Abstract
Root parasitic plants of the Orobanchaceae, broomrapes and witchweeds, pose a severe problem to agriculture in Europe, Asia and especially Africa. These parasites are totally dependent on their host for survival, and therefore, their germination is tightly regulated by host presence. Indeed, their seeds remain dormant in the soil until a host root is detected through compounds called germination stimulants. Strigolactones (SLs) are the most important class of germination stimulants. They play an important role in planta as a phytohormone and, upon exudation from the root, function in the recruitment of symbiotic arbuscular mycorrhizal fungi. Plants exude mixtures of various different SLs, possibly to evade detection by these parasites and still recruit symbionts. Vice versa, parasitic plants must only respond to the SL composition that is exuded by their host, or else risk germination in the presence of non-hosts. Therefore, parasitic plants have evolved an entire clade of SL receptors, called HTL/KAI2s, to perceive the SL cues. It has been demonstrated that these receptors each have a distinct sensitivity and specificity to the different known SLs, which possibly allows them to recognize the SL-blend characteristic of their host. In this review, we will discuss the molecular basis of SL sensitivity and specificity in these parasitic plants through HTL/KAI2s and review the evidence that these receptors contribute to host specificity of parasitic plants.
Keywords: Broomrapes, Host specificity, HTL/KAI2, Receptor-ligand specificity, Strigolactones, Witchweeds
Root Parasitic Plants Are Detrimental to Agriculture
Root parasitic plants of the family Orobanchaceae (from here on referred to as parasitic plants) are a scourge that profoundly affects agriculture across Southern Europe, Africa and Asia (Parker 2012, Spallek et al. 2013). The family contains a large number of obligate parasites that require a host to grow on and reproduce. These include broomrapes of the genera Orobanche and Phelipanche and witchweeds of the genus Striga (Parker 2012). In particular, witchweeds pose a critical threat to food security in Africa (Parker 2009). Up to 50 million ha of agricultural land of the continent is affected by Striga, threatening 300 million farmers and causing damage ranging upward of 7 billion USD annually (Ejeta and Gressel 2007). Although parasitic plants can infect many plant species, staple crops such as cereals and legumes are particularly susceptible (Parker 2012, Spallek et al. 2013). For example, annual damage occurred to rice alone reaches 200 million USD, which is predicted to increase by an additional 30 million each year (Rodenburg et al. 2016). Finally, Striga seed production ranges from 10,000 to >200,000 seeds per individual, which can stay dormant in the soil for up to 20 years (Bebawi et al. 1984, Hearne 2009). These seeds are minuscule, weighing approximately 7 µg, and are easily transmitted by wind, water, animals, humans and agricultural equipment (Berner 1995, Hearne 2009), making it difficult to control the spread of these parasites. Moreover, Striga has been found to infect crops that were previously thought to be non-host species (Ejeta and Gressel 2007, Parker 2012).
Losses caused by parasitic plants are especially threatening to poor farmers that do not have access to the proper means to control infestations (Berner 1995). In the USA, an infestation by Striga asiatica was successfully eradicated by the use of various pesticides, ethylene, methyl bromide and comprehensive quarantining and monitoring (Parker 2012). These solutions are inaccessible or too expensive for African farmers, who have instead resorted to simpler methods such as pulling weeds by hand, intercropping and extra manuring, which are only partially effective (Parker 2012). In some cases, certain crops or infested patches of land are abandoned altogether, which is not a viable option when there are no alternatives (Parker 2009). Therefore, it is of utmost importance to find new, more accessible, methods to combat these parasitic weeds. Here, we review how host specificity in parasitic plants depends on germination in response to host signals, and the molecular mechanism behind this specificity, and how this knowledge may improve our understanding of this intriguing interaction and possibilities to control an important agricultural problem.
Root Parasitic Plant Germination Depends on the Presence of a Host
The obligate parasitic Orobanchaceae are fully dependent on a host for growth and reproduction. Via a structure called the haustorium, the parasite attaches to the roots of the host plant and connects to the host’s vasculature through which it drains water, nutrients and assimilates (Yoshida et al. 2016). Since the seeds of these parasites are tiny, the energy available for germination and haustorium formation is limited and seedlings can only survive for 3–7 d if they do not attach to a host (Berner 1995, Hearne 2009).
Germination of parasitic plant seeds encompasses two steps (Brun et al. 2018): preconditioning and recognition of germination stimulants exuded by the host. Together, this ensures that seeds only germinate under favorable conditions and in the presence of a host. Preconditioning, or dormancy relief, entails exposure to moist and warm conditions (Matusova et al. 2004), during 3 days–3 weeks at an optimal temperature of 18–30°C, depending on the species (Gibot-Leclerc et al. 2004, Matusova et al. 2004, Song et al. 2005, Lechat et al. 2012). After preconditioning, seeds become responsive to host-derived signaling molecules called germination stimulants (Matusova et al. 2004, Brun et al. 2018, Bouwmeester et al. 2021). Strigolactones (SLs) were the first germination stimulants to be discovered and are thought to be the most prominent germination stimulants (Bouwmeester et al. 2021).
SLs: A Role in Host Specificity?
Canonical SLs are composed of a tricyclic lactone called the ABC-rings, which are connected to a butenolide D-ring via an enol ether bridge (Waters et al. 2017, Wang and Bouwmeester 2018, Aliche et al. 2020). The canonical SLs can be divided into strigol- and orobanchol-type SLs, depending on the stereochemistry of the junction between the B- and C-rings. The D-rings of natural SLs are always attached in 2ʹR orientation (Fig. 1). Non-canonical SLs lack the classic ABC-rings of the canonical SLs and instead have a more complex structure (Waters et al. 2017, Wang and Bouwmeester 2018, Aliche et al. 2020). Up until today, approximately 35 different types of SL or SL-like compounds have been isolated from plants (Bouwmeester et al. 2021). The first SL was discovered in 1966 and coined strigol. It was isolated from cotton root exudate and determined to be a potent germination stimulant for Striga lutea (Cook et al. 1966). Subsequently, many other SLs were discovered, and in 1995 the term ‘strigolactones’ was coined (Butler 1995). SLs that are commonly used in experiments include strigol, orobanchol, 5-deoxystrigol (5DS) and 4-deoxyorobanchol (4DO) (Fig. 1). Besides these, the synthetic SL rac-GR24 is also commonly used. However, rac-GR24 is a racemate of two optical isomers: GR245DS which is configured like natural 5DS, with a 2ʹR oriented D-ring, and its mirror image GR24ent−5DS with an unnatural 2ʹS-oriented D-ring (Fig. 1). This mixture could therefore induce responses that natural SLs cannot. Note that other GR24 analogs exist, GR244DO and GR24ent−4DO (Fig. 1), but these are usually discarded during synthesis and therefore not commonly used (Flematti et al. 2016).
Fig. 1.
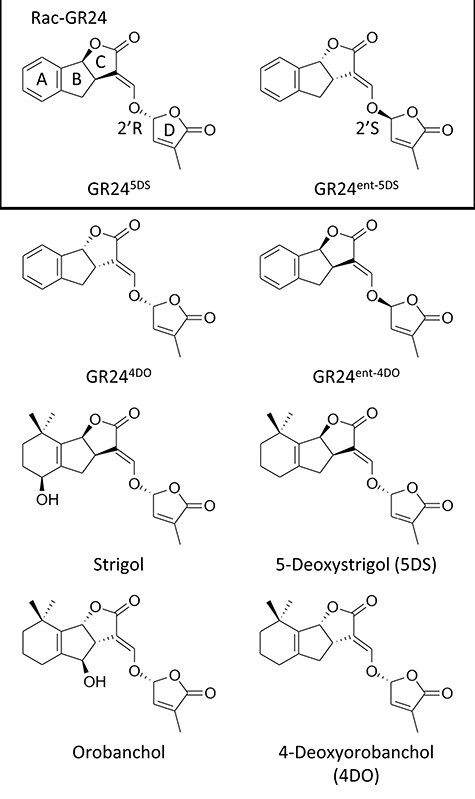
Chemical structures of SLs commonly used in experiments. GR24 is a commonly used synthetic SL that is usually supplied as a racemate of two optical isomers: rac-GR24, which consists of equal amounts of GR245DS and GR24ent−5DS (see the box). Other GR24 analogs exist, GR244DO and GR24ent−4DO, but are usually discarded during production. ABCD-rings are marked on GR245DS, and 2ʹR and 2ʹS configurations of the D-ring are marked on the two stereoisomers in rac-GR24. Strigol, Orobanchol, 5DS and 4DO are naturally occurring SLs that have been used in several studies. These can be divided into strigol- and orobanchol-type SLs, depending on the stereochemical configuration of the junction between the B- and C-rings. Importantly, all natural SLs have 2ʹR-configured D-rings, whereas rac-GR24 consists of both naturally and unnaturally configured GR24. Structures were drawn using the program ChemDraw.
It is important to note that most plant species, so not just hosts of parasitic plants, commonly exude SLs (Wang and Bouwmeester 2018). Moreover, most plant species investigated to date do not exude just one SL, but rather a mixture of SLs (Yoneyama et al. 2009, Wang and Bouwmeester 2018). Interestingly, the composition of these mixtures varies considerably between species and sometimes between varieties or developmental stages within a species. Some species such as petunia and pea only seem to produce orobanchol-type SLs, while others produce both orobanchol- and strigol-type SLs (Wang and Bouwmeester 2018). Furthermore, some species exude around 10 different SLs, while others exude <5. Even though, therefore, most plant species exude SLs, only some of them are a compatible host for a certain parasitic plant species (Nelson 2021). A germination response to the incorrect mixture of SLs could result in suicidal germination, that is, the seeds germinate but cannot attach to the (incompatible) host and therefore cannot survive. Since most parasitic members of the Orobanchaceae are specialized to a relatively narrow host range, it is critical that they only respond to the root exudate that corresponds to that host. Unraveling the mechanism by which they perceive host signals could help to develop targeted control strategies and more efficient induction of suicidal germination.
Indeed, germination of some parasitic plant species displays a strong specificity for the root exudates of their host, while generalist parasitic species seem to respond less specifically (Fernández-Aparicio et al. 2009, 2011b). For example, Orobanche cumana, O. hederae, O. densiflora and O. gracilis germinate almost exclusively with root exudate of their respective hosts. Conversely, Phelipanche aegyptica, P. ramosa and O. minor germinate with root exudate of a broader range of plant species (Fernández-Aparicio et al. 2009, 2011b). These different responses are considered to be at least partially caused by SLs, since with single SLs a similar pattern in the germination response was found. That is, O. minor, P. aegyptiaca and P. nana responded strongly to each of the three SLs that were tested, whereas the other species only responded to one SL or not at all (Fernández-Aparicio et al. 2011b). Similarly, Striga hermonthica and Striga gesnerioides show distinct responses to various stereoisomers of strigol, sorgolactone, orobanchol, sorgomol and 5DS (Nomura et al. 2013). Of the two species, S. hermonthica is most sensitive, germinating in response to all SL variants and stereoisomers, albeit at strongly varying rates. It responds most strongly to those with the same stereochemistry as 5DS. In contrast, S. gesnerioides germinated only in response to a small subset of SLs. Interestingly, SLs that strongly induce S. hermonthica only weakly induce S. gesnerioides germination, and vice versa. Interestingly, some SLs even suppress S. gesnerioides germination (Nomura et al. 2013). Moreover, Orobanche and Phelipanche species mainly germinate in response to orobanchol-type SLs, whereas S. hermonthica mainly responds to strigol-type SLs (Bouwmeester et al. 2021). In some parasitic plant species, other compounds in the root exudate seem to have taken on a role similar to that of SLs. Orobanche cumana seeds, for example, germinate in response to dehydrocostus lactone, which is present in root exudate of sunflower (Joel et al. 2011, Cala et al. 2017), and the glucosinolate derivative 2-phenylethyl isothiocyanate (2-PEITC) in the root exudate of rapeseed induces germination in P. ramosa (Auger et al. 2012, de Saint Germain et al. 2021).
Certain parasitic plant species exhibit ecotype-specific variation in host specificity (Thorogood et al. 2009, Huang et al. 2012, Gibot-Leclerc et al. 2013). Striga hermonthica isolated from sorghum germinated more often in response to sorghum root exudate than a different line collected from millet (Dafaallah 2020). Although there is as yet no evidence that this variation is caused by differences in sensitivity to different SLs, these results do suggest that perception of exuded chemicals may play a role.
Although there is therefore ample evidence that SLs play a role in the host specificity of these parasitic plants, relatively little is known about how this works at the molecular level. Next, we will discuss what is known about the mechanism by which a parasitic plant can perceive the signals of its host amidst a multitude of non-host signals. However, it should first be considered why SLs are exuded in the first place, what their endogenous functions are, and how they are perceived.
Parasitic Plants Have Repurposed HTL/KAI2 to Detect SLs
With the discovery of the hormonal role of SLs and the identification of their receptor, D14 (Box 1), D14 was a logical candidate for the SL receptor in parasitic plants. However, transcriptomic data showed that parasitic plants express at most one D14 homolog (Conn et al. 2015, Das et al. 2015), likely involved in the perception of its endogenous, but not the host-exuded SLs (Conn et al. 2015, Xu et al. 2018, Nelson 2021). Instead, the focus shifted toward a homolog of D14 called HYPOSENSITIVE TO LIGHT (HTL)/KARRIKIN INSENSITIVE2 (KAI2). Both proteins are part of a larger family of α/β-fold hydrolase receptors, and both contain the same Ser-His-Asp catalytic triad (Waters et al. 2012). Much like D14 signaling, HTL/KAI2 signaling depends on the interaction with MAX2 (Nelson et al. 2011, Waters et al. 2012, Bunsick et al. 2020). However, the downstream targets of this pathway are different; Arabidopsis HTL/KAI2 (AtKAI2) interacts with MAX2 to induce the degradation of SMAX1 and SMXL2 (Stanga et al. 2013, 2016, Khosla et al. 2020, Wang et al. 2020). Importantly, a mutation in D14 does not affect seed germination, whereas a mutation of AtKAI2 does. Arabidopsis kai2 mutants show impaired seed germination, and D14 expressed under the KAI2 promoter cannot rescue this phenotype. Thus, in non-parasitic plants, HTL/KAI2 is thought to function in the germination response to karrikin (KAR), a chemical released from burning plant material, which promotes germination of the seeds of fire-following species, but also many other species (Flematti 2004, Sun and Ni 2011, Nelson et al. 2012, Waters et al. 2012, Kochanek et al. 2016). Although KAR seems to bind to HTL/KAI2 and activate signaling, most studies show that binding is relatively weak (Nelson 2021). In addition, crystal structures are inconsistent in determining the orientation of bound KAR, and several in vitro and in vivo experiments show that KAR by itself cannot activate HTL/KAI2 (Nelson 2021). Therefore, it was proposed that HTL/KAI2 binds an unidentified compound called KAI2-ligand (KL), which is probably a metabolized form of KAR that may function as a phytohormone (Waters et al. 2012, Conn and Nelson 2016, Swarbreck et al. 2019, Villaécija-Aguilar et al. 2019). Recently, a study in sunflowers has suggested that sesquiterpene lactones 8-epixanthatin and tomentosin could constitute KL, at least in the Asteraceae (Rahimi and Bouwmeester 2021). Finally, KAR and possibly KL are chemically similar to SL since they (presumably) contain a butenolide D-ring-like structure. This similarity suggests that certain HTL/KAI2s may be able to bind SL, which could result in germination.
