assessment of AD severity is usually considered essential to standardize comparisons and reduce detection biases for interventions that cannot be blinded. The international Harmonising Outcome Measures for Eczema initiative has recently established a core outcome set for clinical trials for AD that includes objective measurement of signs using Eczema Area and Severity Index (Williams et al., 2022). Objective assessment of AD severity usually requires assessment by trained clinical staff for signs such as redness or lichenification graded from none (=0) to severe (=3) for each sign.
Complete skin examination in a face-to-face environment is desirable for objective assessment of AD severity. However, it is resource-intensive for both the patient/study participant and the trained assessor. Studies that require repeated visits to the clinic for assessments are especially challenging and can contribute to large quantities of missing data. The recent COVID-19 pandemic has placed additional constrain on such face-to-face visits. It is also known that inter- and intra-observer variation can be a significant challenge when assessing AD severity objectively (Schmitt et al., 2013).
A form of automated remote assessment of AD severity using digital images is desirable because it could enable and standardize the remote assessment of AD severity and reduce the inter- and intra-observer variability. However, the methods published to date are often not fully automated to detect AD lesions and assess AD severity. They require manual intervention in their pipeline. For example, Bang et al. (2021) trained and tested a severity assessment algorithm using fixed-size image crops manually prepared to cover only AD areas. Automatic detection of AD lesions in real-world images and design of end-to-end pipelines to assess AD severity from digital camera images are needed to minimize human intervention.
Pan et al. (2020) recently developed a convolutional neural network–based computer vision pipeline called EczemaNet that first detects AD regions and then assesses the severity of seven disease signs (dryness, erythema, excoriation, cracking, exudation, lichenification, and edema). The pipeline was trained using real-world images taken with digital cameras in a published clinical trial so that EczemaNet can be used in real-world situations where the images are of different sizes, taken under various imaging conditions (e.g., resolution, lighting, focus, and blur), or include non-AD skin and non-skin background. However, training data for the AD region detection model in EczemaNet was obtained from non-experts, and the model’s detection quality was considered a bottleneck for better severity assessment.
This study aims to develop more accurate and robust computer vision algorithms for AD region detection to enable reliable assessment of AD severity from digital images. We use pixel-level AD segmentation data obtained from clinicians that provide granular information on the extent of AD regions in the images. We evaluate the quality and robustness of eczema segmentation and its effects on the severity prediction using skin segmentation as a benchmark. Previously, Nisar et al. (2021) attempted automatic segmentation of AD lesions in 84 images that contain only AD regions and neighboring pixels and Son et al. (2021) explored the segmentation and classification of erythema lesions. In comparison, we introduce EczemaNet2 in this study that segments the areas of AD (not only erythema) lesions using 1,345 real-world images that include non-AD skin and non-skin background. We also apply data augmentation techniques and quantify the robustness of AD region detection to varying imaging conditions.
Results
Overview of EczemaNet1
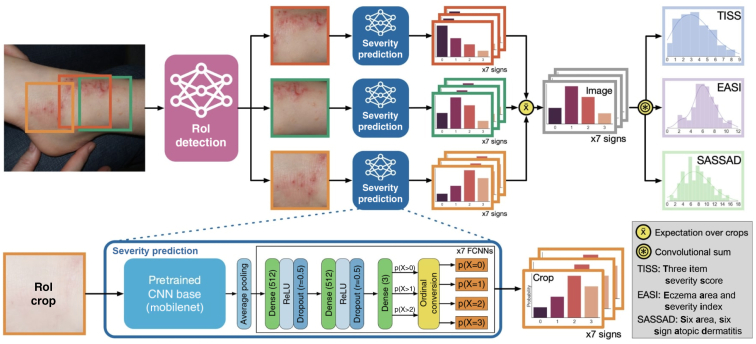
The published EczemaNet model (Pan et al., 2020) has two main components (Figure 1): the region of interest (RoI) detection model and the severity prediction model. The latter makes probabilistic predictions of seven disease signs of AD for each crop produced by the RoI detection model. The predicted severity of each disease sign is aggregated to produce regional severity scores for the whole image, such as regional versions of Six Area, Six Sign Atopic Dermatitis score (Berth-Jones, 1996); Three-Item Severity score (Wolkerstorfer et al., 1999); and Eczema Area and Severity Index (Hanifin et al., 2001).
