Abstract
Epiphytes with crassulacean acid metabolism (CAM) photosynthesis are widespread among vascular plants, and repeated evolution of CAM photosynthesis is a key innovation for micro-ecosystem adaptation. However, we lack a complete understanding of the molecular regulation of CAM photosynthesis in epiphytes. Here, we report a high-quality chromosome-level genome assembly of a CAM epiphyte, Cymbidium mannii (Orchidaceae). The 2.88-Gb orchid genome with a contig N50 of 22.7 Mb and 27 192 annotated genes was organized into 20 pseudochromosomes, 82.8% of which consisted of repetitive elements. Recent expansions of long terminal repeat retrotransposon families have made a major contribution to the evolution of genome size in Cymbidium orchids. We reveal a holistic scenario of molecular regulation of metabolic physiology using high-resolution transcriptomics, proteomics, and metabolomics data collected across a CAM diel cycle. Patterns of rhythmically oscillating metabolites, especially CAM-related products, reveal circadian rhythmicity in metabolite accumulation in epiphytes. Genome-wide analysis of transcript and protein level regulation revealed phase shifts during the multifaceted regulation of circadian metabolism. Notably, we observed diurnal expression of several core CAM genes (especially βCA and PPC) that may be involved in temporal fixation of carbon sources. Our study provides a valuable resource for investigating post-transcription and translation scenarios in C. mannii, an Orchidaceae model for understanding the evolution of innovative traits in epiphytes.
Key words: epiphytes, CAM photosynthesis, multi-omics, phase shifts, rhythmic metabolites
This study reports a high-quality chromosome-level genome assembly of a CAM epiphyte, Cymbidium mannii (Orchidaceae). The high-resolution transcriptomics, proteomics, and metabolomics data across a CAM diel cycle reveal circadian rhythmicity in metabolite accumulation in epiphytes. Genome-wide characterization of transcript and protein level regulation reveals phase shifts during the multifaceted regulation of circadian metabolism.
Introduction
Epiphytes, plants that grow on other plants only for physical support, represent a distinct group in terrestrial carbon-cycling ecosystems (Zotz, 2016). In vascular plants, about 28 000 (∼9%) known epiphyte species are distributed in 73 families, including 18 families of lycopods and ferns, one gymnosperm, and many angiosperms (Zotz, 2013). However, most vascular epiphytes are from the particularly species-rich orchid family (Orchidaceae), with approximately 70% of Orchidaceae species being epiphytes, totaling ∼70% of all known epiphytic flowering plants (Pridgeon, 2009; Stevens, 2017). Orchidaceae are thus an important subject for understanding the innovative traits of epiphytes and their associations with adaptation and rapid diversification.
Although two photosynthesis types—C3 and crassulacean acid metabolism (CAM)—exist in epiphytes (Motomura et al., 2008; Martin et al., 2010), the widespread occurrence of CAM photosynthesis in epiphytes is of particular interest (Zotz, 2004; Bone et al., 2015; Crayn et al., 2015). CAM photosynthesis is a water-conserving carbon dioxide (CO2) fixation pathway predominantly found in desert succulents and tropical epiphytes that temporarily or permanently inhabit water-deficit environments (Nobel, 1991; Donoghue et al., 2014; Bone et al., 2015). Through use of a temporally separated carbon-concentrating mechanism, a CAM plant is able to photosynthesize during the day and exchange gases at night to minimize water loss. The CAM process has been divided into four phases across a diel cycle according to dynamic changes in CO2 fixation rate, malic acid content, and stomatal resistance (Osmond, 1978). CO2 is converted to a four-carbon acid that is stored in the vacuoles at night, then decarboxylated to release CO2 during the day. CAM has been documented in 37 families, including Crassulaceae, Bromeliaceae, Cactaceae, Agavaceae, and Orchidaceae (Benzing et al., 1985; Luttge, 2004; Silvera et al., 2009; Gamisch et al., 2021). Previous studies of Bromeliaceae and Orchidaceae have shown that CAM photosynthesis evolved convergently in epiphytes (Benzing et al., 1985; Silvera et al., 2009). Five subfamilies (Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae, and Epidendroideae) are recognized within Orchidaceae (Ramirez et al., 2007; Chase et al., 2015; Govaerts et al., 2021). Except for Neottieae and a few early diverging groups, the core Epidendroideae are highly speciose, including more than 19 500 species, and largely comprise epiphytic CAM plants, accounting for ∼70% of all epiphytic flowering plants and ∼60% of all CAM plants (Winter and Smith, 1996; Pridgeon, 2009; Zotz, 2013).
Our understanding of the genetic basis and molecular modules of CAM photosynthesis–mediated metabolic physiology remains incomplete, particularly for epiphytes. Previous studies of molecular regulation in CAM plants have focused mainly on terrestrial CAM plants at the genomic or transcriptional regulation level: identification of cis-regulatory elements (CREs) of CAM pathway genes associated with circadian clock genes in Ananas comosus (Ming et al., 2015), diel shifts and altered transcript abundance between CAM and C3 plants of Sedum album (Wai et al., 2019) and Erycina (Heyduk et al., 2018), convergent evolution of genes involved in CO2 fixation and stomatal movement (Yang et al., 2017), and gene toolkits directly involved in CO2 fixation metabolic pathways in representative orchids (Cai et al., 2015; Zhang et al., 2016). A more recent study of the aquatic CAM lycopod Isoetes revealed its distinction from terrestrial CAM plants at the genome and transcriptome levels (Wickell et al., 2021). However, metabolic and proteomic contributions to the physiological adaptations of CAM photosynthesis remain to be fully clarified. Proteomic changes in guard and mesophyll cells were analyzed in the obligate CAM plant Kalanchoë fedtschenkoi (Abraham et al., 2020) and the facultative CAM plant Mesembryanthemum crystallinum (Guan et al., 2021). A multi-omics study of terrestrial CAM was performed in Agave (Abraham et al., 2016), but whether the findings apply to epiphytes is largely unknown. Because transcriptome changes translate into protein fluctuations and then into metabolite rhythms, multifaceted regulation of the CAM pathway is worthy of further study.
The genus Cymbidium belongs to Epidendroideae (Orchidaceae) and comprises 60–80 perennial plants and many horticultural cultivars (Liu et al., 2006; Puy and Cribb, 2007; Pridgeon, 2009). Members of the genus Cymbidium can be epiphytic or terrestrial and are found in diverse vegetation types, from tropical rainforests to subtropical grasslands (Motomura et al., 2008; Ogura-Tsujita et al., 2012). CAM photosynthesis has been documented to occur frequently in Cymbidium (Motomura et al., 2008; Zhang et al., 2015); thus, minor genetic changes might enable the evolution of such photosynthetic traits. Diverse ecological adaptations make Cymbidium an ideal model for understanding the association between epiphytes and photosynthetic types. In Cymbidium, three genomes, all from terrestrial C3 species, have recently been reported (Ai et al., 2021; Sun et al., 2021; Yang et al., 2021), but the genomes of epiphytic CAM plants remain to be explored. Although the genome of the epiphytic orchid Phalaenopsis equestris has been reported, this work mainly emphasized the evolution of CAM-associated genes (Cai et al., 2015). In the present study, we assembled a high-quality chromosome-level genome of the CAM epiphyte C. mannii and investigated genome-wide molecular regulation of its metabolic physiology by integrating transcriptomics, proteomics, and metabolomics data across a 24-h diel cycle. Our work provides a high-quality genome for a CAM epiphyte and broadens our understanding of the temporal regulation of CAM photosynthesis–mediated metabolic physiology.
Results
Physiological characteristics and chromosome-level genome assembly and annotation of C. mannii
We surveyed the day–night patterns of physiological parameters in the CAM plant C. mannii, including net carbon uptake, enzyme activities, and metabolites involved in carboxylation. Nocturnal CO2 uptake was generally synchronous with stomatal opening in C. mannii leaves (Supplemental Figure 1). During the day–night shift, the content of oxaloacetic acid increased until dusk; this was followed by a dramatic increase in pyruvic acid at night, which may reflect the corresponding increase in phosphoenolpyruvate carboxylase (PPC) activity (Supplemental Figure 1). During the day, stored malate was decarboxylated by malic enzyme (ME), likely resulting in high concentrations of CO2 around Rubisco.
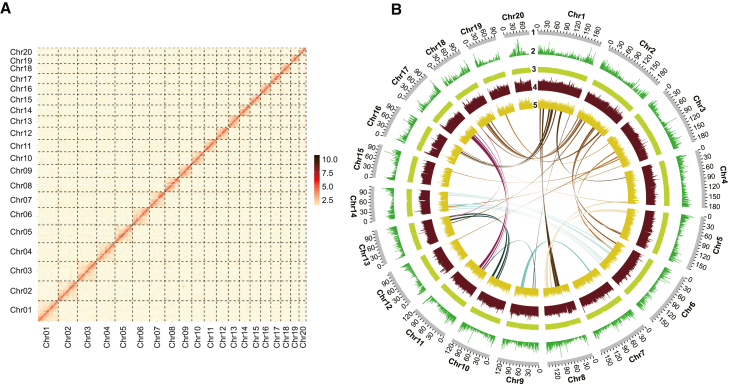
We then sequenced the C. mannii genome using a combination of platforms (Supplemental Table 1). The assembly was 2.88 Gb in length, comprising 1892 contigs with a contig N50 of 22.7 Mb (Supplemental Tables 2 and 3). Using chromatin interaction signals from Hi-C data, 20 pseudochromosomes were constructed, anchoring 2.73 Gb (94.79%) of the genome sequences; the longest pseudochromosome was 202.7 Mb and the shortest 79.1 Mb (Figure 1; Supplemental Table 4). The assembled genome size was consistent with predictions based on flow cytometry (2.84 Gb) and k-mer frequency distribution (2.84 Gb) (Supplemental Figure 2), and the continuous evaluation contig N50 was higher than that of other reported genome assemblies in Orchidaceae (Supplemental Table 3).
Figure 1.
Genome characteristics of the CAM plant Cymbidium mannii.
(A) Genome-wide all-by-all Hi-C interactions of the intra-chromosomal contact matrix. The interaction heatmap represents the normalized counts of Hi-C links in 500-kb bins on a logarithmic scale.
(B) Genomic features of C. mannii. Outer to inner circles show (1) chromosome length, (2) gene density, (3) GC content, (4) LTR sequences, and (5) DNA transposons. Homologous regions among chromosomes are displayed with connecting lines colored according to the chromosome number.
