Abstract
Strigolactones (SLs) are a class of plant hormones that regulate many aspects of plant growth and development. SLs also improve symbiosis with arbuscular mycorrhizal fungi (AMF) in the rhizosphere. Recent studies have shown that the DWARF14-LIKE (D14L)/KARRIKIN-INSENSITIVE2 (KAI2) family, paralogs of the SL receptor D14, are required for AMF colonization in several flowering plants, including rice. In this study, we found that (−)-GR5, a 2′S-configured enantiomer of a synthetic SL analog (+)-GR5, significantly activated SL biosynthesis in rice roots via D14L. This result is consistent with a recent report, showing that the D14L pathway positively regulates SL biosynthesis in rice. In fact, the SL levels tended to be lower in the roots of the d14l mutant under both inorganic nutrient-deficient and -sufficient conditions. We also show that the increase in SL levels by (−)-GR5 was observed in other mycorrhizal plant species. In contrast, the KAI2 pathway did not upregulate the SL level and the expression of SL biosynthetic genes in Arabidopsis, a non-mycorrhizal plant. We also examined whether the KAI2 pathway enhances SL biosynthesis in the liverwort Marchantia paleacea, where SL functions as a rhizosphere signaling molecule for AMF. However, the SL level and SL biosynthetic genes were not positively regulated by the KAI2 pathway. These results imply that the activation of SL biosynthesis by the D14L/KAI2 pathway has been evolutionarily acquired after the divergence of bryophytes to efficiently promote symbiosis with AMF, although we cannot exclude the possibility that liverworts have specifically lost this regulatory system.
Keywords: Arabidopsis thaliana, Arbuscular mycorrhizal fungi, Biosynthesis, Oryza sativa, Strigolactone, Symbiosis
Introduction
Strigolactones (SLs), carotenoid-derived small compounds, have been initially isolated as germination stimulants for root parasitic plants (Xie et al. 2010). SLs were later identified as root-derived symbiotic signals for arbuscular mycorrhizal fungi (AMF). SLs stimulate hyphal branching, hyphal growth and spore germination in AMF (Akiyama et al. 2005, Besserer et al. 2006). The rhizosphere signaling molecule for AMF has been recently shown as the ancestral function of SLs using the liverwort Marchantia paleacea (Kodama et al. 2022). In addition, SLs were identified as a class of plant hormones that inhibit shoot branching (Gomez-Roldan et al. 2008, Umehara et al. 2008). Many studies have shown that SLs regulate not only shoot branching but also various other developmental processes and environmental responses (Waters et al. 2017, Wu et al. 2022).
SLs are biosynthesized from all-trans-β-carotene or related carotenoids. The sequential action of DWARF27 (D27), carotenoid cleavage dioxygenase 7 (CCD7) and CCD8 converts all-trans-β-carotene to carlactone (CL) that contains an enol ether bond and the D ring, both of which are critical for the biological activity of SLs (Alder et al. 2012, Seto et al. 2014). Then, CYP711A, a subfamily of cytochrome P450 oxygenases, catalyzes the conversion from CL to carlactonoic acid (CLA) (Abe et al. 2014, Yoneyama et al. 2018). Recently, some members of CYP711A and CYP722C subfamilies have been shown to convert CLA into canonical SLs in several plant species (Zhang et al. 2014, Wakabayashi et al. 2019, Mori et al. 2020). In rice, CYP711A2/Os900 converts CL into 4-deoxyorobanchol (4DO) and CYP711A3/Os1400 converts 4DO into orobanchol (ORO) (Zhang et al. 2014). Furthermore, a SABATH methyltransferase (CLAMT) has been reported to convert CLA into a non-canonical SL, methyl carlactonoate (MeCLA) in Arabidopsis (Wakabayashi et al. 2021, Mashiguchi et al. 2022). MeCLA was shown to be converted to CLA and hydroxymethyl carlactonoate (1ʹ-OH-MeCLA) by the LATERAL BRANCHING OXIDOREDUCTASE (LBO) family of 2-oxoglutarate-dependent dioxygenases in several plant species (Yoneyama et al. 2020).
Various endogenous and external stimuli regulate SL biosynthesis (Mashiguchi et al. 2021). The phosphate (P) deficiency signal is one of the environmental factors that drastically influence SL production in many plant species (Lopez-Raez et al. 2008, Umehara et al. 2010, Liu et al. 2011, Yoneyama et al. 2012). The nitrogen (N) deficiency also increases SL production, but this response is not universally conserved among plant species (Liu et al. 2011, Yoneyama et al. 2012). The expression of SL biosynthetic genes such as D27 and CYP711A is increased under the P-deficient condition (Umehara et al. 2010, Liu et al. 2011, Mori et al. 2020). Because SLs are symbiotic signals that contribute to the colonization with AMF, which enables host plants to take up inorganic nutrients, it has been proposed that the enhancement of SL biosynthesis under the P-deficient condition is the nutrient acquisition strategy of plants in the rhizosphere (Andreo-Jimenez et al. 2015). In addition, it has been proposed that endogenous SLs accumulated under P deficiency play a hormonal role in inhibiting tiller bud outgrowth and regulating leaf senescence to efficiently utilize the available P in rice (Umehara et al. 2010, Yamada et al. 2014).
Karrikins (KARs) are butenolide molecules isolated from smoke as seed germination stimulants (Flematti et al. 2004). The DWARF14-LIKE (D14L)/KARRIKIN-INSENSITIVE2 (KAI2) family of α/β-fold hydrolases, which is homologous to the D14 family of SL receptors, has been shown to act as a receptor for KARs and an as-yet-unknown endogenous KAI2 ligand (KL) (Waters et al. 2012, 2015, Conn and Nelson 2016). The D14L/KAI2 and D14 signaling pathways are highly similar in which both D14L/KAI2 and D14 receptors form ternary complexes with a common F-box protein, D3/MORE AXILLARY GROWTH2 (MAX2) and D53/SUPPRESSOR OF MAX2 1-LIKE (SMXL) repressors upon ligand binding. Then, the SCFD3/MAX2 ubiquitin ligase polyubiquitinates D53/SMXLs, which are further degraded by the proteasome (Temmerman et al. 2022). It has been shown that D14L and KAI2 receptors target OsSMAX1 and SMAX1/SMXL2 repressors in rice (Zheng et al. 2020) and Arabidopsis (Stanga et al. 2016), respectively. In addition to the similarity of protein structures and signaling pathways of D14L/KAI2 and D14 receptors, the D14L/KAI2 family has been shown to have selectivity for synthetic SL derivatives with a C-2′S configuration, which possess opposite stereochemistry to natural SLs with a C-2′R configuration. Among four GR24 stereoisomers, 2′S-configured (−)-ent-GR24 (also known as GR24ent-5DS) is preferentially perceived by D14L/KAI2 proteins as well as KARs (or their metabolites) in several plant species including rice (Scaffidi et al. 2014, Carbonnel et al. 2020, Zheng et al. 2020). Furthermore, KAI2 proteins have been shown to interact directly with (−)-ent-GR24 in Arabidopsis, Lotus japonicus, Marchantia polymorpha and pea (Waters et al. 2015, Carbonnel et al. 2020, Mizuno et al. 2021, Guercio et al. 2022).
In rice, the D14L pathway is involved in mesocotyl elongation in the dark (Kameoka and Kyozuka 2015). It has also been shown that the D14L pathway is essential for symbiosis with AMF in rice because D14L- and D3-defective mutants could not establish AMF colonization (Yoshida et al. 2012, Gutjahr et al. 2015). The phenotype of these mutants is unique as the responsiveness to AMF was severely impaired, and this is different from that of SL biosynthetic mutants in rice, in which AMF colonization is reduced because of the attenuated hyphopodium formation, but most colonization processes are normal (Yoshida et al. 2012, Kobae et al. 2018). Recent studies have also demonstrated that the rice SMAX1, OsSMAX1, functions as a repressor of the D14L pathway in mesocotyl elongation and symbiosis with AMF (Choi et al. 2020, Zheng et al. 2020). Interestingly, the expression of SL biosynthetic genes and the levels of SLs [4DO and methoxy-5-deoxystrigol (methoxy-5DS)] were upregulated by the smax1 mutation (Choi et al. 2020). These findings suggest that the D14L pathway positively regulates SL biosynthesis in rice. However, the detailed mechanism of this regulatory system needs to be clarified. It is also unknown whether the interaction between the D14L/KAI2 pathway and SL biosynthesis is observed in other plant species.