Figure 1.
Overview of the EczemaNet1 pipeline. Reproduced with permission from Pan et al. (2020). The Region-of-Interest (RoI) detection model generates AD crops of the input image that contain AD regions. The severity prediction model makes probabilistic predictions of seven disease signs in each crop. The AD severity scores for each disease sign are integrated to give the regional severity scores for the whole image. AD, atopic dermatitis; EASI, Eczema Area and Severity Index; SASSAD, Six Area, Six Sign Atopic Dermatitis; RoI, region of interest.
The RoI detection model for EczemaNet1 was trained on crops obtained from non-experts. Its outputs are rectangular bounding boxes of the detected AD regions in an image that may also include background pixels and do not provide information about the particular shape of the AD regions, limiting the performance of AD severity assessment in EczemaNet1.
Pixel-level segmentation data
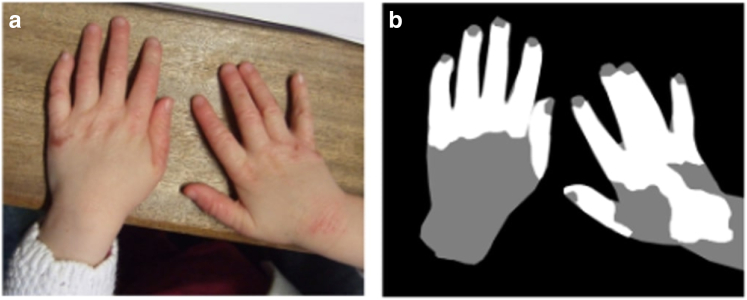
We obtained 1,345 photographs of AD regions from 287 children previously collected in the Softened Water Eczema Trial (Thomas et al., 2011). The photographs had varying image quality and resolution. Four dermatologists segmented the AD regions from each of the 1,345 photographs at the pixel level, providing finer resolution than the crops used in EczemaNet1. The pixel-level segmentation data included skin segmentation masks for background, non-AD skin, and AD skin (Figure 2).
Figure 2.
Illustration of pixel-level segmentation masks. (a) A sample image and (b) the corresponding pixel-level segmentation masks of background (black), non-AD skin (gray), and AD skin (white). AD, atopic dermatitis.
An RoI detection model using pixel-level segmentation for EczemaNet2
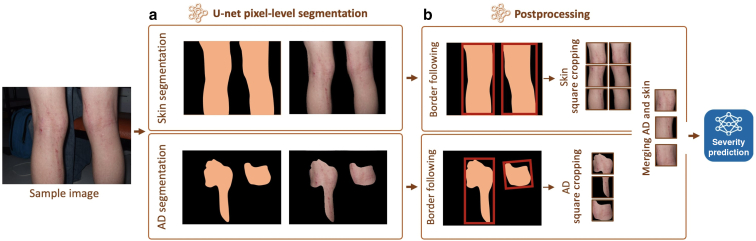
We introduced EczemaNet2, which is an improved version of EczemaNet1. EczemaNet2 uses a modified RoI detection model with a standard pixel-level segmentation U-Net (Ronneberger et al., 2015) to produce skin and AD segmentation masks (Figure 3a). We added three postprocessing steps (Figure 3b) to extract crops for the subsequent severity prediction model. The postprocessing steps include a border-following algorithm to generate rectangular crops, square cropping to extract square crops without distortion, and adding surrounding non-AD skin pixels to the AD skin pixels in the square crops. This step was applied only when both skin and AD segmentation masks were available.
Figure 3.
Overview of the RoI detection part of EczemaNet2. (a) U-Net pixel-level segmentation for skin or AD and (b) postprocessing steps to produce crops that are inputs for the subsequent severity prediction model. Merging non-AD skin pixels with AD pixels is applied when both AD and skin segmentation are available. AD, atopic dermatitis; RoI, region of interest.
Systematic evaluation of the quality and robustness of RoI detection
We compared the performance of the RoI models with various configurations of training data and data augmentation (Table 1). For a consistent and fair comparison of all models, the same data were used with a split ratio of 6:2:2 for training, validation, and test sets. The performance was evaluated in terms of the quality and the robustness of RoI detection, two essential aspects of computer vision methods to be used in clinical practices.
Table 1.