Box 1.
Other functions of SLs
For several decades after the discovery of strigol and other SLs, it remained a mystery why plants exude cues for parasitic plants. In 2005, it was discovered that SLs exuded by the plant root facilitate the beneficial interaction with AM fungi through the induction of hyphal branching (Akiyama et al. 2005, Akiyama and Hayashi 2006, López‐Ráez et al. 2008), hyphopodium formation (Kobae et al. 2018) and boosting of metabolism (Besserer et al. 2006, 2008). Besides symbiosis with AM fungi, SLs are also thought to regulate interactions with other microorganisms (Peláez-Vico et al. 2016, Lanfranco et al. 2018, Kim et al. 2022). Together, the benefits of these interactions appear to outweigh the risk of infection by parasitic plants. The interaction with both parasitic plants and AM fungi implies that there are two selective pressures at work (Wang and Bouwmeester 2018): (I) to produce and exude SLs to recruit AM fungi and (II) to prevent detection by parasitic plants by producing SL variants that do not trigger germination. The interplay of these pressures is thought to have contributed to the large variety and different mixtures of SLs that are exuded (Brun et al. 2018, Wang and Bouwmeester 2018). This is illustrated by the mutation of the Low Germination Stimulant 1 (LGS1) locus in sorghum, which resulted in Striga resistance in sorghum (Gobena et al. 2017). Lgs varieties have low germination stimulant activity and show significantly less parasitization by Striga in field experiments (Vogler et al. 1996, Gobena et al. 2017). Intriguingly, lgs1 sorghum produces considerably less 5DS than susceptible LGS1 lines and exudes orobanchol instead (Gobena et al. 2017). Because Striga species are generally more responsive to strigol- than orobanchol-type SLs (Nomura et al. 2013, Bouwmeester et al. 2021), this change in the SL palette reduces detection by Striga. This was corroborated by another study where high 5DS and low orobanchol production significantly correlated with Striga germination and parasitization (Mohemed et al. 2018). Importantly, this change in the SL profile did not seem to produce negative side effects and the degree of symbiosis with AM fungi remained unchanged (Gobena et al. 2017).
Besides their function in the rhizosphere, it was later shown that the SLs in planta are also plant hormones. SLs inhibit shoot branching, and SL mutants display excessive branching (Gomez-Roldan et al. 2008, Umehara et al. 2008). Over the past decade, a wide variety of additional functions have been identified for the SL hormone, including the regulation of lateral root outgrowth, root hair growth, root and stem elongation, leaf shape and senescence, secondary stem growth and other developmental processes (Al-Babili and Bouwmeester 2015, Waters et al. 2017). SL production is increased in phosphate (P)-deficient plants (Yoneyama et al. 2007, López‐Ráez et al. 2008). This is considered to improve AM fungi recruitment to alleviate the P deficiency (Bouwmeester et al. 2007, Czarnecki et al. 2013). However, also some of the shoot and root phenotypes regulated by SL play a role in the mitigation of P deficiency (Koltai et al. 2010, Umehara et al. 2010, Kohlen et al. 2011, Ruyter-Spira et al. 2011, Umehara 2011, Czarnecki et al. 2013). Hence, SL is an important player in the P deficiency response of plants.
Perception of endogenous SL
The plant hormone SL is perceived by a receptor, the α/β-fold hydrolase DWARF14 (D14)/DECREASED APICAL DOMINANCE2 (DAD2)/RAMOSUS3 (Arite et al. 2009, Hamiaux et al. 2012, Waters et al. 2012, 2017, Nakamura et al. 2013, de Saint Germain et al. 2016, Yao et al. 2016, 2018). Interestingly, the D14 signaling pathway works similarly to that of auxin, GA and jasmonic acid (JA) (Blázquez et al. 2020). Each of these pathways transduces signals by interacting with the Skp1/Cullin/F-box (SCF)-type E3 ubiquitin ligase complex. In response to SLs, D14 interacts with the F-box protein MORE AXILLARY GROWTH 2 (MAX2)/DWARF3 (D3) (Hamiaux et al. 2012, Zhao et al. 2015, Yao et al. 2016). The resulting D14–SCFMAX2 complex then targets SUPPRESSOR OF MAX2 (SMAX2)-LIKE (SMXL) proteins for degradation (Soundappan et al. 2015, Wang et al. 2015, 2020, Waters et al. 2017, Li et al. 2022). SMXLs are negative regulators of transcription factors, and their degradation induces the expression of SL-responsive genes. In Arabidopsis, SMAX1 and SMXL2/6/7/8 are targeted by SL signaling, affecting various developmental processes.
Unlike most other receptors in plants, D14 possesses catalytic activity toward its substrate (Hamiaux et al. 2012, de Saint Germain et al. 2016, Yao et al. 2016, Waters et al. 2017). A conserved Ser-His-Asp catalytic triad is thought to hydrolyze bound SL at the junction between the ABC- and D-rings. Subsequently, the D-ring is bound to the catalytic His residue, forming a CLIM (Hamiaux et al. 2012, de Saint Germain et al. 2016, Yao et al. 2016, 2018, Nelson 2021). CLIM formation is thought to induce conformational changes that allow the binding of D14 to MAX2 and the subsequent degradation of SMXLs. At some point, the D-ring is released, allowing for the binding of new SLs. It is still unclear if SL hydrolysis and CLIM formation are strict requirements for signal transduction, since D14 can activate downstream signaling without hydrolysis in some cases (Shabek et al. 2018, Seto et al. 2019). A few hours after SL detection, D14 is degraded in a MAX2-dependent manner (Chevalier et al. 2014, Hu et al. 2017).
Irrespective of its true ligand, HTL/KAI2 is thought to be the ancestral form from which D14 has evolved (Waters et al. 2012, Conn et al. 2015, Bythell-Douglas et al. 2017, Xu et al. 2018). This means that the SL-sensing ability of D14 is a derived trait and that HTL/KAI2s had neo-functionalized to perceive SLs (Bythell-Douglas et al. 2017). Combined with the fact that Arabidopsis HTL/KAI2 plays a role in seed germination, this led to the hypothesis that parasitic plants use HTL/KAI2s to detect host-derived SLs. This was confirmed in three 2015 Science papers (Conn et al. 2015, Toh et al. 2015, Tsuchiya et al. 2015): in lieu of D14, parasitic plants perceive SLs via a large, diversified clade of HTL/KAI2 proteins. Parasitic plants contain many HTL/KAI2s, and a subset of these has neo-functionalized to perceive host-exuded SLs.
Parasitic Plants Contain Many SL Receptors
Transcriptomic data show that parasitic plant species express on average five to six HTL/KAI2 paralogs, which generally have a high-sequence identity of 54–80% (Conn et al. 2015). Since these numbers are based on transcriptomes, the actual amounts of paralogs encoded in the genomes could be much higher. Indeed, genome sequences showed that especially Striga species are rich in HTL/KAI2 paralogs (Nelson 2021). For example, S. hermonthica contains 13 paralogs and S. asiatica 21 (Conn et al. 2015, Yoshida et al. 2019). HTL/KAI2s form a monophyletic clade that can be divided into three subclades: conserved (KAI2c), intermediate (KAI2i) and divergent (KAI2d). KAI2c and KAI2i are present in most angiosperms, whereas the largest clade, KAI2d, occurs exclusively in parasitic plants. Most parasitic HTL/KAI2s belong to KAI2d, which is the clade that can perceive SLs (Conn et al. 2015, Toh et al. 2015, Tsuchiya et al. 2015).
Fitting with their role as SL receptors, KAI2d paralogs have binding pockets that are significantly larger than those of other clades and are more similar to D14 (Conn et al. 2015). Conversely, KAI2c paralogs have the smallest binding pocket, similar to that of AtKAI2. Paralogs of the KAI2i clade have binding pockets intermediate between AtKAI2 and D14. Although the binding pocket of KAI2d superficially seems to be most similar to D14, phylogenetic analysis shows that D14 actually diverged from HTL/KAI2 before KAI2c, KAI2i and KAI2d diverged from each other (Conn et al. 2015, Xu et al. 2018). Nevertheless, the large size of the binding pocket and its similarity to that of D14 indicate that KAI2c has undergone convergent evolution and can now detect SLs (Conn et al. 2015).
Also at the mechanistic level, SL perception by parasitic HTL/KAI2 proceeds analogous to D14 (Yao et al. 2017). The conserved Ser-His-Asp catalytic triad hydrolyzes SLs at the junction between the ABC- and D-rings by forming an intermediate molecule that is covalently bound to the Ser and His residues. Afterward the D-ring remains covalently bound to only the His residue and is called the Covalently Linked Intermediate Molecule (CLIM) (Yao et al. 2017, Uraguchi et al. 2018, Xu et al. 2018, de Saint Germain et al. 2021). CLIM formation is thought to trigger conformational changes that facilitate the interaction with MAX2 (Yao et al. 2017). The binding interface between HTL/KAI2 and MAX2 is strikingly similar between Arabidopsis and S. hermonthica orthologs (Shahul Hameed et al. 2018) and is likely conserved across land plants (Bythell-Douglas et al. 2017). Indeed, downstream signaling in parasitic plants is comparable to Arabidopsis, involving MAX2 and SMXL orthologs (Bunsick et al. 2020, Nelson 2021).
Striga hermonthica expresses one MAX2 ortholog, which can complement most of the Arabidopsis max2 mutant phenotypes, including the SL response and root and shoot phenotypes (Liu et al. 2014). In addition, when S. hermonthica HTL/KAI2s (ShHTLs) are expressed in Arabidopsis they require SMAX1 and MAX2 to function (Bunsick et al. 2020). Finally, direct interactions between several ShHTLs and ShMAX2 or AtMAX2 have also been shown in vitro (Xu et al. 2018, Wang et al. 2021, 2022). In yeast-two-hybrid and pull-down assays, the presence of rac-GR24 triggers ShHTL7–ShMAX2 and ShHTL4–ShMAX2 interactions, while ShHTL1–ShMAX2 interactions occur regardless of the presence of ligand (Yao et al. 2017, Xu et al. 2018). Another study showed that ShHTL5/7/8/9 interact with AtMAX2 and ShMAX2 in response to several types of natural SL, mainly 5DS and 4DO (Wang et al. 2021). Parasitic HTL/KAI2s presumably also interact with parasitic SMXL orthologs (Nelson 2021); however, there is no direct evidence for this since parasitic SMXLs have not yet been characterized.
Parasitic HTL/KAI2 Clades Have Functionally Diversified
A common way of measuring the SL response of parasitic HTL/KAI2 proteins is by performing cross-species complementation assays (Conn et al. 2015, Toh et al. 2015, Uraguchi et al. 2018, de Saint Germain et al. 2021, Nelson 2021). Upon exposure to heat, Arabidopsis seed becomes dormant in a process called thermoinhibition, which can be alleviated in an AtKAI2-dependent manner (Toh et al. 2012). Consequently, kai2 mutants germinate very poorly, and parasitic HTL/KAI2 proteins heterologously expressed in the kai2 mutant can be assayed for their ability to complement this phenotype. Such complementation assays have been employed to explore the specificities of AtKAI2, KAI2c, KAI2i and KAI2d (Conn et al. 2015, Toh et al. 2015, Takei et al. 2023). AtKAI2 induces germination in response to both KAR and rac-GR24, likely triggered by the unnaturally configured enantiomer GR24ent−5DS (Conn et al. 2015, Arellano-Saab et al. 2021). Despite having binding pockets similar to AtKAI2, KAI2c proteins are generally unable to confer a germination response to KAR and rac-GR24 (Conn et al. 2015, Toh et al. 2015). KAI2i paralogs show a response to KAR and rac-GR24, but not to natural SLs (Conn et al. 2015, Toh et al. 2015). Finally, KAI2d proteins generally do not respond to KAR, but most do strongly respond to rac-GR24 and SLs. Importantly, this response seems to be specific to natural SLs or naturally configured GR24 enantiomers, such as GR245DS and GR244DO (Conn et al. 2015, de Saint Germain et al. 2021). Together, these findings show that the different clades of parasitic HTL/KAI2 have functionally diversified.
Additional cross-species complementation experiments have made it clear that the SL sensitivity of parasitic HTL/KAI2 proteins can be exceptionally high. S. hermonthica KAI2d clade proteins generally trigger Arabidopsis germination in response to micro- and nanomolar levels of SLs (Toh et al. 2015, Tsuchiya et al. 2015). However, some receptors in particular are even more sensitive. For example, S. hermonthica HTL7 (ShHTL7) is able to confer a response to natural SL at picomolar levels (Toh et al. 2015), or in certain cases even at femtomolar levels, as observed with natural 5DS or the synthetic SL-like germination stimulant spironolactone-7 (SPL7) (Uraguchi et al. 2018). Curiously enough, the potent response to SPL7 seems to be exclusive to ShHTL7, as it binds poorly to other receptors (Uraguchi et al. 2018). This signifies that even small changes in the structure of the binding pocket can dramatically alter ligand affinity. Although one HTL/KAI2 may be highly sensitive to a certain SL, another receptor may not be and instead prefer a different SL. Indeed, parasitic receptors vary considerably in their specificity to different SLs (Toh et al. 2012, Tsuchiya et al. 2015, Uraguchi et al. 2018, Xu et al. 2018, Wang et al. 2021).