A total of 27 192 protein-coding genes were annotated in the C. mannii genome, at least 21 960 (79.1%) of which were supported with transcriptome data. InterProScan analysis revealed that 26 313 (96.4%) genes could be functionally annotated, 10 785 of which encoded metabolic enzymes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Supplemental Table 5). Nine hundred forty-three protein kinases and 1660 transcription factors (TFs) were identified in the C. mannii genome (Supplemental Table 6). The coding regions were typical in size, but the average gene size was expanded compared with that of basal Orchidaceae species and selected monocots, probably because of an increase in noncoding sequences (Supplemental Table 7). The annotation quality was assessed using Benchmarking Universal Single-Copy Orthologs (BUSCO); 1566 complete gene models out of 1614 (97.0%) were recovered, consisting of 95.4% single-copy genes and 1.6% duplicates, suggesting the high quality and completeness of the gene annotations (Supplemental Figure 3; Supplemental Table 8).
Transposable element–mediated genome size expansion
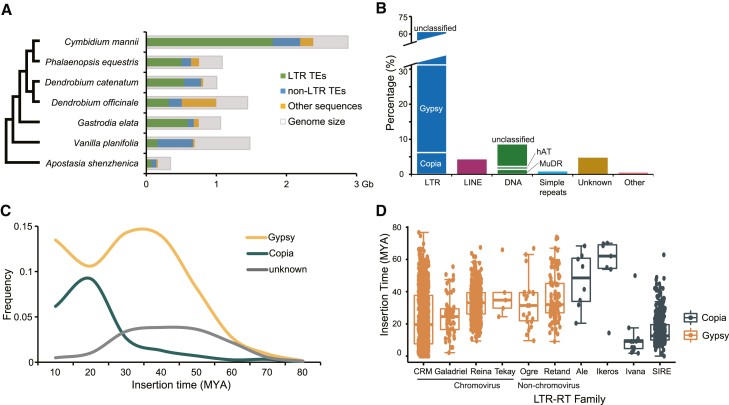
Approximately 82.8% of the C. mannii genome was composed of repetitive elements, 92.2% of which were transposable elements (TEs) (Figure 2A; Supplemental Table 9). Long terminal repeats (LTRs) represented the largest proportion (75.8%) of repetitive elements, of which Gypsy (30.2%) was the most abundant, followed by Copia (7.5%) (Figure 2B). The Gypsy:Copia ratio was approximately 4:1, falling within the general range of most angiosperm LTR ratios (Zhang et al., 2020). The main bursts of Gypsy retrotransposons (RTs) occurred about 30–40 million years ago (MYA), whereas those of Copia retrotransposons occurred approximately 10–20 MYA (Figure 2C). LTRs were classified into 10 families, and their abundance and insertion times were investigated (Figure 2D). CRM and Reina families in Gypsy, which are present only in seed plants (Gorinsek et al., 2004; Neumann et al., 2019), were markedly expanded. The non-chromovirus group Retand, which is only found in angiosperms (Neumann et al., 2019), was the third most abundant LTR group. A similar expansion was observed for the angiosperm-specific group SIRE in the Copia superfamilies. Thus, specific LTR groups had a major effect on C. mannii genome size during evolution.
Figure 2.
TE-mediated genome size expansion.
(A) Comparison of repetitive sequences in orchids.
(B) Relative contents of the different repetitive element classes. Types lower than 0.5% are not shown.
(C) Insertion time distribution of intact LTRs in the C. mannii genome.
(D) LTR-RT families in C. mannii. The 10 LTR-RT families with ≥5 LTRs are plotted. Dots in each boxplot represent the identified LTR elements. Box boundaries indicate 25% and 75% of the distribution of insertion times.
Genome and gene family evolution
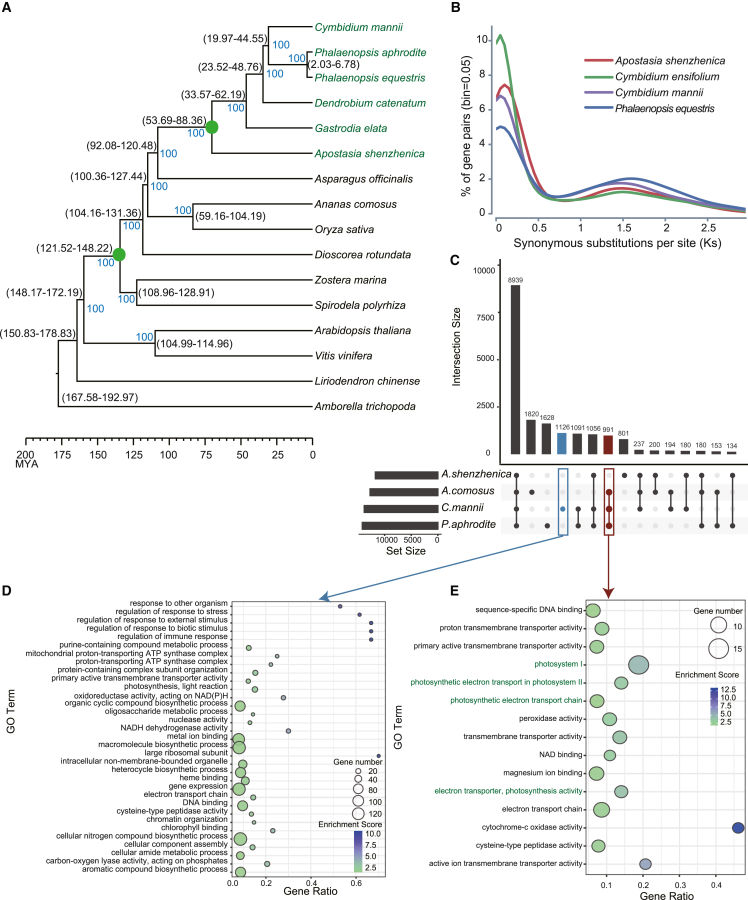
The phylogeny of C. mannii and other Orchidaceae species, along with 10 other representative plant species, was inferred using a concatenated dataset of 423 single-copy genes (Figure 3A). The divergence time between Cymbidium (represented by C. mannii) and its sister clade Phalaenopsis could be dated back to 30.67 MYA or the Early Oligocene (Figure 3A), which was also close to the timing of the LTR-Gypsy burst.
Figure 3.
Comparative and evolutionary genomics of C. mannii and other representative plants.
(A) Phylogenetic relationships among orchids and other selected angiosperms. The phylogeny is constructed from single-copy orthologs among selected taxa. Species names in green belong to the Orchidaceae family. Green circles on the internodes indicate WGD events. Lineage divergence time (black) and bootstrap support value (blue) are marked at each branch node.
(B) Distribution of Ks analysis among selected orchids. Gene pairs were based on paralogs within a given genome.
(C) UpSet plot displaying the orthologous groups present (black circles) or missing (gray circles) across genomes of the selected C3 orchids, the representative terrestrial CAM plant A. comosus, and the CAM epiphyte P. aphrodite. Highlighted blue and dark red boxes and circles indicate C. mannii-specific genes and genes shared by selected CAM plants, respectively.
(D) Significant (FDR < 0.05) GO terms annotated in C. mannii-specific orthologous groups.
(E) Significant GO terms derived from shared CAM plant orthologous groups.
Consistent with previous studies (Zhang et al., 2017; Hasing et al., 2020), our collinearity analysis showed that all orchids, including the early diverging lineages, shared a single whole-genome duplication (WGD) event, but there were no genomic signals of additional WGD events in the formation of the core Cymbidium species (Supplemental Figure 4). Few syntenic blocks (42 blocks containing 416 genes) were identified in the C. mannii genome, further confirming the lack of segmental or chromosome-level duplication (Figure 1B). A lack of recent WGD events was also revealed by the distribution of synonymous substitutions per synonymous site (Ks), in which the selected orchids shared a similar peak (Figure 3B), and by the small proportion of duplicated BUSCO genes in the genomes of C. mannii and other orchids (Supplemental Figure 3).
To obtain a broader picture of the dynamics of gene families, C. mannii was compared with four sequenced orchids and four other angiosperms, including typical CAM species (Supplemental Figure 5). A total of 8353 and 6588 gene families were shared among orchids and the selected angiosperms, respectively. Because our focus was on functional toolkits related to CAM photosynthesis, gene families shared by the selected CAM plants (Phalaenopsis aphrodite and A. comosus) were identified and compared with those of the early diverging C3 orchid Apostasia shenzhenica (Figure 3C). Gene Ontology (GO) enrichment analysis indicated that C. mannii-specific genes were enriched in functional categories related to stress response and internal and external stimuli, as well as energy-related components such as NADH dehydrogenase and ATP synthase complex (Figure 3D). Compared with the terrestrial CAM plant A. comosus and another epiphytic CAM orchid P. aphrodite, C. mannii shared many gene families related to photosystems, photosynthetic electron transport, cytochrome-c oxidase activity, and so forth (Figure 3E). We also compared the assembled C. mannii genome with the recently published genome of Cymbidium ensifolium (a C3 plant), and they largely shared gene clusters enriched in photosynthesis and light reactions (Supplemental Figure 6).
Rhythmic oscillations of the C. mannii metabolome
Temporal variations in the C. mannii leaf metabolome were examined using untargeted liquid chromatography–mass spectrometry (LC–MS) with six biological replicates at 4-h intervals within one diel cycle (starting from zeitgeber time [ZT] 0) (Supplemental Figure 7). A total of 7364 metabolite features were measured, and analysis of coefficients of variation across replicates revealed that 80% of the metabolites had values <0.4, suggesting that the metabolome data were generally of high quality. Principal-component analysis clearly separated the metabolites into different time points, especially metabolic profiles for the day and night (Supplemental Figure 8).