Summary of the AD Detection Pipelines
| Configuration Name | Pipeline | RoI Model | Training Data | Data Augmentation |
|---|---|---|---|---|
| Base | EczemaNet1 | A faster R-CNN | AD crops | No |
| Skin | EczemaNet2 | A U-Net | Skin segmentation masks | No |
| AD | AD segmentation masks | No | ||
| AD/skin | 2 U-Nets | AD and skin segmentation masks | No | |
| AD/skin+Pix2Pix | AD and skin segmentation masks | Pix2Pix | ||
| AD/skin+DA | AD and skin segmentation masks | Traditional |
Abbreviations: AD, atopic dermatitis; CNN, convolutional neural network; RoI, region of interest.
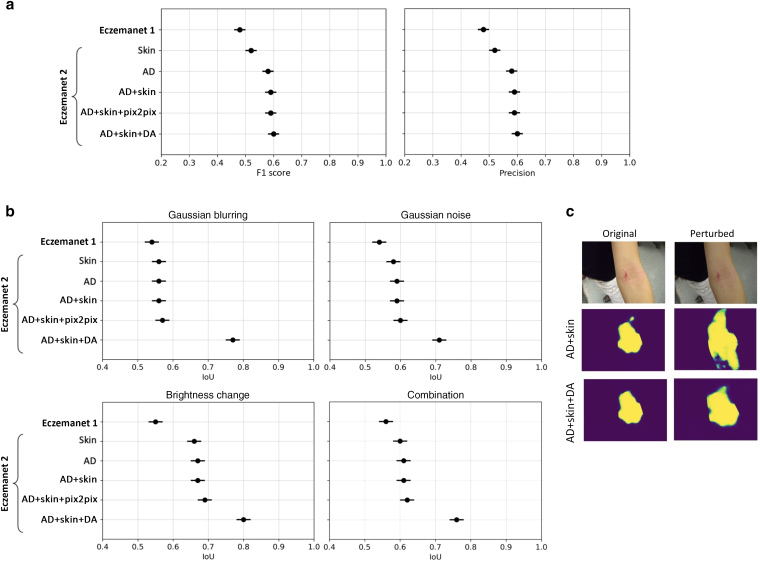
First, we evaluated the quality of RoI detection for each image on the basis of the classification of each pixel into AD region or not in all relevant crops (Figure 4a). Pixel-level segmentation of AD regions obtained from four dermatologists was used as a reference. The quality of RoI detection was evaluated using F1 score and precision, as metrics for classification compared with ground truth. F1 score is the harmonic mean of precision and recall, whereas precision is the fraction of correctly classified pixels in the predicted mask, and recall is the fraction of correctly classified pixels in the ground truth mask. F1 score ranges from 0 to 1, where 1 indicates perfect segmentation accuracy.
Figure 4.
Quality and robustness of RoI detection. (a) Quality measured by F1 score and precision (mean ± SE; the higher, the better) on the test set of 271 photographs. (b) Robustness of RoI detection against image perturbations measured by IoU between predictions made from unperturbed and perturbed images (mean ± SE; the higher, the better) on the test set of 271 photographs, for different perturbations (Gaussian blurring, Gaussian noise, brightness change, and combinations). (c) Example AD segmentation of unperturbed and perturbed images by the RoI models trained with data augmentation (AD+Skin+DA) and with the nonaugmented dataset (AD+Skin). AD+Skin+DA generates consistent masks even in the presence of image perturbations but not AD+Skin. AD, atopic dermatitis; DA, data augmentation; IoU, intersection over union; RoI, region of interest; SE, standard error.
The RoI detection model of EczemaNet2 trained on pixel-level skin segmentation masks (“Skin” in Table 1 and Figure 4a) achieved a better detection quality than the RoI detection model of EczemaNet1. The RoI detection model of EczemaNet2 trained on pixel-level AD segmentation masks (“AD”) achieved a slightly better quality than that on skin segmentation masks. This was expected because training the model with skin segmentation masks leads to many false positives as all predicted skin pixels are considered AD. The RoI quality was not improved by adding surrounding non-AD skin pixels to AD skin segmentation masks (“AD+Skin”) and data augmentation with traditional methods (“AD+Skin+DA”) and Pix2Pix (“AD+Skin+Pix2Pix”). The improvement in AD detection brought by EczemaNet2 from EczemaNet1 is statistically significant (with P = 0.021 in paired t-test), but the detection performance remains moderate, with an average precision and F1 score of approximately 60% across images.