The specificity of HTL/KAI2 proteins can be dissected into two aspects: activation efficiency and binding affinity. These aspects are not necessarily equivalent since activation efficiency is not solely dependent on binding affinity. Instead, it is also influenced by factors such as CLIM formation, ABC-ring binding after hydrolysis and interactions with MAX2 and SMXLs (Uraguchi et al. 2018, Nelson 2021, Wang et al. 2021). Activation efficiency can be measured as the effective concentration of stimulant at which 50% of Arabidopsis plants germinate in complementation assays (EC50), or more easily measurable, as the percentage of plants that germinate in response to a set concentration of SL (Conn et al. 2015, Toh et al. 2015, Tsuchiya et al. 2015, Uraguchi et al. 2018). HTL/KAI2 binding affinity can be measured in several ways: either directly via methods such as isothermal titration calorimetry (ITC) (Xu et al. 2018, Wang et al. 2021) or indirectly via ligand competition assays with fluorescent probes (Tsuchiya et al. 2015, Uraguchi et al. 2018). An example of such probes are Yoshimulactone Green (YLG) and Yoshimulactone Green Double (YLGW) (Tsuchiya et al. 2015). Hydrolysis of YLG or YLGW by D14 or KAI2 results in fluorescence. YLG can therefore be used to detect SL perception and signaling in real-time. In ligand competition assays, an SL is added to compete with YLG after which the median inhibitory concentration (IC50), at which fluorescence has diminished by 50%, can be determined.
HTL/KAI2 Proteins Differ in SL Specificity
HTL/KAI2 specificity has been studied in most detail in S. hermonthica (Conn et al. 2015, Toh et al. 2015, Tsuchiya et al. 2015, Xu et al. 2018, Wang et al. 2021). KAI2c, KAI2i and KAI2d clades of S. hermonthica are comprised of ShHTL1, ShHTL2&3 and ShHTL4–11, respectively. Notably, the germination-stimulating activity of rac-GR24 varies considerably between individual KAI2d proteins (Toh et al. 2015). ShHTL7 causes germination in response to a broad variety of SLs and synthetic agonists, whereas others, such as ShHTL8 and ShHTL9, only confer a response to a more specific set of stimulants (Toh et al. 2015). These differences are reflected in the binding affinities of these receptors (Tsuchiya et al. 2015, Wang et al. 2021). ShHTL6 and ShHTL7 show a high affinity toward all tested SLs, while the rest shows a preference for specific SLs (Tsuchiya et al. 2015). Overall 5DS seems to be the most potent and broadly perceived SL (Toh et al. 2012, Tsuchiya et al. 2015). Together, this shows that different KAI2d proteins provide distinct responses to different SLs. It should be noted that the abundance of the receptor proteins in S. hermonthica is unknown. For instance, ShHTL7 may be more sensitive than ShHTL8; however, the expression of the latter may be much higher. This means that ShHTL8 could confer a stronger SL response, despite its lower sensitivity. Moreover, the interaction partners of the receptors may also further alter the strength of the SL response. Therefore, we have to be careful with the interpretation of the sensitivity of a given ShHTL to a certain SL, as analyzed in Arabidopsis.
Even though ShHTL10 and ShHTL11 are biochemically active, they do not seem to trigger a response to any of the SLs or synthetic agonists that were tested (Toh et al. 2015). Curiously, they do display a quite high affinity for various SLs (Tsuchiya et al. 2015, Wang et al. 2021), suggesting that they can bind SLs but not respond to them, at least in Arabidopsis. Finally, none of these KAI2d clade proteins binds or responds to KAR (Toh et al. 2015, Xu et al. 2018). Together, this shows that the KAI2d clade in S. hermonthica can be divided into an SL-sensitive group comprising ShHTL4-9 and a group that is insensitive, in the Arabidopsis test system, comprising ShHTL10-11 (Toh et al. 2015). The implications of this are not yet understood.
Although the conserved clade receptor ShHTL1 triggers a weak response to rac-GR24, it does not bind with or react to natural SLs (Toh et al. 2015, Wang et al. 2021). Moreover, ShHTL1 is able to bind KAR but does not trigger germination in response to it (Toh et al. 2015, Xu et al. 2018). This suggests that ShHTL1 is not involved in either SL or KAR perception. Instead, KAI2c may function as a potential KL receptor (Conn et al. 2015, Conn and Nelson 2016, Xu et al. 2018). Another explanation could be that it perceives undiscovered and/or untested SLs. Intermediate clade receptors ShHTL2 and ShHTL3 caused germination in response to both rac-GR24 and KAR (Toh et al. 2015), but only ShHTL3 was able to bind KAR (Xu et al. 2016, 2018). In addition, ShHTL2 and ShHTL3 show little to no affinity for natural SLs (Tsuchiya et al. 2015, Wang et al. 2021). Importantly, KAR does not trigger germination of Striga seed, suggesting that activation of ShHTL2 and ShHTL3 is not sufficient to induce germination (Toh et al. 2015). Together, these findings suggest that KAI2i is not involved in SL perception but could potentially function as receptors of exogenous KAR (Conn et al. 2015, Conn and Nelson 2016, Xu et al. 2018).
Also, the broomrapes contain a largely expanded HTL clade. Phelipanche ramosa contains one receptor belonging to the conserved clade, PrKAI2c, and four belonging to the divergent clade, PrKAI2d1–4 (de Saint Germain et al. 2021). However, KAI2i paralogs are absent. Unfortunately, only PrKAI2d3 recombinant protein could be further characterized since the others were not water soluble in vitro. PrKAI2d3 and mutant PrKAI2d3S98A, which replaces the catalytic Ser with Ala, were characterized in Arabidopsis cross-species complementation assays (de Saint Germain et al. 2021). PrKAI2d3 conferred a germination response to both GR245DS and GR24ent−5DS, whereas PrKAI2d3S98A was unresponsive to both. In addition, the researchers tested whether PrKAI2d3 and PrKAI2d3S98A could confer morphological responses to GR24. Unexpectedly, lines expressing either PrKAI2d3 or PrKAI2d3S98A showed a reduction in hypocotyl length in response to GR245DS. This suggests that the catalytic Ser98 is somehow only required for the germination response, but not for seedling morphogenesis.
Next, de Saint Germain et al. (2021) investigated P. ramosa germination sensitivity to various stereoisomers and modified forms of GR24. Seeds were 100-fold more sensitive to GR24 stereoisomers that resembled natural SLs than to those with unnatural configurations. PrKAI2d3 could not hydrolyze these unnatural stereoisomers. Likewise, the addition or removal of methyl groups on the D-ring also results in a large reduction of sensitivity compared to unmodified GR245DS, even though hydrolysis of de-methylated GR24 was more efficient than GR24. This pattern of sensitivity was reflected by the interactions between these ligands and PrKAI2d3. Differential scanning fluorimetry (DSF) revealed that only naturally configured, and to a lesser degree methylated/demethylated, forms of GR24 could bind and destabilize PrKAI2d3. Using a tryptophan intrinsic fluorescence assay, the researchers estimated binding affinity, which was consistent with the DSF results. Together, this shows that PrKAI2d3 preferentially binds naturally configured GR24, which fits with its role as a germination stimulant receptor. Finally, the hydrolytic activity of PrKAI2d3 was monitored using fluorescent probes. Hydrolysis plateaued when the product reaches the concentration of PrKAI2c3, rather than the concentration of the substrate. This signifies that PrKAI3d3 is a single-use protein. Finally, like other parasitic SL receptors, PrKAI2d3 could form a CLIM upon rac-GR24 hydrolysis, with the D-ring covalently bound to the catalytic His residue.
Besides SLs, P. ramosa is known to germinate in response to 2-PEITC, which is a non-SL germination stimulant. Although P. ramosa is more sensitive to SLs than 2-PEITC, the latter is still an important contributor to its host specificity to Brassica napus, since this host does not exude SLs (Auger et al. 2012). Therefore, de Saint Germain et al. (2021) measured P. ramosa seed germination and PrKAI2d3 interaction with 2-PEITC. Seeds germinated at nanomolar levels of both 2-PEITC and a related compound benzyl isothiocyanate, but at picomolar levels of rac-GR24. In addition, 2-PEITC could slightly thermally destabilize PrKAI2d3, indicating that a weak interaction occurs. This fits with the observation that 2-PEITC is a weaker germination stimulant than SL. Interestingly, DSF showed that PrKAI2d3 had different melting temperatures when 2-PEITC or GR24 was bound, suggesting that the two compounds trigger different conformational changes. Mass-spectrometry revealed that 2-PEITC can also form a CLIM by covalently binding to PrKAI2d3. However, unlike SLs, it binds only to the catalytic Ser and not to the catalytic His. Altogether de Saint Germain et al. (2021) showed that PrKAI2d3 is a germination stimulant receptor that detects naturally configured SLs and isothiocyanates.
More recently, five KAI2d proteins were identified and characterized in O. minor. Two of these, OmKAI2d3 and OmKAI2d4, were found to bind various SLs and rescue the kai2 germination phenotype in Arabidopsis (Takei et al. 2023). OmKAI2d3 was found to bind SL more strongly and confer a stronger germination response, whereas OmKAI2d4 was able to bind a wider variety of SLs. Intriguingly, both receptors were also demonstrated to interact with sesquiterpene lactones, another class of germination stimulants known to induce germination of some Orobanche species (de Luque et al. 2000, Joel et al. 2011, Raupp and Spring 2013, Ueno et al. 2014, Takei et al. 2023), albeit to a much lesser degree than to SLs. OmKAI2d1, 2 and 5, on the other hand, were mostly unable to bind and confer a response to SLs or sesquiterpene lactones.
To summarize, parasitic plants possess a large arsenal of HTL/KAI2s, which can be very variable in their sensitivity and specificity. However, it remains largely elusive how this gives rise to specificity at the level of parasite and host. This mystery can be broken down into four important questions: (I) What causes specificity at the level of the receptor? (II) How are the signals triggered by each receptor integrated? (III) Which receptors are expressed, and when and where? Finally, (IV) how do these aspects differ between parasitic plant species? Unfortunately, the latter three questions are far from being answered, in part because of the intractability of parasitic plants. There is currently no single ‘model parasitic plant’, and parasitic plants in general are difficult to reliably and consistently grow, let alone transform, since they require a living host. Therefore, most research has focused on the first question, how SL specificity works on the level of the receptor (Conn et al. 2015, Toh et al. 2015, Xu et al. 2016, Shahul Hameed et al. 2018, Uraguchi et al. 2018, Zhang et al. 2020, Arellano-Saab et al. 2021, Wang et al. 2021).
Binding Pocket Shape, Size and Composition Are Important for SL Recognition
Research into the mechanism behind receptor specificity for SLs started with the investigation of homology models (Conn et al. 2015). Since parasitic HLT/KAI2 proteins have relatively high-sequence identity with AtKAI2 (54–80%), structural models could be predicted based on previously existing crystal structures of KAR-bound AtKAI2. These models show that KAI2c, KAI2i and KAI2d proteins of various parasitic plant species display clear differences in binding pocket size (Conn et al. 2015, de Saint Germain et al. 2021). The binding pockets of KAI2c proteins are the smallest and most similar to AtKAI2, whereas the pockets of SL-binding KAI2d proteins are the largest and most similar to D14. It was therefore hypothesized that the larger, more D14-like, binding pockets of KAI2d proteins are what enables them to recognize SLs (Conn et al. 2015). A generally important amino acid for determining pocket size is Y124, which is present in AtKAI2 and all members of the KAI2c clade in lamiids (Conn et al. 2015). This bulky Tyr residue is thought to separate the main binding pocket from an adjacent cavity. Many KAI2i paralogs contain the less bulky Y124F substitution, while most KAI2d paralogs have a variety of substitutions with even smaller residues. Small residues allow for the fusion of the cavities, thereby increasing the size of the binding pocket. Other residues that were hypothesized to be important for pocket size and shape are W153, F157 and F194, which are all conserved in AtKAI2, KAI2c and KAI2i but are highly variable in KAI2d (Conn et al. 2015).
Since 2015, several proper crystal structures have been determined for parasitic HTL/KAI2s, the first of which was that of the KAI2c protein ShHTL5 (Toh et al. 2015). This revealed that ShHTL5 and AtKAI2 are structurally similar enough to allow for direct comparative analysis. Both receptors have active sites that are enclosed by a cap domain that consists of four α-helices that form a binding pocket. The binding pocket of ShHTL5 is more than twice as large as that of AtKAI2 (Toh et al. 2015). Eight out of the 16 residues covering the interior of the ShHTL5-binding pocket are different from those of AtKAI2, including the Y124 that was identified by Conn et al. (2015). Modeling of GR24 binding indicates that the changes of Y124 to V124 and S196 to Y196 in ShHTL5 are especially important to accommodate the D-ring of GR24 (Toh et al. 2015). Additional key differences in ShHTL5 include mutations of amino acids W153, F194 and A219 (two of which were also found by Conn et al. 2015), which are involved in KAR positioning in AtKAI2. When binding pocket amino acids are compared across all ShHTLs, it is observed that the KAI2c protein ShHTL1 is most similar to AtKAI2, while the KAI2d protein ShHTL7 is most distinct (Toh et al. 2015). Importantly, the degree of similarity to AtKAI2 correlates inversely with SL sensitivity. Therefore, it is postulated that these structural differences change the chemical environment in a way that facilitates SL recognition (Toh et al. 2015).