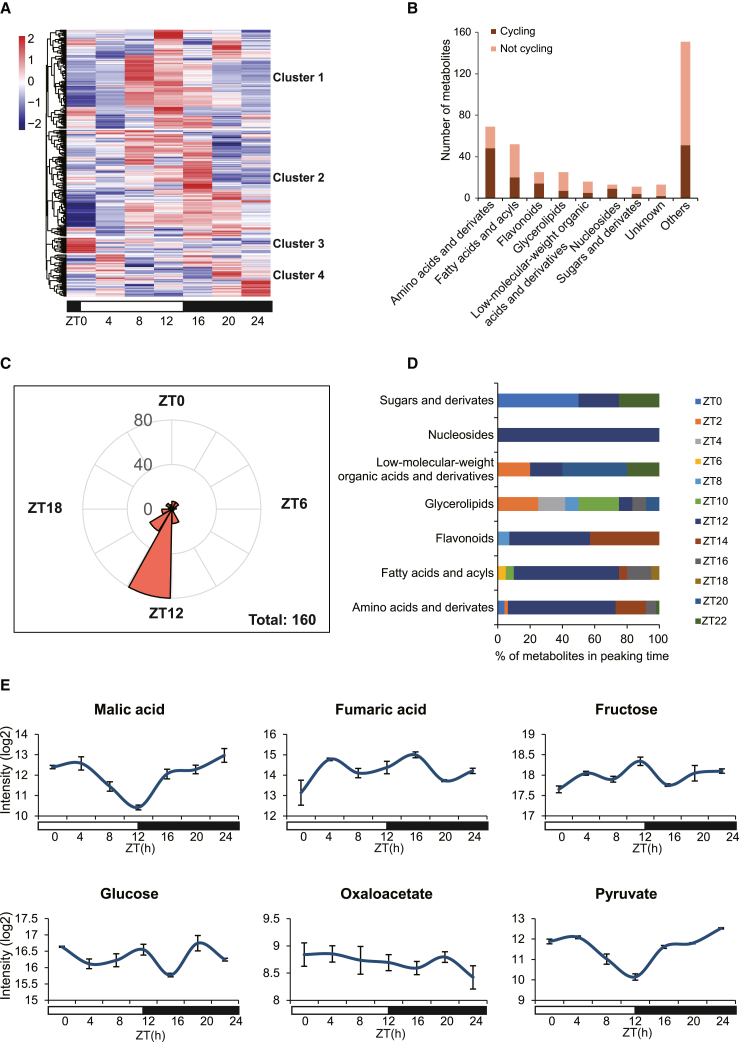
By comparing the mass-to-charge ratio (m/z), retention time, and fragmentation pattern data with the KEGG and Human Metabolome Database, 375 metabolites (from secondary mass spectrometry) were annotated or structurally identified, including 16 low-molecular-weight organic acids and derivatives, 11 sugar-related compounds, 69 amino acids and derivatives, 13 nucleosides, 25 flavonoids, 38 glycerolipids and glycerophospholipids, and 52 fatty acids and acyls (Supplemental Table 10). The 375 metabolites were grouped into four clusters according to their relative abundance at different time points: cluster 1 (141 metabolites) showed higher abundance late in the day (dusk), cluster 2 (152 metabolites) generally showed higher abundance in the middle of the night, cluster 3 (22 metabolites) peaked at the start of the day, and cluster 4 (60 metabolites) displayed higher abundance in the early morning (dawn) (Figure 4A). Metabolic pathway enrichment analysis (false-discovery rate [FDR]-adjusted p < 0.05) indicated that the aminoacyl-tRNA biosynthetic pathway was enriched in clusters 1 and 2, whereas metabolites from glutathione and glycerophospholipid metabolic pathways were enriched in cluster 4. No metabolic pathway enrichment was detected in cluster 3.
Figure 4.
Rhythmic and selected CAM photosynthesis-related metabolites in C. mannii.
(A) Hierarchical clustering metabolic abundance profiles throughout the diel cycle. The abundance values (log2) were averaged from six replicates, and the Z score was transformed. The metabolite annotation is summarized in Supplemental Table 10.
(B) Rhythmic metabolites and their functional categories.
(C) Peak times of rhythmic metabolites across the 24-h diel cycle.
(D) Distribution of rhythmic metabolites in the seven major categories.
(E) Abundance profiles of selected metabolites from secondary mass spectrometry (malic and fumaric acids) and primary mass spectrometry (potentially fructose, glucose, oxaloacetic acid, and pyruvate). Standard errors of six replicates are indicated by error bars.
To further characterize rhythmic metabolites, JTK_CYCLE analysis was performed, and 160 metabolites with rhythmically oscillating patterns of abundance were identified (FDR-adjusted p < 0.05) (Figure 4B). Most detected amino acids and derivatives (69.56%), nucleosides (69.23%), and flavonoids (56.0%) displayed circadian rhythmicity. Most rhythmic metabolites showed peak abundance at the end of the day to early night, accounting for 75.61% of all rhythmic metabolites (Figure 4C). Moreover, 77.78% of nucleosides, 68.97% of organic acids and derivatives (e.g., amino acids), 42.11% of organic oxygen compounds (e.g., sugars, alcohols), and 29.59% of lipids showed rhythmic abundance (Figure 4D).
As a CAM epiphyte, C. mannii can take up CO2 at night as a carbon source. We therefore investigated levels of central metabolites in the CAM pathway (e.g., malic acid, fumaric acid, and soluble sugars) (Figure 4E) and the glycolysis/gluconeogenesis pathway (Supplemental Figure 9). Malic acid abundance decreased during the day and increased at night, whereas fumaric acid fluctuated over the diel cycle but tended to increase at night. Possible soluble sugars, oxaloacetate, and pyruvate were detected by primary mass spectrometry. Glucose and fructose displayed a biphasic pattern in which they accumulated in the middle of the night and in the morning. The dynamic synthesis and decomposition of pyruvate during the day were consistent with the reduced intensity in the day. To compare the fluctuations of these metabolites with those in other CAM and C3 plants, the dynamic patterns of malic acid, fumaric acid, and soluble sugar levels were compared with those in the terrestrial CAM plant Agave and the C3 model plant Arabidopsis (Supplemental Figure 10; Supplemental Table 11). The oscillation phase of malic acid was consistent with CO2 fixation in terrestrial CAM plants and unlike that in the C3 model plant. Fumaric acid tended to show a pattern similar to that in Agave, with higher intensity in the evening. Glucose and fructose displayed similar fluctuations in Agave and Arabidopsis, indicating more than one carbon source for malate synthesis (Arrizon et al., 2010; Abraham et al., 2016). Glycolysis/gluconeogenesis-related metabolites were also investigated (Supplemental Figure 9), and several metabolites from this pathway showed relevant changes. However, because some of the glycolysis/gluconeogenesis-related metabolites were detected from primary mass spectrometry, further identification and verification may be required.
Rhythmic time differences between transcriptome and proteome
To investigate the potential roles of transcription- and translation-level regulation in the day–night metabolic physiology of an epiphyte, transcriptomic and proteomic profiling were performed in triplicate and quadruplicate, respectively, using the same time points sampled for metabolomic analyses. After excluding low-expressed genes (transcripts per million [TPM] < 5), 17 648 transcripts (64.63%) (Supplemental Table 12) were used for circadian analysis, 7568 (27.72%) of which were rhythmically transcribed (Supplemental Table 13; Supplemental Figure 11). Protein abundance was also quantified to evaluate the potential role of post-transcriptional modifications on functional state changes in C. mannii, and 40 721 peptide sequences and 5218 protein groups were obtained (Supplemental Table 14; Supplemental Figure 12A). Among all detected proteins, the abundance of 508 (9.74%) proteins exhibited rhythmic patterns across the diel cycle (FDR-adjusted p < 0.05). Among these, 235 proteins were related to metabolic pathways. Enrichment analysis showed that nucleotide biosynthesis pathways and CAM-related pathways (e.g., carbohydrate metabolism and photosynthesis pathways) were enriched (Supplemental Figure 12B). Moreover, the functions of protein processing in the endoplasmic reticulum, plant hormone signal transduction, and carbohydrate metabolism were enriched among the cycling proteins (Supplemental Figure 12C).
When transcriptome and proteome data were integrated, multifaceted regulation of metabolic physiology was observed in C. mannii leaves (Figure 5). Specifically, the rhythmic transcripts mainly showed peak expression at the beginning of the night (ZT12) (25.17%) (Figure 5A), whereas most proteins peaked at dawn (ZT20–ZT22) (49.80%) (Figure 5B). This phase shift pattern suggests that the molecular regulation of circadian physiology of metabolism occurred at or above post-transcriptional levels. To gain insight into this pattern, we examined 182 genes that displayed rhythmicity at both transcriptional and post-transcriptional levels (Figure 5C). The peak protein abundance was hysteretic. Approximately 66.67% of the genes displayed a lag time of 2–12 h, of which 52 genes (47.27%) lagged for 6–8 h at the protein level relative to their expression level (Figure 5D). To test whether this molecular pattern resembled or differed from that in terrestrial CAM plants, published transcriptomic and proteomic data were reanalyzed using the same pipeline, and rhythmic transcripts and proteins were identified in Agave (Supplemental Figure 13). The percentage of rhythmic transcripts (33.82%) and proteins (9.70%) in Agave was similar to that in C. mannii. The phase-shift pattern could be clearly observed in Agave, and 49.64% of the genes displayed a lag time of 2–12 h, of which 56.52% lagged for 8–10 h. The total proportion of genes that lagged by 8–10 h in Agave (28.06%) is close to that of genes that lagged by 6–8 h in C. mannii (31.51%). However, 34.53% of the genes lagged by 20–22 h in Agave compared with only 16.36% in C. mannii. Our results indicate that the transcript–protein phase shift pattern was shared by different species, but the peak times of transcripts and proteins may be species specific.
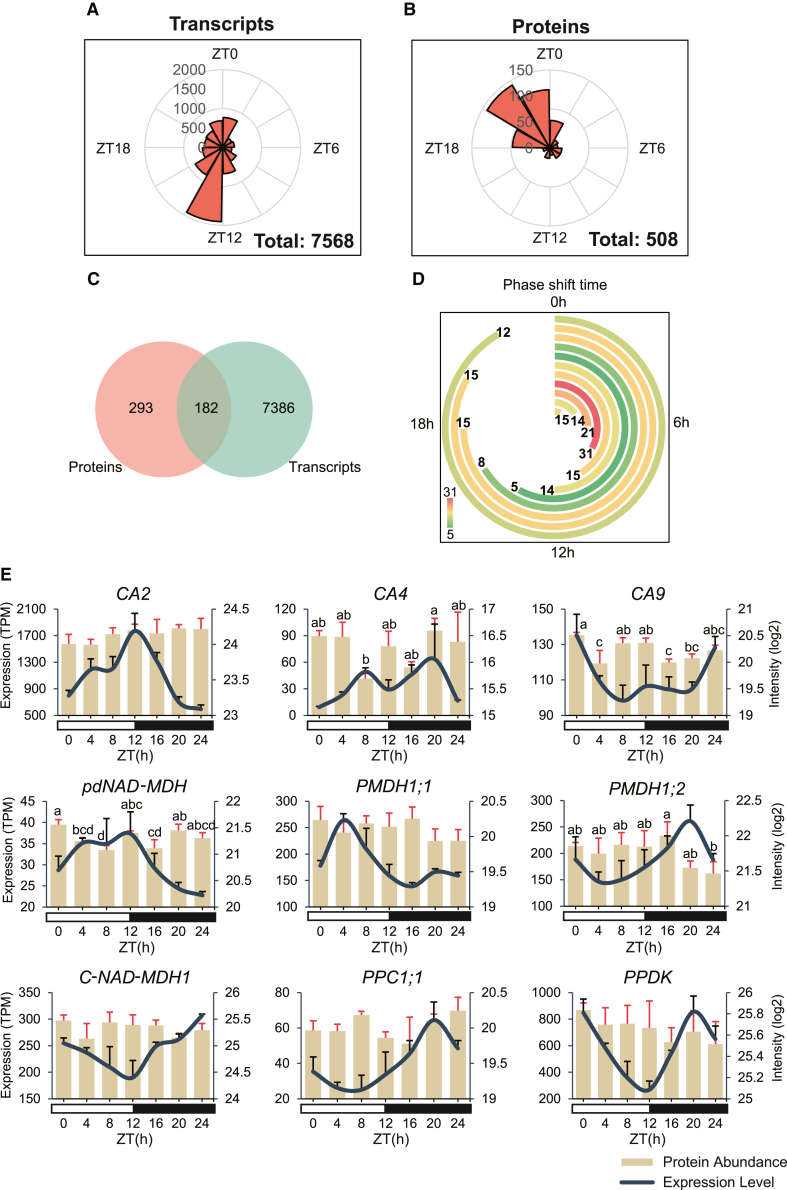
Figure 5.