In this study, following our finding that (−)-GR5, an enantiomer of the synthetic SL analog (+)-GR5, induced SL biosynthesis in rice, we performed a series of experiments on the D14L-mediated activation of SL biosynthesis in rice. Furthermore, we analyzed the relationship between the D14L/KAI2 pathway and SL biosynthesis in mycorrhizal- and non-mycorrhizal plant species.
Results and Discussion
Positive regulation of SL biosynthesis by 2′S-configured stereoisomers of synthetic SL derivatives is mediated by the D14L pathway in rice
We performed a chemical screen for compounds that alter endogenous SL levels using a commercially available chemical library and SL-related compounds. We used the rice SL receptor–defective d14-1 mutant (cv. Shiokari) grown under P-deficient conditions for two reasons. First, the d14 mutant grown under P-deficient conditions strongly increases SL levels (Seto et al. 2014), which is advantageous for analyzing SLs. Second, the chemicals with SL activities that reduce SL levels by negative feedback regulation of SL biosynthesis can be excluded by using the d14 mutant.
In the process of screening, we found that (−)-GR5, a 2′S-configured enantiomer of a synthetic SL analog (+)-GR5 (Johnson et al. 1976), exhibited stronger activities than 2′R -configured (+)-GR5 in increasing 4DO levels in root exudates and extracts (Fig. 1A, B). The increase in endogenous 4DO levels by (−)-GR5 was also observed in the wild type (WT) and under a P-sufficient condition (Supplementary Fig. S1A, B). Furthermore, similar responses to the GR5 stereoisomers were observed in root exudates and extracts of the d14-1N mutant (cv. Nipponbare), in which not only 4DO but also ORO is produced (Supplementary Fig. S1C). We also found that 2′S-configured stereoisomers of GR7 and GR24 [(+)-2′-epi-GR7, (−)-ent-GR7 and (−)-ent-GR24] showed stronger activities than 2′R -configured compounds in increasing 4DO levels, but none of the dihydroGR24 stereoisomers, in which the double bond in the enol ether bridge is reduced to a single bond, showed the activity (Supplementary Fig. S2). These results indicate that both the S configuration at the C-2′ position and the enol ether bridge are important structures in increasing SL levels. It is noted that 2′S-configured (+)-2′-epi-GR24 did not show the activity, suggesting that the stereochemistry of the B–C ring junction also influences the activity of GR24 which has a larger molecular size than GR5 and GR7.
Fig. 1.
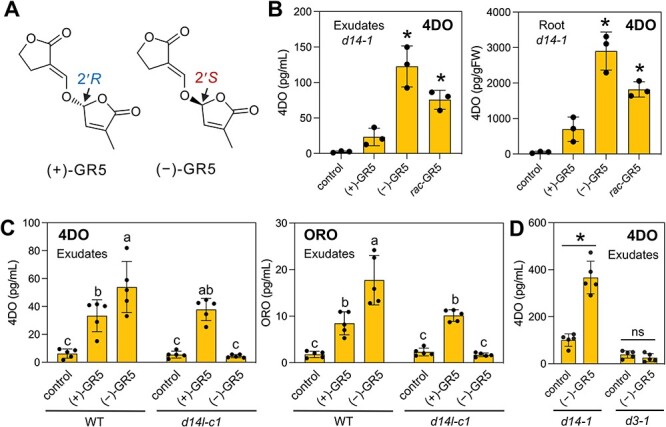
D14L-dependent upregulation of SL levels by (−)-GR5 in rice. (A) Chemical structures of GR5 stereoisomers. (B) The effect of GR5 stereoisomers on 4DO levels in root exudates and roots of d14-1 plants. rac-GR5 is a racemic mixture of GR5 stereoisomers. (C) The effect of GR5 stereoisomers on 4DO and ORO levels in root exudates of WT and d14l-c1 seedlings. (D) The effect of GR5 stereoisomers on 4DO levels in root exudates of d14-1 and d3-1 seedlings. In (B–D), rice seedlings grown hydroponically under P-deficient conditions were treated with 10 μM of each compound for 24 h. Root exudates analyzed contain SLs released into the medium for 24 h. Values are means ± SDs [n = 3 (B) and n = 5 (C, D)]. Significant differences (P < 0.05) compared to control are indicated with asterisks [one-way ANOVA followed by Dunnett’s test in (B) or the t-test in (D)]. Different letters indicate significant differences in C (P < 0.05, Tukey’s Honestly Significant Difference (HSD)).
We speculated that other factors than D14 are involved in the phenomena mentioned earlier because these responses were observed in the d14 mutants. Then, we focused on D14L, a paralog of D14 in rice. The D14 and D14L/KAI2 families have been shown to perceive preferentially synthetic SL derivatives with the 2′R and 2′S configurations, respectively (Scaffidi et al. 2014, Umehara et al. 2015). Among four GR24 stereoisomers, the D14L/KAI2 family preferentially uses (−)-ent-GR24 as a ligand, as observed in Supplementary Fig. S2A (Scaffidi et al. 2014, Mizuno et al. 2021). From these pieces of evidence, we speculated that D14L is responsible for the response to 2′S -configured SL derivatives. So, we analyzed the D14L-defective mutant, d14l-c1, generated using the CRISPR/Cas9 system (Supplementary Fig. S3A). Although we used only one d14l allele in which off-target mutation(s) was not analyzed in this study, the d14l-c1 mutant and the d3 mutant lost the ability to interact with AMF as previously reported (Yoshida et al. 2012, Gutjahr et al. 2015) (Supplementary Fig. S3B). As expected, we found that the d14l-c1 mutant was completely insensitive to (−)-GR5 in increasing both 4DO and ORO production (Fig. 1C), consistent with a recent report that the D14L pathway positively regulates SL biosynthesis in rice (Choi et al. 2020). The importance of the D14L pathway in inducing SL biosynthesis is supported by the observation that the 4DO level was not increased after (−)-GR5 treatment in the D3-deficient mutant (Fig. 1D). Interestingly, the 4DO-inducing activity of (+)-GR5 was observed in the d14l mutant as in the d14 mutant (Fig. 1B, C). Future analysis using the d3 mutant will provide a clue to determine whether D3 is involved in the response to (+)-GR5. It will also be important to examine whether other D14-related proteins, D14L2a and D14L2b, have a role in the response to (+)-GR5 in rice (Waters et al. 2012, Sisaphaithong et al. 2021).
We next investigated the expression of SL biosynthetic genes in the roots of d14 mutants treated with (−)-GR5 by quantitative RT-PCR (qRT-PCR). We first confirmed that (−)-GR5 treatment drastically increased the expression of D14L2a (36.9-fold), a marker gene of the D14L pathway (Zheng et al. 2020), supporting the idea that (−)-GR5 mimics KL and activates D14L-mediated signaling (Supplementary Fig. S4). The (−)-GR5 treatment also significantly induced the expression levels of D27, D17/CCD7, CYP711A2 and CYP711A3 genes, whereas D10/CCD8 expression was not increased. In addition, a CLAMT homolog, Os01g0700300, was upregulated among rice homologs of Arabidopsis CLAMT and LBO genes (Brewer et al. 2016, Mashiguchi et al. 2022) (Fig. 2A, Supplementary Fig. S4). These results matched well with our observation that endogenous levels of both canonical SLs (4DO and ORO) and a non-canonical SL (MeCLA), as well as those of biosynthetic intermediates (CL and CLA), were elevated in (−)-GR5-treated WT, but not in the d14l-c1 mutant (Fig. 2B). In conclusion, our results revealed that 2′S -configured (−)-GR5 could enhance SL biosynthesis via D14L-mediated transcriptional upregulation of its biosynthetic genes in rice roots. Because the activation of SL biosynthesis was observed even in the d14 mutant, D14-mediated feedback inhibition and D14L-mediated activation of SL biosynthesis likely use independent mechanisms.