More recently, the crystal structures for ShHTL1/3/4/7/8 have also been determined (Xu et al. 2016, 2018, Shahul Hameed et al. 2018, Zhang et al. 2020, Wang et al. 2021). Based on these crystal structures, the molecular basis for ligand specificity can be analyzed in greater detail. Comparison of ShHTL1/3/4/5/7 and ShD14 shows that all receptors are structurally similar, constituting an α/β hydrolase domain, and a flexible cap domain composed of four helices αD1, αD2, αD3 and αD4 (Xu et al. 2018). However, structural differences are apparent in the cap domain, especially the αD1 and αD2 helices. Differences seem to be caused primarily by a change of Y150 to F150 in KAI2d clade proteins ShHTL4/5/7, resulting in the loss of a hydrogen bond between helices αD1 and αD3. Consequently, αD1 and αD2 can move away from αD3, thereby enlarging the binding pocket (Xu et al. 2018). It was hypothesized that this mutation contributes to the SL sensitivity and specificity of ShHTL4/5/7. Altogether, the KAI2d proteins ShHTL4/5/7 have binding pockets that are larger than that of ShD14, while those of ShHTL1/3 are slightly smaller than that of AtKAI2. Additional subtle differences in αD1 cause it to further tilt away from the binding pocket, increasing the size of the pocket even further. This phenomenon is especially notable in ShHTL7, which has the largest binding pocket and is generally the most sensitive SL receptor. Therefore especially helix αD1 is thought to be an important contributor to ligand specificity (Xu et al. 2018).
The crystal structure of ShHTL8 is similar to the other receptors (Zhang et al. 2020). Differences mostly occur in the cap domain, especially in helices αD1 and αD2 (Zhang et al. 2020), which were shown to be important for determining binding pocket size (Xu et al. 2018). Interestingly, ShHTL8 contains four amino acids, Y27, L179, S187 and Y151, that form a hydrogen-bond network that may be essential for cap domain integrity (Zhang et al. 2020). The estimation of binding pocket sizes was in line with Xu et al. (2018): ShHTL4/5/7/8 have large binding pockets, while those of ShHTL1/3 are small.
Further analysis within the KAI2d clade (ShHTL4/5/7/8) shows that the amino acid residues inside the binding pocket also contribute to the observed size differences (Xu et al. 2018, Zhang et al. 2020). In particular, the binding pocket of ShHTL7 is coated with small residues, including T142, L153 and T157 (Xu et al. 2018). Xu et al. (2018) postulated that smaller residues allow for a further increase in pocket size without impacting enzymatic activity. Consequently, a reduction in steric interference may facilitate ShHTL7’s high sensitivity to various SLs (Xu et al. 2018). In contrast, binding pockets of AtD14 and OsD14 contain slightly more bulky residues, resulting in smaller pockets. Residues of KAI2c protein ShHTL1 and KAI2i protein ShHTL3 were even bulkier. This may explain why these receptors can only bind KAR and not SLs (Xu et al. 2018). Overlaying ShHTL and AtKAI2 with a GR245DS-bound OsD14 or KAR-bound AtKAI2 crystal structure reveals additional residues that may be blocking ligand access. Residues L142, L/F190 and F194 of ShHTL1/2 and AtKAI2 may physically block the binding of GR245DS, whereas amino acids 157, 218 and 219 of ShHTL4/5/7 and ShD14 possibly interfere with KAR binding (Xu et al. 2018).
To test whether experimental changes in binding pocket residue bulkiness can affect ligand recognition, Xu et al. (2018) created ShHTL7 mutants that partially resemble ShHTL3, by double T190F/C194F and triple L124F/T190F/C194F mutations. The binding affinity of these mutants with rac-GR24 was 2- and 100-fold lower, respectively, but the mutations did enable weak binding to KAR. Together, this shows that modification of residue bulkiness directly affects ligand interaction. Likewise, Zhang et al. (2020) generated various single mutants of ShHTL8, substituting binding pocket residues with bulkier ones. L125F, L125Y and M154W mutations completely abolished YLG hydrolysis, whereas others (I140F, V143F, M147F, S158F, A191F, I194F and M220F) reduced activity by 30–50% (Zhang et al. 2020). L125F and L125Y also reduced hydrolysis of rac-GR24 from c. 60% to near background levels of c. 20%, while M147F and M154W reduced hydrolysis of rac-GR24 to c. 45%, and I194F to c. 35% (Zhang et al. 2020). Modeling of GR245DS binding to ShHTL8 shows that the D-ring is oriented toward the Ser-His-Asp catalytic triad, where it forms hydrophobic interactions with residues L125 and I194, while the ABC-ring is turned toward the opening forming hydrophobic interactions with M147 and M154 (Zhang et al. 2020). Together, these results indicate that M147, M154, I194 and especially L125 are important amino acids for GR24/YLG positioning. In conclusion, replacing certain binding pocket amino acids with bulkier counterparts can block the SL interaction and hydrolysis.
The alignment of ShHTL sequences makes it clear that amino acid L124 of ShHTL7 corresponds with L125 of ShHTL8 (Xu et al. 2018, Zhang et al. 2020). Therefore, combining the results of Xu et al. (2018) and Zhang et al. (2020) suggests that these amino acids play an important role in SL perception, especially since non-SL-perceiving ShHTL1/2/3 and AtKAI2 contain bulky residues at this position (Xu et al. 2018). To investigate the role of these residues in other ShHTLs, Zhang et al. (2020) generated corresponding mutants in ShHTL4 and ShHTL6. Mutation I124F in ShHTL4 did not affect YLG or rac-GR24 hydrolysis, but it did significantly reduce the hydrolysis of both compounds in ShHTL6. This experiment can also be conducted the other way around, where the bulky residues of ShHTL1/2/3 are replaced by smaller ones. Fascinatingly, this increases YLG and rac-GR24 hydrolysis in all three cases (Zhang et al. 2020). Together, these results indicate that L124 plays an important role in ligand positioning and hydrolysis. Overall, this fits well with the earlier research of Toh et al. (2015) and Conn et al. (2015), which already highlighted that amino acid 124 is an important determinant of binding pocket structure.
Another interesting example where receptor specificity was experimentally altered is that of ShHTL7 (Uraguchi et al. 2018). ShHTL7 shows a femtomolar sensitivity toward both the natural 5DS and the synthetic SPL7. Interestingly, 5DS is also detected with high affinity by other ShHTLs, while SPL7 is exclusively detected by ShHTL7. The authors showed that seven key amino acids facilitate the unique SPL7 interaction of ShHTL7: M139, T142, T157, L161, Y174, C194 and M219 (Uraguchi et al. 2018). The specific combination of these seven amino acids was unique to ShHTL7, across all known parasitic HTL/KAI2 proteins. In single to septuple mutants where these amino acids are gradually replaced with the ones found in ShHTL5, affinity to SPL7 and the related H-SPL7 decreased, while affinity for 5DS was unaffected, showing that SPL7 and 5DS specificity can be altered independently (Uraguchi et al. 2018). Although SPL7 is admittedly a synthetic compound, this research shows that specificity toward different SL-like compounds can be encoded within HTL/KAI2-binding sites at least partially independently.
Since SPL7 and 5DS (and other natural SLs) all contain the same D-ring, ligand specificity must be based on the ABC-ring (Uraguchi et al. 2018). Further support for this comes from the varying SL specificities of ShHTLs in general (Conn et al. 2015, Toh et al. 2015, Tsuchiya et al. 2015). Not just chemical composition but also stereochemistry plays a role, as seen for PrKAI2d3 which preferentially binds configurations that mimic certain natural SLs (de Saint Germain et al. 2021). Finally, witchweeds and broomrapes germinate mainly in response to strigol- or orobanchol-type SLs, respectively (Bouwmeester et al. 2021). Therefore, it can be hypothesized that their receptors must also encode a preference for one of either stereochemical conformations. It may be interesting to generate ‘chimeric’ SLs that combine the structures/stereochemistry of different types in order to test what effects different parts of the molecule have on receptor specificity. A study with a similar concept has been performed earlier, testing the effects of 36 artificial configurations of strigol, sorgolactone, orobanchol, sorgomol and 5DS, on S. gesnerioides germination (Nomura et al. 2013).
Molecular dynamic (MD) modeling of AtD14, OsD14, PhDAD2 (D14 ortholog in petunia), ShD14 and ShHTL1/4/5/7/8 crystal structures revealed that their binding pocket entrances are all large enough to allow all 20 SLs that were investigated to enter (Bürger and Chory 2020, Chen et al. 2021). However, further inward, the pockets narrow into a stiff ‘bottleneck’, blocking SLs that are larger in diameter from entering the active site (Bürger and Chory 2020). In contrast, the area surrounding the binding site itself is quite flexible. Slight conformational changes facilitate ligand positioning by increasing pocket volume, allowing SLs to rotate into the correct orientation in which the D-ring faces the active site. Importantly, reorientation appears to be dependent on the Ser and His of the catalytic triad (Bürger and Chory 2020). For example, when the Ser and His of ShHTL8 are replaced by Ala, SL can no longer reorient. Each of the investigated receptors seems to use a common subset of amino acids to bind SL. Amino acids corresponding to AtD14 F159 interact with all bound SLs, while F136, V144 and V194 interact with >80% of bound SLs. In addition, F28 and F126 interact with half of the SLs (Bürger and Chory 2020). Overall, modeling showed that ShHTLs had significantly varying SL preferences, which is in line with earlier wet lab research. Strangely, ShHTL8 and to a lesser extent ShHTL7 were predicted to mainly bind orobanchol-type SLs, contrary to most other studies where they readily bind strigol-type SLs. Therefore, it is imperative that these modeling results are verified in vivo before any conclusions are drawn.
Specificity Evolved via Keystone Mutations
As seen in the previous sections, a lot of amino acid residues have been implicated to be involved or required for ligand specificity. Therefore, one may speculate that adaptation to new ligands requires a series of mutations that gradually alter ligand binding. However, this does not necessarily seem to be the case, as recently demonstrated by gain-of-function analysis where numerous AtKAI2 amino acids were substituted with their ShHTL7 counterparts (Arellano-Saab et al. 2021). In essence, the study screened many gain-of-function mutants in order to identify keystone mutations that allow HTL/KAI2s to detect SLs. Eight binding pocket amino acids were identified that were less bulky or hydrophobic in ShHTL7 than in AtKAI2 (Y26, L124, T142, L153, T157, Y174, T190 and C194). Different combinations of these ShHTL7 amino acids were substituted in AtKAI2, generating 92 hybrid variants bearing single, double or triple mutations. Subsequently, yeast-two-hybrid assays were performed to screen for rac-GR24-dependent interactions between hybrid variants and MAX2. This screen identified seven receptor variants that gained the ability to interact with MAX2 (Arellano-Saab et al. 2021). These had substitutions at position 190 in six variants and at positions 124 and/or 157 in four variants each. This suggests that these amino acids are important for SL-dependent interactions with MAX2. In particular, the importance of amino acid 124 is in line with earlier studies (Toh et al. 2015, Xu et al. 2018, Zhang et al. 2020). To further investigate the functions of these AtKAI2 hybrid receptor variants, Arellano-Saab et al. (2021) screened 87 of them by using them to complement an Arabidopsis kai2 mutant. This led to the identification of the triple mutant Var64 (W153L F157T G190T), which conferred a germination response to nanomolar levels of rac-GR24 with 100-fold higher sensitivity to GR245DS than GR24ent−5DS. The response to GR24ent−5DS was similar to that of native AtKAI2, but the increase in sensitivity toward GR245DS was highly significant. Therefore, the substitution of the ‘ShHTL7-like’ amino acids L153, T157 and T190 in AtKAI2 can broaden its sensitivity to GR245DS, without impairing GR24ent−5DS sensitivity (Arellano-Saab et al. 2021). DSF and hydrolysis assays confirmed that both AtKAI2 and the triple mutant bind and hydrolyze GR24ent−5DS, but that only the latter can bind and hydrolyze GR245DS (Arellano-Saab et al. 2021). These results show that the triple mutant, Var64, gained the ability to respond to naturally configured GR245DS, suggesting that SL sensitivity can evolve with as little as three mutations.
The prevalence of these ShHTL7-like mutations at positions 153, 157 and 190 was investigated across a large number of land plant HTL/KAI2 paralogs (Arellano-Saab et al. 2021). Divergent clade HTL/KAI2 (especially parasitic KAI2d) paralogs are enriched in amino acids structurally related to those of ShHTL7, especially at position 153, whereas D14 paralogs are enriched with such amino acids mainly at position 190. Conversely, the conserved HTL/KAI2 clade was depleted of ShHTL7-like amino acids (Arellano-Saab et al. 2021). Together, this supports the role of L153, T157 and T190 as keystone mutations in the evolution of SL detection.
KL or KAR signaling can be indirectly measured by investigating the elongated leaf phenotype of Arabidopsis kai2 mutants, which is thought to be specific to KL/KAR and not SL signaling (Waters et al. 2012, Arellano-Saab et al. 2021). Leaves of Arabidopsis plants cross-complemented with hybrid receptor variants are more likely to be rescued by single mutants, which are more similar to AtKAI2, than by double or triple mutants. However, this pattern is broken by triple mutants carrying the G190T substitution, which rescues the leaf defect more frequently than mutants lacking it (Arellano-Saab et al. 2021). It is hypothesized that T190 somehow stabilizes the receptor, permitting additional mutations that alter ligand recognition, without directly affecting its activity. This is consistent with the prevalence of T190 in D14 paralogs and SL-detecting HTL/KAI2s (Arellano-Saab et al. 2021). Importantly, Var64 also contains the G190T mutation and fully rescues the leaf defect, showing that although it can detect SLs, it retains its function in KL/KAR signaling (Arellano-Saab et al. 2021). These findings show that one of the keystone mutations that enabled SL perception is a stabilizing mutation that enabled further evolution of the binding affinity.
Structural comparison of Var64 and AtKAI2 confirms the importance of the α-helices constituting the lid domain in determining pocket size, as previously shown by Xu et al. (2018) and Zhang et al. (2020). Mutations W153L and F157T resulted in the loss of hydrogen bonds between helices αD2, αD3 and αD4, consequently increasing lid flexibility and binding pocket size (Arellano-Saab et al. 2021). MD simulations support this increase in flexibility. Furthermore, simulations of ligand binding indicate that Var64 binds GR245DS more effectively than AtKAI2, since the ligand remains in closer proximity to the binding site and fluctuates less in Var64 (Arellano-Saab et al. 2021). The binding of GR245DS also triggers a conformational change in the αD1 helix, making it more flexible, which is thought to promote interactions with downstream targets (Arellano-Saab et al. 2021).