Next, we evaluated the robustness of RoI detection, that is, the sensitivity of the model’s predictions to perturbations in the model’s inputs, using intersection over union (IoU), a metric that measures the similarity of two sets without referring to ground truth. IoU was computed by the area of overlap between predicted RoIs on the original image and on the perturbed version of the image, divided by the area of union between them. It reflects how robust the model prediction is on different imaging conditions. The IoU metric ranges between 0 and 1, and a score of 1 indicates a perfect overlap, whereas a score of 0 indicates no overlap between them.
The perturbations applied are similar to those applied for data augmentation (Table 2), including blurring (e.g., due to incorrect focus), brightness changes (e.g., due to incorrect exposure), noise (e.g., due to poor lighting conditions), and their combinations that could realistically occur in images taken with a smartphone camera.
Table 2.
Traditional Data Augmentation Methods Applied to Create an Augmented Dataset
| Method | Range: Increment Size | Number of Generated Images |
|---|---|---|
| Blur | 2.5–12.5: 2.5 | 5X |
| Noise | 0.01–0.05: 0.01 | 5X |
| Brightness | 0.5–1.5: 0.2 | 6X |
| Zoom | 0.25–1.75: 0.25 | 6X |
| Rotation | −90 to 90 | 2X |
| Flip | Horizontal, vertical | 2X |
| Mix | Combination of two randomly chosen methods | 2X |
RoI detection in EczemaNet2 is significantly (with P = 0.014 in paired t-test) more robust than EczemaNet1 in terms of IoU for all perturbations considered (Figure 4b). Data augmentation of training data using traditional methods (“DA”) improved the robustness of RoI detection (Figure 4c), but data augmentation by Pix2Pix did not (“Pix2Pix”). These results suggest that complex augmentation methods are not necessarily required to improve the robustness of AD detection models. The IoU of the most robust configuration ("AD+Skin+DA") still did not reach a perfect score of 1, suggesting that the AD segmentation remains sensitive to irrelevant features of the images.
Accuracy of severity prediction
Finally, we investigated the impact of the different RoI model configurations on the downstream severity prediction task (Figure 5). We conducted 10-fold cross-validation with a 90:10 train/test split, stratified on patients, and computed the root mean square error of the mean prediction across test images.
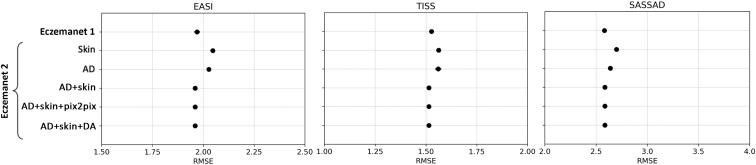
Figure 5.
Accuracy of severity prediction evaluated with RMSE. Points show the mean SE over 10-fold cross-validation with the test set of 136 photographs for EASI (in [0, 12]), TISS (in [0, 9]), and SASSAD (in [0, 18]). EASI, Eczema Area and Severity Index; RMSE, root mean square error; SASSAD, Six Area, Six Sign Atopic Dermatitis; SE, standard error; TISS, Three-Item Severity score.
We evaluated the performance in predicting the regional scores for Eczema Area and Severity Index (erythema, excoriation, lichenification, and edema); Three-Item Severity score (erythema, excoriation, and edema); and Six Area, Six Sign Atopic Dermatitis (cracking, dryness, erythema, excoriation, exudation, and lichenification). The RoI detection models in EczemaNet2 trained with AD or skin segmentation masks (Skin, AD) achieved a better performance than that in EczemaNet1 (Figure 4). It resulted in a slightly higher root mean square error for AD severity assessment (Figure 5), though the practical significance of the difference in root mean square error is debatable. Merging AD segmentation masks with the neighboring skin pixels (“AD+Skin”) provided the downstream severity model with informative and discriminative pattern features between the healthy skin and the lesion (e.g., brightness color, gradient, and texture changes) and led to marginal improvement (P = 0.091 in paired t-test) in the accuracy of severity prediction compared with EczemaNet1 and EczemaNet2 (AD or Skin). Data augmentation, either with traditional methods (“DA”) or Pix2Pix, did not impact the average predictive performance.