Phase shifts between proteomic and expression profiles and in core CAM genes.
(A and B) Rose plots representing the number of rhythmic transcripts (A) and proteins (B) with different peak times across the diel cycle.
(C) Unique and shared gene numbers between cycling transcripts and proteins.
(D) Phase shift time between transcripts and proteins. The full circle represents 24 h, and the angle of each bar represents the shift time. Genes with a 24-h shift are not shown in the chart. Gene numbers at each time point are labeled at the end of each bar, with color indicating high (red) or low (green).
(E) Selected core CAM pathway genes with cycling expression levels. Error bars for transcript expression data represent the standard errors of three replicates. Significant differences in protein abundance were identified using Fisher’s least significant difference and are indicated with lowercase letters. Expression levels were quantified as transcript abundance (TPM), and protein abundance was quantified as log2-transformed peptide intensity.
CA, carbonic anhydrase; NAD-MDH, NAD-malate dehydrogenase; PPC, phosphoenolpyruvate carboxylase; PPDK, pyruvate orthophosphate dikinase.
To gain further insight into the post-transcriptional regulation of epiphytic CAM plants, we analyzed the expression levels and protein abundance of core genes involved in CO2 fixation in the CAM pathway, including carbonic anhydrase (CA), ME, malate dehydrogenase (MDH), PPC, phosphoenolpyruvate carboxykinase (PEPCK), phosphoenolpyruvate carboxylase kinase (PPCK), and pyruvate phosphate dikinase (PPDK) (Supplemental Table 15; Supplemental Figure 14A). The mRNA expression profiles of the nine rhythmically expressed CAM pathway genes (three CAs, four MDHs, one PPC, and one PPDK) were collected and compared with their protein abundance (Figure 5E). CA catalyzes the conversion of CO2 to bicarbonate, playing a crucial role in CO2 fixation. Three CA subfamilies (α, β, and γ) were present in C. mannii. βCA is an upstream regulator of CO2-controlled stomatal movements, and plants that overexpress βCA in guard cells exhibit enhanced water use efficiency (WUE) (Hu et al., 2010). Two rhythmically expressed βCAs (CA2 and CA4) were identified in the CAM orchid C. mannii, and one βCA (CA5) showed a high expression level. CA2 and CA5 exhibited increased expression levels during the day, followed by a decrease at night, whereas CA4 and CA9 displayed the opposite pattern. Therefore, CA2 and CA5 might correlate with stomatal movements, but more evidence is needed to support this possibility. The four cycling MDH genes showed contrasting expression patterns, indicating that the different enzymes had their own regulatory schema and coordinated together in the CAM pathway. Because PPC is a key enzyme for CO2 fixation in the CAM pathway, gene expression and protein abundance of all three gene copies were examined. One copy of PPC showed rhythmic expression (PPC1;1), but another copy (PPC1;3) had a much higher expression level, at least 10 times that of PPC1;1. Moreover, only PPC1;3 displayed significant day–night differences in protein abundance (Supplemental Figure 14B), implying that PPC1;3 might play a dominant role in catalyzing HCO3-dependent carboxylation. Overall, the protein abundance of these genes changed asynchronously with their temporal expression levels, suggesting important roles of post-transcriptional regulation in epiphyte physiology.
Similarities and differences in CAM-related gene expression between epiphytic and terrestrial plants
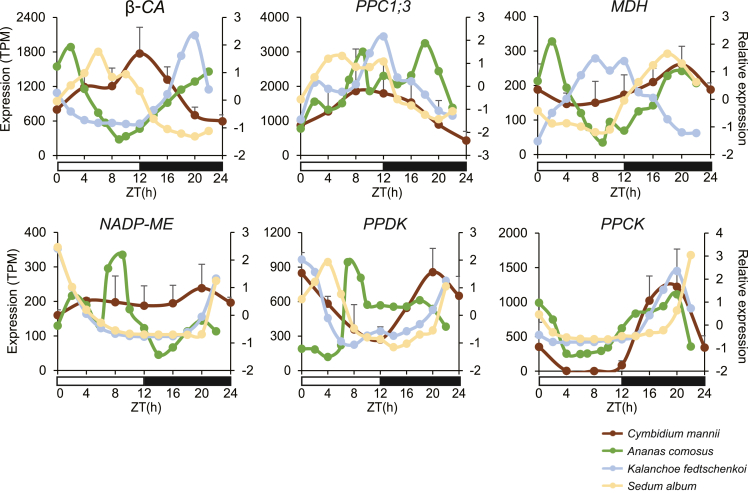
We next compared the expression patterns of core genes related to the CAM pathway in C. mannii and three terrestrial CAM plants (A. comosus, K. fedtschenkoi, and S. album) using publicly available cycling data. The genes showed generally coincident expression patterns with shifted phases (Figure 6). The rhythmic βCA (CA2) in C. mannii and its S. album homolog displayed a similar expression pattern, distinct from that of the A. comosus and K. fedtschenkoi homologs, which showed increased expression under dark conditions. The expression pattern (peaking at dawn) of the other highly expressed βCA gene (CA5) was consistent with that of homologs in A. comosus and S. album, but its peak time was roughly between those of the A. comosus and S. album genes (Supplemental Figure 15A), displaying a phase shift. The other two relatively weakly expressed βCA genes exhibited fluctuation patterns that did not match those in terrestrial CAM plants (Supplemental Figure 15B). Likewise, the dominantly expressed PPC1;3 in C. mannii appeared to show an expression pattern similar to its K. fedtschenkoi and S. album homologs (Figure 6), and PPCK showed conserved nighttime expression in both epiphytic and terrestrial plants. The expression levels of NADP-ME and PPDK in C. mannii were highly consistent with those in K. fedtschenkoi and S. album, suggesting that these species all preferred to utilize NADP-ME and PPDK pathways for decarboxylation (Yang et al., 2017; Wai et al., 2019). The intermediate product (pyruvic acid) of these pathways was present at consistently lower levels during the day (Supplemental Figure 1). In addition, the extremely lowexpression level of PEPCK (Supplemental Figure 14A) and the expression pattern of MDH (Figure 6, Supplemental Figure 15D) did not support the alternative MDH and PEPCK decarboxylation pathways reported in A. comosus and the aquatic CAM lycopod Isoetes (Ming et al., 2015; Wickell et al., 2021).
Figure 6.
Expression of selected cycling core CAM genes in C. mannii and terrestrial CAM plants.
TPM expression data for C. mannii are plotted with standard error bars (from three replicates). Relative expression patterns in selected terrestrial CAM plants were plotted for comparison. Source gene IDs used for comparison are provided in Supplemental Table 16. PPCK, phosphoenolpyruvate carboxylase kinase.
Circadian clock–associated regulation in CAM and C3Cymbidium plants
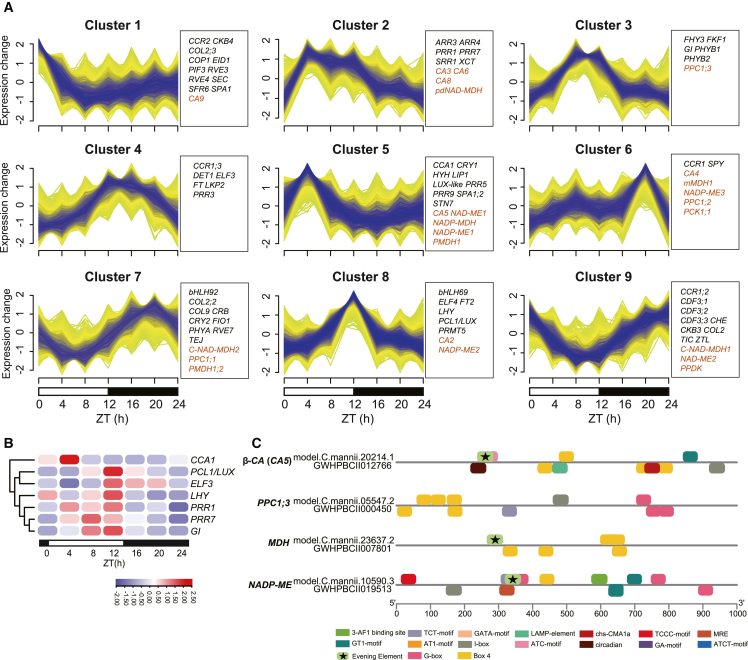
Circadian clock–associated genes and CREs play a crucial role in regulating the CAM pathway (Ming et al., 2015; Wai et al., 2019; Wickell et al., 2021). To explore the circadian clock and CAM gene expression pattern during the diel cycle, we performed a temporal clustering analysis using Mfuzz (Figure 7A). We also examined the expression of circadian clock genes (Supplemental Table 17; Supplemental Figure 16), particularly CCA1 (CIRCADIAN CLOCK ASSOCIATED 1), LUX (LUX ARRHYTHMO), ELF3 (EARLY FLOWERING 3), LHY (LATE ELONGATED HYPOCOTYL), PRR1 (PSEUDO-RESPONSE REGULATOR 1), PRR7 (PSEUDO-RESPONSE REGULATOR 7), and GI (GIGANTEA) (Figure 7B). These circadian oscillator genes were largely similar in expression level, reaching peak abundance near dusk, with the exception of CCA1. CCA1 was in the same cluster as CA5, NAP-MDH, and NADP-ME1, whereas the possibly dominant CO2-fixation gene PPC1;3 was in cluster 3, which displayed a later peak time.