Keystone mutations seem to play an important role in the evolution of parasitic HTL/KAI2 ligand specificity. In animals, keystone mutations have also been shown to be important for steroid hormone receptors, where two mutations are enough to trigger a 70,000-fold change in receptor specificity (Harms et al. 2013). The fact that this phenomenon occurs in both plants and animals suggests that such high-impact mutations may be a more widespread property of receptor evolution (Harms et al. 2013, Arellano-Saab et al. 2021). These findings are important, as they suggest that it is possible for existing KAI2/HTL proteins to relatively easily and rapidly evolve new ligand specificity. This would allow parasitic plants to quickly adapt to a new host species or co-evolve with changes in the SL composition of their host. Further fine-tuning may occur through diversifying selection, which is facilitated by the large amount of parasitic KAI2/HTL paralogs.
HTL/KAI2 SL-Affinity and Germination Induction Are Inconsistent
Molecular mechanisms behind receptor specificity have been investigated in quite some detail. However as mentioned earlier, the activation of these receptors does not solely depend on ligand binding affinity. There exists a considerable discrepancy between SL-binding affinity and seed germination sensitivity (Shahul Hameed et al. 2018, de Saint Germain et al. 2021, Nelson 2021, Wang et al. 2021). For example, ShHTLs generally show IC50-values in the micromolar range, which vary at most a single order of magnitude (Tsuchiya et al. 2015). These values are consistent with dissociation constants obtained for YLG or 5DS (Tsuchiya et al. 2015, Wang et al. 2021). However, when looking at germination sensitivity in cross-species complementation assays, ShHTLs readily confer germination responses to nanomolar and picomolar levels of SL (Toh et al. 2015), and even femtomolar levels in the case of 5DS and ShHTL7 (Uraguchi et al. 2018). ShHTLs seem to confer responses that are several orders of magnitude stronger than what would be expected based on their binding affinities. A similar phenomenon is also observed in P. ramosa. Both P. ramosa and Arabidopsis seeds expressing PrKAI2d3 germinate in response to picomolar levels of naturally configured GR24 analogs, even though PrKAI2d3 only has a micromolar affinity for these compounds (de Saint Germain et al. 2021). These unexplained ‘increases in sensitivity’ are furthermore inconsistent between different paralogs. For example, ShHTL6 and ShHTL7 have roughly equal affinities for 5DS, but the latter confers an upward of 1,000-fold more sensitive germination response. Moreover, receptors like ShHTL10&11 were found to bind 5DS more strongly than ShHTL7 (Wang et al. 2021) but could at best only confer a weak germination response (Toh et al. 2015). Because of these discrepancies, it has been hypothesized that there must be additional mechanisms that amplify HTL/KAI2 activation (Shahul Hameed et al. 2018, de Saint Germain et al. 2021, Nelson 2021), such as CLIM formation, ligand hydrolysis, interactions with the cleaved ABC-ring and interactions with downstream targets (Yao et al. 2017, Uraguchi et al. 2018, de Saint Germain et al. 2021, Wang et al. 2021).
Indeed, CLIM formation has been suggested to be involved in the activation of both D14 and HTL/KAI2, resulting in conformational changes that allow for interactions with downstream targets such as MAX2 (Yao et al. 2016, 2017, 2018). However, it is still controversial what role this exactly plays. In certain cases, D14 activation can occur without hydrolysis or CLIM formation (Seto et al. 2019), or hydrolysis occurs after signal transduction (Shabek et al. 2018). Likewise, experiments on ShHTL7 also show that hydrolysis and CLIM formation are not necessary for activation per se but do somehow contribute to attaining high germination sensitivity (Uraguchi et al. 2018, Wang et al. 2022). Indeed, hydrolysis-resistant SPL7 analogs can induce germination at concentrations of 100 nM. Although this concentration is quite low, it is still almost a million times higher than the concentration at which the hydrolyzable SPL7 can induce germination. Uraguchi et al. (2018) investigated whether this increase can be attributed to the rate at which CLIM forms. Kinetic analysis shows that a lower CLIM formation speed is weakly associated with lower sensitivity. However, GR24 forms CLIM at a higher rate than SPL7, while it is less effective at activating ShHTL7, suggesting that other factors likely also play a role (Uraguchi et al. 2018). Since CLIM consists of the hydrolyzed D-ring bound to the catalytic His residue, it was postulated that it must be the ABC-ring of the hydrolyzed ligand which somehow increases sensitivity (Uraguchi et al. 2018). It is possible that the ABC-ring remains bound to HTL/KAI2 where it could fine-tune the conformational change caused by the CLIM, thereby enabling interactions with different downstream targets depending on the ligand. However, this remains to be tested.
Complex formation with MAX2 and SMXLs plays a more clear role in boosting HTL/KAI2 ligand sensitivity (Wang et al. 2021). As mentioned before, parasitic D14 and KAI2d proteins interact with MAX2 and SMXLs in an SL-dependent manner in vitro (Yao et al. 2017, Xu et al. 2018, Nelson 2021, Wang et al. 2021). This interaction triggers SMXL ubiquitination and degradation, activating downstream signaling. In yeast-two-hybrid and pull-down assays, ShHTL5/7/8/9 could form SL-dependent complexes with either AtMAX2 on its own, AtSMAX2 on its own, or form a tripartite complex with both AtMAX2 and AtSMAX1 (Wang et al. 2021). Conversely, ShHTL1/2/3/10/11 did not interact with AtMAX2 or AtSMAX1 in an SL-dependent manner. ShHTL4 and 6 displayed autoactivation in yeast; however, they do show weak SL-dependent interactions in the pull-down assay (Wang et al. 2021).
SL concentrations at which ShHTL5/7/8/9 begin to form complexes with AtMAX2 and AtSMAX1 correlate well with their ability to confer a germination response (Wang et al. 2021). That is to say, ShHTLs that form complexes in response to lower levels of SL also trigger germination responses at lower levels of SL, and vice versa. For example, ShHTL7 confers the strongest germination response and also interacts with AtMAX2 and AtSMAX1 at the lowest SL concentrations. Importantly, during the formation of this tripartite ShHTL–AtMAX2–AtSMAX1 complex, SMAX1 and MAX2 appear to act synergistically in their SL-dependent binding with ShHTL. Indeed, when both AtMAX2 and AtSMAX2 are present, interactions occur at 100-fold lower SL concentrations than when either partner is present on its own (Wang et al. 2021). These synergistic interactions occur at SL concentrations that are seemingly lower than what would be expected when only considering receptor affinity (e.g. 100 pM 5DS for ShHTL7, which has an affinity of Kd = 0.91 µM). Therefore, these observations led Wang et al. (2021) to hypothesize that the exceptionally high sensitivity of ShHTLs may be rooted in their interactions with downstream targets. It should be noted that all of the findings mentioned earlier are also achieved with ShMAX2 instead of AtMAX2, further supporting this hypothesis (Wang et al. 2021). Finally, an important domain for ShHTL–AtMAX2–AtSMAX1 complex affinity is the D2-domain of SMAX1, which significantly boosts the interaction strength between ShHTL7 and AtMAX2 (Shabek et al. 2018, Wang et al. 2021).
When forming SL-dependent complexes, ShHTL5/7/8/9 show varying preferences for different types of natural SL (Wang et al. 2021), which is consistent with the distinct germination responses that they confer (Toh et al. 2015). Complex formation in the presence of strigol, orobanchol, 5DS, 4DO and rac-GR24 indicates that ShHTL7 is most sensitive, responding to each of the tested SLs. Whereas other ShHTLs are more specific, responding mainly to 5DS, 4DO or GR24. To investigate the molecular mechanism behind these SL-dependent interactions with AtMAX2 and AtSMAX1, Wang (2021) determined and analyzed a crystal structure of ShHTL7. Comparison of the ShHTL7 crystal structure with other ShHTLs showed differences in the αD1 and αD2 helices that result in a larger and more flexible binding pocket that may facilitate SL recognition (Wang et al. 2021), corroborating the numerous other comparative studies mentioned earlier (Xu et al. 2018, Zhang et al. 2020, Arellano-Saab et al. 2021).
ShHTL7 interacts with AtMAX2 and AtSMAX1 in response to strigol and orobanchol, but ShHTL5/8/9 do not. Differences in their binding pocket structure may in part explain this phenomenon (Wang et al. 2021). Notably, ShHTL7 contains Leu at positions 146 and 153, whereas the others contain Met at these positions. When substitutions L146M and L153M are introduced into ShHTL7, its ability to interact with AtMAX2 or AtSMAX2 in response to strigol and orobanchol is abolished, while its response to 5DS and 4DO is unaffected (Wang et al. 2021). Admittedly, this mutant likely does not directly affect complex formation, but rather does so indirectly by altering ligand recognition. Nevertheless, these results are interesting since L146M and L153M seem to reduce specificity without impacting sensitivity. Besides, these substitutions decrease the binding pocket size (Wang et al. 2021) and therefore support the notion that larger binding pockets may contribute to broader ligand recognition (Conn et al. 2015, Toh et al. 2015, Xu et al. 2018, Zhang et al. 2020, Arellano-Saab et al. 2021).
Bulkiness of the ShHTL7-binding pocket residues also seems to affect the SL-dependent interaction with AtMAX2 and AtSMAX1. Wang et al. (2021) generated 10 mutants where single amino acids were substituted with Ala to decrease bulkiness, and L146F and L153F were created to increase bulkiness. ITC shows the majority of substitutions that decrease bulkiness either negatively affect, or do not affect, 5DS binding affinity. On the other hand, substitutions L146F and L153F that increase bulkiness caused a slight increase in 5DS affinity (Wang et al. 2021). This is in contrast with the notion that less bulkiness should decrease steric hindrance and therefore increase SL-affinity (Xu et al. 2018, Zhang et al. 2020) and suggests that decreasing bulkiness, by itself, does not improve affinity and that the specific functions of these amino acids must play a role (Wang et al. 2021). Yeast-two-hybrid assays revealed that modifying binding pocket bulkiness does not necessarily have the same effect on complex formation with AtMAX2 and AtSMAX1 as on 5DS binding affinity. Some mutants show both decreased 5DS affinity and interaction strength. However, other mutants actually bind 5DS more strongly but still interact more weakly with AtMAX2/AtSMAX1. Two other mutants show equal interactions with AtMAX2/AtSMAX1 but differ 20-fold in 5DS affinity (Wang et al. 2021). It is clear that 5DS affinity and sensitivity of complex formation are not inherently linked. This is supported by the fact that ShHTL8/9/10/11 have high 5DS affinity but fairly low affinity for SL-dependent interactions with AtMAX2/AtSMAX1 in pull-down assays (Wang et al. 2021). Therefore, these results suggest that while changes in binding pocket bulkiness can indeed affect complex formation, this is not necessarily a consequence of altered SL-affinity (Wang et al. 2021).
One of the aforementioned ShHTL7 mutants, L153F, interacts with AtMAX2 in response to lower 5DS concentrations than the wild type (WT) ShHTL7, even though its 5DS binding affinity is equal (Wang et al. 2021). Its response to 4DO, strigol and orobanchol also remains unchanged. In essence, this mutation affects affinity toward AtMAX2 without affecting SL sensitivity or specificity, making it a good subject to study the mechanism behind ShHTL7–AtMAX2 binding. Modeling of the interaction between ShHTL7 and AtMAX2 shows that the αD2 helix folds in toward the binding pocket entrance upon interactions, closing it off and exposing a binding interface, including L153, to AtMAX2 (Wang et al. 2021). In the L153F mutant, the exposed Leu is replaced with Phe, which engages in a π–π interaction with F646 of AtMAX2. This extra interaction is thought to increase the overall binding strength with AtMAX2 (Wang et al. 2021). The binding interface between HTL/KAI2s and SMXLs has not yet been investigated in detail. However, in Arabidopsis D14, SL-induced conformational changes are slightly more clear (Seto et al. 2019). First, the binding pocket is closed off, exposing the MAX2-binding domain, just as it is in ShHTL7. Next, the loop containing the catalytic Asp moves out toward the surface and then putatively binds SMXLs (Seto et al. 2019). It is possible that the ShHTL–SMAX1 interaction works in a similar fashion.
Further analysis of the AtMAX2-binding interface across different ShHTLs indicates that it is conserved across all paralogs except ShHTL10 and 11. More specifically, ShHTL10 and 11 contain significant differences in amino acids located on the αD2 and αD3 helices (Wang et al. 2021). ShHTL10 and 11 can bind SLs, but do not interact with AtMAX2 or AtSMAX1 nor confer a germination response to them (Toh et al. 2015, Wang et al. 2021). Therefore, the amino acids at the binding interface may be particularly important for the interaction with AtMAX2 (Wang et al. 2021). A subset of the differences observed in ShHTL10 and 11 are also observed in ShHTL6 and 7. ShHTL6 and 7 bind 5DS at roughly equal affinities and have similar hydrolytic properties; however, ShHTL6 interacts more weakly with AtMAX2 in response to SL than ShHTL7, supporting that differences in the binding interface affect the interaction strength with AtMAX2 (Wang et al. 2021). This can be confirmed with gain-of-function mutations in which the amino acids corresponding to ShHTL7 are substituted into ShHTL6 (Wang et al. 2021). Indeed, a quintuple ShHTL6 mutant bearing F157T, M161L, G163A, S180N and I181M binds AtMAX2 with the same strength as ShHTL7. Similarly, a double mutant bearing S180N and I181M shows an intermediate binding strength (Wang et al. 2021). In short, these findings support the notion that amino acids at the surface of ShHTLs determine the strength of SL-dependent interactions with AtMAX2. Consequently, this may contribute to the SL sensitivity of ShHTL–AtMAX2–AtSMAX1 complex formation, which in turn enables a highly sensitive germination response (Wang et al. 2021).