Skin of color subpopulation
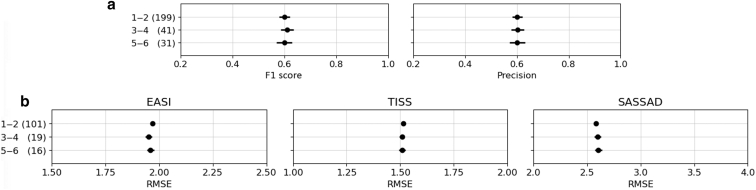
In addition to our primary analysis, we investigated the model’s performance on the skin of color images. We stratified the subjects on the basis of the Fitzpatrick skin phototype and evaluated the model's performance on the quality and robustness of RoI detection and the accuracy of severity prediction. A total of 226 patients (with 1,031 photographs) were types 1–2, 41 patients (with 182 photographs) were types 3–4, and 20 patients (132 photographs) were types 5–6. We found no significant differences in model performance across different Fitzpatrick skin phototypes (Figure 6). This reinforces the potential generalizability and applicability of our model across diverse skin tones, though the result needs to be treated carefully because the number of photographs with types 3–6 was rather small.
Figure 6.
Performance of EczemaNet2 on skin of color subpopulation. Points show the mean ± SE of the metrics across Fitzpatrick bins 1–2, 3–4, and 5–6 with the number of photographs (in parenthesis). (a) Quality measured by F1 score and precision. (b) Accuracy of severity prediction evaluated with RMSE. EASI, Eczema Area and Severity Index; RMSE, root mean square error; SASSAD, Six Area, Six Sign Atopic Dermatitis; SE, standard error; TISS, Three-Item Severity score.
Discussion
Main findings
This study presents EczemaNet2, in which we propose an algorithm to detect AD regions (RoI) (Figure 3) to enable reliable assessment of AD severity from digital camera images. It builds on and improves previously published EczemaNet1 (Figure 1), capable of detecting AD regions from camera images and subsequently assessing the severity of seven AD disease signs without any manual intervention. The RoI model of EczemaNet2 was trained on pixel-level AD segmentation masks provided by four dermatologists (Figure 2). EczemaNet2 could detect and extract AD regions from digital images more accurately than EczemaNet1 (Figure 4a). Data augmentation for the training set boosted the model’s robustness to poor imaging conditions and external noise that is often found in real-life images (Figure 4b and c).
Strengths and limitations of this study
The originality of this study lies in thorough and systematic evaluation of the performance and robustness of AD detection algorithms, the contribution of RoI training data and data augmentation (Pix2Pix and traditional augmentation methods) in terms of detection quality, robustness to poor imaging conditions, and severity prediction performance. By improving the detection quality of the RoI model in EczemaNet2, we improve the interpretability of the entire pipeline because we can be more confident that the severity of AD is assessed from relevant image features. Interpretability of models is relevant to patients’ “right to an explanation” for automated decision making, highlighted in existing regulations, such as the European General Data Protection Regulation (Goodman and Flaxman, 2017).
We used privileged information with 1,345 pixel-level segmentation masks provided by dermatologists and further expanded the dataset with data augmentation techniques. Data augmentation improved the robustness of the RoI model, which is essential to maintain the users’ trust in automated AD severity assessment and ensure the models provide consistent predictions without being sensitive to perturbations in input images.
We recognize that AD segmentation masks may be unreliable because poor agreement among dermatologists was found in detecting AD regions in digital images (Hurault et al., 2022). Poor inter-rater reliability of AD segmentation could explain why the detection accuracy of AD regions remains low in EczemaNet2 (Figure 5). It could further explain why the U-Net did not perform much better for AD detection when it was trained to delineate AD regions as opposed to delineating skin and why merging AD segmentation with surrounding skin pixels achieved only slightly better performance in severity prediction. We believe skin segmentation may be a reasonable alternative to AD segmentation that would require specialist labeling whose quality may be debatable. By definition, skin segmentation does not identify AD lesions. Still, it may sufficiently restrict the inputs to the severity assessment model without excluding potentially informative regions in the images (therefore achieving perfect sensitivity/recall), assuming the images contain a priori representative sites of AD. It could nonetheless be interesting to explore AD segmentation algorithms that can deal with noisy segmentation labels (Karimi et al., 2020).