Fig. 2.
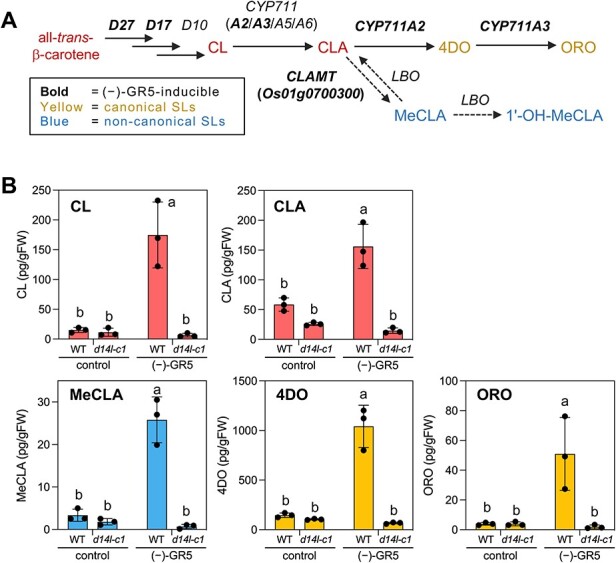
The upregulation of SL biosynthetic genes is linked with the D14L-mediated increase of endogenous SL levels by (−)-GR5 in rice. (A) The proposed SL biosynthesis pathway in rice. The dotted arrows indicate metabolic conversions that have not been experimentally clarified in rice. The genes whose expression was induced by (−)-GR5 are shown in bold (Supplementary Fig. S4). (B) The endogenous levels of SLs and their biosynthetic intermediates in the roots of the WT and d14l-c1 after (−)-GR5 treatment. Rice seedlings grown hydroponically under P-deficient conditions were treated with 10 μM of (−)-GR5 for 24 h. Values are means ± SDs (n = 3). Different letters indicate significant differences at P < 0.05, Tukey’s HSD.
SL levels are moderately decreased in the D14L-defective mutant in rice
In the experiments mentioned earlier, we observed the response of SL biosynthesis using exogenously applied synthetic SL derivatives. In addition, the smax1 mutation that constitutively activates the downstream of the D14L pathway increased the levels of 4DO and methoxy-5DS in root exudates of rice grown under low P conditions (Choi et al. 2020). However, it was unclear whether D14L regulates SL biosynthesis under normal and nutrient-deficient conditions. So, we next examined the SL levels in the roots of the d14l-c1 mutant grown under P-deficient or -sufficient conditions. As a result, the levels of SLs and their biosynthetic intermediates showed a tendency to decrease in root exudates and extracts of the d14l mutant grown under both P-sufficient (Fig. 3A, Supplementary Fig. S5) and P-deficient conditions (Figs. 2B, 3B). The decrease in SL levels in P-sufficient conditions was clearer than that in P-deficient conditions. Moreover, we analyzed the involvement of D14L in SL biosynthesis in the response to the N deficiency (Sun et al. 2014). We found that N deficiency upregulated the 4DO and ORO levels in WT root exudates, but these increases were repressed in the d14l mutant (Supplementary Fig. S6). These observations indicate that the D14L pathway positively regulates SL biosynthesis under both nutrient-deficient and -sufficient conditions in rice roots. They are also consistent with a previous study that CYP711A2/Os900 is downregulated in the roots of the d14l (hebibaAOC) mutant (Gutjahr et al. 2015). However, D14L may have a moderate role in activating SL biosynthesis in the absence of specific signals like (−)-GR5 because the levels of SLs and their intermediates are not always significantly different between WT and d14l plants (e.g. controls of Figs. 1C, 2B). Moreover, D14L is dispensable for the nutrient-deficient response of SL production because SL biosynthesis still tended to increase in the d14l mutant grown under P- or N-deficient conditions versus sufficient conditions (Fig. 3, Supplementary Fig. S6). Our results suggest that the D14L regulation of SL biosynthesis is at least in part independent of the nutrient-deficiency signal in rice.
Fig. 3.
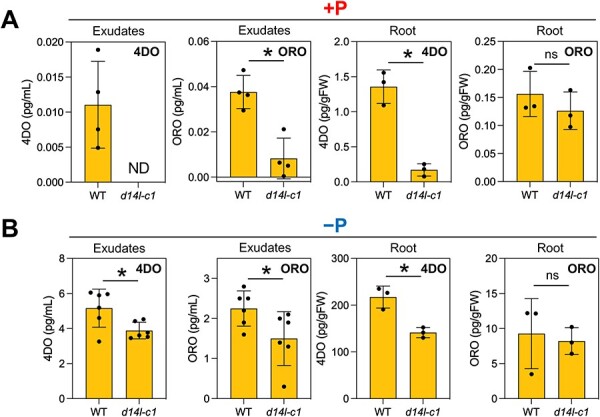
SL levels in the rice d14l mutant. 4DO and ORO levels in root exudates of the WT and d14l-c1 grown under (A) P-sufficient (+P) and (B) P-deficient (−P) conditions. Rice seedlings grown hydroponically under each condition were analyzed. Values are means ± SDs (n = 3–6). ND, not detected. Significant differences (P < 0.05) between the WT and d14l-c1 are indicated with asterisks (t-test; ns, not significant).
(−)-GR5-induced SL biosynthesis in mycorrhizal plants
Because the rice D14L pathway is essential for symbiosis with AMF (Gutjahr et al. 2015), we speculated that the D14L/KAI2 pathway–stimulated SL biosynthesis is related to the symbiotic ability with AMF of analyzed plants. In the experiments mentioned earlier, we showed that (−)-GR5 can mimic natural ligand(s) of D14L because the d14l mutant was insensitive to (−)-GR5 and (−)-GR5 induced the D14L2a expression. Generally, synthetic mimics of plant hormones are useful because these compounds can stimulate the target pathway in many plant species even if these hormonal molecules have been unknown or the signaling factors of the pathway have not been identified. We investigated (−)-GR5-induced SL production in several mycorrhizal plant species, which were grown hydroponically under P-deficient conditions. We measured ORO and solanacol in tomato (Fig. 4A, Supplementary Fig. S7A), zealactone in maize (Fig. 4B, Supplementary Fig. S7B) and 5DS in L. japonicus (Fig. 4C). In all these plant species, (−)-GR5 was effective in upregulating SL levels. These results suggest that the D14L/KAI2 pathway positively regulates SL biosynthesis not only in rice but also in other mycorrhizal angiosperms.
Fig. 4.
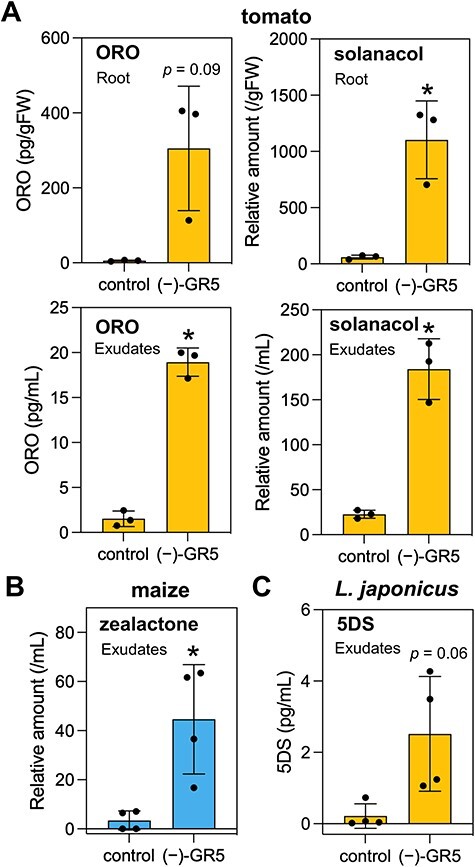
The induction of SL levels by (−)-GR5 in tomato, maize and Lotus japonicus. (A) ORO and solanacol levels in root exudates and roots in tomato. (B) Zealactone levels in root exudates in maize. (C) 5DS levels in root exudates in L. japonicus. WT seedlings grown hydroponically under P-deficient conditions were treated with (−)-GR5 (10 µM) for 24 h. For the quantification of solanacol and zealactone, the area ratio (endogenous/internal standard) was normalized with the quantity of internal standard added before purification and divided by root weight (or root exudate volume). Labeled ORO and 4DO were used as internal standards for the quantification of solanacol and zealactone, respectively. Values are means ± SDs (n = 3–4). Significant differences between control and (−)-GR5 treatment are indicated with asterisks (P < 0.05, t-test). Growth conditions for each experiment are described in Supplementary Table S2.