Integration of HTL/KAI2 Signaling in Parasitic Plants
Parasitic HTL/KAI2s have been studied in fairly great detail, as shown earlier; however, most of the research was done using cross-species or in vitro assays. An obvious drawback of cross-species complementation is that the genetic context of the parasitic plant is lost and is replaced with that of Arabidopsis. Likewise, in vitro studies strip a given protein from its in vivo environment. This leads to a bias toward the proteins themselves and away from how the system works as a whole. Consequently, we know a lot about the proteins themselves, and how they may function in Arabidopsis, but very little of the actual pathway as it exists within parasitic plants. Tight regulation of germination is of critical importance to parasitic plants, germination in response to the correct cue determines whether the plant survives or dies. In contrast, KAR-induced germination in Arabidopsis is not of critical concern. Therefore, it can be hypothesized that the germination control mechanism in parasitic plants is more complex, or at least more tightly regulated. Such complexity may become apparent if additional downstream signaling partners (e.g. MAX2, SMAX1 homologs) are discovered in parasitic plants. In Arabidopsis, D14 and KAI2 together interact with five different SMXLs (SMAX1 and SMXL2/6/7/8). Therefore, it is plausible that parasitic plants also contain several homologs. Overall, it is imperative to sequence a wide variety of parasitic plants to identify these putative interactors.
Conditions that cause germination in Arabidopsis may not necessarily trigger germination in parasitic plants, and vice versa. For example, when expressed in Arabidopsis ShHTL2 and 3 confer a germination response to KAR, even though the plant from which these receptors originate, S. hermonthica, is not responsive to KAR (Toh et al. 2015, Xiong et al. 2016). If S. hermonthica does contain additional MAX2 or SMAX1 homologs, it stands to reason that alternative complexes could form that mediate different downstream responses (Fig. 2). There could be complexes that negatively affect germination, for example, which could turn KAR or perhaps non-host SLs into germination inhibiting signals. One way in which this may work is by interaction and degradation of (unidentified) positive regulators of germination (Fig. 2B). Such an idea is supported by the fact that S. gesnerioides germination is inhibited by certain strigol-type SLs (Nomura et al. 2013). Another way in which negative regulation could be achieved is by sequestration of signaling partners (Nelson 2021). This is accomplished by an HTL/KAI2 variant that can interact with either MAX2 or SMAX1, but not with both, thus blocking downstream signaling while at the same time sequestering the partner that does bind (Fig. 2C) (Nelson 2021). Several mutations have already been discovered that change the way D14 interacts with MAX2 and SMXLs (Zhao et al. 2015, Yao et al. 2016, Seto et al. 2019, Lee et al. 2020). The petunia D14/DAD2 mutant D166A can no longer interact with PhMAX2A but retains its interactions with PhD53A (Lee et al. 2020). This suggests that a similar mutation may be possible in parasitic HTL/KAI2s. ShHTL10 and 11 are candidates that could potentially act as negative regulators, since they strongly bind certain SLs, but do not interact with MAX2 and SMAX1 nor trigger germination (Toh et al. 2015, Tsuchiya et al. 2015, Nelson 2021, Wang et al. 2021). Furthermore, the significant differences in their ‘MAX2’ binding surfaces may imply that they can interact with unidentified partners (Wang et al. 2021).
Fig. 2.
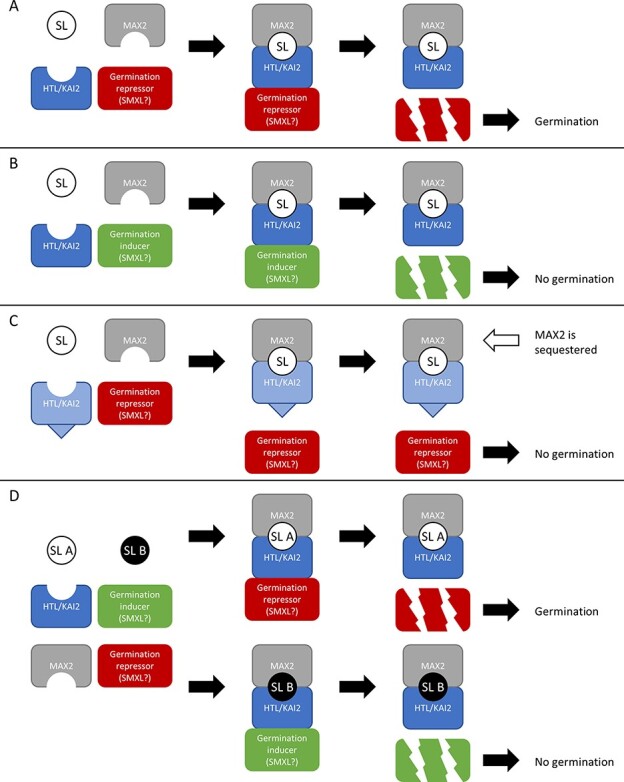
Hypothetical mechanisms of parasitic plant seed germination in response to SLs. Parasitic plants of the Orobanchaceae germinate in response to host-exuded SLs, which are detected using a diverse clade of HTL/KAI2 receptors. It is essential that parasites only respond to the signals corresponding with their host; therefore, signal integration is an important part of host detection. Shown are different models by which HTL/KAI2 paralogs may transduce signals upon binding SL, by interacting with MAX2 and degrading (presumably) SMXLs. Each panel represents a plausible way in which a given HTL/KAI2 could interact with its partners. (A) Positive regulation by degradation of a germination-inhibiting regulator. (B) Negative regulation by degradation of a germination-inducing regulator. (C) Negative regulation by sequestration of a signaling partner, as postulated by Nelson (2021). Certain modifications of HTL/KAI2 may prevent the interaction with either SMXL (as shown here) or MAX2, thereby sequestrating HTL/KAI2 and its signaling partner and preventing further signaling. (D) Positive or negative regulation based on which type of SL is present. Different kinds of SLs may trigger distinct conformational changes, e.g. SL 1 may promote the degradation of regulators that inhibit germination, whereas SL 2 might promote the degradation of regulators that induce germination.
Finally, it is possible that different SLs trigger distinct conformational changes. If this is the case, HTL/KAI2s may exist that bind multiple types of SLs, but only respond to a specific type, or interact with different partners depending on which type is bound (Fig. 2D). When non-host SLs bind the receptor may become ‘blocked’ by preventing the interaction altogether or by sequestering signaling components. As a consequence, other SLs that can induce germination can no longer bind and activate the blocked HTL/KAI2. Alternatively, non-host SLs could induce different conformational changes than host SLs, triggering degradation of positive, instead of negative, regulators of germination (Fig. 2D), in which case the receptor functions as both a negative and positive regulator, depending on the type of SL that is present. One candidate for this is ShHTL6, which binds orobanchol more strongly than strigol-type SLs, even though S. hermonthica is much less sensitive to the former (Tsuchiya et al. 2015). Unfortunately, the effect of orobanchol has not been studied in cross-species complementation assays, so evidence remains weak. Also in ShHTL7 there is evidence for blocking of the binding pocket. Triton X-100 binds but prevents conformational changes in ShHTL7 (Shahul Hameed et al. 2018). To check whether certain SLs have similar effects, future research should focus on screening for SLs with inhibiting effects, rather than activating effects like most previous studies. Interestingly, parallels can be drawn between gibberellic acid (GA) and SL signaling (Box 2), and in GA signaling negative regulation by a receptor has already been shown experimentally.
Box 2.
Parallels between SL and GA signaling
Since auxin, JA, GA and SL signaling are all mediated by SCF complexes, parallels can be drawn between these pathways (Blázquez et al. 2020). SL signaling is especially similar to that of GA, since both consist of separate receptor, F-box and TF-repressors, whereas auxin and JA make use of proteins with combined receptor and F-box functionalities. In addition, HTL/KAI2 and the GA receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) are both members of the α/β-fold hydrolase superfamily (Ueguchi-Tanaka et al. 2005, 2007, Shimada et al. 2008, Blázquez et al. 2020). GID1 interacts with DELLA proteins upon binding with GA and recruits the F-box protein GIBBERELLIN INSENSITIVE DWARF2 (GID2)/SLEEPY1, which causes the degradation of DELLA. It should be noted, however, that GID1 does not possess catalytic activity toward its substrate in contrast to HTL/KAI2. Nevertheless, because of the similarities in the signaling of these hormones, investigating the GA pathway may provide new insights into SL signaling.
Among numerous other functions, GA and GID1 play an important role in seed germination (Voegele et al. 2011, Ge and Steber 2018, Hauvermale and Steber 2020). Arabidopsis contains three GID1 paralogs, GID1a, b and c. The fact that Arabidopsis contains multiple GID1 receptors is interesting, since they may interact with each other and behave in ways that are analogous to the behavior of HTL/KAI2s in parasitic plants. Because of this, GID1a/b/c may be good targets to test what kinds of positive/negative/synergistic interactions can occur when multiple α/β-fold hydrolase receptors are present, especially since Arabidopsis is much more tractable than parasitic plants.
Knockout mutants gid1a and gid1c show significantly slower and less germination, whereas the gid1b mutant germinates similarly to the WT (Ge and Steber 2018). Interestingly, mutant and epistasis analyses indicate that GID1b acts as a negative regulator of germination, while GID1a and c are additive positive regulators: double mutant gid1a gid1c was unable to germinate, showing that the negative effects of gid1a and gid1c are additive. Next, the gid1a gid1b double mutant has a germination phenotype that is intermediate of WT and gid1a, indicating that gid1b can partially rescue gid1a and that GID1b therefore acts as a negative regulator of seed germination. On the other hand, the phenotype of double mutant gid1b gid1c was identical to gid1c, suggesting that the negative regulation by GID1b likely acts upstream of GID1c (Ge and Steber 2018). Application of exogenous GA to GA-biosynthesis inhibited seeds reflect these results: double mutation of gid1a gid1c impacts GA induced germination significantly more strongly than gid1a and gid1c single mutants. Importantly, gid1a gid1b rescues the gid1a single mutant, and gid1b gid1c does not affect the gid1c phenotype (Ge and Steber 2018). GID1 receptors seem to function slightly differently when seeds are plated under dim or green light and incubated under dark conditions. Mutations gid1a and gid1b both seem to increase germination in the dark, whereas gid1c has little effect (Ge and Steber 2018). Like before, gid1a gid1c is unable to germinate at all, but intriguingly, double mutation gid1a gid1b germinates at higher efficiency than both single mutants. This shows that GID1a can act as both a positive and negative regulator, depending on the circumstances. Together, these results show that GID1 paralogs can function as positive and negative regulators and that they can act up- or downstream and in parallel with each other. Because of the similarity of GA and SL signaling, it seems plausible that HTL/KAI2s can interact in similar ways. Once new genetic techniques have been developed for parasitic plants, it will be interesting to replicate these experiments on SL receptors.
Localization of Parasitic Plant HTL/KAI2 Expression and Activity Are Still Unclear
Another important part of the genetic context of parasitic plants is when and where receptors and interactors are expressed and/or activated. Unfortunately, research on this aspect is virtually missing. HTL/KAI2 expression has been investigated to some extent in S. hermonthica, S. asiatica and P. ramosa, whereas the expression of MAX2 is a mystery, and parasitic SMXLs remain unidentified (Tsuchiya et al. 2015, Yoshida et al. 2019, de Saint Germain et al. 2021).
Expression patterns of individual parasitic HTL/KAI2s have been investigated at multiple developmental stages in S. asiatica (Yoshida et al. 2019). Interestingly, only a subset of SaKAI2d receptors are expressed in the seed (10 of the 16 that were tested), whereas all of them were expressed at the early seedling stage, suggesting that some SaKAI2d paralogs still have other functions after germination. Indeed, it has been shown that P. japonicum KAI2d mediates chemotropism toward host SLs to ensure swift parasitization (Ogawa et al. 2022). Similar chemotropism has also been shown for certain arbuscular mycorrhizal (AM) fungi (Sbrana and Giovannetti 2005). Moreover, Around six SaKAI2d proteins remain expressed up to 3 d after germination, while none are expressed 7 d after germination (Yoshida et al. 2019), which fits with the timeframe for parasitization. Finally, SaKAI2i is expressed in the seed and seedling, while SaKAI2c is only expressed in the seedling (Yoshida et al. 2019). In short, these results suggest that S. asiatica HTL/KAI2s are functional in seed, as expected, but also remain active in the early stages of seedling growth.
In P. ramosa, PrKAI2c expression in the seed seems to be affected by both rac-GR24 and 2-PEITC (de Saint Germain et al. 2021). Six hours after treatment with these compounds, PrKAI2d1 and PrKAI2d2 are upregulated, and PrKAI2d3 and PrKAI2d4 transcripts are downregulated compared to control. In contrast, PrKAI2c expression is not affected. It seems counterintuitive that PrKAI2d3 is downregulated, since its function as SL/isothiocyanate receptor has been shown in relatively great detail (de Saint Germain et al. 2021). It could be argued that 6 h is enough to induce the germination process, after which PrKAI2d3 is no longer necessary. Oppositely, PrKAI2d1 and PrKAI2d2 may have other functions after germination, like some of the S. asiatica and P. japonicum receptors mentioned earlier, which would explain their upregulation.
HTL expression in S. hermonthica has been investigated indirectly by measuring fluorescence produced by YLGW hydrolysis in germinating seeds (Tsuchiya et al. 2015). As a consequence no differentiation could be made between different ShHTLs, the observed ‘expression’ pattern is essentially the pooled activity of all SL-hydrolyzing proteins that are present. Nevertheless, fluorescence starts within 20 min of YLGW addition, in the root tip of the plant embryo (Tsuchiya et al. 2015), suggesting that at least one SL-sensitive ShHTL is already expressed within the seed. Over a period of 6 h, called the wake-up wave, fluorescence spreads up toward the cotyledons (Tsuchiya et al. 2015). After the wake-up wave, fluorescence diminished in a period called the pre-gemination pause. Completion of the wake-up wave seems to be important, since YLGW is required for at least 6 h to efficiently induce seed germination, corresponding to the wake-up wave duration (Tsuchiya et al. 2015). At the 15-h mark, the root begins to elongate and fluorescence spreads across the entire root, which is called the elongation tide. These patterns suggest that either the ShHTL expression domain becomes larger or existing proteins somehow become hydrolytically active. The latter option is more likely since RT-PCR shows that the expression of ShHTLs is only weakly, if at all, upregulated upon the addition of rac-GR24 (Tsuchiya et al. 2015). This is in contrast with PrKAI2d expression. PrKAI2d expression changes were within 50% of control, indicating that the effect on ShHTLs may have been missed through a lack of replicates. Alternatively, these species may simply differ in this aspect.