Implications for clinical practice and research
This study proposed a robust method to detect AD regions from digital images while highlighting the challenges of this endeavor. Accurate and robust detection of AD regions is necessary for developing end-to-end pipelines that automatically assess AD severity from real-world digital images. Although there is considerable promise for remote assessment of AD, the performance of the downstream severity prediction remained unchanged in EczemaNet2, highlighting the difficulty of assessing AD severity from real-world digital images. We believe that collecting more and better-quality data (images and labels) would surpass the gains in performance from using cleverer algorithms. In particular, we emphasize the importance of ensuring that the training dataset covers images for various skin tones to limit skin color bias (Daneshjou et al., 2021).
Materials and Methods
Pixel-level segmentation data
We used 1,345 photographs of representative AD regions from 287 children with AD aged 6 months to 16 years collected as a part of the Softened Water Eczema Trial (Thomas et al., 2011). In terms of the Fitzpatrick skin phototypes, 226 patients (with 1,031 photographs) were types 1–2, 41 patients (with 182 photographs) were types 3–4, and 20 patients (132 photographs) were types 5–6. In the Softened Water Eczema Trial, a clinical staff took photographs and recorded the corresponding severity of seven disease signs (dryness, erythema, excoriation, cracking, exudation, lichenification, and edema) from none (=0) to severe (=3) assessed in person. The photographs vary in resolution and image quality, including focus, lighting, and blur.
Four dermatologists delineated (segmented) AD regions at the pixel level in the 1,345 images (307 images by BO, 308 images by EE, 308 images by LS, and 422 images by HCW). This pixel-level AD segmentation data offers a finer resolution of AD regions than the 1,748 AD crops used to train the RoI detection model in EczemaNet1, where the AD crops were manually extracted by three non-medical students rather than by dermatologists. In addition to the pixel-level AD segmentation, a non-medical student (RM) provided skin segmentation at the pixel level, resulting in pixel-level segmentation masks of background, non-AD skin, and AD skin (Figure 2).
An RoI detection model using pixel-level segmentation for EczemaNet2
For EczemaNet2, we modified the RoI detection model of EczemaNet1 with a standard pixel-level segmentation U-Net (Ronneberger et al., 2015). We trained two U-Nets with pixel-level segmentation masks: one produces skin segmentation masks and another produces AD segmentation masks, respectively, for every image (Figure 3a). We also added three sequential postprocessing steps to extract square crops (Figure 3b) required for the subsequent severity prediction model. The first step was to use the border-following algorithm (Suzuki and be, 1985) to generate rectangular crops. The second step was square cropping to extract square crops of the same dimension without distortion. It avoids image distortion and changes in the appearance and proportion of AD regions that occurred in the resizing step of EczemaNet1, in which image crops were squashed or stretched. The final postprocessing step was to add surrounding non-AD skin pixels to the AD skin pixels in the square crops. This step was applied only when both skin and AD segmentation masks were available.
Data augmentation
Data augmentation refers to the artificial expansion of a dataset by adding synthetic data to increase its size and variation (Shorten and Khoshgoftaar, 2019). Data augmentation is often applied to mitigate the adverse effects of training on a small dataset, such as overfitting to training data, producing an improved and more robust model that can better generalize to unseen data.
We created an augmented dataset to train the segmentation model of EczemaNet2. We applied Gaussian blurring and Gaussian noise to the images, brightened, darkened, zoomed in and out, rotated, and flipped the images. The range and increment size of the scaling parameters were chosen to ensure that the augmented dataset includes various images that may be encountered during image acquisition realistically (Table 2). Those values determined the number of generated images for each augmentation method. For example, five new images were produced by blurring every image in the original dataset with five levels of blurriness. We also applied a combination of two randomly chosen augmentation methods to synthesize two images per original image (Mix in Table 2). The final augmented dataset contains all the synthesized images (28 images in total per original image) and the original images.
We also created another augmented dataset by applying a more complex augmentation method to compare the effects of the data augmentation method on the performance of RoI detection. We used Pix2Pix (Isola et al., 2017), which is based on conditional Generative Adversarial Networks (Mirza and Osindero, 20141) and can generate synthetic images from arbitrary segmentation masks (i.e., images with non-AD/AD/background labels in our case) that describe the precise location of RoI in images.