The KAI2 pathway does not positively regulate SL biosynthesis in Arabidopsis
We next examined whether the KAI2 pathway regulates SL biosynthesis in Arabidopsis, a nonhost plant of AMF (Wang and Qiu 2006). The KAI2 pathway inhibits hypocotyl elongation in Arabidopsis seedlings (Nelson et al. 2011, Waters et al. 2012). We first confirmed whether (−)-GR5 acts through the KAI2 pathway by analyzing the inhibitory activity of (+)-GR5, (−)-GR5 and KAR1 on hypocotyl elongation of WT, atd14-2 and kai2-4 mutants. We found that (−)-GR5 and KAR1 treatments for 7 d suppressed hypocotyl elongation in a KAI2-dependent manner (Supplementary Fig. S8A). However, (−)-GR5 treatment for 24 h as we did for other plant species did not induce the expression of DLK2, a marker gene for KAI2-mediated signaling in Arabidopsis (Waters et al. 2012, Scaffidi et al. 2014), in 13-day-old seedlings grown on the P-deficient or -sufficient agar media (Supplementary Fig. S8B). So, we next analyzed 7-d-old seedlings grown on (−)-GR5-containing half-strength Murashige–Skoog (MS) agar media, which were grown in the same manner as plants used for the hypocotyl elongation assay mentioned earlier (Supplementary Fig. S9A). In these seedlings, the transcript level of DLK2 was moderately increased (4.3-fold), indicating that the KAI2 pathway was activated (Supplementary Fig. S9B). However, it was not conclusive whether SL biosynthesis is enhanced by (−)-GR5 because AtD27 and MAX1/CYP711A1 were slightly induced 2.0-fold and 1.6-fold, respectively, but MAX4/CCD8 was downregulated 0.53-fold (Supplementary Fig. S9C).
Because there were many possibilities when the exogenously applied compound was ineffective or showed weaker activity (e.g. growth conditions, treatment methods and tissues analyzed), we investigated the SMAX1-related mutants. It was previously shown that SMAX1 and SMXL2 redundantly function downstream of KAI2 in Arabidopsis seedlings, and the KAI2 pathway is constitutively activated in the smax1 smxl2 double mutant (Stanga et al. 2016). In fact, the expression of DLK2 was significantly increased in the smax1 smxl2 mutant under P-deficient or -sufficient conditions (Fig. 5A, Supplementary Fig. S8B).
Fig. 5.
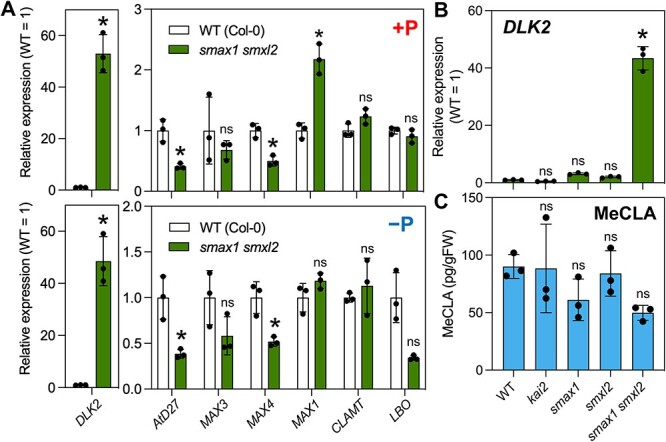
The effect of the KAI2 pathway on SL biosynthesis in Arabidopsis. (A) Expression analysis of DLK2 and SL biosynthesis genes using 13-day-old WT and smax1 smxl2 seedlings grown under P-sufficient (+P) and P-deficient (−P) conditions. (B) Expression levels of DLK2 and (C) MeCLA levels in 2-week-old WT, kai2, smax1, smxl2 and smax1 smxl2 seedlings. Values are means ± SDs (n = 3). Significant differences (P < 0.05) compared to the WT are indicated with asterisks [t-test (A) and one-way ANOVA followed by Dunnett’s test (B); ns, not significant]. Growth conditions for each experiment are described in Supplementary Table S2.
So, we next examined the expression of SL biosynthetic genes in the smax1 smxl2 mutant under these conditions. The transcript levels of AtD27 and MAX4 were reduced under both P conditions and only MAX1 expression was increased 2.2-fold in the smax1 smxl2 mutant under P-sufficient condition (Fig. 5A). The decrease in MAX4 expression in the smax1 smxl2 mutant (Fig. 5A) and (−)-GR5-treated seedlings (Supplementary Fig. S9C) agrees with a previous observation that MAX4 expression was reduced in KARs-treated seedlings in a MAX2-dependent manner (Nelson et al. 2011). These results indicate that the constitutive activation of the KAI2 pathway does not have a great impact on the expression of SL biosynthetic genes.
We further analyzed the endogenous levels of MeCLA, a bioactive SL in Arabidopsis, in the KAI2 pathway–related mutants (kai2-4, smax1, smxl2 and smax1 smxl2). The DLK2 expression was highly induced in the smax1 smxl2 mutant (Fig. 5B), but the endogenous MeCLA levels of the KAI2 pathway–related mutants were not significantly changed compared with those of the WT (Fig. 5C). These results demonstrate that both deactivation (kai2) and activation (smax1 smxl2) of the KAI2 pathway do not affect the endogenous MeCLA levels in Arabidopsis. In conclusion, our results suggest that the KAI2 pathway does not activate SL biosynthesis in Arabidopsis seedlings.
The liverwort KAI2 pathway does not actively regulate SL biosynthesis
Our results using angiosperms suggest that SL biosynthesis induced by the D14L/KAI2 pathway plays a role in symbiosis with AMF because SLs are important molecules in this biological interaction (Akiyama et al. 2005, Besserer et al. 2006, Kobae et al. 2018). We next analyzed the liverwort M. paleacea, in which the biosynthetic pathway for bryosymbiol (BSB), an ancestral SL for AMF symbiosis, has recently been clarified (Kodama et al. 2022). It has also been demonstrated that the KAI2 pathway regulates thalli growth and gemma cup initiation in M. paleacea, like M. polymorpha (Mizuno et al. 2021, Kodama et al. 2022). Moreover, there is one SMAX1-like protein named MpSMXL and MpaSMXL in M. polymorpha and M. paleacea, respectively, and the smxl mutants have recently been shown to increase the number of gemmae in a cup due to the activation of the KAI2 pathway in both species (Komatsu et al. 2023).
To analyze the relationship between the KAI2 pathway and SL biosynthesis, we first examined whether (−)-GR5 can activate the MpaKAI2A pathway in M. paleacea. We found that the transcript level of MpaDLP1, a marker gene for the MpaKAI2A-dependent signaling, was increased 5.0-fold after (−)-GR5 treatment for 24 h under the P-deficient condition (Fig. 6A). However, the endogenous BSB levels were not significantly changed after (−)-GR5 treatment (Fig. 6B, Supplementary Fig. S10).
Fig. 6.