Figure 7.
Core circadian clock gene expression level and CREs in selected CAM genes.
(A) Mfuzz clusters showing gene expression patterns in C. mannii throughout the 24-h diel cycle. Circadian clock (black) and core CAM genes (red) in each cluster are labeled on the right.
(B) Expression of selected core circadian clock genes in C. mannii.
(C) Predicted CREs in the region 1 kb upstream of the selected core CAM genes in C. mannii and the comparable C3 plant C. ensifolium.
Because many circadian oscillator genes can regulate gene expression by binding to specific motifs, we searched for light-responsive CREs in promoter regions (1 kb upstream) of the C. mannii genome and that of the closely related C3 plant C. ensifolium to detect possible factors involved in photoperiodism (Supplemental Table 18). The upstream sequences of these two species were significantly diverse compared with their protein-coding regions but did not differ significantly from the housekeeping genes (Supplemental Figure 17). The evening element (AAATATCT), which can be recognized by CCA1, was present in CA5, MDH, and NADP-ME genes of C. mannii but absent from those of C. ensifolium (Figure 7C), indicating that it may be a possible regulatory factor. The well-known morning element G-box (CACGTG) was found in many gene promoters in both CAM and C3 species, as was the widespread Box 4 (ATTAAT), indicating that they may not directly modulate timing.
Discussion
We generated a chromosome-level genome assembly of the epiphytic CAM orchid C. mannii, one of the higher quality Orchidaceae genome assemblies reported to date (Cai et al., 2015; Zhang et al., 2017; Ai et al., 2021; Sun et al., 2021; Yang et al., 2021). The 2.88-Gb C. mannii genome is rich in repetitive sequences and is the largest percentage of repetitive sequences (82.8%) among all sequenced orchids (Supplemental Table 3). The genome size expansion in Cymbidium is mainly due to LTR insertions, and their insertion time is consistent with the timing of Cymbidium species diversification (Figures 2 and 3). The rapid radiation of Cymbidium orchids in the mountains of southwest China may have been triggered by the uplift and subsequent fluvial incision of the Qinghai–Tibetan Plateau (Wang et al., 2012; Lai et al., 2021). No recent WGD events have occurred within Orchidaceae, suggesting that many innovative features, such as the CAM photosynthesis that evolved in Cymbidium, were unlikely to be caused by WGD (Deng et al., 2016).
We investigated day–night metabolite profiles as an indicator of CAM photosynthesis-mediated metabolic physiology and observed obvious diurnal regulation in C. mannii. The observed rhythmic metabolites and their peak times provide valuable information for epiphytic Cymbidium species and can be used for future research on orchids and other CAM plants. The rhythmic metabolites showed marked temporal differences from those of C3 (Zhou et al., 2022) and C4 (Li et al., 2020) species, and more diversity remains to be explored in other plants. Using metabolite abundance data, we found that trends in accumulation of malic acid and glucose were consistent with features of the tricarboxylic acid cycle in CAM plants (Heyduk et al., 2019). Studies on tissue-specific rhythmic regulation are needed to further clarify mechanisms of CAM physiology in epiphytes. In addition, although oscillating metabolites in the photosynthetic pathway were related to glycolysis/gluconeogenesis to some extent, only a limited number of such metabolites were identified, and further targeted metabolomics of this pathway may reveal more detailed mechanisms.
Integrated analysis of the transcriptome and proteome data revealed the diurnal synchronization of carbon-processing pathways, but a phase shift pattern was observed. Analysis of genes encoding core CAM enzymes (CA, ME, MDH, PPC, and PPDK) identified specific βCA genes that may function mainly in the epiphytic CAM pathway. Because guard cell βCA regulation could directly affect WUE, our study provides solid evidence for core CAM gene expression and regulation in adaptation to CAM photosynthesis and enhancing WUE. Expression and protein abundance data indicated that PPC1;3 may have been co-opted during evolution to perform a key role in CO2 fixation, suggesting that one isoform of PPC has a critical function (Winter and Smith, 2022). Suppression of PPC expression can result in core circadian-clock gene expression changes (Boxall et al., 2020), indicating that the dominant PPC copy detected here could be used for future engineering; its protein abundance alteration is worthy of further study. The other documented CAM genes and their protein abundance data provide valuable information on unique post-transcriptional patterns in CAM plants. We also observed that βCA, PPC, and other core CAM genes have strong correlations with most cycling metabolites (Supplemental Figure 18), suggesting that they play an important role in regulation of the C. mannii leaf metabolome.
When C. mannii was compared with terrestrial CAM plants, we found a highly coincident gene expression pattern but a shifted circadian phase (Figure 6). Comparison with a closely related C3 Cymbidium plant suggested that circadian CREs may affect the cycling genes in C. mannii, and they have potential for future regulation to create a more water-use efficient pathway. Overall, C. mannii represents a valuable model plant in Orchidaceae for understanding the evolution of epiphytic CAM plants and for future breeding of ornamental traits. We elucidated the genomic evolution and CAM pathway of C. mannii using multi-omics datasets, offering an important resource for utilization and modification of water-use efficient plants in a changing climate.
Methods
Plant materials for DNA sequencing
C. mannii plants were collected from the fields of Puer, Yunnan, southwest China, and were grown in Kunming Botanical Garden as previously described (Yang et al., 2013). Genomic DNA extracted from healthy leaves of a single individual with the Qiagen Genomic DNA Extraction Kit (Qiagen, Valencia, CA, USA) was used for both short-read and long-read DNA sequencing. A BGI-seq library was constructed using the MGIEasy Library Prep Kit (MGI, Shenzhen, China) and sequenced on the MGISEQ-2000 platform, generating 292 Gb of high-quality paired-end short reads for genome size estimation and error correction. For long-read DNA sequencing, 43 nanopore libraries were constructed according to the manufacturer’s instructions and sequenced using a GridION X5 or PromethION sequencer (Oxford Nanopore Technologies, Oxford, UK), producing 411 Gb of nanopore raw reads for genome assembly (Supplemental Table 1).
Genome size estimation
The C. mannii genome size was estimated with Jellyfish v2.2.10 (Marcais and Kingsford, 2011) using a 17-k-mer frequency distribution and was experimentally validated by flow cytometry.
Genome assembly, polishing, and evaluation
Raw nanopore long reads were trimmed for adaptor sequences and quality control (QC). Filtered reads were corrected and used for de novo assembly performed with NextDenovo (v2.2.0; https://github.com/Nextomics/NextDenovo) with the parameters: read cutoff = 1000, seed cutoff = 25 000, block size = 1 g. After 50 iterative rounds, the assembled genome with the highest continuity was selected and polished with Nextpolish v1.1.0 (Hu et al., 2020).
Assembly completeness was assessed by mapping all paired-end short reads to the assembly using Burrows–Wheeler Aligner v0.7.17 (Li, 2013). The base-level error rates of the assembly were evaluated with the assembly tool of the Pomoxis toolkit (https://github.com/nanoporetech/pomoxis). The gene detection rate was measured with BUSCO v5.2.2 (Seppey et al., 2019) against the Embryophyta (odb10) dataset.
Pseudochromosome construction with Hi-C data
Five Hi-C libraries were constructed as previously described (Belton et al., 2012) and sequenced on the Illumina HiSeq platform (Illumina, San Diego, CA, USA), generating 721.3 Gb of paired-end reads (2 × 150 bp) with ∼250× coverage for pseudochromosome construction. The Hi-C data were mapped to the polished assembly using Bowtie 2 v2.3.2 (Langmead and Salzberg, 2012), and only uniquely mapped paired-end reads were retained. Assembled contigs were anchored and ordered using LACHESIS (Burton et al., 2013). The chromatin interaction signals were visually inspected using heatmaps at a 100-kb resolution, and any mis-assembled contigs were split and re-anchored with the Hi-C data.
RNA sequencing for genome annotation
Mixed samples from six healthy tissues (root, young leaf, mature leaf, flower bud, mature flower, and fruit) were collected for RNA extraction using the TRIzol Universal Reagent (Tiangen Biotech, Beijing, China). RNA sequencing (RNA-seq) libraries were prepared using the TruSeq Sample Preparation Kit (Illumina) and sequenced on the HiSeq X Ten platform. Clean Illumina data were obtained by removing adapters and low-quality reads from the raw data. For Isoform-sequencing, cDNA products were amplified using KAPA HiFi PCR kits (Kapa Biosystems, Wilmington, MA, USA), followed by purification using the SMRTbell Template Prep Kit (Pacific Biosciences, Menlo Park, CA, USA). Libraries were sequenced on the PacBio Sequel platform.
Genome annotation
The identification of repetitive sequences, including tandem repeats and TEs, was performed before the C. mannii genome annotation. Tandem repeats were identified using Tandem Repeats Finder v4.07b (Benson, 1999), and TEs were identified by a combination of homology-based and de novo approaches. RepeatMasker v4.0.6, RepeatModeler v1.0.4, and RepeatProteinMask (in RepeatMasker) were used to build the repeat library (Tarailo-Graovac and Chen, 2009). LTR insertion time was calculated as described by Hu et al. (2011).
C. mannii gene models were constructed by integrating de novo prediction and RNA-seq- and homology-based evidence. In brief, RNA-seq data were mapped to the genome for expression evidence using TopHat (Trapnell et al., 2009). For homology-based annotation, protein sequences from five sequenced plants, Asparagus officinalis (Harkess et al., 2017), Arabidopsis thaliana (Cheng et al., 2017), Oryza sativa (Ouyang et al., 2007), P. aphrodite (Chao et al., 2018), and Zea mays (Schnable et al., 2009), were downloaded and mapped onto the C. mannii genome using tBlastn (Camacho et al., 2009), followed by inferring the exon–intron boundaries using Exonerate v2.2.0 (Slater and Birney, 2005). Gene structures were also predicted using Augustus v3.2.2 (Stanke et al., 2006). All annotated information was integrated using EVidenceModeler v1.1.1 (Haas et al., 2008) to generate the final gene models. Functional annotation was performed using InterProScan (Jones et al., 2014) by searching against the Swiss-Prot, Eukaryotic Orthologous Groups, KEGG, GO, and non-redundant protein databases. TFs and protein kinases were identified using the iTAK v1.7 pipeline (Zheng et al., 2016). Gene-encoded metabolic enzymes were identified using E2P2 v4.0 as previously described (Schlapfer et al., 2017). CREs were predicted using PlantCARE (Lescot et al., 2002) and plotted using TBtools (Chen et al., 2020).