These complex expression patterns are likely caused by feedback loops triggered by other endogenous signals (Tsuchiya et al. 2018). The addition of ethylene biosynthesis inhibitor aminoethoxyvinylglycine (AVG) or protein translation inhibitor cycloheximide (CHX) impairs SL-mediated seed germination. Furthermore, AVG reduces YLGW fluorescence, while CHX slows the wake-up phase (Tsuchiya et al. 2015). Based on these results, a working model was hypothesized (Tsuchiya et al. 2015, 2018): initial perception of SL in the root tip triggers a local amplification loop by inducing ethylene production, which in turn increases ShHTL catalytic activity. Then an unknown mobile factor ‘X’ is produced and transported up the root, activating additional ShHTLs. The production of mobile factor ‘X’ presumably requires protein translation. Next, the pre-germination pause is possibly caused by the degradation of ShHTL after SL perception, since ligand-dependent degradation occurs for AtD14 and AtKAI2 in a similar timeframe (Chevalier et al. 2014, Waters et al. 2015, Tsuchiya et al. 2018). Finally, SL-hydrolyzing activity returns, presumably through newly expressed ShHTLs. Crucially, Tsuchiya et al. hypothesize that the two waves of ShHTL activity and intermediate pause serve as an additional signal integration mechanism, which allows for further processing of exogenous SLs and ‘double check’ for the correct SLs (Tsuchiya et al. 2018). Since fluorescence assays cannot differentiate between ShHTL paralogs, it is tempting to speculate if they play different roles: could the wake-up phase be triggered by the highly sensitive ShHTL7, after which the more specific ShHTLs take over to fine-tune the response? More research is necessary to validate such hypotheses. Ideally, fluorescent tagging of individual ShHTLs could unravel their precise expression patterns. However, as of yet parasitic plants are too genetically intractable to achieve this. Development of more specific probes or reconstruction of the system in Arabidopsis are more achievable (Tsuchiya et al. 2018).
Conclusions and Future Research
It is clear that SLs are important determinants for parasitic plant germination and therefore host specificity. A lot of progress has been made toward understanding how this specificity works at the level of individual SLs and receptors. However, much more research is needed to elucidate how these receptors fit into the bigger picture: How are the signals generated by host- and non-host-associated SLs differentiated from each other, and on what basis is germination induced? Do all HTL/KAI2s act at the same time, or do they function in a certain order? Can HTL/KAI2s function as negative regulators? What signaling components lay downstream of MAX2 and SMXLs? Are SL-signaling pathways similar across different species in the Orobanchaceae, or is there a large variation? Important requirements to move this research forward are improvement of the regeneration of parasitic plants (in vitro) and making them more genetically tractable. Genetic modification of certain parasitic plants has been possible for >10 years (Tomilov et al. 2007, Ishida et al. 2011, Fernández-Aparicio et al. 2011a, Bandaranayake and Yoder 2018). However, only last year researchers managed to regenerate fertile plants (Maher et al. 2020, Nelson 2021). Another requirement is to have more genome sequences of parasitic plants to identify additional receptors and downstream signaling components, such as additional parasitic SMXLs, which have stayed elusive as of yet. It would be fascinating to see how the HTL/KAI2 receptor functioning in their natural background compares with all the heterologous studies reported so far.
Contributor Information
Sjors Huizinga, Plant Hormone Biology Group, Green Life Sciences Cluster, Swammerdam Institute for Life Science, University of Amsterdam, Science Park 904, Amsterdam 1098 XH, The Netherlands.
Harro J Bouwmeester, Plant Hormone Biology Group, Green Life Sciences Cluster, Swammerdam Institute for Life Science, University of Amsterdam, Science Park 904, Amsterdam 1098 XH, The Netherlands.
Data Availability
No new datasets were generated or analyzed in this study.
Funding
European Research Council Advanced grant CHEMCOMRHIZO 670211 (to H.J.B.); Dutch Research CouncilNederlandse Organisatie voor Wetenschappelijk Onderzoek(NWO/OCW) Gravitation program Harnessing the second genome of plants (MiCRop) 024.004.014 (to H.J.B.); University of Amsterdam Research Priority Area Systems Biology.
Disclosures
The authors have no conflicts of interest to declare.
References
- Akiyama K. and Hayashi H. (2006) Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann. Bot. 97: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K. and Hayashi H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. [DOI] [PubMed] [Google Scholar]
- Al-Babili S. and Bouwmeester H.J. (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66: 161–186. [DOI] [PubMed] [Google Scholar]
- Aliche E.B., Screpanti C., De Mesmaeker A., Munnik T. and Bouwmeester H.J. (2020) Science and application of strigolactones. New Phytol. 227: 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano-Saab A., Bunsick M., Al Galib H., Zhao W., Schuetz S., Bradley J.M., et al. (2021) Three mutations repurpose a plant karrikin receptor to a strigolactone receptor. Proc. Natl. Acad. Sci. USA 118: e2103175118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T., Umehara M., Ishikawa S., Hanada A., Maekawa M., Yamaguchi S., et al. (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Auger B., Pouvreau J.-B., Pouponneau K., Yoneyama K., Montiel G., Le Bizec B., et al. (2012) Germination stimulants of Phelipanche ramosa in the rhizosphere of Brassica napus are derived from the glucosinolate pathway. Mol. Plant-Microbe Interact. 25: 993–1004. [DOI] [PubMed] [Google Scholar]
- Bandaranayake P.C.G. and Yoder J.I. (2018) Factors affecting the efficiency of Rhizobium rhizogenes root transformation of the root parasitic plant Triphysaria versicolor and its host Arabidopsis thaliana. Plant Methods 14: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebawi F.F., Eplee R.E., Harris C.E. and Norris R.S. (1984) Longevity of witchweed (Striga asiatica) seed. Weed Sci. 32: 494–497. [Google Scholar]
- Berner O.K. (1995) Striga research and control: a perspective from Africa. Plant Dis. 79: 652–660. [Google Scholar]
- Besserer A., Bécard G., Jauneau A., Roux C. and Séjalon-Delmas N. (2008) GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 148: 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A., Puech-Pagès V., Kiefer P., Gomez-Roldan V., Jauneau A., Roy S., et al. (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez M.A., Nelson D.C. and Weijers D. (2020) Evolution of plant hormone response pathways. Annu. Rev. Plant Biol. 71: 327–353. [DOI] [PubMed] [Google Scholar]
- Bouwmeester H., Li C., Thiombiano B., Rahimi M. and Dong L. (2021) Adaptation of the parasitic plant lifecycle: germination is controlled by essential host signaling molecules. Plant Physiol. 185: 1292–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester H.J., Roux C., Lopez-Raez J.A. and Bécard G. (2007) Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 12: 224–230. [DOI] [PubMed] [Google Scholar]
- Brun G., Braem L., Thoiron S., Gevaert K., Goormachtig S. and Delavault P. (2018) Seed germination in parasitic plants: what insights can we expect from strigolactone research? J. Exp. Bot. 69: 2265–2280. [DOI] [PubMed] [Google Scholar]
- Bunsick M., Toh S., Wong C., Xu Z., Ly G., McErlean C.S.P., et al. (2020) SMAX1-dependent seed germination bypasses GA signalling in Arabidopsis and Striga. Nat. Plants 6: 646–652. [DOI] [PubMed] [Google Scholar]
- Bürger M. and Chory J. (2020) In‐silico analysis of the strigolactone ligand‐receptor system. Plant Direct 4: e00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler L. (1995) Chemical communication between the parasitic weed Striga and its crop host. InAllelopathy. Edited by Inderjit, A., Dakshini, K. and Einhellig, F. pp. 158–168. American Chemical Society: Washington DC, ACS Symposiun Series 582. [Google Scholar]
- Bythell-Douglas R., Rothfels C.J., Stevenson D.W.D., Graham S.W., Wong G.K.-S., Nelson D.C., et al. (2017) Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biol. 15: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cala A., Molinillo J.M.G., Fernández-Aparicio M., Ayuso J., Álvarez J.A., Rubiales D., et al. (2017) Complexation of sesquiterpene lactones with cyclodextrins: synthesis and effects on their activities on parasitic weeds. Org. Biomol. Chem. 15: 6500–6510. [DOI] [PubMed] [Google Scholar]
- Chen J., White A., Nelson D.C. and Shukla D. (2021) Role of substrate recognition in modulating strigolactone receptor selectivity in witchweed. J. Biol. Chem. 297: 101092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F., Nieminen K., Sánchez-Ferrero J.C., Rodríguez M.L., Chagoyen M., Hardtke C.S., et al. (2014) Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26: 1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn C.E., Bythell-Douglas R., Neumann D., Yoshida S., Whittington B., Westwood J.H., et al. (2015) Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349: 540–543. [DOI] [PubMed] [Google Scholar]
- Conn C.E. and Nelson D.C. (2016) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 6: 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C.E., Whichard L.P., Turner B., Wall M.E. and Egley G.H. (1966) Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154: 1189–1190. [DOI] [PubMed] [Google Scholar]
- Czarnecki O., Yang J., Weston D., Tuskan G. and Chen J.-G. (2013) A dual role of strigolactones in phosphate acquisition and utilization in plants. Int. J. Mol. Sci. 14: 7681–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafaallah A.B. (2020) Variability and host specificity of Striga hermonthica (Del.) Benth. in response to in-situ root exudates of Sorghum bicolor (L.) Moench. J. Res. Weed Sci. 3: 238–253. [Google Scholar]
- Das M., Fernández-Aparicio M., Yang Z., Huang K., Wickett N.J., Alford S., et al. (2015) Parasitic plants Striga and Phelipanche dependent upon exogenous strigolactones for germination have retained genes for strigolactone biosynthesis. Am. J. Plant Sci. 06: 1151–1166. [Google Scholar]
- de Luque A.P., Galindo J.C.G., Macías F.A. and Jorrín J. (2000) Sunflower sesquiterpene lactone models induce Orobanche cumana seed germination. Phytochemistry 53: 45–50. [DOI] [PubMed] [Google Scholar]
- de Saint Germain A., Clavé G., Badet-Denisot M.-A., Pillot J.-P., Cornu D., Le Caer J.-P., et al. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol. 12: 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A., Jacobs A., Brun G., Pouvreau J.-B., Braem L., Cornu D., et al. (2021) A Phelipanche ramosa KAI2 protein perceives strigolactones and isothiocyanates enzymatically. Plant Commun. 2: 100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejeta G. and Gressel J. (2007) Integrating New Technologies for Striga Control: Towards Ending the Witch-Hunt. World Scientific: Singapore. [Google Scholar]
- Fernández-Aparicio M., Flores F. and Rubiales D. (2009) Recognition of root exudates by seeds of broomrape (Orobanche and Phelipanche) species. Ann. Bot. 103: 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Aparicio M., Rubiales D., Bandaranayake P.C., Yoder J.I. and Westwood J.H. (2011a) Transformation and regeneration of the holoparasitic plant Phelipanche aegyptiaca. Plant Methods 7: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Aparicio M., Yoneyama K. and Rubiales D. (2011b) The role of strigolactones in host specificity of Orobanche and Phelipanche seed germination. Seed Sci. Res. 21: 55–61. [Google Scholar]
- Flematti G.R. (2004) A compound from smoke that promotes seed germination. Science 305: 977. [DOI] [PubMed] [Google Scholar]
- Flematti G.R., Scaffidi A., Waters M.T. and Smith S.M. (2016) Stereospecificity in strigolactone biosynthesis and perception. Planta 243: 1361–1373. [DOI] [PubMed] [Google Scholar]
- Ge W. and Steber C.M. (2018) Positive and negative regulation of seed germination by the Arabidopsis GA hormone receptors, GID1a, b, and c. Plant Direct 2: e00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibot-Leclerc S., Corbineau F., Sallé G. and Côme D. (2004) Responsiveness of Orobanche ramosa L. seeds to GR 24 as related to temperature, oxygen availability and water potential during preconditioning and subsequent germination. Plant Growth Regul. 43: 63–71. [Google Scholar]
- Gibot-Leclerc S., Dessaint F., Reibel C. and Le Corre V. (2013) Phelipanche ramosa (L.) Pomel populations differ in life-history and infection response to hosts. Flora - Morphol. Distrib. Funct. Ecol. Plants 208: 247–252. [Google Scholar]
- Gobena D., Shimels M., Rich P.J., Ruyter-Spira C., Bouwmeester H., Kanuganti S., et al. (2017) Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc. Natl. Acad. Sci. USA114: 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V., Fermas S., Brewer P.B., Puech-Pagès V., Dun E.A., Pillot J.-P., et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194. [DOI] [PubMed] [Google Scholar]
- Hamiaux C., Drummond R.S.M., Janssen B.J., Ledger S.E., Cooney J.M., Newcomb R.D., et al. (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22: 2032–2036. [DOI] [PubMed] [Google Scholar]
- Harms M.J., Eick G.N., Goswami D., Colucci J.K., Griffin P.R., Ortlund E.A., et al. (2013) Biophysical mechanisms for large-effect mutations in the evolution of steroid hormone receptors. Proc. Natl. Acad. Sci. USA 110: 11475–11480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale A.L. and Steber C.M. (2020) GA signaling is essential for the embryo-to-seedling transition during Arabidopsis seed germination, a ghost story. Plant Signal. Behav. 15: 1705028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearne S.J. (2009) Control-the Striga conundrum. Pest Manag. Sci. 65: 603–614. [DOI] [PubMed] [Google Scholar]
- Huang K., Whitlock R., Press M.C. and Scholes J.D. (2012) Variation for host range within and among populations of the parasitic plant Striga hermonthica. Heredity 108: 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., He Y., Wang L., Liu S., Meng X., Liu G., et al. (2017) DWARF14, a receptor covalently linked with the active form of strigolactones, undergoes strigolactone-dependent degradation in rice. Front. Plant Sci. 8: 1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida J.K., Yoshida S., Ito M., Namba S. and Shirasu K. (2011) Agrobacterium rhizogenes-mediated transformation of the parasitic plant Phtheirospermum japonicum. PLoS One 6: e25802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel D.M., Chaudhuri S.K., Plakhine D., Ziadna H. and Steffens J.C. (2011) Dehydrocostus lactone is exuded from sunflower roots and stimulates germination of the root parasite Orobanche cumana. Phytochemistry 72: 624–634. [DOI] [PubMed] [Google Scholar]