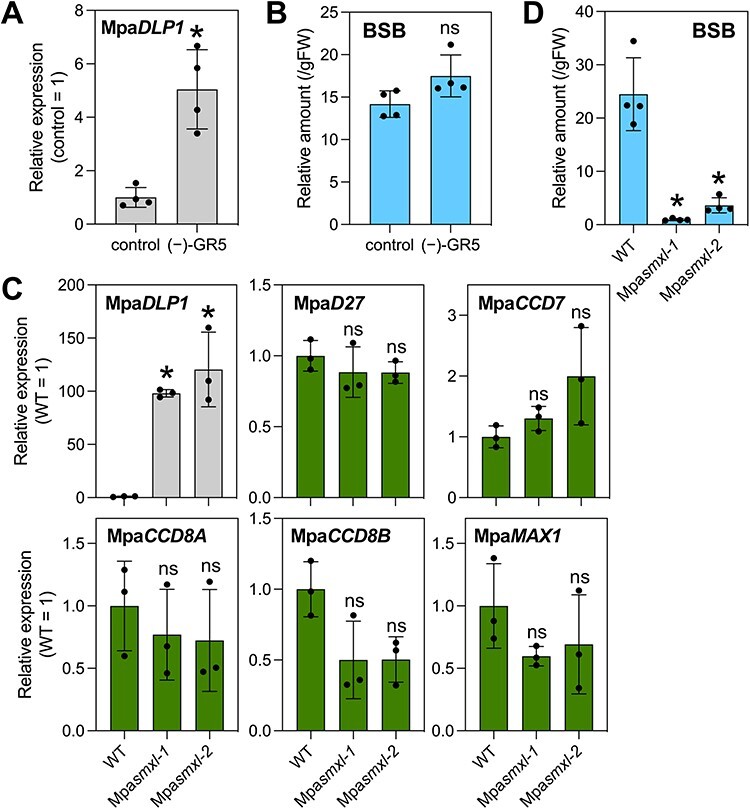
The effect of the KAI2 pathway on SL biosynthesis in M. paleacea. (A, B) Analyses of MpaDLP1 expression (A) and BSB levels (B) in (−)-GR5-treated 6-week-old WT plants. (−)-GR5 (10 µM) was treated for 24 h under the P-deficient condition. Values are means ± SDs (n = 4). (C) Expression analysis of MpaDLP1 and BSB biosynthesis genes using 4-week-old WT and Mpasmxl plants grown under P-sufficient conditions. Values are means ± SDs (n = 3). (D) BSB levels in 4-week-old WT and Mpasmxl plants grown under P-deficient conditions. Values are means ± SDs (n = 4). To quantify BSB in (B) and (D), rac-GR24 was added to each sample as an internal standard before purification. The area ratio (endogenous BSB/rac-GR24) was normalized by dividing by plant fresh weight. Significant differences (P < 0.05) compared to the WT or control are indicated with asterisks [t-test (A, B) and one-way ANOVA followed by Dunnett’s test (C, D); ns, not significant].
We next generated two independent lines of the Mpasmxl mutants by CRISPR/Cas9-based genome editing to analyze SL biosynthesis when the KAI2 pathway is constitutively active (see Materials and Methods). The expression of BSB biosynthetic genes was examined by qRT-PCR using plants grown on P-sufficient (half-strength B5) agar media. MpaDLP1 was drastically upregulated: 98.1-fold and 121-fold in the Mpasmxl-1 and Mpasmxl-2 mutants, respectively, but no BSB biosynthetic genes were upregulated in the Mpasmxl mutants (Fig. 6C). We further measured the endogenous BSB levels in the Mpasmxl-1 and Mpasmxl-2 mutants under the P-deficient condition because we were unable to analyze them under the P-sufficient condition due to low abundance. Interestingly, the BSB levels were not increased but significantly decreased in these mutants (Fig. 6D). These results suggest that constitutive activation of the KAI2 pathway does not upregulate BSB biosynthesis but instead reduces BSB levels in M. paleacea.
We also performed RNA-seq analysis using WT, Mpakai2a-1 and Mpakai2a-2 plants. We confirmed that MpaDLP1 was downregulated in the Mpakai2a mutants (Supplementary Fig. S11A, Supplementary Table S1). In these samples, BSB biosynthetic genes were not significantly downregulated, whereas MpaMAX1 was slightly upregulated in the Mpakai2a mutants (Supplementary Fig. S11B). These results indicate that deactivation of the KAI2 pathway does not downregulate SL biosynthesis. These pieces of evidence demonstrate that SL biosynthesis is not positively regulated by the KAI2 pathway in M. paleacea. Previous observations that the BSB levels in exudates were not reduced in Mpakai2a/2b and Mpamax2 mutants support this idea (Kodama et al. 2022).
Interestingly, it has been shown that the BSB-deficient Mpaccd8a/8b mutant cannot establish symbiosis with AMF, but Mpakai2a and Mpamax2 mutants can (Kodama et al. 2022). These results indicate that the KAI2 pathway and SL biosynthesis are not closely linked in M. paleacea, unlike in mycorrhizal angiosperms described earlier. However, because BSB levels were severely decreased in the Mpasmxl-1 and Mpasmxl-2 mutants (Fig. 6D), excessive activation of the KAI2 pathway may negatively regulate SL levels in M. paleacea. The molecular mechanism underlying this negative regulation needs to be clarified for further understanding of the connection between the KAI2 pathway and SL biosynthesis in M. paleacea.
Conclusion
Our results in this study indicate that the D14L/KAI2 pathway could stimulate SL biosynthesis in several mycorrhizal angiosperms. However, Arabidopsis, a non-mycorrhizal angiosperm, and the liverwort M. paleacea do not seem to have this regulatory mechanism. These findings support the idea that activation of SL biosynthesis by the D14L/KAI2 pathway is evolutionarily acquired in mycorrhizal plants to improve AMF symbiosis. Recently, it has been shown that the D14L-defective mutants showed decreased AMF colonization in barley and Medicago truncatula as previously observed in rice (Choi et al. 2020, Li et al. 2022). The positive transcriptional regulation of HvRLK10, a receptor candidate for AMF-derived lipochitooligosaccharides, by the D14L pathway has been considered as one of the reasons for defects in AMF symbiosis in the d14l mutants in barley (Li et al. 2022). Interestingly, AMF colonization has been shown to increase the expression of D14L2a as well as SL biosynthetic genes in rice (Sisaphaithong et al. 2021), suggesting that AM symbiosis itself activates the D14L pathway which leads to the activation of SL biosynthesis. It will be important to analyze SL levels in the d14l mutants in barley and M. truncatula to ask whether SL biosynthesis is also involved in the D14L-mediated interaction with AMF.
Because the D14L/KAI2 family is highly conserved and widely distributed in land plants (Machin et al. 2020), future studies will clarify whether the D14L/KAI2 pathway–mediated positive regulation of SL biosynthesis can be observed beyond angiosperms and whether it is related to AMF symbiosis. Elucidation of key signaling factor(s) downstream of SMAX1 would be essential for fully understanding the evolution of cross-talk between SL biosynthesis and the D14L/KAI2 signaling pathway. The transcription factor NSP2 has recently been shown to function upstream of the D14L/KAI2 pathway in promoting AMF colonization in M. truncatula (Li et al. 2022). Moreover, overexpression of M. truncatula NSP2 increased SL levels under both low P and high P conditions in barley (Li et al. 2022). Because the NSP1- or NSP2-deficient mutants are severely impaired in P starvation–induced SL biosynthesis in barley, M. truncatula and rice, these transcription factors may play an important role in connecting the nutrient-deficient response of SL production with the D14L/KAI2-mediated regulation of SL biosynthesis (Liu et al. 2011, Li et al. 2022). However, as discussed earlier, because the nutrient deficiency could induce SL biosynthesis in the d14l mutant in rice (Fig. 3), the D14L pathway may not be the only factor that functions downstream of NSP1 and NSP2 in stimulating SL biosynthesis. We also found that NSP2, but not NSP1, was significantly upregulated after (−)-GR5 treatment (Supplementary Fig. S4), which is consistent with a previous report that the NSP2 expression was elevated in the smax1 mutant in rice (Choi et al. 2020). These observations suggest that NSP2 functions not only upstream but also downstream of D14L signaling to enhance SL biosynthesis in rice. To clarify this hypothesis, it will be important to examine whether NSP2 overexpression can stimulate SL biosynthesis in not only barley but also other plant species. Further studies will elucidate the relationship between the D14L/KAI2 pathway and the nutrient signaling, both of which are crucial for AMF symbiosis in mycorrhizal plants.