Synteny analysis
Synteny analysis was performed between the C. mannii genome and two other orchid genomes, A. shenzhenica (Zhang et al., 2017) and Vanilla planifolia (Hasing et al., 2020), using MCScan in the JCVI package (https://github.com/tanghaibao/jcvi) (Tang et al., 2008) with default parameters.
Phylogenetic analyses and evolutionary time estimation
Phylogenetic relationships between C. mannii and 15 other species were analyzed using orthologs identified by OrthoMCL v2.0.9 (https://github.com/apetkau/orthomcl-pipeline) with percentMatchCutoff = 50 and evalueExponentCutoff = −5. Multiple Sequence Comparison by Log-Expectation (Edgar, 2004) was used to align the sequences, and RAxML v8.2.10 (Stamatakis, 2014) was used to construct the phylogeny. The PAML package (Yang, 2007) was used to estimate the species divergence times with the MCMCTree model, and the calibrations for Amborella trichopoda–Liriodendron chinense, A. thaliana–P. aphrodite, A. thaliana–Vitis vinifera, and Spirodela polyrhiza–Zostera marina were obtained from TimeTree (Kumar et al., 2017).
Estimation of WGD events
Ortholog divergence (Ks) was calculated between gene pairs using the Nei–Gojobori method implemented in the JCVI package as previously described (Hasing et al., 2020).
Physiological data acquisition
Plant materials
Healthy, mature C. mannii plants growing in the greenhouse of Kunming Institute of Botany were used for imaging and physiological data acquisition. Data were collected at two night time points (2:00 and 20:00) and two day time points (8:00 and 14:00).
Net CO2 uptake measurement
An LI-6400 portable gas exchange system (Li-Cor Biosciences, Lincoln, NE, USA) was used to measure the net CO2 uptake. Measurements were taken at the middle of the leaf, avoiding the leaf vein. At each time point, four repeated measurements were obtained. All measurements were taken under standard conditions with photosynthetic photon flux density set at 100 μmol m−2 s−1 during the day.
Measurement of organic acids
For measurement of each organic acid, 0.15 g of fresh sample was ground and homogenized with 1.5 ml of water. For preparation of oxaloacetic acid, the homogenized mixture was ultrasonically crushed for 20 min at 60°C and centrifuged at 12 000 g for 10 min. For preparation of pyruvic acid, the homogenized mixture was ultrasonically crushed for 40 min at 50°C and centrifuged at 12 000 g for 15 min. For preparation of malic acid, the homogenized mixture was ultrasonically crushed for 30 min at 40°C and centrifuged at 12 000 g for 15 min. The supernatant of each organic acid preparation was extracted using a Millipore filter (0.22 μm; Merck Millipore, Burlington, MA, USA) for testing. The abundance of the organic acids was estimated by high-performance liquid chromatography (LC-20AT, Shimadzu, Japan). Standard curves were drawn, and a regression equation and linear coefficients were calculated to estimate the content of each organic acid.
Measurement of NAD-ME and PPC activities
NAD-ME and PPC activities were determined according to the kit instructions (Beijing Solarbio Science & Technology, Beijing, China). For extraction of enzyme solution, 1 ml of extraction buffer was added to 0.1 g of tissue for homogenization in an ice bath. For NAD-ME activity measurement, the supernatant was centrifuged at 12 000 rpm at 4°C for 10 min. The total volume of the reaction solution was 170 μl, the sample solution was 10 μl, and the working solution was 160 μl. The NAD-ME activity was calculated by measuring the increased rate of NADH at 340 nm. For the PPC activity measurement, the supernatant was centrifuged at 8000 rpm at 4°C for 10 min. According to the kit instructions, 90 μl working solution was prepared and mixed with 90 μl reagent solution and 20 μl sample solution. PPC activity was calculated on the basis of absorption at 340 nm.
Multi-omics data analysis
Experimental setup
C. mannii plants were maintained in a growth chamber with 12:12 h of light:dark with temperatures of 25°C (light) (Jarvis et al., 2017) and 18°C (dark). Light was provided beginning at 8:00 (ZT0), and dark began at 20:00 (ZT12). Leaf tip zones (∼10 cm) were collected at a 4-h interval after the beginning of the light period over the 24-h time course (ZT0, ZT4, ZT8, ZT12, ZT16, ZT20, and ZT24) for the multi-omics study (Supplemental Figure 6). Three, four, and six biological replicates were prepared for transcriptomic, proteomic, and metabolomic profiling, respectively. All samples were frozen in liquid nitrogen immediately after harvest and stored at −80°C until further use.
RNA-seq and analysis
Total RNA was extracted from leaf tissues as described above. Libraries were sequenced on the Illumina NovaSeq 6000 platform (Illumina) in paired-end mode (2 × 150 bp). To remove adapters and low-quality bases, raw reads were trimmed using Trimmomatic v0.39 (Bolger et al., 2014). The trimmed reads were aligned to the C. mannii genome to quantify gene expression levels using Kallisto v0.44.0 (Bray et al., 2016) with default parameters, and the abundance was defined as TPM. The Mfuzz R package (Kumar and Futschik, 2007) was used to perform gene clustering according to the time courses using normalized read counts.
Proteomics profiling and analysis
Protein extraction and digestion
Approximately 2 g of ground tissue was suspended in SDT lysis buffer (4% SDS, 100 mM Tris–HCl, pH 7.6) and transferred to lysing matrix tubes. Cells were disrupted using the FastPrep-24 system (MP Biomedicals, Shanghai, China) (speed 6.0 m/s, 60 s, twice), followed by boiling for 10 min. The protein was extracted by centrifugation at 14 000 g for 15 min and quantified by bicinchoninic acid assay. For all samples, a filter-aided sample preparation method was used for converting proteins to peptides suitable for proteome analysis (Wisniewski et al., 2009). Dithiothreitol was added to 200 μg of the extracted crude protein to reach a final concentration of 100 mM. The mixture was placed in a boiling water bath for 5 min and cooled to room temperature, followed by addition of 200 μl UA buffer (8 M urea, 150 mM Tris–HCl, pH 8.5), and completely mixed before centrifugation at 12 500 g for 25 min. The pelleted proteins were resuspended in 100 μl of 0.05 M iodoacetamide buffer (100 mM IAA in UA), incubated at room temperature for 30 min, and centrifuged again at 12 500 g for 25 min. The denatured protein was diluted with 100 μl of UA buffer twice (centrifuged at 12 500 g for 15 min) and washed with 100 μl of 100 mM triethylammonium bicarbonate (TEAB) buffer twice (centrifuged at 12 500 g for 15 min). The concentrated proteins were digested with 40 μl of trypsin buffer (4 μg trypsin in 40 μl of 100 mM TEAB buffer) at 37°C for 16–18 h. The concentrated proteins were then centrifuged at 12 500 g for 15 min, followed by addition of 20 μl 100 mM TEAB buffer, and centrifuged at 12 500 g for 15 min. After digestion, the combined filtrates were desalted through a C18 cartridge and solubilized in 40 μl of 0.1% formic acid. The resulting peptides were quantified by bicinchoninic acid assay.
High-performance LC–MS and peptide identification
The Agilent 1260 Infinity II high-performance liquid chromatography system was used for peptide separation. The peptide mixture was automatically loaded (1 ml/min) into the reversed phase Xbridge Peptide BEH C18 column (Waters, Wexford, Ireland). Peptide sequencing analysis was performed using the Easy-nLC system (Thermo Fisher Scientific) and Q Exactive HF-X hybrid quadrupole-orbitrap mass spectrometer (Thermo Fisher Scientific). The data-independent acquisition method (Chaw et al., 2019) was used to analyze the spectra. The settings were as follows: (1) mass spectrometry, scan range (m/z) = 350–1500, resolution = 60 000, automatic gain control (AGC) target = 3e6, maximum injection time = 50 ms; (2) Data-independent acquisition (DIA), resolution = 15 000, AGC target = 2e5, maximum injection time = 45 ms, normalized collision energy = 28%.
Raw DIA data were analyzed with Spectronaut Pulsar X (v12, Biognosys AG, Zurich, Switzerland) with default settings. A local regression normalization (default) strategy was applied (Callister et al., 2006). The spectral data were searched against Orchidaceae species from the UniProt database (http://www.uniprot.org) and a customized C. mannii database. The FDR threshold was 1.0% for precursors, proteins, and peptides.
Metabolite profiling and data analysis
Metabolite extraction
Approximately 100 mg of fast-frozen leaf tissue was ground into fine powder with liquid nitrogen, and the metabolites were extracted with 120 μl of 50% pre-cooled methanol by incubating the well-mixed samples at room temperature for 10 min. The extracted mixture was then stored at −20°C overnight. To isolate the metabolites, the mixture was centrifuged at 4000 g for 20 min, and the supernatants were transferred to new 96-well plates. The samples were stored at −80°C before the LC–MS analysis. Chromatographic separation was performed using an ultra-performance liquid chromatography (UPLC) system (SCIEX, UK). An Acquity UPLC T3 column (100 mm × 2.1 mm × 1.8 μm; Waters) was used for the reversed phase separation by maintaining the temperature at 35°C. The flow rate was 0.4 ml/min, and the mobile phase consisted of solvent A (0.1% formic acid in deionized water) and solvent B (0.1% formic acid in acetonitrile).
LC–MS
A high-resolution tandem mass spectrometer TripleTOF 5600 Plus (SCIEX) was used to detect metabolites eluted from the column. The quadrupole time-of-flight analysis was performed in both positive (Electrospray ionization, ESI+) and negative (ESI−) ion modes using the following parameters: ion spray voltage of 5.0 kV in ESI+ and −4.5 kV in ESI−, curtain gas of 30 psi, ion source gas 1 of 60 psi, ion source gas 2 of 60 psi, and an interface heater temperature of 650°C. The mass spectrometry data were acquired in the data-dependent acquisition mode, and the mass range was set from 60.0 to 1200.0 Da. Mass accuracy was calibrated every 20 samples during acquisition. To evaluate LC–MS stability during the whole acquisition, a QC sample was acquired after every 10 samples.
Data processing
The acquired LC–MS raw data files were converted to the mzXML format using MSConvert in ProteoWizard (v3.0.6150) (Chambers et al., 2012) and processed by the XCMS (Smith et al., 2006), CAMERA (Kuhl et al., 2012), and metaX (Wen et al., 2017) packages in R. Each ion was identified by combining retention time and m/z data.