Model implementation and training
All algorithms tested in this study were developed using libraries and scripts in Python 3.7 and TensorFlow 1.15. The RoI model of EczemaNet2 was trained using Adam for optimizing the cross-entropy loss function through 50 epochs with a learning rate of 0.0001 and batch size of 2 samples, all of which were determined empirically.
Ethics statement
We used 1,345 photographs of representative AD regions from 287 children with AD aged 6 months to 16 years collected as a part of the Softened Water Eczema Trial (Thomas et al., 2011). The softened Water Eczema Trial study was approved by North West Research Ethics Committee (reference 06/MRE08/77), and written informed consent was provided by the parents/caregivers of participating children. The secondary use of the data was approved by Science Engineering Technology Research Ethics Committee at Imperial College London (London, United Kingdom) (SETREC number 22IC7801).
Data availability statement
The source code is publicly available at https://github.com/Tanaka-Group/EczemaNet2. The original images were collected during the Softened Water Eczema Trial for which HCW was the chief investigator. The images cannot be shared publicly owing to restrictions on the participants’ consent.
ORCIDs
Rahman Attar: http://orcid.org/0000-0002-5786-6433
Guillem Hurault: http://orcid.org/0000-0002-1052-3564
Zihao Wang: http://orcid.org/0000-0001-8715-2952
Ricardo Mokhtari: http://orcid.org/0000-0002-7940-6489
Kevin Pan: http://orcid.org/0000-0002-2834-605X
Bayanne Olabi: http://orcid.org/0000-0002-4786-7838
Eleanor Earp: http://orcid.org/0000-0002-8316-0223
Lloyd Steele: http://orcid.org/0000-0003-4745-1338
Hywel C. Williams: http://orcid.org/0000-0002-5646-3093
Reiko J. Tanaka: http://orcid.org/0000-0002-0769-9382
Conflict of Interest
The authors state no conflict of interest.
Acknowledgments
The authors would like to thank Kim S. Thomas and the Softened Water Eczema Trial study team for sharing Softened Water Eczema Trial dataset. The Softened Water Eczema Trial was funded by the National Institute for Health and Care Research (NIHR) Health Technology Assessment Programme. This work was supported by British Skin Foundation.
Author Contributions
Conceptualization: RA, GH, RJT; Data Curation: RA, ZW, RM, KP, BO, EE, LS, HCW; Formal Analysis: RA, GH, ZW, RM; Funding Acquisition: RJT; Investigation: RA, GH, ZW, RM; Methodology: RA, GH, ZW, RM, KP; Software: RA, ZW, RM; Supervision: RJT, HCW; Validation: RA, GH, RJT; Visualization: RA, GH; Writing – Original Draft Preparation: RA, GH; Writing – Review and Editing: RJT, HCW
accepted manuscript published online XXX; corrected proof published online XXX
Cite this article as: JID Innovations 2023.100213
Mirza M, Osindero S. Conditional generative adversarial nets. arXiv 2014.
References
- Bang C.H., Yoon J.W., Ryu J.Y., Chun J.H., Han J.H., Lee Y.B., et al. Automated severity scoring of atopic dermatitis patients by a deep neural network. Sci Rep. 2021;11:6049. doi: 10.1038/s41598-021-85489-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berth-Jones J. Six Area, Six Sign Atopic Dermatitis (SASSAD) seventy score: a simple system for monitoring disease activity in atopic dermatitis. Br J Dermatol Suppl. 1996;135:25–30. doi: 10.1111/j.1365-2133.1996.tb00706.x. [DOI] [PubMed] [Google Scholar]
- Daneshjou R., Smith M.P., Sun M.D., Rotemberg V., Zou J. Lack of transparency and potential bias in artificial intelligence data sets and algorithms: a scoping review. JAMA Dermatol. 2021;157:1362–1369. doi: 10.1001/jamadermatol.2021.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman B., Flaxman S. European Union regulations on algorithmic decision making and a “right to explanation”. AI Mag. 2017;38:50–57. [Google Scholar]
- Hanifin J.M., Thurston M., Omoto M., Cherill R., Tofte S.J., Graeber M. The eczema area and severity index (EASI): assessment of reliability in atopic dermatitis. EASI Evaluator Group. Exp Dermatol. 2001;10:11–18. doi: 10.1034/j.1600-0625.2001.100102.x. [DOI] [PubMed] [Google Scholar]
- Hurault G., Pan K., Mokhtari R., Olabi B., Earp E., Steele L., et al. Detecting eczema areas in digital images: an impossible task? JID Innov. 2022;2 doi: 10.1016/j.xjidi.2022.100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. Paper presented at: 2017 Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 21–26 July 2017; Honolulu, HI.