Materials and Methods
Chemicals
The synthetic rac-GR5 was purchased from Fuji Molecular Planning (Yokohama, Japan), and (+)-GR5 and (−)-GR5 were separated by chiral HPLC with a CHIRALPAK AD-H column (1.0 cm × 25 cm × 5 µm) (DAICEL, Osaka, Japan). The isocratic elution was carried out with n-hexane/2-propanol/methyl tert-butyl ether (80:10:10) at a flow rate of 4.8 ml/min. The synthetic rac-GR24 was purchased from Chiralix (Nijmegen, Netherlands). KAR1 was purchased from Toronto Research Chemicals (Toronto, Canada). The other synthetic SL derivatives, stable isotope-labeled SLs, natural SLs (solanacol and zealactone) and enzymatically produced BSB using MpaMAX1 are described earlier (Xie et al. 2007, Akiyama et al. 2010, Abe et al. 2014, Seto et al. 2014, Xie et al. 2017, Kodama et al. 2022).
Plant materials, plant growth conditions and treatment of SL-related compounds
We used rice cultivars (cv. Nipponbare and cv. Shiokari), an Arabidopsis ecotype Col-0, a tomato cultivar (cv. Micro-Tom), an L. japonicus ecotype Gifu B-129, a maize cultivar (cv. Caroline 86) and an M. paleacea subspecies (ssp. diptera) as the WT. The SL- and KL-related mutants of rice (d14-1, d14-1N, d3-1 and d3-2), Arabidopsis (atd14-2, kai2-4, smax1, smxl2 and smax1 smxl2) and liverwort (Mpakai2a-1 and Mpakai2a-2) were previously described (Ishikawa et al. 2005, Yoshida et al. 2012, Umehara et al. 2015, Stanga et al. 2016, Seto et al. 2019, Kodama et al. 2022). Kamachi’s hydroponic nutrient solution (N = 1 mM and P = 0.6 mM) was used in rice experiments (Kamachi et al. 1991). Norén’s hydroponic nutrient solution (N = 9 mM and P = 1 mM) and the half-strength MS nutrient solution (N = 19.7 mM and P = 0.62 mM) were used to make agar media in Arabidopsis experiments (Norén et al. 2004). The half-strength Hoagland nutrient solution (N = 8 mM and P = 0.5 mM) was used in tomato, maize and L. japonicus experiments. The half-strength Gamborg’s B5 (B5) agar/liquid media (N = 13.4 mM and P = 0.54 mM) was used in M. paleacea experiments. When making a specific nutrient-deficient media, the corresponding nutrient was excluded.
Rice plants were hydroponically grown as previously described (Umehara et al. 2015). Briefly, 1-week-old rice seedlings grown on 0.6% agar media of hydroponic nutrient solution under a 16-h light/8-h dark photoperiod were transferred to glass vials containing 4 or 13 mL of hydroponic nutrient solution (pH 5.7). The hydroponic nutrient solution was replaced with a new medium 1 d before sampling roots and/or root exudates simultaneously with chemical treatment. In all chemical treatment experiments, acetone [final concentration 0.1% (v/v)] was used as a control because acetone was used to dissolve compounds. The details of growth and nutrient conditions of rice and other plant species used in this study are summarized in Supplementary Table S2.
Establishment of the d14l mutant and analysis of AMF colonization phenotype in rice
The annealed oligo DNAs (5′-GTTGTCGAAGTAGTCCGGGTTGGT-3′ and 5′-AAACACCAACCCGGACTACTTCGA-3′) containing the target sequence were subcloned into pU6gRNA-oligo and then cloned into pZH_OsU3gYSA_MMCas9 (Mikami et al. 2015). The resulting binary vector was transformed into the WT (cv. Nipponbare) as previously described (Nakagawa et al. 2002). Because the 175th adenine of D14L ORF (816 bp) is duplicated in the d14l-c1 mutant, a frameshift occurs after the 58th Thr and a premature stop codon is generated (Supplementary Fig. S3A). The transgene was eliminated by segregation in the d14l-c1 mutant. To analyze the interaction with AMF, Rhizophagus irregularis DAOM197198 was inoculated to rice plants for 14 d. Fungal cell walls were stained with wheat germ agglutinin Alexa Fluor 488, and the number of infection sites was counted. Detailed conditions were previously described (Kobae et al. 2018) (Supplementary Fig. S3B).
Establishment of the smxl mutants in M. paleacea
The Mpasmxl-1 mutant was previously described (Komatsu et al. 2023). To make the Mpasmxl-2 mutant, the annealed oligo DNAs (5′-CTCGACAGATTACCATCGAACC-3′ and 5′-AAACGGTTCGATGGTAATCTGT-3′) containing the target sequence were subcloned into pMpGE_En03 (GenBank LC090755) and then cloned into pMpGE011 (GenBank LC090757) (Sugano et al. 2018) by Gateway LR reaction (Thermo Fisher Scientific, Waltham, MA, USA). The resulting binary vector was transformed into the WT as previously described (Kodama et al. 2022). Because the 15-bp sequence (the 261st to 275th) is deleted and the 4-bp sequence (GTTC) is inserted into MpaSMXL ORF (3,861 bp) in the Mpasmxl-2 mutant, a frame-shifting change after the 87th Pro occurs and a premature stop codon is generated.
Inhibition assay of Arabidopsis hypocotyl elongation
After imbibition at 4°C for 3 d under dark conditions, surface-sterilized seeds were germinated on half-strength MS agar media (1% sucrose, pH 5.7) containing each compound dissolved in acetone (1:1,000 dilution). The plates were placed vertically at 22°C under short-day conditions (8-h light/16-h dark, 64 µmol m−2 s−1) for 7 d.
qRT-PCR analysis in rice roots
Total RNA was isolated and purified using the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The QuantiTect Reverse Transcription Kit (QIAGEN) was used for cDNA synthesis from total RNA. The Mx3000P system (Agilent, Santa Clara, CA, USA) was used to perform qRT-PCR by using the THUNDERBIRD Probe qPCR Mix (TOYOBO, Osaka, Japan) and TaqMan probes (for absolute quantification) or the KOD SYBR qPCR Mix (TOYOBO) (for relative quantification). The Ubiquitin gene (Os05g0160200) was used as the housekeeping gene. The primers and TaqMan probes are described in Supplementary Table S3.
qRT-PCR analysis in Arabidopsis
Total RNA was isolated and purified using the Cica Geneus RNA Prep Kit (Kanto Chemical, Tokyo, Japan) or the Total RNA Extraction Kit Mini (Plant) (RBC Bioscience, New Taipei City, Taiwan). The ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO) was used for cDNA synthesis from total RNA. The Mx3000P system (Agilent) was used to perform qRT-PCR by using the KOD SYBR qPCR Mix (TOYOBO). The CACS gene (At5g46630) was used as the housekeeping gene. The primers are described in Supplementary Table S3.
LC–MS/MS analysis of SL-related compounds
LC–MS/MS analyses of SL-related compounds were carried out using an ultra HPLC (Nexera, Shimadzu, Kyoto, Japan) and a quadrupole/time-of-flight tandem mass spectrometer (TripleTOF 5600) (AB SCIEX, Framingham, MA, USA). To analyze SL levels in root exudates, the hydroponic culture solution was collected from each plant sample and extracted using ethyl acetate (EtOAc) twice with stable isotope-labeled SLs as internal standards. The EtOAc fraction was then evaporated to dryness under N2 gas, dissolved in acetonitrile or 50% (vol/vol) acetonitrile and subjected to LC–MS/MS. Calibration curves generated using labeled and non-labeled compounds were used to quantify SLs. MeCLA analysis using Arabidopsis seedlings (0.25–0.57 gFW) was performed mostly according to our previous study (Ramírez et al. 2018). Purifications of SLs and LC–MS/MS conditions are shown in Supplementary Tables S4 and S5, respectively.
qRT-PCR and BSB analysis in M. paleacea
For qRT-PCR and BSB analyses of (−)-GR5-treated M. paleacea, gemmae of the WT were incubated on half-strength B5 agar medium (1% sucrose) at 22°C for 4 weeks under continuous light. Then, plants were transferred to the agar medium (1% sucrose) without P and grown for 2 weeks. Six-week-old plants were treated with (−)-GR5 (10 µM) for 24 h in half-strength B5 liquid medium (1% sucrose) without P. In BSB analysis, rac-GR24 (0.8 pmol) was added to each sample (0.18–0.25 gFW) as an internal standard.