The intensity of peak data were further filtered by removing data detected in less than 50% of QC samples or 80% of biological samples, and the remaining missing values were imputed with the k-nearest neighbor algorithm to further improve the data quality. All retained peaks were normalized with the probabilistic quotient normalization method, followed by QC-robust spline batch corrections. The relative standard deviations of the metabolic features in QC samples were set at a threshold of 30%, and values higher than the threshold were removed.
Student’s t-test was performed to detect significantly changed metabolites among samples, and the p value was adjusted for multiple tests by FDR. Multivariate statistical analysis (principal-component analysis, Partial least squares discriminant analysis) was performed to discriminate the different variables between groups. The variable importance in projection (VIP) values were calculated, and a VIP cutoff value of 1.0 was used to select important features.
Metabolite database search and identification
The online KEGG database (Kanehisa and Goto, 2000) and Human Metabolome Database (Wishart et al., 2018) were used to annotate the metabolites by matching the exact molecular mass data (m/z) of samples with those from the database. If the mass difference between the observed and database value was less than 10 ppm, the metabolites were annotated and the molecular formulae of metabolites were further identified and validated by isotopic distribution measurements. An in-house fragment spectrum library of metabolites was used to validate metabolite identification.
KEGG enrichment analysis
The annotated metabolites were mapped to KEGG pathways, and enrichment analysis was performed for metabolites in each cluster. Pathways with p < 0.05 were considered significantly enriched.
Statistical analyses
Diurnal analysis
The JTK_CYCLE algorithm (Hughes et al., 2010) was used to detect the rhythmic transcripts, proteins, and metabolites, providing the optimal phase, amplitude, and period estimates for each dataset. The cutoff score of FDR-adjusted p < 0.05 was used to determine the cycling data.
Funding
This study was funded by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB31000000) to D.-Z.L. and J.-B.Y., CAS Pioneer Hundred Talents Program to A.Z., the Project for Innovation Team of Yunnan Province (grant no. 202105AE160012) to S.-B.Z., and the Science and Technology Basic Resources Investigation Program of China (grant no. 2021FY100200) to J.-B.Y.
Author contributions
D.-Z.L., A.Z., J.-B.Y., and S.-B.Z. conceived the project and designed the research. J.-B.Y., S.-B.Z., J.-D.Y., and J.-L.H. collected the plant materials. A.Z., Z.-S.H., M.Z., J.F., L.Z., Y.H., and F.L. designed and carried out the sample collection across the diel cycle and collected the physiological data. W.F., Z.-S.H., and A.Z. designed and coordinated computational analyses. W.F., A.Z., and Z.-S.H. wrote the manuscript with input from all co-authors.
Acknowledgments
The authors thank the Molecular Biology Experimental Center and the Service Center for Experimental Biotechnology at the Kunming Institute of Botany, CAS, for plant care and experimental support. We thank the iFlora HPC Center (iFlora High Performance Computing Center) of the Germplasm Bank of Wild Species for support with data analysis. No conflict of interest is declared.
Published: February 21, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information can be found online at Plant Communications Online.
Contributor Information
Jun-Bo Yang, Email: jbyang@mail.kib.ac.cn.
Andan Zhu, Email: zhuandan@mail.kib.ac.cn.
De-Zhu Li, Email: dzl@mail.kib.ac.cn.
Accession numbers
All raw data generated in the current study are available at the National Genomic Data Center of the China National Center for Bioinformation under project number PRJCA006911. The Agave multi-omics data were obtained from Dr. Xiaohan Yang’s laboratory and were published previously (Abraham et al., 2016). Core CAM gene expression data for selected terrestrial CAM plants were obtained from previous publications (Wai et al., 2019; Wickell et al., 2021).
Supplemental information
References
- Abraham P.E., Hurtado Castano N., Cowan-Turner D., Barnes J., Poudel S., Hettich R., Flutsch S., Santelia D., Borland A.M. Peeling back the layers of crassulacean acid metabolism: functional differentiation between Kalanchoe fedtschenkoi epidermis and mesophyll proteomes. Plant J. 2020;103:869–888. doi: 10.1111/tpj.14757. [DOI] [PubMed] [Google Scholar]
- Abraham P.E., Yin H., Borland A.M., Weighill D., Lim S.D., De Paoli H.C., Engle N., Jones P.C., Agh R., Weston D.J., et al. Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat Plants. 2016;2:16178. doi: 10.1038/nplants.2016.178. [DOI] [PubMed] [Google Scholar]
- Ai Y., Li Z., Sun W.H., Chen J., Zhang D., Ma L., Zhang Q.H., Chen M.K., Zheng Q.D., Liu J.F., et al. The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 2021;8:255. doi: 10.1038/s41438-021-00683-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrizon J., Morel S., Gschaedler A., Monsan P. Comparison of the water-soluble carbohydrate composition and fructan structures of Agave tequilana plants of different ages. Food Chem. X. 2010;122:123–130. doi: 10.1016/j.foodchem.2010.02.028. [DOI] [Google Scholar]
- Belton J.M., McCord R.P., Gibcus J.H., Naumova N., Zhan Y., Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzing D.H., Givnish T.J., Bermudes D. Absorptive trichomes in Brocchinia reducta (Bromeliaceae) and their evolutionary and systematic significance. Syst. Bot. 1985;10:81–91. [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone R.E., Smith J.A.C., Arrigo N., Buerki S. A macro-ecological perspective on crassulacean acid metabolism (CAM) photosynthesis evolution in Afro-Madagascan drylands: eulophiinae orchids as a case study. New Phytol. 2015;208:469–481. doi: 10.1111/nph.13572. [DOI] [PubMed] [Google Scholar]
- Boxall S.F., Kadu N., Dever L.V., Kneřová J., Waller J.L., Gould P.J.D., Hartwell J. Kalanchoe PPC1 is essential for crassulacean acid metabolism and the regulation of core circadian clock and guard cell signaling genes. Plant Cell. 2020;32:1136–1160. doi: 10.1105/tpc.19.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O., Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Liu X., Vanneste K., Proost S., Tsai W.C., Liu K.W., Chen L.J., He Y., Xu Q., Bian C., et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015;47:65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- Callister S.J., Barry R.C., Adkins J.N., Johnson E.T., Qian W.J., Webb-Robertson B.J.M., Smith R.D., Lipton M.S. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res. 2006;5:277–286. doi: 10.1021/pr050300l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers M.C., Maclean B., Burke R., Amodei D., Ruderman D.L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y.T., Chen W.C., Chen C.Y., Ho H.Y., Yeh C.H., Kuo Y.T., Su C.L., Yen S.H., Hsueh H.Y., Yeh J.H., et al. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 2018;16:2027–2041. doi: 10.1111/pbi.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase M.W., Cameron K.M., Freudenstein J.V., Pridgeon A.M., Salazar G., van den Berg C., Schuiteman A. An updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015;177:151–174. doi: 10.1111/boj.12234. [DOI] [Google Scholar]
- Chaw S.M., Liu Y.C., Wu Y.W., Wang H.Y., Lin C.Y.I., Wu C.S., Ke H.M., Chang L.Y., Hsu C.Y., Yang H.T., et al. Stout camphor tree genome fills gaps in understanding of flowering plant genome evolution. Nat. Plants. 2019;5:63–73. doi: 10.1038/s41477-018-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Cheng C.Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- Crayn D.M., Winter K., Schulte K., Smith J.A.C. Photosynthetic pathways in Bromeliaceae: phylogenetic and ecological significance of CAM and C3 based on carbon isotope ratios for 1893 species. Bot. J. Linn. Soc. 2015;178:169–221. doi: 10.1111/boj.12275. [DOI] [Google Scholar]
- Deng H., Zhang L.S., Zhang G.Q., Zheng B.Q., Liu Z.J., Wang Y. Evolutionary history of PEPC genes in green plants: implications for the evolution of CAM in orchids. Mol. Phylogenet. Evol. 2016;94:559–564. doi: 10.1016/j.ympev.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Donoghue M.J., Edwards E.J. Biome shifts and niche evolution in plants. Annu. Rev. Ecol. Evol. Syst. 2014;45:547–572. [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamisch A., Winter K., Fischer G.A., Comes H.P. Evolution of crassulacean acid metabolism (CAM) as an escape from ecological niche conservatism in Malagasy Bulbophyllum (Orchidaceae) New Phytol. 2021;231:1236–1248. doi: 10.1111/nph.17437. [DOI] [PubMed] [Google Scholar]
- Gorinsek B., Gubensek F., Kordis D. Evolutionary genomics of chromoviruses in eukaryotes. Mol. Biol. Evol. 2004;21:781–798. doi: 10.1093/molbev/msh057. [DOI] [PubMed] [Google Scholar]
- Govaerts R., Bernet P., Kratochvil K., Gerlach G., Carr G., Alrich P., Pridgeon A.M., Pfahl J., Campacci M.A., Baptista D.H., et al. Facilitated by the Royal Botanic Gardens; Kew: 2021. World Checklist of Orchidaceae. [Google Scholar]
- Guan Q., Kong W., Zhu D., Zhu W., Dufresne C., Tian J., Chen S. Comparative proteomics of Mesembryanthemum crystallinum guard cells and mesophyll cells in transition from C(3) to CAM. J. Proteomics. 2021;231:104019. doi: 10.1016/j.jprot.2020.104019. [DOI] [PubMed] [Google Scholar]
- Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., White O., Buell C.R., Wortman J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A., Zhou J., Xu C., Bowers J.E., Van der Hulst R., Ayyampalayam S., Mercati F., Riccardi P., McKain M.R., Kakrana A., et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Commun. 2017;8:1279. doi: 10.1038/s41467-017-01064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasing T., Tang H., Brym M., Khazi F., Huang T., Chambers A.H. A phased Vanilla planifolia genome enables genetic improvement of flavour and production. Nat. Food. 2020;1:811–819. doi: 10.1038/s43016-020-00197-2. [DOI] [PubMed] [Google Scholar]
- Heyduk K., Moreno-Villena J.J., Gilman I.S., Christin P.A., Edwards E.J. The genetics of convergent evolution: insights from plant photosynthesis. Nat. Rev. Genet. 2019;20:485–493. doi: 10.1038/s41576-019-0107-5. [DOI] [PubMed] [Google Scholar]