- Khosla A., Morffy N., Li Q., Faure L., Chang S.H., Yao J., et al. (2020) Structure–function analysis of SMAX1 reveals domains that mediate its karrikin-induced proteolysis and interaction with the receptor KAI2. Plant Cell 32: 2639–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Westerhuis J.A., Smilde A.K., Floková K., Suleiman A.K.A., Kuramae E.E., et al. (2022) Effect of strigolactones on recruitment of the rice root-associated microbiome. FEMS Microbiol. Ecol. 98: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae Y., Kameoka H., Sugimura Y., Saito K., Ohtomo R., Fujiwara T., et al. (2018) Strigolactone biosynthesis genes of rice are required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant Cell Physiol. 59: 544–553. [DOI] [PubMed] [Google Scholar]
- Kochanek J., Long R.L., Lisle A.T. and Flematti G.R. (2016) Karrikins identified in biochars indicate post-fire chemical cues can influence community diversity and plant development. PLoS One 11: e0161234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W., Charnikhova T., Liu Q., Bours R., Domagalska M.A., Beguerie S., et al. (2011) Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 155: 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai H., Dor E., Hershenhorn J., Joel D.M., Weininger S., Lekalla S., et al. (2010) Strigolactones’ effect on root growth and root-hair elongation may be mediated by auxin-efflux carriers. J. Plant Growth Regul. 29: 129–136. [Google Scholar]
- Lanfranco L., Fiorilli V., Venice F. and Bonfante P. (2018) Strigolactones cross the kingdoms: plants, fungi, and bacteria in the arbuscular mycorrhizal symbiosis. J. Exp. Bot. 69: 2175–2188. [DOI] [PubMed] [Google Scholar]
- Lechat -M.-M., Pouvreau J.-B., Peron T., Gauthier M., Montiel G., Veronesi C., et al. (2012) PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J. Exp. Bot. 63: 5311–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.W., Sharma P., Janssen B.J., Drummond R.S.M., Luo Z., Hamiaux C., et al. (2020) Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners. J. Biol. Chem. 295: 4181–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Martín-Fontecha E.S., Khosla A., White A.R.F., Chang S., Cubas P., et al. (2022) The strigolactone receptor D14 targets SMAX1 for degradation in response to GR24 treatment and osmotic stress. Plant Commun. 3: 100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhang Y., Matusova R., Charnikhova T., Amini M., Jamil M., et al. (2014) striga hermonthicamax2 restores branching but not the very low fluence response in the arabidopsis thaliana max2 mutant. New Phytol. 202: 531–541. [DOI] [PubMed] [Google Scholar]
- López‐Ráez J.A., Charnikhova T., Gómez‐Roldán V., Matusova R., Kohlen W., De Vos R., et al. (2008) Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 178: 863–874. [DOI] [PubMed] [Google Scholar]
- Maher M.F., Nasti R.A., Vollbrecht M., Starker C.G., Clark M.D. and Voytas D.F. (2020) Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38: 84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusova R., Mourik van T. and Bouwmeester H.J. (2004) Changes in the sensitivity of parasitic weed seeds to germination stimulants. Seed Sci. Res. 14: 335–344. [Google Scholar]
- Mohemed N., Charnikhova T., Fradin E.F., Rienstra J., Babiker A.G.T. and Bouwmeester H.J. (2018) Genetic variation in Sorghum bicolor strigolactones and their role in resistance against Striga hermonthica. J. Exp. Bot. 69: 2415–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Xue Y.-L., Miyakawa T., Hou F., Qin H.-M., Fukui K., et al. (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4: 2613. [DOI] [PubMed] [Google Scholar]
- Nelson D.C. (2021) The mechanism of host-induced germination in root parasitic plants. Plant Physiol. 185: 1353–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.C., Flematti G.R., Ghisalberti E.L., Dixon K.W. and Smith S.M. (2012) Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu. Rev. Plant Biol. 63: 107–130. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Scaffidi A., Dun E.A., Waters M.T., Flematti G.R., Dixon K.W., et al. (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura S., Nakashima H., Mizutani M., Takikawa H. and Sugimoto Y. (2013) Structural requirements of strigolactones for germination induction and inhibition of Striga gesnerioides seeds. Plant Cell Rep. 32: 829–838. [DOI] [PubMed] [Google Scholar]
- Ogawa S., Cui S., White A.R.F., Nelson D.C., Yoshida S. and Shirasu K. (2022) Strigolactones are chemoattractants for host tropism in Orobanchaceae parasitic plants. Nat. Commun. 13: 4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. (2009) Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag. Sci. 65: 453–459. [DOI] [PubMed] [Google Scholar]
- Parker C. (2012) Parasitic weeds: a world challenge. Weed Sci. 60: 269–276. [Google Scholar]
- Peláez-Vico M.A., Bernabéu-Roda L., Kohlen W., Soto M.J. and López-Ráez J.A. (2016) Strigolactones in the Rhizobium-legume symbiosis: stimulatory effect on bacterial surface motility and down-regulation of their levels in nodulated plants. Plant Sci. 245: 119–127. [DOI] [PubMed] [Google Scholar]
- Rahimi M. and Bouwmeester H. (2021) Are sesquiterpene lactones the elusive KARRIKIN-INSENSITIVE2 ligand? Planta 253: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raupp F.M. and Spring O. (2013) New sesquiterpene lactones from sunflower root exudate as germination stimulants for Orobanche cumana. J. Agric. Food Chem. 61: 10481–10487. [DOI] [PubMed] [Google Scholar]
- Rodenburg J., Demont M., Zwart S.J. and Bastiaans L. (2016) Parasitic weed incidence and related economic losses in rice in Africa. Agric. Ecosyst. Environ. 235: 306–317. [Google Scholar]
- Ruyter-Spira C., Kohlen W., Charnikhova T., van Zeijl A., van Bezouwen L., de Ruijter N., et al. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones?. Plant Physiol. 155: 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrana C. and Giovannetti M. (2005) Chemotropism in the arbuscular mycorrhizal fungus Glomus mosseae. Mycorrhiza 15: 539–545. [DOI] [PubMed] [Google Scholar]
- Seto Y., Yasui R., Kameoka H., Tamiru M., Cao M., Terauchi R., et al. (2019) Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat. Commun. 10: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N., Ticchiarelli F., Mao H., Hinds T.R., Leyser O. and Zheng N. (2018) Structural plasticity of D3–D14 ubiquitin ligase in strigolactone signalling. Nature 563: 652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahul Hameed U., Haider I., Jamil M., Kountche B.A., Guo X., Zarban R.A., et al. (2018) Structural basis for specific inhibition of the highly sensitive Sh htl 7 receptor. EMBO Rep. 19: e45619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Nakatsu T., Nakajima M., Naoe Y., Ohmiya H., et al. (2008) Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523. [DOI] [PubMed] [Google Scholar]
- Song W.J., Zhou W.J., Jin Z.L., Cao D.D., Joel D.M., Takeuchi Y., et al. (2005) Germination response of Orobanche seeds subjected to conditioning temperature, water potential and growth regulator treatments. Weed Res. 45: 467–476. [Google Scholar]
- Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J.P., Abbas A., et al. (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27: 3143–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek T., Mutuku M. and Shirasu K. (2013) The genus Striga: a witch profile: profile of Striga, the witchweed. Mol. Plant Pathol. 14: 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga J.P., Morffy N. and Nelson D.C. (2016) Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243: 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga J.P., Smith S.M., Briggs W.R. and Nelson D.C. (2013) SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 163: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.-D. and Ni M. (2011) HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Mol. Plant 4: 116–126. [DOI] [PubMed] [Google Scholar]
- Swarbreck S.M., Guerringue Y., Matthus E., Jamieson F.J.C. and Davies J.M. (2019) Impairment in karrikin but not strigolactone sensing enhances root skewing in Arabidopsis thaliana. Plant J. 98: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei S., Uchiyama Y., Bürger M., Suzuki T., Okabe S., Chory J., et al. (2023) A divergent clade KAI2 protein in the root parasitic plant Orobanche minor is a highly sensitive strigolactone receptor and is involved in the perception of sesquiterpene lactones. Plant Cell Physiol. pcad026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorogood C.J., Rumsey F.J. and Hiscock S.J. (2009) Host-specific races in the holoparasitic angiosperm Orobanche minor: implications for speciation in parasitic plants. Ann. Bot. 103: 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S., Holbrook-Smith D., Stogios P.J., Onopriyenko O., Lumba S., Tsuchiya Y., et al. (2015) Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350: 203–207. [DOI] [PubMed] [Google Scholar]
- Toh S., Kamiya Y., Kawakami N., Nambara E., McCourt P. and Tsuchiya Y. (2012) Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant Cell Physiol. 53: 107–117. [DOI] [PubMed] [Google Scholar]
- Tomilov A., Tomilova N. and Yoder J.I. (2007) Agrobacterium tumefaciens and Agrobacterium rhizogenes transformed roots of the parasitic plant Triphysaria versicolor retain parasitic competence. Planta 225: 1059–1071. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Yoshimura M. and Hagihara S. (2018) The dynamics of strigolactone perception in Striga hermonthica: a working hypothesis. J. Exp. Bot. 69: 2281–2290. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Yoshimura M., Sato Y., Kuwata K., Toh S., Holbrook-Smith D., et al. (2015) Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349: 864–868. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Motoyuki A. and Matsuoka M. (2007) Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58: 183–198. [DOI] [PubMed] [Google Scholar]
- Ueno K., Furumoto T., Umeda S., Mizutani M., Takikawa H., Batchvarova R., et al. (2014) Heliolactone, a non-sesquiterpene lactone germination stimulant for root parasitic weeds from sunflower. Phytochemistry 108: 122–128. [DOI] [PubMed] [Google Scholar]
- Umehara M. (2011) Strigolactone, a key regulator of nutrient allocation in plants. Plant Biotechnol. 28: 429–437. [Google Scholar]
- Umehara M., Hanada A., Magome H., Takeda-Kamiya N. and Yamaguchi S. (2010) Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 51: 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200. [DOI] [PubMed] [Google Scholar]
- Uraguchi D., Kuwata K., Hijikata Y., Yamaguchi R., Imaizumi H., Am S., et al. (2018) A femtomolar-range suicide germination stimulant for the parasitic plant Striga hermonthica. Science 362: 1301–1305. [DOI] [PubMed] [Google Scholar]
- Villaécija-Aguilar J.A., Hamon-Josse M., Carbonnel S., Kretschmar A., Schmidt C., Dawid C., et al. (2019) SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet. 15: e1008327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele A., Linkies A., Müller K. and Leubner-Metzger G. (2011) Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination. J. Exp. Bot. 62: 5131–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler R.K., Ejeta G. and Butler L.G. (1996) Inheritance of low production of Striga germination stimulant in sorghum. Crop Sci. 36: 1185–1191. [Google Scholar]
- Wang Y. and Bouwmeester H.J. (2018) Structural diversity in the strigolactones. J. Exp. Bot. 69: 2219–2230. [DOI] [PubMed] [Google Scholar]
- Wang D., Pang Z., Yu H., Thiombiano B., Walmsley A., Yu S., et al. (2022) Probing strigolactone perception mechanisms with rationally designed small-molecule agonists stimulating germination of root parasitic weeds. Nat. Commun. 13: 3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang B., Jiang L., Liu X., Li X., Lu Z., et al. (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27: 3128–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xu Q., Yu H., Ma H., Li X., Yang J., et al. (2020) Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis. Plant Cell 32: 2251–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yao R., Du X., Guo L., Chen L., Xie D., et al. (2021) Molecular basis for high ligand sensitivity and selectivity of strigolactone receptors in Striga. Plant Physiol. 185: 1411–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Gutjahr C., Bennett T. and Nelson D.C. (2017) Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68: 291–322. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Nelson D.C., Scaffidi A., Flematti G.R., Sun Y.K., Dixon K.W., et al. (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Scaffidi A., Flematti G. and Smith S.M. (2015) Substrate-induced degradation of the α/β-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol. Plant 8: 814–817. [DOI] [PubMed] [Google Scholar]
- Xiong G., Li J. and Smith S.M. (2016) Evolution of strigolactone perception by seeds of parasitic plants: reinventing the wheel. Mol. Plant 9: 493–495. [DOI] [PubMed] [Google Scholar]
- Xu Y., Miyakawa T., Nakamura H., Nakamura A., Imamura Y., Asami T., et al. (2016) Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica. Sci. Rep. 6: 31386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Miyakawa T., Nosaki S., Nakamura A., Lyu Y., Nakamura H., et al. (2018) Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat. Commun. 9: 3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R., Ming Z., Yan L., Li S., Wang F., Ma S., et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536: 469–473. [DOI] [PubMed] [Google Scholar]
- Yao R., Wang L., Li Y., Chen L., Li S., Du X., et al. (2018) Rice DWARF14 acts as an unconventional hormone receptor for strigolactone. J. Exp. Bot. 69: 2355–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R., Wang F., Ming Z., Du X., Chen L., Wang Y., et al. (2017) ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res. 27: 838–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K., Xie X., Yoneyama K. and Takeuchi Y. (2009) Strigolactones: structures and biological activities. Pest Manag. Sci. 65: 467–470. [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Yoneyama K., Takeuchi Y. and Sekimoto H. (2007) Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 225: 1031–1038. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Cui S., Ichihashi Y. and Shirasu K. (2016) The haustorium, a specialized invasive organ in parasitic plants. Annu. Rev. Plant Biol. 67: 643–667. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Kim S., Wafula E.K., Tanskanen J., Kim Y.-M., Honaas L., et al. (2019) Genome sequence of Striga asiatica provides insight into the evolution of plant parasitism. Curr. Biol. 29: 3041–3052.e4. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang D., Shen Y. and Xi Z. (2020) Crystal structure and biochemical characterization of Striga hermonthica HYPO-SENSITIVE TO LIGHT 8 (ShHTL8) in strigolactone signaling pathway. Biochem. Biophys. Res. Commun. 523: 1040–1045. [DOI] [PubMed] [Google Scholar]
- Zhao L.-H., Zhou X.E., Yi W., Wu Z., Liu Y., Kang Y., et al. (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 25: 1219–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new datasets were generated or analyzed in this study.