For qRT-PCR analysis of the WT and Mpasmxl mutants, gemmae of the WT, Mpasmxl-1 and Mpasmxl-2 were incubated on half-strength B5 agar medium (1% sucrose) at 22°C for 4 weeks under continuous light.
For BSB analysis of the WT and the Mpasmxl mutants, gemmae of the WT, Mpasmxl-1 and Mpasmxl-2 were incubated on half-strength B5 agar medium (1% sucrose) at 22°C for 3 weeks under continuous light. Then, plants were transferred to the agar medium (1% sucrose) without P and grown for 1 week to induce BSB biosynthesis. As an internal standard, rac-GR24 (0.4 pmol) was added to each sample (0.058–0.12 gFW).
Total RNA was isolated and purified using the RNeasy Plant Mini Kit (QIAGEN). The ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO) was used for cDNA synthesis. The Mx3000P system (Agilent) was used to perform qRT-PCR by using the KOD SYBR qPCR Mix (TOYOBO). The MpaACTIN gene was used as the housekeeping gene. The primers are described in Supplementary Table S3.
BSB was analyzed by an X500R QTOF system (AB SCIEX). Purification of BSB and LC–MS/MS conditions are shown in Supplementary Tables S4 and S5, respectively.
RNA-seq analysis in M. paleacea
Gemmae of the WT, Mpakai2a-1 and Mpakai2a-2 were incubated on half-strength B5 media with 1.0% agar at 22°C for 14 d under continuous light. After 14-d culture, the thalli were transferred to the new medium and incubated for another 7 d. Total RNA was extracted from the harvested plants using the NucleoSpin RNA Plant kit (MACHEREY-NAGEL, Düren, Germany). Library preparation and 100-bp paired-end read sequencing were performed by BGI (Shenzhen, China) with the DNBseqTM platform. Raw data with adapter sequences or low-quality sequences were filtered by SOAPnuke software developed by BGI (Chen et al. 2017). Mapping onto the M. paleacea genome using HISAT2 and calculating the read counts using featureCounts were performed as described previously (Kodama et al. 2022). A summary of RNA-seq analysis is shown in Supplementary Table S1.
Data collection and analysis
Most of the experiments were performed more than twice. The 4DO analyses using GR24, GR7 and dhGR24 stereoisomers were performed once. BSB, qRT-PCR and RNA-seq analyses in M. paleacea were performed once, but two independent mutant lines were analyzed. qRT-PCR analysis of (−)-GR5-treated Arabidopsis seedlings was also performed once. Statistical analyses were performed using Prism 9 (GraphPad Software, Boston, MA, USA).
Supplementary Material
Acknowledgments
We would like to thank Dr. Kohki Akiyama for providing SL-related compounds. We also thank Dr. David Nelson for providing smax1, smxl2 and smax1 smxl2 seeds. We are grateful to Dr. Xiaonan Xie and Dr. Takahito Nomura for providing natural SLs (solanacol and zealactone) and a standard of BSB prepared using the recombinant MpaMAX1 protein, respectively. We are also grateful to the System for Development and Assessment of Sustainable Humanosphere (Research Institute for Sustainable Humanosphere, Kyoto University) for providing a greenhouse.
Contributor Information
Kiyoshi Mashiguchi, Institute for Chemical Research, Kyoto University, Gokasho, Uji, Kyoto, 611-0011 Japan; Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi, 980-8577 Japan.
Ryo Morita, Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi, 980-8577 Japan.
Kai Tanaka, Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi, 980-8577 Japan.
Kyoichi Kodama, Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi, 980-8577 Japan.
Hiromu Kameoka, Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi, 980-8577 Japan.
Junko Kyozuka, Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi, 980-8577 Japan.
Yoshiya Seto, Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi, 980-8577 Japan; School of Agriculture, Meiji University, 1-1-1 Higashi-mita, Tama-ku, Kawasaki, Kanagawa, 214-8571 Japan.
Shinjiro Yamaguchi, Institute for Chemical Research, Kyoto University, Gokasho, Uji, Kyoto, 611-0011 Japan; Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi, 980-8577 Japan.
Supplementary Data
Supplementary data are available at PCP online.
Data Availability
All data underlying this article are available in the article and the online supplementary data.
Funding
Ministry of Education, Culture, Sports, Science and Technology Grants-in-Aid for Scientific Research (KAKENHI) (JP24114010 to S.Y. and JP17H06474 to S.Y.), Japan Society for the Promotion of Science KAKENHI (JP20H05684 to H.K., J.K., Y.S. and S.Y., JP23H05409 to J.K., Y.S. and S.Y., JP19H02892 and JP23H02149 to K.M.), Japan Science and Technology Agency Core Research for Evolutional Science and Technology (CREST) (JPMJCR13B1 to S.Y.), International Collaborative Research Program of Institute for Chemical Research, Kyoto University (2023-132 to Y.S.).
Disclosures
The authors have no conflicts of interest to declare.
References
- Abe S., Sado A., Tanaka K., Kisugi T., Asami K., Ota S., et al. (2014) Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. U.S.A. 111: 18084–18089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K. and Hayashi H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. [DOI] [PubMed] [Google Scholar]
- Akiyama K., Ogasawara S., Ito S. and Hayashi H. (2010) Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 51: 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., et al. (2012) The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351. [DOI] [PubMed] [Google Scholar]
- Andreo-Jimenez B., Ruyter-Spira C., Bouwmeester H.J. and Lopez-Raez J.A. (2015) Ecological relevance of strigolactones in nutrient uptake and other abiotic stresses, and in plant-microbe interactions below-ground. Plant Soil 394: 1–19. [Google Scholar]
- Besserer A., Puech-Pages V., Kiefer P., Gomez-Roldan V., Jauneau A., Roy S., et al. (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer P.B., Yoneyama K., Filardo F., Meyers E., Scaffidi A., Frickey T., et al. (2016) LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113: 6301–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnel S., Torabi S., Griesmann M., Bleek E., Tang Y., Buchka S., et al. (2020) Lotus japonicus karrikin receptors display divergent ligand-binding specificities and organ-dependent redundancy. PLoS Genet. 16: e1009249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.X., Chen Y.S., Shi C.M., Huang Z.B., Zhang Y., Li S.K., et al. (2017) SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7: gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Lee T., Cho J., Servante E.K., Pucker B., Summers W., et al. (2020) The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nat. Commun. 11: 2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn C.E. and Nelson D.C. (2016) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front Plant Sci. 6: 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flematti G.R., Ghisalberti E.L., Dixon K.W. and Trengove R.D. (2004) A compound from smoke that promotes seed germination. Science 305: 977–977. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., Fermas S., Brewer P.B., Puech-Pages V., Dun E.A., Pillot J.P., et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194. [DOI] [PubMed] [Google Scholar]
- Guercio A.M., Torabi S., Cornu D., Dalmais M., Bendahmane A., Le Signor C., et al. (2022) Structural and functional analyses explain Pea KAI2 receptor diversity and reveal stereoselective catalysis during signal perception. Commun. Biol. 5: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Gobbato E., Choi J., Riemann M., Johnston M.G., Summers W., et al. (2015) Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350: 1521–1524. [DOI] [PubMed] [Google Scholar]
- Ishikawa S., Maekawa M., Arite T., Onishi K., Takamure I. and Kyozuka J. (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46: 79–86. [DOI] [PubMed] [Google Scholar]
- Johnson A.W., Rosebery G. and Parker C. (1976) A novel approach to Striga and Orobanche control using synthetic germination stimulants. Weed Res. 16: 223–227. [Google Scholar]
- Kamachi K., Yamaya T., Mae T. and Ojima K. (1991) A role for glutamine synthetase in the remobilization of leaf nitrogen during natural senescence in rice leaves. Plant Physiol. 96: 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka H. and Kyozuka J. (2015) Downregulation of rice DWARF 14 LIKE suppress mesocotyl elongation via a strigolactone independent pathway in the dark. J. Genet. Genomics 42: 119–124. [DOI] [PubMed] [Google Scholar]
- Kobae Y., Kameoka H., Sugimura Y., Saito K., Ohtomo R., Fujiwara T., et al. (2018) Strigolactone biosynthesis genes of rice are required for the punctual entry of arbuscular mycorrhizal fungi into the roots. Plant Cell Physiol. 59: 544–553. [DOI] [PubMed] [Google Scholar]