- Heyduk K., Hwang M., Albert V., Silvera K., Lan T., Farr K., Chang T.H., Chan M.T., Winter K., Leebens-Mack J. Altered gene regulatory Networks are associated with the transition from C3 to crassulacean acid metabolism in Erycina (oncidiinae: Orchidaceae) Front. Plant Sci. 2018;9:2000. doi: 10.3389/fpls.2018.02000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Boisson-Dernier A., Israelsson-Nordström M., Böhmer M., Xue S., Ries A., Godoski J., Kuhn J.M., Schroeder J.I. Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat. Cell Biol. 2010;12:87–93. doi: 10.1038/ncb2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Fan J., Sun Z., Liu S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2020;36:2253–2255. doi: 10.1093/bioinformatics/btz891. [DOI] [PubMed] [Google Scholar]
- Hu T.T., Pattyn P., Bakker E.G., Cao J., Cheng J.F., Clark R.M., Fahlgren N., Fawcett J.A., Grimwood J., Gundlach H., et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M.E., Hogenesch J.B., Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis D.E., Ho Y.S., Lightfoot D.J., Schmöckel S.M., Li B., Borm T.J.A., Ohyanagi H., Mineta K., Michell C.T., Saber N., et al. The genome of Chenopodium quinoa. Nature. 2017;542:307–312. doi: 10.1038/nature21370. [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl C., Tautenhahn R., Böttcher C., Larson T.R., Neumann S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012;84:283–289. doi: 10.1021/ac202450g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L., Futschik M.E. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Lai Y.J., Han Y., Schuiteman A., Chase M.W., Xu S.Z., Li J.W., Wu J.Y., Yang B.Y., Jin X.H. Diversification in Qinghai-Tibet Plateau: orchidinae (Orchidaceae) clades exhibiting pre-adaptations play critical role. Mol. Phylogenet. Evol. 2021;157:107062. doi: 10.1016/j.ympev.2020.107062. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. Preprint at. [DOI] [Google Scholar]
- Li Z., Zhu A., Song Q., Chen H.Y., Harmon F.G., Chen Z.J. Temporal regulation of the metabolome and proteome in photosynthetic and photorespiratory pathways contributes to maize heterosis. Plant Cell. 2020;32:3706–3722. doi: 10.1105/tpc.20.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Chen S., Ru Z. Science Press; Beijing: 2006. The Genus Cymbidium in China. [Google Scholar]
- Lüttge U. Ecophysiology of crassulacean acid metabolism (CAM) Ann. Bot. 2004;93:629–652. doi: 10.1093/aob/mch087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C.E., Mas E.J., Lu C., Ong B.L. The photosynthetic pathway of the roots of twelve epiphytic orchids with CAM leaves. Photosynthetica. 2010;48:42–50. doi: 10.1007/s11099-010-0007-6. [DOI] [Google Scholar]
- Ming R., VanBuren R., Wai C.M., Tang H., Schatz M.C., Bowers J.E., Lyons E., Wang M.L., Chen J., Biggers E., et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015;47:1435–1442. doi: 10.1038/ng.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura H., Yukawa T., Ueno O., Kagawa A. The occurrence of crassulacean acid metabolism in Cymbidium (Orchidaceae) and its ecological and evolutionary implications. J. Plant Res. 2008;121:163–177. doi: 10.1007/s10265-007-0144-6. [DOI] [PubMed] [Google Scholar]
- Neumann P., Novák P., Hoštáková N., Macas J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA. 2019;10:1. doi: 10.1186/s13100-018-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel P.S. Achievable productivities of certain CAM plants: basis for high values compared with C3 and C4 plants. New Phytol. 1991;119:183–205. doi: 10.1111/j.1469-8137.1991.tb01022.x. [DOI] [PubMed] [Google Scholar]
- Ogura-Tsujita Y., Yokoyama J., Miyoshi K., Yukawa T. Shifts in mycorrhizal fungi during the evolution of autotrophy to mycoheterotrophy in Cymbidium (Orchidaceae) Am. J. Bot. 2012;99:1158–1176. doi: 10.3732/ajb.1100464. [DOI] [PubMed] [Google Scholar]
- Osmond C.B. Crassulacean acid metabolism: a curiosity in context. Annu. Rev. Plant Physiol. 1978;29:379–414. doi: 10.1146/annurev.pp.29.060178.002115. [DOI] [Google Scholar]
- Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M., Childs K., Thibaud-Nissen F., Malek R.L., Lee Y., Zheng L., et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridgeon A.M. In: Genera Orchidacearum. Cribb P.J., Chase M.W., Rasmussen F.N., editors. Oxford University Press; Oxford: 2009. [Google Scholar]
- Puy D.D., Cribb P. Royal Botanic Garden; 2007. The Genus Cymbidium. [Google Scholar]
- Ramirez S.R., Gravendeel B., Singer R.B., Marshall C.R., Pierce N.E. Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature. 2007;448:1042–1045. doi: 10.1038/nature06039. [DOI] [PubMed] [Google Scholar]
- Schlapfer P., Zhang P., Wang C., Kim T., Banf M., Chae L., Dreher K., Chavali A.K., Nilo-Poyanco R., Bernard T., et al. Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 2017;173:2041–2059. doi: 10.1104/pp.16.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., Ware D., Fulton R.S., Stein J.C., Wei F., Pasternak S., Liang C., Zhang J., Fulton L., Graves T.A., et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Seppey M., Manni M., Zdobnov E.M. BUSCO: Assessing genome assembly and annotation completeness. Methods Mol. Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- Silvera K., Santiago L.S., Cushman J.C., Winter K. Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the Orchidaceae. Plant Physiol. 2009;149:1838–1847. doi: 10.1104/pp.108.132555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater G.S.C., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinf. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.A., Want E.J., O'Maille G., Abagyan R., Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens P.F. Angiosperm Phylogeny Website. 14 edition. 2017. http://www.mobot.org/MOBOT/research/APweb/ [Google Scholar]
- Sun Y., Chen G.-Z., Huang J., Liu D.-K., Xue F., Chen X.-L., Chen S.-Q., Liu C.-G., Liu H., Ma H., et al. The Cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornam. Plant Res. 2021;1:1–13. doi: 10.48130/opr-2021-0010. [DOI] [Google Scholar]
- Tang H., Bowers J.E., Wang X., Ming R., Alam M., Paterson A.H. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M., Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2009;Chapter 4:4.10.1–4.10.14. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai C.M., Weise S.E., Ozersky P., Mockler T.C., Michael T.P., VanBuren R. Time of day and network reprogramming during drought induced CAM photosynthesis in Sedum album. PLoS Genet. 2019;15:e1008209. doi: 10.1371/journal.pgen.1008209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Kirby E., Furlong K.P., van Soest M., Xu G., Shi X., Kamp P.J.J., Hodges K.V. Two-phase growth of high topography in eastern Tibet during the Cenozoic. Nat. Geosci. 2012;5:640–645. doi: 10.1038/ngeo1538. [DOI] [Google Scholar]
- Wen B., Mei Z., Zeng C., Liu S. metaX: a flexible and comprehensive software for processing metabolomics data. BMC Bioinf. 2017;18:183. doi: 10.1186/s12859-017-1579-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickell D., Kuo L.Y., Yang H.P., Dhabalia Ashok A., Irisarri I., Dadras A., de Vries S., de Vries J., Huang Y.M., Li Z., et al. Underwater CAM photosynthesis elucidated by Isoetes genome. Nat. Commun. 2021;12:6348. doi: 10.1038/s41467-021-26644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K., Smith J. In: Crassulacean Acid Metabolism Biochemistry Ecophysiology & Evolution. Winter K., Smith J.A.C., editors. Springer-Verlag; Berlin: 1996. An introduction to crassulacean acid metabolism. Biochemical Principles and Ecological Diversity; pp. 1–13. [Google Scholar]
- Winter K., Smith J.A.C. CAM photosynthesis: the acid test. New Phytol. 2022;233:599–609. doi: 10.1111/nph.17790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vázquez-Fresno R., Sajed T., Johnson D., Li C., Karu N., et al. Hmdb 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Yang F.X., Gao J., Wei Y.L., Ren R., Zhang G.Q., Lu C.Q., Jin J.P., Ai Y., Wang Y.Q., Chen L.J., et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021;19:2501–2516. doi: 10.1111/pbi.13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.B., Tang M., Li H.T., Zhang Z.R., Li D.Z. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013;13:84. doi: 10.1186/1471-2148-13-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Hu R., Yin H., Jenkins J., Shu S., Tang H., Liu D., Weighill D.A., Cheol Yim W., Ha J., et al. The Kalanchoe genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat. Commun. 2017;8:1899. doi: 10.1038/s41467-017-01491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Paml 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang G.Q., Liu K.W., Li Z., Lohaus R., Hsiao Y.Y., Niu S.C., Wang J.Y., Lin Y.C., Xu Q., Chen L.J., et al. The Apostasia genome and the evolution of orchids. Nature. 2017;549:379–383. doi: 10.1038/nature23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen F., Zhang G.Q., Zhang Y.Q., Niu S., Xiong J.S., Lin Z., Cheng Z.M.M., Liu Z.J. Origin and mechanism of crassulacean acid metabolism in orchids as implied by comparative transcriptomics and genomics of the carbon fixation pathway. Plant J. 2016;86:175–185. doi: 10.1111/tpj.13159. [DOI] [PubMed] [Google Scholar]
- Zhang L., Chen F., Zhang X., Li Z., Zhao Y., Lohaus R., Chang X., Dong W., Ho S.Y.W., Liu X., et al. The water lily genome and the early evolution of flowering plants. Nature. 2020;577:79–84. doi: 10.1038/s41586-019-1852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.B., Dai Y., Hao G.Y., Li J.W., Fu X.W., Zhang J.L. Differentiation of water-related traits in terrestrial and epiphytic Cymbidium species. Front. Plant Sci. 2015;6:260. doi: 10.3389/fpls.2015.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Jiao C., Sun H., Rosli H.G., Pombo M.A., Zhang P., Banf M., Dai X., Martin G.B., Giovannoni J.J., et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant. 2016;9:1667–1670. doi: 10.1016/j.molp.2016.09.014. [DOI] [PubMed] [Google Scholar]
- Zhou J., Liu C., Chen Q., Liu L., Niu S., Chen R., Li K., Sun Y., Shi Y., Yang C., et al. Integration of rhythmic metabolome and transcriptome provides insights into the transmission of rhythmic fluctuations and temporal diversity of metabolism in rice. Sci. China Life Sci. 2022;65:1794–1810. doi: 10.1007/s11427-021-2064-7. [DOI] [PubMed] [Google Scholar]
- Zotz G. How prevalent is crassulacean acid metabolism among vascular epiphytes? Oecologia. 2004;138:184–192. doi: 10.1007/s00442-003-1418-x. [DOI] [PubMed] [Google Scholar]
- Zotz G. The systematic distribution of vascular epiphytes – a critical update. Bot. J. Linn. Soc. 2013;171:453–481. [Google Scholar]
- Zotz G. Springer; 2016. Plants on Plants – the Biology of Vascular Epiphytes. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.