- Karimi D., Dou H., Warfield S.K., Gholipour A. Deep learning with noisy labels: exploring techniques and remedies in medical image analysis. Med Image Anal. 2020;65 doi: 10.1016/j.media.2020.101759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan S.M., Irvine A.D., Weidinger S. Atopic dermatitis. Lancet. 2020;396:345–360. doi: 10.1016/S0140-6736(20)31286-1. [published correction appears in Lancet 2020;396:758] [DOI] [PubMed] [Google Scholar]
- Leshem Y.A., Chalmers J.R., Apfelbacher C., Furue M., Gerbens L.A.A., Prinsen C.A.C., et al. Measuring atopic eczema symptoms in clinical practice: the first consensus statement from the Harmonising Outcome Measures for Eczema in clinical practice initiative. J Am Acad Dermatol. 2020;82:1181–1186. doi: 10.1016/j.jaad.2019.12.055. [DOI] [PubMed] [Google Scholar]
- National Institute for Health and Care Excellence. Dupilumab for treating moderate to severe atopic dermatitis, https://www.nice.org.uk/guidance/ta534; 2018 (accessed August 5, 2023).
- Nisar H, Tan YR, Ho YK. Segmentation of eczema skin lesions using U-net. Paper presented at: 2020 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES). 1–3 March 2021; Langkawi Island, Malaysia.
- Pan K., Hurault G., Arulkumaran K., Williams H., Tanaka R.J. In: Mach Learn Med Imaging. Midtown Manhattan. Liu M., Yan P., Lian C., Cao X., editors. Springer International Publishing; NY: 2020. EczemaNet: automating detection and assessment of atopic dermatitis; pp. 220–230. [Google Scholar]
- Ronneberger O., Fischer P., Brox T. In: Medical image computing and computer-assisted intervention. Midtown Manhattan. Navab N., Hornegger J., Wells W., Frangi A., editors. Springer International Publishing; NY: 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- Schmitt J., Langan S., Deckert S., Svensson A., von Kobyletzki L., Thomas K., et al. Assessment of clinical signs of atopic dermatitis: a systematic review and recommendation. J Allergy Clin Immunol. 2013;132:1337–1347. doi: 10.1016/j.jaci.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J Big Data. 2019;6:1–48. doi: 10.1186/s40537-021-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H.M., Jeon W., Kim J., Heo C.Y., Yoon H.J., Park J.U., et al. AI-based localization and classification of skin disease with erythema. Sci Rep. 2021;11:5350. doi: 10.1038/s41598-021-84593-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., be K. Topological structural analysis of digitized binary images by border following. Comput Vis Graph Image Process. 1985;30:32–46. [Google Scholar]
- Thomas K.S., Koller K., Dean T., o’Leary C.J., Sach T.H., Frost A., et al. A multicentre randomised controlled trial and economic evaluation of ion-exchange water softeners for the treatment of eczema in children: the Softened Water Eczema Trial (SWET) Health Technol Assess. 2011;15:1–156. doi: 10.3310/hta15080. [DOI] [PubMed] [Google Scholar]
- Williams H.C., Schmitt J., Thomas K.S., Spuls P.I., Simpson E.L., Apfelbacher C.J., et al. The HOME Core outcome set for clinical trials of atopic dermatitis. J Allergy Clin Immunol. 2022;149:1899–1911. doi: 10.1016/j.jaci.2022.03.017. [DOI] [PubMed] [Google Scholar]
- Wolkerstorfer A., de Waard Van Der Spek F.B., Glazenburg E.J., Mulder P.G.H., Oranje A.P. Scoring the severity of atopic dermatitis: three item severity score as a rough system for daily practice and as a pre-screening tool for studies. Acta Derm Venereol. 1999;79:356–359. doi: 10.1080/000155599750010256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The source code is publicly available at https://github.com/Tanaka-Group/EczemaNet2. The original images were collected during the Softened Water Eczema Trial for which HCW was the chief investigator. The images cannot be shared publicly owing to restrictions on the participants’ consent.