- Kodama K., Rich M.K., Yoda A., Shimazaki S., Xie X., Akiyama K., et al. (2022) An ancestral function of strigolactones as symbiotic rhizosphere signals. Nat. Commun. 13: 3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu A., Kodama K., Mizuno Y., Fujibayashi M., Naramoto S. and Kyozuka J. (2023) Control of vegetative reproduction in Marchantia polymorpha by the KAI2-ligand signaling pathway. Curr. Biol. 33: 1196–1210.e4. [DOI] [PubMed] [Google Scholar]
- Li X.R., Sun J., Albinsky D., Zarrabian D., Hull R., Lee T., et al. (2022) Nutrient regulation of lipochitooligosaccharide recognition in plants via NSP1 and NSP2. Nat. Commun. 13: 6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Kohlen W., Lillo A., Op den Camp R., Ivanov S., Hartog M., et al. (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23: 3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Raez J.A., Charnikhova T., Gomez-Roldan V., Matusova R., Kohlen W., De Vos R., et al. (2008) Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 178: 863–874. [DOI] [PubMed] [Google Scholar]
- Machin D.C., Hamon-Josse M. and Bennett T. (2020) Fellowship of the rings: a saga of strigolactones and other small signals. New Phytol. 225: 621–636. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., Seto Y., Onozuka Y., Suzuki S., Takemoto K., Wang Y., et al. (2022) A carlactonoic acid methyltransferase that contributes to the inhibition of shoot branching in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 119: e2111565119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K., Seto Y. and Yamaguchi S. (2021) Strigolactone biosynthesis, transport and perception. Plant J. 105: 335–350. [DOI] [PubMed] [Google Scholar]
- Mikami M., Toki S. and Endo M. (2015) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol. Biol. 88: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y., Komatsu A., Shimazaki S., Naramoto S., Inoue K., Xie X., et al. (2021) Major components of the KARRIKIN INSENSITIVE2-dependent signaling pathway are conserved in the liverwort Marchantia polymorpha. Plant Cell 33: 2395–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N., Nomura T. and Akiyama K. (2020) Identification of two oxygenase genes involved in the respective biosynthetic pathways of canonical and non-canonical strigolactones in Lotus japonicus. Planta 251: 40. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Shimamoto K. and Kyozuka J. (2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29: 743–750. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Scaffidi A., Dun E.A., Waters M.T., Flematti G.R., Dixon K.W., et al. (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108: 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norén H., Svensson P. and Andersson B. (2004) A convenient and versatile hydroponic cultivation system for Arabidopsis thaliana. Physiol. Plant 121: 343–348. [Google Scholar]
- Ramírez V., Xiong G., Mashiguchi K., Yamaguchi S. and Pauly M. (2018) Growth- and stress-related defects associated with wall hypoacetylation are strigolactone-dependent. Plant Direct 2: e00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi A., Waters M.T., Sun Y.M.K., Skelton B.W., Dixon K.W., Ghisalberti E.L., et al. (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165: 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y., Sado A., Asami K., Hanada A., Umehara M., Akiyama K., et al. (2014) Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. U.S.A. 111: 1640–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y., Yasui R., Kameoka H., Tamiru M., Cao M., Terauchi R., et al. (2019) Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat. Commun. 10: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisaphaithong T., Yanase M., Mano T., Tanabe S., Minami E., Tanaka A., et al. (2021) Localized expression of the Dwarf14-like2a gene in rice roots on infection of arbuscular mycorrhizal fungus and hydrolysis of rac-GR24 by the encoded protein. Plant Signal Behav. 16: 2009998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga J.P., Morffy N. and Nelson D.C. (2016) Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243: 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S.S., Nishihama R., Shirakawa M., Takagi J., Matsuda Y., Ishida S., et al. (2018) Efficient CRISPR/Cas9-based genome editing and its application to conditional genetic analysis in Marchantia polymorpha. PLoS One 13: e0205117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Tao J., Liu S., Huang S., Chen S., Xie X., et al. (2014) Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 65: 6735–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmerman A., Guillory A., Bonhomme S., Goormachtig S. and Struk S. (2022) Masks start to drop: suppressor of MAX2 1-like proteins reveal their many faces. Front Plant Sci. 13: 887232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M., Cao M., Akiyama K., Akatsu T., Seto Y., Hanada A., et al. (2015) Structural requirements of strigolactones for shoot branching inhibition in rice and Arabidopsis. Plant Cell Physiol. 56: 1059–1072. [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Magome H., Takeda-Kamiya N. and Yamaguchi S. (2010) Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 51: 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200. [DOI] [PubMed] [Google Scholar]
- Wakabayashi T., Hamana M., Mori A., Akiyama R., Ueno K., Osakabe K., et al. (2019) Direct conversion of carlactonoic acid to orobanchol by cytochrome P450 CYP722C in strigolactone biosynthesis. Sci. Adv. 5: eaax9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi T., Yasuhara R., Miura K., Takikawa H., Mizutani M. and Sugimoto Y. (2021) Specific methylation of (11R)-carlactonoic acid by an Arabidopsis SABATH methyltransferase. Planta 254: 88. [DOI] [PubMed] [Google Scholar]
- Wang B. and Qiu Y.L. (2006) Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16: 299–363. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Gutjahr C., Bennett T. and Nelson D.C. (2017) Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68: 291–322. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Nelson D.C., Scaffidi A., Flematti G.R., Sun Y.K., Dixon K.W., et al. (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Scaffidi A., Moulin S.L., Sun Y.K., Flematti G.R. and Smith S.M. (2015) A selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27: 1925–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Gao Y., Yang W., Sui N. and Zhu J. (2022) Biological functions of strigolactones and their crosstalk with other phytohormones. Front Plant Sci. 13: 821563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Kisugi T., Yoneyama K., Nomura T., Akiyama K., Uchida K., et al. (2017) Methyl zealactonoate, a novel germination stimulant for root parasitic weeds produced by maize. J. Pestic. Sci. 42: 58–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Kusumoto D., Takeuchi Y., Yoneyama K., Yamada Y. and Yoneyama K. (2007) 2ʹ-epi-orobanchol and solanacol, two unique strigolactones, germination stimulants for root parasitic weeds, produced by tobacco. J. Agric. Food Chem. 55: 8067–8072. [DOI] [PubMed] [Google Scholar]
- Xie X., Yoneyama K. and Yoneyama K. (2010) The strigolactone story. Annu. Rev. Phytopathol. 48: 93–117. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Furusawa S., Nagasaka S., Shimomura K., Yamaguchi S. and Umehara M. (2014) Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240: 399–408. [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Akiyama K., Brewer P.B., Mori N., Kawano-Kawada M., Haruta S., et al. (2020) Hydroxyl carlactone derivatives are predominant strigolactones in Arabidopsis. Plant Direct 4: e00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K., Mori N., Sato T., Yoda A., Xie X., Okamoto M., et al. (2018) Conversion of carlactone to carlactonoic acid is a conserved function of MAX1 homologs in strigolactone biosynthesis. New Phytol. 218: 1522–1533. [DOI] [PubMed] [Google Scholar]
- Yoneyama K., Xie X., Kim H.I., Kisugi T., Nomura T., Sekimoto H., et al. (2012) How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 235: 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Kameoka H., Tempo M., Akiyama K., Umehara M., Yamaguchi S., et al. (2012) The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol. 196: 1208–1216. [DOI] [PubMed] [Google Scholar]
- Zhang Y., van Dijk A.D., Scaffidi A., Flematti G.R., Hofmann M., Charnikhova T., et al. (2014) Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 10: 1028–1033. [DOI] [PubMed] [Google Scholar]
- Zheng J., Hong K., Zeng L., Wang L., Kang S., Qu M., et al. (2020) Karrikin signaling acts parallel to and additively with strigolactone signaling to regulate rice mesocotyl elongation in darkness. Plant Cell 32: 2780–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data underlying this article are available in the article and the online supplementary data.


